Abstract
The role and functional domain of hepatitis B virus (HBV) X protein (HBx) in regulating HBV transcription and replication were investigated with a transient transfection system in the human hepatoma cell line HepG2 using wild-type or HBx-minus HBV genome constructs and a series of deletion or mutation HBx expression plasmids. We show here that HBx has augmentation effects on HBV transcription and replication as a HBV mutant genome with defective X gene led to decreased levels of 3.5-kb HBV RNA and HBV replication intermediates and that these decreases can be restored by either transient ectopic expression of HBx or a stable HBx expression cell line. The C-terminal two-thirds (amino acids [aa] 51 to 154), which contain the transactivation domain, is required for this function of HBx; the N-terminal one-third (aa 1 to 50) is not required. Using the alanine scanning mutagenesis strategy, we demonstrated that the regions between aa 52 to 65 and 88 to 154 are important for the augmentation function of HBx in HBV replication. By the luciferase reporter gene analysis, we found that the transactivation and coactivation activities of HBx coincide well with its augmentation function in HBV transcription and replication. These results suggest that HBx has an important role in stimulating HBV transcription and replication and that the transcriptional transactivation function of HBx may be critical for its augmentation effect on HBV replication.
Hepatitis B virus (HBV) is the prototypic member of the Hepadnavirdae family, which contains a group of closely related hepatotropic small DNA viruses that infect their respective animal hosts (13, 29). The HBV genome is a 3.2-kb, circular, partially double-stranded DNA molecule with four overlapping open reading frames (ORFs) named PC-C, PS-S, P, and X (35, 38). Upon HBV infection of the hepatocyte, the HBV genome is converted to covalently closed circular DNA in the nucleus. The covalently closed circular DNA serves as the template for transcription by the host RNA polymerase II, generating the 3.5-, 2.4-, 2.1-, and 0.7-kb viral transcripts that encode the HBV core and polymerase polypeptides, the large surface antigen polypeptide, the middle and major surface antigen polypeptides, and the HBx polypeptide. The expression of those transcripts are directed by four HBV promoters (Cp, PS1p, Sp, and Xp), respectively, and influenced by two HBV enhancers (Enh I and Enh II) (14, 29, 35, 38, 43). HBV replicates by reverse transcription of the viral pregenomic 3.5-kb RNA (pgRNA) using the HBV polymerase that catalyzes RNA-dependent DNA synthesis and DNA-dependent DNA synthesis (36, 40). Besides encoding for the HBV core polypeptide and HBV DNA polymerase that compose the viral capsid, the greater-than-genome-length 3.5-kb pgRNA is also encapsidated and serves as the template for reverse transcription. The encapsidated pgRNA is converted into the 3.2-kb partially double-stranded genomic DNA inside the viral capsid in the cytoplasm of the hepatocytes (35, 36, 38, 40). There are several putative regulatory steps for HBV replication, e.g., pgRNA synthesis, encapsidation of pgRNA, and reverse transcription of pgRNA. As the pgRNA encodes both the HBV polymerase and core polypeptides and serves an additional function as the replication template, regulation of the synthesis of this RNA is therefore a critical step in the viral life cycle (35, 38, 39).
HBV infection is a worldwide health problem and is one of the major causes of hepatocellular carcinoma (HCC). The crucial role of HBV in hepatocarcinogenesis is established, while the mechanism by which HBV causes transformation of hepatocytes remains unclear (1, 2, 6). HBV X protein (HBx) has long been suspected of playing a positive role in hepatocarcinogenesis, as avian hepadnaviruses missing the X ORF seem not to be associated with HCC, and some HBx transgenic mice appear to develop HCC (15, 44) or be more sensitive to a carcinogenic environment. However, the exact molecular mechanism remains to be elucidated. HBx, a 154-amino-acid (154-aa) protein with a N-terminal negative regulatory domain and a C-terminal transactivation or coactivation domain, is a multifunctional protein that exhibits effects on gene transcription, signaling pathways, genotoxic stress responses, protein degradation, cell-cycle control, cell proliferation, and apoptosis (22, 23). These activities may affect viral replication and viral proliferation directly or indirectly and may also be relevant to HBV-associated pathogenesis, especially hepatocarcinogenesis (22, 23).
The biological role(s) of X protein in the viral life cycle has been addressed by several experimental systems. X protein has been shown to be essential for woodchuck hepatitis virus (WHV) replication in woodchucks, as an intact X gene is required for the successful establishment of WHV infection in vivo (7, 48). However, its essential role in the HBV life cycle is somewhat controversial. HBV mutant genomes with a defective X gene are replication competent after being transfected into hepatoma cell lines (3, 42) or HBV transgenic mice models (31, 41). The differences in these experiments may suggest a critical role of X protein for the establishment of virus infection, not for the establishment of viral replication. However, several lines of evidence suggest that HBx can enhance HBV replication in both cell culture and the transgenic mice model ( 5, 21,41, 42, 45), although the mechanism remains uncertain. As HBx has been shown to transactivate a variety of viral and cellular promoters, including the HBV promoters (22, 23, 26, 32), whether the transcriptional transactivation function of HBx is involved in the effect in HBV life cycle is still an open question and needs to be explored.
In an attempt to further address the role, functional domain, and possible mechanism(s) of HBx in the life cycle of HBV, we performed HBV replication assays of the wild-type or HBx-minus HBV genome constructs. A series of truncated and mutated HBx proteins were examined for their capacity to modulate the levels of HBV transcription and replication. We found that HBx has an augmentation effect on HBV 3.5-kb RNA and DNA replication intermediate synthesis. The C-terminal transactivation domain of HBx is necessary and sufficient for this stimulation function. Mutagenesis studies have defined the precise regions that are critical for the regulating effect of HBx on HBV replication. The important sequences for augmentation of HBV replication in the viral replication assay correlated well with those required for the transcriptional transactivation function in the reporter gene analysis.
MATERIALS AND METHODS
Plasmid constructions.
The plasmid payw1.2 is a replication-competent construct that contains 1.2 copies of the wild-type HBV genome (subtype ayw) and expresses HBV pregenomic 3.5-kb RNA under the control of the endogenous promoters of HBV (34). The HBx-minus mutant vector payw*7, which contains an ochre termination signal (CAA to UAA) after codon 7 (at codon 8) in the HBx ORF, was derived from payw1.2 by site-directed mutagenesis (21). In this construct, no mutation was introduced in the overlapping HBV polymerase ORF (21).
The mammalian expression plasmid pSG5UTPL was derived from pSG5 (Stratagene) (20, 25). With pSG5UTPL, a FLAG-tagged expression vector, pNKFLAG, was generated as previously reported (11) and used to express amino-terminally FLAG-tagged proteins. All of the HBx (subtype adr) mammalian expression constructs were derived from pNKFLAG. The pNKF-HBx, pNKF-HBxD1, and pNKF-HBxD5 vectors express full-length HBx (aa 1 to 154), truncated HBx (aa 51 to 154), and truncated HBx (aa 1 to 50), respectively, have been previously described (8, 17, 24). An alanine scanning method was applied to construct a series of HBx clustered alanine substitution mutants (designated Cm) by site-directed mutagenesis. The mutagenesis was carried out by a splicing PCR method with the mutated oligonucleotide primer sets (4). The target sequence of 7-aa residues was changed to AAASAAA, and all of the HBx-encoding DNA fragments bearing the clustered mutations were introduced into the EcoRI and BamHI sites of pNKFLAG, generating the pNKF-Xcm1 to pNKF-Xcm21 constructs. All of the constructs were sequenced by the dideoxy method using the Tag sequencing primer kit and a DNA sequencer (370A; Applied Biosystems, Inc, Ltd.). The mammalian expression vector pSG5UTPL-Gal-VP16 was constructed by inserting the sequence of Gal4-VP16, which encodes the transactivation domain of VP16 fused to the Gal4 DNA binding domain, into the EcoRI and BamHI sites of pSG5UTPL (18).
The luciferase (LUC) reporter gene vector pAP1-Luc contains four tandem copies of the AP1 enhancer fused to a TATA-like promoter (PTAL) region from the herpes simplex virus-thymidine kinase promoter, which drives the expression of the firefly luciferase gene (Clontech). The plasmid pFR-Luc was constructed by cloning the luciferase reporter gene sequence downstream of a basic promoter element (TATA box) and joined to five tandem repeats of the GAL4 binding element (Stratagene). HBV DNA sequences in the HBV promoter reporter gene constructs were derived from the plasmid pCP10, which contains two copies of the HBV genome (subtype ayw) cloned into the EcoRI site of pBR322 (12). The firefly LUC reporter gene in these constructs was derived from the plasmid p19DLUC (30). The plasmid CpLUC contains one complete HBV genome located directly 5′ to the promoterless LUC reporter gene, such that the expression of the LUC gene is governed by the HBV nucleocapsid promoter (30). Similarly, the plasmids XpLUC, SpLUC, and PS1pLUC contain one complete HBV genome located directly 5′ to the promoterless LUC reporter gene, such that the expression of the LUC gene is governed by the HBV enhancer 1/X gene, major surface antigen, and large surface antigen promoters, respectively (30).
Cells and transfections.
The human hepatoma cell line HepG2 and HepG2/HBx cell line were grown in RPMI 1640 medium and 10% fetal bovine serum at 37°C in 5% CO2 in air. The HepG2/HBx cell line is a monoclonal HepG2 cell line with stable HBx expression, which was established by introducing HBx expression retrovirus derived from pBabeBlas-FlagHBx and selected and maintained in the presence of 4 μg of blasticidin S (Funakoshi Co., Ltd)/ml. The expression of HBx in this cell line was confirmed by Western blot analysis (see below). Cells were transfected with purified plasmids by the standard calcium phosphate method. The total DNA used for each transfection experiment was adjusted to an equal amount with the appropriate plasmid vector.
Transfections for viral RNA and DNA analysis were performed using 10-cm plates, containing approximately 106 cells. DNA and RNA isolation was performed 3 days posttransfection. Except where indicated in the figure legends, the transfected DNA mixture was composed of 5 μg of payw1.2 or payw*7 plus 0.5 μg of the HBx expression vectors. Controls were derived from cells transfected with HBV DNA and the pNKFLAG expression vector lacking the HBx coding sequence.
Transfections for HBx protein analysis were performed using 10-cm plates containing approximately 106 cells and 20 μg of wild-type or mutated HBx expression plasmids. Cell lysates were prepared 2 days posttransfection and used for Western blot analysis.
Transfections using LUC reporter gene constructs were performed in six-well plates containing approximately 3 × 105 HepG2 cells per well. The transfected DNA mixture comprised 5 μg of a LUC plasmid and 0.25 μg of pRL-CMV, which served as an internal control for transfection efficiency. pRL-CMV directs the expression of the Renilla LUC gene with the cytomegalovirus immediate-early promoter (Promega). When appropriate, the DNA mixture also included 0.5 μg of the HBx expression vectors or the control expression vector. In the coactivation experiment, 1 ng of pSG5UTPL-Gal-VP16 was also added to the mixture as indicated in the figure legends.
Characterization of HBV transcripts and viral replication intermediates.
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described previously (37) with some modifications. Total cellular RNA was isolated with the RNeasy Mini Kit (QIAGEN) as instructed by the manufacturer. For the isolation of viral DNA replication intermediates, the cells were lysed in 0.4 ml of 100 mM Tris hydrochloride (pH 8.0,) and 0.2% (vol/vol) NP-40. The lysate was centrifuged for 1 min at 14,000 rpm in a microcentrifuge to pellet the nuclei. The supernatant was adjusted to 6.75 mM magnesium acetate plus 200 μg of DNase I/ml and incubated for 1 h at 37°C to remove the transfected plasmid DNA. The supernatant was readjusted to 100 mM NaCl, 10 mM EDTA, 0.8% (wt/vol) sodium lauryl sulfate, and 1.6 mg of pronase/ml; it was incubated for an additional 1 h at 37°C. The supernatant was extracted twice with phenol, precipitated with 2 volumes of ethanol, and resuspended in 100 μl of 10 mM Tris hydrochloride (pH 8.0)-1 mM EDTA.
RNA (Northern) and DNA (Southern) filter hybridization analyses were performed with 10 μg of total cellular RNA and 30 μl of viral DNA replication intermediates, respectively, as previously described (33). The 32P-labeled full-length HBV DNA probe was used in both Northern and Southern filter hybridization analyses. Results of filter hybridization were quantitated with a phosphorimaging analyzer (BAS2000; Fuji Film Co., Ltd.).
Cell extracts and Western blot analysis.
Transfected HepG2 cells or HepG2/HBx cells were washed with phosphate-buffered saline; harvested in LAC buffer, which contained 20 mM HEPES (pH7.9), 50 mM KCl, 400 mM NaCl, 1 mM EDTA, 9 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 10% glycerol, 1 mM dithiothreitol, 10 mM aprotinin and leupetin, and 1 mM phenylmethylsulfonyl fluoride; and lysed by sonication. Twenty micrograms of protein from the cell lysates was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto a nitrocellulose membrane, and subjected to Western blot analysis with the anti-FLAG M2 antibody (Sigma). The proteins were visualized with the enhanced chemiluminescence (ECL) kit according to the manufacturer's instructions (Amersham Pharmacia Biotech).
Luciferase assay.
Total cell lysates were prepared from cells harvested 40 to 48 h after transfection. Luciferase assays were performed with the Dual-Luciferase Reporter Assay system (Promega) as instructed by the manufacturer. The firefly luciferase activities were normalized to the level of Renilla luciferase activity in each transfection experiment.
RESULTS
HBx augments HBV transcription and replication in HepG2 cells.
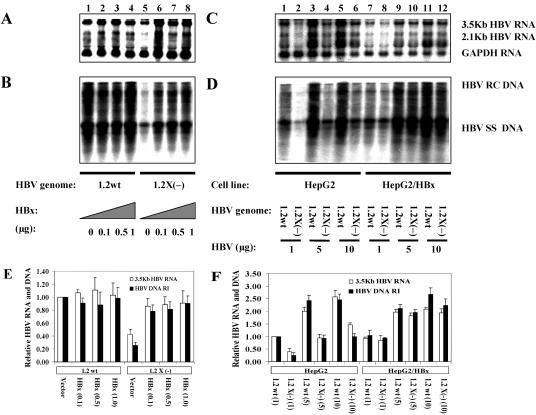
To study the effect of HBx on HBV replication, the HBV RNA transcripts and HBV DNA replication intermediates synthesized from a wild-type HBV genome and a HBx-minus HBV genome were initially characterized and compared in the human hepatoma cell line HepG2 (Fig. 1). Both wild-type and HBx-minus HBV DNAs were transcribed and replicated in HepG2 cells (Fig. 1), suggesting that both were replication-competent genomes and that the HBx protein was not essential for the establishment of HBV replication, as the HBx-minus HBV construct could still support HBV replication while lacking the expression of HBx. However, the level of viral replication intermediates synthesized from the HBx-minus HBV DNA construct was two- to fourfold lower than that of the replication intermediates synthesized from the wild-type HBV construct (Fig. 1B, lanes 1 and 5; 1D, lanes 1 and 2, 3 and 4, 5 and 6; 1E and F). The finding is consistent with previous observations (5, 21, 41, 45) although the reduction fold was lower than that described in some reports. In addition, about a 1.5- to 2.5-fold decrease in the level of 3.5-kb HBV RNA was also observed from the HBx-minus construct compared with the wild-type construct, and this correlated with the reduction in HBV replication (Fig. 1A, lanes 1 and 5; 1C, lanes 1 and 2, 3 and 4, 5 and 6; 1E and F). These results indicate that HBx has an augmentation role on HBV transcription and replication, as the lack of HBx expression led to decreased HBV RNA and HBV DNA synthesis in the HepG2 cells.
FIG. 1.
Augmentation effects of HBx on HBV transcription and replication. (A and B) Human hepatoma HepG2 cells were transiently transfected with the wild-type HBV construct payw1.2 (1.2wt, lanes 1 to 4) or the HBx-minus HBV construct payw*7 [1.2X(−), lanes 5 to 8], plus the control vector pNKFLAG (lanes 1 and 5), or various amount of HBx expression vector pNKF-HBx (0.1, 0.5, and 1 μg) (lanes 2 to 4 and 6 to 8, respectively). (C and D) Human hepatoma HepG2 cells (HepG2, lanes 1 to 6) and stable HBx expression HepG2 cells (HepG2/HBx, lanes 7 to 12) were transiently transfected with the wild-type HBV construct payw1.2 (1.2wt, lanes 1, 3, 5, 7, 9, and 11) or the HBx-minus HBV construct payw*7 [1.2X(−), lanes 2, 4, 6, 8, 10, and 12]. Various amounts of HBV DNA constructs were used in this transfection experiment (1 μg, lanes 1, 2, 7, and 8; 5 μg, lanes 3, 4, 9, and 10; and 10 μg, lanes 5, 6, 11, and 12). (A and C) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B and D) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (E and F) Quantitative analysis of the 3.5-kb HBV RNA and HBV DNA replication intermediates. The levels of the 3.5-kb HBV RNA and HBV DNA replication intermediates (HBV DNA RI) are reported relative to those of the wild-type HBV construct payw1.2 plus the control vector (lane 1 of panels A and B) (E) or to those of the HepG2 cells transfected with 1 μg of payw1.2 (panels C and D, lanes 1) (F), which are set at 1.0. The mean RNA and DNA levels plus standard deviation (indicated by error bars) from two independent analyses are shown. The constructs and cells used in the study are shown below the graph.
To confirm that the reduction of HBV transcription and replication was really caused by the absence of HBx expression, we tested whether these decreases could be restored by ectopic expression of HBx (Fig. 1). By cotransfection of the HBx expression plasmid, the levels of 3.5-kb HBV RNA and HBV replication intermediates synthesized from the wild-type HBV construct were not significantly affected (Fig. 1A and B, lanes 1 and 2 to 4; 1E), while the decreased levels of 3.5-kb HBV RNA and HBV replication intermediates synthesized from the HBx-minus HBV construct were restored to levels similar to that observed with the wild-type construct (Fig. 1A and B, lanes 5 and 6 to 8; 1E). Consistent with this result, no significant differences in the 3.5-kb HBV RNA and HBV replication intermediates were observed between wild-type and HBx-minus HBV constructs when transfected into a stable HBx expression HepG2 cell line (Fig. 1C and D, lanes 7 and 8, 9 and 10, 11 and 12; 1F), indicating that the effect of HBx was complemented by the stable expression of HBx in the cells. These results clearly show that not only transiently but also stable expressed HBx can complement in trans to stimulate HBV RNA synthesis and HBV DNA replication.
The C-terminal transactivation domain of HBx is responsible for its augmentation effect on HBV transcription and replication.
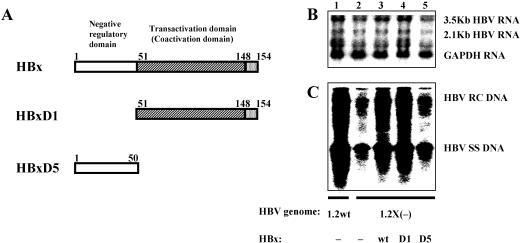
The full-length HBx protein composed by 154 aa has an N-terminal negative regulatory domain and a C-terminal transactivation or coactivation domain (Fig. 2A). In an attempt to determine the functional domain(s) of HBx needed for its stimulatory effect on HBV replication, viral transcription and replication were examined in HepG2 cells with full-length (HBx) and two truncated HBx proteins lacking either the N-terminal domain (HBxD1) or C-terminal domain (HBxD5) (Fig. 2A). Similar to the results showed above (Fig. 1), the reduction of HBV transcription and replication from the HBx-minus HBV construct could be restored by the full-length HBx (Fig. 2B and C, lanes 1 to 3). It was apparent that the N-terminal negative regulatory domain of HBx was not required for its stimulation function on HBV RNA synthesis and HBV DNA replication, as the truncated HBx protein (HBxD1) lacking the N-terminal one-third (aa 1 to 50) activated HBV replication to an extent similar to that of the full-length HBx (Fig. 2B and C, lanes 3 and 4). In contrast, the augmentation effect on HBV transcription and replication was defected when the C-terminal two-thirds (aa 51 to 154), which contains the transactivation domain, was deleted (Fig. 2B and C, lanes 5). The functional deficiency of HBxD5 is not caused by the lack of protein expression, as a similar level of this truncated HBx protein was detected by Western blot analysis compared with that of the full-length HBx (24). This indicates that the C-terminal transactivation domain of HBx is necessary and sufficient for its augmentation function in HBV replication as well as 3.5-kb HBV RNA synthesis.
FIG. 2.
The C-terminal transactivation domain of HBx is sufficient for the stimulation effect on HBV transcription and replication. (A) Schematic representations of the HBx protein showing the locations of the amino-terminal negative regulatory domain and the carboxyl-terminal transactivation (also called the coactivation) domain (22, 23). The amino acids of the full-length HBx (154 aa residues) and the truncated HBx (HBxD1 and HBxD5, spanning aa residues 51 to 154 and 1 to 50, respectively) are shown. (B and C) HepG2 cells were transiently transfected with the wild-type HBV construct payw1.2 (1.2wt, lane 1) or the HBx-minus HBV construct payw*7 [1.2X(−), lanes 2 to 5] plus the empty vector control (−, lanes 1 and 2) or full-length HBx expression vector (wt, lane 3) or different truncated HBx expression vectors (D1, lane 4; D5, lane 5). (B) RNA (Northern) filter hybridization analysis of HBV transcripts. The GAPDH transcript was used as an internal control for RNA loading per lane. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA.
Identification of the HBx regions involved in regulating HBV transcription and replication.
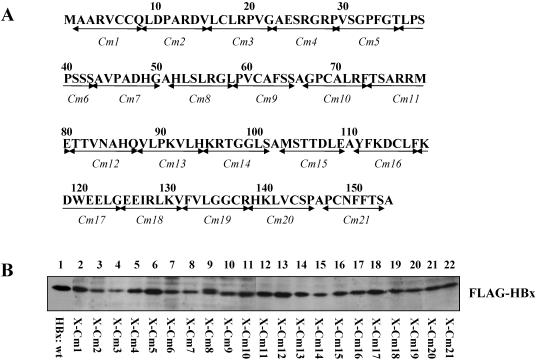
To further investigate the precise regions of HBx involved in the stimulation role in HBV transcription and replication, we constructed a series of HBx clustered alanine substitution mutants. For each mutant, seven consecutive amino acids were replaced by the sequence AAASAAA. Twenty-one of these clustered mutants (Cm1 to Cm21) were made to cover the whole sequence of HBx, with Cm1 to Cm7 covering the N-terminal negative regulatory domain and Cm8 to Cm21 covering the C-terminal transactivation or coactivation domain (Fig. 3A). Initially, the expression levels of these HBx mutants were checked in HepG2 cells. Western blot analysis demonstrated that all of the mutated HBx proteins were expressed, although with slightly different levels of expression (Fig. 3B). However, the mutated proteins HBx-Cm3 and HBx-Cm5 displayed the lowest and the highest expression levels, respectively (Fig. 3B, lanes 4 and 6), and were completely functional with respect to modulating HBV transcription and replication (Fig. 4 and 5). Therefore, it appears that any differences in the level of expression of the mutated HBx proteins used in this study did not influence greatly the function of these HBx proteins.
FIG. 3.
Expression of mutated HBx proteins in HepG2 cells. (A) Schematic representations of a series of clustered alanine substitution mutants (Cm1 to Cm21) of HBx. The amino acid locations of the clustered mutations are shown. (B) Detection of expression of the mutated HBx proteins. Total cell lysates prepared from HepG2 cells transfected with an expression vector encoding the wild-type HBx (HBx:wt, lane 1), and mutant HBx expression vectors (X-Cm1 to X-Cm21, lanes 2 to 22, respectively) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western blot analysis with anti-FLAG M2 antibody.
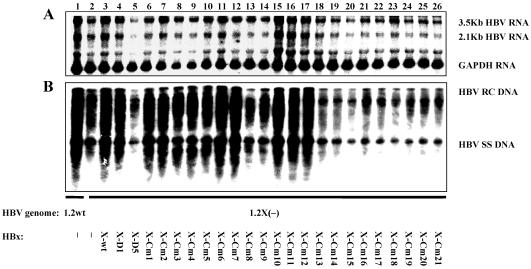
FIG. 4.
Mapping of regions of HBx required for the activation effect on HBV transcription and replication. HepG2 cells were transiently transfected with the wild type HBV construct payw1.2 (1.2wt, lane 1) or the HBx-minus HBV construct payw*7 [1.2X(−), lanes 2 to 26] plus empty vector control (−, lanes 1 and 2), full-length HBx expression vector (X-wt, lane 3), different truncated HBx expression vectors (X-D1, lane 4; X-D5, lane 5), or a series of clustered mutant HBx expression vectors (X-Cm1 to X-Cm21, lanes 6 to 26, respectively). (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The GAPDH transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA.
FIG. 5.
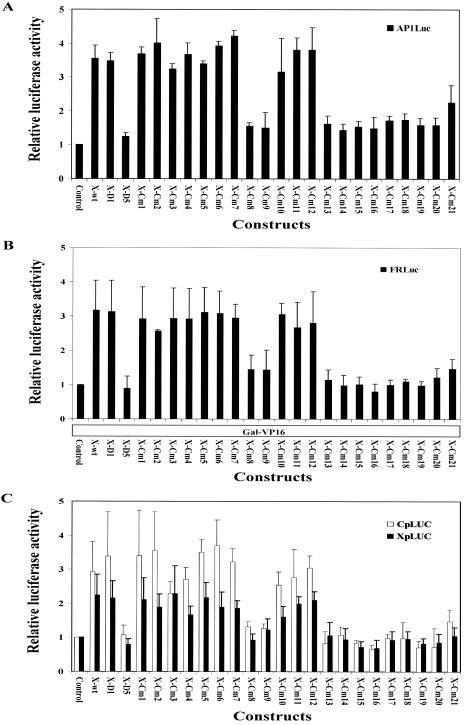
Analysis of the transactivation and coactivation activities of truncated and clustered mutated HBx proteins. (A and C) The effects of truncated and mutated HBx on transcription from the AP1 enhancer-herpes simplex virus-thymidine kinase promoter construct pAP1-Luc (A) and from the HBV nucleocapsid and enhancer 1/X gene promoter constructs CpLUC and XpLUC, respectively (C), were examined. Relative luciferase activities of the LUC constructs in HepG2 cells in the absence or presence of ectopically expressed wild-type, truncated, and mutated HBx proteins are indicated. The expression vectors for the HBx proteins are indicated below the graph. The luciferase activities are reported relative to those of the AP1-Luc, CpLUC, and XpLUC constructs in the absence of HBx expression (control), with a relative activity set at 1.0. The mean luciferase activities plus standard deviations (indicated by error bars) from three independent experiments are shown. (B) The coactivation activities of truncated and mutated HBx. HepG2 cells were transfected with the GAL4 binding element containing LUC construct pFR-Luc and the Gal4-VP16 activator expression construct pSG5UTPL-Gal4-VP16, plus wild-type, truncated, or mutated HBx expression vectors (as indicated below the graph). HBx coactivation activity was determined by luciferase assay. The luciferase activities are reported relative to the pFR-Luc plus Gal-VP16 constructs in the absence of HBx expression (control), with relative activity set at 1.0. The mean luciferase activities plus standard deviations (indicated by the error bars) from three independent analyses are shown.
Using this series of HBx cm mutants, we next addressed what sequence(s) is critical for the activation function of HBx by examining the complementation effect of these mutated HBx proteins on HBV transcription and replication in HepG2 cells (Fig. 4). The HBx mutants Cm1 to Cm7, which contain mutations spanning aa 2 to 50, retained the ability to complement the augmentation effect to a similar extent as the wild-type HBx (Fig. 4, lanes 3 and 6 to 12). This is consistent with the mapping result using the truncated form of HBx (Fig. 2) and confirms that the N-terminal negative regulatory region is dispensable for the activation effect of HBx. The HBx mutants Cm10 to Cm12, with mutations between aa 67 and 87, also retained the ability to complement the augmentation effect of HBx (Fig. 4, lanes 15 to 17). This result suggests that the sequences between aa 67 and 87 are not necessary for the activation function of HBx, although this region is located within the transactivation domain. In contrast, HBx mutants Cm8 to Cm9 and Cm13 to Cm21, which span aa 52 to 65 and 88 to 154, respectively, and cover the majority of the transactivation domain, were unable to restore the augmentation function of HBx (Fig. 4, lanes 13, 14, and 18 to 26). These observations indicate that the regions between aa 52 to 65 and 88 to 154, which are located within the C-terminal transactivation domain, are important for the augmentation function of HBx in HBV replication.
The transcriptional transactivation function(s) of HBx may be critical for the augmentation role in HBV replication.
Based on the observations showed above, the results implied that HBx augments HBV replication through modulation of HBV transcription. To examine whether the function of HBx in HBV replication is mediated through its transactivation activities, a luciferase assay was performed to determine if the stimulatory effect of HBx on HBV replication is correlated with its transactivation and coactivation function (Fig. 5). Initially, the transactivation activities of the wild-type, truncated, and mutated HBx were analyzed using a pAP1-Luc reporter gene construct (Fig. 5A). The results showed that ectopic expression of wild-type HBx activates luciferase activity about fourfold, the truncated HBx D1 and mutated HBx Cm1 to Cm7 and Cm10 to Cm12 retained transactivation ability, mutated HBx Cm21 showed partially transactivation activity, and the truncated HBx D5 and all the other mutated HBx had no transactivation function (Fig. 5A). It is important to note that the pattern of the transactivation activity of HBx mutants correlates with its function in stimulating HBV replication (Fig. 4). The HBx proteins with transactivation activity are those that can augment HBV replication. In addition, a similar pattern of the coactivation function was also observed with those mutated HBx proteins when pFR-Luc was used as a reporter and Gal4-VP16 was used as an activator (Fig. 5B).
Since analysis with the artificial promoters may not precisely reflect the real nature of HBx in modulating HBV transcription, in an attempt to characterize further the effect of these mutated HBx proteins on HBV promoter activity, we performed a luciferase assay using HBV promoter reporter gene constructs, which contain the complete HBV genome located upstream of the LUC gene (Fig. 5C). Ectopic expression of HBx activates transcription from the nucleocapsid and enhancer 1/X gene promoter approximately four- and threefold in constructs CpLUC and XpLUC, respectively (Fig. 5C). With all of the HBx proteins used in this study, no significant effects were observed on the HBV major surface antigen and large surface antigen promoter activities by using SpLUC and PS1pLUC reporter gene constructs (H. Tang and S. Murakami, unpublished data). The patterns of activation are similar to those observed with pAP1-Luc and pFR-Luc and most importantly are correlated with the augmentation function of HBx in the HBV replication assay (Fig. 4 and 5). Therefore, it is apparent that the regions between aa 52 to 65 and 88 to 154 located within the C-terminal transactivation domain of HBx mediated both the activation of transcription from the nucleocapsid and enhancer 1/X gene promoters of HBV (by a reporter gene assay) and the activation of viral transcription and replication (by the HBV replication assay) in HepG2 cells. These observations supported the suggestion that the transcriptional transactivation function(s) of HBx may be important for its augmentation role in HBV replication, and HBx may augment HBV replication by activating the HBV promoter(s).
DISCUSSION
HBx has been the focus of much attention in recent years, because it has been suspected to be an important factor in hepatocarcinogenesis. Although many effects have been reported to be related to HBx, including transactivation of many viral and cellular genes, stimulation of various signal transduction pathways, binding to a variety of protein targets, modulation of protein degradation, and DNA repair and apoptosis (22, 23, 35), the physiological role of X protein during a natural viral infection is not well understood. Despite a well-established function of the X protein in the WHV life cycle (7, 48), there are conflicting reports about the relevance of HBx in the life cycle of HBV (3, 31, 41, 42). In this study, the effect of HBx on HBV transcription and replication was initially characterized by a HBV replication assay (Fig. 1). This analysis demonstrated that HBx has an augmentation role in HBV transcription and replication, which can be complemented by ectopically expressed HBx in trans (Fig. 1). These observations are consistent with several reports regarding the regulation of HBV DNA replication (5, 21, 41, 42, 45) but distinct from some reported results that suggested the positive role of HBx on virus replication could be distinguished from its effect on HBV transcription (5, 45). The reasons for these differences are unclear. However, by comparison with the wild-type construct, we reproducibly observed a decreased level of 3.5-kb HBV RNA that is associated with reduced HBV DNA replication for the HBx-minus HBV construct, and this reduction can be recovered by providing HBx in trans. Also, this result is consistent with a recent study of transgenic mice (41), suggesting that the augmentation of HBV DNA replication by HBx was most likely the result of activation of HBV RNA transcription. In addition, the effect of HBx on HBV replication appears not to be mediated by altering of the precore to pgRNA ratio, as the precore/core RNA ratio was not significantly affected by HBx expression (Tang and Murakami, unpublished).
Although HBx is not essential for the establishment of HBV replication in the transient transfection system (Fig. 1), it is possible that HBx is needed to initiate the infection. In fact, the augmentation effect of HBx on HBV replication may be important for the early step of natural infection, as very low levels of virus may be more easily eliminated by the immune system. This speculation is supported by a recent report that WHV defective in the X gene may replicate at a low level in vivo (47); the inability of the WHV genome with a defect of the X gene to initiate infection in woodchucks may due to its lower replication rate. As the initial infection step of the viral life cycle cannot be analyzed by the transient transfection system, additional studies with HBV infection in cell culture or animal models will be necessary to address this possibility.
HBx comprises a negative regulatory domain at its N terminus and a transactivation domain (also called the coactivation domain) at its C terminus. As HBx has no known direct DNA binding property, therefore, it is not a typical transactivator. Its effects on transcription are thought to be mediated through protein-protein interaction with endogenous cellular proteins, transcription factors and cofactors, or basal transcription machinery (22, 23, 35). Analysis with the truncated HBx proteins indicated the C-terminal transactivation domain is required and that the N-terminal domain is dispensable for the augmentation effects of HBx on HBV transcription and replication (Fig. 2). Mutagenesis studies with the clustered alanine substitution mutant library further confirmed this result and demonstrated that two regions within the C-terminal domain covering aa 52 to 65 and 88 to 154 are critical for the stimulation of HBV replication (Fig. 4). These results are not surprising, as most of the protein targets of HBx are reported to interact through the C-terminal domain, which is responsible for the transactivation and coactivation properties of HBx (18, 22, 23), while the role of the N-terminal regulatory domain remains elusive (24, 27). Importantly, HBx showed stimulation effect not only on HBV replication but also on 3.5-kb HBV RNA under our conditions, although the stimulation varied to some extent between experiments (Fig. 1, 2, and 4). Furthermore, exactly the same sequences of HBx are critical for its transactivation and coactivation ability, as examined by reporter gene analysis (Fig. 5). These results strongly support the notion that the stimulatory effect on HBV replication is due to the transcriptional modulatory activity of HBx. However, it may also be possible that the same surface of HBx serves for the different functions or that the clustered substitution analysis is too crude to identify the diverse functions. Further studies with a point mutant library will be needed in future to define the important amino acid residues of HBx that mediate the transactivation activity and the augmentation effect on HBV transcription and replication.
The molecular mechanisms by which HBx attains its biological functions in HBV replication remain largely unresolved. Several groups have reported a positive role(s) of HBx in HBV replication with different molecular mechanisms, including activation of the calcium signaling cascade (5), proteasome modulation (45), and transcriptional modulation (32, 41), but the exact mechanism(s) of HBx activation on HBV replication is controversial. HBV has a distinct process of viral replication among DNA viruses, since its genetic information is converted to RNA and replicates through reverse transcription. HBV replication can be regulated at many steps and by a variety of factors. The initial step in HBV replication is synthesis of pgRNA transcribed by host RNA polymerase II, controlled by distal and proximal promoter cis elements where a variety of ubiquitous and liver-enriched transcriptional factors have been shown to be involved (19, 28, 38, 39, 46). HBx has been shown to act as a transcriptional coactivator but not as an activator, and the previously described transactional function of HBx may in fact be a reflection of its coactivator function (18). This interpretation is clearly supported by the present results, demonstrating that the same sequences in the C-terminal domain are required for coactivation and transactivation function (Fig. 4 and 5). Also, there are several reports showing the direct interaction of HBx with some transcription factors that can bind to the HBV core promoter, such as RXR (16), PPARγ (9), and C/EBPα (10). Therefore, it is possible that the step of pgRNA synthesis would be modulated by the presence of HBx. Functional analysis using HBV promoter reporter gene constructs demonstrated that the same sequences within the C-terminal transactivation domain were responsible for stimulating HBV transcription and replication in the viral replication analysis and for activating transcription from the nucleocapsid promoter in the reporter gene analysis (Fig. 4 and 5). These results argue that one possible mechanism of HBx action on HBV transcription and replication is mediated through activation of the HBV core promoter. However, the possibility that HBx activates the transcription of a cellular gene(s) that affects the rate of turnover of the HBV transcripts cannot be excluded.
Regardless of the mechanisms, the data clearly demonstrate that HBx has an augmentation role on HBV transcription and replication and that the transcriptional transactivation function of HBx is important for the augmentation effect. Further studies will be needed to define the detailed factor(s) and step(s) that HBx works through or that work through HBx to regulate the transcription and replication of HBV.
Acknowledgments
We are grateful to Margherita Melegari (Massachusetts General Hospital Cancer Center, Charlestown, Mass.) for plasmids payw1.2 and payw*7; Alan McLachlan (The Scripps Research Institute, La Jolla, Calif.) for plasmids CpLUC, XpLUC, SpLUC, and PS1pLUC; and Y. Hirose (The Cancer Research Institute of Kanazawa University, Kanazawa, Japan) for plasmid pFR-Luc. We thank Alan McLachlan for many helpful discussions and critical reading of the manuscript. We thank M. Yasukawa and K. Kuwabara for their technical assistance.
This work was supported by a National Science Fund for Distinguished Young Scholars (grant no. 30325036) from the National Natural Science Foundation of China and a research fellowship from the Japan Society for the Promotion of Science to H.T. and by Grants-in-aid for Scientific Research (B) and Development and a Grant-in-Aid for Scientific Research on Priority Areas (C) in oncogenesis from the Ministry of Education, Sports, Culture, and Technology.
REFERENCES
- 1.Arbuthnot, P., and M. Kew. 2001. Hepatitis B virus and hepatocellular carcinoma. Int. J. Exp. Pathol. 82:77-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beasley, R. P. 1988. Hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer 61:1942-1956. [DOI] [PubMed] [Google Scholar]
- 3.Blum, H. E., Z. S. Zhang, E. Galun, F. von Weizsacker, B. Garner, T. J. Liang, and J. R. Wands. 1992. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 66:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordo, D., and P. Argos. 1991. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 217:721-729. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 6.Buendia, M. A. 2000. Genetics of hepatocellular carcinoma. Semin. Cancer Biol. 10:185-200. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H. S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheong, J. H., M. Yi, Y. Lin, and S. Murakami. 1995. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, Y. H., H. Kim, J. K. Seong, D. Y. Yu, H. Cho, M. O. Lee, J. M. Lee, Y. H. Ahn, S. J. Kim, and J. H. Park. 2004. Hepatitis B virus X protein modulates peroxisome proliferator-activated receptor γ through protein-protein interaction. FEBS Lett. 557:73-80. [DOI] [PubMed] [Google Scholar]
- 10.Choi, B. H., G. T. Park, and H. M. Rho. 1999. Interaction of hepatitis B virus X protein and CCAAT/enhancer-binding protein α synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J. Biol. Chem. 274:2858-2865. [DOI] [PubMed] [Google Scholar]
- 11.Dorjsuren, D., Y. Lin, W. Wei, T. Yamashita, T. Nomura, N. Hayashi, and S. Murakami. 1998. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol. Cell. Biol. 18:7546-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois, M. F., C. Pourcel, S. Rousset, C. Chany, and P. Tiollais. 1980. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc. Natl. Acad. Sci. USA 77:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 3087. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 14.Ganem, D., and H. E. Varmus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 15.Kim, C. M., K. Koike, I. Saito, T. Miyamura, and G. Jay. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351:317-320. [DOI] [PubMed] [Google Scholar]
- 16.Kong, H. J., S. H. Hong, M. Y. Lee, H. D. Kim, J. W. Lee, and J. Cheong. 2000. Direct binding of hepatitis B virus X protein and retinoid X receptor contributes to phosphoenolpyruvate carboxykinase gene transactivation. FEBS Lett. 483:114-118. [DOI] [PubMed] [Google Scholar]
- 17.Lin, Y., T. Nomura, J. H. Cheong, D. Dorjsuren, K. Iida, and S. Murakami. 1997. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcriptional factor IIB and the RNA polymerase II subunit 5. J. Biol. Chem. 272:7132-7139. [DOI] [PubMed] [Google Scholar]
- 18.Lin, Y., H. Tang, T. Nomura, D. Dorjsuren, N. Hayashi, W. Wei, T. Ohta, R. Roeder, and S. Murakami. 1998. The hepatitis B virus X protein is a co-activator of activated transcription that modulates the transcription machinery and distal binding activators. J. Biol. Chem. 273:27097-27103. [DOI] [PubMed] [Google Scholar]
- 19.López-Cabrera, M., J. Letovsky, K.-Q. Hu, and A. Siddiqui. 1990. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc. Natl. Acad. Sci. USA 87:5069-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahe, Y., N. Mukaida, K. Kuno, M. Akiyama, N. Ikeda, K. Matsushima, and S. Murakami. 1991. Hepatitis B virus X protein transactivates human interleukin-8 gene through acting on nuclear factor kB and CCAAT/enhancer-binding protein-like cis-elements. J. Biol. Chem. 266:13759-13763. [PubMed] [Google Scholar]
- 21.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Cloning and characterization of a novel hepatitis B virus X binding protein that inhibits viral replication. J. Virol. 72:1737-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami, S. 1999. Hepatitis B virus X protein: structure, function and biology. Intervirology 42:81-99. [DOI] [PubMed] [Google Scholar]
- 23.Murakami, S. 2001. Hepatitis B virus X protein: multifunctional viral regulator. J. Gastroenterol. 36:651-660. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, S., J. H. Cheong, and S. Kaneko. 1994. Human hepatitis virus X gene encodes a regulatory domain that represses transactivation of X protein. J. Biol. Chem. 269:15118-15123. [PubMed] [Google Scholar]
- 25.Murakami, S., J. H. Cheong, S. Ohno, K. Matsushima, and S. Kaneko. 1994. Transactivation of human hepatitis B virus X protein, HBx, operates through a mechanism distinct from protein kinase C and okadaic acid activation pathways. Virology 199:243-246. [DOI] [PubMed] [Google Scholar]
- 26.Nakatake, H., O. Chisaka, S. Yamamoto, K. Matsubara, and R. Koshy. 1993. Effect of X protein on transactivation of hepatitis B virus promoters and on viral replication. Virology 195:305-314. [DOI] [PubMed] [Google Scholar]
- 27.Noh, E. J., H. J. Jung, G. Jeong, K. S. Choi, H. J. Park, C. H. Lee, and J. S. Lee. 2004. Subcellular localization and transcriptional repressor activity of HBx on p21 (WAF1/Cip1) promoter is regulated by ERK-mediated phosphorylation. Biochem. Biophys. Res. Commun. 319:738-745. [DOI] [PubMed] [Google Scholar]
- 28.Raney, A. K., J. L. Johnson, C. N. A. Palmer, and A. McLachlan. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 71:1058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raney, A. K., and A. McLachlan. 1991. The biology of hepatitis B virus, p. 1-37. In A. McLachlan (ed.), Molecular biology of the hepatitis B virus. CRC Press, Inc., Boca Raton, Fla.
- 30.Raney, A. K., D. R. Milich, A. J. Easton, and A. McLachlan. 1990. Differentiation specific transcriptional regulation of the hepatitis B virus large surface antigen gene in human hepatoma cell lines. J. Virol. 64:2360-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reifenberg, K., P. Nusser, J. Lohler, G. Spindler, C. Kuhn, F. von Weizsacker, and J. Kock. 2002. Virus replication and virion export in X-deficient hepatitis B virus transgenic mice. J. Gen. Virol. 83:991-996. [DOI] [PubMed] [Google Scholar]
- 32.Reifenberg, K., H. Wilts, J. Lohler, P. Nusser, R. Hanano, L. G. Guidotti, F. V. Chisari, and H. J. Schlicht. 1999. The hepatitis B virus X protein transactivates viral core gene expression in vivo. J. Virol. 73:10399-10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Scaglioni, P. P., M. Melegari, and J. R. Wands. 1997. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J. Virol. 71:345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 37.Summers, J., P. M. Smith, M. J. Huang, and M. S. Yu. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 65:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, H., K. E. Banks, A. L. Anderson, and A. McLachlan. 2001. Hepatitis B virus transcription and replication. Drug News Perspect. 14:325-334. [PubMed] [Google Scholar]
- 39.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 98:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Will, H., W. Reiser, T. Weimer, E. Pfaff, M. Buscher, R. Sprengel, R. Cattaneo, and H. Schaller. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, Z., T. S. B. Yen, L. Wu, C. R. Madden, W. Tan, B. L. Slagle, and J. H. Ou. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaginuma, K., Y. Shirakata, M. Kobayashi, and K. Koike. 1987. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc. Natl. Acad. Sci. USA 84:2678-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen, T. S. B. 1993. Regulation of hepatitis B virus gene expression. Semin. Virol. 4:33-42. [Google Scholar]
- 44.Yu, D. Y., H. B. Moon, J. K. Son, S. Jeong, S. L. Yu, H. Yoon, Y. M. Han, C. S. Lee, J. S. Park, C. H. Lee, B. H. Hyun, S. Murakami, and K. K. Lee. 1999. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J. Hepatol. 31:123-132. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Z., U. Protzer, Z. Hu, J. Jacob, and T. J. Liang. 2004. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J. Virol. 78:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, P., A. K. Raney, and A. McLachlan. 1993. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J. Virol. 67:1472-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Z., N. Torii, Z. Hu, J. Jacob, and T. J. Liang. 2001. X-dificient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Investig. 108:1523-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]