Summary
Background
Simian immunodeficiency viruses (SIV) have been jumping between non-human primates in West/Central Africa for thousands of years and yet, the HIV-1 epidemic only originated from a primate lentivirus over 100 years ago.
Methods
This study examined the replicative fitness, transmission, restriction, and cytopathogenicity of 22 primate lentiviruses in primary human lymphoid tissue and both primary human and chimpanzee peripheral blood mononuclear cells.
Findings
Pairwise competitions revealed that SIV from chimpanzees (cpz) had the highest replicative fitness in human or chimpanzee peripheral blood mononuclear cells, even higher fitness than HIV-1 group M strains responsible for worldwide epidemic. The SIV strains belonging to the “HIV-2 lineage” (including SIVsmm, SIVmac, SIVagm) had the lowest replicative fitness. SIVcpz strains were less inhibited by human restriction factors than the “HIV-2 lineage” strains. SIVcpz efficiently replicated in human tonsillar tissue but did not deplete CD4+ T-cells, consistent with the slow or nonpathogenic disease observed in most chimpanzees. In contrast, HIV-1 isolates and SIV of the HIV-2 lineage were pathogenic to the human tonsillar tissue, almost independent of the level of virus replication.
Interpretation
Of all primate lentiviruses, SIV from chimpanzees appears most capable of infecting and replicating in humans, establishing HIV-1. SIV from other Old World monkeys, e.g. the progenitor of HIV-2, replicate slowly in humans due in part to restriction factors. Nonetheless, many of these SIV strains were more pathogenic than SIVcpz. Either SIVcpz evolved into a more pathogenic virus while in humans or a rare SIVcpz, possibly extinct in chimpanzees, was pathogenic immediately following the jump into human.
Funding
Support for this study to E.J.A. was provided by the NIH/NIAID R01 AI49170 and CIHR project grant 385787. Infrastructure support was provided by the NIH CFAR AI36219 and Canadian CFI/Ontario ORF 36287. Efforts of J.A.B. and N.J.H. was provided by NIH AI099473 and for D.H.C., by VA and NIH AI AI080313.
Keywords: HIV-1, Zoonosis, Primate lentiviruses, Pathogenesis
Research in context.
Evidence before this study
Invasion of wild animal habitats by humans can have devastating consequences for the human population as evident by the HIV-1 and SARS-CoV-2 pandemics. Simian immunodeficiency virus (SIV) jumped into humans over 100 years ago from chimpanzees. There have been numerous studies on different evolutionary pathways of primate lentiviruses and the potential for cross-species transmission based primarily on viral genomic sequences and inhibition of various restriction factors in nonhuman primates. A comprehensive, phenotypic analyses of the replication efficiency and pathogenicity of available primate lentiviruses as model for human zoonoses has not been performed.
Added value of this study
We examined replicative fitness and pathogenesis of 26 different primate lentiviruses in human and chimpanzee primary lymphoid cells from blood and within tonsils. SIV from specific chimpanzee sub-species and lowland gorillas were the most capable of infecting and replicating in human and Ptv lymphoid cells, but they did not result in the pathogenesis related to disease in humans. In contrast, SIV from Sooty Mangabeys resulting in the HIV-2 epidemic were pathogenic, but they replicated poorly in primary human cells compared to the SIVcpz lineages leading to the HIV-1 pandemic. Previous studies have shown direct correlations between rate of disease progression within infected individuals with the relative HIV-1 replicative fitness coupled to cytopathogenicity. Low levels of replicative fitness/cytopathogenicity of HIV-1 is typically related to slow disease progression and even, human who are elite suppressors of the HIV-1.
Implications of all the available evidence
Despite robust SIVcpz replication in chimpanzees, this virus appears less virulent in many chimpanzees. Based on the studies herein, SIVcpz strains had similar replicative fitness in human and chimpanzee T cells but with lower cytopathogenicity than HIV-1 strains. There is a possibility that an SIVcpz from chimpanzees jumped into humans circa 1920s and circulated among humans resulting slow or minimal disease but eventually evolved into a pathogenic virus. If true, this SIVcpz evolution leading to pathogenesis in humans must have occurred early in the pandemic considering HIV-1 group M rapidly diverged into distinct lineages, all of which are pathogenic. In general, restriction factors have evolved at fast rates in monkeys/apes likely to adapt and prevent cross-family/species zoonoses over millions of years primate evolution. Jumps of retroviruses between Hominini (African apes) and with other primates, even with low frequency, likely occurred on many occasions during the evolution of humans, chimpanzees, and gorillas over 4 million years but successful introduction and spread of the lentiviruses in Africa apes only dates to hundreds of years ago. As such, various restriction factors in African apes (e.g. humans, gorillas, and chimpanzees) effectively inhibit retroviruses of Old World but are poor inhibitors of SIV/HIV/lentiviruses in African apes, which may explain for frequent jumps and recombination of the lentiviruses between African apes over the past few hundred years. The notion that the HIV-1 group M epidemic in humans emerged from a jump of a very rare SIV from chimpanzee(s) is indirectly supported by this study but proof would require extensive surveillance of wild chimpanzees, with the assumption that the SIV and infected chimpanzees still remain. Greater knowledge of the viral genotypic/phenotypic factors and events that lead to a new zoonoses resulting in a global pandemic (e.g. HIV-1 or SARS-CoV-2) is essential to predicting and possibly mitigating future pandemics.
Introduction
Simian immunodeficiency viruses (SIV) are a collection of primate lentiviruses differing as much as 50% (in nucleotide diversity) and infecting a genetically diverse population of non-human primates.1,2 In contrast, the distinction between “human” immunodeficiency virus type 1 (HIV-1) and “simian”IV from chimpanzees (cpz) are more so in their assigned names, considering SIVcpz and HIV-1 share more homology than SIVs that infected different Old World monkeys.1, 2, 3, 4 Zoonotic transmissions of SIV occurred through several independent jumps from chimpanzees and gorillas to humans, leading to the establishment of 4 groups within HIV-1 (M, N) and (O, P) respectively, and from sooty mangabeys to humans for the 9 groups (A-I) within HIV-2.1, 2, 3, 4, 5, 6, 7, 8 HIV-1 group M is the most prevalent human lentivirus, accounting for >95% of the 36 million infections in humans, while HIV-2 and HIV-1 group O are responsible for 1–2 million and <30,000 infections.2,9 The rare HIV-1 groups N and P are found in less than a hundred individuals residing in or visiting the Equatorial rainforest of West and Central Africa,2,10, 11, 12, 13, 14 which is also the origin of the HIV-1 group M zoonotic jump. West Central African countries, with Cameroon at its epicenter, represent the oldest epidemic illustrated by high HIV genetic variability in the current infected population.2,14
There are more than 40 different SIV lineages infecting more than 45 species of African non-human primates, most commonly in the African apes (Hominini), e.g. chimpanzees, gorillas, and humans and other Old World monkeys.3, 4, 5,8,15,16 As an example, SIVsmm infects sooty mangabeys (smm) (or Cercocebus atys) in West Africa and is the likely source of HIV-2.17, 18, 19 SIVcpz infects the Pan troglodytes troglodytes (Ptt) and Pan troglodytes schweinfurthii (Pts) sub-species of chimpanzees common in central and west central Africa.7,20, 21, 22 While SIVcpz from Ptt is the origin of HIV-1 groups M and N, Pts-related SIV lineages have not been found in humans.7,23 Likewise, SIVcpz has not jumped to other subspecies of chimpanzees (i.e. Pt verus, Pt vellerosus) and bonobos (Pan paniscus), which raises the possibility cross-species transmission is not observed due to incompatibility, lack of opportunity, or simple lack of virus replication in these Pan species.24, 25, 26, 27 It is still unclear if SIV infecting lowland gorillas (Gorilla gorilla or gor) inhabiting the Equatorial forest of Central Africa8,16 were the direct link for the primate lentiviral jump to humans to establish HIV-1 groups O and P, or if this jump involved chimpanzees as an intermediate host.
A cross-species lentiviral inoculation establishing a new, successful zoonosis may be governed by higher replicative capacity2,22 and reduced sensitivity to innate restriction factors in the new primate host.28, 29, 30, 31, 32, 33, 34 As an example, increased sensitivity to human restriction factors or low replicative fitness of SIVcpz from Pts chimpanzees in susceptible human CD4+ cells may have prevented this cross-species transmission. Given the deforestation and exploitation in sub-Saharan Africa after the turn of the 18th century,35 a time that coincided with the introduction and rapid expansions of the HIV-1 and -2 epidemics, it is unlikely, but not impossible, that humans were only exposed to SIV from Ptt chimpanzees, and not Pts chimpanzees.25 Primate lentiviruses have been jumping and circulating through Old World primate populations, including humans, for hundreds of years.36,37 Cross-species transmission and variable host adaptions likely resulted in extinction of many primate lentiviral lineages, attenuations, and even the birth of novel lineages through recombination events that occur during super-infections with SIVs from different primates.38, 39, 40 In fact, SIVcpz are recombinants between two lentiviruses originating from the red-capped mangabey and greater spot-nosed monkey or a closely related species.41
Following the zoonotic transmission to humans around the 1920s, HIV-1 group M rapidly evolved into 10 different subtypes within the Congo Basin but with increased human migration and trade with sub-Saharan Africa, many HIV-1 subtypes founded new epidemics across the globe and even recombined to generate new circulating recombinant forms (CRFs).1,2 This collection of group M subtypes have evolved almost equidistant from the root of the HIV tree but despite the similar evolutionary rates, considerable differences in virulence and transmission efficiency may still be shaping subtype spread.42, 43, 44 HIV-1 subtype D may be the most virulent in humans42,45,46 but has decreased in prevalence in the past 30 years, whereas the rapid expansion and predominance of HIV-1 subtype C worldwide may be related to its lower virulence and average transmissibility.2 Several studies now suggest that in the absence of treatment, people infected HIV-1 subtype C compared to those infected with other HIV-1 subtypes, thus, increasing the opportunity for transmission with subtype C HIV-1 which may help to explain how HIV-1 subtype C has outpaced the spread of other subtypes around the world.2,42,47 Although HIV-1 replicative fitness in primary T cells and macrophages is a strong correlate of disease progression in humans,42,48,49 this relationship is not absolute, especially with HIV-2 in humans and some SIV strains in African green monkeys (agm) and chimpanzees.50, 51, 52, 53, 54, 55 In some primates such as Rhesus macaques, zenotropic infection and pathogenesis by SIVsmm was similar to HIV-1 pathogenesis in humans, which may be related to direct infection of the gut tissues in early infection. During chronic disease, primary lymphoid organs like tonsillar tissue, in which viral pathogenicity, T-cell depletion, and viral propagation rates can be measured.56, 57, 58
In the present study, we utilized various primary human cell types to determine the ex vivo fitness and pathogenicity of a diverse set of 21 primate lentiviral isolates that included HIV-1 groups M, N, O, HIV-2, and SIVs of cpz, gor, mac, smm, and agm. We have performed over 300 pairwise dual infections using the 21 primate lentivirus isolates in peripheral blood mononuclear cells (PBMCs) from both human (Hu) and chimp (Pan troglodytes verus; Ptv) donors, along with monoinfection of PMBCs and cell lines, and dendritic cell + T cell co-cultures. We determined the relative repression of primate lentivirus replication in human cells by various restriction factors and correlated this to replicative fitness. Finally, we accessed the cytopathogenicity on CD4+ T cells in human tonsillar explant tissue following infection with the HIV-1, HIV-2 and SIV isolates. The results described herein indicate that the SIVcpz from Ptt chimpanzees have high replicative fitness, are not significantly inhibited by a subset of human restriction factors as previously reported,28, 29, 30, 31, 32, 33, 34 but showed very low virulence in primary human lymphoid tissue. In contrast, SIV from other primates were highly susceptible to inhibition by human restriction factors, had low replicative fitness, and yet displayed high cytopathogenicity in primary human lymphoid tissue.
Methods
Ethics
Human tonsils were removed during routine tonsillectomy (provided by the Human Tissue Procurement Facility of the University Hospitals Case Medical Center Hospital, Cleveland, Ohio). Use of anonymous surgical waste for experimental HIV infections was approved by the IRB under UHIRB 01-02-45. Human PBMCs were obtained from uninfected, HIV negative donors approved by the IRB under UHIRB 01-02-45. No other human subjects enrolled in this study to collect samples or viruses. All investigations have been conducted according to the principles expressed in the Declaration of Helsinki. Sex and gender were not applicable to this study as it was focused on in vitro primate lentivirus infections in human and chimpanzee cells.
Cells
Primary and cell lines
HIV negative subjects provided whole blood from which peripheral blood mononuclear cells (PBMC) were separated using the Ficoll-Hypaque density centrifugation technique. Whole blood from an HIV negative Chimpanzee (Pan troglodytes verus–Ptv) was made available and purchased for this study from the SNPRC as part of their Primate and Primate Services core (https://snprc.org/primates/). Ptv blood was shipped overnight to Case Western Reserve University, Cleveland and PBMCs were separated as described above. All PBMCs were stimulated for 3–4 days with 2 μg/ml of phytohemagglutinin (PHA; Gibco BRL) and 1 μg/ml of interleukin-2 (IL-2; Gibco BRL) in complete RPMI 1640 containing 2 mM l-glutamine. U87 human glioma cells expressing CD4 and CCR5 (U87.CD4.CCR5–RRID:CVCL_X630 and U87.CD4.CXCR4–RRID:CVCL_X632) were obtained through the AIDS Research and Reference Reagent Program (validated testing reports provided at https://www.hivreagentprogram.org/) and maintained in DMEM supplemented with 10% FBS, Fetal Bovine Serum (FBS; Mediatech, Inc.), Penicillin (100 U/ml), Streptomycin (100 μg/ml), Puromycin (1 μg/ml) and G418 (300 μg/ml). 293 T-cells (human embryonic kidney cells) (RRID:CVCL_0063) were also grown in DMEM media.
Human tonsils
Human tonsils removed during routine tonsillectomy on healthy young adults in the absence of infection but with history frequent, reoccurring strep infections (see above) was received within 5 h of excision. Use of anonymous surgical waste for experimental HIV infections was approved by the IRB under UHIRB 01-02-45. To prepare human lymphocyte aggregate cultures (HLACs), we removed the cauterized tissue from the tonsil as well as the capsule and the fatty parts surrounding the tonsil. Bloody or inflamed parts were also discarded from the tissue. The tonsil tissue was dissected into small pieces by hand with a surgical scissors and the pieces were squeezed using a flat surface. Cells were dispersed and washed in Phosphate-Buffered Saline (PBS) and passed through a 70 μm cell strainer (BD Falcon). Finally, isolated HLACs were plated in 96-well U-bottomed plates (Corning, Inc.) at a concentration of 2 × 106 cells per well in 200 μl RPMI 1640 media containing 10% FCS, 100 U/mL penicillin, 100 μg/ml streptomycin sulfate, 2.5 μg/ml fungizone, 2 mM l-glutamine, 10 mM HEPES, 1 mM sodium pyruvate and 1% non-essential amino acids mixture. One day after HLACs preparation, cells were inoculated with virus in triplicate (0.002 multiplicity of infection; (MOI) per 2 × 106 cells per well in 200 μl). After overnight infection, cells were washed, and supernatants harvested at days 4, 7, and 10 post infection without dispersing the pellet. Lentiviral replication was assessed using an RT assay as described previously.59
Monocyte-derived dendritic cells (MDDCs)
MDDCs were isolated as described previously.60 Briefly, CD14+ beads (Miltenyi Biotech) were used to isolate CD14+ monocytes from healthy donor human PBMCs. These were then cultured in the typical 10% FBS/RPMI media and supplemented with 10 ng/ml IL-4 and 50 ng/ml GM-CSF (Miltenyi Biotech). MDDCs were matured by overnight stimulation with 100 ng/ml LPS (Sigma) 5–6 days after initiation of the cultures. Mature MDDCs were pelleted, washed and resuspended in fresh medium ready for co-culture with PBMCs and infection with viruses.
Viruses
Twenty six primate lentiviruses, primary isolates or molecular clones were used for this study and included HIV-1 group M (n = 10), N (n = 1) and O (n = 3), and HIV-2 (n = 2) and from non-human primates; SIVcpz (n = 6), SIVgor (n = 1), SIVagm (n = 1), smm (n = 1), and mac (n = 1). A full list of the viruses, their origins, phylogenetic relations in env and gag, and titers are outlined in Fig. 1A and B. The HIV-1 group M and group O isolates have been used previously47 and along withSIVcpz, SIVagm, SIVsmm, and SIVmac isolates were obtained from the AIDS Research and Reference Reagent Program of the National Institute of Health. The well characterized group molecular O clone CMO2.41 was also used as one of the representatives of HIV-1 group O.61 SIVgor (CP684) and three of the SIVcpz strains (EK505, Gab1-1, MB897-1) were gifts from Beatrice Hahn. The remaining SIVcpz strains Nico and Noah, were analysed in collaboration with the Institute of Tropical Medicine in Antwerp, Belgium (Drs. Jonathan Heeney and Pascale Ondoa, authors). The SIVcpz strains Nico and Noah were obtained from a chimpanzee infected in the wild, maintained in captivity, and whose virus was then used to infect another chimpanzee.52
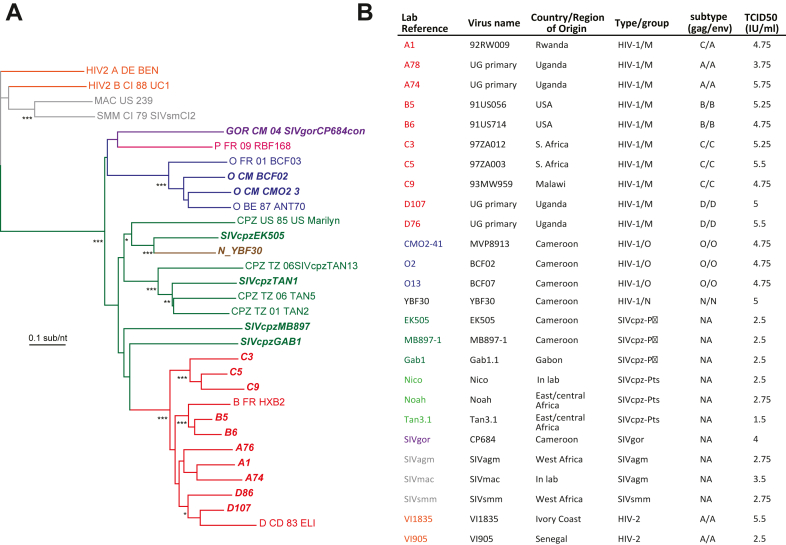
Fig. 1.
Characteristics of lentiviruses used in this study. (A) Neighbor-joining phylogenetic tree of the env sequence of primate lentiviruses with reference strains. (B) List of study viruses showing country/region of origin, genotype, and virus titres (tissue culture infectious dose for 50% infectivity; TCID50). Accession numbers are as follows: A1–AY669700; A74–FJ541298; A78–FJ541299; B5–AY669719; B6–AY669719; C3–AY669741; C5 -AY118165; C9–AY669739; D107–FJ541288; D76–FJ541297; CMO2-41–AY618998; YBF30–AJ006022; EK505–DQ373065; MB897-1–EF535994; Gab1–AF382829; Tan3.1–DQ374658; SIVgor–FJ424871; SIVagm–M58410; SIVmac–EF070330; SIVsmm–AF077017.
As compared to the primary SIV and HIV isolates described above, we also employed proviral clones, plasmids of the proviral clones (CMO2.4, CP684, EK505, Gab1-1, MB897-1) were used to generate infectious virus by transfecting 293T cells using the Effectene transfection reagent (QIAGEN, Valencia, CA). Supernatant containing virus was collected 2 days post transfection, clarified by centrifugation at 2500 rpm for 10 min and purified through a 0.45 μm pore-size filter (Millipore, Billerica, MA). Transfection derived supernatants were subsequently briefly passaged in human PBMC while all other primary isolates were briefly passaged in PBMCs to generate virus stocks. The tissue culture infectious dose for 50% infectivity (TCID50 expressed as IU/ml) of propagated viruses (Fig. 1B) was calculated by the Reed and Muench method as described previously.47 Virus productions in supernatant was determined by a positive RT activity (two standard deviations above the negative control). RT activity is the preferred assay considering the enzyme is present in all HIV or SIV strains.
Assessment of CD4+ T-cell depletion and RT activity in human tonsil histocultures
Twelve days post infection, HLACs were analysed by fluorescence-activated cell sorting (FACS). Uninfected and infected HLACs were stained with Live/dead® violet (Invitrogen, Grand Island, NY), Annexin V-APC (no RRID/not an antibody; e-biosciences, San Diego, CA) and with cell surface markers α- human CD4 PE (RRID: AB_571954; Biolegend, San Diego, CA), α-human CD3 PerCP (RRID: AB_2033956; BD Biosciences) and α- humanCD8 FITC (RRID: AB_11154582; BD Pharmigen). Cells were acquired in a LSRII flow cytometer (BD Biosciences, San Jose, CA) and data analysed with FloJo software (Tree Star, Inc. Ashland, OR). Supernatants from each tonsil infection was also assayed for RT activity.59
Ex-vivo growth competition assay
Ex vivo pathogenic fitness for the isolates was determined by infecting human and chimpanzee PBMCs using methods described previously.44,47,48,62,63 In human PBMC, pathogenic fitness of HIV/SIV isolates was performed by full pair wise competition in triplicate using 9 group M (2 each of subtypes A, B, D; and 3 C), 3 group O, 1 group N, 5 SIVcpz (3 from Ptt and 2 from Pts), 2 HIV-2 and 1 each of SIVsmm, agm and mac. For chimpanzee PBMC the competition was restricted to a selected number of isolates due to the limitation of cell number. All competitions were performed in 48 well plate containing 2 × 105 PBMCs and equal multiplicity of infection (0.005) for both viruses, i.e. 1000 IUs. Virus production was monitored by measuring the RT activity until peak production at day 12 when cells were harvested and stored at −80 °C. Duplicate competitions were analysed resulting in less than 10% variation. The triplicate was analysed with any failure in extraction or PCR in the duplicate. No single, dual virus competition was compared to another. Instead, these analyses involved a comparison of replicative fitness of any one primate lentivirus against all others in direct competition. All statistical analyses reflect this pairwise competition.
PCR and sequence analyses
Due to the closer genetic relationship between HIV-1 group M, DNA extracted from intra-HIV-1 group M competitions were PCR amplified using envelope primers described previously.44,47,48,62,63 Briefly, C2–C3 region of envelope was amplified by PCR using the primers E80 and E125 primers.48 The env primers employed for intra HIV-1 group M competitions were 100% conserved to the target sequences of all HIV-1 group M strains employed. For the DNA extracts of the competitions using the more divergent HIV-1/HIV-2/SIV viruses, conserved primers that spanned the primer binding site pbsdt-F AAAATCTCTAGCAGTGGCGCCCGAACAG (position 622–649 in HxB2; 806–835 in SMM239) and the 5′ end of gag (MAp17) GAGdt-R TTTCCAGCTCCCTGCTTGCCCATACTA (position 890–916 in HxB2 and 1153–1179 in SMM239) was employed for PCR. The 5′UTR/gag primers used for the intergroup/type competitions were 100% conserved to the viruses used herein. However, there are HIV-2 and SIV strains (not employed in this study) that do not possess an exact matched sequence to these primers. The efficiency of PCR amplification of these env and 5′UTR/gag primers have been tested with cloned HIV-1 group M, O, and HIV-2 sequences in the past.44 The NGS protocol to amplify and sequence the 300 competitions using 454 and MiSeq has been recently described. It is important to stress that the same results ( ± 2% variance) was observed with both NGS platforms despite the increased error rates with 454. The alignments and analysis pipeline described below “count” the number of reads that cluster with one versus the other lentiviruses in the competition. All quasispecies of lentiviruses are genetically distinct using this amplified and sequenced gag or env region.
Competition sequence data extraction and analyses
Sequence data was extracted and analysed using SeekDeep, a software suit that enables de novo clustering thereby avoiding potential artifacts that may occur due to alignment directly to reference sequences in situations of divergent sequence populations. It provides start-to-finish workflow from raw sequences files to population-level clustering, tabular and graphical summaries.64 The program has three components namely: extractor, qluster and process Clusters. SeekDeep analyses counts phylogenetically-related clones within 10 nts of the of the most abundant viral clones in the quasispecies/swarm of each lentiviral isolate in the competition and thereby avoid biases that may be engendered by direct mapping to a singular reference/strain sequence.
Sequence reads were first separated at the sample level based on their MIDs/bar codes followed by the detection and removal of forward and reverse PCR primers to leave the targeted region. Additional filtering was performed to ensure expected target length and to remove low quality reads as indicated by low quality or ambiguous (N) bases in the sequences. Next, was the clustering phase which involved the collapsing of unique reads except for high quality differences. Clusters that were up to 2 bases different from a more abundant haplotype were collapsed. Clusters composed of a single read were removed. During the SeekDeep process clustering phase, a final filtering was performed to remove low abundance artifacts and chimeric haplotypes, determined as a haplotype that was a combination of two other more abundant haplotype clusters (>2-fold relative to potential chimera), low frequency chimeric haplotypes that represented combinations of and low abundance artifacts. These final haplotype clusters were than mapped to known input strain sequences allowing up to 5 mismatches and renamed appropriately. The relative abundance of the strains in each competition was then calculated and interactive graphs of the results were created to assess quality and view the data.64
Phylogenetic and evolutionary analysis by Maximum Likelihood method
All group M and O strains used in the study had been partially sequenced in the env gene (C2–V3; 480 nucleotides) and gag (MA p17; 350 nucleotides) as described previously.47 The group N and some of the SIV strains are available as full or partial genome sequences in the HIV Los Alamos database. Sequences were aligned using Clustal X incorporated in the MEGA software. Reference sequences of various HIV types and groups were also included. The evolutionary history was inferred by using the Maximum Likelihood method and Kimura 2-parameter model.65 Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA X.66
SiRNA inhibition of restriction factors
U87.CD4.CCR5 cells were seeded with 5 × 104 per well in DMEM and incubated in 48 well plates overnight at 37 °C. On the next day, cells were washed with PBS and DMEM media replaced with 210ul of Opti-MEM (Thermofisher). siRNA against Trim 5α, APOBEG 3F and G, and Tetherin obtained from Santa Cruz Biotechnology Inc. was dissolved respectively and resuspended in an accompanying diluent. Cells were then transfected with siRNA (10 nM final concentration) using Lipofectamin RNAi Max reagent (Invitrogen) as follows: 0.6 μl Lipofectamin in 20 μl Opti-MEM and 0.25 μl of the respective siRNA in 20 μl Opti-MEM.67 These were then mixed and added dropwise in each well and incubated for 36 h. Cells were then washed with PBS, diluted virus added at a MOI of 0.001 and incubated at 37 °C for 5 h in DMEM. Cells were washed and resuspended in 400 μl DMEM and supernatants were harvested on days 3, 5, 7 and 9 post infection and tested for RT activity. All experiments were done in triplicates; and included virus + scrambled siRNA, virus + siRNA, as well as negative controls (siRNA alone and cells alone). Endogenous expression of these restriction factors is rarely detected by Western blots using antibodies. The reduction of the restriction factor mRNA levels with these optimized siRNA was between 65 and 95%. Please note the same U87 cell culture treated with one specific siRNA was split and utilized for infections with all lentiviruses.
Statistical analyses
We employed the Pearson Product Moment Correlation (and where indicated with fewer data points, Spearman Rank Correlation) to determine if there was any relationship between the genetic distance and the fitness in human PBMC, as well as fitness and CD4+ T-cell depletion in the tonsil by the isolates. One way ANOVA analyses were performed to compare all differences in replicative fitness. All statistical analyses were performed using the GraphPad Prism program.
Role of funders
The funders of this research had no input in the concepts, implementation, data analyses or interpretation of results.
Results
HIV-1, HIV-2 and SIV replication in human PBMCs and U87 cell line
Twenty-six HIV and SIV strains representing 10 HIV-1 group M, 3 group O, 1 group N, 6 SIVcpz, 1 SIVgor, 2 HIV-2, and one each of SIVsmm, SIVmac and SIVagm (Fig. 1A and B) were propagated and tittered on human PBMCs. Each SIV/HIV strain was classified and/or confirmed as R5 based on their replication on U87.CD4.CCR5 but not U87.CD4.CXCR4 cell lines. Unfortunately, all SIV strains had been previously propagated and expanded in human PBMCs, regardless of primate species of origin. The SIV isolation process, preceding this study, was the first evidence of human cell tropism, at least at the level of replication in human PBMCs. Previous phylogenetic analyses of these SIV and HIV isolates identified clusters and provided the nomenclature based on their closest sequence relations and also consistent with their primate hosts in the wild (except SIVmac). In this study, we provide simple, trimmed phylogenetic trees with specific SIV and HIV reference sequences only to highlight their closest relations within the gag and env sequences (Fig. 1A and Supplementary Figure S1). Specifically, the group M strains clustered more closely with SIVcpz MB897 (Fig. 1A), while SIVgor_CP684 grouped with the group O strains, as previously described (Fig. 1A and B). SIVcpz EK505 is more closely related to the HIV-1 group N strain, YBF30. The HIV-2 strains VI1835 and VI1905 (both subtype A) clustered with SIVsmm, the source of HIV-2 (Fig. 1A; Supplementary Figure S1). Strains were continually sequenced in the gag and env genes using Ilumina MiSeq during this study, i.e. prior and post propagation and for strain quantitation/identification in the pairwise competitions (see below). Mutations related to in vitro sequence drift in this SIV and HIV-1 strains was very rare and within the MiSeq error rate of ∼0.1%.
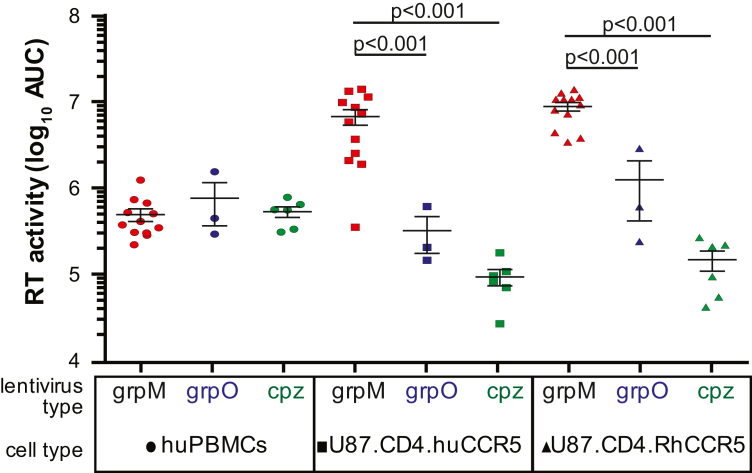
Simple replication kinetics of all the primate lentiviruses were determined on human PBMCs and the brain glioma cell line U87 expressing huCD4 along with either human (Hu) CCR5 or rhesus macaque (Rh) CCR5. All cell types were infected with the same multiplicity of infection (MOI) and monitored for virus production (and cell viability, see below). Virus replication kinetics over a course of a 12-day infection in PHA/IL2-treated human PBMCs as measured by the reverse transcriptase activity were not significantly different between the HIV-1 group M, O and SIVcpz strains (area under curve; Fig. 2). In the human brain glioma cell line expressing HuCD4 with HuCCR5 or MmCCR5, the HIV-1 group M strains had 10-fold and nearly 100-fold greater replication kinetics than HIV-1 group O and SIVcpz strains (one way ANOVA), respectively (Fig. 2) consistent with previous reports.2,44 Many studies have repeated shown that monoinfections in cell lines (e.g. U87) are very poor surrogate of virus replicative fitness when compared to monoinfections and even more so to pairwise competitions in primary cells (e.g. PBMCs, DC-T cell co-cultures, CD4+ T cells, and various tissue explants).44,49,68 In cell lines like U87, monoinfections are less reliable due to inherent variations between separate infections, logarithmic growth of viruses, and variability in viral monitoring assays, i.e. HIV-1 have been shown to have more efficient RT activity than HIV-1 group O, HIV-2, and other lentiviruses.69,70 Finally, many cell lines including U87s have lower, non-physiological levels of various restriction factors as compared to primary cells (e.g. ABOBEC 3G).71
Fig. 2.
Replication of primate lentiviruses in human PBMCs or CD4+ U87 cells expressing human or rhesus macaque CCR5, as measured by reverse transcriptase activity in supernatant. Panel (A) shows the production of HIV-1 group M, group O, and SIVcpz strains over a 12-day infection and plotted as the area-under-the curve of the RT activity in supernatant. All infections were done in triplicate. Standard deviations at each time point were within 20% of the value on average and are not shown.
Competitions of HIV and SIV isolates in human and chimpanzee PBMCs
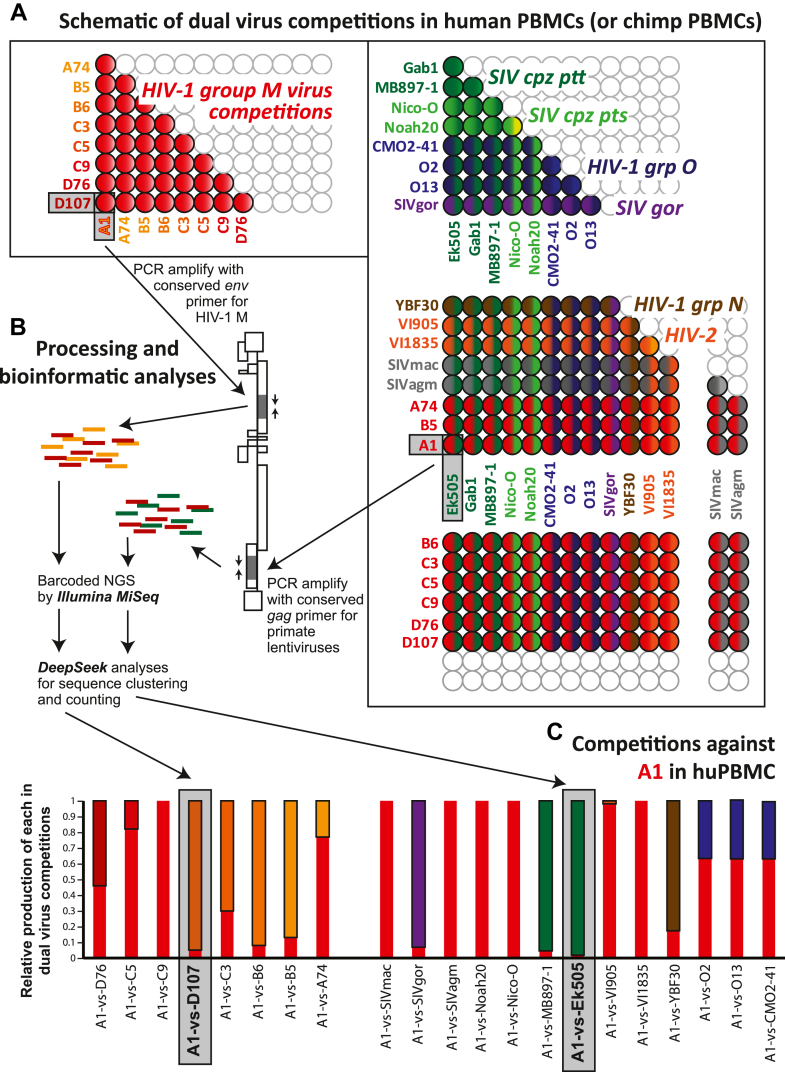
Direct dual virus competitions at low multiplicity of infection in the same contained culture provides a relative, reproducible measure of replicative fitness with minimal variance.44,47, 48, 49,62 Thus, we performed pairwise competitions with 21 lentiviruses in both human and chimpanzee PBMCs in triplicate (schematically illustrated in Fig. 3A). A total of 253 separate pairwise competitions (in triplicate) in HuPBMC at an MOI of 0.005 were harvested after 12 days post-infection, i.e. peak virus production. Dual infections at this MOI and in this timeframe limits recombination to <0.1% of the observed dual virus replication.72,73 We obtained a limited supply of blood/PBMCs from P.t. verus (Ptv) to perform 82 pairwise competitions involving inter-lentiviral pairs (e.g. HIV-1 M strain versus SIVcpz rather than HIV-1 M subtype A versus HIV-1 M subtype B). Ptv was available for PBMCs. Testing the competitive replicative fitness of the SIVcpz originating from Pts against Ptt in PtsPBMCs might give SIVcpz-Pts a competitive advantage and visa versa. As such, use of PBMCs from Ptv might reduce some host adaptation advantage. Also, this Ptz chimpanzee subspecies lacked infection by SIVcpz (as tested to date).24, 25, 26, 27 PCR products of env and gag regions for the group/type competitions44,47,48,62,63 were barcoded and sequenced by next generation deep sequencing, and analyses were obtained through the SeekDeep pipeline, which performs de novo clustering at a single base resolution (Fig. 3B).64 Fig. 3C provides an example of HIV-1 group M A1 virus competed against each of 20 primate lentiviruses in human PBMCs and plotted as relative production of the HIV-1 A1 and the competing virus. Detailed results from 335 competitions involving each pair of primate lentiviruses in human and PtvPBMCs (over 1000 competitions in triplicate) is summarized in Fig. 4 with the statistical one way ANOVA analyses in Supplementary Tables S1–S5.
Fig. 3.
(A) Schematic illustration of the pairwise dual virus competitions in human and chimpanzee PBMC involving 21 primate lentvirus. (D) Processing and bioinformatic analyses following the completion of the competition/infections. (C) Results of the pairwise infections involving competitions of the HIV-1 group M A1 strain against all other primate lentiviruses. Relative production of both viruses as measured by NGS and SeekDeep is shown.
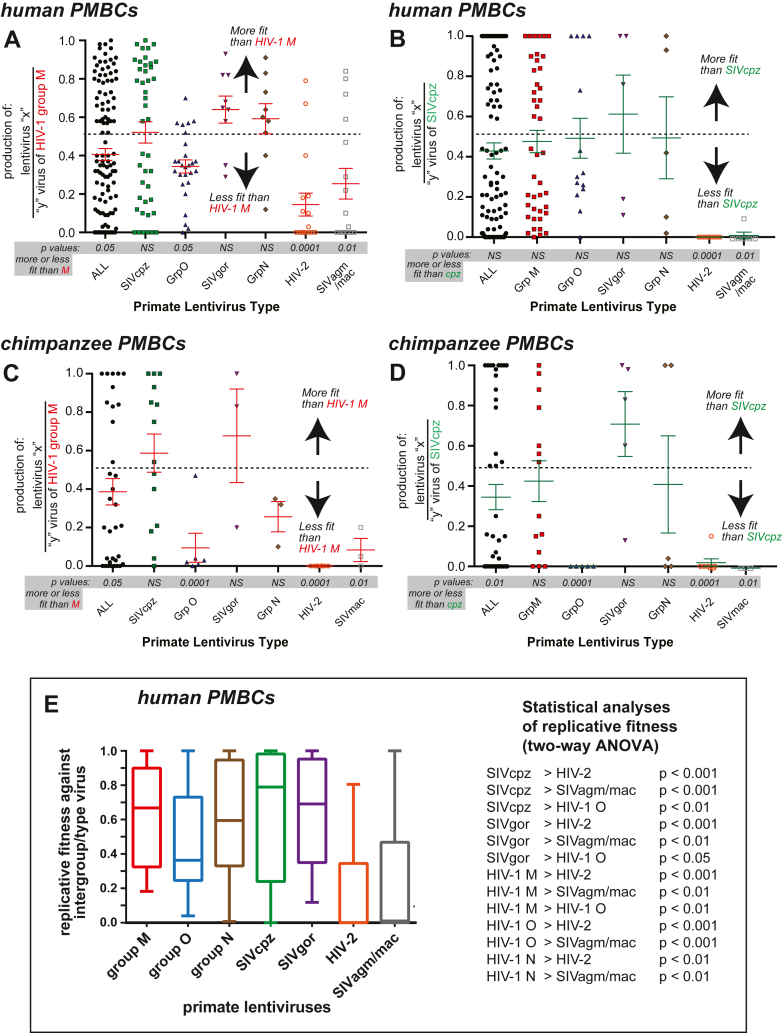
Fig. 4.
Replicative fitness of HIV-1, -2, and SIV isolates in human and chimpanzee PBMCs. As described in Fig. 3, pairwise competitions were performed in human PBMCs and a subset in chimpanzee PBMCs. The production from these dual virus competitions were PCR-amplified, subject to NGS, and analysed by SeekDeep to obtain the relative production of each virus in each dual virus competition. These values are plotted as the production of any lentivirus relative to group M (A and C) or SIVcpz (B and D) in human (Hu) PBMCs (A and B) or chimpanzee (Ptv) PBMCs (C and D). (E) For this comparison, the replicative fitness of members of one primate lentivirus group/type (e.g. HIV-1 group M) competed against members of all other groups/types was analysed for an intergroup/type replicative fitness. These analyses exclude all of the intragroup/type dual virus competitions (e.g. HIV-1 group M A74 versus B5). The geometric mean and box of 95% of the relative fitness values with outlier whiskers is plotted for each group/type of primate lentivirus. Statistical analyses of the date in (E) is presented on the right. Statistical analyses of all panels in Figure 4 is provided in Supplementary Tables S1–S5, respectively.
Fig. 4 shows the competition results of each lentivirus within primate lentivirus groups/types against each HIV-1 group M isolate (4A and C) and SIVcpz isolate (4B and D) in human (Hu) HuPBMCs (4A and B) or in chimpanzee (Ptv) PtvPBMCs (4C and D). In direct competitions, the human lentiviruses HIV-1 group O and HIV-2 were significantly less fit than SIVcpz and HIV-1 group M isolates in either HuPBMCs (Fig. 4A) or PtvPBMCs (Fig. 4C) (ANOVA; Supplementary Tables). SIVgor CP684 and SIVcpz strainscould outcompete most HIV-1 group M isolates in HuPBMCs, suggesting an SIVcpz/gor ancestor may have been capable of establishing an infection in humans (Fig. 4A). Interestingly, HIV-2 and SIVsmm, had the lowest replicative fitness despite ancestor of SIVsmm being responsible for the HIV-2 outbreak. We were unable to propagate sufficient HIV-1 P RBF168 for this study but the SIVgor CP684 was over 90% identical in Gag, Pol and Env sequence to HIV-1 group P.8
SIVcpz strains trended to slightly higher replicative fitness than HIV-1 group M isolates in PtvPBMCs (Fig. 4B and D). Again, the SIV strains infecting other Old orld monkeys (not chimpanzees) or the HIV-2 strains (originating from SIVsmm from sooty mangabeys) were less fit than SIVcpz or group M HIV-1 in both Hu and PtvPBMCs. HIV-2 or SIVsmm have not been identified in chimpanzees. SIVcpz was more fit than HIV-1 group O in PtvPBMCs (Fig. 4B) but not in HuPBMCs (Fig. 4D). The path of zoonotic jumps responsible for the origin of HIV-1 group O may involve a jump from chimpanzees but with an SIV that originated in gorillas.8,74
The competition results are also presented as the replicative fitness values of each primate lentivirus within a group/type competed against the primate lentiviruses outside of this group/type in HuPBMCs (e.g. HIV-1 group M isolates competed against HIV-1 O, N, HIV-2, SIVcpz, gor, agm, and mac; first bar in Fig. 4E). HIV-1 groups M, N, SIVcpz and gor are significantly more fit than HIV-1 group O, HIV-2, SIVagm and mac/smm (Fig. 4E; ANOVA analyses in Supplementary Tables).
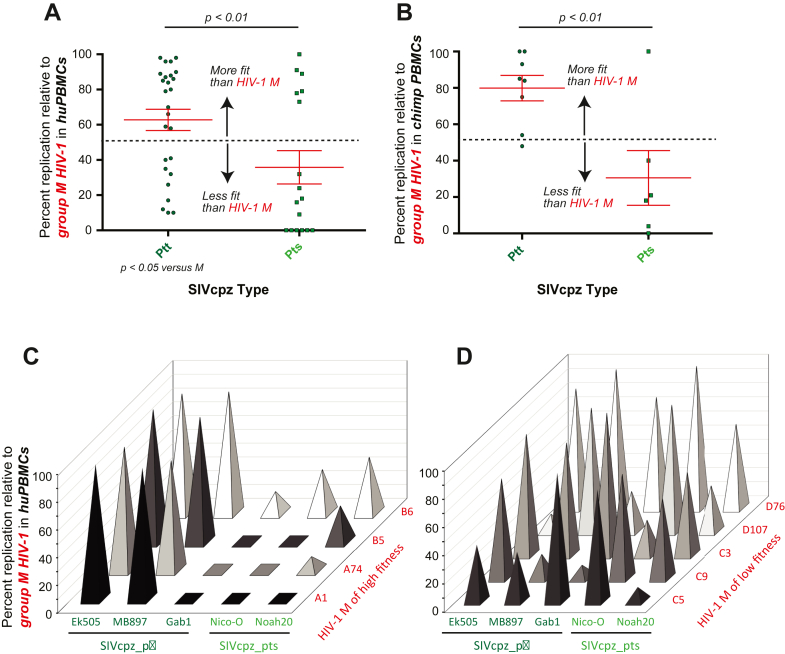
Possible replicative fitness differences among SIVcpz strains derived from different chimpanzee subspecies
When comparing the replicative fitness of SIVcpz versus HIV-1 group M in HuPBMCs and PtvPBMCs, there were SIVcpz strains/viruses with high and low fitness. The competitions included SIVcpz_ptt isolated from the Pan troglodytes troglodytes subspecies and SIVcpz_pts found in the P. troglodytes schweinfurthii. SIVcpz_ptt strains were significantly more fit than SIVcpz_pts compared to HIV-1 group M in both Hu (Fig. 5A) and PtvPBMCs (Fig. 5B) (ANOVA; Supplementary Tables). The SIVcpz_ptt strains, EK505 and MB897 could compete equally with the more fit group M isolates, such as A1, A74, B5, and B6, high replicative fitness also previously described.44,47 The SIVcpz_pts strains, Nico-0 and Noah-20, were out-competed in HuPBMCs (Fig. 5C) by these group M HIV-1 of higher replicative fitness. Gab1, the least fit SIVcpz_ptt strain, could only compete with the HIV-1 group M isolates of the lowest replicative fitness44,47 (Fig. 5D).
Fig. 5.
Fitness of various SIVcpz strains of Ptt and Pts competed against HIV-1 group M strains in human PBMC (A) and chimpanzee PBMC (B). SIVcpz_ptt and _pts strains competed in human PBMCs against HIV-1 group M of high fitness (C) and group M of low fitness (D).
Dendritic cells enhance HIV and SIV infection in PBMCs
Within a human host, the delivery of HIV-1 by dendritic cells to activated CD4+ T-cells in the gut, lymph nodes, and other tissues may be responsible for seeding the majority of HIV-1 replication during systemic infection. The replication of primate lentiviruses in PBMCs mediated by human MDDC trans-infection was significantly greater (by 2- to 3-fold) than that observed in direct lentivirus exposure of PHA/IL-2 treated PBMCs of the same donor (Supplementary Figure S2). The highest fold change (ca. 3-fold) in replication mediated by DCs was observed with the HIV-1 group M A74 virus (Supplementary Figure S2B). In using the MDDC and PBMCs of a different donor, we observed a similar level of enhancement of lentiviral replication with MDDC-mediated trans infection.
Cytopathic phenotype of HIV-1, HIV-2 and SIV in ex-vivo lymphoid tissues
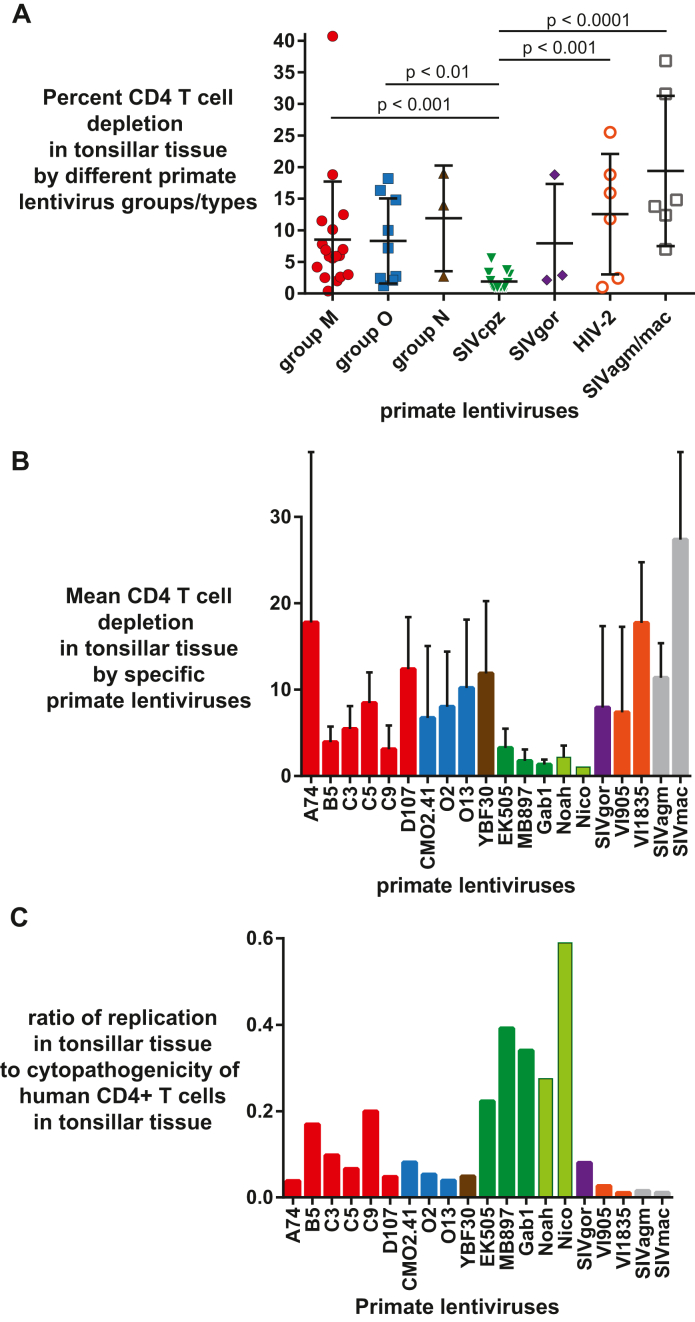
Unlike PBMCs, human tonsils are permissive cells that do not require stimulation with mitogens or IL-2 for HIV propagation.75 To determine the cytopathic effect and CD4+ T-cell depletion rate by these primate lentiviruses, human tonsils were infected with the same infectious titers and monitored for virus production and CD4+ T-cell depletion due to virus exposure. All data presented were relative to uninfected controls, which showed less than <5–8% CD4 T cell depletion after 12 days in culture.56,57 It is important to note that depletion of CD4+ T cells in human tonsils by R5-using HIV-1 subtype B strains (∼10% CD4 T cell depletion) is lower than reported with X4-using HIV-1 strains, typically reaching a 65% loss in tonsillar CD4+ T cells.57 Again, all the primate lentiviruses used in this study were CCR5-using for host cell entry. The highest CD4+ T cell depletion was observed with SIVmac (average 27%), followed by A74 (18%), VI1835 (16%) and D107 (12%) (Fig. 6A). Despite the relatively low replication in CD4+ T cells, HIV-2 and SIVmac/agm still depleted significant levels of CD4+ T cells in human tonsils (Fig. 6B). Exposure to the human tonsillar tissue to SIVcpz strains (Ptt and Pts) resulted in nominal CD4+ T cell depletion (1–3.5%; Fig. 6A) despite high levels of replication in these tissues (Fig. 6B). SIVcpz_Pts Nico had the highest ratio of virus production to CD4+ T-cell depletion in the tonsillar tissues. All the primate lentiviruses within HIV-1 groups (M, N, O), HIV-2, SIVgor and SIVagm/mac were better at depleting CD4+ T-cells than SIVcpz, significantly so when compared to all but HIV-1 group N and SIVgor (limited data points). In contrast, there was no difference in the levels of CD4+ T-cell depletion in the tonsillar tissues between any of the types/groups when SIVcpz was not included in the ANOVA analyses.
Fig. 6.
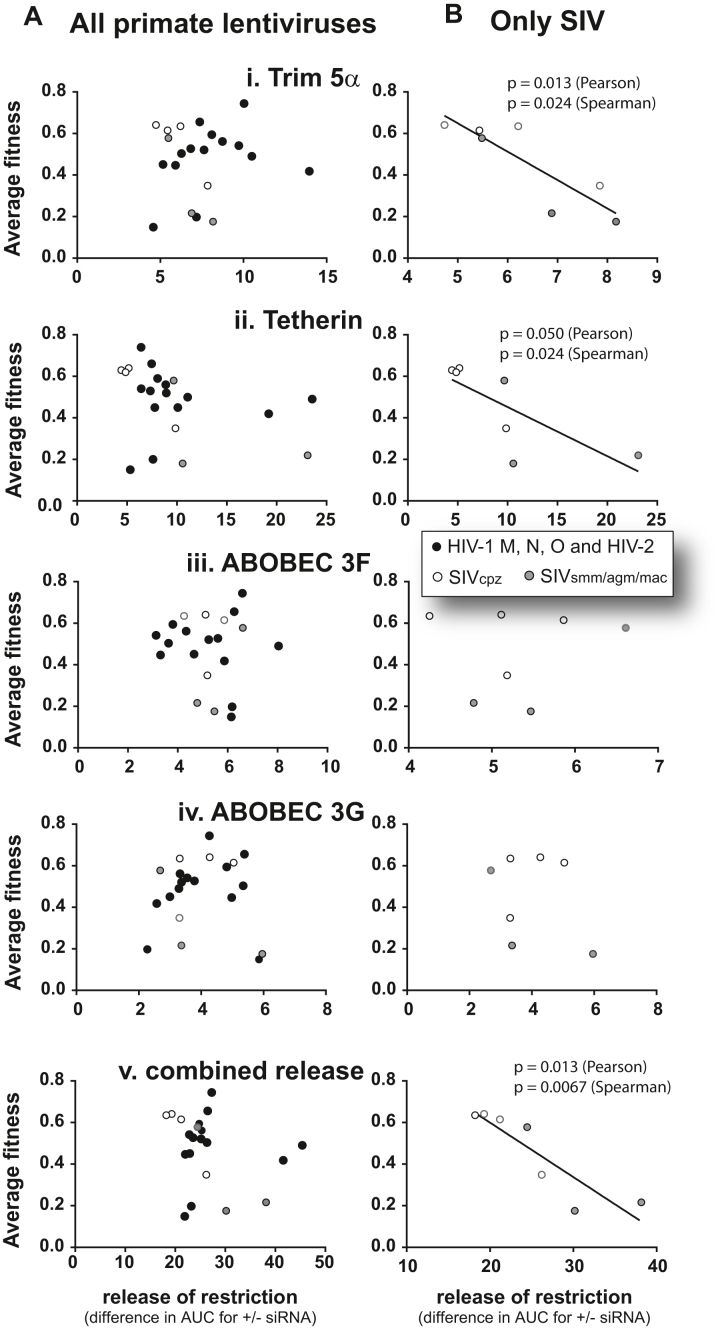
Comparing the average replicative fitness to the release of restriction (mediated by siRNAs). The average replicative fitness for each primate lentivirus was determined from the pairwise competitions in HuPBMCs as described in Supplementary Figure S3. Each primate lentivirus was also used to infect cells in the presence of a restriction factor-specific siRNA or a scrambled siRNA. The area under the curve for the 9-day virus replication (measured by RT activity) with specific siRNA was subtracted from that with scrambled siRNA. (A) For each primate lentivirus, plots of average replicative fitness versus the relative release of replication due to Trim 5α (i), Tetherin/BST2 (ii), ABOBEC 3G (iii), 3F (iv), and all four combined (v). (B) The same plots as in (A) but without the human lentiviruses.
No correlation was observed between the relative replication in the tonsillar tissue and the level of CD4+ T cell depletion by these viruses. SIVcpz had the highest replicative fitness in human PBMCs and in the tonsillar tissues. In contrast, SIVcpz infection was associated with minimal cytopathogenicity in human tonsillar tissue resulting in the greatest ratio of virus production to CD4+ T cell death of any primate lentivirus (Fig. 7C). In contrast, the HIV-2 isolates, SIVagm and SIVmac, showed an average depletion of CD4+ T cells, i.e. comparable to that observed with HIV-1 group M, O, and all other primate lentiviruses (except SIVcpz) (Fig. 7A and B). Based on the poor virus production in human tonsillar tissue (as well as average fitness in human PBMCs), the ratio of replication to CD4+ T cell depletion of HIV-2 and SIVagm/mac was the lowest for all primate lentiviruses (Fig. 7C). As described below, these findings might suggest that pathogenicity/virulence could have evolved as a trait in humans following a zoonotic jump.
Fig. 7.
Primate lentiviral replication in tonsillar tissue and huPBMC fitness (A) Percentage of CD4+ T cell depletion within a group or type of primate relative to CD4 T cell depletion in the mock infection/control; (B) CD4 T-cell depletion levels based on infection of tonsillar tissue in triplicate; (C) Comparing the replication in tonsillar tissue to cytopathogenicity.
Impact of human restriction factors on primate lentivirus replication
Host factors such as Trim5a, APOBEC 3F/G/H, tetherin/BST2, SERINC5, and SamHD1 have all been shown to restrict xenotropic retrovirus replication, and most have been associated with preventing cross species transmission of primate lentiviruses.28, 29, 30, 31, 32, 33, 34,76,77 We explored the ability of these restriction factors in human cells to reduce replication kinetics of NHP lentiviruses as one mechanism to prevent a jump into humans. For these studies, we utilized the cell line U87.CD4.HuCCR5—the cell line most supportive of HIV-1 group M replication and most restrictive to NHP lentiviral replication (Fig. 2). In cells treated with siRNAs to reduce endogenous mRNA of Trim5α, tetherin/BST2, APOBEC 3F and 3G (by 80–90% for 5 days), we observed release of restriction and increase in lentivirus replication (Fig. 6 and Supplementary Figure S3). Reduction of human APOBEC3F and 3G levels resulted in only minimal increases in lentiviral replication over 7 days (Supplementary Figure S3C and D), which is expected considering restriction/inhibition requires an accumulation of deleterious mutations through multiple rounds of replication. siRNA treatment to reduce human Trim5α resulted in only low level “rescue” of the replication of all primate lentiviruses (Supplementary Figure S3A). The strongest restriction of these NHP lentiviruses in these human cell lines was mediated by tetherin/BST2 (Fig. 6 and Supplementary Figure S3B). SIVmac and SIVsmm replication was increased (or “rescued”) by 5–6 fold with a >80% knockdown of tetherin mRNA over a 9-day infection. Tetherin inhibits even HIV-1 group M and O isolates at a low level. In contrast, there was no apparent inhibition of SIVcpz strains by human tetherin.
Next, the average replicative fitness of the 21 primate lentiviruses in human PBMCs was compared to the individual restriction by human Trim 5α (i), Tetherin (ii), APOBEC 3G (iii), 3F (iv), and cumulative restriction by all four. There was no significant correlation between inhibition by human restriction factors and the average replicative fitness of these primate lentiviruses, which included the human immunodeficiency viruses (Fig. 6A). However, the successful jump of primate lentiviruses from chimpanzee to humans was likely mediated in part by the ability of this now “human” lentivirus to resist inhibition by these restriction factors in human cells and survive in the human population. Thus, we removed the human lentivirus viruses from the analyses in Fig. 6B. A significant inverse correlation was now observed between the average replicative fitness of these “non-human” primate lentiviruses (in human PBMCs) and level of restriction by Trim 5α (Fig. 6B i) and Tetherin/BST2 (7B ii). The cumulative restriction by all Trim 5α, Tetherin, and APOBEC3G/3F showed the strongest correlation with average replicative fitness. The SIVcpz strains (especially cpz_Ptt) appeared to be the least inhibited by these restriction factors in human cells which related to the highest replicative fitness in human PBMCs.
Discussion
Lentiviruses have a complex history of interspecies jumps in primates Based on the most recent colonization of Madagascar by mammals from Africa, the discovery of an endogenous primate lentivirus in the gray mouse lemurs of Madagascar but absent in all African primates places the introduction of lentivirus into primates at a minimum of 14 million years ago, which would extend to over 80 million years78 when considering introduction of lentiviruses at the point of divergence of Strepsirrhini (lemurs) and Haplorhini (monkeys and apes)79 or reduced less than 4 million years if an aerial lentivirus introduction by bats occurred.37,80 Despite evidence that lentiviruses many have been introduced into primates in the millions of years ago,81 the existing, unstable lentiviral lineages in various African apes may have appeared much more recently, estimated at hundreds of years.37 HIV-1 group M emerged in humans with a jump from chimpanzees (Ptt) in the Congo basin in the early 20th century, an event that has now resulted in over 40 million deaths with another 37 million infected humans.37,82,83 The jump of SIV from sooty mangabeys into humans in the 1930s37 resulted in HIV-2 with a peak epidemic of 1–2 million and even before the introduction of antiretroviral treatment, there was progressive loss in HIV-2 prevalence in the same regions where HIV-1 group M was expanding rapidly.84,85 HIV-1 group O, N, and P had even less success surviving in humans following introductions from the gorilla and/or chimpanzees; jumps dated to sometime in the 1920s for group O and 1960s for N.21,37,82 As such, convention for SIV/HIV classification has been unstable due to continuous transitions between related apes, changing phylogenetic classifications, and mounting evidence of recombination among primate lentiviruses.37,43,82
This study was designed to compare the competitive replicative fitness and virulence of different lentiviral strains isolated from various primate species using all available replicating primate lentivirus available at the time of study. Exposure of a primate to a xenotropic lentivirus is unlikely to result in a cross-species transmission event due in part to protective barriers, e.g. innate defenses and various restrictions factors. Nonetheless, these restriction factor/innate defenses appear to show variable protection against different, foreign lentiviruses. To understand the overall restriction of primate lentiviruses in primary human cells susceptible to HIV, we compared the replicative fitness of primate lentiviruses in both human and chimpanzee PBMCs by performing over 250 competitions with 21 lentiviral isolates of HIV-1 groups M, N, O, HIV-2, SIVcpz, gor, smm, and agm. To determine relative virulence upon a possible jump to humans, we compared the relative replicative fitness of these primate lentiviruses to the inhibition by various human restriction factors and the relative depletion of CD4+ T cells in human tonsillar tissue.
To summarize, SIV strains from Ptt and Gor and HIV-1 group N appeared slightly more fit than even HIV-1 group M replicating in both human and Ptv PBMCs. However, as previously published, there was a wide range in replicative fitness among HIV-1 group M subtypes with subtype A and C strains, again, being consistently less fit.42,44,47,62 With HIV-1, the replicative fitness of HIV-1 subtypes in human PBMCs directly and significantly correlated with the rate of disease progression in people infected by the same subtypes. However, unlike SIVcpz strains, these HIV-1 isolates also proved to be pathogenic in vitro and within infected individuals.42,43,46,86 When these HIV-1 group M strains of lower fitness were removed from the analyses, there was no significant difference in the replicative fitness of SIV from chimpanzees and gorillas and the HIV-1 group M and group N isolates found in humans. SIVcpz or SIVgor (lineage related to past zoonosis of HIV-1 group P) are significantly more fit than HIV-1 group O in human PBMCs. Finally, SIV from African green monkeys and Sooty mangabey had the lowest replicative fitness in both human and Ptv PBMCs, similar to the low replicative fitness observed with HIV-2. We determined that the relative level of replication of these non-human primate lentiviruses in human and Ptv PBMCs reflects the cumulative inhibition by the various human restriction factors, with tetherin/BST2 being the most restrictive. Based on the cumulative data presented herein, it is clear that SIVcpz and SIVgor have the greatest ability to replicate in human PBMCs.
Based on the similar overlap of humans into the Pts and Ptt habitats in the Rift Valley of East Africa, Congo Basin of Central Africa, and the tropical south of West Africa, it is unlikely that humans would experience less exposure to Pts than to Ptt chimpanzees. Findings presented herein on just six SIVcpz isolates suggest that the SIVcpz of Ptt compared to SIVcpz of Pts was more compatible for cross-species transmission to humans and gained the foothold necessary for transmission and human adaptation. Approximately 45% amino acid sequence diversity were found in the Gag, Pol, and Env alignments of SIVcpz from Ptt and Pts (Supplementary Figure S4), more than diversity than between an HIV-1 group M and O strain. Thus, key substitutions leading to zoonotic transmission and increased pathogenesis of Ptt over Pts in humans will be difficult to define. Nearly 20,000 full HIV-1 genome sequences with 1000s of primary HIV-1 isolates are publicly available as compared to only 30 SIVcpz genome sequences and six primary SIVcpz isolates (at the time of this study). For this study, we utilized replication competent SIVcpz strains available. In past studies involving over 100 primary HIV-1 group M, N, and O isolates,42,44,47,62 the replicative fitness results between human lentivirus types and groups have been highly consistent in human PBMCs and other susceptible cell types. With only five SIVcpz isolates, we cannot assume that all SIVcpz isolates will reflect the results presented herein, but the findings from all five SIVcpz isolates were quite consistent. We have attempted to generate SIVcpz strains based on de novo genome constructions from 25 published sequences (www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). Unfortunately, none of these constructions resulted in replication-competent virus that could be rescued. For now, this study remains the most comprehensive in determining replicative fitness of primate lentiviruses in primary human T cells, chimpanzee T cells, and primary human lymphoid tissue in comparison to relative inhibition by human restriction factors.
The ability of a lentivirus to replicate in the primary susceptible cells of a host does not always correlate with pathogenesis. A strong example is the lack of disease in African Green Monkeys despite the high viral loads of SIVagm produced by an abundance of infected T-cells in these animals50,51 Only one study has characterized disease progression of a chimpanzee infected by SIVcpz (a Pts strain) raised in captivity after being infected in the wild. With this animal, there was little evidence of disease despite an increasing viral load during the length of infection.52 In captured chimpanzees infected with the same “wild” SIVcpz52 via an intra-rectal, -vaginal or -venous routes, lentiviral pathogenesis and disease progression was somewhat similar to HIV infections in humans including some evidence of initial hyperimmune activation.87 In observational studies of wild chimpanzees from western Tanzania, SIVcpz pathogenesis was evident88 with clear T cell depletion in lymph nodes53 along with increased mortality rates in infected infant and adult chimpanzees.87 Consistent with end-stage disease during AIDS, post-mortem histopathology of lymphoid tissue provided evidence of pathogenesis in SIVcpz-infected chimpanzees.89 Thus, it is quite possible that other mortality risk factors in wild chimpanzees leading to reduced human-age corrected life expectancies90 may mask a slow SIVcpz pathogenesis, quite similar to that observed in HIV-infected humans.
In humans, several studies have described a direct correlation between the rate of disease progression and the replicative fitness of the infecting HIV-1 isolates in primary T-cells.42,48,49 This is best illustrated by the low replicative fitness of HIV-1 isolates derived from patients with slow or non-progressing disease versus the high replicative HIV fitness among rapid progressors.48,91 In this study, we compared the cytopathogenicity of human and non-human primate lentiviruses in primary human lymphoid tissue, i.e. resected tonsillar tissue. There was a clear disconnect between the ability of non-human primate lentiviruses to propagate in human tonsillar tissue and the pyroptosis/cell death of the CD4+ T cells within the same tissue. HIV-2, SIVagm, and SIVsmm, with low replicative fitness, mediated similar levels of T cell death as HIV-1 group M/N and SIVgor with high replicative fitness. In contrast, SIVcpz, having the highest replicative fitness in tonsillar tissue (as well as in human and ptv PBMCs), was associated with minimal pyroptosis and all manner of T-cell death in this tonsillar tissue. The inability of SIVcpz to mediate cytopathogenicity in human T-cells may correspond to the minimal pathogenesis or lack of disease described in most SIV-infected chimpanzees.52 Again, in contrast to the most fit SIVcpz's, SIVsmm (the progenitor of HIV-2) and HIV-2 were highly cytopathogenic in these tonsillar tissues despite having the lowest replicative capacity. Although cytopathology of primate lentiviruses is clearly not the only factor associated with pathogenesis, these findings are consistent with slow disease progression associated with HIV-2 infected humans.54,55 HIV-1 group O and SIVgor (the progenitor of HIV-1 group O) had high cytopathogenicity and moderate replicative fitness in these tonsillar tissues but not as low as HIV-2/SIVsmm. People infected with HIV-1 group O progress faster to disease than those infected with HIV-2, but slower than those infected with HIV-1 group M.74
The relative virulence of a virus in their host species is impacted by a multitude of host factors (e.g. innate and acquired immune responses, restriction factors). SIVcpz strains from Ptt and Pts had similar replicative fitness in human cells than HIV-1 group M strains but the SIVcpz strains did not result in cytopathogenicity. This inconsistency between SIVcpz_Ptt and HIV-1 could relate to an adaptation/evolution of SIVcpz in humans to escape restriction factors/barriers following a jump. However, there is little evidence that SIVcpz is restricted from replication in human PBMCs as compared to other SIVs from non-human primates. It is feasible that an increased cytopathogenicity evolved early in the human epidemic during a period of high viral burden and efficient transmission. As HIV-1 group M diversified during the past 100 years, HIV-1 subtypes may have evolved with differences in virulence and transmission efficiency. Increased virulence of HIV-1 subtype D over subtypes A and C has been well documented within infected cohorts and using similar ex vivo studies described herein.42,44,45,47,62
As a closing hypothesis, we can speculate, based on these in vitro findings, that the HIV-1 epidemic was caused by a cross-species transmission of a very rare SIVcpz strain resulting in exceptionally high virulence (replicative fitness + cytopathogenicity) in humans. The rarity of the SIVcpz strain with human tropism and pathogenesis could also explain why a lentiviral epidemic did not appear earlier in human history considering the clear interaction of chimpanzees and humans in central Africa. Clearly, neither of these hypotheses can be proven without the availability of human lentiviral samples closer to the cross-specific transmission event responsible for the HIV-1 epidemic.
Contributors
Denis M. Tebit contributed to figures, writing, study design, data collection, analysis, and interpretation.
Myriam Rodriguez, Nicolas J. Hathaway, Katie Bain, Jennifer Bongorno, Gabrielle Nickel, Jonathan Heeney, Pascale Ondoa, and Angel L. Reyes-Rodriguez contributed to data collection, analysis, and interpretation.
Richard Gibson and Yue Li contributed to coordination of experimentation and supervision of the laboratory-based studies.
David Canaday, David McDonald, and Jeffrey A. Bailey provided supervision, study design, data interpretation, writing, and research funding.
Eric J. Arts provided overall study coordination, supervision, study design, data interpretation, writing, research funding, and final manuscript submission.
All authors have read and approved the final version of the manuscript.
Denis M. Tebit and Jeffrey A. Bailey have verified the underlying data.
Data sharing statement
All data underlying the results herein is either available in supplementary material or upon request, including the data presented as summaries and averages in the article. Data will be shared as Excel Spreadsheets and graphs upon request. Please email Eric Arts at earts@uwo.ca with any data request. For source codes for SeekDeep, please refer https://github.com/bailey-lab/SeekDeep, to reference 65, or email Jeff Bailey at jeffrey_bailey@brown.edu.
Declaration of interests
None of the authors have competing financial interest related to this study.
Acknowledgements
We thank Drs. Beatrice Hahn for providing primate lentiviruses for this study. Support for this study to E.J.A. was provided by the NIH/NIAID R01 AI49170 and CIHR project grant 385787. Infrastructure support was provided by the NIH CFAR AI36219 and Canadian CFI/Ontario ORF 36287. Efforts of J.A.B. and N.J.H. was provided by NIH AI099473. D.H.C. was funded by VA and NIH AI AI080313 for this study.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104965.
Appendix A. Supplementary data
Neighbor joining phylogenetic tree of the gag p24 sequences of primate lentiviruses with reference strains.
Enhancement of HIV and SIV replication in PBMC+dendritic cell cultures. A) Comparison of the replication kinetics of some isolates in PBMC (P, black filled circles) and PBMC+DC (DC, gold filled squares) co-cultures. B) Fold difference showing enhancement in replication by DCs.
Primate lentivirus replication in human CD4+ U87 cells expressing HsCCR5 and pre-treated with siRNA to human restiriction factors Trim 5a (A), Tetherin/BST2 (B), APOBEC3F (C), and APOBEC3G (D). Replication is plotted as the fold change in increased lentivirus replication on days 3, 5, and 7 in the presence of siRNA to specific human restriction factors.
Amino acid sequence alignment of the predicted Gag proteins of SIVcpz GAB2 of chimpanzee Ptt and SIVcpz TAN3 of chimpanzee Ptts.
Amino acid sequence alignment of the predicted Pol proteins of SIVcpz GAB2 of chimpanzee Ptt and SIVcpz TAN3 of chimpanzee Ptts.
Amino acid sequence alignment of the predicted Env proteins of SIVcpz GAB2 of chimpanzee Ptt and SIVcpz TAN3 of chimpanzee Ptts.
Statisical analyses of the data presented in Figure 4A comparing the replicative fitness of the SIVcpz viruses competed against each type and group of primate lentiviruses in pairwise compeititons using human PBMCs.
Statisical analyses of the data presented in Figure 4B comparing the replicative fitness of the SIVcpz viruses competed against each type and group of primate lentiviruses in pairwise compeititons using human PBMCs.
Statisical analyses of the data presented in Figure 4C comparing the replicative fitness of the HIV-1 group M viruses competed against each type and group of primate lentiviruses in pairwise compeititons using chimpanzee PBMCs.
Statisical analyses of the data presented in Figure 4D comparing the replicative fitness of the SIVcpz viruses competed against each type and group of primate lentiviruses in pairwise compeititons using chimpanzee PBMCs.
Statisical analyses of the data presented in Figure 4E comparing the replicative fitness of the SIVcpz viruses competed against all other primate lentiviruses in pairwise compeititons using human PBMCs.
Statistics on SIVcpz_Ptt and SIVcpz_Pts pairwise competitions in human PBMCs using HIV-1 M as the comparator in chimpanzee PBMCs (data from Figure 5A) and in chimp PBMCs using HIV-1 M as the comparator in chimpanzee PBMCs (data from Figure 5B), respectively.
References
- 1.Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Tebit D.M., Arts E.J. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011;11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 3.Hahn B.H., Shaw G.M., De Cock K.M., Sharp P.M. AIDS as a zoonosis: scientific and public health implications. Science (1979) 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 4.Peeters M., D'Arc M., Delaporte E. Origin and diversity of human retroviruses. AIDS Rev. 2014;16:23–34. [PMC free article] [PubMed] [Google Scholar]
- 5.Keele B.F., Van H.F., Li Y., et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science (1979) 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Heuverswyn F., Li Y., Neel C., et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 7.Gao F., Bailes E., Robertson D.L., et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes [see comments] Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 8.D'Arc M., Ayouba A., Esteban A., et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci U S A. 2015;112:E1343–E1352. doi: 10.1073/pnas.1502022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb G.S., Raugi D.N., Smith R.A. 90-90-90 for HIV-2? Ending the HIV-2 epidemic by enhancing care and clinical management of patients infected with HIV-2. Lancet HIV. 2018;5:e390–e399. doi: 10.1016/S2352-3018(18)30094-8. [DOI] [PubMed] [Google Scholar]
- 10.Simon F., Mauclere P., Roques P., et al. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 11.Gürtler L.G., Hauser P.H., Eberle J., et al. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol. 1994;68:1581–1585. doi: 10.1128/jvi.68.3.1581-1585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallari A., Holzmayer V., Harris B., et al. Confirmation of putative HIV-1 group P in Cameroon. J Virol. 2011;85:1403–1407. doi: 10.1128/JVI.02005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aghokeng A.F., Liu W., Bibollet-Ruche F., et al. Widely varying SIV prevalence rates in naturally infected primate species from Cameroon. Virology. 2006;345:174–189. doi: 10.1016/j.virol.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 14.Leoz M., Feyertag F., Kfutwah A., et al. The two-phase emergence of non pandemic HIV-1 group O in Cameroon. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VandeWoude S., Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van H.F., Li Y., Neel C., et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 17.Clavel F., Mansinho K., Chamaret S., et al. Human immunodeficiency virus type 2 infection associated with AIDS in West Africa. N Engl J Med. 1987;316:1180–1185. doi: 10.1056/NEJM198705073161903. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch V.M., Olmsted R.A., Murphey-Corb M., Purcell R.H., Johnson P.R. An African primate lentivirus (SIVsmm) closely related to HIV-2. Nature (London) 1989;339:389–391. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 19.Santiago M.L., Range F., Keele B.F., et al. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79:12515–12527. doi: 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nerrienet E., Santiago M.L., Foupouapouognigni Y., et al. Simian immunodeficiency virus infection in wild-caught chimpanzees from Cameroon. J Virol. 2005;79:1312–1319. doi: 10.1128/JVI.79.2.1312-1319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tebit D.M., Arts E.J. Elsevier Inc.; 2013. From simian to human immunodeficiency viruses (SIV to HIV): emergence from nonhuman primates and transmission to humans. The role of animals in emerging viral diseases; pp. 201–234. [DOI] [Google Scholar]
- 23.Paraskevis D., Lemey P., Salemi M., Suchard M., Van De P.Y., Vandamme A.M. Analysis of the evolutionary relationships of HIV-1 and SIVcpz sequences using bayesian inference: implications for the origin of HIV-1. Mol Biol Evol. 2003;20:1986–1996. doi: 10.1093/molbev/msg207. [DOI] [PubMed] [Google Scholar]
- 24.Li Y., Ndjango J.-B., Learn G.H., et al. Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J Virol. 2012;86:10776–10791. doi: 10.1128/JVI.01498-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Switzer W.M., Parekh B., Shanmugam V., et al. The epidemiology of simian immunodeficiency virus infection in a large number of wild- and captive-born chimpanzees: evidence for a recent introduction following chimpanzee divergence. AIDS Res Hum Retroviruses. 2005;21:335–342. doi: 10.1089/aid.2005.21.335. [DOI] [PubMed] [Google Scholar]
- 26.Corbet S., Muller-Trutwin M.C., Versmisse P., et al. Env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince A.M., Brotman B., Lee D.H., Andrus L., Valinsky J., Marx P. Lack of evidence for HIV type 1-related SIVcpz infection in captive and wild chimpanzees (Pan troglodytes verus) in West Africa. AIDS Res Hum Retroviruses. 2002;18:657–660. doi: 10.1089/088922202760019356. [DOI] [PubMed] [Google Scholar]
- 28.Sauter D., Kirchhoff F. Key viral adaptations preceding the AIDS pandemic. Cell Host Microbe. 2019;25:27–38. doi: 10.1016/j.chom.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Etienne L., Bibollet-Ruche F., Sudmant P.H., Wu L.I., Hahn B.H., Emerman M. The role of the antiviral APOBEC3 gene family in protecting chimpanzees against lentiviruses from monkeys. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaddis N.C., Sheehy A.M., Ahmad K.M., et al. Further investigation of simian immunodeficiency virus vif function in human cells. J Virol. 2004;78:12041–12046. doi: 10.1128/jvi.78.21.12041-12046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaba A., Flath B., Chelico L. Examination of the apobec3 barrier to cross species transmission of primate lentiviruses. Viruses. 2021;13 doi: 10.3390/v13061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F., Wilson S.J., Landford W.C., et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C.-Y., Shingai M., Welbourn S., et al. Antagonism of BST-2/tetherin is a conserved function of the env glycoprotein of primary HIV-2 isolates. J Virol. 2016;90:11062–11074. doi: 10.1128/jvi.01451-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song B., Javanbakht H., Perron M., Park D.H., Stremlau M., Sodroski J. Retrovirus restriction by TRIM5α variants from Old World and New World Primates. J Virol. 2005;79:3930–3937. doi: 10.1128/jvi.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldwin B. University of Central Florida; 2015. "An examination of the instability and exploitation in Congo from king " by Baldwin Beal. [Google Scholar]
- 36.Chesney M.A., Ickovics J.R., Chambers D.B., et al. Self-reported adherence to antiretroviral medications among participants in HIV clinical trials: the AACTG adherence instruments. Patient care committee & adherence working group of the outcomes committee of the adult AIDS clinical trials group (AACTG) AIDS Care. 2000;12:255–266. doi: 10.1080/09540120050042891. [DOI] [PubMed] [Google Scholar]
- 37.Wertheim J.O., Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liégeois F., Schmidt F., Boué V., et al. Full-length genome analyses of two new simian immunodeficiency virus (SIV) strains from mustached monkeys (C. Cephus) in Gabon illustrate a complex evolutionary history among the SIVmus/Mon/gsn lineage. Viruses. 2014;6:2880–2898. doi: 10.3390/v6072880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin M.J., Hui H., Robertson D.L., et al. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courgnaud V., Abela B., Pourrut X., et al. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J Virol. 2003;77:12523–12534. doi: 10.1128/JVI.77.23.12523-12534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailes E., Gao F., Bibollet-Ruche F., et al. Hybrid origin of SIV in chimpanzees. Science (1979) 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 42.Venner C.M., Nankya I., Kyeyune F., et al. Infecting HIV-1 subtype predicts disease progression in women of sub-saharan Africa. eBioMedicine. 2016;13:305–314. doi: 10.1016/j.ebiom.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arien K.K., Vanham G., Arts E.J. Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol. 2007;5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arien K.K., Abraha A., Quinones-Mateu M.E., Kestens L., Vanham G., Arts E.J. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J Virol. 2005;79:8979–8990. doi: 10.1128/JVI.79.14.8979-8990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaleebu P., Ross A., Morgan D., et al. Relationship between HIV-1 Env subtypes A and D and disease progression in a rural Ugandan cohort. AIDS. 2001;15:293–299. doi: 10.1097/00002030-200102160-00001. [DOI] [PubMed] [Google Scholar]
- 46.Neilson J.R., John G.C., Carr J.K., et al. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J Virol. 1999;73:4393–4403. doi: 10.1128/jvi.73.5.4393-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abraha A., Nankya I.L., Gibson R., et al. CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic. J Virol. 2009;83:5592–5605. doi: 10.1128/JVI.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinones-Mateu M.E., Ball S.C., Marozsan A.J., et al. A dual infection/competition assay shows a correlation between Ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol. 2000;74:9006–9018. doi: 10.1128/JVI.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Troyer R.M., Collins K.R., Abraha A., et al. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol. 2005;79:9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartung S., Boller K., Cichutek K., Norley S.G., Kurth R. Quantitation of a lentivirus in its natural host: simian immunodeficiency virus in African green monkeys. J Virol. 1992;66:2143–2149. doi: 10.1128/jvi.66.4.2143-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandrea I., Ribeiro R.M., Gautam R., et al. Simian immunodeficiency virus SIVagm dynamics in African green monkeys. J Virol. 2008;82:3713–3724. doi: 10.1128/JVI.02402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ondoa P., Kestens L., Davis D., et al. Longitudinal comparison of virus load parameters and CD8 T-cell suppressive capacity in two SIVcpz-infected chimpanzees. J Med Primatol. 2001;30:243–253. doi: 10.1034/j.1600-0684.2001.d01-56.x. [DOI] [PubMed] [Google Scholar]
- 53.Keele B.F., Jones J.H., Terio K.A., et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azevedo-Pereira J.M., Santos-Costa Q. HIV interaction with human host: HIV-2 as a model of a less virulent infection. AIDS Rev. 2016;18:44–53. [PubMed] [Google Scholar]
- 55.Whittle H., Morris J., Todd J., et al. HIV-2-infected patients survive longer than HlV-1-infected patients. AIDS. 1994;8:1617–1620. doi: 10.1097/00002030-199411000-00015. [DOI] [PubMed] [Google Scholar]
- 56.Grivel J.-C., Malkevitch N., Margolis L. Human immunodeficiency virus type 1 induces apoptosis in CD4+ but not in CD8+ T cells in ex vivo-infected human lymphoid tissue. J Virol. 2000;74:8077–8084. doi: 10.1128/jvi.74.17.8077-8084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geuenich S., Kaderali L., Allespach I., Sertel S., Keppler O.T. Biological signature Characteristics of primary isolates from human immunodeficiency virus type 1 group O in ex vivo human tonsil histocultures. J Virol. 2009;83:10494–10503. doi: 10.1128/jvi.00928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pandrea I.V., Gautam R., Ribeiro R.M., et al. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J Immunol. 2007;179:3035–3046. doi: 10.4049/jimmunol.179.5.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marozsan A.J., Fraundorf E., Abraha A., et al. Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates. J Virol. 2004;78 doi: 10.1128/JVI.78.20.11130-11141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reyes-Rodriguez A.L., Reuter M.A., McDonald D. Dendritic cells enhance HIV infection of memory CD4(+) T cells in human lymphoid tissues. AIDS Res Hum Retroviruses. 2016;32:203–210. doi: 10.1089/AID.2015.0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tebit D.M., Zekeng L., Kaptue L., et al. Construction and characterization of an HIV-1 group O infectious molecular clone and analysis of vpr- and nef-negative derivatives. Virology. 2004;326:329–339. doi: 10.1016/j.virol.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 62.Ball S.C., Abraha A., Collins K.R., et al. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J Virol. 2003;77:1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubio A.E., Abraha A., Carpenter C.A., et al. Similar replicative fitness is shared by the subtype B and unique BF recombinant HIV-1 isolates that dominate the epidemic in Argentina. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hathaway N.J., Parobek C.M., Juliano J.J., Bailey J.A. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res. 2018;46:e21. doi: 10.1093/nar/gkx1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 66.Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 67.Gao Y., Lobritz M.A., Roth J., et al. Targets of small interfering RNA restriction during human immunodeficiency virus type 1 replication. J Virol. 2008;82 doi: 10.1128/JVI.02126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arts E.J., Quinones-Mateu M.E. Sorting out the complexities of HIV-1 fitness. AIDS. 2003;17:780–781. doi: 10.1097/00002030-200303280-00026. [DOI] [PubMed] [Google Scholar]
- 69.Arts E.J., Stetor S.R., Li X., et al. Initiation of (-) strand DNA synthesis from tRNALys3 on lentiviral RNAs: implications of specific HIV-1 RNA-tRNALys3 interactions inhibiting primer utilization by retroviral reverse transcriptases. Proc Natl Acad Sci U S A. 1996;93 doi: 10.1073/pnas.93.19.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menendez-Arias L., Abraha A., Quinones-Mateu M.E., Mas A., Camarasa M.J., Arts E.J. Functional characterization of chimeric reverse transcriptases with polypeptide subunits of highly divergent HIV-1 group M and O strains. J Biol Chem. 2001;276:27470–27479. doi: 10.1074/jbc.M104342200. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y.J., Wang X., Zhang H., et al. Expression and regulation of antiviral protein APOBEC3G in human neuronal cells. J Neuroimmunol. 2009;206:14–21. doi: 10.1016/j.jneuroim.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quinones-Mateu M.E., Gao Y., Ball S.C., Marozsan A.J., Abraha A., Arts E.J. In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J Virol. 2002;76:9600–9613. doi: 10.1128/JVI.76.19.9600-9613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baird H.A., Gao Y., Galetto R., et al. Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination. Retrovirology. 2006;3:91. doi: 10.1186/1742-4690-3-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bush S., Tebit D.M. HIV-1 group O origin, evolution, pathogenesis, and treatment: unraveling the complexity of an outlier 25 years later. AIDS Rev. 2015;17:147–158. [PubMed] [Google Scholar]
- 75.Eckstein D.A., Penn M.L., Korin Y.D., et al. HIV-1 actively replicates in naive CD4+ T cells residing within human lymphoid tissues. Immunity. 2001;15:671–682. doi: 10.1016/S1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 76.Malim M.H., Bieniasz P.D. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tavakoli-Tameh A., Janaka S.K., Zarbock K., et al. Loss of tetherin antagonism by Nef impairs SIV replication during acute infection of rhesus macaques. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gifford R.J., Katzourakis A., Tristem M., Pybus O.G., Winters M., Shafer R.W. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci U S A. 2008;105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marciniak S., Mughal M.R., Godfrey L.R., et al. Evolutionary and phylogenetic insights from a nuclear genome sequence of the extinct, giant, “subfossil” koala lemur Megaladapis edwardsi. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2022117118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gilbert C., Maxfield D.G., Goodman S.M., Feschotte C. Parallel germline infiltration of a lentivirus in two malagasy lemurs. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gifford R.J. Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 2012;28:89–100. doi: 10.1016/j.tig.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 82.Lemey P., Pybus O.G., Rambaut A., et al. The molecular population genetics of HIV-1 group O. Genetics. 2004;167:1059–1068. doi: 10.1534/genetics.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Korber B., Muldoon M., Theiler J., et al. Timing the ancestor of the HIV-1 pandemic strains. Science (1979) 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 84.Kanki P.J., Travers K.U., Mboup S., et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343:943–946. doi: 10.1016/s0140-6736(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 85.van der Loeff M.F., Awasana A.A., Sarge-Njie R., et al. Sixteen years of HIV surveillance in a West African research clinic reveals divergent epidemic trends of HIV-1 and HIV-2. Int J Epidemiol. 2006;35:1322. doi: 10.1093/ije/dyl037. [DOI] [PubMed] [Google Scholar]
- 86.Kaleebu P., French N., Mahe C., et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–1250. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- 87.Greenwood E.J.D., Schmidt F., Kondova I., et al. Simian immunodeficiency virus infection of chimpanzees (Pan troglodytes) shares features of both pathogenic and non-pathogenic lentiviral infections. PLoS Pathog. 2015;11:1–26. doi: 10.1371/journal.ppat.1005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudicell R.S., Jones J.H., Wroblewski E.E., et al. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Terio K.A., Kinsel M.J., Raphael J., et al. Pathologic lesions in chimpanzees (Pan trogylodytes schweinfurthii) from Gombe National Park, Tanzania, 20042010. J Zoo Wildl Med. 2011;42:597–607. doi: 10.1638/2010-0237.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hill K., Boesch C., Goodall J., Pusey A., Williams J., Wrangham R. Mortality rates among wild Chimpanzees. J Hum Evol. 2001;40:437–450. doi: 10.1006/jhev.2001.0469. [DOI] [PubMed] [Google Scholar]
- 91.Lassen K.G., Lobritz M.A., Bailey J.R., et al. Elite suppressor-derived hiv-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor joining phylogenetic tree of the gag p24 sequences of primate lentiviruses with reference strains.
Enhancement of HIV and SIV replication in PBMC+dendritic cell cultures. A) Comparison of the replication kinetics of some isolates in PBMC (P, black filled circles) and PBMC+DC (DC, gold filled squares) co-cultures. B) Fold difference showing enhancement in replication by DCs.
Primate lentivirus replication in human CD4+ U87 cells expressing HsCCR5 and pre-treated with siRNA to human restiriction factors Trim 5a (A), Tetherin/BST2 (B), APOBEC3F (C), and APOBEC3G (D). Replication is plotted as the fold change in increased lentivirus replication on days 3, 5, and 7 in the presence of siRNA to specific human restriction factors.
Amino acid sequence alignment of the predicted Gag proteins of SIVcpz GAB2 of chimpanzee Ptt and SIVcpz TAN3 of chimpanzee Ptts.
Amino acid sequence alignment of the predicted Pol proteins of SIVcpz GAB2 of chimpanzee Ptt and SIVcpz TAN3 of chimpanzee Ptts.
Amino acid sequence alignment of the predicted Env proteins of SIVcpz GAB2 of chimpanzee Ptt and SIVcpz TAN3 of chimpanzee Ptts.
Statisical analyses of the data presented in Figure 4A comparing the replicative fitness of the SIVcpz viruses competed against each type and group of primate lentiviruses in pairwise compeititons using human PBMCs.
Statisical analyses of the data presented in Figure 4B comparing the replicative fitness of the SIVcpz viruses competed against each type and group of primate lentiviruses in pairwise compeititons using human PBMCs.
Statisical analyses of the data presented in Figure 4C comparing the replicative fitness of the HIV-1 group M viruses competed against each type and group of primate lentiviruses in pairwise compeititons using chimpanzee PBMCs.
Statisical analyses of the data presented in Figure 4D comparing the replicative fitness of the SIVcpz viruses competed against each type and group of primate lentiviruses in pairwise compeititons using chimpanzee PBMCs.
Statisical analyses of the data presented in Figure 4E comparing the replicative fitness of the SIVcpz viruses competed against all other primate lentiviruses in pairwise compeititons using human PBMCs.
Statistics on SIVcpz_Ptt and SIVcpz_Pts pairwise competitions in human PBMCs using HIV-1 M as the comparator in chimpanzee PBMCs (data from Figure 5A) and in chimp PBMCs using HIV-1 M as the comparator in chimpanzee PBMCs (data from Figure 5B), respectively.