Abstract
Target of rapamycin (TOR) proteins are members of the phosphatidylinositol kinase-related kinase (PIKK) family and are highly conserved from yeast to mammals. TOR proteins integrate signals from growth factors, nutrients, stress, and cellular energy levels to control cell growth. The ribosomal S6 kinase 1 (S6K) and eukaryotic initiation factor 4E binding protein 1(4EBP1) are two cellular targets of TOR kinase activity and are known to mediate TOR function in translational control in mammalian cells. However, the precise molecular mechanism of TOR regulation is not completely understood. One of the recent breakthrough studies in TOR signaling resulted in the identification of the tuberous sclerosis complex gene products, TSC1 and TSC2, as negative regulators for TOR signaling. Furthermore, the discovery that the small GTPase Rheb is a direct downstream target of TSC1-TSC2 and a positive regulator of the TOR function has significantly advanced our understanding of the molecular mechanism of TOR activation. Here we review the current understanding of the regulation of TOR signaling and discuss its function as a signaling nexus to control cell growth during normal development and tumorigenesis.
INTRODUCTION
Rapamycin is an antifungal agent originally purified from Streptomyces hygroscopicus (4). Structural analysis of rapamycin reveals that it is an analogue of the macrolide antibiotic FK506. Similar to FK506, rapamycin also has immunosuppressive effects (4). Rapamycin analogs with improved pharmaceutical properties have been used clinically to inhibit both host rejection following organ transplantation and the restenosis of coronary arteries after angioplasty (77). Importantly, recent studies found that rapamycin has potent growth inhibitory activity against the development of various types of tumors and has potential for being used as anticancer treatment (115, 210).
Rapamycin inhibits cell growth in many types of cells including the budding yeast Saccharomyces cerevisiae (106). The yeast TOR1 and TOR2 genes were originally identified as the targets of rapamycin (143). Mutations in the yeast TOR1 or TOR2 gene confer resistance to the growth-inhibitory properties of rapamycin. However, rapamycin does not directly inhibit TOR; instead, it forms a complex with FKBP12 (FK506 binding protein). It is the FKBP12-rapamycin complex that binds to TOR and inhibits its activity (4). Subsequently, the structurally and functionally conserved mammalian counterpart of yeast TOR, mTOR (also known as FKBP-rapamycin-associated protein [28], rapamycin and FKBP12 target [206], or rapamycin target [43]) was discovered based on its ability to bind to the FKBP12-rapamycin complex. Recently, TOR homologs have also been discovered in Drosophila (dTOR) (181, 278), Caenorhabditis elegans (CeTOR) (155), fungus (TOR1 in Cryptcococcus neoformans) (54), and plants (AtTOR in Arabidopsis thaliana) (167). Thus, TOR is an evolutionarily conserved protein.
The TOR proteins have a C-terminal region with high homology to the catalytic domain of phosphatidylinositol 3-kinase (PI3K) and thus have been termed PI kinase-related kinases (PIKKs). In addition to TOR, the PIKK family includes DNA-PK, ataxia telangiectasia mutated (ATM), and ataxia telangiectasia and Rad-3-related (ATR) (111). The PIKKs are involved in a diverse array of cellular processes, including cell growth control, cell cycle regulation, and DNA damage checkpoint regulation. Interestingly, although the protein sequences of PIKKs demonstrate significant homology to those of lipid kinases, no PIKKs have to date been shown to have lipid kinase activity. Consistently, studies indicate that both yeast and mammalian TOR proteins are protein kinases that preferentially phosphorylate serine or threonine residues (32, 33, 125). Although few physiological substrates of yeast TOR are currently known, biochemical studies have indicated that mTOR can directly phosphorylate a (Ser/Thr)-Pro motif, as seen in the substrates 4EBP1 and STAT3, or a threonine flanked by bulky hydrophobic residues, as seen in the substrate S6K. However, as discussed later in this review, it is also possible that mTOR may indirectly phosphorylate these substrates by regulating a yet to be identified mTOR-associated kinase.
Studies of TOR signaling in yeasts, Drosophila, and mammals have demonstrated that TOR is involved in cellular growth control (where cell growth refers to an increase in both cell size and cell number) by regulating several processes such as transcription, translation, protein degradation, and ribosome biogenesis. Disruption of TOR genes leads to early embryonic death in both flies and mammals, indicating that TOR plays an essential role in development (171, 181). In flies and mammals, TOR plays a critical role in regulating cell size in response to extracellular growth factors and nutrient availability (72, 73, 181, 278). These topics have been previously discussed extensively in several excellent reviews (141, 195). In this review, we focus on recent progress in TOR research. Biochemical purification has identified a high-molecular-weight complex associated with TOR in vivo. The identification of TOR-interacting proteins, such as Kog1p (Raptor) and Lst8p (mLst8/GβL), has significantly advanced our understanding of how TOR is regulated and how it recruits its target substrates (95, 135, 136, 154). Furthermore, recent Drosophila genetic studies have identified two tumor suppressor proteins, TSC1 and TSC2, as negative regulators of TOR signaling (75). The identification of a small G protein, Rheb, as a target of TSC1-TSC2 and an activator of TOR signaling has generated new interest in the function and regulation of the TOR pathway (209, 233, 279). The TSC1-TSC2-Rheb-TOR pathway integrates a wide variety of signals from growth factors, nutrients (amino acids), lipids (phophatidic acid), osmotic stress, and energy (ATP) to control cell growth (76, 121, 243). We discuss in detail how TOR protein function is regulated by TSC1-TSC2 and Rheb.
TORs AND THEIR INTERACTING PROTEINS
TOR Complexes in Yeast
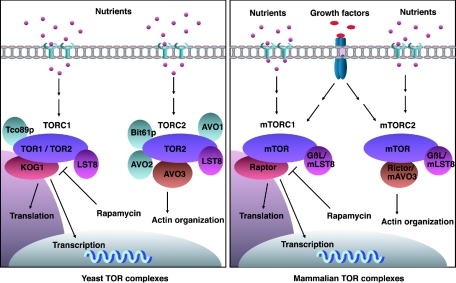
S. cerevisiae has two different TOR genes, termed TOR1 and TOR2, which encode proteins with molecular masses of approximately 280 kDa. Tor1p and Tor2p have 67% sequence identity and are partially redundant in function (107). Both Tor1p and Tor2p play a critical role in translation initiation and cell cycle progression in response to nutrients (14) (Fig. 1), and both of these functions are inhibited by rapamycin. An important function unique to Tor2p is its ability to regulate the polarization of the actin cytoskeleton during cell cycle progression (215, 217, 218). Interestingly, this function of Tor2p is rapamycin insensitive because the rapamycin-FKBP12 complex does not bind to the TOR complex 2 (see below).
FIG. 1.
TOR complexes in yeast and mammalian cells. TOR complex 1 (TORC1) contains either Tor1p or Tor2p with Kog1p, Lst8p, and Tco89p. TORC1 regulates a variety of functions including transcription, mRNA turnover, protein turnover, and translation. All of these TORC1 functions are rapamycin sensitive. TORC2 consists of Tor2p, Avo1-3p, Bit61p, and Lst8p. TORC2 regulates cytoskeleton organization. This function is specific for TORC2 and rapamycin insensitive (left panel). Mammalian TOR also forms two complexes. mTORC1 consists of mTOR, Raptor, and GβL (mLst8). The identified function of mTORC1 complex is similar to that of yeast TORC1. Recently, Rictor/mammalian AVO3 has been identified as a component of mTORC2. Rictor/mAVO3 and GβL are involved in regulating cytoskeleton organization in a rapamycin-insensitive manner.
Recently, Hall and coworkers have successfully purified and identified the components of two distinct TOR-associated complexes in yeast (Fig. 1) (154). TOR complex 1 (TORC1) contains either Tor1p or Tor2p, together with Kontroller of growth 1 (Kog1p) and Lethal with sec thirteen (Lst8p). TORC1 modulates translation initiation, inhibits protein turnover, and represses the transcription of specific genes that are induced by nutrient starvation. It has been found that the functions of TORC1 are inhibited by the rapamycin-FKBP complex. Furthermore, disruption of TORC1 in yeast mimics the phenotype seen after rapamycin treatment, suggesting that TORC1 is the physiological target of rapamycin. Similar studies with mammalian cells have demonstrated that TORC1 is structurally and functionally conserved in high eukaryotes (95, 135). TOR complex 2 (TORC2) consists of Tor2p, Avo1p (Adheres voraciously to Tor2), Avo2p, Avo3p, and Lst8p (Fig. 2). TORC2 mediates the reorganization of the actin cytoskeleton (154) (Fig. 1). Notably, in contrast to TORC1, the rapamycin-FKBP12 complex does not bind to or inhibit TORC2. Studies of the individual functions of the various components of TORC1 and TORC2 will provide a better understanding of mechanisms underlying the distinct functions of TORC1 and TORC2 in yeast. Also, the mechanisms by which rapamycin selectively inhibits nutrient signaling but not actin cytoskeleton reorganization are clarified. Interestingly, the TOR-interacting proteins, Avo1p and Avo3p, are conserved both in the fission yeast (Sin1p and Ste20p, respectively) and in mammals (hSIN1 and Rictor, respectively) (128, 208, 267). hSin1 has an mRNA expression profile similar to that of mTOR (154), although an interaction between mTOR and hSIN1 has not been reported. Recently, two groups have discovered that mammalian AVO3, also known as rapamycin-insensitive companion of mTOR (Rictor), interacts with mTOR (128, 208). Therefore, it is very likely that a functional equivalent of TORC2 might exist in mammalian cells. Recently, Powers's group has also purified additional proteins, Tco89p and Bit61p, as binding partners of TORC1 and TORC2, respectively. Notably, Tco89p was proposed as another important component of TORC1 and participates in cellular integrity (201).
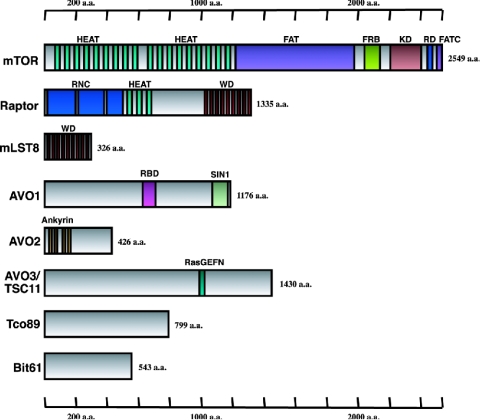
FIG. 2.
Structural domains of mTOR and TOR-associated proteins. mTOR consists of HEAT, FAT, FATC, FRB, and kinase domains. All of them are evolutionarily conserved in TOR orthologs. Raptor-Kog1p consists of RNC, HEAT, and WD40 domains. All domains are also conserved in raptor orthologs. Avo1p has homologies to the Ras-binding domain in the middle of the protein (amino acids [a.a.] 843 to 919) and to the SIN1 gene product. Sin1p is implicated in regulating transcriptional processes and chromatin assembly and is also characterized as the Sapkp (stress activated-protein kinase)-interacting protein. Avo2p has four ankyrin repeats in the N-terminal region. Rictor-Avo3p, also known as Tsc11p, has homology to the Ras GEF N terminus in the middle of the protein (a.a. 990 to 1046). Tco89p and Bit61p are novel yeast TOR-interacting proteins. Abbreviations: HEAT, Huntingtin, elongation factor 3, A subunit of PP2A, TOR; FAT, FRAP, ATM, TRRAP; FATC, FAT carboxyl terminal; FRB, FKBP12-rapamycin binding; RNC, raptor N-terminal conserved; WD, WD40.
mTOR Complex in Mammalian Cells
Mammals contain a single TOR gene, termed mTOR. Recent studies have indicated that like yeast TOR, mTOR forms a functional protein complex with at least two proteins: Raptor (regulatory associated protein of mTOR), a mammalian counterpart of Kog1p in yeast, and mLst8/GβL, a mammalian ortholog of the yeast Lst8p (95, 135, 136) (Fig. 1), indicating the existence of mammalian TORC1 (mTORC1). Raptor is a 150-kDa protein, containing a conserved N-terminal domain, three HEAT domains, and seven WD40 motifs; mLst8 is composed of seven WD40 motif repeats and has sequence homology to the β subunit of heterotrimeric G proteins (Fig. 2). The mTOR-Raptor-GβL complex possesses a total of 14 WD40 motifs and 23 HEATs motifs, two domains that are important for protein-protein interactions, suggesting that this complex could serve as a central nexus for TOR signaling. The structural features of this complex support the notions that, like its homologs in yeast and in Drosophila, mTOR functions in the context of a larger signaling complex. Both biochemical and genetic studies of mammalian cells indicate that both Raptor and mLst8 play important roles in regulating mTOR function.
The expression pattern of Raptor in different mammalian tissues is strikingly similar to that of mTOR, suggesting that it may function as an important subunit in the mTOR-associated complex (135). When Raptor is present in the mTOR complex, immunopreciptiated mTOR can efficiently phosphorylate both S6K and 4EBP1, two well-known targets of mTOR. However, whereas when Raptor is absent from the mTOR complex, mTOR shows decreased ability to phosphorylate its substrates, especially 4EBP1 (95). This suggests that Raptor is required for mTOR to efficiently phosphorylate its substrates. Consistently, increasing Raptor levels in the cell by overexpression promoted the kinase activity of mTOR in vitro (95). Furthermore, knock down of Raptor expression caused a decrease in the expression of mTOR as well, indicating that Raptor may contribute to maintaining mTOR protein stability (135). In mammalian cells, knock down of Raptor expression using small interfering RNA (siRNA) caused a decrease in S6K phosphorylation as well as a decrease in cell size, just as in cells in which mTOR expression was knocked down, highlighting the close relationship between Raptor and mTOR functionally (135). Similarly, knock down of the Raptor homolog in C. elegans caused a phenotype similar to that associated with ceTOR knock down (95).
These studies have led to a conclusion that complex formation between mTOR and Raptor is critical for mTOR function. Several models have been proposed to explain how the complex containing Raptor and mTOR works. One model proposed by Hara et al. suggests that Raptor serves as a scaffolding protein to bridge the interaction between mTOR and its substrates (95). This idea is supported by the fact that both S6K and 4EBP1 physically interact with Raptor (95). Schalm and Blenis have discovered that S6K contains a TOR signaling motif (TOS motif), mutation of which abolishes the phosphorylation of S6K by mTOR; the TOS motif is also found in 4EBP1 (211). Mutation of the TOS motif diminishes both S6K and 4EBP1 binding to Raptor, and such mutants displayed a relatively low rate of phosphorylation by mTOR in vitro (22, 46, 178, 212). In addition, several groups more recently have shown that, distinct from the TOS motif, the 4-amino-acid RAIP motif in 4EBP1 is also important for mTOR-dependent phosphorylation of 4EBP1 (22, 46). Mutation of this motif disrupts the binding of 4EBP1 with Raptor in vivo, although the first 24 amino acids in 4EBP1 containing an intact RAIP motif is not sufficient to bind Raptor, suggesting that the RAIP motif is necessary but not sufficient for 4EBP1 to engage with Raptor. Taken together, these biochemical analyses have demonstrated that the TOS motifs of S6K and 4EBP1 are used for binding to Raptor and suggest that Raptor plays a role as an adaptor to recruit substrates to be phosphorylated by mTOR.
Alternatively, it has also been proposed that, instead of merely presenting substrates to mTOR, Raptor directly influences mTOR kinase activity (135). Kim et al. reported that several agents that inhibit mTOR signaling by affecting mitochondrial function or energy metabolism, such as antimycin and 2-deoxyglucose (2-DG), enhanced the Raptor-mTOR association, whereas agents that stimulate mTOR activity, such as amino acids (e.g., leucine), decreased affinity in the binding between Raptor and mTOR (135). Based on these observations, Kim et al. proposed that Raptor could bind mTOR in either a high-affinity state or a low-affinity state. When binding of Raptor to mTOR is in a high-affinity state, mTOR activity is inhibited, whereas when the Raptor-mTOR binding is in a low-affinity state, mTOR kinase activity is enhanced (135). These observations were made with endogenous Raptor and mTOR. This phenomenon, however, has not been successfully reproduced with overexpressed mTOR and Raptor (95). For example, amino acids failed to affect the affinity of association between ectopically expressed mTOR and Raptor (95). Further studies are needed to confirm whether nutrient signals affect the binding between Raptor and mTOR. Intriguingly, Kim et al. also noted that rapamycin efficiently disrupted the Raptor-mTOR complex under phosphate-rich conditions. This finding may suggest one mechanism whereby rapamycin inhibits mTOR function and is consistent with the notion that rapamycin does not directly inhibit intrinsic mTOR kinase as mTOR autophosphorylation is insensitive to rapamycin (189).
Third, it is possible that mTOR does not directly phosphorylate S6K or 4EBP1. Instead, Raptor may bring in yet to be identified mTOR-dependent kinases to phosphorylate S6K and 4EBP1 at rapamycin-sensitive sites. To test this idea, it will be important to ask whether recombinant Raptor can rescue the kinase activity of mTOR purified under high-nonionic-detergent conditions, which easily disrupt the Raptor-mTOR complex (95, 135). In this situation, any rescued kinase activity should reflect the activity of other, perhaps novel, Raptor-associated kinases, rather than that of mTOR. Indeed, Proud's group has shown that the mutation in TOS motif in 4EBP1 resulted in dephosphorylation of 4EBP1 on its rapamycin-sensitive as well as insensitive sites. These observations suggest that other Raptor-associated kinases, rather than mTOR, are responsible for phosphorylation on rapamycin-insensitive sites of 4EBP1 (22).
GβL (mLst8), a counterpart of Lst8p in yeast, is another essential component of the functional mTORC1 (39, 154, 262). Sabatini and colleagues first identified mLst8/GβL as an important binding protein of mTOR (136). GβL interacts with mTOR within the kinase domain, and GβL mutants with reduced binding affinity to mTOR show decreased ability to activate mTOR. Unlike the Raptor-mTOR complex, the mTOR-GβL complex is not sensitive to nonionic-detergent conditions. Knock down of GβL expression using siRNA inhibits the phosphorylation of S6K in response to amino acids and serum. GβL knock down also results in a decrease in cell size, a phenotype similar to that associated with knock down of either mTOR or Raptor. Consistent with this finding, GβL-depleted conditions prevented mTOR-mediated activation of S6K. These observations led to the conclusion that GβL, like Raptor, positively regulates mTOR function and is critical for targeting the mTOR to its substrates. However, in contrast to Raptor, GβL does not directly interact with mTOR substrates or regulate the mTOR protein level (136).
The obvious question that arises from these observations is that of how GβL affects mTOR activity. Some evidence suggests that GβL is required for formation of the Raptor-mTOR complex. For instance, knock down of GβL decreased the association between mTOR and Raptor (136). In contrast, overexpression of GβL stabilizes the interaction between coexpressed mTOR and Raptor and enhances mTOR kinase activity (136). Furthermore, GβL is postulated to mediate cellular signals, such as that from nutrients, to modulate the formation of the mTOR and Raptor complex (136). The dissociation of overexpressed mTOR and Raptor by leucine can be observed only when GβL is expressed in sufficient quantities. A rapamycin-induced decrease in the interaction between overexpressed mTOR and Raptor is also dependent on GβL expression levels. In view of the model proposed by Kim and colleagues, where Raptor enhances mTOR activity under low-binding-affinity conditions and inhibits mTOR activity under a high-binding-affinity conditions, caution must be exercised in interpreting the role of GβL and one must acknowledge the possibility that the regulation of the Raptor-mTOR complex by GβL could be far more complex than what it appears to be. Nevertheless, evidence is rapidly accumulating to strongly support the current consensus that GβL appears to play a positive role in mTOR signaling.
More recently, important progress was made in identifying a rapamycin-insensitive mTOR complex. Both Sabatini's and Hall's groups have identified a new component of mTOR complex, termed Rictor, a mammalian counterpart of Avo3p in yeast (128, 208). Like yeast TORC2, the Rictor-mTOR complex does not possess Raptor and therefore is referred to as mammalian TORC2 (mTORC2) (Fig. 1). The studies demonstrate that the Rictor-mTOR complex plays an important role in the regulation of cytoskeleton organization in mammalian cells. Similar to the yeast TORC2, mTORC2 seems to function upstream of Rho GTPases and protein kinase Cα (PKCα) to control cytoskeleton organization, although the precise mechanism of how mTORC2 regulates those effectors has not been determined (154, 208). Sabatini and coworkers have confirmed that the association between Rictor and mTOR is resistant to rapamycin under phosphate-rich conditions (208). Given the crucial role of Rictor in organizing the cytoskeleton, this may explain the previous observation that in yeast, rapamycin fails to disrupt TORC2 regulation of cytoskeleton. Taken together, these studies have shown that mTOR binds either Raptor or Rictor to form two distinct structural and functional complexes, mTORC1 and mTORC2, respectively, which are conserved between yeast and mammals.
FUNCTIONS AND EFFECTORS OF TOR
Downstream Effectors
Identification of the targets of TOR is critical for obtaining a complete picture of the cellular responses mediated by TOR. This process has been facilitated by studying rapamycin-induced cellular responses. Several downstream targets of mTOR have been found by this approach, including insulin receptor substrate 1 (IRS-1) (101, 102, 238), glycogen synthase (11, 228), cytoplasmic linker protein 170 (CLIP-170) (44, 45), eukaryotic elongation factor 2 (eEF2) kinase (200), hypoxia-inducible factor 1α (117, 247), p27 and p21 cyclin-dependent kinase inhibitors (116, 119), retinoblastoma protein (252), eukaryotic initiation factor 4E-binding protein (4EBP1) (86, 152), ribosomal S6 kinase 1 (S6K-1) (48, 197), STAT3 (274), Lipin (118), PKCα, δ, and ɛ (183, 208), and protein phosphatase 2A (PP2A) (17, 190) (Table 1). However, many of the proteins are likely indirect downstream targets of the mTOR signaling pathway, with only a few convincingly demonstrated as direct targets of TOR.
TABLE 1.
Downstream targets of mTOR
| Target | Related function | Phosphorylation by:
|
Phosphorylation site(s) | Consequences of regulation by mTOR | Reference(s) | |
|---|---|---|---|---|---|---|
| mTOR pathway present | mTOR, direct or indirect | |||||
| CLIP-170 | Cytoskeleton organization | + | NDa | ND | Activation | 44, 45 |
| eEF2 kinase | Protein kinase | + | Indirect | ND | Inhibition | 193 |
| Glycogen synthase | Glycogen synthesis | ND | ND | ND | Activation | 11, 218 |
| HIF-1α | Transcription | ND | ND | ND | Activation | 115, 237 |
| lipin | Adipose tissue development | + | ND | ND | Activation | 116 |
| 4EBP-1 | Translation inhibitor | + | Direct | T37, T46, S65, T70 | Inhibition | 85, 148 |
| PKCδ | Protein kinase | + | ND | S662 | Activation | 176 |
| PKCɛ | Protein kinase | + | ND | S729 | Activation | 176 |
| IRS-1 | Adaptor protein | + | ND | S302 | Inhibition | 99, 100, 228 |
| PP2A | Phosphatase | + | Direct | ND | Inhibition | 17, 183 |
| p21 CDKI | Protein kinase | ND | ND | ND | Inhibition | 114 |
| p27 CDKI | Protein kinase | ND | ND | ND | Inhibition | 114, 117 |
| S6K-1 | Protein kinase | + | Direct | S371, T389 | Activation | 48, 190 |
| Rb | Transcription factor | + | ND | S780, S795, S807, S811 | Inhibition | 241 |
| STAT3 | Transcription factor | + | Direct | S727 | Activation | 263 |
| URI | Component of transcriptional complex | + | ND | ND | Activation | 90 |
| TIF-IA | Component of transcriptional complex | + | Indirect | S44 | Activation | 159 |
| S199 | Inhibition | |||||
ND, not determined.
TOR in Regulation of PP2A
In both yeast and mammalian cells, TOR regulates PP2A (213). PP2A is a multimetric serine/threonine phosphatase that is highly conserved from budding yeast to humans. It is involved in a variety of cellular functions such as cellular metabolism, DNA replication, transcription, translation, cell proliferation, and cell transformation (166, 266). The PP2A holoenzyme is composed of a catalytic subunit C in complex with a scaffolding subunit A and a regulatory subunit B (82). The regulatory subunits affect PP2A substrate specificity and localization. In yeast, Pph21p and Pph22p are the homologs of mammalian PP2A catalytic subunits and are associated with an A subunit, Tpd3p, and one of two regulatory B subunits, Cdc55p or Rts1p (130). Sit4p is a yeast PP2A-related catalytic subunit that associates with one of its four regulatory proteins, Sit4p-associated proteins: Sap4p, Sap155p, Sap185p, or Sap190p. Loss of three of the SAPs, SAP155, SAP185, and SAP190, leads to a phenotype similar to that caused by SIT4 deletion, indicating that these regulatory SAPs are required for Sit4p function (156, 235). Tap42p is a yeast PP2A-interacting protein with a molecular mass of 42 kDa, which plays a critical role in regulating the phosphatase activity of Pphs and Sit4p (266).
Genetic screens of S. cerevisiae have identified TOR as an essential regulator of the Pphs/Sit4p and Tap42p complexes (64, 130). Under nutrient-rich conditions, phosphorylated Tap42p binds and inhibits Pph21/22p and Sit4p (15, 64) (Fig. 3). On nutrient starvation or in the presence of rapamycin, dephosphorylated Tap42p dissociates from Pph21/22p and/or Sit4p and relieves Tap42p inhibition on Pphs/Sit4p. Most recently, it has been shown that dephosphorylated Tap42p can also activate Pph21/22p and Sit4p (68). The activated Pphs/Sit4p subsequently dephosphorylates downstream effectors, such as nitrogen permease reactivator kinase (Npr1p) and GATA transcription factor Gln3p (55) (Fig. 4). Although the details of the mechanism remain unclear, two models have been postulated to explain how TOR regulates the Tap42p-Pphs/Sit4p complex.
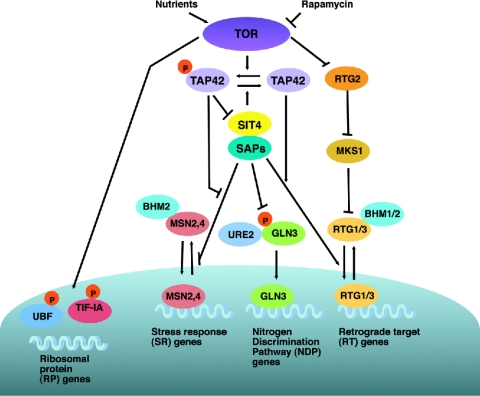
FIG. 3.
Regulation of PP2A by TOR in yeast.
FIG. 4.
Transcriptional regulation by TOR. In yeast, TOR regulates multiple transcription factors through its inactivation of PP2A activity. Recently, a study of yeast has indicated that the regulation of ribosomal protein (RP) genes expression is independent of Tap42p. TOR-dependent regulation of UBF, TIF-1A, and URI has been demonstrated in mammalian system.
The first model postulates that TOR phosphorylates Tap42p and increases Tap42p binding to Sit4p and/or Pph21/22p, thereby inactivating their phosphatase activities (Fig. 3). This is supported by the finding that TOR phosphorylates Tap42p both in vivo and in vitro in a rapamycin-sensitive manner (130). Interestingly, Tap42p is also subjected to Pphs/Sit4p-dependent dephosphorylation: inactivation of Pphs, through mutation of Cdc55p and Tpd3p regulatory subunits (Fig. 3), leads to rapamycin resistance, enhanced phosphorylation of Tap42p, and increased binding between Tap42p and Pphs/Sit4p. These data suggest that the phosphorylation status of Tap42p is determined by reciprocal regulation by both TOR and Pphs/Sit4p (130) (Fig. 3).
Alternatively, it has been suggested that TOR indirectly controls the binding of Tap42p to Sit4p by regulating the Tap42p-interacting protein Tip41p, an inhibitor of the interaction between Tap42p and Sit4p (126) (Fig. 3). TOR-dependent phosphorylation of Tip41p causes dissociation of Tip41p from Tap42p and consequently promotes the binding of Tap42p to Sit4p, whereas rapamycin stimulates the binding between Tip41p and Tap42p via Sit4p-dependent dephosphorylation of Tip41p, suggesting that Tip41p plays a role in the amplification of Sit4p phosphatase activity in a feedback loop under conditions where TOR is inactive (126) (Fig. 3). These lines of evidence suggest that TOR might regulate Tap42p both directly and indirectly to inhibit Pphs/Sit4p phosphatase activity. Although the details of TOR regulation of the Tap42p-Pphs/Sit4p complex need to be further clarified, it has been generally accepted that TOR inhibits the function of the phosphatase Pph21/22p and Sit4p via promoting the binding between Tap42p and Pphs/Sit4p (Fig. 3).
α4 is mammalian ortholog of yeast Tap42p that interacts with the mammalian catalytic subunit(s) of PP2A (124, 172). Recombinant α4 represses PP2A activity in vitro (173). In α4 knockout B cells, S6K activation induced by anti-immunoglobulin M antibody was defective, suggesting involvement of α4 in the mTOR-S6K pathway (172). In addition, it has been observed that mTOR directly phosphorylates PP2A and represses its functions in vitro whereas rapamycin treatment activates PP2A activity in vivo (190), indicating that mTOR negatively regulates PP2A through direct phosphorylation. However, it remains unclear whether mTOR regulates PP2A by affecting the association between α4 and PP2A, as seen in yeast, where TOR regulates the binding between Tap42p and Pphs/Sit4p. Further studies are required to elucidate the mechanisms by which mTOR regulates PP2A in mammalian cells.
Given a relatively broad spectrum of substrates targeted by PP2A, the regulation of this highly conserved phosphatase by TOR links TOR signaling to a variety of cellular activities. Indeed, in yeast most of the readouts of TOR signaling are mediated through TOR-dependent regulation of Pphs/Sit4p. TOR globally inhibits transcription of stress-responsive genes, starvation-specific genes, nitrogen discrimination pathway associated genes, and retrograde target genes by sequestering several responsive transcription factors in the cytoplasm, such as Gln3p, Msn2p-Man4p (15), and Rtg1p-Rtg3p (140) (Fig. 4). These processes are reportedly dependent on regulation of the Tap42p-Pph21-Pph22p-Sit4p system by Tor (see below). Similarly, in mammalian cells, the negative regulation of PP2A by mTOR is regarded as one of the mechanisms utilized by mTOR to maintain the phosphorylation status of several of its downstream effectors, such as S6K, 4EBP1, and PKC (183, 189, 190).
TOR in Regulation of Protein Translation
TOR targets S6K and 4EBP to control protein translation initiation and cell size.
In Drosophila, mutation of dTOR results in a dramatic effect on cell size. Drosophila cells containing nonfunctional dTOR are approximately four times smaller than wild-type cells (75, 181, 278). Similarly, in mammalian systems, rapamycin treatment significantly reduces the size of cultured mammalian cells (73, 123). Collectively, these results demonstrate that TOR is an important cell size regulator in both Drosophila and mammals. Moreover, an increasing amount of data has been accumulated indicating that S6K and 4EBP1, two of the most well-characterized TOR targets involved in protein translation, are key TOR pathway elements that mediate the regulation of cell size (73).
S6K is the well-known ribosomal protein S6 kinase in mammalian cells (10, 246). Activating the phosphorylation of S6K contributes to increased kinase activity and results in an elevated phosphorylation of ribosomal S6 polypeptide (198). It has been reported that S6 phosphorylation selectively increases the translation of mRNA transcripts containing a tract of pyrimidine (TOP) motif. These TOP-containing mRNAs often code for ribosomal proteins and other translational regulators (168). In this way, S6K enhances the translational capacity of cells. A significant amount of evidence indicates that S6K is a key downstream target of TOR (129, 245). Overexpression of rapamycin-resistant mTOR activates S6K even in the presence of rapamycin (29), whereas knock down of mTOR expression via siRNA represses the activation of S6K (135). Furthermore, rapamycin treatment also blocks S6K activation by a variety of growth-stimulating signals (48, 197).
TOR phosphorylation of S6K involves multiple sites. Among them, Thr389 and Ser371 are the major mTOR-targeting sites in vitro (33, 207). Phosphorylation on these two sites by mTOR is essential for S6K activation (186), since rapamycin was shown to inhibit phosphorylation on these sites (93). In addition, five amino acid residues (Phe-Asp-Ile-Asp-Ile) located close to the N terminus of S6K form the TOS motif (211). Deletion or mutation of the TOS motif significantly blocked insulin-induced activation of S6K (211). Similarly, deletion of the N terminus of S6K rendered it insensitive to rapamycin treatment (38, 265), suggesting that the TOS motif is critical for mTOR-mediated activation of S6K.
Importantly, S6K and 4EBP1 are two key elements of the TOR pathway that mediate regulation of cell size by TOR. Fruit flies deficient in S6K are smaller than wild-type flies and display decreased cell size (169). Similarly, S6K1 knockout mice have a body size approximately 80% of that of their wild-type littermates (230). This phenotype is similar to those seen in Drosophila with dTOR mutations, as well as mice treated with rapamycin (24, 108). Furthermore, S6K overexpression rescues TOR knockout phenotypes in Drosophila (278). These data identify S6K as a major TOR target that mediates the effects of TOR on cell size regulation.
4EBP1 is another well-characterized TOR target (79, 86, 152). Hypophosphorylated 4EBP1 acts as a translational repressor by binding and inhibiting the eukaryotic translation initiation factor 4E (eIF4E), which recognizes the 5′-end cap of the majority of eukaryotic mRNAs (81, 92, 152). Phosphorylation of 4EBP1 by mTOR results in the dissociation of 4EBP1 from eIF4E, thereby relieving the inhibitory effect of 4EBP1 on eIF4E-dependent translation initiation (151, 185). Evidence for mTOR-mediated phosphorylation of 4EBP1 came from studies demonstrating that rapamycin reduces the phosphorylation of 4EBP1 and partially prevents its dissociation from eIF4E (19, 86, 152). Subsequently, it was shown that overexpression of rapamycin-insensitive mTOR resulted in 4EBP1 phosphorylation even in the presence of rapamycin (32). Several phosphorylation sites of 4EBP1 have been identified, including Thr37, Thr46, Ser65, Thr70, Ser83, Ser101, and Ser112 (70, 105, 260). Phosphorylation of Thr70 and Ser65 is more sensitive to rapamycin treatment than is phosphorylation of Thr37 and Thr46 (80, 170).
Besides the direct phosphorylation sites, two other motifs are important for efficient phosphorylation of 4EBP in vivo. One of these is a TOS motif located at the end of its C terminus (Phe-Glu-Met-Asp-Ile) (211), and the other is a RAIP motif (Arg-Als-Ile-Pro) generated by caspase cleavage of the Asp24-Gly-25 bond in 4EBP1 (244). 4EBP1 also plays a role as a cell size regulator. Overexpression of 4EBP carrying a TOS motif mutation results in a decreased size of mammalian cells (73). Thus, mTOR-dependent phosphorylation of both S6K and 4EBP might represent one of the underlying mechanisms by which mTOR positively regulates cell growth and especially cell size in mammalian cells.
In yeast, loss of TOR function also results in a rapid and strong inhibition of translation initiation (14). In contrast to mammalian cells, a rapamycin-sensitive S6K homolog has yet to be found (6, 131), nor have yeast ribosomal protein mRNAs containing a 5′-TOP sequence been found (131). Moreover, the phosphorylation of S6 protein does not correlate with yeast cell growth (131). These observations indicate that mTOR-S6K-dependent translation control is unique in metazoans and that yeast TOR might use distinct mechanisms to control protein translation. One of the postulated mechanisms involves TOR-dependent phosphorylation of eIF4E. First, genetic studies demonstrate that eIF4E (Cdc33p) and TOR mutants have remarkably similar phenotypes (14, 58), indicating that eIF4E plays an important role in mediating the effects of yeast TOR. Second, biochemical analysis indicates that TOR maintains the eIF4E-eIF4G complex by both stabilizing eIF4G, the scaffolding component in the eIF4F complex, and recruiting other critical initiation factors such as eIF4A and eIF3 (21) into the initiation complex. Finally, Eap1p protein has recently been identified as interacting with eIF4E in S. cerevisiae (51). Eap1p contains a canonical eIF4E-binding motif and inhibits cap-dependent translation. Eap1p competes with eIF4G for binding to eIF4E in vivo, suggesting that Eap1p plays a role similar to that of mammalian 4EBP1. Targeted disruption of the EAP1 gene confers partial resistance to growth inhibition by rapamycin, indicating the involvement of EAP1 in the TOR signaling pathway (51). Therefore, the yeast eIF4E system might be the major player that mediates TOR regulation of translational machinery in yeast.
TOR regulates ribosome biogenesis.
Ribosome biogenesis is an energy- and nutrient-consuming process, which involves more than 100 genes and all three RNA polymerases (pol) (I, II, and III) (261). To regulate cell growth and proliferation on the basis of cellular energy levels, nutrient availability, and metabolic status, cells must tightly control the production and abundance of the ribosomes. In both yeast and mammals, TOR regulates ribosome biogenesis at multiple levels, including transcription, rRNA processing, and translation. Inhibition of the TOR pathway with rapamycin or by nutrient starvation results in a downregulation of transcription of ribosomal protein mRNAs (pol II dependent) (35, 99, 196), tRNA, and rRNA (pol I and pol III dependent) (196, 276). Several mechanisms have been proposed to explain how TOR regulates ribosome biogenesis.
One mechanism by which mTOR regulates ribosome biogenesis and translation involves the action of S6K and S6 (also see the previous section). As described above, phosphorylation of S6 selectively enhances the translation of mRNA transcripts that contains a 5′-TOP sequence in their 5′ untranslated region (5′-UTR) (168). Many mRNA transcripts encoding ribosomal proteins and translational regulators contain TOP sequence in their 5′-UTR. Therefore, phosphorylation of S6 may increase the global translation capacity of a cell. However, this model is perhaps oversimplified. Recent studies have indicated that S6K activity does not exactly correlate with increased translation of 5′-TOP-containing mRNAs (231, 234, 239). In addition, it was reported that S6 phosphorylation and translation of 5′-TOP-containing mRNAs are still responsive to mitogens in a rapamycin-dependent fashion in S6K-1 S6K-2 double-knockout murine cells (187). These studies indicate that S6K may not be the sole mediator of transmission of TOR signaling to S6 protein.
rDNA is DNA that specifically codes for components of the ribosome, and TOR is a conserved regulator of rDNA transcription. Nutrient starvation and rapamycin treatment rapidly inhibit rDNA transcription in both yeast and mammalian cells (146, 159, 196, 276). Zheng's group recently reported that TOR regulates the structure of the nucleolus, the region of the nucleus where rDNA transcription and ribosome assembly occurs (248). During nutrient starvation or rapamycin treatment, both RPD3 and SIN3, members of the histone deacetylase family, bound rDNA chromatin, resulting in histone H4 deacetylation, rDNA chromatin condensation, and nucleolar size reduction. These authors also found that this chain of events further leads to the delocalization of RNA pol I, the polymerase used for rDNA transcription, from the nucleolus and subsequently results in rDNA transcriptional inhibition. These observations indicate that TOR regulates rDNA transcription through multiple chromatin-dependent mechanisms to control ribosome biogenesis (248). In addition to these observations, Grummt's group has shown that the activity of TIF-IA, an essential initiation factor associated with the initiation-competent form of pol I, is also regulated in a mTOR-dependent manner (165). Rapamycin inhibits TIF-IA activity by decreasing Ser44 phosphorylation and increasing Ser199 phosphorylation on TIF-IA. The phosphorylation of Ser44 and Ser199 is mediated by both mTOR-dependent kinases including S6K as well as phosphatase PP2A activity. Furthermore, inactivation of TIF-IA by rapamycin causes a dissociation of TIF-IA from the pol I complex and leads to translocation of TIF-IA into the cytoplasm, thereby suppressing pol I complex activity. Another mechanism, proposed by Hannan's group, is that mTOR activates 45S ribosomal transcription by activating nucleolar transcription upstream binding factor (UBF) (94). mTOR-dependent activation of UBF goes through S6 kinase activation. Although S6K failed to phosphorylate UBF as well as TIF-IA in vitro, phosphorylated UBF binds to SL-1 (basal rDNA transcription factor) and stimulates rDNA transcription to enhance ribosomal biogenesis (94). Thus, mTOR activates pol I-dependent rDNA transcription by multiple mechanisms.
It has been reported that TOR also regulates ribosome biogenesis by modulating rRNA processing, including processing of the 35S precursor rRNA (5, 196). Rapamycin destabilized these mRNAs in wild-type yeast cells but failed to destabilize them in rapamycin-resistant Tor1p-containing yeast, indicating that TOR not only stimulates transcription but also enhances mRNA stability to increase ribosomal biogenesis. Interestingly, the effect of rapamycin on mRNA stability is not limited to ribosomal mRNAs. Indeed, rapamycin treatment of mammalian cells destabilizes the interleukin-3 and cyclin D1 mRNAs (13, 103).
How does TOR regulate such a variety of processes to control ribosome biogenesis? It has been proposed that TOR regulation of the Tap42p-Pphs-Sit4p system in yeast is critical in these processes. A mutation in TAP42 inhibits polyribosome formation, suggesting that TAP42 functions upstream of translation initiation (64). Moreover, TPD3 (A subunit of PP2A) and SIT4 function upstream of RNA pol III and pol II, respectively (258). From these observations, Powers and Walter have proposed that the Tap42p-Pphs-Sit4p system plays a major role in mediating TOR signaling to control ribosomal biogenesis (196).
TOR in Regulation of Localization of Transcription Factors
On nutrient starvation, yeast cells become stationary, a phenotype featured by a global reduction of protein synthesis and an altered gene expression pattern. While a few pol II-transcribed genes that are required for adapting to the “low-nutrient crisis conditions” are turned on, RNA pol I-, II-, and III-associated gene transcription is down-regulated. When nutrients are abundant, TOR inhibits starvation-specific gene expression by sequestering several nutrient-responsive transcription factors within the cytoplasm (15, 35, 99) (Fig. 4). For instance, GLN3 is a GATA family transcription factor responsible for expression of nitrogen discrimination pathway genes that promote the utilization of less-favorable nitrogen sources when nutrients are limited (15). Under nutrient-rich conditions, Gln3p is retained in the cytoplasm by binding with Ure2p, a cytoplasmic retention protein that is phosphorylated by TOR (35, 99) (Fig. 4). Rapamycin-induced dephosphorylation of Ure2p disrupts its interaction with Gln3p and results in subsequent nuclear translocation of Gln3p (Fig. 4). Therefore, TOR inhibits the transcription activity of Gln3p by cytoplasmic sequestration of this transcription factor.
TOR also inhibits the transcription of stress-responsive genes by sequestering the general stress transcription factors Msn2p and Msn4p (Zn2+ transcription factor) in the cytoplasm (15). TOR sequesters Msn2p and Msn4p possibly by promoting their association with the cytoplasmic 14-3-3 proteins Bmh1p and Bmh2p (Fig. 4). Another example comes from the fact that TOR negatively regulates the heterodimeric basic helix-loop-helix/Zip transcription factors Rtg1p and Rtg3p (53, 140). RTG1 and RTG3 regulate the expression of tricarboxylic acid and glyoxylate cycle genes that are involved primarily in α-ketoglutarate synthesis used for generating several amino acids, such as glutamine and glutamate. The mechanism by which TOR inhibits Rtg1p-Rtg3p is unknown, but epistasis analysis indicates that this inhibition might occur through Rtg2p and Mks1p (65, 221, 224). TOR seems to antagonize Rtg2p, which is a negative regulator of Mks1p (65), and Mks1p inhibits Rtg1p-Rtg3p (65, 221). It has also been reported that 14-3-3 proteins induce Rtg3p to adopt an inactive configuration (254) (Fig. 4). Since Gln3p and Rtg1p-Rtg3p respond to intracellular levels of glutamine, the TOR pathway appears to sense glutamine, among other unknown nutrient compounds, to regulate cellular nutrient status by modulating metabolism and transporters (53). Taken together, these studies indicate that TOR regulates the intracellular localization of transcription factors to regulate gene transcription.
Another mechanism used by that TOR to regulate transcription involves regulating RNA polymerase. Krek's group purified a 1-MDa protein complex from mammalian cells (91), which includes a member of the prefoldin (PFD) family, termed URI (for “unconventional prefolding RPB5 interactor”) and Rpb5, a subunit shared by RNA pol I, II, and III (269). Sequence comparison indicates that URI is an evolutionarily conserved protein (269). Yeast genetics indicate that the majority of the genes whose expression is affected by loss of URI overlap with those genes that have been identified previously to be affected by amino acid starvation or rapamycin treatment (91). Interestingly, biochemical analyses of mammalian cells show that URI phosphorylation is regulated by mTOR, suggesting that TOR regulates nutrient-dependent transcription at least partially through URI phosphorylation and that this mechanism might be conserved from yeast to humans (91).
In mammalian cells, mTOR is also involved in mitogen-induced transcription regulation. For example, it phosphorylates and activates the transcriptional activator STAT3 on Ser727 (274). It has been reported that Ser727 phosphorylation contributes to STAT3 activity (264). Interestingly, primary sequence analysis shows that STAT3 has both a TOS motif (FDMEL) and a RAIP motif (RAIL). Thus, like S6K and 4EBP1, STAT3 appears to be recruited by Raptor to be phosphorylated by mTOR.
Effect of TOR on Cellular Nutrient Levels by Controlling Both Autophagy and Amino Acid Transporters
Autophagy is a catabolic membrane-trafficking phenomenon observed in yeast and mammalian cells (2, 138). Nutrient starvation and serum deprivation induce autophagy, which is defined as the hydrolysis of cytoplasmic contents to generate nutrients for cellular adaptation under nutrient starvation conditions. The observation that rapamycin treatment also induces autophagy indicates the involvement of TOR in the process (177). In both yeast and mammalian cells, TOR inhibits autophagy. APG1 is reportedly an essential kinase gene required for autophagy and is negatively regulated by TOR in yeast (164). Under nutrient-poor conditions or after rapamycin treatment, Apg1p forms a complex with Apg13p and turns on autophagy. Under nutrient-rich conditions, TOR enhances Apg13p phosphorylation and prevents its association with Apg1p, thereby inhibiting the autophagy (23, 132, 229).
It has been reported that inhibition of autophagy by mTOR is implicated in the pathogenesis of neurodegenerative disorders such as Huntington's disease (199). Treatment with the rapamycin ester CCI-779 enhanced the clearance of aggregates of the mutant Huntingtin protein in the brain and attenuated pathological symptoms in a mouse model of Huntington's disease.
It is well known that TOR activity is largely regulated by the intracellular amino acid concentration. Interestingly, TOR also regulates intracellular amino acid levels by modulating the activities of amino acid permeases. It has been reported that TOR regulates two types of amino acid permeases. One type is the general amino acid permease (GAP1), and the other includes those that are specific for single amino acids, such as the histidine permease HIP1 and tryptophan permease TAT2 (16, 214). Interestingly, TOR exerts opposite effects on HIP1, TAT2, and the general permease GAP1 in response to nutrient availability. For example, under nutrient-poor conditions, TOR inhibits Tat2p by enhancing its ubiquitination and consequent degradation (16, 214). In contrast, under the same conditions, TOR activates Gap1p by preventing its ubiquitination. Both effects of TOR are mediated by the same Ser/Thr nitrogen permease reactivator kinase, Npr1p (61, 87, 88, 253). It has been reported that Npr1p is a phosphoprotein and that its phosphorylation is controlled by TOR. Under nutrient-rich conditions, TOR maintains Npr1p phosphorylation, thereby inhibiting it and leading to Gap1p degradation. On the other hand, under nutrient-poor conditions, Npr1p is dephosphorylated and activated, thereby resulting in an activation of the general amino acids permease Gap1p. Therefore, regulation of Npr1p by TOR provides a molecular mechanism by which TOR regulates amino acid transport. It has been reported that the activation of Npr1p kinase is also mediated by Sit4p-Pphs phosphatase activity (126).
REGULATION OF TOR
TOR and Growth Factors
Mammalian cell growth is regulated by both growth factors and the availability of nutrients. mTOR is involved in both growth factor and nutrient signaling in mammalian cells. One of the important signaling pathways that controls cell growth is the PI3K pathway (141, 180). Many growth factors, including insulin, activate PI3K, which phosphorylates phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] at the 3′ position to generate PtdIns(3,4,5)P3 (27). The 3-phosphoinositide-dependent protein kinase 1 (PDK1) and PKB (also known as Akt) bind with PtdIns(3,4,5)P3 through their pleckstrin homology domains and are recruited to the membrane. This recruitment results in phosphorylation of Akt by PDK1 and activation of Akt. It has been shown that, in mammals, stimulation of growth factors such as insulin results in the phosphorylation of both S6K and 4EBP1 via the PI3K pathway (48, 151, 197). The phosphorylation of S6K and 4EBP1 could be inhibited by either a low concentration of wortmannin, a specific inhibitor of PI3K, or rapamycin. These studies strongly suggest that both the mTOR pathway and PI3K signaling are involved in the regulation of the phosphorylation of S6K and 4EBP1.
The question that naturally arises from these observations is whether TOR functions downstream of insulin-PI3K-Akt or whether it represents a parallel pathway that impinges on S6K and 4EBP1. Treatment with rapamycin does not interfere with any steps between binding of insulin to its receptor and activation of PI3K-Akt (85), suggesting that PI3K-Akt is upstream of or parallel to mTOR. The sequence around Ser2448 of mTOR matches the consensus Akt phosphorylation site, making mTOR an attractive target for Akt phosphorylation. Insulin stimulation or Akt overexpression in cells enhances the Ser2448 phosphorylation of mTOR in vivo (174, 220). In addition, the purified active Akt directly phosphorylates mTOR in vitro (222), suggesting that mTOR acts downstream of PI3K. Furthermore, increased phosphorylation of Ser2448 on mTOR in response to PI3K-Akt correlates with enhancement of S6K and 4EBP1 phosphorylation (202), suggesting that PI3K-Akt-induced Ser2448 phosphorylation of mTOR is important for its kinase activity toward S6K and 4EBP1 in vivo. Consistent with this observation, the phosphorylation of Ser2448 of mTOR is also controlled by amino acid availability and the intracellular ATP levels (123, 174). However, it has also been reported that phosphorylation of Ser2448 in mTOR may not be required for mTOR kinase activity (222). Furthermore, the phosphorylation of Ser2448 on mTOR by PI3K-Akt activation represents only one of the mechanisms by which growth factors such as insulin stimulate S6K and 4EBP1. However, it is also possible that PI3K-Akt can regulate S6K independently of the mTOR pathway. S6K mutants with deletion of the TOS motif and/or N- and C-terminal regions become resistant to rapamycin treatment yet remain responsive to both insulin and the PI3K inhibitor (85, 211). Thus, the PI3K pathway can regulate S6K in both an mTOR-dependent and -independent manner. Since TOR controls cell growth in both yeast and plants, which lack an equivalent PI3K signaling pathway, the PI3K pathway may have been added to the TOR pathway later in evolution.
TOR and Nutrients
In yeast, the TOR pathway can be activated by both nitrogen and carbon sources (19, 53, 144). Although the exact nature of this regulation remains unclear, Hall and colleagues found that the amino acid glutamine is particularly important as a nitrogen source to regulate TOR signaling (53). In mammalian cells, amino acid deprivation causes rapid dephosphorylation of both 4EBP1 and S6K (96, 259, 270) whereas amino acid supplementation stimulates 4E-BP1 and S6K phosphorylation. Rapamycin abrogates these responses, suggesting that mTOR may also be involved in amino acid signaling in mammalian cells. Glucose is another nutrient source that stimulates 4EBP1 and S6K phosphorylation in an mTOR-dependent manner (137, 184). However, whether glucose itself, glucose metabolites, or glucose-derived products directly stimulate mTOR remains unclear. In addition to affecting protein synthesis, both amino acids and glucose regulate gene transcription. A recent transcriptional profiling of mammalian cells has revealed that rapamycin treatment mimics glutamine or leucine starvation more than it mimics glucose starvation (188). Moreover, Dennis et al. have observed that depletion of ATP levels by the glucose analog 2-deoxyglucose or by the mitochondrial uncoupler rotenone inhibited S6K and 4EBP phosphorylation in an mTOR-dependent manner even in the presence of amino acids (62). They also found that in vitro, mTOR has a Km for ATP of around 1 mM, which is significantly higher than the Km for most kinases. These observations led them to propose that mTOR is an ATP sensor under physiological conditions to regulate translation initiation. However, the intracellular ATP concentration is normally higher than the Km of mTOR, implying that a drastic change of ATP levels within the cells is necessary for altering mTOR kinase activity.
The question that logically arises from these observations is whether mTOR is the primary physiological ATP sensor in vivo. For instance, several groups recently have shown that 5′-AMP-activated protein kinase (AMPK), a kinase that is activated by a high AMP/ATP ratio in response to ATP depletion, also modulates S6K activity in an mTOR-dependent manner (25, 137, 142). AMPK is thought to be a much more sensitive ATP sensor than mTOR because the intracellular AMP concentration is much lower than that of ATP (98). Thus, a small change in ATP levels could greatly increase the AMP/ATP ratio, suggesting that AMPK may function as a primary physiological sensor of the intracellular energy level to inhibit the mTOR signaling pathway. In summary, TOR may serve as a highly conserved signaling nexus for a variety of nutrient signals.
How does TOR transmit nutrient signals? Although the details remain mysterious, a few studies suggest possible mechanisms involved. For example, it has been reported that TOR controls the expression of nutrient-regulated genes by modulating the subcellular localization of several nutrient-responsive transcription factors in the cytoplasm, including Gln3p, Gat1p, Rtg1p, Rtg3p, and Msn2p-Msn4p (Fig. 4). Interestingly, glutamine starvation activates only Gln3p and Rtg1p-Rtg3p but not Gat1p and Msn2p-Msn4p, suggesting that nutrient sources other that glutamine target TOR to regulate nutrient-responsive transcription factors such as Gat1p and Msn2p-Msn4p (53). These lines of evidence suggest that different nutrient signals may impinge on TOR to control distinct transcription factors. In addition, it has been reported that TOR affects its downstream kinases to transmit nutrient signals.
One of these kinases is GCN2 (42). In yeast, the Gcn2p kinase senses intracellular amino acid availability through a tRNA-binding domain that exhibits similarity to histidyl-tRNA synthetase (263). On amino acid limitation, the levels of uncharged tRNA increase significantly, resulting in an increase in binding to the Gcn2p kinase. The binding of uncharged tRNA activates Gcn2p kinase, which consequently phosphorylates and activates Gcn4p, a transcriptional activator responsible for the induction of several genes involved in amino acid biosynthesis, thus leading to increased synthesis of amino acids (109, 110). Recently, it was reported that rapamycin treatment reduced Ser577 phosphorylation of Gcn2p through the phosphatase complex Tap42p-Sit4p, thus activating Gcn2p kinase and ultimately leading to inhibition of translation initiation. This inhibition of translation is mediated through phosphorylation of Ser51 on elF2α by Gcn2p (42). Activation of Gcn2p by dephosphorylation of Ser577 also requires the binding of uncharged tRNA to the C-terminal histidyl-tRNA synthetase-related domain of Gcn2p. These data suggest that both TOR activity and amino acid signaling impinge on Gcn2p kinase to influence protein synthesis in yeast cells. The Gcn2p protein is conserved in mammals (20, 134, 232). Hence, it would be interesting to determine whether the same regulatory mechanism is also conserved in mammals.
TSC1 and TSC2 as Negative Regulators of mTOR Functions
The recent identification of the TSC1-TSC2 complex and its function significantly advances the understanding of how TOR cooperatively receives signals from both nutrient and growth factor pathways. Tuberous sclerosis is a relatively common autosomal dominant disorder, occurring in approximately 1 in 6,000 to 10,000 of the population and characterized by the development of benign tumors called hamartomas in a variety of organs. Common clinical symptoms include seizures, mental retardation, autism, kidney failure, facial angiofibromas, and cardial rhabdomyomas (83). Two independent genes are responsible for TSC disease. TSC1 encodes a 130-kDa protein, also known as hamartin, with several coiled-coil domains but no obvious catalytic domains (256) (Fig. 5). TSC2 encodes a 200-kDa protein, also known as tuberin, with a coiled-coil domain and a C-terminal region with homology to the Rap GTPase-activating protein (GAP) (69a) (Fig. 5).
FIG. 5.
Structural domains and phosphorylation sites of TSC1 and TSC2. TSC1 (hamartin) has transmembrane and coiled-coil domains at the N- and C-terminal regions, respectively. Thr447, Ser584, and Thr1047 are cdc2-dependent phosphorylation sites, and these phosphorylations negatively regulate TSC1-TSC2 complex activity (8). TSC2 (tuberin) consists of a leucine zipper domain, two coiled-coils domains, and a Rheb-GAP domain. It has been reported that the N-terminal coiled-coil domain is critical for its association with TSC1. Ser939, Ser1130, Ser1132, and Thr1462 are Akt-dependent phosphorylation sites (57, 122, 161, 194), and Ser1254 is a MAPKAP-K2 phophorylation site (149). Phosphorylation of TSC2 by Akt and MAPKAP-K2 inactivates TSC1-TSC2 complex activity. Thr1271 and Ser1387 are sites of phosphorylation by AMPK, and AMPK phosphorylation activates the TSC1-TSC2 complex (123).
TSC1 and TSC2 form a physical and functional complex that is most stable as a heterodimer (175, 191, 257). Homozygous inactivation of either TSC1 or TSC2 in mice is lethal to embryos, while heterozygous animals are tumor prone (9, 139, 182), indicating that both TSC1 and TSC2 function as tumor suppressors. In Drosophila, inactivation of either dTSC1 or dTSC2 increases cell size and cell proliferation while overexpression of both dTSC1 and dTSC2 together decrease cell size, supporting the fact that TSC1 and TSC2 form a functional complex to regulate cell growth (74, 193, 240). Biochemical studies demonstrated that the TOR pathway is highly activated in either TSC1 or TSC2 mutant cells (84, 145). Furthermore, the high-level phosphorylation of S6K and 4EBP1 can be effectively inhibited by rapamycin (133). Overexpression of TSC1 and TSC2 suppress both S6K and 4EBP1 phosphorylation, and transient knock down of TSC2 by RNAi enhances S6K phosphorylation (122, 242). These results provide strong evidence that TSC1-TSC2 negatively regulates TOR activity. Furthermore, a rapamycin-resistant S6K mutant is also resistant to inhibition by overexpression of TSC1-TSC2, further supporting a role for TSC1-TSC2 in TOR regulation (122, 242).
TSC2 in growth factor signaling: effects of Akt.
It is thought that activation of mTOR requires integration of signals from at least four different sources including PI3K activation, amino acids, and adequate intracellular ATP levels, as well as lipids, especially phosphatidic acid (PA) (40). Lack of any one of these inputs inhibits the ability of mTOR to fully phosphorylate and activate S6K. However, none of these signals, except PA, has been found to directly target TOR. Therefore, an important question in the field is that of which molecules function between the upstream signals and TOR and how different signaling pathways are integrated at TOR. Recent genetic analyses using Drosophila have indicated that dTSC1 and dTSC2 act downstream of the insulin/insulinlike growth factor receptor in the control of cell growth (74, 195, 240). The functional position of the TSC complex is between dAkt and dTOR (75). Biochemical studies by several groups have shown that Akt directly phosphorylates TSC2 and inhibits its function (57, 122, 161, 194) (Fig. 6). Several mechanisms of TSC2 inactivation by Akt phosphorylation have been proposed, including (i) mislocalization of TSC2, (ii) dissociation between TSC2 and TSC1, and (iii) degradation of TSC2 mediated by ubiquitination (18, 122, 194). Knock down of TSC2 by RNAi causes an enhancement of S6K phosphorylation and activity, while overexpression of TSC1 and TSC2 inhibits S6K phosphorylation and activity in both Drosophila and mammalian cells (75, 122, 194, 242). Taken together, a favored model of how insulin activates TOR signaling is that Akt phosphorylates TSC2 and relieves TSC2 inhibition of TOR signaling. Interestingly, it has been reported that loss of the PTEN phosphatase, a negative regulator of the Akt pathway, is associated with Cowden's disease and the Bannayan-Riley-Ruvalcaba syndrome, two dominantly inherited hamartoma syndromes (150, 162). In PTEN−/− cells, the Akt-dependent phosphorylation of Ser939 and T1462 on TSC2 is constitutive (161). Furthermore, both S6K and 4EBP1 phosphorylations are highly enhanced in PTEN−/− cells (176). Thus, it is appealing to propose that one of the mechanisms underlying these PTEN-related hamartomas is due to inactivation of TSC2 function. However, a recent study by Dong and Pan challenges the current model in which TSC2 functions as a downstream target of Akt and mediates Akt effects on mTOR (66). Using a transgenic strategy in flies, Dong and Pan demonstrated that mutation of the Akt phosphorylation sites did not significantly inhibit the biological activity of dTSC2 supporting fly development, indicating that dTSC2 is not the major target of dAkt during normal Drosophila development (66). Whether this conclusion is applicable to the mammalian system remains to be determined.
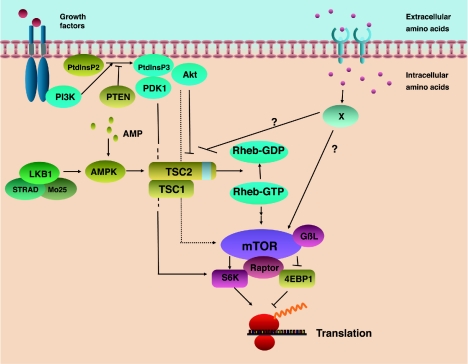
FIG. 6.
TOR functions as a multiple-channel sensor for a variety of upstream signals. Multiple signaling cascades impinge on the TSC1-TSC2 complex-Rheb-mTOR complex to regulate translational machinery. LKB1 complex consists of LKB1 kinase, STRAD, and Mo25. STRAD and Mo25 (12, 26) are required for full activation of LKB1 kinase and are important for cytosolic localization of LKB1. It has been reported that LKB1 complex activates not only AMPK but also another 12 AMPK-related kinases (153). Whether these AMPK members participate in the regulation of translation machinery has not yet been determined. Protein X represents a putative amino acid-sensing molecule.
TSC2 activity is also regulated by the PKC and mitogen-activated protein kinase (MAPK) pathways (241). Blenis and collegues have recently reported that phorbol 12-myristate 13-acetate (PMA)- and epidermal growth factor-induced activation of the mTOR pathway are also suppressed by TSC1 and TSC2 overexpression (241). They have shown that activation of the PKC-MAPK pathway also leads to phosphorylation of TSC2 at multiple sites. Interestingly, the Akt phosphorylation sites of TSC2 (S939 and T1462) are also enhanced by PMA without affecting Akt activity itself. These studies suggest that PMA-dependent kinase may phosphorylate Akt phosphorylation sites on TSC2 and inhibit TSC2 activity. Subsequently, the same group has demonstrated that one of the Akt phosphorylation sites is also phosphorylated by RSK1 (57, 204). They have also shown that PA-induced S6K activation is inhibited by overexpressed TSC1 and TSC2 (241). Although PA is thought to activate mTOR via direct binding (71), it will be interesting to test whether it affects TSC1-TSC2 activity. Taken together, these studies demonstrate that pathways from multiple growth factors integrate on TSC2 to regulate the mTOR activity.
TSC2 in nutrient-sensing pathways: effects of amino acids.
It is widely accepted that TOR is in a pathway linking intracellular amino acid levels to the regulation of cell growth control. Recent studies have revealed that TSC1-TSC2 may mediate amino acid signals to regulate TOR function. It was recently reported that amino acid depletion causes TSC2 phosphorylation in vivo and may contributes to TSC2 activation (123). Amino acid-induced S6K and 4EBP1 phosphorylation is inhibited by overexpressed TSC1 and TSC2 (122, 242). More importantly, loss of TSC1 or TSC2 confers cellular resistance to S6K dephosphorylation induced by amino acid depletion in Drosophila and in mammalian cells (75). Further studies are required to elucidate detailed mechanisms by which TSC1-TSC2 mediates amino acid signals, including identifying the kinases and/or phosphatases that are responsible for TSC2 phosphorylation in response to amino acid signaling.
TSC2 in energy-sensing pathways: effects of AMPK.
Intracellular energy (ATP) is one of the most important inputs regulating mTOR activity (62). Protein synthesis consumes approximately 20% of intracellular ATP (219). ATP depletion by 2-deoxyglucose (d-glucose analog) drastically inhibits S6K and 4EBP1 phosphorylation in cells without affecting PI3K activation and intracellular amino acid concentrations (62). Although mTOR has been proposed to be a cellular energy sensor, it has been suggested that the Km of mTOR for ATP is still considerably lower than normal cellular ATP levels and that therefore a drastic decrease in ATP would be required to affect the activity of mTOR (198). On the other hand, it has been reported recently that the 5′AMP-activated protein kinase (AMPK), an intracellular energy sensor, inhibits translation in response to changes in the intracellular ATP/AMP ratio (97, 98). AMPK plays a pivotal role in maintaining energy homeostasis and regulating cell growth and viability under energy deprivation conditions. Mutation of the γ2 subunit of AMPK is responsible for familial cardiac hypertrophy (59). It has been shown that AMPK-induced inhibition of S6K seems to be mTOR dependent, because mutant S6K lacking a TOS motif is resistant to both active AMPK overexpression and AICAR (an AMPK stimulator) treatment (137).
Our laboratory has investigated whether TSC1-TSC2 involved in the cellular energy response and has found that AMPK activates TSC2 by direct phosphorylation (123). AMPK phosphorylates multiple serine and threonine sites on TSC2 and inhibits the mTOR pathway. Direct phosphorylation sites by AMPK mainly involve T1271 and S1387 (residue numbers are for the human TSC2 long variant) on TSC2. ATP depletion-induced S6K dephosphorylation is compromised in TSC2-deficient cells or cells treated with TSC2 siRNA. More importantly, the phosphorylation of TSC2 by AMPK has critical roles in intracellular energy-dependent control of cell size and viability. For instance, reduced cellular energy levels by glucose starvation results in smaller TSC2 wild-type cells but not smaller TSC2-deficient cells. Furthermore, TCS2−/− cells expressing a TSC2 mutant that is unable to be phosphorylated by AMPK fails to fully restore the cellular energy response. Moreover, under energy depletion conditions, TSC2-expressing cells survive longer than both TSC2-deficient cells and mutant TSC2-expressing cells, indicating that AMPK phosphorylation of TSC2 plays a pivotal role not only in cell size regulation but also in cell viability under the energy limitation conditions. However, TSC2-deficient cells and mutant TSC2-expressing cells can survive energy starvation in the presence of rapamycin. Thus, loss of TSC1-TSC2 results in uncontrolled mTOR activity, which may cause inappropriate translation initiation even though the environmental energy supply is not sufficient. However, in these cells, AMPK could block translation elongation via eEF2 kinase phosphorylation in response to energy starvation (30, 113). Therefore, such a disruption in the coordination between translation initiation and elongation under energy starvation conditions might induce apoptosis. Alternatively, it is also plausible that energy starvation-induced apoptosis in TSC2−/− cells may be due to the lack of an autophagy response. It has been reported that nutrient starvation or rapamycin treatment induces autophagy in mammalian cells (23, 229). However, it has not been studied whether TSC2−/− cells lacks autophagy in response to various stress conditions. Taken together, these studies suggest that TSC1-TSC2 is a physiological regulators of the energy-sensing pathway upstream of mTOR. Further, they indicate that three important inputs (PI3K, amino acids, and ATP) for mTOR activation seem to be mediated by TSC1-TSC2, supporting a potential model where TSC1-TSC2 functions as a signal integration point to regulate cell growth control (Fig. 6).
Interestingly, several groups have recently reported that LKB1 kinase, the product of the gene inactivated in Peutz-Jeghers syndrome, is an upstream kinase of AMPK (104, 112, 268). Peutz-Jeghers syndrome is another dominantly inherited genetic disorder that is characterized by the formation of gastrointestinal hamartomas histologically similar to those observed in TSC patients (63). The LKB1 kinase is required for activation of AMPK. We have recently observed that overexpressed LKB1 inhibits the phosphorylation of both S6K1 and 4EBP1, two targets of mTOR (50). In addition, LKB1 plays an important role in inhibiting the mTOR pathway in response to energy starvation. Under energy starvation or stress conditions, LKB1 is necessary to protect cells from death triggered by these stresses (50, 226, 227). Furthermore, LKB1 enhances TSC2 phosphorylation of T1271 and S1387, which are AMPK-dependent phosphorylation sites, suggesting that TSC and Peutz-Jeghers syndrome may have a common pathomechanism, namely, dysregulation of mTOR signaling (50, 226).
Rheb as an Upstream Activator of mTOR
Rheb is a small GTP-binding protein that was first identified by a differential screen of mRNAs induced in neurons by agents that provoke seizures. It is ubiquitously expressed in many tissues but particularly abundant in both muscles and the brain (271). Rheb is highly conserved from yeast to mammals and has greatest homology to the Rap subfamily of small GTPases; however, close sequence analysis suggests that it occupies its own separate phylogenetic branch (250). It has been reported that Rheb is overexpressed in several tumor cell lines (90) and that ectopic expression of Rheb can induce the transformation of mouse fibroblast (273), indicating the involvement of Rheb in cell growth. In yeast, loss of Rheb results in a phenotype similar to those due to nitrogen starvation, suggesting that Rheb might be involved in nutrient signaling (158). Genetic screens for genes that are involved in controlling Drosophila cell growth have also led to the identification of Rheb as an important regulator of cell growth (209, 233). Mutation in Rheb decreased the cell size, while overexpression of Rheb increased it. Genetic epistasis analyses and biochemical studies indicate that Rheb possibly functions downstream of TSC1-TSC2 and upstream of TOR (37, 76, 121, 209, 233, 243, 279). Loss of Rheb suppresses the cell growth and S6K phosphorylation caused by loss of TSC2. In contrast, inhibition of cell growth by loss of dTOR is not rescued by Rheb overexpression. Drosophila cells with a mutation in Rheb show a much lower phosphorylation of S6K. Using an RNA interference-based screen, Pan and colleagues also discovered an important role of Rheb in S6K phosphorylation in Drosophila S2 cells (279). This screen showed that RNAi-mediated inhibition of Rheb, but not any of the other 17 GTPases examined, including Rap1, Rab5, Rac1, and cdc42, abolished S6K phosphorylation in S2 cells (279). These data established Rheb as a positive regulator of S6K phosphorylation.
Several groups showed that, in mammalian cells, Rheb potently activates S6K and 4EBP1 phosphorylation (37, 76, 121, 243). Furthermore, Rheb-induced S6K phosphorylation was blocked by either overexpression of an mTOR kinase inactive mutant or rapamycin treatment (76, 121). In addition, Rheb stimulates mTOR phosphorylation and cannot activate mutant S6K with a TOS motif mutation, indicating that Rheb is upstream of mTOR (121, 243), consistent with the notion that TSC mediates signals from growth factors, amino acids, intracellular ATP levels, and possibly PA. Rheb overexpression rescued S6K dephosphorylation caused by PI3K inactivation and amino acid depletion, mild ATP depletion, and inhibition of PA production (76, 121). These results suggest that Rheb may be a key molecule that relays upstream inputs to the regulation of mTOR. However, there are several studies that argue against this notion. For instance, if Rheb is a sensor to regulate TOR activity, upstream signals should affect Rheb GTP levels. In contrast to this prediction, Rheb GTP levels were not altered by cellular conditions known to affect mTOR activity (279). However, most of the experiments were performed with overexpressed Rheb, which may not be regulated by cellular conditions that affect the activity of endogenous Rheb. Indeed, it was reported that insulin was found to increase GTP loading of endogenous Rheb in A14 NIH 3T3 cells (76). Therefore, it is likely that Rheb is a key mediator for the inputs to modulate TOR activities.
TSC2 is a GAP Specific for Rheb
Genetic epistatic analyses of Drosophila have placed Rheb function downstream of dTSC1-dTSC2. Several groups performed biochemical analyses and demonstrated that, in both Drosophila and mammalian cells, Rheb is a direct target of TSC2 GAP activity. TSC2 stimulates Rheb GTP hydrolysis both in vivo and in vitro (37, 76, 121, 243, 279). It is worth noting that most of the missense mutations identified in TSC patients occur in the GAP domain of TSC2 (9, 56). Many TSC disease-associated TSC2 mutants are unable to stimulate Rheb GTP hydrolysis in vivo, resulting in uncontrolled Rheb activity (76, 121, 243, 279). These data strongly imply role for uncontrolled Rheb activity in TSC. Taken together, a simplified model is that TSC1-TSC2 inhibits the TOR pathway by stimulating Rheb GTP hydrolysis through TSC2 GAP activity and that Rheb positively stimulates mTOR activity (Fig. 6).
CONCLUSIONS AND FUTURE DIRECTIONS
A decade after the discovery of TOR as the target of rapamycin, we now realize that TOR is a central regulator for cell growth, which integrates signaling from both growth factors and nutrients. Recent important discoveries include the finding of a negative regulator, the TSC1-TSC2 complex, and positive regulators, such as Rheb, Raptor, and mLst8. Furthermore, another emerging concept is that similar to yeast TORC2, mTOR also has rapamycin-independent functions to regulate cytoskeletal reorganization. Although these findings provide critical insight into the upstream and downstream components of the TOR signaling pathway, many gaps in our knowledge of the TOR signaling pathway remain to be filled. For example, how does TSC1-TSC2 sense upstream signals from nutrients? How does Rheb regulate TOR activity? What are other downstream substrates of TOR? Furthermore, is there any cross talk between the TSC1-TSC2-mTOR signaling pathway and other signaling pathways that are involved in oncogenesis and metabolic diseases? All these information will be important for constituting a complete picture of TSC1-TSC2-mTOR biology.
How Rheb Is Regulated
Convincing evidence has placed Rheb as downstream of nutrient signaling and upstream of mTOR. Thus, it is logical to anticipate that Rheb activity is subjected to nutrient regulation. Although several groups failed to detect any significant regulation of the GTP level of overexpressed Rheb by amino acid depletion (76, 121, 279), these studies used overexpressed Rheb, and overexpressed Rheb may not accurately mimic the endogenous Rheb in the responsiveness to amino acid depletion. Further studies aimed at determining the regulation of endogeneous Rheb by amino acids will be required to address this question. Furthermore, establishment of TSC2 as a Rheb GAP makes it legitimate to hypothesize that the activity of Rheb is determined by the balance of the activity of its GAP (TSC2) and its yet unidentified guanine exchange factor (GEF), if any exists. Any signal such as amino acids, which regulates Rheb, should work through these two proteins. Identifying a Rheb-GEF and elucidating how TSC2 and Rheb-GEF are regulated by upstream signals will shed light on the issue of how Rheb responds to upstream signals.
Sequence analyses of Rheb and TSC2 show that Rheb and TSC2 are a unique pair of small G protein and GAP (147). It has been reported that some of the residues in the GAP domain of TSC2 (1638K, 1643N, 1651N; residue numbers are for the human TSC2 long variant) are critical for TSC2 GAP activity (76, 243, 279). Interestingly, TSC2 does not carry an arginine finger, which is a crucial moiety for stabilizing the basal catalytic activity of small G proteins. Therefore, TSC2 increases the GTPase activity of Rheb by a different arginine or by a mechanism different from that employed by Ras-GAP (237) toward its substrates. Supporting this idea, recent structural and biochemical studies of Rap1GAP showed that Rap1GAP used a catalytic asparagine (N290) instead of an arginine (60). Interestingly, the catalytic N290 on Rap1GAP is conserved in TSC2 GAP (N1643), and this asparagine mutation on TSC2 has been found in TSC and leads to loss of its GAP activity toward Rheb, suggesting that, like N290 on Rap1GAP, the N1643 on TSC2 may be a critical catalytic residue for Rheb (60).
Given the observations that many upstream signals, including growth factors and nutrients, both regulate TSC2 phosphorylation and affect the TOR signaling pathway that is dependent on Rheb function, it would be logical to postulate that TSC2 GAP activity and TSC2 phosphorylation should be two tightly coupled events. The question that naturally arises from these observations is whether TSC2 GAP activity is directly related to its phosphorylation status. It is possible that TSC2 phosphorylation affect its stability, trafficking, or conformational change, which could indirectly relate to Rheb activity. Although our own preliminary studies using an overexpression strategy in mammalian cells failed to detect any difference of GAP activity between TSC2 wild type and TSC2 phosphorylation mutants, including mutation of Akt and certain AMPK phosphorylation sites (unpublished results). Hopefully, further studies focusing on the endogenous Rheb GTP will provide critical information about this issue.
GEFs often receive upstream signals that trigger the various small G-protein-associated signal transduction cascades (219) and constitute an important force in opposition of a GAP to determine the activity of downstream effectors (216). It is therefore reasonable to speculate that, in the case of Rheb regulation, there might exist a Rheb-GEF and that its function, just like that of Rheb GAP (TSC2), is also regulated by signals affecting the TOR pathway, such as nutrients. However, whether Rheb is regulated by a GEF is an open question because Rheb exists in a highly GTP-bound state (120). Furthermore, Rheb-N20, which is equivalent to Ras-N17, a dominant negative mutant of Ras able to bind and sequester Ras-GEF but defective in binding guanine nucleotides (41), does not function as a dominant negative Rheb (121, 158). This suggests that Rheb might not require a GEF for its activation or that Rheb-GEF catalyzes Rheb by a different mechanism from that employed by Ras-GEF. It has been reported that Rheb-K60 and Rheb-V60, a Rheb mutant lacking the ability to bind GTP, act as dominant negative forms in yeast and mammalian cells (236). Expression of Rheb-K60 inhibits the basal level of S6K phosphorylation in HEK293 cells. However, we did not detect a dominant negative effect of either Rheb-K60 or Rheb-V60 in mammalian cells. In fact, Rheb-V60 stimulates S6K phosphorylation (T389) in our assays, although it is less active than the wild-type Rheb (147, 148, 160). The discrepancy of these results could be due to different expression levels of overexpressed Rheb in cells. Therefore, the issue of a dominant negative Rheb remains open.
How Does Rheb Activate mTOR?
Currently, there are two proposed mechanisms for how Rheb could regulate TOR. First, Rheb may be a component of the TOR complex, thereby directly regulating TOR function. Like other Ras family members, Rheb is modified posttranslationally by farnesyltransferase (49, 272). The enzyme adds farnesyl isoprenoids covalently to a cysteine residue at the C terminus of Rheb, which is important for Rheb to optimally function as an activator of mTOR signaling (37, 243). Like other farnesylated small G proteins (277), farnesylated Rheb preferentially associates with the membrane. Since TOR is also found in the membrane fraction, including those of the mitochondria and endoplasmic reticulum (67, 205), the membrane-associated Rheb may recruit the TOR signaling complex to the membrane. However, until now, no convincing direct association between Rheb and TOR has been reported. Therefore, it is possible that Rheb may bind to and activate mTOR-interacting proteins such as Raptor or GβL, rather than interacting with and activating TOR directly. It will be interesting to examine whether wild-type Rheb or Rheb harboring mutations in the effector domain affect the association between mTOR and raptor or GβL. Furthermore, with the identification of the Rictor-mTOR complex, it will be also important to determine whether Rheb activates TORC2. Those investigations will provide new insights into the mechanisms by which Rheb activates TOR complexes. A different but not mutually exclusive hypothesis is that Rheb may regulate TOR function by affecting the transport of upstream signals such as amino acids (158, 209, 251, 255). For instance, Saucedo et al. have shown that overexpressed Rheb functions to promote cell growth and animal viability under conditions of protein starvation (209). In addition, it has been reported that the cells depend on Rheb to grow normally under situations with a limited amount of nitrogen in fission yeast (158). However, there are several caveats associated with this model. First, Rheb has also been linked to inhibit amino acid uptake, in this case arginine in both Schizosaccharomyces pombe (fission yeast) and S. cerevisiae (budding yeast) (251, 255). Second, it has been reported that disruption of either Tsc1p or Tsc2p in fission yeast results in a mislocalization of amino acid permease and reduction of permeases expression (163). Third, it remains unclear whether Tsc1p-Tsc2p and/or Rhebp regulation of nutrient uptake requires Tor function, since Tor has been known for its role in regulating amino acids uptake. Fourth, overexpressed Rheb can efficiently activate mTOR pathway under amino acid-free conditions in both mammalian and Drosophila cells (76, 121, 209). Therefore, it still remains open whether TSC1-TSC2 and Rheb stimulate mTOR function by regulating amino acid uptake.
mTOR and Hamartomas
Many studies during the past decade have revealed that TOR plays a central role in controlling cell growth, proliferation, and metabolism in a variety of organisms. Multiple signals impinge on TOR, and a variety of effects emanate from TOR (52, 127). It is not surprising that dysregulation of the TOR signaling pathway has been associated with disturbed organ development and tissue homeostasis and thus ultimately with the formation of tumors. Several pieces of evidence now point to a promising linkage between the mTOR signaling pathway and tumor development, although oncogenic mutations of mTOR have not yet been found in tumors. First, some neuroblastomas and glioblastomas are particularly sensitive to rapamycin treatment, implicating TOR function in the development of these tumors (114). Second, the mTOR signaling pathway is constitutively activated in pancreatic cancer cells (89). Third, mTOR promotes the translation of c-Myc, a transcription factor that is frequently dysregulated in many tumor types, whereas mTOR destabilizes p27kip1, an inhibitor of the cyclin-dependent kinase (78, 157, 179). Fourth, many proteins that are related to the mTOR signaling pathway possess transforming potential, including elF4E (47, 203). Fifth, mTOR regulates proto-oncogenic factors, such as hypoxia-inducible factor and vascular endothelial growth factor (3, 7, 31, 69, 117). All these studies highlight a critical role for the TOR signal pathway in tumor formation.
Importantly, recent genetic analysis and biochemical studies have linked mTOR to the PTEN-PI3K-Akt-TSC1-TSC2 (225) and LKB1-AMPK-TSC1-TSC2 (36, 123) pathways, which are dysfunctional in many tumors and organ hypertrophy (Fig. 6). Inactivation of the TSC1-TSC2 complex leads to TSC. Loss of function of PTEN is also held responsible for hamartoma syndromes such as Cowden's disease (34). Inactivation of LKB results in Peutz-Jeghers syndrome (268). Mutation of the γ2 subunit of AMPK, leading to inactivation of AMPK, is responsible for familial cardiac hypertrophy (59) (Fig. 6). These are seemingly distinct and independent clinical disease entities. However, from the molecular etiology point of view, all of them might have the same root, i.e., the dysfunction of mTOR-related signaling pathways (36, 123, 225). The results of these studies are of great clinical significance. We propose that dysregulation of the mTOR pathway might represent a common molecular mechanism that contributes to the pathogenesis of many hamartoma syndromes. More generally, they suggest that in the age of molecular medicine, the clinical diagnosis of various hamartomas should include more molecular-etiology-based criteria rather than merely reflecting on the clinical features of the diseases. Such a molecular-etiology-based diagnostic method would not only prove to be more instrumental and meaningful for differential diagnoses for various hamartomas but also, perhaps more importantly, provide a solid mechanistic foundation leading to increased treatment efficiency and thus to more successful and predictable treatment outcomes. From a therapeutic point of view, the appreciation of dysfunction of mTOR signaling as the underlying molecular mechanism of several hamartomas immediately suggests that rapamycin, as a specific TOR inhibitor, would have a potential therapeutic impact on treating hamartoma syndromes associated with TOR activation, such as Cowden's disease, Peutz-Jeghers syndrome, and TSC (133, 176, 192, 275). However, caution should be exercised in considering the potential application of rapamycin on benign tumors. Several groups have reported that there is a negative-feedback system from S6K to the molecules upstream of Akt, such as insulin receptor substrates (IRSs) (100, 102, 223, 249). IRS is a physiological target of S6K, and S6K phosphorylation of IRSs inhibits their functions, including activating the PI3K-Akt signaling pathway. Akt, besides regulating the mTOR pathway, also has many mTOR-independent functions that are involved in oncogenic signaling pathways. The existence of this negative-feedback loop may explain why tumors associated with the dysfunction of the mTOR pathway are virtually benign. Moreover, since rapamycin could disrupt this negative-feedback loop, the safety of mTOR inhibitors as therapeutic agents for benign hamartoma syndromes needs to be carefully evaluated.
Given the important roles of mTOR in the normal life of mammalian cells and the severe consequences associated with disturbed TOR signals, it is conceivable that mTOR is subjected to additional regulation other than the PTEN-PI3K-Akt and LKB1-AMPK and that mTOR might have a target spectrum much broader than the spectrum we know about at present. To further elucidate the involvement of TOR in oncogenesis, it would be important to identify these additional linkages as well as other novel mTOR effectors. These efforts would broaden our knowledge of mTOR biology and potentially expand the therapeutic spectrum of rapamycin.
Acknowledgments
We thank Eric Tang, Michael N. Corradetti, and Chong-Han Lee for critical reading of the review.
This work is supported by grants from the National Institutes of Health (to K.-L.G.).
REFERENCES
- 1.Reference deleted.
- 2.Abeliovich, H., and D. J. Klionsky. 2001. Autophagy in yeast: mechanistic insights and physiological function. Microbiol. Mol. Biol. Rev. 65:463-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abraham, R. T. 2004. mTOR as a positive regulator of tumor cell responses to hypoxia. Curr. Top. Microbiol. Immunol. 279:299-319. [DOI] [PubMed] [Google Scholar]
- 4.Abraham, R. T., and G. J. Wiederrecht. 1996. Immunopharmacology of rapamycin. Annu. Rev. Immunol. 14:483-510. [DOI] [PubMed] [Google Scholar]
- 5.Albig, A. R., and C. J. Decker. 2001. The target of rapamycin signaling pathway regulates mRNA turnover in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 12:3428-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altmann, M., and H. Trachsel. 1997. Translation initiation factor-dependent extracts from yeast Saccharomyces cerevisiae. Methods 11:343-352. [DOI] [PubMed] [Google Scholar]
- 7.Arsham, A. M., J. J. Howell, and M. C. Simon. 2003. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 278:29655-29660. [DOI] [PubMed] [Google Scholar]
- 8.Astrinidis, A., W. Senapedis, T. R. Coleman, and E. P. Henske. 2003. Cell cycle-regulated phosphorylation of hamartin, the product of the tuberous sclerosis complex 1 gene, by cyclin-dependent kinase 1/cyclin B. J. Biol. Chem. 278:51372-51379. [DOI] [PubMed] [Google Scholar]
- 9.Au, K. S., J. A. Rodriguez, J. L. Finch, K. A. Volcik, E. S. Roach, M. R. Delgado, E. Rodriguez, Jr., and H. Northrup. 1998. Germ-line mutational analysis of the TSC2 gene in 90 tuberous-sclerosis patients. Am. J. Hum. Genet. 62:286-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avruch, J., C. Belham, Q. Weng, K. Hara, and K. Yonezawa. 2001. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 26:115-154. [DOI] [PubMed] [Google Scholar]
- 11.Azpiazu, I., A. R. Saltiel, A. A. DePaoli-Roach, and J. C. Lawrence. 1996. Regulation of both glycogen synthase and PHAS-I by insulin in rat skeletal muscle involves mitogen-activated protein kinase-independent and rapamycin-sensitive pathways. J. Biol. Chem. 271:5033-5039. [DOI] [PubMed] [Google Scholar]
- 12.Baas, A. F., J. Boudeau, G. P. Sapkota, L. Smit, R. Medema, N. A. Morrice, D. R. Alessi, and H. C. Clevers. 2003. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 22:3062-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banholzer, R., A. P. Nair, H. H. Hirsch, X. F. Ming, and C. Moroni. 1997. Rapamycin destabilizes interleukin-3 mRNA in autocrine tumor cells by a mechanism requiring an intact 3′ untranslated region. Mol. Cell. Biol. 17:3254-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 16.Beck, T., A. Schmidt, and M. N. Hall. 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146:1227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begum, N., and L. Ragolia. 1996. cAMP counter-regulates insulin-mediated protein phosphatase-2A inactivation in rat skeletal muscle cells. J. Biol. Chem. 271:31166-31171. [DOI] [PubMed] [Google Scholar]
- 18.Benvenuto, G., S. Li, S. J. Brown, R. Braverman, W. C. Vass, J. P. Cheadle, D. J. Halley, J. R. Sampson, R. Wienecke, and J. E. DeClue. 2000. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene 19:6306-6316. [DOI] [PubMed] [Google Scholar]
- 19.Beretta, L., A. C. Gingras, Y. V. Svitkin, M. N. Hall, and N. Sonenberg. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 15:658-664. [PMC free article] [PubMed] [Google Scholar]
- 20.Berlanga, J. J., J. Santoyo, and C. De Haro. 1999. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur. J. Biochem. 265:754-762. [DOI] [PubMed] [Google Scholar]
- 21.Berset, C., H. Trachsel, and M. Altmann. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beugnet, A., X. Wang, and C. G. Proud. 2003. Target of rapamycin (TOR)-signaling and RAIP motifs play distinct roles in the mammalian. TOR-dependent phosphorylation of initiation factor 4E-binding protein 1. J. Biol. Chem. 278:40717-40722. [DOI] [PubMed] [Google Scholar]
- 23.Blommaart, E. F., J. J. Luiken, P. J. Blommaart, G. M. van Woerkom, and A. J. Meijer. 1995. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270:2320-2326. [DOI] [PubMed] [Google Scholar]
- 24.Bodine, S. C., T. N. Stitt, M. Gonzalez, W. O. Kline, G. L. Stover, R. Bauerlein, E. Zlotchenko, A. Serimgeour, J. C. Lawrence, D. J. Glass, and G. D. Yancopoulos. 2001. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3:1014-1019. [DOI] [PubMed] [Google Scholar]
- 25.Bolster, D. R., S. J. Crozier, S. R. Kimball, and L. S. Jefferson. 2002. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J. Biol. Chem. 277:23977-23980. [DOI] [PubMed] [Google Scholar]
- 26.Boudeau, J., A. F. Baas, M. Deak, N. A. Morrice, A. Kieloch, M. Schutkowski, A. R. Prescott, H. C. Clevers, and D. R. Alessi. 2003. MO25alpha/beta interact with STRADalpha/beta enhancing their ability to bind, activate and localize LKB1 in the cytoplasm. EMBO J. 22:5102-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brazil, D. P., and B. A. Hemmings. 2001. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem. Sci. 26:657-664. [DOI] [PubMed] [Google Scholar]
- 28.Brown, E. J., M. W. Albers, T. B. Shin, K. Ichikawa, C. T. Keith, W. S. Lane, and S. L. Schreiber. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369:756-758. [DOI] [PubMed] [Google Scholar]
- 29.Brown, E. J., P. A. Beal, C. T. Keith, J. Chen, T. B. Shin, and S. L. Schreiber. 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377:441-446. [DOI] [PubMed] [Google Scholar]
- 30.Browne, G. J., S. G. Finn, and C. G. Proud. 2004. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J. Biol. Chem. 279:1220-1231. [DOI] [PubMed] [Google Scholar]
- 31.Brugarolas, J. B., F. Vazquez, A. Reddy, W. R. Sellers, and W. G. Kaelin, Jr. 2003. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell 4:147-158. [DOI] [PubMed] [Google Scholar]
- 32.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 33.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Synder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantley, L. C., and B. G. Neel. 1999. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. USA 96:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carling, D. 2004. The AMP-activated protein kinase cascade—a unifying system for energy control. Trends Biochem. Sci. 29:18-24. [DOI] [PubMed] [Google Scholar]
- 37.Castro, A. F., J. F. Rebhun, G. J. Clark, and L. A. Quilliam. 2003. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 278:32493-32496. [DOI] [PubMed] [Google Scholar]
- 38.Cheatham, L., M. Monfar, M. M. Chou, and J. Blenis. 1995. Structural and functional analysis of pp70S6k. Proc. Natl. Acad. Sci. USA 92:11696-11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, E. J., and C. A. Kaiser. 2003. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161:333-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, J., and Y. Fang. 2002. A novel pathway regulating the mammalian target of rapamycin (mTOR) signaling: Biochem. Pharmacol. 64:1071-1077. [DOI] [PubMed] [Google Scholar]
- 41.Cherfils, J., and P. Chardin. 1999. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24:306-311. [DOI] [PubMed] [Google Scholar]
- 42.Cherkasova, V. A., and A. G. Hinnebusch. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 17:859-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiu, M. I., H. Katz, and V. Berlin. 1994. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 91:12574-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi, J. H., N. R. Adames, T. F. Chan, C. Zeng, J. A. Cooper, and X. F. Zheng. 2000. TOR signaling regulates microtubule structure and function. Curr. Biol. 10:861-864. [DOI] [PubMed] [Google Scholar]
- 45.Choi, J. H., P. G. Bertram, R. Drenan, J. Carvalho, H. H. Zhou, and X. F. Zheng. 2002. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 3:988-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi, K. M., L. P. McMahon, and J. C. Lawrence, Jr. 2003. Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor. J. Biol. Chem. 278:19667-19673. [DOI] [PubMed] [Google Scholar]
- 47.Chung, J., R. E. Bachelder, E. A. Lipscomb, L. M. Shaw, and A. M. Mercurio. 2002. Integrin (alpha 6 beta 4) regulation of eIF-4E activity and VEGF translation: a survival mechanism for carcinoma cells. J. Cell Biol. 158:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung, J., C. J. Kuo, G. R. Crabtree, and J. Blenis. 1992. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69:1227-1236. [DOI] [PubMed] [Google Scholar]
- 49.Clark, G. J., M. S. Kinch, K. Rogers-Graham, S. M. Sebti, A. D. Hamilton, and C. J. Der. 1997. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J. Biol. Chem. 272:10608-10615. [DOI] [PubMed] [Google Scholar]
- 50.Corradetti, M. N., K. Inoki, N. Bardeesy, R. A. DePinho, and K. L. Guan. 2004. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18:1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cosentino, G. P., T. Schmelzle, A. Haghighat, S. B. Helliwell, M. N. Hall, and N. Sonenberg. 2000. Eaplp, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crespo, J. L., and M. N. Hall. 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66:579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crespo, J. L., T. Powers, B. Fowler, and M. N. Hall. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99:6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cruz, M. C., L. M. Cavallo, J. M. Gorlach, G. Cox, J. R. Perfect, M. E. Cardenas, and J. Heitman. 1999. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell. Biol. 19:4101-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cutler, N. S., X. Pan, J. Heitman, and M. E. Cardenas. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dabora, S. L., S. Jozwiak, D. N. Franz, P. S. Roberts, A. Nieto, J. Chung, Y. S. Choy, M. P. Reeve, E. Thiele, J. C. Egelhoff, J. Kasprzyk-Obara, D. Domanska-Pakiela, and D. J. Kwiatkowski. 2001. Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am. J. Hum. Genet. 68:64-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dan, H. C., M. Sun, L. Yang, R. I. Feldman, X. M. Sui, R. S. Yeung, D. J. Halley, S. V. Nicosia, W. J. Pledger, and J. Q. Cheng. 2002. PI3K/AKT pathway regulates TSC tumor suppressor complex by phosphorylation of tuberin. J. Biol. Chem. 277:35364-35370. [DOI] [PubMed] [Google Scholar]
- 58.Danaie, P., M. Altmann, M. N. Hall, H. Trachsel, and S. B. Helliwell. 1999. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem. J. 340:135-141. [PMC free article] [PubMed] [Google Scholar]
- 59.Daniel, T., and D. Carling. 2002. Functional analysis of mutations in the gamma 2 subunit of AMP-activated protein kinase associated with cardiac hypertrophy and Wolff-Parkinson-White syndrome. J. Biol. Chem. 277:51017-51024. [DOI] [PubMed] [Google Scholar]
- 60.Daumke, O., M. Weyand, P. P. Chakrabarti, I. R. Vetter, and A. Wittinghofer. 2004. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429:197-201. [DOI] [PubMed] [Google Scholar]
- 61.De Craene, J. O., O. Soetens, and B. Andre. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939-43948. [DOI] [PubMed] [Google Scholar]
- 62.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 63.Devroede, G., B. Lemieux, S. Masse, J. Lamarche, and P. S. Herman. 1988. Colonic hamartomas in tuberous sclerosis. Gastroenterology 94:182-188. [DOI] [PubMed] [Google Scholar]
- 64.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 65.Dilova, I., C. Y. Chen, and T. Powers. 2002. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr. Biol. 12:389-395. [DOI] [PubMed] [Google Scholar]
- 66.Dong, J., and D. Pan. 2004. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 18:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drenan, R. M., X. Liu, P. G. Bertram, and X. F. Zheng. 2004. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J. Biol. Chem. 279:772-778. [DOI] [PubMed] [Google Scholar]
- 68.Duvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11:1467-1478. [DOI] [PubMed] [Google Scholar]
- 69.El-Hashemite, N., V. Walker, H. Zhang, and D. J. Kwiatkowski. 2003. Loss of Tsc1 or Tsc2 induces vascular endothelial growth factor production through mammalian target of rapamycin. Cancer Res. 63:5173-5177. [PubMed] [Google Scholar]
- 69a.European Chromosome 16 Tuberous Sclerosis Consortium. 1993. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 75:1305-1315. [DOI] [PubMed] [Google Scholar]
- 70.Fadden, P., T. A. Haystead, and J. C. Lawrence, Jr. 1997. Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J. Biol. Chem. 272:10240-10247. [DOI] [PubMed] [Google Scholar]
- 71.Fang, Y., M. Vilella-Bach, R. Bachmann, A. Flanigan, and J. Chen. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294:1942-1945. [DOI] [PubMed] [Google Scholar]
- 72.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151-3171. [DOI] [PubMed] [Google Scholar]
- 73.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao, X., and D. Pan. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15:1383-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao, X., Y. Zhang, P. Arrazola, O. Hino, T. Kobayashi, R. S. Yeung, B. Ru, and D. Pan. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4:699-704. [DOI] [PubMed] [Google Scholar]
- 76.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11:1457-1466. [DOI] [PubMed] [Google Scholar]
- 77.Garza, L., Y. W. Aude, and J. F. Saucedo. 2002. Can we prevent in-stent restenosis? Curr. Opin. Cardiol. 17:518-525. [DOI] [PubMed] [Google Scholar]
- 78.Gera, J. F., I. K. Mellinghoff, Y. Shi, M. B. Rettig, C. Tran, J. H. Hsu, C. L. Sawyers, and A. K. Lichtenstein. 2004. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J. Biol. Chem. 279:2737-2746. [DOI] [PubMed] [Google Scholar]
- 79.Gingras, A. C., S. G. Kennedy, M. A. O'Leary, N. Sonenberg, and N. Hay. 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 12:502-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 82.Goldberg, Y. 1999. Protein phosphatase 2A: who shall regulate the regulator? Biochem. Pharmacol. 57:321-328. [DOI] [PubMed] [Google Scholar]
- 83.Gomez, M. R. 1991. Phenotypes of the tuberous sclerosis complex with a revision of diagnostic criteria. Ann. N. Y. Acad. Sci. 615:1-7. [DOI] [PubMed] [Google Scholar]
- 84.Goncharova, E. A., D. A. Goncharov, A. Eszterhas, D. S. Hunter, M. K. Glassberg, R. S. Yeung, C. L. Walker, D. Noonan, D. J. Kwiatkowski, M. M. Chou, R. A. Panettieri, Jr., and V. P. Krymskaya. 2002. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. J. Biol. Chem. 277:30958-30967. [DOI] [PubMed] [Google Scholar]
- 85.Grammer, T. C., L. Cheatham, M. M. Chou, and J. Blenis. 1996. The p70S6K signalling pathway: a novel signalling system involved in growth regulation. Cancer Surv. 27:271-292. [PubMed] [Google Scholar]
- 86.Graves, L. M., K. E. Bornfeldt, G. M. Argast, E. G. Krebs, X. Kong, T. A. Lin, and J. C. Lawrence, Jr. 1995. cAMP- and rapamycin-sensitive regulation of the association of eukaryotic initiation factor 4E and the translational regulator PHAS-I in aortic smooth muscle cells. Proc. Natl. Acad. Sci. USA 92:7222-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grenson, M. 1983. Inactivation-reactivation process and repression of permease formation regulate several ammonia-sensitive permeases in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 133:135-139. [DOI] [PubMed] [Google Scholar]
- 88.Grenson, M. 1983. Study of the positive control of the general amino-acid permease and other ammonia-sensitive uptake systems by the product of the NPR1 gene in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 133:141-144. [DOI] [PubMed] [Google Scholar]
- 89.Grewe, M., F. Gansauge, R. M. Schmid, G. Adler, and T. Seufferlein. 1999. Regulation of cell growth and cyclin D1 expression by the constitutively active FRAP-p70s6K pathway in human pancreatic cancer cells. Cancer Res. 59:3581-3587. [PubMed] [Google Scholar]
- 90.Gromov, P. S., P. Madsen, N. Tomerup, and J. E. Celis. 1995. A novel approach for expression cloning of small GTPases: identification, tissue distribution and chromosome mapping of the human homolog of rheb. FEBS Lett. 377:221-226. [DOI] [PubMed] [Google Scholar]
- 91.Gstaiger, M., B. Luke, D. Hess, E. J. Oakeley, C. Wirbelauer, M. Blondel, M. Vigneron, M. Peter, and W. Krek. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302:1208-1212. [DOI] [PubMed] [Google Scholar]
- 92.Haghighat, A., S. Mader, A. Pause, and N. Sonenberg. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14:5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han, J. W., R. B. Pearson, P. B. Dennis, and G. Thomas. 1995. Rapamycin, wortmannin, and the methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J. Biol. Chem. 270:21396-21403. [DOI] [PubMed] [Google Scholar]
- 94.Hannan, K. M., Y. Brandenburger, A. Jenkins, K. Sharkey, A. Cavanaugh, L. Rothblum, T. Moss, G. Poortinga, G. A. McArthur, R. B. Pearson, and R. D. Hannan. 2003. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 23:8862-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 96.Hara, K., K. Yonezawa, Q. P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 97.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 98.Hardie, D. G., and S. A. Hawley. 2001. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays 23:1112-1119. [DOI] [PubMed] [Google Scholar]
- 99.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harrington, L. S., G. M. Findlay, A. Gray, T. Tolkacheva, S. Wigfield, H. Rebholz, J. Barnett, N. R. Leslie, S. Cheng, P. R. Shepherd, I. Gout, C. P. Downes, and R. F. Lamb. 2004. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hartman, M. E., M. Villela-Bach, J. Chen, and G. G. Freund. 2001. Frap-dependent serine phosphorylation of IRS-1 inhibits IRS-1 tyrosine phosphorylation. Biochem. Biophys. Res. Commun. 280:776-781. [DOI] [PubMed] [Google Scholar]
- 102.Haruta, T., T. Uno, J. Kawahara, A. Takano, K. Egawa, P. M. Sharma, J. M. Olefsky, and M. Kobayashi. 2000. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol. Endocrinol. 14:783-794. [DOI] [PubMed] [Google Scholar]
- 103.Hashemolhosseini, S., Y. Nagamine, S. J. Morley, S. Desrivieres, L. Mercep, and S. Ferrari. 1998. Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J. Biol. Chem. 273:14424-14429. [DOI] [PubMed] [Google Scholar]
- 104.Hawley, S. A., J. Boudeau, J. L. Reid, K. J. Mustard, L. Udd, T. P. Makela, D. R. Alessi, and D. G. Hardie. 2003. Complexes between the LKB1 tumor suppressor, STRADalpha/beta and MO25alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heesom, K. J., M. B. Avison, T. A. Diggle, and R. M. Denton. 1998. Insulin-stimulated kinase from rat fat cells that phosphorylates initiation factor 4E-binding protein 1 on the rapamycin-insensitive site (serine-111). Biochem. J. 336:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 107.Helliwell, S. B., P. Wagner, J. Kunz, M. Deuter-Reinhard, R. Henriquez, and M. N. Hall. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hentges, K., K. Thompson, and A. Peterson. 1999. The flat-top gene is required for the expansion and regionalization of the telencephalic primordium. Development 126:1601-1609. [DOI] [PubMed] [Google Scholar]
- 109.Hinnebusch, A. G. 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA 81:6442-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hinnebusch, A. G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem. 272:21661-21664. [DOI] [PubMed] [Google Scholar]
- 111.Hoekstra, M. F. 1997. Responses to DNA damage and regulation of cell cycle checkpoints by the ATM protein kinase family. Curr. Opin. Genet. Dev. 7:170-175. [DOI] [PubMed] [Google Scholar]
- 112.Hong, S. P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Horman, S., G. Browne, U. Krause, J. Patel, D. Vertommen, L. Bertrand, A. Lavoinne, L. Hue, C. Proud, and M. Rider. 2002. Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Curr. Biol. 12:1419-1423. [DOI] [PubMed] [Google Scholar]
- 114.Hosoi, H., M. B. Dilling, L. N. Liu, M. K. Danks, T. Shikata, A. Sekulic, R. T. Abraham, J. C. Lawrence, Jr., and P. J. Houghton. 1998. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol. Pharmacol. 54:815-824. [DOI] [PubMed] [Google Scholar]
- 115.Huang, S., and P. J. Houghton. 2003. Targeting mTOR signaling for cancer therapy. Curr. Opin. Pharmacol. 3:371-377. [DOI] [PubMed] [Google Scholar]
- 116.Huang, S., L. N. Liu, H. Hosoi, M. B. Dilling, T. Shikata, and P. J. Houghton. 2001. p53/p21 (CIP1) cooperate in enforcing rapamycin-induced G(1) arrest and determine the cellular response to rapamycin. Cancer Res. 61:3373-3381. [PubMed] [Google Scholar]
- 117.Hudson, C. C., M. Liu, G. G. Chiang, D. M. Otterness, D. C. Loomis, F. Kaper, A. J. Giaccia, and R. T. Abraham. 2002. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 22:7004-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huffman, T. A., I. Mothe-Satney, and J. C. Lawrence, Jr. 2002. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. USA 99:1047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ilyin, G. P., D. Glaise, D. Gilot, G. Baffet, and C. Guguen-Guillouzo. 2003. Regulation and role of p21 and p27 cyclin-dependent kinase inhibitors during hepatocyte differentiation and growth. Am. J. Physiol. 285:G115-G127. [DOI] [PubMed] [Google Scholar]
- 120.Im, E., F. C. von Lintig, J. Chen, S. Zhuang, W. Qui, S. Chowdhury, P. F. Worley, G. R. Boss, and R. B. Pilz. 2002. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene 21:6356-6365. [DOI] [PubMed] [Google Scholar]
- 121.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Inoki, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4:648-657. [DOI] [PubMed] [Google Scholar]
- 123.Inoki, K., T. Zhu, and K. L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577-590. [DOI] [PubMed] [Google Scholar]
- 124.Inui, S., H. Sanjo, K. Maeda, H. Yamamoto, E. Miyamoto, and N. Sakaguchi. 1998. Ig receptor binding protein 1 (alpha4) is associated with a rapamycin-sensitive signal transduction in lymphocytes through direct binding to the catalytic subunit of protein phosphatase 2A. Blood 92:539-546. [PubMed] [Google Scholar]
- 125.Isotani, S., K. Hara, C. Tokunaga, H. Inoue, J. Avruch, and K. Yonezawa. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274:34493-34498 [DOI] [PubMed] [Google Scholar]
- 126.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017-1026. [DOI] [PubMed] [Google Scholar]
- 127.Jacinto, E., and M. N. Hall. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 128.Jacinto, E., R. Loewith, A. Schmidt, S. Lin, M. A. Ruegg, A. Hall, and M. N. Hall. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122-1128. [DOI] [PubMed] [Google Scholar]
- 129.Jefferies, H. B., C. Reinhard, S. C. Kozma, and G. Thomas. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA 91:4441-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Johnson, S. P., and J. R. Warner. 1987. Phosphorylation of the Saccharomyces cerevisiae equivalent of ribosomal protein S6 has no detectable effect on growth. Mol. Cell. Biol. 7:1338-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kenerson, H. L., L. D. Aicher, L. D. True, and R. S. Yeung. 2002. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 62:5645-5650. [PubMed] [Google Scholar]
- 134.Kilberg, M. S., R. G. Hutson, and R. O. Laine. 1994. Amino acid-regulated gene expression in eukaryotic cells. FASEB J. 8:13-19. [DOI] [PubMed] [Google Scholar]
- 135.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 136.Kim, D. H., D. Sarbassov dos, S. M. Ali, R. R. Latek, K. V. Guntur, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11:895-904. [DOI] [PubMed] [Google Scholar]
- 137.Kimura, N., C. Tokunaga, S. Dalal, C. Richardson, K. Yoshino, K. Hara, B. E. Kemp, L. A. Witters, O. Mimura, and K. Yonezawa. 2003. A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells 8:65-79. [DOI] [PubMed] [Google Scholar]
- 138.Klionsky, D. J., and Y. Ohsumi. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1-32. [DOI] [PubMed] [Google Scholar]
- 139.Kobayashi, T., O. Minowa, Y. Sugitani, S. Takai, H. Mitani, E. Kobayashi, T. Noda, and O. Hino. 2001. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc. Natl. Acad. Sci. USA 98:8762-8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kozma, S. C., and G. Thomas. 2002. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24:65-71. [DOI] [PubMed] [Google Scholar]
- 142.Krause, U., L. Bertrand, and L. Hue. 2002. Control of p70 ribosomal protein S6 kinase and acetyl-CoA carboxylase by AMP-activated protein kinase and protein phosphatases in isolated hepatocytes. Eur. J. Biochem. 269:3751-3759. [DOI] [PubMed] [Google Scholar]
- 143.Kunz, J., R. Henriquez, U. Schneider, M. Deuter-Reinhard, N. R. Movva, and M. N. Hall. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73:585-596. [DOI] [PubMed] [Google Scholar]
- 144.Kuruvilla, F. G., A. F. Shamji, and S. L. Schreiber. 2001. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc. Natl. Acad. Sci. USA 98:7283-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kwiatkowski, D. J., H. Zhang, J. L. Bandura, K. M. Heiberger, M. Glogauer, N. el-Hashemite, and H. Onda. 2002. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11:525-534. [DOI] [PubMed] [Google Scholar]
- 146.Leicht, M., A. Simm, G. Bertsch, and J. Hoppe. 1996. Okadaic acid induces cellular hypertrophy in AKR-2B fibroblasts: involvement of the p70S6 kinase in the onset of protein and rRNA synthesis. Cell Growth Differ. 7:1199-1209. [PubMed] [Google Scholar]
- 147.Li, Y., M. N. Corradetti, K. Inoki, and K. L. Guan. 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29:32-38. [DOI] [PubMed] [Google Scholar]
- 148.Li, Y., K. Inoki, and K. L. Guan. 2004. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24:7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li, Y., K. Inoki, P. Vacrasis, and K. L. Guan. 2003. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 (TSC2) gene product, and enhances its interaction with 14-3-3. J. Biol. Chem. 278:13663-13671. [DOI] [PubMed] [Google Scholar]
- 150.Liaw, D., D. J. Marsh, J. Li, P. L. Dahia, S. I. Wang, Z. Zheng, S. Bose, K. M. Call, H. C. Tsou, M. Peacocke, C. Eng, and R. Parsons. 1997. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 16:64-67 [DOI] [PubMed] [Google Scholar]
- 151.Lin, T. A., X. Kong, T. A. Haystead, A. Pause, G. Belsham, N. Sonenberg, and J. C. Lawrence, Jr. 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266:653-656. [DOI] [PubMed] [Google Scholar]
- 152.Lin, T. A., X. Kong, A. R. Saltiel, P. J. Blackshear, and J. C. Lawrence, Jr. 1995. Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J. Biol. Chem. 270:18531-18538. [DOI] [PubMed] [Google Scholar]
- 153.Lizcano, J. M., O. Goransson, R. Toth, M. Deak, N. A. Morrice, J. Boudeau, S. A. Hawley, L. Udd, T. P. Makela, D. G. Hardie, and D. R. Alessi. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 155.Long, X., C. Spycher, Z. S. Han, A. M. Rose, F. Muller, and J. Avruch. 2002. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 12:1448-1461. [DOI] [PubMed] [Google Scholar]
- 156.Luke, M. M., F. Della Seta, C. J. Di Como, H. Sugimoto, R. Kobayashi, and K. T. Arndt. 1996. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16:2744-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Luo, Y., S. O. Marx, H. Kiyokawa, A. Koff, J. Massague, and A. R. Marks. 1996. Rapamycin resistance tied to defective regulation of p27Kip1. Mol. Cell. Biol. 16:6744-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mach, K. E., K. A. Furge, and C. F. Albright. 2000. Loss of Rhb1, a Rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mahajan, P. B. 1994. Modulation of transcription of rRNA genes by rapamycin. Int. J. Immunopharmacol. 16:711-721. [DOI] [PubMed] [Google Scholar]
- 160.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28:573-576. [DOI] [PubMed] [Google Scholar]
- 161.Manning, B. D., A. R. Tee, M. N. Logsdon, J. Blenis, and L. C. Cantley. 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10:151-162. [DOI] [PubMed] [Google Scholar]
- 162.Marsh, D. J., P. L. Dahia, Z. Zheng, D. Liaw, R. Parsons, R. J. Gorlin, and C. Eng. 1997. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 16:333-334. [DOI] [PubMed] [Google Scholar]
- 163.Matsumoto, S., A. Bandyopadhyay, D. J. Kwiatkowski, U. Maitra, and T. Matsumoto. 2002. Role of the Tsc1-Tsc2 complex in signaling and transport across the cell membrane in the fission yeast Schizosaccharomyces pombe. Genetics 161:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Matsuura, A., M. Tsukada, Y. Wada, and Y. Ohsumi. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245-250. [DOI] [PubMed] [Google Scholar]
- 165.Mayer, C., J. Zhao, X. Yuan, and I. Grummt. 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mayer-Jaekel, R. E., and B. A. Hemmings. 1994. Protein phosphatase 2A—a menage a trois'. Trends. Cell Biol. 4:287-291. [DOI] [PubMed] [Google Scholar]
- 167.Menand, B., T. Desnos, L. Nussaume, F. Berger, D. Bouchez, C. Meyer, and C. Robaglia. 2002. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99:6422-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Meyuhas, O. 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267:6321-6330. [DOI] [PubMed] [Google Scholar]
- 169.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 170.Mothe-Satney, I., D. Yang, P. Fadden, T. A. Haystead, and J. C. Lawrence, Jr. 2000. Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol. Cell. Biol. 20:3558-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Murakami, M., T. Ichisaka, M. Maeda, N. Oshiro, K. Hara, F. Edenhofer, H. Kiyama, K. Yonezawa, and S. Yamanaka. 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24:6710-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Murata, K., J. Wu, and D. L. Brautigan. 1997. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 94:10624-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nanahoshi, M., T. Nishiuma, Y. Tsujishita, K. Hara, S. Inui, N. Sakaguchi, and K. Yonezawa. 1998. Regulation of protein phosphatase 2A catalytic activity by alpha4 protein and its yeast homolog Tap42. Biochem. Biophys. Res. Commun. 251:520-526. [DOI] [PubMed] [Google Scholar]
- 174.Nave, B. T., M. Ouwens, D. J. Withers, D. R. Alessi, and P. R. Shepherd. 1999. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem. J. 344:427-431. [PMC free article] [PubMed] [Google Scholar]
- 175.Nellist, M., B. Verhaaf, M. A. Goedbloed, A. J. Reuser, A. M. van den Ouweland, and D. J. Halley. 2001. TSC2 missense mutations inhibit tuberin phosphorylation and prevent formation of the tuberin-hamartin complex. Hum. Mol. Genet. 10:2889-2898. [DOI] [PubMed] [Google Scholar]
- 176.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98:10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963-3966. [DOI] [PubMed] [Google Scholar]
- 178.Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, N. Tanaka, J. Avruch, and K. Yonezawa. 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278:15461-15464. [DOI] [PubMed] [Google Scholar]
- 179.Nourse, J., E. Firpo, W. M. Flanagan, S. Coats, K. Polyak, M. H. Lee, J. Massague, G. R. Crabtree, and J. M. Roberts. 1994. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature 372:570-573. [DOI] [PubMed] [Google Scholar]
- 180.Oldham, S., and E. Hafen. 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13:79-85. [DOI] [PubMed] [Google Scholar]
- 181.Oldham, S., J. Montagne, T. Radimerski, G. Thomas, and E. Hafen. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14:2689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Onda, H., A. Lueck, P. W. Marks, H. B. Warren, and D. J. Kwiatkowski. 1999. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J. Clin. Investig. 104:687-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Parekh, D., W. Ziegler, K. Yonezawa, K. Hara, and P. J. Parker. 1999. Mammalian TOR controls one of two kinase pathways acting upon nPKCdelta and nPKCepsilon. J. Biol. Chem. 274:34758-34764. [DOI] [PubMed] [Google Scholar]
- 184.Patel, J., X. Wang, and C. G. Proud. 2001. Glucose exerts a permissive effect on the regulation of the initiation factor 4E binding protein 4E-BP1. Biochem. J. 358:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pause, A., G. J. Belsham, A. C. Gingras, O. Donze, T. A. Lin, J. C. Lawrence, Jr., and N. Sonenberg. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762-767. [DOI] [PubMed] [Google Scholar]
- 186.Pearson, R. B., P. B. Dennis, J. W. Han, N. A. Williamson, S. C. Kozma, R. E. Wettenhall, and G. Thomas. 1995. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 14:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pende, M., S. H. Um, V. Mieulet, M. Sticker, V. L. Goss, J. Mestan, M. Mueller, S. Fumagalli, S. C. Kozma, and G. Thomas. 2004. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Peng, T., T. R. Golub, and D. M. Sabatini. 2002. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol. Cell. Biol. 22:5575-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Peterson, R. T., P. A. Beal, M. J. Comb, and S. L. Schreiber. 2000. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 275:7416-7423. [DOI] [PubMed] [Google Scholar]
- 190.Peterson, R. T., B. N. Desai, J. S. Hardwick, and S. L. Schreiber. 1999. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin-associated protein. Proc. Natl. Acad. Sci. USA 96:4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Plank, T. L., R. S. Yeung, and E. P. Henske. 1998. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 58:4766-4770. [PubMed] [Google Scholar]
- 192.Podsypanina, K., R. T. Lee, C. Politis, I. Hennessy, A. Crane, J. Puc, M. Neshat, H. Wang, L. Yang, J. Gibbons, P. Frost, V. Dreisbach, J. Blenis, Z. Gaciong, P. Fisher, C. Sawyers, L. Hedrick-Ellenson, and R. Parsons. 2001. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− mice. Proc. Natl. Acad. Sci. USA 98:10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Potter, C. J., H. Huang, and T. Xu. 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105:357-368. [DOI] [PubMed] [Google Scholar]
- 194.Potter, C. J., L. G. Pedraza, and T. Xu. 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4:658-665. [DOI] [PubMed] [Google Scholar]
- 195.Potter, C. J., and T. Xu. 2001. Mechanisms of size control. Curr. Opin. Genet. Dev. 11:279-286. [DOI] [PubMed] [Google Scholar]
- 196.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Price, D. J., J. R. Grove, V. Calvo, J. Avruch, and B. E. Bierer. 1992. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science 257:973-977. [DOI] [PubMed] [Google Scholar]
- 198.Proud, C. G. 2002. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 269:5338-5349. [DOI] [PubMed] [Google Scholar]
- 199.Ravikumar, B., C. Vacher, Z. Berger, J. E. Davies, S. Luo, L. G. Oroz, F. Scaravilli, D. F. Easton, R. Duden, C. J. O'Kane, and D. C. Rubinsztein. 2004. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36:585-595. [DOI] [PubMed] [Google Scholar]
- 200.Redpath, N. T., E. J. Foulstone, and C. G. Proud. 1996. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. EMBO J. 15:2291-2297. [PMC free article] [PubMed] [Google Scholar]
- 201.Reinke, A., S. Anderson, J. M. McCaffery, J. Yates, 3rd, S. Aronova, S. Chu, S. Fairclough, C. Iverson, K. P. Wedaman, and T. Powers. 2004. TOR complex 1 (TORC1) includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in S. cerevisiae. J. Biol. Chem. 279:14752-14762. [DOI] [PubMed] [Google Scholar]
- 202.Reynolds, T. H. IV, S. C. Bodine, and J. C. Lawrence, Jr. 2002. Control of Ser2448 phosphorylation in the mammalian target of rapamycin by insulin and skeletal muscle load. J. Biol. Chem. 277:17657-17662. [DOI] [PubMed] [Google Scholar]
- 203.Rhoads, R. E. 1991. Protein synthesis, cell growth and oncogenesis. Curr. Opin. Cell Biol. 3:1019-1024. [DOI] [PubMed] [Google Scholar]
- 204.Roux, P. P., B. A. Ballif, R. Anjum, S. P. Gygi, and J. Blenis. 2004. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 101:13489-13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sabatini, D. M., R. K. Barrow, S. Blackshaw, P. E. Burnett, M. M. Lai, M. E. Field, B. A. Bahr, J. Kirsch, H. Betz, and S. H. Snyder. 1999. Interaction of RAFT1 with gephyrin required for rapamycin-sensitive signaling. Science 284:1161-1164. [DOI] [PubMed] [Google Scholar]
- 206.Sabatini, D. M., H. Erdjument-Bromage, M. Lui, P. Tempst, and S. H. Snyder. 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78:35-43. [DOI] [PubMed] [Google Scholar]
- 207.Saitoh, M., N. Pullen, P. Brennan, D. Cantrell, P. B. Dennis, and G. Thomas. 2002. Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J. Biol. Chem. 277:20104-20112. [DOI] [PubMed] [Google Scholar]
- 208.Sarbassov dos, D., S. M. Ali, D. H. Kim, D. A. Guertin, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296-1302. [DOI] [PubMed] [Google Scholar]
- 209.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566-571. [DOI] [PubMed] [Google Scholar]
- 210.Sawyers, C. L. 2003. Will mTOR inhibitors make it as cancer drugs? Cancer Cell 4:343-348. [DOI] [PubMed] [Google Scholar]
- 211.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 212.Schalm, S. S., D. C. Fingar, D. M. Sabatini, and J. Blenis. 2003. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13:797-806. [DOI] [PubMed] [Google Scholar]
- 213.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 214.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schmidt, A., M. Bickle, T. Beck, and M. N. Hall. 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88:531-542. [DOI] [PubMed] [Google Scholar]
- 216.Schmidt, A., and A. Hall. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587-1609. [DOI] [PubMed] [Google Scholar]
- 217.Schmidt, A., and M. N. Hall. 1998. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:305-338. [DOI] [PubMed] [Google Scholar]
- 218.Schmidt, A., J. Kunz, and M. N. Hall. 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 93:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schmidt, E. V. 1999. The role of c-myc in cellular growth control. Oncogene 18:2988-2996. [DOI] [PubMed] [Google Scholar]
- 220.Scott, P. H., G. J. Brunn, A. D. Kohn, R. A. Roth, and J. C. Lawrence, Jr. 1998. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc. Natl. Acad. Sci. USA 95:7772-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sekito, T., Z. Liu, J. Thornton, and R. A. Butow. 2002. RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3]. Mol. Biol. Cell 13:795-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sekulic, A., C. C. Hudson, J. L. Homme, P. Yin, D. M. Otterness, L. M. Karnitz, and R. T. Abraham. 2000. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 60:3504-3513. [PubMed] [Google Scholar]
- 223.Shah, O. J., Z. Wang, and T. Hunter. 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14:1650-1656. [DOI] [PubMed] [Google Scholar]
- 224.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 225.Shamji, A. F., P. Nghiem, and S. L. Schreiber. 003. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol. Cell 12:271-280. [DOI] [PubMed] [Google Scholar]
- 226.Shaw, R. J., N. Bardeesy, B. D. Manning, L. Lopez, M. Kosmatka, R. A. DePinho, and L. C. Cantley. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6:91-99. [DOI] [PubMed] [Google Scholar]
- 227.Shaw, R. J., M. Kosmatka, N. Bardeesy, R. L. Hurley, L. A. Witters, R. A. DePinho, and L. C. Cantley. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 101:3329-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shepherd, P. R., B. T. Nave, and K. Siddle. 1995. Insulin stimulation of glycogen synthesis and glycogen synthase activity is blocked by wortmannin and rapamycin in 3T3-L1 adipocytes: evidence for the involvement of phosphoinositide 3-kinase and p70 ribosomal protein-S6 kinase. Biochem. J. 305:25-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shigemitsu, K., Y. Tsujishita, K. Hara, M. Nanahoshi, J. Avruch, and K. Yonezawa. 1999. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J. Biol. Chem. 274:1058-1065. [DOI] [PubMed] [Google Scholar]
- 230.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sollner-Webb, B., and J. Tower. 1986. Transcription of cloned eukaryotic ribosomal RNA genes. Annu. Rev. Biochem. 55:801-830. [DOI] [PubMed] [Google Scholar]
- 232.Sood, R., A. C. Porter, D. A. Olsen, D. R. Cavener, and R. C. Wek. 2000. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics 154:787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Stocker, H., T. Radimerski, B. Schindelholz, F. Wittwer, P. Belawat, P. Daram, S. Breuer, G. Thomas, and E. Hafen. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5:559-565. [DOI] [PubMed] [Google Scholar]
- 234.Stolovich, M., H. Tang, E. Hornstein, G. Levy, R. Cohen, S. S. Bae, M. J. Birnbaum, and O. Meyuhas. 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22:8101-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sutton, A., D. Immanuel, and K. T. Arndt. 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11:2133-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tabancay, A. P., Jr., C. L. Gau, I. M. Machado, E. J. Uhlmann, D. H. Gutmann, L. Guo, and F. Tamanoi. 2003. Identification of dominant negative mutants of Rheb GTPase and their use to implicate the involvement of human Rheb in the activation of p70S6K. J. Biol. Chem. 278:39921-39930. [DOI] [PubMed] [Google Scholar]
- 237.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 238.Takano, A., I. Usui, T. Haruta, J. Kawahara, T. Uno, M. Iwata, and M. Kobayashi. 2001. Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptor substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol. Cell. Biol. 21:5050-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tang, H., E. Hornstein, M. Stolovich, G. Levy, M. Livingstone, D. Templeton, J. Avruch, and O. Meyuhas. 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21:8671-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tapon, N., N. Ito, B. J. Dickson, J. E. Treisman, and I. K. Hariharan. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105:345-355. [DOI] [PubMed] [Google Scholar]
- 241.Tee, A. R., R. Anjum, and J. Blenis. 2003. Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase (PI3K)/Akt-dependent and -independent phosphorylation of tuberin. J. Biol. Chem. 278:37288-37296. [DOI] [PubMed] [Google Scholar]
- 242.Tee, A. R., D. C. Fingar, B. D. Manning, D. J. Kwiatkowski, L. C. Cantley, and J. Blenis. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 99:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13:1259-1268. [DOI] [PubMed] [Google Scholar]
- 244.Tee, A. R., and C. G. Proud. 2002. Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol. Cell. Biol. 22:1674-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Terada, N., H. R. Patel, K. Takase, K. Kohno, A. C. Nairn, and E. W. Gelfand. 1994. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl. Acad. Sci. USA 91:11477-11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Thomas, G. 2002. The S6 kinase signaling pathway in the control of development and growth. Biol. Res. 35:305-313. [DOI] [PubMed] [Google Scholar]
- 247.Treins, C., S. Giorgetti-Peraldi, J. Murdaca, G. L. Semenza, and E. Van Obberghen. 2002. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J. Biol. Chem. 277:27975-27981. [DOI] [PubMed] [Google Scholar]
- 248.Tsang, C. K., P. G. Bertram, W. Ai, R. Drenan, and X. F. Zheng. 2003. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 22:6045-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Um, S. H., F. Frigerio, M. Watanabe, F. Picard, M. Joaquin, M. Sticker, S. Fumagalli, P. R. Allegrini, S. C. Kozma, J. Auwerx, and G. Thomas. 2004. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431:200-205. [DOI] [PubMed] [Google Scholar]
- 250.Urano, J., C. Ellis, G. J. Clark, and F. Tamanoi. 2001. Characterization of Rheb functions using yeast and mammalian systems. Methods Enzymol. 333:217-231. [DOI] [PubMed] [Google Scholar]
- 251.Urano, J., A. P. Tabancay, W. Yang, and F. Tamanoi. 2000. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J. Biol. Chem. 275:11198-11206. [DOI] [PubMed] [Google Scholar]
- 252.Usui, I., T. Haruta, M. Iwata, A. Takano, T. Uno, J. Kawahara, E. Ueno, T. Sasaoka, and M. Kobayashi. 2000. Retinoblastoma protein phosphorylation via PI 3-kinase and mTOR pathway regulates adipocyte differentiation. Biochem. Biophys. Res. Commun. 275:115-120. [DOI] [PubMed] [Google Scholar]
- 253.Vandenbol, M., J. C. Jauniaux, S. Vissers, and M. Grenson. 1987. Isolation of the NPR1 gene responsible for the reactivation of ammonia-sensitive amino-acid permeases in Saccharomyces cerevisiae. RNA analysis and gene dosage effects. Eur. J. Biochem. 164:607-612. [DOI] [PubMed] [Google Scholar]
- 254.van Heusden, G. P., and H. Y. Steensma. 2001. 14-3-3 Proteins are essential for regulation of RTG3-dependent transcription in Saccharomyces cerevisiae. Yeast 18:1479-1491. [DOI] [PubMed] [Google Scholar]
- 255.Van Slegtenhorst, M., E. Carr, R. Stoyanova, W. Kruger, and E. P. Henske. 2004. Tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 279:12706-12713. [DOI] [PubMed] [Google Scholar]
- 256.van Slegtenhorst, M., R. de Hoogt, C. Hermans, M. Nellist, B. Janssen, S. Verhoef, D. Lindhout, A. van den Ouweland, D. Halley, J. Young, M. Burley, S. Jeremiah, K. Woodward, J. Nahmias, M. Fox, R. Ekong, J. Osborne, J. Wolfe, S. Povey, R. G. Snell, J. P. Cheadle, A. C. Jones, M. Tachataki, D. Ravine, D. J. Kwiatkowski, et al. 1997. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science 277:805-808. [DOI] [PubMed] [Google Scholar]
- 257.van Slegtenhorst, M., M. Nellist, B. Nagelkerken, J. Cheadle, R. Snell, A. van den Ouweland, A. Reuser, J. Sampson, D. Halley, and P. van der Sluijs. 1998. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum. Mol. Genet. 7:1053-1057. [DOI] [PubMed] [Google Scholar]
- 258.van Zyl, W., W. Huang, A. A. Sneddon, M. Stark, S. Camier, M. Werner, C. Marck, A. Sentenac, and J. R. Broach. 1992. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4946-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wang, X., L. E. Campbell, C. M. Miller, and C. G. Proud. 1998. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 334:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wang, X., W. Li, J. L. Parra, A. Beugnet, and C. G. Proud. 2003. The C terminus of initiation factor 4E-binding protein 1 contains multiple regulatory features that influence its function and phosphorylation. Mol. Cell. Biol. 23:1546-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 262.Wedaman, K. P., A. Reinke, S. Anderson, J. Yates III, J. M. McCaffery, and T. Powers. 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14:1204-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wek, R. C., B. M. Jackson, and A. G. Hinnebusch. 1989. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. USA 86:4579-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 265.Weng, Q. P., K. Andrabi, M. T. Kozlowski, J. R. Grove, and J. Avruch. 1995. Multiple independent inputs are required for activation of the p70 S6 kinase. Mol. Cell. Biol. 15:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wera, S., and B. A. Hemmings. 1995. Serine/threonine protein phosphatases. Biochem. J. 311:17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wilkinson, M. G., T. S. Pino, S. Tournier, V. Buck, H. Martin, J. Christiansen, D. G. Wilkinson, and J. B. Millar. 1999. Sin1: an evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 18:4210-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Woods, A., S. R. Johnstone, K. Dickerson, F. C. Leiper, L. G. Fryer, D. Neumann, U. Schlattner, T. Wallimann, M. Carlson, and D. Carling. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13:2004-2008. [DOI] [PubMed] [Google Scholar]
- 269.Woychik, N. A., S. M. Liao, P. A. Kolodziej, and R. A. Young. 1990. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 4:313-323. [DOI] [PubMed] [Google Scholar]
- 270.Xu, G., G. Kwon, C. A. Marshall, T. A. Lin, J. C. Lawrence, Jr., and M. L. McDaniel. 1998. Branched-chain amino acids are essential in the regulation of PHAS-1 and p70 S6 kinase by pancreatic beta-cells. A possible role in protein translation and mitogenic signaling. J. Biol. Chem. 273:28178-28184. [DOI] [PubMed] [Google Scholar]
- 271.Yamagata, K., L. K. Sanders, W. E. Kaufmann, W. Yee, C. A. Barnes, D. Nathans, and P. F. Worley. 1994. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J. Biol. Chem. 269:16333-16339. [PubMed] [Google Scholar]
- 272.Yang, W., A. P. Tabancay, Jr., J. Urano, and F. Tamanoi. 2001. Failure to farnesylate Rheb protein contributes to the enrichment of G0/G1 phase cells in the Schizosaccharomyces pombe farnesyltransferase mutant. Mol. Microbiol. 41:1339-1347. [DOI] [PubMed] [Google Scholar]
- 273.Yee, W. M., and P. F. Worley. 1997. Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol. Cell. Biol. 17:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Yokogami, K., S. Wakisaka, J. Avruch, and S. A. Reeves. 2000. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 10:47-50. [DOI] [PubMed] [Google Scholar]
- 275.Yu, K., L. Toral-Barza, C. Discafani, W. G. Zhang, J. Skotnicki, P. Frost, and J. J. Gibbons. 2001. mTOR, a novel target in breast cancer: the effect of CCI-779, an mTOR inhibitor, in preclinical models of breast cancer. Endocr. Relat. Cancer 8:249-258. [DOI] [PubMed] [Google Scholar]
- 276.Zaragoza, D., A. Ghavidel, J. Heitman, and M. C. Schultz. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhang, F. L., and P. J. Casey. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65:241-269. [DOI] [PubMed] [Google Scholar]
- 278.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumor suppressor proteins. Nat. Cell Biol. 5:578-581. [DOI] [PubMed] [Google Scholar]