Abstract
Biomass conversion to ethanol as a liquid fuel by the thermophilic and anaerobic clostridia offers a potential partial solution to the problem of the world's dependence on petroleum for energy. Coculture of a cellulolytic strain and a saccharolytic strain of Clostridium on agricultural resources, as well as on urban and industrial cellulosic wastes, is a promising approach to an alternate energy source from an economic viewpoint. This review discusses the need for such a process, the cellulases of clostridia, their presence in extracellular complexes or organelles (the cellulosomes), the binding of the cellulosomes to cellulose and to the cell surface, cellulase genetics, regulation of their synthesis, cocultures, ethanol tolerance, and metabolic pathway engineering for maximizing ethanol yield.
INTRODUCTION
Need for Alternative Energy Source
Of the total amount of gasoline used in the United States as a transportation fuel and by industry, about half has to be imported (358). The amount of solar energy received at the earth's surface is 2.5 × 1021 Btu/year, which far exceeds the present human usage of 2.0 × 1017 Btu/year. The amount of energy from the sun which is stored as carbon via photosynthesis is 10 times the world usage. Lignocellulose is the most abundant renewable natural resource and substrate available for conversion to fuels. It is inexpensive, plentiful, and renewable. On a worldwide basis, terrestrial plants produce 1.3 × 1010 metric tons (dry weight basis) of wood per year, which is equivalent to 7 × 109 metric tons of coal or about two-thirds of the world's energy requirement. Available cellulosic feedstocks from agriculture and other sources are about 180 million tons per year (203). Furthermore, tremendous amounts of cellulose are available as municipal and industrial wastes which today contribute to our pollution problems. Thus, there is great interest in the use of cellulosic biomass as a renewable source of energy via breakdown to sugars that can then be converted to liquid fuel.
With the hike in oil prices around the world in the 1970s and the realization that the world's oil supply is finite, the quest for alternative fuels began in 1975, and researchers looked for economical ways to produce ethanol, preferably from abundantly available, biodegradable, and renewable raw materials. Ethanol is an excellent transportation fuel, in some respects superior to gasoline (199, 200). In particular, with respect to gasoline, neat (unblended) ethanol burns more cleanly, has a higher octane rating, can be burned with greater efficiency, is thought to produce smaller amounts of ozone precursors (thus decreasing urban air pollution), and is particularly beneficial with respect to low net CO2 put into the atmosphere. Furthermore, ethanol by fermentation offers a more favorable trade balance, enhanced energy security, and a major new crop for a depressed agricultural economy. Ethanol is considerably less toxic to humans than is gasoline (or methanol). Ethanol also reduces smog formation because of low volatility; its photochemical reactivity and that of its combustion products are low, and only low levels of smog-producing compounds are formed by its combustion (359). Its high heat of vaporization, high octane rating, and low flame temperature yield good engine performance. Cellulosics, at $42 per dry ton, cost the same as petroleum, at 6 to $7 per barrel based on equivalent mass or 12 to $13 per barrel based on equivalent energy content (203, 356); today, the price of petroleum is much higher. It appears probable that ethanol from cellulose could become competitive with gasoline if the processing costs for the former could be lowered. Ethanol is also used as an oxygenate to reduce automobile emissions. With the current and impending phase-out of methyl tert-butyl ether as an oxygenate in many states in the United States, ethanol will fill the void. Useful reviews on the biological conversion of lignocellulosic biomass to ethanol have been published (183, 196, 198, 200, 201, 203, 204, 206, 224, 356).
The potential quantity of ethanol that could be produced from cellulose is over an order of magnitude larger than that producible from corn. In contrast to the corn-to-ethanol conversion, the cellulose-to-ethanol route involves little or no contribution to the greenhouse effect and has a clearly positive net energy balance (five times better). As a result of such considerations, microorganisms that metabolize cellulose have gained prominence in recent years (38, 329).
Lignocellulose is difficult to hydrolyze because (i) it is associated with hemicellulose, (ii) it is surrounded by a lignin seal which has a limited covalent association with hemicellulose, and (iii) much of it has a crystalline structure with a potential formation of six hydrogen bonds, four intramolecular and two intermolecular, giving it a highly ordered, tightly packed structure (344). Pretreatments aim at increasing the surface area of cellulose by (i) removing the lignin seal, (ii) solubilizing hemicellulose, (iii) disrupting crystallinity, and/or (iv) increasing pore volume. The value of a cellulase system that attacks crystalline cellulose lies in the observation that many of the pretreatments which increase surface area also increase crystallinity. These include dilute sulfuric acid, alkali, and ethylenediamine.
The rate-limiting step in the conversion of cellulose to fuels is its hydrolysis, especially the initial attack on the highly ordered, insoluble structure of crystalline cellulose, since the products of this attack are readily solubilized and converted to sugars. A great deal of effort has gone into the development of methods for conversion of cellulose to sugars. Most of this work has emphasized the biochemistry, genetics, and process development of fungi (especially Trichoderma reesei) coupled to the further conversion of the sugars produced to ethanol by yeast (Saccharomyces cerevisiae). After many years of study, it is becoming apparent that such a process is not the only potential solution.
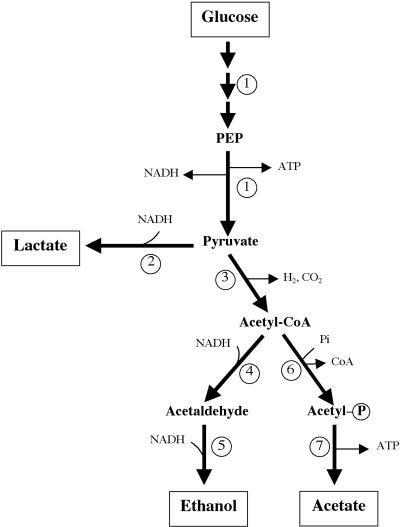
The large amounts of attention and resources devoted over the past 30 years to the Trichoderma-Saccharomyces concept have perhaps interfered in the industrial development of the potential of the cellulolytic, thermophilic anaerobic bacteria such as Clostridium thermocellum (49, 51, 66, 81, 216) and their cellulases and hemicellulases (28, 32, 78, 195, 204, 274, 301, 315). C. thermocellum breaks down cellulose, with the formation of cellobiose and cellodextrins as main products. Cellobiose, a disaccharide of two glucose moieties held together by a β-1,4 linkage, can be further utilized by the organism, and the final end products are ethanol, acetic acid, lactic acid, hydrogen, and carbon dioxide (181) (Fig. 1). Small cellodextrins can also be taken into the cell, broken down further, and metabolized (203). The interest in this organism is due to several factors (Table 1). First, C. thermocellum can utilize lignocellulosic waste and generate ethanol, a rare property among living organisms. Indeed, the cellulase system of C. thermocellum mediates hydrolysis of lignin-containing materials such as hardwood pretreated with dilute acid as well as model substrates not containing lignin (196, 198). Second, for large-scale culture, anaerobiosis is an advantage because one of the most expensive steps in industrial fermentations is that of providing adequate oxygen transfer, e.g., for cellulase production. Third, since the optimum temperature for the cultivation of the organism is 60°C, the problems of contamination are lessened and the cooling of large fermentors is much simplified. Fourth, growth at a high temperature facilitates the recovery of ethanol. Fifth, thermophiles are thought to be robust microorganisms and contain stable enzymes. Sixth, anaerobes generally have a low cell yield, and hence more of the substrate is converted to ethanol. Experience suggests that even more advantageous are in situ cellulase production and the high rates of growth on and metabolism of cellulose and hemicellulose. Since the carbon dioxide produced during fermentation and fuel use of ethanol is recycled by growth of plants, ethanol does not contribute to carbon dioxide accumulation in the atmosphere and possible global warming (359).
FIG. 1.
The clostridial coculture process in which C. thermocellum serves as the cellulase and hemicellulase producer. The hemicellulose-derived pentoses can be utilized by C. thermosaccharolyticum but not C. thermocellum. C. thermosaccharolyticum uses cellobiose faster and is a better ethanol producer. In addition to cellobiose, cellodextrins are also produced from cellulose and can be utilized directly.
TABLE 1.
Advantages of using C. thermocellum for ethanol fermentation from biomass
| The cellulolytic and ethanogenic nature, allowing saccharification and fermentation in a single step |
| The anaerobic nature, negating the need for expensive oxygen transfer |
| Low cell growth yield, favoring ethanol conversion |
| The thermophilic nature, facilitating ethanol removal and recovery |
| The thermophilic nature, reducing cooling cost |
| Thermophilic fermentation being less prone to contamination |
| Thermophilic biomass-degrading enzymes enhancing protein stability |
| Amenability to coculture with other ethanol-producing and pentose-fermenting organisms |
In 1990, 90% of new cars in Brazil used neat ethanol, while the remainder utilized a blend of 20 to 22% ethanol and 80 to 78% gasoline. Three to four billion gallons are produced in Brazil from sugar per year. The United States in 1987 was using 8.6 metric tons (340 million bushels) of corn to make 850 million gallons of anhydrous ethanol for blending with gasoline at a level of 10%. The ethanol-gasoline blend was supplying about 8% of the U.S. gasoline market. Although small, this figure had risen from less than 1% in 1981 (355). In 1990, the United States consumed 112 billion gallons of gasoline. By 1993, the United States was producing 1.0 billion gallons of ethanol from corn (358); by 1996, the amount had risen to 1.3 billion gallons (197, 247); by 2000, it was 1.6 billion gallons; and by 2001, it was 1.8 billion gallons (84, 349). If all cellulosic feedstocks in the United States could be used for ethanol, 20 billion gallons could be produced (203, 224), which would be more than enough to add 10% ethanol to all gasoline used in the United States.
Cellulosic biomass in the early 1980s, at 20 to $30/dry ton, was much cheaper than corn, at $110/dry ton, yet cellulose hydrolysis was not competitive with starch hydrolysis to produce sugar. The problems included cellulose crystallinity and lignin content, requiring expensive pretreatment. Also, corn usage provided oil and protein by-products, which were of much higher value than lignin (247). By 1990, the cost of ethanol production from cellulose (by Trichoderma plus yeast) was thought to be approaching that of production from corn (by yeast) on an unsubsidized basis (130, 196, 200). However, neither ethanol produced from corn nor ethanol produced from cellulose is currently competitive with conventional liquid fuels. A state-of-the-art process designed for ethanol production from cellulose had a selling price of U.S. $1.35 per gallon in 1988 (200), $1.18 per gallon in 1996 (201), and $1.20 in 2003 (356). For production from corn, the price in 1994 was $1.22 per gallon and that in 2001 was $1.65 (355, 357). However, a price of 50 cents per gallon would have been required for ethanol to compete with gasoline in the early 1990s, 67 cents per gallon would have been required in 1994, and 70 cents per gallon would have been required in 2000. With improved technology applied to the microbial biomass-to-ethanol technology, it was projected that the selling price of pure ethanol could be 50 cents per gallon and in the best case could be as low as 34 cents per gallon (201). Thus, substantial cost reductions are possible and could make biological ethanol a competitive neat transportation fuel (206).
The cost of the cellulase in the Trichoderma-yeast process is still prohibitively high, whereas for a direct clostridial coculture process, the enzymes cost very little because they are produced by the fermenting organism in the course of ethanol production. The direct fermentation of cellulose to ethanol could save 50 cents per gallon compared to a state-of-the-art Trichoderma-yeast simultaneous saccharification and fermentation process, since the former process combines cellulase production, hydrolysis, and fermentation in a single bioreactor (130). Conversion of mixed hardwood flour to ethanol in a continuous fermentor was 2.5 times higher with C. thermocellum than with the simultaneous saccharification and fermentation process using Trichoderma cellulase, β-glucosidase, and S. cerevisiae (310); furthermore, the rate of conversion was four times higher. Mixed cultures of thermophilic anaerobic bacteria offer the further potential of decreasing the production costs of lignocellulose conversion to ethanol by twofold (198). With resources dedicated to the exploitation of these bacteria, the conversion of agricultural, forest, and urban resources into ethanol could become an economic substitute for petroleum fuels when oil prices are about U.S. $30 or more per barrel. This new technology could provide a profitable outlet for renewable resources sooner than would occur by waiting for the Trichoderma-yeast process to become economical by gradual increases in the price of petroleum. The primary advantages of a direct clostridial conversion include elimination of capital or operating costs for enzyme production, greatly reduced diversion of substrate for enzyme production, and compatible enzyme and fermentation systems. Moreover, with the increased use of ethanol as an octane enhancer replacing tetraethyl lead and as an oxygenate replacing methyl tert-butyl ether, the development of an anaerobic bacterial process using waste feedstocks is likely to be justified at current petroleum prices.
THE CLOSTRIDIUM THERMOCELLUM CELLULASE SYSTEM
Strains and Media
Because of the need to develop alternative forms of liquid energy, in the late 1970s we began studies at the Massachusetts Institute of Technology (M.I.T.) involving C. thermocellum strain ATCC 27405. This microbe had been isolated by Viljoen et al. (336) from manure and later described by McBee (218). The Japanese strain F1 and Russian strain F7 are very similar to ATCC 27405 DSM 1237, NCIB 10682) (110). Our group was chiefly concerned with its nutrition, regulation of enzyme synthesis, and enzymology. We found that at a low yeast extract concentration (0.2%), only cellulose and cellobiose were readily utilized, out of approximately 50 carbon sources. However, upon increasing yeast extract to 0.6%, the organism assimilated glucose (304). We developed a chemically defined medium in which the yeast extract was replaced by para-aminobenzoic acid, vitamin B12, pyridoxamine, and biotin (140) and found that C. thermocellum could use ammonium, nitrate, urea, or certain amino acids as major nitrogen sources (93). The organism was found to break down xylan but could not use the resulting xylose or xylose oligomers. Another strain of C. thermocellum (YM4) requires para-aminobenzoic acid, vitamin B12, vitamin B6, and folic acid (237).
Properties of the Cellulase System
Our initial enzymatic studies employed a cellulase assay with the chromogenic substrate trinitrophenyl-carboxymethylcellulose (TNP-CMC), which measured endo-β-glucanase activity. We found the TNP-CMCase activity of C. thermocellum to be constitutive, neither repressed nor inhibited by cellobiose or glucose (304), and over 95% of the extracellular enzymatic activity on TNP-CMC appeared to be unaffected by oxygen, thiol compounds, thiol reagents, and mono- or divalent cations.
Unlike fungal cellulases, the C. thermocellum cellulase complex has very high activity on crystalline cellulose (140); i.e., it has “true cellulase activity” (also called Avicelase) which is characterized by its ability to completely solubilize crystalline forms of cellulose such as cotton and Avicel (141). This unique cellulase system has been studied by a number of groups biochemically, immunologically, and via molecular biological techniques. The complex is comprised of: (i) numerous endo-β-glucanases which are responsible for the random breakdown of amorphous types of cellulose, including CMC and TNP-CMC (4, 30, 31, 41, 62, 80, 93, 122, 143-145, 162, 221, 239, 240, 249, 267, 268, 276, 295, 296, 304, 307, 312); (ii) at least four exoglucanases (152, 222, 234, 308, 342, 372); (iii) a cellobiose phosphorylase that breaks down cellobiose to glucose and glucose-1-phosphate (5); (iv) a cellodextrin phosphorylase that phosphorylyzes β-1,4-oligoglucans (300); and (v) two β-glucosidases that hydrolyze cellobiose to glucose (3, 4, 104). C. thermocellum also possesses at least six xylanases (107, 232), two lichenases (367), two laminarinases (332), and minor activities of β-xylosidase, β-galactosidase, and β-mannosidase (164). A strain of C. thermocellum has been shown to degrade pectin and probably produces pectin lyase, polygalacturonate hydrolase, and pectin methylesterase (314). We found the activity responsible for hydrolysis of crystalline cellulose, unlike the endoglucanases, to be inhibited by cellobiose (141); glucose had no such inhibitory effect.
In our experiments (141), filter paper was found to be the preferred substrate for Trichoderma cellulase, whereas cotton was the best substrate for the clostridial enzyme complex. This high activity on a highly ordered substrate reflects C. thermocellum's ability to proliferate under thermophilic, anaerobic conditions on partially digested plant tissues. As an anaerobe depending solely on glycolysis for its cellular energy, C. thermocellum cannot afford to produce large quantities of extracellular proteins. Thus, it makes a cellulase complex with about 50-fold-higher specific activity than that of Trichoderma under the assay conditions used; the latter is excreted at a high extracellular concentration but is rather weak in activity.
For a long time, it was puzzling that C. thermocellum could grow well on crystalline cellulose whereas its extracellular fluid only poorly degraded crystalline cellulose (e.g., Avicel). When the organism was grown on agar containing Avicel, we found clear zones around the colonies, indicating production of an active, extracellular enzyme. We finally were able to solve this problem. We found the extracellular cellulase activity from C. thermocellum to have three unusual properties that had not been previously seen with cellulase preparations from other bacteria or fungi and which made it a uniquely active enzyme complex. (i) This thermophilic cellulase complex contains sulfhydryl groups that are essential either for the saccharification of crystalline celluloses such as cotton fibers, filter paper, or Avicel or for its structural stability (142). This property requires that the cellulase complex be used under reducing conditions and renders the activity susceptible to oxidation and sulfhydryl inactivation. When the cellulase activity is protected by reducing agents such as dithiothreitol (DTT), cysteine, sodium dithionate, glutathione, or mercaptoethanol, it displays high specific activity. (ii) The complex has a requirement for Ca2+. Cotton, Avicel, and filter paper are completely solubilized under reducing conditions in the presence of Ca2+. Although its activity was stimulated by 10 mM DTT, it was inhibited by a lower DTT concentration (0.1 to 1 mM). We found this to be due to autooxidation of low DTT producing H2O2, which inactivates the enzyme under aerobic or anaerobic conditions (139). The inactivation was prevented by catalase but not by superoxide dismutase or hydroxyl radical scavengers. As expected, sulfhydryl oxidizing agents [e.g., o-iodosobenzoate and 5,5′-dithio-bis-(2 nitrobenzoic acid)] inhibited the activity, and the inhibition was reversed by 10 mM DTT. Furthermore, nonoxidizing SH reagents such as N-ethylmaleimide, iodoacetate, and p-chloromercuribenzoate were also inhibitory. Since oxidation of thiol groups involves metals, it was not surprising that a low concentration of EDTA (1 mM) inhibited inactivation by low DTT concentration whereas H2O2 and Cu2+ stimulated inactivation. The component of the cellulase complex that was susceptible to sulfhydryl inactivation appeared not to be an endoglucanase, since endoglucanase activity is unaffected by oxidation or thiol reagents. It probably is exoglucanase CelS (also known as SS, equivalent to component S8 of the complex) whose stability is increased by Ca2+ and thiols and whose activity is inhibited by cellobiose (234). Another exoglucanase (CBH3) has a molecular mass (78 kDa) similar to that of CelS but some different properties, such as substrate specificity and less inhibition by cellobiose; it also is protected by Ca2+ and DTT from high temperature inactivation (308). It should be noted, however, that some endoglucanases (e.g., CelD) are stimulated and stabilized at high temperature by Ca2+ (55). (iii) The third unusual property of the C. thermocellum true cellulase system is the possible involvement of a transition metal ion, presumably iron (139). Cellulose saccharification under anaerobic (reducing) conditions was inhibited by the chelators o-phenanthroline and dipyridyl; this was reversed by preincubation with Fe2+ plus Fe3+. The presence of a catalytic metal ion and/or sulfhydryl groups could provide an explanation for the high specific activity of clostridial cellulase on crystalline cellulose.
Breakdown products of cellulose are both cellodextrins and cellobiose. When these enter the cell, they can be broken down extracytoplasmically via phosphorolytic cleavage by cellodextrin phosphorylase and/or cellobiose phosphorylase or via hydrolytic cleavage by β-glucosidase. Two intracellular β-glucosidases have been purified, characterized, and cloned (3, 103, 148, 153, 294). Rates of phosphorolytic cleavage have been found to be 20-fold higher than those of hydrolytic cleavage (365). From a bioenergetic standpoint, phosphorolytic cleavage is more beneficial because it provides a route to ATP synthesis. Cellodextrins appear to be more favorable to phosphorolysis than hydrolysis; the apparent Km for cellodextrin phosphorylase (measured with cellopentaose) was found to be considerably lower (0.61 mM) than that for cellobiose phosphorylase (3.3 mM). It appears that the mean cellodextrin length assimilated during growth on cellulose is equal to or larger than four hexose units (Y.-H. P. Zhang, and L. R. Lynd, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. I-002, 2002).
C. thermocellum cells as well as its extracellular cellulase complex completely solubilize the model crystalline substrate Avicel and a more realistic substrate, dilute-acid-pretreated mixed hardwoods (198, 202). With either cells or extracellular filtrate, the performances on the two substrates were similar, indicating that insoluble lignin does not interfere with growth or hydrolysis. The pretreated hardwood supports growth not only in batch culture but also in continuous culture. A new procedure, an enzyme-linked immunosorbent assay, is now available for quantitation of cell and cellulose mass concentrations during batch fermentations of C. thermocellum (364).
THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME
Components SL and SS
Early studies in our laboratory on purification of the cellulase complex from C. thermocellum (137) indicated that the true cellulase activity was part of a large aggregate with a molecular weight of over 1.5 × 106. Workers in Israel (16, 21, 179) purified a multisubunit complex from the culture supernatant that they named the cellulosome (18, 20, 174, 175).
The highly ordered arrangement of the cellulosome gives it stability. This resistance to environmental insults correlates with its resistance to dissociation into individual components even in the presence of urea or nonionic detergent; hence, purification of individual proteins was extremely difficult (174, 175). However, we accomplished a breakthrough in the purification of this complex aggregate of cellulolytic proteins (353, 354). Using Avicel breakdown as a turbidimetric assay for true cellulase activity and CMC hydrolysis as an assay for endoglucanases, we found the aggregate to contain at least four endoglucanases of different molecular weights accompanying true cellulase activity. We dissociated the aggregate by mild sodium dodecyl sulfate (SDS) treatment plus EDTA and DTT, but the resulting individual fractions exhibited only endoglucanase activity, the true cellulase activity being lost. However, we were able to reconstitute true cellulase activity by combining two of the major components, which we called SS (Mr = 82,000) and SL (Mr = 250,000). SS and SL are more abundant than any other cellulosomal components (179, 351). They were purified by gel filtration chromatography and by elution from an SDS-polyacrylamide gel, respectively. The reconstituted true cellulase activity yielded cellobiose as the predominant product of hydrolysis, was inhibited by cellobiose, required Ca2+ and reducing conditions, and thus behaved like the crude cell-free supernatant. In 1984, we proposed that an exoglucanase was the component subject to oxidation (139). SS, an exoglucanase, appeared to be that component responsible for cellobiose inhibition, calcium dependency, and oxidation sensitivity of the true cellulase activity (169, 170, 231, 234). These results indicate that SS plays an important role in the cellulolytic activity of the cellulosome and that it may be the rate-limiting cellulosomal component. SL is glycosylated, a rare modification for a bacterial extracellular protein. As many as 13 cellulosomal proteins may be glycosylated (164, 236, 249), but SL has the major part of the sugar, with about 40% of this component being carbohydrate (97, 99). The oligosaccharides that have been characterized are O-glycosidically attached via galactopyranose to threonine residues of SL (98). These threonine residues are in the Thr/Pro-rich regions which link the cellulose-binding domain (CBD) to the enzyme receptor regions (i.e., cohesins; see below) of SL. The major carbohydrate has (i) a basic tetrasaccharide structure containing two galactose units, one galactitol unit, and one 3-O-methyl-N-acetylglucosamine unit and (ii) a disaaccharide structure containing d-galactose.
SS alone acted on CMC, but SL alone had little to no enzymatic activity (179, 354). The enzymatic activity of SS on CMC was not enhanced by SL, but its adsorption to crystalline cellulose was (354). We hypothesized that the cooperative degradation of crystalline cellulose involves an interaction between SS (and presumably other cellulases), SL, and the insoluble substrate. SL (an anchorage subunit) would function to bind SS (and other catalytic proteins of the complex) to the cellulose surface in a manner optimal for hydrolysis (351, 353). As discussed below, the DNA sequence of the SL gene (cipA) reveals that SL indeed contains a CBD and multiple enzyme receptor domains, consistent with the anchor-enzyme hypothesis. The anchor-enzyme model has been further confirmed by using recombinant forms of SS and SL (168). The anchorage function of SL is the basis of our current understanding of the cellulosome structure (see next section).
We and others devoted considerable efforts to the isolation and purification of a number of proteins of the C. thermocellum system as well as the cloning, expression, and sequencing of their relevant genes in Escherichia coli and other hosts (80, 96, 148, 163, 276, 277). Of particular interest was our gene sequencing of SL (96, 277) and the gene sequencing of SS (341).
Protein SL, which is equivalent to component S1 described by Lamed et al. (179), is now called the cellulosome-integrating protein (CipA), the scaffolding protein, or scaffoldin (18). It contains approximately 1,850 amino acid residues and is the most important protein of the cellulosome. In addition to its function of binding other members of the cellulase complex to itself, it also binds to cellulose (33). Our first cloning and sequencing of cipA involved a truncated 5′ region (277). Later, the remainder of the gene was cloned by chromosome walking and the entire sequence was determined (96). Its nucleotide sequence revealed a deduced protein size of 196,800 Da, a CBD, and nine domains of about 150 to 166 amino acid residues each. The CBD is of type 3 (28, 96). The nine repeated sequences, called cohesins by Bayer et al. (18), are quite similar to each other, i.e., exhibiting between 60 and 100% identity, with six of the nine domains being at least 90% identical. They are the receptors that bind the individual cellulases, xylanases, and other enzymes to CipA (88, 96, 327) (Fig. 2).
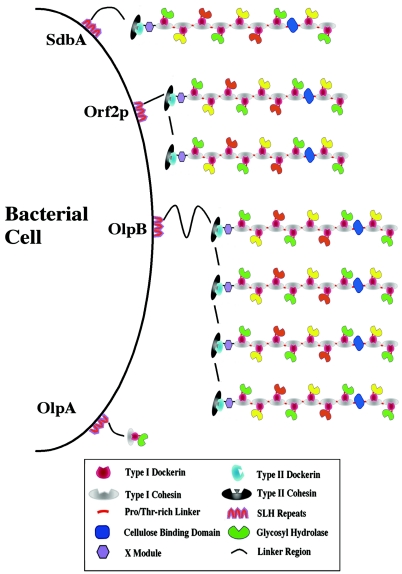
FIG. 2.
A schematic diagram of the C. thermocellum cellulosome. The type I dockerins mediate attachment of the catalytic subunits to the scaffoldin, which is comprised of nine cohesins, a CBD, a hydrophilic domain of unknown function (X) and a type II dockerin. The scaffoldin likewise binds through its type II dockerin domain to a type II cohesin-containing protein on the bacterial cell surface that is thought to anchor the complex through a series of three SLH domains (see Fig. 3 for details). Reproduced from reference 337 with permission of John Wiley & Sons.
Protein SS, also called CelS (343), is the major catalytic subunit of the cellulosome and is equivalent to component S8 described by the Israeli group (231). It is an exoglucanase (168-170, 231, 234). Sequencing of celS revealed an open reading frame of 2,241 bp encoding 741 amino acid residues with a predicted molecular weight of 80,670 (341, 343). The sequence revealed that CelS belonged to a new cellulase family (341). It was later classified as a member of family 48 glycosyl hydrolases. Although it is the most abundant catalytic subunit of the cellulosome, its low or complete lack of activities on CMC and other synthetic substrates explains why it had been difficult to clone its gene. In fact, relatively few enzymes in this family have been identified since we reported the celS sequence. Family 48 enzymes are found mostly in bacterial cellulase systems (P. M. Coutinho and B. Henrissat, 1999, Carbohydrate-Active Enzymes server at http://afmb.cnrs-mrs.fr/CAZY/index.html). They are now considered a key component in the bacterial scheme for breaking down cellulosic materials (133). A recent study on Ruminococcus albus mutants that are defective in cellulose degradation found that all three mutants failed to produce family 48 and family 9 enzymes (67), again indicating that the family 48 enzymes are crucial for bacterial cellulase systems. CelS is the only family 48 enzyme found in the C. thermocellum genome (http://genome.jgi-psf.org/draft_microbes/cloth/cloth.home.html).
CelS contains a duplicated 24-amino-acid-residue dockerin, the site of binding to scaffoldin. The recombinant enzyme produced in E. coli behaves like an exoglucanase, hydrolyzing phosphoric acid-swollen cellulose faster than Avicel and hydrolyzing Avicel more rapidly than CMC (169, 342). Like the cellulosome itself, recombinant CelS is inhibited by cellobiose and only marginally so by glucose (167). Inhibition by cellobiose was found to be competitive.
In collaboration with Alzari's group in the Pasteur Institute, we determined the crystal structure of CelS (111). The overall structure resembles that of C. cellulolyticum CelF (262, 263, 271). The protein folds into an (α/α)6 barrel with a tunnel-shaped substrate-binding region. The most salient feature of the CelS structure is that its tunnel-shaped substrate-binding site, which is capable of binding seven glucose moieties, is adjacent to an open cleft that accommodates two glucose moieties. It is proposed that the cellulose chain threads through the tunnel and that the glycosidic bond between the second and third glucose residues is hydrolyzed to produce cellobiose as the product. Upon the release of cellobiose from the open cleft, the cellulose chain slides forward through the tunnel with a distance of two glucose units. This “processivity,” consisting of alternating steps of hydrolysis and sliding-threading, explains the cellobiohydrolase nature of CelS. Structural comparisons with other (α/α)6 barrel glycosidases indicate that CelS and endoglucanase CelA, a family 8 glycosidase whose sequence is unrelated to that of CelS and which has a groove-shaped substrate-binding region, use the same catalytic machinery to hydrolyze the glycosidic linkage, despite a low sequence similarity and a different endo-exo mode of action. CelS and CelA can therefore be classified in a new clan of glycoside hydrolases. For the substrate-binding site, CelA has an open cleft while CelS has a closed tunnel, explaining their different endo-exo modes of action, as also observed in other endo-exo pairs (see reference 324 for a review).
Thus, an association is formed by a synergistic cassette of catalytic proteins, which is optimal for hydrolysis of insoluble polymers to the level of soluble oligosaccharides. Synergism between two cloned C. thermocellum endoglucanases and one cloned exoglucanase has been observed in vitro (333). The proximity of these synergistic enzymes to their cellulosic substrate as mediated by the scaffolding protein CipA may provide the structural basis for the high specific activity of the cellulosome.
Cellulosome Structure
Cellulosomes are crucially important for the efficient breakdown and utilization of crystalline cellulose (22, 23, 29, 73, 174, 175, 293, 305). The cellulosome is a macromolecular machine (multienzyme complex) which, like a ribosome, is dedicated to organized, concerted, synergistic, and efficient catalysis of cellular activities (22). Cellulosomes are unique in that no other extracellular protein complexes with the size and complexity of the cellulosomes have been reported. They have molecular weights of 2 × 106 to 6 × 106, have diameters of about 18 nm, and contain 14 to 50 polypeptides ranging in size from 37 to 210 kDa (164, 174, 175, 215). Over 95% of the endoglucanase activity of C. thermocellum is associated with the cellulosome (37). Cellulosomes contain 5 to 7% carbohydrate (99).
Researchers at the University of Georgia (81, 131) found even larger aggregates, of ca. 108 × 106 Da (polycellulosomes). Such protuberances covering the surface of the cell are packed with (poly)cellulosomes; each protuberance seems to contain several hundred cellulosomes (176). A mutant which did not bind cellulose was found to lack cellulosomes and protuberances (21). When cells are grown on cellobiose, cellulosome complexes are packed into discrete exocellular structures. When grown on cellulose, these polycellulosome-containing organelles (protuberances with diameters of 60 to 200 nm) undergo extensive structural modification (17). After attachment to the insoluble substrate, the protuberances rapidly aggregate into “contact corridors” that physically mediate between the cellulosome, which is attached to the cellulose, and the bacterial cell surface. Protuberances are not produced when the organism is grown under cellulase-repressing conditions (178). The proteins of the cellulosome are arranged in a highly ordered chain-like array (215). The cellulose-bound cellulosome clusters appear to be the sites of active cellulolysis, and the products may be channeled down the fibrous structures to the cell. Cellulosomes also contain lipids with a high concentration of unsaturated fatty acids (44). The lipid material is thought to be localized mainly at the contact point between cellulosomes and crystalline cellulose. Both the cellulosomes described by the Israeli group (177) and the polycellulosomes described by the Georgia group display the same requirement for reducing agents and Ca2+ that we had found for true cellulase activity of the C thermocellum culture supernatant (139).
The most important component of the cellulosome is the nonenzymatic scaffoldin (96, 233). It is a unique scaffolding protein subunit, which assembles cellulases and related enzyme subunits (Fig. 2). The catalytic subunits, on the other hand, contain different modules (dockerins) which are responsible for their attachment to the scaffold (96, 327). Important in this relationship are (i) cohesin domains on scaffoldin, (ii) dockerin domains on the enzymes, and (iii) a CBD on the scaffoldin, binding the complex to cellulose. Cellulosomes and scaffoldin have been found in many bacteria, such as Clostridium cellulovorans (306), Clostridium cellulolyticum (258), Clostridium josui (149), Clostridium acetobutylicum (254, 279), Acetovibrio cellulolyticus (360), Bacteroides cellulosolvens, R. albus, Ruminococcus flavefaciens (273), Vibrio sp., and the anaerobic fungal genera Neocallimastix, Piromyces, and Orpinomyces (305). At least eight scaffoldin genes from cellulolytic bacteria have been sequenced (74). These include cipA from C. thermocellum, cbpA from C. cellulovorans (306), cipC from C. cellulolyticum (258), cipA from C. josui (149), cipA from C. acetobutylicum (254, 280), scaB from R. flavefaciens (71), cipBc from B. cellulosolvens (70), and cipV from A. cellulolyticus (69). The last two named organisms are closely related to the clostridia (191). Quite similar to the C. thermocellum cellulosomal complex are those of the mesophilic anaerobes C. cellulovorans (72) and C. cellulolyticum (90). In C. cellulovorans, the cellulosome contains a large, nonenzymatic scaffoldin, called CbpA, which has a signal peptide, a CBD, a hydrophilic domain (HLD) present four times, and a hydrophobic domain present nine times. The hydrophobic domains are the cohesins of this species. Although C. acetobutylicum is not known to degrade cellulose, the genome sequence reveals the presence of a large cellulosome gene cluster (254). This cluster contains the genes encoding the scaffolding protein CipA, the processive endocellulase Cel48A, several endoglucanases of families 5 and 9, the mannanase Man5G, and a hydrophobic protein, OrfXp. The genetic organization of this large cluster is very similar to that of C. cellulolyticum. An inactive cellulosome with an apparent molecular mass of 665 kDa has been subsequently reported (279). Recently, the entire scaffoldin (CipA) of this bacterium has been cloned and successfully expressed in E. coli (280). Chimeric miniscaffoldins consisting of domains derived from C. cellulolyticum and C. thermocellum have been expressed in C. acetobutylicum (266). In C. papyrosolvens, a mesophilic anaerobe, the cellulase system (53) can be fractionated by ion-exchange chromatography into seven high-molecular-weight multiprotein complexes, with the molecular weights ranging from 500,000 to 600,000. Each complex has a different ultrastructure (269) and a unique profile of enzymatic activities (270). The common protein appearing in each fraction is a glycoprotein (Mr = 125,000) that lacks any enzymatic activity. Whether this protein serves as the scaffoldin of the complexes and how these complexes are assembled remain to be investigated.
A number of studies on the dissociation of the cellulosome have been done. We used a mild SDS-EDTA-DTT treatment (352, 354) to separate the components of the cellulosome. Morag and coworkers (235) found that cellulosomes were dissociated under nondenaturing conditions by incubation at 60°C in the presence of EDTA and crystalline cellulose. During this dissociation, scaffoldin remained tightly bound to the cellulose but the enzyme subunits were released. A mixture of the dissociated free subunits minus scaffoldin had activity equal to that of undissociated cellulosomes on soluble or acid-swollen cellulose but had only 25 to 30% of the activity on crystalline cellulose. Bhat and Bhat (39) reported that cellulosomes can be disassociated without much loss of the ability to degrade crystalline cellulose by use of 50 mM Na acetate buffer (pH 5.0) containing 10 mM DTT, 10 mM EDTA, and 0.2% SDS at 30°C for 25 min.
Cohesins and Dockerins
Two types of cohesin exist: type I cohesins bind specifically to type I dockerin domains on the catalytic subunits, and type II cohesins are on some cell surface proteins which bind the dockerin of scaffoldin to the cell wall.
Cloning and DNA sequencing showed that genes encoding at least nine cellulosomal endoglucanases, one exoglucanase (169), one xylanase, and one lichenase (367) from C. thermocellum contain a highly conserved, noncatalytic region of 50 to 60 residues which is usually found at the carboxy terminus. These duplicated sequences, now called docking sequences or dockerins (18), are not essential for catalytic activity (113) but are responsible for the binding of the respective cellulosomal enzymes, e.g., endoglucanase D (CelD) and xylanase Z (XynZ), to one or more of the nine cohesins of CipA (186). Dockerin domains consist of two very similar segments of 22 to 24 residues connected by a peptide containing 8 to 17 amino acid residues; they are over 65% identical among the different subunits of the cellulosome (27, 28, 292, 327). Two types of dockerin exist: type I (186), which anchors catalytic subunits to scaffoldin, and type II, which anchors scaffoldin and free enzymes to the cell surface. Both depend on Ca2+. All cellulosomal enzymes contain dockerin modules. Based on current understanding, if an enzyme does not contain a dockerin, it is not part of a cellulosome. For example, endoglucanase CelC does not contain dockerins, does not bind to scaffoldin, and is not cellulosomal. When the dockerin of CelD was grafted onto CelC, it gained the ability to bind to CipA (325).
The anchor-enzyme model that we proposed was verified by data obtained from recombinant proteins (168), showing that (i) recombinant CelS, via its dockerin, forms a stable complex with cohesin 3 (also known as R3) of CipA but not with the CBD of CipA and (ii) the attachment of recombinant CelS to cellulose is dependent on the presence of a protein sequence containing both cohesin 3 and CBD but not on either alone. In both of these cases, the binding of CelS was dependent on its dockerin, since removal of the dockerin eliminated binding. The binding of endoglucanase CelD to a recombinant “mini-CipA” containing a cohesin and a CBD enhanced catalytic activity, as did the binding of CelS to CipA (89). CelD is the most active endoglucanase of the C. thermocellum cellulosome (92). Scaffoldin enhanced the activity of CelD by at least 10-fold on Avicel but had no effect on the activity of a truncated CelD lacking an intact dockerin. Similarly, the activity enhancement of an endoglucanase from C. thermocellum against Avicel by the presence of scaffoldin was found to be the result of the attachment of the enzyme to a structure bearing a CBD (151). The anchorage function of the scaffoldin has also been demonstrated by using the truncated forms of CipC of C. cellulolyticum (260). In C. cellulovorans, mini-CbpA could help cellulase components degrade insoluble cellulose but not soluble cellulose (243), as would be expected from the anchor-enzyme model. In this work, it was also demonstrated that an endoglucanase initiates the attack on cellulose, followed by ExgS, which is homologous to CelS.
The CelS dockerin can bind any of the cohesins of CipA (207). In C. thermocellum, the draft genome sequence indicates that there are at least 72 dockerin-containing proteins (see “Dockerin-Containing Proteins” below) but only nine cohesins per scaffoldin molecule. These cohesins are highly homologous, and five of them have more than 90% identity. The cohesins of a strain recognize nearly all of the dockerins of the same strain. These observations indicate that, at least in individual species, binding between catalytic dockerins and scaffoldin cohesins is relatively nonspecific (32, 168, 207, 362; P. Beguin, Abstr. Pasteur Symp., p. 37-40, 1995), with the exception of one report on binding efficiency or specificity for the C. cellulovorans CbpA (261). In C. josui, the affinity of a dockerin for various cohesins can vary, and up to a 34-fold difference has been observed (136). Although cellulosomes in a particular species appear to be heterogeneous and their assembly does not appear to follow a single pattern, there is specificity between species. For example, there is no interaction between the cohesin domain of C. thermocellum and dockerins of C. cellulolyticum and vice versa (257) (see details in “Assembly of the Cellulosome” below).
Calcium is the main metal of the cellulosome, and the reaction between dockerins and cohesins requires calcium in C. thermocellum (59, 362) and C. cellulolyticum (259); dockerins bind calcium (see “Assembly of the Cellulosome” below). This is the reason that the cellulosome can be dissociated by mild conditions if EDTA is present (25, 37, 352-354). EDTA inhibits cellulosomal activity due to its ability to chelate Ca2+ (131, 137, 139). Ca2+ is tightly bound in the cellulosome once it is taken up (58). If incubated in 50 mM Tris buffer (pH 7.5), 0.1 M NaCl, and 5 mM EDTA at 37°C, the cellulosome breaks up into polypeptides and the Ca2+ is released. Bands with masses of 160, 98, 76, and 54 kDa are lost, and new bands of 150, 132, 91, 71, 57, and 46 kDa appear. The 91- and 71-kDa polypeptides represent CelS and truncated CelS, respectively. Cleavage occurs at asparagine residue 681, eliminating 60 residues from the C terminus. All catalytic subunits examined contain a similar asparagine residue which is part of their dockerin regions. Thus, the dissociated catalytic subunits may be susceptible to proteolytic degradation at this position.
In view of the calcium requirement for optimal true cellulase activity, it is of interest that endoglucanase D has three binding sites for calcium and that calcium lowers the dissociation constant (KD) for CMC and increases thermal stability (81, 147). This endoglucanase also has one possible binding site for zinc (147). Calcium also increases the thermostability of exoglucanase CelS, the most abundant enzyme of the cellulosome (170). Finally, protein folding of the dockerin is dependent on Ca2+, explaining why Ca2+ is essential for the cohesin-dockerin interaction and hence the structural stability of the cellulosome (210, 211).
Cellulose-Binding Domain
As mentioned above, the enhancement of activity of CelS, CelD, and an endoglucanase from C. thermocellum against Avicel by the presence of scaffoldin was found to be the result of the attachment of the enzymes to a structure bearing a CBD (89, 151, 168). Different CBDs appear to target different sites on crystalline cellulose (50). In comparing the C. thermocellum CipA CBD with three others (two from T. reesei and one from Cellulomonas fimi), the CipA CBD appeared to target many more sites of the cellulose molecule than did the other three.
Individual domains of CipA, obtained by protease or spontaneous degradation, bind to cellulose, to cellulosomal enzymes, or both (288). Since certain fragments as large as 200 kDa which failed to bind cellulose were obtained, it was concluded that the CBD was at one of the termini of CipA and distinct from the catalytic domain (301). The experimental observation is therefore consistent with the CipA domain structure deduced from its DNA sequence. Although CBDs are also found in some but not all catalytic subunits, the CBD on CipA binds cellulose much more tightly than CBDs on individual cellulosomal enzymes that have been studied. The C. thermocellum CipA CBD belongs to family IIIa (19, 329) as do the CBDs on all other known scaffoldins except ScaB of R. flavefaciens, which does not have a CBD. Purified C. thermocellum CipA CBD binds crystalline cellulose with a KD of 0.4 μM and a maximum binding capacity of 10 mg of CBD per g of microcrystalline cellulose (0.54 μmol/g) (230). The capacity for amorphous cellulose is 20-fold higher. The KDs of other clostridial scaffoldin family IIIa CBDs are as follows: 0.6 μM for CbpA of C. cellulovorans (102), 0.038 μM for CipA of C. acetobutylicum (280), and 0.14 μM for C. cellulolyticum (91).
The C. thermocellum CipA CBD has been cloned and overexpressed in E. coli, and its crystal structure has been determined (330). As the cohesin, the CBD assumes a nine-stranded β sandwich with a jelly-roll topology. Cellulose binding likely involves interactions between a planar strip of aromatic amino acid residues on a surface of the CBD and glucose moieties on a cellulose chain and between polar amino acids and two adjacent glucose chains of crystalline cellulose. The CBD binds a calcium ion whose function is unknown. The crystal structure of the C. cellulolyticum CipC CBD has also been determined (302). It is very similar to that of the CBD from the CipA, with minor differences. It includes a well-conserved calcium-binding site, a putative cellulose-binding surface, and a conserved shallow groove of unknown function. It is clear that the function of the scaffoldin CBD is to anchor the catalytic subunits to the substrate surface. An additional function of the CBD in modifying the cellulose surface to facilitate enzymatic hydrolysis has also been suggested (68, 260).
Each of the known scaffoldins except the R. flavefaciens ScaB has one internal or N-terminal CBD. The catalytic subunit may also have its own CBD. The CBD in a nonscaffoldin subunit may further enhance binding of the cellulosome to cellulose. However, in some family 9 cellulases, their respective catalytic domains are immediately adjacent to a family IIIc CBD, of which many aromatic amino acid residues on the planar strip thought to be crucial for binding to cellulose are not conserved (19). Examples include CelI (101, 123, 373), CelN (373), CelQ (9), CelF of C. thermocellum (19), CelG of C. cellulolyticum (90), EngH of C. cellulovorans (194, 321), CelZ of C. stercorarium (134), and cellulase E4 of Thermomonospora fusca (132). Family IIIc CBDs are considered to play a very different role from that of the family IIIa CBDs. The crystal structure of cellulase E4 revealed the novel feature of the catalytic domain and adjacent family IIIc CBD interacting with each other (285). Sakon et al. (285) first proposed that the CBD may act by binding to a single cellulose chain and feeding it into the active-site cleft of the catalytic domain and thus that it participates directly in the catalytic function of the enzyme (285, 348). Thus, family IIIc CBDs are better considered as a “cellulose-binding subsite” of the catalytic domain (19, 132). Indeed, deletion of the family IIIc CBD from these enzymes rendered the enzyme almost completely inactive (9, 90, 101, 132, 285). It is interesting that each cellulosomal catalytic subunit (CelF, CelN, CelG, and EngH) has a family IIIc CBD only but each noncellulosomal enzyme (CelI, CelZ, and E4) has an additional family IIIa CBD. These cellulosomal proteins may thus depend on the scaffoldin's family IIIa CBD for binding to cellulose. It is also interesting that all except 2 of 13 identified or putative cellulosomal cellulases containing a family 9 glycosyl hydrolase domain have a family IIIc CBD (7 enzymes) or an immunoglobulin (Ig)-like domain (4 enzymes) immediately adjacent to the catalytic domain (370). The Ig-like domain likely participates in the catalytic function, as does the family IIIc CBD. Bayer et al. (23) have classified family 9 glycosyl hydrolases into four themes of molecular architecture: (i) the theme A enzymes lack any accessory module, (ii) the theme B enzymes possess a family IIIc CBD fused to the C-terminal end of the catalytic domain, (iii) the theme C enzymes possess an N-terminal Ig-like domain, and (iv) the theme D enzymes contain both an Ig-like domain and a family 4 CBD. Recently, a new type of CBD (i.e., the family 30 CBD) was found to be N terminal to the family 9 catalytic domain of C. thermocellum CelJ, both being linked by an Ig-like domain (8). The family 30 CBD binds to cellulose and is crucial for catalytic activity. Although the involvement of the CBD in the catalytic function remains to be characterized, it is clear that CelJ belongs to a new theme of family 9 enzymes (8). The family 4 CBD is generally not essential for catalytic function, except for C. cellulolyticum CelE (94). The family 4 CBD of the C. thermocellum CelK has a binding capacity of ∼4 mmol/g of cellulose (154), similar to those of the family 3 CBDs which have been characterized. It is interesting that LicA, a noncellulosomal C. thermocellum β-1,3-glucanase, has four family 4 CBDs at its C-terminal end (86).
An interesting application of the CBD is its use in cloning cellulosomal genes in E. coli (242). The recombinant products of such genes from C. cellulovorans unfortunately are expressed as insoluble inclusion bodies. However, when the catalytic domain contained the CBD of the noncellulosomal EngD enzyme, the recombinant protein was produced in E. coli in soluble form.
Attachment of Cellulosomes to the Cell Surface
Cellulosomes are attached to the cell during early log phase, they start to become free in late exponential growth, and most are in the medium and attached to cellulose in the stationary phase (20, 64, 215). All cellulosomal proteins are excreted to the outside of the cell, possessing leader peptides which are removed during excretion (305). The complex is constructed extracellularly, probably at the cell surface. As protein folding of the dockerin is dependent on Ca2+ (210), it is possible that assembly of the cellulosome is facilitated by the presence of Ca2+ outside the cell (337).
CipA itself contains, at its COOH terminus, a dockerin, which suggested that it may self-associate or play a role in anchoring the cellulosome to the surface of the cell (28, 88, 96). Gel scanning densitometry indicated the cellulosome to contain at least two CipA components per 2.1-MDa complex (179, 180). Evidence against CipA self-association (287) was obtained by finding that the seventh cohesin of CipA did not bind to the dockerin of CipA. Indeed, the dockerin of CipA does not bind to any of the CipA cohesins (207).
Immediately downstream from cipA in the C. thermocellum genome are three open reading frames (ORFs) encoding cell surface proteins, forming a three-gene cluster on DNA. These four genes form two operons (87). All three ORFs encode C-terminal domains (three repeats of about 65 amino acid residues) responsible for the cell surface layer (S-layer) location of the encoded proteins. These are called S-layer homology (SLH) repeats, i.e., modules which are present in polypeptides associated with bacterial cell surfaces and promote a noncovalent attachment of the protein to peptidoglycan of the cell wall (214). The three genes are called olpA (olp for outer layer protein; previously known as ORF3p), olpB (previously ORF1p), and orf2p. A fourth gene, sdbA, encoding another cell surface protein, is in another part of the genome (186). The proteins encoded by these genes anchor cellulosomes or free enzymes to the cell.
The gene furthest from cipA, i.e., olpA, encodes a protein containing a cohesin sequence in its NH2-terminal region which can bind the dockerins of the catalytic subunits (88, 286). Protein OlpA has been localized to the cell surface of C. thermocellum (286). It was hypothesized that this region may anchor the cellulosome to the cell surface (87). However, there was no binding found between the dockerin of CipA and the receptor domain of OlpA (287). Thus, CipA does not appear to anchor itself to cell surface protein OlpA. OlpA has a single type I cohesin molecule binding cellulosomal enzymes via their type I dockerins. Since the OlpA receptor binds to the dockerin of CelD, it has been suggested that the catalytic proteins bind to OlpA prior to the binding to CipA of the cellulosome. Gene olpB encodes OlpB, which is also located at the cell surface (189).
Protein SdbA of C. thermocellum has a type II cohesin domain in its NH2-terminal domain which specifically binds the type II CipA dockerin domain (187); its carboxy terminus contains SLH repeats. A model of the attachment of CipA to the cell involving use of its type II dockerin binding to a cohesin domain in the SdbA cell surface protein was proposed by Beguin and Alzari (27). SdbA is anchored via its SLH domain to the S-layer of the cell, which is external to the peptidoglycan layer of the cell wall. Actually, the binding between CipA and the cell surface probably occurs between the carboxy-terminal dockerin domain of CipA and the type II cohesin domains at the amino-terminal ends of OlpB, ORF2p, and SdbA (32). SdbA has one cohesin domain, Orf2p has two, and OlpB has four cohesins, presumably binding one, two, and four molecules of scaffoldin, respectively. The binding is a type II interaction (186), which involves calcium, as does the type I interaction. Binding of CipA dockerin to SdbA as a function of Ca2+ concentration is sigmoid, corresponding to a Hill coefficient of 2, suggesting the presence of two cooperatively bound Ca2+ ions in the cohesin-dockerin complex. Western blotting of C. thermocellum subcellular fractions and electron microscopy of immunocytochemically labeled cells indicated that SdbA is not only a cell surface protein but also a cellulosome component (187). It has been determined that OlpB binds to the C. thermocellum cell wall with a dissociation constant on the order of 10−7 M (56).
Figure 3 summarizes our current understanding of how the cellulosome is attached to the cell surface in C. thermocellum. Some noncellulosomal enzymes, may bind directly to the cell surface, e.g., xylanase XynX. It contains no dockerin domain but does have SLH segments (32). Similarly, LicA contains three SLH domains at its N-terminal end that have been shown to mediate its association with cells (86). Many scaffoldins, including that of C. cellulovorans (CbpA), do not have a dockerin. It has been postulated that the four HLDs of CbpA, each having partial homology with the SLH domain, play a role in binding the cellulosome to the cell surface (320). Furthermore, EngE, one of the three major subunits of the C. cellulovorans cellulosome, has three SLH domains at the N-terminal half, a family 5 glycosyl hydrolase domain, and a dockerin at the C terminus (320). It has been shown that EngE bridges the cellulosome and the cell by binding to the cellulosome via its dockerin and to the cell surface via its SLH domains (166). Thus, a cellulosomal enzyme may serve as an anchor for the cellulosome on the cell surface. Most scaffoldins carry HLDs with copy numbers ranging from one to six (74). The HLDs are sometimes called the X domains (19). The solution structure of one of such domain, the X2-1 domain of C. cellulolyticum, has been determined (238). It has an immunoglobulin-like fold with two β-sheets packed against each other.
FIG. 3.
Schematic drawing depicting cellulosome attachment to the cell surface through the type II dockerin-cohesin interaction with SLH domain-containing cell surface proteins, SdbA, Orf2p, and OlpB (22). On the other hand, OlpA, which contains a type I cohesin, is presumed to anchor an enzyme or protein containing a type I dockerin. Adapted from reference 337 with permission of John Wiley & Sons.
Assembly of the Cellulosome
The three-dimensional structures of two type I cohesin domains from C. thermocellum and one from C. cellulolyticum have been solved by X-ray crystallography (303, 313, 323). Cohesin 2 of C. thermocellum CipA forms a nine-stranded β-sandwich with an overall jelly-roll topology, similar to that of a bacterial cellulose-binding domain (303, 330). The structure of cohesin 7 of CipA is similar to that of cohesin 2, although minor differences were observed. These differences may be due to amino acid sequence deviations, since the cohesin domains at both ends of CipA (in particular, cohesins 1, 2, and 9) are less conserved than the central domains, which have >95% identity with one another (136, 207, 323). Despite low sequence identity (32%), the C. cellulolyticum cohesin domain (cohesin 1 of CipC) has a fold similar to those of the C. thermocellum cohesins. However, the putative dockerin binding surface is notably much less polar. This difference may explain the cross-species specificity of cohesin in recognizing the cognate dockerin (313). Structures of these three cohesins suggest that all cohesins may be similar in their overall folding.
The type I cohesin-dockerin interaction is extremely strong, with dissociation constants on the order of 10−10 M in the presence of calcium (82, 136, 219, 290). This high affinity explains the stability of the quaternary structure of the cellulosome in the extracellular environment. Our study (208) and that of Fierobe and coworkers (82) showed that both duplicated segments of the dockerin domain are required for binding to cohesin. As mentioned, it has also been demonstrated that within a given species, the interaction between the various dockerins and cohesins is nonselective (207, 362). However, the interaction displays species specificity between dockerins and cohesins from C. thermocellum and the mesophile C. cellulolyticum (257). Based on sequence comparisons of dockerins from these two species, two residues in each duplicated segment, i.e., positions 10 and 11 of each calcium-binding loop, were pinpointed as being likely recognition determinants in the binding of dockerin to cohesin (257). This prediction has been corroborated by experiments in which the species specificity of the interaction was altered through site-directed mutagenesis of these residues (219, 220). However, the results also indicated that additional residues are involved in the interaction, since binding between the mutated dockerins and the complementary cohesins from the same species was not disrupted.
As a further step toward understanding the cohesin-dockerin interaction, we undertook a solution nuclear magnetic resonance (NMR) study to determine the structure of the dockerin domain from the C. thermocellum CelS. To this date, CelS is the only cellulosomal protein with three-dimensional structures of both the catalytic and dockerin domains determined. Interestingly, two-dimensional 15N-1H heteronuclear single quantum correlation NMR spectroscopy revealed that Ca2+ induces folding of the dockerin into its tertiary structure (210). The calcium-dependent folding may be a mechanism that evolved to prevent folding into an active form in the cytoplasm, where the free Ca2+ concentration is only on the order of 1 mM. The Ca2+ requirement could potentially safeguard against binding of the catalytic subunits to scaffoldin prior to secretion (210). It is remarkable that the unfolded dockerin remains soluble at the concentration typically used for NMR analysis (∼1 mM). The solution structure of the folded CelS dockerin in the presence of Ca2+ has been determined by protein NMR spectroscopy (209, 337). Although the highly conserved dockerin domain is characterized by two Ca2+-binding sites with sequence similarity to the EF-hand motif, the dockerin domain assumes a completely different structure. The structure consists of two Ca2+-binding loop-helix motifs connected by a linker; the E helices entering each loop of the classical EF-hand motif are absent from the dockerin domain. This result agrees with the secondary structure prediction reported by Pages et al. (257). Each dockerin Ca2+-binding subdomain is stabilized by a cluster of buried hydrophobic side chains. Structural comparisons reveal that, in its noncomplexed state, the dockerin fold displays a dramatic departure from that of Ca2+-bound EF-hand domains and represents a novel Ca2+-binding domain.
The dockerin structure is symmetric, as expected from its duplicated primary sequence. How the symmetric dockerin docks to the nonsymmetric cohesin had been a puzzle. While experimental data indicate that the type I cohesin and dockerin form a 1:1 complex, results of site-directed mutagenesis on positions 10 and 11 of the dockerin on either segment suggest that either segment of the dockerin is capable of binding to the cohesin (290). The notion that only one of the two halves of the symmetric dockerin is used to bind to cohesin was confirmed when the cohesin-dockerin complex structure was determined by coexpressing cohesin 2 of CipA and the dockerin domain of a xylanase (Xyn 10B) in E. coli (52). While the cohesin structure remains essentially unchanged in the complex, the dockerin undergoes conformational change and ordering compared with its solution structure to display a near-perfect internal twofold symmetry. On the other hand, the classical 12-residue EF-hand coordination to two calcium ions is maintained. Significantly, the cohesin binds predominantly to the second segment of the dockerin, suggesting that the first segment of the dockerin may provide the second binding site, enabling the formation of a 1:2 complex (one dockerin to two cohesins) and thus a higher order of the cellulosome structure. On the other hand, a few amino acid residues on the first segment also participate in the complex formation, through either hydrophobic interactions or hydrogen bonding. Therefore, both segments of the dockerin participate in binding even in a 1:1 complex. The results agree with a report demonstrating that the two segments (subdomains) of the CelS dockerin are both required for interaction with a cohesin (208). Similar results were obtained for the C. cellulolyticum dockerin, the two segments of which are homologous enough to replace each other (82). The structure of the complex further revealed that protein-protein recognition is mediated by hydrophobic interactions between one face of the cohesin and the helices of the dockerin. Many of these hydrophobic amino acids of the dockerin have been predicted to be involved in protein docking, since they are solvent accessible in the dockerin solution structure (209). Isothermal titration calorimetry also showed that the cohesin-dockerin binding is mainly hydrophobic (290). Five hydrogen bonds between the two proteins are found, which are dominated by a Ser-Thr pair (positions 10 and 11 of the Ca2+-binding loop). Notably, the Ser-Thr pairs are strictly conserved in the C. thermocellum dockerins but not in the C. cellulolyticum dockerins. Other amino acid residues forming direct hydrogen bonds with the cohesin, such as Arg (or Lys) and Leu (or Ile), are also well conserved. Conservation of the amino acid residues involved in hydrophobic interactions and hydrogen bonding in both the cohesin and dockerin explains the lack of binding selectivity in cohesin-docking binding in C. thermocellum (207, 362). On the other hand, replacements of the hydrogen bonding residues (Ser, Thr, and Arg) with other residues (Ala, Ile, and Lys) in C. cellulolyticum dockerins provide a partial explanation for the cross-species specificity of the dockerin in recognizing the cognate cohesin. Bioinformatic analysis and site-specific mutagenesis have previously indicated the important roles of the dockerin's Ser-Thr pair and Arg (position 22 based on numbering of the Ca2+-binding loop) in complex formation (219, 220, 257). The cohesin amino acids previously identified to be involved in dockerin-cohesin interaction by site-directed mutagenesis (226) were found to be present in the interface of the complex (52).
The structures of the type I dockerin, cohesin, and their complex shed light on how the cellulosome is assembled and provide a blueprint for reengineering the cellulosome for biotechnological explorations. Unfortunately, to date, no three-dimensional structures of type II components have been reported. The dissociation constant for a type II cohesin-dockerin pair (i.e., the CipA dockerin and the SdbA cohesin) is on the order of 10−9 M (135). Secondary-structure assignments of the SdbA cohesin have been made by 1H, 13C, and 15N protein NMR (309). It was found that the positioning of the secondary structural elements is very similar to that observed in the type I cohesin except that an α-helix was present between β-strands 6 and 7 of SdbA, which may contribute to the type I-type II specificity. Further elucidation of the structures of type II components is essential for understanding the molecular basis of this specificity.
Although species specificity is generally observed, exceptions have been reported. For example, in C. thermocellum, the dockerin of Cel9D-Cel44A (formerly CelJ [1]) does not seem to bind to cohesins 1, 4, and 7 (136) or to cohesins 2 and 3 (362) of CipA. Its binding specificity remains to be determined. Furthermore, in C. josui, the affinities of binding of a dockerin to various cohesins can vary as much as 34-fold (136). Another exception to species specificity was reported in the same study, revealing that the Xyn11A dockerin of C. thermocellum binds to various cohesins of C. josui with high affinities (KD of ∼10−8 M) (136). Thus, binding specificity and affinity may be influenced by subtle differences in the primary or tertiary structures of the dockerin and cohesin. Further studies are needed to completely understand these subtleties.
Genes and Enzymes
Genes cloned from C. thermocellum in E. coli, S. cerevisiae, Bacillus subtilis, Bacillus stearothermophilus, and Lactobacillus plantarum include cipA and genes for over 30 endoglucanases, 4 exoglucanases (also known as cellobiohydrolases), 2 cellular β-glucosidases, 6 xylanases, 2 lichenases (β-1,3-1,4-glucanases), 1 putative xylan esterase, 2 laminarinases (β-1,3-glucanases), 1 mannanase, 1 chitinase, and 1 pectate lyase (15, 22, 27, 34, 46, 62, 63, 83, 96, 103, 104, 106, 107, 120, 124, 143, 145, 148, 160, 162, 163, 221, 222, 225, 239, 240, 267, 268, 275-278, 281, 284, 291, 292, 295-297, 299, 305, 312, 332, 341, 343, 366, 367, 369, 371).
Four cellulosomal cellobiohydrolases have been reported in C. thermocellum. They are CbhA (formerly Cbh3; component S3) (222, 308, 332, 375), CelS (S8) (343), CelK (S5) (152), and CelO (372). Genes cbhA and celK have been cloned and sequenced. Gene celK is upstream of cbhA with an intervening sequence of 524 bp (374). The two genes are highly homologous, both encoding a family 4 CBD, an Ig-like domain, a family 9 glycosyl hydrolase, and a dockerin. The only difference is that CbhA contains two fibronectin-like domains and a family 3b CBD, both N terminal to the dockerin. These two genes are likely the result of gene duplication and recombination. CelO consists of a family 3b CBD, a family 5 glycosyl hydrolysis domain, and a dockerin. It produces cellobiose as the only product from cellulose. The crystal structures of CelS in complex with its substrate or inhibitor indicate that CelS hydrolyzes the cellulose chain from the reducing end (111). CelO activities on various substrates indicate that it also attacks the cellulose chain from the same end (372). On the other hand, the same study revealed that CbhA hydrolyzes the cellulose chain from the nonreducing end. The facts that the catalytic domain of CelK is highly homologous to that of CbhA and that CelK is active on p-nitrophenyl-β-d-cellobioside indicate that its mode of action is like that of CbpA (13). Why the bacterium needs two sets of exocellulases remains to be explained. It appears that CbhA and CelO are capable of reducing the viscosity of a CMC solution (332, 372), whereas CelS is not (170, 234), indicating that the former two enzymes have some endoglucanase activity. A recent reexamination of the mode of action of CbhA showed that it had a very high activity on CMC relative to that on ball-milled crystalline cellulose, it rapidly reduced the viscosity of CMC, and it produced 40% insoluble reducing sugars from filter paper. Thus, in these tests CbhA behaved like an endocellulase (C. McGrath and D. B. Wilson, personal communication), and it should be reclassified as such. The results illustrate the importance of using multiple criteria to evaluate an exoglucanase. CelS behaved like an exoglucanase in these same tests (170, 342). The behaviors of CelK and CelO in these tests remain to be studied. CelF, the CelS equivalent in C. cellulolyticum, also displays significant activity in reducing the viscosity of a CMC solution (271).
Some of the cloned endoglucanase genes are celA (26), celB (105), celC (298), celD (143, 326), celE (113), celF (245), celG (188), celH (361), celI (101, 123, 373), celJ (encoding component S2 of the complex) (1, 8), celM (162), celN (373), celQ (9), celT (172), and celX (113).
Six of the xylanases present in the C. thermocellum cellulosome are XynA, XynB, XynC, XynX, XynY, and XynZ (107), the genes of which have all been cloned (121, 146). Despite the large number of xylanases, the organism cannot grow on xylan or xylose.
The cellulosomal pectate lyase gene in C. cellulovorans has an ORF containing 2,742 bp and encoding a protein of 914 amino acid residues with a molecular mass of 94,458 Da. The protein contains a dockerin at its C terminus. It breaks down polygalacturonic acid into di- and trigalacturonic acids. This is the first reported pectate lyase in cellulolytic clostridia (319). The mannanase gene man26B of the C. thermocellum cellulosome has been cloned and sequenced (171).
Sequencing of celS has been described above (see “Components SL and SS”). Another important catalytic component is CelJ (2), the largest catalytic subunit, which is an endoglucanase equivalent to component S2 described by Lamed et al. (179, 180). Like CelS, it has some activity on Avicel, and it also attacks CMC, lichenan, and xylan.
In the early 1990s, we reported on the isolation of three new endoglucanases. These included one having a molecular mass of 83 kDa, an isoelectric point of 3.55, an optimum pH of 6.6 and an optimum temperature of 70°C (80). It hydrolyzed CMC and, at a higher rate, cellotetraose and cellopentaose; Avicel, cellotriose, and cellobiose were not attacked. It was originally thought to be CelS due to its similar size and its reaction with CelS antibodies; however, CelS has been found to be an exoglucanase (234). Thus, our designation of this protein as CelS was erroneous. A second new endonuclease had the following properties: an Mr of 76,000, an isoelectric point of 5.05, an optimum pH of 7.0, and an optimum temperature of 70°C (276). Both of these endoglucanases were inhibited by Hg2+ and p-chloromercuribenzoate, suggesting the existence of thiol groups essential for activity or stability. Ca2+ stimulated both endoglucanases, whereas Mg2+ stimulated the 76-kDa enzyme but inhibited the 83-kDa enzyme. NaCl inhibited the larger enzyme but not the smaller one. The substrate specificities of the two enzymes were similar. A third new endoglucanase was CelM (162). Its nucleotide sequence indicated a molecular mass of 35 kDa. The enzyme had an optimum pH of 5.5 to 6.5 and a temperature optimum of 60°C. It lacked the dockerin and thus appeared to be noncellulosomal. It attacked CMC but not Avicel or xylan. It was not affected by Ca2+, Mg2+, or NaCl. Its inhibition by p-chloromercuribenzoate suggested the presence of thiol groups; the deduced amino acid sequence indicated the presence of seven cysteine residues.
A simplified cellulosomal fraction which was more active on Avicel (8-fold) and on CMC (15-fold) than the crude extracellular protein was isolated from the cellulase complex of C. thermocellum (161); it was called the subcellulosome preparation. The isolation utilized a lectin (Jacalin) preferentially binding to galactosyl carbohydrates. Subcellulosome activity was inactivated by SH reagents and inhibited by the chelators EDTA and ο-phenanthroline. This subcellulosome was composed of six major proteins, including CipA and CelS, ranging in molecular mass from 58 to 210 kDa. These were cloned and expressed in E. coli (163). A similar subpopulation of cellulosomes was isolated by cellulose affinity chromatography followed by anion-exchange chromatography (6). Its activity was slightly higher than that of unfractionated cellulosomes and was similarly affected by calcium, DTT, and cellobiose. Of the six subunits in this subpopulation, three were known cellulosomal proteins, but the other three were not. Another subcellulosomal preparation (minicellulosome) was constructed by using CipA, exoglucanases S5 and S8 (CelS), and endoglucanase S11. Adding CipA to the other three enzymes increased Avicel hydrolysis sevenfold (39).
Dockerin-Containing Proteins
Proteins containing a dockerin domain are considered components of the cellulosome. To date, a large number of the genes encoding dockerin-containing proteins have been cloned and characterized. However, the intriguing questions concerning how many exist in a clostridial genome and the nature of these gene products remain unresolved. These questions are now partially answered, thanks to the availability of the C. thermocellum draft genome sequence. We have submitted the C. thermocellum ATCC 27405 genomic DNA to the U.S. Department of Energy Joint Genome Institute for sequencing, and a draft genome sequence is now available (http://genome.jgi-psf.org/draft_microbes/cloth/cloth.home.html). Searching of the draft genome sequence for genes encoding dockerin-containing proteins yielded at least 72 entries, with only 28 of them having been previously identified through genetic cloning (368, 370). These dockerin-containing proteins are listed in Table 2. Besides CipA, there are enzymes of various glycosyl hydrolase families, including families 5 (5 entries, including CelO, CelB, and CelG), 8 (1 entry [CelA]), 9 (14 entries, including CbhA, CelK, CelD, CelN, CelR, CelQ, CelF, and CelT), and 48 (1 entry [CelS]) and multifunctional proteins (8 entries, including CelJ, CelH, XynZ, XynY, and CelE). In addition, there are at least four xylanases (XynA, XynB, XynC, and XynD), one lichenase (LicB), one chitinase (ChiA), one mannanase (ManA), one xyloglucanase (XghA), four putative hemicellulases, five putative glycosidases, four putative carbohydrate esterases, and five putative pectinases. Also interesting is that the list contains 1 putative protease, 2 putative protease inhibitors, and 13 proteins with unknown functions (including CseP).
TABLE 2.
Dockerin-containing proteins found in the draft genome of C. thermocellum
| Protein family or putative function group | Gene producta | Putative function and/or reading frameb | Mol mass (kDa) | Reference(s) | Module structurec |
|---|---|---|---|---|---|
| Structural component | CipA (+) | Scaffoldin, Chte02002467, ZP_00312244 | 197 | 88, 96, 374 | 2(Coh1)-CBM3a-7(Coh1)-UN-Doc2 |
| Cellulase/β-glucanase | CelO | Cellobiohydrolase, Chte02003367, ZP_00311446 | 75 | 372 | CBM3b-GH5-Doc1 |
| — | Chte02003044, ZP_00311743 | 103 | GH5-CBM6-Fn3-Doc1 | ||
| CelB | Endoglucanase, Chte02000960, ZP_00313593 | 64 | 105 | GH5-Doc1 | |
| CelG (+) | Endoglucanase, Chte02001813, ZP_00312831 | 63 | 188 | GH5-Doc1 | |
| — | Chte02001497, ZP_00313148 | 58 | GH5-Doc1 | ||
| CelA (+) | Endoglucanase, Chte02000171, ZP_00314359 | 53 | 30 | GH8-Doc1 | |
| CbhA (+) | Cellobiohydrolase, Chte02001488, ZP_00313140 | 138 | 375 | CBM4-Ig-GH9-2(Fn3)-CBD3b-Doc1 | |
| CelK (+) | Cellobiohydrolase, Chte02001490, ZP_00313141 | 101 | 152, 374 | CBM4-Ig-GH9-Doc1 | |
| CelD | Endoglucanase, Chte02001777, ZP_00312895 | 72 | 143 | Ig-GH9-Doc1 | |
| — | Chte02001890, ZP_00312800 | 110 | GH9-CBM3c-CBM3b-Doc1 | ||
| — | Chte02001086, ZP_00313565 | 105 | GH9-CBM3c-CBM3b-Doc1 | ||
| CelN (+) | Endoglucanase, Chte02000571, ZP_00314035 | 82 | 373 | GH9-CBM3c-Doc1 | |
| CelR (+) | Endoglucanase, Chte02001003, ZP_00313635 | 85 | GH9-CBM3c-Doc1 | ||
| CelQ (+) | Endoglucanase, Chte02001251, ZP_00313301 | 80 | 9 | GH9-CBM3c-Doc1 | |
| CelF | endoglucanase, Chte02000967, ZP_00313600 | 82 | 245 | GH9-CBM3c-Doc1 | |
| — | Chte02001891, ZP_00312801 | 80 | GH9-CBM3c-Doc1 | ||
| — | Chte02001467, ZP_00313120 | 89 | GH9-CBM3c-Doc1 | ||
| — | Chte02002786, ZP_00311957 | 82 | GH9-CBM3c-Doc1 | ||
| — | Chte02000166, ZP_00314354 | 63 | GH9-Doc1 | ||
| CelT (+) | Endoglucanase, Chte02001327, ZP_00313235 | 69 | 172 | GH9-Doc1 | |
| CelS (+) | Exoglucanase, Chte02001227, ZP_00313420 | 83 | 341 | GH48-Doc1 | |
| Xylanases | XynD (+) | Xylanase, Chte02000364, ZP_00314122 | 72 | CBM22-GH10-Doc1 | |
| XynC (+) | Xylanase, Chte02001156, ZP_00313489 | 70 | 120 | CBM22-GH10-Doc1 | |
| XynA, XynU (+) | Xylanase, Chte02001934, ZP_00312745 | 74 | 121 | GH11-CBM4-Doc1-NodB | |
| XynB, XynV (+) | Xylanase connecting error? | 50 | 121 | GH11-CBM4-Doc1 | |
| Other hemicellulases | LicB (+) | Lichenase, Chte02000203, ZP_00314391 | 38 | 367 | GH16-Doc1 |
| ChiA (+) | Chitinase, Chte02000170, ZP_00314358 | 55 | 366 | GH18-Doc1 | |
| ManA (+) | Mannanase, Chte02001325, ZP_00313234 | 67 | 114 | CBM-GH26-Doc1 | |
| — | Chte02000583, ZP_00314046 | 67 | GH26-Doc1 | ||
| — | Chte02002707, ZP_00312042 | 71 | GH30-CBM6-Doc1 | ||
| — | Chte02002259, ZP_00312443 | 47 | GH53-Doc1 | ||
| — | Chte02002200, ZP_00312458 | 86 | GH81-Doc1 | ||
| Putative glycosidases | — | Chte02003048, ZP_00311747 | 104 | GH2-CBM6-Doc1 | |
| — | Chte02003396, ZP_00311430 | 88 | GH39-2(CBM6)-Doc1 | ||
| — | Chte02003047, ZP_00311746 | 59 | GH43-CBM6-Doc1 | ||
| — | Chte02002202, ZP_00312459 | 64 | GH43-CBM13-Doc1 | ||
| — | Chte02000090, ZP_00314291 | 75 | GH43-2(CBM6)-Doc1 | ||
| Xyloglucan hydrolase | XghA (+) | Xyloglucanase, Chte02002261, ZP_00312445 | 92 | GH74-CBM2-Doc1 | |
| Putative carbohydrate esterases | — | Chte02001642, ZP_00312970 | 91 | Fn3-CE12-Doc1-CBM6-CE12 | |
| — | Chte02001749, ZP_00312868 | 55 | CE3-CE3-Doc1 | ||
| — | Chte02001822, ZP_00312838 | 55 | Doc1-CE6 | ||
| /PICK> | |||||
| — | Chte02003045, ZP_00311744 | 54 | CE1-CBM6-Doc1 | ||
| Putative pectinases | — | Chte02001171, ZP_00313369 | 92 | GH28-Doc1 | |
| — | Chte02002538, ZP_00312205 | 60 | PL1-Doc1-CBM6 | ||
| — | Chte02000923, ZP_00313704 | 98 | PL1-Doc1-CBM6-PL9 | ||
| — | Chte02002537, ZP_00312204 | 42 | PL10-UN-Doc1 | ||
| — | Chte02002767, ZP_00311987 | 89 | Doc1-CBM6-PL11 | ||
| Multifunctional components | CelJ (+) | Cellulase, Chte02001252, ZP_00313302 | 178 | 1 | CBM30-Ig-GH9-GH44-Doc1-UN |
| CelH | Endoglucanase, Chte02003109, ZP_00311690 | 102 | 361 | GH26-GH5-CBM9-Doc1 | |
| — | Chte02003393, ZP_00311428 | 111 | GH30-GH54-GH43-Doc1 | ||
| — | Chte02001578, ZP_00313008; Chte02001577, ZP_00313007, frame shift? | 79 | GH54-Doc1-GH43 | ||
| — | Chte02003394, ZP_00311429 | 66 | GH54-GH43-Doc1 | ||
| ZynZ (+) | Xylanase, Chte02002412, ZP_00312306 | 92 | 106 | CE1-CBM6-Doc1-GH10 | |
| XynY | Xylanase, Chte02002344, ZP_00312390 | 120 | 83 | CBM22-GH10-CBM22-Doc1-CE1 | |
| CelE (+) | Endoglucanase, Chte02001748, ZP_00312867 | 90 | 122 | GH5-Doc1-CE2 | |
| Putative protease and protease inhibitors | — | Chte02001648, ZP_00312975 | 40 | Subtilisin-like serine protease-Doc1 | |
| — | Chte02000228, ZP_00314411 | 64 | Fn3-Doc1-serpin | ||
| — | Chte02000229, ZP_00314412 | 68 | Doc1-serpin | ||
| Components with unknown function | — | Chte02002760, ZP_00311980 | 117 | 2(UN)-UN-UN(CelP 550-870)-Doc1 | |
| — | Chte02003046, ZP_00311745 | 105 | UN-CBM6-Doc1 | ||
| CseP (+) | Chte02000570, ZP_00314034 (structural component) | 62 | 373 | UN-Doc1 | |
| — | Chte02002500, ZP_00312225 | 58 | Doc1-UN | ||
| — | Chte02000182, ZP_00314370 | 51 | Doc1-U | ||
| — | Chte02001566, ZP_00313102 | 76 | Doc1-UN | ||
| — | Chte02002663, ZP_00312096 | 65 | 2(UN)-Doc1 | ||
| — | Chte02003357, ZP_00311454 | 135 | UN-UN-UN-Doc1 | ||
| Chte02001465, ZP_00313118 | 40 | Doc1-UN | |||
| Chte02001653, ZP_00312979 | 47 | UN-Doc1 | |||
| Chte02000318, ZP_00314080 | 37 | UN-Doc1 | |||
| Chte02001119, ZP_00313455 | 236 | 2(UN)-UN-Doc2 | |||
| Chte02002121, ZP_00312553 | 19 | Doc1-Doc1 |
The ORFs in the draft genome sequence, each containing at least a dockerin molecule, are listed. A gene designation indicates that the gene encoding the component has been cloned and the encoded protein has been biochemically characterized; a dash indicates that this is not the case. +, the existence of the component in the cellulosome has been experimentally verified.
The enzymatic activity or function (putative or known) and the ORF number (Chte) from REFSEQ accession number AABG00000000 on the NCBI website (http://www.ncbi.nlm.nih.gov/RefSeq/; database as of 17 June 2004) are provided. The Chte number indicates the locus tag in the genome sequence (nucleotide search RefSeq_DNA); the ZP number indicates the protein identification code (protein search RefSeq_Prot).
Module classification according to P. M. Coutinho and B. Henrissat (Carbohydrate- Active Enzymes server at http://afmb.cnrs-mrs.fr/CAZY/index.html, 1999). Coh, cohesion; Doc, dockerin; CBM, carbohydrate-binding module; GH, glycosyl hydrolase family; Fn3, fibronectin III; Ig, Ig-like fold; CE, carbohydrate esterase; PL, pectin lyase; UN, module with unknown function; serpin, serine protease inhibitor homologous module. Adapted from reference 368 with permission of Wiley-VCH Verlag.
It is thus clear that in C. thermocellum alone, there are a surprisingly large number of proteins which can potentially be “inserted” into the cellulosome through their dockerin domains. The cellulosome may therefore be a versatile extracellular organelle whose functions can be tailored by incorporating different dockerin-containing subunits. The roles of the cellulosome in clostridial physiology must be complicated, and a clear picture awaits further studies. Inevitably, these studies will have to reveal how the organism coordinates the expression of these dockerin-containing proteins in concert with the expression of noncellulosomal proteins. As described below, we know only very little about the regulatory processes at this time.
CARBON SOURCE NUTRITION
C. thermocellum has been reported to utilize a number of different sugars, but quite a bit of disagreement is seen in various reports. McBee (217) reported growth of strain 651 with cellulose, hemicellulose, cellobiose, and xylose but not with glucose, fructose, sorbitol, or mannitol. Growth on xylose by strain ATCC 27405 (the designation given by the American Type Culture Collection to the 651 strain) was not confirmed by us (304). We also did not observe growth on xylan, a component of hemicellulose (93). Patni and Alexander (264, 265) found C. thermocellum 651 to grow on mannitol, glucose, xylose, and fructose. Ng et al. (248) obtained no growth of strain LQ8 on glucose and xylose but later obtained growth on glucose of strain LQ8Rl, derived from LQ8 (181). Strain YS has been reported to grow only on cellulose or cellobiose (76). C. thermocellum I-1-B grows on esculin in addition to cellulose, cellobiose, glucose, and fructose (289). Recently, we found that C. thermocellum ATCC 27405 grows well on laminaribiose, a β-1,3-linked glucose dimer, with a growth rate and extent comparable to those obtained on cellobiose (M. Newcomb and J. H. D. Wu, unpublished data). The growth of a number of strains on glucose in complex medium after a lag of 30 h has been reported (125, 126, 212, 346). We later obtained growth of strain ATCC 27405 on glucose, fructose, and sorbitol (but not on mannitol) in chemically defined medium, but only after an exceptionally long lag of over 100 h (138). With cellobiose as the sole carbon source, C. thermocellum achieved its maximum cell density within 20 to 24 h. When a cellobiose-grown inoculum was transferred to chemically defined medium with either glucose, fructose, or sorbitol as the sole carbon source, there followed a long lag of over 100 h. At the end of this time period, growth occurred on these three carbon sources, and subsequent transfers into medium with the same carbon source allowed growth to occur within 24 h. Freier et al. (85) reported growth of strain JW20 on cellulose, cellobiose, and xylooligomers and, after long-term adaptation, on glucose, fructose, and xylose. We thus felt that the use of carbon sources other than cellulose and cellobiose often depends on the presence of high levels of yeast extract and/or requires long periods of adaptation.
One possible explanation considered for the long lag period was physiological adaptation; i.e., the pathways for the utilization of glucose, fructose, and sorbitol could require induction or deregulation, and thus the cells would not grow immediately on these carbon sources because they could not internalize and utilize these carbon sources immediately. To understand the mode of sugar utilization in this organism, we felt it essential to study the differences observed when cellobiose-grown cells were fed cellobiose, glucose, and fructose separately and to compare these results with those for glucose-grown cells fed glucose and fructose-grown cells fed fructose (253). One of the key factors related to growth is the energy status of the cell. In anaerobic bacteria, the proton motive force is generated through the activity of the proton-translocating ATPase, which hydrolyzes ATP (generated via glycolysis) and causes the extrusion of protons. According to the chemiosmotic theory (229), the transmembrane electrical and chemical potentials generated in this manner create a driving force to perform work, and a number of cellular processes can be driven in this manner (119). The three major components related to the energetics of a cell are (i) the chemical potential (ΔpH), (ii) the electrical potential (ΔY) (which together constitute the proton motive force, Δp), and (iii) ATP, which is the driving force, especially in anaerobic bacteria. Cell bioenergetics have been linked to different phenomena, including product formation (43, 45, 241), chemotaxis (156, 316), and the growth status of the cell (150). Although there were a few reports on the metabolism of sugars by C. thermocellum, aspects such as the rate of utilization of sugars, formation of sugar phosphates, modes of sugar phosphorylation, and bioenergetics had not been examined in any detail.
We carried out a study of the bioenergetics of C. thermocellum; the mode by which cellobiose, glucose, and fructose were metabolized; the mechanism of the lag observed when a cellobiose-grown inoculum was transferred to a medium with glucose or fructose as the sole carbon source; and correlations between energy-related parameters and cellulase production (253). Long lags of 180 to 200 h were observed, followed by rapid growth on glucose or fructose. During the lag on fructose, true cellulase production increased markedly, but during the lag on glucose, production was poor. These observations were similar to those previously reported by Johnson et al. (138). When cells began to grow on fructose, cellulase levels were still elevated compared to those in cellobiose-grown cultures. The final cell densities reached with glucose and fructose were similar to that achieved on cellobiose. Repeated transfers on glucose or fructose resulted in growth of the culture within 24 h, and repeated transfers on fructose (but not glucose) resulted in maintenance of high cellulase titers. The growth rates of C. thermocellum growing on cellobiose, glucose, and fructose were comparable.
Data on the energetics of C. thermocellum, obtained with in vivo 13C-NMR measurements (251, 328), revealed that fructose was transported into cellobiose-grown cells at a higher rate than cellobiose, which in turn was transported faster than glucose. When these rates were compared to those of glucose transport in glucose-adapted cells and fructose transport in fructose-adapted cells, it became clear that the rate of sugar transport was not the cause of the long lags on glucose and fructose. The rates of sugar phosphate formation were comparable in all cases, leading to the conclusion that the initial steps in glycolysis were not limiting growth. The mode of sugar phosphorylation was found to be ATP dependent for all three sugars (251). Thus, there was no apparent correlation between the growth lag and the mode of sugar phosphorylation. ATP measurements on cellobiose-grown cells fed cellobiose, glucose, or fructose and on fructose-grown and glucose-grown cells fed fructose and glucose, respectively, revealed that in all cases there was no lack of ATP for carrying out cellular functions. In fact, the ATP levels were higher in cellobiose-grown cells fed glucose and fructose than in such cells fed cellobiose. The values of ΔY, the transmembrane electrical potential, were comparable irrespective of the carbon source fed to cellobiose-grown cells. Thus, the ΔY values did not offer an explanation for the initial lags on glucose and fructose from a cellobiose inoculum. It was obvious that the hypothesis described above, i.e., that growth lags were due to a lack of ability to take up or metabolize fructose and glucose, was incorrect. Two other hypotheses were considered next: (i) that the lag represents the time taken for a mutant cell (capable of utilizing glucose or fructose), which was present in the cellobiose-grown inoculum, to grow, and (ii) that eventual growth is due to a mutation(s) during the extended lag phase. Plating experiments with cellobiose-grown cells showed that colonies appeared suddenly at 180 to 200 h and not before. Thus, the lag was not due to mutant cells carried over from the inoculum (253). Instead, the occurrence of mutations at a high frequency (2 × 10−4) was found to be the mechanism involved. The mutations were stable in that after mutant cells were grown on fructose or glucose and put through a transfer on cellobiose, they still grew rapidly on fructose or glucose. These findings (138, 253) constituted one of the early reports of adaptive mutation (48, 112, 350).
Another interesting finding was that during the lag in glucose or fructose, the cells have a much lower ΔpH (or internal pH) than do cells actively growing in cellobiose, glucose, or fructose (250). This low pH gradient and the lack of growth is terminated by the mutation which consistently occurs. We postulated that ΔpH (or internal pH) might be the critical factor for the lag in growth. Two alternative hypotheses might explain the relationship between ΔpH and the initial lack of growth on glucose and fructose. Either (i) the breakdown and internalization of the disaccharide cellobiose contribute to the formation of the essential pH gradient or (ii) the transport of glucose and fructose via proton symport dissipates the pH gradient and effectively prevents growth until the mutation(s) somehow corrects the problem.
REGULATION OF CELLULASE PRODUCTION
Carbon Source Regulation
Growth conditions markedly affect the cellulolytic activity of C. thermocellum (85, 138, 177, 253) as well as the profile of cellulosomal components (21). The overall cellulase activity is higher when cells are grown on cellulose than when they are grown on cellobiose (21, 177, 340, 352). Although total endoglucanase activity in C. thermocellum is constitutive and not subject to carbon source repression (93, 115, 304), transcription of endoglucanase genes celA, celD, and celF appears to be controlled temporally (227). Transcripts were not detected until the late exponential growth stage, probably at the time that the cellobiose concentration had markedly decreased. We observed that C. thermocellum carefully regulates its production of true cellulase activity, with enzyme production being dependent upon the source and availability of carbon and energy (138). True cellulase activity was formed when the bacterium was grown with cellulose, cellobiose, glucose, fructose, or sorbitol, but specific titers varied more than 200-fold between the different substrates. Unexpectedly, marked derepression resulting in very high transient rates of synthesis was observed when C. thermocellum was adapting, during long-term (over 100-h) lag phases, to growth on fructose or sorbitol but not when it was adapting long term to growth on glucose. High-level enzyme production was also observed when carbon and energy availability was limited by growth on crystalline cellulose. High cellulase production on fructose was repressed rapidly by addition of cellobiose (138). Superior enzyme production on fructose and sorbitol is thought to reflect their inactivity as carbon source repressors, in contrast to the repressive activity of cellobiose and glucose (340; J. H. Wang and J. H. D. Wu, Abstr. Annu. Meet. Soc. Ind. Microbiol., abstr. p. 25, 1989). There is no evidence that a specific inducer is involved in regulation of cellulase synthesis (138), despite a claim that cellobiose is an inducer (40). At present, it appears that carbon source repression and/or growth rate (see below) is the principal mechanism of regulation of cellulase synthesis in C. thermocellum. It is worth noting, however, that laminaribiose was recently shown to serve as an inducer for genes encoding cellulases which are also active on β-1,3-glucan (246) (for details, see “Negative regulation” below).
As mentioned above, when C. thermocellum was grown on the different carbon sources, the amount of true cellulase activity varied. The true cellulase titer was highest in cells grown on fructose, intermediate in cells grown on glucose, and lowest in cells grown on cellobiose. Due to the fact that the mode of sugar phosphorylation is ATP dependent for all three sugars studied (251), no correlation could be drawn between sugar transport and the (de)regulation of cellulase. Since the intracellular ATP level was highest on fructose, lower on glucose, and lowest on cellobiose, there was a direct (but not linear) relationship between cellular ATP and true cellulase production. There was no obvious relationship between ΔpH, rates of uptake of inorganic phosphate and of sugars, and the true cellulase titer. However, Δp, the proton motive force, and, more directly, its electrical potential component, ΔY, showed a nonlinear inverse relationship with true cellulase titer (252).
Recent studies by Dror et al. (76), using batch cultures as well as continuous culture, have shown that CelS transcription is controlled by growth rate under conditions of limitation of the carbon or nitrogen source. Expression under carbon restriction was higher than that under nitrogen limitation, suggesting some effect of carbon source on control of transcription. Transcriptional activity was found to be inversely proportional to growth rate. Expression of celS was threefold higher in the mid-exponential growth phase on cellulose (growth rate of 0.23 h−1) than on cellobiose (growth rate of 0.35 h−1). These data confirmed those of Johnson et al. (138), who observed higher levels of cellulase with cells growing slowly on crystalline cellulose or during adaptation to growth on fructose or sorbitol. Dror et al. (76) also observed two major transcriptional start sites at positions −140 and −145 bp upstream of the translational start site of celS. Potential promoters showed homology to σA (σ70) and σD in B. subtilis. Earlier, the Beguin group in Paris, France, had proposed that regulation of cellulase synthesis may be accomplished, at least in part, by sigma factors directing RNA polymerase to certain promoters. They found similarities between the promoter sequences of celA and celD and promoters preferred by σA and σD in B. subtilis (35, 227).
The control of CipA and cell surface anchoring proteins OlpB and Orf2p was similarly found to be a function of growth rate (77). Thirty-eight to 66 transcripts of these genes per cell were observed under cellobiose limitation at the low growth rate of 0.04 h−1 in continuous culture, and 2 to 13 transcripts were observed at an exponential growth rate of 0.35 h−1 in batch culture on cellobiose. Under nitrogen limitation in the chemostat, transcript levels were lower, reaching 25 to 31 per cell, again indicating a contribution to regulation by carbon source repression. Transcription of the cell surface gene sdbA was not influenced by growth rate. Two transcriptional start sites were located at −81 and −50 bp upstream of cipA's translational start site. The potential promoters showed homology to σA and σL (σ54) of B. subtilis. Although transcription from the σA-like promoter was observed only under carbon limitation, transcription from the σL-like promoter occurred under all growth conditions examined.
In the mesophile C. cellulovorans, crystalline cellulose promotes assembly of cellulosomes (213). When grown with cellobiose, the cellulosomal proteins were not organized but were present free in the medium. Upon addition of crystalline cellulose to the extracellular broth, cellulosomes formed. Growth on cellulose also induces production of cellular protuberances (42), but these are not observed during growth on glucose, fructose, cellobiose, or CMC. Addition of soluble carbohydrate to cellulose-grown cells led to a rapid disappearance of the protuberances within minutes. The cellulosome of C. cellulovorans contains enzymes not only for the degradation of cellulose but also for the breakdown of xylan, lichenan, pectin, and mannan (318), like the cellulosome of C. thermocellum. The cellulosomal enzyme mannanase, encoded by manA, is repressed by cellobiose compared to acid-swollen cellulose as revealed by Western blot analysis. Studies at the protein level by zymograms and sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed that the composition of xylanases and their quantities depend on the carbon source used (165). A recent study using Northern dot blot technique to semiquantify expression of various cellulase, hemicellulase, and pectin lyase genes revealed that cellulose, xylan, or pectin, when used as the carbon source, gave rise to high-level expression of most genes (117). Moderate expression was observed with cellobiose or fructose. Low-level expression was obtained with lactose, mannose, and locust bean gum, and the lowest or no expression was observed with glucose, galactose, maltose, and sucrose. These results provide evidence of coordinate expression of cellulase and hemicellulase genes, the existence of catabolite repression, and influence of hemicellulose on cellulose degradation. On the other hand, an enzyme preparation from pectin-grown cells was found to be more effective in generating protoplasts from plant cells than preparations from glucose-, cellobiose-, xylan-, or locust bean gum-grown cells (322). Cellulosomal proteome profiles were found to be more affected by the carbon source than noncellulosomal enzymes (116). As in C. thermocellum, the promoter regions (of cbpA, engE, manA, and hbpA) harbor a conserved sequence exhibiting strong similarity to the σA consensus promoter sequences of gram-positive bacteria (118).
Cellulase Gene Clusters
Despite the fact that cellulase and xylanase genes of C. thermocellum are mostly scattered over the chromosome (110), several gene clusters have been found, suggesting the existence of operons as units of gene regulation (Fig. 4). The first two operons found are in the cipA cluster as two tandem operons, including cipA plus olpB and orfp2 plus olpA (“Attachment of cellulosomes to the cell surface” above). More recently, a noncellulosomal endoglucanase, CelI, was shown to be clustered with cellulosomal cellulase CelN and a possible cellulosomal structural, component CseP (for cellulosomal element protein) (373). cseP codes for a type I dockerin-containing protein and is in the opposite strand and likely transcribed independently from celI and celN. The recombinant CseP displayed no detectable enzymatic activities. Although the exact function of CseP is unknown, Western blot analysis revealed that the cellulosome contains a substantial amount of this protein (373). Another gene cluster consists of the genes encoding the noncellulosomal endoglucanase CelC, a LacI-like protein, the lichenan-degrading enzyme LicA, and a potential membrane protein with eight putative transmembrane regions (86). We (246) reported that this gene cluster is immediately upstream of the genes encoding ManB (171) and CelT (172). The cluster therefore has a total of six genes (246). The gene cluster is preceded by a putative transposase gene and orf2. All of these genes are on the same strand. As described below, our data indicate that the LacI-like protein regulates the expression of the gene cluster by binding to the promoter region. The fourth gene cluster consists of celA, chiA, and orfZ (366). CelA is a noncellulosomal endoglucanase. ChiA (family 18) is the only chitinase in the cellulosome. OrfZ has a domain of unknown function and a dockerin. Finally, a five-gene cluster consisting of genes encoding five dockerin-containing proteins was found in analyzing the C. thermocellum genome (M. Newcomb, T. Dabral, and J. H. D. Wu, unpublished data). Interestingly, none of these five genes have been previously cloned. The first gene product is a putative family 2 glycosyl hydrolase. The second encoded protein is a putative family 43 glycosyl hydrolase, which contains a family 4 CBD. The third encoded protein contains a domain of unknown function and a family 6 CBD. The fourth encoded protein is a putative esterase with a family 6 CBD. The last gene product is a putative family 5 cellulase with a family 4 CBD and a fibronectin-like domain. Gene clusters that presumably resulted from gene duplication include the gene pairs of celK-cbhA (374) and xynB-xynA (121). As the genome sequencing of C. thermocellum is being completed, more cellulase and hemicellulase gene clusters may be detected.
FIG. 4.
Cellulase gene clusters found in C. thermocellum. The arrow indicates the transcription direction and the approximate size of the gene. GH, glycosyl hydrolase family.
In mesophilic clostridia, large cellulase gene clusters have been found (Fig. 5). Interestingly, these gene clusters all start with a scaffoldin gene, followed by a gene encoding a family 48 enzyme (exoglucanase working from the reducing end), one to two endoglucanase genes, and another gene encoding an exoglucanase working from the nonreducing end (293). Similarities among these gene clusters indicate that these clostridia are closely related. Alternatively, the similarities may be the results of lateral gene transfer. In C. cellulovorans, the nine-gene cluster contains the genes encoding the scaffolding protein CbpA, the exoglucanase ExgS, several family 9 endoglucanases, the mannanase ManA, and the hydrophobic protein HbpA containing a surface layer homology domain and a hydrophobic (or cohesin) domain (318, 321). The sequence of the nine-gene cluster is cbpA-exgS-engH-engK-hbpA-engL-manA-engM-engN and spans about 22 kb in length. Several possible transcription terminators have been found between some of the genes. Northern hybridization revealed that the cellulosomal cbpA gene cluster is transcribed as polycistronic mRNAs of 8 and 12 kb. The 8-kb mRNA codes for CbpA and ExgS, and the 12-kb mRNA codes for CbpA, ExgS, EngH, and EngK (118). On the other hand, manA is transcribed as a monocistronic messenger.
FIG. 5.
Cellulase gene clusters found in mesophilic clostridia. The arrow indicates the transcription direction and the approximate size of the gene. The sequence of the C. josui cluster is incomplete, and additional genes of the cluster may exist.
In C. cellulolyticum, a similar gene cluster has been reported. The sequence of the 10-gene, 20-kb cluster is cipC-celF-celC-celG-celE-orfXp-celH-celJ-manK-celM. Results of Northern blot analysis revealed that celC and celG are expressed as a polycistronic transcriptional unit which possibly includes a third gene (12).
The genome sequence of C. acetobutylicum revealed at least 11 genes encoding dockerin- or cohesin-containing proteins (254). Ten of these genes form a large cluster, similar to the C. cellulovorans and C. cellulolyticum gene clusters, with the sequence cipA-celA-celB-celC-orfXp-celD-celE-celF-manG-celH (279). The existence of these sequences suggests that the bacterium is capable of producing a cellulosome-like structure, and such a structure has been identified (279), as mentioned above. However, the bacterial strain is incapable of hydrolyzing amorphous or crystalline cellulose, although it hydrolyzes CMC. In addition, most of the xylanase genes in this bacterium are located in a predicted operon (254). In C. josui, a gene cluster containing at least four genes has been reported (149). The scaffoldin gene cipA is the first member of this cluster, followed by celD, celB, and celE.
Negative Regulation
The task of elucidating the mechanisms controlling the biosynthesis of biomass-degrading enzymes in clostridia is obviously complicated by the large number of genes and proteins involved. In C. thermocellum, the long list of the cellulosomal genes is further complicated by many noncellulosomal enzyme components. The large number of genes involved necessitates the use of a genomics approach. By searching the genome, we have identified three C. thermocellum proteins, GlyR1, GlyR2, and GlyR3 (formerly CelR1, CelR2, and CelR3, respectively), that are homologous to E. coli LacI (246). Each of these putative regulatory proteins contains two major domains. A helix-turn-helix DNA-binding domain is N terminal to a sugar-binding domain. The domain structure of the GlyR proteins is thus similar to the LacI structure and suggests that they belong to the Lac I family of negative regulators. Among them, GlyR3 is encoded by a member of the celC gene cluster mentioned above, including the genes encoding CelC, GlyR3, LicA, a putative membrane protein, ManB, and CelT, respectively. The DNA-binding domain of GlyR3 shows a helix-turn-helix structure very similar to that of LacI. On the other hand, the folding of the sugar-binding domain of GlyR3, as predicted by homology modeling, is substantially different from that of its equivalent in LacI (246). The results are consistent with our expectation that GlyR3 is a DNA-binding protein regulated by a sugar other than lactose. Our experimental evidence indicates that GlyR3 binds to the celC promoter region and that such binding is specifically inhibited by laminaribiose (246; M. Newcomb and J. H. D. Wu, unpublished data). Thus, laminaribiose appears to serve as an inducer of the gene cluster by inactivating binding of GlyR3 to the promoter region. This is the first demonstration, after a long search for transcription regulators of the C. thermocellum cellulase system, that the cellulase genes can be regulated by negative control. The demonstration will undoubtedly prompt efforts to find additional transcription factors that regulate cellulase formation in clostridia. Thorough understanding of the cellulase and hemicellulase regulatory mechanisms will be crucial for deregulating their production through rational genetic manipulations.
CLOSTRIDIAL COCULTURES
In addition to our basic interest in the cellulolytic complex of C. thermocellum, we are intrigued by the possibility that the anaerobic and thermophilic clostridia will someday represent a practical means of directly converting cellulose and hemicellulose to fuel ethanol. In principle, the concept of a thermophilic ethanol fermentation is very simple, i.e., a fermentation at high temperature (60 to 70°C), eliminating or reducing the need for power-consuming cooling and aeration of large reactor vessels and facilitating the constant removal of the volatile ethanol by evaporation and distillation, thus maintaining a low ethanol concentration in the fermentor and circumventing the need for an organism that is resistant to very high ethanol concentrations; only moderately resistant cultures would be necessary. Research on the optimization of ethanol production by clostridia has focused on substrate utilization (61, 283; D. I. C. Wang and G. C. Avgerinos, Abstr. Am. Inst. Chem. Eng. Annu. Meet., 1983), substrate pretreatment (79, 205), manipulation of fermentation conditions (140, 335), mutation and screening or selection for better cellulase producers (93), ethanol-tolerant strains (10, 127, 317), and strains which do not produce acids (Wang and Avgerinos, Abstr. AIChE Annu. Mtg., 1983).
No microbial system that simultaneously synthesizes a cellulase system at the high activities required and produces ethanol at the required high selectivity (i.e., without acetate and lactate as side products) is presently known. One could conceivably convert an excellent ethanol producer, such as S. cerevisiae, into a cellulase producer, but this would be extremely difficult because of the complexity of cellulase systems; indeed, such efforts have failed in the past to yield commercially interesting cultures. On the other hand, it would be much simpler to genetically modify an excellent cellulase producer that produces ethanol, acetate, and lactate so that mainly ethanol is produced. This approach has the added benefit of addressing the more serious economic problem, that of product selectivity, rather than the less important problem (i.e., for a thermophile) of ethanol sensitivity. C. thermocellum is the organism of choice for such an effort, since it is the most thoroughly described cellulolytic ethanol-producing thermophile. Studies (198) revealed that continuous cultures of C. thermocellum grown on pretreated hardwood can achieve essentially complete hydrolysis in a 12-h residence time. Pretreatment of lignocellulose has been discussed by Grethlein (109), Grethlein and Converse (108), and Khan et al. (155).
Because C. thermocellum is capable of utilizing only the hexoses and not the pentose sugars generated from cellulose and hemicellulose, the use of mixed-culture (i.e., dual-culture) systems is of great interest. C. thermocellum has been cultivated with thermophilic, anaerobic bacteria that are capable of utilizing pentose as well as hexose sugars, i.e., Clostridium thermosaccharolyticum (282, 335, 338), Clostridium thermohydrosulfuricum (95, 247, 282), Thermoanaerobacter ethanolicus (347) and Thermoanaerobium brockii (181). One such direct process was studied at M.I.T. (11, 335, 338), and involved a mixture of C. thermocellum and C. thermosaccharolyticum (78). This combination forms closely associated, syntrophic, and very stable dual cultures (345). Cellulose is broken down by the cellulase complex of C. thermocellum to cellobiose and cellodextrins, which are then utilized by the organisms to produce ethanol; unfortunately, acetate and lactate are also formed.
C. thermocellum is unable to utilize the pentose sugars, mainly xylose and xylobiose, formed by its breakdown of hemicellulose. The other anaerobic thermophile in the dual culture, C. thermosaccharolyticum, uses the pentoses to form ethanol, acetate, and lactate. The latter organism also uses the cellobiose resulting from cellulase action faster than C. thermocellum, thus preventing buildup of this inhibitory and repressive disaccharide. Residual lignin formed from the breakdown of lignocellulosic materials could be sold for adhesive formulations, burned as fuel, or converted into steam and electricity (47).
The dual-culture process has great potential, but the problem of side products such as acetate and lactate that decrease the yield of ethanol and can act as weak uncouplers and slow cell growth (129) must also be addressed. The method of acid production is clear: acetate production involves the activity of phosphotransacetylase and acetate kinase, whereas lactate formation involves lactate dehydrogenase (Fig. 6). These enzymes should be genetically removable without disrupting growth of the organism. Physiological manipulations can also be important; growth of C. thermosaccharolyticum during transient states in a continuous-culture reactor resulted in an ethanol selectivity (moles of ethanol per mole of other products) in excess of 11 (199). The purification and characterization of acetate kinase from C. thermocellum have been described (192, 193).
FIG. 6.
Ethanol, lactate, and acetate fermentation by C. thermocellum. 1, enzymes of Embden-Meyerhof pathway; 2, lactate dehydrogenase; 3, pyruvate-ferredoxin oxidoreductase; 4, acetaldehyde dehydrogenase; 5, alcohol dehydrogenase; 6, phosphotransacetylase; 7, acetate kinase.
The other problem with the thermoanaerobes is the buildup of ethanol which can inhibit cell growth (128). The basis and extent of ethanol sensitivity in thermoethanologenic bacteria are still unclear. Proposed mechanisms include ethanol-induced changes in the cell membrane, overreduction of the pyridine nucleotide pool, and inhibition of alcohol dehydrogenase. Although clostridia have lower ethanol tolerance than yeast, certain developments indicate the difference not to be as great as generally thought. In batch culture, it had been shown that growth of C. thermocellum ATCC 27405 was 50% inhibited at 4 to 16 g of ethanol per liter (127, 289). However, ethanol-resistant strains have which are 50% inhibited only at higher levels, e.g., at 25 and 48 g per liter, have been obtained (see the table in reference 14). Another isolated strain, C. thermocellum strain I-1-B is 50% inhibited by 27g of ethanol per liter, compared to 16g per liter for ATCC 27405 (289). It produced 23.6g of ethanol per liter from 80g of cellulose per liter; other products were 8.5g of lactate per liter, 2.9g of acetate per liter, and 0.9g of formate per liter. Such an ethanol titer is high for a monoculture.
Growth of C. thermosaccharolyticum strain HG-8 in continuous culture was not inhibited by 21g of ethanol (endogenous plus exogenous) per liter and was 50% inhibited by 28 to 29 g/liter at its optimum temperature of 60°C (159). At 55°C, 36 to 40 g of ethanol per liter was required to inhibit growth by 50% (14, 159). These data indicate a much greater ethanol tolerance than previously thought (130, 199). At 30°C, 7°C below the optimum temperature for S. cerevisiae growth, the ethanol concentration causing 50% inhibition in continuous culture of yeast was about 50 g/liter (24, 100). Thus, at temperatures 5 to 7°C below those for optimum growth (lowering the temperature decreases the toxicity of ethanol), the difference in levels causing 50% growth inhibition in continuous culture between S. cerevisiae and C. thermosaccharolyticum is rather small, i.e., ca. 50 versus 40 g/liter, respectively. Of course, ethanol sensitivity becomes even less important in a thermophilic process where ethanol can continuously be removed by evaporation and distillation. True cellulase activity of C. thermocellum is rather resistant to ethanol, with 50% inhibition requiring 8% ethanol (by weight) (36).
Although strains which show decreased acid production or increased ethanol tolerance have been obtained by mutagenesis followed by screening or selection (60, 127, 173, 317, 338), genetic stability has been a problem (60, 78, 228) and further genetic work is needed. It is encouraging that a hypercellulolytic strain isolated from C. thermocellum Ym4 showed stability through five subcultures (236). Since the number of variables and mutant strains which can be examined at one time in fermentors is limited, we scaled down the M.I.T. coculture process from fermentors into flasks by using our simple chemically defined medium with Solka Floc as a carbon source (335). Prior to these studies, flask experiments worked effectively only at low substrate concentrations, e.g., 5 g of cellulose per liter; higher cellulose concentrations yielded no additional ethanol. We were able to raise the substrate concentration to 40g/liter and obtain increased ethanol production by raising the iron concentration in the medium. Ethanol titers of 12 to 16 g/liter were attained with carbon balances of 91 to 97%. The flask system can be used to attack the problems mentioned above in order to remove the bottlenecks to economic progress.
APPLICATION OF RECOMBINANT DNA TECHNOLOGY TO THE SELECTIVITY PROBLEM
A major obstacle to using a clostridial coculture for ethanol production is the branched fermentation pathway shared by members of this genus, which results in a reduced ethanol yield due to formation of other fermentation products, notably acetate and lactate (Fig. 6). One potential solution is the elimination of metabolic branches, which would result in ethanol formation being the sole means for the cell to rid itself of excess reducing equivalents. In doing so, factors which are likely to affect gene transfer in the two thermophilic organisms, C. thermocellum and C. thermosaccharolyticum, have to be investigated. Genetic manipulation with the goal of increasing ethanol yield has been hampered by the paucity of information on the molecular genetics of C. thermocellum and C. thermosaccharolyticum. One obvious approach is to knock out the genes (encoding acetate kinase and/or phosphotransacetylase and lactate dehydrogenase) that are responsible for the branched metabolic pathways. The DNA sequences of these target genes can be obtained by cloning the respective genes from C. thermocellum and C. thermosaccharolyticum into E. coli or by searching the genomic sequences, if available, for these genes. Thereafter, a knockout vector can be constructed by using a nonreplicative, temperature-sensitive or otherwise unstable plasmid containing a fragment(s) of the target gene, to be introduced into the respective organism by a method such as electroporation. Insertional inactivation (single crossover) or deletion (double crossover) of the endogenous, functional genes in the chromosome via homologous recombination would yield stable knockout strains directing the substrate mainly to ethanol.
A prerequisite for any such manipulation would be the ability to introduce foreign DNA into these bacteria. Although many genes from thermophilic clostridia have been cloned, reports on the introduction of DNA into these organisms are almost nonexistent, except for a recent breakthrough (334) (see below). In addition to one initial report of polyethylene glycol-mediated transformation of the thermophile C. thermohydrosulfuricum (synonymous with Thermoanaerobacter thermohydrosulfuricus [185]) with plasmid pUB110 (311), some progress was made in a joint Dartmouth College-M.I.T. investigation of C. thermocellum and C. thermosaccharolyticum dealing with restriction endonuclease systems (157). Restriction-modification systems are usually composed of an endonuclease that digests DNA at a specific base sequence and a methylase that protects the sequence from digestion by its action at a specific position on one or more bases in that same sequence. Restriction-modification systems can also be obstacles to transformation, as they digest incoming DNA that is not properly methylated. The action of endonucleases had been shown to be the primary barrier to achieving efficient transformation of C. acetobutylicum with DNA that is not protected by appropriate methylation (223). Accordingly, Klapatch et al. (157) first investigated endonuclease activity, the use of interspecific methylase expression to protect DNA in situations where endonuclease attack would otherwise occur, and the identity of the restriction site.
Cell extracts were used to determine whether a restriction system was operative in C. thermocellum. It was found that extracts prepared by high pressure disruption or protoplast extraction exhibited sequence-specific restriction endonuclease activity with little nonspecific exonuclease activity. Next to be examined was the question of whether an interspecific methylase could protect DNA from endonuclease cleavage. The Dam methylation system of E. coli completely protected all DNAs tested (totaling >100 kb, ensuring that most base pair combinations were exposed). Based on both the Dam recognition sequence and the similarity in action of cell extracts and MboI DNA digests, the C. thermocellum restriction enzyme recognition sequence appeared to be 5′-GATC-3′. Since plasmids prepared from Dam− E. coli strains or from B. subtilis, which does not exhibit Dam methylation, were digested by the cell extract, it was concluded that the genome of C. thermocellum exhibits a Dam+ phenotype. These results suggested that E. coli shuttle plasmids used to transform C. thermocellum should be prepared from a Dam+ E. coli host to escape digestion at this commonly occurring (on average, once every 256 bp) site. In C. thermocellum ATCC 27405, isoschizomers of BclI (TGATCA) and EcoRII (CC[A/T]GG) had been reported (363), but digestion by extracts at these sequences was not observed, despite the fact that their respective restriction sites were present in the DNA studied. The finding that C. thermocellum DNA prepared in a dam+ host is resistant to endonuclease attack has been confirmed (256).
The action of C. thermosaccharolyticum extracts against methylated and unmethylated DNA was also investigated (157). Two independently prepared cell extracts that were exposed to >100 kb of DNA did not exhibit restriction endonuclease activity at 55°C. In contrast to that of C. thermocellum, the genome of C. thermosaccharolyticum was digested by MboI, indicating that it has a Dam− phenotype. It is noteworthy that a third thermophile, C. thermohydrosulfuricum, exhibits restriction activity at GATC which is prevented by Dam methylation found in that organism (272). Nonspecific degradation of both linear and circular DNA was observed by Klapatch et al. (157) upon incubation with cell extracts from either C. thermocellum or C. thermosaccharolyticum at 63°C but not at 55 or 60°C. DNase activity had been reported previously for both organisms (184), but the effect of temperature on this activity had not been addressed.
Further work was aimed at creating a stable strain by using homologous recombination-mediated targeted mutagenesis (65). As mentioned above, one key prerequisite for carrying out this approach is the development of a transformation system to allow introduction of modified target genes into the cell for homologous recombination with the endogenous genes. A preliminary report on successful transformation of C. thermocellum had appeared (331), but the efficiency of gene transfer was not disclosed. Recently, electrotransformation of C. thermocellum was achieved by using plasmid pIKm1 with selection based on resistance to erythromycin and lincomycin (334). Transformation efficiency was optimized. Dam methylation increased the transformation efficiency by 7- or 40-fold, depending on the host strain.
C. thermosaccharolyticum was successfully transformed by electrotransformation, and the foreign erythromycin resistance character was expressed (65, 158). Cells were prepared in 20% glycerol and electroporated using a high field strength (10 kV/cm) with a time constant of approximately 16 ms. The glycerol electroporation buffer, which those authors successfully used for C. thermosaccharolyticum, had been used with Clostridium perfringens (7) to increase transformation efficiency over that obtained with phosphate-buffered sucrose. Evidence supporting the reproducible transformation with pCTC1 included a positive hybridization signal on a Southern blot of total genomic DNA from transformed C. thermosaccharolyticum when probed with pCTC1, visualization on agarose gels of pCTC1 in 100% of the tested transformed C. thermosaccharolyticum clones, correlation of resistance to erythromycin with the presence of the plasmid, transformation of E. coli with pCTC1 isolated from C. thermosaccharolyticum, reisolation of the plasmid from E. coli, and a dose response of the number of C. thermosaccharolyticum transformants with increasing amounts of DNA. A 95% survival frequency was observed during electroporation with C. thermosaccharolyticum. The average transformation frequency was low but reproducible and was similar to the initial frequencies seen with other pioneering transformation studies using various organisms. The transformation efficiency was low when the plasmid was prepared in E. coli, but it was two orders of magnitude higher when plasmid produced in C. thermosaccharolyticum was used, i.e., 52 transformants per μg of DNA. The preferred growth temperature of C. thermosaccharolyticum is 60°C, but in the experiments of Klapatch et al. (158), cells were grown at 45°C. A lower growth temperature was used because increased plasmid segregational instability of pAMb1 replicons with increasing temperature had been reported (311). Klapatch et al. (158) also obtained indirect evidence of this in the form of lowered plasmid yields of alkaline lysis maxipreparations of transformed C. thermosaccharolyticum when cells were grown at 60°C. Isolation of pCTC1 from E. coli following passage through C. thermosaccharolyticum and examination of the restriction pattern did not produce any changes from the expected pattern. Since C. thermosaccharolyticum HG8 does not exhibit restriction endonuclease activity and lacks DNA methylation activity, proper methylation of the incoming DNA does not appear to be the reason why C. thermosaccharolyticum-prepared DNA transformed 100-fold better that E. coli-prepared DNA. One possible explanation is that the topology of DNA affects gene expression (75, 339) and may affect replicon recognition. Thermophilic bacteria have been reported to contain unique topoisomerase activity, and this proper “twisting” of the DNA molecule may be reflected in increased recognition of the replicon by host cell replication factors and increased transformation efficiency (54).
The Dartmouth-M.I.T. group also attempted the isolation of the clostridial acetate kinase and phosphotransacetylase genes, ack and pta, respectively, in E. coli (65). We used strains of E. coli that lack ack, pta, or both. Such mutants grow more slowly as colonies on acetate minimal plates than does the wild-type parent (190). This constituted a convenient screening system, since the recipient mutants should grow slowly unless the correct gene has been inserted and expressed. To test the system, we transformed overexpression plasmids (pT7-7) containing ack (designated pML703) and pta (designated pML702) from the thermophile Methanosarcina thermophila (182) into the E. coli mutants and examined growth on minimal acetate plates. Transformants TA3514(pML702) and TA3515(pML703) produced colonies at 24 h. The mutants without inserts, the mutants with the “wrong” insert, and the double mutant with either insert failed to produce colonies until 72 h. Activity tests showed TA3515(pML703) to contain 7.7 U of acetate kinase per mg, Ack− TA3515 had 0.1 U/mg, and Pta− TA3514 contained 6.0 U/mg. Thus, the screening procedure was effective. In addition, we showed for the first time that an acetate kinase gene from a thermophile could complement an acetate kinase deficiency in the mesophile E. coli K-12. The deduced amino acid sequences of the ack genes from M. thermophila and E. coli show 60% similarity and 40% identity (182). This work was important since the M. thermophila genes provide an alternative path in case of failure to isolate the clostridial genes; i.e., one could inactivate the M. thermophila genes in E. coli and incorporate these deletion-bearing constructs into the clostridial chromosomes via homologous recombination by using the electrotransformation methodology described above.
Ozcengiz et al. (255) investigated three different strategies for cloning ack from C. thermocellum into E. coli: (i) heterologous complementation, (ii) heterologous probing, and (iii) PCR-based amplification of a C. thermocellum ack fragment and homologous probing. The use of a PCR-based approach was found to be best.
Recently, success has been achieved by the Lynd laboratory at Dartmouth College, where an efficient gene transfer system was developed for C. thermocellum (334). Genes ldh and ack have been individually eliminated from Thermoanaerbacterium saccharolyticum, resulting in markedly decreased production of lactate and acetate, respectively. Plans are in place for eliminating both genes in a single strain and repeating this exciting work in C. thermocellum.
The DNA-shuffling technique for evolution of improved enzymes has been applied to genes engB and engD of C. cellulovorans to increase thermostability (244).
Another exciting development is the cloning and expression of the genes encoding the EngB endoglucanase and the mini-CbpA1 scaffoldin of C. cellulovorans in B. subtilis and the isolation of minicellulosomes from the gram-positive aerobe (57). This could potentially lead to the production of “designer cellulosomes” in B. subtilis.
CLOSING COMMENTS
Today the world is being inundated with urban, agricultural, and industrial waste. Landfill space to handle this waste is becoming unavailable, and its cost is rising rapidly. A potential solution to this problem is the conversion of lignocellulosic biomass into motor fuel, i.e., ethanol, by a coculture of thermophilic, anaerobic microorganisms (i.e., a cellulolytic strain such as C. thermocellum and a saccharolytic strain such as C. thermosaccharolyticum). Together, these strains attack cellulose and hemicellulose and convert the sugars produced to ethanol.
Attention has focused on anaerobic thermophiles as “ethanologens” for the following reasons: (i) thermophiles are thought to be robust and contain stable enzymes; (ii) anaerobes generally have a low cellular growth yield, and hence more of the substrate is converted to ethanol; (iii) thermophilic fermentations are less prone to detrimental effects of contamination; and (iv) growth at higher temperatures may facilitate the removal and recovery of volatile products such as ethanol. Our experience suggests that even more important are the advantages of cellulase production in situ and the high rates of metabolism of cellulose and hemicellulose.
In addition to addressing a pollution problem, the clostridial coculture system is potentially capable of dramatically increasing the use of ethanol as a major liquid fuel with renewable photosynthetic biomass as feedstock. The major obstacle to an economic process is the production of the side products, acetate and lactate, which limits conversion yield. In principle, the concept of a thermophilic ethanol fermentation is a very simple one involving a high-temperature fermentation with a reduced need for power-consuming cooling and agitation or aeration of reactor vessels and with the four biologically mediated events involved in ethanol production (cellulase and hemicellulase formation, cellulose and hemicellulose hydrolysis, hexose fermentation, and pentose fermentation) consolidated in a single process step. A combination of recombinant DNA technology and metabolic engineering research is sure to overcome the bottleneck described above. The selectivity problem is being addressed by using a molecular genetics approach involving inactivation of the genes encoding acetate- and lactate-producing enzymes of both members of the coculture system.
Acknowledgments
We thank Lee R. Lynd and Mark Laser of Dartmouth College for their advice on ethanol as a fuel and Sara Ladd for proofreading the manuscript.
The cellulase work by J.H.D.W.'s group was supported by grants from DOE (DE-FG02-94ER20155) and USDA. We also thank Link Foundation for providing a graduate fellowship.
Footnotes
This review is dedicated to the late Marek Romaniec, who brought the light of molecular biology to our M.I.T. laboratory but whose own light went out much too soon.
REFERENCES
- 1.Ahsan, M. M., T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1996. Cloning, DNA sequencing, and expression of the gene encoding Clostridium thermocellum cellulase CelJ, the largest catalytic component of the cellulosome. J. Bacteriol. 178:5732-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahsan, M. M., M. Matsumoto, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1997. Purification and characterization of the family J catalytic domain derived from the Clostridium thermocellum endoglucanase CelJ. Biosci. Biotechnol. Biochem. 61:427-431. [DOI] [PubMed] [Google Scholar]
- 3.Ait, N., N. Creuzet, and J. Cattaneo. 1982. Properties of β-glucosidase purified from Clostridium thermocellum. J. Gen. Microbiol. 128:569-577. [Google Scholar]
- 4.Ait, N., N. Creuzet, and P. Forget. 1979. Partial purification of cellulase from Clostridium thermocellum. J. Gen. Microbiol. 113:399-402. [Google Scholar]
- 5.Alexander, J. K. 1968. Purification and specificity of cellobiose phosphorylase from Clostridium thermocellum. J. Biol. Chem. 243:2899-2904. [PubMed] [Google Scholar]
- 6.Ali, B. R., M. P. Romaniec, G. P. Hazlewood, and R. B. Freedman. 1995. Characterization of the subunits in an apparently homogeneous subpopulation of Clostridium thermocellum cellulosomes. Enzyme Microb. Technol. 17:705-711. [DOI] [PubMed] [Google Scholar]
- 7.Allen, S. P., and H. P. Blaschek. 1990. Factors involved in the electroporation-induced transformation of Clostridium perfringens. FEMS Microbiol. Lett. 58:217-220. [DOI] [PubMed] [Google Scholar]
- 8.Arai, T., R. Araki, A. Tanaka, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2003. Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J. Bacteriol. 185:504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arai, T., H. Ohara, S. Karita, S. Kimura, K. Sakka, and K. Ohmiya. 2001. Sequence of celQ and properties of CelQ, a component of the Clostridium thermocellum cellulosome. Appl. Microbiol. Biotechnol. 57:660-666. [DOI] [PubMed] [Google Scholar]
- 10.Avgerinos, G. C., H. Y. Fang, I. Biocic, and D. I. C. Wang. 1981. A novel single step conversion of cellullosic biomass to ethanol, p. 119-124. In M. Moo-Young and W. Robinson (ed.), Advances in biotechnology, vol. 2. Pergamon Press, Oxford, United Kingdom.
- 11.Avgerinos, G. C., and D. I. C. Wang. 1980. Direct microbiological conversion of cellulosics to ethanol. Ann. Rep. Ferm. Proc. 4:165-191. [Google Scholar]
- 12.Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J.-P. Belaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 13.Barr, B. K., Y.-L. Hsieh, B. Ganem, and D. B. Wilson. 1996. Identification of two functionally different classes of exocellulases. Biochemistry 35:586-592. [DOI] [PubMed] [Google Scholar]
- 14.Baskaran, S., H.-J. Ahn, and L. R. Lynd. 1995. Investigation of the ethanol tolerance of Clostridium thermosaccharolyticum in continuous culture. Biotechnol. Prog. 11:276-281. [Google Scholar]
- 15.Bates, E. E., H. J. Gilbert, G. P. Hazlewood, J. Huckle, J. I. Laurie, and S. P. Mann. 1989. Expression of a Clostridium thermocellum endoglucanase gene in Lactobacillus plantarum. Appl. Environ. Microbiol. 55:2095-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayer, E. A., R. Kenig, and R. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayer, E. A., and R. Lamed. 1986. Ultrastructure of the cell surface cellulosome of Clostridium thermocellum and its interaction with cellulose. J. Bacteriol. 167:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:379-386. [DOI] [PubMed] [Google Scholar]
- 19.Bayer, E. A., E. Morag, R. Lamed, S. Yaron, and Y. Shoham. 1998. Cellulosome structure: four-proned attack using biochemistry, molecular biology, crystallography and bioinformatics, p. 39-65. In M. Claeyssens, W. Nerinckx, and K. Piens (ed.), Carbohydrases from Trichoderma reesei and other microorganisms. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 20.Bayer, E. A., E. Morag, Y. Shoham, J. Tormo, and R. Lamed. 1996. The cellulosome: a cell surface organelle for the adhesion to and degradation of cellulose, p. 155-182. In M. Fletcher (ed.), Bacterial adhesion: molecular and ecological diversity. Wiley-Liss, Inc., New York, N.Y.
- 21.Bayer, E. A., E. Setter, and R. Lamed. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayer, E. A., L. J. W. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes—structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 23.Bayer, E. A., Y. Shoham, and R. Lamed. 2000. Cellulose-decomposing prokaryotes and their enzyme systems. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer, New York, N.Y.
- 24.Bazua, C. D., and C. R. Wilke. 1977. Ethanol effects on the kinetics of a continuous fermentation with Saccharomyces cerevisiae. Biotechnol. Bioeng. Symp. 7:105-118. [PubMed] [Google Scholar]
- 25.Beattie, L., K. M. Bhat, and T. M. Wood. 1994. The effect of cations on reassociation of the components of the cellulosome cellulase complex synthesized by the bacterium Clostridium thermocellum. Appl. Microbiol. Biotechnol. 40:740-744. [Google Scholar]
- 26.Beguin, P. 1990. Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 44:219-248. [DOI] [PubMed] [Google Scholar]
- 27.Beguin, P., and P. M. Alzari. 1998. The cellulosome of Clostridium thermocellum. Biochem. Soc. Trans. 26:178-185. [DOI] [PubMed] [Google Scholar]
- 28.Beguin, P., and J. P. Aubert. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25-58. [DOI] [PubMed] [Google Scholar]
- 29.Beguin, P., S. Chauvaux, M.-K. Chaveroche, G. Guglielmi, I. Kataeva, E. Leibovitz, and I. Miras. 1998. The cellulosome: a versatile system for coupling cellulolytic enzymes and attaching them to the cell surface, p. 66-72. In M. Claeyssens, W. Nerinckx, and K. Piens (ed.), Carbohydrases from Trichoderma reesei and other microorganisms. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 30.Beguin, P., P. Cornet, and J. P. Aubert. 1985. Sequence of a cellulase gene of the thermophilic bacterium Clostridium thermocellum. J. Bacteriol. 162:102-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beguin, P., P. Cornet, and J. Millet. 1983. Identification of the endoglucanase encoded by the celB gene of Clostridium thermocellum. Biochimie 65:495-500. [DOI] [PubMed] [Google Scholar]
- 32.Beguin, P., and M. Lemaire. 1996. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31:201-236. [DOI] [PubMed] [Google Scholar]
- 33.Beguin, P., J. Millet, and J.-P. Aubert. 1992. Cellulose degradation by Clostridium thermocellum: from manure to molecular biology. FEMS Microbiol. Lett. 100:523-528. [DOI] [PubMed] [Google Scholar]
- 34.Beguin, P., J. Millet, and J. P. Aubert. 1987. The cloned cel (cellulose degradation) genes of Clostridium thermocellum and their products. Microbiol. Sci. 4:277-280. [PubMed] [Google Scholar]
- 35.Beguin, P., M. Rocancourt, M. C. Chebrou, and J. P. Aubert. 1986. Mapping of mRNA encoding endoglucanase A from Clostridium thermocellum. Mol. Gen. Genet. 202:251-254. [DOI] [PubMed] [Google Scholar]
- 36.Bernardez, T. D., K. Lyford, and L. R. Lynd. 1994. Kinetics of the extracellular cellulose complex of Clostridium thermocellum for pretreated mixed hardwood and avicel. Appl. Microbiol. Biotechnol. 41:620-625. [Google Scholar]
- 37.Bhat, K. M., and T. M. Wood. 1992. The cellulase of the anaerobic bacterium Clostridium thermocellum: isolation, dissociation, and reassociation of the cellulosome. Carbohydr. Res. 227:293-300. [Google Scholar]
- 38.Bhat, M. K., and S. Bhat. 1997. Cellulose degrading enzymes and their potential industrial applications. Biotechnol. Adv. 15:585-620. [DOI] [PubMed] [Google Scholar]
- 39.Bhat, M. K., and S. Bhat. 1998. Clostridium thermocellum cellulosome: dissociation, isolation, and characterization of subunits and potential biotechnological implications. Recent Res. Dev. Biotechnol. Bioeng. 1:59-84. [Google Scholar]
- 40.Bhat, S., P. W. Goodenough, E. Owen, and M. K. Bhat. 1993. Cellobiose: a true inducer of cellulosome in different strains of Clostridium thermocellum. FEMS Microbiol. Lett. 111:73-78. [Google Scholar]
- 41.Bhat, S., E. Owen, and M. K. Bhat. 2001. Isolation and characterization of a major cellobiohydrolase (S8) and a major endoglucanase (S11) subunit from the cellulosome of Clostridium thermocellum. Anaerobe 7:171-179. [Google Scholar]
- 42.Blair, B. G., and K. L. Anderson. 1999. Regulation of cellulose-inducible structures of Clostridium cellulovorans. Can. J. Microbiol. 45:242-249. [DOI] [PubMed] [Google Scholar]
- 43.Blaut, M., V. Muller, K. Fiebig, and G. Gottschalk. 1985. Sodium ions and an energized membrane required by Methanosarcina barkeri for the oxidation of methanol to the level of formaldehyde. J. Bacteriol. 164:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolobova, A. V., A. V. Zhukov, and A. A. Klyosov. 1994. Lipids and fatty acids in cellulosomes of Clostridium thermocellum. Appl. Microbiol. Biotechnol. 42:128-133. [Google Scholar]
- 45.Buchanan, R. L., S. B. Jones, W. V. Gerasimowicz, L. L. Zaika, H. G. Stahl, and L. A. Ocker. 1987. Regulation of aflatoxin biosynthesis: assessment of the role of cellular energy status as a regulator of the induction of aflatoxin production. Appl. Environ. Microbiol. 53:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bumazkin, B. K., G. A. Velikodvorskaya, K. Tuka, M. A. Mogutov, and A. Strongin. 1990. Cloning of Clostridium thermocellum endoglucanase genes in Escherichia coli. Biochem. Biophys. Res. Commun. 167:1057-1064. (Erratum, 168:1326-1327.) [DOI] [PubMed] [Google Scholar]
- 47.Bungay, R. R. 1982. Biomass refining. Science 218:643-646. [DOI] [PubMed] [Google Scholar]
- 48.Cairns, J., J. Overbaugh, and S. Miller. 1988. The origin of mutants. Nature 335:142-145. [DOI] [PubMed] [Google Scholar]
- 49.Canganella, F., and J. Wiegel. 1993. The potential of thermophilic clostridia in biotechnology, p. 393-429. In D. R. Woods (ed.), The clostridia and biotechnology. Butterworth-Heinemann, Boston, Mass.
- 50.Carrard, G., A. Koivula, H. Soderlund, and P. Beguin. 2000. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA 97:10342-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carreira, L. H., and L. G. Ljungdahl. 1993. Production of ethanol from biomass using anaerobic thermophilic bacteria, p. 1-28. In D. L. Wise (ed.), Liquid fuel developments. CRC Press, Boca Raton, Fla.
- 52.Carvalho, A. L., F. M. V. Dias, J. A. M. Prates, T. Nagy, H. J. Gilbert, G. J. Davies, L. M. A. Ferreira, M. J. Romao, and C. M. G. A. Fontes. 2003. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc. Natl. Acad. Sci. USA 100:13809-13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavedon, K., S. B. Leschine, and E. Canale-Parola. 1990. Characterization of the extracellular cellulase from a mesophilic clostridium (strain C7). J. Bacteriol. 172:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charbonnier, F., and P. Forterre. 1994. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J. Bacteriol. 176:1251-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chauvaux, S., P. Beguin, J.-P. Aubert, K. M. Bhat, L. A. Gow, T. M. Wood, and A. Bairoch. 1990. Calcium-binding affinity and calcium-enhanced activity of Clostridium thermocellum endoglucanase D. Biochem. J. 265:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chauvaux, S., M. Matuschek, and P. Beguin. 1999. Distinct affinity of binding sites for S-layer homologous domains in Clostridium thermocellum and Bacillus anthracis cell envelopes. J. Bacteriol. 181:2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho, H.-Y., H. Yukawa, M. Inui, R. H. Doi, and S.-L. Wong. 2004. Production of minicellulosomes from Clostridium cellulovorans in Bacillus subtilis WB800. Appl. Environ. Microbiol. 70:5704-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi, S. K., and L. G. Ljungdahl. 1996. Dissociation of the cellulosome of Clostridium thermocellum in the presence of ethylenediaminetetraacetic acid occurs with the formation of truncated polypeptides. Biochemistry 35:4897-4905. [DOI] [PubMed] [Google Scholar]
- 59.Choi, S. K., and L. G. Ljungdahl. 1996. Structural role of calcium for the organization of the cellulosome of Clostridium thermocellum. Biochemistry 35:4906-4910. [DOI] [PubMed] [Google Scholar]
- 60.Chuvil'skaya, N. A., Y. Y. Atakishieva, and V. K. Akimenko. 1991. Production of ethanol-resistant mutants of Clostridium thermocellum. Mikrobiologiya 60:107-115. (English translation, 60:78-84.) [Google Scholar]
- 61.Cooney, C. L., D. I. C. Wang, S.-D. Wang, J. Gordon, and M. Jiminez. 1978. Simultaneous cellulose hydrolysis and ethanol production by a cellulolytic anaerobic bacterium. Biotechnol. Bioeng. Symp. 8:103-114. [Google Scholar]
- 62.Cornet, P., J. Millet, P. Beguin, and J.-P. Aubert. 1983. Characterization of two cel (cellulose degradation) genes of Clostridium thermocellum coding for endo-glucanases. Bio/Technology 1:589-594. [Google Scholar]
- 63.Cornet, P., D. Tronik, J. Millet, and J.-P. Aubert. 1983. Cloning and expression in Escherichia coli of Clostridium thermocellum genes coding for amino acid synthesis and cellulose hydrolysis. FEMS Microbiol. Lett. 16:137-141. [Google Scholar]
- 64.Coughlan, M. P., K. Hon-Nami, H. Hon-Nami, L. G. Ljungdahl, J. J. Paulin, and W. E. Rigsby. 1985. The cellulolytic enzyme complex of Clostridium thermocellum is very large. Biochem. Biophys. Res. Commun. 130:904-909. [DOI] [PubMed] [Google Scholar]
- 65.Demain, A. L., T. R. Klapatch, K. H. Jung, and L. R. Lynd. 1996. Recombinant DNA technology in development of an economical conversion of waste to liquid fuel. Ann. N. Y. Acad. Sci. 782:402-412. [Google Scholar]
- 66.Demain, A. L., and J. H. D. Wu. 1989. The cellulase complex of Clostridium thermocellum, p. 68-86. In T. K. Ghose (ed.), Bioprocess engineering, the first generation. Ellis Horwood, Chichester, United Kingdom.
- 67.Devillard, E., D. B. Goodheart, S. K. R. Karnati, E. A. Bayer, R. Lamed, J. Miron, K. E. Nelson, and M. Morrison. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J. Bacteriol. 186:136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Din, N., N. R. Gilkes, B. Tekant, R. C. Miller, R. A. J. Warren, and D. G. Kilburn. 1991. Non-hydrolytic disruption of cellulose fibres by the binding domain of a bacterial cellulose. Bio/Technology 9:1096-1099. [Google Scholar]
- 69.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 1999. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J. Bacteriol. 181:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 2000. A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J. Bacteriol. 182:4915-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ding, S.-Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doi, R. H., M. Goldstein, S. Hashida, J.-S. Park, and M. Takagi. 1994. The Clostridium cellulovorans cellulosome. Crit. Rev. Microbiol. 20:87-93. [DOI] [PubMed] [Google Scholar]
- 73.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell wall degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 74.Doi, R. H., A. Kosugi, K. Murashima, Y. Tamaru, and S. O. Han. 2003. Cellulosomes from mesophilic bacteria. J. Bacteriol. 185:5907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drlica, K. 1992. Control of bacterial DNA supercoiling. Mol. Microbiol. 6:425-433. [DOI] [PubMed] [Google Scholar]
- 76.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dror, T. W., A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of expression of scaffoldin-related genes in Clostridium thermocellum. J. Bacteriol. 185:5109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duong, C. T. V., E. A. Johnson, and A. L. Demain. 1983. Thermophilic, anaerobic and cellulolytic bacteria. Enzyme Ferm. Biotechnol. 7:156-195. [Google Scholar]
- 79.Fan, L. T., Y. Lee, and D. H. Breadmore. 1980. Mechanism of the enzymatic hydrolysis of cellulose: effects of major structural features of cellulose on enzymatic hydrolysis. Biotechnol. Bioeng. Symp. 22:177-199. [Google Scholar]
- 80.Fauth, U., M. P. Romaniec, T. Kobayashi, and A. L. Demain. 1991. Purification and characterization of endoglucanase SS from Clostridium thermocellum. Biochem. J. 279:67-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Felix, C. R., and L. G. Ljungdahl. 1993. The cellulosome: the extracellular organelle of Clostridium. Annu. Rev. Microbiol. 47:791-819. [DOI] [PubMed] [Google Scholar]
- 82.Fierobe, H. P., S. Pages, A. Belaich, S. Champ, D. Lexa, and J. P. Belaich. 1999. Cellulosome from Clostridium cellulolyticum: molecular study of the dockerin/cohesin interaction. Biochemistry 38:12822-12832. [DOI] [PubMed] [Google Scholar]
- 83.Fontes, C. M. G. A., G. P. Hazlewood, E. Morag, J. Hall, B. H. Hirst, and H. J. Gilbert. 1995. Evidence for a general role for non-catalytic thermostabilizing domains in xylanases from thermophilic bacteria. Biochem. J. 307:151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fox, J. L. 2002. Legislation, technology boosting renewable fuel, materials uses. ASM News 68:480-481. [Google Scholar]
- 85.Freier, D., C. F. Mothershed, and J. Wiegel. 1988. Characterization of Clostridium thermocellum JW20. Appl. Environ. Microbiol. 54:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fuchs, K.-P., V. V. Zverlov, G. A. Velikodvorskaya, F. Lottspeich, and W. H. Schwarz. 2003. Lic16A of Clostridium thermocellum, a non-cellulosomal, highly complex endo-beta-1,3-glucanase bound to the outer cell surface. Microbiology 149:1021-1031. [DOI] [PubMed] [Google Scholar]
- 87.Fujino, T., P. Beguin, and J.-P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fujino, T., S. Karita, and K. Ohmiya. 1992. Cloning of the gene cluster encoding putative cellulosomal catalytic component from Clostridium josui, p. 67-75. In K. Shimada, S. Hosino, K. Ohmiya, K. Sakka, Y. Kobayashi, and S. Karita (ed.), Genetics, biochemistry and ecology of lignocellulose degradation. Uni Publishers, Tokyo, Japan.
- 89.Fukumura, M., A. Begum, K. Kruus, and J. H. D. Wu. 1997. Interactions and synergism between the recombinant CelD, an endoglucanase, and the cellulosome-integrating protein (CipA) of Clostridium thermocellum. J. Ferm. Bioeng. 83:146-151. [Google Scholar]
- 90.Gal, L., C. Gaudin, A. Belaich, S. Pages, C. Tardif, and J. P. Belaich. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gal, L., S. Pages, C. Gaudin, A. Belaich, C. Reverbel-Leroy, C. Tardif, and J. P. Belaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garcia-Campayo, V., and P. Beguin. 1997. Synergism between the cellulosome-integrating protein CipA and endoglucanase CelD of Clostridium thermocellum. J. Biotechnol. 57:39-47. [DOI] [PubMed] [Google Scholar]
- 93.Garcia-Martinez, D. V., A. Shinmyo, A. Madia, and A. L. Demain. 1980. Studies on cellulase production by Clostridium thermocellum. Eur. J. Appl. Microbiol. Biotechnol. 9:189-197. [Google Scholar]
- 94.Gaudin, C., A. Belaich, S. Champ, and J.-P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Germain, P., F. Toukourou, and L. Donaduzzi. 1986. Ethanol production by anaerobic thermophilic bacteria: regulation of lactate dehydrogenase activity in Clostridium thermohydrosulfuricum. Appl. Microbiol. Biotechnol. 24:300-305. [Google Scholar]
- 96.Gerngross, U. T., M. P. M. Romainiec, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 97.Gerwig, G. J., P. de Waard, J. P. Kamerling, J. F. Vliegenthart, E. Morgenstern, R. Lamed, and E. A. Bayer. 1989. Novel O-linked carbohydrate chains in the cellulase complex (cellulosome) of Clostridium thermocellum. 3-O-Methyl-N-acetylglucosamine as a constituent of a glycoprotein. J. Biol. Chem. 264:1027-1035. [PubMed] [Google Scholar]
- 98.Gerwig, G. J., J. P. Kamerling, J. F. Vliegenthart, E. Morag, R. Lamed, and E. A. Bayer. 1993. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J. Biol. Chem. 268:26956-26960. [PubMed] [Google Scholar]
- 99.Gerwig, G. J., J. P. Kamerling, J. F. Vliegenthart, E. Morag, R. Lamed, and E. A. Bayer. 1991. Primary structure of O-linked carbohydrate chains in the cellulosome of different Clostridium thermocellum strains. Eur. J. Biochem. 196:115-122. [DOI] [PubMed] [Google Scholar]
- 100.Ghose, T. K., and R. D. Tyagi. 1979. Rapid ethanol fermentation of cellulose hydrolysate. 2. Product and substrate inhibition and optimization of fermentor design. Biotechnol. Bioeng. Symp. 21:1400-1420. [Google Scholar]
- 101.Gilad, R., L. Rabinovich, S. Yaron, E. A. Bayer, R. Lamed, H. J. Gilbert, and Y. Shoham. 2003. CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goldstein, M. A., M. Takagi, S. Hashida, O. Shoseyov, R. H. Doi, and I. H. Segel. 1993. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J. Bacteriol. 175:5762-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grabnitz, F., M. Seiss, K. P. Rucknagel, and W. L. Staudenbauer. 1991. Structure of the beta-glucosidase gene bglA of Clostridium thermocellum. Sequence analysis reveals a superfamily of cellulases and beta-glycosidases including human lactase/phlorizin hydrolase. Eur. J. Biochem. 200:301-309. [DOI] [PubMed] [Google Scholar]
- 104.Grabnitz, F., and W. L. Staudenbauer. 1988. Characterization of two β-glucosidase genes from Clostridium thermocellum. Biotechnol. Lett. 10:73-78. [Google Scholar]
- 105.Grepinet, O., and P. Beguin. 1986. Sequence of the cellulase gene of Clostridium thermocellum coding for endoglucanase B. Nucleic Acids Res. 14:1791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grepinet, O., M.-C. Chebrou, and P. Beguin. 1988. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J. Bacteriol. 170:4582-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grepinet, O., M.-C. Chebrou, and P. Beguin. 1988. Purification of Clostridium thermocellum xylanase Z expressed in Escherichia coli and identification of the corresponding product in the culture medium of Clostridium thermocellum. J. Bacteriol. 170:4576-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grethlein, H., and A. O. Converse. 1983. Flow reactor for acid hydrolysis or pretreatment of cellulosic biomass, p. 97-128. In D. L. Wise (ed.), Liquid fuel developments. CRC Press, Boca Raton, Fla.
- 109.Grethlein, H. E. 1984. Pretreatment for enhanced hydrolysis of cellulosic biomass. Biotechnol. Adv. 2:43-62. [DOI] [PubMed] [Google Scholar]
- 110.Guglielmi, G., and P. Beguin. 1998. Cellulase and hemicellulase genes of Clostridium thermocellum from five independent collections contain few overlaps and are widely scattered across the chromosome. FEMS Microbiol. Lett. 161:209-215. [DOI] [PubMed] [Google Scholar]
- 111.Guimaraes, B. G., H. Souchon, B. L. Lytle, J. H. David Wu, and P. M. Alzari. 2002. The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J. Mol. Biol. 320:587-596. [DOI] [PubMed] [Google Scholar]
- 112.Hall, B. G. 1990. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics 126:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hall, J., G. P. Hazlewood, P. J. Barker, and H. J. Gilbert. 1988. Conserved reiterated domains in Clostridium thermocellum endoglucanases are not essential for catalytic activity. Gene 69:29-38. [DOI] [PubMed] [Google Scholar]
- 114.Halstead, J. R., P. E. Vercoe, H. J. Gilbert, K. Davidson, and G. P. Hazlewood. 1999. A family 26 mannanase produced by Clostridium thermocellum as a component of the cellulosome contains a domain which is conserved in mannanases from anaerobic fungi. Microbiology 145:3101-3108. [DOI] [PubMed] [Google Scholar]
- 115.Hammerstrom, R. A., K. D. Claus, J. W. Cughlan, and R. H. McBee. 1955. The constitutive nature of bacterial cellulase. Arch. Biochem. Biophys. 56:123-129. [DOI] [PubMed] [Google Scholar]
- 116.Han, S. O., H.-Y. Cho, H. Yukawa, M. Inui, and R. H. Doi. 2004. Regulation of expression of cellulosomes and noncellulosomal (hemi)cellulolytic enzymes in Clostridium cellulovorans during growth on different carbon sources. J. Bacteriol. 186:4218-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans. J. Bacteriol. 185:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J. Bacteriol. 185:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harold, F. M. 1986. The bacterial paradigm: useful work, p. 143-144. In F. M. Harold (ed.), In the vital force: a study of bioenergetics. W. H. Freeman and Co., New York, N.Y.
- 120.Hayashi, H., K.-I. Takagi, M. Fukumura, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1997. Sequence of xynC and properties of XynC, a major component of the Clostridium thermocellum cellulosome. J. Bacteriol. 179:4246-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hayashi, H., M. Takehara, T. Hattori, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1999. Nucleotide sequences of two contiguous and highly homologous xylanase genes xynA and xynB and characterization of XynA from Clostridium thermocellum. Appl. Microbiol. Biotechnol. 51:348-357. [DOI] [PubMed] [Google Scholar]
- 122.Hazlewood, G. P., K. Davidson, J. H. Clarke, A. J. Durrant, J. Hall, and H. J. Gilbert. 1990. Endoglucanase E, produced at high level in Escherichia coli as a lacZ′ fusion protein, is part of the Clostridium thermocellum cellulosome. Enzyme Microb. Technol. 12:656-662. [DOI] [PubMed] [Google Scholar]
- 123.Hazlewood, G. P., K. Davidson, J. I. Laurie, N. S. Huskisson, and H. J. Gilbert. 1993. Gene sequence and properties of CelI, a family E endoglucanase from Clostridium thermocellum. J. Gen. Microbiol. 139:307-316. [DOI] [PubMed] [Google Scholar]
- 124.Hazlewood, G. P., M. P. M. Romaniec, K. Davidson, O. Grepinet, P. Beguin, J. Millet, O. Raynaud, and J. P. Aubert. 1988. A catalogue of Clostridium thermocellum endoglucanase, β-glucosidase and xylanase genes cloned in Escherichia coli. FEMS Microbiol. Lett. 51:231-236. [Google Scholar]
- 125.Hernandez, P. E. 1981. Carbohydrate utilization by Clostridium thermocellum. M.S. thesis. Massachussetts Institute of Technology, Cambridge, Mass.
- 126.Hernandez, P. E. 1982. Transport of d-glucose in Clostridium thermocellum ATCC 27405. J. Gen. Appl. Microbiol. 28:469-477. [Google Scholar]
- 127.Herrero, A. A., and R. F. Gomez. 1980. Development of ethanol tolerance in Clostridium thermocellum: effect of growth temperature. Appl. Environ. Microbiol. 40:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Herrero, A. A., R. F. Gomez, and M. F. Roberts. 1985. 35P NMR studies of Clostridium thermocellum. Mechanism of end product inhibition by ethanol. J. Biol. Chem. 260:7442-7451. [PubMed] [Google Scholar]
- 129.Herrero, A. A., R. F. Gomez, B. Snedecor, C. J. Tolman, and M. F. Roberts. 1985. Growth inhibition of Clostridium thermocellum by carboxylic acids: a mechanism based on uncoupling by weak acids. Appl. Microbiol. Biotechnol. 22:536-563. [Google Scholar]
- 130.Hogsett, D. A., H.-J. Ahn, T. D. Bernardez, C. R. South, and L. R. Lynd. 1992. Direct microbial conversion. Prospects, progress, and obstacles. Appl. Biochem. Biotechnol. 34.35:527-541. [Google Scholar]
- 131.Hon-nami, K., M. P. Coughlan, H. Hon-nami, and L. G. Ljungdahl. 1986. Separation and characterization of the complexes constituting the cellulolytic enzyme system of Clostridium thermocellum. Arch. Microbiol. 145:13-19. [Google Scholar]
- 132.Irwin, D., D.-H. Shin, S. Zhang, B. K. Barr, J. Sakon, P. A. Karplus, and D. B. Wilson. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 134.Jauris, S., K. P. Rucknagel, W. H. Schwarz, P. Kratzsch, K. Bronnenmeier, and W. L. Staudenbauer. 1990. Sequence analysis of the Clostridium stercorarium celZ gene encoding a thermoactive cellulase (Avicelase I): identification of catalytic and cellulose-binding domains. Mol. Gen. Genet. 223:258-267. [DOI] [PubMed] [Google Scholar]
- 135.Jindou, S., T. Kajino, M. Inagaki, S. Karita, P. Beguin, T. Kimura, K. Sakka, and K. Ohmiya. 2004. Interaction between a type-II dockerin domain and a type-II cohesin domain from Clostridium thermocellum cellulosome. Biosci. Biotechnol. Biochem. 68:924-926. [DOI] [PubMed] [Google Scholar]
- 136.Jindou, S., A. Soda, S. Karita, T. Kajino, P. Beguin, J. H. D. Wu, M. Inagaki, T. Kimura, K. Sakka, and K. Ohmiya. 2004. Cohesin-dockerin interactions within and between Clostridium josui and Clostridium thermocellum: binding selectivity between cognate dockerin and cohesin domains and species specificity. J. Biol. Chem. 279:9867-9874. [DOI] [PubMed] [Google Scholar]
- 137.Johnson, E. A. 1983. Regulation of cellulase activity and synthesis in Clostridium thermocellum. Ph.D. thesis. Massachussetts Institute of Technology, Cambridge, Mass.
- 138.Johnson, E. A., F. Bouchot, and A. L. Demain. 1985. Regulation of cellulase formation in Clostridium thermocellum. J. Gen. Microbiol. 131:2303-2308. [Google Scholar]
- 139.Johnson, E. A., and A. L. Demain. 1984. Probable involvement of sulfhydryl groups and a metal as essential components of the cellulase of Clostridium thermocellum. Arch. Microbiol. 137:135-138. [Google Scholar]
- 140.Johnson, E. A., A. Madia, and A. L. Demain. 1981. Chemically defined minimal medium for growth of the anaerobic cellulolytic thermophile Clostridium thermocellum. Appl. Environ. Microbiol. 41:1060-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Johnson, E. A., E. T. Reese, and A. L. Demain. 1982. Inhibition of Clostridium thermocellum cellulase by end products of cellulolysis. J. Appl. Biochem. 4:64-71. [Google Scholar]
- 142.Johnson, E. A., M. Sakajoh, G. Halliwell, A. Madia, and A. L. Demain. 1982. Saccharification of complex cellulosis substrates by the cellulase system from Clostridium thermocellum. Appl. Environ. Microbiol. 43:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joliff, G., P. Beguin, and J.-P. Aubert. 1986. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum. Nucleic Acids Res. 14:8605-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Joliff, G., P. Beguin, M. Juy, J. Millet, A. Ryter, R. Poljak, and J.-P. Aubert. 1986. Isolation, crystallization and properties of a new cellulase of Clostridium thermocellum, overproduced in Escherichia coli. Bio/Technology 4:896-900. [Google Scholar]
- 145.Jung, K. H., J.-H. Lee, Y.-T. Yi, H.-K. Kim, and M.-Y. Pack. 1992. Properties of a novel Clostridium thermocellum endo-β-1,4-glucanase expressed in Escherichia coli. Kor. J. Appl. Microbiol. Biotechnol. 20:505-510. [Google Scholar]
- 146.Jung, K. H., K. M. Lee, H. Kim, K.-H. Yoon, S.-H. Park, and M. Y. Pack. 1998. Cloning and expression of a Clostridium thermocellum xylanase gene in Escherichia coli. Biochem. Mol. Biol. Int. 44:283-292. [DOI] [PubMed] [Google Scholar]
- 147.Juy, M., A. G. Amit, P. M. Alzari, R. J. Poljak, M. Claeyssens, P. Beguin, and J.-P. Aubert. 1992. Crystal structure of a thermostable bacterial cellulose-degrading enzyme. Nature 357:89-91. [Google Scholar]
- 148.Kadam, S., A. L. Demain, J. Millet, P. Beguin, and J. P. Aubert. 1988. Molecular cloning of a gene for a thermostable β-glucosidase from Clostridium thermocellum into Escherichia coli. Enzyme Microb. Technol. 10:9-14. [Google Scholar]
- 149.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kashket, E. R., A. G. Blanchard, and W. C. Metzger. 1980. Protonmotive force during growth of Streptococcus lactis cells. J. Bacteriol. 143:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kataeva, I., G. Guglielmi, and P. Beguin. 1997. Interaction between Clostridium thermocellum endoglucanase CelD and polypeptides derived from the cellulosome-integrating protein CipA: stoichiometry and cellulolytic activity of the complexes. Biochem. J. 326:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kataeva, I., X. L. Li, H. Chen, S. K. Choi, and L. G. Ljungdahl. 1999. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J. Bacteriol. 181:5288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kataeva, I. A., N. Chuvil'skaya, N. P. Golochenko, and V. K. Akimenko. 1990. Two β-glucosidases of Clostridium thermocellum; isolation and properties. Biokhimiya 56:1429-1438. [Google Scholar]
- 154.Kataeva, I. A., R. D. Seidel III, X.-L. Li, and L. G. Ljungdahl. 2001. Properties and mutation analysis of the CelK cellulose-binding domain from the Clostridium thermocellum cellulosome. J. Bacteriol. 183:1552-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Khan, A. W., M. Asther, and C. Giuliano. 1984. Utilization of steam and explosion decompressed aspen wood by some anaerobes. J. Ferm. Technol. 62:335-339. [Google Scholar]
- 156.Khan, S., and R. M. Macnab. 1980. Proton chemical potential, proton electrical potential and bacterial motility. J. Mol. Biol. 138:599-614. [DOI] [PubMed] [Google Scholar]
- 157.Klapatch, T. R., A. L. Demain, and L. R. Lynd. 1996. Restriction endonuclease activity in Clostridium thermocellum and Clostridium thermosaccharolyticum. Appl. Microbiol. Biotechnol. 45:127-131. [DOI] [PubMed] [Google Scholar]
- 158.Klapatch, T. R., M. L. Guerinot, and L. R. Lynd. 1996. Electrotransformation of Clostridium thermosaccharolyticum. J. Ind. Microbiol. 16:342-347. [DOI] [PubMed] [Google Scholar]
- 159.Klapatch, T. R., D. A. Hogsett, S. Baskaran, and S. Pal. 1994. Organism development and characterization for ethanol production using thermophilic bacteria. Appl. Biochem. Biotechnol. 45/46:209-223. [Google Scholar]
- 160.Klesov, A. A. 1990. Biochemistry and enzymology of cellulose hydrolysis. Biokhimiya 55:1731-1765 (English translation, 55:1295-1318.) [Google Scholar]
- 161.Kobayashi, T., M. P. Romaniec, U. Fauth, and A. L. Demain. 1990. Subcellulosome preparation with high cellulase activity from Clostridium thermocellum. Appl. Environ. Microbiol. 56:3040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kobayashi, T., M. P. M. Romaniec, P. J. Barker, U. T. Gerngross, and A. L. Demain. 1993. Nucleotide sequence of gene celM encoding a new endoglucanase (CelM) of Clostridium thermocellum and purification of the enzyme. J. Ferm. Bioeng. 76:251-256. [Google Scholar]
- 163.Kobayashi, T., M. P. M. Romaniec, U. Fauth, P. J. Barker, and A. L. Demain. 1992. Cloning and expression in Escherichia coli of Clostridium thermocellum DNA encoding subcellulosomal proteins. Enzyme Microb. Technol. 14:447-453. [Google Scholar]
- 164.Kohring, S., J. Wiegel, and F. Mayer. 1990. Subunit composition and glycosidic activities of the cellulase complex from Clostridium thermocellum JW 20. Appl. Environ. Microbiol. 56:3798-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on growth substrates. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kosugi, A., K. Murashima, Y. Tamaru, and R. H. Doi. 2002. Cell-surface-anchoring role of N-terminal surface layer homology domains of Clostridium cellulovorans EngE. J. Bacteriol. 184:884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kruus, K., A. Andreacchi, W. K. Wang, and J. H. D. Wu. 1995. Product inhibition of the recombinant CelS, an exoglucanase component of the Clostridium thermocellum cellulosome. Appl. Microbiol. Biotechnol. 44:399-404. [DOI] [PubMed] [Google Scholar]
- 168.Kruus, K., A. C. Lua, A. L. Demain, and J. H. D. Wu. 1995. The anchorage function of CipA (CeL), a scaffolding protein of the Clostridium thermocellum cellulosome. Proc. Natl. Acad. Sci. USA 92:9254-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kruus, K., W. K. Wang, P.-C. Chiu, J. Ching, T.-Y. Wang, and J. H. D. Wu. 1994. CelS: a major exoglucanase component of Clostridium thermocellum cellulosome, p. 84-99. In M. E. Himmel, J. O. Baker, and R. P. Overend (ed.), Enzymatic conversion of biomass for fuels production. American Chemical Society, Washington, D.C.
- 170.Kruus, K., W. K. Wang, and J. H. D. Wu. 1995. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J. Bacteriol. 177:1641-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kurokawa, J., E. Hemjinda, T. Arai, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2001. Sequence of the Clostridium thermocellum mannanase gene man26B and characterization of the translated product. Biosci. Biotechnol. Biochem. 65:548-554. [DOI] [PubMed] [Google Scholar]
- 172.Kurokawa, J., E. Hemjinda, T. Arai, T. Kimura, K. Sakka, and K. Ohmiya. 2002. Clostridium thermocellum cellulase CelT, a family 9 endoglucanase without an Ig-like domain or family 3c carbohydrate-binding module. Appl. Microbiol. Biotechnol. 59:455-461. [DOI] [PubMed] [Google Scholar]
- 173.Kurose, N., S. Kinoshita, J. Yagyu, M. Uchida, S. Hanai, and A. Obayashi. 1988. Improvement of ethanol production of thermophilic Clostridium sp. by mutation. J. Ferm. Technol. 66:467-472. [Google Scholar]
- 174.Lamed, R., and E. A. Bayer. 1988. The cellulosome concept: exocellular/extracellular enzyme reactor centers for efficient binding and cellulolysis, p. 101-116. In J.-P. Aubert, P. Beguin, and J. Millet (ed.), Biochemistry and genetics of cellulose degradaation. Academic Press, London, United Kingdom.
- 175.Lamed, R., and E. A. Bayer. 1988. The cellulosome of Clostridium thermocellum. Adv. Appl. Microbiol. 33:1-46. [Google Scholar]
- 176.Lamed, R., and E. A. Bayer. 1986. Contact and cellulolysis in Clostridium thermocellum via extensile surface organelles. Experientia 42:72-73. [Google Scholar]
- 177.Lamed, R., R. Kenig, E. Setter, and E. A. Bayer. 1985. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb. Technol. 7:37-41. [Google Scholar]
- 178.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Specialized cell surface structures in cellulolytic bacteria. J. Bacteriol. 169:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lamed, R., E. Setter, R. Kenig, and E. A. Bayer. 1983. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol. Bioeng. Symp. 13:163-181. [Google Scholar]
- 181.Lamed, R., and J. G. Zeikus. 1980. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J. Bacteriol. 144:569-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Latimer, M. T., and J. G. Ferry. 1993. Cloning, sequence, analysis, and hyperexpression of the genes encoding phosphotransacetylase and acetate kinase from Methanosarcina thermophila. J. Bacteriol. 175:6822-6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lee, J. 1997. Biological conversion of lignocellulosic biomass to ethanol. J. Biotechnol. 56:1-24. [DOI] [PubMed] [Google Scholar]
- 184.Lee, S. Y., L. D. Mermelstein, G. N. Bennett, and E. T. Popoutsakis. 1992. Vector construction, transformation, and gene amplification in Clostridium acetobutylicum ATCC 824. Ann. N. Y. Acad. Sci. Biochem. Eng. 665:39-51. [DOI] [PubMed] [Google Scholar]
- 185.Lee, Y.-E., M. K. Jain, C. Lee, S. E. Lowe, and J. G. Zeikus. 1993. Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xyanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int. J. Syst. Bacteriol. 43:41-51. [Google Scholar]
- 186.Leibovitz, E., and P. Beguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Leibovitz, E., H. Ohayon, P. Gounon, and P. Beguin. 1997. Characterization and subcellular localization of the Clostridium thermocellum scaffolding dockerin binding protein SdbA. J. Bacteriol. 179:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lemaire, M., and P. Beguin. 1993. Nucleotide sequence of the celG gene of Clostridium thermocellum and characterization of its product, endoglucanase CelG. J. Bacteriol. 175:3353-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Beguin. 1995. OlpA, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.LeVine, S. M., F. Ardeshir, and G. Ferro-Luzzi Ames. 1980. Isolation and characterization of acetate kinase and phophotransacetylase mutants of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 143:1081-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lin, C., J. W. Urbance, and D. A. Stahl. 1994. Acetivibrio cellulolyticus and Bacteroides cellulosolvens are members of the greater clostridial assemblage. FEMS Microbiol. Lett. 124:151-155. [DOI] [PubMed] [Google Scholar]
- 192.Lin, W. R., C. C. Lee, J. J. Hsu, J.-F. Hamel, and A. L. Demain. 1998. Properties of acetate kinase activity in Clostridium thermocellum cell extracts. Appl. Biochem. Biotechnol. 69:137-145. [DOI] [PubMed] [Google Scholar]
- 193.Lin, W. R., Y. Peng, S. Lew, C. C. Lee, J. J. Hsu, J.-F. Hamel, and A. L. Demain. 1998. Purification and characterization of acetate kinase from Clostridium thermocellum. Tetrahedron 54:15915-15925. [Google Scholar]
- 194.Liu, C.-C., and R. H. Doi. 1998. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene 211:39-47. [DOI] [PubMed] [Google Scholar]
- 195.Ljungdahl, L. G., and K.-E. Eriksson. 1985. Ecology of microbial cellulose degradation, p. 237-299. In K. C. Marshall (ed.), Advances in microbial ecology, vol. 8. Plenum, New York, N.Y.
- 196.Lynd, L. R. 1990. Large-scale fuel ethanol from lignocellulose. Potential, economics, and research priorities. Appl. Biochem. Biotechnol. 24.25:695-719. [Google Scholar]
- 197.Lynd, L. R. 1996. Overview and evaluation of fuel ethanol from cellulosic biomass: technology, economics, the environment, and policy. Annu. Rev. Energy Environ. 21:403-465. [Google Scholar]
- 198.Lynd, L. R. 1989. Production of ethanol from lignocellulosic material using thermophilic bacteria: critical evaluation of potential and review. Adv. Biochem. Eng. Biotechnol. 38:1-52. [Google Scholar]
- 199.Lynd, L. R., H.-J. Ahn, G. Anderson, P. Hill, D. S. Kersey, and T. R. Klapatch. 1991. Thermophilic ethanol production. Investigation of ethanol yield and tolerance in continuous culture. Appl. Biochem. Biotechnol. 28.29:549-570. [Google Scholar]
- 200.Lynd, L. R., J. H. Cushman, R. J. Nichols, and C. E. Wyman. 1991. Fuel ethanol from cellulosic biomass. Science 251:1318-1323. [DOI] [PubMed] [Google Scholar]
- 201.Lynd, L. R., R. T. Elander, and C. E. Wyman. 1996. Likely features and costs of mature biomass ethanol technology. Appl. Biochem. Biotechnol. 57/58:741-761. [Google Scholar]
- 202.Lynd, L. R., and H. G. Grethlein. 1987. Hydrolysis of dilute acid pretreated hardwood and purified microcrystalline cellulose by cell-free broth from Clostridium thermocellum. Biotechnol. Bioeng. Symp. 29:92-100. [DOI] [PubMed] [Google Scholar]
- 203.Lynd, L. R., H. Jin, J. G. Michels, C. E. Wyman, and B. Dale. 2003, posting date. Bioenergy: background, potential, and policy. Center for Strategic and International Studies, Washington, D.C. [Online.] http://agriculture.senate.gov/Hearings/hearings.cfm?hearingid=1161&witnessId=3320
- 204.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lynd, L. R., R. H. Wolkin, and H. E. Grethlein. 1986. Continuous fermentation of Avicel and pretreated mixed hardwood by Clostridium thermocellum. Biotechnol. Bioeng. Symp. 17:265-274. [Google Scholar]
- 206.Lynd, L. R., C. E. Wyman, and T. U. Gerngross. 1999. Biocommodity engineering. Biotechnol. Prog. 15:777-793. [DOI] [PubMed] [Google Scholar]
- 207.Lytle, B., C. Myers, K. Kruus, and J. H. D. Wu. 1996. Interactions of the CelS binding ligand with various receptor domains of the Clostridium thermocelum cellulosomal scaffolding protein, CipA. J. Bacteriol. 178:1200-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lytle, B., and J. H. D. Wu. 1998. Involvement of both dockerin subdomains in assembly of the Clostridium thermocellum cellulosome. J. Bacteriol. 180:6581-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lytle, B. L., B. F. Volkman, W. M. Westler, M. P. Heckman, and J. H. D. Wu. 2001. Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain. J. Mol. Biol. 307:745-753. [DOI] [PubMed] [Google Scholar]
- 210.Lytle, B. L., B. F. Volkman, W. M. Westler, and J. H. D. Wu. 2000. Secondary structure and calcium-induced folding of the Clostridium thermocellum dockerin domain determined by NMR spectroscopy. Arch. Biochem. Biophys. 379:237-244. [DOI] [PubMed] [Google Scholar]
- 211.Lytle, B. L., W. M. Westler, and J. H. D. Wu. 1999. Molecular assembly of the Clostridium thermocellum cellulosome, p. 444-449. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry, and ecology of cellulose degradation. Uni Publishers, Tokyo, Japan.
- 212.Madia, A. M., J. M. Townsend, and A. L. Demain. 1979. Microbial studies on cellulase production by Clostridium thermocellum. Abstr. Annu. Meet. Am. Soc. Microbiol. abstr. O(H)21, p. 205, 1979.
- 213.Matano, Y., J.-S. Park, M. Goldstein, and R. Doi. 1994. Cellulose promotes extracellular assembly of Clostridium cellulovorans cellulosomes. J. Bacteriol. 176:6952-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Matuschek, M., G. Burchhardt, K. Sahm, and H. Bahl. 1994. Pullulanase of Thermoanaerobacter thermosulfurigenes EM1 (Clostridium thermosulfurogenes): molecular analysis of the gene, composite structure of the enzyme, and the common model for its attachment to the cell surface. J. Bacteriol. 176:3295-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mayer, F., M. P. Coughlan, Y. Mori, and L. G. Ljungdahl. 1987. Macromolecular organization of the cellulolytic complex of Clostridium thermocellum as revealed by electron microscopy. Appl. Environ. Microbiol. 53:2785-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.McBee, R. H. 1950. The anaerobic thermophilic cellulolytic bacteria. Bacteriol. Rev. 14:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.McBee, R. H. 1954. The characteristics of Clostridium thermocellum. J. Bacteriol. 67:505-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.McBee, R. H. 1948. The culture and physiology of a thermophilic cellulose-fermenting bacterium. J. Bacteriol. 56:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mechaly, A., H.-P. Fierobe, A. Belaich, J.-P. Belaich, R. Lamed, Y. Shoham, and E. A. Bayer. 2001. Cohesin-dockerin interaction in cellulosome assembly. A single hydroxyl group of a dockerin domain distinguishes between nonrecognition and high affinity recognition. J. Biol. Chem. 276:9883-9888. [DOI] [PubMed] [Google Scholar]
- 220.Mechaly, A., S. Yaron, R. Lamed, H. P. Fierobe, A. Belaich, J. P. Belaich, Y. Shoham, and E. A. Bayer. 2000. Cohesin-dockerin recognition in cellulosome assembly: experiment versus hypothesis. Proteins Struct. Funct. Genet. 39:170-177. [DOI] [PubMed] [Google Scholar]
- 221.Mel'nik, M. S., D. V. Kapkov, M. A. Mogutov, M. L. Rabinovich, and A. A. Klesov. 1989. A new type of Clostridium thermocellum endoglucanase produced by the recombinant strain of E. coli. Some properties and identification in donor cells. Biokhimiia 54:387-395. [PubMed] [Google Scholar]
- 222.Mel'nik, M. S., M. L. Rabinovich, and Y. V. Voznyi. 1991. Cellobiohydrolase of Clostridium thermocellum produced by a recombinant E. coli strain. Biokhimiya 56:1787-1797. (English translation.) [PubMed] [Google Scholar]
- 223.Mermelstein, L. D., N. E. Welker, G. N. Bennett, and E. T. Papoutsakis. 1992. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Bio/Technology 10:190-195. [DOI] [PubMed] [Google Scholar]
- 224.Mielenz, J. R. 2001. Ethanol production from biomass: technology and commercialization status. Curr. Opin. Microbiol. 4:324-329. [DOI] [PubMed] [Google Scholar]
- 225.Millet, J., D. Petre, P. Beguin, O. Raynaud, and J.-P. Aubert. 1985. Cloning of ten distinct DNA fragments of Clostridium thermocellum coding for cellulases. FEMS Microbiol. Lett. 29:145-149. [Google Scholar]
- 226.Miras, I., F. Schaeffer, P. Beguin, and P. M. Alzari. 2002. Mapping by site-directed mutagenesis of the region responsible for cohesin-dockerin interaction on the surface of the seventh cohesin domain of Clostridium thermocellum CipA. Biochemistry 41:2115-2119. [DOI] [PubMed] [Google Scholar]
- 227.Mishra, S., P. Beguin, and J. P. Aubert. 1991. Transcription of Clostridium thermocellum endoglucanase genes celF and celD. J. Bacteriol. 173:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mistry, F. R. 1986. Production of ethanol by Clostridium thermosaccharolyticum in a continuous culture cell recycle system. Ph.D. thesis. Massachusetts Institute of Technology, Cambridge, Mass.
- 229.Mitchell, P. 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature 191:144-148. [DOI] [PubMed] [Google Scholar]
- 230.Morag, E., A. Lapidot, D. Govorko, R. Lamed, M. Wilchek, E. A. Bayer, and Y. Shoham. 1995. Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl. Environ. Microbiol. 61:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Morag, E., E. A. Bayer, G. P. Hazlewood, H. J. Gilbert, and R. Lamed. 1993. Cellulase Ss (CelS) is synonymous with the major cellobiohydrolase (subunit S8) from the cellulosome of Clostridium thermocellum. Appl. Biochem. Biotechnol. 43:147-151. [DOI] [PubMed] [Google Scholar]
- 232.Morag, E., E. A. Bayer, and R. Lamed. 1990. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J. Bacteriol. 172:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Morag, E., E. A. Bayer, and R. Lamed. 1992. Unorthodox intra-subunit interactions in the cellulosome of Clostridium thermocellum. Appl. Biochem. Biotechnol. 33:205-217. [Google Scholar]
- 234.Morag, E., I. Halevy, E. A. Bayer, and R. Lamed. 1991. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J. Bacteriol. 173:4155-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Morag, E., S. Yaron, R. Lamed, R. Kenig, Y. Shoham, and E. A. Bayer. 1996. Expression, purification and crystallization of a cohesin domain from the cellulosome of Clostridium thermocellum. J. Biotechnol. 51:235-242. [DOI] [PubMed] [Google Scholar]
- 236.Mori, Y. 1990. Isolation of mutants of Clostridium thermocellum with enhanced cellulase production. Agric. Biol. Chem. 54:825-826. [Google Scholar]
- 237.Mori, Y. 1995. Nutritional interdependence between Thermoanaerobacter thermohydrosulfuricus and Clostridium thermocellum. Arch. Microbiol. 164:152-154. [Google Scholar]
- 238.Mosbah, A., A. Belaich, O. Bornet, J.-P. Belaich, B. Henrissat, and H. Darbon. 2000. Solution structure of the module X2_1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum. J. Mol. Biol. 304:201-217. [DOI] [PubMed] [Google Scholar]
- 239.Mosolova, T. P., S. V. Kaliuzhnyi, and G. A. Velikodvorskaia. 1993. Purification and some properties of Clostridium thermocellum endoglucanase produced in a recombinant strain Escherichia coli. Biokhimiia 58:1213-1220. [PubMed] [Google Scholar]
- 240.Mosolova, T. P., S. V. Kalyuzhnyi, S. D. Varfolomeyev, and G. A. Velikodvorskaya. 1993. Purification and properties of Clostridium thermocellum endoglucanase 5 produced in Escherichia coli. Appl. Biochem. Biotechnol. 42:9-18. [DOI] [PubMed] [Google Scholar]
- 241.Muller, V., M. Blaut, and G. Gottschalk. 1987. Oxidation of trimethylamine to the level of formaldehyde by Methanosarcina barkeri is dependent on the protonmotive force. FEMS Microbiol. Lett. 43:183-186. [Google Scholar]
- 242.Murashima, K., A. Kosugi, and R. H. Doi. 2003. Solubilization of cellulosomal cellulases by fusion with cellulose-binding domain of noncellulosomal cellulase EngD from Clostridium cellulovorans. Proteins Struct. Funct. Genet. 50:620-628. [DOI] [PubMed] [Google Scholar]
- 243.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. J. Bacteriol. 184:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Thermostabilization of cellulosomal endoglucanase EngB from Clostridium cellulovorans by in vitro DNA recombination with non-cellulosomal endoglucanase EngD. Mol. Microbiol. 45:617-626. [DOI] [PubMed] [Google Scholar]
- 245.Navarro, A., M.-C. Chebrou, P. Beguin, and J.-P. Aubert. 1991. Nucleotide sequence of the cellulase gene celF of Clostridium thermocellum. Res. Microbiol. 142:927-936. [DOI] [PubMed] [Google Scholar]
- 246.Newcomb, M., and J. H. D. Wu. 2004. Transcription regulators of the Clostridium thermocellum cellulase system, p. 148-154. In K. Ohmiya, K. Sakka, S. Karita, T. Kimura, M. Sakka, and Y. Onishi (ed.), Biotechnology of lignocellulose degradation and biomass utilization. Uni Publishers, Tokyo, Japan.
- 247.Ng, T. K., A. Ben-Bassat, and J. G. Zeikus. 1981. Ethanol production by thermophilic bacteria: fermentation of cellulosic substrates by cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl. Environ. Microbiol. 41:1337-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ng, T. K., T. K. Weimer, and J. G. Zeikus. 1977. Cellulolytic and physiological properties of Clostridium thermocellum. Arch. Microbiol. 114:1-7. [DOI] [PubMed] [Google Scholar]
- 249.Ng, T. K., and J. G. Zeikus. 1981. Purification and characterization of an endoglucanase (1,4-beta-d-glucan glucanohydrolase) from Clostridium thermocellum. Biochem. J. 199:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Nochur, S. V., A. L. Demain, and M. F. Roberts. 1992. Carbohydrate utilization by Clostridium thermocellum: importance of internal pH in regulating growth. Enzyme Microb. Technol. 14:338-349. [Google Scholar]
- 251.Nochur, S. V., G. R. Jacobson, M. F. Roberts, and A. L. Demain. 1992. Mode of sugar phosphorylation in Clostridium thermocellum. Appl. Biochem. Biotechnol. 33:33-41. [Google Scholar]
- 252.Nochur, S. V., M. F. Roberts, and A. L. Demain. 1993. True cellulase production by Clostridium thermocellum grown on different carbon sources. Biotechnol. Lett. 15:641-646. [Google Scholar]
- 253.Nochur, S. V., M. F. Roberts, and A. L. Demain. 1990. Mutation of Clostridium thermocellum in the presence of certain carbon sources. FEMS Microbiol. Lett. 71:199-204. [Google Scholar]
- 254.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ozcengiz, G., J.-H. Kim, W. R. Lin, E. Ozcengiz, D. Westenberg, L. R. Lynd, and A. L. Demain. 1998. Superiority of the PCR-based approach for cloning the acetate kinase gene of Clostridium thermocellum. J. Ind. Microbiol. Biotechnol. 21:145-149. [Google Scholar]
- 256.Ozkan, M., S. G. Desai, Y. Zhang, D. M. Stevenson, J. Beane, E. A. White, M. L. Guerinot, and L. R. Lynd. 2001. Characterization of 13 newly isolated strains of anaerobic, cellulolytic, thermophilic bacteria. J. Ind. Microbiol. Biotechnol. 27:275-280. [DOI] [PubMed] [Google Scholar]
- 257.Pages, S., A. Belaich, J.-P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 258.Pages, S., A. Belaich, H. P. Fierobe, C. Tardif, C. Gaudin, and J. P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Pages, S., A. Belaich, C. Tardif, C. Reverbel-Leroy, C. Gaudin, and J.-P. Belaich. 1996. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 178:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Pages, S., L. Gal, A. Belaich, C. Gaudin, C. Tardif, and J. Belaich. 1997. Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. J. Bacteriol. 179:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Park, J.-S., Y. Matano, and R. H. Doi. 2001. Cohesin-dockerin interactions of cellulosomal subunits of Clostridium cellulovorans. J. Bacteriol. 183:5431-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Parsiegla, G., M. Juy, C. Reverbel-Leroy, C. Tardif, J.-P. Belaïch, H. Driguez, and R. Haser. 1998. The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 Å resolution. EMBO J. 17:5551-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Parsiegla, G., C. Reverbel-Leroy, C. Tardif, J. P. Belaich, H. Driguez, and R. Haser. 2000. Crystal structures of the cellulase Cel48F in complex with inhibitors and substrates give insights into its processive action. Biochemistry 39:11238-11246. [DOI] [PubMed] [Google Scholar]
- 264.Patni, N. J., and J. K. Alexander. 1971. Catabolism of fructose and mannitol in Clostridium thermocellum: presence of phosphoenolpyruvate:fructose phosphotransferase, fructose-1-phosphate kinase, phosphoenol-pyruvate:mannitol phosphotransferase, and mannitol-1-phosphate dehydrogenase in cell extracts. J. Bacteriol. 105:226-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Patni, N. J., and J. K. Alexander. 1971. Utilization of glucose by Clostridium thermocellum: presence of glucokinase and other glycolytic enzymes in cell extracts. J. Bacteriol. 105:220-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Perret, S., L. Casalot, H.-P. Fierobe, C. Tardif, F. Sabathe, J.-P. Belaich, and A. Belaich. 2004. Production of heterologous and chimeric scaffoldins by Clostridium acetobutylicum ATCC 824. J. Bacteriol. 186:253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Petre, D., J. Millet, R. Longin, P. Beguin, H. Girard, and J. P. Aubert. 1986. Purification and properties of the endoglucanase C of Clostridium thermocellum produced in Escherichia coli. Biochimie 68:687-695. [DOI] [PubMed] [Google Scholar]
- 268.Petre, J., R. Longin, and J. Millet. 1981. Purification and properties of an endo-beta-1,4-glucanase from Clostridium thermocellum. Biochimie 63:629-639. [DOI] [PubMed] [Google Scholar]
- 269.Pohlschroder, M., E. Canale-Parola, and S. Leschine. 1995. Ultrastructural diversity of the cellulase complexes of Clostridium papyrosolvens C7. J. Bacteriol. 177:6625-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Pohlschroder, M., S. B. Leschine, and E. Canale-Parola. 1994. Multicomplex cellulase-xylanase system of Clostridium papyrosolvens C7. J. Bacteriol. 176:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Reverbel-Leroy, C., S. Pages, A. Belaich, J. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Richards, D. F., P. E. Linnett, F. D. Oultram, and M. Young. 1988. Restriction endonucleases in Clostridium pasteurianum ATCC 6013 and C. thermohydrosulfuricum DSM 568. J. Gen. Microbiol. 134:3151-3157. [DOI] [PubMed] [Google Scholar]
- 273.Rincon, M. T., S. Y. Ding, S. I. McCrae, J. C. Martin, V. Aurilia, R. Lamed, Y. Shoham, E. A. Bayer, and H. J. Flint. 2003. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 185:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Robson, L. M., and G. H. Chambliss. 1989. Cellulases of bacterial origin. Enzyme Microb. Technol. 11:626-644. [Google Scholar]
- 275.Romaniec, M. P., N. G. Clarke, and G. P. Hazlewood. 1987. Molecular cloning of Clostridium thermocellum DNA and the expression of further novel endo-beta-1,4-glucanase genes in Escherichia coli. J. Gen. Microbiol. 133:1297-1307. [DOI] [PubMed] [Google Scholar]
- 276.Romaniec, M. P., U. Fauth, T. Kobayashi, N. S. Huskisson, P. J. Barker, and A. L. Demain. 1992. Purification and characterization of a new endoglucanase from Clostridium thermocellum. Biochem. J. 283:69-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Romaniec, M. P., T. Kobayashi, U. Fauth, U. T. Gerngross, and A. L. Demain. 1991. Cloning and expression of a Clostridium thermocellum DNA fragment that encodes a protein related to cellulosome component SL. Appl. Biochem. Biotechnol. 31:119-134. [DOI] [PubMed] [Google Scholar]
- 278.Romaniec, M. P. M., K. Davidson, and G. P. Hazlewood. 1987. Cloning and expression in Escherichia coli of Clostridium thermocellum DNA encoding β-glucosidase activity. Enzyme Microb. Technol. 9:474-478. [Google Scholar]
- 279.Sabathe, F., A. Belaich, and P. Soucaille. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217:15-22. [DOI] [PubMed] [Google Scholar]
- 280.Sabathe, F., and P. Soucaille. 2003. Characterization of the CipA scaffolding protein and in vivo production of a minicellulosome in Clostridium acetobutylicum. J. Bacteriol. 185:1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sacco, M., J. Millet, and J. P. Aubert. 1984. Cloning and expression in Saccharomyces cerevisiae of a cellulase gene from Clostridium thermocellum. Ann. Microbiol. (Paris) 135A:485-488. [DOI] [PubMed] [Google Scholar]
- 282.Saddler, J. N., and M. K.-H. Chan. 1984. Conversion of pretreated lignocellulosic substrates to ethanol by Clostridium thermosaccharolyticum and Clostridium thermohydrosulphuricum. Can. J. Microbiol. 30:212-220. [Google Scholar]
- 283.Saddler, J. N., and M. K.-H. Chan. 1982. Optimization of Clostridium thermocellum growth on cellulose and pretreated wood substrates. Eur. J. Appl. Microbiol. Biotechnol. 16:99-104. [Google Scholar]
- 284.Sakka, K., S. Furuse, and K. Shimada. 1989. Cloning and expression in Escherichia coli of the thermophilic Clostridium sp. F1 genes related to cellulose hydrolysis. Agric. Biol. Chem. 53:905-911. [Google Scholar]
- 285.Sakon, J., D. Irwin, D. B. Wilson, and P. A. Karplus. 1997. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4:810-818. [DOI] [PubMed] [Google Scholar]
- 286.Salamitou, S., M. Lemaire, T. Fujino, H. Ohayon, P. Gounon, P. Beguin, and J.-P. Aubert. 1994. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J. Bacteriol. 176:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Salamitou, S., O. Raynaud, M. Lemaire, M. Coughlan, P. Beguin, and J.-P. Aubert. 1994. Recognition specificity of the duplicated segments present in Clostridium thermocellum endoglucanase CelD and in the cellulosome-intergrating protein CipA. J. Bacteriol. 176:2822-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Salamitou, S., K. Tokatlidis, P. Beguin, and J. P. Aubert. 1992. Involvement of separate domains of the cellulosomal protein S1 of Clostridium thermocellum in binding to cellulose and in anchoring of catalytic subunits to the cellulosome. FEBS Lett. 304:89-92. [DOI] [PubMed] [Google Scholar]
- 289.Sato, K., M. Tomita, S. Yonemura, S. Goto, K. Sekine, E. Okuma, Y. Takagi, K. Hon-nami, and T. Saiki. 1993. Characterization of and ethanol hyper-production by Clostridium thermocellum I-1-B. Biosci. Biotechnol. Biochem. 57:2116-2121. [Google Scholar]
- 290.Schaeffer, F., M. Matuschek, G. Guglielmi, I. Miras, P. M. Alzari, and P. Beguin. 2002. Duplicated dockerin subdomains of Clostridium thermocellum endoglucanase CelD bind to a cohesin domain of the scaffolding protein CipA with distinct thermodynamic parameters and a negative cooperativity. Biochemistry 41:2106-2114. [DOI] [PubMed] [Google Scholar]
- 291.Schimming, S., W. H. Schwarz, and W. L. Staudenbauer. 1991. Properties of a thermoactive β-1,3-1,4-glucanase (lichenase) from Clostridium thermocellum expressed in Escherichia coli. Biochem. Biophys. Res. Commun. 177:447-452. [DOI] [PubMed] [Google Scholar]
- 292.Schimming, S., W. H. Schwarz, and W. L. Staudenbauer. 1992. Structure of the Clostridium thermocellum gene licB and the encoded beta-1,3-1,4-glucanase. A catalytic region homologous to Bacillus lichenases joined to the reiterated domain of clostridial cellulases. Eur. J. Biochem. 204:13-19. [DOI] [PubMed] [Google Scholar]
- 293.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 294.Schwarz, W. H., K. Bronnemeier, and W. L. Staudenbauer. 1985. Molecular cloning of Clostridium thermocellum genes involved in β-glucan degradation in bacteriophage lambda. Biotechnol. Lett. 7:859-864. [Google Scholar]
- 295.Schwarz, W. H., F. Grabnitz, and W. L. Staudenbauer. 1986. Properties of a Clostridium thermocellum endoglucanase produced in Escherichia coli. Appl. Environ. Microbiol. 51:1293-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Schwarz, W. H., S. Schimming, and W. L. Staudenbauer. 1988. Degradation of barley β-glucan by endoglucanase C of Clostridium thermocellum. Appl. Microb. Biotechnol. 29:25-31. [Google Scholar]
- 297.Schwarz, W. H., S. Schimming, and W. L. Staudenbauer. 1988. Isolation of a Clostridium thermocellum gene encoding a thermostable β-1,3-glucanase (laminarinase). Biotechnol. Lett. 10:225-230. [Google Scholar]
- 298.Schwarz, W. H., S. Schimming, K. P. Rucknagel, S. Burgschwaiger, G. Kreil, and W. L. Staudenbauer. 1988. Nucleotide sequence of the celC gene encoding endoglucanase C of Clostridium thermocellum. Gene 63:23-30. [DOI] [PubMed] [Google Scholar]
- 299.Schwarz, W. H., S. Schimming, and W. L. Staudenbauer. 1987. High-level expression of Clostridium thermocellum cellulase genes in Escherichia coli. Appl. Microbiol. Biotechnol. 27:50-56. [Google Scholar]
- 300.Sheth, K., and J. K. Alexander. 1969. Purification and properties of β-1,4-oligoglucan:orthophosphate glucosyltransferase from Clostridium thermocellum. J. Biol. Chem. 244:457-464. [PubMed] [Google Scholar]
- 301.Shimada, K., S. Karita, K. Sakka, and K. Ohmiya. 1994. Cellulases, xylanases, and their genes from bacteria. Bioprocess Technol. 19:395-429. [PubMed] [Google Scholar]
- 302.Shimon, L. J., S. Pages, A. Belaich, J. P. Belaich, E. A. Bayer, R. Lamed, Y. Shoham, and F. Frolow. 2000. Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 A resolution. Acta Crystallogr. D 56:1560-1568. [DOI] [PubMed] [Google Scholar]
- 303.Shimon, L. J. W., E. A. Bayer, E. Morag, R. Lamed, S. Yaron, Y. Shoham, and F. Frolow. 1997. A cohesin domain from Clostridium thermocellum: the crystal structure provides new insights into cellulosome assembly. Structure 5:381-390. [DOI] [PubMed] [Google Scholar]
- 304.Shinmyo, A., D. V. Garcia-Martinez, and A. L. Demain. 1979. Studies on the extracellular cellulolytic enzyme complex produced by Clostridium thermocellum. J. Appl. Biochem. 1:202-209. [Google Scholar]
- 305.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 306.Shoseyov, O., M. Takagi, M. A. Goldstein, and R. H. Doi. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc. Natl. Acad. Sci. USA 89:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Singh, R. N., and V. K. Akimenko. 1994. Isolation and characterization of a complex forming hydrophilic endoglucanase of Clostridium thermocellum. Biochem. Mol. Biol. Int. 32:409-417. [PubMed] [Google Scholar]
- 308.Singh, R. N., and V. K. Akimenko. 1993. Isolation of a cellobiohydrolase of Clostridium thermocellum capable of degrading natural crystalline substrates. Biochem. Biophys. Res. Commun. 192:1123-1130. [DOI] [PubMed] [Google Scholar]
- 309.Smith, S. P., P. Beguin, P. M. Alzari, and K. Gehring. 2002. 1H, 13C, 15N NMR sequence-specific resonance assignment of a Clostridium thermocellum type II cohesin module. J. Biomol. NMR 23:73-74. [DOI] [PubMed] [Google Scholar]
- 310.South, C. R., D. A. Hogsett, and A. R. Lynd. 1993. Continuous fermentation of cellulosic biomass to ethanol. Appl. Biochem. Biotechnol. 39/40:587-600. [Google Scholar]
- 311.Soutschek-Bauer, E., L. Hartl, and W. L. Staudenbauer. 1985. Transformation of Clostridium thermohydrosulfuricum DSM 586 with plasmid DNA. Biotechnol. Lett. 7:705-710. [Google Scholar]
- 312.Soutschek-Bauer, E., and W. L. Staudenbauer. 1987. Synthesis and secretion of a heat-stable carboxymethylcellulase from Clostridium thermocellum in Bacillus subtilis and Bacillus stearothermophilus. Mol. Gen. Genet. 208:537-541. [DOI] [PubMed] [Google Scholar]
- 313.Spinelli, S., H.-P. Fierobe, A. Belaich, J.-P. Belaich, B. Henrissat, and C. Cambillau. 2000. Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition. J. Mol. Biol. 304:189-200. [DOI] [PubMed] [Google Scholar]
- 314.Spinnier, H. E., B. Lavigne, and H. Blachere. 1986. Pectinolytic activity of Clostridium thermocellum: its use for anaerobic fermentation of sugar beet pulp. Appl. Microbiol. Biotechnol. 23:434-437. [Google Scholar]
- 315.Stutzenberger, F. 1990. Bacterial cellulases, p. 37-70. In W. M. Fogarty and C. T. Kelly (ed.), Microbial enzymes and biotechnology. Elsevier Applied Science, London, United Kingdom.
- 316.Szmelcman, S., and J. Adler. 1976. Change in membrane potential during bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 73:4387-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Tailliez, P., H. Girard, J. Millet, and P. Beguin. 1989. Enhanced cellulose fermentation by an asporogenous and ethanol-tolerant mutant of Clostridium thermocellum. Appl. Environ. Microbiol. 55:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Tamaru, Y., and R. H. Doi. 2000. The engL gene cluster of Clostridium cellulovorans contains a gene for cellulosomal ManA. J. Bacteriol. 182:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tamaru, Y., and R. H. Doi. 2001. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. USA 98:4125-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Tamaru, Y., S. Ui, K. Murashima, A. Kosugi, H. Chan, R. H. Doi, and B. Liu. 2002. Formation of protoplasts from cultured tobacco cells and Arabidopsis thaliana by the action of cellulosomes and pectate lyase from Clostridium cellulovorans. Appl. Environ. Microbiol. 68:2614-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Tavares, G. A., P. Beguin, and P. M. Alzari. 1997. The crystal structure of a type I cohesin domain at 1.7 A resolution. J. Mol. Biol. 273:701-713. [DOI] [PubMed] [Google Scholar]
- 324.Teeri, T. T. 1997. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 15:160-167. [Google Scholar]
- 325.Tokatlidis, K., P. Dhurjati, and P. Beguin. 1993. Properties conferred on Clostridium thermocellum endoglucanase CelC by grafting the duplicated segment of endoglucnase CelD. Protein Eng. 6:947-952. [DOI] [PubMed] [Google Scholar]
- 326.Tokatlidis, K., P. Dhurjati, J. Millet, P. Beguin, and J. P. Aubert. 1991. High activity of inclusion bodies formed in Escherichia coli overproducing Clostridium thermocellum endoglucanase D. FEBS Lett. 282:205-208. [DOI] [PubMed] [Google Scholar]
- 327.Tokatlidis, K., S. Salamitou, P. Beguin, P. Dhurjati, and J. P. Aubert. 1991. Interaction of the duplicated segment carried by Clostridium thermocellum cellulases with cellulosome components. FEBS Lett. 291:185-188. [DOI] [PubMed] [Google Scholar]
- 328.Tolman, C. J., S. Kanodia, and M. F. Roberts. 1986. 31P and 13C NMR analyses of the energy metabolism of the thermophilic anaerobe Clostridium thermocellum. J. Biol. Chem. 262:11088-11096. [PubMed] [Google Scholar]
- 329.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 330.Tormo, J., R. Lamed, A. J. Chirino, E. Morag, E. A. Bayer, Y. Shoham, and T. A. Steitz. 1996. Crystal structure of a bacterial family III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15:5739-5751. [PMC free article] [PubMed] [Google Scholar]
- 331.Tsoi, T. V., N. A. Chuvil'skaya, Y. Y. Atakishieva, T. D. Dzhavakhishvili, V. K. Akimento, and A. M. Boronin. 1987. Clostridium thermocellum—a new object of genetic investigations. Mol. Genet. Mikrobiol. Virusol. 11:18-23. [PubMed] [Google Scholar]
- 332.Tuka, K., V. V. Zverlov, B. K. Bumazkin, G. A. Velikodvorskaya, and A. Strongin. 1990. Cloning and expression of Clostridium thermocellum genes coding for thermostable exoglucanases (cellobiohydrolases) in Escherichia coli cells. Biochem. Biophys. Res. Commun. 169:1055-1060. [DOI] [PubMed] [Google Scholar]
- 333.Tuka, K., V. V. Zverlov, and G. A. Velikodvorskaya. 1992. Synergism between Clostridium thermocellum cellulases cloned in Escherichia coli. Appl. Biochem. Biotechnol. 37:201-207. [DOI] [PubMed] [Google Scholar]
- 334.Tyurin, M. V., S. G. Desai, and L. R. Lynd. 2004. Electrotransformation of Clostridium thermocellum. Appl. Environ. Microbiol. 70:883-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Venkateswaren, S., and A. L. Demain. 1986. The Clostridium thermocellum-Clostridium thermosaccharolyticum ethanol production process: nutritional studies and scale-down. Chem. Eng. Commun. 45:53-60. [Google Scholar]
- 336.Viljoen, J. A., E. B. Fred, and W. H. Peterson. 1926. The fermentation of cellulose by thermophilic bacteria. J. Agric. Sci. 16:1-17. [Google Scholar]
- 337.Volkman, B. F., B. L. Lytle, and J. H. D. Wu. 2004. Dockerin domains, p. 617-628. In A. Messerschmidt, W. Bode, and M. Cygler (ed.), Metalloproteins, vol. 3. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 338.Wang, D. I. C., G. C. Avgerinos, I. Biocic, S.-D. Wang, and H.-Y. Fang. 1983. Ethanol from cellulosic biomass. Phil. Trans. R. Soc. London 300:323-333. [Google Scholar]
- 339.Wang, J.-Y., and M. Syvanen. 1992. DNA twist as transcriptional sensor for environmental changes. Mol. Microbiol. 6:1861-1866. [DOI] [PubMed] [Google Scholar]
- 340.Wang, J. H. 1989. Sugar utilization and cellulase formation by Clostridium thermocellum. M.S. thesis. University of Rochester, Rochester, N.Y.
- 341.Wang, W. K., K. Kruus, and J. H. D. Wu. 1993. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS), a major cellulosome component. J. Bacteriol. 175:1293-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Wang, W. K., K. Kruus, and J. H. D. Wu. 1994. Cloning and expression of the Clostridium thermocellum celS gene in Escherichia coli. Appl. Microbiol. Biotechnol. 42:346-352. [DOI] [PubMed] [Google Scholar]
- 343.Wang, W. K., and J. H. D. Wu. 1993. Structural features of the Clostridium thermocellum cellulase SS gene. Appl. Biochem. Biotechnol. 39-40:149-158. [DOI] [PubMed] [Google Scholar]
- 344.Weil, J., P. Westgate, K. Kohlmann, and M. R. Ladisch. 1994. Cellulose pretreatments of lignocellulosic substrates. Enzyme Microb. Technol. 16:1002-1004. [DOI] [PubMed] [Google Scholar]
- 345.Wiegel, J. 1980. Formation of ethanol by bacteria. A pledge for the use of extreme thermophilic anaerobic bacteria in industrial ethanol fermentation processes. Experientia 36:1434-1446. [Google Scholar]
- 346.Wiegel, J., and L. G. Ljungdahl. 1979. Ethanol as fermentation product of extreme thermophilic, anaerobic bacteria, p. 117-127. In H. Dellweg (ed.), In Klassische Prozesse mit Neuen Aussichten. Verlag Versuchs und Lehranstalt fuer Spiritusfabrikation und Fermentations Technologie im Insituut fuer Gaerungsgewerbe und Biotechnologie, Berlin, Germany.
- 347.Wiegel, J., and L. G. Ljungdahl. 1981. Thermoanaerobacter ethanolicus gen. nov., a new extreme thermophilic, anaerobic bacterium. Arch. Microbiol. 128:343-348. [Google Scholar]
- 348.Wilson, D. B., D. Irwin, J. Sakon, and P. A. Karplus. 1998. Thermomonospora fusca cellulase E4: a processive endocellulase, p. 133-138. In M. Claeyssens, W. Nerinckx, and K. Piens (ed.), Carbohydrases from Trichoderma reesei and other microorganisms. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 349.Wirth, T. E., C. B. Gay, and J. D. Podesta. 2003. The future of energy policy. Foreign Affairs 82:132-155. [Google Scholar]
- 350.Wright, B. E. 1997. Does selective gene activation direct evolution? FEBS Lett. 402:4-8. [DOI] [PubMed] [Google Scholar]
- 351.Wu, J. H. D. 1993. Clostridium thermocellum cellulosome: new mechanistic concept for cellulose degradation, p. 251-264. In M. E. Himmel and G. Georgiou (ed.), Biocatalyst design for stability and specificity. American Chemical Society, Washington, D.C.
- 352.Wu, J. H. D. 1987. Cooperative degradation of crystalline cellulose by the Clostridium thermocellum cellulase components. Ph.D. thesis. Massachusetts Institute of Technology, Cambridge, Mass.
- 353.Wu, J. H. D., and A. L. Demain. 1988. Proteins of the Clostridium thermocellum complex responsible for degradation of crystallise cellulose, p. 117-131. In J. P. Aubert, P. Beguin, and J. Millet (ed.), Biochemistry and genetics of cellulose degradation. Academic Press, New York, N.Y.
- 354.Wu, J. H. D., W. H. Orme-Johnson, and A. L. Demain. 1988. Two components of an extracellular protein aggregate of Clostridium thermocellum together degrade crystalline cellulose. Biochemistry 27:1703-1709. [Google Scholar]
- 355.Wyman, C. E. 1994. Alternative fuels from biomass and their impact on carbon dioxide accumulation. Appl. Biochem. Biotechnol. 45/46:897-915. [Google Scholar]
- 356.Wyman, C. E. 2003. Potential synergies and challenges in refining cellulosic biomass to fuels, chemicals, and power. Biotechnol. Prog. 19:254-262. [DOI] [PubMed] [Google Scholar]
- 357.Wyman, C. E. 2001. Twenty years of trials, tribulations, and research progress in bioethanol technology: selected key events along the way. Appl. Biochem. Biotechnol. 91-93:5-21. [DOI] [PubMed] [Google Scholar]
- 358.Wyman, C. E., and B. J. Goodman. 1993. Biotechnology for production of fuels, chemicals, and materials from biomass. Appl. Biochem. Biotechnol. 39/40:41-59. [Google Scholar]
- 359.Wyman, C. E., and N. D. Hinman. 1990. Ethanol. Fundamentals of production from renewable feedstocks and use as a transportation fuel. Appl. Biochem. Biotechnol. 24/25:735-753. [Google Scholar]
- 360.Xu, Q., W. Gao, S. Y. Ding, R. Kenig, Y. Shoham, E. A. Bayer, and R. Lamed. 2003. The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell surface anchoring protein. J. Bacteriol. 185:4548-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Yague, E., P. Beguin, and J. P. Aubert. 1990. Nucleotide sequence and deletion analysis of the cellulase-encoding gene celH of Clostridium thermocellum. Gene 89:61-67. [DOI] [PubMed] [Google Scholar]
- 362.Yaron, S., E. Morag, E. A. Bayer, R. Lamed, and Y. Shoham. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121-124. [DOI] [PubMed] [Google Scholar]
- 363.Young, M., N. P. Minton, and W. L. Staudenbauer. 1989. Recent advances in the genetics of the clostridia. FEMS Microbiol. Rev. 5:301-325. [DOI] [PubMed] [Google Scholar]
- 364.Zhang, Y., and L. R. Lynd. 2003. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: development of an enzyme-linked immunosorbent assay-based method with application to Clostridium thermocellum batch cultures. Anal. Chem. 75:219-227. [DOI] [PubMed] [Google Scholar]
- 365.Zhang, Y.-H. P., and L. R. Lynd. 2004. Kinetics and relative importance of phosphorylytic and hydrolytic cleavage of cellodextrins and cellobiose in cell extracts of Clostridium thermocellum. Appl. Environ. Microbiol. 70:1563-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Zverlov, V. V., K.-P. Fuchs, and W. H. Schwarz. 2002. Chi18A, the endochitinase in the cellulosome of the thermophilic, cellulolytic bacterium Clostridium thermocellum. Appl. Environ. Microbiol. 68:3176-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Zverlov, V. V., K.-P. Fuchs, W. H. Schwarz, and G. A. Velikodvorskaya. 1994. Purification and cellulosomal localization of Clostridium thermocellum mixed linkage β-glucanase LicB (1,3-1,4-β-d-glucanase). Biotechnol. Lett. 16:29-44. [Google Scholar]
- 368.Zverlov, V. V., J. Kellermann, and W. H. Schwarz. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics, in press. [DOI] [PubMed]
- 369.Zverlov, V. V., D. A. Laptev, V. I. Tishkov, and G. A. Velikodvorskaya. 1991. Nucleotide sequence of the Clostridium thermocellum laminarinase gene. Biochem. Biophys. Res. Commun. 181:507-512. [DOI] [PubMed] [Google Scholar]
- 370.Zverlov, V. V., and W. H. Schwarz. 2004. The Clostridium thermocellum cellulosome—the paradigm of a multienzyme complex, p. 137-147. In K. Ohmiya, K. Sakka, S. Karita, T. Kimura, M. Sakka, and Y. Onishi (ed.), Biotechnology of lignocellulose degradation and biomass utilization. Uni Publishers, Tokyo, Japan.
- 371.Zverlov, V. V., and G. A. Velikodvorskaya. 1990. Cloning of the Clostridium thermocellum thermostable laminarinase gene in Escherichia coli: the properties of the enzyme thus produced. Biotechnol. Lett. 12:811-816. [Google Scholar]
- 372.Zverlov, V. V., G. A. Velikodvorskaya, and W. H. Schwarz. 2002. A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum: investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose. Microbiology 148:247-255. [DOI] [PubMed] [Google Scholar]
- 373.Zverlov, V. V., G. A. Velikodvorskaya, and W. H. Schwarz. 2003. Two new cellulosome components encoded downstream of celI in the genome of Clostridium thermocellum: the non-processive endoglucanase CelN and the possibly structural protein CseP. Microbiology 149:515-524. [DOI] [PubMed] [Google Scholar]
- 374.Zverlov, V. V., G. A. Velikodvorskaya, W. H. Schwarz, J. Kellermann, and W. L. Staudenbauer. 1999. Duplicated Clostridium thermocellum cellobiohydrolase gene encoding cellulosomal subunits S3 and S5. Appl. Microbiol. Biotechnol. 51:852-859. [DOI] [PubMed] [Google Scholar]
- 375.Zverlov, V. V., G. V. Velikodvorskaya, W. H. Schwarz, K. Bronnenmeier, J. Kellermann, and W. L. Staudenbauer. 1998. Multidomain structure and cellulosomal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J. Bacteriol. 180:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]