Abstract
The ascendancy of metallo-β-lactamases within the clinical sector, while not ubiquitous, has nonetheless been dramatic; some reports indicate that nearly 30% of imipenem-resistant Pseudomonas aeruginosa strains possess a metallo-β-lactamase. Acquisition of a metallo-β-lactamase gene will invariably mediate broad-spectrum β-lactam resistance in P. aeruginosa, but the level of in vitro resistance in Acinetobacter spp. and Enterobacteriaceae is less dependable. Their clinical significance is further embellished by their ability to hydrolyze all β-lactams and by the fact that there is currently no clinical inhibitor, nor is there likely to be for the foreseeable future. The genes encoding metallo-β-lactamases are often procured by class 1 (sometimes class 3) integrons, which, in turn, are embedded in transposons, resulting in a highly transmissible genetic apparatus. Moreover, other gene cassettes within the integrons often confer resistance to aminoglycosides, precluding their use as an alternative treatment. Thus far, the metallo-β-lactamases encoded on transferable genes include IMP, VIM, SPM, and GIM and have been reported from 28 countries. Their rapid dissemination is worrisome and necessitates the implementation of not just surveillance studies but also metallo-β-lactamase inhibitor studies securing the longevity of important anti-infectives.
INTRODUCTION
The increase in antibiotic resistance among gram-negative bacteria is a notable example of how bacteria can procure, maintain, and express new genetic information that can confer resistance to one or several antibiotics. This genetic plasticity can occur both inter- and intragenerically. Gram-negative bacterial resistance possibly now equals or usurps that of gram-positive bacterial resistance and has prompted calls for similar infection control measures to curb their dissemination (134). Reports of resistance vary, but a general consensus appears to prevail that quinolone and broad-spectrum β-lactam resistance is increasing in members of the family Enterobacteriaceae and Acinetobacter spp. and that treatment regimes for the eradication of Pseudomonas aeruginosa infections are becoming increasingly limited (90, 106). For example, a 5-year longitudinal study involving many centers from Latin America indicated that year after year, P. aeruginosa resistance has continually risen to the point where approximately 40% are resistant to “antipseudomonal” drugs, including carbapenems (3). While the advent of carbapenems in the 1980s heralded a new treatment option for serious bacterial infections, carbapenem resistance can now be observed in Enterobacteriaceae and Acinetobacter spp. and is becoming commonplace in P. aeruginosa.
Gram-negative bacteria have at their disposal a plethora of resistance mechanisms that they can sequester and/or evince, eluding the actions of carbapenems and other β-lactams. The common form of resistance is either through lack of drug penetration (i.e., outer membrane protein [OMP] mutations and efflux pumps), hyperproduction of an AmpC-type β-lactamase, and/or carbapenem-hydrolyzing β-lactamases. Based on molecular studies, two types of carbapenem-hydrolyzing enzymes have been described: serine enzymes possessing a serine moiety at the active site, and metallo-β-lactamases (MBLs), requiring divalent cations, usually zinc, as metal cofactors for enzyme activity (20, 23, 24, 45).
The serine carbapenemases are invariably derivatives of class A or class D enzymes and usually mediate carbapenem resistance in Enterobacteriaceae or Acinetobacter spp. The enzymes characterized from Enterobacteriaceae include NmcA, Sme1-3, IMI-1, KPC1-3, and GES-2 (107, 133, 139, 142, 197, 200). Despite the avidity of these enzymes for carbapenems, they do not always mediate high-level resistance and not all are inhibited by clavulanic acid (108). In contrast, the oxacillinases have been characterized from Acinetobacter baumannii only and include OXA 23 to 27 (1, 16), OXA-40 (61), and OXA-48 (128). These enzymes hydrolyze carbapenems poorly but are able to confer resistance and are only partially inhibited by clavulanic acid. The class A and class D carbapenemases are encoded by genes that have been procured by the bacterium and can be chromosomally encoded (sometime associated with integrons) or carried on plasmids (108).
MBLs, like all β-lactamases, can be divided into those that are normally chromosomally mediated and those that are encoded by transferable genes. The early studies on chromosomally mediated MBLs mainly centered around Bacillus cereus (BC II) (83), and Stenotrophomonas maltophilia (L1) (178). However, primarily due to genomic sequencing, increasingly more chromosomally mediated genes are being discovered but are often found in obscure nonclinical bacteria (89, 103, 146, 149, 161).
Over the last decade there have been several articles summarizing the levels of MBLs in the bacterial community (20, 22, 23, 85, 86, 108, 117). However, in the past 3 to 4 years many new transferable types of MBLs have been studied and appear to have rapidly spread. In some countries, P. aeruginosa possessing MBLs constitute nearly 20% of all nosocomial isolates, whereas in other countries the number is still comparatively small (46, 80). In recent years MBL genes have spread from P. aeruginosa to Enterobacteriaceae, and a clinical scenario appears to be developing that could simulate the global spread of extended-spectrum β-lactamases. Moreover, given that MBLs will hydrolyze virtually all classes of β-lactams and that we are several years away from the implementation of a therapeutic inhibitor, their continued spread would be a clinical catastrophe. This review will focus on the biochemical and genetic characterization of MBLs and in particular transferable MBLs from Pseudomonas spp., Acinetobacter spp., and Enterobacteriaceae, including their epidemiology and methods for detecting them, and review those MBL experimental inhibitors studied thus far.
CLASSIFICATION OF MBLs
MBLs were first formally categorized from serine β-lactamases in 1980 in the classification scheme proposed by Ambler (2). At the time, very few MBLs had been sufficiently studied to merit inclusion, the most notable exceptions being L1 (from S. maltophilia) and BCII (from B. cereus). In 1989, Bush further classified MBLs into a separate group (group 3) according to their functional properties and remains the recommended referencing system for β-lactamases generally (21). This scheme was primarily based on substrate profiles (in particular imipenem hydrolysis), their sensitivity to EDTA, and their lack of inhibition by serine β-lactamase inhibitors. This scheme was updated in 1995 and further modified in 1997 to accommodate the growing number of group 3 enzymes continually being classified (24, 141). At the time, only two transferable types of MBLs had been studied, Bacteroides fragilis CcrA and IMP-1 from P. aeruginosa.
All MBLs hydrolyze imipenem, but their ability to achieve this varies considerably and the rate of hydrolysis may or may not correlate with the bacterium's level of resistance to carbapenems (92). Accordingly, this classification further segregated these enzymes into subgroups primarily on the basis of imipenem and other β-lactam hydrolysis (141). Essentially, group 3a enzymes possess a broad spectrum of activity; group 3b enzymes possess a preferential avidity for carbapenems; and group 3c enzymes hydrolyze carbapenems poorly compared to other β-lactam substrates. However, these enzymes possess the characteristic hallmark of being universally inhibited by EDTA as well as other chelating agents of divalent cations, a quintessential feature of MBLs that correlates with their mechanistic function (141).
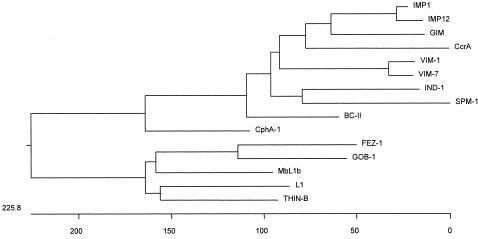
At a molecular level, the MBLs are a disparate group of proteins that make classifying and standardizing their structures virtually impossible (Fig. 1). Attempts have been made to subdivide class B enzymes based on sequence identity and other structural features (141). The phylogenetic tree (Fig. 1) indicates the relatedness of one enzyme to another as judged by nucleotide sequences. The rationale of class B1 is that the enzymes possess the key zinc coordinating residues of three histidines and one cysteine and accommodates the transferable MBLs IMP, VIM, GIM, and SPM-1. Class B2 include those that possess an asparagine instead of the histidine at the first position of the principal zinc-binding motif, NXHXD, and derive from Aeromonas spp. and the Serratia fonticola enzyme SFH-1. MBL L1 is the sole occupant of the class B3 enzymes, as it is singularly unique among all β-lactamases in being functionally represented as a tetramer (140).
FIG. 1.
Phylogeny of chromosomally encoded MBLs. Represented sequences of various MBLs were obtained from GenBank. IMP-1 and IMP-12 (most divergent from IMP-1) and VIM-1 and VIM-7 (most divergent from VIM-1) were also added for comparison. Signal peptides were removed prior to alignment. Sequences were aligned and phylogeny trees were constructed with Clustal W (PAM250 matrix; DNA Star) using the neighbor joining method.
Standardization of MBLs has been proposed based on a numbering scheme used to standardize class A β-lactamases; however, the class B enzymes are far more problematic due to their disparate nature, not least, their variability in the number of amino acids (48). This numbering scheme is primarily based on the alignment of key zinc-coordinating residues but unfortunately does not accommodate new sequences comfortably when they have substantial inserts in the middle of the protein, as has been recently shown for the P. aeruginosa enzyme SPM-1 loop (167). The numbering scheme has been recently updated to accommodate newly discovered MBLs (50).
CHROMOSOMALLY ENCODED MBLs
Some bacteria, usually from environmental habitats, ubiquitously carry MBLs, although there is much debate as to why this is the case. One argument is that over a considerable period of time, the bacteria have been exposed to β-lactams or β-lactam-type compounds and the bacteria have been conscripted to acquire and maintain these genes and their products. Another argument is that these enzymes perform a normal cellular function that is yet to be fully elucidated. Regardless of the viewpoint, a number of these MBL genes are inducible, and the majority of the bacteria carrying them are or can become highly resistant to β-lactams. Fortunately, these organisms are opportunistic pathogens, and with the arguable exception of S. maltophilia and Bacillus anthracis, seldom cause serious infections. Chromosomally encoding MBL bacteria include B. cereus (BC II) (83), Bacillus anthracis (30), S. maltophilia (L1) (178), Aeromonas hydrophilia (CphA) (92), Chryseobacterium meningosepticum (BlaB or GOB-1) (147), Chryseobacterium indologenes (IND-1) (13), Legionella gormannii (FEZ-1) (15), Caulobacter crescentus (Mbl1B) (161), Myroides spp. (TUS-1, MUS-1) (89), and Janthinobacterium lividium (THIN-B) (146) Flavobacterium johnsoniae (JOHN-1) (103) and S. fonticola (SFH-1) (149) are listed in Table 1.
TABLE 1.
Chromosomally encoded MBLs
| Subgroup | Organism | Enzyme name | Reference |
|---|---|---|---|
| B1 | Bacillus cereus | BCII-5/B/6 | 83 |
| BCII-569/H | 65 | ||
| Bacillus anthracis | Bla2 | 30 | |
| Alkalophilic Bacillus spp. | Bce 170 | 65 | |
| Chryseobacterium indologenes | IND-1 | 10 | |
| IND-2, 2a, 3, 4 | 13 | ||
| Chryseobacterium meningosepticum | BlaB | 148 | |
| BlaB2, BlaB3 | 188 | ||
| BlaB4-8 | 12 | ||
| Chryseobacterium gleum | CGB-1 | 12 | |
| Myroides odoratus | TUS-1 | 89 | |
| Myroides odoratimimus | MUS-1 | 89 | |
| Flavobacterium johnsoniae | JOHN-1 | 103 | |
| B2 | Aeromonas hydrophilia | CphA | 92 |
| Aeromonas veronii | ImiS | 180 | |
| AsbM1 | 196 | ||
| Serratia fonticola | SFH-1 | 149 | |
| B3 | Caulobacter crescentus | Mb11B | 161 |
| CAU-1 | 42 | ||
| Janthinobacterium lividium | THIN-B | 146 | |
| Legionella gormanii | FEZ-1 | 15 | |
| Chryseobacterium meningosepticum | GOB-1-7 | 13 | |
| Stenotrophomonas maltophilia | L1a | 178 | |
| L1-BlaS | 151 | ||
| L1c, L1d, L1e | 7 |
Generally speaking, the chromosomal MBLs from a particular species or genus vary little from one another. The most notable exception to this is the MBLs from Chryseobacterium meningosepticum, where the BlaB and GOB-type enzymes vary considerably (11% identity) and accordingly have been divided into separate subclasses (Table 1). The chromosomally mediated enzymes are also often coregulated with serine β-lactamases. For instance, both A. hydrophila and Aeromonas veronii bv. sobia produce three β-lactamases, a penicillinase, a cephalosporinase, and an MBL, all of which are overexpressed when high-level (derepressed) β-lactam-resistant mutants are selected. Similar phenomena are found with other bacteria, not least S. maltophilia, where high-level β-lactam resistance is primarily due to overexpression of the L1 MBL and its chromosomal class A enzyme L2 (179). This derepression usually occurs at a frequency of 1 in 105 to 107 and can happen in vivo under β-lactam therapy (179).
One group of MBL genes that are often described as chromosomal but are in fact transferable are those associated with Bacteroides spp. Compared to other anaerobic bacteria, B. fragilis is relatively resistant to β-lactams, not least due to the potential production of its MBL, designated CfiA or sometimes CcrA (143, 164). The cfiA MBL gene was first genetically characterized in 1990 and is one of the most intensely studied with respect to its mechanism of catalysis and structure-function properties, often providing a paradigm for similar enzymes (34, 37, 183, 185). cfiA is often quiescent and requires a surrogate sequence to provide an adequate promoter, thereby potentiating expression of the structural gene. Insertion elements, such as IS942, IS1186, and IS4351, have been shown to embed immediately upstream of the ribosome-binding site, providing enhanced transcriptional capabilities for the cfiA gene (125, 126, 143). Studies have shown that in most countries, the silent cfiA gene is present in B. fragilis at approximately 2 to 4% of strains (125, 191).
GENETIC APPARATUS OF TRANSFERABLE MBLs
The dissemination of MBL genes is thought to be driven by the regional consumption of extended-spectrum cephalosporins or carbapenems (80, 87). Most, if not all, genes encoding IMP- and VIM-type as well as GIM-1 are found as gene cassettes in class 1 integrons (29, 79, 129, 131, 156, 192), although IMP MBL genes are also found on class 3 integrons (5, 32). Integrons are capable of procuring gene cassettes via a site-specific recombination event between two DNA sites, one in the integron and one in the gene cassette. Integrons consist of three regions: the 5′ conserved region, the 3′conserved region, and a variable region. The 5′ region consists of the integrase gene (intl), its adjacent recombination site (attI), and a promoter, which facilitates expression of the procured gene cassettes in the variable region. The 3′ conserved region often consists of a partially deleted qac gene (qacEΔ1) fused to a sul gene and, correspondingly, confers resistance to antiseptics and sulfonamide, respectively.
Gene cassettes are small pieces of circular DNA, approximately 1 kb in size, comprising a single gene together with a recombination site termed a 59-base element (14). blaVIM genes from some European counties have been found with a truncated 59-base element and the gene cassettes are likely to be “fused” (114, 181). In most instances, this involves the MBL gene and an aacA4 gene that encodes kanamycin, neomycin, amikacin, and streptomycin resistance. Therefore, both aminoglycosides and β-lactams will select clinical bacteria harboring this fused gene cassette, further compromising these antibiotic regimens (181).
While gene cassettes carrying aminoglycoside and β-lactam resistance genes can freely move from one integron to another, they cannot by themselves move from one organism to another and require the assistance of other genetic elements such as plasmids and transposons (14). The majority of MBL genes are found on plasmids usually between 120 and 180 kb; however some, such as blaVIM-7 from the United States, are carried on a conjugative plasmid of 24 kb (166). The in vivo transfer of large native plasmids that carry MBL genes probably involves the promiscuity of the accommodating bacteria as much as the size of the plasmid or what other genes, besides the MBL gene, are carried on it. At present, there is very little information as to what other genes are carried on these plasmids or whether the functions they encode will aid or hinder their bacterial host. A recent study evaluated the possibility for an increase in virulence afforded by the blaIMP gene using cell monolayers (4). While there was no increase in virulence, the study examined the effect of the MBL gene only and not the native plasmids normally accommodating these genes.
The genetic context of MBLs not only represents a measure of its plasticity but also enables reasonable speculation as to how transferable MBL genes spread internationally. In 2003, the first account of an MBL gene (blaIMP-13) and its integron were reported to be embedded in a Tn5051-type transposon from an Italian P. aeruginosa isolate (165). The insertion of the elements containing blaIMP-13 and blaVIM-2 from a P. aeruginosa from Poland are in an identical site. Moreover, the tnpR genes of the transposon from both sites are identical, suggesting that the transposon is responsible for dissemination of the class 1 integron, which then procured the different MBL genes (165). These data are corroborated by the fact that the P. aeruginosa isolates possess different pulsed-field gel electrophoresis patterns and that there was no evidence of plasmid carriage. Some of the Japanese integron mediated blaVIM-2 have also been speculated to be embedded in transposons (199).
Not all MBL genes are associated with integrons or transposons. The genetic context of blaSPM-1 was shown to be unique, being adjacent to genes closely related to Salmonella enterica serovar Typhimurium and not associated with an integron or transposon (167). Interestingly, blaSPM-1 and its surrounding genes are part of a mobile genomic pathogenicity island and found on a plasmid of approximately 180 kb. Salmonella pathogenicity islands have been associated with mobile common regions that have also been associated with other mobile elements called SXT regions. These regions can be mobilized under bacterial stress, as has been shown recently when resistant elements linked to SXT increased 300-fold when the bacterium was exposed to fluoroquinolones (9, 67). Further analysis of the Brazilian P. aeruginosa isolates demonstrated that the upstream DNA contained common regions or CR elements, in this case CR4 (130). Compared to integrons and transposons, very little is known about CR elements, particularly how they facilitate the mobilization of genes despite the fact that they have been associated with antibiotic resistance genes (115). SPM-1 genes in P. aeruginosa from Sao Paolo also contain common regions, although these are very different from the isolates from Recife, suggesting a different genetic origin (M. A. Toleman, unpublished data). At present, the genetic region immediately surrounding blaSPM-1 is not associated with coresistance to other antibiotics.
BIOCHEMISTRY OF MBLs
MBLs and serine β-lactamase both mediate resistance to β-lactams by cleaving the amide bond of the β-lactam ring; however, the way in which the two groups of enzymes achieve this differs considerably (45). MBLs possess a distinct set of amino acids that define the finite architecture of the active site which coordinates the zinc ions. The zinc ions in turn usually coordinate two water molecules necessary for hydrolysis (186). The principal zinc-binding motif is histidine-X- histidine-X-aspartic acid (HXHXD), which is common to most MBLs apart from the class B2 enzymes (141). Without exception, the preferred metal is zinc, and while most MBLs accommodate two zinc ions in their active site, the class B2 enzymes possess just a single zinc ion (141). The proposed mechanism of hydrolysis suggests that the active site orients and polarizes the β-lactam bond to facilitate nucleophilic attack by zinc-bound water/hydroxides (94, 162, 184).
The MBL mechanism of hydrolysis is complex and varies from one MBL to another (162). Elucidation of the crystal structure of MBLs has offered invaluable insights into their catalytic mechanisms. Despite the fact that MBLs may share less than 25% amino acid identity with one another, they all share the unique αββα fold and their active site architecture is virtually superimposable. While there are many crystal structures of MBLs to facilitate our understanding of their hydrolytic mechanism, there are only two structures of MBLs complexed to β-lactams (J. Spencer, personal communication) (49), leaving much room for speculation about their catalytic steps (27, 28, 33, 34, 51, 52, 175). It appears that most MBLs have a loop that is flexible and this is thought to facilitate binding and hydrolysis of the β-lactam substrates.
Unlike serine β-lactamases, MBLs possess a wide plastic active-site groove and accordingly can accommodate most β-lactam substrates, facilitating their very broad spectrum of activity. They are also impervious to the impeding effects of serine inhibitors such as clavulanic acid and sulbactum that are often treated as poor substrates (111, 112). Interestingly, none of the MBLs hydrolyze aztreonam particularly well, and it has been speculated that it could be considered a therapeutic MBL inhibitor (see section on inhibitors). However, in studies of animals with pneumonia caused by P. aeruginosa producing VIM-2, infection could not be eradicated with aztreonam even when the animals were given high drug doses (11).
The affinity of an enzyme for a substrate is reported as the Km, the enzyme's ability to turn over the substrate is the kcat, and kcat/Km is a measure of the enzyme's overall catalytic efficiency. Table 2 shows the Km, kcat and kcat/Km values of the MBLs GIM-1, IMP-1, VIM-1, VIM-2, and SPM-1 derived under similar experimental conditions.
TABLE 2.
Steady-state kinetic values of GIM-1 (29), IMP-1 (78), VIM-1 (41), VIM-2 (41), and SPM-1 (102) against a range of β-lactams
| Antibiotic | GIM-1
|
IMP-1
|
VIM-1
|
VIM-2
|
SPM-1
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | |
| Penicilllin | 6.6 | 46 | 0.14 | 320 | 520 | 0.62 | 29 | 841 | 0.034 | 55.8 | 49 | 1.14 | 108 | 38 | 2.8 |
| Ampicillin | 3.3 | 20 | 0.16 | 950 | 200 | 4.8 | 37 | 917 | 0.04 | 117 | 72 | 1.6 | |||
| Carbenicillin | 4.1 | 170 | 0.02 | NDa | ND | 0.02 | 167 | 75 | 2.2 | 74 | 814 | 0.09 | |||
| Azlocillin | ND | ND | ND | 1,525 | 123 | 12 | 53 | 147 | 0.35 | ||||||
| Piperacillin | 6.9 | 69 | 0.10 | ND | ND | 0.72 | 1,860 | 3,500 | 0.53 | 32.7 | 72 | 0.45 | 117 | 59 | 2 |
| Ticarcillin | 2.3 | 57 | 0.04 | 1.1 | 740 | 0.0015 | 452 | 1,117 | 0.41 | 31.7 | 46 | 0.69 | ND | <0.35 | ND |
| Nitrocefin | 5.8 | 12 | 0.47 | 63 | 27 | 2.3 | 95 | 17 | 5.6 | 0.53 | 4 | 0.12 | |||
| Cephalothin | 16 | 22 | 0.72 | 48 | 21 | 2.4 | 281 | 53 | 5.3 | 56.2 | 44 | 1.28 | 43 | 4 | 11.7 |
| Cefuroxime | 5.9 | 7 | 0.80 | 8 | 37 | 0.22 | 324 | 42 | 7.7 | 12.1 | 22 | 0.55 | 37 | 4 | 8.8 |
| Cefoxitin | 8.3 | 206 | 0.04 | 16 | 8b | 2 | 26 | 131 | 0.2 | 3 | 24 | 0.12 | 8 | 2 | 4 |
| Ceftazidime | 18 | 31 | 0.58 | 8 | 44 | 0.18 | 60 | 794 | 0.076 | 89 | 98 | 0.90 | 28 | 46 | 0.6 |
| Cefotaxime | 1.1 | 4 | 0.24 | 1.3 | 4b | 0.35 | 169 | 247 | 0.68 | 27.5 | 32 | 0.86 | 16 | 9 | 1.9 |
| Cefepime | 17 | 431 | 0.04 | 7 | 11b | 0.66 | 549 | 145 | 3.8 | 4.7 | 184 | 0.03 | 18 | 18 | 1 |
| Imipenem | 27 | 287 | 0.09 | 46 | 39 | 1.2 | 2.0 | 1.5 | 1.3 | 9.9 | 10 | 0.99 | 33 | 37 | 1 |
| Meropenem | 2.7 | 25 | 0.11 | 50 | 10 | 0.12 | 13 | 48 | 0.27 | 1.4 | 5 | 0.28 | 63 | 281 | 0.22 |
| Moxalactam | 14 | 1,035 | 0.01 | 88 | 10b | 8.8 | 14.8 | 80 | 0.18 | 13 | 97 | 0.13 | |||
| Aztreonam | ND | ND | ND | >0.01 | >1,000 | <1 × 10−5 | <0.01 | >1,000 | <1 × 10−5 | <0.5 | ND | ND | ND | <0.3 | ND |
| Clavulanic acid | ND | ND | ND | ND | >0.1 | ND | |||||||||
| Tazobactam | ND | ND | ND | >1,000 | >3.98 | 0.0039 | 5.3 | 337 | 0.016 | 0.6 | 3 | 0.2 | |||
ND, data could not be determined.
Km was obtained as the Ki value.
The data in Table 2 demonstrate that while these enzymes share common features in fold and active site architecture, their ability to bind and hydrolyze β-lactams varies considerably. The most noticeable example of this is the difference between VIM-1 and VIM-2, which are structurally very similar. For instance, VIM-2 tends to bind most β-lactams more tightly than VIM-1 and possesses significantly lower Km values for benzylpenicillin, ampicillin, piperacillin, mezocillin, ticarcillin, cefalothin, cefoxitin, cefotaxime, ceftazidime, cefpirome, moxalactam, and meropenem. The most notable exception is imipenem, where VIM-1 and VIM-2 possess Km values of 1.5 μM and 10 μM, respectively (41). However, VIM-1 is capable of hydrolyzing most β-lactams (piperacillin, azlocillin, ticarcillin, cefaloridine, cefalothin, cefuroxime, cefotaxime, ceftazidime, cepirome, and meropenem) more efficiently than VIM-2. Again, the most notable exception is imipenem, where VIM-1 and VIM-2 possess kcat values of 0.2/s and 34/s, respectively. Docquier et al. speculate that these substantial kinetic differences are due to amino acid substitutions near or at the active site, namely, histidine/tyrosine at position 224 and serine/arginine at position 228 (41).
Table 2 indicates that SPM-1 hydrolyzes most therapeutic β-lactams well and is generally a more efficient enzyme (higher kcat/Km values) than IMP-1 (78) and GIM-1 (29), the exceptions being ampicillin, imipenem, and moxalactam for IMP-1. SPM-1 also binds cephalosporins, particularly cefoxitin, more tightly (lower Km values) than penicillins and carbapenems. GIM-1 primarily functions as a penicillinase with moderate activity against narrow-spectrum cephalosporins and carbapenems, although it does bind most β-lactams tightly with the notable exceptions of imipenem (Km 287 μM), cefepime (Km 431 μM), and moxalactam (Km 1,035 μM).
The kinetic values demonstrated by the MBLs raise many questions about why there should be such variation in binding and hydrolysis when these enzymes are very similar. Transient kinetic studies have tried to probe the catalytic mechanism of MBLs in binding and hydrolyzing β-lactam substrates. However, most of these studies have utilized the chromogenic substrate nitrocefin, which was shown to be atypical for some enzymes (162). Furthermore, many studies have centered on the MBLs BCII and CcrA rather than the more clinically relevant enzymes, those encoded by highly transferable genes (18, 95, 163).
TRANSFERABLE MBLs
IMP-Type MBLs
The origin and bacterial hosts of blaIMP genes are summarized in Table 3. The first indication of mobile MBLs was with the discovery of P. aeruginosa strain GN17203 in Japan in 1988 (187). The isolate possessed an imipenem MIC of 50 μg/ml as well as resistance to extended-spectrum cephalosporins e.g., a ceftazidime MIC of >400 μg/ml. The resistance allele was found on a transferable conjugative plasmid that could be readily mobilized to other Pseudomonas strains. Three years later an identical gene was found in Serratia marcescens strain Tn9106 isolated from a urinary tract infection at Aichi Hospital in Okazaki, Japan (110). Two years later, the same gene was characterized from S. marcescens (AK9373) from a hospital in the city of Anjyo, situated next to the city of Okazaki (5). This IMP-1 allele, blaIMP-1. was found within a class 3 integron adjacent to a aac(6′)Ib-like gene and was harbored on a large plasmid (120 kb).
TABLE 3.
Origins and bacterial hosts of the IMP-type MBLs
| IMP-type MBL | Host | Origin | Integron | Reference(s) or accession no. |
|---|---|---|---|---|
| IMP-1 | Pseudomonas aeruginosa | Japan | Class 1 | 158, 187 |
| Japan | Class 3? | 157 | ||
| Japan | ? | 62, 190 | ||
| Brazil | Class 1 | Toleman, unpublished data | ||
| Korea | ? | 80 | ||
| Pseudomonas putida | Japan | 158 | ||
| Japan | Class 3? | 156, 158 | ||
| Japan | ? | 62, 201 | ||
| Singapore | ? | 75 | ||
| Serratia marcescens | Japan | Class 3? | 5, 156 | |
| Japan | Class 1 | 66, 68, 158 | ||
| Japan | ? | 190, 62 | ||
| Acinetobacter baumannii | Korea | ? | 80 | |
| Japan | Class 1 | 158 | ||
| England? | ? | 172 | ||
| Pseudomonas fluorescens | Singapore | ? | 75 | |
| Japan | Class 1 | 158 | ||
| Japan | ? | 62 | ||
| Pseudomonas stutzeri | Japan | Class 1 | 158 | |
| Klebsiella pneumoniae | Japan | Class 3? | 156 | |
| Japan | Class 1 | 158 | ||
| Klebsiella oxytoca | Japan | Class 3? | 156 | |
| Achromobacter xylosoxidans | Japan | Class 1 | 158 | |
| Japan | Class 1 | 158 | ||
| Alcaligenes xylosoxidans | Japan | Class 1 | 158 | |
| Alcaligenes faecalis | Japan | ? | 62 | |
| Citrobacter freundii | Japan | Class 1 | 158 | |
| Japan | Class 1 | 158 | ||
| Enterobacter aerogenes | Japan | Class 1 | 158 | |
| Enterobacter cloacae | Japan | ? | 6 | |
| Escherichia coli | Japan | Class 1 | 158 | |
| Proteus vulgaris | Japan | Class 1 | 158 | |
| Providencia rettgeri | Japan | Class 1 | 158 | |
| Acinetobacter spp. | England | ? | 174 | |
| IMP-2 | Acinetobacter baumannii | Italy | Class 1 | 144 |
| Japan | Class 1 | 158 | ||
| Acinetobacter lwoffii | Japan | Class 1 | 158 | |
| Pseudomonas aeruginosa | Japan | Class 1 | 158 | |
| IMP-3 | Shigella flexneri | Japan | Class 1 | 68 |
| IMP-4 | Acinetobacter baumannii | Hong Kong | Class 1 | 31 |
| Citrobacter freundii | Class 1 | 60 | ||
| Australia | Class 1 | 132 | ||
| Australia | Class 1 | 132 | ||
| Pseudomonas aeruginosa | China | Class 1 | 60 | |
| Australia | ? | 123 | ||
| IMP-5 | Acinetobacter baumannii | Portugal | Class 1 | 38 |
| IMP-6 | Acinetobacter baumannii | Brazil | ? | 47 |
| Serratia marcescens | Japan | Class 1 | 198 | |
| IMP-7 | Pseudomonas aeruginosa | Canada | Class 1 | 54 |
| Malaysia | Class 1 | 63 | ||
| IMP-8 | Enterobacter cloacae | Taiwan | Class 1 | 193 |
| Klebsiella pneumoniae | Taiwan | Class 1 | 194 | |
| IMP-9 | Pseudomonas aeruginosa | China | Class 1 | Accession no. AY033653 |
| IMP-10 | Pseudomonas aeruginosa | Japan | Class 1 | 69 |
| Alcaligenes xylosoxidans | Japan | Class 1 | 69 | |
| IMP-11 | Pseudomonas aeruginosa | Japan | ? | Accession no. AB074437 |
| Acinetobacter baumannii | Japan | ? | Accession no. AB074436 | |
| IMP-12 | Pseudomonas putida | Italy | Class 1 | 43 |
| IMP-13 | Pseudomonas aeruginosa | Italy | Class 1 | 165 |
| IMP-14 | ? | ? | ? | Assigned (http://www.lahey.org/studies/) |
| IMP-15 | ? | ? | ? | Assigned (http://www.lahey.org/studies/) |
| IMP-16 | Pseudomonas aeruginosa | Brazil | Class 1 | Accession no. AJ584652 |
| IMP-17 | ? | ? | ? | Assigned (http://www.lahey.org/studies/) |
| IMP-18 | Pseudomonas aeruginosa | USA | ? | 59; accession no. AY780674 |
A further study from seven general hospitals from Japan in 1993 identified four S. marcescens which carried blaIMP-1 (66). A further pan-Japanese hybridization study screened 3,700 P. aeruginosa isolates collected between 1992 and 1994 from 17 general hospitals with blaIMP-1 probes (157). Fifteen strains from five hospitals from different geographical areas probed positive with blaIMP-1. Interestingly, when the imipenem MICs of the MBL-positive isolates were tested, they varied from 2 mg/liter to 128 mg/liter, which suggests that acquisition of MBLs alone does not ubiquitously confer resistance to enems. In a further study of 54 isolates possessing ceftazidime resistance (MIC > 128 mg/liter) from 18 hospitals in Japan (157), 22 additional blaIMP-1-positive isolates were detected by PCR. These positive bacterial isolates included nine S. marcescens, two Achromobacter xylosoxidans, one Pseudomonas putida, and one Klebsiella pneumoniae. PCR detected the intI3 gene in 33 of the 42 (78.5%) blaIMP-positive strains, implicating the involvement of class 3 integrons with blaIMP-1. Further Japanese reports denote the wide dispersion of MBL genes principally carried on class 3 integrons in 16 different species of gram-negative bacteria (Table 3) (158), which has also been indicated in a more local setting (62, 190). Studies have shown that MBL-positive P. putida strains possessing identical genotypes have remained in Japanese hospital environments for extended periods of time (62, 201).
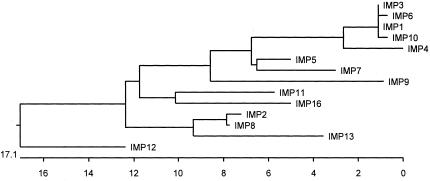
During a survey of clinical isolates of Shigella flexneri, S. marcescens, P. aeruginosa, and Alcaligenes spp. for the presence of MBLs, three minor variants of IMP-1 have been identified in Japan, IMP-3 (68), IMP-6 (198), and IMP-10 (69) (Fig. 2). IMP-3 has two amino acid changes from IMP-1 and was identified in an S. flexneri isolate. Genetic and kinetic studies determined that the substitution of glycine for serine at position 196 caused a reduction in the activity against penicillin (68). This same amino acid change is observed in IMP-6, which displays not only reduced activity against penicillin G and piperacillin but also a higher level of meropenem hydrolysis compared to imipenem, the opposite to IMP-1 (198) (Fig. 2).
FIG. 2.
Phylogeny of IMP-type MBLs. Signal peptides were removed prior to alignment. Sequences were aligned and phylogeny trees were constructed with Clustal W (PAM250 matrix; DNA Star) using the neighbor-joining method.
IMP-10 was discovered as a result of a study of IMP-producing isolates of P. aeruginosa and Alcaligenes spp. collected from 1995 to 2001 (69). The position of the changed amino acid found in both IMP-3 and IMP-6 corresponds to the same amino acid (glycine) in other innate MBLs such as BCII, leading to the hypothesis that IMP-3 may actually be the progenitor of IMP-1 rather than just being a variant of IMP-1 (Fig. 2) (68). The blaIMP-10 gene was found to be plasmid mediated in one P. aeruginosa isolate and chromosomally mediated in one P. aeruginosa and one Achromobacter xylosoxidans isolate. The blaIMP-10 gene differs from blaIMP-1 by a single base change responsible for a change of phenylalanine for valine at position 49. This amino acid change caused a marked reduced hydrolysis of penicillins but, unlike the changes responsible for IMP-3 and IMP-6, did not alter the carbapenem hydrolysis profile.
The belief that mobile MBLs genes were solely a distant Japanese problem was negated with the advent of blaIMP-2 in 1997 and blaIMP-5 in 1998 from Italy and Portugal, respectively (36, 38). Subsequent reports described in detail the blaIMP genes, their chromosomal location, and the details of their genetic environments (40, 144). IMP-2 had 36 amino acid changes relative to IMP-1, and IMP-5 had 17 amino acid changes relative to IMP-1, and both were different in terms of their genetic context, blaIMP-2 being found next to two aminoglycoside resistance-conferring alleles (aacA4 followed by aadA1) and blaIMP-5 being the sole gene cassette.
Subsequent to these reports, two other IMP variants have been described from Italy. IMP-12 was produced by a P. putida clinical isolate from Varesse in 2000 and its allele, blaIMP-12, was located on a 50-kb nontransferable plasmid (43). IMP-12 is highly divergent from IMP-1, possessing 36 different amino acids and displaying poor activity against penicillin. blaIMP-13 was cloned from a P. aeruginosa clinical specimen isolated in Rome and was 19 amino acids different from IMP-1 (165) and chromosomally encoded. The blaIMP-13 allele has subsequently been found on a plasmid (M. A. Toleman, unpublished results) and has been associated with a minor epidemic in a hospital in the south of Italy (98).
The differences between the European IMPs and those from Southeast Asia cannot be reconciled with global dissemination of IMP alleles from Japan. It is more likely that these alleles represent local emergence. However, IMP-1 has very recently been found in England (172, 174) in isolates of Acinetobacter junii and A. baumannii. The A. junii isolate carrying blaIMP-1 is identical in amino acid sequence, although the nucleotide sequence contains several silent changes. The A. baumanii strain was isolated from a female who had recently been on holiday in Spain and hospitalized there for 4 weeks before being transferred to Britain, and the origin of the resistant isolate and its genetic details have yet to be fully described.
Retrospective studies on resistant isolates collected as early as 1994 in Hong Kong and 1995 in Canada determined that the carbapenem resistance was due to IMP-7 (P. aeruginosa) in Canada (54) and IMP-4 (Acinetobacter spp.) in Hong Kong (31). The MBL gene blaIMP-4 was detected in 66% of imipenem-resistant strains collected between 1994 and 1998 at the Prince of Wales hospital in Hong Kong (31). IMP-4 was 10 amino acids different from IMP-1 and 37 amino acids different from IMP-2. blaIMP-4 was harbored on a plasmid and integron encoded along with three other resistance genes (qacG2, aacA4, and catB3) (64). Acinetobacter strains harboring IMP-4 were found in 1997 and 1998 at a prevalence of ≈14% of all Acinetobacter strains and disappeared in 1999, but the reasons for this is not clear (64). blaIMP-4 was subsequently (1998 and 2000) found in Citrobacter youngae and P. aeruginosa isolates from Guangzhou in mainland China. Guangzhou is close to Hong Kong, which suggests local dissemination of this IMP allele (60). IMP-4 has now been found in Australia in Escherichia coli, Klebsiella pneumoniae, and P. aeruginosa, possibly “imported” from Southeast Asia (123, 132).
IMP-7 was responsible for a clonal P. aeruginosa outbreak in Canada at two hospitals. The integron was cloned, and blaIMP-7 was found in the third gene cassette position with other gene cassettes encoding aminoglycoside resistance (orf1, aacC4, and aacC1) (54). Subsequently, an identical blaIMP-7 gene was found in 1999 in a carbapenem-resistant P. aeruginosa isolate in Malaysia (63).
Further examination for MBL-positive isolates in Japan revealed that there is very little evidence of spread of these resistance alleles from Japan. In Taiwan, the IMP variant IMP-8 has been found in a single institution the National Cheung Kung Hospital. Here, the blaIMP-8 gene cassette was found in a class 1 integron next to aacA4 in a K. pneumoniae specimen isolated in 1998 (194). IMP-8 is more similar to IMP-2 (two amino acids different) than IMP-1, suggesting no clear link with the Japanese IMP-1-producing isolates. Furthermore, hybridization studies with a blaIMP-8 probe found blaIMP-8 genes in 28.5% of 140 ceftazidime-resistant K. pneumoniae strains isolated from 1999 to 2000, although only 12.5% of these were carbapenem resistant (194). The blaIMP-8 gene was also used to probe 9,082 enteric isolates, and 36 of 1,261 Enterobacter cloacae isolates were positive (193). Recently, IMP-1 and IMP-1-like MBLs have been reported from Singapore and Korea, respectively. IMP-1-producing K. pneumoniae was reported in a large tertiary-care hospital in Singapore in 2001 (74). Since then, one other blaIMP-1-harboring isolate was detected in a Pseudomonas fluorescens isolate from 2001 (75). However, genetic analysis found that this IMP-1-producing allele was, like the blaIMP-1 allele found in A. junii from Britain, more similar in nucleotide sequence to blaIMP-3.
In Korea, two studies have revealed that Korea has a considerable problem with carbapenem-resistant isolates due to MBLs. One study found IMP-like MBL genes in 35% of 130 carbapenem- or ceftazidime-resistant isolates (including two P. aeruginosa) (109). Another study examined isolates from 28 hospitals located in six cities and provinces across Korea in 2000 to 2001 (80). MBLs were present in 60% of all Korean hospitals tested, and IMP-like MBLs in 41.7% of all hospitals, accounting for 28.9% of all Acinetobacter isolates. Imipenem resistance has risen in Korea from 6% of all isolates in 1996 to 19% in 2001. Unfortunately, the lack of genetic data on these strains does not give us the necessary information to say whether these IMP-like genes are actually evidence of dissemination from Japan.
The only information in the scientific literature on IMP-like MBLs in the Americas has principally come from Brazil, where there is a serious problem with isolates of multidrug-resistant Acinetobacter spp. Sequencing of a PCR product amplified with IMP-specific primers gave 100% identity with blaIMP-6. However, the PCR product represented only partial gene sequence (47). Further recent studies of isolates collected through the SENTRY worldwide antimicrobial surveillance program have identified five further isolates from Brazil harboring IMP-1 and a new divergent IMP allele, blaIMP-16 (Fig. 2) (97). The most recent IMP-like MBL (IMP-18) has been found in a P. aeruginosa isolate from Las Cruces, New Mexico (59).
VIM-Type MBLs
The second dominant group of acquired MBLs is the VIM-type enzymes (Table 4). VIM-1 was described first in Verona, Italy, from a P. aeruginosa isolate (79). This clinical isolate, recovered in 1997, was resistant to a series of β-lactams, including piperacillin, ceftazidime, imipenem, and aztreonam. In particular, the MIC of imipenem was >128 μg/ml. Biochemical analysis performed from a crude extract of a culture of this strain revealed a carbapenem-hydrolyzing activity that was inhibited by EDTA and restored upon addition of Zn2+. These observations strongly suggested production of a metalloenzyme. The β-lactamase gene was cloned, and the deduced amino acid sequence revealed a 266-amino-acid preprotein with a pI of 5.3. VIM-1 (Veronese imipenemase) is distantly related to other metalloenzymes. It is most closely related to BCII from B. cereus, sharing only 39% amino acid identity (Fig. 1) (79). The hydrolytic profile of VIM-1 analyzed from a culture of a recombinant E. coli strain expressing this enzyme is typical of class B enzymes, hydrolyzing most β-lactams except aztreonam. Resistance to the monobactam aztreonam, in the original P. aeruginosa isolate, was likely due to a plethora of resistance mechanisms such as efflux and cephalosporinase hyperproduction. As found for blaIMP genes, the blaVIM-1 gene was integrated as a gene cassette into a class 1 integron (79). This integron carried an integrase gene typical of class 1 integrons and, in addition to the blaVIM-1 gene cassette, an aacA4 gene cassette encoding resistance to aminoglycosides. In this P. aeruginosa isolate, the blaVIM-1-containing integron was probably located on the chromosome (79).
TABLE 4.
Origins and bacterial hosts of the VIM-type MBLs
| VIM-type MBL | Host | Origin | Integron present | Reference or accession no. |
|---|---|---|---|---|
| VIM-1 | Pseudomonas aeruginosa | Italy | + | 79 |
| Achromobacter xylosoxidans | Italy | + | 145 | |
| Pseudomonas putida | Italy | + | 87 | |
| Escherichia coli | Greece | + | 154 | |
| France | + | Personal data | ||
| Klebsiella pneumoniae | Greece | + | 53 | |
| VIM-2 | Pseudomonas aeruginosa | France | + | 127 |
| Greece | + | 135 | ||
| Italy | + | 114 | ||
| Japan | + | 199 | ||
| Korea | 81 | |||
| Portugal | + | 26 | ||
| Spain | ? | 137 | ||
| Croatia | + | 152 | ||
| Poland | + | 181 | ||
| Chile | + | 96 | ||
| Venezuela | + | 96 | ||
| Argentina | ? | 113 | ||
| USA | + | 150 | ||
| Acinetobacter baumannii | Korea | + | 203 | |
| Enterobacter cloacae | Korea | + | 70 | |
| Serratia marcescens | Korea | + | 203 | |
| Pseudomonas putida | Korea | ? | 81 | |
| Japan | + | 158 | ||
| Pseudomonas fluorescens | Chile | + | 96 | |
| Pseudomonas stutzeri | Taiwan | ? | 192 | |
| Acinetobacter genomosp. 3 | Korea | + | 203 | |
| Achromobacter xylosoxidans | Japan | + | 158 | |
| Citrobacter freundii | Taiwan | ? | 193 | |
| VIM-3 | Pseudomonas aeruginosa | Taiwan | ? | 192 |
| VIM-4 | Pseudomonas aeruginosa | Greece | + | 136 |
| Sweden | ? | 55 | ||
| Poland | + | 116 | ||
| Enterobacter cloacae | Italy | ? | 88 | |
| Klebsiella pneumoniae | Italy | ? | 88 | |
| VIM-5 | Klebsiella pneumoniae | Turkey | + | Unpublished data |
| Pseudomonas aeruginosa | Turkey | + | 8 | |
| VIM-6 | Pseudomonas putida | Singapore | ? | 75 |
| VIM-7 | Pseudomonas aeruginosa | USA | + | 167 |
| VIM-8 | Pseudomonas aeruginosa | Columbia | ? | AY524987.1 |
| VIM-9 | Pseudomonas aeruginosa | United Kingdom | ? | AY534988.1 |
| VIM-10 | Pseudomonas aeruginosa | United Kingdom | ? | AY524989.1 |
| VIM-11a | Pseudomonas aeruginosa | Argentina | ? | AY605049.1 |
| VIM-11b | Pseudomonas aeruginosa | Italy | ? | AY635904.1 |
Subsequently, a blaVIM-1 gene was found in Achromobacter xylosoxidans in the same hospital in Verona (145). This isolate exhibited resistance to all β-lactams, including carbapenems, and harbored a 30-kb nonconjugative plasmid carrying a class 1 integron. This integron, In70, contained four gene cassettes and three different aminoglycoside resistance genes (aacA4, aphA15, and aadA1) located downstream of the blaVIM-1 gene cassette. As observed in In31 carrying the blaIMP-1 gene cassette, In70 was flanked by inverted repeats and a truncated tni module was detected in its 3′ part. Thus, In70 can be also considered a member of the group of class 1 integrons associated with defective transposon derivatives originating from Tn402-like elements.
Additionally, VIM-1 has been detected in three clonally related P. putida isolates in Italy as a source of nosocomial infections, underlining that environmental isolates are either the source or at least vectors of MBLs (87). These isolates were recovered in the same hospital in Varese, Italy. The MICs of imipenem and meropenem were >32 μg/ml, whereas that of aztreonam was 32 μg/ml. These isolates harbored a ca. 52-kb plasmid that encoded the VIM-1 determinant (87).
VIM-1 has been also detected in E. coli (154) and in several K. pneumoniae isolates in Greece (53). A similar VIM-1-positive K. pneumoniae strain has been detected very recently in France (P. Nordmann, unpublished data) that was associated with the extended-spectrum β-lactamase SHV-5. Interestingly, the carbapenem resistance level among the enterobacterial isolates was variable. In a study performed by Scoulica et al., the MICs of imipenem and meropenem for the E. coli clinical isolates were below the proposed breakpoint definition for resistance (154).
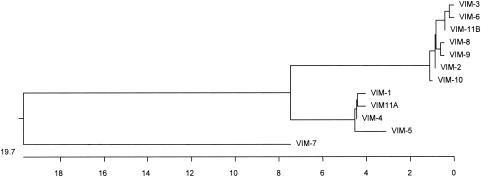
blaVIM-2 was first identified in southern France from a P. aeruginosa isolate in a blood culture from a neutropenic patient in 1996 (127). This isolate was resistant to most β-lactams, including ceftazidime, cefepime, and imipenem, but remained susceptible to aztreonam. VIM-2 is closely related to VIM-1 (90% amino acid identity) and was encoded by a gene cassette, the only resistance gene identified in the blaVIM-2-positive class 1 integron in that isolate (Fig. 3). The blaVIM-2 gene was located on a nonconjugative ca. 45-kb plasmid. However, this plasmid was transferable by electroporation to P. aeruginosa. β-Lactamases VIM-1 and VIM-2 have identical amino acid residues that may be involved near or in the active site of these enzymes (41). Sequence heterogeneity is mostly observed in the NH2- and carboxy-terminal regions of VIM-1 and VIM-2.
FIG. 3.
Phylogeny of VIM-type MBLs. Signal peptides were removed prior to alignment. Sequences were aligned and phylogeny trees were constructed with Clustal W (PAM250 matrix; DNA Star) using the neighbor-joining method.
Subsequently, two other P. aeruginosa isolates had been identified in Paris, France, that harbored the same blaVIM-2 gene cassette (129). Both isolates had similar resistance patterns compared to the P. aeruginosa COL-1 isolate, with a high level of resistance to all β-lactams except aztreonam. In these two isolates, the blaVIM-2 gene cassettes were embedded in different class 1 integrons, In58 and In59. The blaVIM-2-positive integrons carried a variety of aminoglycoside resistance genes in addition to the sulfonamide resistance gene usually found in the 3′ element.
In addition, a retrospective epidemiological study in the hospital in Marseilles, France, where the first VIM-2-producing P. aeruginosa strain was isolated, revealed that from 1995 to 1999, 10 other VIM-2-positive P. aeruginosa isolates were identified in patients hospitalized in different units. These isolates had indistinguishable genotypic patterns, but the class 1 integrons containing the blaVIM-2 gene may vary in size and structure (P. Nordmann, upublished data).
Similarly, VIM-2-producing P. aeruginosa isolates were found to be the source of outbreaks in two university hospitals in Italy and Greece during the same period (77, 135). VIM-2-producing P. aeruginosa strains have also been reported from other countries, such as Japan, South Korea, Portugal, Spain, Poland, Croatia, Chile, Venezuela, Argentina, Belgium, and most recently in the United States (26, 96, 137, 152, 181, 199, 203). The U.S. “outbreak” involved four patients in an intensive care unit and typically the P. aeruginosa harboring VIM-2 was sensitive to aztreonam only (150). The VIM-2-producing P. aeruginosa isolates from the other cases were often involved in serious infections, such as septicemia and pneumonia in different patients, and they exhibited a high level of resistance to imipenem. In addition, VIM-2 has been detected in Citrobacter freundii in Taiwan, in S. marcescens in South Korea, and in Enterbacter cloacae in South Korea (70, 193, 203). In the last strain, the MICs of imipenem and meropenem were 4 μg/ml.
Recently, VIM-2 and a novel variant of the VIM series, VIM-3, have been identified in P. aeruginosa isolates in Taiwan (192). The amino acid sequence of VIM-3 differs from that of VIM-2 by two amino acid substitutions. The precise genetic environment of the blaVIM-3 gene remains unknown, even if a chromosomal location was noticed (192).
VIM-4 was reported from a P. aeruginosa isolate from Larissa, Greece (136). This strain was recovered in 2001 from a patient who had received imipenem. This strain was resistant to all β-lactams but kept some antibacterial activity for aztreonam (MIC of 16 μg/ml) (82). VIM-4 differs from VIM-1 by a single amino acid change (Ser175Arg) that also differs between VIM-2 and VIM-3. Interestingly, a carbapenem-resistant VIM-4-producing P. aeruginosa isolate was also identified in Sweden but from a patient that was transferred from Greece (55). Nothing is known about its genetic support. Very recently, Luzzaro et al. identified the same MBL gene in K. pneumoniae and E. cloacae isolates of a single patient hospitalized in May 2002 in Varese, Italy (88). This patient had received a carbapenem-containing therapy that likely contributed to selection of the VIM producers. The MICs of imipenem and meropenem for the K. pneumoniae isolate were 2 and 0.5 μg/ml, respectively, whereas those of the E. cloacae clinical isolate were 0.25 and 0.12 μg/ml, respectively, the latter having abnormally low carbapenem MICs. Thus, since it was demonstrated that blaVIM-4 was encoded by the same plasmid in both isolates, it is interesting that carbapenem MIC levels may vary significantly among enterobacterial isolates despite a common resistance mechanism (88).
A survey performed on carbapenem-resistant P. aeruginosa isolates recovered from children in Warsaw, Poland, evidenced that several clonally distinguishable VIM-4-producing P. aeruginosa strains were present in that country, one of the clones now being considered an endemic strain (116). All the isolates possessed an identical class 1 integron with the aminoglycoside resistance gene cassette aacA4 at the first position and the blaVIM-4 gene cassette at the second and last position. This allele has now been found in P. putida from the same institute (T. R. Walsh, unpublished data).
VIM-5 differs from VIM-1 by five amino acid changes (Fig. 3) (8). It has been identified in K. pneumoniae (P. Nordmann, unpublished data) and in P. aeruginosa isolates from Ankara, Turkey (8). The P. aeruginosa isolate in which blaVIM-5 has been identified was resistant to all β-lactams, including aztreonam. β-Lactamase VIM-6 was identified from two P. putida isolates from Singapore (75). These isolates were highly resistant to β-lactams, with MICs of >32 μg/ml for imipenem and meropenem, >256 μg/ml for ceftazidime, and 128 μg/ml for aztreonam. VIM-6 differs from VIM-2 by two amino acid changes (glutamine/arginine at position 59 and asparagine/serine at position 165) and from VIM-3 by only one amino acid (75).
The latest VIM-type β-lactamase to be fully characterized is VIM-7, which has been characterized from a carbapenem-resistant P. aeruginosa isolate from Houston, Texas (166). It shares only 77% identity with VIM-1 and 74% with VIM-2 and therefore constitutes a third subgroup among the VIM-type β-lactamases (Fig. 3). The blaVIM-7 gene was located on a ca. 24-kb plasmid and likely to be integron borne. It was identified from a clinical isolate that was resistant to all β-lactams, including aztreonam, and to all other available antibiotics except polymyxin B.
Whereas reports indicate that VIM-type β-lactamases may be identified in distantly related geographical areas, several studies have been performed to evaluate the spread of such enzymes in certain areas. Although VIM-1 and -2 have been identified in several enterobacterial species, P. aeruginosa remains the most important known reservoir of these enzymes (Table 4). Thus, Lagatolla et al. evaluated the occurrence of MBL-encoding genes in P. aeruginosa isolates at Trieste University Hospital in Italy and found that 20% of P. aeruginosa isolates and 70% of the carbapenem-resistant P. aeruginosa isolates produced the VIM-1 and -2 enzymes. They demonstrated a clonal diversity among VIM-positive isolates and showed heterogeneity of the coresistance determinants. In addition, they identified VIM-positive P. aeruginosa isolates from outpatients (77).
Similarly, a retrospective survey has been performed in a tertiary-care hospital in Korea since 1995. Imipenem resistance reached 16% of all P. aeruginosa isolates, and 9% of the resistant isolates were producing VIM-2 β-lactamase (81). These isolates were mostly clonally related, and their carbapenem resistance determinant was transferable by conjugation. The authors noted that the MIC range of β-lactams for these VIM-2-producing isolates was quite large. For example, the MICs of aztreonam, imipenem, and meropenem all varied from 8 to 128 μg/ml and that of ceftazidime from 32 to 128 μg/ml. Thus, identification of VIM producers on the sole basis of the β-lactam susceptibility profile remains uncertain.
Another study has been performed in Greece in which all carbapenem-resistant P. aeruginosa isolates recovered from separate patients during a 1-year period at the University Hospital of Thessaly, Larissa, were studied for MBL production. blaVIM-like genes were detected in 47 of the 53 (88.7%) carbapenem-resistant P. aeruginosa isolates that corresponded to seven genotypes. Four genotypes possessed blaVIM-2 and three possessed blaVIM-4. They were carried as single gene cassettes or along with an aminoglycoside resistance gene (aacA29a) in class 1 integrons. In addition, the spread of VIM-producing-P. aeruginosa isolates in Greece was confirmed (135). In France, VIM-producing strains have recently been reported in a series of P. aeruginosa that are scattered throughout the territory (Poirel and Nordmann, unpublished results). Thus, it seems that repeated reports of VIM-producing strains in southern Europe and in Southeast Asia correspond to a true spread of these isolates rather than to a special interest of research teams located in theses areas. An extended North American survey was performed from 1999 to 2002, studying 1,111 P. aeruginosa and 236 A. baumannii strains from 23 medical centers (72) and found only a single VIM-positive isolate. This was denoted VIM-7, although it sequence is very different from that of the other VIMs and it probably arose from a different ancestral source (166).
Several sequences of new blaVIM genes have been submitted to the EMBL database but have not been formally published (Table 4). The first of these is blaVIM-8 which was isolated from P. aeruginosa in Colombia, adding to the growing number of blaVIM genes discovered in South America (96). Two others, blaVIM-9 and balVIM-10 (accession numbers AY534988 and AY534989, respectively), are from the United Kingdom and differ by just two amino acids. At present, there exist two blaVIM-11 EMBL submissions isolated from P. aeruginosa in Argentina and Italy (AY605049 and AY635904, respectively). This is the second incidence of an MBL from Argentina (113).
SPM-1
A clinical P. aeruginosa isolate from 1997 from Sao Paulo, Brazil, was analyzed as part of the SENTRY surveillance program and shown to contain a novel gene, designated blaSPM-1 (Sao Paulo MBL) (167). The strain, 48-1997A, was a bloodstream isolate from a 4-year-old leukemic girl who eventually succumbed to the infection. The isolate was shown to be highly resistant to all standard anti-gram-negative anti-infectives except colistin (46).
When the sequence of SPM-1 was compared to that of other MBLs, the highest identities were seen with to IMP-1 (35.5%), ImiS (32.2%), CphA (32.1%), BCII (30.0%), and CcrA (27.0%) (167) (Fig. 1). The sequence of SPM-1 differs significantly from that of both IMP and VIM, not least due to the presence of an “insertion” of 24 amino acids just after the active site, HFHLD. This insertion has been shown to be very flexible and acts as a “loop,” probably augmenting the binding and hydrolysis of β-lactams (T. R. Walsh, unpublished data). The genetic context of blaSPM-1 is unique in that it is immediately associated with common region elements and not with transposons or integrons (130). Interestingly, these common elements differ significantly in P. aeruginosa strains collected from different areas of Brazil even though the blaSPM genes are identical (M. A. Toleman, unpublished results).
The ability of SPM-1 to hydrolyze various β-lactams is summarized in Table 2. As judged by kcat values, the preferred substrates of SPM-1 are penillins: penicillin (108), ampicillin (117), piperacillin (117), carbencillin (74), azlocillin (53), and cephalothin (43). Generally, SPM-1 binds cephalosporins more tightly than penicillins, which give relatively large Km values (38 to 814 μM). Like IMP-1 and VIM-1, SPM-1 does not hydrolyze clavulanic acid or aztreonam particularly efficiently, which can act as competitive inhibitors (Km of >0.1 and <0.3, respectively; Table 2) (102).
GIM-1
In 2002, five P. aeruginosa isolates were recovered from different patients from a medical site in Dusseldorf, Germany, and shown to possess a novel class B β-lactamase designated GIM-1 (German imipenemase) (29). Typical of most P. aeruginosa isolates possessing MBLs, the five isolates were susceptible only to polymyxin B. By pulsed-field gel electrophoresis analysis, the five P. aeruginosa isolates were indistinguishable. These strains were compared with six carbapenem-susceptible isolates recovered at the same time from the same medical site in Germany. All susceptible isolates were, by pulsed-field gel electrophoresis analysis, significantly different from the carbapenem-resistant isolates and also distinct from each other.
The amino acid sequence of GIM-1 displayed most identity with IMP variants IMP-6, IMP-1, and IMP-4 (43.5, 43.1, and 43.1%, respectively), with identity to VIM variants ranging from a high of 31.2% compared to VIM-7, 28.8% compared with VIM-1, VIM-4, and VIM-5, and only 28.0% similarity with SPM-1 (Fig. 1). GIM-1 possesses the major consensus features of the MBL class B1 family, such as the principal zinc-binding motif (HXHXD), and has been shown to contain two zincs at its active site (141). GIM-1 demonstrates a hydrolytic profile similar to that of IMP-1 but is arguably a weaker enzyme, as denoted by its lower kcat and higher Km values (Table 2).
Similar to the majority of MBL genes, blaGIM-1 was found on a class 1 integron that is carried on relatively small plasmid of 45 kb. This integron also harbors three other resistance genes, two aminoglycoside resistance genes, aacA4 and aadA1, and a β-lactamase gene, blaOXA-2. Unusually, the blaGIM-1 and aacA4 genes appeared to be accommodated in a single gene cassette that has probably been generated from individual cassettes by deletion of most of the intervening 59-base element. Gene fusions have also been seen with blaVIM-type and aacA4 genes as well. This convoluted genetic arrangement implies that either aminoglycoside or β-lactam therapy will select for blaGIM-1.
EXPERIMENTAL INHIBITORS OF MBLs
The introduction of amoxicillin-clavulanate in the 1980s set a paradigm for therapeutic potentiation between a β-lactam (ampicillin) and a β-lactamase inhibitor (clavulanic acid). Imbued by the success of amoxicillin-clavulanate, other combinations have been studied with inhibitors directed against all classes of β-lactamases, including MBLs (100, 168). Theoretically, the panoply of β-lactamases could be inhibited with a similar approach; however, in the case of MBLs, there are additional obstacles to circumvent (121). First, MBLs possess subtle but significant variations in their active site architecture, so that designing a single inhibitor efficacious against even the transferable MBLs will be problematic (38). Moreover, many inhibitor-screening studies have not included the more clinically important enzymes, i.e., IMP, VIM, and SPM, but used the older, better-characterized enzymes as models, which may or may not be appropriate. Second, unlike clavulanic acid, which interacts directly with class A enzymes and forms a stable covalent intermediate, MBLs do not form highly populated metastable reaction intermediates. Therefore, given their very broad spectrum of activity, attempting to inhibit MBLs with β-lactam-like derivatives may not meet with the same success.
Third, while many studies have used an array of compounds to inhibit these enzymes at a kinetic level, few have examined the potentiation of these compounds with potent β-lactams on P. aeruginosa containing MBL genes. Demonstrating adequate affinity of the inhibitor for the enzyme does not necessarily correlate to lower MICs in the presence of an antipseudomonal β-lactam. Fourth, part of the success of clavulanic acid was due to the fact that there was no homologous mammalian target, i.e., relatively low toxicity. Unfortunately, MBLs have active site motifs similar to those for mammalian enzymes that are highly likely to be quintessential for cellular functions. For instance, human glyoxalase II has a similar protein fold and shares most of the key zinc binding residues of MBLs and accordingly possesses a similar active site architecture (38). Glyoxalase II is a thioesterase and is crucial for the catabolism of toxic 2-oxoaldehydes (153). Therefore, it is likely to be extremely taxing to design compounds that inhibit IMP, VIM, and SPM but do not interact with, for instance, human glyoxalase II. Comparative studies have occasionally used carboxypeptidase A (58); however, these enzymes possess only a single zinc ion and their overall fold is substantially different (189). Studies on MBL inhibitors have hitherto not included human glyoxalase II or other mammalian binuclear enzymes for toxicity screening.
A variety of structurally disparate compounds have been examined as MBL inhibitors, including thioester derivatives (44, 58, 118, 119, 121), trifluoromethyl alcohols and ketones (182), thiols (6, 18, 44, 56, 57, 71, 76, 101, 155, 159), sulfonyl hydrazones (160), tricyclic natural products (122), succinic acid derivatives (171), biphenyl tetrazoles (169, 170), cysteinyl peptides (17), mercaptocarboxylates (57, 121), 1-β-methylcarbapenem (104, 105) cefotetan (140), thioxocephalosporins (173), and penicillin derivatives (25). The activities of these compounds against selected MBLs are summarized in Table 5.
TABLE 5.
Characteristics of experimental MBL inhibitors
| Inhibitor type | Representative compound | Enzyme tested | Affinitya (μM) | Demonstrable potentiation | Reference |
|---|---|---|---|---|---|
| Thioester derivative | Morpholinoethanesulfonic acid | CcrA | Ki 23 | NDb | 44 |
| SB217782/8018/9158 | L1 | IC50 <1.9 | ND | 119 | |
| SB214752 | L1 | IC50 2 | ND | 119 | |
| Biphenylmethyl derivatives | IMP-1 | IC50 0.0004 | Potentiation with E. coli expressing IMP-1 | 170 | |
| CcrA | IC50 180 | ND | |||
| Trifluoromethyl alcohol | d-Alanine derivative | L1 | Ki 1.5 | ND | 182 |
| BCII | Ki 300 | ND | |||
| Thiol | Mercaptoacetic acid | IMP-1 | Ki 0.23 | ND | 56 |
| Mercaptopropionic acid | IMP-1 | Ki 0.19 | ND | ||
| 2′-Mercaptoethyl-derivative | BCII | Ki 70 | ND | 18 | |
| Thiobenzoate derivative | IMP-1 | IC50 0.0004 | ND | 57 | |
| CcrA | IC50 180 | ND | |||
| 2-para-Thiomandelic acid | BCII | Ki 0.21 | ND | 101 | |
| Quinoline C45H | IMP-1 | IC50 1.2 | ND | 71 | |
| VIM-2 | IC50 1.1 | ND | |||
| Sulfonyl hydrazone | 2-Naphthyl derivatives | IMP | IC50 1.6 | ND | 160 |
| Tricyclic product | SB238569 | BCII | Ki 79 | ND | 122 |
| IMP-1 | Ki 17 | No potentiation found with P. aeruginosa (IMP-1) | |||
| CcrA | Ki 3.4 | 8-fold synergistic effect with B. fragilis (CcrA) | |||
| 2S-3S disubstitute | IMP-1 | IC50 >0.21 | ND | 171 | |
| Biphenyl tetrazole | L161, 189 | CcrA | IC50 0.30 | Possessed activity alone and potentiation with imipenem against B. fragilis (CcrA) | 170 |
| Cysteinyl peptide | d-Phenylalanine derivative | BCII | Ki 3.0 | ND | 17 |
| 1-β-Methyl-carbapenem | J-110, 441 | IMP-1 | Ki 0.0037 | Potentiation with imipenem for S. marcescens (IMP-1) and for some P. aeruginosa (IMP-1) strains | 104 |
| CcrA | Ki 0.23 | ND | |||
| L1 | Ki 1.0 | ND | |||
| BCII | Ki 0.83 | ND | |||
| J111, 225 | IMP-1 | Ki 0.18 | 8-fold potentiation with imipenem for P. aeruginosa (IMP-1) | 105 | |
| Penicillin derivative | Penicillinate sulfone | L1 | IC50 0.10 | Some potentiation evident with piperacillin/tazobactam against E. coli (IMP-1) | 25 |
| BCII | IC50 1.4 | See above | |||
| Thioxocephalosporin | Thioacid | BCII | Ki 96 | ND | 173 |
IC50, concentration of inhibitor required to inhibit 50% of MBL activity.
ND, no data.
As Table 5 illustrates, inhibition studies on MBLs have used different enzymes, making direct comparison between compounds difficult, and where studies have used a number of disparate MBLs, their avidity for a given inhibitor varies markedly (122). The other notable aspect of Table 5 is the comparatively small number of bacterial whole-cell assays. While some groups have used transferable MBLs in a P. aeruginosa background to evaluate their inhibitors and demonstrated a degree of potentiation, most have not (104, 105). Perhaps the most worrisome aspect of these studies is that there are few lead compounds with efficacious activity at the submicromolar level. The more recent compounds being developed are being synthesized around a β-lactam scaffold (25, 173), which may be pharmacokinetically more promising. Therapeutically, clinically available β-lactams (e.g., aztreonam) have also been suggested as being potential inhibitors of MBLs due to their competitive inhibition (Table 3) (161). However, this phenomenon appears to be restricted to only a few compounds, and where such β-lactam “inhibitors” of MBLs have been used in animal studies, their efficacy is still to be substantially verified (11).
DETECTION OF MBLs
Rather like the accepted ethos for the early detection of extended-spectrum β-lactamases, it is judicious to detect MBLs for precisely the same reasons. Unfortunately, there are no standardized phenotypic methods available and the testing criteria are likely to depend on whether the gene is carried by P. aeruginosa or a member of the Enterobacteriaceae, i.e., the evincible level of resistance. For example, most Enterobacteriaceae and some Acinetobacter spp. carrying MBL genes will appear sensitive, with imipenem MICs of between 1 and 2 μg/ml (154, 194). Therefore, the implementation of a screening plate to detect MBLs, as has been advocated for extended-spectrum β-lactamases, must take account of the genus of the bacterium, i.e., pseudomonads intrinsically have higher carbapenem MICs than Enterobacteriaceae. It is plausible that for screening Enterobacteriaceae for the presence of MBLs, a plate could contain ceftazidime with and without EDTA, but this would only be effective if the bacterium did not also produce an extended-spectrum β-lactamase, which cannot be assumed.
The identification of some β-lactamases has been aided by isoelectric focusing with the aid of counterstaining the gel with the chromogenic substrate nitrocefin to determine the enzyme's isoelectric point. This technique is based on the surface charge properties of these enzymes, which are neutralized at a certain pH. For closely related enzymes e.g., TEM and SHV, the isoelectric point represents a valuable tool in the identification process. However, MBLs, even the transferable types, differ considerably from one another, and thus, isoelectric focusing is not recommended as a tool to identify them, although it can provide useful information as to the isoelectric point of unknown MBLs by using EDTA inhibition (preincubated with the enzyme prior to electrophoresis or soaking the gel with EDTA after electrophoresis) as part of the isoelectric focusing process (120).
Given the fact that all MBLs are affected by the removal of zinc from the active site, in principle, their detection should be straightforward, and studies have seized upon this principle and used a variety of inhibitor-β-lactam combinations to detect strains possessing these clinically important enzymes (Table 6) (195). However, MBLs vary in their level of inhibition with certain compounds and also vary in their ability to confer resistance to ceftazidime or imipenem, two substrates commonly used in screening MBLs. As previously mentioned, Enterobacteriaceae carrying MBLs are often carbapenem intermediate or susceptible and can be missed when using imipenem or meropenem in the detection method. Consequently, there is no perfect inhibitor-β-lactam combination to detect all transferable MBLs.
TABLE 6.
MBL detection techniques
| Technique | Test | Substrate-inhibitor combination | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|
| Clinical microbiology | Disk approximation | Ceftazidime and 2-mercaptoproprionic acid | Easy to use | Disk and distance of disk placement not standardized and not always easy to interpret | 6 |
| Disk diffusion | Imipenem and EDTA | Easy to use and relatively easy to interpret | Disk not standardized. MBL-producing bacteria can be imipenem sensitive | 202 | |
| Microdilution test | Imipenem and EDTA and 1,10-phenanthroline | Based on reduction in MICs, easy to interpret | Specialized and labor intensive, MBL-producing bacteria can be imipenem sensitive | 99 | |
| Etest | Imipenem and EDTA | Easy to use and relatively easy to interpret | MBL-producing bacteria can be imipenem sensitive and borderline cases may be missed | 177 | |
| Carbapenem hydrolysis | Meropenem and EDTA | Very sensitive and deemed to be the gold standard | Highly specialized, labor intensive, and interpretation not straightforward | 180 | |
| Molecular detection | PCR for genes for IMP, VIM, etc. | Easy to perform, specific for gene family | Requires tailor-made DNA primers, cannot differentiate between variants, may not detect new variants | 158 | |
| DNA probes | Specialized, labor intensive | Probe required for each gene family, cannot differentiate between variants | |||
| Cloning and sequencing | Molecular gold standard | Labor intensive, intepretation of data requires experience | 167 |
The nonmolecular “gold standard” is well established in research laboratories where bacterial crude cell extracts are examined for their ability to hydrolyze carbapenems and whether this hydrolysis is EDTA sensitive. These data indicate that the enzyme is being produced regardless of its genotype. However, this technique utilizes specialized spectrophotometric equipment that precludes its implementation in a routine diagnostic laboratory.
The problem of using EDTA in combination with imipenem is further complicated by the fact that a minority of MBL-negative P. aeruginosa produced reduced imipenem MICs in the presence of EDTA. This is due, in part, to the effect of zinc on OprD and the newly described CzcR-CzcS system (35, 124). With the advent of new and increasing numbers of MBLs, the phenotypic methods listed in Table 6 must be continually evaluated for sensitivity and specificity, particularly for detection in Enterobacteriaceae.
For clinical laboratories concerned about implementing a reasonable screening system, we suggest the following. First, target key isolates based on ceftazidime and carbapenem MIC data. For example, P. aeruginosa isolates with an imipenem MIC of ≥16 μg/ml may be considered appropriate candidates. For Acinetobacter spp. isolates, an imipenem MIC of ≥8 μg/ml, whereas for Enterobacteriaceae, an MIC of ≥2 μg/ml may be appropriate. For ease of application for most microbiology laboratories, the Etest MBL strip is recommended (177), where one half of the strip is impregnated with an imipenem gradient across seven dilutions and the other half with another imipenem gradient overlaid with a constant concentration of EDTA (177) (Fig. 4). However, the current strip will not detect all MBL-positive Enterobacteriaceae due to the low level of “resistance” and will need to be supplemented by the disk approximation test for some Enterobacteriaceae isolates. However, to increase the sensitivity of this technique, several substrates (imipenem, ceftazidime, and meropenem) should be used, preferably with more than one inhibitor (EDTA and mercaptopropionic acid) (6, 202). Positive isolates should be forwarded to a state or national reference laboratory where molecular techniques can verify the phenotypic observations. The excellence of a screening program is dictated by the sensitivity and specificity of the methods it employs; the screening program is also not perfect, for instance, some P. aeruginosa strains are likely to give false-positive results due to altered OprD levels and not the presence of an MBL (35).
FIG. 4.
Etest MBL strip (reprinted with permission from AB BIODISK, Solna, Sweden) and an Acinetobacter sp. expressing a VIM-2 MBL. The intersection of the ellipses at the strip is read from two halves, i.e., at the section with imipenem alone (IP) and imipenem plus EDTA (IPI). A reduction in the MIC of imipenem of ≥3 dilutions in the presence of EDTA is interpreted as a positive test.
The genetic techniques used to detect MBLs are similar to those that have also been used to molecularly characterize countless other β-lactamase, not least extended-spectrum β-lactamases. PCR and DNA probing, while sensitive, are based on the presumption that the clinical isolates produce a related MBL gene, which may or may not be the case. These methods will not indicate the type of variant that is present, which will require sequencing. As most MBLs are strongly linked to class 1 integrons, the genetic elements themselves could be amplified and sequenced, thereby giving information on the structural gene and its adjacent DNA. However, these methods, rather like gene cloning, are highly specialized and beyond most clinical laboratories.
TREATMENT OF INFECTIONS WITH MBL-POSITIVE GRAM-NEGATIVE BACTERIA
The MBL producers that are most clinically significant are primarily those where the gene encoding the enzyme is transferable and include P. aeruginosa and Acinetobacter spp. and to a lesser extent enterobacterial species. While S. maltophilia is also clinically important, its MBL carriage can be predicted, as can its β-lactam resistance profile. Thus, these broad-spectrum β-lactamases are mostly identified from bacterial species that already have a high degree of natural resistance to many antibiotic classes. Concerning β-lactam resistance, these species express a cephalosporinase, have efficient efflux pumps, and have low intrinsic outer membrane permeability to many hydrophilic molecules. Thus, multidrug resistance may be easily observed in those species as a result of combined mechanisms of resistance. The unique problem with MBLs is their unrivalled broad-spectrum resistance profile. In addition, in many cases the MBL genes may be located on plasmids with genes encoding other antibiotic resistance determinants, i.e., aminoglycoside resistance genes. These MBL-positive strains are usually resistant to β-lactams, aminoglycosides, and fluoroquinolones. However, they usually remain susceptible to polymyxins.
No extended survey with a series of human infections with MBL-positive isolates has been performed to determine the optimal treatment. Thus, suitable therapy for treating those infections remains unknown. Using an animal model of pneumonia infection with a VIM-2-positive P. aeruginosa isolate, it was shown that aztreonam at a high dose reduced the bacterial load and may be a useful drug. Although carbapenems retained some activity in the same study, its clinical usefulness under these conditions remains doubtful (11).
One of the main interests for detecting MBL producers would be to contraindicate the use of carbapenems. Although metalloenzyme inhibitors may be used in vitro, no MBL inhibitors are available for treating patients. The association of other antibiotic molecules such as aminoglycosides may be limited due to the coresistance mechanisms cited above (108). The only therapeutic alternative may be the therapeutic administration of polymyxins, which have recently been shown to be efficient for treating multidrug-resistant gram-negative bacilli (73, 84). It has been claimed recently that polymyxins are not as toxic as previously thought (91). In any case, these molecules should not be used in monotherapy, and rapid determination of MICs of aminoglycosides by MIC methods (not disk diffusion) may help to choose an aminoglycoside molecule that may have kept some activity. In addition, rifampin may be an interesting agent for treating multidrug-resistant P. aeruginosa infections.
Clearly, in the absence of novel agents in the near future, the spread of MBL producers may lead to therapeutic dead ends. This would be particularly the case if these MBL genes spread in gram-negative isolates to outpatients. Taking into account the current distribution of these enzymes in different geographical areas (South America, southern Europe, Southeast Asia), early detection of MBL producers in patients from these areas should be undertaken. It would also be prudent to detect colonization with MBL producers when patients are admitted to clinical wards, in particular intensive care units and oncology units. Early detection may avoid spread of these multidrug-resistant isolates and may help maintain first- and second-line therapies (176).
The occurrence of an MBL-positive isolate in a localized hospital environment poses not only a therapeutic problem but also a serious concern for infection control management (109). The microbiology laboratory should promptly inform infection control management, the patient should be regarded as high-risk, and appropriate isolation measures should be enforced. If necessary, patient medical forms should indicate the high-risk nature of the infection, informing clinicians and other health care workers that may come into contact with the patient.
EPIDEMIOLOGY OF MBLs: NOW AND IN THE FUTURE
The spread of extended-spectrum β-lactamases such as TEM, SHV, and CTX M-types should provide a salutary lesson from which we should draw experience to aid us in combating the spread of MBLs (19). However, even with universal infection control policies, the MBL gene pool in some countries may already be firmly established. The recently reported situation in Korea, where 11.4% of imipenem-resistant P. aeruginosa and 14.2% of imipenem-resistant A. baumanii isolates produced MBLs, is a disturbing revelation (80). Moreover, this survey encompassed 28 hospitals, and MBL-producing isolates were found in 60.7% of Korean hospitals. Unsubstantiated reports from Brazilian hospitals indicate that the incidence of imipenem-resistant P. aeruginosa possessing SPM-1 is approaching 20% (unpublished data).
It is impossible to predict what impact MBL genes will have on future antimicrobial regimens. There is little doubt that in some countries the numbers of MBL possessing P. aeruginosa and Acinetobacter spp. infections are such that the mainstay antibiotic regimens used to eradicate these bacteria can no longer be relied upon. For example, the first characterized P. aeruginosa isolate possessing blaSPM-1 was fully resistant to all antibiotics except colistin, which was deemed inappropriate due to the clinical presentation of the patient, who subsequently died of the infection. This case is now mirrored elsewhere in Brazilian hospitals, such that it now precludes the consideration of any β-lactam or aminoglycoside treatment.
At present, it would appear that blaVIM-2 first came from a patient residing in Portugal in 1995 (26). Typically, the isolate was a P. aeruginosa. Since then, blaVIM-2-positive P. aeruginosa has appeared in France, Italy, Greece, Spain, Poland, Croatia, Germany, and Belgium and has asserted itself as the dominant European MBL (93, 114, 127, 137, 152, 181). Outside Europe, it has been found in Venezuela, Chile, Argentina, Korea, Japan, Taiwan, Saudi Arabia, and most recently from an outbreak in the United States (81, 96, 113, 193, 199). It is a mute point whether blaVIM-2 was “created” in these countries or was introduced, and only by studying the mobile genetic elements of the MBL genes and their site of insertion can this be elucidated. It is noteworthy that the first Scandinavian P. aeruginosa blaVIM (blaVIM-4) isolate may have been introduced into Sweden via a Greek patient, highlighting the important role of human traffic in spreading MBL genes (55).
However, it is highly likely that the gene pool is more extensive and either in a quiescent state or not being detected or both. In Taiwan, where 140 multiresistant K. pneumoniae isolates were examined for IMP and VIM genes using hybridization studies, 40 were positive for IMP yet only 5 of 40 were carbapenem resistant (194). Similar findings have been reported from Korea, where P. aeruginosa and Acinetobacter spp. isolates were selected on the premise of imipenem or ceftazidime resistance; however, some carrying the balVIM-2 gene were susceptible to imipenem (109). These hybridization studies provide a useful insight into the extent of the gene pool within a hospital; however, despite the potential problem of MBL genes in Europe, no uniform European study examining the frequency or the spread of MBL genes has been established.
It would appear that MBL genes are first propagated in pseudomonads (usually P. aeruginosa) before appearing in Enterobacteriaceae, including S. marcescens, K. pneumoniae, C. freundii, E. coli, and Enterobacter spp. Based on circumstantial evidence only, it is likely that pseudomonads have transferred their plasmids to Enterobacteriaceae, probably within a clinical environment. The extent of this phenomenon is difficult to ascertain, as the majority of MBL genes in Enterobacteriaceae do confer a resistant phenotype, as judged by in vitro MICs (154, 194). No animal efficacy studies have been undertaken to exam β-lactam eradication in MBL-positive Enterobacteriaceae infections, the data from which would provide a useful benchmark of their clinical importance.
CONCLUSION
The clinical significance of bacteria possessing an MBL is ultimately judged by their ability to confer in vivo β-lactam resistance and, ultimately, whether the patient fails β-lactam therapy. Many isolates possessing transferable MBL genes, particularly Acinetobacter spp. and Enterobacteriaceae, are sensitive to the carbapenems, and doubts have been raised about their in vivo resistance. Accordingly, further in vivo studies need to be undertaken to evaluate the significance of MBL genes is a range of different bacterial hosts. However, there is little doubt that these enzymes contribute significantly to β-lactam resistance even if not singularly responsible for it.
MBL detection by clinical laboratories is hindered by the fact that the presence of the MBL gene does not always confer resistance. There also exists a constant debate as to which of the published methods are more appropriate, and thus, there ensues a compelling need for large multicenter double-blind studies to be undertaken using a variety of transferable MBL genes in a disparate group of clinically important bacteria.
MBLs also represent a clinical threat due to their unrivalled spectrum of activity and their resistance to therapeutic serine β-lactamase inhibitors. The fact that MBL and aminoglycoside resistance genes are genetically linked merely compounds this problem. The problem of an appropriate treatment regimen is also amplified by the lack of new antimicrobials that will possess broad-spectrum potency against clinically significant P. aeruginosa and Acinetobacter spp. (138).
In 1993, Payne deliberated whether MBLs were or could be a serious clinical threat (117). Since that time, a number of events have emerged that command reevaluation of these deliberations. In 1993, there existed just the single publication of a transferable MBL reported from P. aeruginosa. Twelve years hence, there exist good scientific studies reporting the presence of bacteria possessing transferable MBLs from more than 28 countries. In part, some of this increase has arisen through better awareness and clinical laboratories utilizing appropriate methods to identify and evaluate them. However, it is now generally acknowledged that the first blaVIM gene is blaVIM-2 and it is possible that the index case occurred in 1995 in Portugal. Given that blaVIM-2 has now appeared in 18 different countries on five different continents, it is likely that their spread is both human and bacteriological. All blaVIM-type genes are associated with integrons that can be embedded in transposons, which in turn can be accommodated on plasmids, thereby resulting in a highly mobile genetic apparatus. The spread of MBL genes is likely to escalate and further highlights the importance of international resistance surveillance programs such as SENTRY, MYSTIC, the Alexander Project, and EARSS in reporting the emergence and epidemic spread of these remarkable but menacing enzymes.
Acknowledgments
We thank James Spencer for reviewing parts of the manuscript.
Mark Toleman is funded by JMI and by the EC through COBRA contract LSHM-CT-2003-503335. Laurent Poirel is also funded by the same EU COBRA contract.
REFERENCES
- 1.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler, R. P. 1980. The structure of β-lactamases. Phil. Trans. R. Soc. Lond. B. Biol. Sci. 289:321-331. [DOI] [PubMed] [Google Scholar]
- 3.Andrade, S. S., R. N. Jones, A. C. Gales, and H. S. Sader. 2003. Increasing prevalence of antimicrobial resistance among Pseudomonas aeruginosa isolates in Latin American medical centres: 5 year report of the SENTRY Antimicrobial Surveillance Program (1997-2001). J. Antimicrob. Chemother. 52:140-U7. [DOI] [PubMed] [Google Scholar]
- 4.Aoki, S., Y. Hirakata, A. Kondoh, N. Gotoh, K. Yanagihara, Y. Miyazaki, K. Tomono, Y. Yamada, S. Kohno, and S. Kamihira. 2004. Virulence of metallo-β-lactamase-producing Pseudomonas aeruginosa in vitro and in vivo. Antimicrob. Agents Chemother. 48:1876-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avison, M. B., C. S. Higgins, C. J. von Heldreich, P. M. Bennett, and T. R. Walsh. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahar, G., A. Mazzariol, R. Koncan, A. Mert, R. Fontana, G. M. Rossolini, and G. Cornaglia. 2004. Detection of VIM-5 metallo-β-lactamase in a Pseudomonas aeruginosa clinical isolate from Turkey. J. Antimicrob. Chemother. 54:282-283. [DOI] [PubMed] [Google Scholar]
- 9.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72-74. [DOI] [PubMed] [Google Scholar]
- 10.Bellais, S., S. Leotard, L. Poirel, T. Naas, and P. Nordmann. 1999. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol. Lett. 171:127-132. [DOI] [PubMed] [Google Scholar]
- 11.Bellais, S., O. Mimoz, S. Leotard, A. Jacolot, O. Petitjean, and P. Nordmann. 2002. Efficacy of β-lactams for treating experimentally induced pneumonia due to a carbapenem-hydrolyzing metallo-β-lactamase-producing strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:2032-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellais, S., T. Naas, and P. Nordmann. 2002. Genetic and biochemical characterization of CGB-1, an ambler class B carbapenem-hydrolyzing β-lactamase from Chryseobacterium gleum. Antimicrob. Agents Chemother. 46:2791-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellais, S., L. Poirel, S. Leotard, T. Naas, and P. Nordmann. 2000. Genetic diversity of carbapenem-hydrolyzing metallo-β-lactamases from Chryseobacterium (Flavobacterium) indologenes. Antimicrob. Agents Chemother. 44:3028-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett, P. M. 1999. Integrons and gene cassettes: a genetic construction kit for bacteria. J. Antimicrob. Chemother. 43:1-4. [PubMed] [Google Scholar]
- 15.Boschi, L., P. S. Mercuri, M. L. Riccio, G. Amicosante, M. Galleni, J. M. Frere, and G. M. Rossolini. 2000. The Legionella (Fluoribacter) gormanii metallo-β-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 β-lactamases. Antimicrob. Agents Chemother. 44:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bou, G., A. Oliver, and J. Martinez-Beltran. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bounaga, S., M. Galleni, A. P. Laws, and M. I. Page. 2001. Cysteinyl peptide inhibitors of Bacillus cereus zinc β-lactamase. Bioorg. Med. Chem. 9:503-510. [DOI] [PubMed] [Google Scholar]
- 18.Bounaga, S., A. P. Laws, M. Galleni, and M. I. Page. 1998. The mechanism of catalysis and the inhibition of the Bacillus cereus zinc-dependent β-lactamase. Biochem. J. 331:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bush, K. 1999. β-Lactamases of increasing clinical importance. Curr. Pharm. Des. 5:839-845. [PubMed] [Google Scholar]
- 21.Bush, K. 1989. Classification of β-lactamases—group-2c, group-2d, group-2e, group-3, and group-4. Antimicrob. Agents Chemother. 33:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bush, K. 1998. Metallo-β-lactamases: a class apart. Clin. Infect. Dis. 27:S48-53. [DOI] [PubMed] [Google Scholar]
- 23.Bush, K. 2001. New β-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085-1089. [DOI] [PubMed] [Google Scholar]
- 24.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buynak, J. D., H. Chen, L. Vogeti, V. R. Gadhachanda, C. A. Buchanan, T. Palzkill, R. W. Shaw, J. Spencer, and T. R. Walsh. 2004. Penicillin-derived inhibitors that simultaneously target both metallo- and serine-β-lactamases. Bioorg. Med. Chem. Lett. 14:1299-1304. [DOI] [PubMed] [Google Scholar]
- 26.Cardoso, O., R. Leitao, A. Figueiredo, J. C. Sousa, A. Duarte, and L. V. Peixe. 2002. Metallo-β-lactamase VIM-2 in clinical isolates of Pseudomonas aeruginosa from Portugal. Microb. Drug Resist. 8:93-97. [DOI] [PubMed] [Google Scholar]
- 27.Carfi, A., E. Duee, M. Galleni, J. M. Frere, and O. Dideberg. 1998. 1.85 A resolution structure of the zinc (II) β-lactamase from Bacillus cereus. Acta Crystallogr. D Biol. Crystallogr. 54:313-323. [DOI] [PubMed] [Google Scholar]
- 28.Carfi, A., E. Duee, R. Paul-Soto, M. Galleni, J. M. Frere, and O. Dideberg. 1998. X-ray structure of the ZnII β-lactamase from Bacteroides fragilis in an orthorhombic crystal form. Acta Crystallogr. D Biol. Crystallogr. 54:45-57. [DOI] [PubMed] [Google Scholar]
- 29.Castanheira, M., M. A. Toleman, R. N. Jones, F. J. Schmidt, and T. R. Walsh. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob. Agents Chemother. 48:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, Y. H., J. Succi, F. C. Tenover, and T. M. Koehler. 2003. β-Lactamase genes of the penicillin-susceptible Bacillus anthracis Sterne strain. J. Bacteriol. 185:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu, Y. W., M. Afzal-Shah, E. T. Houang, M. I. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collis, C. M., M. J. Kim, H. W. Stokes, and R. M. Hall. 2002. Integron-encoded Intl integrases preferentially recognize the adjacent cognate attl site in recombination with a 59-be site. Mol. Microbiol. 46:1415-1427. [DOI] [PubMed] [Google Scholar]
- 33.Concha, N. O., C. A. Janson, P. Rowling, S. Pearson, C. A. Cheever, B. P. Clarke, C. Lewis, M. Galleni, J. M. Frere, D. J. Payne, J. H. Bateson, and S. S. Abdel-Meguid. 2000. Crystal structure of the IMP-1 metallo β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39:4288-4298. [DOI] [PubMed] [Google Scholar]
- 34.Concha, N. O., B. A. Rasmussen, K. Bush, and O. Herzberg. 1996. Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure 4:823-836. [DOI] [PubMed] [Google Scholar]
- 35.Conejo, M. C., I. Garcia, L. Martinez-Martinez, L. Picabea, and A. Pascual. 2003. Zinc eluted from siliconized latex urinary catheters decreases OprD expression, causing carbapenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:2313-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornaglia, G., M. L. Riccio, A. Mazzariol, L. Lauretti, R. Fontana, and G. M. Rossolini. 1999. Appearance of IMP-1 metallo-β-lactamase in Europe. Lancet 353:899-900. [DOI] [PubMed] [Google Scholar]
- 37.Crowder, M. W., Z. Wang, S. L. Franklin, E. P. Zovinka, and S. J. Benkovic. 1996. Characterization of the metal-binding sites of the β-lactamase from Bacteroides fragilis. Biochemistry 35:12126-12132. [DOI] [PubMed] [Google Scholar]
- 38.Daiyasu, H., K. Osaka, Y. Ishino, and H. Toh. 2001. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 503:1-6. [DOI] [PubMed] [Google Scholar]
- 39.Da Silva, G. J., M. Correia, C. Vital, G. Ribeiro, J. C. Sousa, R. Leitao, L. Peixe, and A. Duarte. 2002. Molecular characterization of blaIMP-5, a new integron-borne metallo-β-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 215:33-39. [DOI] [PubMed] [Google Scholar]
- 40.Da Silva, G. J., R. Leitao, and L. Peixe. 1999. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 37:2109-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Docquier, J. D., J. Lamotte-Brasseur, M. Galleni, G. Amicosante, J. M. Frere, and G. M. Rossolini. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 42.Docquier, J. D., F. Pantanella, F. Giuliani, M. C. Thaller, G. Amicosante, M. Galleni, J. M. Frere, K. Bush, and G. M. Rossolini. 2002. CAU-1, a subclass B3 metallo-β-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. Antimicrob. Agents Chemother. 46:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Docquier, J. D., M. L. Riccio, C. Mugnaioli, F. Luzzaro, A. Endimiani, A. Toniolo, G. Amicosante, and G. M. Rossolini. 2003. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47:1522-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzgerald, P. M., J. K. Wu, and J. H. Toney. 1998. Unanticipated inhibition of the metallo-β-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution. Biochemistry 37:6791-6800. [DOI] [PubMed] [Google Scholar]
- 45.Frere, J. M. 1995. β-lactamases and bacterial resistance to antibiotics. Mol. Microbiol. 16:385-395. [DOI] [PubMed] [Google Scholar]
- 46.Gales, A. C., L. C. Menezes, S. Silbert, and H. S. Sader. 2003. Dissemination in distinct Brazilian regions of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM metallo-β-lactamase. J. Antimicrob. Chemother. 52:699-702. [DOI] [PubMed] [Google Scholar]
- 47.Gales, A. C., M. C. Tognim, A. O. Reis, R. N. Jones, and H. S. Sader. 2003. Emergence of an IMP-like metallo-enzyme in an Acinetobacter baumannii clinical strain from a Brazilian teaching hospital. Diagn. Microbiol. Infect. Dis. 45:77-79. [DOI] [PubMed] [Google Scholar]
- 48.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, and J. M. Frère. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garau, G., C. Bebrone, C. Anne, M. Galleni, J. M. Frere, and O. Dideberg. 2005. A metallo-β-lactamase enzyme in action: Crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 345:785-795. [DOI] [PubMed] [Google Scholar]
- 50.Garau, G., I. Garcia-Saez, C. Bebrone, C. Anne, P. Mercuri, M. Galleni, J. M. Frere, and O. Dideberg. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48:2347-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Saez, I., J. Hopkins, C. Papamicael, N. Franceschini, G. Amicosante, G. M. Rossolini, M. Galleni, J. M. Frere, and O. Dideberg. 2003. The 1.5-A structure of Chryseobacterium meningosepticum zinc β-lactamase in complex with the inhibitor, D-captopril. J. Biol. Chem. 278:23868-23873. [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Saez, I., P. S. Mercuri, C. Papamicael, R. Kahn, J. M. Frere, M. Galleni, G. M. Rossolini, and O. Dideberg. 2003. Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-β-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril. J. Mol. Biol. 325:651-660. [DOI] [PubMed] [Google Scholar]
- 53.Giakkoupi, P., A. Xanthaki, M. Kanelopoulou, A. Vlahaki, V. Miriagou, S. Kontou, E. Papafraggas, H. Malamou-Lada, L. S. Tzouvelekis, N. J. Legakis, and A. C. Vatopoulos. 2003. VIM-1 metallo-β-lactamase-producing Klebsiella pneumoniae strains in Greek hospitals. J. Clin. Microbiol. 41:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giske, C. G., M. Rylander, and G. Kronvall. 2003. VIM-4 in a carbapenem-resistant strain of Pseudomonas aeruginosa isolated in Sweden. Antimicrob. Agents Chemother. 47:3034-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goto, M., T. Takahashi, F. Yamashita, A. Koreeda, H. Mori, M. Ohta, and Y. Arakawa. 1997. Inhibition of the metallo-β-lactamase produced from Serratia marcescens by thiol compounds. Biol. Pharm. Bull. 20:1136-1140. [DOI] [PubMed] [Google Scholar]
- 57.Greenlee, M. L., J. B. Laub, J. M. Balkovec, M. L. Hammond, G. G. Hammond, D. L. Pompliano, and J. H. Epstein-Toney. 1999. Synthesis and SAR of thioester and thiol inhibitors of IMP-1 metallo-β-lactamase. Bioorg. Med. Chem. Lett. 9:2549-2554. [DOI] [PubMed] [Google Scholar]
- 58.Hammond, G. G., J. L. Huber, M. L. Greenlee, J. B. Laub, K. Young, L. L. Silver, J. M. Balkovec, K. D. Pryor, J. K. Wu, B. Leiting, D. L. Pompliano, and J. H. Toney. 1999. Inhibition of IMP-1 metallo-β-lactamase and sensitization of IMP-1-producing bacteria by thioester derivatives. FEMS Microbiol. Lett. 179:289-296. [DOI] [PubMed] [Google Scholar]
- 59.Hanson, N. D., A. Hossain, L. L. Buck, E. S. Moland, and K. S. Thomson. 2004. Program and abstracts of the 44th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, D.C., abstr. C1-291, 2004.
- 60.Hawkey, P. M., J. Xiong, H. Ye, H. Li, and F. H. M′Zali. 2001. Occurrence of a new metallo-β-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People's Republic of China. FEMS Microbiol. Lett. 194:53-57. [DOI] [PubMed] [Google Scholar]
- 61.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirakata, Y., K. Izumikawa, T. Yamaguchi, H. Takemura, H. Tanaka, R. Yoshida, J. Matsuda, M. Nakano, K. Tomono, S. Maesaki, M. Kaku, Y. Yamada, S. Kamihira, and S. Kohno. 1998. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 42:2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ho, S. E., G. Subramaniam, S. Palasubramaniam, and P. Navaratnam. 2002. Carbapenem-resistant Pseudomonas aeruginosa in Malaysia producing IMP-7 β-lactamase. Antimicrob. Agents Chemother. 46:3286-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houang, E. T. S., Y. W. Chu, W. S. Lo, K. Y. Chu, and A. F. B. Cheng. 2003. Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and metallo-β-lactamase (blaIMP-4) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrob. Agents Chemother. 47:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hussain, M., A. Carlino, M. J. Madonna, and J. O. Lampen. 1985. Cloning and sequencing of the metallothioprotein β-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J. Bacteriol. 164:223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito, H., Y. Arakawa, S. Ohsuka, R. Wacharotayankun, N. Kato, and M. Ohta. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 39:824-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwanaga, M., C. Toma, T. Miyazato, S. Insisiengmay, N. Nakasone, and M. Ehara. 2004. Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob. Agents Chemother. 48:2364-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iyobe, S., H. Kusadokoro, J. Ozaki, N. Matsumura, S. Minami, S. Haruta, T. Sawai, and K. O′Hara. 2000. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:2023-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iyobe, S., H. Kusadokoro, A. Takahashi, S. Yomoda, T. Okubo, A. Nakamura, and K. O'Hara. 2002. Detection of a variant metallo-β-lactamase, IMP-10, from two unrelated strains of Pseudomonas aeruginosa and an Alcaligenes xylosoxidans strain. Antimicrob. Agents Chemother. 46:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeong, S. H., K. Lee, Y. Chong, J. H. Yum, S. H. Lee, H. J. Choi, J. M. Kim, K. H. Park, B. H. Han, S. W. Lee, and T. S. Jeong. 2003. Characterization of a new integron containing VIM-2, a metallo-β-lactamase gene cassette, in a clinical isolate of Enterobacter cloacae. J. Antimicrob. Chemother. 51:397-400. [DOI] [PubMed] [Google Scholar]
- 71.Jin, W. C., Y. Arakawa, H. Yasuzawa, T. Taki, R. Hashiguchi, K. Mitsutani, A. Shoga, Y. Yamaguchi, H. Kurosaki, N. Shibata, M. Ohta, and M. Goto. 2004. Comparative study of the inhibition of metallo-β-lactamases (IMP-1 and VIM-2) by thiol compounds that contain a hydrophobic group. Biol. Pharm. Bull. 27:851-856. [DOI] [PubMed] [Google Scholar]
- 72.Jones, R. N., L. M. Deshpande, T. R. Fritsche, and H. S. Sader. 2004. Determination of epidemic clonality among multidrug-resistant strains of Acinetobacter spp. and Pseudomonas aeruginosa in the MYSTIC Programme (UAS 1999-2003). Diagn. Microbiol. Infect. Dis. 49:211-216. [DOI] [PubMed] [Google Scholar]
- 73.Karabinis, A., E. Paramythiotou, D. Mylona-Petropoulou, A. Kalogeromitros, N. Katsarelis, F. Kontopidou, I. Poularas, and H. Malamou-Lada. 2004. Colistin for Klebsiella pneumoniae-associated sepsis. Clin. Infect. Dis. 38:E7-E9. [DOI] [PubMed] [Google Scholar]
- 74.Koh, T. H., L. H. Sng, G. S. Babini, N. Woodford, D. M. Livermore, and L. M. Hall. 2001. Carbapenem-resistant Klebsiella pneumoniae in Singapore producing IMP-1 β-lactamase and lacking an outer membrane protein. Antimicrob. Agents Chemother. 45:1939-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koh, T. H., G. C. Y. Wang, and L. H. Sng. 2004. IMP-1 and a novel metallo-β-lactamase, VIM-6, in fluorescent pseudomonads isolated in Singapore. Antimicrob. Agents Chemother. 48:2334-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kurosaki, H., H. Yasuzawa, Y. Yamaguchi, W. C. Jin, Y. Arakawa, and M. Goto. 2003. Detection of a metallo-β-lactamase (IMP-1) by fluorescent probes having dansyl and thiol groups. Org. Biomol. Chem. 1:17-20. [DOI] [PubMed] [Google Scholar]
- 77.Lagatolla, C., E. A. Tonin, C. Monti-Bragadin, L. Dolzani, F. Gombac, C. Bearzi, E. Edalucci, F. Gionechetti, and G. M. Rossolini. 2004. Endemic carbapenem-resistant Pseudomonas aeruginosa with acquired metallo-β-lactamase determinants in European hospital. Emerg. Infect. Dis. 10:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laraki, N., N. Franceschini, G. M. Rossolini, P. Santucci, C. Meunier, E. de Pauw, G. Amicosante, J. M. Frere, and M. Galleni. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee, K., W. G. Lee, Y. Uh, G. Y. Ha, J. Cho, and Y. Chong. 2003. VIM- and IMP-type metallo-β-lactamase-producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emerg. Infect. Dis. 9:868-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Libisch, B., M. Gacs, K. Csiszar, M. Muzslay, L. Rokusz, and M. Fuzi. 2004. Isolation of an integron-borne blaVIM-4 type metallo-β-lactamase gene from a carbapenem-resistant Pseudomonas aeruginosa clinical isolate in Hungary. Antimicrob. Agents Chemother. 48:3576-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lim, H. M., J. J. Pene, and R. W. Shaw. 1988. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 β-lactamase II structural gene. J. Bacteriol. 170:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Linden, P. K., S. Kusne, K. Coley, P. Fontes, D. J. Kramer, and D. Paterson. 2003. Use of parenteral colistin for the treatment of serious infection due to antimicrobial-resistant Pseudomonas aeruginosa. Clin. Infect. Dis. 37:E154-E160. [DOI] [PubMed] [Google Scholar]
- 85.Livermore, D. M. 2002. The impact of carbapenemases on antimicrobial development and therapy. Curr. Opin. Investig. Drugs 3:218-224. [PubMed] [Google Scholar]
- 86.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 87.Lombardi, G., F. Luzzaro, J. D. Docquier, M. L. Riccio, M. Perilli, A. Coli, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2002. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-β-lactamase. J. Clin. Microbiol. 40:4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luzzaro, F., J. D. Docquier, C. Colinon, A. Endimiani, G. Lombardi, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2004. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-β-lactamase encoded by a conjugative plasmid. Antimicrob. Agents Chemother. 48:648-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mammeri, H., S. Bellais, and P. Nordmann. 2002. Chromosome-encoded β-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (Formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymes. Antimicrob. Agents Chemother. 46:3561-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maniatis, A. N., S. Pournaras, S. Orkopoulou, P. T. Tassios, and N. J. Legakis. 2003. Multiresistant Acinetobacter baumannii isolates in intensive care units in Greece. Clin. Microbiol. Infect. 9:547-553. [DOI] [PubMed] [Google Scholar]
- 91.Markou, N., H. Apostolakos, C. Koumoudiou, M. Athanasiou, A. Koutsoukou, I. Alamanos, and L. Gregorakos. 2003. Intravenous colistin in the treatment of sepsis from multiresistant Gram-negative bacilli in critically ill patients. Crit. Care 7:R78-R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Massidda, O., G. M. Rossolini, and G. Satta. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J. Bacteriol. 173:4611-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mavroidi, A., A. Tsakris, E. Tzelepi, S. Pournaras, V. Loukova, and L. S. Tzouvelekis. 2000. Carbapenem-hydrolysing VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 46:1041-1042. [DOI] [PubMed] [Google Scholar]
- 94.McManus-Munoz, S., and M. W. Crowder. 1999. Kinetic mechanism of metallo-β-lactamase L1 from Stenotrophomonas maltophilia. Biochemistry 38:1547-1553. [DOI] [PubMed] [Google Scholar]
- 95.Melino, S., C. Capo, B. Dragani, A. Aceto, and R. Petruzzelli. 1998. A zinc-binding motif conserved in glyoxalase II, β-lactamase and arylsulfatases. Trends Biochem. Sci. 23:381-382. [DOI] [PubMed] [Google Scholar]
- 96.Mendes, R. E., M. Castanheira, P. Garcia, M. Guzman, M. A. Toleman, T. R. Walsh, and R. N. Jones. 2004. First isolation of blaVIM-2 in Latin America: Report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 48:1433-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mendes, R. E., M. A. Toleman, J. Ribeiro, H. S. Sader, R. N. Jones, and T. R. Walsh. 2004. Genetic characterization of a novel metallo-β-lactamase gene, blaIMP-16: a highly divergent blaIMP with a unique genetic context. Report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 48:4654-4661.15561840 [Google Scholar]
- 98.Migliavacca, R., C. Colinon, J. D. Docquier, M. Spalla, E. Nucleo, M. Li Bergoli, M. Labonia, G. M. Rossolini, and L. Pagani. 2004. Abstr. 14th European Congress on Clinical Microbiology and Infectious Diseases, abstr. P1876, 2004.
- 99.Migliavacca, R., J. D. Docquier, C. Mugnaioli, G. Amicosante, R. Daturi, K. W. Lee, G. M. Rossolini, and L. Pagani. 2002. Simple microdilution test for detection of metallo-β-lactamase production in Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4388-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miller, L. A., K. Ratnam, and D. J. Payne. 2001. β-lactamase-inhibitor combinations in the 21st century: current agents and new developments. Curr. Opin. Pharm. 1:451-458. [DOI] [PubMed] [Google Scholar]
- 101.Mollard, C., C. Moali, C. Papamicael, C. Damblon, S. Vessilier, G. Amicosante, C. J. Schofield, M. Galleni, J. M. Frere, and G. C. Roberts. 2001. Thiomandelic acid, a broad spectrum inhibitor of zinc β-lactamases: kinetic and spectroscopic studies. J. Biol. Chem. 276:45015-45023. [DOI] [PubMed] [Google Scholar]
- 102.Murphy, T. A., A. M. Simm, M. A. Toleman, R. N. Jones, and T. R. Walsh. 2003. Biochemical characterization of the acquired metallo-β-lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Naas, T., S. Bellais, and P. Nordmann. 2003. Molecular and biochemical characterization of a carbapenem-hydrolysing β-lactamase from Flavobacterium johnsoniae. J. Antimicrob. Chemother. 51:267-273. [DOI] [PubMed] [Google Scholar]
- 104.Nagano, R., Y. Adachi, T. Hashizume, and H. Morishima. 2000. In vitro antibacterial activity and mechanism of action of J.-111,225, a novel 1β-methylcarbapenem, against transferable IMP-1 metallo-β-lactamase producers. J. Antimicrob. Chemother. 45:271-276. [DOI] [PubMed] [Google Scholar]
- 105.Nagano, R., Y. Adachi, H. Imamura, K. Yamada, T. Hashizume, and H. Morishima. 1999. Carbapenem derivatives as potential inhibitors of various β-lactamases, including class B metallo-β-lactamases. Antimicrob. Agents Chemother. 43:2497-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neuhauser, M. M., R. A. Weinstein, R. Rydman, L. H. Danziger, G. Karam, and J. P. Quinn. 2003. Antibiotic resistance among gram-negative bacilli in US intensive care units—implications for fluoroquinolone use. JAMA 289:885-888. [DOI] [PubMed] [Google Scholar]
- 107.Nordmann, P., S. Mariotte, T. Naas, R. Labia, and M. H. Nicolas. 1993. Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 37:939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 109.Oh, E. J., S. Lee, Y. J. Park, J. J. Park, K. Park, S. I. Kim, M. W. Kang, and B. K. Kim. 2003. Prevalence of metallo-β-lactamase among Pseudomonas aeruginosa and Acinetobacter baumannii in a Korean university hospital and comparison of screening methods for detecting metallo-β-lactamase. J. Microbiol. Methods 54:411-418. [DOI] [PubMed] [Google Scholar]
- 110.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Page, M. I. 1999. The reactivity of β-lactams, the mechanism of catalysis and the inhibition of β-lactamases. Curr. Pharm. Des. 5:895-913. [PubMed] [Google Scholar]
- 112.Page, M. I. 2002. Understanding metallo-β-lactamases. ASM News 68:217-221. [Google Scholar]
- 113.Pagniez, G., M. Radice, A. Amoroso, A. Famiglietti, and G. Gutkind. 2004. Abstr. 44th Interscience Conference on Antimicrobial Agents and Chemotherapy, abstr. C1-293, Washington, D.C.
- 114.Pallecchi, L., M. L. Riccio, J. D. Docquier, R. Fontana, and G. M. Rossolini. 2001. Molecular heterogeneity of blaVIM-2-containing integrons from Pseudomonas aeruginosa plasmids encoding the VIM-2 metallo-β-lactamase. FEMS Microbiol. Lett. 195:145-150. [DOI] [PubMed] [Google Scholar]
- 115.Partridge, S. R., and R. M. Hall. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185:6371-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Patzer, J., M. A. Toleman, L. M. Deshpande, W. Kaminska, D. Dzierzanowska, P. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Pseudomonas aeruginosa strains harbouring an unusual blaVIM-4 gene cassette isolated from hospitalized children in Poland (1998-2001). J. Antimicrob. Chemother. 53:451-456. [DOI] [PubMed] [Google Scholar]
- 117.Payne, D. J. 1993. Metallo-β-lactamases—a new therapeutic challenge. J. Med. Microbiol. 39:93-99. [DOI] [PubMed] [Google Scholar]
- 118.Payne, D. J., J. H. Bateson, B. C. Gasson, T. Khushi, D. Proctor, S. C. Pearson, and R. Reid. 1997. Inhibition of metallo-β-lactamases by a series of thiol ester derivatives of mercaptophenylacetic acid. FEMS Microbiol. Lett. 157:171-175. [DOI] [PubMed] [Google Scholar]
- 119.Payne, D. J., J. H. Bateson, B. C. Gasson, D. Proctor, T. Khushi, T. H. Farmer, D. A. Tolson, D. Bell, P. W. Skett, A. C. Marshall, R. Reid, L. Ghosez, Y. Combret, and J. Marchand-Brynaert. 1997. Inhibition of metallo-β-lactamases by a series of mercaptoacetic acid thiol ester derivatives. Antimicrob. Agents Chemother. 41:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Payne, D. J., R. Cramp, J. H. Bateson, J. Neale, and D. Knowles. 1994. Rapid identification of metallo- and serine β-lactamase. Antimicrob. Agents Chemother. 38:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Payne, D. J., W. Du, and J. H. Bateson. 2000. β-Lactamase epidemiology and the utility of established and novel β-lactamase inhibitors. Expert Opin. Investig. Drugs 9:247-261. [DOI] [PubMed] [Google Scholar]
- 122.Payne, D. J., J. A. Hueso-Rodriguez, H. Boyd, N. O. Concha, C. A. Janson, M. Gilpin, J. H. Bateson, C. Cheever, N. L. Niconovich, S. Pearson, S. Rittenhouse, D. Tew, E. Diez, P. Perez, J. De La Fuente, M. Rees, and A. Rivera-Sagredo. 2002. Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-β-lactamases. Antimicrob. Agents Chemother. 46:1880-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peleg, A. Y., C. Franklin, J. Bell, and D. W. Spelman. 2004. Emergence of IMP-4 metallo-β-lactamase in a clinical isolate from Australia. J. Antimicrob. Chemother. 54:699-700. [DOI] [PubMed] [Google Scholar]
- 124.Perron, K., O. Caille, C. Rossier, C. van Delden, J. L. Dumas, and T. Kohler. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279:8761-8768. [DOI] [PubMed] [Google Scholar]
- 125.Podglajen, I., J. Breuil, I. Casin, and E. Collatz. 1995. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J. Bacteriol. 177:5270-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Podglajen, I., J. Breuil, and E. Collatz. 1994. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol. Microbiol. 12:105-114. [DOI] [PubMed] [Google Scholar]
- 127.Poirel, L., L. Collet, and P. Nordmann. 2000. Carbapenem-hydrolyzing metallo-β-lactamase from a nosocomial isolate of Pseudomonas aeruginosa in France. Emerg. Infect. Dis. 6:84-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Poirel, L., C. Heritier, and P. Nordmann. 2004. Chromosome-encoded ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Poirel, L., M. Magalhaes, M. Lopes, and P. Nordmann. 2004. Molecular analysis of metallo-β-lactamase gene blaSPM-1-surrounding sequences from disseminated Pseudomonas aeruginosa isolates in Recife, Brazil. Antimicrob. Agents Chemother. 48:1406-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Poirel, L., J. N. Pham, L. Cabanne, B. J. Gatus, S. M. Bell, and P. Nordmann. 2004. Carbapenem-hydrolysing metallo-β-lactamases from Klebsiella pneumoniae and Escherichia coli isolated in Australia. Pathology (Philadelphia) 36:366-367. [DOI] [PubMed] [Google Scholar]
- 133.Poirel, L., G. F. Weldhagen, T. Naas, C. De Champs, M. G. Dove, and P. Nordmann. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Poole, K. 2003. Overcoming multidrug resistance in gram-negative bacteria. Curr. Opin. Investig. Drugs 4:128-139. [PubMed] [Google Scholar]
- 135.Pournaras, S., M. Maniati, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 51:1409-1414. [DOI] [PubMed] [Google Scholar]
- 136.Pournaras, S., A. Tsakris, M. Maniati, L. S. Tzouvelekis, and A. N. Maniatis. 2002. Novel variant (blaVlM-4) of the metallo-β-lactamase gene blaVIM-1 in a clinical strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:4026-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Prats, G., E. Miro, B. Mirelis, L. Poirel, S. Bellais, and P. Nordmann. 2002. First isolation of a carbapenem-hydrolyzing β-lactamase in Pseudomonas aeruginosa in Spain. Antimicrob. Agents Chemother. 46:932-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Projan, S. J. 2003. Why is big Pharma getting out of antibacterial drug discovery? Curr. Opin. Microbiol. 6:427-430. [DOI] [PubMed] [Google Scholar]
- 139.Queenan, A. M., C. Torres-Viera, H. S. Gold, Y. Carmeli, G. M. Eliopoulos, R. C. Moellering, J. P. Quinn, J. Hindler, A. A. Medeiros, and K. Bush. 2000. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44:3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Quiroga, M. I., N. Franceschini, G. M. Rossolini, G. Gutkind, G. Bonfiglio, L. Franchino, and G. Amicosante. 2000. Interaction of cefotetan and the metallo-β-lactamases produced in Aeromonas spp. and in vitro activity. Chemotherapy 46:177-183. [DOI] [PubMed] [Google Scholar]
- 141.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rasmussen, B. A., K. Bush, D. Keeney, Y. J. Yang, R. Hare, C. Ogara, and A. A. Medeiros. 1996. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 40:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rasmussen, B. A., and E. Kovacs. 1991. Identification and DNA sequence of a new Bacteroides fragilis insertion sequence-like element. Plasmid 25:141-144. [DOI] [PubMed] [Google Scholar]
- 144.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rossolini, G. M., M. A. Condemi, F. Pantanella, J. D. Docquier, G. Amicosante, and M. C. Thaller. 2001. Metallo-β-lactamase producers in environmental microbiota: new molecular class B enzyme in Janthinobacterium lividum. Antimicrob. Agents Chemother. 45:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rossolini, G. M., N. Franceschini, L. Lauretti, B. Caravelli, M. L. Riccio, M. Galleni, J. M. Frere, and G. Amicosante. 1999. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaACME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamases. Antimicrob. Agents Chemother. 43:2193-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rossolini, G. M., N. Franceschini, M. L. Riccio, P. S. Mercuri, M. Perilli, M. Galleni, J. M. Frere, and G. Amicosante. 1998. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem. J. 332:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Saavedra, M. J., L. Peixe, J. C. Sousa, I. Henriques, A. Alves, and A. Correia. 2003. Sfh-1, a subclass B2 metallo-β-lactamase from a Serratia fonticola environmental isolate. Antimicrob. Agents Chemother. 47:2330-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sahud, A., K. Lolans, A. M. Queenan, and J. P. Quinn. 2004. Presented at the 44th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, D.C.
- 151.Sanschagrin, F., J. Dufresne, and R. C. Levesque. 1998. Molecular heterogeneity of the L-1 metallo-β-lactamase family from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42:1245-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sardelic, S., L. Pallecchi, V. Punda-Polic, and G. M. Rossolini. 2003. Carbapenem-resistant Pseudomonas aeruginosa-carrying VIM-2 metallo-β-lactamase determinants, Croatia. Emerg. Infect. Dis. 9:1022-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schilling, O., N. Wenzel, M. Naylor, A. Vogel, M. Crowder, C. Makaroff, and W. Meyer-Klaucke. 2003. Flexible metal binding of the metallo-β-lactamase domain: glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry 42:11777-11786. [DOI] [PubMed] [Google Scholar]
- 154.Scoulica, E. V., I. K. Neonakis, A. I. Gikas, and Y. J. Tselentis. 2004. Spread of blaVIM-1-producing E. coli in a university hospital in Greece. Genetic analysis of the integron carrying the blaVIM-1 metallo-β-lactamase gene. Diagn. Microbiol. Infect. Dis. 48:167-172. [DOI] [PubMed] [Google Scholar]
- 155.Scrofani, S. D., J. Chung, J. J. Huntley, S. J. Benkovic, P. E. Wright, and H. J. Dyson. 1999. NMR characterization of the metallo-β-lactamase from Bacteroides fragilis and its interaction with a tight-binding inhibitor: role of an active-site loop. Biochemistry 38:14507-14514. [DOI] [PubMed] [Google Scholar]
- 156.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shibata, N., Y. Doi, K. Yamane, T. Yagi, H. Kurokawa, K. Shibayama, H. Kato, K. Kai, and Y. Arakawa. 2003. PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J. Clin. Microbiol. 41:5407-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Siemann, S., A. J. Clarke, T. Viswanatha, and G. I. Dmitrienko. 2003. Thiols as classical and slow-binding inhibitors of IMP-1 and other binuclear metallo-β-lactamases. Biochemistry 42:1673-1683. [DOI] [PubMed] [Google Scholar]
- 160.Siemann, S., D. P. Evanoff, L. Marrone, A. J. Clarke, T. Viswanatha, and G. I. Dmitrienko. 2002. N-Arylsulfonyl hydrazones as inhibitors of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 46:2450-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Simm, A. M., C. S. Higgins, S. T. Pullan, M. B. Avison, P. Niumsup, O. Erdozain, P. M. Bennett, and T. R. Walsh. 2001. A novel metallo-β-lactamase, Mbl1b, produced by the environmental bacterium Caulobacter crescentus. FEBS Lett. 509:350-354. [DOI] [PubMed] [Google Scholar]
- 162.Spencer, J., A. R. Clarke, and T. R. Walsh. 2001. Novel mechanism of hydrolysis of therapeutic β-lactams by Stenotrophomonas maltophilia L1 metallo-β-lactamase. J. Biol. Chem. 276:33638-33644. [DOI] [PubMed] [Google Scholar]
- 163.Spencer, J., A. M. Simm, A. R. Clarke, R. B. Sessions, and T. R. Walsh. 2003. Abstr. 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., abstr. C1-568.
- 164.Thompson, J. S., and M. H. Malamy. 1990. Sequencing of the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus β-lactamase BCII. J. Bacteriol. 172:2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Toleman, M. A., D. Biedenbach, D. Bennett, R. N. Jones, and T. R. Walsh. 2003. Genetic characterization of a novel metallo-β-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase genes in Europe: report from the SENTRY worldwide antimicrobial surveillance programme. J. Antimicrob. Chemother. 52:583-590. [DOI] [PubMed] [Google Scholar]
- 166.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 168.Toney, J. H. 2003. Metallo-β-lactamase inhibitors: could they give old antibacterials new life? Curr. Opin. Investig. Drugs 4:115-116. [PubMed] [Google Scholar]
- 169.Toney, J. H., K. A. Cleary, G. G. Hammond, X. Yuan, W. J. May, S. M. Hutchins, W. T. Ashton, and D. E. Vanderwall. 1999. Structure-activity relationships of biphenyl tetrazoles as metallo-β-lactamase inhibitors. Bioorg. Med. Chem. Lett. 9:2741-2746. [DOI] [PubMed] [Google Scholar]
- 170.Toney, J. H., P. M. Fitzgerald, N. Grover-Sharma, S. H. Olson, W. J. May, J. G. Sundelof, D. E. Vanderwall, K. A. Cleary, S. K. Grant, J. K. Wu, J. W. Kozarich, D. L. Pompliano, and G. G. Hammond. 1998. Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-β-lactamase. Chem. Biol. 5:185-196. [DOI] [PubMed] [Google Scholar]
- 171.Toney, J. H., G. G. Hammond, P. M. Fitzgerald, N. Sharma, J. M. Balkovec, G. P. Rouen, S. H. Olson, M. L. Hammond, M. L. Greenlee, and Y. D. Gao. 2001. Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-β-lactamase. J. Biol. Chem. 276:31913-31918. [DOI] [PubMed] [Google Scholar]
- 172.Towner, K. J., T. Gee, and T. Boswell. 2002. An unwanted import to the UK: a carbapenem-resistant clinical isolate of Acinetobacter baumannii producing metallo-β-lactamase. J. Antimicrob. Chemother. 50:1092-1093. [DOI] [PubMed] [Google Scholar]
- 173.Tsang, W. Y., A. Dhanda, C. J. Schofield, J. M. Frere, M. Galleni, and M. I. Page. 2004. The inhibition of metallo-β-lactamase by thioxo-cephalosporin derivatives. Bioorg. Med. Chem. Lett. 14:1737-1739. [DOI] [PubMed] [Google Scholar]
- 174.Tysall, L., M. W. Stockdale, P. R. Chadwick, M. F. Palepou, K. J. Towner, D. M. Livermore, and N. Woodford. 2002. IMP-1 carbapenemase detected in an Acinetobacter clinical isolate from the UK. J. Antimicrob. Chemother. 49:217-218. [DOI] [PubMed] [Google Scholar]
- 175.Ullah, J. H., T. R. Walsh, I. A. Taylor, D. C. Emery, C. S. Verma, S. J. Gamblin, and J. Spencer. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution. J. Mol. Biol. 284:125-136. [DOI] [PubMed] [Google Scholar]
- 176.Urban, C., S. Segal-Maurer, and J. J. Rahal. 2003. Considerations in control and treatment of nosocomial infections due to multidrug-resistant Acinetobacter baumannii. Clin. Infect. Dis. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 177.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, and A. Gales. 2002. Evaluation of a new Etest for detecting metallo-β-lactamases in routine clinical testing. J. Clin. Microbiol. 40:2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Walsh, T. R., L. Hall, S. J. Assinder, W. W. Nichols, S. J. Cartwright, A. P. MacGowan, and P. M. Bennett. 1994. Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1218:199-201. [DOI] [PubMed] [Google Scholar]
- 179.Walsh, T. R., A. P. MacGowan, and P. M. Bennett. 1997. Sequence analysis and enzyme kinetics of the L2 serine β-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1460-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Walsh, T. R., W. A. Neville, M. H. Haran, D. Tolson, D. J. Payne, J. H. Bateson, A. P. MacGowan, and P. M. Bennett. 1998. Nucleotide and amino acid sequences of the metallo-β-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob. Agents Chemother. 42:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Walsh, T. R., M. A. Toleman, W. Hryniewicz, P. M. Bennett, and R. N. Jones. 2003. Evolution of an integron carrying blaVIM-2 in Eastern Europe: report from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 52:116-119. [DOI] [PubMed] [Google Scholar]
- 182.Walter, M. W., A. Felici, M. Galleni, R. P. Soto, R. M. Adlington, J. E. Baldwin, J.-M. Frère, M. Gololobov, and C. J. Schofield. 1996. Trifluoromethyl alcohol and ketone inhibitors of metallo-β-lactamases. Bioorg. Med. Chem. Lett. 6:2455-2458. [Google Scholar]
- 183.Wang, Z., and S. J. Benkovic. 1998. Purification, characterization, and kinetic studies of a soluble Bacteroides fragilis metallo-β-lactamase that provides multiple antibiotic resistance. J. Biol. Chem. 273:22402-22408. [DOI] [PubMed] [Google Scholar]
- 184.Wang, Z., W. Fast, and S. J. Benkovic. 1998. Direct observation of an enzyme-bound intermediate in the catalytic cycle of the metallo-β-lactamase from Bacteroides fragilis. J. Am. Chem. Soc. 120:10788-10789. [Google Scholar]
- 185.Wang, Z., W. Fast, and S. J. Benkovic. 1999. On the mechanism of the metallo-β-lactamase from Bacteroides fragilis. Biochemistry 38:10013-10023. [DOI] [PubMed] [Google Scholar]
- 186.Wang, Z., W. Fast, A. M. Valentine, and S. J. Benkovic. 1999. Metallo-β-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 3:614-622. [DOI] [PubMed] [Google Scholar]
- 187.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Woodford, N., M. F. Palepou, G. S. Babini, B. Holmes, and D. M. Livermore. 2000. Carbapenemases of Chryseobacterium (Flavobacterium) meningosepticum: distribution of blaB and characterization of a novel metallo-β-lactamase gene, blaB3, in the type strain, NCTC 10016. Antimicrob. Agents Chemother. 44:1448-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wouters, M. A., and A. Husain. 2001. Changes in zinc ligation promote remodeling of the active site in the zinc hydrolase superfamily. J. Mol. Biol. 314:1191-1207. [DOI] [PubMed] [Google Scholar]
- 190.Yamasaki, K., M. Komatsu, T. Yamashita, K. Shimakawa, T. Ura, H. Nishio, K. Satoh, R. Washidu, S. Kinoshita, and M. Aihara. 2003. Production of CTX-M-3 extended-spectrum β-lactamase and IMP-1 metallo β-lactamase by five Gram-negative bacilli: survey of clinical isolates from seven laboratories collected in 1998 and 2000, in the Kinki region of Japan. J. Antimicrob. Chemother. 51:631-638. [DOI] [PubMed] [Google Scholar]
- 191.Yamazoe, K., N. Kato, H. Kato, K. Tanaka, Y. Katagiri, and K. Watanabe. 1999. Distribution of the cfiA gene among Bacteroides fragilis strains in Japan and relatedness of cfiA to imipenem resistance. Antimicrob. Agents Chemother. 43:2808-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yan, J. J., P. R. Hsueh, W. C. Ko, K. T. Luh, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yan, J. J., W. C. Ko, C. L. Chuang, and J. J. Wu. 2002. Metallo-β-lactamase-producing Enterobacteriaceae isolates in a university hospital in Taiwan: prevalence of IMP-8 in Enterobacter cloacae and first identification of VIM-2 in Citrobacter freundii. J. Antimicrob. Chemother. 50:503-511. [DOI] [PubMed] [Google Scholar]
- 194.Yan, J. J., W. C. Ko, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Outbreak of infection with multidrug-resistant Klebsiella pneumoniae carrying blaIMP-8 in a university medical center in Taiwan. J. Clin. Microbiol. 39:4433-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yan, J. J., J. J. Wu, S. H. Tsai, and C. L. Chuang. 2004. Comparison of the double-disk, combined disk, and Etest methods for detecting metallo-β-lactamases in gram-negative bacilli. Diagn. Microbiol. Infect. Dis. 49:5-11. [DOI] [PubMed] [Google Scholar]
- 196.Yang, Y., and K. Bush. 1996. Biochemical characterization of the carbapenem-hydrolyzing β-lactamase AsbM1 from Aeromonas sobria AER 14M: a member of a novel subgroup of metallo-β-lactamases. FEMS Microbiol. Lett. 137:193-200. [DOI] [PubMed] [Google Scholar]
- 197.Yang, Y. J., P. J. Wu, and D. M. Livermore. 1990. Biochemical characterization of a β-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob. Agents Chemother. 34:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yano, H., A. Kuga, R. Okamoto, H. Kitasato, T. Kobayashi, and M. Inoue. 2001. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob. Agents Chemother. 45:1343-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yatsuyanagi, J., S. Saito, S. Harata, N. Suzuki, Y. Ito, K. Amano, and K. Enomoto. 2004. Class 1 integron containing metallo-β-lactamase gene blaVIM-2 in Pseudomonas aeruginosa clinical strains isolated in Japan. Antimicrob. Agents Chemother. 48:626-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yomoda, S., T. Okubo, A. Takahashi, M. Murakami, and S. Iyobe. 2003. Presence of Pseudomonas putida strains harboring plasmids bearing the metallo-β-lactamase gene blaIMP in a hospital in Japan. J. Clin. Microbiol. 41:4246-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yong, D., K. Lee, J. H. Yum, H. B. Shin, G. M. Rossolini, and Y. Chong. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 40:3798-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yum, J. H., D. Yong, K. Lee, H. S. Kim, and Y. Chong. 2002. A new integron carrying VIM-2 metallo-β-lactamase gene cassette in a Serratia marcescens isolate. Diagn. Microbiol. Infect. Dis. 42:217-219. [DOI] [PubMed] [Google Scholar]