Abstract
Vibrio cholerae has multiple survival strategies which are reflected both in its broad distribution in many aquatic environments and its high genotypic diversity. To obtain additional information regarding the content of the V. cholerae genome, suppression subtractive hybridization (SSH) was used to prepare libraries of DNA sequences from two southern California coastal isolates which are divergent or absent in the clinical strain V. cholerae O1 El Tor N16961. More than 1,400 subtracted clones were sequenced. This revealed the presence of novel sequences encoding functions related to cell surface structures, transport, metabolism, signal transduction, luminescence, mobile elements, stress resistance, and virulence. Flanking sequence information was determined for loci of interest, and the distribution of these sequences was assessed for a collection of V. cholerae strains obtained from southern California and Mexican environments. This led to the surprising observation that sequences related to the toxin genes toxA, cnf1, and exoY are widespread and more common in these strains than those of the cholera toxin genes which are a hallmark of the pandemic strains of V. cholerae. Gene transfer among these strains could be facilitated by a 4.9-kbp plasmid discovered in one isolate, which possesses similarity to plasmids from other environmental vibrios. By investigating some of the nucleotide sequence basis for V. cholerae genotypic diversity, DNA fragments have been uncovered which could promote survival in coastal environments. Furthermore, a set of genes has been described which could be involved in as yet undiscovered interactions between V. cholerae and eukaryotic organisms.
Although it is best known as the causative agent of the human disease cholera, Vibrio cholerae is also an autochthonous inhabitant of many aquatic environments, including estuarine and coastal waters (13). Indeed, the great majority of its more than 200 serogroups, excluding O1 and O139, are not associated with epidemic disease. V. cholerae has been isolated routinely from many aquatic environments throughout the world, often in association with plankton, plants, invertebrates, and fish, and there are some reports of its presence in water birds, seals, and diseased farm animals (2, 26, 27, 33, 43, 56, 66). The prevalence of V. cholerae in the environment is influenced by temperature and salinity (reviewed in reference 42) as well concentrations of dissolved organic carbon (44). A number of genes have been previously implicated in environmental survival, and this has already led to a better understanding of the genetic basis for V. cholerae adaptations. Genes have been uncovered which are important for biofilm formation (for examples, see references 68 and 73), zooplankton association (9), survival with filamentous blue cyanobacteria (28), and the degradation of nonbiting midge (Chironomos sp.) egg masses (24). These genes clearly provide V. cholerae with mechanisms to avoid environmental stresses and obtain nutrients in aquatic environments.
The evolution of pandemic, virulent strains of V. cholerae has required the acquisition of large islands of genes encoding pathogenicity functions, including the vibrio pathogenicity island (32) and the cholera toxin phage genome (67). Additionally, human disease potential can be affected by other loci, including prophage K139, the superintegron, the sxt conjugative integrating element, O-antigen genes (reviewed in reference 18), and cryptic plasmid pTLC (59). While horizontal gene transfer has played an important role in the emergence of V. cholerae strains with altered human virulence, little is known about genome alterations (i.e., insertions or deletions) which influence this species' environmental distribution. By exploring the gene content of multiple strains within V. cholerae, it will be possible to discover new metabolic capabilities and survival strategies not represented in the single V. cholerae strain sequenced to date. To investigate this, we have undertaken a search for sequences present in two environmental V. cholerae strains which are lacking in the sequenced clinical O1 El Tor strain N16961 (25). The hypothesis underlying this effort is that additional genes encoding new survival strategies are likely to be found in nonpathogenic environmental V. cholerae strains, and the identification and characterization of these genes could provide useful information about factors that influence the environmental reservoir of V. cholerae. Consistent with this view, previous molecular fingerprinting studies have suggested a high level of genomic diversity among non-O1/non-O139 strains, suggesting that genomic flexibility enables environmental survival in this species (for examples, see references 4, 31, and 75).
A number of methods exist to characterize genetic differences among closely related bacteria. In the case of V. cholerae, representational difference analysis has previously been used by Calia et al. to uncover genes distinguishing pathogenic serotypes and biotypes (7), and this method was useful in the identification of an RTX toxin gene locus (41). In addition, comparative microarray analysis has revealed genes shared among pathogenic V. cholerae or unique to the El Tor biotype (15). Genetic differences among strains can also be characterized by suppression subtractive hybridization (SSH) (14), a technique that selectively amplifies DNA segments present in one bacterial genome (the tester) but not in another (the driver). These segments have arisen through one of two processes: recent acquisition in the strain of interest relative to the driver strain or loss in the driver strain while being retained in the strain of interest. This and other subtractive hybridization approaches have been used to discover virulence loci and pathogenicity islands (for examples, see references 53 and 74) and to explore genomic diversity in Thermotoga maritima (47). Here we report the use of SSH to isolate and subsequently characterize V. cholerae sequences present in two environmental isolates.
MATERIALS AND METHODS
Strains.
The Vibrio mimicus and V. cholerae strains used in this study are listed in the supplemental material (see Table S1 in the supplemental material). Briefly, 48 environmental V. cholerae isolates from Newport Beach, Calif., were kindly provided by Sunny Jiang, University of California, Irvine (30). These isolates were obtained over a 1-year period from several points along the San Diego Creek with different temperature and salinity profiles. Two environmental non-O1 V. cholerae isolates from Mission Bay, San Diego, Calif., were kindly provided by Barbara Hemmingsen, San Diego State University. Clinical V. cholerae strains O1 El Tor N16961 and O1 Classical O395 were kindly provided by John Mekalanos, Harvard University. An additional five V. cholerae strains were provided by M. Lizarraga-Partida, Centro de Investigacion Cientifica y de Educacion Superior de Ensenada, Ensenada, Mexico. V. mimicus was obtained from the American Type Culture Collection (ATCC 33653).
The two environmental isolates of V. cholerae that were the focus of this study were obtained on 13 July 2000, following an enrichment regimen similar to that which has been used by others (for examples, see references 10, 30, and 65) coupled with a diagnostic PCR screen. In particular, strain SIO was isolated from seawater collected off the Scripps Institution of Oceanography (SIO) pier during a bloom of the dinoflagellate Lingulodinium polyedrum, and V. cholerae strain TP was obtained from plankton collected in the Torrey Pines Beach State Preserve estuary. Additional information concerning the isolation of these strains is presented in the supplemental material.
Additional characterization of strains.
The V. cholerae isolates were examined to determine if they were members of the O1 serogroup by using a direct fluorescent antibody assay (cholera DFA; New Horizons Diagnostics Corporation, Columbia, Md.) and an Olympus BX51 fluorescence microscope. Substrate utilization preferences were scored by using the Microstation ML3 system from Biolog (Hayward, Calif.).
Comparison of V. cholerae strains SIO and TP by locus sequencing.
The V. cholerae strains were compared by sequencing rRNA genes (16S, 23S, and V. cholerae-V. mimicus-specific 16S-23S intergenic spacer region [ISR]) (12) and protein markers (deoxyribodipyrimidine photolyase-1 [phrB-1], deoxyribodipyrimidine photolyase-2 [phrB-2], DNA helicase II [uvrD], DNA helicase IV [helD], integrase/recombinase [xerC], and site-specific recombinase [intl4]) (Table 1; see Table S2 in the supplemental material). Primers and detailed methods used to amplify and sequence these genes are presented in the supplemental material. Furthermore, specific amplification of the ompW gene (46) and analysis of seven simple sequence repeat loci was performed to further establish V. cholerae identity (Y. Danin-Poleg, L. A. Cohen, H. Gancz, E. Malul, Y. Y. Broza, H. Goldschmidt, M. Halpern, M. Broza, and Y. Kashi, unpublished data).
TABLE 1.
Percent nucleotide sequence identity of phylogenetic marker genes to published N16961 sequences among V. cholerae strains used in this study
| Gene | % Identity to V. cholerae strain:
|
|
|---|---|---|
| TPb | SIOc | |
| 16S rRNA | 99.9 | 99.5 |
| 23S rRNA | 99 | 99 |
| Intergenic spacer region type 2a | 99.0 | 99.0 |
| phrB-1 | 96 | 96 |
| phrB-2 | 98 | 97 |
| uvrD | 94 | 93 |
| helD | 99 | 95 |
| xerC | 98 | 97 |
| intl4 | 97 | 90 |
Intergenic spacer region type 2 comparisons were made to the N16961 type 2 ISR amplified and sequenced for this study. All other comparisons were made to the sequenced N16961 genome.
Isolated from Torrey Pines Beach State Preserve estuary.
Isolated from SIO pier during dinoflagellate bloom.
Subtractive hybridizations.
Two subtracted libraries were constructed by using the PCR-select bacterial genome subtraction kit (Clontech, San Diego, Calif.). In both libraries, V. cholerae strain N16961 was used as the driver DNA to remove sequences shared between strain N16961 and the two environmental V. cholerae strains, SIO and TP. The manufacturer′s protocol was followed except that the final products were cloned into pCR4.0-Topo (Invitrogen, Carlsbad, Calif.). Clones were picked into 96-well microtiter plates and sequenced with M13F and M13R primers at the San Diego State University Microchemical Core Facility.
Computer analyses.
The ABI traces were analyzed by using the PHRED/PHRAP package (http://www.phrap.org/) to remove the vector sequence and mask a poor quality sequence as well as a sequence that was 85% identical to N16961 sequence over 20 bp or more. The sequence was masked and trimmed based on BLASTN searches using custom PERL scripts (http://bartlettlab.ucsd.edu). A masked sequence present at the ends of fragments was removed. SIO- and TP-specific masked and trimmed sequences were compared against the GenBank nonredundant database with BLASTX. The automated comparisons were performed by using the BLASTCL3 client available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The data were parsed with scripts available at http://bartlettlab.ucsd.edu to provide a best hit to the Vibrio sequence and a best hit to the database. The BLASTX results of matches to the entire database were used as the basis of the annotation described herein.
Contig assemblies of libraries of unique sequences in SIO and TP were performed by using DNASTAR (Madison, Wis.). Contigs were generated with sequences that underwent more stringent screening for adaptor sequences and other short repeated sequences that are artifacts of the SSH procedure. Contig assembly was dependent upon finding a match of 20 bp with 90% identity, and all other parameters of the DNASTAR program were maintained on default settings. The resulting contig spectra were used as the basis to approximate the completeness of the libraries and compute novel genome sizes for SIO and TP. The single sequences used to generate the contigs, as well as the assembled contig sequences, are available at http://bartlettlab.ucsd.edu.
To describe the DNA segments shared between SIO and TP, the sets of sequences used for contig assembly (as described above) were compared to each other by using the TBLASTX algorithm in two separate analyses (SIO versus TP and TP versus SIO). Sequences with an e value of less than 0.001 were considered significantly similar to one another. The categories of similar sequences were determined by matching each sequence to its top BlastX match and comparing the results between SIO and TP. These sequences were not masked for the N16961-like sequence (as described above); therefore, DNA segments shared between SIO, TP, and N16961 were found from this analysis. The proportion of similar sequences found to be unique to and shared between SIO and TP was determined by manually filtering out sequences with top BLASTX hits to the N16961 genome.
Panhandle PCR and sequencing of selected genes.
Panhandle PCR was performed to obtain additional genomic sequence data upstream and downstream of short reads of genes of interest in the SIO and TP environmental strains (16, 64). Detailed methods are presented in the supplemental material.
Colony hybridization.
To analyze the distribution of genes among other environmental strains of V. cholerae, we obtained strains isolated from southern California and Mexico from a number of laboratories (see Table S1 in the supplemental material). These strains, in addition to SIO, TP, and N16961, were streaked and patched onto L agar plates and incubated overnight at 30°C. Colonies were blotted onto Immobilon-NY+ 82-mm-pore-size nylon filters (Millipore, Bedford, Mass.) according to the method of Sambrook et al. (61). DNA was immobilized on the filters by UV cross-linking (Stratalinker) and/or baking.
Hybridization probes were synthesized by PCR and purified with the UltraClean PCR clean-up DNA purification kit (Mo Bio Laboratories, Solana Beach, Calif.). Primers were designed within or including regions of BlastX similarity to the genes of interest, with the following product sizes (in base pairs): toxA, 1,084; cnf, 704; cya, 427; luxA, 1,194; rnr, 1,258; rtx-ox, 324. Probes to the SIO plasmid are as follows: pSIO1-1 includes orf3, with similarity to virD4 genes (823 bp); pSIO1-2 includes orf2, with similarity to a gene from the plasmid pOM1 from Francisella tularensis (1,292 bp); pSIO1-3 includes a region similar to a 135-bp region from a plasmid from Vibrio shiloi (reclassified as Vibrio mediterranei) (1,042 bp); pSIO-4 includes orf1 (1,073 bp). Probe labeling, hybridization, washing, and detection were performed with the AlkPhosDirect labeling kit and ECF detection substrate (Amersham) according to the manufacturer's instructions, with modifications detailed in the supplemental material.
V. cholerae strain SIO plasmid cloning and sequencing.
Plasmid pSIO1 was isolated by using a QIAprep spin miniprep kit (QIAGEN). Restriction digestion with KpnI resulted in a linearized fragment, which was cloned into the KpnI site of the pUC18 vector and sequenced with primers derived from the pUC18 sequence or from prior rounds of pSIO1 sequencing.
Bioluminescence assay.
V. cholerae isolates demonstrating positive hybridization with the luxA probe were assayed for bioluminescence with a luminometer. Individual strains were grown for 14 h in Luria-Bertani at 30°C. Measurements were made on 1 ml of culture transferred to a scintillation vial by using an ATP photometer model 2000 (SAI Technology Company).
RESULTS AND DISCUSSION
Isolation and characterization of strains SIO and TP.
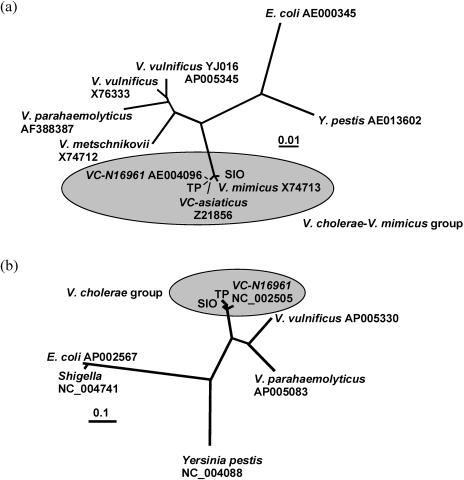
Two environmental strains of V. cholerae were isolated from a coastal and an estuary environment near SIO by following established enrichment techniques coupled with a PCR screen (see Materials and Methods). The strain obtained off of the SIO pier was designated SIO, and the strain recovered from the nearby Torrey Pines Beach State Preserve was designated TP. To confirm the identity of these isolates as V. cholerae, sequence information was obtained and analyzed for 16S rRNA genes, 23S rRNA genes, the ISR, phrB-1, phrB-2, uvrD, helD, xerC, and intl4. The results clearly demonstrated that SIO and TP are closely related to V. cholerae (Table 1). Phylogenetic trees based upon 16S rRNA genes and xerC sequences placed both SIO and TP within the V. cholerae-V. mimicus cluster (Fig. 1). Further comparisons of specific ISR regions from SIO, TP, assorted V. cholerae strains, and V. mimicus were performed in an attempt to assign SIO and TP to one or the other of these closely related species (12). Neighbor-joining phylogenetic trees deduced from alignments of ISR type 1 and 2 sequences always produced distinct V. cholerae and V. mimicus clusters. SIO and TP never grouped within the V. mimicus cluster, but in some percentage of cases, SIO also did not group with V. cholerae (data not shown). Further identification of SIO and TP was accomplished by specifically amplifying the ompW gene with primers documented to yield a PCR product in V. cholerae but not in V. mimicus (46). This gene was amplified in both SIO and TP, thus confirming that both isolates are V. cholerae strains (Y. Danin-Poleg and Y. Kashi, unpublished data). Simple sequence repeat analysis demonstrated that SIO and TP are not related to strains of the O1 or O139 serogroups (Danin-Poleg and Kashi, unpublished). These results are consistent with the direct fluorescence antibody assay and phylogenetic placement obtained with the BIOLOG GN substrate utilization assay. In light of these results, both SIO and TP were assigned as non-O1/non-O139 strains of V. cholerae.
FIG. 1.
Phylogeny of Vibrio cholerae strains SIO and TP based upon 16S rRNA gene sequence (A) and xerC sequence (B). Unrooted trees were generated by the neighbor-joining method. The scale bars indicate 0.01 (A) and 0.1 (B) substitutions per position.
Identification of genes unique to strains SIO and TP.
SSH was used to isolate loci present in the environmental strains (tester DNA sources) that were missing in strain N16961 (driver DNA source). The complete genome sequence of clinical isolate N16961 is available (25), which makes it a useful reference strain for comparative analyses. Subtracted clones were obtained and sequenced from the SIO and TP libraries (892 and 521, respectively). These single reads were trimmed of vector sequence and then masked for N16961-like sequence greater than 85% identical over more than 20 bp and compared against the nonredundant database by using BLASTX (3). It was also possible to prepare contigs from some of the single reads. This resulted in 114 SIO and 54 TP contigs with an average size of 472 bp (SIO) and 508 bp (TP). These sequences are available at http://bartlettlab.ucsd.edu.
When the masked SIO SSH single-read fragments were compared to the nonredundant database with the BlastX algorithm, about 45% returned no match. After removing fragments with insignificant (e value greater than 0.001) matches and those which duplicated previous matches, roughly 24% of the total number of fragments originally obtained remained for further analysis. Of these, 25% were most similar to V. cholerae N16961 proteins and 35% were similar to proteins found in other Vibrio species. Of the remainder, only two fragments were similar to the Archaea and Eukarya and the rest were similar to sequences in other Bacteria. However, roughly one-third of the sequences with a significant match in the database were most similar to hypothetical proteins in the Bacteria (Table 2). Performing the same analysis with the SSH fragments obtained from the TP strain showed that roughly 57% of the masked sequences returned no match to the nonredundant database. After removing fragments with insignificant matches and those which duplicated previous matches, there remained about 20% of the total number of fragments. Of these, only 7% were similar to N16961 proteins, 45% were similar to proteins found in other Vibrio species, and 46% were similar to other Bacteria. Only one match was to a protein found in the Eukarya, and there were no matches to archaeal proteins. Of the total number of sequences with significant matches to the database, roughly 20% were to hypothetical proteins (Table 2).
TABLE 2.
Functional categories of genes that are significantly similar to SIO and TP sequence fragments using the BlastX algorithm
| Functional category of genes | No. of genes (% of strain total) in strain:
|
|
|---|---|---|
| SIO | TP | |
| Hypothetical and unknown function | 75 (35.0) | 20 (21.1) |
| Intracellular functions | 7 (3.3) | 10 (10.5) |
| Bioluminescence | 3 | 6 |
| Cell division | 1 | 0 |
| Nonribosomal peptide synthase | 2 | 4 |
| Flagellar biosynthesis | 1 | 0 |
| Mobile DNA | 15 (7.0) | 10 (10.5) |
| Phage and plasmid encoded | 6 | 5 |
| Super-integron-related elements | 2 | 0 |
| Transposases and integrases | 7 | 3 |
| Retroelement and addiction module | 0 | 2 |
| DNA modification and structure | 9 (4.2) | 5 (5.3) |
| Restriction-modification systems | 6 | 4 |
| Helicases, Dam methylase, binding protein | 3 | 1 |
| Metabolism | 24 (11.2) | 14 (14.7) |
| Mannitol utilization | 0 | 4 |
| Starch utilization | 6 | 0 |
| DNA/RNA synthesis and processing | 2 | 0 |
| Other anabolism/catabolism | 16 | 10 |
| Transporters/uptake/secretion | 20 (9.3) | 5 (5.3) |
| ABC transporters | 4 | 0 |
| PTS permease | 1 | 1 |
| Secretion | 1 | 1 |
| Iron uptake | 2 | 1 |
| Other transporters | 12 | 2 |
| EPS/OM/O-antigen/murein biosynthesis | 16 (7.5) | 11 (11.6) |
| Extracellular polysaccharide synthesis/transport | 3 | 1 |
| O-antigen and LPS synthesis | 10 | 8 |
| Murein recycling | 3 | 0 |
| Other | 0 | 2 |
| Virulence | 12 (5.6) | 5 (5.3) |
| Toxins and hemolysins | 9 | 3 |
| Virulence-associated factors | 3 | 2 |
| Resistance to stress | 14 (6.5) | 6 (6.3) |
| Heat shock/protein folding | 1 | 1 |
| Efflux of toxics out of cell | 3 | 3 |
| Other | 10 | 2 |
| Regulation and signal transduction | 17 (7.9) | 8 (8.4) |
| Two-component regulatory systems | 4 | 0 |
| LysR-like, GGDEF domain, and other regulators | 10 | 7 |
| Chemotaxis | 3 | 1 |
| Archaea | 3 (1.4) | 1 (1.1) |
| Eukarya | 2 (0.9) | 0 (0.0) |
| Total | 214 (100) | 95 (100) |
V. cholerae genomic diversity.
Although SSH is not a quantitative procedure, it was possible to estimate the amount of novel sequence present in the genomes of SIO and TP. Calculating the total number of unique base pairs in the consensus sequences of the contigs and the remaining single reads provides one such estimate. After removing N16961-like sequence also recovered by SSH, there could be 0.20 Mb of novel sequence in SIO and 0.14 Mb in TP. To account for the incomplete recovery of DNA segments by SSH, a Chao2 estimator (8), typically applied to estimate species richness in ecological studies, was also used, indicating 0.8 Mb and 0.7 Mb of novel sequence in SIO and TP, respectively. One issue with these estimates is that the SSH library was not constructed from randomly generated DNA segments, since a restriction enzyme, RsaI, was used in the first step to generate fragments of DNA. Thus, some sequences are more likely to be amplified in this procedure simply due to the proximity of RsaI restriction sites. With this caveat in mind, it can be concluded that the amount of novel sequence is in the range of 3 to 20% of the N16961 genome size for these bacteria.
To determine the set of DNA segments shared between SIO and TP but absent from N16961, the SSH libraries from SIO and TP were compared to one another by using the TBLASTX algorithm after removing adaptor sequences but prior to masking for the N16961-like sequence. A significant proportion of the shared segments were most similar to the N16961 sequence and were not considered further. Of the 214 types of unique gene segments found in SIO (Table 2), only 41 were shared with TP. This result suggests that a minority of gene fragments are shared and that each strain has retained a significant number of unique genes. There was no tendency for one category of metabolic function to be represented more than any other. Preliminary microarray analyses comparing the number of SIO and TP genes shared with N16961 indicate that SIO and TP strains are missing 14 and 9% of N16961 genes, respectively (M. Miller and G. Schoolnik, unpublished data). Roughly 6% of N16961 genes are missing in SIO but are present in TP, while only 1% of N16961 genes are missing in TP but are present in SIO. Approximately 8% of N16961 genes are missing in both SIO and TP. In contrast, clinical, pandemic, prepandemic, and environmental strains of the O1 or O139 serogroups or the El Tor or Classical biotypes lacked only ∼1% of N16961 genes (15).
Other studies have estimated the genomic variation among strains of the same species, and most have found percentages of strain-specific genes similar to the values reported above for SIO, TP, and N16961. The percentages can vary from less than 4% in Pseudomonas aeruginosa PAO1 compared to clinical and environmental strains (70), to 2.6 to 16.3% among Campylobacter jejuni strains isolated from different sources (51), to 12 to 18% in Helicobacter pylori (60), and up to 20% in T. maritima (47). In general, genome variation within a single species can be as high as 26%, with many isolates showing variation between 5 and 15% (summarized in reference 47).
Categories of genes present in SIO and TP but not in N16961.
The DNA segments recovered from the SSH were compared to the nonredundant database with the BLASTX algorithm, and they were placed in 12 categories based upon the cellular function of the most similar sequence in the database (Table 2). Similar categories of novel sequences were present within the two environmental V. cholerae strains. In both cases, a large percentage of the genes was associated with metabolism, mobile elements, restriction-modification systems, and the cell surface. However, strain SIO possessed a larger fraction of sequences related to transporters and stress resistance genes, whereas TP had more unique genes related to intracellular pathways and DNA mobilization. These categories of genes are typical of those that appear to vary between closely related strains of the same species in other bacteria. For example, extensive differences in O-antigen genes have previously been detected within strains of a number of species (55, 74), including within other V. cholerae strains (for an example, see reference 39). The V. cholerae examined in this study shared differences with Aeromonas hydrophila and H. pylori among related strains in restriction-modification systems (1, 74), with T. maritima in sugar transport and utilization genes (47), and with Escherichia coli in phage sequences (52). Specific findings, highlighting DNA segments of particular interest, are presented below.
Lipopolysaccharide structure.
Several SIO segments were similar to genes within the lipopolysaccharide core oligosaccharide biosynthesis gene cluster (wav gene cluster) of two O6 serogroup strains (48). The TP strain had a sequence related to a O-antigen ligase present in a non-O1/non-O139 strain (48). Several other segments in TP were most similar to unknown or hypothetical proteins encoded within the O-antigen synthesis cluster of O139 and O22 serogroups (72).
Nutrient uptake.
SIO and TP both had genes that appeared to be indicative of iron uptake, including a segment similar to a heme receptor present in V. mimicus. SIO was also found to possess many sequences related to carbohydrate utilization, including a starch utilization operon in Klebsiella oxytoca (19) and glycosidases in Vibrio vulnificus.
DNA transfer.
Top BlastX matches to genes indicative of DNA transfer and rearrangement included transposases present in the genomes of other gram-negative bacteria. In the SIO strain, there were segments similar to cassettes associated with the super integrons in Vibrionaceae (58). In TP, a segment significantly similar to the retroelement EC67 originally described for clinical strains of E. coli was found, although retrons have been identified previously in V. cholerae (63).
Plasmid pSIO1.
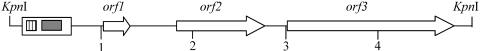
The presence of a number of SSH sequences showing similarity to plasmid genes prompted further study of the possibility of extrachromosomal DNA in the environmental V. cholerae strains. High-copy plasmid preparations from strain TP did not yield any DNA, but those from strain SIO resulted in the isolation of a 4.9-kbp element which was designated pSIO1. The complete nucleotide sequence of pSIO1 was obtained, revealing three open reading frames of approximately 300, 900, and 1,800 bp in length. Intriguingly, ORF2 and ORF3 were similar to genes involved in DNA transfer, and there is a region very similar to a sequence present in plasmids from V. shiloi and another environmental V. cholerae isolate. These results are summarized in Fig. 2 and Table 3.
FIG. 2.
Structure of plasmid pSIO1 (4,906 bp). The white box indicates a 337-bp region 96% identical to a region in the cryptic plasmid pVC isolated from a Vibrio cholerae strain in Hong Kong. The shaded box indicates a 135-bp region 88% identical to similar region in the coral pathogen Vibrio shiloi (reclassified as Vibrio mediterranei). The striped box designates a 27-bp region identical to a portion of the p15A replicon. Size markings (in kbp) are indicated.
TABLE 3.
Top BlastX matches of genes of interest
| Putative gene name | GenBank accession no. | ORF length (aa)a | Top match(es) after BLASTX search | e value | Accession no. of similar protein |
|---|---|---|---|---|---|
| toxA | AY876053 | 656 | Exotoxin A precursor from P. aeruginosa PA01 | 2e-84 | NP_249839.1 |
| cnf | AY876054 | 846 | Cytotoxic necrotizing factor type 1 from V. fischeri | 1e-122 | JE0362 |
| Cytotoxic necrotizing factor 1 from E. coli | 3e-64 | AF483829.1 | |||
| cya | AY876055 | 1,046 | Probable inner membrane protein VC0822 from N16961 | 5e-42 | D82276 |
| 126,-000-molecular-weight pathogenicity-associated protein from V. cholerae | 5e-42 | T09441 | |||
| Adenylate cyclase ExoY from P. aeruginosa PA01 | 1e-19 | NP_250881.1 | |||
| rtx | Unknown protein from Synechocystis sp. strain PCC 6803 | 2e-31 | NP_442018.1 | ||
| Autotransporter adhesin from V. vulnificus CMCP6 | 1e-30 | NP_761533.1 | |||
| RTX protein from A. salmonicida | 3e-22 | AF218037.1 | |||
| luxA | AY876056 | Bacterial luciferase alpha chain from V. harveyi | 1e-149 | P07740 | |
| rnr | Putative VacB/RNase R from V. parahaemolyticus | 0.0 | NP_798269.1 | ||
| pSIO1 orf1 | AY876057 | 104 | No significant matches in database | ||
| pSIO1 orf2 | AY876057 | 312 | Integrase (COG0582) from Azotobacter vinelandii | 3e-72 | ZP_00091084.1 |
| Orf3 on plasmid pOM1 from Francisella tularensis | 3e-66 | NP_052245.1 | |||
| Putative integrase from Bordetella bronchiseptica | 7e-56 | NP_887463.1 | |||
| pSIO1 orf3 | AY876057 | 595 | VirD4-like protein encoded by gene on plasmid pMD136 from Pediococcus pentosaceus | 1e-19 | NP_037557.1 |
| TraG from Pseudomonas aeruginosa | 2e-13 | AAP22624 |
ORF, open reading frame; aa, amino acids.
Additional characterization of selected genes.
Surprisingly, several DNA fragments were recovered from strains SIO and TP which had strong similarity to genes encoding virulence toxins in the bacterial pathogens P. aeruginosa, uropathogenic E. coli, and others. Since these sequences had not been previously observed in V. cholerae, additional sequence information surrounding these fragments was obtained to ascertain the following: (i) whether each of these sequences was part of an uninterrupted open reading frame, (ii) whether the genes were present in a single island of recently transferred DNA rather than scattered throughout the genome, (iii) the GC content of these genes, as a means of estimating whether they have been recently transferred, and (iv) the conservation of amino acid residues in the encoded protein required for its putative enzymatic function. Finally, DNA fragments related to genes involved in bioluminescence were recovered, and as very few V. cholerae strains are bioluminescent (54), additional sequence information was obtained. In all cases, additional information was obtained by panhandle PCR. The results are summarized in Table 3.
Exotoxin A.
A total of 3.7 kbp of sequence was obtained surrounding an open reading frame of 1,968 bp encoding the putative toxA gene from TP. It is located between sequences homologous to VC1644 and VC1645 on chromosome I of the V. cholerae N16961 genome. P. aeruginosa toxA encodes exotoxin A, which is a member of a family of ADP-ribosylating enzymes, including cholera, diphtheria, and pertussis toxins, E. coli heat-labile enterotoxin, and Salmonella enterica SpvB (34, 38, 49). The TP-deduced enzyme sequence includes most of the conserved residues involved with receptor binding (with a number of insertions) and all of the residues important for disulfide bond formation and localization to the endoplasmic reticulum as well as a glutamic acid residue within the catalytic domain which is absolutely conserved among members of this enzyme family (37, 69).
Cytotoxic necrotizing factor.
A 4.4-kbp segment of TP sequence was obtained surrounding a 2.5-kbp open reading frame encoding a protein similar to cytotoxic necrotizing factor type 1 (CNF1) and CNF2 found in E. coli (6) and to the putative cnf gene of Vibrio fischeri (40). In E. coli, CNF1 is encoded within a pathogenicity island and is associated with extraintestinal infections (5). The toxin causes reorganization of actin microfilaments, formation of stress fibers, and membrane ruffling in eukaryotic cells (20, 62). The TP protein sequence shows strong conservation of the amino-terminal receptor binding domain and membrane spanning domain, but the remainder of the protein, including the catalytic domain, has sustained significant deletions.
Adenylate cyclase.
A 4.4-kbp segment of TP DNA was obtained surrounding an open reading frame encoding a protein similar in its amino terminus to the deduced 126-kDa pathogenicity-associated protein encoded on the V. cholerae vibrio pathogenicity island I and in its carboxy terminus to a secreted adenylate cyclase from P. aeruginosa (exoY) (71), Bacillus anthracis (cya) (36), and Bordetella pertussis (cyaA) (22). The function of the V. cholerae 126-kDa protein gene is unknown (32). Like other secreted adenylate cyclases, the TP protein lacks a signal peptide sequence and encodes all of the residues documented to be important for nucleotide binding (23, 71).
Repeat in structural toxin.
A 5.5-kbp segment of SIO DNA sequence was obtained as two contigs which included a region of BLASTX similarity to RTX (repeat in structural toxin) proteins present in Aeromonas salmonicida and E. coli with 55% identity over 251 amino acids along one portion of the Aeromonas sequence. No RTX consensus could be found in the protein sequence, but repetitive sequence was evident: 18 amino acids were repeated five times within a total stretch of 160 amino acids. The RTX gene family comprises a group of diverse proteins present in a number of gram-negative bacteria (35). These include some hemolysins, leukotoxins, and metalloproteases, which can insert themselves into target membranes and form pores (35). An RTX gene cluster has previously been found to contribute to virulence and to cross-link actin in V. cholerae O1 El Tor strains N16961 and E7946 and to depolymerize the actin cytoskeleton in a variety of mammalian cell lines (21, 41).
RNase R.
A 2.38-kbp segment was obtained from strain SIO which included a region with similarity to rnr, an exoribonuclease which is involved in the control of virulence gene expression in Shigella flexneri and enteroinvasive E. coli (11, 72).
LuxA.
A 2.0-kbp segment was obtained from TP which included open reading frames related to the luxA and luxD products of Vibrio harveyi. The luxA and luxD genes encode the alpha subunit of the luciferase enzyme and an acyl transferase, or myristoyl-ACP-specific thioesterase, respectively. luxA sequences have previously been detected in other V. cholerae environmental strains (50, 54), and at least one bioluminescent V. cholerae strain has been described previously (50). TP and SIO were evaluated for bioluminescence with a luminometer. Neither strain was visibly luminescent, but low-level light emission was detected from TP.
Distribution of selected genes among additional V. cholerae strains.
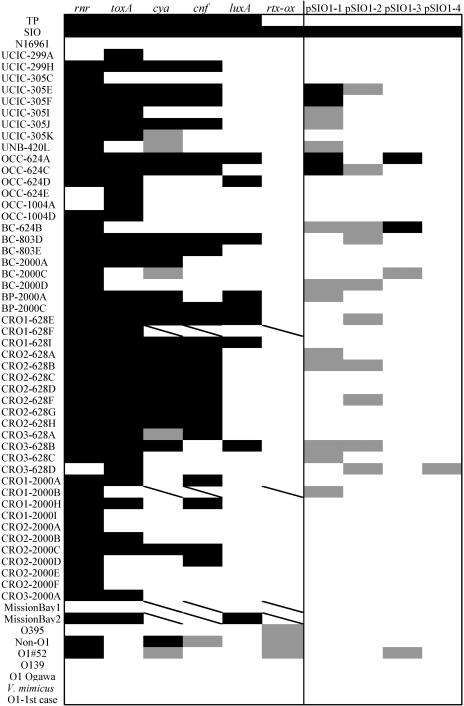
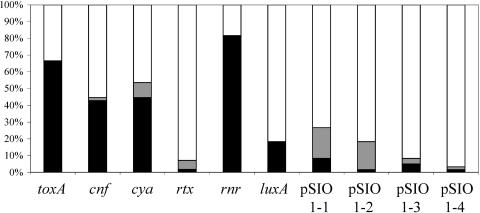
The distribution of all of the above selected gene sequences, as well as segments of plasmid pSIO1, was determined by colony hybridization for more than 50 V. cholerae isolates. Most of these isolates were obtained by Jiang and Fu from coastal and river samples obtained in southern California near Newport Beach (30). Several additional San Diego County coastal isolates, several strains from Mexico, a Classical O1 strain (O395), an El Tor O1 strain (N16961), and a V. mimicus control were also examined. The results for each strain are presented in Fig. 3, and a compilation of the results for each probe is presented in Fig. 4.
FIG. 3.
Distribution of selected environmental genes among individual Vibrio isolates. Colony hybridization results for a panel of environmental genes of interest (listed along the top) against a panel of Vibrio isolates obtained primarily from the southwestern United States (listed along the left side) are shown. Positive signal is indicated by black shading, no signal is indicated by white shading, and a faint positive signal is indicated by grey shading. A slash indicates that no data were obtained. All strains are Vibrio cholerae, unless otherwise designated. See Materials and Methods for additional descriptions of probes and strains used.
FIG. 4.
Percent distribution of selected environmental genes among Vibrio strains. For the putative cnf and toxA genes and the rtx-like region, a total of 55 Vibrio cholerae isolates and 1 Vibrio mimicus strain were included. For all other genes, a total of 59 Vibrio cholerae isolates and 1 Vibrio mimicus strain were included. Black shading indicates the percentage of isolates which are hybridization positive for the gene of interest; grey shading indicates the percentage of isolates which exhibit weak hybridization to the gene probe.
Most of the putative virulence genes identified in the SSH were present in many of the environmental isolates collected off the southern California coast. The rnr gene was widely distributed among the isolates, with greater than 80% yielding a positive signal. The putative toxA gene, encoding a protein similar to exotoxin A from P. aeruginosa, was present in more than 60% of the isolates. Roughly half of the isolates contained the putative cya or cnf gene. In contrast, the RTX probe only hybridized to SIO, the strain from which it was derived, although previous results have shown that V. cholerae rtxA genes are widely distributed among O1, O139, and non-O1 environmental and clinical isolates (11). This suggests that the RTX-like region in SIO may be a narrowly distributed sequence feature. The plasmid pSIO1 sequences were also present in only a few isolates. The luxA probe hybridized to 18% of isolates examined, and this hybridization frequency is similar to that previously reported by Palmer and Colwell (50). In their study, 15% of environmental and clinical V. cholerae strains strongly hybridized to a luxA probe, and another 15% weakly hybridized to the probe. SIO and TP shared all putative virulence genes, except for the gene hybridizing to the RTX probe.
We hypothesize that the genes encoding proteins with significant similarity to toxins could contribute to a pathogenic phenotype with an as yet unknown host organism(s), since the only known function of these genes is virulence, the sequences do not appear to be pseudogenes (i.e., containing frameshift mutations), and they are widespread. By comparison, genes encoding the cholera toxin have been found in 17% of non-O1/non-O139 strains in southern California (29), 10% of non-O1/non-O139 strains from Brazil (57), and 21% of non-O1/non-O139 isolates from Calcutta, India (45). Thus, the intriguing possibility exists that the toxin-like genes uncovered here could actually be more widespread within environmental V. cholerae than are the ctx toxin genes. Furthermore, these genes do not appear to be recently horizontally transferred, based upon %GC content analysis, and they are not present in a single region in the chromosome (data not shown).
This observation may be especially timely, as environmental non-O1/non-O139 strains lacking ctxA and tcpA were recently shown to be virulent in mammalian models of V. cholerae (17). Virulence was not necessarily correlated with the presence of known virulence factors, suggesting that environmental strains of V. cholerae may have additional undiscovered virulence genes. It remains to be determined whether the virulence-like gene sequences uncovered here contribute to mammalian virulence or virulence against aquatic organisms present in the environment from which strains TP and SIO were obtained.
In summary, the data presented here have provided information about the amount and the nature of DNA sequence differences existing between non-O1/non-O139 environmental V. cholerae and the sequenced O1 El Tor strain. Much additional information is needed, including the nature of more subtle gene sequence variation among strains and genome variation in V. cholerae from other environments including those associated with cholera outbreaks. Furthermore, the effects of these chromosomal alterations on V. cholerae phenotypic diversity, including environmental fitness and biogeography, must be determined.
Supplementary Material
Acknowledgments
We are grateful to Sunny Jiang, Marcial Lizarraga-Partida, John Mekalanos, and Barbara Hemmingsen for generously providing V. cholerae strains. We gratefully acknowledge Michael Miller and Gary Schoolnik for generously sharing unpublished results, and Yael Danin-Poleg and Yechezkel Kashi for performing the simple sequence repeat analysis and sharing their technique with us prior to publication. We also thank Suhelen Egan for assistance with panhandle PCR and Hideto Takami for assistance with plasmid annotation. Finally, we also thank three anonymous reviewers for insightful comments.
This work was supported by grant AI46600-01 from the National Institute of Allergy and Infectious Disease to F.A. and D.H.B. A.P. is supported by a Howard Hughes Medical Institute Predoctoral Fellowship.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D., A. DePaola, S. Chirtel, and R. B. Young. 1998. Detection of Vibrio cholerae in oyster (Crassostrea virginica) homogenate based on centrifugal removal of antimicrobial agents. J. Microbiol. Methods 33:237-244. [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bik, E. M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum, G., V. Falbo, A. Caprioli, and J. Hacker. 1995. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin from the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol. Lett. 126:189-196. [DOI] [PubMed] [Google Scholar]
- 6.Boquet, P. 2001. The cytotoxic necrotizing factor 1 (CNF1) from Escherichia coli. Toxicon 39:1673-1680. [DOI] [PubMed] [Google Scholar]
- 7.Calia, K. E., M. K. Waldor, and S. B. Calderwood. 1998. Use of representational difference analysis to identify genomic differences between pathogenic strains of Vibrio cholerae. Infect. Immun. 66:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 9.Chiavelli, D. A., J. W. Marsh, and R. K. Taylor. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choopun, N., V. Louis, A. Huq, and R. R. Colwell. 2002. Simple procedure for rapid identification of Vibrio cholerae from the aquatic environment. Appl. Environ. Microbiol. 68:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow, K. H., T. K. Ng, K. Y. Yuen, and W. C. Yam. 2001. Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, J., A. Huq, and R. R. Colwell. 1999. Analysis of 16S-23S intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl. Environ. Microbiol. 65:2202-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 14.Diatchenko, L., Y.-F. C. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egan, S., S. James, C. Holmstrom, and S. Kjelleberg. 2002. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 4:433-442. [DOI] [PubMed] [Google Scholar]
- 17.Faruque, S. M., N. Chowdhury, M. Kamruzzaman, M. Dziejman, M. H. Rahman, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. USA 101:2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruque, S. M., and J. J. Mekalanos. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11:505-510. [DOI] [PubMed] [Google Scholar]
- 19.Fiedler, G., M. Pajatsch, and A. Bock. 1996. Genetics of a novel starch utilization pathway present in Klebsiella oxytoca. J. Mol. Biol. 256:279-291. [DOI] [PubMed] [Google Scholar]
- 20.Flatau, G., E. Lemichez, M. Gauthier, P. Chardin, S. Paris, C. Fiorentini, and P. Boquet. 1997. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature 387:729-733. [DOI] [PubMed] [Google Scholar]
- 21.Fullner, K. J., and J. J. Mekalanos. 2000. In vivo cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 19:5315-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glaser, P., D. Ladant, O. Sezer, F. Pichot, A. Ullmann, and A. Danchin. 1988. The calmodulin-sensitive adenylate cyclase of Bordetella pertussis: cloning and expression in Escherichia coli. Mol. Microbiol. 2:19-30. [PubMed] [Google Scholar]
- 23.Glaser, P., H. Sakamoto, J. Bellalou, A. Ullman, and A. Danchin. 1988. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 7:3997-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halpern, M., H. Gancz, and Y. Kashi. 2003. Vibrio cholerae hemagglutinin/protease degrades chironomid egg masses. Appl. Environ. Microbiol. 69:4200-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. Mcdonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam, M. S., B. S. Drasar, and D. J. Bradley. 1990. Long-term persistence of toxigenic Vibrio cholerae 01 in the mucilaginous sheath of a blue-green alga, Anabaena variabilis. J. Trop. Med. Hyg. 93:133-139. [PubMed] [Google Scholar]
- 27.Islam, M. S., B. S. Drasar, and R. B. Sack. 1993. The aquatic environment as a reservoir of Vibrio cholerae: a review. J. Diarrhoeal Dis. Res. 11:197-206. [PubMed] [Google Scholar]
- 28.Islam, M. S., M. M. Goldar, M. G. Morshed, M. N. H. Khan, M. R. Islam, and R. B. Sack. 2002. Involvement of the hap gene (mucinase) in the survival of Vibrio cholerae O1 in association with the blue-green alga, Anabaena sp. Can. J. Microbiol. 48:793-800. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, S., W. P. Chu, and W. X. Fu. 2003. Prevalence of cholera toxin genes (ctxA and zot) among non-O1/O139 Vibrio cholerae strains from Newport Bay, California. Appl. Environ. Microbiol. 69:7541-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, S. C., and W. Fu. 2001. Seasonal abundance and distribution of Vibrio cholerae in coastal waters quantified by a 16S-23S intergenic spacer probe. Microb. Ecol. 42:540-548. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, S. C., V. Louis, N. Choopun, A. Sharma, A. Huq, and R. R. Colwell. 2000. Genetic diversity of Vibrio cholerae in Chesapeake Bay determined by amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 66:140-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karaolis, D. K. R., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaysner, C. A., C. Abeyta, Jr., M. M. Wekell, A. DePaula, Jr., R. F. Stott, and J. M. Leitch. 1987. Incidence of Vibrio cholerae from estuaries of the United States west coast. Appl. Environ. Microbiol. 53:1344-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krueger, K. M., and J. T. Barbieri. 1995. The family of bacterial ADP-ribosylating exotoxins. Clin. Microbiol. Rev. 8:34-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lally, E. T., R. B. Hill, I. R. Kieba, and J. Korostoff. 1999. The interaction between RTX toxins and target cells. Trends Microbiol. 7:356-361. [DOI] [PubMed] [Google Scholar]
- 36.Leppla, S. H. 1982. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cAMP concentration in eukaryotic cells. Proc. Natl. Acad. Sci. USA 79:6532-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesnick, M. L., and D. G. Guiney. 2001. The best defense is a good offense—Salmonella deploys an ADP-ribosylating toxin. Trends Microbiol. 9:2-5. [DOI] [PubMed] [Google Scholar]
- 38.Lesnick, M. L., N. E. Reiner, J. Fierer, and D. G. Guiney. 2001. The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol. Microbiol. 39:1464-1470. [DOI] [PubMed] [Google Scholar]
- 39.Li, M. R., T. Shimada, J. G. Morris, A. Sulakvelidze, and S. Sozhamannan. 2002. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect. Immun. 70:2441-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, J.-W., L.-M. Chen, H.-Y. Chen, and S.-F. Weng. 1998. Identification and analysis of the regulatory region R&R* with the cnf1 gene encoding the cytotoxic necrotizing factor type 1 that closely links to the lux regulon of Vibrio fischeri. Biochim. Biophys. Acta 250:462-465. [DOI] [PubMed] [Google Scholar]
- 41.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 96:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipp, E. K., I. N. G. Rivera, A. I. Gil, E. M. Espeland, N. Choopun, V. R. Louis, E. Russek-Cohen, A. Huq, and R. R. Colwell. 2003. Direct detection of Vibrio cholerae and ctxA in Peruvian coastal water and plankton by PCR. Appl. Environ. Microbiol. 69:3676-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mourino-Perez, R. R., A. Z. Worden, and F. Azam. 2003. Growth of Vibrio cholerae O1 in red tide waters off California. Appl. Environ. Microbiol. 69:6923-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nandi, B., R. K. Nandy, S. Mukhopadhyay, G. B. Nair, T. Shimada, and A. C. Ghose. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38:4145-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nesbo, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nesper, J., A. Kraib, S. Schild, J. Blab, K. E. Klose, J. Bockemuhl, and J. Reidl. 2002. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect. Immun. 70:2419-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otto, H., D. Tezcan-Merdol, R. Girisch, F. Haag, M. Rhen, and F. Koch-Nolte. 2000. The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Mol. Microbiol. 37:1106-1115. [DOI] [PubMed] [Google Scholar]
- 50.Palmer, L., and R. R. Colwell. 1991. Detection of luciferase gene sequence in nonluminescent Vibrio cholerae by colony hybridization and polymerase chain reaction. Appl. Environ. Microbiol. 57:1286-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 52.Pradel, N., S. Leroy-Setrin, B. Joly, and V. Livrelli. 2002. Genomic subtraction to identify and characterize sequences of Shiga toxin-producing Escherichia coli O91:H21. Appl. Environ. Microbiol. 68:2316-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 54.Ramaiah, N., J. Chun, J. Ravel, W. L. Straube, R. T. Hill, and R. R. Colwell. 2000. Detection of luciferase gene sequences in nonluminescent bacteria from the Chesapeake Bay. FEMS Microbiol. Ecol. 33:27-34. [DOI] [PubMed] [Google Scholar]
- 55.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: Identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddacliff, G. L., M. Hornitzky, J. Carson, R. Petersen, and R. Zelski. 1993. Mortalities of goldfish, Carassius-auratus (L), associated with Vibrio cholerae (non01) infection. J. Fish Dis. 16:517-520. [Google Scholar]
- 57.Rivera, I. N. G., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowe-Magnus, D. A., A.-M. Guerout, and D. Mazel. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 13:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubin, E. J., W. Lin, J. Mekalanos, and M. K. Waldor. 1998. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 28:1247-1254. [DOI] [PubMed] [Google Scholar]
- 60.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and F. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Schmidt, G., P. Sehr, M. Wilm, J. Selzer, M. Mann, and K. Aktories. 1997. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387:725-729. [DOI] [PubMed] [Google Scholar]
- 63.Shimamoto, T., M. Kobayashi, T. Tsuchiya, S. Shinoda, H. Kawakami, S. Inouye, and M. Inouye. 1999. A retroelement in Vibrio cholerae. Mol. Microbiol. 34:631-632. [DOI] [PubMed] [Google Scholar]
- 64.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.U.S. Food and Drug Administration. 1998. Bacteriological analytical manual, 8th ed. AOAC International, Gaithersburg, Md.
- 66.Visser, I. J. R., P. Vellema, H. van Dokkum, and T. Shimada. 1999. Isolation of Vibrio cholerae from diseased farm animals and the surface water in the Netherlands. Vet. Rec. 144:451-452. [DOI] [PubMed] [Google Scholar]
- 67.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 68.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wedekind, J. E., C. B. Trame, M. Dorywalska, P. Koehl, T. M. Raschke, M. McKee, D. FitzGerald, R. J. Collier, and D. B. McKay. 2001. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J. Mol. Biol. 314:823-837. [DOI] [PubMed] [Google Scholar]
- 70.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamasaki, S., H. Shimizu, K. Hoshino, S.-T. Ho, T. Shimada, G. B. Nair, and Y. Takeda. 1999. The genes responsible for O-antigen synthesis of Vibrio cholerae O139 are closely related to those of Vibrio cholerae O22. Gene 237:321-332. [DOI] [PubMed] [Google Scholar]
- 73.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, Y. L., C. T. Ong, and K. Y. Leung. 2000. Molecular analysis of genetic differences between virulent and avirulent strains of Aeromonas hydrophila isolated from diseased fish. Microbiology 146:999-1009. [DOI] [PubMed] [Google Scholar]
- 75.Zo, Y. G., I. N. G. Rivera, E. Russek-Cohen, M. S. Islam, A. K. Siddique, M. Yunus, R. B. Sack, A. Huq, and R. R. Colwell. 2002. Genomic profiles of clinical and environmental isolates of Vibrio cholerae O1 in cholera endemic areas of Bangladesh. Proc. Natl. Acad. Sci. USA 99:12409-12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.