Abstract
Bacteriophage S-PM2 infects several strains of the abundant and ecologically important marine cyanobacterium Synechococcus. A large lytic phage with an isometric icosahedral head, S-PM2 has a contractile tail and by this criterion is classified as a myovirus (1). The linear, circularly permuted, 196,280-bp double-stranded DNA genome of S-PM2 contains 37.8% G+C residues. It encodes 239 open reading frames (ORFs) and 25 tRNAs. Of these ORFs, 19 appear to encode proteins associated with the cell envelope, including a putative S-layer-associated protein. Twenty additional S-PM2 ORFs have homologues in the genomes of their cyanobacterial hosts. There is a group I self-splicing intron within the gene encoding the D1 protein. A total of 40 ORFs, organized into discrete clusters, encode homologues of T4 proteins involved in virion morphogenesis, nucleotide metabolism, gene regulation, and DNA replication and repair. The S-PM2 genome encodes a few surprisingly large (e.g., 3,779 amino acids) ORFs of unknown function. Our analysis of the S-PM2 genome suggests that many of the unknown S-PM2 functions may be involved in the adaptation of the metabolism of the host cell to the requirements of phage infection. This hypothesis originates from the identification of multiple phage-mediated modifications of the host's photosynthetic apparatus that appear to be essential for maintaining energy production during the lytic cycle.
Strains of unicellular cyanobacteria of the genera Synechococcus and Prochlorococcus are abundant in the world's oceans and constitute the prokaryotic component of the picophytoplankton. Together, these photosynthetic bacteria contribute a significant proportion of primary production in oligotrophic regions of the oceans (21, 35, 37, 68). Viral infection of marine unicellular cyanobacteria was first reported in 1990 (53, 63), and cyanovirus isolates were first characterized in the laboratory in 1993 (62, 69, 74). The majority of these phages belong to the myoviruses. Myoviruses are physically robust and remarkably versatile; this virion design can apparently be easily adapted to a variety of different ecological niches (64). S-PM2 is a lytic cyanomyovirus with an icosahedral head and long contractile tail that infects marine Synechococcus strains. The genome has been shown to have a size of ∼194 kb (27). Bacteriophage T4 that infects Escherichia coli is the archetype myovirus, and S-PM2 was shown to have a genetic module that encodes distant homologues of most of the major virion proteins of T4 (27). T4 has been extensively studied and is extremely well understood; it serves as a superb, if somewhat complex, model for S-PM2.
A previous phylogenetic analysis of the sequences of the major head and tail genes of a wide range of T4-type bacteriophages indicated at least three distinct phylogenetic subgroups of these phages (64). There is a large cluster of phages, termed the T-evens, members of which are all closely related to T4, the archetype of the Myoviridae. The second subgroup is surprisingly phylogenetically divergent from the T-evens, but morphologically similar; these are called the pseudoT-evens (47), and they includes phages such as RB49 and RB42 that infect E. coli. The third cluster includes Aeromonas phages and vibriophages such as nt-1, KVP20, KVP40, 65, and Aeh1. Such phages have heads that are more elongated than those of both T-evens and the pseudoT-evens and thus are called the schizoT-evens (64). Phylogenetic analysis based on the major capsid protein gp23 has shown that S-PM2 and the related cyanomyovirus S-PWM3 are quite distinct from the other characterized T4-like phages and form a new discrete group, the exoT-evens (27). These marine T4-type phages have apparently diverged significantly from the T4 archetype. Beyond the fact that they have a contractile tail, these phages have little morphological resemblance to the other T4-type phages. Among the many differences between the exoT-evens and the other T-type phages are those that relate to the photosynthetic physiology of their hosts. It is clear that S-PM2 (41) and several other marine cyanomyoviruses (36, 43) encode homologues of the D1 and D2 proteins of the host photosystem II that presumably become associated with the bacterial photosynthetic apparatus. In order to ascertain the extent to which S-PM2 shares additional modules with the other T-type phages or its host, we sequenced and characterized the complete genome of this novel “photosynthetic” phage.
MATERIALS AND METHODS
Propagation of phage S-PM2 and DNA extraction.
Phage S-PM2 was originally isolated from the English Channel (74) and was subsequently reassigned as a myovirus (73). It was propagated on Synechococcus sp. strain WH7803 grown in artificial seawater (ASW), as previously described (74) (7). Cyanophages were purified by polyethylene glycol precipitation and CsCl gradient centrifugation, and cyanophage DNA was then extracted by phenol extraction and alcohol precipitation as described previously (74). Briefly, 1 liter of exponentially growing Synechococcus culture (optical density at 750 nm of between 3.5 and 4) was infected with S-PM2 at a multiplicity of infection of approximately 1. Following lysis cell, debris was then removed by adding 58.4 g of NaCl and incubating on ice for 1 h, followed by centrifugation at 11,000 × g for 10 min. Phages were then precipitated from the supernatant by adding PEG 6000 (Sigma) to a final concentration of 10% (wt/vol) and incubating in ice water for 1 h, followed by a further centrifugation at 11,000 × g for 10 min. The precipitated cyanophages were resuspended in approximately 2 ml of ASW. This was layered onto a CsCl step gradient (made up in ASW) and centrifuged at 4°C in an SW40 Beckman rotor at 22,000 × g. Concentrated cyanophages were removed and dialyzed against ASW prior to DNA extraction by using 1 volume of Tris.HCl buffer (pH 8.0)-saturated phenol, followed by 1:1 Tris-HCl buffer (pH 8.0)-saturated phenol-chloroform, and finally with chloroform-isoamyl alcohol (24:1). The DNA from the resulting aqueous layer was precipitated by adding 2 volumes of isopropanol and 0.4 volumes of 7.5 M ammonium acetate. After incubation for 10 min at room temperature, the precipitated DNA was collected by centrifugation and redissolved in Tris-EDTA buffer.
DNA sequencing.
The initial phase of sequencing the S-PM2 genome was carried out commercially by Agowa GmbH, Berlin. The bacteriophage DNA was shotgun cloned into the plasmid pUC19. For this purpose, 20 μg of bacteriophage DNA was sonicated, and the resulting fragments were purified by agarose gel electrophoresis. The fraction of 1,200 to 1,800 bp was eluted from the gel, and the DNA fragments were blunted with T4 DNA polymerase. These fragments were subcloned into an SmaI-digested, alkaline phosphatase-treated pUC19 sequencing vector. The subclones were sequenced by using Big Dye-terminator chemistry (Applied Biosystems). Data were collected by using ABI 3730 automated sequencers (Applied Biosystems). A total of approximately 600 clones were sequenced to cover the complete bacteriophage genome six- to eightfold. Sequence data were further processed by the program PHRED (http://www.phrap.org/phredphrapconsed.html) and assembled by using the program GAP4 (http://staden.sourceforge.net/staden_home.html). This approach yielded 14 contigs totaling 170.9 kb of sequence.
The contigs were ordered by PCR with multiple primers and joined by primer walking. This was done by designing PCR primers (Primer Designer 3.0; Scientific and Educational Software, Durham, N.C.) to extend outwards from the contigs. PCR was then performed by using purified S-PM2 DNA as a template. If the two contigs were adjacent, a PCR product was obtained. This product was then gel purified (with QIAGEN gel extraction kits) and used as a template in a sequencing reaction. The product was sequenced in both directions. This process was continued until the PCR product was fully sequenced. Primers were then designed to the reverse of the sequenced strand to ensure each region was sequenced in both directions. Approximately 25 kb of the genome was sequenced by using primer walking. Contigs and new sequence data were assembled and ambiguities were identified by using SEQMAN (DNASTAR, Inc. Madison, Wis.). Primers were designed to the ends of the contigs and to any equivocal regions of the genome. The resulting PCR products were sequenced. This work involved over 100 different primers sets, and optimization was typically done by using the following thermal cycler conditions: 1 cycle for 1 min at 94°C, linked to 34 cycles of 20s at 94°C, then for 20s at 50 to 65°C, and then for 30 s to 4 min at 72°C, depending on the size of the product (30 s was added for every 500 bp of product). Optimal conditions were then used to prepare the template for sequencing. Each sequencing reaction was analyzed in an ABI Prism 3100 sequencer and used 100 ng of DNA and Big Dye version 3.1.
Computer analysis of DNA and protein sequences.
The completed genomic sequence of S-PM2 was analyzed by both Glimmer (14) and GenemarkS (3) to predict protein coding regions and by tRNAscan-SE version 1.21 (38) to predict tRNA genes. Genome annotation was carried out by using Artemis (56), and genome comparison was made using ACT (www.sanger.ac.uk/Software/ACT/). The similarity of putative S-PM2 proteins with proteins in the NCBI nr database and at the T4-like Genome website (phage.bioc.tulane.edu/) was detected by using BLAST (2). Prediction of homologies was also carried out by using the GTOP database (spock.genes.nig.ac.jp/∼genome/gtop.html). PSORT-B (20), TMHMM (34), SOSUI (30), and MaxH (5) were used to predict the localization of proteins, and SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) was used to detect possible signal peptide cleavage sites. Putative right-handed beta-helix folds in proteins were detected by using BetaWrap (6). The presentation of the genome was produced with our own custom software written in the Q script language version 6 (http://www.star.le.ac.uk/∼rw). Promoter consensus features were generated by using WebLogos (12).
The completed S-PM2 genome sequence was deposited in the EMBL database under the accession number AJ630128.
RESULTS AND DISCUSSION
Genetic organization of the S-PM2 genome and its similarities to T4.
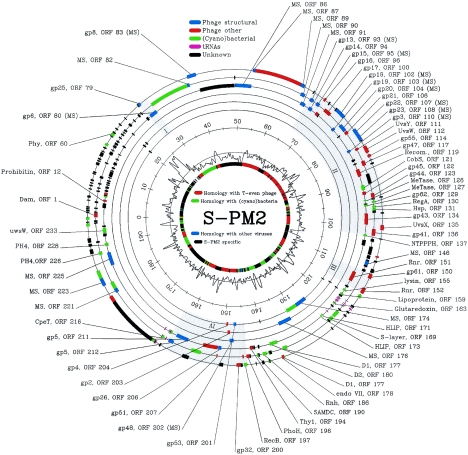
The circularly permuted genome of phage S-PM2 is 196,280 bp in size (Fig. 1) (EMBL accession number AJ630128). Its G+C content (37.8%) differs significantly from the value of 59.4% (51) of Synechococcus sp. WH8102, a laboratory host. Interestingly, the mol% GC of the T4 genome is also substantially lower than that of its host (46). Bioinformatics methods (see Materials and Methods) identified 239 probable protein-coding sequences and 25 tRNA genes in this genome. Table 1 shows all the open reading frames (ORFs) that encode proteins with clear homologues in other phages or cellular organisms. A complete list of all ORFs and tRNA genes and their coordinates in the S-PM2 genome is found in the EMBL database (AJ630128). There is only one potential intron in the S-PM2 genome, interrupting ORF 177 (psbA); although it is only 212 nucleotides long, it has the canonical features of a group I self-splicing intron (43). There is a marked asymmetry in the distribution of the genes on the two phage DNA strands, with 248 on the plus strand and only 16 on the minus strand. The S-PM2 genome was previously shown to contain blocks of genes homologous to coliphage T4 (27) (A. Letarov and H. M. Krisch, unpublished observations). This limited initial observation of homology to a portion of the T4 genome is now considerably extended. A total of at least 40 ORFs have convincing homology to sequences of the T4-type phages. The vast majority these T4-type ORFs are grouped together in four clusters. The smallest one, cluster I (coordinates 26866 to 29066), consists of only two contiguous ORFs encoding homologues of the baseplate subunits gp25 and gp6; in the T4 genome these genes are not adjacent, but they are in the schizoT-even phage KVP40 (45). Cluster II (coordinates 66833 to 102295) is much larger, ∼35 kb, and encodes 24 homologues of T4 genes. This genomic region had been only partially characterized previously (27). It starts with an ORF encoding a gp13 homologue and extends as a largely contiguous block to gp23, essentially maintaining the same gene order as in T4. Thus, this module contains most of the structural components of the phage head and the contractile tail. The synteny, however, ceases after ORF 108 (the major capsid protein g23), which is followed by g3, the tail sheath terminator gene. The T4-type genes further downstream in cluster II encode homologues of many of the T4 replication and repair proteins (UvsY, UvsW, UvsX, gp41, gp43, gp44, gp45 gp46, and gp62). Hence, cluster II encodes the virion structural proteins as well as other T4-like proteins involved in DNA replication, recombination, transcription, and translational control. Cluster III (coordinates 110575 to 115044) contains just three T4-type ORFs, those encoding the α- and β-subunits of ribonucleotide reductase and DNA primase (gp61). Cluster IV (coordinates 144631 to 162664) includes several genes encoding baseplate hub subunits (gp5, gp26, gp48, and gp53), the head completion protein (gp4), and proteins involved with DNA replication (gp32 and gp59), DNA end protection (gp2), and late transcription (gp33).
FIG. 1.
Organization of the genome of phage S-PM2. The circles from outside to inside indicate the following: 1 to 6 represent the six reading frames, 7 is the scale bar (in kilobases), 8 is G+C content (smoothed with a sigma = 200 bp; Gaussian; range is 28.2 to 60.4%, with a mean of 37.8%), and 9 shows homology with other organisms. Labels show ORF numbers and gene designations where a putative homologue in T4 has been identified. MS indicates that presence of protein in the virion has been established by mass spectrometry and is shown in parentheses if the ORF has already been shown to encode a virion structural protein on the basis of proposed homology. The following color scheme has been used: green, ORFs encoding proteins exhibiting similarity to cyanobacterial proteins; blue, phage structural proteins; red, other phage proteins; purple, tRNA genes; and black, unidentified ORFs. The four clusters of T4-like genes are shaded in gray.
TABLE 1.
ORFs with putative homologues in other phages or cellular organisms or identified as a virion structural protein by mass spectroscopya
| ORF | Size (aa) | T4 similarity | E-value | Comment |
|---|---|---|---|---|
| 1 | 295 | 2e-66 | Possible Dam methylase | |
| 12 | 134 | 3e-25 | Possible prohibitin. | |
| 16 | 136 | 5e-06 | Similar to KVP40 CDS 229 | |
| 53 | 170 | 4e-31 | Similar to Gloeobacter violaceus hypothetical protein | |
| 60 | 200 | 4e-07 | Similar to Trichodesmium hypothetical protein; contains a prolyl 4-hydroxylase domain | |
| 79 | 130 | gp25 | 2e-09 | Baseplate wedge subunit |
| 80 | 602 | gp6 | 3e-40 | Baseplate wedge subunit |
| 81 | 3048 | 3e-13 | Similar to Novosphingobium aromaticivorans hypothetical protein | |
| 82 | 175 | Virion structural protein | ||
| 83 | 634 | gp8 | 4e-15 | Baseplate wedge subunit, virion structural protein |
| 86 | 1251 | Virion structural protein | ||
| 87 | 168 | Virion structural protein | ||
| 88 | 3338 | 9e-5 | Similar to phage Aeh1 hypothetical protein | |
| 89 | 306 | Virion structural protein, contains a fibrinogen domain | ||
| 90 | 327 | 9e-5 | Virion structural protein, contains a pentraxin domain | |
| 91 | 379 | Virion structural protein | ||
| 93 | 267 | gp13 | 2e-07 | Neck protein, virion structural protein |
| 94 | 292 | gp14 | 4e-16 | Neck protein |
| 95 | 266 | gp15 | 7e-26 | Proximal tail sheath stabilizer, virion structural protein |
| 96 | 139 | gp16 | 3e-05 | Terminase subunit |
| 100 | 548 | gp17 | 1e-97 | Terminase large subunit |
| 102 | 743 | gp18 | 5e-62 | Contractile tail sheath protein, virion structural protein |
| 103 | 204 | gp19 | 4e-17 | Tail tube protein, virion structural protein |
| 104 | 564 | gp20 | 0 | Portal vertex protein, virion structural protein |
| 106 | 214 | gp21 | e-105 | Prohead protease |
| 107 | 392 | gp22 | e-112 | Prohead core protein, virion structural protein |
| 108 | 468 | gp23 | 0 | Major capsid protein, virion structural protein |
| 110 | 169 | gp3 | 0.073 | Tail completion protein, virion structural protein |
| 111 | 157 | UvsY | 0.039 | Recombination and repair |
| 112 | 487 | UvsW | 6e-61 | DNA helicase |
| 114 | 164 | gp55 | 7e-13 | Sigma factor for late transcription |
| 117 | 349 | gp47 | 1e-36 | Recombination endonuclease |
| 119 | 576 | gp46 | 2e-75 | Recombination protein |
| 121 | 372 | 5e-26 | Partial similarity to Salmonella enterica cobalt insertion protein CobS; has a MoxR-like ATPase domain | |
| 122 | 221 | gp45 | 8e-11 | Sliding clamp |
| 123 | 315 | gp44 | 2e-45 | Sliding clamp loader subunit |
| 126 | 59 | 6e-06 | Similar to N-terminal portion of Nostoc methyl transferase | |
| 127 | 132 | 7e-13 | Similar to C-terminal portion of Nostoc methyl transferase | |
| 129 | 128 | gp62 | 1e-12 | Sliding clamp loader subunit |
| 130 | 142 | RegA | 2e-39 | Translation repressor of early genes, |
| 131 | 137 | 1e-19 | Small heat shock protein (hsp 20 family), | |
| 134 | 830 | gp43 | 1e-136 | DNA polymerase |
| 135 | 343 | UvsX | 3e-35 | RecA-like recombination protein |
| 136 | 470 | gp41 | 3e-90 | DNA primase-helicase subunit, |
| 137 | 135 | 5e-10 | Similar to conserved hypothetical bacterial protein family; contains a putative nucleotide pyrophosphohydrolase domain | |
| 146 | 316 | Virion structural protein | ||
| 150 | 329 | gp61 | 1e-33 | DNA primase subunit |
| 151 | 776 | NrdA | 1e-158 | Ribonucleotide reductase alpha subunit |
| 152 | 391 | NrdB | 1e-108 | Ribonucleotide reductase beta subunit |
| 155 | 136 | 1e-39 | Similar to Nostoc lysin; contains a phage lysozyme domain | |
| 159 | 120 | 1e-20 | Possesses a rare lipoprotein A-2 domain, | |
| 163 | 81 | 7e-06 | Glutaredoxin | |
| 167 | 75 | 1e-08 | Similar to N-terminal portion of putative tryptophan halogenase; contains a tryptophan halogenase domain | |
| 168 | 410 | 1e-08 | Similar to C-terminal portion of tryptophan halogenase; contains a tryptophan halogenase domain | |
| 169 | 279 | 33-13 | Possesses an S-layer homology domain | |
| 171 | 162 | 5e-04 | High-light inducible protein | |
| 173 | 39 | 1e-4 | High-light inducible protein | |
| 174 | 1095 | Virion structural protein, possible adhesin | ||
| 175 | 1058 | 2e-07 | Similar to putative Leptospira interrogans outer membrane protein | |
| 176 | 1177 | Virion structural protein, possible adhesin | ||
| 177 | 359 | 9e-65 | Photosynthetic reaction centre protein D1, contains a 212 bp intron | |
| 178 | 143 | 3e-67 | Similar to bacteriophage phiYeO3-12 ORF; contains an endonuclease VII domain | |
| 180 | 353 | 2e-58 | Photosynthetic reaction centre protein D2 | |
| 182 | 83 | 7e-04 | Partial similarity to glutaredoxin | |
| 185 | 78 | 1e-19 | Similar to a conserved family of cyanobacterial proteins | |
| 186 | 287 | Rnh | 9e-49 | RNase H |
| 190 | 110 | 1e-23 | S-adenosylmethionine decarboxylase | |
| 194 | 213 | 2e-61 | Thymidylate synthase-complementing protein (homologue in phage P60) | |
| 196 | 250 | 3e-21 | Partial similarity to PhoH, ATPase related to phosphate starvation (homologue in phage P60). | |
| 197 | 229 | 7e-14 | Similar to hypothetical Nostoc protein; contains a RecB domain | |
| 198 | 249 | gp33 | 0.003 | gp33 phage 44RR |
| 200 | 295 | gp32 | 5e-52 | Single-strand DNA binding protein |
| 201 | 220 | gp53 | 1e-08 | Base plate wedge component. |
| 202 | 332 | gp48 | 2e-08 | Base plate, tail tube associated; virion structural protein |
| 203 | 220 | gp2 | 8e-12 | DNA end protector protein |
| 204 | 145 | gp4 | 8e-33 | Head completion protein. |
| 206 | 238 | gp26 | 3e-19 | Base plate hub subunit |
| 207 | 57 | 6e-05 | Possible base plate hub assembly catalyst | |
| 208 | 1167 | 9e-10 | Partial similarity to bacteriophage CP-1 orf18 | |
| 209 | 749 | Contains a domain found in membrane proteins related to metalloendopeptidases | ||
| 211 | 981 | gp5? | 4e-08 | Partial similarity baseplate hub subunit and tail lysozyme |
| 212 | 367 | gp5? | 3e-04 | Partial similarity baseplate hub subunit and tail lysozyme |
| 213 | 114 | 4e-11 | Similar to marine Synechococcus and Prochlorococcus conserved hypothetical proteins | |
| 216 | 175 | 3e-20 | Similar to cyanobacterial CpeT | |
| 218 | 193 | 2e-06 | Partially similar to prolyl 4-hydroxylase alpha subunit | |
| 221 | 295 | Virion structural protein | ||
| 222 | 567 | 1e-15 | Partially similar to Methanosarcina hypothetical protein; shares a similar C-terminal domain with ORFs 223 and 224 | |
| 223 | 560 | 5e-07 | Partially similar to Methanosarcina hypothetical protein; shares a similar C-terminal domain with ORFs 222 and 224; virion structural protein | |
| 224 | 582 | 3e-11 | Partially similar to Methanosarcina hypothetical protein; shares a similar C-terminal domain with ORFs 222 and 223 | |
| 225 | 1037 | 5e-07 | Possible short tail fiber; virion structural protein | |
| 226 | 238 | 2e-16 | Similar to a conserved bacterial family of proteins; contains a prolyl 4-hydroxylase domain | |
| 228 | 231 | 2e-21 | Similar to a conserved bacterial family of proteins; contains a prolyl 4-hydroxylase domain | |
| 229 | 169 | Contains a prolyl 4-hydroxylase domain | ||
| 233 | 415 | UvsW | 2e-04 | DNA or RNA helicase. |
| 235 | 202 | 9e-13 | Similar to Ralstonia hypothetical protein | |
| 239 | 186 | 5e-15 | Similar to Helicobacter hypothetical protein |
Mass spectroscopy data are from our laboratory (unpublished results).
A prominent and puzzling feature of the S-PM2 genome is the region extending from ORF 2 to ORF 78 (∼26 kb), in which only a few of the predicted proteins (e.g., ORF 12) are not database orphans. The large majority (69 out of 76) of these ORFs encode small proteins of fewer than 150 amino acids (aa). Most of the uncharacterized proteins of T4 are also quite small; 62 predicted T4 ORFs encode proteins of fewer than 100 aa, and the smallest known functional protein in T4, Stp, contains of only 29 aa. The shortest ORF in the S-PM2 genome is ORF 166 with 30 amino acids. Similarly, there are many small ORFs in the T4-type Vibriophage KVP40 (45). The role that these numerous and often conserved small proteins play in the life cycle of large virulent phages is unclear. We speculate that they may have a crucial role as accessory factors that bind to and subtly modify the specificity of host proteins so that they function appropriately during phage infection.
Paradoxically, S-PM2 also encodes extremely large proteins; 10 ORFs encode more than 1,000 aa, and one is a giant sequence of nearly 3,800 aa. There is a notable clustering of seven of these large ORFs. Three ORFs (174, 175, and 176) are contiguous and are transcribed from the “wrong” negative strand of S-PM2. Four other large ORFs (81, 84, 86, and 88), while not contiguous, are in propinquity. In terms of nucleotide composition, there is only one region of the S-PM2 genome that has a clearly anomalous G+C composition. This region, from 21.2 to 25.00 kb, has a 55.2% GC content, 2.5 standard deviations above the average for the genome. Interestingly, this region is particularly sparsely populated with predicted ORFs (Fig. 1).
Transcription.
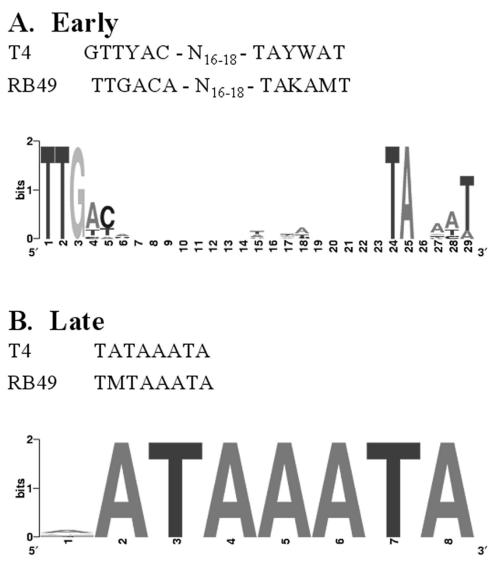
An intriguing feature of T4 infection is the complex choreography of phage gene expression (49). Much of the temporal control is mediated by a cascade of modifications in the specificity of the host RNA polymerase. In T4 infections, the earliest phage transcription involves the recognition of a consensus promoter sequence that differs from the sequence recognized by the host RNA polymerase with an associated σ70 (71). The sequence of σ70-dependent promoters in both cyanobacteria and E. coli are very similar to each other (13). Since the S-PM2 genome has no homologues of the characterized T4 early genes (Alt, ModA, and ModB) that modify the specificity of the host RNA polymerase, the S-PM2 early promoters would, a priori, be expected to be quite similar in sequence to the σ70 promoters of their host. Visual sequence analysis of the region of the early regions of the S-PM2 genome revealed a series of sequences that contained −35 and −10 boxes characteristic of the σ70 promoter recognition sequence. Almost without exception, these phage sequences were on the correct strand and in a context that was perfectly compatible with an early promoter function. Among the genes with putative early promoters (Table 2) are those encoding the PSII reaction center proteins D1 and D2 together with the σ-factor required for late gene transcription. The consensus sequence (TTGHH-N18-TANNHW) for the putative early S-PM2 promoters is shown in Fig. 2 and compared to those of T4 and RB49.
TABLE 2.
ORFs associated with putative early promoters
| Promoter coordinatesa | ORF start coordinate | ORF | Comment |
|---|---|---|---|
| 885-913 | 925 | 2 | |
| 1042-1070 | 1049 | 3 | |
| 3515-3543 | 3550 | 11 | |
| 5322-5350 | 5370 | 16 | Similar to phage KVP40 CDS 229 |
| 15773-15801 | 15769 | 53 | Similar to Gloeobacter violaceus hypothetical protein |
| 17468-17496 | 17526 | 57 | |
| 19880-19908 | 19921 | 64 | |
| 22290-22318 | 22340 | 72 | |
| 84480-84508 | 84532 | 114 | gp55 sigma factor for late transcription |
| 93713-93741 | 93757 | 124 | |
| 96625-96653 | 96674 | 132 | |
| 102150-102178 | 102267 | 137 | Similar to conserved hypothetical bacterial protein family; contains a putative nucleotide pyrophosphohydrolase domain |
| 103581-103609 | 103581 | 140 | |
| 110519-110547 | 110575 | 150 | DNA primase subunit |
| 116293-116321 | 116307 | 155 | |
| 124309-124337 | 124362 | 171 | |
| 128351-128379R | 128316R | 174 | possible adhesin |
| 135214-135242 | 135236 | 177 | D1 |
| 137766-137794 | 137820 | 180 | D2 |
| 145229-145257 | 145298 | 198 | |
| 145887-145915 | 145969 | 200 | |
| 154887-154915 | 154952 | 209 | |
| 162737-162765 | 162825 | 213 | |
| 163183-163211 | 163191 | 214 |
R, reverse strand.
FIG. 2.
Consensus features of S-PM2 putative early (A) and late (B) promoters. The features were calculated by using WebLogo (12). The height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of each symbol within the stack reflects the relative frequency of the corresponding base at that position. The consensus promoter features of phages T4 and RB49 (15) are shown for comparison.
Later during a T4 infection, the MotA gene encodes a transcription factor that replaces the host σ70, which is now inactivated by the phage Asi function. Such a middle mode transcription does not appear to occur in S-PM2. This phage lacks homologues of both MotA and Asi. In this respect S-PM2 resembles the only other well-characterized T4-type phage, RB49, where the absence of a T4-like middle mode of transcription has also been noted (15). As in RB49, the middle mode of transcription in S-PM2 appears to have been largely replaced by overlapping transcription that initiates from early and late promoters (15). All the S-PM2 homologues of the T4 genes whose expression is MotA dependent in T4 have both early and late promoters in the S-PM2 genome. In this way transcription of these genes can be assured during different periods of the phage cycle (15).
Regardless of these striking differences in the transcription of S-PM2 and T4 during the first part of infection, there is a strong similarity between them with regard to late transcription. S-PM2 has a homologue of T4 g55, an alternative σ-factor required for the transcription of late genes including those encoding head, tail, and fiber proteins (72). Two other phage proteins, gp33 and gp45 (sliding clamp), are important in late gene transcription in T4, and S-PM2 has homologues of both these genes. The consensus sequence of the −10 region of late promoters in T4 is TATAAATA, though the first T is sometimes replaced by an A (46, 72). In S-PM2 the late consensus sequence, NATAAATA, is slightly less rigid, although the sequence is most frequently AATAAATA (Fig. 2). Nearly all of these promoters are found in a context that is fully consistent with a late promoter function (Table 3). For example, the sequence NATAAATA is found upstream of the various homologues of virion proteins (gp8, gp17, gp18, etc.) that are only expressed late in T4 (39, 72). Thus, it seems that S-PM2, like RB49 and KVP40 (45), resembles T4 in the way in which it initiates late gene expression. It is interesting that the two genes psbA and psbD encoding the two photosystem II proteins D1 and D2 (see below) are also preceded by both putative early and late T4-like late promoter sequences, suggesting that they may be transcribed throughout the infection cycle. In contrast, there is a large block of viral genes extending from ORF 17 to ORF 52 that lack detectable consensus early or late promoters. Are these genes only transcribed under certain environmental conditions and require a novel σ-factor? Intriguingly, we have identified a conserved promoter-like sequence motif (MCNCCRNARNNNNNNNTNNWRNNNTAWNMTA) located at several positions in the S-PM2 genome just after potential transcription termination signals. These motifs are invariably located at the 5′ end of what appears to be a long, densely packed, polycistronic operon composed exclusively, or nearly so, of S-PM2 database orphan genes. Since these putative S-PM2 operons lack both consensus early or late phage promoter sequences, we are now investigating a possible role of the motif in the transcription of these large blocks of unknown phage genes.
TABLE 3.
ORFs associated with putative late promoters
| Promoter coordinatesa | ORF start coordinate | ORF | Comment |
|---|---|---|---|
| 4136-4142 | 4219 | 13 | |
| 25168-25174 | 25268 | 75 | |
| 26835-26841 | 26866 | 79 | gp25 base plate wedge |
| 38767-38773 | 38809 | 83 | gp8 base plate wedge |
| 49118-49124 | 49179 | 86 | Virion structural protein |
| 53450-53456 | 53491 | 88 | Similar to hypothetical phage Aeh1 protein |
| 66560-66566 | 66595 | 92 | |
| 66798-66804 | 66833 | 93 | gp13 neck protein |
| 69733-69739 | 69759 | 97 | Similar to hypothetical phage Aeh1 protein |
| 70827-70833 | 70856 | 100 | gp17 terminase large subunit |
| 72774-72780 | 72817 | 102 | gp18 contractile tail sheath protein |
| 75701-75707 | 75737 | 104 | gp20 portal vertex protein |
| 77424-77430 | 77454 | 105 | |
| 78284-78290 | 78328 | 107 | gp22 prohead core |
| 81533-81539 | 81564 | 110 | gp3 tail completion protein |
| 96601-96607 | 96674 | 132 | |
| 102999-103005 | 103025 | 139 | |
| 106774-106780 | 106808 | 143 | |
| 107814-107820 | 107846 | 145 | |
| 108090-108096 | 108134 | 146 | Virion structural protein |
| 115170-115176 | 115200 | 153 | |
| 117312-117318 | 117438 | 159 | Rare lipoprotein A-2 domain |
| 117800-117806 | 117831 | 160 | |
| 119712-119718 | 119815 | 162 | |
| 128355-128361R | 128316R | 174 | Virion structural protein, possible adhesin |
| 135130-135136R, 135148-135154R | 135094R | 176 | Virion structural protein, possible adhesin |
| 135241-135247 | 135326 | 177 | D1 |
| 136654-136660 | 136684 | 178 | gp49 endonuclease VII |
| 137793-137799 | 137820 | 180 | D2 |
| 143623-143629 | 143633 | 195 | |
| 149642-149648, 149648-149654 | 149677 | 205 | |
| 154865-154871 | 154952 | 209 | Metalloendopeptidase domain |
| 163661-163667 | 163747 | 215 | |
| 165885-165891 | 165928 | 219 | |
| 188463-188469 | 188506 | 227 |
R, reverse strand.
Translation. (i) Alternative start codons.
Of the predicted ORFs of S-PM2, 84.1% have an AUG start codon; however, the remaining ORFs start with either GUG or UUG start codons, each at a 7.9% frequency. These results are in contrast to the situation in T4, where alternative start codons are rare (46). However, high frequencies of unusual start codons have been reported for phage KVP40 (45). Codon usage in the S-PM2 genome is constrained by the low mol% GC content, and there are marked differences in preferences for synonymous codons compared to those of one its hosts, Synechococcus sp. WH8102. S-PM2 encodes 23 apparently functional tRNA genes and 2 tRNA pseudo-genes organized almost entirely into two large adjacent blocks. This situation is similar to that of KVP40, which has a total of 30 tRNA genes (including five pseudo-genes) organized in a single block (45). By contrast T4 has only eight tRNA genes (48). Although S-PM2 has a low mol% GC content, only 13 out of 23 of the tRNAs recognize codons with A or U in the third position. Fourteen of the codons recognized by the phage tRNAs are among the least frequently used codons in the Synechococcus sp. WH8102 genome (51).
(ii) Translational coupling.
When the translation initiation of a downstream gene is dependent on the translation of the gene immediately upstream, the translation of these genes is said to be coupled. In phage genomes such as T4, where the protein coding capacity is extremely densely packed, transcriptional coupling is commonplace (46). Such coupling has been shown to be a major factor in maintaining the correct stoichiometry of the subunits in protein assemblies (66). Bacteriophage T4 genes have been inferred to be translationally coupled when the downstream initiation codon is close to, or overlaps, the upstream stop codon, and there are 52 clusters of genes where this situation applies (46). Applying the same diagnostic methods to S-PM2 indicates that there are 129 genes in 47 clusters that may be translationally coupled.
Translational repression plays a significant role in posttranscriptional regulation of gene expression in T4 (44). RegA, in particular, binds and translationally represses more than a dozen T4 early mRNAs. A homologue of the T4 RegA is encoded by S-PM2, and it is very likely that this protein plays a similar regulatory role during S-PM2 infection as does the T4 RegA protein. Two of the central proteins of T4 DNA replication apparatus, gp32 and gp43, are each self-regulatory at the level of translation, binding to sequences near the ribosomal binding site of their respective messages and thus inhibiting their own translation initiation. The sequences and structural features implicated in the translational repression of both genes 32 and 43 in T4 (52, 60) are not obviously conserved in the homologues of these genes in S-PM2. Either these S-PM2 genes have no translational control, or translational control may be functionally conserved, but in such a case the regulatory sites involved must have massively diverged.
DNA replication, repair, recombination, and nucleotide metabolism.
The T4 replisome consists of seven proteins: DNA polymerase (gp43), sliding clamp loader (gp44 and gp62), sliding clamp (gp45), DNA helicase (gp41), DNA primase (gp61), and single-strand DNA binding (gp32) proteins (46). Additionally, RNase H and DNA ligase are required to join Okazaki fragments. Potential homologues of all these proteins, except DNA ligase (which is not actually critical in T4 infection since the host-encoded enzyme can, under certain circumstances, substitute for it) are encoded by S-PM2. Thus, it seems that replisome structure is generally conserved in the two phages. T4 recombination is intimately coupled to replication because phage recombinational intermediates are often used to initiate chromosome replication. The key proteins involved in T4 homologous recombination are gp32, UvsX, UvsY, gp46, and gp47, and all of these proteins have homologues in S-PM2. Thus, like the situation in T4, recombination apparently has an important role in the S-PM2 life cycle. S-PM2 encodes a homologue of T4 UvsW, a key component in T4 replication and recombination during the later stages of infection. In general the proteins involved in T4 recombination are also involved in DNA repair. Broken or damaged T4 replication forks can be repaired by join-copy recombination, and homologues of the key T4 repair proteins such as UvsX are encoded by S-PM2.
T4 encodes a nucleotide precursor complex that takes both cellular nucleoside diphosphates and the deoxynucleotide monophosphates from host-DNA breakdown and converts them into deoxyribonucleotide triphosphates for T4 DNA synthesis (25). S-PM2 encodes at least two enzymes of nucleotide metabolism, aerobic ribonucleotide diphosphate reductase and thymidylate synthase. Thus, S-PM2 is potentially capable of scavenging ribonucleotides for DNA synthesis. The marine cyanopodovirus P60 also encodes ribonucleotide diphosphate reductase (10), and the thymidylate synthase gene was found in the marine phage roseophage S101, which infects strains of Roseobacter (55). The majority of enzymes required for phage DNA synthesis would appear to be phage encoded. However, there is no evidence that S-PM2 utilizes any modified nucleotides such as hydroxymethyl cytosine or that it glycosylates its DNA as does T4.
Virion structural proteins.
There are 27 genes in the S-PM2 genome that are predicted to encode either T4-type virion structural proteins on the basis of sequence similarity between their predicted protein products and known virion components of T4 and other phages or those identified as virion structural proteins by mass spectroscopy. Many of the genes associated with head and contractile tail assembly are found in cluster II. Cluster I contains just two genes encoding baseplate subunit proteins; however, 7 of the 12 ORFs between clusters I and II have been identified as virion structural proteins by a proteomic approach (our unpublished data). Consequently, this whole region of 55.2 kb (26866 to 82073) extending from the beginning of cluster I to the middle of cluster II may be associated with virion assembly. Cluster IV encodes several genes encoding baseplate hub subunits, the head completion protein, and DNA end protection protein.
The long T4 tail fibers with their associated adhesins mediate the initial recognition and attachment of the phage to its host. In T4 the adsorption specificity is largely determined by the C-terminal domain of the 1,026-aa gp37 component of the distal tail fiber (76). This adhesin domain recognizes the specific receptor molecules on the surface of the bacteria host such as lipopolysaccharide (LPS) (22). It is critical for our understanding of the phage-host interaction in the oceanic picophytoplankton to identify both the phage adhesins and their targets on the Synechococcus cell surface. There is good evidence of the extensive lateral transfer of genes involved in LPS and surface polysaccharide biosynthesis from the analysis of Prochlorococcus and Synechococcus genomes (51, 54). This is consistent with the idea that the hyperplasticity of bacterial cell surfaces is a response to the selective pressures exerted by phage infection and/or grazing by protists, thought to be the two major causes of picophytoplankton mortality. There is also evidence from one phage infecting a freshwater cyanobacterium Anabaena sp. strain PCC 7120 that sensitivity to infection depends on LPS (77). However, no homologue of g37 was detectable in the S-PM2 genome. Several proteins involved in viral adhesion have been shown to share structural features with proteins involved in the recognition or metabolism polysaccharides or LPSs (70). These viral adhesion proteins were predominantly fibrous, elongated homotrimers with β-sheet topologies containing unusual repetitive folds including triple β-helices and triple β-spirals. Among the ORFs identified by proteomic studies and mass spectroscopy (our unpublished results) as virion structural proteins are ORF 174 and ORF 176. These ORFs together with the intervening ORF 175 form a highly unusual contiguous block of genes that encode large proteins ranging between 1,058 and 1,177 aa and are found on the negative strand of the virus. Analysis of the proteins encoded by these ORFs using the BetaWrap program (6), which assesses compatibility with the right-handed beta-helix fold, gave very good scores for the two virion structural proteins encoded by ORFs 174 and 176. Thus, on the basis of structural predictions these two proteins are candidates for components of the S-PM2 adhesin(s), but there is no detectable similarity between ORFs 174 and 176 and the tail fiber adhesins of T4 or T4-like phages (65). Nevertheless, the S-PM2 distal tail fibers could be folded into another type of fiber structure, such as an α-helical coiled-coil.
Cell envelope proteins.
S-PM2 encodes 19 proteins that could be associated with the cell envelope; 15 of these proteins are strongly predicted by at least two algorithms to be in the cytoplasmic or thylakoid membranes (Table 4). Interestingly, the PSORT prediction for the T4 genome also gives 19 cytoplasmic membrane proteins (46). P-SORT-B (the successor to PSORT) only predicts two S-PM2 inner membrane proteins, and these are the D1 and D2 proteins of PSII. There is some degree of clustering in the S-PM2 genome among the presumed envelope-associated ORFs. One cluster (ORFs 7, 11, 12, and 17) encodes four of the inner membrane proteins, while another cluster (ORFs 170, 171, 173, 177, 180, and 185) encodes at least four proteins that are thylakoid associated.
TABLE 4.
Potential S-PM2 inner membrane/thylakoid proteins
| ORF | Prediction of inner membrane localizationa | No. of trans-membrane domains | Signal peptide cleavage sitesb | Size (aa) | Comment |
|---|---|---|---|---|---|
| 7 | H, S, T, M | 1 | 28-29 | 59 | |
| 11 | H, S, T | 1 | 42 | ||
| 12 | H, S, T, M | 1 | 134 | Putative prohibitin | |
| 17 | H, S, T, M | 2 | 68 | ||
| 73 | H, S, T, M | 1 | 23-24 | 48 | |
| 98 | H, S, T | 1 | 18-19 | 76 | |
| 137 | H, S | 1 | 135 | ||
| 149 | H, S, T | 1 | 154 | ||
| 161 | H, T | 1 | 117 | ||
| 170 | H, S, T, M | 2 | 116 | ||
| 171 | H, S, T | 1 | 162 | Putative HLIP | |
| 173 | H, S, T | 1 | 39 | Putative HLIP | |
| 177 | P, H, S, T, M | 8 | 359 | PsbA | |
| 180 | P, H, S, T, M | 6 | 353 | PsbD | |
| 185 | H, S, T, M | 1 | 78 | Member of a conserved family of cyanobacterial proteins |
P, PSORT-B prediction score of >7.5 (20); H, HMTOP (67) prediction as a component of PSORT-B; S, SOSUI prediction (30); T, TMHMM prediction (34); M, MaxH prediction score of >0.900.
SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) prediction.
There are other lines of evidence that point to a localization of several phage proteins in the periplasm or outer membrane. The 120-aa polypeptide encoded by ORF 159 was found to contain a rare lipoprotein A domain. Furthermore, LipoP (31) predicts the protein to have a signal peptidase cleavage site between residues 23 and 24. ORF 169 (279 aa) potentially encodes a protein with an S-layer homology domain (protein family database accession no. 00395) (17). S-layers are little understood but very common surface structures of bacteria that are monomolecular quasi-crystalline arrays of proteinaceous substrates external to the outer membrane (59). Several bacterial proteins are noncovalently anchored to the cell surface via an S-layer homology domain, and in some cases this may involve pyruvylation of the polysaccharide (42). The presence of S-layers in cyanobacteria is well established (61) and has also been reported for a marine Synechococcus strain (57). It is known that an S-layer-associated protein (ORF 169) is expressed during the course of infection (our unpublished results), but there is no evidence suggesting a function for this phage-encoded protein. One can speculate that it is either involved in preventing the grazing of infected Synechococcus cells by protozoa or may prevent superinfection. Another possibility is that an alteration in the surface properties of infected cells allows them to aggregate with uninfected cells and thus ensure available hosts for the released progeny phages.
Cellular homologues of S-PM2 proteins and the photosynthetic apparatus.
There are 29 ORFs with significant similarities to proteins encoded by cyanobacteria (19) or other bacteria (9). With the exception of the five ORFs associated with the photosynthetic apparatus, only a few of these homologues have known function. For example ORF 12 encodes a prohibitin domain that may somehow be involved in proteolysis. ORF 126/7 encodes a putative cyanobacterial-type cytosine-specific DNA methyl transferase. ORF 196 encodes a probable thymidylate synthase, and ORF 131 encodes a probable small heat shock protein.
Among the phage ORFs with cyanobacterial homology, a function can be most easily attributed for the proteins associated with the photosynthetic apparatus. The discovery that S-PM2 encoded the D1 and D2 proteins of photosystem II (41) suggests that a component of the phage's replicative strategy is to maintain the structural and functional integrity of at least part of the photosynthetic apparatus in order to provide energy for phage replication (41). All cyanobacteria studied to date contain multigene psbA families, the expression of which has been extensively studied in Synechococcus sp. PCC 7942 and is clearly regulated in response to environmental conditions (for a review, see reference 23). The cyanobacterium Synechococcus sp. PCC 7942 encodes three psbA genes encoding two distinct forms of D1 (24). Normally growing cells express the psbA1 gene and D1.1 is produced, but cells exposed to “excitation stress” predominantly express psbAII and psbAIII, leading to production of D1.2. Cells with D1.2 appear to be more resistant to excess excitation pressure than those possessing D1.1, and this in part derives from the higher intrinsic resistance of PSII containing D1.2 to photoinhibition (8, 9) that may be due to an alteration in the redox behavior of the reaction center (58). When the phage-encoded D1 protein is compared with D1.1 and D1.2, it shares all the amino acid features in the transmembrane helices of D1.2, suggesting that it is a high-light form. Furthermore, in the context of photosystem II, there is the phage ORF 190 to consider, which potentially encodes an S-adenosylmethionine decarboxylase homologue, a key enzyme in the biosynthesis of spermidine and spermine. Polyamines have been implicated in photoadaptation and photoinhibition in other oxygenic phototrophs (33). Furthermore, a mutant of the cyanobacterium Synechocystis sp. PCC6803 having a reduced spermidine content was found to exhibit reduced psbA2 transcript stability (50).
The characterization of the complete S-PM2 genome has revealed other ORFs encoding proteins that are likely to interact with the photosynthetic apparatus. In vascular plants the major light-harvesting complex (LHC) is primarily composed of integral membrane proteins with three transmembrane helices, the LHC polypeptides, that bind chlorophylls a and b. Genes have been identified in cyanobacteria that encode single-helix members of an extended LHC family (HLIPs) and have been designated hli (high-light inducible) (16, 19). Deletion of all four hli genes in the cyanobacterium Synechocystis sp. PCC6803 led to the creation of a strain unable to adapt to, or survive, high light intensities (28). There are two ORFs (171 and 173) upstream from the psbA gene (D1) encoding proteins of 162 and 39 residues that exhibit significant similarity to HLIPs, and both have a single predicted transmembrane helix. An analysis of 73 hli genes from marine and freshwater cyanobacteria defined 24 clusters, with a strong divergence between marine and freshwater species (4). The protein encoded by ORF 171 is most similar to HLIPs from Prochlorococcus rather than Synechococcus. The same is true for the putative HLIP encoded by ORF 173, which also includes the TGQIIPGF motif that is strongly conserved in the C terminus of Prochlorococcus HLIPs (4). It seems possible that these genes were acquired from Prochlorococcus rather than Synechococcus, which suggests that the host range of S-PM2 extends to Prochlorococcus or that the genes were acquired by recombination with a phage that infects Prochlorococcus. The physiological function of these viral HLIPs is presumably the same as that of the host HLIPs and permits photosynthesis to provide the energy for the viral life cycle even under conditions of high light stress.
In cyanobacteria the major peripheral light-harvesting antenna are the phycobilisomes, and their component proteins, including the chromophore-carrying phycobiliproteins, represent a significant fraction of total cellular protein. ORF 216 encodes a polypeptide that exhibits considerable similarity to CpeT from the Fremyella diplosiphon and other cyanobacteria (11). Typically cpeT is part of a cluster of genes cpeESTR with a conserved order, and the product of one of these genes, cpeR, has been implicated in the control of expression of the genes encoding the α- and β-subunits of phycoerythrin (11), which is the primary light-harvesting component of the phycobilisomes of S-PM2 hosts. Thus, it is possible that S-PM2 is capable of modulating the expression of the phycoerythrin genes in infected cells and thereby controls the size of the overall photosynthetic antenna, which is consistent with its possession of hli genes.
Conclusions.
Until quite recently the only phage genomes that have been characterized were those that infect heterotrophic hosts. The present analysis of a phage that infects an obligately phototrophic host has revealed unexpected, novel, and potentially important interactions between this phage and its host. There is evidence from other host-virus systems to suggest numerous lateral gene transfer events in both directions (18), and one of the most interesting features of the S-PM2 genome is the presence of numerous phage-encoded genes that are actually homologues of host functions. The most exciting example of such a presumed phage recruitment of host genes involves functions that become part of the photosynthetic apparatus. The psbA and psbD photosynthesis genes presumably provide a significant adaptive advantage to the phage by allowing photosynthesis to continue throughout infection and thus augment the phage's burst size. Similarly, the S-PM2 genes encoding homologues of the host's HLIPs and S-adenosylmethionine decarboxylase could allow the infected cell to adapt to increased light intensity and to avoid photoinhibition. Recently, partial analyses of the genomes of three myoviruses and a podovirus infecting Prochloroccus strains were described (36). Given the close phylogenetic relationship between these two groups of marine cyanobacteria, it is not surprising that their phages share several features, in particular the possession of photosynthesis-related genes. All of the Prochloroccus phages, including the podovirus, carried copies of psbA (D1 protein), and two of the myoviruses also carried psbD (D2 protein). Two of the myoviruses encoded potential HLIPs and one, P-SSM2, encoded two photosynthetic electron transport genes coding for plastocyanin (petE) and ferredoxin (petF) that are not encoded by S-PM2. However, the possession of photosynthesis-related genes is not a universal feature of phages infecting marine cyanobacteria, as was shown by the analysis of the cyanopodovirus P60 (10). The survival advantage for phage to carry such host functions is self-evident. Another similar example is the phage-encoded homologue of the host cpeT gene. This function could modulate the expenditure of the infected cell's resources in carbon and photosynthetic energy that are directed toward phycoerythrin synthesis. The production of this major photopigment could thus be finely adjusted to the ambient light conditions in order to optimize phage multiplication. Unfortunately, the majority of the S-PM2 sequences that are homologous to host genes have completely unknown function. Nevertheless, it should be noted that in their natural environment (e.g., oligotrophic central gyres of the oceans) S-PM2-type phages often infect hosts growing under conditions of acute nutrient stress. In such situations, the infected cell's physiology must be vastly different than in nutrient-replete laboratory cultures (40). For example, S-PM2 can enter a so-called pseudolysogenic state when it infects a phosphate-limited host (73). Pseudolysogeny is a little studied, and even less understood, type of phage of infection that probably occurs frequently in nature. Pseudolysogeny may be viewed as a sort of viral hibernation, where under unfavorable conditions the phage infection is suspended at an early stage of developmental cycle (H. M. Krisch and F. Tétart, unpublished data). Such arrested infections can either end by cell death occurring prior to the production of viable phage or devolve into a normal virulent infection. This choice depends on a complex set of unknown environmental factors. Future experiments will be directed at looking at S-PM2 gene expression in pseudolysogenic infections and under conditions of environmental stress to determine if some of the phage-encoded cellular homologues are either preferentially expressed or repressed in such situations. In the context of such experiments, it is particularly worth drawing attention to the large block of S-PM2 genes extending from ORF 17 to ORF 52 that lack both early and late phage promoters. How is a contiguous group of viral genes of largely unknown function expressed? It is tempting to speculate that they are transcribed by a phage-encoded σ-factor that functions only under specific environmental conditions. In this context an analysis of the genomes of the several cyanobacteria so far sequenced revealed that all σ70-like groups are represented, but no σ54-like factor could be identified, and a characteristic feature is the presence of multiple group 2 σ-factors (26). A group 2 σ-factor has been shown to be required for the lytic development of the temperate cyanophage A4-L that infects the freshwater cyanobacterium Anabaena sp. strain PCC7120 (32).
Clearly much remains to be done to understand the complex regulatory interactions between marine cyanophages and their phototrophic hosts. The S-PM2-Synechococcus system provides us with an excellent experimental model system to accomplish such a task. Potentially the most exciting outcome of this future work will be the blurring of the distinction between the phage and host functions. The bipartite evolution of these shared host and phage functions now appears perfectly plausible. If this proves true, it means there was a direct and potentially important role of phage in the evolutionary history of some nontrivial fraction of the “cellular” genes in the biosphere. Phages are both unbelievably abundant and unbelievably diverse in nature (for a review, see reference 75). These characteristics, coupled with their unusually promiscuous genetic exchange (29), may make phages the preferred testing ground for evolutionary experimentation. Successful innovations that emerge from phage evolution would then rapidly be transferred to the host. It seems likely that cellular systems would not have ignored the chance to develop some means to efficiently exploit such a cheap, plentiful source of highly relevant genetic diversity.
Acknowledgments
This work was supported by grants from the Natural Environment Research Council. A.C. and A.M. were supported by Natural Environment Research Council and Biotechnology and Biological Sciences Research Council studentships, respectively. The Toulouse contribution to this work was supported by the CNRS and by grants from the Ministère de la recherche (PRFMMIP and ACI-Microbiologie). The CNRS-IFR109 and the Toulouse Genopole provided funding for the DNA sequencing facilities.
We thank the laboratory of Ken Nishikawa for providing the GTOP analysis. We greatly appreciate the advice and assistance generously given by Jim Karam, Hans Ackermann, Françoise Tétart, E. Peter Geiduschek, and Yvette de Preval.
REFERENCES
- 1.Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes. Bacteriophage taxonomy, p. 13-44. CRC Press, Boca Raton, Fla.
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaya, D., A. Dufresne, D. Vaulot, and A. Grossman. 2002. Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiol. Lett. 215:209-219. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D., C. Schierle, and J. Beckwith. 1998. How many membrane proteins are there? Protein Sci. 7:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley, P., M. Cowen, J. Menke, J. King, and B. Berger. 2001. Predicting the beta-helix fold from protein sequence data. Presented at the RECOMB 2001, the Fifth Annual International Conference on Computational Molecular Biology, Montreal, Canada. [DOI] [PubMed]
- 7.Bratbak, G., W. Wilson, and M. Heldal. 1996. Viral control of Emiliania huxleyi blooms? J. Mar. Syst. 9:75-81. [Google Scholar]
- 8.Campbell, D., D. Bruce, C. Carpenter, P. Gustafsson, and G. Oquist. 1996. Two forms of the photosystem II D1 protein alter energy dissipation and state transitions in the cyanobacterium Synechococcus sp. PCC 7942. Photosynth. Res. 47:131-144. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, D., M. J. Eriksson, G. Oquist, P. Gustafsson, and A. K. Clarke. 1998. The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc. Natl. Acad. Sci. USA 95:364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobley, J. G., A. C. Clark, S. Weerasurya, F. A. Queseda, J. Y. Xiao, N. Bandrapali, I. D'Silva, M. Thounaojam, J. F. Oda, T. Sumiyoshi, and M. H. Chu. 2002. CpeR is an activator required for expression of the phycoerythrin operon (cpeBA) in the cyanobacterium Fremyella diplosiphon and is encoded in the phycoerythrin linker-polypeptide operon (cpeCDESTR). Mol. Microbiol. 44:1517-1531. [DOI] [PubMed] [Google Scholar]
- 12.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis, S. E., and J. A. Martin. 1994. The transcription apparatus and the regulation of transcription initiation, p. 613-639. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 14.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desplats, C., C. Dez, F. Tetart, H. Eleaume, and H. M. Krisch. 2002. Snapshot of the genome of the pseudo-T-even bacteriophage RB49. J. Bacteriol. 184:2789-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolganov, N. A. M., D. Bhaya, and A. R. Grossman. 1995. Cyanobacterial protein with similarity to the chlorophyll a/b-binding proteins of higher plants: evolution and regulation. Proc. Natl. Acad. Sci. USA 92:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer cell wall interactions. J. Struct. Biol. 124:276-302. [DOI] [PubMed] [Google Scholar]
- 18.Filee, J., P. Forterre, and J. Laurent. 2003. The role played by viruses in the evolution of their hosts: a view based on informational protein phylogenies. Res. Microbiol. 154:237-243. [DOI] [PubMed] [Google Scholar]
- 19.Funk, C., and W. Vermaas. 1999. A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38:9397-9404. [DOI] [PubMed] [Google Scholar]
- 20.Gardy, J. L., C. Spencer, K. Wang, M. Ester, G. E. Tusnady, I. Simon, S. Hua, K. deFays, C. Lambert, K. Nakai, and F. S. L. Brinkman. 2003. PSORT-B: improving protein subcellular localization prediction for gram-negative bacteria. Nucleic Acids Res. 31:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goericke, R., and N. A. Welschmeyer. 1993. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep Sea Res. Part I 40:2283-2294. [Google Scholar]
- 22.Golberg, E., L. Grinius, and L. Letellier. 1994. Recognition, attachment and injection, p. 347-356. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 23.Golden, S. S. 1994. Light-responsive gene expression and the biochemistry of the photosystem II reaction center, p. 693-714. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer, Dordrecht, The Netherlands.
- 24.Golden, S. S., J. Brusslan, and R. Haselkorn. 1986. Expression of a family of psbA genes encoding a photosystem-II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 5:2789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenberg, G. R., P. He, J. Hilfinger, and M.-J. Tseng. 1994. Deoxyribonucleoside triphosphate synthesis and T4 DNA replication, p. 14-25. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 26.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 27.Hambly, E., F. Tétart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, Q. F., N. Dolganov, O. Bjorkman, and A. R. Grossman. 2001. The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J. Biol. Chem. 276:306-314. [DOI] [PubMed] [Google Scholar]
- 29.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 31.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khudyakov, I. Y., and J. W. Golden. 2001. Identification and inactivation of three group 2 sigma factor genes in Anabaena sp. strain PCC 7120. J. Bacteriol. 183:6667-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotzabasis, K., B. Strasser, E. Navakoudis, H. Senger, and D. Dornemann. 1999. The regulatory role of polyamines in structure and functioning of the photosynthetic apparatus during photoadaptation. J. Photochem. Photobiol. B 50:45-52. [Google Scholar]
- 34.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 35.Li, W. K. W. 1995. Composition of ultraphytoplankton in the central north-Atlantic. Mar. Ecol. Prog. Ser. 122:1-8. [Google Scholar]
- 36.Lindell, D., M. B. Sullivan, Z. I. Johnson, A. C. Tolonen, F. Rohwer, and S. W. Chisholm. 2004. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 101:11013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, H. B., H. A. Nolla, and L. Campbell. 1997. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 12:39-47. [Google Scholar]
- 38.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luke, K., A. Radek, X. P. Liu, J. Campbell, M. Uzan, R. Haselkorn, and Y. Kogan. 2002. Microarray analysis of gene expression during bacteriophage T4 infection. Virology 299:182-191. [DOI] [PubMed] [Google Scholar]
- 40.Mann, N. H. 2003. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 27:17-34. [DOI] [PubMed] [Google Scholar]
- 41.Mann, N. H., A. Cook, A. Millard, S. Bailey, and M. Clokie. 2003. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424:741. [DOI] [PubMed] [Google Scholar]
- 42.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millard, A., M. R. Clokie, D. A. Shub, and N. H. Mann. 2004. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. USA 101:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, E. S. 1994. Control of translation initiation: mRNA structure and protein repressors, p. 193-205. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 45.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monod, C., F. Repoila, M. Kutateladze, F. Tetart, and H. M. Krisch. 1997. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J. Mol. Biol. 267:237-249. [DOI] [PubMed] [Google Scholar]
- 48.Mosig, G. 1994. Synthesis and maturation of T4-encoded tRNAs, p. 182-185. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 49.Mosig, G., and D. H. Hall. 1994. Gene expression: a paradigm of integrated circuits, p. 127-131. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 50.Mulo, P., T. Eloranta, E. M. Aro, and P. Maenpaa. 1998. Disruption of a spe-like open reading frame alters polyamine content and psbA-2 mRNA stability in the cyanobacterium Synechocystis sp. PCC 6803. Botanica Acta 111:71-76. [Google Scholar]
- 51.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 52.Pavlov, A. R., and J. D. Karam. 2000. Nucleotide-sequence-specific and non-specific interactions of T4 DNA polymerase with its own mRNA. Nucleic Acids Res. 28:4657-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine-bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 54.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 55.Rohwer, F., A. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 56.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 57.Samuel, A. D., J. D. Petersen, and T. S. Reese. 2001. Envelope structure of Synechococcus sp. WH8113, a nonflagellated swimming cyanobacterium. BMC Microbiol. 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sane, P. V., A. G. Ivanov, D. Sveshnikov, N. P. A. Huner, and G. Oquist. 2002. A transient exchange of the photosystem II reaction center protein D1: 1 with D1: 2 during low temperature stress of Synechococcus sp. PCC 7942 in the light lowers the redox potential of Q(B). J. Biol. Chem. 277:32739-32745. [DOI] [PubMed] [Google Scholar]
- 59.Sara, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shamoo, Y., A. Tam, W. H. Konigsberg, and K. R. Williams. 1993. Translational repression by the bacteriophage-T4 gene 32 protein involves specific recognition of an RNA pseudoknot structure. J. Mol. Biol. 232:89-104. [DOI] [PubMed] [Google Scholar]
- 61.Smarda, J., D. Smajs, J. Komrska, and V. Krzyzanek. 2002. S-layers on cell walls of cyanobacteria. Micron 33:257-277. [DOI] [PubMed] [Google Scholar]
- 62.Suttle, C. A., and A. M. Chan. 1993. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth-characteristics. Mar. Ecol. Prog. Ser. 92:99-109. [Google Scholar]
- 63.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467-469. [Google Scholar]
- 64.Tétart, F., C. Desplats, M. Kutateladze, C. Monod, H. W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tétart, F., F. Repoila, C. Monod, and H. M. Krisch. 1996. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J. Mol. Biol. 258:726-731. [DOI] [PubMed] [Google Scholar]
- 66.Torgov, M. Y., D. M. Janzen, and M. K. Reddy. 1998. Efficiency and frequency of translational coupling between the bacteriophage T4 clamp loader genes. J. Bacteriol. 180:4339-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 68.Veldhuis, M. J. W., G. W. Kraay, J. D. L. VanBleijswijk, and M. A. Baars. 1997. Seasonal and spatial variability in phytoplankton biomass, productivity and growth in the northwestern Indian Ocean: the southwest and northeast monsoon, 1992-1993. Deep Sea Res. Part I 44:425-449. [Google Scholar]
- 69.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weigele, P. R., E. Scanlon, and J. King. 2003. Homotrimeric, beta-stranded viral adhesins and tail proteins. J. Bacteriol. 185:4022-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilkens, K., and W. Ruger. 1994. Transcription from early promoters, p. 132-141. In J. D. Karam (ed.), Molecular biology of phage T4. American Society for Microbiology, Washington, D.C.
- 72.Williams, K. P., G. A. Kassavetis, D. R. Herendeen, and E. P. Geiduschek. 1994. Regulation of late gene expression, p. 161-175. In J. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington D.C.
- 73.Wilson, W. H., N. G. Carr, and N. H. Mann. 1996. The effect of phosphate status on the kinetics of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. WH7803. J. Phycol. 32:506-516. [Google Scholar]
- 74.Wilson, W. H., I. R. Joint, N. G. Carr, and N. H. Mann. 1993. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl. Environ. Microbiol. 59:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wood, W. B., F. A. Eiserling, and R. A. Crowther. 1994. Long tail fibers: proteins, structure and assembly, p. 282-290. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 77.Xu, X., I. Khudyakov, and C. P. Wolk. 1997. Lipopolysaccharide dependence of cyanophage sensitivity and aerobic nitrogen fixation in Anabaena sp. strain PCC 7120. J. Bacteriol. 179:2884-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]