Abstract
Background
Tumor-associated antigen (TAA)-specific CD8(+) T cells are essential for nivolumab therapy, and irradiation has been reported to have the potential to generate and activate TAA-specific CD8(+) T cells. However, mechanistic insights of T-cell response during combinatorial immunotherapy using radiotherapy and nivolumab are still largely unknown.
Methods
Twenty patients included in this study were registered in the CIRCUIT trial (ClinicalTrials.gov, NCT03453164). All patients had multiple distant metastases and were intolerance or had progressed after primary and secondary chemotherapy without any immune checkpoint inhibitor. In the CIRCUIT trial, eligible patients were treated with a total of 22.5 Gy/5 fractions/5 days of radiotherapy to the largest or symptomatic lesion prior to receiving nivolumab every 2 weeks. In these 20 patients, T-cell responses during the combinatorial immunotherapy were monitored longitudinally by high-dimensional flow cytometry-based, multiplexed major histocompatibility complex multimer analysis using a total of 46 TAAs and 10 virus epitopes, repertoire analysis of T-cell receptor β-chain (TCRβ), together with circulating tumor DNA analysis to evaluate tumor mutational burden (TMB).
Results
Although most TAA-specific CD8(+) T cells could be tracked longitudinally, several TAA-specific CD8(+) T cells were detected de novo after irradiation, but viral-specific CD8(+) T cells did not show obvious changes during treatment, indicating potential irradiation-driven antigen spreading. Irradiation was associated with phenotypical changes of TAA-specific CD8(+) T cells towards higher expression of killer cell lectin-like receptor subfamily G, member 1, human leukocyte antigen D-related antigen, T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain, CD160, and CD45RO together with lower expression of CD27 and CD127. Of importance, TAA-specific CD8(+) T cells in non-progressors frequently showed a phenotype of CD45RO(+)CD27(+)CD127(+) central memory T cells compared with those in progressors. TCRβ clonality (inverted Pielou’s evenness) increased and TCRβ diversity (Pielou’s evenness and Diversity Evenness score) decreased during treatment in progressors (p=0.029, p=0.029, p=0.012, respectively). TMB score was significantly lower in non-progressors after irradiation (p=0.023).
Conclusion
Oligo-fractionated irradiation induces an immune-modulating effect with potential antigen spreading and the combination of radiotherapy and nivolumab may be effective in a subset of patients with gastric cancer.
Keywords: gastric cancer, immune modulatory, combination therapy, radiotherapy/radioimmunotherapy, immune checkpoint inhibitor
WHAT IS ALREADY KNOWN ON THIS TOPIC.
It was reported that radiotherapy has the potential to generate and activate tumor-associated antigen (TAA)-specific CD8(+) T cells. However, the mechanism of T-cell response in combinatorial immunotherapy with irradiation in patients with gastric cancer is poorly understood.
WHAT THIS STUDY ADDS
We revealed that oligo-fractionated irradiation (22.5 Gy/5 fractions/5 days) induces TAA-specific CD8(+) T cells and that these CD8(+) T cells express a phenotype of CD45RO(+)CD27(+)CD127(+) central memory T cells in patients with unresectable advanced or recurrent gastric cancer who respond to the combination of radiotherapy and nivolumab.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our results suggest that the combinatory treatment of oligo-fractionated irradiation (22.5 Gy/5 fractions/5 days) with anti-programmed cell death protein-1 therapy may be effective in a subset of patients with gastric cancer.
Background
Immune checkpoint inhibitors (ICIs) targeting programmed cell death protein-1 (PD-1) axis have become a standard therapy for patients with advanced gastric cancer (GC). Although response rate to monotherapy of PD-1 inhibitor is still limited in patients with GC, combination therapy with chemotherapy was shown to prolong overall survival and progression-free survival compared with chemotherapy alone.1–4 Currently, combinatorial immunotherapy with chemotherapy is being developed to enhance the therapeutic efficacy of ICIs and is considered to be the first line treatment for patients with advanced GC.5 6
It was reported that irradiation activates the cancer immunity cycle and results in the expansion of tumor-associated antigen (TAA)-specific CD8(+) T cells, which are essential for anti-PD-1 therapy.7–9 We also reported that cancer-testis antigen-specific CD8(+) T cells were induced by chemoradiation in 38% of patients with advanced esophageal squamous cell carcinoma.10 In several clinical trials, a combination therapy of radiotherapy with ICIs targeting PD-1 axis induced the clinical benefit in patients with non-small-cell lung cancer (PACIFIC trial), metastatic triple negative breast cancer (TONIC trial).11–13 We recently conducted a single-arm, phase I/II trial in 41 patients with unresectable advanced or recurrent GC treated with a combination of oligo-fractionated irradiation and nivolumab (CIRCUIT trial) (ClinicalTrials.gov, NCT03453164), in which radiotherapy of total 22.5 Gy/5 fractions/5 days was given to the largest or symptomatic lesion, followed by nivolumab administration every 2 weeks.14 As a result, we reported that this combinatorial immunotherapy has a promising clinical effect of median survival time of 230 days without obvious additional adverse events.14 Thus, our and other reports suggest that combinatorial immunotherapy using radiotherapy and ICI targeting PD-1 axis is an attractive treatment strategy in any type of cancer, including GC.
Although previous articles indicated that radiotherapy generates and activates TAA-specific CD8(+) T cells and modifies diversification of T-cell repertoire and T-cell exhaustion, mechanistic insights of T-cell response during combinatorial immunotherapy using irradiation are still largely unknown.9 10 15 16 In order to gain a better understanding of T-cell responses during the combinatorial immunotherapy using radiotherapy and ICIs targeting PD-1 axis in patients with GC, we performed comprehensive immunological and molecular profiling in patients with GC enrolled in the CIRCUIT trial. In this study, peripheral blood samples were monitored longitudinally by high-dimensional flow cytometry-based, multiplexed major histocompatibility complex (MHC) multimer analysis using a total of 46 TAAs, next-generation sequencing-based repertoire analysis of T-cell receptor β-chain (TCRβ), and tumor mutational burden (TMB) analysis by using plasma-derived circulating tumor DNA (ctDNA). Our findings show that several TAA-specific CD8(+) T cells were detected de novo after irradiation and those in non-progressors frequently showed a phenotype of CD45RO(+)CD27(+)CD127(+) central memory T cells compared with those in progressors, and TCRβ clonality significantly increased and TCRβ diversity significantly decreased during this combination therapy in progressors.
Methods
Patient cohorts and sample collection
Forty-one patients with unresectable advanced or recurrent GC were enrolled in the CIRCUIT trial (ClinicalTrials.gov, NCT03453164) and all enrolled patients were treated with a total of 22.5 Gy/5 fractions/5 days of radiotherapy to the largest or symptomatic lesion prior to receiving nivolumab every 2 weeks.14 The human leukocyte antigen (HLA) genotype test was performed in 30 out of enrolled patients and 24 patients were expressing HLA-A*02:01 and/or HLA-A*24:02. Of these 24 patients, 20 patients were included in this study for whom peripheral blood samples were completely collected at three time points; before treatment (Pre), 10–17 days after the completion of radiotherapy and before nivolumab administration (radiotherapy (RT)), and after nivolumab administrations (Nivo).14 All peripheral blood samples were analyzed to longitudinally assess treatment-induced immune modulation. The CIRCUIT trial ended on January 31, 2021, and overall survival was defined as the time from the start date of radiotherapy until the date of death from any cause and the confirmation of survival in this study was conducted on December 31, 2022.
For each blood collection, a tube of BD PAXgene Blood ccfDNA Tube (cat. no. 768165; Becton, Dickinson and Company, Franklin Lakes, New Jersey, USA) was used for the collection of plasma and two tubes of BD Vacutainer CPT Cell Preparation Tube with Sodium HeparinN (cat no. 362753, Becton, Dickinson and Company) were used for the collection of peripheral blood mononuclear cell (PBMC). Plasma samples were stored in a −80° freezer and used for ctDNA analysis, and PBMC samples were stored in a liquid nitrogen tank and were used for highly multiplexed flow cytometric analysis and TCR repertoire analysis.
Highly multiplexed flow cytometric analysis
Highly multiplexed flow cytometric analysis was conducted by ImmunoScape Private Limited (Singapore). We used 16 different fluorescently labeled antibodies for immune cell subset discrimination and phenotypic profiling (online supplemental table S1), and a total of 56 epitopes restricted to HLA-A*02:01 or HLA-A*24:02, which included 46 TAAs and 10 virus-derived peptides (online supplemental table S2). PBMCs were stained with triple-coded, fluorescently labeled peptide-MHC tetramer cocktails containing 56 epitope antigens followed by staining with 16 different fluorescently labeled antibodies.17 For the generation of a triple-coded tetramer staining cocktail, three out of eight different fluorescently labeled streptavidins were randomly combined by using an automated pipetting device (Tecan Group, Männedorf, Switzerland) resulting in a total of 56 unique possible combinations to encode a single peptide candidate. All samples were run in technical replicates, with the same peptide in each replicate labeled by a different combination of fluorescently labeled streptavidin molecules. Tetramer and surface antibody labeled cells were further stained for live cells using Live/Dead Fixable Blue (Thermo Fisher Scientific, Waltham, Massachusetts, USA). As positive control for antigen-specific T-cell identification, one healthy donor PBMC sample that matched at least one of the patients’ HLA alleles was included in each experiment. All samples were acquired on an FACSymphony flow cytometer (BD Biosciences, San Jose, California, USA).
jitc-2023-008385supp001.pdf (773.5KB, pdf)
All data were exported from FACSDiva software (BD Biosciences) and imported into FlowJo software V.10.7.1 (Tree Star, Ashland, Oregon, USA). After the compensation matrix was adjusted, immune cell subsets were identified by gating on live lymphocytes using a classical gating strategy (online supplemental figure S1A). CD8(+) T cells from each sample underwent a specific manual tetramer gating sequence for the detection of triple-coded antigen-specific CD8(+) T cells. Bona fide antigen-specific T cells were further validated based on the following criteria18: (1) the detection cut-off threshold (≥2 events to be detected in each technical replicate, ie, a total of at least four positive events), (2) the background noise (the frequencies of specific CD8(+) T-cell events must be greater than events from the CD4(+) T-cell population), (3) the difference between the frequencies of a hit in the two technical replicates must be less than 2.5-fold (online supplemental figure S1B).
Antigen-specific CD8(+) T cells were further phenotypically profiled using high-dimensional analysis methods and assessed for phenotypic changes in response to treatment through ImmunoScape’s cloud based analytical pipeline tool Cytographer. For high-dimensional analysis, we included all samples and downsampled to a maximum of 10,000 cells per sample. Uniform Manifold Approximation and Projection (UMAP) was used as dimensionality reduction technique for data visualization.19 Data plots were generated using GraphPad Prism V.8 software (GraphPad Software, La Jolla, California, USA).
TCR repertoire analysis
Purification of RNA from PBMC samples and next-generation sequencing-based repertoire analysis of TCRβ was conducted by Repertoire Genesis Incorporation (Osaka, Japan).20 Briefly, total RNA was extracted from each patient’s PBMC sample using the RNeasy mini kit (QIAGEN, Valencia, California, USA) and converted to complementary DNA (cDNA) with SuperScript III reverse transcriptase (Thermo Fisher Scientific). The double-stranded cDNA (ds-cDNA) was synthesized and P10EA/P20EA adaptor was ligated to the 5’ end of the ds-cDNA, and then the adaptor-ligated ds-cDNA was digested with a Not I restriction enzyme. After removal of the primer and adaptor with MinElute Reaction Cleanup Kit (QIAGEN), PCR was performed with TCRβ constant region-specific primer and P20EA, and the second PCR was performed using the first PCR product with nested primer and P20EA. After amplification, high-throughput sequencing was performed using amplicons with the Illumina MiSeq paired-end platform (2×300 base pairs). Each sequence read was analyzed by the bioinformatics software created by Repertoire Genesis Incorporation, and each sequence read having identical TCRβ-V-gene (TRBV) and TCRβ-J-gene (TRBJ), and deduced amino acid sequence of complementarity-determining region three was defined as a unique read. Out-of-frame sequences were excluded from the analyses. The diversity of TCRβ repertoires was described by Pielou’s evenness and Diversity Evenness score (DE50), and the clonality was described by inverted Pielou’s evenness (1 – Pielou’s evenness).
Assessment of TMB using plasma derived ctDNA
Briefly, ctDNA was extracted from 1 mL of plasma sample by using QIAamp MinElute ccfDNA kit (QIAGEN) as per manufacturer’s instruction. After isolation of ctDNA the samples were subjected for library preparation by using QIAseq TMB Panel (QIAGEN). This commercial panel has covered the variants in 486 genes related to tumor and immune biology, covering a total 1.3 Mb of DNA. After library preparation paired end sequencing was performed using Illumina NovaSeq 6000 platform. After sequencing run TMB score was calculated for each sample by counting the number of synonymous and non-synonymous mutations across 1.3 Mb region spanning 486 genes with computational germline status and oncogenic driver filtering, which were performed by CLC Genomics Workbench V.22 (QIAGEN) using QIAseq TMB ready-to-use bioinformatics workflows in Biomedical Genomics Analysis plugin.
Statistical analysis
Statistical analyses were performed using the R software (V.4.0.3.). Two groups were compared by Wilcoxon’s rank-sum test. Comparisons between Pre and RT, Pre and Nivo, RT and Nivo in each target were performed with Wilcoxon’s sign-rank test, followed by the adjustment using Bonferroni method. Due to the small number of subjects, no multiplicity adjustment for testing across categorical variables in each data set was performed. All p values<0.05 were considered to be statistically significant.
Results
Patient’s characteristics
In the present study, in order to perform the comprehensive immunological monitoring including TAA-specific CD8(+) T cells, we enrolled 20 patients with HLA-A*02:01 or HLA-A*24:02 that are major HLA-A types in Japanese from the phase I/II trial (CIRCUIT trial) in 41 patients with unresectable advanced or recurrent GC treated with a combination of oligo-fractionated irradiation and nivolumab.14 The baseline characteristics of 20 patients in this study are presented in table 1.
Table 1.
Patients and tumor characteristics
| No | Age | HLA-A type | OS | BOR | HER2 score | Pathology | Laurén classification | cStage |
| 1 | 50–59 | A*24:02, A*31:01 | 34 | PD | 2+ | por, sig, tub2 | Diffuse type | IVB |
| 2 | 60–69 | A*02:01, A*31:01 | 73 | PD | 0 | por | Diffuse type | IVB |
| 3 | 70–79 | A*02:07, A*24:02 | 83 | PD | Unknown | Unknown | Not determined | IVB |
| 4 | 30–39 | A*24:02, A*24:02 | 95 | PD | 3+ | tub2>tub1 | Intestinal type | IVB |
| 5 | 60–69 | A*02:06, A*24:02 | 157 | PD | 0 | tub1>tub2 | Intestinal type | IVB |
| 6 | 60–69 | A*24:02, A*24:02 | 158 | PD | 0 | tub2 | Intestinal type | IVB |
| 7 | 70–79 | A*24:02, A*26:01 | 167 | PD | 2+ | tub2>>por1 | Intestinal type | IVB |
| 8 | 60–69 | A*11:01, A*24:02 | 174 | PD | 0 | tub1, tub2 | Diffuse type | IVB |
| 9 | 70–79 | A*24:02, A*31:01 | 202 | PD | 0 | tub1, tub2 | Intestinal type | IVB |
| 10 | 70–79 | A*02:01, A*33:03 | 290 | PD | 3+ | tub2, por | Intestinal type | IVB |
| 11 | 60–69 | A*02:06, A*24:02 | 303 | SD | 2+ | tub1, tub2 | Intestinal type | IVB |
| 12 | 60–69 | A*11:01, A*24:02 | 330 | SD | 0 | tub2>por | Intestinal type | IVB |
| 13 | 70–79 | A*02:01, A*24:02 | 342 | SD | 1+ | tub1>tub2 | Intestinal type | IVB |
| 14 | 80–89 | A*24:02, A*26:01 | 435 | PR | 1+ | Unknown | Not determined | IVB |
| 15 | 70–79 | A*24:02, A*24:02 | 651 | CR | 3+ | tub | Intestinal type | IVB |
| 16 | 70–79 | A*24:02, A*33:03 | 888 | PR | Unknown | tub2>tub1 | Intestinal type | IVB |
| 17 | 70–79 | A*02:01, A*02:06 | 1111 | SD | 0 | por1 | Diffuse type | IVB |
| 18 | 70–79 | A*24:02, A*26:01 | 1118 | CR | 2+ | tub1>tub2>por1 | Intestinal type | IVB |
| 19 | 70–79 | A*02:06, A*24:02 | 1160 | CR | 0 | por2 | Diffuse type | IVB |
| 20 | 70–79 | A*11:01, A*24:02 | 1489 | PR | 0 | tub2 | Intestinal type | IVB |
Cases with italicized case number were survivors as of the date of confirmation of survival. Clinical stage (cStage) was determined by the eighth Edition of the UICC tumor, node, metastases Classification of Malignant Tumors for the stomach.
BOR, best overall response; CR, complete response; HER2, human epidermal growth factor receptor 2; HLA, human leukocyte antigen; OS, overall survival; PD, progressive disease; por1, poorly differentiated adenocarcinoma, solid type; por2, poorly differentiated adenocarcinoma, non-solid type; por, poorly differentiated adenocarcinoma; PR, partial response; SD, stable disease; sig, signet-ring cell carcinoma; tub1, tubular adenocarcinoma, well differentiated; tub2, tubular adenocarcinoma, moderately differentiated; tub, tubular adenocarcinoma; UICC, Union for International Cancer Control.
Since all of the patients in this study had multiple distant metastases at the registration of CIRCUIT trial, they were all at clinical stage IVB determined by eighth Edition of the Union for International Cancer Control tumor, node, metastasis Classification of Malignant Tumors for the stomach (table 1). The summary of treatments administered prior to the treatment protocol of CIRCUIT trial was shown in online supplemental table S3. All patients in this study were intolerant or had progressed after primary and secondary chemotherapy, and no patient was administered an immune checkpoint inhibitor in previous treatments. The treatment protocol of CIRCUIT trial was initiated 29 days after the completion of the previous treatment.
The best overall response was evaluated according to Response Evaluation Criteria in Solid Tumors guideline V.1.1. The complete response (CR) rate was 15.0% (3/20), partial response (PR) rate was 15.0% (3/20), stable disease (SD) rate was 20.0% (4/20), and progressive disease (PD) rate was 50% (10/20), and three patients were alive as of the date of confirmation of survival (table 1). Patients with PD were classified as progressors and patients with CR or PR or SD were classified as non-progressors in this study.
Longitudinal analysis of lymphocyte subpopulations
Peripheral blood samples were collected from all enrolled patients before treatment (Pre), after radiotherapy (RT), and after nivolumab administration (Nivo), details of which are described in “METHODS”. We used 16 different fluorescently labeled antibodies for immune cell subset discrimination and phenotypic profiling of PBMC collected on Pre, RT, and Nivo (online supplemental table S1).
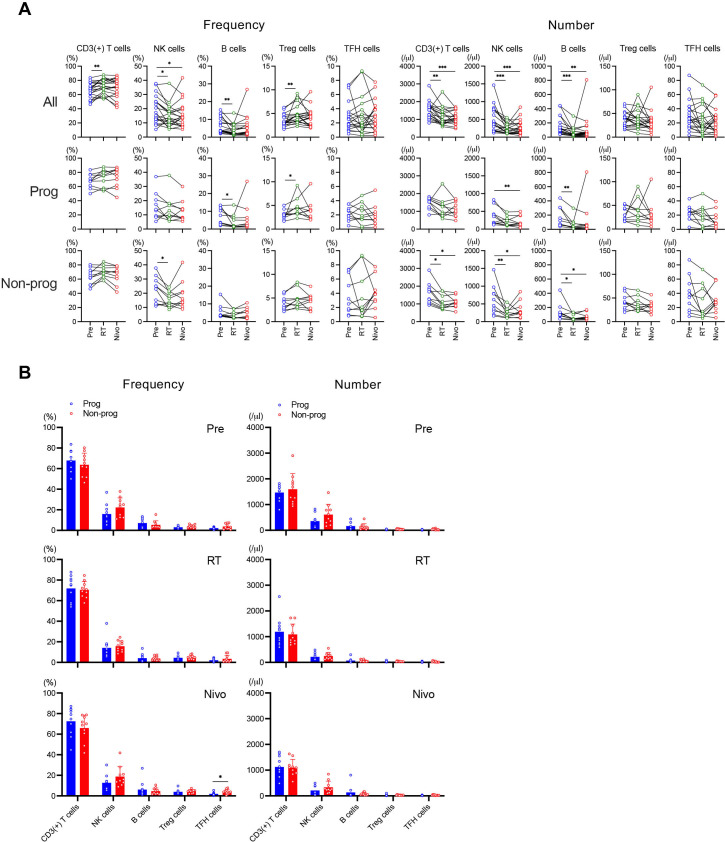
In all patients, the frequency of CD3(+) T cells and regulatory T (Treg) cells significantly increased on RT compared with Pre, while that of natural killer (NK) cells and B cells significantly decreased on RT compared with Pre, and a significant decrease compared with Pre in NK cells frequency continued on Nivo (figure 1A, left top). The frequency of B cells significantly decreased and that of Treg cells significantly increased on RT compared with Pre in progressors (figure 1A, left middle), and that of NK cells significantly decreased on RT compared with Pre in non-progressors (figure 1A, left bottom). The number of CD3(+) T cells, NK cells, and B cells significantly decreased on RT compared with Pre, which further continued on Nivo in all patients (figure 1A, right top). In progressors, the number of NK cells significantly decreased on Nivo compared with Pre and that of B cells significantly decreased on RT compared with Pre (figure 1A, right middle). On the other hand, the number of CD3(+) T cells, NK cells, and B cells significantly decreased on RT compared with Pre, which further continued on Nivo in non-progressors (figure 1A, right bottom).
Figure 1.
Immune cell subsets in response to treatment. Peripheral blood mononuclear cell samples were evaluated by the highly multiplexed flow cytometric analysis. The frequency (%) and number (/μL) of immune cell subsets were analyzed among treatments (A) and between progressors (Prog) and non-progressors (Non-prog) (B). *p<0.05, **p<0.01, ***p<0.001. Error bars represent±SD. Nivo, after nivolumab administrations; NK, natural killer; Pre, before treatment; RT, after radiotherapy; TFH, T follicular helper; Treg, regulatory T.
Next, we compared the frequency and number of each immune cell subset between progressors and non-progressors at each time point. The frequency of T follicular cells, which are involved in the humoral response, significantly increased in non-progressors on Nivo (figure 1B). Overall, it is likely that there was a minor difference between progressors and non-progressors in the dynamic change of immune cell subset during this combination therapy.
Immune cell activation and exhaustion parameters
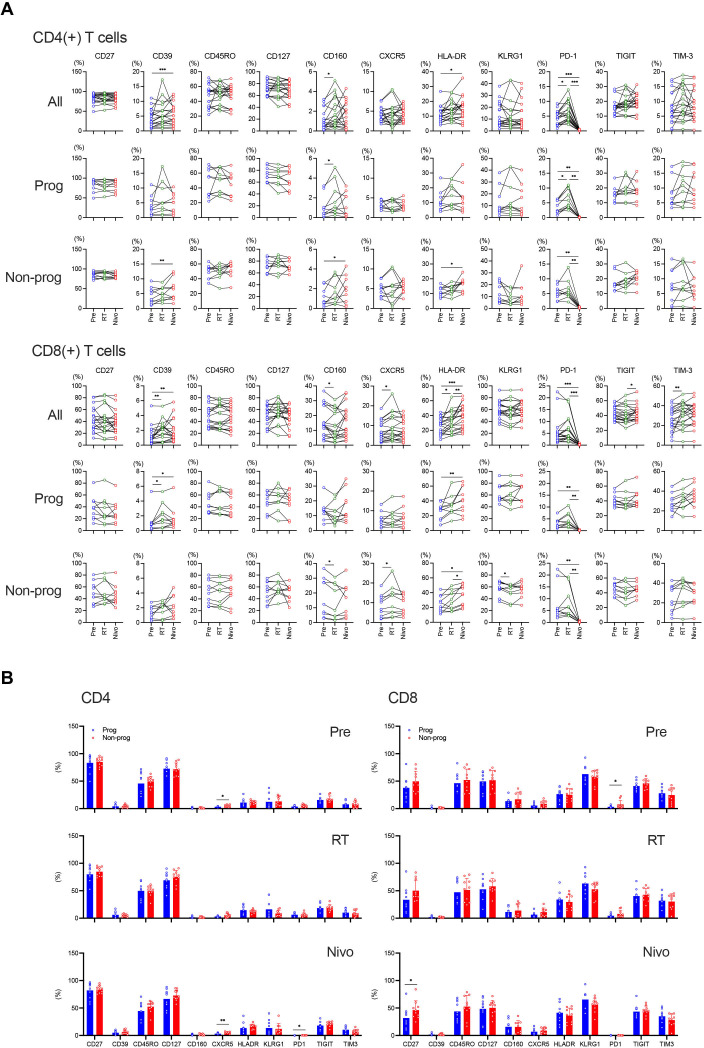
T-cell activation during this combination therapy was assessed based on the expression of HLA D-related antigen (HLA-DR), CD39 and CD160. CD4(+) T cells showed a higher activation status on Nivo compared with Pre in all patients and non-progressors (figure 2A). On the other hand, CD8(+) T cells were characterized by a higher activation phenotype on RT compared with Pre, which was further maintained on Nivo in all patients (figure 2A). This tendency was also observed in both progressors and non-progressors (figure 2A). Interestingly, the prevalence of CD160(+)CD8(+) T cells decreased, but the frequency of CD8(+) T cells expressing C-X-C chemokine receptor type 5 (CXCR5) was upregulated on RT compared with Pre in non-progressors (figure 2A, bottom).
Figure 2.
Modulation of T-cell phenotypes in response to treatment. The frequency of CD4(+) and CD8(+) T cells expressing activation and exhaustion markers was analyzed during treatment (A) and between progressors (Prog) and non-progressors (Non-prog) (B). *p<0.05, **p<0.01, ***p<0.001. Error bars represent±SD. CXCR5, C-X-C chemokine receptor type 5; HLA, human leukocyte antigen; HLA-DR, HLA D-related antigen; ITIM, immunoreceptor tyrosine-based inhibitory motif; KLRG1, killer cell lectin-like receptor subfamily G, member 1; Nivo, after nivolumab administrations; PD-1, programmed cell death protein-1; Pre, before treatment; RT, after radiotherapy; TIGIT, T-cell immunoglobulin and ITIM domain; TIM-3, T-cell immunoglobulin-3.
PD-1, T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT), and T-cell immunoglobulin-3 (TIM-3) expression were assessed to measure T-cell exhaustion. Irradiation induced upregulation of PD-1 expression on CD4(+) T cells in all patients and progressors, however, PD-1 expression on both CD4(+) and CD8(+) T cells was not detectable on Nivo, most likely because of competition of nivolumab with the antibody used for staining (figure 2A). Of note, the frequency of TIGIT(+)CD8(+) T cells increased on Nivo compared with RT and TIM-3(+)CD8(+) T cells increased on RT compared with Pre in all patients (figure 2A).
Subsequently, the frequency of CD4(+) and CD8(+) T cells expressing each activation and exhaustion parameter at each time point was compared between progressors and non-progressors. The frequency of CXCR5(+)CD4(+) T cells significantly increased in non-progressors on both Pre and Nivo (figure 2B, left). Although there was a significant difference in PD-1(+)CD4(+) T cells on Nivo, their frequencies were marginal (figure 2B, left). Regarding CD8(+) T cells, the frequency of PD-1(+) T cells on Pre and CD27(+) T cells on Nivo significantly increased in non-progressors (figure 2B, right).
Diversity and clonality of T cells by longitudinal TCR repertoire analysis
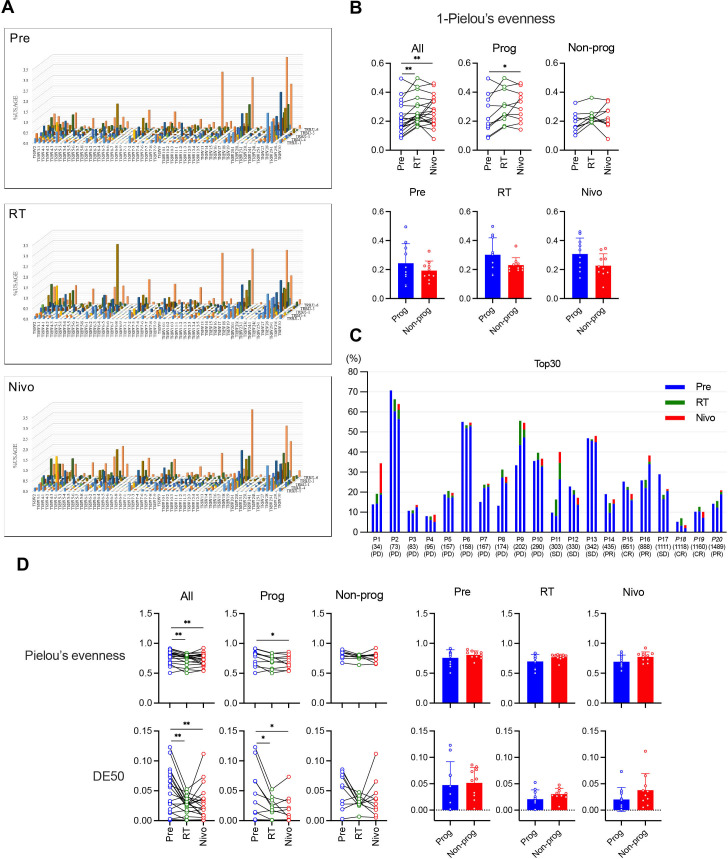
To study T-cell clonality, we performed a next-generation sequencing-based repertoire analysis of TCRβ and evaluated expression of TRBV and TRBJ as markers of T-cell repertoire diversity (figure 3A). The basic stat of TCR repertoire analysis was presented in online supplemental table S4. The clonality was evaluated by the inverted Pielou’s evenness (1 – Pielou’s evenness) and the frequency of top 30 most frequent clonotypes.21 22 In the analysis of the inverted Pielou’s evenness, we observed a significant increase of clonality on RT compared with Pre, which was further maintained on Nivo in all patients (figure 3B, upper). Furthermore, the clonality significantly increased on Nivo compared with RT in progressors but not in non-progressors (figure 3B, upper). There was no significant difference in the clonality between progressors and non-progressors at each time point (figure 3B, lower). Analysis of the top 30 most frequent clonotypes showed that many sequence-reads, which were not found on Pre, appeared on RT and Nivo (figure 3C, green and orange bars).
Figure 3.
Clonality and diversity of T cells evaluated by T-cell receptor (TCR) repertoire analysis. TCR β-chain (TCRβ) was evaluated using next-generation sequencing-based repertoire analysis. (A) Expression of TCRβ-J-gene and TCRβ-V-gene in representative samples shows the diversity of the entire TCR repertoire. (B) Modulation of clonality in response to treatment was evaluated by the inverted Pielou’s evenness (1 – Pielou’s evenness). The clonality was analyzed among treatments (upper) and between progressors (Prog) and non-progressors (Non-prog) (lower). (C) The frequency of the top 30 most frequent clonotypes was evaluated at each time point. P represents each patient’s identification number in this study. For each patient, the left bar shows the frequency before treatment (Pre), the middle bar shows the frequency after radiotherapy (RT), and the right bar shows the frequency after nivolumab administrations (Nivo). Overall survival for each patient is shown below the patient’s identification number and best overall response for each patient is also shown below overall survival. Cases with italicized case number were survivors as of the date of confirmation of survival. (D) Modulation of repertoire diversity in response to treatment was evaluated by Pielou’s evenness (upper) and Diversity Evenness score (DE50) (lower). The diversity was analyzed among treatments and between Prog and Non-prog. *p<0.05, **p<0.01, ***p<0.001. Error bars represent±SD. CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease.
Repertoire diversity was then evaluated by Pielou’s evenness and DE50. In contrast to clonality, repertoire diversity was reduced on RT compared with Pre and this trend was maintained on Nivo in all patients and progressors but not in non-progressors (figure 3D, left). There was no significant difference in the diversity between progressors and non-progressors at each time point (figure 3D, right). Taken together, the clonality significantly increased and diversity significantly decreased during this combination therapy in progressors.
TMB score reduced on RT in non-progressors
Next, we performed the TMB analysis using ctDNA derived from plasma samples. The concentration of ctDNA at each time point was presented in online supplemental table S5. We observed no significant changes in TMB score during this combination therapy (online supplemental figure S2A). However, it was observed that a significantly lower TMB score in non-progressors on RT (online supplemental figure S2B), suggesting that TMB score might reflect tumor volume during the treatment.
Characterization of antigen-specific CD8(+) T cells during this combination therapy using multiplexed MHC multimer analysis
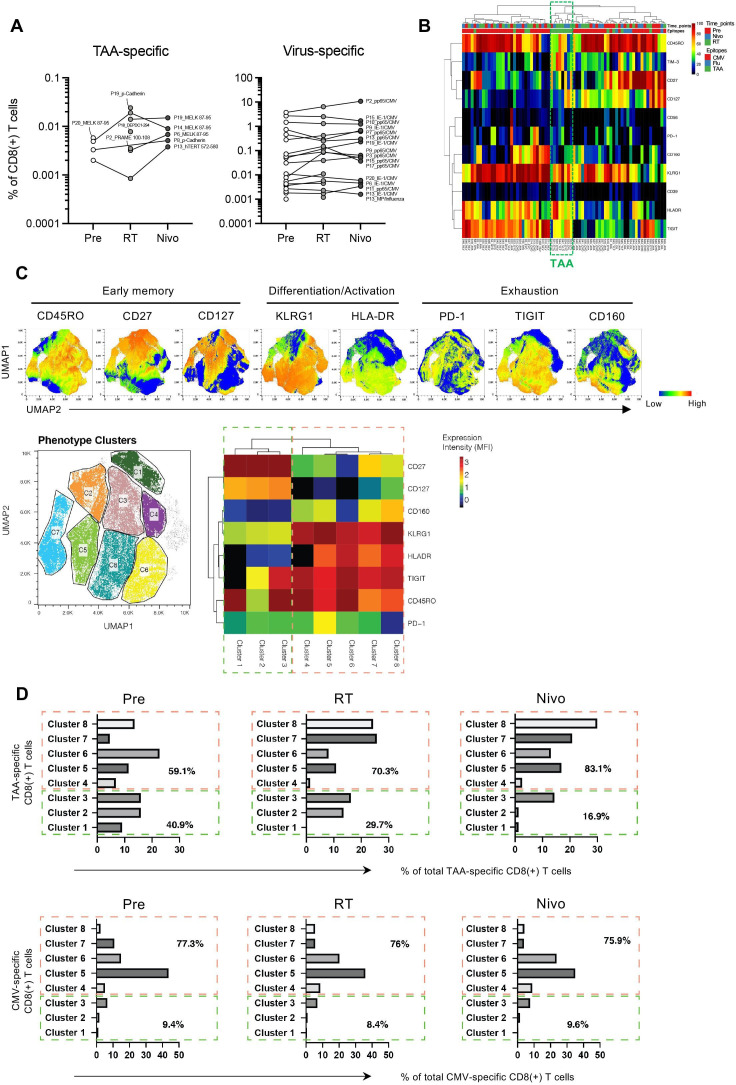
Antigen-specific CD8(+) T-cell responses in PBMC were assessed using a total of 56 peptide epitopes restricted to HLA-A*02:01 or HLA-A*24:02, which included 46 TAAs and 10 virus epitopes according to the previously published data (online supplemental table S2). Across the three time points, we detected a total of 16 TAA-specific CD8(+) T cells in 8 of the 20 patients analyzed (figure 4A and online supplemental table S6). Detected TAA HLA-A*24:02 epitopes were MELK 87–95 (EYCPGGNLF), p-Cadherin (DYLNEWGSRF), and DEPDC1-294 (EYYELFVNI), and detected TAA HLA-A*02:01 epitopes were hTERT 572–580 (YLFFYRKSV) and PRAME 100–108 (VLDGLDVLL). Of note, after irradiation, five de novo TAA-specific CD8(+) T cells were detected in five of the patients (figure 4A and online supplemental table S6), indicating that irradiation may be inducing antigen-spreading with novel TAA-specific CD8(+) T-cell responses. In addition, we also detected T cells specific for viral epitopes including a total of 54 reactivities against cytomegalovirus (CMV) and influenza-derived epitopes in the same cohorts (figure 4A and online supplemental table S6). Of importance, frequencies of virus-specific CD8(+) T cells did not show obvious changes during this combination therapy (figure 4A and online supplemental table S6). Taken together, these results suggest that irradiation may be inducing tumor antigen spreading, but does not affect pre-existing T-cell responses against viral antigens.
Figure 4.
Characteristics of tumor-associated antigen (TAA)-specific CD8(+) T cells. (A) In 20 patients analyzed in highly multiplexed flow cytometric analysis, 5 different TAA specificities and a total of 54 virus reactivities were detected across 8 and 14 out of 20 patients, respectively (online supplemental table S6). P represents each patient’s identification number in this study and antigens were presented in online supplemental table S6. (B) Levels of immune marker expression in TAA-specific (mostly clustered in the green dashed line box) and virus-specific CD8(+) T cells are shown in the heatmap. (C) Uniform Manifold Approximation and Projection (UMAP) plots display the expression intensity of all phenotypic markers on TAA-specific CD8(+) T cells (upper). We further analyzed the phenotypes of TAA-specific CD8(+) T cells and showed eight distinct phenotypic cell clusters (lower left) and heat map presenting the phenotypes for each cluster (lower right). (D) The distribution of TAA-specific and cytomegalovirus (CMV)-specific CD8(+) T cells across these clusters before treatment (Pre), after radiotherapy (RT), and after nivolumab administrations (Nivo). We downsampled the numbers of CMV-specific T cells here to a maximum of 100 cells/hit to avoid phenotypic bias caused by large frequency hits. Flu, influenza; HLA, human leukocyte antigen; HLA-DR, HLA D-related antigen; ITIM, immunoreceptor tyrosine-based inhibitory motif; KLRG1, killer cell lectin-like receptor subfamily G, member 1; MFI, mean fluorescence intensity; PD-1, programmed cell death protein-1; TIGIT, T-cell immunoglobulin and ITIM domain; TIM-3, T-cell immunoglobulin-3.
We further analyzed the phenotypes of TAA-specific and virus-specific CD8(+) T cells identified by multiplexed MHC multimer analysis. TAA-specific CD8(+) T cells showed a unique phenotype with a higher expression of HLA-DR and TIGIT as compared with influenza-specific CD8(+) T cells, and with higher frequencies of HLA-DR and TIM-3 as compared with CMV-specific CD8(+) T cells (figure 4B). Subsequently, UMAP was used as dimensionality reduction technique for data visualization and to display the expression intensity of all phenotypic markers analyzed (figure 4C, upper).19 This was apparent when we visually delineated distinct cell clusters of the possible phenotypes observed across all TAA-specific CD8(+) T cells and displayed the phenotypes of these clusters (figure 4C, lower left), based on the mean fluorescence intensity of each marker assessed, as heat map for each cluster (figure 4C, lower right). Quantitative analysis showed a substantial decrease of cells present in clusters 1–3 (mainly CD27(+) and CD127(+)), but an increase of cells distributed across clusters 4–8 (mainly killer cell lectin-like receptor subfamily G, member 1 (KLRG1)(+), CD160(+), HLA-DR(+), TIGIT(+), and CD45RO(+)) following irradiation and nivolumab treatment in TAA-specific CD8(+) T cells (figure 4D, upper). In contrast, CMV-specific CD8(+) T cells did not change their phenotypes following treatment and were largely distributed at similar frequencies across the same clusters at the different time points assessed (figure 4D, lower).
Phenotypes of TAA-specific CD8(+) T cells in non-progressors
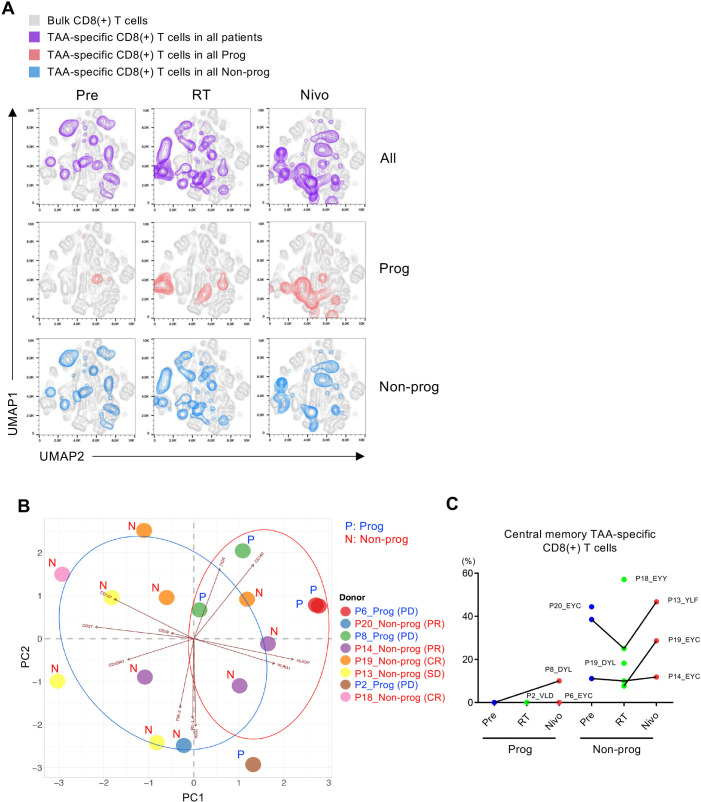
Next, we focused on eight patients whom we were able to identify the TAA-specific CD8(+) T cells (online supplemental table S6). Across all eight analyzed patients, irradiation induced an enrichment of TAA-specific CD8(+) T cells with a high expression of KLRG1, HLA-DR, TIGIT and CD160, while driving lower expression of CD27 and CD127 (figure 5A, purple contour plots). Moreover, these phenotypic patterns on TAA-specific CD8(+) T cells are maintained during nivolumab treatment. Interestingly, TAA-specific CD8(+) T cells in progressors (n=3) revealed a relatively homogenous profile with a late differentiated phenotype characterized by a high expression of KLRG1 and a low expression of CD45RO (figure 5A, red contour plot), while those in non-progressors (n=5) showed a more heterogeneous phenotype including an early memory effector phenotype characterized by a high expression of CD27, CD127 and low expression of CD160 (figure 5A, blue contour plot).
Figure 5.
Phenotypical change of tumor-associated antigen (TAA)-specific CD8(+) T cells in response to treatment. (A) Uniform Manifold Approximation and Projection (UMAP) plots display the expression intensity of all phenotypic markers on TAA-specific CD8(+) T cells analyzed before treatment (Pre), after radiotherapy (RT), and after nivolumab administrations (Nivo) in all patients (All), progressors (Prog), and non-progressors (Non-prog). (B) Principal component analysis shows a distinct differentiation profile of TAA-specific CD8(+) T cells on Pre, RT, and Nivo in Prog and Non-prog. P represents each patient’s identification number in this study and the best overall response for each patient is also shown. (C) Frequency of CD45RO(+)CD27(+)CD127(+) central memory TAA-specific CD8(+) T cells on Pre, RT, and Nivo in Prog and Non-prog (online supplemental table S6). CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease.
Consistent with the above observations, principal component analysis (PCA) showed a distinct differentiation profile of TAA-specific CD8(+) T cells in non-progressors as compared with progressors, which was mainly driven by different expressions of CD45RO, CD27, CD127 and KLRG1 (figure 5B). Moreover, most TAA-specific CD8(+) T cells in progressors clustered at the right upper quadrant of the PCA plot (figure 5B). In contrast, TAA-specific CD8(+) T cells in non-progressors tended to be distributed across different quadrants in the PCA representation (figure 5B). Interestingly, TAA-specific CD8(+) T cells in non-progressors frequently showed a phenotype of CD45RO(+)CD27(+)CD127(+) central memory T cells compared with those in progressors (figure 5C).
Discussion
In the present study, in order to perform the comprehensive immunological monitoring including TAA-specific CD8(+) T cells, we enrolled 20 patients with HLA-A*02:01 or HLA-A*24:02 that are major HLA-A types in Japanese from the phase I/II trial (CIRCUIT trial) (ClinicalTrials.gov, NCT03453164) in 41 patients with unresectable advanced or recurrent GC treated with a combination of oligo-fractionated irradiation and nivolumab.14
Multiplexed MHC multimer and phenotypic analysis was performed to longitudinally evaluate the immune modulating effect induced by radiotherapy and nivolumab treatment. Post-irradiation, we detected the presence of several new TAA-specific CD8(+) T cells which were not detected before irradiation, suggesting that irradiation drove some degree of antigen spreading and resulted in expansion of de novo TAA-specific CD8(+) T cells or pre-existing T-cell clones that were below sensitivity of detection in pretreatment samples (figure 4A and online supplemental table S6). Previous report by Huang et al indicated that major tumor infiltrating T cells can also be found in peripheral blood and we have been able to detect TAA-specific CD8(+) T cells in PBMC of patients with cancer, in which expansion of novel TAA-specific CD8(+) T cells in the periphery may have been induced by tumor cell death and generation of novel antigens taken up, processed and presented by dendritic cells.18 23 Moreover, in the present study, irradiation was associated with phenotypic changes in TAA-specific CD8(+) T cells including higher expression of KLRG1, HLA-DR, TIGIT, CD160, and CD45RO, together with lower expression of CD27 and CD127 (figure 4, C-D). In particular, we observed that the prevalence of CD8(+) T cells expressing CD160 was reduced and that of T cells expressing CXCR5 increased following irradiation in non-progressors (figure 2A, bottom). Since CD160 is a marker associated with the impairment of functional CD8(+) T cells and CXCR5 enables T cells to migrate to lymph nodes, irradiation may activate CD8(+) T cells and accelerate the generation of a functional antitumor immune response.24 25 Although it is necessary to prove the correlation between these T-cell markers and effector function by a functional assay, irradiation may be inducing immunological modulation with antigen spreading to some degree.
The stereotactic body radiation therapy for renal cell carcinoma reported that intratumoral T-cell clonality was modulated by radiotherapy, and that pre-existing T-cell clones within the tumor microenvironment expand into peripheral blood.26 Moreover, it has been reported that increased CD8(+) T-cell effector function and increased TCR diversity with extended activation of selective tumor infiltrating CD8(+) T cells were associated with antitumor effects, and the degree of expansion and contraction of peripheral blood T-cell clones has been shown to be the strongest predictors of clinical responses to radiotherapy combined with ICI.26 27 It has also been reported that responders to anti-PD-1 therapy have less clonality and more diversity of TCRβ repertoire at treatment baseline.21 28 In the present study, we evaluated the modulation of TCRβ repertoire in response to the combination treatment and found that the TCRβ clonality significantly increased and TCRβ diversity significantly decreased during this combination therapy in progressors (figure 3, B and D). It has been reported that radiotherapy and ICIs targeting PD-1 axis augmented the diversity of the TCR repertoire of tumor infiltrating lymphocytes,29–31 and it was also reported that TCR repertoires of tumor infiltrating T cells were different from those in peripheral T cells, when responded to treatment.32 33 Therefore, there is still controversy regarding the relationship between TCR repertoires and efficacy of the treatment, and the relationship between TCR repertoires in tumor infiltrating T cells and those in peripheral T cells.
In association with previous papers describing potential biomarkers for ICI treatments, the present study indicated that the frequencies of both PD-1(+)CD8(+) T cells and CXCR5(+)CD4(+) T cells on Pre were significantly higher in non-progressors (figure 2B) and TMB score was significantly lower in non-progressors on RT (online supplemental figure S2B).28 34–36 In addition, we also found that TAA-specific CD8(+) T cells in non-progressors frequently showed a phenotype of CD45RO(+)CD27(+)CD127(+) central memory T cells compared with those in progressors (figure 5C). Furthermore, we recently reported that the frequency of peripheral CD45RA(+)CD27(+)CD127(+) central memory CD4(+) and CD8(+) T cells was significantly reduced during the treatment course in non-responders to nivolumab therapy for advanced esophageal squamous cell carcinoma.16 Although further investigation is necessary, the frequency of peripheral central memory CD8(+) T cells expressing CD27 and CD127 may correlate with a favorable response to anti-PD-1 therapy and/or combinatory treatment of irradiation with anti-PD-1 therapy.
As for the optimization of radiation-induced immunogenicity, there is still controversy between non-ablative oligo-fractionated irradiation and definitive irradiation conditions to enhance the synergistic effect of irradiation with ICIs. Of importance, it has been reported that activation of cyclic guanosine monophosphate–adenosine monophosphate synthase (cGAS)-stimulator of interferon genes (STING) (cGAS-STING) pathway and its related chemokine profile is strongly associated with synergistic effect of irradiation with ICI.37 For example, comparison of oligo-fractionated irradiation with a single high-dose of irradiation showed a completely different profile for immune response including cGAS-STING pathway and its downstream recruitment of dendritic cells and activation of CD8(+) T cells. Moreover, we have recently reported that irradiation can induce remodeling of the tumor microenvironment through tumor cell-intrinsic expression of cGAS-STING.38 Therefore, better understanding for immunological remodeling of tumor microenvironment induced by irradiation would be necessary, in order to further enhance the synergistic effect of irradiation with ICI.
There are several limitations in the present study. First, the total number of analysis population was small and the power of statistical analysis was weak. Second, although we have shown that some immune and exhaustion markers of peripheral T cells were statistically significant between non-progressors and progressors, the differences were not so marked. Therefore, it would be difficult to directly apply these markers for predictive biomarkers for the present combinatory treatment. Further analysis of peripheral blood T cells using liquid biopsy samples obtained from phase III clinical trials would be desirable in the future.
Taken together, the present study suggests that oligo-fractionated irradiation (22.5 Gy/5 fractions/5 days) may induce an immune-modulating effect with antigen spreading. Furthermore, we found that TAA-specific CD8(+) T cells in non-progressors frequently showed a phenotype of CD45RO(+)CD27(+)CD127(+) central memory T cells, and TCRβ clonality significantly increased and TCRβ diversity significantly decreased during this combination therapy in progressors.
Acknowledgments
We are grateful to Masayo Sugeno from Fukushima Medical University School of Medicine and Rika Takahashi from Kanagawa Cancer Center for their administrative assistance.
Footnotes
Contributors: KM, YS and KK contributed to the conception of this study. KM and KK designed the experiments. KM, TO, YY, DY, SN, HS, NM, TY, YW, TT, HF, YI, SH, HH, ZS, HK, TO, YS and KK acquired samples and patient clinical information. FT analyzed the data. PHDN, HK, MF, and AN performed the highly multiple flow cytometric analysis. SR, Y-CY, and AG performed the tumor mutational burden analysis. KM and KK interpreted the analyzed data. KM, SN, and KK contributed to generating figures and tables. KM and KK wrote the manuscript. AG and AN reviewed and edited the manuscript. All authors read and approved the latest version of the article. KK is responsible for the overall content as guarantor.
Funding: This study was funded by ONO Pharmaceutical Co., Ltd. and Bristol-Myers Squibb.
Competing interests: PHDN (former), HK (former), MF, and AN are shareholders or employees of ImmunoScape Pte Ltd. or ImmunoScape Inc. AN is on the board of directors of ImmunoScape Pte Ltd. KK reports speaker fee from ONO Pharmaceutical Co., Ltd. and Bristol-Myers Squibb. The remaining authors have declared no conflict of interest exists.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Fukushima Medical University Research Ethics Committee (Reference No. 2021-321). Written informed consent was received from all participants prior to participation. This study was conducted in accordance with the 1964 Declaration of Helsinki principles and its later amendments.
References
- 1.Kang Y-K, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:2461–71. 10.1016/S0140-6736(17)31827-5 [DOI] [PubMed] [Google Scholar]
- 2.Muro K, Chung HC, Shankaran V, et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. Lancet Oncol 2016;17:717–26. 10.1016/S1470-2045(16)00175-3 [DOI] [PubMed] [Google Scholar]
- 3.Janjigian YY, Shitara K, Moehler M, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40. 10.1016/S0140-6736(21)00797-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang Y-K, Chen L-T, Ryu M-H, et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): a randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2022;23:234–47. 10.1016/S1470-2045(21)00692-6 [DOI] [PubMed] [Google Scholar]
- 5.Kono K, Nakajima S, Mimura K. Current status of immune checkpoint inhibitors for gastric cancer. Gastric Cancer 2020;23:565–78. 10.1007/s10120-020-01090-4 [DOI] [PubMed] [Google Scholar]
- 6.Japanese Gastric Cancer Association . Japanese Gastric Cancer Treatment Guidelines 2021 (6th edition). Gastric Cancer 2023;26:1–25. 10.1007/s10120-022-01331-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 8.Weichselbaum RR, Liang H, Deng L, et al. Radiotherapy and immunotherapy: a beneficial liaison? Nat Rev Clin Oncol 2017;14:365–79. 10.1038/nrclinonc.2016.211 [DOI] [PubMed] [Google Scholar]
- 9.Hwang WL, Pike LRG, Royce TJ, et al. Safety of combining radiotherapy with immune-checkpoint inhibition. Nat Rev Clin Oncol 2018;15:477–94. 10.1038/s41571-018-0046-7 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki Y, Mimura K, Yoshimoto Y, et al. Immunogenic tumor cell death induced by chemoradiotherapy in patients with esophageal squamous cell carcinoma. Cancer Res 2012;72:3967–76. 10.1158/0008-5472.CAN-12-0851 [DOI] [PubMed] [Google Scholar]
- 11.Antonia SJ, Villegas A, Daniel D, et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:1919–29. 10.1056/NEJMoa1709937 [DOI] [PubMed] [Google Scholar]
- 12.Faivre-Finn C, Vicente D, Kurata T, et al. Four-Year Survival With Durvalumab After Chemoradiotherapy in Stage III NSCLC-an Update From the PACIFIC Trial. J Thorac Oncol 2021;16:860–7. 10.1016/j.jtho.2020.12.015 [DOI] [PubMed] [Google Scholar]
- 13.Voorwerk L, Slagter M, Horlings HM, et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat Med 2019;25:920–8. 10.1038/s41591-019-0432-4 [DOI] [PubMed] [Google Scholar]
- 14.Mimura K, Ogata T, Yoshimoto Y, et al. Phase I/II clinical trial of nivolumab in combination with oligo-fractionated irradiation for unresectable advanced or recurrent gastric cancer. Commun Med (Lond) 2023;3:111. 10.1038/s43856-023-00343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takehara Y, Mimura K, Suzuki Y, et al. Anti‐PD‐1 monoclonal antibody‐resistant esophageal squamous cell carcinoma showing the abscopal effect: A case report with T‐cell receptor/B‐cell receptor repertoire analysis. Cancer Reports 2023;6. 10.1002/cnr2.1832 Available: https://onlinelibrary.wiley.com/toc/25738348/6/7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakuma M, Mimura K, Nakajima S, et al. A Potential Biomarker of Dynamic Change in Peripheral CD45RA−CD27+CD127+ Central Memory T Cells for Anti-PD-1 Therapy in Patients with Esophageal Squamous Cell Carcinoma. Cancers 2023;15:3641. 10.3390/cancers15143641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newell EW, Klein LO, Yu W, et al. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods 2009;6:497–9. 10.1038/nmeth.1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehlings M, Jhunjhunwala S, Kowanetz M, et al. Late-differentiated effector neoantigen-specific CD8+ T cells are enriched in peripheral blood of non-small cell lung carcinoma patients responding to atezolizumab treatment. J Immunother Cancer 2019;7:249. 10.1186/s40425-019-0695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol December 3, 2018. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- 20.Kitaura K, Shini T, Matsutani T, et al. A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) α and β repertoires and identifying potential new invariant TCR α chains. BMC Immunol 2016;17:38. 10.1186/s12865-016-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gros A, Robbins PF, Yao X. PD-1 identifies the patient-specific CD8. J Clin Invest 2014;124:2246–59. 10.1172/JCI73639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60–5. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viganò S, Banga R, Bellanger F, et al. CD160-associated CD8 T-cell functional impairment is independent of PD-1 expression. PLoS Pathog 2014;10:e1004380. 10.1371/journal.ppat.1004380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawano S, Mitoma H, Inokuchi S, et al. TNFR2 Signaling Enhances Suppressive Abilities of Human Circulating T Follicular Regulatory Cells. The Journal of Immunology 2022;208:1057–65. 10.4049/jimmunol.2100323 [DOI] [PubMed] [Google Scholar]
- 26.Chow J, Hoffend NC, Abrams SI, et al. Radiation induces dynamic changes to the T cell repertoire in renal cell carcinoma patients. Proc Natl Acad Sci U S A 2020;117:23721–9. 10.1073/pnas.2001933117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosoi A, Takeda K, Nagaoka K, et al. Increased diversity with reduced “diversity evenness” of tumor infiltrating T-cells for the successful cancer immunotherapy. Sci Rep 2018;8:1058. 10.1038/s41598-018-19548-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon M, An M, Klempner SJ, et al. Determinants of Response and Intrinsic Resistance to PD-1 Blockade in Microsatellite Instability-High Gastric Cancer. Cancer Discov 2021;11:2168–85. 10.1158/2159-8290.CD-21-0219 [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Wang W, Hu S, et al. Radiotherapy remodels the tumor microenvironment for enhancing immunotherapeutic sensitivity. Cell Death Dis 2023;14:679. 10.1038/s41419-023-06211-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Twyman-Saint Victor C, Rech AJ, Maity A, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015;520:373–7. 10.1038/nature14292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spassova I, Ugurel S, Terheyden P, et al. Predominance of Central Memory T Cells with High T-Cell Receptor Repertoire Diversity is Associated with Response to PD-1/PD-L1 Inhibition in Merkel Cell Carcinoma. Clin Cancer Res 2020;26:2257–67. 10.1158/1078-0432.CCR-19-2244 [DOI] [PubMed] [Google Scholar]
- 32.Bai X, Zhang Q, Wu S, et al. Characteristics of Tumor Infiltrating Lymphocyte and Circulating Lymphocyte Repertoires in Pancreatic Cancer by the Sequencing of T Cell Receptors. Sci Rep 2015;5:13664. 10.1038/srep13664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui C, Tian X, Wu J, et al. T cell receptor β-chain repertoire analysis of tumor-infiltrating lymphocytes in pancreatic cancer. Cancer Sci 2019;110:61–71. 10.1111/cas.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumagai S, Togashi Y, Kamada T, et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat Immunol 2020;21:1346–58. 10.1038/s41590-020-0769-3 [DOI] [PubMed] [Google Scholar]
- 35.Si H, Kuziora M, Quinn KJ, et al. A Blood-based Assay for Assessment of Tumor Mutational Burden in First-line Metastatic NSCLC Treatment: Results from the MYSTIC Study. Clin Cancer Res 2021;27:1631–40. 10.1158/1078-0432.CCR-20-3771 [DOI] [PubMed] [Google Scholar]
- 36.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. 10.1038/s41591-018-0101-z [DOI] [PubMed] [Google Scholar]
- 37.Dewan MZ, Galloway AE, Kawashima N, et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clin Cancer Res 2009;15:5379–88. 10.1158/1078-0432.CCR-09-0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima S, Mimura K, Kaneta A, et al. Radiation-Induced Remodeling of the Tumor Microenvironment Through Tumor Cell-Intrinsic Expression of cGAS-STING in Esophageal Squamous Cell Carcinoma. Int J Radiat Oncol Biol Phys 2023;115:957–71. 10.1016/j.ijrobp.2022.10.028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-008385supp001.pdf (773.5KB, pdf)
Data Availability Statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.