Abstract
Despite recent advancements in machine learning (ML) applications in health care, there have been few benefits and improvements to clinical medicine in the hospital setting. To facilitate clinical adaptation of methods in ML, this review proposes a standardized framework for the step-by-step implementation of artificial intelligence into the clinical practice of radiology that focuses on three key components: problem identification, stakeholder alignment, and pipeline integration. A review of the recent literature and empirical evidence in radiologic imaging applications justifies this approach and offers a discussion on structuring implementation efforts to help other hospital practices leverage ML to improve patient care.
Clinical trial registration no. 04242667
© RSNA, 2024
Summary
Implementing machine learning algorithms in clinical settings is achievable through a generalizable framework to integrate artificial intelligence–empowered software into the clinical practice of radiology.
Essentials
■ Real-world, representative data acquisition enables effective machine learning (ML).
■ Radiologists and other key stakeholders must work together for iterative and clinically informed use cases for ML in radiology.
■ Clinical implementation strategies involve the coordination of multidisciplinary teams and alignment with key institutional values.
■ Effectively characterizing the success of implementation of artificial intelligence in the clinical practice of radiology requires using user-centered metrics in real-world environments integrated with existing software and clinical workflows.
Introduction
Recent successes in artificial intelligence (AI) methods have enabled the impressive performance of machine learning (ML) models on health care–related tasks, including radiology report analysis (1,2), medical image acquisition and reconstruction (3–5), image lesion detection (6,7), and disease classification (8,9). However, real-world adaptation of such methods remains sparse. As of November 2022, there are 521 U.S. Food and Drug Administration–approved ML algorithms for clinical use, with 392 (75.2%) proposed for radiology applications (10). However, when compared with the progress made in other high-stakes domains including financial analytics and autonomous driving, practical AI implementation in health care settings lags behind (11,12).
Extensive prior work (13–16) has analyzed these trends and offers both empirical and qualitative evidence for the slow-moving headway in clinical AI implementation. Reyes et al (17) and Topol (18) highlight the need for increased transparency in ML model training and interpretability of prediction outputs. Kahn (19) emphasizes the importance of bias mitigation efforts in radiologic AI applications. Rowell and Sebro (20) examine AI use in the context of complex insurance cost practices and patient privacy standards. Whereas these works have introduced the challenges facing AI implementation in health care, there has been much less discussion on exactly how such implementation efforts should be structured to benefit clinical medicine.
A standardized and effective framework that answers this question remains an active area of exploration. Proctor et al (21) proposed guidelines for defining and operationalizing implementation strategies across multiple facets of health care innovation. Such structured frameworks are already in place for domains such as drug discovery (22,23), telemedicine (24), and global health (25). Whereas similar regulatory initiatives for clinical AI applications have progressed (26), methods for clinical AI are still in their infancy with the potential for further improvement and expansion (27).
This review addresses this limitation in a stepwise approach, as follows: first, by analyzing existing health care data sources important for implementation of clinical AI; second, by introducing empirical end-to-end standardized guidelines for how clinical teams can approach clinical implementation of AI based on both evidence from recent literature and firsthand experience of members of a radiology department; third, by considering insights and constraints from stakeholders; and finally, by addressing research questions relevant to future work before scalable AI implementation in health care is achievable.
The Role of Data Curation and Data Set Acquisition in Implementation of Clinical AI
The availability of health care data dictates the success of method development within ML research and its applications, particularly in scaling deep neural networks. However, patient data is important in the implementation of clinical AI models.
Public Data Sets
Publicly released data sets enabled the development of so-called foundation models, which are AI algorithms trained on large amounts of data extracted from a large, diverse population (28,29). Foundation models can be subsequently tuned and optimized for specific tasks for commercial AI products. For tasks related to the health sciences, data sets such as the UK Biobank, ISARIC-COVID-19, and fastMRI+, among others, enabled work in disease prediction from genomic, health record, epidemiologic, and image data (30–32). These data sets facilitate the reproducibility of ML research and the development of both open-source and commercial AI technologies for implementation into patient care.
Publicly available data sets are not sufficient for the implementation of these algorithms into existing radiology workflows. This is because it is crucial to test prediction models on in-domain patient data (ie, in-house data) to mitigate the effects of bias and data distribution shift (ie, when training and test data sets are not from the same patient population) that are well-studied in existing literature (8,19). Despite ongoing regulatory efforts by the U.S. Food and Drug Administration (26), existing ML models continue to generalize poorly to previously unseen patient populations, as documented by Belbasis and Panagiotou (33). Thus, even if commercial AI models demonstrate excellent performance on the large population on which they were initially trained, they may not exhibit similar performance on the smaller patient populations.
To overcome these challenges, it is beneficial for radiologists to test implemented AI solutions on their own patients and scans. Hesitancies with personal data collection may lead to recruitment bias (34), which de Man et al (35) show can be effectively mitigated by implementing opt-out consent protocols. Patient-physician conversations surrounding data collection can also indirectly influence downstream care, introducing potential performance bias. These sources of bias may adversely impact both data acquisition and AI performance with respect to at-risk patient groups. Recognizing and attenuating the impact of these imperfections in data collection is therefore crucial.
Practice-specific Data Curation
In community clinics and resource-constrained settings, AI algorithms are poised to assist in patient care through radiographic image interpretation (6,7,36), patient triage (37), and other applications (1). To enable effective clinical AI implementation, we strongly encourage radiologists to emphasize in-house data acquisition as much as allowable given pre-existing resource constraints.
Multiple commercial and open-source tools are increasingly available to facilitate data curation at both academic and community radiology practices (38). Of note, Micard et al (39) and Kuhn Cuellar et al (40) have introduced data management systems for medical image storage. Demirer et al (41) introduced a radiologist-friendly, no-code interface to streamline physician annotation of medical images, whereas Scherer et al (42) proposed an imaging data platform specifically designed for distributed data sharing across multiple hospital sites.
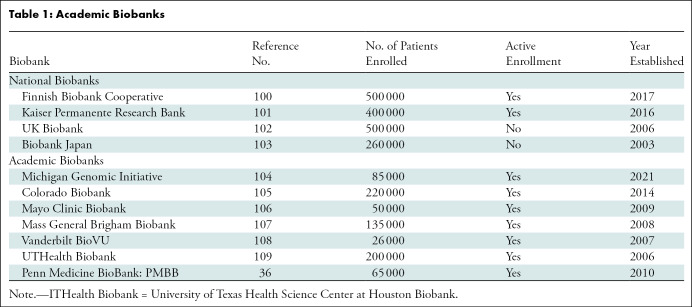
Academic Biobanks
Academic biobanks are patient data sets collected by academic hospitals and affiliated research institutions (43). Example biobanks include the Penn Medicine BioBank, Mayo Clinic Biobank, and University of Texas Health Science Center at Houston Biobank (UTHealth Biobank) (Table 1, Appendix S1). Figure 1 shows an overview of the imaging data available in the Penn Medicine BioBank. There are many potential applications of the patient imaging data acquired from an academic biobank. For example, clinically informed image analysis can be used to identify patients with different common diseases (Fig 2).
Table 1:
Academic Biobanks
Figure 1:
Overview of Penn Medicine BioBank (PMBB) imaging data. (A) Bar graph shows the number of studies within the Penn Medicine BioBank by imaging modality. The number of studies per Penn Medicine BioBank capita is the average number of studies per patient within the Penn Medicine BioBank. (B) Line graph shows the number of imaging studies acquired per year contained within the Penn Medicine BioBank by imaging modality. (C) Line graph of 1–CDF, where CDF is the cumulative distribution function. 1–CDF corresponds to the proportion of patients (by modality) according to number of examinations. (D) Histogram shows the time between sequential repeat imaging studies by patient for the four most common imaging modalities.
Figure 2:
Comparisons of principal component distributions of six artificial intelligence–extracted image-derived phenotype (IDP) metrics (36), calculated from abdominal CT in 1276 anonymized patients from the Penn Medicine BioBank. These image-derived phenotypes included liver CT attenuation, spleen CT attenuation, liver volume, spleen volume, visceral fat volume, and subcutaneous fat volume. Using principal component analysis (PCA), the principal component of these image-derived phenotypes was extracted and its distribution was plotted as a histogram for patients stratified by different clinical diagnoses. Bar graphs show different image-derived phenotype principal component distributions in patients without diagnoses (gray bars) versus in patients diagnosed with (A) obesity (n = 91), (B) obstructive sleep apnea (n = 201), and (C) hypertension (n = 1082). Image-derived phenotype principal component distributions in patients without diagnoses (gray bars) versus in patients diagnosed with (D) nonalcoholic fatty liver disease (NAFLD; n = 429) and (E) diabetes (n = 790). (F) Genitourinary diseases (n = 1202), which are not clinically associated with these image-derived phenotype metrics, were not associated with a statistically significant different principal component distribution compared with healthy patients (P = .12). P values were calculated with statistical analyses comparing distributions of patients with and without disease diagnoses, with a two-sample Kolmogorov-Smirnov test for goodness of fit.
In U.S. facilities, academic biobanks are practice-specific and capture the attributes and variance of the patient population of the hospital. Missing or repeated values in these data sets also mirror the messiness of real-world patient data, for which implemented algorithms and applications must be equipped to handle for any practical use (44).
Whereas biobanks have proven useful for certain academic hospitals, they are by no means necessary for clinical AI implementation in the average radiology practice. For satellite hospitals and community practices that share similar patient populations with the parent academic hospital, academic biobanks and their subsets can serve as “pseudo-in-house” data sets. This can prove particularly useful in settings used for implementation without the resources or infrastructure to systematically collect their own in-house test data.
A Proposed Framework for Clinical AI Implementation
A proposed three-step framework for clinical AI implementation in an academic research hospital environment focuses on the following central considerations: problem identification, stakeholder alignment, and pipeline integration. Table 2 summarizes this framework.
Table 2:
A Proposed Framework for Clinical AI Implementation
Step 1: Problem Identification
Over the past decade, the potential use cases of ML methods for multimodal breast nodule detection and early detection of breast cancer has been explored (45,46). Given these studies, the Department of Radiology at the University of Pennsylvania developed the infrastructure necessary for the AI-assisted detection of breast cancer. However, it became clear that practicing radiologists had no interest in using the proposed tools even at the onset of clinical implementation. Through subsequent discussions with physician partners, this underuse was because ML-enabled image interpretation performed no better than radiologists, and there was no excess demand for image interpretation that could not already be met by radiologists. In addition to the risks and liability associated with automated performance and difficulty in widespread adaptation, these barriers limited the use of AI tools in this clinical setting.
This experience reinforced the importance of first identifying and naming the specific problem that a proposed ML approach will solve, which is a key step in implementation science that extends beyond clinical AI implementation alone. Problem identification refers to the determination of real problems from the perspective of individuals in the target population for use. The ostensible problem addressed was to automate the detection of breast cancer with human expert–level accuracy. However, from the perspective of the radiologists that diagnose breast masses daily, there was no problem with the accuracy of diagnoses, number of patient cases, or time of diagnoses. Thus, the addressed problem was no practical problem at all for the target population (ie, the breast radiologists).
Common guiding principles exist for problem identification regarding clinical AI implementation. The U.S. Centers for Disease Control and Prevention introduced its Planning and Operations Language for Agent-based Regional Integrated Simulation framework for problem identification in public health (47), and Stannard (48) highlighted case studies on effective problem identification in evidence-based practices. Cumulatively, these studies raise important questions that inform our own institutional practices: Who is impacted by the problem at hand? And what evidence exists to quantify and characterize the impact? The process of answering these questions through relevant interviews, analysis, and literature reviews provides a generalizable framework for robust problem identification.
From experience at the University of Pennsylvania, clarifying the problem that AI implementation will solve revolves around one of the following four objective tenets: unsatisfactory diagnostic accuracy or therapeutic efficacy (18), long time to diagnosis or low therapeutic efficiency (8), high costs or low revenue (3), and inefficiencies in patient care and cumbersome legacy systems (37). Most AI implementation solutions should attempt to improve on at least one of these pressure points. For adequate progress tracking, quantitative data are necessary, including time scales, patient surveys, changes in relative value units, and application-specific efficacy metrics.
The task of problem identification is inherently tied to interdisciplinary communication and collaboration. Communication must be both documented and frequent to allow for iterative problem identification. Similarly, if proposed solutions are intended for patients, it is equally important to communicate with patients. Effective problem identification is a task that should evolve to ensure problem-solution alignment for researchers, clinicians, and other relevant stakeholders.
Step 2: Stakeholder Alignment
Parallel to problem identification is aligning proposed solutions with the goals and values of key stakeholders: institutional leaders, radiologists, and the referring clinicians (49). The stakeholder alignment step is similar to problem identification: To gain tractable support for clinical AI implementation, it is important to convince both clinicians and hospital leadership, who often have different needs and goals.
Institutional leaders are important stakeholders behind any plan for implementation. For example, conversations with departmental directors and chairs at the University of Pennsylvania allowed for a better understanding of the shared guiding principles for institutional stakeholders. We found that framing the proposed implementation strategies through the lens of revenue generation and cost reduction at the level of the health care enterprise was crucial in getting stakeholder buy-in. Any initiative, whether related to AI implementation or not, must be sustainable without excessive consumption of the hospital’s limited resources. Of note, larger academic institutions often provide many clinical services that enable resource pooling and opportunities for adaptation of technologies that could yield net negative revenue, particularly in the short term. It is also crucial to investigate algorithm implementation strategies in community-based hospitals and practices in terms that make financial sense, cognizant that there may not be complementary institutional initiatives to provide a financial buffer.
Although revenue generation is a priority for many institutional stakeholders, many departmental leaders are also interested in secondary community benefits to either the health care system or the greater scientific community. Most academic biobanks provide operable data to institution-affiliated researchers. It is equally important to align implementation efforts with nonsupervisory parties. Of course, clinicians and users of implemented AI strategies seek to improve the quality, ease, and efficiency of health care. In many radiology practices with large volumes of imaging studies, radiologist users seek to optimize the practical user experience to avoid additional user-facing computer applications, mouse clicks, and other extra steps in workflows. To streamline patient care, users also seek to minimize downtime and technical errors with respect to software latency and service outages. Adapted from our own experience, risk stratification questions for stakeholders to better characterize these potential sources of risk are in Table 3.
Table 3:
Risk Stratification Questions to Ask Potential Commercial AI Vendors
Accomplishing these goals requires the expertise of a multidisciplinary team of engineers and product managers to use clinical data from hospital sources and implement AI algorithms into clinical practice. Initial efforts often require rapid expansion of clinical implementation and oversight teams when establishing roles for project managers, directors, support staff, and steering committees.
Finally, whereas the primary focus of this article is on a subgroup of key stakeholders in clinical AI implementation, AI implementation will impact technologists, equipment operators, support staff, and hospital administrators. For example, patients and patient advocacy groups also seek to use AI algorithms that are trustworthy, equitable, and easy for patients to understand (50). The influence of AI solutions on these roles emphasizes the importance of interdisciplinary collaboration (21,51).
Step 3: Pipeline Integration to Enable Algorithm Validation in Hospital Systems
Clinical pipelines are necessary for the proper validation of proposed ML algorithms. Prior studies by Argent et al (52) and Yu et al (53) have argued for real-world validation from a data perspective because model inputs in practical clinical scenarios may be imprecise and have missing data values.
Our empirical experiences suggest that the greatest value added from validation in existing clinical pipelines is the ability to define and quantify user-centered metrics for successful implementation. Metrics optimized during training and reported after testing do not necessarily correspond to metrics that best reflect human expert objectives, as explored in detail by Park et al (54). For example, Zhao et al (7) reported that maximizing the structural similarity index measure to train models in computer vision applications can excessively smooth images and therefore reduce the ability for clinical detection of smaller lesions at imaging. Classifier models in ML are also typically scored using threshold-agnostic functions, such as the area under the receiver operating characteristic curve. However, real-world applications are more interested in fine-tuning thresholds so that trade-offs between model sensitivity, specificity, and predictive power are in accordance with clinician priorities and error tolerances as assessed through direct human-computer interfacing. Exact user-centered metrics used to assess the impact of implemented AI solutions will largely depend on the specific application and iterative conversations with end users.
Another important consideration in real-world validation is determining how ML models will interface with medical imaging data. Whereas the specific implementation details largely vary from site to site, there are generally three conserved frameworks for pipeline design: on-scanner, cloud-based pre–picture archiving and communication system, and cloud-based co–picture archiving and communication system (Fig 3). At a high level, on-scanner implementation allows ML models to have easy access to raw data, enabling physics-based image reconstruction (55) and improvements to scanner outputs by using information from both data and environmental variables. On-scanner applications are typically offline and therefore often feature faster inference times and more secure data transfer. However, they are largely limited by an intrinsic difficulty in scaling ML models over time and keeping software up to date with vendor-led changes to acquisition parameters, hardware, and internal software. This issue is addressed by moving AI data interfacing to cloud-based environments, where models trade access to hardware data for scalability, easier interfacing for researchers, and the ability to work with more interpretable medical imaging data alongside additional data types such as laboratories, medical records, and patient demographic information. However, cloud-based environments need to be able to process data from any scanner in an unbiased manner, which is often difficult in practice. Whereas scanner-based implementation has been studied, the cloud-based co–picture archiving and communication system ML framework has the greatest empirical success according to Elahi et al (56).
Figure 3:
Overview of common frameworks for integrating machine learning and medical imaging. (A) Artificial intelligence (AI) models interface directly with raw acquired data and are implemented on a per-scanner basis. (B) Machine learning models have access to the processed image outputs from the individual scanners before they are sent to the central picture archiving and communication system (PACS) server. (C) Models communicate with the picture archiving and communication system server to obtain relevant inputs for AI interpretation.
In most practical cases, the space of possible interfacing opportunities is limited by both who is developing an ML model and what is its intended use case. For example, MRI reconstruction algorithms that aim to reconstruct anatomic images from sparsely sampled data take raw scanner data and parameters as input and therefore are exclusively implemented via scanner-based ML (4,5). Whereas general open-source frameworks and application programming interfaces have become increasingly available (57,58), optimizing on-scanner ML relies on an understanding of vendor-specific software and hardware and requires close collaboration with manufacturers. Conversely, methods in medical image segmentation often do not require access to raw scanner data and could therefore benefit from cloud-based implementation frameworks (36).
In addition to the engineering efforts of effective pipeline integration, this step of clinical AI implementation offers an opportunity for internal validation and is where data acquisition proves useful. Whereas AI solutions may be U.S. Food and Drug Administration–approved, externally validated performance rarely translates into clinical practice (33). The human-in-the-loop interactions between clinical users and AI models may differ across practices. Therefore, hospital-specific qualification of these algorithms is necessary (59). The extent to which independent evaluation of these workflows is possible will depend on the institution-specific availability of resources, and it remains an open problem in the field of clinical AI implementation.
In many cases, pipeline integration can be accomplished with the aid of commercial vendor teams after the purchase of AI systems. We have found that these teams can work together with existing information technology staff at clinical sites to help overcome the technologic barriers associated with implementation of AI.
Data Curation
There are several approaches that do not require large amounts of data curation to validate and adapt commercial AI tools for successful implementation. One popular method in radiographic applications is image harmonization, which projects medical imaging data from community hospitals into the image space used to train an ML model (60,61). However, such methods require access to the data used to train and test the model of interest. A more practical solution is model fine-tuning, which takes an existing public model previously trained on a large corpus of data and optimizes its performance on a substantially smaller data set derived from the community hospital. Model fine-tuning can enable strong predictive performance in settings with limited data and resources (1,5,36), with Candemir et al already offering a promising solution (62). Affordable, resource-optimized methods for model fine-tuning were viable in a variety of potential practice settings (63,64). These techniques require access to transparent model parameters and careful documentation of available data and models to tune available AI models for use in settings where they are most needed. Whereas model parameters are often not released publicly, the U.S. Food and Drug Administration and other regulatory parties have strongly advocated for this practice in recent years (26). Finally, if fine-tuning the model is not possible, recording test-time performance on patient data may offer valuable empirical metrics to quantify the impact of implemented AI solutions after pipeline integration. Whereas many hospital practices may be unable to devote large volumes of resources toward institutional validation, any degree of validation is better than no validation at all.
The Role of AI in Medicine
Recent advancements in AI performance and implementation in subsets of consumer markets have sparked speculation that AI may replace health care workers in the future. Instead, we argue that clinical AI implementation is more likely to augment patient health care and should be designed with the goal of working together with clinicians and their patients. Longoni (65) and Richardson et al (66) show that patients generally distrust AI compared with human experts for their medical care because of a lack of personalized care and a desire for human empathy and understanding. Shuaib et al (67) discuss the ethical and liability concerns unique to high-stakes AI applications, such as patient care, and recommend against the adaptation of AI models that operate independently of human supervision.
Kundu (68) and Juluru et al (69) proposed several potential use cases in remote patient and prescription adherence monitoring among other related tasks. In accordance with Park et al (70) and Paranjape et al (71), the same generative models that power advancements in protein folding can also generate practice imaging cases for clinician trainees, and even augment training data sets for other ML applications (72,73). AI may also be readily used for supportive oversight and lower-stakes tasks such as automated resource allocation (74), triage (37,70,71,75), clinical pipeline supervision (76), and patient follow-up (8) that may not require direct human-algorithm interfacing.
Finally, clinical AI implementation in radiology practices is especially poised for high-value impact in resource-limited settings, such as in rural communities across the globe with few radiologists. Firsthand experiences from Elahi et al (56) and Ciecierski-Holmes et al (77) demonstrate that the major challenges to clinical AI implementation unique to these environments are more related to establishing appropriate digital infrastructure and support networks to enable AI-assisted clinical medicine. In the settings where human experts are limited, AI diagnostic tools have crucial roles in interpreting patient data that is either read by machines or not read at all.
Research Directions for Future Work
The U.S. Food and Drug Administration recently proposed a set of guidelines and important strategic directions for implementing AI tools in clinical practice (26). Such directions include improving both the AI compatibility and capabilities for internal validation of radiology digital infrastructure (Fig 4).
Figure 4:
Robust Detection of Data Distribution Shift
Successful implementation necessitates detecting input data outside of the training data distribution. Allen et al (78) reported that over 90% of ML users found U.S. Food and Drug Administration–cleared AI algorithms to have unexpectedly poor predictive performance on their own data compared with the data used for model training and U.S. Food and Drug Administration clearance. In medical imaging applications, harmonization techniques have attempted to project data sets from out-of-domain to in-domain, although such methods inherently alter input images and therefore the information content (60,61,79,80). Other methods have been proposed (81,82), but evidence for their adaptation into clinical workflows remains limited. Detecting data distribution shift is important for understanding algorithmic limitations and determining model lifetime among others. Furthermore, in global health applications where there is often insufficient in-domain training data and an overall lack of resources, researchers need to be able to characterize and potentially improve out-of-domain generalization of model performance. For example, MacLean et al (36) demonstrated that data augmentation with patient subsets identified by unique clinical features could improve segmentation model performance in data with the same clinical features without sacrificing overall performance.
Interpretability of ML Methods
Despite recent advancements in model reasoning by Wei et al (83) and Lample et al (84), trade-offs between model complexity and explainability are well-documented in related work (85–87). Explainability may take the form of explicit human-level reasoning (83,84) or providing model uncertainty quantification (88–90) based on current directions in ML research.
Clinical data can be unclear and conflicting, and laboratory values outside the so-called normal range are not always causes for concern in medicine. The learned expertise of clinicians to synthesize information in the context of a clinical picture is not replicable in existing ML methods. For broad clinical adaptation, physicians must be able to interpret ML outputs and understand the associated reasoning process to contextualize them with the available patient information (8).
A Standardized Framework for Machine Intelligence Reporting
Existing black-box approaches in ML are not conducive to continual learning or to effectively mitigate bias (19,91,92). In addition to improving the explainability of ML models, AI algorithms in health care need standardized, interinstitutional reporting frameworks that detail important attributes of each implemented AI algorithm (26). Many companies have implemented such internal organizational standards, and the recent introduction of so-called model cards, which detail important metadata about ML models (and their associated training data) to improve ML documentation, have gained tractable adaptation among ML researchers (93–96). Using model cards and other successful software documentation frameworks from other fields may help improve existing guidelines for radiology.
Fortunately, adapting existing infrastructure in health care may also assist in standardization. For example, Rubin et al (97) introduced the common data element in clinical radiology. Common data elements may be used to define queryable metadata that can describe an ML model’s training process, pertinent features of its training data, key dependencies, test evaluation, and performance. Together with associated work by Belbasis et al (33) and Heil et al (98), Mongan et al (99) proposed a framework of core common data elements to AI reporting in clinical medicine. In the immediate future, our goal is to implement such a framework for better reproducibility standards and trustworthiness of ML applications in patient health care.
Data and Code Availability
Data related to the Penn Medicine BioBank are available to investigators on request. The code used to generate Figures 1 and 2 is available on a Github repository (https://github.com/michael-s-yao/PennMedBiobankAIDeployment).
Conclusion
Implementation of clinical artificial intelligence (AI) in radiology practices is a challenging process with important considerations in a rapidly evolving space. Nonetheless, there are common principles to guide implementation efforts derived from both empirical analysis and insights from other medical fields. Empowered by the increased availability of different forms of radiologic data, AI algorithm implementation involves identifying the problems in health care for which there may exist machine learning–based solutions, aligning these solutions with key clinical stakeholders, and verifying that these solutions work seamlessly in real-world clinical practices. Ultimately, these steps can serve to standardize the implementation of AI technologies and consequently improve patient care in radiologic practices.
A.C. and M.S.Y. contributed equally to this work.
W.R.W. and J.C.G. are co-senior authors.
M.S.Y. is supported by the National Institutes of Health (NIH) Training Program in Biomedical Imaging and Informational Sciences (T32 EB009384) at the University of Pennsylvania. N.C. is supported by an institutional training grant from the NIH (5T32EB004311). W.R.W. is supported by the NIH (R01 HL137984, P41 EB029460). J.C.G. is supported by the NIH (R01 HL133889).
Data sharing: Data generated or analyzed during the study are available from the corresponding author by request.
Disclosures of conflicts of interest: A.C. No relevant relationships. M.S.Y. No relevant relationships. H.S. Scholar Grant from RSNA; Society of Radiologists in Ultrasound Early Career Award, Penn-Calico Collaborative grant (pending); patent planned in the future for aspects of an artificial intelligence pipeline; leadership role in MDPI Computers Special Issue editor, SIIM Clinical Data Informaticist Task Force. A.D.G. No relevant relationships. N.C. No relevant relationships. M.T.M. Research stipend from Sarnoff Cardiovascular Research Foundation; meeting and travel support from Sarnoff Cardiovascular Research Foundation. J.D. No relevant relationships. A.E. No relevant relationships. A.B. No relevant relationships. M.D.R. No relevant relationships. D.R. No relevant relationships. C.E.K. Consulting fees from Daccan Partners; travel support and honorarium from American Society of Gastrointestinal Endoscopy; Editor for Radiology: Artificial Intelligence. W.R.W. No relevant relationships. J.C.G. Grants to author’s institution from NIH, Siemens Healthineers; stock/stock options from Merck.
Abbreviations:
- AI
- artificial intelligence
- ML
- machine learning
References
- 1. Yan A , McAuley J , Lu X , et al . RadBERT: Adapting transformer-based language models to radiology . Radiol Artif Intell 2022. ; 4 ( 4 ): e210258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jaiswal A , Tang L , Ghosh M , Rousseau JF , Peng Y , Ding Y . RadBERT-CL: Factually-Aware Contrastive Learning For Radiology Report Classification . Proc Mach Learn Res 2021. ; 158 : 196 – 208 . [PMC free article] [PubMed] [Google Scholar]
- 3. Lee JE , Choi SY , Hwang JA , et al . The potential for reduced radiation dose from deep learning-based CT image reconstruction: A comparison with filtered back projection and hybrid iterative reconstruction using a phantom . Medicine (Baltimore) 2021. ; 100 ( 19 ): e25814 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sriram A , Zbontar J , Murrell T , et al . End-to-End Variational Networks for Accelerated MRI Reconstruction . In: MICCAI 2020. . Springer International Publishing; , 2020. ; 64 – 73 . [Google Scholar]
- 5. Yao MS , Hansen MS . A Path Towards Clinical Adaptation of Accelerated MRI . Proc Mach Learn Res 2022. ; 193 : 489 – 511 . [PMC free article] [PubMed] [Google Scholar]
- 6. Rajpurkar P , Irvin J , Zhu K , et al . CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning . arXiv 1711.05225 [preprint] https://arxiv.org/abs/1711.05225. Updated December 25, 2017. Accessed August 11, 2023.
- 7. Zhao R , Zhang Y , Yaman B , Lungren MP , Hansen MS . End-to-End AI-based MRI Reconstruction and Lesion Detection Pipeline for Evaluation of Deep Learning Image Reconstruction . arXiv 2109.11524 [preprint] https://arxiv.org/abs/2109.11524. Published September 23, 2021. Accessed August 11, 2023.
- 8. Yao MS , Chae A , MacLean MT , et al . SynthA1c: Towards Clinically Interpretable Patient Representations for Diabetes Risk Stratification . arXiv 2209.10043 [preprint] https://arxiv.org/abs/2209.10043. Updated July 28, 2023. Accessed August 11, 2023. [DOI] [PMC free article] [PubMed]
- 9. Fung DLX , Liu Q , Zammit J , Leung CK , Hu P . Self-supervised deep learning model for COVID-19 lung CT image segmentation highlighting putative causal relationship among age, underlying disease and COVID-19 . J Transl Med 2021. ; 19 ( 1 ): 318 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices . U.S. Food & Drug Administration . https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices. Updated October 5, 2022. Accessed November 1, 2022.
- 11. Matheny ME , Whicher D , Thadaney Israni S . Artificial intelligence in health care: A report from the national academy of medicine . JAMA 2020. ; 323 ( 6 ): 509 – 510 . [DOI] [PubMed] [Google Scholar]
- 12. Khunte M , Chae A , Wang R , et al . Trends in clinical validation and usage of US Food and Drug Administration-cleared artificial intelligence algorithms for medical imaging . Clin Radiol 2023. ; 78 ( 2 ): 123 – 129 . [DOI] [PubMed] [Google Scholar]
- 13. Loh E . Medicine and the rise of the robots: A qualitative review of recent advances of artificial intelligence in health . BMJ Leader 2018. ; 2 ( 2 ): 59 – 63 . [Google Scholar]
- 14. Sun TQ . Adopting artificial intelligence in public healthcare: The effect of social power and learning algorithms . Int J Environ Res Public Health 2021. ; 18 ( 23 ): 12682 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thomas LB , Mastorides SM , Viswanadhan NA , Jakey CE , Borkowski AA . Artificial intelligence: Review of current and future applications in medicine . Fed Pract 2021. ; 38 ( 11 ): 527 – 538 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petersson L , Larsson I , Nygren JM , et al . Challenges to implementing artificial intelligence in healthcare: a qualitative interview study with healthcare leaders in Sweden . BMC Health Serv Res 2022. ; 22 ( 1 ): 850 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reyes M , Meier R , Pereira S , et al . On the interpretability of artificial intelligence in radiology: Challenges and opportunities . Radiol Artif Intell 2020. ; 2 ( 3 ): e190043 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Topol EJ . High-performance medicine: the convergence of human and artificial intelligence . Nat Med 2019. ; 25 ( 1 ): 44 – 56 . [DOI] [PubMed] [Google Scholar]
- 19. Kahn CE Jr . Hitting the mark: Reducing bias in AI systems . Radiol Artif Intell 2022. ; 4 ( 5 ): e220171 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rowell C , Sebro R . Who will get paid for artificial intelligence in medicine? Radiol Artif Intell 2022. ; 4 ( 5 ): e220054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Proctor EK , Powell BJ , McMillen JC . Implementation strategies: recommendations for specifying and reporting . Implement Sci 2013. ; 8 ( 1 ): 139 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kiriiri GK , Njogu PM , Mwangi AN . Exploring different approaches to improve the success of drug discovery and development projects: A review . Futur J Pharm Sci 2020. ; 6 ( 1 ): 1 – 12 . [Google Scholar]
- 23. Hughes JP , Rees S , Kalindjian SB , Philpott KL . Principles of early drug discovery . Br J Pharmacol 2011. ; 162 ( 6 ): 1239 – 1249 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Dyk L . A review of telehealth service implementation frameworks . Int J Environ Res Public Health 2014. ; 11 ( 2 ): 1279 – 1298 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Villalobos Dintrans P , Bossert TJ , Sherry J , Kruk ME . A synthesis of implementation science frameworks and application to global health gaps . Glob Health Res Policy 2019. ; 4 ( 1 ): 25 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Artificial Intelligence and Machine Learning in Software as a Medical Device . U.S. Food & Drug Administration . https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device. Updated September 22, 2021. Accessed February 15, 2023.
- 27. Hwang TJ , Kesselheim AS , Vokinger KN . Lifecycle regulation of artificial intelligence– and machine learning–based software devices in medicine . JAMA 2019. ; 322 ( 23 ): 2285 – 2286 . [DOI] [PubMed] [Google Scholar]
- 28. Willemink MJ , Roth HR , Sandfort V . Toward foundational deep learning models for medical imaging in the new era of transformer networks . Radiol Artif Intell 2022. ; 4 ( 6 ): e210284 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sellergren AB , Chen C , Nabulsi Z , et al . Simplified transfer learning for chest radiography models using less data . Radiology 2022. ; 305 ( 2 ): 454 – 465 . [DOI] [PubMed] [Google Scholar]
- 30. Bycroft C , Freeman C , Petkova D , et al . The UK Biobank resource with deep phenotyping and genomic data . Nature 2018. ; 562 ( 7726 ): 203 – 209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia-Gallo E , Merson L , Kennon K , et al. ; ISARIC Clinical Characterization Group. ISARIC-COVID-19 dataset: A prospective, standardized, global dataset of patients hospitalized with COVID-19 . Sci Data 2022. ; 9 ( 1 ): 454 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao R , Yaman B , Zhang Y , et al . fastMRI+, Clinical pathology annotations for knee and brain fully sampled magnetic resonance imaging data . Sci Data 2022. ; 9 ( 1 ): 152 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Belbasis L , Panagiotou OA . Reproducibility of prediction models in health services research . BMC Res Notes 2022. ; 15 ( 1 ): 204 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaba P , Bhatt DL . Recruitment practices in multicenter randomized clinical trials: Time for a relook . J Am Heart Assoc 2021. ; 10 ( 22 ): e023673 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Man Y , Wieland-Jorna Y , Torensma B , et al . Opt-in and opt-out consent procedures for the reuse of routinely recorded health data in scientific research and their consequences for consent rate and consent bias: Systematic review . J Med Internet Res 2023. ; 25 : e42131 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MacLean MT , Jehangir Q , Vujkovic M , et al . Quantification of abdominal fat from computed tomography using deep learning and its association with electronic health records in an academic biobank . J Am Med Inform Assoc 2021. ; 28 ( 6 ): 1178 – 1187 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yao LH , Leung KC , Tsai CL , Huang CH , Fu LC . A novel deep learning-based system for triage in the emergency department using electronic medical records: Retrospective cohort study . J Med Internet Res 2021. ; 23 ( 12 ): e27008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boch M , Gindl S , Barnett A , et al . A Systematic Review of Data Management Platforms . In: Rocha A , Adeli H , Dzemyda G , Moreira F (eds). Information Systems and Technologies . WorldCIST 2022. Lecture Notes in Networks and Systems , vol 469 . Springer; , Cham: . [Google Scholar]
- 39. Micard E , Husson D , Felblinger J . ArchiMed: A data management system for clinical research in imaging . Front ICT 2016. ; 3 . [Google Scholar]
- 40. Kuhn Cuellar L , Friedrich A , Gabernet G , et al . A data management infrastructure for the integration of imaging and omics data in life sciences . BMC Bioinformatics 2022. ; 23 ( 1 ): 61 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Demirer M , Candemir S , Bigelow MT , et al . A user interface for optimizing radiologist engagement in image data curation for artificial intelligence . Radiol Artif Intell 2019. ; 1 ( 6 ): e180095 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scherer J , Nolden M , Kleesiek J , et al . Joint imaging platform for federated clinical data analytics . JCO Clin Cancer Inform 2020. ; 4 ( 4 ): 1027 – 1038 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Henderson GE , Cadigan RJ , Edwards TP , et al . Characterizing biobank organizations in the U.S.: results from a national survey . Genome Med 2013. ; 5 ( 1 ): 3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Willemink MJ , Koszek WA , Hardell C , et al . Preparing medical imaging data for machine learning . Radiology 2020. ; 295 ( 1 ): 4 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gastounioti A , Desai S , Ahluwalia VS , Conant EF , Kontos D . Artificial intelligence in mammographic phenotyping of breast cancer risk: a narrative review . Breast Cancer Res 2022. ; 24 ( 1 ): 14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Witowski J , Heacock L , Reig B , et al . Improving breast cancer diagnostics with deep learning for MRI . Sci Transl Med 2022. ; 14 ( 664 ): eabo4802 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Office of the Associate Director for Policy and Strategy: POLARIS . Centers for Disease Control and Prevention . https://www.cdc.gov/policy/polaris/index.html. Accessed February 21, 2023.
- 48. Stannard D . Problem identification: The first step in evidence-based practice . AORN J 2021. ; 113 ( 4 ): 377 – 378 . [DOI] [PubMed] [Google Scholar]
- 49. Daye D , Wiggins WF , Lungren MP , et al . Implementation of clinical artificial intelligence in radiology: Who decides and how? Radiology 2022. ; 305 ( 3 ): 555 – 563 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Z , Citardi D , Wang D , Genc Y , Shan J , Fan X . Patients’ perceptions of using artificial intelligence (AI)-based technology to comprehend radiology imaging data . Health Informatics J 2021. ; 27 ( 2 ): 14604582211011215 . [DOI] [PubMed] [Google Scholar]
- 51. Patel B , Usherwood T , Harris M , et al . What drives adoption of a computerised, multifaceted quality improvement intervention for cardiovascular disease management in primary healthcare settings? A mixed methods analysis using normalisation process theory . Implement Sci 2018. ; 13 ( 1 ): 140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Argent R , Bevilacqua A , Keogh A , Daly A , Caulfield B . The importance of real-world validation of machine learning systems in wearable exercise biofeedback platforms: A case study . Sensors (Basel) 2021. ; 21 ( 7 ): 2346 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu AC , Mohajer B , Eng J . External validation of deep learning algorithms for radiologic diagnosis: A systematic review . Radiol Artif Intell 2022. ; 4 ( 3 ): e210064 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park SH , Sul AR , Han K , Sung YS . How to determine if one diagnostic method, such as an artificial intelligence model, is superior to another: Beyond performance metrics . Korean J Radiol 2023. ; 24 ( 7 ): 601 – 605 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yaman B , Hosseini SAH , Moeller S , Ellermann J , Uğurbil K , Akçakaya M . Self-supervised learning of physics-guided reconstruction neural networks without fully sampled reference data . Magn Reson Med 2020. ; 84 ( 6 ): 3172 – 3191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elahi A , Dako F , Zember J , et al . Overcoming challenges for successful PACS installation in low-resource regions: Our experience in Nigeria . J Digit Imaging 2020. ; 33 ( 4 ): 996 – 1001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hansen MS , Sørensen TS . Gadgetron: an open source framework for medical image reconstruction . Magn Reson Med 2013. ; 69 ( 6 ): 1768 – 1776 . [DOI] [PubMed] [Google Scholar]
- 58. Veldmann M , Ehses P , Chow K , Nielsen JF , Zaitsev M , Stöcker T . Open-source MR imaging and reconstruction workflow . Magn Reson Med 2022. ; 88 ( 6 ): 2395 – 2407 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silcox C , Dentzer S , Bates DW . AI-enabled clinical decision support software: A “Trust and value checklist” for clinicians . NEJM Catal 2020. ; 1 ( 6 ): CAT.20.0212. [Google Scholar]
- 60. Guan H , Liu Y , Yang E , Yap PT , Shen D , Liu M . Multi-site MRI harmonization via attention-guided deep domain adaptation for brain disorder identification . Med Image Anal 2021. ; 71 : 102076 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mali SA , Ibrahim A , Woodruff HC , et al . Making radiomics more reproducible across scanner and imaging protocol variations: A review of harmonization methods . J Pers Med 2021. ; 11 ( 9 ): 842 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Candemir S , Nguyen XV , Folio LR , Prevedello LM . Training strategies for radiology deep learning models in data-limited scenarios . Radiol Artif Intell 2021. ; 3 ( 6 ): e210014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ding N , Qin Y , Yang G , et al . Parameter-efficient fine-tuning of large-scale pre-trained language models . Nat Mach Intell 2023. ; 5 ( 3 ): 220 – 235 . [Google Scholar]
- 64. Duval A , Lamson T , de Kerouara GdL , Gallé M . Breaking Writer's Block: Low-cost Fine-tuning of Natural Language Generation Models . arXiv 2101.03216 [preprint] https://arxiv.org/abs/2101.03216. Updated March 2, 2021. Accessed August 11, 2023.
- 65. Longoni C . Resistance to medical artificial intelligence . J Consum Res 2019. ; 46 ( 4 ): 629 – 650 . [Google Scholar]
- 66. Richardson JP , Smith C , Curtis S , et al . Patient apprehensions about the use of artificial intelligence in healthcare . NPJ Digit Med 2021. ; 4 ( 1 ): 140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shuaib A , Arian H , Shuaib A . The increasing role of artificial intelligence in health care: Will robots replace doctors in the future? Int J Gen Med 2020. ; 13 : 891 – 896 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kundu S . How will artificial intelligence change medical training? Commun Med (Lond) 2021. ; 1 ( 1 ): 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Juluru K , Shih HH , Keshava Murthy KN , et al . Integrating Al Algorithms into the Clinical Workflow . Radiol Artif Intell 2021. ; 3 ( 6 ): e210013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Park SH , Do KH , Kim S , Park JH , Lim YS . What should medical students know about artificial intelligence in medicine? J Educ Eval Health Prof 2019. ; 16 : 18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Paranjape K , Schinkel M , Panday RN , Car J , Nanayakkara P . Introducing Artificial Intelligence Training in Medical Education . JMIR Med Ed 2019. ; 5 ( 2 ): e16048 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pinaya WHL , Tudosiu P , Dafflon J , et al . Brain imaging generation with latent diffusion models . In: Deep Generative Models . Springer Nature Switzerland; , 2022. ; 117 – 126 . [Google Scholar]
- 73. Duong MT , Rauschecker AM , Rudie JD , et al . Artificial intelligence for precision education in radiology . Br J Radiol 2019. ; 92 ( 1103 ): 20190389 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Żbikowski K , Ostapowicz M , Gawrysiak P . Deep reinforcement learning for resource allocation in business processes . In: Process Mining Workshops . Lecture Notes in Business Information Processing; 2023. ; 468 . [Google Scholar]
- 75. Miles J , Turner J , Jacques R , Williams J , Mason S . Using machine-learning risk prediction models to triage the acuity of undifferentiated patients entering the emergency care system: a systematic review . Diagn Progn Res 2020. ; 4 ( 1 ): 16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li X , Wang L , Xin Y , Yang Y , Chen Y . Automated vulnerability detection in source code using minimum intermediate representation learning . Appl Sci (Basel) 2020. ; 10 ( 5 ): 1692 . [Google Scholar]
- 77. Ciecierski-Holmes T , Singh R , Axt M , Brenner S , Barteit S . Artificial intelligence for strengthening healthcare systems in low- and middle-income countries: a systematic scoping review . NPJ Digit Med 2022. ; 5 ( 1 ): 162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Allen B , Agarwal S , Coombs L , Wald C , Dreyer K . 2020 ACR data science institute artificial intelligence survey . J Am Coll Radiol 2021. ; 18 ( 8 ): 1153 – 1159 . [DOI] [PubMed] [Google Scholar]
- 79. Bashyam VM , Doshi J , Erus G , et al. ; iSTAGING and PHENOM consortia . Deep generative medical image harmonization for improving cross‐site generalization in deep learning predictors . J Magn Reson Imaging 2022. ; 55 ( 3 ): 908 – 916 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Keller H , Shek T , Driscoll B , et al . Noise-based image harmonization significantly increases repeatability and reproducibility of radiomics features in PET images: A phantom study . Tomography 2022. ; 8 ( 2 ): 1113 – 1128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Baek C , Jiang Y , Raghunathan A , Kolter Z . Agreement-on-the-Line: Predicting the Performance of Neural Networks under Distribution Shift . arXiv.2206.13089 [preprint] https://arxiv.org/abs/2206.13089. Updated May 11, 2023. Accessed August 11, 2023.
- 82. Hulkund N , Fusi N , Vaughan JW , Alvarez-Melis D . Interpretable distribution shift detection using optimal transport . arXiv 2208.02896 [preprint] https://arxiv.org/abs/2208.02896. Published August 4, 2022. Accessed August 11, 2023.
- 83. Wei J , Wang X , Schuurmans D , Bosma M , Ichter B , Xia F , Chi E , Le Q , Zhou D . Chain-of-thought prompting elicits reasoning in large language models . arXiv 2201.11903 [preprint] https://arxiv.org/abs/2201.11903. Updated January 10, 2023. Accessed August 11, 2023.
- 84. Lample G , Lachaux M , Lavril T , Martinet X , Hayat A , Ebner G , Rodriguez A , Lacroix T . HyperTree proof search for neural theorem proving . arXiv 2205.11491 [preprint] https://arxiv.org/abs/2205.11491. Published May 23, 2022. Accessed August 11, 2023.
- 85. Herm L , Heinrich K , Wanner J , Janiesch C . Stop ordering machine learning algorithms by their explainability! A user-centered investigation of performance and explainability . Int J Inf Manage 2023. ; 69 : 102538 . [Google Scholar]
- 86. Dziugaite GK , Ben-David S , Roy DM . Enforcing interpretability and its statistical impacts: Trade-offs between accuracy and interpretability . arXiv 2010.13764 [preprint] https://arxiv.org/abs/2010.13764. Updated October 28, 2020. Accessed August 11, 2023.
- 87. Sidey-Gibbons JAM , Sidey-Gibbons CJ . Machine learning in medicine: a practical introduction . BMC Med Res Methodol 2019. ; 19 ( 1 ): 64 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Psaros AF , Meng X , Zou Z , Guo L , Karniadakis GE . Uncertainty quantification in scientific machine learning: Methods, metrics, and comparisons . J Comput Phys 2023. ; 477 : 111902 . [Google Scholar]
- 89. Abdar M , Pourpanah F , Hussain S , et al . A review of uncertainty quantification in deep learning: Techniques, applications and challenges . Inf Fusion 2021. ; 76 : 243 – 297 . [Google Scholar]
- 90. Park S , Bastani O , Matni N , Lee I . PAC confidence sets for deep neural networks via calibrated prediction . arXiv 2001.00106 [preprint] https://arxiv.org/abs/2001.00106. Updated February 15, 2020. Accessed August 11, 2023.
- 91. Caruana R , Nori H . Why data scientists prefer glassbox machine learning: Algorithms, differential privacy, editing and bias mitigation . Proc. ACM SIGKDD Conference on Knowledge Discovery and Data Mining 2022. ; 4776 – 7 . [Google Scholar]
- 92. Adam H , Balagopalan A , Alsentzer E , Christia F , Ghassemi M . Mitigating the impact of biased artificial intelligence in emergency decision-making . Commun Med (Lond) 2022. ; 2 ( 1 ): 149 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crisan A, Drouhard M, Vig J, Rajani N. Interactive model cards: A human-centered approach to model documentation. In: ACM Conference on Fairness, Accountability, and Transparency (FAccT) 2022;427–39. [Google Scholar]
- 94. Wadhwani A , Jain P . Machine learning model cards transparency review: Using model card toolkit . In: IEEE Pune Section International Conference (PuneCon) 2020. ; 133 – 37 . [Google Scholar]
- 95. Pushkarna M , Zaldivar A , Kjartansson O . Data Cards: Purposeful and Transparent Dataset Documentation for Responsible AI . arXiv 2204.01075 [preprint] https://arxiv.org/abs/2204.01075. Published April 3, 2022. Accessed August 11, 2023.
- 96. Mitchell M , Wu S , Zaldivar A , et al . Model Cards for Model Reporting . In: Proceedings of the Conference on Fairness, Accountability, and Transparency 2019. ; 220 – 9 . [Google Scholar]
- 97. Rubin DL , Kahn CE Jr , Charles E . Common data elements in radiology . Radiology 2017. ; 283 ( 3 ): 837 – 844 . [DOI] [PubMed] [Google Scholar]
- 98. Heil BJ , Hoffman MM , Markowetz F , Lee SI , Greene CS , Hicks SC . Reproducibility standards for machine learning in the life sciences . Nat Methods 2021. ; 18 ( 10 ): 1132 – 1135 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mongan J , Moy L , Kahn CE Jr . Checklist for artificial intelligence in medical imaging (CLAIM): A guide for authors and reviewers . Radiol Artif Intell 2020. ; 2 ( 2 ): e200029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lähteenmäki J , Vuorinen AL , Pajula J , et al . Integrating data from multiple Finnish biobanks and national health-care registers for retrospective studies: Practical experiences . Scand J Public Health 2022. ; 50 ( 4 ): 482 – 489 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Feigelson HS , Clarke CL , Van Den Eeden SK , et al . The Kaiser Permanente Research Bank Cancer Cohort: a collaborative resource to improve cancer care and survivorship . BMC Cancer 2022. ; 22 ( 1 ): 209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sudlow C , Gallacher J , Allen N , et al . UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age . PLoS Med 2015. ; 12 ( 3 ): e1001779 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nagai A , Hirata M , Kamatani Y , et al. ; BioBank Japan Cooperative Hospital Group . Overview of the BioBank Japan Project: Study design and profile . J Epidemiol 2017. ; 27 ( 3S 3S ): S2 – S8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fritsche LG , Gruber SB , Wu Z , et al . Association of polygenic risk scores for multiple cancers in a phenome-wide study: Results from the Michigan genomics initiative . Am J Hum Genet 2018. ; 102 ( 6 ): 1048 – 1061 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wiley L , Shortt J , Roberts E , et al. Colorado Center for Personalized Medicine . Building a vertically-integrated genomic learning health system: The Colorado Center for Personalized Medicine Biobank . MedRxiv [preprint] 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Olson JE , Ryu E , Johnson KJ , et al . The Mayo Clinic Biobank: a building block for individualized medicine . Mayo Clin Proc 2013. ; 88 ( 9 ): 952 – 962 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Boutin NT , Schecter SB , Perez EF , et al . The evolution of a large biobank at Mass General Brigham . J Pers Med 2022. ; 12 ( 8 ): 1323 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Roden DM , Pulley JM , Basford MA , et al . Development of a large-scale de-identified DNA biobank to enable personalized medicine . Clin Pharmacol Ther 2008. ; 84 ( 3 ): 362 – 369 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sanner JE , Nomie KJ . The biobank at the University of Texas Health Science Center at Houston . Biopreserv Biobank 2015. ; 13 ( 3 ): 224 – 225 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data related to the Penn Medicine BioBank are available to investigators on request. The code used to generate Figures 1 and 2 is available on a Github repository (https://github.com/michael-s-yao/PennMedBiobankAIDeployment).