Abstract
Sneathia vaginalis is a Gram-negative vaginal species that is associated with pregnancy complications. It produces cytopathogenic toxin A (CptA), a pore-forming toxin. To determine whether CptA is expressed in vivo and to examine the mucosal Ab response to the toxin, we examined human midvaginal swab samples obtained during pregnancy for IgM, IgA, and IgG Abs with CptA affinity. This subcohort study included samples from 93 pregnant people. S. vaginalis relative abundance was available through 16S rRNA survey. There were 22 samples from pregnancies that resulted in preterm birth in which S. vaginalis relative abundance was <0.005%, 22 samples from pregnancies that resulted in preterm birth with S. vaginalis ≥0.005%, 24 samples from pregnancies that resulted in term birth with S. vaginalis <0.005%, and 25 samples from pregnancies that resulted in term birth with S. vaginalis ≥0.005%. IgM, IgA, and IgG with affinity for CptA were assessed by ELISA. The capacity for the samples to neutralize CptA was quantified by hemolysis assay. All three Ab isotypes were detectable within different subsets of the samples. There was no significant association between relative abundance of S. vaginalis and the presence of any Ab isotype. The majority of vaginal swab samples containing detectable levels of anti-CptA Abs neutralized the hemolytic activity of CptA, with the strongest correlation between IgA and neutralizing activity. These results demonstrate that S. vaginalis produces CptA in vivo and that CptA is recognized by the host immune defenses, resulting in the production of Abs with toxin-neutralizing ability.
Introduction
The etiology of preterm birth varies, with differing levels of influence from both intrinsic and external factors (1, 2). Infectious agents are an important trigger of preterm birth, and, among infectious triggers, bacterial species that normally reside in the vagina are likely responsible in a substantial proportion of the cases (3). However, the roles of specific bacterial taxa in the pathogenesis of infectious preterm birth need to be defined more precisely to promote advances in treatment modalities (4). Identification of virulence determinants that are expressed by various vaginal taxa during colonization and the host response to those factors are key to understanding pathogenesis.
Sneathia vaginalis, formerly known as Sneathia amnii (5), is a Gram-negative streptobacillus. Sneathia species, of which S. vaginalis is the most common, are frequently detected in the vaginal microbiome (6, 7). S. vaginalis is present at an abundance level of ≥0.1% in more than 40% of vaginal microbiomes (8) and tends to be more abundant in high-diversity community state types. However, S. vaginalis is the dominant species in less than 2% of vaginal microbiomes among nonpregnant people, and the relative abundance of this taxon tends to decrease during pregnancy (9). Evidence supports a greater virulence potential for S. vaginalis relative to other normal vaginal species. It is consistently associated with preterm birth (10–14), is frequently detected in amniotic fluid (15–17), and has been identified as the causative agent in cases of neonatal and maternal postpartum bacteremia and sepsis (18–20). Ex vivo studies have shown that S. vaginalis damages and traverses the fetal membrane, and cell culture models reveal that it is cytotoxic (21). The cytotoxic effect is due to the cytopathogenic toxin A (CptA), an ∼230 kDa pore-forming exotoxin that permeabilizes human RBCs and epithelial cells (21). CptA is the only S. vaginalis virulence factor that has been characterized to date. It is the effector component of what appears to be a two-partner secretion system, but it remains associated with the bacterial cell surface rather than being secreted into the extracellular milieu in vitro (21). Ostensibly, it would seem that CptA plays a role in vivo, because S. vaginalis does not appear to have a reservoir outside the human body; however, all prior experimental analyses of the toxin and its expression have been performed in vitro, and in vivo expression has yet to be demonstrated. The goals of this study were to determine whether CptA is expressed in vivo and to investigate the mucosal Ab response against the toxin during vaginal colonization. We found IgA, IgM, and IgG with affinity for CptA in vaginal swab samples, demonstrating that CptA is produced in vivo and that it elicits the production of Abs capable of neutralizing its hemolytic activity.
Materials and Methods
Participant enrollment, informed consent, and sample collection
Participants in this study were enrolled under the Multiomic Microbiome Study Pregnancy Initiative (MOMS-PI) study (10). All participants in this study were enrolled from visitors to women’s clinics in Virginia. All study procedures involving human subjects were reviewed and approved by the Virginia Commonwealth University Institutional Review Board (IRB no. HM15527). The study was performed in compliance with all relevant ethical regulations. Written informed consent was obtained from all participants, and parental permission and assent were obtained for participating minors at least 15 y of age. Participants were informed that they could withdraw from the study at any time and that providing samples was at their sole discretion. Those who were incapable of understanding the informed consent or assent forms or who were incarcerated were excluded from the study. Comprehensive demographic, health history, and dietary assessment surveys were administered, and relevant clinical data (for example, gestational age, height, weight, blood pressure, vaginal pH, diagnosis, and so forth) were recorded. Women with any of the following conditions were excluded from sampling at a given visit: incapable of self-sampling due to mental, emotional, or physical limitations; more than minimal vaginal bleeding as judged by the clinician; ruptured membranes before 37 wk of gestation; and active herpes lesions in the vulvovaginal region.
From the MOMS-PI cohort, 93 samples from 93 participants were chosen for this study solely on the basis of S. vaginalis abundance and gestational age at delivery. Group 1 comprised 22 samples in which S. vaginalis relative abundance was <0.005% from pregnancies that resulted in birth at <37 gestational weeks at delivery. Group 2 comprised 22 samples with S. vaginalis ≥0.005% relative abundance from pregnancies that resulted in birth at <37 gestational weeks at delivery. Group 3 comprised 24 samples in which S. vaginalis relative abundance was <0.005% from pregnancies that resulted in birth at ≥37 gestational weeks at delivery. Group 4 comprised 25 samples with S. vaginalis ≥0.005% relative abundance from pregnancies that resulted in birth at ≥37 gestational weeks at delivery. The abundance level of 0.005% was chosen because it is between the 75th percentile for the term birth and preterm birth cohorts (10).
Sample collection and processing
Samples were collected and processed as previously described (9). Midvaginal samples were collected either by a clinician during a pelvic examination or by self-sampling using a double-tipped CultureSwab EZ swab. Swabs for preservation of Abs and other proteins were immediately immersed in PBS and stored at −80°C. Swabs for DNA isolation were immersed in MoBio PowerSoil Bead Solution and processed using the MoBio PowerSoil DNA Isolation Kit, as described by the manufacturer. DNA samples were stored at −80°C.
16S rRNA taxonomic surveys of the vaginal microbiome
Analysis of 16S rRNA profiles was performed as previously described (9, 10). DNA samples were amplified with barcoded primers targeting the V1–V3 region of the 16S rRNA and validated for vaginal taxa essentially as previously reported (22). The samples were randomized at the PCR stage and again at the sequencing stage. Samples from the MOMS-PI study were multiplexed (384 samples per run) and sequenced using 2 × 300 base (b) PE technology on an Illumina MiSeq sequencer to generate a depth of coverage of at least 50,000 reads per sample. Briefly, the raw paired-end sequence data were demultiplexed into sample-specific paired-end fastq files based on unique barcode sequences using custom python scripts. The merging of overlapping pairs and quality filtering were performed using the MeFiT¯pipeline (23), with a maximum expected error cutoff of 1.0. Each high-quality amplicon sequence was taxonomically classified to the species level using STIRRUPS (22) by alignment to a custom reference database of vaginally relevant species. Samples were filtered on the basis of sequencing depth; samples having fewer than 1000 high-quality amplicons were removed from further analysis. Taxa assigned below threshold by STIRRUPS and less than 0.01% in abundance were also filtered out, and relative proportions were recomputed.
Determination of S. vaginalis absolute abundance
Absolute abundance levels of S. vaginalis in the ELISA subcohort samples were determined by quantitative PCR (qPCR). Standards of S. vaginalis strain SN35 ranging in concentration from 17 to 1,714,000 CFU/ml and Escherichia coli 120,000 CFU/ml were prepared in PBS. The bacteria in the standards were enumerated by plating and counting colonies. Real-time PCR reactions contained 1 μl of each standard, PBS alone, or the E. coli–negative control, 500 nmol SNRTFWD 5′-TGATCCAGCAATTCTGTGTG-3′, and 500 nmol SNRTREV 5′-TAGGCAAGCCTATGGTTGAG-3′ in a final concentration of 1× iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories). The RT-PCR reaction was carried out on a Bio-Rad iQ5. The optimal reaction conditions were determined to be initial denaturation for 30 s at 94°C, followed by 35 cycles of 94°C for 20 s, 55.2°C for 20 s, and 72°C for 20 s. The melting temperature of the target product was 79–80.5°C. Off-target products appeared in the negative controls after 35 cycles and had melting temperatures outside the target range. A standard curve plotting CFU/ml in the 1,714–1,714,000 CFU/ml standards (lower quantities of SN35 did not fit within the linear range) against the 2−Ct from the RT-PCR reaction was generated. To test the vaginal swab samples, 1 μl was tested by RT-PCR, and the absolute abundance was determined from the 2−Ct using the standard curve. Samples with a Ct ≥35 or with significant peaks with melting temperatures outside of the acceptable range (79–80.5°C) were labeled below threshold.
Recombinant CptA protein expression, extraction, and purification
A portion of CptA extending from aa 24 to 1349 was expressed in E. coli and purified as previously described (21). The pET32a plasmid containing the gene fragment was transformed into E. coli BL21-CodonPlus (DE3)-RIL cells (Agilent) for expression. Bacteria were grown in Luria-Bertani broth containing 100 µg ampicillin and 50 µg chloramphenicol per milliliter at 37°C, and expression was induced, when the OD595nm reached 0.5, for 2 h with 1 mM isopropyl β-d-thiogalactoside at 21°C with shaking at 225 rpm. Bacteria were collected and lysed using a French press set at 2000 ψ in 50 mM NaH2PO4, pH 8.0, containing 300 mM NaCl, EDTA-free cOmplete protease inhibitors (Roche), 5 mM EDTA, and 0.1% Triton X-100. Insoluble material was removed from the lysate by centrifugation at 30,000 × g, and the proteins were purified by histidine affinity using cOmplete His-Tag Purification Resin (Sigma-Aldrich) according to the manufacturer’s instructions and eluted with 200 mM imidazole. The fractions were concentrated to ∼4–5 mg/ml using 50,000 Da molecular mass cutoff Amicon Ultra-0.5 centrifugal filters, and 0.5 ml was loaded onto a Superose 6 Increase 10/300 GL within an AKTA Pure FPLC system. The system collected 0.5-ml fractions, and all fractions were tested for hemolytic activity and analyzed by SDS-PAGE. Fractions containing proteins of the correct size were pooled and concentrated and stored at −80°C in 50 mM NaH2PO4, pH 8.0, containing 300 mM NaCl and 5% glycerol.
ELISA
For ELISA, 96-well polystyrene plates were coated overnight at 4°C with 200 μl per well of 0.186 mg CptA24-1349/ml PBS. After 16 h, plates were inverted to remove the protein and washed three times with PBST. After three washes, 200 μl PBS containing 5% skim milk was incubated in each well for 1 h at 37°C. The blocking agent was removed, and 90 μl PBST was added to each well followed by 10 μl of a 1:10 dilution of vaginal swab sample in PBS. PBS alone was used as a negative control, and 10 μl of a 1:1000 dilution of anti-CptA rabbit antiserum was used as a positive control. After 1 h at room temperature (RT), plates were inverted and washed three times. Any remaining liquid in the wells was removed by striking the plate on a stack of paper towels. The secondary Ab was 100 μl of 1:200 anti-human IgA, IgG, or IgM-HRP in PBST per well. After 1 h at RT, plates were inverted and washed six times, striking the plate after the last two washes to ensure removal of secondary Ab. To detect Ab, 50 μl of tetramethylbenzidine substrate solution was added to each well for 15 min, and 50 μl of 0.2 mM sulfuric acid was added to stop the reaction. The OD at 450 nm of each well was determined using an ELISA plate reader.
Hemolysis assay
RBCs from human whole blood (purchased from Biochemed Services) collected in EDTA were collected by centrifugation at 500 × g for 8 min and washed twice in PBS. After the third wash, RBCs were resuspended in three times the original blood volume of PBS. Vaginal swab sample (5 μl) was combined with 5.3 μg CptA24-1349 (23 pmol) and incubated for 15 min at RT. After incubation, a 100-μl washed RBC suspension was added to each tube and incubated for 2 h in a 37°C water bath. The tubes were gently agitated to resuspend the RBCs and dislodge free hemoglobin and centrifuged for 8 min at 500 × g. In a microtiter plate, 90 μl deionized water was added to each well, and 10 μl supernatant from each test reaction was added. Optical densities at 405 nm of the samples were determined by spectrophotometry. Neutralization assays were performed in duplicate, and the average of the two values is reported.
Statistical analysis
Parametric ANOVA was used to compare the means of sample sets with assumed equal variance, and the Kruskal–Wallis test was used to compare means of sample sets with unequal variance. The association between two qualitative variables was tested using Fisher’s exact test. Correlations between ELISA values and percentage neutralization were calculated as Pearson’s coefficient.
Results
The two known CptA protein sequences display high identity
There are currently genome sequences for two S. vaginalis strains available, the reference genome, strain SN35 (accession number NZ_CP011280; https://www.ncbi.nlm.nih.gov/nuccore/808076794) and the whole-genome shotgun sequence for CCUG strain 52976 (DSM 16630) (accession number JASSPO000000000 version JASSPO010000000; https://www.ncbi.nlm.nih.gov/nuccore/JASSPO000000000.1). Comparing the CptA amino acid sequences from these two strains revealed 1538/1565 identities (98%), 1553/1565 positives (99%), and 1/1565 gaps (0%).
Vaginal microbiome profiles of subjects
Because the goal of this study was to determine whether S. vaginalis CptA is produced during vaginal colonization and not to investigate the association between CptA and gestational age at delivery, it was not designed as a case-control study. Instead, vaginal swab samples with low relative abundance of S. vaginalis, ranging from 0% to <0.005%, or high relative abundance, ranging from ≥0.005% to >25%, from pregnancies that resulted in preterm or term births were chosen from the MOMS-PI study (9, 10). Species-level taxonomic analysis of the samples and the association between taxa and between taxa and preterm birth was performed in a previous study (10), so this analysis was not repeated in this study, but bar plots showing relative abundance levels of bacteria in the participants at different time points during their pregnancies are shown in Supplemental Fig. 1, and the S. vaginalis abundance levels at each visit are shown in Supplemental Table I. Longitudinal analysis demonstrated, as expected, that relative abundance levels of all species detected, including S. vaginalis, varied over time.
Table I lists the four groups and the average, median, and range of the gestational age at delivery and relative abundance of S. vaginalis for each group. Group 1 consisted of 22 samples from pregnancies that resulted in preterm birth in which S. vaginalis relative abundance of the sample chosen was <0.005%. Group 2 consisted of 22 samples from pregnancies resulting in preterm births with S. vaginalis ≥0.005%. Group 3 consisted of 24 samples from pregnancies resulting in term births in which S. vaginalis was <0.005%, and group 4 consisted of 25 samples with ≥0.005% S. vaginalis from term births. Because the bacterial burden in the vagina can vary greatly, absolute abundance can provide additional information about the exposure of the host to a particular species. Absolute abundances in the samples selected for this study were determined by qPCR. Supplemental Table I lists the gestational age at delivery for all samples, S. vaginalis relative abundance at all time points tested in this study, and absolute abundance at the time point tested in this study. All of the participants included in the study were self-reported as Black.
Table I. Gestational age at delivery and S. vaginalis relative abundance per group.
| GAD Range | GAD Average | GAD Median | RA Range | RA Average | RA Median | |
|---|---|---|---|---|---|---|
| Group 1 | 23 wk 2 d–36 wk 6 d | 34 wk 1d | 35 wk 3 d | 0–0.0038 | 0.0007 | 0 |
| Group 2 | 24 wk 5 d–36 wk 4 d | 34 wk 2 d | 35 wk 3 d | 0.0071–25.5093 | 4.96 | 0.8095 |
| Group 3 | 37 wk 1 d–41 wk 1 d | 39 wk 2 d | 39 wk 2 d | 0–0.0038 | 0.0006 | 0 |
| Group 4 | 37 wk 0 d–41 wk 1 d | 39 wk 2 d | 39 wk 6 d | 0.0082–19.1546 | 3.152 | 1.6867 |
The range, average, and medians of the gestational age at delivery (GAD) and percentage relative abundance (RA) for each group are shown.
CptA-specific Abs can be detected in vaginal swab specimens
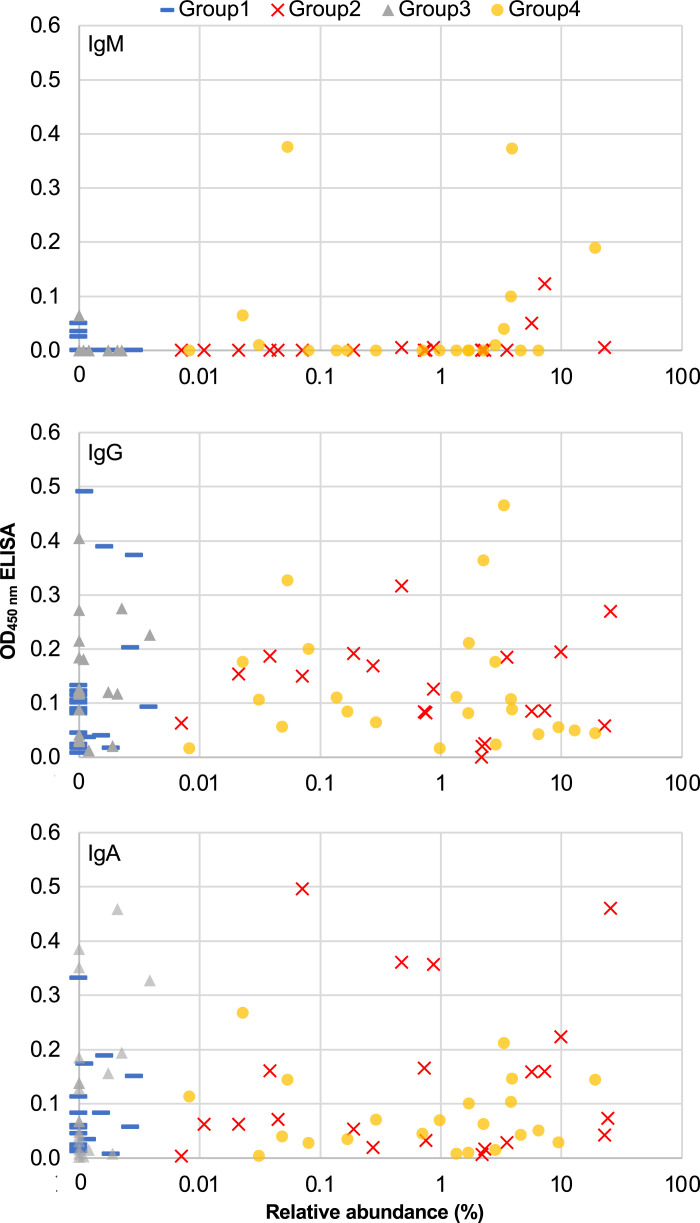
A previously described antigenic portion of the CptA protein (24) was produced and purified and used to coat microtiter plates, and vaginal swab samples were analyzed for the toxin by ELISA. The averages and ranges of all OD450nm values for each of the four groups are shown in Table II. The OD450nm values from for each of the individual samples tested (Supplemental Table II) were plotted against the relative abundance of S. vaginalis in the samples tested, and the resulting scatterplots are shown in Fig. 1. The majority of samples with detectable anti-CptA IgM were from groups 2 and 4, the two groups with S. vaginalis relative abundance ≥0.005%. However, significant correlations between ELISA values and relative abundance were not indicated by Pearson’s coefficients for any of the isotypes [IgM r(92) = 0.09, p = 0.389; IgA r(92) = 0.13, p = 0.227; IgG r(92) = -0.07, p = 0.503].
Table II. Averages and ranges of ELISA and hemolysis values for groups 1–4.
| IgM Average | IgM Range | IgG Average | IgG Range | IgA Average | IgA Range | Hemolysis Average | Range Hemolysis | |
|---|---|---|---|---|---|---|---|---|
| Group 1 | 0.00 | −0.02, 0.05 | 0.12 | 0.01, 0.39 | 0.07 | −0.01, 0.33 | 47.04% | 0, 134.21% |
| Group 2 | 0.01 | −0.03, 0.05 | 0.11 | −0.05, 0.32 | 0.14 | 0, 0.50 | 43.37% | −2.57, 111.76% |
| Group 3 | 0.00 | −0.02, 0.07 | 0.13 | 0.01, 0.4 | 0.11 | −0.01, 0.46 | 50.01% | 0, 126.51% |
| Group 4 | 0.04 | −0.02, 0.38 | 0.12 | −0.01, 0.47 | 0.07 | −0.01, 0.27 | 48.35% | −1.14, 115.44% |
The average and range for all ELISA values obtained for each of the four groups and the averages and ranges of the percentage hemolysis for each group are listed.
FIGURE 1.
Relative abundance of S. vaginalis versus ELISA values. Scatterplots of relative abundance determined by 16S rRNA survey for each of the swab samples (x-axis in log10 scale) relative to the average of two OD450nm values obtained by isotype-specific ELISA (y-axis). Distinct markers were used for group 1 (preterm birth [PTB], low S. vaginalis), group 2 (PTB, high S. vaginalis), group 3 (term birth [TB], low S. vaginalis), and group 4 (TB, high S. vaginalis).
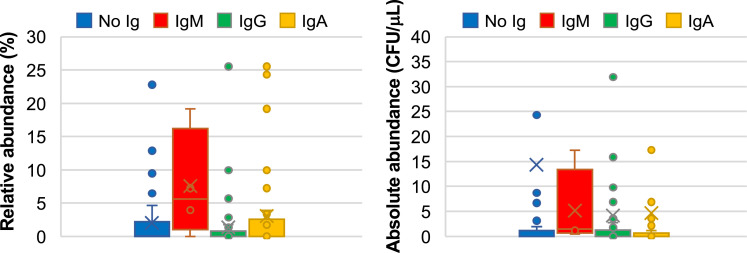
We also used a binomial scoring system to categorize the samples as positive or negative by ELISA. Positive samples had an OD450nm reading ≥0.05 (after subtracting the values of the negative control wells) in two separate ELISAs. Using this scoring system, only 4 of the 93 samples were positive for IgM. IgG was positive in 49 samples, and IgA was positive in 33 samples. At least one of the three isotypes was positive in 59 samples, and no anti-CptA Abs were positive in 35 samples. Among 46 samples with <0.005% S. vaginalis, anti-CptA Ab was positive in 31 (15 from group 1 and 16 from group 3), and among 47 samples with ≥0.005% S. vaginalis, 28 (14 from group 2 and 14 from group 4) were positive. Fisher’s exact test revealed no significant association between low (<0.005%) versus high (≥0.005%) relative abundance (p = 0.3001) or between low (not detectable) versus high (detectable) absolute abundance (p = 0.6544) of S. vaginalis and positive/negative score for anti-CptA IgG. Fisher’s exact test revealed no significant association between low (<0.005%) versus high (≥0.005%) relative abundance (p = 0.5179) or low (not detectable) versus high (detectable) absolute abundance (p = 1) and positive/negative score for anti-CptA IgA. The absolute and relative abundances of S. vaginalis in the samples tested are grouped by Ig detected and plotted in box-and-whisker plots in Fig. 2. A Shapiro–Wilk test showed that the data were not normally distributed. The Kruskal–Wallis test determined that there was no significant association between Ig absence versus presence of IgG, IgA, or presence of any Ig and the relative or absolute abundance of S. vaginalis (p = 0.94432). The IgM group was not large enough to include in the Kruskal–Wallis test. All four IgM-positive samples had relative abundance values ≥0.005% S. vaginalis, but a Fisher’s exact test conducted with a contingency table of categories IgM present and IgM absent and groups ≥0.005% and <0.005% S. vaginalis determined that IgM presence and S. vaginalis abundance are statistically independent (p = 0.0559). All four IgM-positive samples had absolute abundance values >0 and a Fisher’s exact test of IgM presence/absence in samples with absolute abundance of 0 or >0 determined that S. vaginalis abundance and IgM are statistically associated (p = 0.014).
FIGURE 2.
Positive anti-CptA ELISA is not a good predictor of current S. vaginalis presence. ELISA data were used to categorize samples as anti-CptA positive (two OD450nm readings ≥0.05) or negative (one or zero OD450nm readings ≥0.05). The vaginal swab samples were grouped as positive or negative for IgM, IgG, or IgA. Samples that contained anti-CptA Abs of more than one isotype are represented multiple times in the plots. Box-and-whisker plots show range, outliers, means (X), and the median (line). The y-axis indicates relative abundance determined by 16S rRNA survey or absolute abundance determined by qPCR.
Vaginal swab samples containing anti-CptA Ab neutralize hemolytic activity
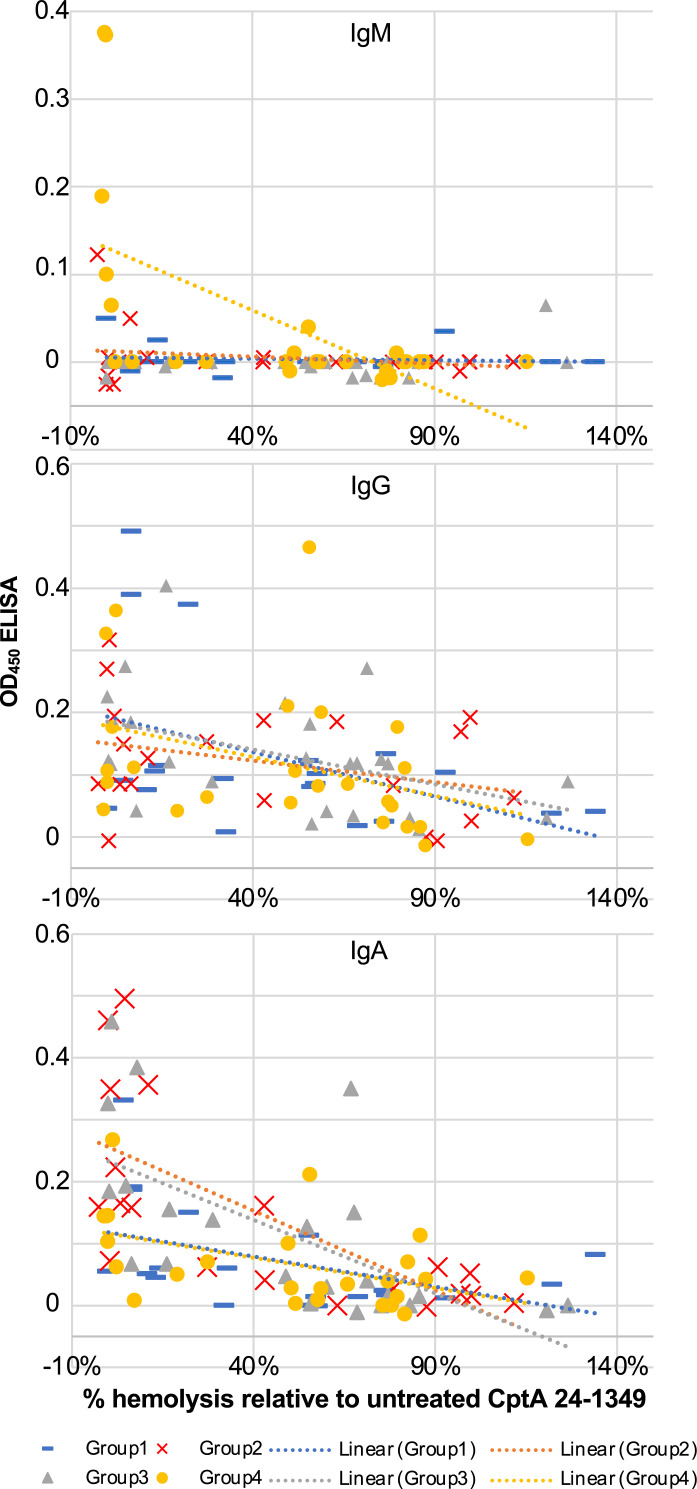
To assess the ability of host Ab to neutralize the pore-forming activity of CptA, we tested the samples using a hemolysis assay (four samples were not tested because of limited sample availability). Purified recombinant CptA24-1349, a fragment of CptA previously shown to have hemolytic activity (24), was pretreated with the samples, and the percentage of hemolytic activity relative to untreated toxin was determined. The range and average of the percentage of hemolysis for each group are shown in Table II, and the values for each sample are listed in Supplemental Table II. Scatterplots suggested a possible linear relationship between ELISA values and hemolysis-neutralizing activity (Fig. 3), and Pearson’s coefficients support negative correlations between anti-CptA ELISA values and remaining hemolytic activity following pretreatment [IgM r(86) = −0.27, p = 0.0119; IgG r(86) = −0.38, p = 0.0002; IgA r(86) = −0.57, p < 0.0001].
FIGURE 3.
Anti-CptA ELISA values correlate with toxin-neutralizing activity. Recombinant purified CptA24-1349 was pretreated with PBS alone (positive hemolysis control) or a vaginal swab sample and then added to washed RBCs. After 2 h at 37°C, released hemoglobin was measured by OD at 450 nm. The negative control (no CptA24-1349) value was subtracted from all test values, and the percentage of hemolysis relative to the positive control was calculated by setting untreated CptA24-1349 as 100%. All hemolysis assays were performed in duplicate, and averages of the two values are shown. Scatterplots showing percentage hemolysis (x-axis) relative to isotype-specific anti-CptA ELISA data (y-axis). Distinct markers were used for group 1 (preterm birth [PTB], low S. vaginalis), group 2 (PTB, high S. vaginalis), group 3 (term birth [TB], low S. vaginalis), and group 4 (TB, high S. vaginalis).
Discussion
The S. vaginalis CptA toxin has been shown in vitro to lyse epithelial cells and to contribute to traversal through chorion, suggesting that it may play a role in pathogenesis. However, it has never been proved that CptA is produced in vivo or that it comes into contact with the host. To investigate this, we examined midvaginal swab samples for the presence of anti-CptA Abs. We compared the CptA protein from the two sequenced strains of S. vaginalis, which were from geographically distinct regions, to investigate the utility of an ELISA to detect Abs against CptA from different strains. The cptA gene in strain SN35 encodes a 2213-aa protein, but the cptA gene sequence from DSM 16630 is 4695 nucleotides, encoding a 1565-aa protein. CptA in DSM 16630 is shorter because the gene encodes fewer carboxy-terminal repeats (21). This is likely an artifact of the assembly because the repeats were compressed in the original version of SN35 (version NZ_CP011280.1). The version described in this study (version NZ_CP011280.2) was produced by Oxford Nanopore sequencing, and the long reads generated using this technique can easily sequence the entire repeat region. Nonetheless, the portion of the toxin that was used as bait in the ELISA was within a region of high identity, as determined using BLASTp, not within the repeat region.
Through ELISA, the presence of IgG, IgA, and IgM in vaginal swab samples was assessed. We examined IgM because it is a marker of recent infection, and we hypothesized that participants might have recently been colonized or infected with S. vaginalis. We examined IgA because the vaginal epithelium is a mucosal surface, and secretory IgA is abundant within mucosae. Additionally, IgA is often efficient at neutralizing toxins, and high levels of IgA against the pore-forming toxin of Gardnerella vaginalis have been shown to tend to be negatively associated with adverse pregnancy outcome (25). We examined IgG because it has been reported to be the most abundant Ab isotype in vaginal secretions (26). We identified samples containing anti-CptA IgM, IgA, and IgG. IgG was the most commonly identified isotype because it occurred in 49 of 93 samples. IgA was the next most common isotype, with 33 of 93 positive samples, and IgM was the least common isotype with only 4 of 93 samples testing positive.
We hypothesized that there would be an association, negative or positive, between anti-CptA Abs and S. vaginalis abundance. Presence and titer of Abs could be positively associated with S. vaginalis abundance because Ab production could be ramped up in response to bacterial proliferation. Alternatively, the Ab response could be negatively associated with S. vaginalis abundance if Abs prevent growth or contribute to eradication of the bacteria. However, we found no significant association or even a trend between the presence of IgA or IgG anti-CptA and S. vaginalis abundance. It is possible that both phenomena happen, and the opposing effects cancel each other out. It is also possible that the anti-CptA Abs have little to no impact on the growth of S. vaginalis or that other immune components play a more important role in controlling its growth. It is also possible that other vaginal bacteria, such as lactobacilli, have a greater impact on the abundance of S. vaginalis than the immune response. Only 4 of 93 samples were positive for IgM, and all 4 had relative abundance values >0.005% and detectable absolute abundance levels of S. vaginalis. Fisher’s exact test determined that IgM was independent of S. vaginalis relative abundance but associated with absolute abundance. The variable results are likely due to the small number of IgM-positive samples in the cohort. Biologically, an association between the levels of S. vaginalis may have been found because these participants were only recently colonized by S. vaginalis, inducing an IgM response, and the affinity of IgM Abs was not high enough or had not been present long enough to eradicate the bacteria. IgM-positive vaginal swab samples did neutralize CptA effectively, but all IgM-positive samples contained either IgG or IgA as well, so it cannot be determined whether the IgM or IgA/IgG was responsible for the neutralizing activity. Furthermore, IgG and IgA against CptA or other bacterial Ags may be superior at opsonizing the bacteria and may play a larger role in eradication, and IgM may not function as well in this regard.
Some samples tested positive for anti-CptA Abs by ELISA but did not have detectable S. vaginalis by 16S survey or qPCR. S. vaginalis was detected in at least one time point in 37 of the 46 low abundance samples (90% or 84 of the 93 total participants in the study), and the study represents less than 1 y of each participant’s life, suggesting that most people are colonized with S. vaginalis at some point. Thus, prior colonization, outside of the time points analyzed through the MOMS-PI study, may explain why some samples from participants with no detectable S. vaginalis had Ab that recognized CptA. It is also possible that Abs present in some of the samples cross-react with CptA, even though the host was naïve to CptA. The samples used for this study were from the MOMS-PI project and were limited in quantity, but a larger, dedicated study, looking at Ab production and S. vaginalis abundance longitudinally could provide more insight into the impact of Ab production on the growth of S. vaginalis during colonization. Additionally, because S. vaginalis abundance decreases during pregnancy (9), a study of anti-CptA Ab levels in nonpregnant participants could yield different results.
We hypothesized that anti-CptA Abs would be capable of neutralizing the toxin, and the results confirmed that anti-CptA Abs did effectively neutralize the hemolytic activity of the toxin. The majority of samples without anti-CptA Abs failed to neutralize the toxin, supporting the hypothesis that the Abs rather than other components of vaginal fluid were responsible. Presence of IgM, IgA, and IgG all correlated with the capacity to neutralize the hemolytic activity of the toxin, suggesting that Abs against CptA present in vaginal secretions could play a role in protecting the host. Serologic Ab responses against other vaginal pathogens, such as Ureaplasma, have been analyzed, and some studies suggest that serum Abs are associated with preterm birth (27). We did not look for serum Abs in this study, but it would be of interest because a systemic response could be a marker of invasion in contrast to the local mucosal response that was analyzed in this study.
Although we hypothesized that CptA neutralizing Abs could play a role in preventing preterm birth, there was not a significant association. We did not look for an association between S. vaginalis abundance and preterm birth, because we chose samples at the high and low extremes of S. vaginalis abundance, which would bias this comparison. However, the larger preterm birth study found a significant association between S. vaginalis and preterm birth (10).
In conclusion, this study confirms that CptA is produced in vivo by S. vaginalis; that CptA provokes a host immune response; and that host Ab, particularly IgA, can neutralize CptA. S. vaginalis is associated with various vaginal and reproductive diseases and complications, so further investigation and study of its pathogenesis is important. Weaknesses of the study are the lack of ELISA data from longitudinal samples, the relatively small sample size, and limitations of sample quantity, which limited the number of replicates for experimental analyses. A larger study that includes participants of different races, a study of nonpregnant participants, and analysis of Abs against other S. vaginalis Ags would contribute to our further understanding of the interaction of this vaginal species with the host immune defenses. Strengths of this study include the determination of both relative and absolute abundance values, the 16S rRNA survey data from longitudinal samples, and the toxin neutralization study.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
The Multi Omic Microbiome Study Pregnancy Initiative was funded by a grant from the National Institutes of Health (8U54HD080784). This subcohort study was funded by National Institutes of Health Grant R21 AI166168 and a “breakthrough award” funded by Virginia Commonwealth University.
The data presented in this article were generated by the Virginia Vaginal Microbiome Consortium (vmc.vcu.edu) in the Multi Omic Microbiome Study Pregnancy Initiative (MOMS-PI). Open-access data, including raw 16S rRNA sequences, cytokine data, and limited metadata from the MOMS-PI study, are available at the Human Microbiome Project Data Coordination Center (https://portal.hmpdacc.org).
- CptA
- cytopathogenic toxin A
- MOMS-PI
- Multiomic Microbiome Study Pregnancy Initiative
- qPCR
- quantitative PCR
- RT
- room temperature
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Romero, R., Dey S. K., Fisher S. J.. 2014. Preterm labor: one syndrome, many causes. Science 345: 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldenberg, R. L., Culhane J. F., Iams J. D., Romero R.. 2008. Epidemiology and causes of preterm birth. Lancet 371: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastek, J. A., Gómez L. M., Elovitz M. A.. 2011. The role of inflammation and infection in preterm birth. Clin. Perinatol. 38: 385–406. [DOI] [PubMed] [Google Scholar]
- 4.Nadeau, H. C. G., Subramaniam A., Andrews W. W.. 2016. Infection and preterm birth. Semin. Fetal Neonatal Med. 21: 100–105. [DOI] [PubMed] [Google Scholar]
- 5.Eisenberg, T., Gronow S., Falgenhauer J., Imirzalioglu C., Mühldorfer K., Rau J., Blom J., Fawzy A., Glaeser S. P., Kämpfer P.. 2019. Sneathia vaginalis sp. nov. (Fusobacteriales, Leptotrichiaceae) as a replacement of the species “Sneathia amnii” Harwich et al. 2012 and “Leptotrichia amnionii” Shukla et al. 2002, and emended description of Sneathia Collins et al. 2001. Int. J. Syst. Evol. Microbiol. 71: 004663. [DOI] [PubMed] [Google Scholar]
- 6.Ravel, J., Gajer P., Abdo Z., Schneider G. M., Koenig S. S. K., McCulle S. L., Karlebach S., Gorle R., Russell J., Tacket C. O., et al. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 108(Suppl 1): 4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma, B., Forney L. J., Ravel J.. 2012. Vaginal microbiome: rethinking health and disease. Annu. Rev. Microbiol. 66: 371–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harwich, M. D., Jr., Serrano M. G., Fettweis J. M., Alves J. M. P., Reimers M. A., Buck G. A., Jefferson K. K.; Vaginal Microbiome Consortium (additional members) . 2012. Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics 13(Suppl 8): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano, M. G., Parikh H. I., Brooks J. P., Edwards D. J., Arodz T. J., Edupuganti L., Huang B., Girerd P. H., Bokhari Y. A., Bradley S. P., et al. 2019. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 25: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fettweis, J. M., Serrano M. G., Brooks J. P., Edwards D. J., Girerd P. H., Parikh H. I., Huang B., Arodz T. J., Edupuganti L., Glascock A. L., et al. 2019. The vaginal microbiome and preterm birth. Nat. Med. 25: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson, D. B., Hanlon A., Nachamkin I., Haggerty C., Mastrogiannis D. S., Liu C., Fredricks D. N.. 2014. Early pregnancy changes in bacterial vaginosis-associated bacteria and preterm delivery. Paediatr. Perinat. Epidemiol. 28: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theis, K. R., Florova V., Romero R., Borisov A. B., Winters A. D., Galaz J., Gomez-Lopez N.. 2021. Sneathia: an emerging pathogen in female reproductive disease and adverse perinatal outcomes. Crit. Rev. Microbiol. 47: 517–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hočevar, K., Maver A., Vidmar Šimic M., Hodžić A., Haslberger A., Premru Seršen T., Peterlin B.. 2019. Vaginal microbiome signature is associated with spontaneous preterm delivery. Front. Med. (Lausanne) 6: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florova, V., Romero R., Tarca A. L., Galaz J., Motomura K., Ahmad M. M., Hsu C.-D., Hsu R., Tong A., Ravel J., et al. 2021. Vaginal host immune-microbiome interactions in a cohort of primarily African-American women who ultimately underwent spontaneous preterm birth or delivered at term. Cytokine 137: 155316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiGiulio, D. B. 2012. Diversity of microbes in amniotic fluid. Semin. Fetal Neonatal Med. 17: 2–11. [DOI] [PubMed] [Google Scholar]
- 16.DiGiulio, D. B., Romero R., Amogan H. P., Kusanovic J. P., Bik E. M., Gotsch F., Kim C. J., Erez O., Edwin S., Relman D. A.. 2008. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One 3: e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero, R., Miranda J., Chaemsaithong P., Chaiworapongsa T., Kusanovic J. P., Dong Z., Ahmed A. I., Shaman M., Lannaman K., Yoon B. H., et al. 2015. Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J. Matern. Fetal Neonatal Med. 28: 1394–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Martino, S. J., Mahoudeau I., Brettes J. P., Piemont Y., Monteil H., Jaulhac B.. 2004. Peripartum bacteremias due to Leptotrichia amnionii and Sneathia sanguinegens, rare causes of fever during and after delivery. J. Clin. Microbiol. 42: 5940–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruwier, L., Sprenkels A., Hulsbosch S., Vankeerberghen A., Cartuyvels R.. 2021. Sneathia amnii bacteraemia and chorioamnionitis leading to second trimester abortion: a case report. Access Microbiol. 3: 000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen, A., Ferrero L., Lazarevic V., Gaia N., Martinez de Tejada B., Schrenzel J., Berkane N.. 2023. Postpartum septic arthritis of pubic symphysis due to Sneathia sanguinegens, Sneathia vaginalis, and Mageeibacillus indolicus: contribution of clinical metagenomics. New Microbes New Infect. 53: 101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentile, G. L., Rupert A. S., Carrasco L. I., Garcia E. M., Kumar N. G., Walsh S. W., Jefferson K. K.. 2020. Identification of a cytopathogenic toxin from Sneathia amnii. J. Bacteriol. 202: e00162-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fettweis, J. M., Serrano M. G., Sheth N. U., Mayer C. M., Glascock A. L., Brooks J. P., Jefferson K. K., Buck G. A.; Vaginal Microbiome Consortium (additional members) . 2012. Species-level classification of the vaginal microbiome. BMC Genomics 13(Suppl 8): S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parikh, H. I., Koparde V. N., Bradley S. P., Buck G. A., Sheth N. U.. 2016. MeFiT: merging and filtering tool for Illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinformatics 17: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Brien, C. K., Raskin J. R., Amankwa Asare I., Wei C., Ma J., McCoy Z. T., Jefferson K. K.. 2023. Identification of the pore-forming and binding domains of the Sneathia vaginalis cytopathogenic toxin A. PLoS One 18: e0284349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cauci, S., Thorsen P., Schendel D. E., Bremmelgaard A., Quadrifoglio F., Guaschino S.. 2003. Determination of immunoglobulin A against Gardnerella vaginalis hemolysin, sialidase, and prolidase activities in vaginal fluid: implications for adverse pregnancy outcomes. J. Clin. Microbiol. 41: 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Usala, S. J., Usala F. O., Haciski R., Holt J. A., Schumacher G. F.. 1989. IgG and IgA content of vaginal fluid during the menstrual cycle. J. Reprod. Med. 34: 292–294. [PubMed] [Google Scholar]
- 27.Ireland, D. J., Keelan J. A.. 2014. The maternal serological response to intrauterine Ureaplasma sp. infection and prediction of risk of pre-term birth. Front. Immunol. 5: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.