Abstract
Amyotrophic lateral sclerosis (ALS) is a complex, fatal neurodegenerative disease. Disease pathophysiology is incompletely understood but evidence suggests gut dysbiosis occurs in ALS, linked to impaired gastrointestinal integrity, immune system dysregulation and altered metabolism. Gut microbiome and plasma metabolome have been separately investigated in ALS, but little is known about gut microbe-plasma metabolite correlations, which could identify robust disease biomarkers and potentially shed mechanistic insight. Here, gut microbiome changes were longitudinally profiled in ALS and correlated to plasma metabolome. Gut microbial structure at the phylum level differed in ALS versus control participants, with differential abundance of several distinct genera. Unsupervised clustering of microbe and metabolite levels identified modules, which differed significantly in ALS versus control participants. Network analysis found several prominent amplicon sequence variants strongly linked to a group of metabolites, primarily lipids. Similarly, identifying the features that contributed most to case versus control separation pinpointed several bacteria correlated to metabolites, predominantly lipids. Mendelian randomization indicated possible causality from specific lipids related to fatty acid and acylcarnitine metabolism. Overall, the results suggest ALS cases and controls differ in their gut microbiome, which correlates with plasma metabolites, particularly lipids, through specific genera. These findings have the potential to identify robust disease biomarkers and shed mechanistic insight into ALS.
Keywords: 16S rRNA sequencing, metabolomics, motor neuron disease, Mendelian randomization, multi-omics
Guo et al. assess gut microbial community structure in ALS and identify differences versus controls in several genera, which correlate with levels of plasma metabolites, especially lipids. Mendelian randomization indicates possible causality from lipids related to fatty acid and acylcarnitine metabolism.
Introduction
Amyotrophic lateral sclerosis (ALS) is a complex, clinically heterogeneous, fatal neurodegenerative disease.1 Patients generally only survive 2–4 years post-diagnosis and treatment primarily focuses on symptom management and palliative care.2 Most ALS cases, around 85%, are sporadic and lack a known genetic aetiology.3 Recent research has focused on the environmental contributions to ALS,4 including the gut microbiome,5,6 which correlates with disease progression in human7 and animal studies.8-10
The human gastrointestinal tract is inhabited by millions of microorganisms, which communicate bidirectionally with the host, including through the gut–brain axis.11 The microbiome and bacterially-derived metabolites exert important regulatory activities in the host, such as immune modulation,5,8 energy and metabolic homeostasis5,9 and intestinal barrier integrity.12 In turn, host lifestyle, dietary habits and exposures e.g. to antibiotics,13 shape the microbiome. During health, the microbiome exists in exquisite balance with the host, but dysbiosis, an imbalance in microbial communities, occurs during disease, including ALS.5 The microbiome intersects with several aspects of ALS pathophysiology. Neuroinflammation and immune system dysregulation are ALS hallmarks,14 which correlate with specific microbiome signatures in rodent studies.8,10 ALS patients also have distinct plasma metabolomic profiles,15,16 which may, in part, be influenced by specific microbes in the gut.9 Moreover, structural, functional and biochemical gastrointestinal abnormalities occur in ALS patients,17 correlating with shifts in the intestinal microbiome.12
Little is known about the associations between the gut microbiome to host plasma metabolites in ALS patients at disease onset and with progression. However, gut microbiome signatures in ALS may suggest potential biomarkers and treatment avenues. We previously performed untargeted metabolomics on cohorts of ALS and control participants, identifying dysregulated circulating metabolites linked to disease status,15 especially lipids.16 Herein, we longitudinally profiled the gut microbiome of ALS versus control participants and integrated the initial microbiome profile with circulating plasma metabolites, in the first study, to our knowledge, to examine microbiome-plasma metabolome correlations in ALS. We found altered gut microbial structure at the phylum level in ALS, along with differential abundance in several distinct genera. Our integrated microbiome-plasma metabolome analysis identified several prominent amplicon sequence variants (ASVs) strongly linked to several metabolites, especially lipids. Finally, we performed Mendelian randomization to examine potential causality from metabolites, identifying lipids, especially species related to fatty acid and acylcarnitine metabolism, as possible candidates. Overall, our findings point to a gut microbiome signature in ALS linked to distinct host plasma lipid profiles, which may provide potential biomarkers and future treatment avenues.
Materials and methods
Study participants and sample processing
Study recruitment is already published.15,16 Briefly, this was a case-control study from samples collected between April 2016 to July 2020. ALS patients 18 years or older and able to communicate in English, seen at the Pranger ALS Clinic at Michigan Medicine, were invited to participate. ALS patients were diagnosed based on original and/or revised El Escorial criteria,18 EMG and clinical and family history. All ALS patients meeting the inclusion criteria of definitive, probable, probable with laboratory support, possible or suspected ALS were eligible to participate in the study. All participants met current Gold Coast Criteria. Control participants 18 years or older without a neurodegenerative condition or family history of ALS were recruited through a university-managed recruitment website at the University of Michigan Institute for Clinical and Health Research. Controls were compensated for participating in the study. Demographics, including sex, age, height, weight and body mass index (BMI), were collected from all participants. ALS disease characteristics, including onset segment and phenotype (with frontotemporal dementia), days from symptom onset to diagnosis, days from diagnosis to stool sample, diagnosis by El Escorial criteria, disease severity by ALS Functional Rating Scale-Revised (ALSFRS-R) and percutaneous endoscopic gastrostomy (PEG) status, were additionally collected from ALS patients. All participants provided informed consent for this research, which was approved by the University of Michigan Medical School Institutional Review Board (HUM00028826).
Microbiome profiling and analysis
Approximately 200 µl faecal samples were seeded into PowerMag Glass Bead plates (MO BIO Laboratories) and bacterial DNA was isolated using a MagAttract PowerMicrobiome DNA/RNA Kit (Qiagen) and an epMotion 5075 liquid handling system. The V4 region of the bacterial 16S rRNA gene was amplified on an Illumina MiSeq at the University of Michigan Microbiome Core, as previously reported.19 Raw FASTQ files were read into a microbial package (https://github.com/guokai8/microbial), which integrated multiple functions from DADA2,20 phyloseq21 and DESeq222 packages. The reads were processed by the ‘processSeq’ function, which de-replicated, de-noised, removed chimeras, generated tables of ASVs and assigned taxonomy based on the SILVA (v138.1) database.23
ASVs of unknown species at the phylum level and present in fewer than three samples were filtered out from subsequent analyses. Richness within samples was measured by alpha diversity and relative abundance was then calculated for principal coordinate analysis (PCoA) based on the Bray-Curtis dissimilarity distance score. Differential abundance analysis was performed with P < 0.05 and |log2(fold-change)|- > 1 to identify significant ASVs. The Tax4Fun2 R package24 was used to predict the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional profiling for each sample using the ‘Ref99NR’ database. Adjusted P-values of < 0.05 were considered significant after multiple testing of the resultant KEGG pathways with the Benjamini-Hochberg (BH) method. Butyrate-producing bacteria in our dataset were identified as previously,25 using a curated literature-derived taxonomy file of commensal butyrate-producing bacterial species.26-29
Logistical regression models
General logistic regression analysis regressed bacterial abundance against the ALS versus control comparison, adjusted for age, sex and BMI. Benjamini-Hochberg correction accounted for multiple testing.
Plasma metabolomics
This study used a subset of a previously published metabolomics dataset.15,16 Briefly, whole blood samples from consented participants were drawn without fasting as it was deemed unethical to request fasting from ALS patients. Blood was collected into EDTA tubes and plasma was obtained. Plasma metabolites were profiled by ultra-high-performance liquid chromatography-tandem mass spectroscopy (Metabolon). A subset of ALS and control participants from the metabolomics cohort, which overlap with participants in the microbiome cohort, was used to determine plasma metabolite-gut microbiome associations using weighted gene co-expression network analysis (WGCNA) and two-way orthogonal partial least square with discriminant analysis (O2PLS-LDA).
Weighted gene co-expression network analysis
Biological interactions between gut bacteria and circulating plasma metabolites in ALS were determined with WGCNA v1.7130,31 to build unsigned co-expression networks. Microbiome (relative abundance of ASVs) and metabolites were profiled for ALS (n = 53) and control (n = 80) participants at the first collection point (T1). To use a homogenous approach for handling both datasets, the microbiome and metabolomics data were combined into a single array. The merged array was filtered to include members, i.e. ASVs and metabolites, which were present in at least 10 participants. The co-expression network was built using the standardized (scaled) microbiome and metabolome data. The WGCNA framework was applied to the merged dataset to identify members, i.e. ASVs and metabolites, with similar profiles suggesting they are strongly correlated. Sample clustering was conducted to detect outliers. A soft power of 3 was chosen by the WGCNA ‘pickSoftThreshold’ function to construct the co-expression network, which was built by computing the Pearson correlation coefficient between any pair of nodes, i.e. ASVs and metabolites. The selected soft threshold guarantees a scale-free network topology of unsigned R-squared > 0.9.
The WGCNA module that most significantly correlated with ALSFRS-R was further analysed for associations to change in ALSFRS-R with time. First, ALSFRS-R scores closest to the time of faecal (microbiome) and blood (metabolome) collection were identified. Second, all subsequent ALSFRS-R scores from the participant database were collected. Next, the rate of change between the anchoring ALSFRS-R score (i.e. closest to faecal and blood collection) and all subsequent ALSFRS-R evaluations was calculated. Finally, Spearman correlation analysis was applied to explore potential associations between this rate of change in ALSFRS-R and the abundance of significant ASVs and metabolites in the WGCNA module.
Two-way orthogonal partial least square with discriminant analysis
O2PLS-DA was performed using the R package o2plsda (https://cran.rstudio.com/web/packages/o2plsda/index.html). O2PLS-DA is a bidirectional multivariate regression and derivative method of partial least squares derivative analyses (PLS-DA). O2PLS-DA separates the covariance between different datasets from the systematic sources of variance specific to each dataset separately. The abundances of microbiome and metabolomics data were scaled and group-balanced Monte Carlo cross-validation was used to determine the optimal number of latent variables for each model structure to avoid overfitting the model. The discriminant analysis was performed based on the systematic predictive variation from the two datasets.
Mendelian randomization
Mendelian randomization32 was performed using the TwoSampleMR v 0.5.7 R package (https://mrcieu.github.io/TwoSampleMR/index.html).33 TwoSampleMR performs Mendelian randomization using genome-wide association studies (GWAS) summary data, which are obtained automatically from the MRC Integrative Epidemiology Unit (IEU) open GWAS database (https://gwas.mrcieu.ac.uk/). Two sample Mendelian randomization (2SMR) estimates the causal effect of an exposure on an outcome using only GWAS summary statistics. For exposure, the metabolite dataset embedded in the TwoSampleMR v 0.5.7 R package was employed, which contains a GWAS of 452 metabolites in whole human blood.34 This database contains many specific metabolites that were linked to ALS in this study. Five ALS GWAS studies were obtained from IEU GWAS database (Supplementary Table 1).
To perform 2SMR of a specific metabolite against ALS, single-nucleotide polymorphisms (SNPs) were identified that influence that specific metabolite (i.e. extracted the instrumental variables) and then extracted those SNPs from ALS GWAS datasets. Extracted SNPs were harmonized to ensure the effect of an SNP on the exposure and the effect of the same SNP on the outcome correspond to the same allele. Next, instrumental variables for each exposure-metabolite were obtained, with per-allele beta coefficient estimates (β) and standard errors. Finally, significant effects in the 2SMR analysis were estimated by inverse variance weighted, MR Egger, weighted median, weighted mode and simple mode.
Statistical methods for demographics
Participant demographics were represented as the mean ± standard deviation (SD) for continuous variables, assessed by two-tailed Student’s t-test, and n (%) for categorical variables, assessed by Fisher’s exact test.
Results
Microbiome and overlapping microbiome-metabolomics participants
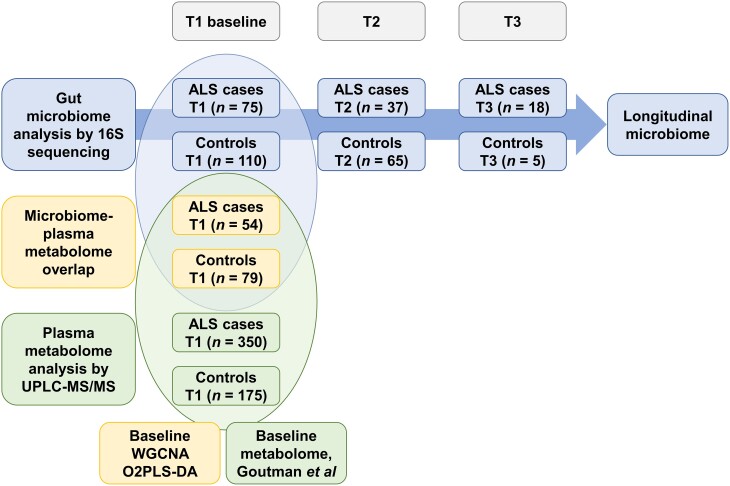
The microbiome cohort consisted of 43 male (57.3%) and 32 female (42.7%) participants in the ALS group (n = 75 total), which differed significantly by per cent from the 44 male (44.0%) and 66 female (60.0%) control participants (n = 110 total; P = 0.025) (Table 1 and Fig. 1). Moreover, ALS participants were significantly older on average than controls (P = 0.010), although they had similar age ranges. Of the ALS participants, 48 (64.0%) had limb onset and 27 (36.0%) had bulbar onset, and 67 (89.3%) reported no family history of ALS. For participants that were both profiled for microbiome and plasma metabolomics (n = 54 total), 33 were male (61.1%) and 21 were female (38.9%), which differed significantly in sex ratio from the control participants with 30 males (38.0%) and 49 females (62.0%) (n = 79 total; P = 0.013) (Table 2 and Fig. 1). Of these ALS participants, 36 (66.7%) had limb onset and 18 (33.3%) had bulbar onset, and six (11.1%) reported a family history of ALS, indicating most cases were sporadic.
Table 1.
Clinical characteristics of participants in the microbiome cohort
| Characteristics | ALS group (n = 75) | Control group (n = 110) | P-value |
|---|---|---|---|
| Age, yearsa | 65.7 ± 10.0 (36.3–84.8) | 62.0 ± 8.6 (38.81–86.26) | 0.010b |
| Sex | – | – | |
| Male | 43 (57.3%) | 44 (40.0%) | 0.025c |
| Female | 32 (42.7%) | 66 (60.0%) | |
| Family history of ALS | – | – | |
| Yes | 7 (9.3%) | 0 (0%) | |
| No | 67 (89.3%) | 110 (100.0%) | |
| Unknown | 1 (1.3%) | 0 (0%) | |
| Race | – | – | |
| White | 75 (100.0%) | 110 (100.0%) | |
| ALS with frontotemporal dementia | 1 (1.33%) | – | |
| Onset segment | – | – | |
| Bulbar | 25 (33.3%) | – | |
| Cervical | 24 (32.0%) | – | |
| Lumbar | 24 (32.0%) | – | |
| Respiratory | 2 (2.67%) | – | |
| El Escorial criteria | – | – | |
| Definite | 16 (21.6%) | – | |
| Probable | 30 (40.5%) | – | |
| Probable, lab supported | 17 (23.0%) | – | |
| Possible | 9 (12.2%) | – | |
| Suspected | 2 (2.70%) | – | |
| Missing | 1 (1%) | – | |
| ALS Functional Rating Scale-Revisedd | 37.0 (34.0–40.0) | – | |
| Days from symptom onset to diagnosisd | 343.0 (216.5–617.5) | – | |
| Feeding tube | – | – | |
| Yes | 6 (8.0%) | – | |
| No | 69 (92.0%) | – | |
| Days from diagnosis to stool sampled | 98.0 (39.5–191.5) | – |
Bold font indicates significant P-values.ALS = amyotrophic lateral sclerosis.
aMean ± standard deviation (range).
bStudent’s t-test.
cFisher’s exact test.
dMedian (interquartile range).
Figure 1.
Study design. Top: Gut microbiome was longitudinally profiled in faecal samples from amyotrophic lateral sclerosis (ALS) (n = 75) versus control (n = 110) participants by 16S rRNA sequencing at baseline (T1), T2 and T3. Bottom: Plasma metabolites were profiled in plasma samples from ALS (n = 350) versus control (n = 175) participants by untargeted metabolomics using ultra-high-performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) at baseline (T1), published in Goutman et al.15,16Middle: Fifty-three ALS cases and 80 controls overlap between the baseline microbiome and metabolome cohorts. Significantly associated bacteria and circulating metabolites were identified by weighted gene co-expression network analysis (WGCNA) and two-way orthogonal partial least square with discriminant analysis (O2PLS-DA).
Table 2.
Clinical characteristics of participants overlapping between microbiome and metabolomics cohorts
| Characteristics | ALS group (n = 54) | Control group (n = 79) | P-value |
|---|---|---|---|
| Age, yearsa | 66.7 ± 9.4 (43.1–84.8) | 61.2 ± 8.3 (38.8–86.3) | 0.004b |
| Sex | – | – | |
| Male | 33 (61.1%) | 30 (38.0%) | 0.013c |
| Female | 21 (38.9%) | 49 (62.0%) | |
| Family history of ALS | – | – | |
| Yes | 6 (11.1%) | 0 (0%) | |
| No | 47 (87.0%) | 79 (100.0%) | |
| Unknown | 1 (1.9%) | 0 (0%) | |
| Race | – | – | |
| White | 54 (100.0%) | 79 (100.0%) | |
| ALS with frontotemporal dementia | 0 (0%) | – | |
| Onset segment | – | – | |
| Bulbar | 17 (31.5%) | – | |
| Cervical | 19 (35.2%) | – | |
| Lumbar | 17 (31.5%) | – | |
| Respiratory | 1 (1.9%) | – | |
| El Escorial criteria | – | – | |
| Definite | 7 (13.0%) | – | |
| Probable | 23 (42.6%) | – | |
| Probable, lab supported | 13 (24.1%) | – | |
| Possible | 8 (14.8%) | – | |
| Suspected | 3 (5.6%) | – | |
| ALS Functional Rating Scale-Revisedd | 38.0 (34.0–40.8) | – | |
| Days from symptom onset to diagnosisd | 370.5 (231.8–588.5) | – | |
| Feeding tube | – | – | |
| Yes | 4 (7.4%) | – | |
| No | 50 (92.6%) | – | |
| Days from diagnosis to stool sampled | 82.5 (44.0–193.2) | – |
Bold font indicates significant P-values.ALS = amyotrophic lateral sclerosis.
aMean ± standard deviation (range).
bStudent’s t-test.
cFisher’s exact test.
dMedian (interquartile range).
Microbial structure differs in ALS cases versus controls
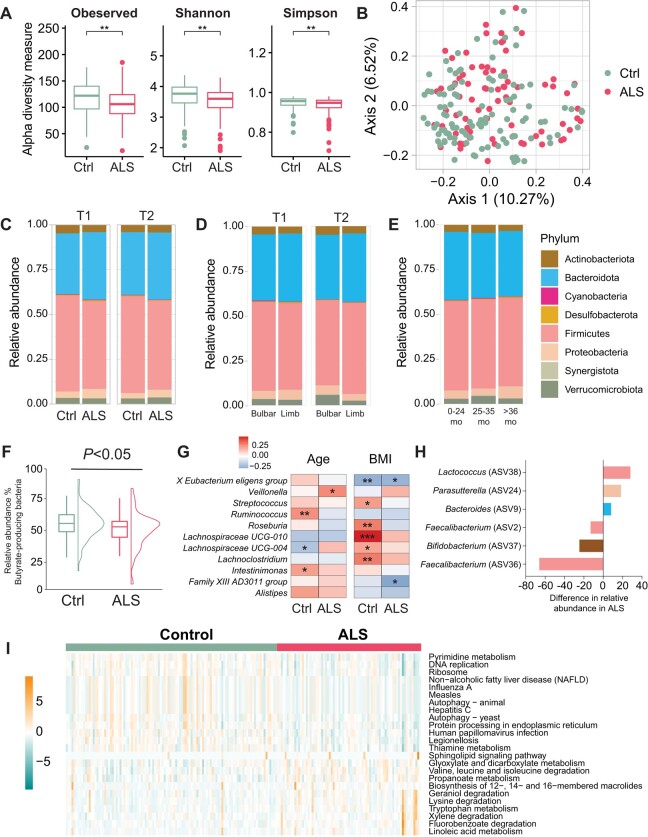
Gut microbiome composition in faecal samples from ALS versus control participants was determined by 16S rRNA sequencing of bacterial DNA, identifying 542 unique ASVs at the genus level after filtering.35 We observed significantly lower intracommunal microbial diversity by alpha diversity at baseline (T1) in ALS cases versus controls using three different metrics (P < 0.01; Fig. 2A), which persisted, albeit non-significantly, at the T2 time point. Inter-group beta-diversity analysis based on the Bray-Curtis distance matrix did not clearly separate ALS cases from controls, although it was statistically significant at T1 (P = 0.005; Fig. 2B), but not at T2.
Figure 2.
Microbial structure differs in ALS cases versus controls. (A) At the first time point (T1), intragroup alpha-diversity was significantly lower in amyotrophic lateral sclerosis (ALS) (n = 75) versus control (Ctrl, n = 110) faecal samples, represented by box plots [median by horizontal line; interquartile range (IQR) by box; maximum/minimum by vertical lines]; *P < 0.01 by t-test. (B) At T1, intergroup beta-diversity differed significantly between ALS versus control faecal samples, represented by principal coordinate analysis based on Bray-Curtis dissimilarity analysis; P = 0.005 by ANOVA. (C) Relative abundance of phyla (Actinobacteria, Bacteroidetes, Cyanobacteria, Desulfobacterota, Firmicutes, Proteobacteria, Synergistota, Verrucomicrobiota) shown as stacked bar plots for ALS versus control; at T1 Firmicutes (P = 4.7 × 1016), Cyanobacteria (P = 0.010), at T2 Firmicutes (P = 7.4 × 1016), in ALS versus control, by Wilcoxon test. (D) Relative abundance of phyla in faecal samples from ALS cases with bulbar (n = 27) versus limb (n = 47) onset at T1 and T2; at T1 Bacteroidetes (P = 0.022), at T2 Firmicutes (P = 0.0014), in ALS versus control, by Wilcoxon test. (E) Relative abundance of phyla in faecal samples from ALS cases from disease onset to sample collection stratified by 0–24 months (mo; n = 87), 25–35 months (n = 26) and >36 months (n = 26); Synergistota (P = 0.0074) in 0–24 months versus 25–35 months, Synergistota (P = 0.0074) in 25–35 months versus >36 months, by ANOVA. (F) At T1, butyrate-producing bacteria abundance was lower in ALS versus control focal samples, represented by box plots with data distribution; P < 0.05 by Wilcoxon test. (G) At T1, relative abundance of faecal ASVs at the genus level in ALS cases and controls by age (left) and BMI (right) represented by heat map; gradient colour scale represents correlation r-value. *P < 0.05, **P < 0.01, ***P < 0.001 by Spearman’s rank correlation. (H) At T1, genera ASVs driving gut microbial differences in ALS versus control faecal samples, assessed by logistic regression adjusted for age, sex and BMI. (I) At T1, significant biological pathways associated with gut microbial relative abundance in ALS cases versus control faecal samples represented by heat map using KEGG functional annotations; gradient colour scale shows over-represented (positive) and under-represented (negative) pathways in ALS; values scaled by row; significant pathways between groups with adjusted P < 0.05, by Wilcoxon test. BMI = body mass index.
Next, we examined the eight most abundant phyla using stacked blot plots of raw relative abundance. Firmicutes (P = 4.7 × 1016) and Cyanobacteria (P = 0.010) differed significantly in cases versus controls at T1, whereas only Firmicutes differed significantly at T2 (P = 7.4 × 1016) (Fig. 2C). When we examined phyla abundance by segment onset, we found that Bacteroidetes (P = 0.022) significantly differed at T1 while Firmicutes (P = 0.0014) differed at T2 in bulbar versus limb onset ALS (Fig. 2D). Lastly, we evaluated phyla abundance differences by the time between symptom onset and time of faecal sample collection (Supplementary Table 2). There were no statistically significant differences between samples collected 0–24 months or >35 months from disease onset. However, Synergistota differed significantly in samples taken 25–35 months from disease onset versus samples collected 0–24 months or over 35 months (Fig. 2E). Thus, overall, microbial structure by phyla differs in ALS cases, even by segment onset, compared to controls.
Next, we investigated more granular microbial information, examining genera and ASV differences by case/control status. We examined butyrate-producing bacteria due to their central role in energy36 and immune tolerance.37 We observed a slightly lower relative abundance of butyrate-producing bacteria in ALS versus controls at T1 (P < 0.05; Fig. 2F). Among genera, Spearman correlation found age and BMI associated differentially with specific genera by case/control status (Fig. 2G). Most correlations occurred within control participants, for both age and BMI. The most significant and most positive correlation by age was to Ruminococcus in control participants, whereas the most significant and most positive correlation by BMI was to Lachnospiraceae UCG-010, also in control participants. The most significant negative correlation was by BMI for X Eubacterium eligens group in control participants.
Consequently, we identified differential genera by case/control status at T1 using logistic regression analysis, adjusted for age, sex and BMI. Six genera differed significantly, with higher relative abundance in ALS of Bacteroides (ASV9; phylum Bacteroidetes), Parasutterella (ASV24; phylum Proteobacteria) and Lactococcus (ASV38; phylum Firmicutes) and higher relative abundance in control of Faecalibacterium (ASV36, ASV2; phylum Firmicutes) and Bifidobacterium (ASV37; phylum Actinobacteria) (Fig. 2H).
Finally, functional analysis of gut microbiota in ALS versus control participants was performed to infer metagenomic pathways by KEGG enrichment. In sum, we identified 24 enriched biological pathways, of which 11 were overrepresented in ALS, related to lipid metabolism (‘linoleic acid metabolism’, ‘propanoate metabolism’, ‘sphingolipid signaling pathway’) and amino acid metabolism and degradation (‘tryptophan metabolism’, ‘lysine degradation’, ‘valine, leucine and isoleucine degradation’) (Fig. 2I). Conversely, 13 biological pathways were under-represented in ALS, related to nucleotide metabolism (‘pyrimidine metabolism’), DNA replication, ribosome, protein metabolism (‘autophagy’, ‘protein processing in endoplasmic reticulum’) and infection.
Lipids are linked to distinct amplicon sequence variants in ALS
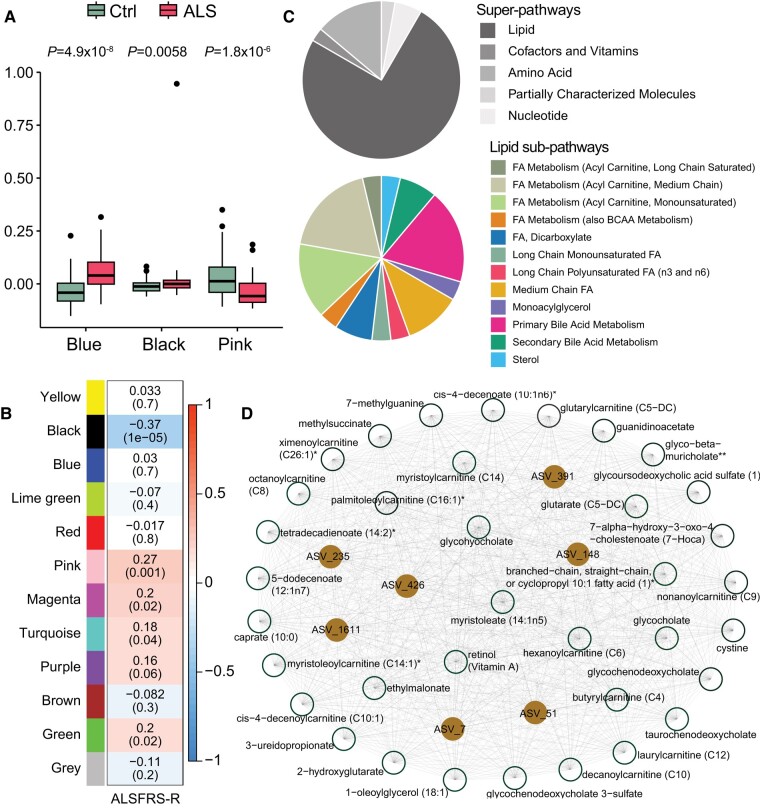
The gut microbiome influences the host plasma metabolome.38 Thus, we next integrated the faecal microbiome dataset with the plasma metabolomics dataset at baseline, i.e. the T1 time point, to identify microbial communities significantly correlated to plasma metabolite abundance in ALS. First, we performed WGCNA, a network-based, unsupervised clustering method.30,31 WGCNA did not identify any sample outliers in the dataset after dendrogram clustering nor any distinct ALS versus control clusters (Supplementary Fig. 1A). We, therefore, reran WGCNA using a soft threshold (power of β = 3) to minimize noise and maximize clustering (Supplementary Fig. 1B). The final network generated 12 modules, which contained 34 to 647 members (Supplementary Fig. 1C and Supplementary Table 3).
The blue module contained 77 members, all plasma metabolites without any ASVs. The pink module comprised 40 members, including five ASVs and 35 plasma metabolites. The black module had seven ASVs and 36 metabolites. Finally, the grey module was the largest and encompassed 647 members, which could not be assigned to any other module due to outlying profiles. The blue (P = 4.9 × 10−8), pink (P = 1.8 × 10−6) and black (P = 0.0058) modules significantly differed between ALS versus controls (Fig. 3A); all other modules were non-significant (Supplementary Fig. 2). Moreover, the black module had a high correlation coefficient (r = 0.7) and low P-value (P = 1.7 × 10−7) of both ASVs and plasma metabolites, suggesting they may contribute to the observed ALS phenotype (Supplementary Fig. 3). Indeed, when we examined the link of modules to the functional status of ALS cases, the ALSFRS-R, the black module was most significant, with a negative correlation (Fig. 3B), i.e. higher levels of black module components with lower ALSFRS-R score and more progressive disease.
Figure 3.
Lipids and ASVs cluster in ALS. (A) Blue, black and pink module expression differed significantly in amyotrophic lateral sclerosis (ALS) cases versus controls (Ctrl), represented by box plots, by Wilcoxon test. (B) Correlation of module colour (P-value in brackets) to functional status of ALS cases, assessed by ALSFRS-R (ALS Functional Rating Scale-Revised); gradient colour scale shows correlations to ALSFRS-R; significant correlations assessed by Pearson correlation. (C) Pie chart of black module metabolites by super-pathway (top) and lipid sub-pathway (bottom) membership. (D) Black module network plot of ASVs (brown circles) connected to metabolites (white circles), generated by Cytoscape. Metabolite membership to super-pathway and sub-pathway outlined in Supplementary Table 2. FA = fatty acid. *A compound that has not been confirmed based on a standard, but that the analytical platform is confident in its identity. **A compound for which a standard is not available, but that the analytical platform is reasonably confident in its identity or the information provided. ASV = amplicon sequence variant.
Network analysis of the black module found that the seven ASVs, corresponding to the families Akkermansiaceae (ASV7), Lachnospiraceae (ASV148, ASV235, ASV391), Rikenellaceae (ASV51), Marinifilaceae (ASV426) and Anaerococcus (ASV1611) strongly correlated to the 36 plasma metabolites, which were mostly (75%), related to and enriched in ‘lipids’ (Fig. 3C and D, Supplementary Fig. 4 and Supplementary Table 3). The lipids belonged to acylcarnitines (37%), medium-chain fatty acids (11%), long-chain fatty acids (7%), various fatty acids (11%), monoacylglycerol (4%), primary (19%) and secondary (7%) bile acids, and sterol (4%). Other biological pathways represented in the black module were ‘amino acid’ (14%), ‘nucleotide’ (6%), ‘cofactors and vitamins’ (3%) and ‘partially characterized molecules’ (3%). Overall, lipids were linked to ALS, mediated by seven ASVs of various families, especially Lachnospiraceae, through the black module.
As the WGCNA module that correlated most significantly with functional status, we also further analysed the association of black module ASVs and metabolites to changes in ALSFRS-R score over time. We found that 26 out of the 36 black module metabolites significantly correlated to decline in ALSFRS-R scores (Supplementary Table 4). Intriguingly, most metabolites exhibited negative correlations, i.e. higher metabolite levels corresponded with a decrease in ALSFRS-R scores over time. This potentially suggests a role for these metabolites in ALS pathogenesis or, alternatively, as biomarkers of disease progression. Specifically, metabolites such as 7-alpha-hydroxy-3-oxo-4-cholestenoate, cystine and glutarate (C5-DC) displayed consistent significant correlations, even at 1-year and 2-year post-profiling intervals (Supplementary Fig. 5). There were also several acylcarnitines, intermediates of fatty acid metabolism, that correlated negatively with ALSFRS-R in the analysis of all scores. Additionally, of the seven ASVs in the black module (Supplementary Table 4), ASV_148 and ASV_1611 notably correlated with a decline in ALSFRS-R scores.
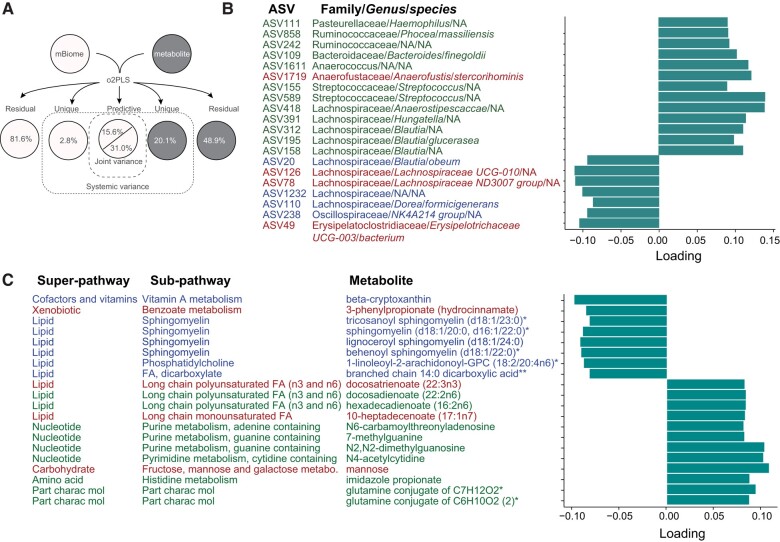
Next, we examined microbiome-metabolome correlations using multivariate regression O2PLS, a derivative of the widely used PLS-DA, which separates groups by maximizing covariance between datasets based on systematic sources of variance within each dataset separately. O2PLS identifies systematic trends across datasets, e.g. microbiome and metabolome. Our O2PLS analysis selected several metabolites and ASVs that were significantly intercorrelated. The optimal model comprised 10 joint variance components between ASV relative abundance and metabolite levels. The unique variance was 2.8% in ASVs and 20.1% in metabolites (Fig. 4A). Noise variance was 81.6% for ASVs and 48.9% for metabolites. Using the joint variance in microbiome to predict metabolites explained 85.5% of variance, while using joint variance in metabolites to predict microbiome explained 88.1% of variance. The joint variance explained 15.6% of variance in ASVs and 31.0% of variance in metabolites. The considerable extent of joint variance suggests that bacterial variation was accompanied by changes in metabolite levels.
Figure 4.
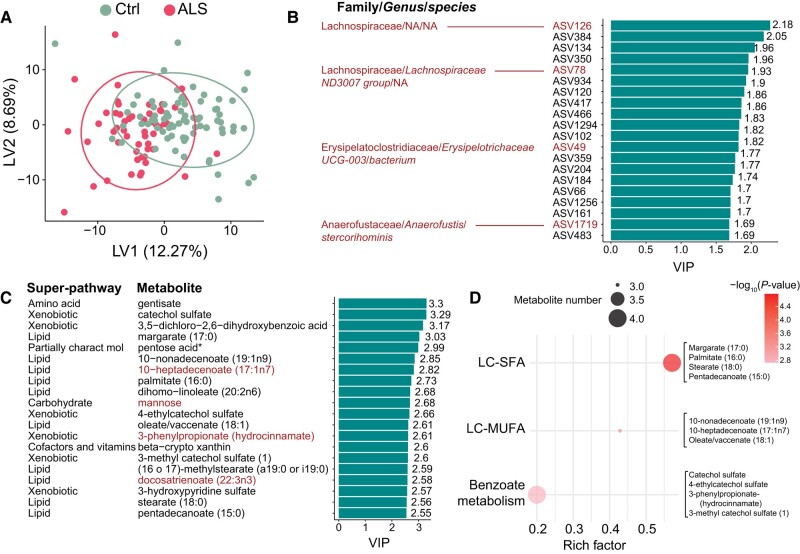
Lipids are inter-correlated with ASVs in ALS. (A) Schematic overview of two-way orthogonal partial least square (O2PLS) integrative analysis showing structure model and corresponding percent of total variance explained by a total of 10 components. Joint variance analysis identified the top 20 highly inter-correlated (B) ASVs and (C) metabolites. ASVs annotated with Family/Genus/species, green font represents positively correlated with metabolites, blue font represents negatively correlated with metabolites, red font represents ASVs contributing to ALS versus control group separation (Fig. 5B). Metabolites annotated with super-pathway and sub-pathway, green font represents positively correlated with ASV, blue font represents negatively correlated with ASVs, red font represents metabolites contributing to ALS versus control group separation (Fig. 5C). FA = fatty acid; NA = not applicable (not identified to the genus and/or species level); part charac mol = partially characterized molecules. *A compound that has not been confirmed based on a standard, but that the analytical platform is confident in its identity. **A compound for which a standard is not available, but that the analytical platform is reasonably confident in its identity or the information provided. ASV = amplicon sequence variant.
The family Lachnospiraceae represented 50% of the top 20 highly correlated ASVs (Fig. 4B), while lipid pathways comprised 50% of highly inter-correlated metabolites (Fig. 4C). Lipid sub-pathways corresponded to sphingomyelins (40%), long-chain polyunsaturated fatty acids (30%), long-chain monounsaturated fatty acids (10%), fatty acid dicarboxylate (10%) and phosphatidylcholine (10%). Notably, long-chain polyunsaturated and monounsaturated fatty acids correlated positively with ASVs, while the other lipids correlated negatively.
Next, we performed the O2PLS discriminatory analysis (O2PLS-DA) based on the joint variance from the O2PLS model. We found that most ALS cases could be separated from control participants based on the model (Fig. 5A). We used the variable importance in projection (VIP) score, a measure of importance to group separation between ALS cases versus controls, with a cut-off value of VIP > 1, identifying 133 significant ASVs (Supplementary Table 5). Most ASVs responsible for group separation belonged to the Lachnospiraceae family (Fig. 5B). In addition, among the top 20 ASVs with VIP >1, ASV126 (Lachnospiraceae family), ASV78 (Lachnospiraceae family), ASV49 (Erysipelatoclostridiaceae family) and ASV1719 (Anaerofustaceae family) were also highly inter-correlated with metabolites (Fig. 4C).
Figure 5.
Lipids and ASVs separate ALS cases from controls. (A) Partial least squares derivative analyses (PLS-DA) score plot separates ALS cases from controls (Ctrl). Top 20 (B) amplicon sequence variants (ASVs) and (C) metabolites contributing to the separation of amyotrophic lateral sclerosis (ALS) cases from controls, as assessed by variable importance in projection (VIP) score plot. ASVs and metabolites present in the joint variance analysis are indicated by red font (Fig. 4), suggesting strong biomarker candidates. ASVs in red font annotated with Family/Genus/species. All metabolites annotated with super-pathway. (D) Dot plot of significant enrichment analysis for the top 20 metabolites from C. LC-SFA = long-chain saturated fatty acid; LC-MUFA = long chain monounsaturated fatty acid; NA = not applicable (not identified to the genus and/or species level). *A compound that has not been confirmed based on a standard, but that the analytical platform is confident in its identity.
There were 247 metabolites with VIP > 1 (Supplementary Table 5) and of the top 20 metabolites contributing to group separation, most were from the ‘lipid’ and ‘xenobiotic’ super-pathways (Fig. 5C). Lipids represented 50% of the top 20 metabolites, of which 40% corresponded to fatty acid branched lipids, 30% to long-chain fatty acids, 20% to long-chain polyunsaturated fatty acids and 10% to long-chain saturated fatty acids. Other identified metabolites belonged to ‘amino acids’, ‘carbohydrates’, ‘cofactors and vitamins’ and ‘partially characterized molecules’ super-pathways. Among the top 20 metabolites with VIP >1, 10-heptadecenoate (17:1n7), mannose, 3-phenylpropinate and docosatrienoate were also highly inter-correlated with ASVs (Fig. 4B). Moreover, functional enrichment analysis for metabolites with VIP > 1, identified long-chain saturated fatty acid pathways as the most significant, along with significant enrichment of long-chain fatty acids and benzoate metabolism (Fig. 5D).
Mendelian randomization suggests a potential causal role of lipids in ALS
Both the WGCNA and O2PLS-DA analyses pinpointed potential links between microbiome and metabolites in ALS, especially through lipids (Supplementary Table 6). However, both analyses are correlative and do not shed light on possible causation. We therefore performed Mendelian randomization of five ALS GWAS datasets of diverse racial and/or ethnic background39-42 against SNPs regulating a panel of 452 blood metabolites.34 The 452-metabolite blood panel examined correlations to broader metabolite and lipid classes, several that we found correlated to ALS in this study. This facilitated a direct comparison of our Mendelian randomization analysis to specific species selected by the black, blue and pink WGCNA modules, as those differing significantly in ALS cases versus controls, and by the O2PLS-DA, which selected metabolites most responsible for the group separation of ALS cases from controls.
Eleven metabolites from the black module out of 36 members (31%) were significant in the Mendelian randomization analysis along with eight metabolites from the blue module out of 77 members (10%) and two in the pink module out of 35 members (6%). The black module was especially of interest as that most correlated to functional status by ALSFRS-R and as containing the most species of any WGNCA module identified by Mendelian randomization. All 11 overlapping metabolites, modulated by 105 SNPs, were lipids, primarily acylcarnitines of fatty acid metabolism, along with a range of fatty acid, monoacylglycerol and primary bile acid species. Between the blue module and Mendelian randomization, seven of the eight overlapping metabolites were lipids, mostly long-chain saturated fatty acids. Twenty-three of the metabolites with VIP > 1 out of 247 (9%) from the O2PLS-DA analysis were significant in our Mendelian randomization analysis. Most were lipids, at eight species primarily of long-chain fatty acids, followed by six amino acids, four cofactors and vitamins and other various super-pathway members. Thus, overall, our Mendelian randomization analysis suggested potential genetically determined causality of blood metabolites, especially lipid species linked to fatty acid and acylcarnitine metabolism, to ALS, warranting further investigation.
Discussion
There is evidence suggesting gut dysbiosis occurs in ALS.5 The microbiome regulates host gastrointestinal integrity, immune system activity and metabolism,38 which are all impacted in ALS.8-10,12,15-17 Although microbiome5,7,43-45 and plasma metabolome15,16,46-50 have been separately investigated in ALS, this is the first study, to our knowledge, to identify specific microbiome-plasma metabolite correlations in ALS, which could strengthen the robustness of potential biomarkers and offer mechanistic insight. Herein, we longitudinally profiled gut microbiome changes in ALS and correlated the initial gut microbiome profile to the plasma metabolome. We found that gut microbial structure at the phylum level differed in ALS versus control participants, with differential abundance of several distinct genera. Unsupervised clustering of microbe and metabolite co-expression levels identified three modules that differed significantly in ALS versus control participants. Network analysis of the module, which was most significantly correlated to ALS functional status, found several prominent ASVs strongly linked to a group of metabolites, primarily lipids. Similarly, examining the microbes and metabolites that most deeply contribute to case versus control separation again identified several ASVs correlated to metabolites, predominantly lipids.
Our earlier investigation into the microbiome of the mutant SOD1G93A ALS mouse model found gut dysbiosis, which evolved with time.8 We launched the present study to examine the microbiome longitudinally in ALS in humans. First, we examined the overall community structure and found lower intra-communal alpha diversity at baseline in ALS cases versus controls, meaning a less diverse gut microbiome in ALS. Inter-group beta-diversity did not clearly separate ALS cases from controls but differed significantly, indicating a distinct gut microbiome in ALS. Trends were maintained but were non-significant at subsequent time points. Lower alpha-diversity in ALS was noted by a human study,43 but not by others.7,44,45,51,52 Generally, although diminishing alpha-diversity is linked to unhealthy ageing, it is actually dependent on the loss or gain of ‘core’ microbes,53 which may account for differences across studies, in addition to study size. Lack of evident group separation but with significant differences in beta-diversity in cases versus controls was usually noted in the literature7,43,44 although not universally.45,52,54
When we broke down community structure by phyla, we noted that Firmicutes and Cyanobacteria differed significantly in ALS cases versus controls at baseline. Cyanobacteria43 abundance was elevated and Firmicutes51 reduced in the ALS microbiome in some studies, but without differences by phyla in some.7 We also found Bacteroidetes significantly differed at T1 and Firmicutes at T2 in bulbar versus limb onset ALS. Few reports have examined microbiome composition differences by ALS onset segment; however, no variation or non-significant variation was observed by studies that did investigate the dependence on onset segment or clinical phenotype.7,52 Dependence of microbiome composition on survival was noted by one study.7 Since the microbiome evolves with ageing or disease course,53 discordances across studies may have arisen from the time of sample collection relative to disease onset. Indeed, we noted differences in phyla abundance by the time of sample collection relative to disease onset, which may, in part, explain study variation. Thus, future investigations of the microbiome in ALS should account for the time of sample collection.
Next, we homed in on microbiome differences in ALS at the genus level to generate finer-grained information. Certain bacterial genera provide energy to host cells through short-chain fatty acids, e.g. butyrate,36 which may be dysregulated in disease.55 Indeed, ALS cases had a slightly lower relative abundance of beneficial butyrate-producing bacteria versus controls. These findings align with previous studies45,52,56 although one report found no difference.44 In our cohort, ALS cases were characterized by a higher relative abundance of Bacteroides (ASV9; phylum Bacteroidetes), Parasutterella (ASV24; phylum Proteobacteria) and Lactococcus (ASV38; phylum Firmicutes) and lower relative abundance of Faecalibacterium (ASV36, ASV2; phylum Firmicutes) and Bifidobacterium (ASV37; phylum Actinobacteria). Other studies have noted ALS cases with elevated Bacteroides45,56 and diminished Faecalibacterium56 and Bifidobacterium7,45; otherwise, distinct ASVs were identified across different studies, which may underlie ALS heterogeneity.52
Since microbiome restructures with ageing53 and metabolic status,57 we examined genera correlations with age and BMI. Additionally, ALS patients are characterized by significant BMI changes58 linked to hypermetabolism,6,59,60 which may potentially reflect in their microbiome. Associations were strongest and most significant among control participants, although three genera correlated significantly in ALS participants, Veillonella positively with age and X Eubacterium eligens group and Family XIII AD3011 group negatively with BMI. Previous studies similarly identified a genetic correlation of Veillonella with age by Mendelian randomization.61 One other study found links between BMI43 and also disease progression52 to specific genera in ALS, but associations were to distinct ASVs.
Our last examination solely of gut community was a functional analysis of the inferred metagenome. Multiple pathways were upregulated in ALS microbiota, including lipid and amino acid metabolism, whereas nucleotide metabolism, DNA replication, ribosome, autophagy, protein processing in endoplasmic reticulum and various infection pathways were downregulated. This is partly aligned with a recent study of faecal microbiome and metabolites from ALS participants, which also highlighted diminished nucleotide metabolism but also decreased amino acid and carbohydrate metabolism.51 Another study identified 41 differential faecal metabolites in ALS cases versus controls, which were primarily lipids along with peptides, nucleic acids and various other metabolite species.62 By contrast, other studies reported no functional or inferred metagenomic differences in ALS versus control microbiome.7,54 The relevance of dysbiosis in ALS presently remains incompletely understood; however, Mendelian randomization studies have examined causality, finding a bidirectional relationship between gut microbiota and ALS.63,64
We next integrated the faecal microbiome and plasma metabolome datasets to seek microbe-plasma metabolite correlations. The first approach was unsupervised clustering of ASV abundance and metabolite levels by WGCNA, which identified three significant modules. Network analysis of the black module, which most strongly correlated with ALS and functional status, found seven ASVs, Akkermansia muciniphila (ASV7), Lachnospiraceae (ASV148, ASV235, ASV391), Rikenellaceae (ASV51), Marinifilaceae (ASV426) and Anaerococcus (ASV1611), correlated to 36 plasma metabolites, 75% of which were lipids. The second integrative method, O2PLS, identified ASVs and metabolites that considerably contributed to the joint variance, suggesting that bacterial variation occurred in tandem with metabolite variation. Of the top 20 candidates, half were of the Lachnospiraceae family, along with Ruminococcaceae, Streptococcaceae and Anaerococcus, among others, and again, lipids represented 50% of metabolites. O2PLS-DA further highlighted Lachnospiraceae UCG-010 (ASV126), Lachnospiraceae ND3007 group (ASV78), Erysipelotrichaceae UCG-003 (ASV49) and Anaerofustaceae Anaerofustis stercorihominis (ASV1719), along with four highly intercorrelated metabolites, which contributed to ALS versus control group separation, advocating them as potential biomarkers.
The ASVs that clustered or correlated with metabolites have documented relevance to ALS and neurogenerative disease. Akkermansia muciniphila is downregulated in the colon of SOD1G93A ALS mice,8 and colonization improves motor function and survival.9 ASV391 is of the genus Hungatella, which is positively associated with Parkinson’s disease65 and gastrointestinal symptoms.66 At the family level, Lachnospiraceae (genus bacterium A4) correlates strongly with disease progression in SOD1G93A ALS mice.67 In human studies, several genera of the Lachnospiraceae family differentiate ALS cases from controls,7,43,45 including Lachnospiraceae Dorea and Lachnospiraceae Blautia.7 Additionally, ASVs of the families Ruminococcaceae and Rikenellaceae also differentiate ALS cases from controls.7,43 ASVs of the Streptococcaceae family encompass the Streptococcus genus; although frequently commensal, a few Streptococcus species are pathogenic. Early reports suggest that infection, such as by virulence factors68 from Streptococcus pneumoniae and immune system activation,69 might aggravate ALS disease course, although this is debatable.70 Marinifilaceae Butyricimonas (ASV426), Peptoniphilaceae Anaerococcus (ASV1611), Erysipelotrichaceae UCG-003 (ASV49) and Anaerofustaceae Anaerofustis stercorihominis (ASV1719) are, to our knowledge, the first instances shown to be potentially linked to ALS at the family and genus level.
Herein, lipids were the plasma metabolites most frequently correlated to ASVs, including acylcarnitines, medium- and long-chain fatty acids of various saturation and branching, sphingomyelins and primary and secondary bile acids. Additional associated metabolites belonged to carbohydrates, nucleotides, amino acids, cofactors and vitamins, partially characterized molecules and benzoate metabolism. Furthermore, correlation of the black module to functional status prompted examination of member ASVs and metabolites to change in ALSFRS-R over time. Several significant candidates emerged; among them was 7-alpha-hydroxy-3-oxo-4-cholestenoate, which is involved in primary bile acid biosynthesis. Cystine is the oxidized form of the amino acid cysteine, the precursor to the antioxidant glutathione, and is essential for maintaining redox balance. Astrocytic expression of the cystine/glutamate antiporter is elevated in ALS71; oxidative stress raises the need for glutathione, stimulating cystine import concomitant with glutamate export,72 which could trigger excitotoxicity, as seen in ALS. However, we found cystine increased with ALSFRS-R decline, indicative of an alternative or compensatory mechanism. Glutarate (C5-DC) is produced from amino acid metabolism, including lysine and tryptophan degradation. Excessive glutarate can deplete carnitine, with implications for energy metabolism and, along with other similar intermediates, is neurotoxic and contributes to the neurodegenerative disease glutaric aciduria type I.73
Changes in plasma lipids are a recurrent theme in ALS participants.15,16,48 Our Mendelian randomization analysis using a rich GWAS dataset of genetically determined blood metabolites narrowed down the lipids potentially causal to ALS to fatty acid and acylcarnitine metabolism. Altered acylcarnitines and fatty acids could point to impaired β-oxidation.16 In our analysis to change in ALSFRS-R, several acylcarnitines increased in level as functional score decreased with time. Primary and secondary bile acids, which correlate with BMI in ALS,58 could indicate maladapted intestinal lipid absorption and dysregulated cholesterol metabolism,74 which may be causal in ALS.75-79 Glycochenodeoxycholate is a primary bile acid that was selected by the black WGNCA module and Mendelian randomization and is a very closely structurally related and derivatized species80 to a recently trialled ALS treatment, tauroursodeoxycholic acid.81 To our knowledge, no study has examined microbiome-plasma correlations in ALS. However, relatively small studies of the faecal microbiome-metabolome noted a few correlations, including between microbes to bile acids51,62 and an acylcarnitine.51
The gut microbiome is intimately linked with host metabolism, including lipids.82 Under homeostatic conditions, specific beneficial gut microbes degrade dietary fibre, generating short-chain fatty acids, such as butyrate. Short-chain fatty acids serve as energy or biosynthesis substrates to the host, sensitize metabolically active tissue to insulin83 and promote host immune tolerance.37 The gut microbiota also metabolize primary bile acids into secondary bile acids. Bile acids primarily aid in lipid digestion within the gastrointestinal tract, but additionally bind to various receptors, stimulating host metabolism.82 Under pathologic conditions, gut dysbiosis occurs and may correlate or potentially causally promote impaired host metabolism, e.g. in obesity.84 Indeed, in addition to changes in plasma lipid levels, ALS is characterized by impaired metabolism,85 such as hypermetabolism and elevated energy expenditure,59,60 along with altered fatty acid β-oxidation.86,87 Overall, dysregulated lipids are a persistent theme in ALS and our findings here suggest that these changes may be potentially mediated by microbiome differences in patients. However, further investigation is needed, especially in animal models to assess causality. Indeed, the evidence to date supports this possibility; modifying lipid metabolism by ablating carnitine palmitoyl transferase 1, the transporter that shuttles fatty acids into mitochondria, from mutant SOD1 ALS mice simultaneously alters microbiome composition.88,89
This study has several strengths, such as the longitudinal design in a relatively large cohort of ALS cases and controls. Moreover, bacteria were identified down to the genus level using ASVs, and metabolomics analysis was untargeted using a highly validated platform, which accounts for instrument and process variability along with data curation. Computationally, we integrated the faecal microbiome and plasma metabolome datasets using two methods, WGCNA and O2PLS-DA. Our Mendelian randomization analysis utilized a large dataset of SNPs for genetically determined levels in 452 blood metabolites to examine possible causality of metabolites to ALS, the most fine-grained analysis, to our knowledge. Our study also has limitations. We did not ask ALS participants to fast as it was deemed unethical. Moreover, since we did not collect dietary information, we could not match dietary habits, which influence microbiome90 and plasma metabolome,91 in ALS cases versus controls. Environment, e.g. dwelling, cohabitation, also impacts gut microbiome composition,92 but we did not recruit controls from the same household. Furthermore, microbiome samples from ALS cases were collected at different time points relative to symptom onset. Finally, our study employed 16S RNA sequencing, which generally only identifies microorganisms to the genus level, and we inferred functional profiles computationally. Thus, our study may have missed important species-level or functional information, such as horizontal gene transfer, which are only captured by higher resolution methods, such as shotgun metagenomics.24
In conclusion, our study revealed significant differences in the gut microbiome structure of ALS cases versus controls, both at the phylum and genus levels. Our integration of the microbiome and plasma metabolomics datasets identified specific ASVs that showed significant correlations to lipids, as revealed by two bioinformatics methods. Moreover, we identified ASVs and metabolites that strongly associated with ALSFRS-R decline, unlocking novel avenues for research into molecular mechanisms underlying ALS, developing potential therapeutic interventions and biomarker discovery. These findings are in line with previous reports on altered plasma lipidome in ALS. Moreover, Mendelian randomization advocates potential causality from particular lipid species, especially fatty acid and acylcarnitine metabolism, and promotes future research avenues into targeted treatment approaches. Given the critical role of the gut microbiome in regulating host immune system health and the occurrence of immune dysregulation in ALS, future integration of the microbiome and/or the plasma metabolome with the inflammasome may constitute a fruitful line of investigation.
Supplementary Material
Acknowledgements
We are grateful to the study participants and their families. We also thank Jayna Duell, R.N., Caroline Piecuch, Amanda Williams, Hasan Farid, Samuel Teener and Ian Webber-Davis for facilitating participant consent and sample collection. This research was supported by work performed by The University of Michigan Microbiome Core. We also thank Dr. Stacey A. Sakowski Jacoby for editorial assistance.
Contributor Information
Kai Guo, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Claudia Figueroa-Romero, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Mohamed H Noureldein, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Benjamin J Murdock, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Masha G Savelieff, Department of Biomedical Sciences, University of North Dakota, Grand Forks, ND 58202, USA.
Junguk Hur, Department of Biomedical Sciences, University of North Dakota, Grand Forks, ND 58202, USA.
Stephen A Goutman, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Eva L Feldman, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Data availability
Anonymized data will be shared by request from any qualified investigator.
Funding
This work was supported by the Michigan Institute for Clinical and Health Research (MICHR, UL1TR000433), the National Center for Advancing Translational Sciences at the National Institutes of Health (UL1TR002240), NINDS R01NS127188, NIEHS R01ES030049 Parent Grant and Diversity Supplement, NIEHS K23ES027221, the Centers for Disease Control and Prevention/Agency for Toxic Substances and Disease Registry/National ALS Registry (R01TS000289), the Centers for Disease Control and Prevention/U.S. Department of Health and Human Services (R01TS000339), the NeuroNetwork Therapeutic Discovery Fund, the Peter R. Clark Fund for ALS Research, the Sinai Medical Staff Foundation, the Scott L. Pranger ALS Clinic Fund, the A. Alfred Taubman Medical Research Institute, and the NeuroNetwork for Emerging Therapies, University of Michigan.
Competing interests
S.A.G. served on a DSMB. The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Feldman EL, Goutman SA, Petri S, et al. Amyotrophic lateral sclerosis. Lancet. 2022;400:1363–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goutman SA, Hardiman O, Al-Chalabi A, et al. Recent advances in the diagnosis and prognosis of amyotrophic lateral sclerosis. Lancet Neurol. 2022;21:480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goutman SA, Hardiman O, Al-Chalabi A, et al. Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 2022;21:465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Chalabi A, Pearce N. Commentary: Mapping the human exposome: Without it, how can we find environmental risk factors for ALS? Epidemiology. 2015;26:821–823. [DOI] [PubMed] [Google Scholar]
- 5. Boddy SL, Giovannelli I, Sassani M, et al. The gut microbiome: A key player in the complexity of amyotrophic lateral sclerosis (ALS). BMC Med. 2021;19:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuraszkiewicz B, Goszczyńska H, Podsiadły-Marczykowska T, et al. Potential preventive strategies for amyotrophic lateral sclerosis. Front Neurosci. 2020;14:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ngo ST, Restuadi R, McCrae AF, et al. Progression and survival of patients with motor neuron disease relative to their fecal microbiota. Amyotroph Lateral Scler Frontotemporal Degener. 2020;21:549–562. [DOI] [PubMed] [Google Scholar]
- 8. Figueroa-Romero C, Guo K, Murdock BJ, et al. Temporal evolution of the microbiome, immune system and epigenome with disease progression in ALS mice. Dis Model Mech. 2019;13:dmm041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blacher E, Bashiardes S, Shapiro H, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572:474–480. [DOI] [PubMed] [Google Scholar]
- 10. Burberry A, Wells MF, Limone F, et al. C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature. 2020;582:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cryan JF, O'Riordan KJ, Cowan CSM, et al. The Microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013. [DOI] [PubMed] [Google Scholar]
- 12. Wu S, Yi J, Zhang YG, Zhou J, Sun J. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol Rep. 2015;3:e12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun J, Zhan Y, Mariosa D, et al. Antibiotics use and risk of amyotrophic lateral sclerosis in Sweden. Eur J Neurol. 2019;26:1355–1361. [DOI] [PubMed] [Google Scholar]
- 14. Beers DR, Appel SH. Immune dysregulation in amyotrophic lateral sclerosis: Mechanisms and emerging therapies. Lancet Neurol. 2019;18:211–220. [DOI] [PubMed] [Google Scholar]
- 15. Goutman SA, Boss J, Guo K, et al. Untargeted metabolomics yields insight into ALS disease mechanisms. J Neurol Neurosurg Psychiatry. 2020;91:1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goutman SA, Guo K, Savelieff MG, et al. Metabolomics identifies shared lipid pathways in independent amyotrophic lateral sclerosis cohorts. Brain. 2022;145:4425–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin S, Battistini C, Sun J. A gut feeling in amyotrophic lateral sclerosis: Microbiome of mice and men. Front Cell Infect Microbiol. 2022;12:839526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brooks BR, Miller RG, Swash M, Munsat TL; World Federation of Neurology Research Group on Motor Neuron Diseases . El escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. [DOI] [PubMed] [Google Scholar]
- 19. Seekatz AM, Theriot CM, Molloy CT, Wozniak KL, Bergin IL, Young VB. Fecal Microbiota transplantation eliminates Clostridium difficile in a murine model of relapsing disease. Infect Immun. 2015;83:3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from illumina amplicon data. Nat Methods. 2016;13:581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wemheuer F, Taylor JA, Daniel R, et al. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ Microbiome. 2020;15:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo K, Figueroa-Romero C, Noureldein M, et al. Gut microbiota in a mouse model of obesity and peripheral neuropathy associated with plasma and nerve lipidomics and nerve transcriptomics. Microbiome. 2023;11:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: An overview using omics data. mSystems. 2017;2:e00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barcenilla A, Pryde SE, Martin JC, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut Microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10:e02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. [DOI] [PubMed] [Google Scholar]
- 30. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. [DOI] [PubMed] [Google Scholar]
- 31. Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hemani G, Zheng J, Elsworth B, et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cani PD, Van Hul M, Lefort C, Depommier C, Rastelli M, Everard A. Microbial regulation of organismal energy homeostasis. Nat Metab. 2019;1:34–46. [DOI] [PubMed] [Google Scholar]
- 37. Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. [DOI] [PubMed] [Google Scholar]
- 38. Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Rheenen W, Shatunov A, Dekker AM, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48:1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benyamin B, He J, Zhao Q, et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun. 2017;8:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nicolas A, Kenna KP, Renton AE, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97:1268–1283 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iacoangeli A, Lin T, Al Khleifat A, et al. Genome-wide meta-analysis finds the ACSL5-ZDHHC6 locus is associated with ALS and links weight loss to the disease genetics. Cell Rep. 2020;33:108323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Di Gioia D, Bozzi Cionci N, Baffoni L, et al. A prospective longitudinal study on the microbiota composition in amyotrophic lateral sclerosis. BMC Med. 2020;18:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hertzberg VS, Singh H, Fournier CN, et al. Gut microbiome differences between amyotrophic lateral sclerosis patients and spouse controls. Amyotroph Lateral Scler Frontotemporal Degener. 2022;23:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nicholson K, Bjornevik K, Abu-Ali G, et al. The human gut microbiota in people with amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2021;22:186–194. [DOI] [PubMed] [Google Scholar]
- 46. Bjornevik K, Zhang Z, O'Reilly ÉJ, et al. Prediagnostic plasma metabolomics and the risk of amyotrophic lateral sclerosis. Neurology. 2019;92:e2089–e2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chang KH, Lin CN, Chen CM, et al. Altered metabolic profiles of the plasma of patients with amyotrophic lateral sclerosis. Biomedicines. 2021;9:1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sol J, Jové M, Povedano M, et al. Lipidomic traits of plasma and cerebrospinal fluid in amyotrophic lateral sclerosis correlate with disease progression. Brain Commun. 2021;3:fcab143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lawton KA, Brown MV, Alexander D, et al. Plasma metabolomic biomarker panel to distinguish patients with amyotrophic lateral sclerosis from disease mimics. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15:362–370. [DOI] [PubMed] [Google Scholar]
- 50. Wuolikainen A, Jonsson P, Ahnlund M, et al. Multi-platform mass spectrometry analysis of the CSF and plasma metabolomes of rigorously matched amyotrophic lateral sclerosis, Parkinson's disease and control subjects. Mol Biosyst. 2016;12:1287–1298. [DOI] [PubMed] [Google Scholar]
- 51. Zeng Q, Shen J, Chen K, et al. The alteration of gut microbiome and metabolism in amyotrophic lateral sclerosis patients. Sci Rep. 2020;10:12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Niccolai E, Di Pilato V, Nannini G, et al. The gut Microbiota-immunity axis in ALS: A role in deciphering disease heterogeneity? Biomedicines. 2021;9:753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeffery IB, Lynch DB, O'Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10:170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brenner D, Hiergeist A, Adis C, et al. The fecal microbiome of ALS patients. Neurobiol Aging. 2018;61:132–137. [DOI] [PubMed] [Google Scholar]
- 55. Mirzaei R, Bouzari B, Hosseini-Fard SR, et al. Role of microbiota-derived short-chain fatty acids in nervous system disorders. Biomed Pharmacother. 2021;139:111661. [DOI] [PubMed] [Google Scholar]
- 56. Fang X, Wang X, Yang S, Meng F, Wei H, Chen T. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using high-throughput sequencing. Front Microbiol. 2016;7:1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pinart M, Dötsch A, Schlicht K, et al. Gut microbiome composition in obese and non-obese persons: A systematic review and meta-analysis. Nutrients. 2021;14:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goutman SA, Boss J, Iyer G, et al. Body mass index associates with amyotrophic lateral sclerosis survival and metabolomic profiles. Muscle Nerve. 2023;67:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jésus P, Fayemendy P, Nicol M, et al. Hypermetabolism is a deleterious prognostic factor in patients with amyotrophic lateral sclerosis. Eur J Neurol. 2018;25:97–104. [DOI] [PubMed] [Google Scholar]
- 60. Steyn FJ, Ioannides ZA, van Eijk RPA, et al. Hypermetabolism in ALS is associated with greater functional decline and shorter survival. J Neurol Neurosurg Psychiatry. 2018;89:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. He D, Liu L, Zhang Z, et al. Association between gut microbiota and longevity: A genetic correlation and Mendelian randomization study. BMC Microbiol. 2022;22:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gong Z, Ba L, Tang J, et al. Gut microbiota links with cognitive impairment in amyotrophic lateral sclerosis: A multi-omics study. J Biomed Res. 2022;37:125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang L, Zhuang Z, Zhang G, Huang T, Fan D. Assessment of bidirectional relationships between 98 genera of the human gut microbiota and amyotrophic lateral sclerosis: A 2-sample Mendelian randomization study. BMC Neurol. 2022;22:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ning J, Huang SY, Chen SD, Zhang YR, Huang YY, Yu JT. Investigating casual associations among gut Microbiota, metabolites, and neurodegenerative diseases: A Mendelian randomization study. J Alzheimers Dis. 2022;87:211–222. [DOI] [PubMed] [Google Scholar]
- 65. Zhang K, Paul KC, Jacobs JP, et al. Parkinson's disease and the gut microbiome in rural California. J Parkinsons Dis. 2022;12:2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen W, Bi Z, Zhu Q, et al. An analysis of the characteristics of the intestinal flora in patients with Parkinson's disease complicated with constipation. Am J Transl Res. 2021;13:13710–13722. [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y, Ogbu D, Garrett S, Xia Y, Sun J. Aberrant enteric neuromuscular system and dysbiosis in amyotrophic lateral sclerosis. Gut Microbes. 2021;13:1996848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goos M, Zech WD, Jaiswal MK, et al. Expression of a Cu, Zn superoxide dismutase typical for familial amyotrophic lateral sclerosis increases the vulnerability of neuroblastoma cells to infectious injury. BMC Infect Dis. 2007;7:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nguyen MD, D'Aigle T, Gowing G, Julien JP, Rivest S. Exacerbation of motor neuron disease by chronic stimulation of innate immunity in a mouse model of amyotrophic lateral sclerosis. J Neurosci. 2004;24:1340–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ebert S, Goos M, Rollwagen L, et al. Recurrent systemic infections with Streptococcus pneumoniae do not aggravate the course of experimental neurodegenerative diseases. J Neurosci Res. 2010;88:1124–1136. [DOI] [PubMed] [Google Scholar]
- 71. Kazama M, Kato Y, Kakita A, et al. Astrocytes release glutamate via cystine/glutamate antiporter upregulated in response to increased oxidative stress related to sporadic amyotrophic lateral sclerosis. Neuropathology. 2020;40:587–598. [DOI] [PubMed] [Google Scholar]
- 72. Albano R, Liu X, Lobner D. Regulation of system x(c)- in the SOD1-G93A mouse model of ALS. Exp Neurol. 2013;250:69–73. [DOI] [PubMed] [Google Scholar]
- 73. Sauer SW, Opp S, Hoffmann GF, Koeller DM, Okun JG, Kölker S. Therapeutic modulation of cerebral L-lysine metabolism in a mouse model for glutaric aciduria type I. Brain. 2011;134:157–170. [DOI] [PubMed] [Google Scholar]
- 74. Di Ciaula A, Garruti G, Lunardi Baccetto R, et al. Bile acid physiology. Ann Hepatol. 2017;16:S4–S14. [DOI] [PubMed] [Google Scholar]
- 75. van Rheenen W, van der Spek RAA, Bakker MK, et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet. 2021;53:1636–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xia K, Klose V, Högel J, et al. Lipids and amyotrophic lateral sclerosis: A two-sample Mendelian randomization study. Eur J Neurol. 2023;30:1899–1906. [DOI] [PubMed] [Google Scholar]
- 77. Zeng P, Zhou X. Causal effects of blood lipids on amyotrophic lateral sclerosis: A Mendelian randomization study. Hum Mol Genet. 2019;28:688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen X, Yazdani S, Piehl F, Magnusson PKE, Fang F. Polygenic link between blood lipids and amyotrophic lateral sclerosis. Neurobiol Aging. 2018;67:202.e1–202.e6. [DOI] [PubMed] [Google Scholar]
- 79. Bandres-Ciga S, Noyce AJ, Hemani G, et al. Shared polygenic risk and causal inferences in amyotrophic lateral sclerosis. Ann Neurol. 2019;85:470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kusaczuk M. Tauroursodeoxycholate-Bile acid with chaperoning activity: Molecular and cellular effects and therapeutic perspectives. Cells. 2019;8:1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Paganoni S, Macklin EA, Hendrix S, et al. Trial of sodium phenylbutyrate-taurursodiol for amyotrophic lateral sclerosis. N Engl J Med. 2020;383:919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schoeler M, Caesar R. Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord. 2019;20:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. [DOI] [PubMed] [Google Scholar]
- 84. Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: Moving beyond associations. Cell Host Microbe. 2017;22:589–599. [DOI] [PubMed] [Google Scholar]
- 85. Blasco H, Lanznaster D, Veyrat-Durebex C, et al. Understanding and managing metabolic dysfunction in amyotrophic lateral sclerosis. Expert Rev Neurother. 2020;20:907–919. [DOI] [PubMed] [Google Scholar]
- 86. Steyn FJ, Li R, Kirk SE, et al. Altered skeletal muscle glucose-fatty acid flux in amyotrophic lateral sclerosis. Brain Commun. 2020;2:fcaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Szelechowski M, Amoedo N, Obre E, et al. Metabolic reprogramming in amyotrophic lateral sclerosis. Sci Rep. 2018;8:3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Trabjerg MS, Mørkholt AS, Lichota J, et al. Dysregulation of metabolic pathways by carnitine palmitoyl-transferase 1 plays a key role in central nervous system disorders: Experimental evidence based on animal models. Sci Rep. 2020;10:15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Trabjerg MS, Andersen DC, Huntjens P, et al. Downregulating carnitine palmitoyl transferase 1 affects disease progression in the SOD1 G93A mouse model of ALS. Commun Biol. 2021;4:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wolter M, Grant ET, Boudaud M, et al. Leveraging diet to engineer the gut microbiome. Nat Rev Gastroenterol Hepatol. 2021;18:885–902. [DOI] [PubMed] [Google Scholar]
- 91. Pourafshar S, Nicchitta M, Tyson CC, et al. Urine and plasma metabolome of healthy adults consuming the DASH (dietary approaches to stop hypertension) diet: A randomized pilot feeding study. Nutrients. 2021;13:1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized data will be shared by request from any qualified investigator.