Abstract
Injuries to pancreatic β-cells are intricately linked to the onset of diabetes mellitus (DM). Metformin (Met), one of the most widely prescribed medications for diabetes and metabolic disorders, has been extensively studied for its antioxidant, anti-aging, anti-glycation, and hepatoprotective activities. N6-methyladenosine (m6A) plays a crucial role in the regulation of β-cell growth and development, and its dysregulation is associated with metabolic disorders. This study aimed to elucidate the mechanistic basis of m6A involvement in the protective effects of Met against oxidative damage in pancreatic β-cells. Hydrogen peroxide (H2O2) was employed to induce β-cell damage. Remarkably, Met treatment effectively increased methylation levels and the expression of the methyltransferase METTL14, subsequently reducing H2O2-induced apoptosis. Knocking down METTL14 expression using siRNA significantly compromised cell viability. Conversely, targeted overexpression of METTL14 specifically in β-cells substantially enhanced their capacity to withstand H2O2-induced stress. Molecular evidence suggests that the anti-apoptotic properties of Met may be mediated through Bcl-xL and Bim proteins. In conclusion, our findings indicate that Met induces METTL14-mediated alterations in m6A methylation levels, thereby shielding β-cells from apoptosis and oxidative damage induced by oxidative stress.
Keywords: Metformin, N6-methyladenosine, Hydrogen peroxide, METTL14, Apoptosis
1. Introduction
Pancreatic β-cell damage stands as a pivotal factor that contributes to the impaired function of β-cells, thereby promoting the pathological progression of diabetes [1,2]. This multifactorial process involves various stressors, including but not limited to metabolic and oxidative stress [3,4], altered growth factors [5], immune attacks [6], and circulating hormones [7]. It has been observed by researchers that the expression levels of catalase and glutathione peroxidase in pancreatic β-cells are comparatively lower than those in other organs [8]. As a consequence, β-cells exhibit a greater susceptibility and sensitivity to oxidative stress in contrast to other cell types [8,9]. The exposure of cells to substantial amounts of hydrogen peroxide (H2O2), a typical reactive oxygen species, often leads to an imbalance between cellular oxidation and antioxidants, ultimately resulting in cell death [10].
Metformin (Met), known as the most conventional antihyperglycemic medication, has been extensively employed in the treatment of type 2 diabetes for a considerable duration [11]. The prominent mechanism underlying Met's hypoglycemic effects lies in its ability to diminish hepatic glucose synthesis [12]. Recent evidence suggests that Met may exert regulatory control over endoplasmic reticulum (ER) stress, oxidative stress, immunological response, and gut microbiota [[13], [14], [15], [16]]. Notably, Met exhibits the capability to reduce the production of reactive oxygen species [17,18]. However, the precise manner in which this ancient medication safeguards pancreatic β-cells against damage still remains elusive.
N6-methyladenosine (m6A) has emerged as a pivotal post-transcriptional mechanism governing gene regulation [19,20], playing a crucial role in numerous physiological and pathological processes [21,22]. The proper development and maintenance of β-cells heavily rely on the modulation of m6A methylation [23]. Notably, evidence suggests that individuals with type 2 diabetes exhibit significantly reduced levels of m6A methylation in their islets and plasma, potentially attributed to the downregulation of m6A methyltransferase METTL3 and upregulation of demethylases such as FTO [[24], [25], [26]]. Further investigation is required to elucidate whether m6A methylation exerts control over H2O2-induced pancreatic apoptosis.
The objective of this study was to investigate the protective potential of Metformin (Met) against oxidative damage in the NIT-1 mouse pancreatic β-cell line. NIT-1 cells were treated with Met in the presence of H2O2 as a direct oxidizing agent. Subsequently, cellular growth and viability were assessed, and the underlying molecular mechanisms of Met-induced m6A methylation were evaluated. Furthermore, we explored the cytoprotective role of the m6A methyltransferase METTL14 in Met-treated β-cells, elucidating the impact of Met on β-cell survival, apoptosis, and the expression of m6A-methylated proteins.
2. Materials and Methods
2.1. Cell culture and reagent
The NIT-1 cell line (Cl-0562, Procell, Wuhan, China), derived from mouse islet β-cells, was cultured in a proliferative medium consisting of DMEM (11,885,084, Gibco, NY, USA) supplemented with 10 % FBS (10,099,141, Gibco, NY, USA) at 37 °C in a humidified atmosphere with 5 % CO2. Cells were treated with hydrogen peroxide (H2O2, S0051, Beyotime, Shanghai, China) or Metformin (Met, D150959, Sigma-Aldrich, St. Louis, USA) for the indicated durations in each experimental group. The concentrations of H2O2 and Met were determined based on previous relevant research studies [27,28], which employed 300 μM H2O2 and 0.5 mM Met treatment for 1 day in NIT-1 cells, respectively.
2.2. SiRNA transfection silenced METTL14 expression
Small interfering RNA (siMETTL14) targeting METTL14 and a siRNA negative control (siNC) were synthesized by RiboBio (Guangzhou, China). Lipofectamine 3000 (L3000015, Invitrogen, USA) was diluted with Opti-MEM and adjusted to the appropriate concentrations. The siRNAs were mixed with the medium to form the transfection complex, followed by incubation at 23 °C for 15 min as instructed, once the cell fusion reached approximately 30 %–50 %. The cells were then cultured in the transfection complex and incubated at 37 °C for 36 h to allow for subsequent experiments. The sequences of the siRNAs targeting mouse METTL14 mRNA were as follows: S1: 5′-CCGGATGTACAGAGGAAAT‐3'; S2: 5′-TTGAAGAATACCCTAAACT-3'; S3: 5′-AGATGAACAGAGGGAGATT-3'.
2.3. The adenovirus METTL14 infects NIT-1 cells
The recombinant adenovirus vectors encoding the empty vector (Ad-NC) and METTL14 overexpression construct (Ad-METTL14) were synthesized by HanBio (Shanghai, China). NIT-1 cells were transfected with Ad-NC or Ad-METTL14 at a multiplicity of infection (MOI) of 100. After incubating in the transfection medium for 6–8 h, the medium was replaced, and subsequent experiments were conducted.
2.4. Cell viability/cytotoxicity testing
To discriminate between living and dead cells, a combination of Calcein Acetoxymethyl Ester (calcein AM) and propidium iodide (PI) staining was used. Briefly, after the initial treatment, the cell culture medium was removed, and NIT-1 cells were rinsed with PBS. Subsequently, 0.1 mL of a diluted solution containing calcein AM and PI was added to each well. The plate was then incubated in darkness at 37 °C for 30 min. Calcein AM stained the living cells, emitting a green fluorescence, while PI stained the dead cells, resulting in a red fluorescence.
2.5. Flow cytometry analysis
To assess apoptosis of NIT-1 cells using flow cytometry, the cells were cultured and treated in 6-well plates. Subsequently, they were dissociated using 0.25 % Trypsin Solution without EDTA (C0205, Beyotime, Shanghai, China) at 37 °C for 3–5 min. The cells were then stained with serial additions of Annexin V-FITC/PI at 4 °C for 15 min, following the protocol provided by the Apoptosis Testing Kit (BestBio, Shanghai, China). During the detection process, flow cytometry analysis was performed within 1 h on the CytoFLEX instrument (Beckman Coulter, USA). Viability was determined based on the rate of double-negative staining cells, while apoptotic cells were identified as Annexin V-positive cells.
2.6. m6A RNA methylation quantification
Total RNA was extracted using TRIzol Reagent (DP424, TIANGEN, Beijing, China), and the concentration of RNA was determined using a Nanodrop 2000 spectrophotometer (Abcam, Cambridge, UK). Subsequently, 200 ng of total RNA from each group was used per well for replicates. The quantification of m6A methylation was analyzed utilizing the m6A RNA Methylation Detection Kit (Abcam, Cambridge, UK), following previously described methods [28].
2.7. qRT-PCR detection
Reverse transcription of RNA into cDNA according to the instructions of the kit (KR123, TIANGEN, Beijing, China). qRT-PCR testing was conducted based on the SYBR Premix ExTaq kit (208,054, QIAGEN, Hilden, Germany). GAPDH primers were used to normalize the relative expression of aim genes, which were determined according to 2−ΔΔCt method. Primer pair sequence: Mouse METTL3: Forward primer 5′-CATCCGTCTTGCCATCTCTACGC-3′, Reverse primer 5′-GCAGACAGCTTGGAGTGGTCAG-3’; Mouse METTL14: Forward primer 5′-TCGACCGAAGTCACCTCCTC-3′, Reverse primer 5′-AGGAGTAAAGCCGCCTCTGT-3’; Mouse FTO: Forward primer 5′-GACACTTGGCTTCCTTACCTGACC-3′, Reverse primer 5′-ACCTCCTTATGCAGCTCCTCTGG-3’; Mouse ALKBH5: Forward primer 5′-GCAAGGTGAAGAGCGGCATCC-3′, Reverse primer 5′-GTCCACCGTGTGCTCGTTGTAC-3’; Mouse GAPDH: Forward primer 5′-GGTTGTCTCCTGCGACTTCA-3′, Reverse primer 5′-TGGTCCAGGGTTTCTTACTCC-3’.
2.8. Western blot analysis
NIT-1 cells were lysed in 100 μL of modified RIPA protein lysis buffer supplemented with 1 × PMSF (Abcam, Cambridge, UK) in each well of the 6-well plates. The proteins were then separated by SDS-PAGE (Abcam, Cambridge, UK) and transferred onto nitrocellulose membranes (CST, Beverly, USA) without delay. Subsequently, the membranes were incubated with primary antibodies at 4 °C for 12 h, followed by appropriate secondary antibodies at room temperature for 2 h. Blots were visualized using a Tanon 5200 visualizer (Tanon, Shanghai, China). The antibodies employed in this experiment included mouse anti-METTL14 (ab220030, 1:1000, Abcam, Cambridge, UK), rabbit anti-Bcl-xL (2764, 1:1000, CST, Beverly, MA, USA), rabbit anti-Bim (2933, 1:1000, CST, Beverly, MA, USA), rabbit anti-cleaved caspase 3 (9664, 1:1000, CST, Beverly, MA, USA), with β-actin (T0022, 1:3000, Affinity, Cincinnati, OH, USA) serving as a reference for total protein levels. Data analysis was performed using ImageJ software.
2.9. Statistical analyses
The data were presented as mean ± SD. For comparisons between different groups, the t-test was utilized. To assess differences among multiple groups, ANOVA followed by the N–K test was applied. Statistical significance was determined by a P-value of less than 0.05.
3. Results
3.1. Met protects against H2O2-Induced NIT-1 cells death
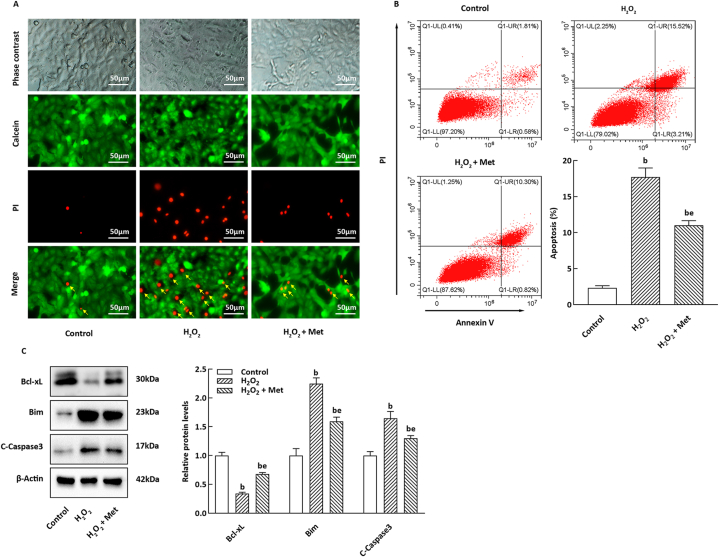
To investigate the potential role of Met in H2O2-induced injury to pancreatic β-cells, we assessed the viability of NIT-1 cells treated with Met. As depicted in Fig. 1A, H2O2 markedly increased the death rate of NIT-1 cells compared to the control group. However, co-treatment with Met significantly ameliorated cell death, as evidenced by calcein AM/PI staining. Moreover, annexin V/PI staining revealed that Met effectively suppressed the apoptosis level of NIT-1 cells induced by H2O2 (Fig. 1B). In NIT-1 cells exposed to H2O2, Met treatment prominently elevated the expression of Bcl-xL protein, while exerting an opposite effect on the protein expression levels of Bim and cleaved caspase 3 (Fig. 1C).
Fig. 1.
Met improved H2O2-induced apoptosis of NIT-1 cells. (A) Calcein AM/PI staining result of NIT-1 cells. Green fluorescence corresponding to living cells; red fluorescence is marked by a yellow arrow due. Scale bar = 50 μm. (B) Representative images of apoptotic NIT-1 cells based on annexin V/PI staining. The histogram refers to the apoptotic NIT-1 cells rate (n = 3). (C) The panel and histogram correspond to the relative levels of apoptosis-associated proteins, the internal control was β-actin (n = 3). bP < 0.05, eP < 0.05, compare to the control and H2O2 group, respectively.
3.2. Met reverses H2O2-Induced m6A RNA modification and METTL14 level reduction in NIT-1 cells
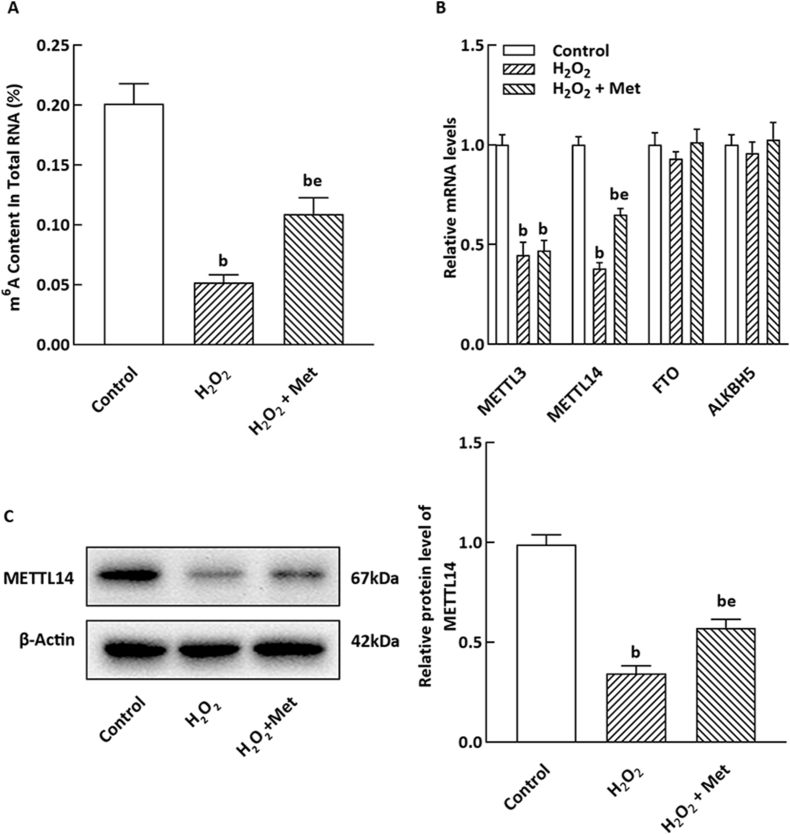
To investigate the potential role of Met in m6A modification in NIT-1 cells, we quantified the m6A content in these cells. As shown in Fig. 2A, H2O2 treatment led to a significant reduction in m6A methylation levels compared to metformin-treated cells. Additionally, mRNA expressions of the m6A methyltransferases METTL3 and METTL14 were decreased in NIT-1 cells treated with H2O2 (P < 0.05), while there were no noticeable changes in the mRNA levels of the m6A demethylases FTO and ALKBH5. Interestingly, treatment with Met increased the degree of m6A methylation in the H2O2-treated group and partially restored the mRNA expression of METTL14, but did not significantly impact the mRNA levels of METTL3 or FTO (Fig. 2B). Furthermore, Western blot analysis demonstrated that Met treatment enhanced the protein level of METTL14 (Fig. 2C).
Fig. 2.
Effect of Met on m6A content and expression of m6A methylated enzyme in NIT-1 cells incubated with H2O2. (A) m6A methylation level in total RNA was detected by ELISA (n = 3). (B) mRNA expression of m6A methyltransferase METTL3 and METTL14 and demethylase FTO and were detected by qRT-PCR (n = 3). (C) METTL14 level was detected by WB method (n = 3). The mean of bP < 0.05, eP < 0.05 are the same to above.
3.3. Effects of METTL14 knockdown in NIT-1 cells
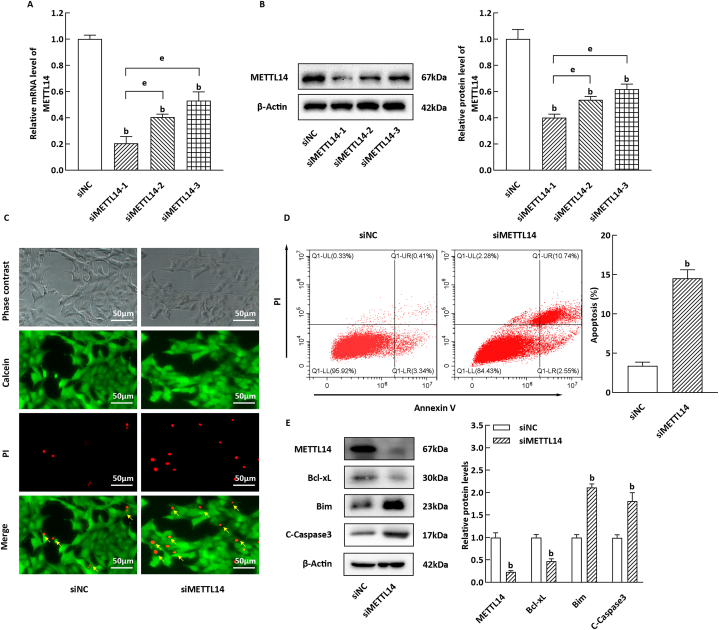
METTL14, a key component of the methyltransferase complex, plays a crucial role in the growth and development of β-cells. Building upon previous studies [29], we sought to further investigate the involvement of METTL14 in the apoptosis of NIT-1 cells. In our experimental procedure, we validated the knockdown efficiency of siMETTL14-1, −2, and −3, ultimately selecting siMETTL14-1 for subsequent experiments (Fig. 2A and B). Notably, as illustrated in Fig. 3C, NIT-1 cells in the siMETTL14 group exhibited significantly higher mortality compared to the siNC group, as determined by calcein-AM/PI staining (P < 0.05). Furthermore, quantitative analysis using flow cytometry demonstrated a significant increase in apoptotic NIT-1 cells in the siMETTL14 group compared to the siNC group (Fig. 3D, P < 0.05). Moreover, the levels of the anti-apoptotic protein Bcl-xL were decreased in NIT-1 cells, while the levels of the pro-apoptotic proteins Bim and cleaved caspase 3 were notably enhanced (Fig. 3E, P < 0.05).
Fig. 3.
Apoptosis of NIT-1 cells induced by METTL14 silencing. (A–B) The silencing efficiency of si-METTL14-1, 2 and 3 was tested based on qRT-PCR and WB method (n = 3). (C) Calcein AM/PI staining result of the relevant silenced NIT-1 cells, Scale bar = 50 μm. (D) Representative images of apoptotic staining NIT-1 cells, the histogram corresponds to apoptotic NIT-1 cells (n = 3). (E) The panel and histogram refer to relative levels of apoptosis-related proteins (n = 3). bP < 0.05, compared to siNC group; eP < 0.05, to si-METTL14-1 group.
3.4. Overexpression of METTL14 inhibited H2O2-Induced apoptosis
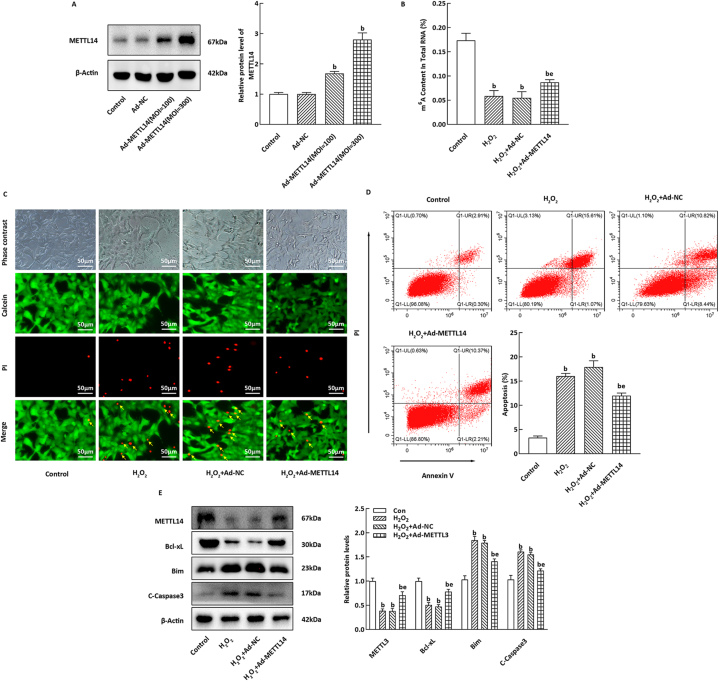
The efficiency of Ad-METTL14 overexpression was assessed using western blotting, and the optimal viral infection concentration was determined (Fig. 4A). In comparison to the Ad-NC group, METTL14 overexpression in NIT-1 cells significantly elevated the m6A methylation level induced by H2O2 (Fig. 4B, P < 0.05), while simultaneously reducing cell mortality and apoptosis (Fig. 4C and D, P < 0.05). Moreover, Ad-METTL14 infection effectively inhibited the H2O2-induced increase in pro-apoptotic protein levels of Bim in NIT-1 cells, and restored the level of Bcl-xL (Fig. 4E, P < 0.05).
Fig. 4.
Effect of METTL14 overexpression on H2O2-induced apoptosis of NIT-1 cells. (A) WB testing result of Ad-METTL14 overexpression in NIT-1 cells (n = 3). (B) m6A methylation level in total RNA was detected by ELISA (n = 3). (C) Calcein AM/PI staining result of the NIT-1 cells. (D) Representative images of apoptotic staining NIT-1 cells, the histogram refers to the apoptotic NIT-1 cells rate (n = 3). (E) The histogram represents the relative expression levels of apoptosis-related proteins (n = 3). bP < 0.05, eP < 0.05, compare to the control and H2O2 group, respectively.
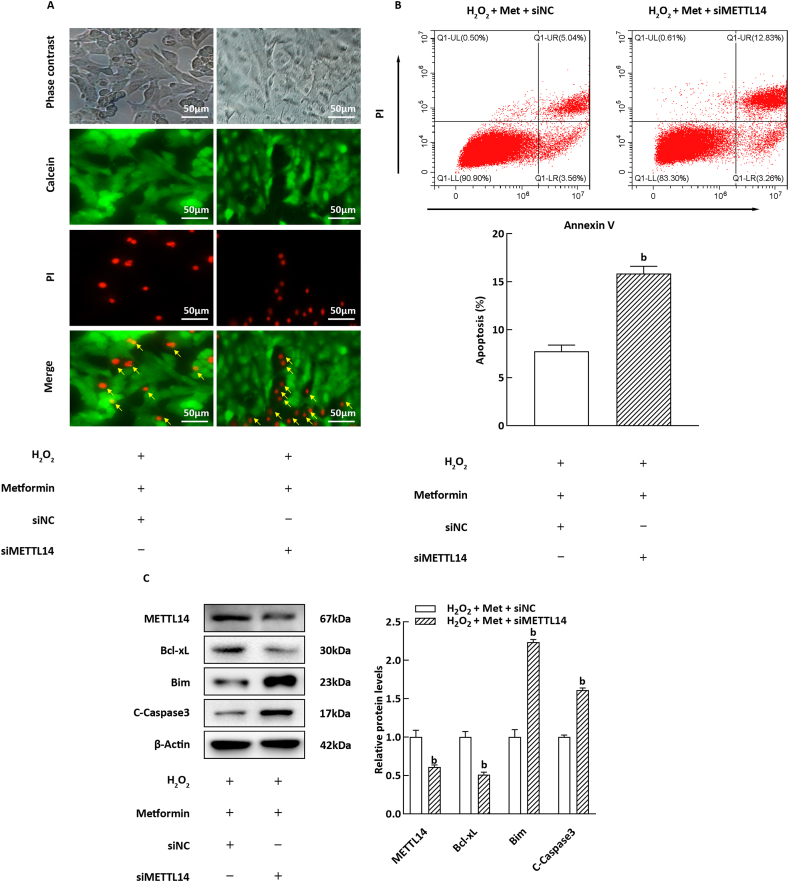
3.5. Influence of METTL14 silencing on the improvement of H2O2-Induced apoptosis by metformin
To elucidate the role of METTL14 in mediating the anti-apoptotic effects of Met, siRNA transfection was employed to inhibit the expression of METTL14 in NIT-1 cells prior to Met treatment. As depicted in Fig. 5A and B, silencing of METTL14 significantly reversed the protective effect of Met against cell death and apoptosis in NIT-1 cells exposed to H2O2 (P < 0.05). Furthermore, upon siMETTL14 transfection, the regulatory impact of Met on the expression of Bcl-xL and Bim was attenuated (Fig. 5C, P < 0.05). These findings suggest that Met enhances H2O2-induced β-cell apoptosis by upregulating METTL14 expression.
Fig. 5.
Influence of METTL14 overexpression on H2O2-induced apoptosis of NIT-1 cells. (A) Calcein AM/PI staining result of the NIT-1 cells. (B) Representative images of apoptotic staining NIT-1 cells, the histogram refers to the apoptotic NIT-1 cells rate (n = 3). (C) The histogram corresponds to the relative expression levels of apoptosis-related proteins (n = 3). bP < 0.05, compared with H2O2 + Met + siNC group.
4. Discussion
Diabetes mellitus (DM), a severe and complex chronic condition, afflicts over 536 million individuals globally [30]. The destruction of β-cells and subsequent impairment of insulin secretion play a significant role in the progression of this disease [31,32]. β-cell injury is a crucial factor contributing to the development of type 2 diabetes mellitus (T2DM) due to β-cell dysfunction, encompassing a multifactorial process. D'Addio et al. have shed light on the cyclic regulation of the IGFBP3/TMEM219 axis in β-cell expansion and function, uncovering the intricate and variable nature of T2DM occurrence and progression [7]. Mounting evidence suggests a close association between oxidative stress-induced dysfunction of pancreatic β-cells and the pathogenesis of DM [33]. Hydrogen peroxide (H2O2) has been employed as a prototype for studying oxidative damage [34]. Various cell types, particularly β-cells with limited antioxidant capacity, are susceptible to oxidative damage induced by H2O2 [8,35]. Previous investigations have demonstrated that higher doses of H2O2 expedite the loss of beta cell viability within 24 h [8]. Given the complexity of the signaling pathways and biochemical processes involved, we intend to delve into the underlying molecular mechanisms.
Metformin (Met) is a frontline hypoglycemic agent employed in clinical practice, renowned for its ability to confer protection upon pancreatic β-cells both in vivo and in vitro. It effectively mitigates β-cell apoptosis and attenuates the decline in insulin secretion induced by lipid toxicity and oxidative stress [36]. Our findings align with earlier studies [37], demonstrating that Met treatment enhances Bcl-xL expression, suppresses Bim protein levels, and ameliorates H2O2-induced apoptosis in NIT-1 cells. Elucidating the precise mechanisms underlying the cellular protective effects of Met is an intriguing avenue that warrants further exploration.
M6A modification is a prevalent form of mRNA methylation that is regulated by methyltransferase complexes and demethylases [38]. Remarkably, T2DM patients exhibit significantly reduced levels of m6A methylation in their islets and plasma, although the underlying mechanism remains unclear due to the influence of various factors [39]. The alterations in m6A methylation levels are primarily associated with decreased expression of METTL3 and METTL14 [17,18], as well as increased expression of FTO [25,26]. Furthermore, there is limited research on the role of ALKBH5 in T2DM, even though it appears to play a crucial role in anti-oxidative stress in gastric cancer cells and myocardial cell lines [40,41]. Notably, the expression of ALKBH5 in blood samples from both T2DM patients and rats does not appear to differ significantly from that of normal groups [42]. Our study indicates that ALKBH5 may have minimal impact on oxidative stress in islet β-cells. We found that the m6A methylation level of total RNA was significantly reduced in H2O2-treated NIT-1 cells, suggesting that increased oxidative stress might contribute to the decrease in m6A methylation levels in diabetic β-cells. However, further analysis revealed that the precise mechanism underlying this phenomenon remains unclear, demanding extensive experimental investigation. In order to assess whether the effect of Met on H2O2-induced apoptosis is related to m6A modification, we examined the m6A methylation content and mRNA levels of METTL3, METTL14, FTO, and ALKBH5 in this study. Interestingly, Met treatment partially restored m6A methylation levels in H2O2-exposed NIT-1 cells, potentially attributed to Met-mediated upregulation of the m6A methyltransferase METTL14 expression.
METTL14 forms a stable heterodimer with METTL3 and serves as a crucial substrate for the methyltransferase complex [43]. Liu et al. demonstrated that pancreatic β-cell-specific knockout of METTL14 in mice leads to increased islet apoptosis. Inhibition of METTL14 expression in the mouse pancreatic β-cell line MIN6 decreased Bcl-xL expression while exerting the opposite effect on Bim, ultimately resulting in enhanced apoptosis of MIN6 cells [29]. Consistent findings were observed in this study when METTL14 was silenced in NIT-1 cells, indicating that inhibition of METTL14 levels can induce β-cell apoptosis under normal culture conditions. Additionally, overexpression of METTL14 significantly reduced H2O2-induced apoptosis in NIT-1 cells and elevated m6A levels. These results strongly suggest that METTL14 indeed regulates the level of apoptosis in β-cells. Furthermore, based on our experimental findings, it can be inferred that inhibition of METTL14 expression in NIT-1 cells led to decreased expression of cleaved caspase 3 and Bim, while increasing Bcl-xL expression mediated by Met. Consequently, this weakened the anti-apoptotic effect of Met against H2O2-induced apoptosis. Overall, our experiments demonstrate that Met, a widely-used antidiabetic drug, safeguards NIT-1 cells from cellular damage and apoptosis via the regulation of m6A methylation, with METTL14 playing a pivotal role in this methylation regulation mechanism.
5. Conclusions
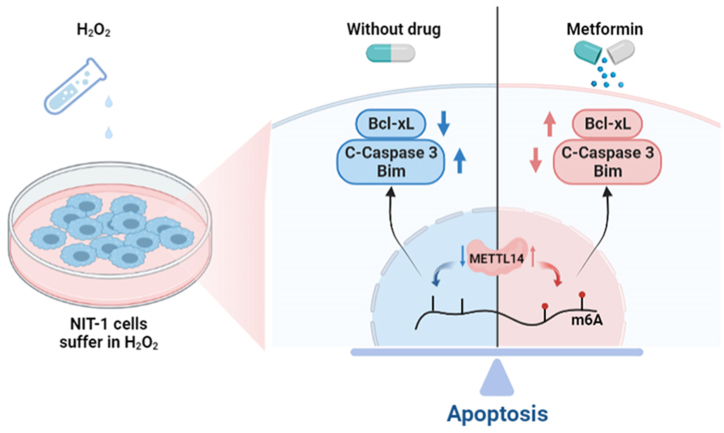
In conclusion, the results lend support to the hypothesis that m6A methylation alterations, regulated by METTL14, mediate H2O2-induced apoptosis in pancreatic β-cells. Moreover, we have demonstrated that Met enhances METTL14 expression and promotes m6A methylation, thereby attenuating H2O2-induced apoptosis in NIT-1 cells. These findings provide novel insights into the protective mechanisms of Met on pancreatic β-cells (Fig. 6). Consequently, our study delineates a potential avenue for investigating the underlying mechanisms of Met in the management of diabetes mellitus.
Fig. 6.
An overview of how Met decreases H2O2-induced oxidative stress apoptosis in vivo.
Data availability statement
No data was used for the research described in the article.
CRediT authorship contribution statement
Si-min Zhou: Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Conceptualization. Xin-ming Yao: Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Conceptualization. Yi Cheng: Resources, Methodology. Yu-jie Xing: Validation, Software. Yue Sun: Resources, Methodology, Data curation. Qiang Hua: Data curation. Shu-jun Wan: Project administration. Xiang-jian Meng: Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank all the authors for their contributions to this study.
References
- 1.Li M., Song L.J., Qin X.Y. Advances in the cellular immunological pathogenesis of type 1 diabetes. J. Cell Mol. Med. 2014;18:749–758. doi: 10.1111/jcmm.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meier J.J., Bonadonna R.C. Role of reduced beta-cell mass versus impaired beta-cell function in the pathogenesis of type 2 diabetes. Diabetes Care. 2013;36(Suppl 2):S113–S119. doi: 10.2337/dcS13-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caturano A., D'Angelo M., Mormone A., Russo V., Mollica M.P., Salvatore T., et al. Oxidative stress in type 2 diabetes: impacts from pathogenesis to lifestyle modifications. Curr. Issues Mol. Biol. 2023;45(8):6651–6666. doi: 10.3390/cimb45080420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halban P.A., Polonsky K.S., Bowden D.W., Hawkins M.A., Ling C., Mather K.J., et al. β-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care. 2014;37(6):1751–1758. doi: 10.2337/dc14-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguayo-Mazzucato C., Zavacki A.M., Marinelarena A., Hollister-Lock J., El Khattabi I., Marsili A., Weir G.C., et al. Thyroid hormone promotes postnatal rat pancreatic β-cell development and glucose-responsive insulin secretion through MAFA. Diabetes. 2013;62(5):1569–1580. doi: 10.2337/db12-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandrup-Poulsen T. beta-cell apoptosis: stimuli and signaling. Diabetes. 2001;50(Suppl 1):S58–S63. doi: 10.2337/diabetes.50.2007.s58. [DOI] [PubMed] [Google Scholar]
- 7.D'Addio F., Maestroni A., Assi E., Ben Nasr M., Amabile G., Usuelli V., et al. The IGFBP3/TMEM219 pathway regulates beta cell homeostasis. Nat. Commun. 2022;13(1):684. doi: 10.1038/s41467-022-28360-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sampson S.R., Bucris E., Horovitz-Fried M., Parnas A., Kahana S., Abitbol G., et al. Insulin increases H2O2-induced pancreatic beta cell death. Apoptosis. 2010;15:1165–1176. doi: 10.1007/s10495-010-0517-5. [DOI] [PubMed] [Google Scholar]
- 9.Eguchi N., Vaziri N.D., Dafoe D.C., Ichii H. The role of oxidative stress in pancreatic beta cell dysfunction in diabetes. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marinho H.S., Cyrne L., Cadenas E., Antunes F. H2O2 delivery to cells: steady-state versus bolus addition. Methods Enzymol. 2013;526:159–173. doi: 10.1016/B978-0-12-405883-5.00010-7. [DOI] [PubMed] [Google Scholar]
- 11.Tabatabaei-Malazy O., Nikfar S., Larijani B., Abdollahi M. Drugs for the treatment of pediatric type 2 diabetes mellitus and related co-morbidities. Expert Opin. Pharmaco. 2016;17:2449–2460. doi: 10.1080/14656566.2016.1258057. [DOI] [PubMed] [Google Scholar]
- 12.Marshall S.M. 60 years of metformin use: a glance at the past and a look to the future. Diabetologia. 2017;60:1561–1565. doi: 10.1007/s00125-017-4343-y. [DOI] [PubMed] [Google Scholar]
- 13.Anisimov V.N. Metformin: do we finally have an anti-aging drug? Cell Cycle. 2013;12:3483–3489. doi: 10.4161/cc.26928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajaei E., Haybar H., Mowla K., Zayeri Z.D. Metformin one in a million efficient medicines for rheumatoid arthritis complications: inflammation, osteoblastogenesis, cardiovascular disease, malignancies. Curr. Rheumatol. Rev. 2019;15:116–122. doi: 10.2174/1573397114666180717145745. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X., Zhao Y., Xu J., Xue Z., Zhang M., Pang X., et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015;5 doi: 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malvandi A.M., Loretelli C., Ben Nasr M., Zuccotti G.V., Fiorina P. Sitagliptin favorably modulates immune-relevant pathways in human beta cells. Pharmacol. Res. 2019;148 doi: 10.1016/j.phrs.2019.104405. [DOI] [PubMed] [Google Scholar]
- 17.Moon J.S., Karunakaran U., Elumalai S., Lee I.K., Lee H.W., Kim Y.W., et al. Metformin prevents glucotoxicity by alleviating oxidative and ER stress-induced CD36 expression in pancreatic beta cells. J. Diabetes Complicat. 2017;31:21–30. doi: 10.1016/j.jdiacomp.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Kim H.I., Lee J.S., Kwak B.K., Hwang W.M., Kim M.J., Kim Y.B., et al. Metformin ameliorates lipotoxic beta-cell dysfunction through a concentration-dependent dual mechanism of action. Diabetes Metab. J. 2019;43:854–866. doi: 10.4093/dmj.2018.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22 doi: 10.1016/j.stem.2017.11.016. 191-205 e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Sun J., Lin Z., Zhang W., Wang S., Wang W., et al. m(6)A mRNA methylation controls functional maturation in neonatal murine beta-cells. Diabetes. 2020;69:1708–1722. doi: 10.2337/db19-0906. [DOI] [PubMed] [Google Scholar]
- 24.De Jesus D.F., Zhang Z., Kahraman S., Brown N.K., Chen M., Hu J., et al. m(6)A mRNA methylation regulates human beta-cell biology in physiological states and in type 2 diabetes. Nat. Metab. 2019;1:765–774. doi: 10.1038/s42255-019-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen F., Huang W., Huang J.T., Xiong J., Yang Y., Wu K., et al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 2015;100:E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Shen F., Huang W., Qin S., Huang J.T., Sergi C., et al. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2019;104:665–673. doi: 10.1210/jc.2018-00619. [DOI] [PubMed] [Google Scholar]
- 27.Kang M.J., Moon J.W., Lee J.O., Kim J.H., Jung E.J., Kim S.J., et al. Metformin induces muscle atrophy by transcriptional regulation of myostatin via HDAC6 and FoxO3a. J. Cachexia Sarcopenia Muscle. 2022;13:605–620. doi: 10.1002/jcsm.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou S., Sun Y., Xing Y., Wang Z., Wan S., Yao X., et al. Exenatide ameliorates hydrogen peroxide-induced pancreatic beta-cell apoptosis through regulation of METTL3-mediated m(6)A methylation. Eur. J. Pharmacol. 2022;924 doi: 10.1016/j.ejphar.2022.174960. [DOI] [PubMed] [Google Scholar]
- 29.Liu J., Luo G., Sun J., Men L., Ye H., He C., et al. METTL14 is essential for beta-cell survival and insulin secretion. Biochim. Biophys. Acta, Mol. Basis Dis. 2019;1865:2138–2148. doi: 10.1016/j.bbadis.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Sun H., Saeedi P., Karuranga S., Pinkepank M., Ogurtsova K., Duncan B.B., et al. IDF Diabetes Atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022;183 doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 32.Prentki M., Nolan C.J. Islet beta cell failure in type 2 diabetes. J. Clin. Invest. 2006;116:1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerber P.A., Rutter G.A. The role of oxidative stress and hypoxia in pancreatic beta-cell dysfunction in diabetes mellitus. Antioxidants Redox Signal. 2017;26:501–518. doi: 10.1089/ars.2016.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M.S., Chyau C.C., Wang C.P., Wang T.H., Chen J.H., Lin H.H. Flavonoids identification and pancreatic beta-cell protective effect of Lotus seedpod. Antioxidants. 2020;9 doi: 10.3390/antiox9080658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nath S., Ghosh S.K., Choudhury Y. A murine model of type 2 diabetes mellitus developed using a combination of high fat diet and multiple low doses of streptozotocin treatment mimics the metabolic characteristics of type 2 diabetes mellitus in humans. J. Pharmacol. Toxicol. Methods. 2017;84:20–30. doi: 10.1016/j.vascn.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J., Massey S., Story D., Li L. Metformin: an old drug with new applications. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19102863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung T.W., Lee M.W., Lee Y.J., Kim S.M. Metformin prevents endoplasmic reticulum stress-induced apoptosis through AMPK-PI3K-c-Jun NH2 pathway. Biochem. Biophys. Res. Commun. 2012;417:147–152. doi: 10.1016/j.bbrc.2011.11.073. [DOI] [PubMed] [Google Scholar]
- 38.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 39.Deglasse J.P., Roma L.P., Pastor-Flores D., Gilon P., Dick T.P., Jonas J.C. Glucose acutely reduces cytosolic and mitochondrial H2O2 in rat pancreatic beta cells. Antioxidants Redox Signal. 2019;30:297–313. doi: 10.1089/ars.2017.7287. [DOI] [PubMed] [Google Scholar]
- 40.Chen C., Zhai E., Liu Y., Qian Y., Zhao R., Ma Y., Liu J., Huang Z., Chen J., Cai S. ALKBH5-mediated CHAC1 depletion promotes malignant progression and decreases cisplatin-induced oxidative stress in gastric cancer. Cancer Cell Int. 2023;23(1):293. doi: 10.1186/s12935-023-03129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D., Li L., Dong S., Yu Y., Zhang L., Jiang S. ALKBH5 regulates N(6)-methyladenosine (m6A) methylation of MG53 to attenuate myocardial infarction by inhibiting apoptosis and oxidative stress. J. Cardiovasc. Pharmacol. 2023;17 doi: 10.1097/FJC.0000000000001515. [DOI] [PubMed] [Google Scholar]
- 42.Shen F., Huang W., Huang J.T., Xiong J., Yang Y., Wu K., Jia G.F., Chen J., Feng Y.Q., Yuan B.F., Liu S.M. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 2015;100(1):E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.