Abstract
Using a genomic approach, we have identified a new Salmonella pathogenicity island, SPI-4, which is the fourth Salmonella pathogenicity island to be identified. SPI-4 was located at 92 min on the chromosome map and was flanked by the ssb and soxSR loci. The DNA sequence covering the entire SPI-4 and both boundaries was determined. The size of SPI-4 was about 25 kb and it contains 18 putative open reading frames (ORFs). Three of these ORFs encode proteins that have significant homology with proteins involved in toxin secretion. Another five ORFs encode proteins that have significant homology with hypothetical proteins from Synechocystis sp. strain PCC6803 or Acinetobacter calcoaceticus. The rest of the ORFs encode novel proteins, one of which has five membrane-spanning domains. SPI-4 is likely to carry a type I secretion system involved in toxin secretion. Furthermore, a previously identified locus (ims98), which is required for intramacrophage survival, was also mapped within the SPI-4 region. These findings suggested that SPI-4 is needed for intramacrophage survival.
The genetic maps of Salmonella enterica and Escherichia coli K-12 are highly conserved (3, 42). However, several large differences in the lengths of gene intervals between the two genomes have been observed (26). Each of these large excess DNA fragments in either bacterium is called a loop (38). The estimated sizes of these loops are from 20 to 70 kb (26). These loops represent a collection of major genomic changes accumulated during the diversification of these two genera from a common ancestor since about 100 million years ago (20, 26, 35, 38). It is estimated that 16 loops are present in S. enterica and 15 are present in E. coli (37).
A large number of genes responsible for the specific biochemical characters of Salmonella spp. are mapped to these loop regions. These genes include the tct genes (57 min) for transporting citrate into the cell (47), the phoN gene (96 min) for a nonspecific acid phosphatase (20), the triMR gene (1 min) for utilization of carballylic acid (47), inlA (92 min) and inlB (55 min) for utilization of inositol (38), and fljA and fljB (56 to 57 min) for flagellar antigens and phase variation (38, 42). Furthermore, two of the three identified Salmonella pathogenic islands, SPI-1 (63 min) and SPI-3 (82 min), which represent segments of the chromosome with clusters of virulence genes, are also located at these loop regions (6, 31). Apparently, major physiological and biochemical differences between E. coli and Salmonella spp. are contributed to by these macroscopic genomic differences, i.e., loops. In this study, we have identified and sequenced a 27-kb chromosomal region covering a Salmonella loop region located at 92 min in the Salmonella enterica chromosome map. Sequence analysis indicated that the cluster of genes located at 92 min may represent a fourth Salmonella pathogenicity island, SPI-4, which is required for survival within murine macrophage. This also represents the first completely sequenced Salmonella pathogenicity island.
MATERIALS AND METHODS
Bacterial strains and lambda DASHII clones.
S. enterica RKS4699 (Dublin), R613 (Minnesota), RKS53 (Enteritidis), RKS4994 (Gallinarum), RKS4993 (Paratyphi A), SA3302 (Paratyphi B), RKS4587 (Paratyphi C), RKS5078 (Pullorum), ty21a (Typhi), and SARA2 (Typhimurium) were obtained from the Salmonella Genetic Stock Center. These strains are 10 of the most common electrophoretic types (ETs) of S. enterica serovars. Clinical isolates of Citrobacter freundii, Klebsiella aerogenes, Enterobacter cloacae, Pseudomonas aeroginosa, Serratia marcescens, and Proteus mirabilis were obtained from Alan Greener. Rhizobium meliloti and Escherichia coli K-12 were from our laboratory collection. Lambda DASHII clones were prepared as described previously (50).
Preparation of DNA from lambda DASHII clones.
Liquid lysates were used for the preparation of DNA. A total of 107 log-phase E. coli SRB(P2) bacteria were infected with lambda phage at a multiplicity of infection of 0.01 in 500 μl of adsorption buffer (10 mM CaCl2, 10 mM MgSO4). Phage adsorption was done at 37°C for 15 min, and then 10 ml of Luria broth containing 10 mM MgSO4–glucose was added. The lysate was grown overnight at 37°C with shaking. Cell debris was removed from the lysate by centrifugation at 3,000 × g. Lambda phage particles were then pelleted by ultracentrifugation with an SW40 rotor at 30,000 rpm for 30 min. Phage pellets were resuspended in 1× universal buffer and digested with DNase and RNase to remove contaminated chromosomal DNA. Lambda phage DNA was then obtained by phenol-chloroform extraction and ethanol precipitation.
PCR, molecular cloning, and nucleotide sequencing.
A long PCR subcloning strategy was developed to sequence the entire SPI-4. The 17- and 14-kb DNA inserts from lambda DASHII clones 12A5 and 980, respectively, were PCR amplified with the TaqPlus PCR system (Stratagene, La Jolla, Calif.) as described previously (49). About 20 μg of purified PCR products was obtained from each lambda clone and was sonicated with an Ultrasonic-60 sonicator (Fisher Scientific, Pittsburgh, Pa.) for 10 s with the power setting at 3. Sonicated fragments were blunt ended by digestion with mung bean nuclease (Boehringer Mannheim, Indianapolis, Ind.). Blunt-ended fragments were fractionated in low-melting-point agarose to obtain 1- to 2-kb fragments, which were then cloned into pCR-blunt (Invitrogen, Carlsbad, Calif.). ABI PRISM dye terminator cycle sequencing ready reaction kits with AmpliTaq FS were used. Sequencing reactions were analyzed with an ABI 377 sequencer (Perkin-Elmer, Foster City, Calif.).
Gel electrophoresis, probe preparation, and hybridization.
EcoRI restriction digests of DNA, extracted from 19 lambda DASHII clones mapped at 91 to 96 min (50), were resolved in a 0.8% agarose gel. The size of the gel was 20 by 24 cm with a thickness of 0.5 cm. After electrophoresis, fragments were transferred to a Duralon membrane (Stratagene) by a capillary method and were UV cross-linked. An [α-32P]dCTP-labeled probe was generated from total genomic DNA extracted from E. coli K-12 with a random priming labeling kit (Stratagene). Hybridization was performed at 55°C in 5× SSC–0.5% blocking reagent (Boehringer Mannheim)–0.1% N-lauroylsarcosine–0.02% sodium dodecyl sulfate solution overnight (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization signals were detected by exposure to Kodak XAR-5 film.
Sequence analysis.
Sequences were assembled into contigs by using AssemblyLign (Oxford Molecular Group, Campbell, Calif.). The coding potential of the SPI-4 sequence was predicted with GeneMark software (9) by using ecoli, ecophage, ecohiexp, and Salm matrices via an E-mail server or a web server (17a) at the Georgia Institute of Technology. Comparison with GenBank data was performed with blastn, blastx, and tblastx analyses through the Internet with the MacVector software package (Oxford Molecular Group) or by using the nucleic acid sequence launcher (51) at the website provided by the Human Genome Center at Baylor College of Medicine (24a). Potential tRNA gene prediction was done by using DNASIS (Hitachi Software, San Bruno, Calif.). Final analysis of amino acid homologies utilized gapped BLAST with the BLASTP2.0 algorithm (2) accessible at the National Center for Biotechnology Information website (32a). In general, homologies were considered significant if P < 0.01 and if the similarity extended over one-third of the sequence. Amino acid sequences of all putative proteins were also searched against the BLOCK database at the BLOCK WWW server (22a).
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was assigned GenBank accession no. AF060869.
RESULTS
Identification of SPI-4 at 91 to 96 min.
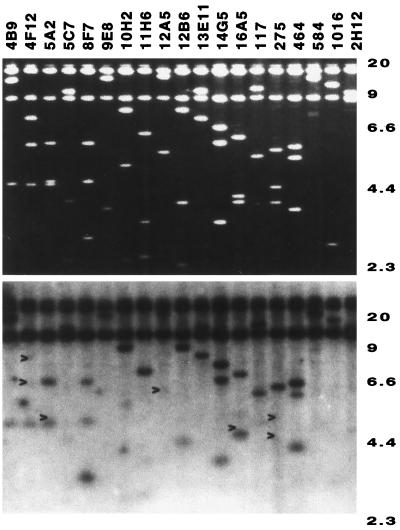
DNAs extracted from 19 lambda clones spanning 91 to 96 min on the map of the S. enterica chromosome (50) were digested with EcoRI and resolved by agarose gel electrophoresis. Figure 1 shows an ethidium-stained gel (top) and the corresponding Southern blot (bottom) probed with total genomic DNA from E. coli K-12. Several EcoRI fragments, derived from the lambda clones 4F12, 5A2, 12A5, 16A5, and 275, did not hybridize with E. coli K-12 DNA (Fig. 1). Thus, these EcoRI fragments represent S. enterica-specific DNA. Lambda clones 4F12, 5A2, and 12A5 were located at 92 min, while lambda clones 16A5 and 275 were located at 94 min. These map locations represent two previously defined loops (38). All the EcoRI fragments from these lambda clones were subcloned and partially sequenced at both ends. Partial sequences obtained from the 92-min lambda clones did not match any entries in the GenBank database, but partial sequences obtained from the 94-min lambda clones matched a previously sequenced Salmonella-specific phosphatase gene, phoN (20), and the insertion element IS200 (5) (data not shown). Moreover, by comparing the physical distances encompassed by the homologous genes of S. enterica (50) and E. coli K-12 (7) around the 91- to 96-min regions of the respective chromosomes, we were able to estimate the size of the loop at 92 min to be about 25 kb and the size of the loop at 94 min to be 8 kb. The discovery of novel sequence at the 92-min region from this initial effort prompted us to determine the complete sequence of this entire region, which is named SPI-4 (Salmonella pathogenicity island 4) in this study.
FIG. 1.
Identification of Salmonella-specific DNA fragment in 91- to 96-min region. (Top) Ethidium bromide staining of EcoRI-digested fragments resolved in a 0.7% agarose gel from 19 overlapping lambda DASHII clones covering the 91- to 96-min region. (Bottom) A Southern blot was generated from the gel shown in panel A and hybridized at low stringency with radiolabelled probe made from E. coli total genomic DNA. Arrows indicate Salmonella-specific DNA fragments that did not hybridize with E. coli total genomic DNA. Molecular sizes are shown on the right in kilobases.
SPI-4 is conserved among the 10 most common serovars of S. enterica.
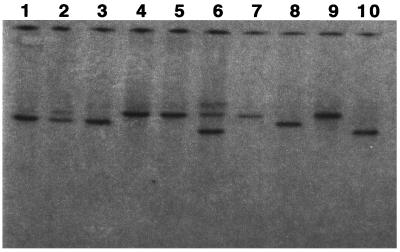
A Southern blot was generated from 10 of the most common serovars of S. enterica and 8 other bacteria as listed in Materials and Methods. A probe was prepared from a 5-kb EcoRI fragment isolated from the lambda clone 12A5 and was hybridized against this collection of bacteria. All the Salmonella strains hybridized strongly with the probe (Fig. 2) but none of the other bacteria tested gave any signal (data not shown). Moreover, a restriction length polymorphism was detected among these 10 Salmonella strains (Fig. 2). Similar results were also obtained with a probe generated from a 5-kb EcoRI fragment isolated from lambda clone 4F12. The locations of these probes within the SPI-4 are indicated in Fig. 5.
FIG. 2.
SPI-4 is present in different ETs of S. enterica serovars. A Southern blot was generated from 10 of the most common ET S. enterica serovars and hybridized with a DNA probe generated from SPI-4 (Fig. 5). Lanes: 1, Dublin; 2, Enteritidis; 3, Gallinarum; 4, Minnesota; 5, Paratyphi A; 6, Paratyphi B; 7, Paratyphi C; 8, Pullorum; 9, Typhi, 10, Typhimurium.
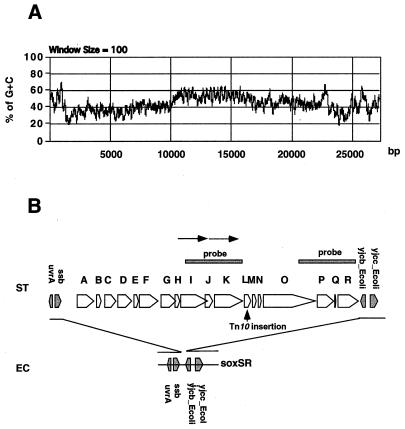
FIG. 5.
Organization and structure of SPI-4. (A) G+C percentages with a window of 100 bp across the entire 27,290-bp sequence. (B) Alignment of SPI-4 with the corresponding regions in E. coli. Coding potential was predicted by the GeneMark program. The 18 putative ORFs are represented by open arrowheads and are assigned a letter from A to R. Shaded arrowheads represent genes present in both species. The region of putative duplication (tandem arrows) and the location of a previous Tn10 insertion affecting intramacrophage survival are indicated.
Sequencing of SPI-4.
Fourteen more lambda clones that hybridized with clones 4A5 and 12A5 (which contain part of SPI-4) were identified from our genomic library, which consists of about 2,000 mapped lambda clones (50). After preliminary sequencing of the ends of the DNA inserts from these lambda clones, two lambda clones that have minimal overlap but which cover the entire SPI-4 region were chosen for complete sequencing. DNA inserts from these two lambda clones were PCR amplified, and the PCR fragments were sonicated to obtain 1- to 2-kb fragments for subcloning into pCR-blunt for sequencing. A total of 90 kb of overlapping sequences was derived from 39 plasmids (subcloned from lambda clone 980), 41 plasmids (subcloned from lambda clone 12A5), and the region adjacent to the vector in 16 lambda clones. This 90-kb total sequence was assembled into five contigs. Three of the sequencing gaps proved to be less than 500 bp and were sequenced by primer walking. One of the gaps was 3.3 kb and was PCR amplified from the chromosome and sequenced by a novel partial digestion method to generate nested deletion clones (48). Finally, a single contig of 27,290 bp, which covers the entire SPI-4 and the adjacent regions, was obtained.
Analysis of the boundaries of the SPI-4.
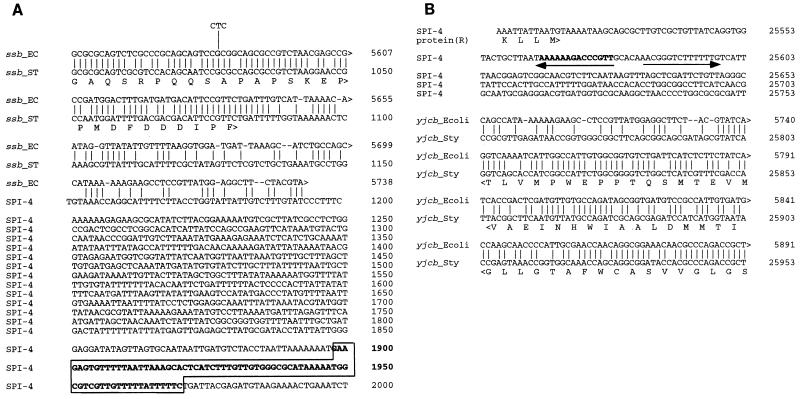
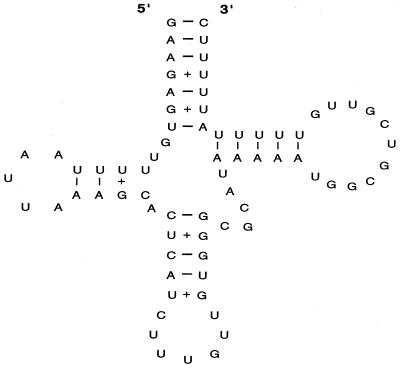
The boundaries of the SPI-4 were identified by aligning the sequence obtained with that of the corresponding regions from the whole E. coli genomic sequence (7). It was found that SPI-4 was located between ssb and yjcb_Ecoli, and the alignment of these boundaries is shown in Fig. 3. The DNA base composition changes dramatically at both boundaries. At the ssb boundary, the average G+C content changes from 52 to 37%. The homology between E. coli and S. enterica ends directly after the stop codon of the ssb coding sequence, which is followed by a 1,119-bp noncoding region (G+C content, ∼30%) before the first codon of the putative open reading frame (ORF) _A. This noncoding region contains a segment of DNA (highlighted in Fig. 3A) which carries a putative tRNA gene sequence as predicted by DNASIS software. The predicted tRNA structure is shown in Fig. 4. At the yjcb_Ecoli boundary, the average G+C content changed from 52 to 44%. A strong terminator structure is present after the last putative ORF_R (Fig. 3B). Further analysis of both boundaries did not reveal sequence repeats of significant length, IS elements, or phage attachment sites.
FIG. 3.
Nucleotide sequence alignments at the boundaries of SPI-4. The DNA sequences of the ssb boundary (A) and the yjcb_Ecoli boundary (B) of SPI-4 (ST) are aligned with the corresponding sequences of the E. coli chromosomal region (EC). Alignments were performed with the MacVector program. The numbering of the nucleotides of SPI-4 corresponds to the 27,290-bp DNA sequence determined in this study. A DNA sequence that forms a tRNA-like structure is boxed and shown in boldface letters in panel A. The hairpin structure is indicated by a pair of inverted arrows in panel B. The numbering of the E. coli sequences is from the published sequence (accession no. ECAE000479). Single-letter symbols are used for the amino acid sequences of Ssb and Yjcb_Ecoli and protein R shown beneath the nucleotide sequences. yjcb_Sty, a hypothetical 13-kDa protein.
FIG. 4.
tRNA structure at the ssb junction. The tRNA structure was predicted by using the DNASIS program. Regular hydrogen bonding for A-U and G-C is represented by a dash and weaker hydrogen bonding for G-U is represented by a plus sign.
Coding potential and homology search of SPI-4 against sequences in database.
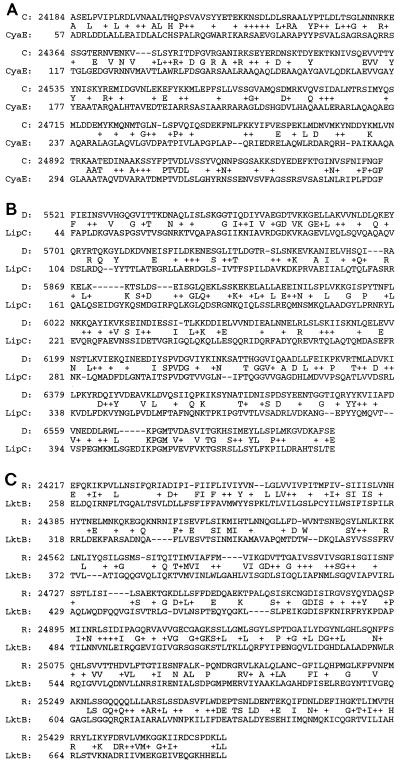
The coding potential of SPI-4 was analyzed with GeneMark software. A total of 18 ORFs were predicted. All these ORFs were transcribed in the same orientation (Fig. 5). Initial similarity searches were performed with the blastn and blastx algorithms via the Internet with the MacVector software package. The final identity and similarity percentages of these 18 putative proteins (A to R) to similar protein sequences were calculated by gapped BLAST using the blastp2.0 algorithm accessible at the NCBI website (32a) or the BCM website (24a). Furthermore, the amino acid sequences of these 18 putative proteins were searched against the BLOCK 10.0 database at the BLOCK WWW server (22a). The results of these analyses are summarized in Table 1. Protein C has significant homology with members of the ABC transporter outer membrane protein family such as CyaE, protein D has significant homology with members of the HlyD secretion protein family such as LipC, and protein R has significant homology with members of the ABC transporter protein family with ATP or GTP binding site motif A such as the LktB toxin secretion proteins (Fig. 6). CyaE is an outer membrane protein involved in the secretion of a cadcalmodulin-sensitive adenylate cyclase toxin, CyaA (related to the RTX family of pore-forming toxins), from Bordetella pertussis (21, 30). LipC is a membrane fusion protein responsible for the secretion of a lipase, LipA, in Serratia marcescens 8000 (1). The amino acid sequence of LipC shows a large degree of sequence similarity with HlyD, which is involved in the secretion of hemolysin in pathogenic E. coli (30). LktB is an ATP-binding protein involved in the secretion of leukotoxin from Pasteurella haemolytica (44). Proteins F, G, I, K, and O have significant homologies with part of a 308.8-kDa hypothetical protein from Synechocystis sp. strain PCC6803 (D63999) and part of a 93.4-kDa hypothetical protein from Acinetobacter calcoaceticus (accession no. AF011339) (Table 1). The rest of the putative proteins are novel proteins. Prediction of the transmembrane regions by Tmpred software at the ISREC website (24b) indicated that proteins A, C, D, E, G, O, and R have one or more transmembrane regions (Table 1). Moreover, a 223-bp sequence derived from a Tn10 insertion at a previously identified locus (ims98), which is needed for intramacrophage survival (4), is within SPI-4. The location of the Tn10 insertion at the ims96 locus is indicated in Fig. 5.
TABLE 1.
Summary of sequence analysis of putative proteins encoded by SPI-4
| Protein | No. of amino acids (predicted mass [kDa]) | Characteristics |
|---|---|---|
| A | 524 (58) | Has five transmembrane helices with one at the N terminus |
| B | 121 (14) | Unknown |
| C | 439 (51) | Shares significant homology with aggA gene product (22% identity, 45% similarity), hasF gene product (18% identity, 45% similarity), prtF gene product (21% identity, 43% similarity), and cyaE gene product (19% similarity, 43% similarity); has one transmembrane helix at N terminus; belongs to ABC exporter outer membrane component family |
| D | 425 (48) | Shares significant homology with lipC gene product (24% identity, 46% similarity), prtE gene product (25% identity, 45% similarity), hlyD gene product (23% identity, 47% similarity), and lktD gene product (21% identity, 43% similarity); has one transmembrane helix at N terminus; belongs to HlyD family of secretion proteins with periplasmic domains |
| E | 130 (14) | Has one transmembrane helix at N terminus |
| F | 526 (56) | Shares significant homology with a 93.4-kDa hypothetical protein of A. calcoaceticus (27% identity and 45% similarity over a stretch of 444 amino acids) |
| G | 431 (46) | Shares significant homology with a 93.4-kDa hypothetical protein of A. calcoaceticus (27% identity and 43% similarity over a stretch of 396 amino acids); has one transmembrane helix |
| H | 116 (13) | Unknown |
| I | 754 (81) | Shares significant homology with part of a hypothetical 308.8-kDa protein of Synechocystis sp. strain PCC6803 (31% identity and 44% similarity over a stretch of 724 amino acids) |
| J | 172 (19) | Unknown |
| K | 820 (88) | Shares significant homology with part of a hypothetical 308.8-kDa protein of Synechocystis sp. strain PCC6803 (32% identity and 46% similarity over a stretch of 780 amino acids) |
| L | 206 (22) | ORF_M was inserted by Tn10 into strain MST2097, which is unable to survive within macrophage |
| M | 123 (13) | Unknown |
| N | 92 (9.6) | Unknown |
| O | 1,512 (162) | Shares significant homology with part of a hypothetical 308.8-kDa protein of Synechocystis sp. strain PCC6803 (25% identity and 42% similarity over a stretch of 1,327 amino acids); has three transmembrane helices with one at the C terminus |
| P | 463 (49) | Unknown |
| Q | 38 (4.5) | Unknown |
| R | 598 (67) | Shares significant homology with lktB gene product (28% identity, 51% similarity), cylB gene product (27% identity, 52% similarity), hlyB gene product (27% identity, 50% similarity), apxIB gene product (26% identity, 50% similarity); has five transmembrane helices (N terminus outside); belongs to ABC transporter family with ATP-GTP-binding site motif A |
FIG. 6.
Alignment of deduced amino acid sequences of proteins C, D, and R of SPI-4 with published amino acid sequences for CyaE (accession no. P11092) (A), LipC (accession no. D49826) (B), and LktB (accession no. P16532) (C). The numbering of the SPI-4-encoded proteins corresponds to the 27,290-bp DNA sequence determined in this study, while the numbering of the amino acid sequences of CyaE, LipC, and LktB is from published data. Conservative substitutions are indicated by plus signs.
DISCUSSION
Pathogenicity islands, which is a term used to describe a large cluster of virulence genes in the bacterial chromosome (19, 27), have been identified in many different bacterial pathogens such as uropathogenic E. coli, enteropathogenic E. coli, Helicobacter pylori, Yersinia enterocolitica, Shigella flexneri, and Salmonella species (6, 8, 11, 12, 16, 34, 36). However, only one or two pathogenicity islands have been identified so far in any one of these bacteria. Even from the completed genomic sequence of H. pylori, only one pathogenicity island has been identified (17). The identification of a fourth SPI in this study further substantiates Salmonella’s elaborate systems for interacting with its infected host. This is probably the result of a long-term interaction with infected hosts since the divergence of Salmonella from E. coli 100 million years ago (35). It has been suggested that the evolution of Salmonella’s pathogenicity is through the acquiring of several pathogenicity islands by lateral transfer. Specific virulence factors are encoded by each SPI and are required for its complex life cycle within the infected host (18).
Using a set of mapped lambda clones from S. enterica (50) for hybridization with E. coli total genomic DNA, we were able to show that unique Salmonella DNA fragments (Fig. 1) were located at the previously defined loops at 92 min and 94 min (37). The loop at 92 min was found to contain a new Salmonella pathogenicity island (SPI-4). SPI-4 is flanked by the gene ssb (encodes the single-stranded DNA binding protein) and the gene yjcb_Ecoli (encodes a hypothetical protein of 13 kDa that is upstream of the superoxide regulatory gene soxSR) at 92 min on the chromosome map of S. enterica (Fig. 5). SPI-4 has a mosaic G+C content as shown in Fig. 5A. A 9-kb region (37% G+C) is followed by a 7-kb region (54% G+C) and then by a 9-kb region (44% G+C). The average G+C content of Salmonella is about 52 to 54%. Thus, the lower G+C contents of the two 9-kb regions suggest that they are likely to have been acquired by lateral transfer. On the other hand, the 7-kb fragment appears to comprise two 3.5-kb similar fragments (71% sequence identity) (Fig. 5). This may be the result of a duplication insertion since the first 3.5-kb region is flanked by a direct repeat of 567 bp at both ends. A 5-kb DNA probe, derived from this 7-kb region, was found to hybridize to a Southern blot generated from the 10 most common ETs of S. enterica serovars (Fig. 2). A similar result was obtained with a 5-kb probe derived from the 9-kb (44% G+C) region (data not shown). The locations of the probes within SPI-4 are indicated in Fig. 5. Thus, both the low G+C and the regular G+C regions within SPI-4 are present in the S. enterica tested. However, further analysis of the distribution and the organization of the mosaic structure of SPI-4 among the phylogenetically defined eight subspecies of S. enterica (10, 33) and related bacteria is needed to better understand the origin and evolution of SPI-4.
The SPI-4 encodes 18 putative proteins as predicted by GeneMark analysis. The letters A to R were assigned to each of the putative ORFs (Fig. 5). Apparently, the ORFs are organized as a single operon and a strong hairpin transcription terminator structure is present after the last ORF, ORF_R. Interestingly, 153 bp upstream from the first codon of ORF_A is a DNA region with a high potential to encode a tRNA-like gene as predicted by DNASIS software (Fig. 4). Pathogenicity islands have often been found to be located next to tRNA genes (19, 22). SPI-2 is adjacent to the tRNA val locus (23) and SPI-3 is adjacent to the selC tRNA locus (6). Since a few phage have been found to use tRNA as a site for integration (19), similar mechanisms may have operated for the acquisition of SPI-4 into Salmonella through specialized transduction. Moreover, it has been previously demonstrated that tRNA genes influence pathogenicity island gene expression in uropathogenic E. coli through specific transcriptional or translational control mechanisms (39, 45). The presence of a putative tRNA-like gene may also be related to the regulation of SPI-4 gene expression. Besides the putative tRNA structure and transcription terminator structure detected at the boundaries of SPI-4, no sequence repeats of significant length, IS elements, or phage attachment sites were found. Similar results have been obtained in the analysis of the boundaries at SPI-1, SPI-2, and SPI-3 (23, 31, 34). This might suggest that SPIs are relatively stable in the contemporary Salmonella spp. and that mutational changes accumulated over a long period of time have obscured the original insertion sites.
The presence of membrane-spanning domains in at least seven of the putative proteins and the high similarities of some of these putative proteins to known toxin secretion proteins, such as CyaE, LipC, and LktB (Fig. 6), suggest that SPI-4 contains a secretory system. In fact, we have found that a Salmonella strain, MS2097 (4), which was found to have a Tn10 insertion in the ORF_L of SPI-4, is defective in the secretion of a large extracellular protein (data not shown). However, ORF_L neither has the capacity to encode this secreted protein nor can it encode a protein with putative transmembrane domains. Since Tn10 insertion may have a polar effect on the expression of all the genes downstream from ORF_L, expression of ORF_M to ORF_R may be affected. In regard to the size of the secreted protein, ORF_O may have the capacity to encode the secreted protein. However, further analysis is required to prove this. We speculate that a type I secretory mechanism (41) is thus likely to be used by SPI-4 because CyaE, LipC, and LktB proteins are all involved in a type I secretory system for the secretion of toxins belonging to the RTX toxin family (46). This is in contrast with SPI-1 and SPI-2 (15, 34), which use a type III secretion mechanism (41).
Several toxins isolated from different pathogenic bacteria have been shown to induce apoptosis in various immune cells (14). These toxins include the leukotoxin isolated from P. haemolytica (43) and adenylate cyclase-hemolysin isolated from B. pertussis (25), whose secretory proteins share significant percentages of identity with proteins encoded by SPI-4 (Fig. 6). It has been shown that Salmonella can induce apoptosis of infected macrophages (13, 32), and the identification of cytotoxins in Salmonella species has also been reported (28, 29, 40). Thus, it is tempting to speculate that a similar mechanism of induction of apoptosis by cytotoxins may be shared by these bacteria and that SPI-4 might be involved in the secretion of a cytotoxin in Salmonella species. The inability of strain MS2097, which has a Tn10 insertion within SPI-4 (Fig. 5), to grow within macrophages may be due to a defective system for cytotoxin secretion. However, further genetic and biochemical studies are required to dissect the functions of SPI-4.
Interestingly, all the SPIs identified so far are directly or indirectly involved in intramacrophage survival or macrophage cytotoxicity. Although SPI-1 contributes mainly to the ability to invade epithelial cells, mutations in several genes within SPI-1 render the bacteria noncytotoxic to macrophages (13, 32). Similarly to SPI-1, SPI-2 contributes to another type III secretion system, and mutations in SPI-2 render the bacteria less cytotoxic to cultured macrophages and less invasive to HEp-2 cells (24). It has also been suggested that SPI-2 is required for intramacrophage survival (34), although it is still a controversial issue (24). SPI-3 is required for intramacrophage survival and contributes to the ability to survive in a Mg2+-limiting environment (6). Here, we have identified a new Salmonella pathogenicity island (SPI-4), and the results of our analysis suggest that SPI-4 is required for intramacrophage survival. Apparently, the ability to surmount the host immune system is the driving force for the acquisition of pathogenicity islands in Salmonella. The functions of individual genes within SPI-4 are currently being investigated.
ACKNOWLEDGMENTS
We thank Susan Varnum and Rita Cheng for their helpful comments on the manuscript.
Nucleotide sequencing, sequencing methodology development, and sequence analysis were done in Wong’s lab and were supported by the Battelle Memorial Institute. Lambda library production, sorting of the initial lambda clones spanning this region, and identification of the existence and location of this loop were done in McClelland’s lab and were supported by grant R01 AI34829 from the NIH.
REFERENCES
- 1.Akatsuka H, Binet R, Kawai E, Wandersman C, Omori K. Lipase secretion by bacterial hybrid ATP-binding cassette exporters: molecular recognition of the LipBCD, PrtDEF, and HasDEF exporters. J Bacteriol. 1997;179:4754–4760. doi: 10.1128/jb.179.15.4754-4760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped Blast and Psi-Blast—a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann B J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisercic M, Ochman H. The ancestry of insertion sequences common to Escherichia coli and Salmonella typhimurium. J Bacteriol. 1993;175:7863–7868. doi: 10.1128/jb.175.24.7863-7868.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc-Potard A B, Groisman E A. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Colladovides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Blum G, Falbo V, Caprioli A, Hacker J. Gene clusters encoding the cytotoxic necrotizing factor type 1, prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. 1995;126:189–195. doi: 10.1111/j.1574-6968.1995.tb07415.x. [DOI] [PubMed] [Google Scholar]
- 9.Borodovsky M, McIninch J D. GeneMark: parallel gene recognition for both DNA strands. Comput Chem. 1993;17:123–133. [Google Scholar]
- 10.Boyd E F, Li J, Ochman H, Selander R K. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J Bacteriol. 1997;179:1985–1991. doi: 10.1128/jb.179.6.1985-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal high-pathogenicity island in biotype 1b Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini S, Lange C, Xiang Z Y, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L M, Kaniga K, Galan J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Zychlinsky A. Apoptosis induced by bacterial pathogens. Microb Pathog. 1994;17:203–212. doi: 10.1006/mpat.1994.1066. [DOI] [PubMed] [Google Scholar]
- 15.Collazo C M, Galan J E. The invasion-associated type-III protein secretion system in Salmonella. Gene. 1997;192:51–59. doi: 10.1016/s0378-1119(96)00825-6. [DOI] [PubMed] [Google Scholar]
- 16.Donnenberg M S, Lai L C, Taylor K A. The locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli encodes secretion functions and remnants of transposons at its extreme right end. Gene. 1997;184:107–114. doi: 10.1016/s0378-1119(96)00581-1. [DOI] [PubMed] [Google Scholar]
- 17.Figura N. Identifiable Helicobacter pylori strains or factors important in the development of duodenal ulcer disease. Helicobacter. 1997;2:S3–S12. doi: 10.1111/j.1523-5378.1997.06b06.x. [DOI] [PubMed] [Google Scholar]
- 17a.GeneMark.http://genemark.biology.gatech.edu/GeneMark.
- 18.Groisman E A, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–9. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 19.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–4. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 20.Groisman E A, Saier M H, Jr, Ochman H. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 1992;11:1309–1316. doi: 10.1002/j.1460-2075.1992.tb05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross R. Domain structure of the outer membrane transporter protein CyaE of Bordetella pertussis. Mol Microbiol. 1995;17:1219–1220. doi: 10.1111/j.1365-2958.1995.mmi_17061219_3.x. [DOI] [PubMed] [Google Scholar]
- 22.Hacker J, Blumoehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria—structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 22a.Henikoff, J., S. Pietrokovski, and S. Henikoff. BLOCKS WWW server. http://www.blocks.fhcrc.org. [DOI] [PubMed]
- 23.Hensel M, Shea J E, Baumler A J, Gleeson C, Blattner F, Holden D W. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hensel M, Shea J E, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden D W. Functional analysis of ssaj and the ssak/u operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 24a.Human Genome Center (Baylor College of Medicine).http://gc.bcm.tmc.edu:8088/search-launcher/launcher.html.
- 24b.ISREC.http://ULREC3.unil.ch/software.
- 25.Khelef N, Zychlinsky A, Guiso N. Bordetella pertussis induces apoptosis in macrophages: role of adenylate cyclase-hemolysin. Infect Immun. 1993;61:4064–4071. doi: 10.1128/iai.61.10.4064-4071.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C A. Pathogenicity islands and the evolution of bacterial pathogens. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 28.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe N, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik P, Sharma V D, Thapliyal D C. Partial purification and characterization of Salmonella cytotoxin. Vet Microbiol. 1996;49:9–11. doi: 10.1016/0378-1135(95)00151-4. [DOI] [PubMed] [Google Scholar]
- 30.Masure H R, Au D C, Gross M K, Donovan M G, Storm D R. Secretion of the Bordetella pertussis adenylate cyclase from Escherichia coli containing the hemolysin operon. Biochemistry. 1990;29:140–145. doi: 10.1021/bi00453a017. [DOI] [PubMed] [Google Scholar]
- 31.Mills D M, Bajaj V, Lee C A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 32.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.National Center for Biotechnology Information.http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast.
- 33.Ochman H, Groisman E A. Distribution of pathogenicity islands in Salmonella spp. Infect Immun. 1996;64:5410–5412. doi: 10.1128/iai.64.12.5410-5412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman H, Wilson A C. Evolutionary history of enteric bacteria. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1649–1654. [Google Scholar]
- 36.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley M, Krawiec S. Genome organization. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 967–981. [Google Scholar]
- 38.Riley M, Sanderson K E. Comparative genetics of Escherichia coli and Salmonella typhimurium. In: Drlica K, Riley M, editors. The bacterial chromosome. Washington, D.C: American Society for Microbiology; 1990. pp. 85–95. [Google Scholar]
- 39.Ritter A, Blum G, Emody L, Kerenyi M, Bock A, Neuhierl B, Rabsch W, Scheutz F, Hacker J. tRNA genes and pathogenicity islands—influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 40.Rumeu M T, Suarez M A, Morales S, Rotger R. Enterotoxin and cytotoxin production by Salmonella enteritidis strains isolated from gastroenteritis outbreaks. J Appl Microbiol. 1997;82:19–31. doi: 10.1111/j.1365-2672.1997.tb03293.x. [DOI] [PubMed] [Google Scholar]
- 41.Salmond G P, Reeves P J. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 42.Sanderson K E, Hessel A, Rudd K E. Genetic map of Salmonella typhimurium, edition VIII. Microbiol Rev. 1995;59:241–303. doi: 10.1128/mr.59.2.241-303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens P K, Czuprynski C J. Pasteurella haemolytica leukotoxin induces bovine leukocytes to undergo morphologic changes consistent with apoptosis in vitro. Infect Immun. 1996;64:2687–2694. doi: 10.1128/iai.64.7.2687-2694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strathdee C A, Lo R Y. Cloning, nucleotide sequence, and characterization of genes encoding the secretion function of the Pasteurella haemolytica leukotoxin determinant. J Bacteriol. 1989;171:916–928. doi: 10.1128/jb.171.2.916-928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Susa M, Kreft B, Wasenauer G, Ritter A, Hacker J, Marre R. Influence of cloned tRNA genes from a uropathogenic Escherichia coli strain on adherence to primary human renal tubular epithelial cells and nephropathogenicity in rats. Infect Immun. 1996;64:5390–5394. doi: 10.1128/iai.64.12.5390-5394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welch R A. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5:521–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 47.Widenhorn K A, Boos W, Somers J M, Kay W W. Cloning and properties of the Salmonella typhimurium tricarboxylate transport operon in Escherichia coli. J Bacteriol. 1988;170:883–888. doi: 10.1128/jb.170.2.883-888.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong K K, Markillie L M, Saffer J D. A novel method for producing partial restriction digestion of DNA fragments by PCR with 5-methyl-CTP. Nucleic Acids Res. 1997;25:4169–4171. doi: 10.1093/nar/25.20.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong K K, Stillwell L C, Dockery C A, Saffer J D. Use of tagged random hexamer amplification (TRHA) to clone and sequence minute quantities of DNA—application to a 180 kb plasmid isolated from Sphingomonas F199. Nucleic Acids Res. 1996;24:3778–3783. doi: 10.1093/nar/24.19.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong K K, Wong R M, Rudd K E, McClelland M. High-resolution restriction map for a 240-kilobase region spanning 91 to 96 minutes on the Salmonella typhimurium LT2 chromosome. J Bacteriol. 1994;176:5729–5734. doi: 10.1128/jb.176.18.5729-5734.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Worley K C, Wiese B A, Smith R F. BEAUTY: an enhanced BLAST-based search tool that integrates multiple biological information resources into sequence similarity search results. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]