Abstract
Brain mural cells regulate development and function of the blood-brain barrier and control blood flow. Existing in vitro models of human brain mural cells have low expression of key mural cell genes, including NOTCH3. Thus, we asked whether activation of Notch3 signaling in hPSC-derived neural crest could direct the differentiation of brain mural cells with an improved transcriptional profile. Overexpression of the Notch3 intracellular domain (N3ICD) induced expression of mural cell markers PDGFRβ, TBX2, FOXS1, KCNJ8, SLC6A12, and endogenous Notch3. The resulting N3ICD-derived brain mural cells produced extracellular matrix, self-assembled with endothelial cells, and had functional KATP channels. ChIP-seq revealed that Notch3 serves as a direct input to relatively few genes in the context of this differentiation process. Our work demonstrates that activation of Notch3 signaling is sufficient to direct the differentiation of neural crest to mural cells and establishes a developmentally relevant differentiation protocol.
Activation of Notch3 signaling in stem cell–derived neural crest is sufficient to yield brain mural cells.
INTRODUCTION
Vascular mural cells, which encompass microvessel-associated pericytes and large vessel–associated vascular smooth muscle cells (VSMCs), regulate blood vessel development, stability, and vascular tone (1, 2). In the brain, mural cells regulate resting cerebral blood flow, neurovascular coupling, blood-brain barrier (BBB) development and maintenance, and neuronal survival (3–15). Several pericyte and VSMC subtypes have been identified in vivo based on morphology, location on the vascular tree, marker expression, and single-cell transcriptome profiles, and their relative contributions to the aforementioned mural cell functions remain the subject of active investigation (8, 16–18). Brain mural cells are also implicated in the pathogenesis of neurological disorders including Alzheimer’s disease (19–21) and cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) (22, 23).
In contrast to the mesodermal origin of most mural cells, those in the face and forebrain are derived from the neural crest (24–27). During embryogenesis, cranial neural crest–derived mesenchyme surrounds the anterior neural tube, and mural cells are specified and invade the developing prosencephalon alongside mesoderm-derived endothelial cells from the perineural vascular plexus (13, 28). This neural crest–derived mesenchyme also forms portions of the meninges, facial cartilage and connective tissue, and skull (27, 29). Molecular signals controlling specification of neural crest–derived mesenchyme to these diverse fates are poorly understood. In vivo loss-of-function experiments have suggested roles for transforming growth factor-β (TGF-β), platelet-derived growth factor (PDGF), and Notch signaling in this process (30–34). Notably, NOTCH3 mutations form the genetic basis of CADASIL (22, 23), and loss-of-function studies in zebrafish have implicated Notch signaling in brain pericyte proliferation (33) and specification of pericytes from naïve mesenchyme (32). Further, global Notch3 knockout mice have reduced arterial VSMC coverage, which correlates with deficits in BBB function (35). Similarly, mice lacking mural cell expression of RBPJ, the DNA binding protein that forms a complex with Notch intracellular domains (NICDs) to regulate transcription, exhibit arteriovenous malformations and defects in pericyte coverage (36). In vitro gain-of-function studies may provide complementary insight into the effects of molecular factors on neural crest differentiation to mural cells (37).
Human pluripotent stem cells (hPSCs) are an in vitro model system well suited for such developmental studies, as they can generate multipotent neural crest cells (38, 39) and potentially account for species differences in mural cell phenotype (40, 41). Further, in contrast to primary cell cultures, hPSCs permit longitudinal analysis of cell fate determination and differentiation. The resulting mural cells could be further used in diverse in vitro modeling applications (42). We recently demonstrated that brain pericyte–like cells could be differentiated from hPSC-derived neural crest via treatment with serum-supplemented E6 medium (43, 44). Other protocols for generating hPSC-derived brain pericyte–like cells via a neural crest intermediate use PDGF-BB and/or fibroblast growth factor 2 (FGF2) in serum-containing medium (45, 46). The sufficiency of serum to cause differentiation of neural crest cells makes interrogation of specific molecular factors difficult, motivating development of a serum-free differentiation scheme. Furthermore, existing hPSC-derived pericyte-like cells lack expression of several key brain mural cell genes and have aberrant expression of some fibroblast-associated genes, features also observed in cultured primary brain pericytes (41). This further motivates development of a new method for differentiation of hPSCs to cells having an improved molecular phenotype. Additional desired features of hPSC-derived brain mural cells include organotypic, central nervous system (CNS)–specific gene expression (e.g., ZIC1) (41), and species–specific attributes observed in vivo (e.g., expression of FN1 and lack of VTN in human brain mural cells) (40, 41).
In this work, we found that expression levels of NOTCH3 and canonical transcriptional targets of Notch signaling were very low in existing hPSC–derived brain pericyte-like cells compared to human brain pericytes in vivo. We therefore tested the hypothesis that activation of Notch3 signaling would direct the differentiation of hPSC-derived neural crest to brain mural cells. We activated Notch signaling by lentiviral overexpression of the human Notch3 intracellular domain (N3ICD) in neural crest cells maintained in serum-free medium. The resulting cells were PDGFRβ+ and displayed robust up-regulation of mural cell markers including Tbx2, HEYL, RGS5, FOXS1, KCNJ8, ABCC9, and endogenous Notch3 (16, 47). The resulting N3ICD-derived mural cells produced extracellular matrix (ECM) that supported endothelial cord formation, self-assembled into compact aggregates with endothelial cells, and had functional adenosine triphosphate–sensitive potassium channel (KATP) channels. Last, we used chromatin immunoprecipitation sequencing (ChIP-seq) to identify the direct transcriptional targets of N3ICD and define the first level of the Notch3-regulated mural cell gene regulatory network during differentiation of neural crest to mural cells. Overall, our work suggests that Notch3 signaling is sufficient to direct differentiation of neural crest to brain mural cells, and establishes a new, serum-free protocol for generation of brain mural cells from hPSCs.
RESULTS
Transcriptome analysis of hPSC-derived brain pericyte–like cells
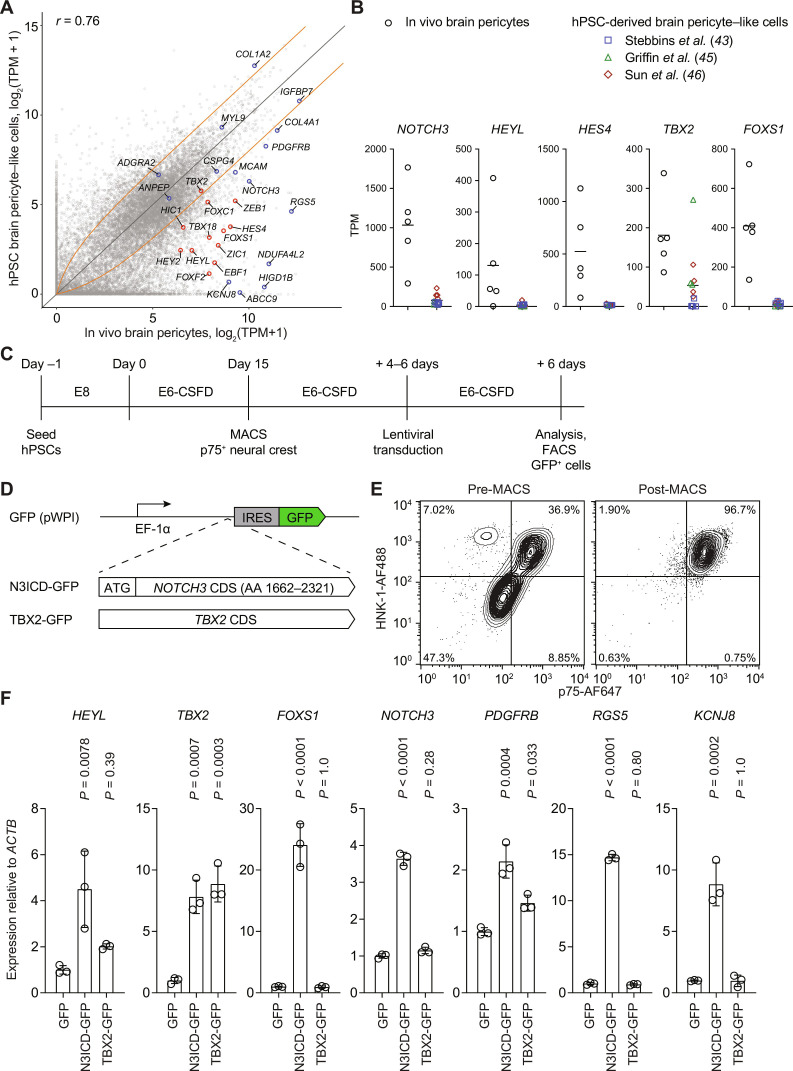
We analyzed RNA-sequencing (RNA-seq) gene expression profiles of brain pericyte–like cells differentiated from hPSCs from three independent studies (table S1) (43, 45, 46). While methodologies differ slightly, all three protocols proceed through a neural crest intermediate and yield cells with molecular and functional characteristics similar to cultured primary brain pericytes. We compared these cells to acutely isolated human brain pericytes, using single-cell/nucleus RNA-seq data from a previous meta-analysis (41). There was modest correlation between the transcriptome of hPSC-derived pericyte-like cells and in vivo pericytes, with similar expression of some canonical markers such as PDGFRB, ANPEP, CSPG4, COL4A1, IGFBP7, and MYL9 (Fig. 1A and file S1). Compared to in vivo pericytes, however, hPSC-derived pericyte-like cells had markedly lower expression of several signaling mediators and transcription factors, including RGS5, NOTCH3, HEYL, HEY2, HES4, TBX2, FOXS1, and FOXF2 (Fig. 1, A and B, and file S1). Critically, many of these genes are expressed not only in pericytes but also in VSMCs (16, 41) and are enriched in these brain mural cells compared to both neural crest and other mesenchymal derivatives (fig. S1A) (48). Furthermore, many of these same genes have reduced expression in cultured primary brain pericytes (fig. S1B) (41). These results therefore suggest that key molecular factors for induction and maintenance of the brain mural cell phenotype are absent under traditional culture conditions, and that augmentation of these factors during hPSC differentiation might yield mural cells with improved phenotype.
Fig. 1. Overview of differentiation strategy.
(A) Comparison of protein-coding transcript abundances in hPSC-derived brain pericyte–like cells versus in vivo human brain pericytes. Data for hPSC-derived brain pericyte–like cells were generated by averaging transcripts per million (TPM) across 11 bulk RNA-seq datasets: 5 datasets from (43), 3 datasets from (45), and 4 datasets from (46) (table S1). Data for in vivo human brain pericytes were obtained from a previous meta-analysis of five single-cell RNA-seq datasets (41). The Pearson correlation coefficient r is shown. Genes of interest are annotated in red (transcription factors) or blue (others). Orange lines represent fold changes of ±2. (B) Transcript abundance of selected genes in the five in vivo human brain pericyte datasets (black circles) and hPSC-derived brain pericyte–like cell datasets [blue squares (43), green triangles (45), and red diamonds (46)]. Bars indicate mean values. (C) Timeline of the differentiation protocol. (D) Schematic of lentiviral overexpression constructs. A fragment of the human NOTCH3 CDS encoding the intracellular domain of Notch3 and the human TBX2 CDS was cloned into the bicistronic lentiviral vector pWPI. The parental pWPI vector was used as a GFP-only control. IRES, internal ribosome entry site; AA, amino acids. (E) Flow cytometry analysis of p75 and HNK-1 expression in day 15 neural crest cells before and after p75 MACS. (F) RT-qPCR analysis of mural cell gene expression 6 days after transduction with GFP, N3ICD-GFP, or TBX2-GFP lentiviruses. Analysis was performed on non-FACS–purified cultures. Expression of each gene is shown relative to ACTB expression and normalized to expression in GFP-transduced cells. Points represent replicate wells from a differentiation of the H9 hPSC line, and bars indicate mean values. P values: analysis of variance (ANOVA) followed by Dunnett’s test versus GFP-transduced cells.
Overexpression of N3ICD as a strategy to derive mural cells
Given low expression of NOTCH3 and Notch target genes (e.g., HEYL, HEY2, and HES4) in existing hPSC-derived pericyte-like cells and the known roles of Notch signaling in mural cell development (32), we asked whether overexpression of the human N3ICD in hPSC-derived neural crest cells could direct mural cell differentiation. We also evaluated overexpression of Tbx2, a mural cell–enriched transcription factor with similarly low expression in existing hPSC models. We cloned the portion of the NOTCH3 coding sequence (CDS) corresponding to the intracellular domain, and the TBX2 CDS, into bicistronic lentiviral vectors also encoding green fluorescent protein (GFP) (Fig. 1, C and D). We differentiated neural crest cells from hPSCs according to a previously established protocol (43). After 15 days of differentiation in E6-CSFD medium, we selected p75+ cells via magnetic-activated cell sorting (MACS), resulting in a homogeneous population of p75+ HNK-1+ neural crest cells that were briefly expanded before transduction (Fig. 1, C and E). We transduced neural crest cells with GFP-only, N3ICD-GFP, or TBX2-GFP lentiviruses and, after 6 days, either analyzed the resulting populations or performed fluorescence-activated cell sorting (FACS) to isolate only the GFP+ cells for further analysis.
Six days after lentiviral transduction, N3ICD-GFP–transduced cells had significantly elevated transcript expression of HEYL and endogenous NOTCH3 (using primers targeting the 3′ untranslated region, which is not present in transgene-derived transcripts) compared to GFP controls (Fig. 1F). Notably, these cells also had increased transcript expression of other transcription factors present in mural cells but not other mesenchymal cell populations in vivo (TBX2 and FOXS1) and canonical mural cell markers (PDGFRB, RGS5, and KCNJ8), suggesting that Notch3 signaling may be sufficient to activate a genetic program for mural cell differentiation (Fig. 1F). In contrast, while TBX2-GFP–transduced cells had elevated expression of TBX2, suggesting successful overexpression, and a slight increase in PDGFRB expression, all other mural cell genes evaluated were unchanged versus GFP controls (Fig. 1F). To confirm that N3ICD-GFP–mediated transcriptional changes occurred via a canonical Notch transactivation mechanism, we used coimmunoprecipitation to assess whether Notch3 was associated with RBPJ, the DNA binding protein in the Notch transactivation complex. RBPJ clearly associated with Notch3 in N3ICD-GFP–transduced but not GFP-transduced cultures (fig. S2, A and B). We also performed a loss-of-function experiment with CB-103, a small-molecule inhibitor of NICD-RBPJ assembly (49), which ablated the effects of N3ICD overexpression on mural cell gene expression signature (fig. S2, C and D). We also asked whether overexpression of an alternative NICD would achieve a similar effect; N1ICD-GFP–transduced neural crest cells underwent similar transcriptional changes to N3ICD-GFP–transduced cells (fig. S3, A and B). We elected, however, to conduct further experiments using cells derived via N3ICD overexpression, given enrichment of Notch3 compared to other Notch receptors in brain mural cells, and loss-of-function studies establishing the necessity of Notch3 for mural cell development (32, 33).
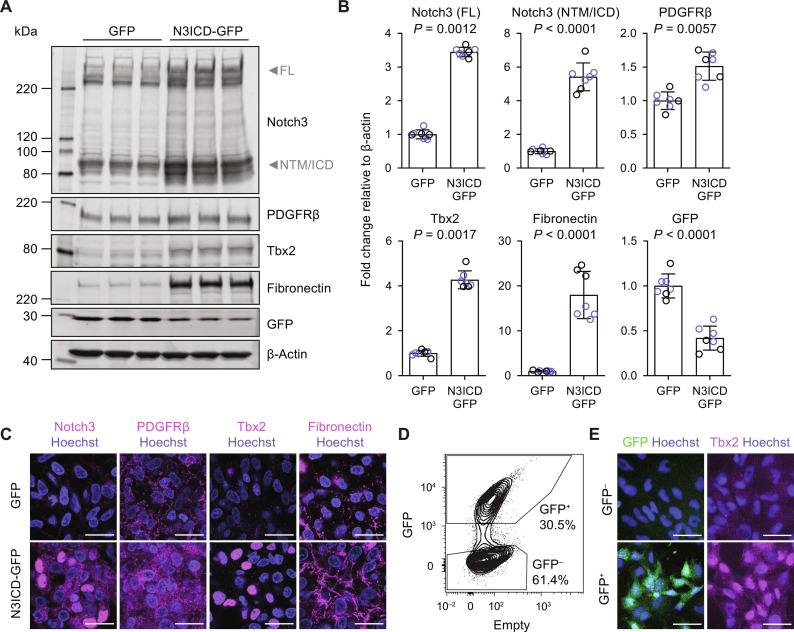
We validated protein-level overexpression of N3ICD via Western blotting with a Notch3 antibody detecting a C-terminal (intracellular domain) epitope. As expected, we observed a significant increase in the intensity of a low molecular weight band corresponding to the Notch transmembrane fragment and intracellular domain in N3ICD-GFP–transduced cultures compared to GFP-transduced cultures (Fig. 2, A and B). A high molecular weight band corresponding to full-length Notch3 also had increased abundance in N3ICD-GFP–transduced cultures, suggestive of positive feedback and consistent with enrichment of Notch3 in brain mural cells compared to neural crest in vivo (16, 48, 50). The canonical mural cell marker PDGFRβ and transcription factor Tbx2 were also upregulated in N3ICD-GFP–transduced cultures (Fig. 2, A and B). We also observed a marked increase in fibronectin abundance in N3ICD-GFP–transduced cultures (Fig. 2, A and B), consistent with human (but not mouse) brain mural cell expression of FN1 in vivo (41). Confocal immunocytochemistry corroborated these findings and revealed clear nuclear localization of Notch3 and Tbx2 in N3ICD-GFP–transduced cells (Fig. 2C). Consistent with transcript-level observations, N1ICD overexpression also achieved similar effects to N3ICD on the protein level (fig. S3, C and D). Together, these results suggest that activation of Notch signaling in neural crest cells is sufficient to drive mural cell differentiation.
Fig. 2. Results of N3ICD overexpression in neural crest cells.
(A) hPSC-derived neural crest cells were transduced with GFP or N3ICD-GFP lentiviruses. After 6 days in culture, Western blots were performed and membranes were probed with Notch3, PDGFRβ, Tbx2, fibronectin, GFP, and β-actin antibodies. On the Notch3 Western blot, arrows indicate the full-length (FL) and Notch transmembrane/intracellular domain (NTM/ICD) bands. The NTM and ICD fragments are similar in molecular weight, and thus, bands are not distinguishable. A representative blot from the H9 hPSC line is shown. (B) Quantification of Western blots. Band intensities were normalized to β-actin band intensities. Points represent replicate wells from two independent differentiations, one in the H9 hPSC line (black circles) and one in the WTC11 hPSC line (blue circles). Bars indicate mean values ± SD, with values normalized within each differentiation such that the mean of the GFP condition equals 1. P values: two-way ANOVA on unnormalized data. (C) Confocal immunocytochemistry analysis of Notch3, PDGFRβ, Tbx2, and fibronectin expression in mixed transduced cultures having both GFP+ and GFP− cells 6 days after transduction with GFP or N3ICD-GFP lentiviruses. Hoechst nuclear counterstain overlaid in all images. Scale bars, 25 μm. (D) Flow cytometry analysis of GFP expression 6 days after transduction of neural crest cells with N3ICD-GFP lentivirus. GFP+ and GFP− gates are representative of those used to isolate populations via FACS. (E) Immunocytochemistry analysis of GFP and Tbx2 expression in cells expanded in E6 medium for 4 days after FACS. Hoechst nuclear counterstain is overlaid in all images. Scale bars, 50 μm.
To confirm that N3ICD-GFP–transduced cells (and not nontransduced cells in the culture) adopt mural cell identity, we used FACS to isolate GFP+ and GFP− cells from cultures 6 days after N3ICD-GFP lentiviral transduction (Fig. 2D). We compared acutely isolated GFP+ and GFP− cells by reverse transcription quantitative polymerase chain reaction (RT-qPCR); GFP+ cells had significantly reduced expression of NGFR, consistent with loss of neural crest identity, and significantly higher expression of markers of mesenchyme (e.g., TBX18) and mural cells (e.g., NOTCH3, TBX2, HEYL, FOXS1, and KCNJ8); the VSMC-enriched gene ACTA2 was not differentially expressed (fig. S4). These results were consistent across multiple hPSC lines. Immunostaining conducted 4 days after FACS confirmed that Tbx2 was selectively expressed in GFP+ cells (Fig. 2E). These results suggest that N3ICD-GFP functions cell-autonomously to direct neural crest-to-mural cell differentiation and that the resulting cells have molecular hallmarks of brain mural cells at this time point. KCNJ8 distinguishes mature pericytes from VSMCs (fig. S1A) (16, 48); Ando and colleagues (47), however, recently demonstrated that brain VSMCs develop from a KCNJ8+ progenitor. We therefore assessed whether our GFP+ (KCNJ8+) cells could be further differentiated to VSMCs. We previously observed induction of the VSMC-enriched protein α-smooth muscle actin (α-SMA) in neural crest cells cultured in minimal E6 medium (44). We therefore asked whether E6 medium would facilitate further specification of N3ICD-overexpressing mural cells to VSMCs. After replating FACS-isolated GFP+ cells in E6 medium for 2 to 10 days, the cells upregulated ACTA2 and downregulated KCNJ8 while maintaining expression of pan-mural cell markers NOTCH3, PDGFRB, RGS5, and the transgene (fig. S5A). The resulting cells had protein-level expression of α-SMA, calponin, and smooth muscle protein-22α after 4 days in E6 medium (fig. S5B). Together, these results support culture of GFP+ cells in E6 medium as a strategy to derive VSMCs.
Molecular properties of cells derived via N3ICD overexpression
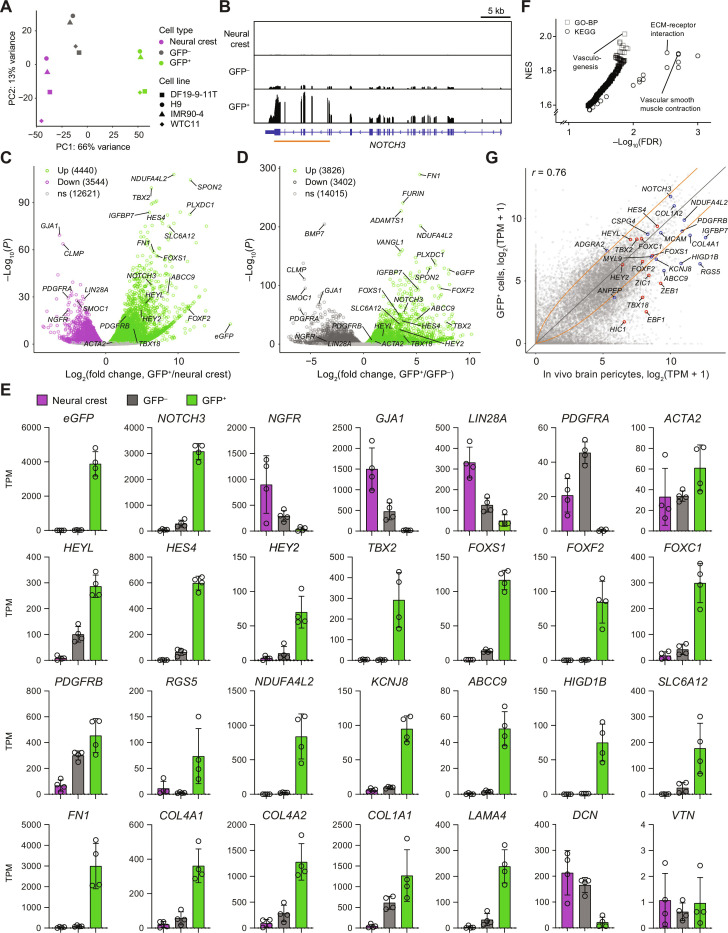
To determine the extent to which N3ICD-overexpressing cells have global molecular similarity to brain mural cells, we next used RNA-seq to obtain transcriptomic profiles of neural crest cells and FACS-isolated GFP− and GFP+ cells from N3ICD-GFP–transduced cultures from four hPSC lines (file S1). In principal component analysis, the three cell types segregated along principal component 1, which explained 66% of the variance, demonstrating global reproducibility across multiple cell lines and differentiations (Fig. 3A). Visualization of RNA-seq reads confirmed that in addition to transgene-derived NOTCH3 transcripts, endogenous NOTCH3 was also upregulated in GFP+ cells (Fig. 3B), consistent with RT-qPCR results. We identified differentially expressed genes in GFP+ cells compared to neural crest (Fig. 3C, fig. S6A, and file S2) and in GFP+ cells compared to GFP− cells (Fig. 3D, fig. S6B, and file S2). In both comparisons, GFP+ cells were enriched for mural cell marker genes, including the key transcription factors HEYL, HES4, TBX2, FOXS1, FOXF2, and FOXC1, some of which have established functional roles in brain mural cell development and function (Fig. 3, C to E, and fig. S6) (51, 52). PDGFRB, RGS5, NDUFA4L2, KCNJ8, ABCC9, HIGD1B, IGFBP7, PLXDC1, CSPG4, and ADAMTS1 were similarly enriched in GFP+ cells (Fig. 3, C to E, and fig. S6). Several of these upregulated genes (KCNJ8, ABCC9, and HIGD1B) have been identified as enriched in adult brain pericytes compared to VSMCs (16). Consistent with results of RT-qPCR, ACTA2 was expressed at moderate levels [~40 to 60 transcripts per million (TPM)] in all cell types but was not differentially expressed in GFP+ cells (Fig. 3, C to E).
Fig. 3. RNA-seq of neural crest, GFP−, and GFP+ cells.
(A) Principal component analysis of whole-transcriptome data. Points are colored by cell type: neural crest (magenta), GFP− cells (gray), GFP+ cells (green). Data are from four independent differentiations, one each from the DF19-9-11T (squares), H9 (circles), IMR90-4 (triangles), and WTC11 (diamonds) hPSC lines. (B) Representative genome browser plots (from DF19-9-11T–derived cells). The orange line indicates the region of NOTCH3 encoding the intracellular domain. (C and D) Differential expression analysis of GFP+ cells compared to neural crest (C) and GFP+ cells compared to GFP− cells (D). Differentially expressed genes (adjusted P < 0.05, DESeq2 Wald test with Benjamini-Hochberg correction) are highlighted, and the numbers of upregulated and downregulated genes are shown. Complete results of differential expression analysis are in file S2. (E) Transcript abundance (TPM) of selected transcripts. The top row displays expression of transgene (eGFP), total NOTCH3 (endogenous and transgene-derived transcripts), neural crest markers (NGFR, LIN28A, and GJA1), nonmural PDGFRA, and VSMC-enriched ACTA2. The second row displays mesenchymal and mural transcription factors; the third row displays mural markers, including those enriched in adult pericytes compared to VSMCs (NDUFA4L2, ABCC9, KCNJ8, HIGD1B, and SLC6A12); the fourth row displays genes encoding components of ECM. Abundance data for all genes are in file S1. (F) Results of Gene Set Enrichment Analysis. Gene sets from the KEGG and GO-BP databases enriched in GFP+ cells compared to neural crest (FDR < 0.05) are shown. NES, normalized enrichment score. Complete results are provided in file S2. (G) Comparison of protein-coding transcript abundances in GFP+ cells versus in vivo human brain pericytes. In vivo data are as described in Fig. 1 (41). The Pearson correlation coefficient r is shown. Genes of interest are annotated in red (transcription factors) or blue (others). Orange lines represent fold changes of ±2.
The canonical neural crest marker NGFR was downregulated in GFP+ cells compared to both neural crest and GFP− cells, as was LIN28A, which plays a role in neural crest multipotency (53). PDGFRA, which is enriched in fibroblasts compared to mural cells in vivo (16, 54), was nearly absent in GFP+ cells, but expressed by GFP− cells (Fig. 3E). Although GFP− cells also expressed PDGFRB, they lacked several other mural cell markers, retained some expression of neural crest genes, and expressed several fibroblast markers (DCN, LUM, ADAMTS2, MMP2, MFAP4, PTGDS, and PODN) (16, 48, 55, 56) (Fig. 3E and fig. S6D), suggesting that interactions with N3ICD-overexpressing GFP+ cells could cause partial differentiation of these cells to a nonmural fate. Hierarchical clustering revealed a gene module with highly enriched expression in GFP+ cells compared to both GFP− cells and neural crest; this module contained known mural transcripts, including FOXD1, GJA4, PTGIR, and MCAM (CD146), in addition to many of those mentioned above (fig. S6, C and D), further supporting the mural identity of cells derived via Notch3 activation. Genes with known enrichment in brain mural cells compared to those of other organs, including PTN, GPER1, and SLC6A17 (16, 41), were also enriched in GFP+ cells (fig. S6, C and D, and file S2), supporting the notion that brain-enriched expression is at least partially attributable to the neural crest origin. Furthermore, the γ-aminobutyric acid (GABA) transporter gene SLC6A12, which we and others recently identified as enriched in human compared to mouse brain pericytes (40, 41, 54), was robustly upregulated in GFP+ cells compared to neural crest and GFP− cells (Fig. 3, C to E). SLC6A1, another human-enriched pericyte gene (41, 54), however, was not expressed (fig. S6D), highlighting that while Notch signaling activates a mural cell transcriptional program, other factors are likely required for complete acquisition of mural cell phenotype. We also observed minimal expression of VTN in all cells (approximately 1 TPM), consistent with observations that human brain pericytes lack VTN, despite robust expression in mouse brain pericytes (40, 41, 57), while other ECM-related genes (FN1, COL4A1, COL4A2, COL1A1, and LAMA4) were upregulated in GFP+ cells (Fig. 3, C to E). Together, these results suggest that cells derived from hPSCs via this strategy (i) are mural cells, (ii) have molecular hallmarks that distinguish brain and nonbrain mural cells, and (iii) can at least partially capture species-specific differences in mural cell gene expression observed in vivo.
We next identified gene sets enriched in GFP+ cells compared to neural crest using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology-Biological Process (GO-BP) databases (Fig. 3F, fig. S7, and file S2). As expected, the Notch Signaling Pathway gene sets from both KEGG and GO-BP databases were enriched (fig. S7 and file S2). Additional enriched gene sets included GO-BP Vasculogenesis, GO-BP Nitric Oxide–Mediated Signal Transduction, and KEGG Vascular Smooth Muscle Contraction, which was driven by enrichment of genes encoding guanylate and adenylate cyclases (e.g., GUCY1B1, GUCY1A2, and ADCY5) and regulators of actomyosin contraction and cytoskeleton (e.g., PPP1R14A, MYLK, and ROCK1), consistent with vascular mural cell identity (Fig. 3F and fig. S7). Other vasculature-related GO-BP gene sets were similarly enriched (file S2). Consistent with previously noted upregulation of ECM-related genes, the KEGG gene set ECM-Receptor Interaction was enriched (Fig. 3F and fig. S7). Notably, we observed enrichment of the KEGG gene set Neuroactive Ligand-Receptor Interaction, which we previously reported as depleted in cultured primary brain pericytes compared to in vivo pericytes (Fig. 3F) (41). Highly enriched genes in this set included PTGIR, PTH1R, EDNRA, and GIPR (fig. S7), but some genes encoding key mural cell receptors such as P2RY14 were not expressed (fig. S7 and file S1), suggesting that other cues may be required to obtain cells with the complete mural cell receptor repertoire. Last, we directly compared the average transcriptome profile of GFP+ cells to that of in vivo human brain pericytes (Fig. 3G). While the overall correlation across all genes was similar to that of existing hPSC-derived brain pericyte–like cells (Fig. 1A), GFP+ cells exhibited a notable improvement in the expression of key mural cell transcription factors and other markers, including NOTCH3, HEYL, HEY2, ZIC1, FOXS1, FOXF2, KCNJ8, ABCC9, RGS5, HIGD1B, and NDUFA4L2 (Fig. 3G and fig. S6, E and F), reflective of the key role of Notch signaling in driving the mural cell genetic program.
Functional attributes of N3ICD-derived brain mural cells
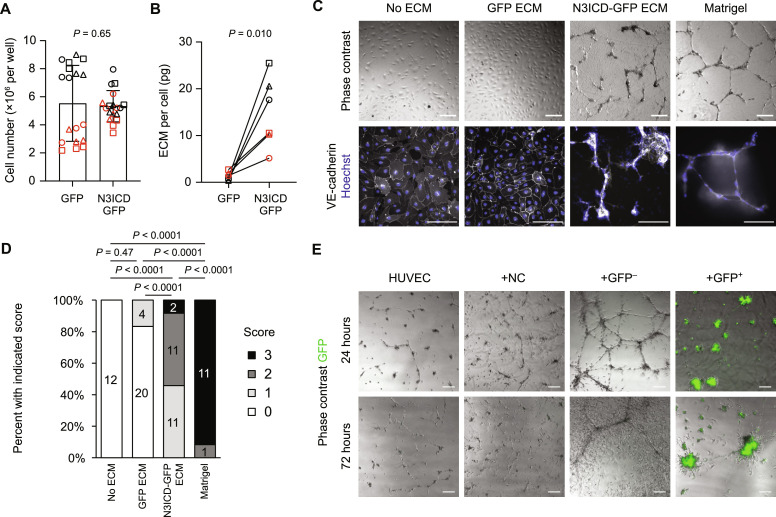
Production of vascular basement membrane is a key function of mural cells. We observed with the naked eye an apparent thick layer of ECM in N3ICD-GFP–transduced cultures but not GFP-transduced cultures. We confirmed this finding by decellularization followed by quantification of remaining total protein. Compared to GFP-transduced cultures, N3ICD-overexpressing cultures generated approximately 10 to 20 times more ECM per cell (Fig. 4, A and B), consistent with the marked up-regulation of ECM-encoding genes in RNA-seq data and protein-level enrichment of fibronectin. Cell yield in N3ICD-transduced cultures represented an approximately 60-fold expansion relative to neural crest input (see Materials and Methods and Fig. 4A). We next evaluated the ability of these decellularized matrices to support formation of endothelial cords, a widely used in vitro proxy for angiogenic potential (43, 46, 58, 59). While human umbilical vein endothelial cells (HUVECs) cultured on ECM from GFP-transduced cultures adopted the same cobblestone morphology as HUVECs cultured directly on tissue culture–treated polystyrene (no ECM), HUVECs cultured on ECM from N3ICD-GFP–transduced cultures formed cords, albeit more variably than on the positive control Matrigel substrate (Fig. 4, C and D, and fig. S8A). We also directly cocultured neural crest cells and GFP+ and GFP− cells 5 days after FACS with HUVECs on the Matrigel substrate. Twenty-four hours after cell seeding, neural crest coculture did not affect HUVEC cord formation (Fig. 4E), while cords in GFP− cell cocultures were longer, consistent with the ability of many mesenchymal cell types to modulate cord formation (59, 60). In GFP+ cell cocultures, however, we observed highly reproducible formation of mural-endothelial aggregates, a phenomenon previously observed in primary pericyte-endothelial cell cocultures at later time points (46) and in cocultures of HUVECs with immature smooth muscle cells derived from hPSCs (Fig. 4E) (59). Aggregate formation was observed with GFP+ cells derived from two different hPSC lines (fig. S8B). Aggregates contained both GFP+ cells and HUVECs, and GFP+ cells did not form such large aggregates in the absence of HUVECs (fig. S8, C and D).
Fig. 4. Mural cell ECM production and endothelial cell interaction.
(A) Number of cells 6 days after transduction of neural crest with GFP or N3ICD-GFP lentiviruses. Points represent replicate wells from three differentiations of the H9 line (black) and three of the IMR90-4 line (red), with each differentiation indicated with a different shape. Bars indicate mean values ± SD. P value: two-way ANOVA. (B) Total ECM production 6 days after transduction of neural crest. Each point represents average ECM quantity from three wells of a differentiation of the H9 line (black) or IMR90-4 line (red), normalized to average cell number from three parallel wells, with each differentiation indicated with a different shape. P value: paired Student’s t test. (C) HUVEC cord formation assay on decellularized ECM from GFP- or N3ICD-GFP–transduced cultures, and from no ECM and Matrigel controls. Scale bars, 200 μm. (D) Quantification of HUVEC cord formation for the conditions described in (C). Blinded images were scored from 0 (no cords) to 3 (virtually all cells associated with cords); see Materials and Methods and fig. S8A. Data from the GFP ECM and N3ICD-GFP ECM conditions are from two differentiations of the H9 line and one of the IMR90-4 line; data from no ECM and Matrigel controls are from two independent experiments. P values: Kruskal-Wallis test followed by Steel-Dwass test. (E) Coculture cord formation assay with HUVECs, HUVECs and neural crest cells (+NC), HUVECs and GFP− cells from an N3ICD-GFP–transduced culture (5 days after FACS; +GFP−), and HUVECs and GFP+ cells from an N3ICD-GFP–transduced culture (5 days after FACS; +GFP+). Representative images are shown from 24 and 72 hours after initiating assay, from a differentiation of the H9 line. Images from a differentiation of the IMR90-4 line are shown in fig. S8D. Scale bars, 200 μm.
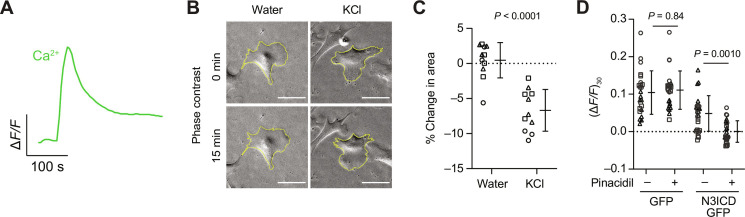
Mural cells regulate vascular tone, and while the relative contributions of different mural cell subtypes to neurovascular coupling remain the subject of debate, virtually all mural cells appear capable of contraction, at least under artificial stimuli (18). Potassium (at concentrations causing depolarization) is widely used to assess contractility of pericytes and smooth muscle cells in vitro and in vivo (45, 61). We first used calcium imaging of an N3ICD-GFP–transduced culture (at 6 days after transduction) to confirm that application of 40 mM KCl led to depolarization and calcium influx (Fig. 5A). Because the cell density at this time point precludes assessment of cell size, we asked whether KCl application would cause contraction of GFP+ cells isolated via FACS and replated at low cell density (Fig. 5B). Two days after FACS, cells underwent an average reduction in area of approximately 7% 15 min after KCl addition, compared to an average 0% change after addition of a water vehicle control, with cells in both conditions extending and withdrawing cellular processes (Fig. 5, B and C). These results support the contractile ability of mural cells derived from neural crest via N3ICD overexpression.
Fig. 5. Mural cell contractility and KATP channel function.
(A) Calcium response (FLIPR Calcium 6 dye) of cells in an N3ICD-GFP–transduced culture (6 days after transduction) to KCl application. KCl (40 mM) was added at t = 50 s. (B) Phase contrast images of GFP+ cells from an N3ICD-GFP–transduced culture (2 days after FACS) at t = 0 min and t = 15 min after treatment with 40 mM KCl. Scale bars, 50 μm. (C) Change in area of GFP+ cells (2 days after FACS) 15 min after treatment with water (vehicle) or 40 mM KCl. Points represent replicate wells from three differentiations of the H9 hPSC line, each differentiation indicated with a different shape. Bars indicate mean values ± SD. P value: two-way ANOVA. (D) Intracellular calcium 30 s after addition of 40 mM KCl to GFP-transduced or N3ICD-GFP–transduced cultures (6 days after transduction) pretreated with pinacidil or DMSO (vehicle). Points represent replicate wells from three differentiations of the H9 hPSC line, each differentiation indicated with a different shape. Bars indicate mean values ± SD. P values: Tukey’s honest significant difference test following ANOVA, P = 0.0011 for interaction (DMSO versus pinacidil) × (GFP versus N3ICD-GFP).
KCNJ8 and ABCC9 encode subunits of the KATP channel, which has been implicated in pericyte propagation of hyperpolarizing signals during neurovascular coupling (62). Our RNA-seq data demonstrated upregulation of KCNJ8 and ABCC9 (but not KCNJ11 or ABCC8, which encode alternative KATP subunits) in acutely isolated GFP+ cells (Fig. 3E). We therefore used the KATP channel opener pinacidil (62) to assess the functionality of this channel in our cells. Hyperpolarization caused by K+ outflow through open KATP channels would be expected to partially counteract KCl-induced depolarization, reducing calcium influx and/or release from intracellular stores. N3ICD-GFP–transduced cultures (at 6 days after transduction) pretreated with pinacidil exhibited reduced intracellular calcium after KCl application compared to control [dimethyl sulfoxide (DMSO)]–treated cells; such a difference was not observed in GFP-transduced cultures (Fig. 5D). These results suggest that KATP channels are functional in mural cells derived via N3ICD overexpression.
CREs underlying Notch3 function in mural cell differentiation
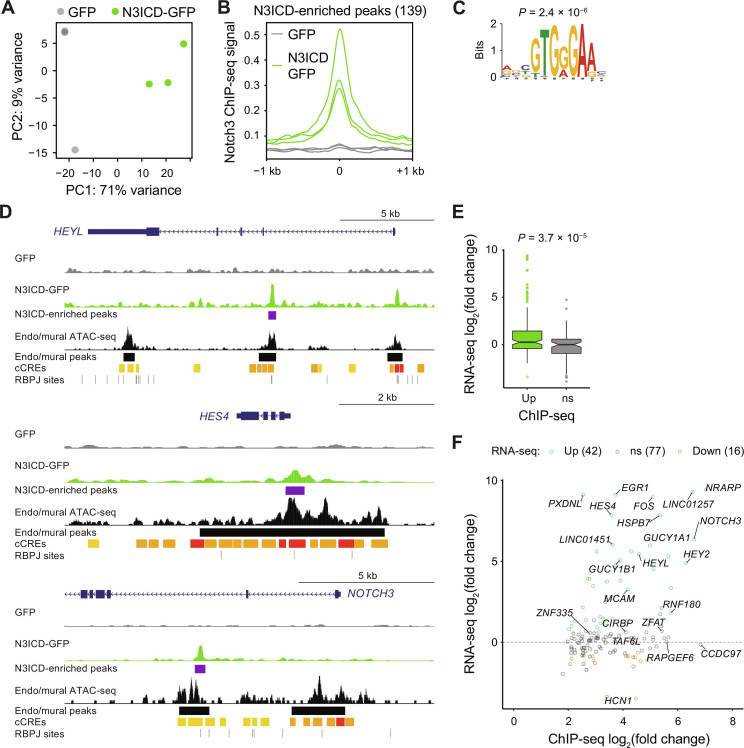
In the canonical model of Notch signaling, NICDs activate transcription of a small number of transcription factors (i.e., HES and HEY family members); however, the genome contains tens of thousands of putative RBPJ binding sites, and NICDs regulate a diverse array of genes in a cell type–specific/tissue-specific context (63). We wondered whether in our system N3ICD serves as a direct input to a large number of the observed upregulated genes, or whether N3ICD directly regulates only a small number of genes that subsequently activate the differentiation cascade. Thus, as a first step toward defining this gene regulatory network, we used Notch3 ChIP-seq to identify cis-regulatory elements (CREs) directly bound by N3ICD in N3ICD-GFP–transduced cultures. In principal component analysis based on all Notch3 peaks (called by comparison of Notch3 IP and matched input controls), N3ICD-GFP–transduced cells clustered distinctly from control GFP-transduced cells along principal component 1, which explained 71% of the variance (Fig. 6A). Peak enrichment analysis using the DiffBind package (64) revealed 139 peaks enriched in N3ICD-GFP–transduced cells (“N3ICD-enriched peaks”), 90 nonenriched peaks, and no Notch3 peaks enriched in GFP-transduced cells (Fig. 6B and fig. S9A), consistent with the lack of Notch3-RBPJ coimmunoprecipitation in GFP-transduced cells (fig. S2, A and B). The RBPJ consensus motif GTGGGAA (65) was overrepresented in N3ICD-enriched peaks (Fig. 6C), and N3ICD-enriched peaks were located proximal to canonical transcriptional targets of Notch3 signaling, including HEYL, HES4, HEY2, and NRARP (Fig. 6D; fig. S9, A and B; and file S3). We also identified a putative enhancer peak within the first intron of the NOTCH3 gene (Fig. 6D), consistent with the positive feedback observed on the transcript and protein levels. Additional N3ICD-enriched peaks were proximal to genes including MCAM, GUCY1A1, GUCY1B1, CIRBP, ZNF335, RNF180, and TAF6L, in addition to a small number of distal intergenic peaks (fig. S9, A and B, and file S3). N3ICD-enriched peaks aligned with single-cell assay for transposase–accessible chromatin (ATAC)–seq peaks from a human cortex mural cell/endothelial cell cluster (66), putative promoter and enhancer signatures from the ENCODE candidate CRE database (67), and putative RBPJ binding sites from the JASPAR database (68) (Fig. 6D and fig. S9B). Together, these results support the potential functional relevance of N3ICD-bound CREs identified in our model system.
Fig. 6. Notch3 ChIP-seq of N3ICD-GFP– and GFP-transduced cells.
(A) Principal component analysis based on all peaks. Points are colored by cell type: GFP-transduced cells (gray) and N3ICD-GFP–transduced cells (green). Data are from three independent differentiations of the H9 hPSC line. (B) Profile plots for the 139 N3ICD-enriched peaks. Each line represents one sample. Profiles are generated by averaging the ChIP-seq signals across the 139 genomic loci containing an N3ICD-enriched peak. Genomic coordinates and annotations for all peaks are provided in file S3. (C) Motif logo for the overrepresented motif in N3ICD-enriched peaks. P value as reported by MEME-ChIP (108). (D) Representative genome browser plots for the HEYL, HES4, and NOTCH3 loci. From top, tracks are Notch3 ChIP-seq signal from GFP-transduced cells, Notch ChIP-seq signal from N3ICD-GFP–transduced cells, N3ICD-enriched peaks as identified by DiffBind (purple bars), ATAC-seq signal from a human cortex endothelial/mural cell cluster (66), endothelial/mural ATAC-seq peaks as identified by MACS (66), ENCODE cCREs (red, promoter-like signature; orange, proximal enhancer-like signature; yellow, distal enhancer-like signature) (67), and putative RBPJ binding sites from the JASPAR database (68). Additional genome browser plots are shown in fig. S5B. (E) RNA-seq log2(fold change) data from comparison of GFP+ (mural) and neural crest cells for genes associated with N3ICD-enriched Notch3 ChIP-seq peaks (Up; n = 134) and genes associated with nonenriched peaks (ns; n = 75). On average, genes associated with (proximal to) N3ICD-enriched peaks have larger fold upregulation in RNA-seq data than genes associated with nonenriched peaks. P value: Student’s t test. (F) Comparison of RNA-seq log2(fold change) data [GFP+ (mural) versus neural crest cells] and ChIP-seq log2(fold change) data (N3ICD-GFP–transduced versus GFP-transduced cells) for genes associated with N3ICD-enriched Notch3 ChIP-seq peaks. Points are colored by differential expression in RNA-seq data, and the numbers of upregulated, downregulated, and nonsignificant genes are shown in the legend.
In RNA-seq differential expression analysis of GFP+ mural cells and neural crest, the average fold change of genes associated with N3ICD-enriched peaks exceeded that of nonenriched peaks (Fig. 6E). Many genes associated with N3ICD-enriched peaks were upregulated in RNA-seq data, including HEYL, HES4, HEY2, NRARP, NOTCH3, GUCY1A1, GUCY1B1, EGR1, and FOS (Fig. 6F). Other peak-associated genes, including the transcriptional regulators ZNF335 and TAF6L, were not differentially expressed and thus likely do not regulate the observed downstream differentiation processes (Fig. 6F). Notably, non-HES/HEY family mesenchymal and mural cell transcription factors such as FOXF2, FOXC1, FOXS1, TBX18, and TBX2, which were upregulated on the transcript level, were not associated with Notch3 ChIP-seq peaks (file S3), suggesting transcriptional regulation downstream of HEYL, HES4, HEY2, and potentially other upstream factors. In summary, these data suggest that Notch3 regulates mural cell differentiation by serving as an input to a relatively small number of transcriptional targets.
DISCUSSION
In vivo loss-of-function experiments have demonstrated that Notch signaling is required for mural cell development from both mesoderm- and neural crest–derived progenitors (32, 33). Here, using hPSC-derived neural crest, we show that Notch signaling is sufficient to direct specification and differentiation of mural cells in vitro. Overexpression of the N3ICD in neural crest cells caused cell-autonomous differentiation to cells with molecular and functional properties of mural cells. Compared to controls, N3ICD-overexpressing cells exhibited increased expression of PDGFRβ, increased expression of full-length Notch3, and induction of mural cell transcription factors such as HEYL, HES4, TBX2, FOXS1, FOXF2, and FOXC1, some of which have previously reported roles in mural cell development (51, 52). Cells derived via N3ICD overexpression produced abundant ECM, which supported endothelial cell cord formation; these cells also self-assembled with endothelial cells and contracted in response to KCl-induced depolarization. Overexpression of the Notch1 intracellular domain had similar effects, consistent with promiscuous binding of intracellular domains from all Notch family members to RBPJ and MAML (49, 63). Other signaling pathways implicated in mural cell development such as PDGF-B and TGF-β (30, 69, 70) were not required for mural cell differentiation in our model system, suggesting that these pathways may regulate other aspects of mural cell development such as recruitment to vessels and/or maturation. PDGF-BB and TGF-β, however, merit future examination for their potential ability to achieve aspects of mural cell phenotype not observed using Notch3.
Using RNA-seq of N3ICD-derived mural cells after 6 days of transduction, we observed Notch-induced upregulation of genes previously identified as enriched in brain mural cells compared to mural cells of other organs, such as PTN, GPER1, SLC6A12, SLC6A17, SLC38A11, and ZIC1 (16, 41). Observation of this brain-enriched mural cell gene signature in our in vitro culture system, which lacks CNS tissue–derived factors, suggests that the neural crest origin at least partially defines the brain-specific molecular profile. Future work evaluating the results of N3ICD overexpression in mesodermal progenitors derived from hPSCs could further inform these findings and permit functional comparisons of mural cells derived from these two distinct lineages. Similarly, we observed up-regulation of genes previously identified as enriched in human compared to mouse mural cells, including FN1 and SLC6A12 (40, 41, 54, 57). In addition, VTN, which is highly expressed by mouse brain mural cells, but not expressed by human brain mural cells in vivo (16, 40, 41, 54, 57, 71), was negligibly expressed in our hPSC-derived cells. These results suggest that species differences in brain mural cell gene expression are at least partially attributable to cell-intrinsic genetic programs.
A common approach for differentiating mural cells from hPSCs, via both mesodermal and neural crest intermediates, has been to use medium supplemented with fetal bovine serum (FBS) and growth factors such as PDGF-BB, TGF-β1, activin A, and/or FGF2 (45, 46, 59, 72–75), but the necessity and/or sufficiency of each factor to drive mural cell differentiation has not been rigorously established. Several studies have used commercially available “pericyte medium,” which includes FBS and a proprietary cocktail of growth factors (45, 46, 75), precluding systematic examination of molecular mechanisms. Our work here establishes a serum-free method for mural cell differentiation that relies on a single, defined molecular factor, which should enable future mechanistic studies. Furthermore, while the molecular profile of existing hPSC-derived brain pericyte–like cells is remarkably similar across studies and to cultured primary brain pericytes (43, 45, 46), this molecular profile has notable departures from the in vivo phenotype. For example, cultured cells coexpress mural and fibroblast-associated markers and lack robust expression of canonical mural and pericyte genes such as HEYL, FOXS1, and KCNJ8. Our approach achieves robust induction of these mural cell transcripts without concomitant up-regulation of fibroblast-associated PDGFRA. We used a lentiviral overexpression system, which achieves moderate-to-high transduction efficiency and offers the flexibility to use the same lentivirus across multiple hPSC lines, which will facilitate future disease modeling applications. Alternative approaches to activate Notch signaling that permit temporal- and dose-control and target nearly 100% of cells (e.g., engineering an hPSC line for inducible N3ICD expression) merit future development. In addition, the duration of the differentiation protocol (~25 days) may be a limitation for some applications. Although hPSC differentiations are constrained to some extent by intrinsic temporal programs, future work could evaluate strategies to accelerate the differentiation process.
Among the genes upregulated by N3ICD overexpression were markers that distinguish adult pericytes from VSMCs in vivo [e.g., KCNJ8, ABCC9, and HIGD1B (16, 41)] (Figs. 1 and 3 and figs. S4 and S6). These results are consistent with rapid expression of abcc9 upon zebrafish mural cell emergence (32) and suggest that pericytes may be the “default” mural cell fate and that additional molecular signals may be required for VSMC specification. An alternative hypothesis is that immature mural cells adopt an intermediate pericyte-VSMC phenotype and that additional factors would be required to fully achieve either fate. This hypothesis is supported by recent data demonstrating that brain VSMCs develop from a KCNJ8+ABCC9+ mural cell progenitor, but the extent to which these common progenitors resemble mature pericytes with respect to function and global transcriptional profile remains unknown (47). We further demonstrated that N3ICD-derived mural cells isolated via FACS and replated in minimal E6 medium downregulated KCNJ8 and acquired expression of α-SMA, calponin, and SM22α, representing a potential strategy to achieve further specification of VSMCs. These results motivate future functional evaluation of these putative VSMCs; for example, given expression of contractile proteins, it will be important to determine whether these cells exhibit increased KCl-induced contraction compared to acutely isolated N3ICD-derived mural cells. Last, our hPSC-based mural cell model is well suited to identify molecular drivers of further pericyte/VSMC differentiation using either candidate factor or genetic/pharmacologic screening approaches. Nonetheless, given expression of many adult pericyte-enriched genes, N3ICD-mural cells may be useful in modeling pericyte functions, as we demonstrated by assessing KATP channel function.
We found that mural cells derived from neural crest via N3ICD overexpression produce ECM capable of supporting endothelial cord formation, consistent with expression data suggesting that brain mural cells produce components of the vascular basement membrane (16, 41), and with a previous study using placental pericyte-derived ECM (76). We also observed that the resulting GFP+ cells self-assemble with endothelial cells into compact aggregates, a phenomenon distinct from that observed in this work with neural crest and GFP− cells from an N3ICD-transduced culture, and from that previously observed with hPSC-derived pericyte-like cells and primary brain pericytes (43). These aggregates permit direct cell-cell contact and three-dimensional cytoarchitecture, features that are not readily achievable in monolayer cultures, and therefore represent a potential system for interrogating mural-endothelial interactions. Such interactions are important in physiological processes, such as BBB development and maintenance, and in disease (4, 11–13, 19, 20, 77). hPSC-based models have been used to advance understanding of neurovascular unit physiology and pathology (42, 78–80), motivating similar use of mural cells derived from hPSCs via N3ICD overexpression and their incorporation into multicellular models. Last, ChIP-seq suggested that Notch3 induces mural cell differentiation through direct binding to a small number of target genes, including the transcription factors HEYL, HES4, and HEY2. These results motivate further characterization of the gene regulatory network controlling mural cell differentiation, for example, by profiling targets of these HEY/HES family members. In summary, we show that activation of Notch signaling in hPSC-derived neural crest is sufficient to direct the differentiation of brain mural cells, and establish an improved in vitro model that should facilitate improved understanding of brain mural cell development and function.
MATERIALS AND METHODS
hPSC maintenance
Matrigel-coated plates were prepared by resuspending a frozen 2.5-mg aliquot of Matrigel, Growth Factor Reduced (Corning, Glendale, AZ) in 1 ml of Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Life Technologies, Carlsbad, CA) and diluting the resulting solution in 29 ml of DMEM/F12. One milliliter of this solution was used to coat each well of five six-well plates. Plates were stored at 37°C for at least 1 hour before use and up to 1 week. The following hPSC lines were used: H9 human embryonic stem cells (hESCs) (81) (WiCell, Madison, WI), IMR90-4 induced pluripotent stem cells (iPSCs) (82) (WiCell), DF19-9-11T iPSCs (83) (WiCell), and WTC11 iPSCs (84) (Gladstone Institutes, San Francisco, CA). hPSCs were maintained at 37°C, 5% CO2 on Matrigel-coated plates in E8 medium (STEMCELL Technologies, Vancouver, Canada) with daily medium changes. When hPSC colonies began to touch, cells were dissociated as colonies using ~7 min of Versene (Life Technologies) treatment and transferred to a new Matrigel-coated plate at a split ratio of 1:12.
Neural crest differentiation
Neural crest was differentiated according to a previously established protocol (43, 44). When hPSC colonies began to touch, three to four wells of cells were dissociated using ~7 min of Accutase (Innovative Cell Technologies, San Diego, CA) treatment, 1 ml per well. The Accutase/single-cell suspension was transferred to 4× volume of DMEM/F12 medium, and cells were counted using a hemocytometer. Cells were centrifuged for 5 min at 200g. The cell pellet was resuspended in 1 ml of E8 medium, and a volume of the resulting suspension containing 2.84 × 106 cells was transferred to a tube containing 6.5 ml of E8 medium supplemented with 10 μM ROCK inhibitor Y-27632 (Tocris, Bristol, UK). The resulting cell suspension was distributed to three wells of a Matrigel-coated six-well plate, 2 ml per well. Cells were incubated at 37°C, 5% CO2. The following day, differentiation was initiated by changing medium to E6-CSFD medium. E6-CSFD medium is E6 medium prepared according to (85) supplemented with 1 μM CHIR 99021 (Tocris), 10 μM SB431542 (Tocris), FGF2 (10 ng/ml; Waisman Biomanufacturing, Madison, WI), 1 μM dorsomorphin dihydrochloride (Tocris), and heparin sodium salt (22.5 μg/ml) from porcine intestinal mucosa (Sigma-Aldrich, St. Louis, MO). E6-CSFD medium was replaced daily for 15 days. Cells were passaged when confluent: One well of cells was dissociated with 1 ml of Accutase for ~5 min. The Accutase/single-cell suspension was transferred to 4× volume of DMEM/F12 medium and centrifuged for 5 min at 200g. The cell pellet was resuspended in 600 μl of E6-CSFD medium, and 100 μl of the resulting suspension was transferred to each of three to six wells of a six-well plate each containing 2 ml of E6-CSFD medium (for a split ratio of 1:6).
MACS of neural crest
On day 15, three to six wells of neural crest cells were dissociated using ~5 min of Accutase treatment, 1 ml per well. The Accutase/single-cell suspension was transferred to 4× volume of DMEM/F12 medium, and cells were counted using a hemocytometer. Cells were centrifuged for 5 min at 180g, 4°C. MACS buffer was prepared by supplementing Dulbecco’s phosphate-buffered saline (DPBS), no calcium, no magnesium (Life Technologies) with 0.5% bovine serum albumin (Sigma-Aldrich) and 2 mM ethylenediaminetetraacetic acid (Sigma-Aldrich). The cell pellet was resuspended in 60 μl of MACS buffer per 107 cells. FcR blocking reagent, human (Miltenyi Biotec, Auburn, CA) and neural crest stem cell microbeads, human (Miltenyi Biotec) were added to the cell suspension at 20 μl each per 107 cells. Cells were incubated for 15 min at 4°C. Cells were sorted through an LS column in a MidiMACS Separator (Miltenyi Biotec) according to the manufacturer’s protocols. Briefly, the column was primed with 3 ml of MACS buffer, cells were loaded onto the column, the column was washed three times with 3 ml of MACS buffer, the column was removed from the MidiMACS Separator, and the cells were eluted with 5 ml of MACS buffer. The eluate was centrifuged for 5 min at 180g, 4°C and sorted through another LS column. Cells in the eluate were counted using a hemocytometer. The eluate was centrifuged for 5 min at 180g, 4°C. The resulting cell pellet was resuspended in a volume of E6-CSFD medium required to achieve a concentration of 105 cells/ml. The resulting cell suspension was distributed to Matrigel-coated six-well plates, 2 ml per well. For some experiments, Matrigel-coated 12-well plates (1 ml of cell suspension per well), 24-well plates (500 μl of cell suspension per well), or 48-well plates (250 μl of cell suspension per well) were used. Cells intended for confocal imaging were seeded on Matrigel-coated #1.5 glass-bottom plates (Cellvis, Sunnyvale, CA). Cells were incubated at 37°C, 5% CO2.
Flow cytometry
On day 15 of the neural crest differentiation, two aliquots of 106 cells were transferred to conical tubes before MACS and kept on ice until MACS was complete. Cells (106) from the final MACS eluate were also transferred to a conical tube. These three cell suspensions were centrifuged for 5 min at 180g, 4°C. One pre-MACS cell pellet and the post-MACS cell pellet were each resuspended in 100 μl of DPBS containing 0.2 μl of p75 antibody and 0.2 μl of HNK-1 antibody (table S2). The other pre-MACS cell pellet was resuspended in 100 μl of DPBS containing the mouse immunoglobulin G1 (IgG1) isotype control antibody and the mouse IgM isotype control antibody (table S2) at concentrations matched to the corresponding p75 and HNK-1 antibodies. Samples were incubated for 30 min on ice, washed by adding 2 ml of DPBS, and centrifuged for 5 min at 180g, 4°C. Each cell pellet was resuspended in 100 μl of DPBS containing 1:500 goat anti-mouse IgG1 Alexa Fluor 647 antibody and 1:500 goat anti-mouse IgM Alexa Fluor 488 antibody. Samples were incubated for 30 min on ice protected from light, washed by adding 2 ml of DPBS, and centrifuged for 5 min at 180g, 4°C. Cell pellets were fixed in 500 μl of 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 15 min at room temperature protected from light. Samples were centrifuged for 5 min at 180g, resuspended in 300 μl of DPBS, transferred to 5-ml flow cytometry tubes, and analyzed on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) with excitation at 488 and 635 nm, Alexa Fluor 488 emission detected with a 530/30 filter and Alexa Fluor 647 emission detected with a 661/16 filter. FlowJo software (BD Biosciences) was used for analysis.
Lentivirus production
The lentiviral plasmids pWPI (Addgene plasmid no. 12254), psPAX2 (Addgene plasmid no. 12260), and pMD2.G (Addgene plasmid no. 12259) were obtained from Addgene (Watertown, MA) as gifts from D. Trono. To generate pWPI-N3ICD, we amplified a cDNA fragment encoding the intracellular domain of Notch3 from a cDNA library generated from hPSC-derived neural crest. This fragment spans nucleotides 5074 to 7056 of National Center for Biotechnology Information (NCBI) Reference Sequence NM_000435.3, corresponding to amino acids 1662 to 2321 of NP_00426.2. To generate pWPI-N1ICD, we amplified a cDNA fragment encoding the intracellular domain of Notch1 from the neural crest cDNA library. This fragment spans nucleotides 5522 to 7930 of NM_017617.5, corresponding to amino acids 1754 to 2556 of NP060087.3. For N3ICD and N1ICD, the forward primers (table S3) contained a Kozak consensus sequence and start codon; forward and reverse primers (table S3) included Pac I restriction enzyme sites. To generate pWPI-TBX2, we amplified the TBX2 CDS from pcDNA3.1-TBX2 (NCBI Reference Sequence NM_005994.4) (GenScript, Piscataway, NJ). The forward primer (table S3) contained a Kozak consensus sequence; forward and reverse primers (table S3) included Pac I restriction enzyme sites. pWPI and the resulting PCR fragments were digested with Pac I. Ligation was performed with Instant Sticky-end Ligase Master Mix (New England Biolabs, Ipswich, MA), and resulting products were transformed into NEB Stable Competent Escherichia coli (New England Biolabs). Single ampicillin-resistant colonies were picked and PCR-screened for the presence of insert using primers annealing to the EF-1α promoter and internal ribosomal entry site (IRES) (table S3). Sanger sequencing was used to identify clones with forward-oriented inserts. pWPI, pWPI-N3ICD, pWPI-N1ICD, and pWPI-TBX2 plasmids were expanded and purified using the EndoFree Plasmid Maxi Kit (Qiagen, Germantown, MD). These plasmids have been deposited to Addgene: pWPI-N3ICD (Addgene no. 185524), pWPI-N1ICD (Addgene no. 185525), and pWPI-TBX2 (Addgene no. 185526).
293TN cells (System Biosciences, Palo Alto, CA) were maintained on uncoated six-well plates in DMEM (Life Technologies) supplemented with 10% FBS (Peak Serum, Wellington, CO), 1 mM sodium pyruvate (Life Technologies), and 0.5× GlutaMAX Supplement (Life Technologies). When 293TN cells reached 90% confluence, psPAX2 (1 μg per well), pMD2.G (0.5 μg per well), and pWPI or pWPI-N3ICD or pWPI-N1ICD or pWPI-TBX2 (1.5 μg per well) were cotransfected using FuGENE HD Transfection Reagent (9 μl per well) (Promega, Madison, WI). Medium was replaced 16 hours after transfection, and virus-containing supernatants were collected 24, 48, and 72 hours later. Supernatants were filtered through a 0.45-μm filter and concentrated 100× using Lenti-X Concentrator (Takara Bio, Mountain View, CA).
Lentiviral transduction
When neural crest cells reached ~40 to 50% confluence (~2 to 3 days after MACS), lentiviral transduction was performed by replacing medium in each well E6-CSFD medium containing 30 to 50 μl of N3ICD-GFP, N1ICD-GFP, or TBX2-GFP lentivirus per milliliter, or 5 to 8 μl of GFP (control) lentivirus per milliliter, which achieved transduction efficiencies of 50 to 80%. E6-CSFD medium was replaced every other day for 6 days. In some experiments, culture medium was supplemented with 10 μM CB-103 (MedChemExpress, Monmouth Junction, NJ). The resulting cultures were either used directly for analysis or sorted to isolate GFP+ and GFP− cells as described below.
Reverse transcription quantitative PCR
RNA extraction was performed using the RNeasy Plus Micro Kit (Qiagen). Cells were lysed with 350 μl of Buffer RLT supplemented with 1% β-mercaptoethanol (Sigma-Aldrich) and transferred to genomic DNA (gDNA) Eliminator spin columns. Three hundred fifty microliters of 70% ethanol was added to each lysate, and lysates were loaded onto RNeasy MinElute spin columns. Columns were washed with Buffer RW1, Buffer RPE, and 80% ethanol according to the manufacturer’s protocols. RNA was eluted with ribonuclease (RNase)–free water, and the concentration was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA). RNA (250 to 1000 ng) was reverse-transcribed for 1 hour at 37°C using the OmniScript RT Kit (Qiagen) and 1 μM Oligo(dT)12–18 primers (Life Technologies). RNaseOUT (1 U/μl; Life Technologies) was included in the reverse transcription reactions. Reaction products were diluted to 10 ng/μl. Twenty microliters of qPCR reactions was carried out with 10 ng of cDNA and 500 nM each forward and reverse primers (table S3) using PowerUp SYBR Green Master Mix (Life Technologies) and an AriaMx Real-Time PCR System (Agilent Technologies, Santa Clara, CA). An annealing temperature of 60°C was used for all reactions.
Immunoprecipitation
Cells were washed once with DPBS and lysed with cell lysis buffer (Cell Signaling Technology, Danvers, MA) supplemented with 1× Halt Protease Inhibitor Cocktail (Thermo Scientific). Lysates were sonicated with three 5-s pulses at 40% power with a 1/8-inch probe and centrifuged at 4°C for 5 min, 14,000g. Supernatants were transferred to new tubes, and protein concentrations were quantified using the Pierce BCA Protein Assay Kit (Thermo Scientific). Lysates were diluted with lysis buffer to 1 mg/ml. Each lysate (420 μl) was precleared by adding 40 μl of prewashed Protein A Magnetic Beads (Cell Signaling Technology) and incubating with rotation for 20 min at room temperature. Beads were removed using a magnetic separation rack. To reduce nonspecific adsorption of DNA to magnetic beads, DNA was fragmented by adding 2 μl (2000 U) of micrococcal nuclease to each lysate and incubating for 30 min at 37°C. Digestion was stopped by adding 10 μl of 0.5 M EDTA to each lysate. A 20-μl aliquot of each lysate was removed and stored at −80°C to serve as a 10% input control for total immunoprecipitation prey (RBPJ) abundance. The remaining 400 μl of each lysate was split between two new tubes, and Notch3 or isotype control antibody (at matched concentration, table S2) was added. Lysates were incubated with rotation overnight at 4°C. Twenty microliters of prewashed Protein A Magnetic Beads was added to each lysate and incubated with rotation for 20 min at room temperature. Beads were pelleted using a magnetic separation rack, supernatant was removed, and beads were washed with 500 μl of lysis buffer. This step was repeated for a total of five washes. After the final wash, beads were pelleted, supernatant was removed, and beads were resuspended in 20 μl of Western blot sample buffer. Western blot sample buffer (20 μl) was also added to each 10% input sample. All samples were heated at 95°C for 5 min. Beads were pelleted via centrifugation, and the resulting supernatants and 10% input samples were processed for anti-RBPJ Western blotting as described below, except that a mouse anti-rabbit IgG conformation-specific secondary antibody (table S2) was used for detection.
Western blotting
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Rockland Immunochemicals, Pottstown, PA) supplemented with 1× Halt Protease Inhibitor Cocktail and centrifuged at 4°C for 5 min, 14,000g. Supernatants were collected and transferred to new tubes, and protein concentrations were quantified using the Pierce BCA Protein Assay Kit. For each sample, ~20 μg of protein was diluted to equal volume with water, mixed with sample buffer, and heated at 95°C for 5 min. Samples were resolved on 4 to 12% tris-glycine gels and transferred to nitrocellulose membranes. Membranes were blocked for 1 hour in tris-buffered saline plus 0.1% Tween 20 (TBST) supplemented with 5% nonfat dry milk. Primary antibodies (table S2) were diluted in TBST supplemented with 5% nonfat dry milk and were added to membranes and incubated overnight at 4°C on a rocking platform. Membranes were washed five times with TBST. Secondary antibodies (table S2) were diluted in TBST supplemented with 5% nonfat dry milk and were added to membranes and incubated for 1 hour at room temperature on a rocking platform, protected from light. Membranes were washed five times with TBST and imaged using an Odyssey 9120 (LI-COR, Lincoln, NE). Band intensities were quantified using Image Studio software (LI-COR).
FACS and post-FACS culture
Six days after lentiviral transduction, cells were dissociated using ~30 min of Accutase treatment, 1 ml per well. The Accutase/single-cell suspension was transferred to 4× volume of DMEM/F12 medium and centrifuged for 5 min at 200g. The cell pellet was resuspended in MACS buffer supplemented with 4′,6-diamidino-2-phenylindole (DAPI; 2 μg/ml; Life Technologies). A FACSAria III cell sorter (BD Biosciences) was used to isolate DAPI−GFP+ cells (live, N3ICD-overexpressing cells) and DAPI−GFP− cells (live, nonoverexpressing cells). Excitation was at 405 and 488 nm, with DAPI emission detected with a 450/50 filter and GFP emission detected with a 502LP dichroic and 530/30 filter. Cells from a nontransduced well were used as a gating control. The resulting cell suspensions were centrifuged for 5 min at 200g, 4°C. Cell pellets were resuspended in E6 medium and seeded on Matrigel-coated plates at 2 × 104 cells/cm2. Medium was replaced daily.
Immunocytochemistry
Cells were washed once with DPBS and fixed with 4% paraformaldehyde for 15 min. Cells were washed three times with DPBS and blocked/permeabilized with DPBS supplemented with 10% goat serum (Life Technologies) and 0.1% Triton X-100 (Sigma-Aldrich) for 1 hour at room temperature. Primary antibodies (table S2) diluted in DPBS supplemented with 10% goat serum were added to cells and incubated overnight at 4°C on a rocking platform. Cells were washed three times with DPBS. Secondary antibodies (table S2) diluted in DPBS supplemented with 10% goat serum were added to cells and incubated for 1 hour at room temperature on a rocking platform, protected from light. Cells were washed three times with DPBS. Cells were incubated for 5 min in DPBS supplemented with 4 μM Hoechst 33342 (Life Technologies). Images were acquired using an Eclipse Ti2-E epifluorescence microscope (Nikon, Tokyo, Japan) with a 20× objective or an A1R-Si+ confocal microscope (Nikon) with a 100× oil objective.
RNA sequencing
RNA-seq was performed on cells from the H9, IMR90-4, DF19-9-11T, and WTC11 hPSC lines. For each line, differentiation-matched samples of neural crest cells and GFP− and GFP+ cells isolated via FACS 6 days after transduction of neural crest with N3ICD-GFP lentivirus were analyzed. Neural crest cells were dissociated with Accutase for ~5 min. The Accutase/single-cell suspension was transferred to 4× volume of DMEM/F12 medium and centrifuged for 5 min at 200g, 4°C. FACS was performed as described above; resulting GFP− and GFP+ populations were centrifuged for 5 min at 200g, 4°C. Supernatants were aspirated, and the resulting cell pellets were immediately lysed with Buffer RLT Plus (Qiagen) supplemented with 1% β-mercaptoethanol and frozen at −80°C. RNA extraction was performed using the RNeasy Plus Micro Kit (Qiagen). Lysates were thawed on ice, processed through gDNA Eliminator spin columns, processed through RNeasy MinElute spin columns per the manufacturer’s instructions, and eluted into RNase-free water.
RNA quality control, library preparation, and sequencing were performed by Novogene (Sacramento, CA). RNA quantity was assessed using a NanoDrop spectrophotometer; RNA quality was assessed using an Agilent 2100 Bioanalyzer. Poly(A) mRNA enrichment was performed using poly(T) oligo–conjugated magnetic beads, first-strand cDNA synthesis was performed using random hexamer primers, second-strand cDNA synthesis was performed, and libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs). Libraries were sequenced on a NovaSeq 6000 (Illumina, San Diego, CA) with approximately 20 million 150–base pair (bp) paired-end reads obtained for each sample.
RNA-seq data analysis
A DNA sequence from lentiviral transfer plasmid pWPI extending from the Pac I site to the 3′ end of the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) (containing the IRES and eGFP CDS) was added to the reference genome (hg38) to permit quantification of transgene-derived transcripts. RNA-seq FASTQ files from the experiment described above and from the literature (obtained from the Gene Expression Omnibus, see table S1) were aligned to the resulting reference genome using STAR (version 2.5.3a) (86). Gene-level counts were generated using the featureCounts function from Subread (version 2.0.3) (87). TPMs were calculated using gene lengths derived from featureCounts as previously described (40).
Transcriptome comparison between hPSC-derived brain pericyte–like cells, GFP+ cells from this work, and in vivo human brain pericytes was performed for protein-coding genes (based on the list at genenames.org/download/statistics-and-files). Data for in vivo brain pericytes were obtained from a previous meta-analysis of single-cell RNA-seq studies (41). In this meta-analysis, a mock bulk RNA-seq dataset was constructed from each source dataset (88–92) by (i) averaging gene counts across the pericyte cluster, (ii) obtaining the subset of protein-coding genes, and (iii) generating mock TPM values by normalizing total counts to 106. Bulk RNA-seq TPM values from hPSC-derived brain pericyte–like cells (from the literature) and GFP+ cells (this work) were similarly renormalized to 106 after obtaining the subset of protein-coding genes. To generate the scatterplots in Figs. 1A and 3G, resulting TPM values for each gene across the 11 hPSC-derived brain pericyte–like cell datasets or 4 GFP+ cell datasets were averaged, and TPM values for each gene across the 5 in vivo human brain pericyte datasets were averaged, followed by log transformation as log2(TPM + 1). The Pearson correlation coefficients were calculated on the basis of the log-transformed average TPM values.
Raw counts from featureCounts were input to DESeq2 (version 1.32.0) (93) for differential expression and principal component analyses. The DESeq2 variance stabilizing transformation was used to generate counts data for input to principal component analysis and hierarchical clustering. Hierarchical clustering on genes and samples (one minus Pearson correlation with average linkage) was performed using Morpheus (software.broadinstitute.org/morpheus/). Differential expression analysis was performed using the DESeq2 Wald test with Benjamini-Hochberg correction. The DESeq2 design included differentiation (hPSC line) matching as described above. Genes with adjusted P values <0.05 were considered differentially expressed. Gene Set Enrichment Analysis (GSEA; version 4.2.3) (94) was performed using DESeq2-normalized counts for neural crest and GFP+ cell samples. GSEA was performed with gene set permutation and otherwise default settings. Genes enriched in GFP+ cells compared to neural crest were tested against the KEGG (95) and GO-BP (96) databases (version 7.5.1). Gene sets with false discovery rates < 0.05 were considered enriched. Visualization of reads aligned to the human genome was performed using Integrative Genomics Viewer (version 2.5.0) (97).
Analysis of single-cell RNA-seq data from developing mouse brain (48) was performed in Scanpy (version 1.9.1) (98). The loom file containing expression data and metadata was obtained from the authors’ website (see table S1). Clusters annotated as neural crest and mesenchymal cell types by the authors were selected for analysis. A complete list of the authors’ subclass identifiers is shown in fig. S1A. A dot plot was used to visualize expression of neural crest, pan-mesenchymal, fibroblast, pan-mural, pericyte, and VSMC markers.
Decellularization and quantification of ECM
Decellularization was performed 6 days after transduction of neural crest cultures in 12-well plates with GFP or N3ICD-GFP lentiviruses as described above. The decellularization protocol was adapted from (99). The following buffers were prewarmed to 37°C: DPBS, wash buffer 1 (100 mM disodium phosphate, 2 mM magnesium chloride, and 2 mM EDTA, pH 9.6), lysis buffer (8 mM disodium phosphate and 1% Triton X-100, pH 9.6), and wash buffer 2 (10 mM disodium phosphate and 300 mM potassium iodide, pH 7.5). Cells were washed twice with 1 ml of DPBS and three times with 1 ml of wash buffer 1. Cells were incubated with 1 ml of lysis buffer for 15 min at 37°C. Lysis buffer was replaced with 1 ml of fresh lysis buffer; cells were incubated for 1 hour at 37°C. Lysis buffer was replaced with 1 ml of fresh lysis buffer; cells were incubated for an additional 1 hour at 37°C. Lysis buffer was removed, and the resulting ECM was washed three times with 1 ml of wash buffer 2 and four times with 1 ml of water. Water was removed, and 250 μl of RIPA buffer was added. ECM was scraped from the bottom of the well. The resulting solution was transferred to a microcentrifuge tube and sonicated with two 10-s pulses at 40% power with a 1/8-inch probe. Protein concentration in the resulting solution was quantified using the bicinchoninic acid (BCA) assay. For normalization of total protein to cell number, cells from a parallel well of the 12-well plate were dissociated using Accutase and counted using a hemocytometer.
Cord formation assays
To assess the ability of ECM from GFP- and N3ICD-GFP–transduced cells to support endothelial cord formation, neural crest cultures in 48-well plates 6 days after transduction with GFP or N3ICD-GFP lentiviruses were decellularized. The decellularization protocol was as described above, except that all wash and incubation steps were performed using 200 μl of solution, and the protocol was terminated after the final wash with water. This procedure was also performed on parallel cell-free wells to serve as a no-ECM control. Additional parallel wells were coated with 200 μl of Matrigel, Growth Factor Reduced, which was allowed to gel at 37°C for 1 hour. HUVECs (2.75 × 104) in 250 μl of EGM-2 medium were added to each well. Phase contrast images were acquired 16 hours after addition of HUVECs using an Eclipse Ti2-E microscope with a 4× objective. Cells were subsequently fixed and processed for VE-cadherin immunocytochemistry as described above. To quantify the extent of cord formation, blinded phase contrast images were scored on the following four-point scale: 0, no cords apparent; 1, few cords apparent, most cells not associated with cords; 2, many cords apparent, most cells associated with cords; 3, virtually all cells associated with cords.
The coculture cord formation assay was performed 5 days after isolation of GFP+ and GFP− cells via FACS from an N3ICD-GFP–transduced culture as described above. Eight-well chamber slides were coated with Matrigel, Growth Factor Reduced, at 250 μl per well. Matrigel was allowed to gel at 37°C for 1 hour. HUVECs (American Type Culture Collection, Manassas, VA) maintained in EGM-2 medium (Lonza, Walkersville, MD) were dissociated using a ~15-min treatment with 0.25% trypsin-EDTA (Gibco). The resulting cell suspension was transferred to a 4× volume of DMEM supplemented with 10% FBS. Neural crest cells, GFP+ cells, and GFP− cells were dissociated using 5 to 15 min of Accutase treatment, and the resulting cell suspensions were transferred to 4× volumes of DMEM/F12 medium. Cells were counted using a hemocytometer and centrifuged for 5 min at 200g. Supernatants were removed, and cell pellets were resuspended in 1 ml of EGM-2 medium. For the HUVEC-only control, HUVEC cell suspension and EGM-2 medium were combined to yield a suspension containing 2.2 × 104 HUVECs per 500 μl. For the coculture conditions, HUVEC cell suspension, coculture cell suspension (neural crest, GFP+, or GFP− cell suspension), and EGM-2 medium were combined to yield suspensions containing 2.2 × 104 HUVECs and 6.6 × 104 coculture cells per 500 μl. Five hundred microliters of the resulting cell suspensions was added to the prepared wells of the eight-well chamber slides. Phase contrast and GFP images were acquired after 24 and 72 hours using an Eclipse Ti2-E microscope with a 4× objective.
Calcium imaging and contraction assay
Calcium imaging was performed 6 days after transduction of neural crest cultures with N3ICD-GFP lentivirus as described above. FLIPR Calcium 6 dye (Molecular Devices, San Jose, CA) was prepared according to the manufacturer’s instructions. The prepared dye (500 μl) was added to the existing 500 μl of culture medium, and cells were incubated at 37°C, 5% CO2 for 2 hours. The plate was transferred to a microscope environmental chamber at 37°C, 5% CO2 and equilibrated for 30 min. Images were acquired every 5 s for 300 s using an Eclipse Ti2-E microscope with a 4× objective. At t = 50 s, a 1:114 dilution of saturated KCl solution (4.56 M, for a final concentration of 40 mM) was added to the well. FIJI/ImageJ (100) was used to quantify mean fluorescence intensity Ft at each time point, and data are displayed as ΔF/F = (Ft – F0)/F0.
To assess function of KATP channels, cells 6 days after transduction with GFP or N3ICD-GFP lentiviruses were labeled with FLIPR Calcium 6 dye as above, and 20 μM pinacidil (Santa Cruz Biotechnology, Dallas, TX) or DMSO was added 1.5 hours after addition of dye. After an additional 30 min of incubation, the plate was transferred to the microscope environmental chamber. Images were acquired every 5 s for 60 s, and 40 mM KCl was added at t = 20 s. Mean fluorescence intensity was quantified as above.
The contraction assay was performed 2 days after isolation of GFP+ cells via FACS from an N3ICD-GFP–transduced culture as described above. The plate was transferred to a microscope environmental chamber at 37°C, 5% CO2 and equilibrated for 30 min. At t = 0 min, a 1:114 dilution of saturated potassium chloride solution, or an equivalent volume of water, was added to the culture medium and the plate was briefly rocked to mix. Phase contrast images were acquired immediately upon addition of potassium chloride or water and 15 min thereafter using an Eclipse Ti2-E microscope with a 20× objective. The Freehand Selection Tool in FIJI/ImageJ was used to trace the outlines of 16 cells per 20× field at times 0 and 15 min, and the Measure function was used to obtain A0 and A15, the cell areas at times 0 and 15 min, respectively. For each cell, the percent change in area was computed as (A15 – A0)/A0 × 100%. These values were averaged across the 16 cells in each field to generate the values shown in Fig. 4.
Chromatin immunoprecipitation sequencing
ChIP-seq was performed on cells 6 days after transduction of neural crest cultures with GFP and N3ICD-GFP lentivirus as described above. Three independent differentiations were performed, with average transduction efficiency of GFP-transduced cultures 82 ± 9% GFP+ and N3ICD-GFP–transduced cultures 87 ± 3% GFP+. Cells were dissociated with Accutase and transferred to DMEM/F12. Cross-linking was performed with 1% formaldehyde for 10 min, and the reaction was quenched with glycine. Cells were centrifuged for 5 min at 2000g, 4°C. Supernatants were removed, and cell pellets were stored at −80°C. Nuclei isolation and chromatin digestion with micrococcal nuclease were performed using the SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads) (Cell Signaling Technology) according to the manufacturer’s protocols. Fifty microliters of resulting chromatin preparation from a GFP-transduced sample was subject to cross-link reversal and DNA purification to confirm appropriate digestion and estimate chromatin concentration. Approximately 60 μg of chromatin was used for each immunoprecipitation. Samples were diluted to 500 μl, and Notch3 antibody (table S2) was added. Samples were incubated overnight with rotation at 4°C. Antibody-bound fragments were isolated using protein G magnetic beads. Resulting chromatin samples and matched 10% input control samples were subjected to cross-link reversal and DNA purification using the SimpleChIP Enzymatic Chromatin IP Kit, per the manufacturer’s protocols. DNA quality control, library preparation, and sequencing were performed by Novogene. Libraries were sequenced on a NovaSeq 6000 with approximately 30 million 150-bp paired-end reads obtained for each sample.
ChIP-seq data analysis
Sequencing reads were aligned to the human genome (hg38) using bowtie2 (version 2.2.1) (101). Sambamba (version 0.8.2) (102) and bedtools (version 2.30.0) (103) were used to remove unmapped reads, reads mapped to blacklist regions (using the database available at https://github.com/Boyle-Lab/Blacklist) (104), and reads mapped to decoys and nonchromosomal assemblies. Peakcalling was performed with MACS (version 3.0.0a7) (105), using IP and matched input control samples, paired-end mode, and a P value cutoff of 0.0005. DiffBind (version 3.2.7) (64) calling DESeq2 (93) was used to identify peaks enriched in N3ICD-GFP–transduced samples compared to GFP-transduced samples (adjusted P < 0.05, DESeq2 Wald test with Benjamini-Hochberg correction). ChIPseeker (version 1.28.3) (106) was used for peak annotation. DeepTools (version 3.5.1) (107) was used to generate bigWig files and peak profile plots. MEME-ChIP (https://meme-suite.org/meme/tools/meme-chip, version 5.4.1) (108) was used for motif enrichment analysis. Genome browser visualizations were created using the UCSC Genome Browser (109).
Statistical analysis
Individual wells of cultured cells that underwent identical experimental treatments are defined as replicates. Details of replication strategy are provided in figure legends. Student’s unpaired or paired t tests were used for comparison of means from two experimental groups. One-way analysis of variance (ANOVA) was used for comparison of means from three or more experimental groups. Two- or three-way ANOVA was used for comparison of means and blocking of differentiation-based variability if data from multiple differentiations were combined [one/two factors being the experimental treatment(s) and one factor being the differentiation]. Following ANOVA, Dunnett’s post hoc test was used for comparison of multiple treatments to a single control, or Tukey’s honest significant difference test was used for multiple pairwise comparisons. For cord formation score data, the nonparametric Kruskal-Wallis test was used followed by the Steel-Dwass test for multiple pairwise comparisons.
Acknowledgments
We thank M. J. Stebbins for advice in the conceptualization of this project, G. Jin for advice related to lentivirus production, M. E. Floy for advice related to decellularization, J. Svaren for advice related to ChIP-seq, and K. L. Foreman for assistance in image analysis. We acknowledge the University of Wisconsin Carbone Cancer Center Flow Cytometry Laboratory (supported by NIH Cancer Center Support Grant P30 CA014520) for use of a flow cytometer and for performing FACS, the University of Wisconsin–Madison Biochemistry Optical Core for use of a confocal microscope, and CyVerse (www.cyverse.org, supported by the National Science Foundation under award numbers DBI-0735191, DBI-1265383, and DBI-1743442) for web-accessible hosting of ChIP-seq data.
Funding: This work was funded by the NIH (NS103844 and NS132441 to E.V.S., S.P.P., and R.D.). B.D.G. was supported by the NIH Biotechnology Training Program (T32 GM008349) and the NSF Graduate Research Fellowship Program (1747503). M.E.S. was supported by a Hilldale Fellowship from the University of Wisconsin–Madison. H.E.H. was supported by The Hartwell Foundation Postdoctoral Fellowship.
Author contributions: B.D.G.: Conceptualization, formal analysis, investigation, methodology, and writing—original draft. M.E.S.: Conceptualization, investigation, methodology, and writing—review and editing. H.E.H.: Investigation and writing—review and editing. R.D.: Conceptualization, funding acquisition, supervision, and writing—review and editing. S.P.P.: Conceptualization, funding acquisition, supervision, and writing–review and editing. E.V.S.: Conceptualization, funding acquisition, supervision, and writing—review and editing.
Competing interests: B.D.G., S.P.P., and E.V.S. are inventors on U.S. patent application US17/933,730 related to this work. The other authors declare no competing interests.
Data and materials availability: RNA-seq and ChIP-seq data have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE205214. ChIP-seq data can additionally be viewed in the UCSC Genome Browser at: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubUrl=https://data.cyverse.org/dav-anon/iplant/home/gastfriend/hub.txt. Plasmids generated in this study have been deposited at Addgene (see Materials and Methods). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 to S3
Figs. S1 to S9
Legends for files S1 to S3
References
Other Supplementary Material for this manuscript includes the following:
Files S1 to S3
REFERENCES AND NOTES
- 1.Armulik A., Abramsson A., Betsholtz C., Endothelial/pericyte interactions. Circ. Res. 97, 512–523 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Armulik A., Genové G., Betsholtz C., Pericytes: Developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Hosford P. S., Christie I. N., Niranjan A., Aziz Q., Anderson N., Ang R., Lythgoe M. F., Wells J. A., Tinker A., Gourine A. V., A critical role for the ATP-sensitive potassium channel subunit KIR6.1 in the control of cerebral blood flow. J. Cereb. Blood Flow Metab. 39, 2089–2095 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikolakopoulou A. M., Montagne A., Kisler K., Dai Z., Wang Y., Huuskonen M. T., Sagare A. P., Lazic D., Sweeney M. D., Kong P., Wang M., Owens N. C., Lawson E. J., Xie X., Zhao Z., Zlokovic B. V., Pericyte loss leads to circulatory failure and pleiotrophin depletion causing neuron loss. Nat. Neurosci. 22, 1089–1098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kisler K., Nikolakopoulou A. M., Sweeney M. D., Lazic D., Zhao Z., Zlokovic B. V., Acute ablation of cortical pericytes leads to rapid neurovascular uncoupling. Front. Cell. Neurosci. 14, 27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kisler K., Nelson A. R., Rege S. V., Ramanathan A., Wang Y., Ahuja A., Lazic D., Tsai P. S., Zhao Z., Zhou Y., Boas D. A., Sakadžić S., Zlokovic B. V., Pericyte degeneration leads to neurovascular uncoupling and limits oxygen supply to brain. Nat. Neurosci. 20, 406–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall C. N., Reynell C., Gesslein B., Hamilton N. B., Mishra A., Sutherland B. A., O’Farrell F. M., Buchan A. M., Lauritzen M., Attwell D., Capillary pericytes regulate cerebral blood flow in health and disease. Nature 508, 55–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill R. A., Tong L., Yuan P., Murikinati S., Gupta S., Grutzendler J., Regional blood flow in the normal and ischemic brain is controlled by arteriolar smooth muscle cell contractility and not by capillary pericytes. Neuron 87, 95–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peppiatt C. M., Howarth C., Mobbs P., Attwell D., Bidirectional control of CNS capillary diameter by pericytes. Nature 443, 700–704 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rungta R. L., Chaigneau E., Osmanski B.-F. F., Charpak S., Vascular compartmentalization of functional hyperemia from the synapse to the Pia. Neuron 99, 362–375.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armulik A., Genové G., Mäe M., Nisancioglu M. H., Wallgard E., Niaudet C., He L., Norlin J., Lindblom P., Strittmatter K., Johansson B. R., Betsholtz C., Pericytes regulate the blood–brain barrier. Nature 468, 557–561 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Bell R. D., Winkler E. A., Sagare A. P., Singh I., LaRue B., Deane R., Zlokovic B. V., Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 68, 409–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daneman R., Zhou L., Kebede A. A., Barres B. A., Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grubb S., Cai C., Hald B. O., Khennouf L., Murmu R. P., Jensen A. G. K. K., Fordsmann J., Zambach S., Lauritzen M., Precapillary sphincters maintain perfusion in the cerebral cortex. Nat. Commun. 11, 395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann D. A., Berthiaume A. A., Grant R. I., Harrill S. A., Koski T., Tieu T., McDowell K. P., Faino A. V., Kelly A. L., Shih A. Y., Brain capillary pericytes exert a substantial but slow influence on blood flow. Nat. Neurosci. 24, 633–645 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanlandewijck M., He L., Mäe M. A., Andrae J., Ando K., Del Gaudio F., Nahar K., Lebouvier T., Laviña B., Gouveia L., Sun Y., Raschperger E., Räsänen M., Zarb Y., Mochizuki N., Keller A., Lendahl U., Betsholtz C., A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Grant R. I., Hartmann D. A., Underly R. G., Berthiaume A. A., Bhat N. R., Shih A. Y., Organizational hierarchy and structural diversity of microvascular pericytes in adult mouse cortex. J. Cereb. Blood Flow Metab. 39, 411–425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmann D. A., Coelho-Santos V., Shih A. Y., Pericyte control of blood flow across microvascular zones in the central nervous system. Annu. Rev. Physiol. 84, 331–354 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell R. D., Winkler E. A., Singh I., Sagare A. P., Deane R., Wu Z., Holtzman D. M., Betsholtz C., Armulik A., Sallstrom J., Berk B. C., Zlokovic B. V., Apolipoprotein e controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halliday M. R., Rege S. V., Ma Q., Zhao Z., Miller C. A., Winkler E. A., Zlokovic B. V., Accelerated pericyte degeneration and blood-brain barrier breakdown in apolipoprotein E4 carriers with Alzheimer’s disease. J. Cereb. Blood Flow Metab. 36, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nortley R., Korte N., Izquierdo P., Hirunpattarasilp C., Mishra A., Jaunmuktane Z., Kyrargyri V., Pfeiffer T., Khennouf L., Madry C., Gong H., Richard-Loendt A., Huang W., Saito T., Saido T. C., Brandner S., Sethi H., Attwell D., Amyloid β oligomers constrict human capillaries in Alzheimer’s disease via signaling to pericytes. Science 365, eaav9518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joutel A., Andreux F., Gaulis S., Domenga V., Cecillon M., Battail N., Piga N., Chapon F., Godfrain C., Tournier-Lasserve E., The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J. Clin. Invest. 105, 597–605 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh M., Balbi M., Hellal F., Dichgans M., Lindauer U., Plesnila N., Pericytes are involved in the pathogenesis of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Ann. Neurol. 78, 887–900 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Etchevers H. C., Vincent C., Le Douarin N. M., Couly G. F., The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development 128, 1059–1068 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Korn J., Christ B., Kurz R., Kurz H., Kurz R., Kurz H., Kurz R., Kurz H., Neuroectodermal origin of brain pericytes and vascular smooth muscle cells. J. Comp. Neurol. 442, 78–88 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Yamanishi E., Takahashi M., Saga Y., Osumi N., Penetration and differentiation of cephalic neural crest-derived cells in the developing mouse telencephalon. Dev. Growth Differ. 54, 785–800 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Le Lièvre C. S., Le Douarin N. M., Mesenchymal derivatives of the neural crest: Analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol. 34, 125–154 (1975). [PubMed] [Google Scholar]
- 28.Etchevers H. C., Couly G., Vincent C., Le Douarin N. M., Anterior cephalic neural crest is required for forebrain viability. Development 126, 3533–3543 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Jiang X., Iseki S., Maxson R. E., Sucov H. M., Morriss-Kay G. M., Tissue origins and interactions in the mammalian skull vault. Dev. Biol. 241, 106–116 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Lindahl P., Johansson B. R., Levéen P., Betsholtz C., Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242–245 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Hellström M., Kalén M., Lindahl P., Abramsson A., Betsholtz C., Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047–3055 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Ando K., Wang W., Peng D., Chiba A., Lagendijk A. K., Barske L., Crump J. G., Stainier D. Y. R. R., Lendahl U., Koltowska K., Hogan B. M., Fukuhara S., Mochizuki N., Betsholtz C., Peri-arterial specification of vascular mural cells from naïve mesenchyme requires Notch signaling. Development 146, dev165589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Pan L., Moens C. B., Appel B., Notch3 establishes brain vascular integrity by regulating pericyte number. Development 141, 307–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wurdak H., Inactivation of TGFβ signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 19, 530–535 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henshall T. L., Keller A., He L., Johansson B. R., Wallgard E., Raschperger E., Mäe M. A., Jin S., Betsholtz C., Lendahl U., Notch3 is necessary for blood vessel integrity in the central nervous system. Arterioscler. Thromb. Vasc. Biol. 35, 409–420 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Nadeem T., Bogue W., Bigit B., Cuervo H., Deficiency of Notch signaling in pericytes results in arteriovenous malformations. JCI Insight 5, e125940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah N. M., Groves A. K., Anderson D. J., Alternative neural crest cell fates are instructively promoted by TGFbeta superfamily members. Cell 85, 331–343 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Menendez L., Yatskievych T. A., Antin P. B., Dalton S., Menendez L., Yatskievych T. A., Antin P. B., Dalton S., Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc. Natl. Acad. Sci. U.S.A. 109, 9220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee G., Kim H., Elkabetz Y., Al Shamy G., Panagiotakos G., Barberi T., Tabar V., Studer L., Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 25, 1468–1475 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Song H. W., Foreman K. L., Gastfriend B. D., Kuo J. S., Palecek S. P., Shusta E. V., Transcriptomic comparison of human and mouse brain microvessels. Sci. Rep. 10, 12358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gastfriend B. D., Foreman K. L., Katt M. E., Palecek S. P., Shusta E. V., Integrative analysis of the human brain mural cell transcriptome. J. Cereb. Blood Flow Metab. 41, 3052–3068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gastfriend B. D., Palecek S. P., Shusta E. V., Modeling the blood–brain barrier: Beyond the endothelial cells. Curr. Opin. Biomed. Eng. 5, 6–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stebbins M. J., Gastfriend B. D., Canfield S. G., Lee M.-S. S., Richards D., Faubion M. G., Li W.-J. J., Daneman R., Palecek S. P., Shusta E. V., Human pluripotent stem cell–derived brain pericyte–like cells induce blood-brain barrier properties. Sci. Adv. 5, eaau7375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gastfriend B. D., Stebbins M. J., Du F., Shusta E. V., Palecek S. P., Differentiation of brain pericyte-like cells from human pluripotent stem cell−derived neural crest. Curr. Protoc. 1, e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffin C., Bajpai R., Neural crest-derived human cranial pericytes model primary forebrain pericytes and predict disease-specific cranial vasculature defects. SSRN Electron. J., (2017). [Google Scholar]
- 46.Sun J., Huang Y., Gong J., Wang J., Fan Y., Cai J., Wang Y., Qiu Y., Wei Y., Xiong C., Chen J., Wang B., Ma Y., Huang L., Chen X., Zheng S., Huang W., Ke Q., Wang T., Li X., Zhang W., Xiang A. P., Li W., Transplantation of hPSC-derived pericyte-like cells promotes functional recovery in ischemic stroke mice. Nat. Commun. 11, 5196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ando K., Tong L., Peng D., Vázquez-Liébanas E., Chiyoda H., He L., Liu J., Kawakami K., Mochizuki N., Fukuhara S., Grutzendler J., Betsholtz C., KCNJ8/ABCC9-containing K-ATP channel modulates brain vascular smooth muscle development and neurovascular coupling. Dev. Cell 57, 1383–1399.e7 (2022). [DOI] [PubMed] [Google Scholar]
- 48.La Manno G., Siletti K., Furlan A., Gyllborg D., Vinsland E., Mossi Albiach A., Mattsson Langseth C., Khven I., Lederer A. R., Dratva L. M., Johnsson A., Nilsson M., Lönnerberg P., Linnarsson S., Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Lehal R., Zaric J., Vigolo M., Urech C., Frismantas V., Zangger N., Cao L., Berger A., Chicote I., Loubéry S., Choi S. H., Koch U., Blacklow S. C., Palmer H. G., Bornhauser B., González-Gaitán M., Arsenijevic Y., Zoete V., Aster J. C., Bourquin J. P., Radtke F., Pharmacological disruption of the Notch transcription factor complex. Proc. Natl. Acad. Sci. U.S.A. 117, 16292–16301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soldatov R., Kaucka M., Kastriti M. E., Petersen J., Chontorotzea T., Englmaier L., Akkuratova N., Yang Y., Häring M., Dyachuk V., Bock C., Farlik M., Piacentino M. L., Boismoreau F., Hilscher M. M., Yokota C., Qian X., Nilsson M., Bronner M. E., Croci L., Hsiao W.-Y. Y., Guertin D. A., Brunet J.-F. F., Consalez G. G., Ernfors P., Fried K., Kharchenko P. V., Adameyko I., Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Siegenthaler J. A., Choe Y., Patterson K. P., Hsieh I., Li D., Jaminet S.-C. S.-C. C., Daneman R., Kume T., Huang E. J., Pleasure S. J., Foxc1 is required by pericytes during fetal brain angiogenesis. Biol. Open 2, 647–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyahi A., Nik A. M., Ghiami M., Gritli-Linde A., Pontén F., Johansson B. R., Carlsson P., Foxf2 is required for brain pericyte differentiation and development and maintenance of the blood-brain barrier. Dev. Cell 34, 19–32 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharya D., Rothstein M., Azambuja A. P., Simoes-Costa M., Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis. eLife 7, e40556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang A. C., Vest R. T., Kern F., Lee D. P., Agam M., Maat C. A., Losada P. M., Chen M. B., Schaum N., Khoury N., Toland A., Calcuttawala K., Shin H., Pálovics R., Shin A., Wang E. Y., Luo J., Gate D., Schulz-Schaeffer W. J., Chu P., Siegenthaler J. A., McNerney M. W., Keller A., Wyss-Coray T., A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muhl L., Genové G., Leptidis S., Liu J., He L., Mocci G., Sun Y., Gustafsson S., Buyandelger B., Chivukula I. V., Segerstolpe Å., Raschperger E., Hansson E. M., Björkegren J. L. M., Peng X., Vanlandewijck M., Lendahl U., Betsholtz C., Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun. 11, 3953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeSisto J., O’Rourke R., Jones H. E., Pawlikowski B., Malek A. D., Bonney S., Guimiot F., Jones K. L., Siegenthaler J. A., Single-cell transcriptomic analyses of the developing meninges reveal meningeal fibroblast diversity and function. Dev. Cell 54, 43–59.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia F. J., Sun N., Lee H., Godlewski B., Mathys H., Galani K., Zhou B., Jiang X., Ng A. P., Mantero J., Tsai L.-H., Bennett D. A., Sahin M., Kellis M., Heiman M., Single-cell dissection of the human brain vasculature. Nature 603, 893–899 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubota Y., Kleinman H. K., Martin G. R., Lawley T. J., Role of laminin and basement membrane in the morphological differentiation of human endothelial cells into capillary-like structures. J. Cell Biol. 107, 1589–1598 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar A., D’Souza S. S., Moskvin O. V., Toh H., Wang B., Zhang J., Swanson S., Guo L. W., Thomson J. A., Slukvin I. I., Specification and diversification of pericytes and smooth muscle cells from mesenchymoangioblasts. Cell Rep. 19, 1902–1916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lilly B., Kennard S., Differential gene expression in a coculture model of angiogenesis reveals modulation of select pathways and a role for Notch signaling. Physiol. Genomics 36, 69–78 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzales A. L., Klug N. R., Moshkforoush A., Lee J. C., Lee F. K., Shui B., Tsoukias N. M., Kotlikoff M. I., Hill-Eubanks D., Nelson M. T., Contractile pericytes determine the direction of blood flow at capillary junctions. Proc. Natl. Acad. Sci. U.S.A. 117, 27022–27033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zambach S. A., Cai C., Helms H. C. C., Hald B. O., Dong Y., Fordsmann J. C., Nielsen R. M., Hu J., Lønstrup M., Brodin B., Lauritzen M. J., Precapillary sphincters and pericytes at first-order capillaries as key regulators for brain capillary perfusion. Proc. Natl. Acad. Sci. U.S.A. 118, e2023749118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray S. J., Gomez-Lamarca M., Notch after cleavage. Curr. Opin. Cell Biol. 51, 103–109 (2018). [DOI] [PubMed] [Google Scholar]
- 64.R. Stark, G. Brown, DiffBind: Differential binding analysis of ChIP-Seq peak data; http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf.
- 65.Castel D., Mourikis P., Bartels S. J. J. J., Brinkman A. B., Tajbakhsh S., Stunnenberg H. G., Dynamic binding of RBPJ is determined by notch signaling status. Genes Dev. 27, 1059–1071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziffra R. S., Kim C. N., Ross J. M., Wilfert A., Turner T. N., Haeussler M., Casella A. M., Przytycki P. F., Keough K. C., Shin D., Bogdanoff D., Kreimer A., Pollard K. S., Ament S. A., Eichler E. E., Ahituv N., Nowakowski T. J., Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo Y., Hitz B. C., Gabdank I., Hilton J. A., Kagda M. S., Lam B., Myers Z., Sud P., Jou J., Lin K., Baymuradov U. K., Graham K., Litton C., Miyasato S. R., Strattan J. S., Jolanki O., Lee J. W., Tanaka F. Y., Adenekan P., O’Neill E., Cherry J. M., New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 48, D882–D889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castro-Mondragon J. A., Riudavets-Puig R., Rauluseviciute I., Lemma R. B., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Pérez N. M., Fornes O., Leung T. Y., Aguirre A., Hammal F., Schmelter D., Baranasic D., Ballester B., Sandelin A., Lenhard B., Vandepoele K., Wasserman W. W., Parcy F., Mathelier A., JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen S., Lechleider R. J., Transforming growth factor-beta-induced differentiation of smooth muscle from a neural crest stem cell line. Circ. Res. 94, 1195–1202 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Hirschi K. K., Rohovsky S. A., D’Amore P. A., PDGF, TGF-beta, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J. Cell Biol. 141, 805–814 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He L., Vanlandewijck M., Raschperger E., Andaloussi Mäe M., Jung B., Lebouvier T., Ando K., Hofmann J., Keller A., Betsholtz C., Analysis of the brain mural cell transcriptome. Sci. Rep. 6, 35108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheung C., Bernardo A. S., Trotter M. W. B., Pedersen R. A., Sinha S., Generation of human vascular smooth muscle subtypes provides insight into embryological origin–dependent disease susceptibility. Nat. Biotechnol. 30, 165–173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheung C., Goh Y. T., Zhang J., Wu C., Guccione E., Modeling cerebrovascular pathophysiology in amyloid-β metabolism using neural-crest-derived smooth muscle cells. Cell Rep. 9, 391–401 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Patsch C., Challet-Meylan L., Thoma E. C., Urich E., Heckel T., O’Sullivan J. F., Grainger S. J., Kapp F. G., Sun L., Christensen K., Xia Y., Florido M. H. C., He W., Pan W., Prummer M., Warren C. R., Jakob-Roetne R., Certa U., Jagasia R., Freskgard P. O., Adatto I., Kling D., Huang P., Zon L. I., Chaikof E. L., Gerszten R. E., Graf M., Iacone R., Cowan C. A., Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 17, 994–1003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faal T., Phan D. T. T. T., Davtyan H., Scarfone V. M., Varady E., Hughes C. C. W. W., Inlay M. A., Blurton-Jones M., Hughes C. C. W. W., Inlay M. A., Induction of mesoderm and neural crest-derived pericytes from human pluripotent stem cells to study blood-brain barrier interactions. Stem Cell Reports 12, 451–460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown L. A., Sava P., Garcia C., Gonzalez A. L., Haven N., Brown L. A., Sava P., Garcia C., Gonzalez A. L., Haven N., Proteomic analysis of the pericyte derived extracellular matrix. Cell. Mol. Bioeng. 8, 349–363 (2015). [Google Scholar]
- 77.Montagne A., Nikolakopoulou A. M., Zhao Z., Sagare A. P., Si G., Lazic D., Barnes S. R., Daianu M., Ramanathan A., Go A., Lawson E. J., Wang Y., Mack W. J., Thompson P. M., Schneider J. A., Varkey J., Langen R., Mullins E., Jacobs R. E., Zlokovic B. V., Pericyte degeneration causes white matter dysfunction in the mouse central nervous system. Nat. Med. 24, 326–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Blanchard J. W., Bula M., Davila-Velderrain J., Akay L. A., Zhu L., Frank A., Victor M. B., Bonner J. M., Mathys H., Lin Y. T., Ko T., Bennett D. A., Cam H. P., Kellis M., Tsai L. H., Reconstruction of the human blood–brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. Nat. Med. 26, 952–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gastfriend B. D., Nishihara H., Canfield S. G., Foreman K. L., Engelhardt B., Palecek S. P., Shusta E. V., Wnt signaling mediates acquisition of blood–brain barrier properties in naïve endothelium derived from human pluripotent stem cells. eLife 10, 70992 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nishihara H., Perriot S., Gastfriend B. D., Steinfort M., Cibien C., Soldati S., Matsuo K., Guimbal S., Mathias A., Palecek S. P., Shusta E. V., Du Pasquier R., Engelhardt B., Intrinsic blood-brain barrier dysfunction contributes to multiple sclerosis pathogenesis. Brain 145, 4334–4348 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomson J. A., Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 82.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A., Induced pluripotent stem cell lines derived from human somatic cells. Science 1917, 154–155 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I. I., Thomson J. A., Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kreitzer F. R., Salomonis N., Sheehan A., Huang M., Park J. S., Spindler M. J., Lizarraga P., Weiss W. A., So P., Conklin B. R., A robust method to derive functional neural crest cells from human pluripotent stem cells. Am. J. Stem Cells 2, 119–131 (2013). [PMC free article] [PubMed] [Google Scholar]
- 85.Chen G., Gulbranson D. R., Hou Z., Bolin J. M., Ruotti V., Probasco M. D., Smuga-Otto K., Howden S. E., Diol N. R., Propson N. E., Wagner R., Lee G. O., Antosiewicz-Bourget J., Teng J. M. C., Thomson J. A., Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Hodge R. D., Bakken T. E., Miller J. A., Smith K. A., Barkan E. R., Graybuck L. T., Close J. L., Long B., Johansen N., Penn O., Yao Z., Eggermont J., Höllt T., Levi B. P., Shehata S. I., Aevermann B., Beller A., Bertagnolli D., Brouner K., Casper T., Cobbs C., Dalley R., Dee N., Ding S. L., Ellenbogen R. G., Fong O., Garren E., Goldy J., Gwinn R. P., Hirschstein D., Keene C. D., Keshk M., Ko A. L., Lathia K., Mahfouz A., Maltzer Z., McGraw M., Nguyen T. N., Nyhus J., Ojemann J. G., Oldre A., Parry S., Reynolds S., Rimorin C., Shapovalova N. V., Somasundaram S., Szafer A., Thomsen E. R., Tieu M., Quon G., Scheuermann R. H., Yuste R., Sunkin S. M., Lelieveldt B., Feng D., Ng L., Bernard A., Hawrylycz M., Phillips J. W., Tasic B., Zeng H., Jones A. R., Koch C., Lein E. S., Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Polioudakis D., de la Torre-Ubieta L., Langerman J., Elkins A. G., Shi X., Stein J. L., Vuong C. K., Nichterwitz S., Gevorgian M., Opland C. K., Lu D., Connell W., Ruzzo E. K., Lowe J. K., Hadzic T., Hinz F. I., Sabri S., Lowry W. E., Gerstein M. B., Plath K., Geschwind D. H., A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103, 785–801.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L. E., Stott S. R. W., Toledo E. M., Villaescusa J. C., Lönnerberg P., Ryge J., Barker R. A., Arenas E., Linnarsson S., Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W., Zhou Y., Ye F., Jiang M., Wu J., Xiao Y., Jia X., Zhang T., Ma X., Zhang Q., Bai X., Lai S., Yu C., Zhu L., Lin R., Gao Y., Wang M., Wu Y., Zhang J., Zhan R., Zhu S., Hu H., Wang C., Chen M., Huang H., Liang T., Chen J., Wang W., Zhang D., Guo G., Construction of a human cell landscape at single-cell level. Nature 581, 303–309 (2020). [DOI] [PubMed] [Google Scholar]
- 92.Zhong S., Ding W., Sun L., Lu Y., Dong H., Fan X., Liu Z., Chen R., Zhang S., Ma Q., Tang F., Wu Q., Wang X., Decoding the development of the human hippocampus. Nature 577, 531–536 (2020). [DOI] [PubMed] [Google Scholar]
- 93.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.The Gene Ontology Consortium, Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G., Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wolf F. A., Angerer P., Theis F. J., SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Floy M. E., Givens S. E., Matthys O. B., Mateyka T. D., Kerr C. M., Steinberg A. B., Silva A. C., Zhang J., Mei Y., Ogle B. M., McDevitt T. C., Kamp T. J., Palecek S. P., Developmental lineage of human pluripotent stem cell-derived cardiac fibroblasts affects their functional phenotype. FASEB J. 35, e21799 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Amemiya H. M., Kundaje A., Boyle A. P., The ENCODE Blacklist: Identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Shirley X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 107.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Machanick P., Bailey T. L., MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D., The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Everson J. L., Fink D. M., Yoon J. W., Leslie E. J., Kietzman H. W., Ansen-Wilson L. J., Chung H. M., Walterhouse D. O., Marazita M. L., Lipinski R. J., Sonic hedgehog regulation of Foxf2 promotes cranial neural crest mesenchyme proliferation and is disrupted in cleft lip morphogenesis. Development 144, 2082–2091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3
Figs. S1 to S9
Legends for files S1 to S3
References
Files S1 to S3