Abstract
The G·U wobble base pair is a fundamental unit of RNA secondary structure that is present in nearly every class of RNA from organisms of all three phylogenetic domains. It has comparable thermodynamic stability to Watson–Crick base pairs and is nearly isomorphic to them. Therefore, it often substitutes for G·C or A·U base pairs. The G·U wobble base pair also has unique chemical, structural, dynamic and ligand-binding properties, which can only be partially mimicked by Watson–Crick base pairs or other mispairs. These features mark sites containing G·U pairs for recognition by proteins and other RNAs and allow the wobble pair to play essential functional roles in a remarkably wide range of biological processes.
Historical perspective
Little did Francis Crick realize 30 years ago that his G·U wobble base pair used in decoding mRNA codons would have such an expansive biological significance (Crick, 1966). When considering codon–anticodon interactions, Crick noticed that the G and U bases would be able to form two hydrogen bonds by interacting through the same face of the base involved in Watson–Crick pairing (Figure 1). This prediction was confirmed when the G·U wobble pair in yeast tRNAPhe was observed at atomic resolution (Ladner et al., 1975; Quigley and Rich, 1975). The history of the G·U pair is, in fact, even older. Yeast tRNAAla, which was the first RNA molecule whose primary sequence was determined, could be folded into a secondary structure containing a single G·U wobble pair (Holley et al., 1965). It was later shown that the G·U pair is a major determinant of that molecule’s amino acid acceptor identity (Hou and Schimmel, 1988; McClain and Foss, 1988). G·U pairs have now been found in virtually every class of functional RNA, and have been shown to play many essential roles that are based upon the unique chemical and structural properties of the wobble pair. Structural features of G·U wobble pairs and other non-Watson–Crick pairs have recently been reviewed elsewhere (Hermann and Westhof, 1999; Masquida and Westhof, 2000). Here, we will review the biological roles of G·U pairs and discuss the specific features that allow them to function in a remarkably wide variety of biological contexts.
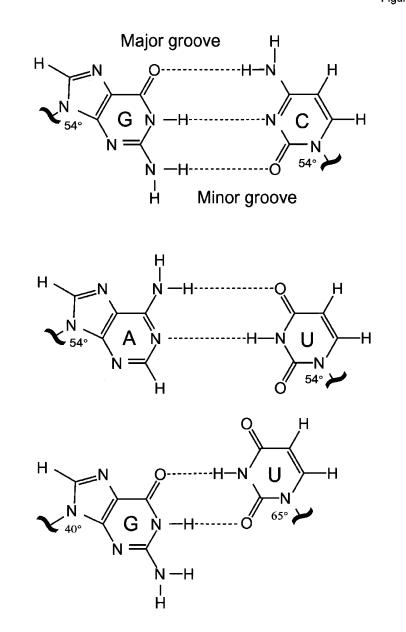
Fig. 1. Watson–Crick G·C and A·U base pairs differ from wobble G·U pairs in the type and location of functional groups that are projected into major and minor grooves. These pairs also differ in the orientation of the bases with respect to the phosphodiester backbone. Whereas the glycosidic angle is similar (~54°) for all nucleosides in Watson–Crick pairs, both angles for G and U differ in the wobble pair.
Ubiquitous building blocks of RNA structure
The functional importance of G·U pairs is underscored by their frequently high evolutionary conservation. For example, the wobble pair at the third position of the acceptor helix of tRNAAla is conserved in nearly all living organisms (Sprinzl et al., 1996). This conservation implies that the G·U pair possesses unique features that cannot be duplicated by any other nucleotide pair. G·U pairs constitute the most common mismatch in the helices of rRNA (Gautheret et al., 1995), and other tRNAs contain as many as four, but typically one, G·U pairs (Sprinzl et al., 1996). The mRNAs coding for ribosomal proteins S15 (Bénard et al., 1998) and L30 (Li et al., 1996) contain G·U pairs that provide recognition signals for autoregulation of protein synthesis. With the discovery of RNA catalysis, G·U pairs were also found to be associated with this ancient biological function of RNA. In fact, the first class of ribozymes discovered, group I self-splicing introns, contain a G·U pair at the site of cleavage that is nearly universally conserved (Doudna et al., 1989; Hur and Waring, 1995). Replacement of this wobble pair with a Watson–Crick pair compromises reactivity and fidelity (Doudna et al., 1989; Pyle et al., 1994). The hepatitis delta virus ribozyme also identifies its self-cleavage site through recognition of a G·U wobble pair (Perrotta and Been, 1996; Ferre-D’Amare et al., 1998). Group II self-splicing introns contain a nearly universally conserved G·U pair within domain 5, surrounded by two base pairs that are also nearly invariant (Peebles et al., 1995; Abramovitz et al., 1996; Konforti et al., 1998). Finally, RNAs that have been selected in vitro from large (1014) pools of randomized RNA sequences to perform various biological functions often contain G·U pairs (Ciesolka and Yarus, 1996; Illangasekare and Yarus, 1999; Khvorova et al., 1999).
The G·U pair functions in so many different contexts because it specifies unique recognition sites for proteins as diverse as aminoacyl-tRNA synthetases and ribosomal proteins, for other RNAs and for divalent metal ions. This ability resides in the distinctive structural, chemical and thermodynamic properties of the G·U pair, and in the unique conformational properties it confers upon RNA double helices. Each of these features can be exploited during recognition by proteins and other ligands.
Distinctive chemical, thermodynamic and structural properties
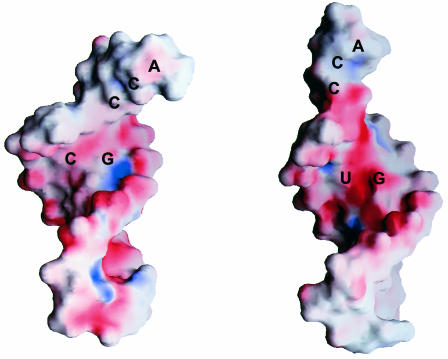
G·U pairs present a unique array of hydrogen bond donors and acceptors in the RNA major and minor grooves. These chemical groups can be recognized by complementary functionalities in a protein, RNA or other ligands. The exocyclic amino group in the minor groove is a distinctive feature of the G·U pair, since it is not base paired and is shifted with respect to the equivalent group in a normal G·C pair (Figure 1). Distinguishing chemical features are distributed in the major grooves well, and the presence of co-planar guanosine N7, guanosine O6 and uridine O4 defines a region of deep negative electrostatic potential (Allain and Varani, 1995a; McDowell and Turner, 1996). In contrast, G·C and A·U pairs both project an NH2 group into the major groove that interrupts the electrostatic field (Figure 2). This steep electrostatic gradient is a major determinant of the ability of divalent metal ions to bind the major groove of G·U pairs, but not that of double helical tracts composed entirely of Watson–Crick pairs (Ott et al., 1993; Allain and Varani, 1995a; Konforti et al., 1998). The affinity for metal ions becomes even stronger when two G·U pairs are present side-by-side (Cate and Doudna, 1996; McDowell and Turner, 1996; Kieft and Tinoco, 1997). Since most RNA enzymes are metalloenzymes in the sense that metal-binding sites generated by unique RNA structures position metals for promoting catalysis (Pyle, 1993), the ability of G·U pairs to bind divalent metal ions is likely to be important for RNA catalysis (Konforti et al., 1998).
Fig. 2. The G·C, A·U and G·U pairs have different functional groups in the major groove, leading to a significantly more electronegative environment at and near the G·U pair. As shown in this image, the surface electrostatic potential (negative is red and positive is blue) of tRNAAla acceptor end minihelices differs between the wild-type molecule containing G·U (right) and a mutant molecule where G·U is replaced by G·C (left).
A second important aspect of G·U pair function is its thermodynamic stability, which approaches that of Watson–Crick pairs and exceeds that of most other mispairs (Jaeger et al., 1989; Mathews et al., 1999; Strazewski et al., 1999). This high thermodynamic stability allows G·U pairs to substitute functionally for Watson–Crick base pairs in phylogenetically conserved double-helical tracts in rRNA, ribozymes and other RNAs. Also, in some sequence contexts, G·U pairs stabilize backbone turns. In tRNA, for example, the G·U pair is most frequently found at the junction of the single-stranded V loop and the T helix where the polynucleotide chain makes a sharp turn (Clark and Klug, 1975). However, thermodynamic stability is not sufficient for G·U function. For example, the aminoacylation capacity of active and inactive mutants of tRNAAla with various substitutions of G·U does not correlate with their respective thermodynamic stability (Strazewski et al., 1999).
G·U wobble pairs embedded within A-form RNA helices have a distinctive structure that results from the displacement of the bases of the G·U pair relative to the bases of Watson–Crick pairs. The glycosidic bond angle between the base and C1′ sugar atom of Watson–Crick pairs in an A-form RNA helix is ~54° for each of the four nucleotides (Figure 1). Thus, the four standard base pairs can be interchanged at a particular site in a double-stranded region without substantially affecting the helical parameters. In contrast, the glycosidic bond angles are dissimilar in the G·U wobble pair (G, 40° and U, 65°, approximately). The substitution of G·U by any Watson–Crick pair or by a U·G pair therefore results in a structural perturbation in any sequence context in which the wobble pair is located. In addition to these sequence-independent properties, other features of the structure of double helical regions containing wobble pairs depend strongly on sequence context (Masquida and Westhof, 2000). As noted both in crystal (Westhof et al., 1985; Masquida et al., 1999; Mueller et al., 1999) and in solution structures (Allain and Varani, 1995b; Ramos and Varani, 1997), G·U pairs introduce a pattern of overtwisting/undertwisting of the RNA double helix. This helical twist is influenced by the identity of the base pairs immediately adjacent to the wobble pair. The conformation of the phosphodiester backbone around the wobble pair also varies between G·U-containing sequences and even between crystallographic environments of a given sequence.
A final distinctive property of G·U base pairs refers to their conformational flexibility. G·U pairs are conformationally soft in the sense that they are found in different conformations in different chemical and structural environments. For example, G·U pairs respond more sharply to sequence context and crystal packing interactions than do Watson–Crick base pairs (Westhof et al., 1985). Because of this property, the conformation of the RNA double helix can be more easily altered at sites containing wobble pairs, allowing for recognition by induced fit (Ramos and Varani, 1997; Chang et al., 1999). As an example, a significant helical kink is observed at a G·U pair in the aspartyl-tRNA synthetase-bound form of tRNAAsp relative to free, unbound tRNAAsp (Ruff et al., 1991).
Specific recognition: chemical identity and structural context
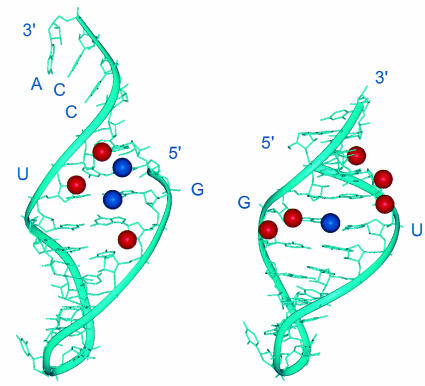
As discussed in the previous section, unique chemical features distinguish G·U pairs from Watson–Crick pairs or other mismatches. The functional role of individual chemical groups of the G·U pair in acceptor minihelix tRNAAla was demonstrated in an in vitro analysis of the enzymic activity of alanyl-tRNA synthetases (Musier-Forsyth et al., 1991; Musier-Forsyth and Schimmel, 1992). These studies identified specific chemical groups in the G·U pair that are critical for alanyl-tRNA synthetase activity. These include the exocyclic amino group of two guanosines, including that of G·U, and several ribose 2′-OH groups, all located in the minor groove of the tRNA acceptor end (Figure 3). A related but not identical ensemble of chemical groups was also demonstrated to be important for the ability of the catalytic core of group I introns to recognize the substrate for splicing or to stabilize the enzyme’s transition state (Strobel and Cech, 1995).
Fig. 3. View into the minor groove of RNA double helices showing the exocyclic amino groups (blue) and 2′-OH groups (red) that are critical for the activity of the G·U pairs in different functional contexts. The figure shows the three-dimensional structure of the tRNAAla acceptor end minihelix (left) and of the P1 helix substrate of group I self-splicing introns (right), highlighting chemical groups in the minor groove that contribute to G·U activity. A set of related but not identical chemical groups at and around the wobble pair is essential for activity in both contexts.
In addition to presenting distinctive chemical groups in the major and minor groove, individual G·U pairs also possess unique conformational features dictated by the sequence context in which the wobble pair is found and by factors that bind to it. The importance of sequence context and of the structural and dynamic features of the wobble pair in its function was demonstrated in genetic and biochemical studies of tRNAAla function conducted in vivo (Gabriel et al., 1996; Chang et al., 1999; McClain et al., 1999). These studies identified C·C and C·A nucleotide substitutions that functionally replace the G·U in tRNAAla even though they contain different chemical features. These results and those from other extensive investigations led to the conclusion that the G·U pair in tRNAAla is recognized through a combination of the direct read-out of its distinctive chemical identity and by the indirect recognition of the structural features of the double helix in which the G·U pair is embedded. G·U pairs in other functional contexts may also be recognized through a similarly complex mechanism. For example, the guanosine unpaired amino group is energetically important in the function of the group I self-splicing intron (Strobel and Cech, 1996), and other mispairs do not function as well as G·U within the upstream splice site for cleavage. However, A·C, which may be a close structural analogue of G·U, provides the best substitute (Doudna et al., 1989).
Since the conformation of an RNA double helix at and around the wobble pair varies from one sequence to another, incorporation of a G·U pair and the subsequent sequence-dependent distortion of the RNA double helix leads to an expansion of the structural diversity of A-form RNA. This property allows G·U pairs to form the main anchor points or fulcrums in networks of three-dimensional contacts with the catalytic core of an RNA enzyme or the RNA recognition domain of a protein. These networks of contacts centered on the wobble pair contribute to the high specificity observed in recognition or catalysis. Indirect recognition of G·U is a particularly powerful discrimination mechanism. It allows for productive interaction with cognate ligands, and thus positive discrimination. It also allows for rejection of non-cognate substrates through non-productive interactions with those substrates that lack a required G·U. In addition, a non-cognate substrate can be rejected as a result of steric conflict resulting from the distinctive structure of a G·U pair. An example of the latter type of discrimination is the non-productive interaction between charged selenocysteine tRNA and elongation factor Tu, which results from a particular G·U pair in that specialized tRNA (Rudinger et al., 1996).
Summary
G·U pairs are used with remarkable frequency in biology to expand the chemical and conformational universe accessible to double-stranded RNA and thereby provide unique recognition sites. Protein and RNA enzymes and RNA-binding proteins bind to unique G·U sites by recognizing chemical features common to all G·U pairs and that differ from Watson–Crick and other mismatched pairs. Conformational features of G·U-containing RNA double helices unique to the specific sequence context of each G·U pair provide diversity and allow a variety of unique interaction sites to be built from this otherwise simple recognition tag.
References
- Abramovitz D.L., Friedman, R.A. and Pyle, A.M. (1996) Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science, 271, 1410–1413. [DOI] [PubMed] [Google Scholar]
- Allain F.H.-T. and Varani, G. (1995a) Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res., 23, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain F.H.-T. and Varani, G. (1995b) Structure of the P1 helix from group I self splicing introns. J. Mol. Biol., 250, 333–353. [DOI] [PubMed] [Google Scholar]
- Bénard L., Mathy, N., Grunberg-Magnago, M., Ehresmann, B., Ehresmann, C. and Portier, C. (1998) Identification in a pseudoknot of a U·G motif essential for the regulation of the expression of ribosomal protein S15. Proc. Natl Acad. Sci. USA, 95, 2564–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H. and Doudna, J.A. (1996) Metal-binding sites in the major groove of a large ribozyme domain. Structure, 4, 1221–1229. [DOI] [PubMed] [Google Scholar]
- Chang K.Y., Varani, G., Bhattacharya, S., Choi, H. and McClain, W.H. (1999) Correlation of deformability at a tRNA recognition site and aminoacylation specificity. Proc. Natl Acad. Sci. USA, 96, 11764–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesolka J. and Yarus, M. (1996) Small RNA-divalent domains. RNA, 2, 785–793. [PMC free article] [PubMed] [Google Scholar]
- Clark B.F.C. and Klug, A. (1975) Structure and function of tRNA with special reference to the three dimensional structure of yeast phenylalanine tRNA. Proc. Tenth FEBS Meet., 39, 183–206. [Google Scholar]
- Crick F.H.C. (1966) Codon–anticodon pairing: The wobble hypothesis. J. Mol. Biol., 19, 548–555. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Cormack, B.P. and Szostak, J.W. (1989) RNA structure, not sequence, determines the 5′ splice-site specificity of a group I intron. Proc. Natl Acad. Sci. USA, 86, 7402–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare A., Zhou, K. and Doudna, J.A. (1998) Crystal structure of a hepatitis delta ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- Gabriel K., Schneider, J. and McClain, W.H. (1996) Functional evidence for indirect recognition of G·U in tRNAAla by alanyl-tRNA synthetase. Science, 271, 195–197. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Konings, D. and Gutell, R.R. (1995) G·U base pairing motifs in ribosomal RNA. RNA, 1, 807–814. [PMC free article] [PubMed] [Google Scholar]
- Hermann T. and Westhof, E. (1999) Non Watson–Crick base pairs in RNA–protein recognition. Chem. Biol., 6, R335–R343. [DOI] [PubMed] [Google Scholar]
- Holley R.W., Apgar, J., Everett, G.A., Madison, J.T., Marquisee, M., Merrill, S.H., Penswick, J.R. and Zamir, A. (1965) Structure of ribonucleic acid. Science, 147, 1462–1465. [DOI] [PubMed] [Google Scholar]
- Hou Y.-M. and Schimmel, P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature, 333, 140–145. [DOI] [PubMed] [Google Scholar]
- Hur M. and Waring, R.B. (1995) Two group I introns with a C·G basepair at the 5′ splice-site instead of the very highly conserved U·G basepair: is selection post-translational? Nucleic Acids Res., 23, 4466–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illangasekare M. and Yarus, M. (1999) A tiny RNA that catalyzes both aminoacyl-RNA and peptidyl-RNA synthesis. RNA, 5, 1482–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J.A., Turner, D.H. and Zuker, M. (1989) Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA, 86, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Kwak, Y.-G., Tamkun, M., Majerfeld, I. and Yarus, M. (1999) RNAs that bind and change the permeability of phospholipid membranes. Proc. Natl Acad. Sci. USA, 96, 10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft J.S. and Tinoco, I.J. (1997) Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexamine. Structure, 5, 713–721. [DOI] [PubMed] [Google Scholar]
- Konforti B.B., Abramowitz, D.L., Duarte, C.M., Karpeisky, A., Beigelman, L. and Pyle, A.M. (1998) Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell, 1, 433–441. [DOI] [PubMed] [Google Scholar]
- Ladner J.E., Jack, A., Robertus, J.D., Brown, R.S., Rhodes, D., Clark, B.F.C. and Klug, A. (1975) Structure of yeast phenylalanine transfer RNA at 2.5 Å resolution. Proc. Natl Acad. Sci. USA, 72, 4414–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Vilardel, J. and Warner, J.R. (1996) An RNA structure involved in feedback regulation of splicing and translation is critical for biological fitness. Proc. Natl Acad. Sci. USA, 93, 1596–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B. and Westhof, E. (2000) On the wobble G·U and related pairs. RNA, 6, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B., Sauter, C. and Westhof, E. (1999) A sulfate pocket formed by the G·U pairs in the 0.97 Å resolution X-ray structure of a nonameric RNA. RNA, 5, 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H., Sabina, J., Zuker, M. and Turner, D.H. (1999) Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- McClain W.H. and Foss, K. (1988) Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science, 240, 793–796. [DOI] [PubMed] [Google Scholar]
- McClain W.H., Jou, Y.-Y., Bhattacharya, S., Gabriel, K. and Schneider, J. (1999) The reliability of in vivo structure-function analysis of tRNA aminoacylation. J. Mol. Biol., 290, 391–409. [DOI] [PubMed] [Google Scholar]
- McDowell J.A. and Turner, D.H. (1996) Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry, 35, 14077–14089. [DOI] [PubMed] [Google Scholar]
- Mueller U., Schübel, H., Sprinzl, M. and Heinemann, U. (1999) Crystal structure of acceptor stem of tRNAAla from Escherichia coli shows unique G·U wobble base pair at 1.16 Å resolution. RNA, 5, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musier-Forsyth K. and Schimmel, P. (1992) Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature, 357, 513–515. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Usman, N., Scaringe, S., Doudna, J.A., Green, R. and Schimmel, P. (1991) Specificity for aminoacylation of an RNA helix: An unpaired exocyclic amino group in the minor groove. Science, 253, 784–786. [DOI] [PubMed] [Google Scholar]
- Ott G., Arnold, L. and Limmer, S. (1993) Proton NMR studies of manganese ion binding to tRNA-derived acceptor arm duplexes. Nucleic Acids Res., 21, 5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C.L., Zhang, M., Perlman, P.S. and Franzen, J.S. (1995) Catalytically critical nucleotides in domain 5 of a group II intron. Proc. Natl Acad. Sci. USA, 92, 4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A.T. and Been, M.D. (1996) Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res., 24, 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A.M. (1993) Ribozymes: A distinct class of metalloenzymes. Science, 261, 709–714. [DOI] [PubMed] [Google Scholar]
- Pyle A.-M., Moran, S., Strobel, S.A., Chapman, T., Turner, D.H. and Cech, T.R. (1994) Replacement of the conserved G·U with a G-C pair at the cleavage site of the Tetrahymena ribozyme decreases binding, reactivity and fidelity. Biochemistry, 33, 13856–13863. [DOI] [PubMed] [Google Scholar]
- Quigley G.J. and Rich, A.R. (1975) Structural domains of transfer RNA molecules. Science, 194, 796–806. [DOI] [PubMed] [Google Scholar]
- Ramos A. and Varani, G. (1997) Structure of the acceptor stem of Escherichia coli tRNAAla: Role of the G3·U70 base pair in synthetase recognition. Nucleic Acids Res., 25, 2083–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J., Hillenbrandt, R., Sprinzl, M. and Giegé, R. (1996) Antideterminants present in minihelixSec hinder its recognition by prokaryotic elongation factor Tu. EMBO J., 15, 650–657. [PMC free article] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy, S., Boeglin, M., Poterszman, A., Mitschler, A., Podjarny, A., Rees, B., Thierry, J.C. and Moras, D. (1991) Class II aminoacyl transfer RNA synthetases: Crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science, 252, 1682–1689. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Horn, C., Brown, M., Ioudovitch, A. and Steinberg, S. (1996) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazewski P., Biala, E., Gabriel, K. and McClain, W.H. (1999) The relationship of thermodynamic stability at a G·U recognition site to tRNA aminoacylation specificity. RNA, 5, 1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S.A. and Cech, T.R. (1995) Minor groove recognition of the conserved G·U pair at the Tetrahymena ribozyme reactive site. Science, 267, 675–679. [DOI] [PubMed] [Google Scholar]
- Strobel S.A. and Cech, T.R. (1996) Exocyclic amine of the conserved G·U pair at the cleavage site of the Tetrahymena ribozyme contributes to 5′-splice site selection and transition state stabilization. Biochemistry, 35, 1201–1211. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas, P. and Moras, D. (1985) Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol., 184, 119–145. [DOI] [PubMed] [Google Scholar]