Abstract
The process of partitioning bacterial sister chromosomes into daughter cells seems to be distinct from chromatid segregation during eukaryotic mitosis. In Escherichia coli, partitioning starts soon after initiation of replication, when the two newly replicated oriCs move from the cell centre to quarter positions within the cell. As replication proceeds, domains of the compact, supercoiled chromosome are locally decondensed ahead of the replication fork. The nascent daughter chromosomes are recondensed and moved apart through the concerted activities of topoisomerases and the SeqA (sequestration) and MukB (chromosome condensation) proteins, all of which modulate nucleoid superhelicity. Thus, genes involved in chromosome topology, once set aside as ‘red herrings’ in the search for ‘true’ partition functions, are again recognized as being important for chromosome partitioning in E. coli.
Introduction
Our view of chromosome partitioning in bacteria has traditionally been based upon our understanding of mitosis in eukaryotic cells. After completion of replication, duplicated chromosomes are condensed, aligned as pairs in the middle of the cell, and pulled apart by contractile fibres that emanate from centrioles near the two poles of the cell, and attach themselves to specific regions on the chromosomes called centromeres. Intuitively, we have been trying to create models for bacterial chromosome partitioning along similar lines, incorporating modifications that accommodate the bacterial replication and cell cycle characteristics that are distinct from those of eukaryotic cells.
A number of essential differences that must be accounted for in any model of bacterial partitioning include the following. First, in contrast to eukaryotic cells where genome duplication (S) and mitosis (M) are distinctly separated by clear gap periods (G1 and G2), bacterial cells undergoing rapid growth may not only abolish the gap G1 (B-period in the bacterial cell cycle), but may even sustain overlapping rounds of replication on the same genome (Helmstetter, 1996). This would result in continuous chromosome replication, precluding the separation of distinct S and M periods. Bacterial cells must therefore have the ability to undergo mitosis while replication is in progress. Secondly, bacterial replication distinguishes itself from eukaryotic replication in that daughter chromosomes do not undergo concerted condensation after replication is complete. Studies with fluorescence in situ hybridization (FISH) and/or green fluorescent protein (GFP) fusions have shown that the chromosomal segments move from origin to terminus (Niki and Hiraga, 1998; Roos et al., 1999; Niki et al., 2000) through the replication machinery localized at mid-cell (Lemon and Grossman, 1998). However, post-replicative condensation has not been detected for whole bacterial chromosomes. Thirdly, bacteria seem to lack a visible spindle apparatus from which contractile fibres could pull the chromosomes apart as during the transition from interphase to prophase for eukaryotic chromatids. Sister chromosomes never even align themselves for post-replicative segregation.
Prokaryotic centromeres
The first evidence for the presence of centromere sites on prokaryotic genomes was obtained for plasmids with active partitioning mechanisms. These include P1, F and R1, all of which possess a trans-acting partition factor that binds at a specific sequence on the plasmid genomes and pairs them through interactions among the DNA-bound proteins. The sequences are considered to be eukaryotic centromere analogues (Abeles et al., 1985; Gerdes and Molin, 1986; Mori et al., 1986). Such sites have recently been detected on the Bacillus subtilis chromosome near its replication origin (Lin and Grossman, 1998), and in Caulobacter crescentus (Mohl and Gober, 1997), although the mechanisms of chromosome positioning in these organisms too have yet to be worked out.
No centromere analogue has yet been detected for the Escherichia coli chromosome, although the demonstration of active partitioning of plasmids in E. coli hosts suggests that they possess the machinery necessary for centromere-mediated segregation. Furthermore, although the E. coli chromosome itself has all the characteristics of active partitioning, oriC minichromosomes do not exhibit the controlled dynamics of an ordered separation (as visualized by GFP or FISH) unless associated with the partition region from a plasmid such as F or P1 (Niki and Hiraga, 1999). Hence, if a chromosomal centromere is present in E. coli, it does not lie close to the replication origin oriC, and oriC itself cannot function as the centromere analogue. Nevertheless, as in B. subtilis and C. crescentus, the E. coli replication origin displays ordered dynamics during replication and cell division: the single chromosomal oriC of the pre-replicative cell moves from its polar location to a mid-cell position just before initiation, and the duplicated oriCs move to quarter-length positions during early stages of replication (Niki et al., 2000). The dynamics of chromosomal oriC positioning thus appears to be independent of centromere site(s), suggesting the possibility of an alternative mechanism for the movement and anchoring of the chromosome domains.
The search for partition functions
The earliest efforts to identify partition genes in bacteria involved screening for conditional lethal mutations that cause defects in the segregation of bacterial nucleoids, the folded nucleoprotein structure of the E. coli genome. A large number of segregation-defective mutants were identified and designated as par (partition) mutants. Most of these mapped in genes coding for gyrases or topoisomerases (parA = gyrase B, parD = gyrase A, parC = topoisomerase IVA, parE = topoisomerase IVB). With the exception of parB, which turned out to be an allele of the DNA primase gene dnaG (Norris et al., 1986), most of the Par products were found to be involved in elongation and/or termination of replication through their influence on the superhelicity of the chromosome (Schaechter, 1990). Loss-of-function of parA and parD resulted in the presence of large nucleoid masses in the middle of filamentous cells, suggesting failure in decatenation of sister chromosomes as the cause of the partition defects (Kato et al., 1990). Similarly, parC and parE loss-of-function mutants gave rise to cells with unseparated bacterial nucleoids. In vitro and in vivo investigations showed that ParC and ParE can function as type II topoisomerases capable of relaxing negative and positive supercoils, and of resolving knotted DNA. Conditional lethality caused by mutations in these genes was also associated with incomplete chromosome replication. Thus, the Par– phenotype actually resulted from defects in replication elongation, replication termination, and/or daughter chromosome resolution by decatenation, all of which were believed to be upstream activities needed to be completed prior to partition. These results were considered unavoidable artifacts that would be generated in any general screen for partition defects. A new genetic approach was therefore thought to be necessary for the identification of genes responsible for positioning of the replicated chromosomes without affecting replication, termination or resolution.
muk genes: positioning nucleoids and plasmids
In the late 1980s, S. Hiraga developed an ingenious genetic screen for the isolation of par genes and was successful in identifying a group of positioning mutants termed muk (Hiraga et al., 1989). These mutations caused a subpopulation of the cells to produce one anucleate and one diploid daughter cell. The mukA gene codes for an outer membrane protein previously known as tolC (Hiraga et al., 1989), and mukB was a hitherto unknown gene that codes for a very large protein (170 kDa) containing both a DNA-binding domain and a domain with ATPase activity (Niki et al., 1991; Yamanaka et al., 1994). Its native form in solution is dimeric, and the homodimer has a rod and hinge structure similar to some of the motor proteins found in eukaryotic cells (Niki et al., 1992). The two other muk genes, mukE and mukF, code for proteins that form complexes with the MukB protein, and it was believed that the Muk proteins form a spindle analogue (Yamazoe et al., 1999). FISH was used to compare the localization of oriC and other chromosomal segments in exponentially or synchronously growing wild-type and muk mutant strains. It was shown that the intracellular positions of chromosome segments follow a reproducible pattern during cell cycle progression in wild-type cells, but that this pattern is seriously perturbed in ΔmukB::kan strains (Niki et al., 2000; Weitao et al., 2000). Nevertheless, plasmid replicons carrying their own partition system were stably maintained in mukB hosts (Ezaki et al., 1991; Funnell and Gagnier, 1995) although their positioning in the cell was affected (Weitao et al., 2000).
The MukB– phenotype: suppression by regulating nucleoid structure
Not all muk genes fulfil the expected criteria for ‘true’ partitioning genes. For example, null mutations in the mukB gene caused conditional lethality associated with filamentous cells and irregular nucleoid distribution. Thus, not only the positioning but also the separation of the nucleoids was affected. Nucleoids in cells of a ΔmukB strain had lost their position as well as compact shape and were spread out through the interior of the cell (S. Dasgupta, unpublished observation; Gullbrand and Nordström, 2000). They were found to sediment more slowly than those from a wild-type strain in a sucrose gradient. When the isolated nucleoids were stained with DAPI and examined by fluorescence microscopy, they looked flaccid and spread-out rather than compact and rounded as was the case for wild-type nucleoids (Weitao et al., 1999). Clearly, inactivation of the mukB gene had a profound effect on the hydrodynamic properties of the nucleoid, consistent with unfolding of the bacterial chromosome. Spontaneously arising suppressors of the temperature-sensitivity phenotype of mukB-null mutant strains were isolated and mapped near the topA locus (Sawitzke and Austin, 2000). This was the first genetic evidence that the loss of MukB function can be compensated by the loss of a topoisomerase activity, which influences nucleoid structure by altering its average superhelical density. It had already been shown that mukB mutation renders cells more sensitive to the gyrase-inhibiting drug novobiocin (Weitao et al., 1999). Also, inactivation of SeqA [a factor involved in sequestration of newly replicated hemi-methylated DNA and in imposition of synchrony in initiation from oriC (Crooke, 1995)], which in itself leads to a significant compaction of E. coli nucleoids, suppressed the segregation defects and conditional lethality of mukB null mutants (Weitao et al., 1999). Direct measurement of the superhelicity of the bacterial nucleoids in the wild-type strain and its mukB and seqA derivatives showed that the chromosomes from the mutant strains have lower and higher negative superhelicity, respectively, compared with that of the wild-type strain (T. Weitao, K. Nordström and S. Dasgupta, in preparation). Plasmid pBR322 was used as a probe for the level of supercoiling and it was confirmed that mukB and seqA alter the general supercoiling potential of the cells. Thus, their effect on nucleoid structure is not necessarily due to direct interaction with the bacterial chromosome. Taken together, these results indicate that phenotypic suppression of mukB can be brought about by any activity that leads to condensation of the bacterial nucleoids, and that such condensation can be generated by increased negative superhelicity of the nucleoids. The idea that MukB-mediated condensation might at least in part derive from altered levels of supercoiling has recently gained broader acceptance (Holmes and Cozzarelli, 2000).
Partitioning: condensation and superhelicity
The data discussed above suggest that MukB, through its effect on the superhelicity of E. coli nucleoids, might provide the tensile force necessary to condense and partition daughter chromosomes. Structural studies have now shown that MukB is an analogue of the SMC (structural maintenance of chromosomes) protein family whose members are involved in promoting DNA condensation and chromosome segregation in both eukaryotes and prokaryotes (Britton et al., 1998; Moriya et al., 1998; Jensen and Shapiro, 1999). Dimeric forms of these gigantic coiled-coil proteins, which have DNA-binding domains at each extremity and a centrally located hinge, could be conceived to bind distant segments of DNA (Melby et al., 1998; Hirano, 1999), bringing them closer and folding the chromosome into a condensed state. It is already known that MukB maintains the condensation state of the nucleoids of stationary cells and also of cells in pre-replicative or replication run-out stages (S. Dasgupta, unpublished observations; Gullbrand and Nordström, 2000). During replication, a large number of SeqA molecules (and possibly additional unknown factors) bind co-operatively to the hemimethylated DNA near the centrally located replication machinery behind the replication fork (Hiraga et al., 1998; Onogi et al., 1999). There, they hold the replicating chromosome in an unfolded state, thereby preventing topoisomerase-mediated rewinding (Torheim and Skarstad, 1999) which would result in a tangled mess. As the replicated DNA is remethylated, the SeqA–DNA complex dissociates and MukB can re-establish the folded, condensed state of the newly replicated domain, perhaps in concert with the histone-like proteins (Jaffé et al., 1997). According to this scenario, partitioning does not wait until replication is complete, but starts soon after initiation and continues in parallel with their replication and cell growth, transporting chromosomal domains to their proper intracellular locations in order of their replication (van Helvoort and Woldringh, 1994; Roos et al., 1999; Niki et al., 2000).
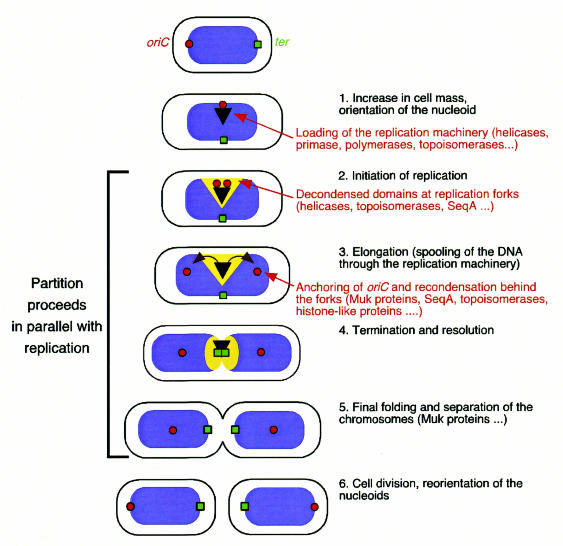
While eukaryotic replication (S) and mitosis (M) are clearly separated events, bacterial replication and partition are parallel processes. As the compartmentalization of replication and partitioning disappears, so do the distinctions between the classical and ‘true’ Par functions, including those performed by the Muk genes. All the par genes, muk genes and seq gene(s) work together in parallel to coordinate not only chromosome replication, but also separation and partitioning of sister chromosomes into the daughter cells (Figure 1).
Fig. 1. Model for nucleoid segregation through decondensation/condensation and supercoiling during the E. coli cell cycle. Red circles represent the origin of replication (oriC), green squares the terminus, and the black triangle the replication machinery. Blue represents the condensed, and yellow the decondensed, domains of the nucleoid or the bacterial chromosome. Refolding and partitioning of the chromosome occur continuously and in parallel with replication. According to this model, DNA decondenses in the vicinity of the replicating segment. Duplicate copies of oriC move rapidly apart from the centre of the cell to quarter positions on opposite sides. The replicated DNA behind the fork recondenses and positions itself within the cell.
Interestingly, all of these genes seem to modulate the superhelical state of the chromosome directly or indirectly. Hence, prokaryotic partitioning distinguishes itself from its eukaryotic counterpart by not having any external partitioning apparatus that pulls the chromosomes apart. The moving force may come from the torsional energy introduced by negatively supercoiling the bacterial chromosome (Pettijohn, 1996). This feature is absent from the linearized histone-coated DNA complexes that are found in eukaryotic nuclei. It is possible that the evolution of the elaborate mitotic apparatus, complete with spindles and microtubules, occurred concomitantly with the transition of genomes from free-floating, naked DNA to DNA–histone complexes confined within a nuclear membrane. It would be interesting to find organisms where remnants of both partitioning mechanisms co-exist. Some extremophiles are known to maintain their chromosomes in either relaxed or positively supercoiled states (Forterre et al., 1996). Examination of the partitioning mechanisms in such organisms might shed further light on the role of superhelicity in partition.

Acknowledgments
Acknowledgements
Our work was supported by The Swedish Natural Science Research Council and The Swedish Cancer Society.
References
- Abeles A.L., Friedman, S.A. and Austin, S.J. (1985) Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J. Mol. Biol., 185, 261–272. [DOI] [PubMed] [Google Scholar]
- Britton R.A., Lin, D.C.-H. and Grossman, A.G. (1998) Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev., 12, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E. (1995) Regulation of chromosomal replication in E. coli: Sequestration and beyond. Cell, 82, 877–880. [DOI] [PubMed] [Google Scholar]
- Ezaki B., Ogura, T., Niki, H. and Hiraga, S. (1991) Partitioning of a mini-F plasmid into anucleate cells of the mukB null mutant. J. Bacteriol., 173, 6643–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P., Bergerat, A. and Lopez-Garcia, P. (1996) The unique DNA topology and DNA topoisomerases of hyperthermophilic archea. FEMS Microbiol. Rev., 18, 237–248. [DOI] [PubMed] [Google Scholar]
- Funnell B.E. and Gagnier, L. (1995) Partition of P1 plasmids in Escherichia coli mukB chromosomal partition mutants. J. Bacteriol., 177, 2381–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K. and Molin, S. (1986) Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J. Mol. Biol., 190, 269–279. [DOI] [PubMed] [Google Scholar]
- Gullbrand B. and Nordström, K. (2000) FtsZ ring formation without subsequent cell division after replication runout in Escherichia coli. Mol. Microbiol., 36, 1349–1359. [DOI] [PubMed] [Google Scholar]
- Helmstetter C. (1996) Timing of synthetic activities in the cell cycle. In Neidhardt, F.C. (ed.), Escherichia coli and Salmonella. ASM Press, Washington, DC, pp. 1627–1639.
- Hiraga S., Niki, H., Ogura, T., Ichinose, C., Mori, H., Ezaki, B. and Jaffé, A. (1989) Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol., 171, 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Ichinose, C., Niki, H. and Yamazoe, M. (1998) Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA–protein complexes in E.coli. Mol. Cell., 1, 381–387. [DOI] [PubMed] [Google Scholar]
- Hirano T. (1999) SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev., 13, 11–19. [DOI] [PubMed] [Google Scholar]
- Holmes V.F. and Cozzarelli, N.R. (2000) Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl Acad. Sci. USA, 97, 1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé A., Vinella, D. and D’Ari, R. (1997) The Escherichia coli histone-like protein HU affects DNA initiation, chromosome partitioning via MukB, and cell division via MinCDE. J. Bacteriol., 179, 3494–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B. and Shapiro, L. (1999) The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl Acad. Sci. USA, 96, 10661–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Nishimura, Y., Imamura, R., Niki, H., Hiraga, S. and Suzuki, H. (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell, 63, 393–404. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman, A.D. (1998) Localization of bacterial DNA polymerase: Evidence for a factory model of replication. Science, 282, 1516–1519. [DOI] [PubMed] [Google Scholar]
- Lin D.C.H. and Grossman, A.D. (1998) Identification and characterization of a bacterial chromosome partition site. Cell, 92, 675–685. [DOI] [PubMed] [Google Scholar]
- Melby T.E., Ciampaglio, C.N., Briscoe, G. and Erickson, H.P. (1998) The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol., 142, 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl D.A. and Gober, J.W. (1997) Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell, 88, 675–684. [DOI] [PubMed] [Google Scholar]
- Mori H., Kondo, A., Ohshima, A., Ogura, T. and Hiraga, S. (1986) Structure and function of the F plasmid genes essential for partitioning. J. Mol. Biol., 192, 1–15. [DOI] [PubMed] [Google Scholar]
- Moriya S., Tsujikawa, E., Hassan, A.K.M., Asai, K., Kodama, T. and Ogasawara, N. (1998) A Bacillus subtilis gene encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol., 29, 179–187. [DOI] [PubMed] [Google Scholar]
- Niki H. and Hiraga, S. (1998) Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev., 12, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H. and Hiraga, S. (1999) Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol. Microbiol., 34, 498–503. [DOI] [PubMed] [Google Scholar]
- Niki H., Jaffé, A., Imamura, R., Ogura, T. and Hiraga, S. (1991) The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E.coli. EMBO J., 10, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Imamura, R., Kitaoka, M., Yamanaka, K., Ogura, T. and Hiraga, S. (1992) E.coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J., 11, 5101–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Yamaichi, Y. and Hiraga, S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev., 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- Norris V., Alliotte, T., Jaffé, A. and D’Ari, R. (1986) DNA replication termination in Escherichia coli parB (a dnaG allele), parA, and gyrB mutants affected in DNA distribution. J. Bacteriol., 168, 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onogi T., Niki, H., Yamazoe, M. and Hiraga, S. (1999) The assembly and migration of SeqA–GFP fusion in living cells of Escherichia coli. Mol. Microbiol., 31, 1775–1782. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. (1996) The nucleoid. In Neidhardt, F.C. (ed.), Escherichia coli and Salmonella. ASM Press, Washington, DC, pp. 158–166.
- Roos M., van Geel, A.B., Aarsman, M.E., Veuskens, J.T., Woldringh, C.L. and Nanninga, N. (1999) Cellular localization of oriC during the cell cycle of Escherichia coli as analyzed by fluorescent in situ hybridization. Biochimie, 81, 797–802. [DOI] [PubMed] [Google Scholar]
- Sawitzke J.A. and Austin, S. (2000) Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl Acad. Sci. USA, 97, 1671–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M. (1990) The bacterial equivalent of mitosis. In Drlica, K. and Riley, M. (eds), The Bacterial Chromosome. ASM Press, Washington, DC, pp. 313–322.
- Torheim N.K. and Skarstad, K. (1999) Escherichia coli SeqA protein affects DNA topology and inhibits open complex formation at oriC. EMBO J., 18, 4882–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helvoort J.M. and Woldringh, C.L. (1994) Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol. Microbiol., 13, 577–583. [DOI] [PubMed] [Google Scholar]
- Weitao T., Nordström, K. and Dasgupta, S. (1999) Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol., 34, 157–168. [DOI] [PubMed] [Google Scholar]
- Weitao T., Dasgupta, S. and Nordström, K. (2000) Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Mitani, T., Feng, J., Ogura, T., Niki, H. and Hiraga, S. (1994) Two mutant alleles of mukB, a gene essential for chromosome partition in Escherichia coli. FEMS Microbiol. Lett., 123, 27–31. [DOI] [PubMed] [Google Scholar]
- Yamazoe M., Onogi, T., Sunako, Y., Niki, H., Yamanaka, K., Ichimura, T. and Hiraga, S. (1999) Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J., 18, 5873–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]