Abstract
Protein unfolding is a key step in the import of some proteins into mitochondria and chloroplasts and in the degradation of regulatory proteins by ATP-dependent proteases. In contrast to protein folding, the reverse process has remained largely uninvestigated until now. This review discusses recent discoveries on the mechanism of protein unfolding during translocation into mitochondria. The mitochondria can actively unfold preproteins by unraveling them from the N-terminus. The central component of the mitochondrial import motor, the matrix heat shock protein 70, functions by both pulling and holding the preproteins.
Introduction
Proteins must fold into well-defined three-dimensional structures to function. Unfolding of proteins, however, is also essential for several processes in the cell. Two examples are protein translocation across membranes and protein degradation by ATP-dependent proteases. Approximately half of all the proteins synthesized in the average eukaryotic cell are translocated into or across a membrane (Schatz and Dobberstein, 1996). Some preproteins fold before translocation and must then be unfolded during import into mitochondria (Eilers and Schatz, 1986; Rassow et al., 1989, 1990), chloroplasts (Walker et al., 1996), or in some cases, even during import into the endoplasmic reticulum (ER) (Paunola et al., 1998). This review focuses on protein unfolding during translocation into the mitochondrial matrix because this is the best understood physiological unfolding process.
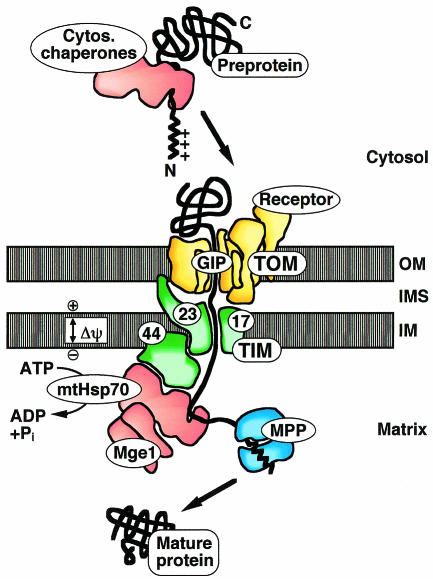
Most mitochondrial matrix proteins are synthesized in the cytosol as preproteins with positively charged N-terminal targeting sequences, and are subsequently imported into mitochondria by the TOM (translocase of the outer mitochondrial membrane) and TIM (translocase of the inner membrane) complexes, macromolecular translocases of the outer and inner membranes, respectively (Figure 1) (Schatz and Dobberstein, 1996; Neupert, 1997; Pfanner et al., 1997; Jensen and Johnson, 1999; Voos et al., 1999; Bauer et al., 2000). Import requires both an electrochemical potential across the inner mitochondrial membrane and the ATP-dependent action of mitochondrial heat shock protein 70 (mtHsp70) in the matrix. mtHsp70 drives import in conjunction with its partner proteins: Tim44, a component of the inner membrane import channel, and the matrix protein Mge1, which functions as nucleotide exchanging co-chaperone. Once the preprotein reaches the matrix, the targeting sequence is usually proteolytically removed and the protein refolds into its native structure.
Fig. 1. The mitochondrial protein import machinery. Shown is the import pathway of a matrix protein. The preprotein is synthesized in the cytosol with an N-terminal positively charged presequence. Cytosolic chaperones can bind to the preprotein. The translocase of the outer mitochondrial membrane (TOM) contains receptors that recognize the presequence and a general import pore (GIP) that mediates translocation across the outer membrane (OM). The translocase of the inner membrane (TIM) includes an import channel formed by Tim23 and Tim17 and the peripheral subunit Tim44. The membrane potential Δψ across the inner membrane (IM) drives translocation of the presequence. Matrix Hsp70 (mtHsp70) binds the preprotein in transit and, together with Tim44 and the co-chaperone Mge1, forms an ATP-dependent import motor. The presequence is cleaved off by the mitochondrial processing peptidase (MPP). IMS, intermembrane space.
Protein unfolding: an essential step for mitochondrial import
It is clear that preproteins are not imported into mitochondria in their native state because stabilizing preproteins by ligand binding blocks their import (Eilers and Schatz, 1986). Proteins are normally unfolded during translocation and are threaded through the import machinery as linear chains (Rassow et al., 1990; Schwartz et al., 1999), although larger structures can be tolerated by the import channels (Vestweber and Schatz, 1989; Schwartz et al., 1999; Schwartz and Matouschek, 1999). The extent of unfolding that is required during translocation is presumably determined by the size of the protein import channels. The channel in the outer mitochondrial membrane has an internal diameter of ~22 Å (Hill et al., 1998; Künkele et al., 1998; Schwartz and Matouschek, 1999) and therefore might allow import of very small folded domains, although typical protein domains must be unfolded before translocation. The channel in the inner membrane is apparently narrower and even a small amount of steric bulk retards translocation into the matrix, indicating that proteins must be unfolded when passing through this channel (Schwartz and Matouschek, 1999).
Although it is known that import into mitochondria can occur post-translationally, both in vitro and in vivo (Schatz and Dobberstein, 1996; Neupert, 1997; Pfanner et al., 1997), it has often been assumed that folding of preproteins before import is generally prevented by cytosolic chaperones. Moreover, some preproteins may also be imported by a co-translational mechanism (Fünfschilling and Rospert, 1999; Lithgow, 2000). However, the two in vivo studies of protein import that have analyzed the folding states of preproteins found them to be in their native forms prior to translocation. First, when a mitochondrially-targeted form of dihydrofolate reductase (DHFR) was expressed in yeast cells, introduction of a substrate analogue into the cytosol inhibited import, indicating that DHFR must have been in its active conformation (Wienhues et al., 1991). Secondly, in vivo import of the heme-binding domain of cytochrome b2 was blocked when the unfolding activity in the mitochondrial inner membrane was disengaged (Bömer et al., 1997). It is likely that other mitochondrial preproteins also fold in the cytosol before import since the N-terminal targeting sequences usually exert only small effects on the folding of the attached mature proteins (Mattingly et al., 1993; Matouschek et al., 1997; Huang et al., 1999) and protein folding in the eukaryotic cytosol occurs very soon after translation, the N-terminal domains of nascent proteins folding even before synthesis of the C-terminal domains is complete (Netzer and Hartl, 1997).
If mitochondrial preproteins fold in the cytosol, why are cytosolic chaperones required for the import of some proteins? The involvement of cytosolic chaperones in import has been studied primarily using cell free assays, which showed that import of a subset of authentic and engineered preproteins requires extramitochondrial ATP (Wachter et al., 1994; Mihara and Omura, 1996). The preproteins with this requirement were either membrane proteins or subunits of multimeric protein complexes (Wachter et al., 1994). This finding suggested that cytosolic chaperones, which require an ATP source, may facilitate the import of preproteins that are unable to fold in the cytosol and are therefore prone to aggregation. In contrast, several preproteins that do not require external ATP for import in vitro are known to fold in a reticulocyte lysate, e.g. cytochrome b2 (Glick et al., 1993; Wachter et al., 1994; Gärtner et al., 1995) and chimeric DHFR or barnase preproteins (Eilers and Schatz, 1986; Rassow et al., 1990; Wachter et al., 1994; Matouschek et al., 1997). Also, it is well established that mitochondria can import chemically pure folded preproteins (Eilers and Schatz, 1986; Bömer et al., 1998). Although the fraction of total preproteins that fold in the cytosol has not been determined, it is evident that mitochondria are presented with at least some folded preproteins and therefore must somehow unfold these proteins during import.
Active unfolding of proteins by mitochondria
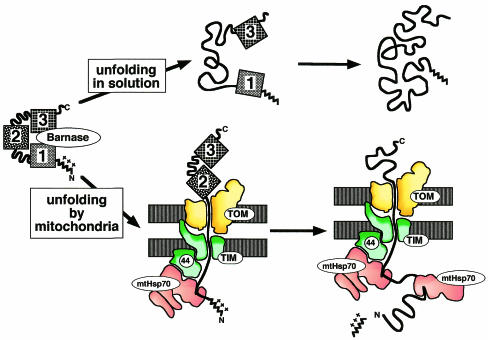
It has been debated if mitochondria play an active role (Matouschek et al., 1997; Huang et al., 1999) or a passive role (Gaume et al., 1998) in protein unfolding. An active role was demonstrated by the observation that import, and thus unfolding, of folded preproteins by mitochondria can be significantly faster than their spontaneous unfolding in solution (Matouschek et al., 1997; Lim, N. Pfanner and W. Voos, unpublished). How do mitochondria actively unfold proteins? The mitochondrial import apparatus can be viewed as a large enzyme that catalyzes the unfolding and translocation of a preprotein. The mechanism has been determined for a form of the ribonuclease barnase that was converted into a mitochondrial preprotein by the attachment of a targeting sequence to its N-terminus (Huang et al., 1999). During import of the model preprotein into isolated yeast mitochondria, it is unraveled from its N-terminus (Figure 2, lower pathway). Once this process has been initiated at the N-terminus, the rest of the preprotein denatures rapidly. In contrast, spontaneous unfolding of wild-type barnase in free solution begins as a global process, with a large part of the structure, particularly the middle portion, unfolding early (Figure 2, upper pathway). Thus mitochondria catalyze unfolding by changing the unfolding pathway (Huang et al., 1999).
Fig. 2. Different unfolding pathways of barnase in solution and by mitochondria. The three portions of barnase are indicated by boxes 1–3 (from N- to C-terminus). In solution, unfolding mainly starts with the middle portion (box 2) of the protein (upper pathway). Mitochondria unfold barnase by unraveling it from its N-terminus (box 1). Barnase was converted to a mitochondrial preprotein by attaching an N-terminal presequence.
The mechanism of unfolding probably depends on structural properties of the preprotein. Barnase is an α+β protein and its N-terminal amino acids form an α-helix at the surface of the protein. Similarly, the heme-binding domain of the mitochondrial intermembrane space protein cytochrome b2 possesses an N-terminal α-helix, as does citrate synthase. It is easy to imagine how these proteins can be unraveled by disruption of these surface structures. Indeed, barnase and the heme-binding domain are efficiently unfolded by mitochondria even when stabilized by ligand binding (Glick et al., 1993; Voos et al., 1993; Huang et al., 1999). However, many other proteins have different architectures. The most frequently occurring domain structure is the α/β fold (Branden and Tooze, 1998), found for example in DHFR, mtHsp70 and aldehyde dehydrogenase. In these structures the N-terminal amino acids are often buried, such as in DHFR whose N-terminus forms an internal β-strand in a β-sheet sandwiched between two layers of α-helices. Presumably, therefore, DHFR must first unfold globally before it can release its N-terminus. Indeed, although mitochondria catalyze the unfolding of DHFR preproteins (Matouschek et al., 1997), ligands can stabilize DHFR to the extent that unfolding and import are completely blocked (Eilers and Schatz, 1986; Rassow et al., 1989; Huang et al., 1999; Voisine et al., 1999). The ability of a protein to be unfolded by mitochondria does not simply depend on global thermodynamic properties because barnase is considerably more stable against unfolding in solution than is DHFR and the respective ligands bind to the two proteins with similar affinities. It therefore appears that some other property of preproteins, presumably the structure of their N-termini, determines their potential to be unraveled by mitochondria (Huang et al., 1999).
Import driving forces of mitochondria
Active unfolding of a protein domain at the mitochondrial surface occurs when its N-terminal part, including the targeting sequence, is long enough to reach the import machinery of the inner membrane (Matouschek et al., 1997; Huang et al., 1999). The simplest mechanism by which the unfolding machinery could unravel the mature domain at a distance is by pulling on its targeting sequence (Kang et al., 1990; Glick, 1995; Huang et al., 1999). Pulling a portion of the polypeptide chain out of a folded structure would collapse the protein because folding is highly cooperative (Neira and Fersht, 1999).
Two import driving forces have been identified (Figure 3A): the electrical component Δψ of the membrane potential across the inner membrane (Martin et al., 1991) and the ATP-dependent action of mtHsp70 in the mitochondrial matrix (Neupert et al., 1990; Simon et al., 1992; Glick, 1995; Pfanner and Meijer, 1995).
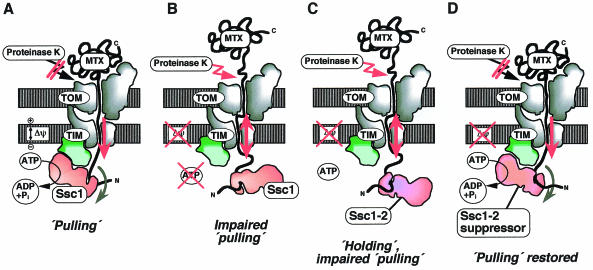
Fig. 3. Import driving forces acting on a mitochondrial preprotein. A preprotein has been arrested in the mitochondrial import machinery in a two-membrane spanning manner by attaching a tightly folded domain to the C-terminus [DHFR with bound methotrexate (MTX) that cannot be unfolded by mitochondria]. (A) Two import driving forces, the membrane potential Δψ and matrix Hsp70 (termed Ssc1 in yeast), pull the preprotein in. Therefore, the folded C-terminal domain is tightly pulled against the outer membrane and cannot be cleaved off by proteinase K added to the mitochondria. (B) When both Δψ and mtHsp70 are inactivated, pulling is impaired. The preprotein slides back in the import channel and the C-terminal domain can be cleaved off by proteinase K. (C) A mutant form of mtHsp70 (Ssc1-2) efficiently holds preproteins, but its interaction with the TIM machinery is impaired. Therefore pulling is impaired and, upon dissipation of Δψ, the preprotein slides back. (D) An intragenic suppressor mutation restores the interaction of mtHsp70 with the TIM machinery and thereby restores pulling of the preprotein.
The membrane potential is positive at the outer (intermembrane space) surface and negative at the inner (matrix) surface of the inner membrane. Since mitochondrial targeting sequences have a net positive charge, a targeting sequence in the import channel will experience a force directed towards the matrix (Martin et al., 1991) and this could lead to unfolding.
The import of both folded and unfolded preproteins into the mitochondrial matrix strictly requires the ATP-dependent action of mtHsp70 (Gambill et al., 1993; Wachter et al., 1994). Moreover, mtHsp70 contributes directly to protein unfolding; the ssc1-2 mutation in mtHsp70 has a severe effect on the import of folded preproteins, but not of unfolded preproteins (Kang et al., 1990; Voisine et al., 1999).
Mechanism of mtHsp70 action: pulling and holding
mtHsp70 is a member of the large family of homologous 70 kD chaperones (Hsp70s), which are involved in a wide range of biological processes that entail unfolding and disassembly of protein complexes (Hartl, 1996; Bukau and Horwich, 1998). Hsp70s consist of an N-terminal ATPase domain, a central peptide-binding domain and a shorter C-terminal segment. mtHsp70 binds the N-terminus of an incoming preprotein via its peptide-binding domain. However, it is unique in that it binds to Tim44 of the inner membrane translocase (Schatz and Dobberstein, 1996; Neupert, 1997; Pfanner et al., 1997). The interaction with Tim44 requires the ATPase domain and is stabilized by the peptide-binding domain and the C-terminal segment (Krimmer et al., 2000).
Two popular models for the action of mtHsp70 in unfolding are the ‘Brownian ratchet’ (‘trapping’ or ‘holding’) and the ‘motor’ (‘pulling’) models. In the ratchet model, mtHsp70 molecules bind to segments of preproteins that emerge from the import channel through spontaneous fluctuations (Neupert et al., 1990; Simon et al., 1992; Ungermann et al., 1994; Pfanner and Meijer, 1995; Bauer et al., 2000). The bound mtHsp70 molecules then block diffusion back into the channel. The motor model is based on the experimental observation that Hsp70s undergo conformational changes in an ATP-dependent manner (Liberek et al., 1991; von Ahsen et al., 1995; Shi et al., 1996). Since mtHsp70 binds to both incoming preprotein and Tim44, an ATP-dependent conformational change could directly generate a pulling force at the N-terminus of a preprotein. If binding of mtHsp70 to preprotein and Tim44, followed by a conformational change, were repeated in an ATP-dependent cycle, mtHsp70 could function as a motor, pulling preproteins into the matrix by a mechanism reminiscent of that whereby myosin functions in muscle contraction (Glick, 1995; Pfanner and Meijer, 1995).
These two models of mtHsp70 action are not as different from each other as it might initially appear. Even classical biological motors, such as myosins and kinesins, are thought by many to function as Brownian ratchets when considered at the single molecule level: they probably act by trapping spontaneous conformational changes within a motor molecule in a nucleotide-dependent manner (Astumian, 1997). And Brownian ratchets can certainly act as motors (Feynman et al., 1963). In the case of protein import, by trapping diffusion of preproteins in one direction only, the molecular ratchet would in effect ‘pull’ at the N-termini of the preproteins. Both models of mtHsp70 action introduced above therefore contain elements of Brownian ratchets and both can describe motors.
It has been assumed that in a ratchet model of mtHsp70 function, protein unfolding by mitochondria cannot be faster than spontaneous global unfolding (Gaume et al., 1998). However, both the ratchet model and the motor model of mtHsp70 action can lead to changes in the unfolding pathway of a preprotein if the spontaneous N-terminal unfolding fluctuations that are trapped by mtHsp70 do not normally lead to global unfolding. Therefore, in both models, protein unfolding by mitochondria can be faster than spontaneous global unfolding (Huang et al., 1999).
A clear-cut difference between the two models of mtHsp70 action is the manner in which spontaneous fluctuations of preproteins are trapped: in the ratchet model by the binding to whole molecules of mtHsp70, without any gross conformational changes of mtHsp70, and in the motor model through substantial conformational changes of the prebound mtHsp70. It may therefore be more appropriate to describe the ratchet model as ‘holding by mtHsp70’, and the motor model as ‘active pulling by mtHsp70’. The active pulling scenario could be more efficient at unfolding proteins than the holding scenario. By conformational changes, mtHsp70 would be able to trap much smaller fluctuations as well as provide additional energy input for unraveling preproteins from the N-terminus when a spontaneous fluctuation is not large enough to overcome the activation energy barrier by itself. It would thereby more actively promote unfolding reactions and may help to overcome restrictions imposed by the interactions of preproteins with the import channels (Chauwin et al., 1998).
Experimental evidence for a pulling function of mtHsp70 has been obtained through the analysis of preproteins that are artificially arrested in the midst of import, in a state spanning both mitochondrial membranes (Schwarz et al., 1993; Voisine et al., 1999). A long presequence was attached to DHFR to promote it’s import, and the conformation of the protein was stabilized by binding of the specific ligand methotrexate. In this system, the N-terminal portion of the preprotein is driven into mitochondria by two forces, the membrane potential and the ATP-dependent action of mtHsp70. The folded DHFR is pulled so tightly against the outer membrane that an externally added protease has no access to the preprotein (Figure 3A). Upon dissipation of the membrane potential and inactivation of mtHsp70 by lowering the ATP level, the preprotein slides back in the import channel, allowing DHFR to be cleaved off by the protease (Figure 3B). Addition of ATP restores protease inaccessibility, even without the force of the membrane potential, demonstrating pulling of the preprotein by mtHsp70 alone. Interestingly, at low ATP concentrations, release of preprotein from mtHsp70 is retarded, in effect leading to a holding of the preprotein by mtHsp70 without a pulling action (Voisine et al., 1999). These results suggest that pulling of the preprotein requires a continuous supply of ATP to drive mtHsp70 through the reaction cycle of binding to Tim44 followed by conformational changes.
Studies with mitochondria from wild-type and mtHsp70 mutant strains (termed ssc1 in yeast) provided evidence that a single mechanism is not sufficient to explain the role of mtHsp70 in import. Instead, they showed that both holding and active pulling cooperate (Pfanner and Meijer, 1995; Voos et al., 1996; Voisine et al., 1999). The ssc1-2 strain carries a mutant mtHsp70 that efficiently holds preproteins, but is impaired in binding to Tim44 of the inner membrane translocase. The mutant mitochondria efficiently import loosely folded preproteins, presumably by a holding mechanism, but are impaired in the import of folded domains and do not function in the pulling assay described above (Figure 3C). Intragenic suppressors of the ssc1-2 mutation restore the interaction of mtHsp70 with Tim44, and thereby re-establish the pulling of the preprotein (Figure 3D) and the import of folded proteins (Voisine et al., 1999). Inactivation of Tim44 leads to a phenotype similar to that of the ssc1-2 mutation and inhibits primarily the import of folded preproteins (Bömer et al., 1998; Merlin et al., 1999). The binding of mtHsp70 to Tim44 thus appears to serve two functions. First, it concentrates mtHsp70 at the exit of the import channel to allow efficient trapping of preproteins (Pfanner and Meijer, 1995; Gaume et al., 1998). Secondly, it promotes a directed conformational change in mtHsp70 that is bound to both preprotein and translocase (Glick, 1995; Pfanner and Meijer, 1995; Voisine et al., 1999). It is likely that two Tim44 and two mtHsp70 molecules are present per import channel, allowing rapid binding of a second mtHsp70 to the preprotein after release of the first mtHsp70 from the import site (Moro et al., 1999). A combination of pulling and holding by mtHsp70 will thus provide an efficient import motor (Pfanner and Meijer, 1995; Voisine et al., 1999).
Perspectives
As with mitochondria, a number of proteins must be unfolded during post-translational import into chloroplasts (Walker et al., 1996). Most proteins destined for the ER are imported co-translationally (Ng et al., 1996). However, at least one protein folds in the cytosol and then unfolds during translocation into the ER (Paunola et al., 1998). In all three translocase systems, an Hsp70 homologue [or possibly other chaperones in the case of chloroplasts (Keegstra and Froehlich, 1999)] binds to the trans side of the translocation channel and associates with the incoming polypeptide chain. In the case of the ER, Matlack et al. (1999) have demonstrated a trapping/holding function for the lumenal Hsp70 homologue, BiP, in the import of a loosely folded preprotein; a folded preprotein has not been analyzed yet. Thus, understanding the mechanism of mitochondrial protein unfolding and the proposed dual role of the mitochondrial import machinery will be relevant to protein unfolding by other organelles. Systems for protein degradation may also take advantage of these machineries for protein unfolding (Horwich et al., 1999). Again, ATP hydrolysis, N- or C-terminal targeting sequences and substrate unfolding coupled to movement of the extended polypeptide chain through a channel have been implicated. Therefore, a characterization of protein unfolding during translocation into organelles also provides a conceptual framework for the analysis of protein unfolding by ATP-dependent proteases.



Acknowledgments
Acknowledgements
We thank Elizabeth Craig, Ben Glick and Jonathan Widom for helpful discussions. Work of the authors’ laboratories was supported by the National Science Foundation, the American Cancer Society, the March of Dimes Foundation, the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388 and the Fonds der Chemischen Industrie.
References
- Astumian R.D. (1997) Thermodynamics and kinetics of a Brownian motor. Science, 276, 917–922. [DOI] [PubMed] [Google Scholar]
- Bauer M.F., Hofmann, S., Neupert, W. and Brunner, M. (2000) Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol., 10, 25–31. [DOI] [PubMed] [Google Scholar]
- Bömer U., Meijer, M., Guiard, B., Dietmeier, K., Pfanner, N. and Rassow, J. (1997) The sorting route of cytochrome b2 branches from the general mitochondrial import pathway at the preprotein translocase of the inner membrane. J. Biol. Chem., 272, 30439–30446. [DOI] [PubMed] [Google Scholar]
- Bömer U., Maarse, A.C., Martin, F., Geissler, A., Merlin, A., Schönfisch, B., Meijer, M., Pfanner, N. and Rassow, J. (1998) Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO J., 17, 4226–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branden C. and Tooze, J. (1998) Introduction to Protein Structure. Garland, New York.
- Bukau B. and Horwich, A.L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell, 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Chauwin J.F., Oster, G. and Glick, B.S. (1998) Strong precursor-pore interactions constrain models for mitochondrial protein import. Biophys. J., 74, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M. and Schatz, G. (1986) Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature, 322, 228–232. [DOI] [PubMed] [Google Scholar]
- Feynman R.P., Leighton, R.B. and Sands, M. (1963) The Feynman Lectures on Physics. Addison Wesley, Reading, ch. 46.
- Fünfschilling U. and Rospert, S. (1999) Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell., 10, 3289–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill B.D., Voos, W., Kang, P.J., Miao, B., Langer, T., Craig, E.A. and Pfanner, N. (1993) A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol., 123, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner F., Bömer, U., Guiard, B. and Pfanner, N. (1995) The sorting signal of cytochrome b2 promotes early divergence from the general mitochondrial import pathway and restricts the unfoldase activity of matrix Hsp70. EMBO J., 14, 6043–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume B., Klaus, C., Ungermann, C., Guiard, B., Neupert, W. and Brunner, M. (1998) Unfolding of preproteins upon import into mitochondria. EMBO J., 17, 6497–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S. (1995) Can hsp70 proteins act as force generating motors? Cell, 80, 11–14. [DOI] [PubMed] [Google Scholar]
- Glick B.S., Wachter, C., Reid, G.A. and Schatz, G. (1993) Import of cytochrome b2 to the mitochondrial intermembrane space: the tightly folded heme-binding domain makes import dependent upon matrix ATP. Protein Sci., 2, 1901–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U. (1996) Molecular chaperones in cellular protein folding. Nature, 381, 571–579. [DOI] [PubMed] [Google Scholar]
- Hill K., Model, K., Ryan, M.T., Dietmeier, K., Martin, F., Wagner, R. and Pfanner, N. (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature, 395, 516–521. [DOI] [PubMed] [Google Scholar]
- Horwich A.L., Weber-Ban, E.U. and Finley, D. (1999) Chaperone rings in protein folding and degradation. Proc. Natl Acad. Sci. USA, 96, 11033–11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Ratliff, K.S., Schwartz, M.P., Spenner, J.M. and Matouschek, A. (1999) Mitochondria unfold precursor proteins by unraveling them from their N-termini. Nature Struct. Biol., 6, 1132–1138. [DOI] [PubMed] [Google Scholar]
- Jensen R.E. and Johnson, A.E. (1999) Protein translocation: is Hsp70 pulling my chain? Curr. Biol., 9, R779–R782. [DOI] [PubMed] [Google Scholar]
- Kang P.-L., Ostermann, J., Shilling, J., Neupert, W., Craig, E.A. and Pfanner, N. (1990) Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature, 348, 137–143. [DOI] [PubMed] [Google Scholar]
- Keegstra K. and Froehlich, J.E. (1999) Protein import into chloroplasts. Curr. Opin. Plant Biol., 2, 471–476. [DOI] [PubMed] [Google Scholar]
- Krimmer T., Rassow, J., Kunau, W.-H., Voos, W. and Pfanner, N. (2000) The mitochondrial protein import motor: the ATPase domain of matrix Hsp70 is crucial for binding to Tim44, while the peptide binding domain and the carboxy-terminal segment play a stimulatory role. Mol. Cell. Biol., 20, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künkele K.P., Heins, S., Dembowski, M., Nargang, F.E., Benz, R., Thieffry, M., Walz, J., Lill, R., Nussberger, S. and Neupert, W. (1998) The preprotein translocation channel of the outer membrane of mitochondria. Cell, 93, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Liberek K., Skowyra, D., Zylicz, M., Johnson, C. and Georgopoulos, C. (1991) The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J. Biol. Chem., 266, 14491–14496. [PubMed] [Google Scholar]
- Lithgow T. (2000) Targeting of proteins to mitochondria. FEBS Lett., 476, 22–26. [DOI] [PubMed] [Google Scholar]
- Martin J., Mahlke, K. and Pfanner, N. (1991) Role of an energized inner membrane in mitochondrial protein import: Δψ drives the movement of presequences. J. Biol. Chem., 266, 18051–18057. [PubMed] [Google Scholar]
- Matlack K.E., Misselwitz, B., Plath, K. and Rapoport, T.A. (1999) BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell, 97, 553–564. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Azem, A., Ratliff, K., Glick, B.S., Schmid, K. and Schatz, G. (1997) Active unfolding of precursor proteins during mitochondrial protein import. EMBO J., 16, 6727–6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly J.R.Jr, Iriarte, A. and Martinez-Carrion, M. (1993) Structural features which control folding of homologous proteins in cell-free translation systems. The effect of a mitochondrial-targeting presequence on aspartate aminotransferase. J. Biol. Chem., 268, 26320–26327. [PubMed] [Google Scholar]
- Merlin A., Voos, W., Maarse, A.C., Meijer, M., Pfanner, N. and Rassow, J. (1999) The J-related segment of Tim44 is essential for cell viability: a mutant Tim44 remains in the mitochondrial import site, but inefficiently recruits mtHsp70 and impairs protein translocation. J. Cell Biol., 145, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara K. and Omura, T. (1996) Cytoplasmic chaperones in precursor targeting to mitochondria: the role of MSF and hsp70. Trends Cell Biol., 6, 104–108. [DOI] [PubMed] [Google Scholar]
- Moro F., Sirrenberg, C., Schneider, H.C., Neupert, W. and Brunner, M. (1999) The TIM17·23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J., 18, 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira J.L. and Fersht, A.R. (1999) Acquisition of native-like interactions in C-terminal fragments of barnase. J. Mol. Biol., 287, 421–432. [DOI] [PubMed] [Google Scholar]
- Netzer W. and Hartl, F. (1997) Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature, 388, 343–349. [DOI] [PubMed] [Google Scholar]
- Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem., 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Neupert W., Hartl, F.U., Craig, E.A. and Pfanner, N. (1990) How do polypeptides cross the mitochondrial membranes? Cell, 63, 447–450. [DOI] [PubMed] [Google Scholar]
- Ng D.T., Brown, J.D. and Walter, P. (1996) Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol., 134, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunola E., Suntio, T., Jamsa, E. and Makarow, M. (1998) Folding of active β-lactamase in the yeast cytoplasm before translocation into the endoplasmic reticulum. Mol. Biol. Cell, 9, 817–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N. and Meijer, M. (1995) Pulling in the proteins. Curr. Biol., 5, 132–135. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Craig, E.A. and Hönlinger, A. (1997) Mitochondrial preprotein translocase. Annu. Rev. Cell. Dev. Biol., 13, 25–51. [DOI] [PubMed] [Google Scholar]
- Rassow J., Guiard, B., Wienhues, U., Herzog, V., Hartl, F.U. and Neupert, W. (1989) Translocation arrest by reversible folding of a precursor protein imported into mitochondria. A means to quantitate translocation contact sites. J. Cell Biol., 109, 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J., Hartl, F.-U., Guiard, B., Pfanner, N. and Neupert, N. (1990) Polypeptides traverse the mitochondrial envelope in an extended state. FEBS Lett., 275, 190–194. [DOI] [PubMed] [Google Scholar]
- Schatz G. and Dobberstein, B. (1996) Common principles of protein translocation across membranes. Science, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Schwartz M.P. and Matouschek, A. (1999) The dimensions of the protein import channels in the outer and inner mitochondrial membranes. Proc. Natl Acad. Sci. USA, 96, 13086–13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.P., Huang, S. and Matouschek, A. (1999) The structure of precursor proteins during import into mitochondria. J. Biol. Chem., 274, 12759–12764. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Seytter, T., Guiard, B. and Neupert, W. (1993) Targeting of cytochrome b2 into the mitochondrial intermembrane space: specific recognition of the sorting signal. EMBO J., 12, 2295–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Kataoka, M. and Fink, A.L. (1996) Conformational characterization of DnaK and its complexes by small-angle X-ray scattering. Biochemistry, 35, 3297–3308. [DOI] [PubMed] [Google Scholar]
- Simon S.M., Peskin, C.S. and Oster, G.F. (1992) What drives the translocation of proteins? Proc. Natl Acad. Sci. USA, 89, 3770–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Neupert, W. and Cyr, D.M. (1994) The role of hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science, 266, 1250–1253. [DOI] [PubMed] [Google Scholar]
- Vestweber D. and Schatz, G. (1989) DNA–protein conjugates can enter mitochondria via the protein import pathway. Nature, 338, 170–172. [DOI] [PubMed] [Google Scholar]
- Voisine C., Craig, E.A., Zufall, N., von Ahsen, O., Pfanner, N. and Voos, W. (1999) The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell, 97, 565–574. [DOI] [PubMed] [Google Scholar]
- von Ahsen O., Voos, W., Henninger, H. and Pfanner, N. (1995) The mitochondrial protein import machinery: role of ATP in dissociation of the Hsp70–Mim44 complex. J. Biol. Chem., 270, 29848–29853. [DOI] [PubMed] [Google Scholar]
- Voos W., Gambill, B.D., Guiard, B., Pfanner, N. and Craig, E.A. (1993) Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol., 123, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W., von Ahsen, O., Müller, H., Guiard, B., Rassow, J. and Pfanner, N. (1996) Differential requirement for the mitochondrial Hsp70–Tim44 complex in unfolding and translocation of preproteins. EMBO J., 15, 2668–2677. [PMC free article] [PubMed] [Google Scholar]
- Voos W., Martin, H., Krimmer, T. and Pfanner, N. (1999) Mechanisms of protein translocation into mitochondria. Biochim. Biophys. Acta, 1422, 235–254. [DOI] [PubMed] [Google Scholar]
- Wachter C., Schatz, G. and Glick, B.S. (1994) Protein import into mitochondria: the requirement for external ATP is precursor-specific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol. Biol. Cell, 5, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D., Chaddock, A.M., Chaddock, J.A., Roberts, L.M., Lord, J.M. and Robinson, C. (1996) Ricin A chain fused to a chloroplast-targeting signal is unfolded on the chloroplast surface prior to import across the envelope membranes. J. Biol. Chem., 271, 4082–4085. [DOI] [PubMed] [Google Scholar]
- Wienhues U., Becker, K., Schleyer, M., Guiard, B., Tropschug, M., Horwich, A.L., Pfanner, N. and Neupert, W. (1991) Protein folding causes an arrest of preprotein translocation into mitochondria in vivo. J. Cell Biol., 115, 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]