Abstract
AKAP450 (also known as AKAP350, CG-NAP or Hyperion) and pericentrin are large coiled-coil proteins found in mammalian centrosomes that serve to recruit structural and regulatory components including dynein and protein kinase A. We find that these proteins share a well conserved 90 amino acid domain near their C-termini that is also found in coiled-coil proteins of unknown function from Drosophila and fission yeast. Fusion of the C-terminal region from either protein to a reporter protein confers a centrosomal localization, and overexpression of the domain from AKAP450 displaces endogenous pericentrin, suggesting recruitment to a shared site. When isolated from transfected cells the C-terminal domain of AKAP450 was associated with calmodulin, suggesting that this protein could contribute to centrosome assembly.
INTRODUCTION
Centrosomes organize microtubules in interphase cells and in the mitotic spindle formed during cell division. They comprise a central pair of centrioles surrounded by amorphous pericentriolar material that appears to be the site of microtubule nucleation. Although the molecular organization of the centrosome is not well understood, progress is being made in identifying its components, and microtubule nucleation has recently been shown to involve complexes containing γ-tubulin (Zimmerman et al., 1999; Schiebel, 2000). Additional components have been identified by the use of centrosomal specific antisera, one such being pericentrin, a large coiled-coil protein of the pericentriolar material (Doxsey et al., 1994; Dictenberg et al., 1998). Pericentrin has been found to interact with protein kinase A, and with cytoplasmic dynein, suggesting that it serves to hold in place a range of centrosomal components (Purohit et al., 1999; Diviani et al., 2000). Another centrosomal coiled-coil protein was recently identified by its ability to bind protein kinase A, and named AKAP450, AKAP350, CG-NAP or Hyperion (Schmidt et al., 1999; Takahashi et al., 1999; Witczak et al., 1999). For simplicity we will refer to it as AKAP450, as this is closest to its predicted molecular weight.
If these large coiled-coil proteins serve to recruit other factors to the centrosome this raises the question of how they themselves are localized. We have recently investigated the intracellular targeting of large coiled-coil proteins found on the Golgi, and extended these studies to include AKAP450, as it had been reported that this protein is found on the Golgi as well as in centrosomes, although other studies had only found a centrosomal location (Munro and Nichols, 1999; Takahashi et al., 1999). In this paper we report that AKAP450 and pericentrin share a conserved domain at their C-termini, outside of their coiled-coil regions and protein kinase A binding sites. This domain is sufficient to confer a centrosomal localization on green fluorescent protein (GFP), and so appears to be responsible for anchoring AKAP450 and pericentrin to the centrosome.
RESULTS
A conserved domain at the C-terminus of AKAP450 and pericentrin
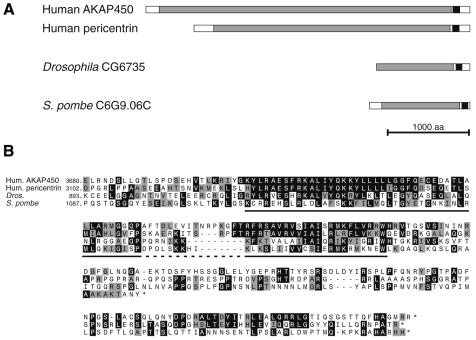
AKAP450 and pericentrin are both very large proteins predicted to form a coiled-coil over most of their length except for N- and C-terminal regions of ∼200 amino acids (Figure 1A). Searching the database with the C-terminal region of AKAP450 revealed a strong homology to the equivalent region of pericentrin, and also with the C-terminus of a putative coiled-coil protein of Drosophila that has not been previously characterized. Using the iterative search program PSI-BLAST (Altschul et al., 1997), with the region shared by these proteins identified a single further match to the C-terminus of an uncharacterized coiled-coil protein from S. pombe (significance score of 1 × e–5). These sequences show homology over a region of ∼90 amino acids, divided into two more highly conserved blocks (Figure 1B). Three of the proteins extend a further c120 residues C-terminal but this region is less well conserved and relatively proline rich.
Fig. 1. A conserved region at the C-terminus of a family of large coiled-coil proteins. (A) Structure of the human centrosomal proteins AKAP450 and pericentrin, and of proteins encoded by open reading frames from the indicated organisms. The related region is shown in black, and regions predicted to be predominantly coiled-coil are shown in grey. Pericentrin was originally cloned from mouse, and the human pericentrin homologue (kendrin) appears larger, but in fact the 3′ untranslated region of the original mouse cDNA ends with a sequence encoding a homologue of a known protein, suggesting it is a fusion of two cDNAs formed during cloning (Doxsey et al., 1994). (B) Alignment of the C-terminal sequences of the proteins in (A). Residues identical (black) or related (grey) in two or more of the sequences are shown, and the two particularly well conserved sections are underlined.
The C-terminus of AKAP450 is sufficient to localize GFP to the centrosome
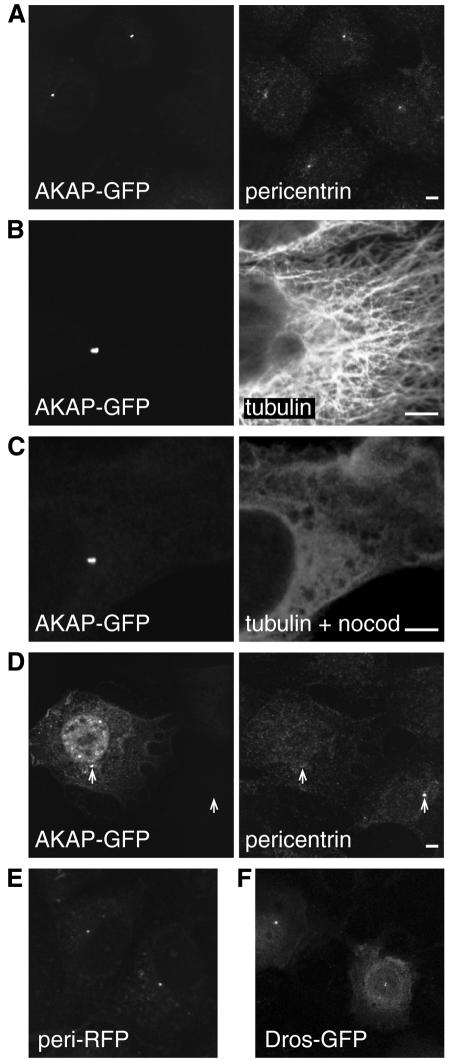
To investigate the function of this conserved C-terminal domain, the last 266 amino acids of AKAP450 were attached to the C-terminus of GFP, and expressed in COS cells. Examination of the transfected cells revealed bright GFP fluorescence in perinuclear spots characteristic of the centrosome (Figure 2A). At moderate expression levels these were the only structures visible, whilst at higher levels diffuse fluorescence was also present throughout the cell. The structures labeled with the GFP chimera were also recognized by antibodies against pericentrin and γ-tubulin, confirming that they were centrosomes (Figure 2A and data not shown). The antisera against pericentrin were raised against part of the coiled-coil region of the protein, and hence should not cross react with the region of AKAP450 present in the GFP fusion. We did not observe any staining of the Golgi apparatus, which is consistent with the notion that AKAP450 is found only on the centrosome. Figure 2C shows that the targeting of the GFP chimera to the centrosome was not affected when microtubules were depolymerized with nocodazole, indicating that the centrosomal location is not simply a reflection of an association with a minus-end-directed motor that concentrates the material near the microtubule organizing centre (Paschal et al., 1993). The C-terminal 226 residues of Drosophila protein CG6735 also targeted GFP to the centrosome in COS cells, indicating that the binding site of the domain has been well conserved in evolution (Figure 2F).
Fig. 2. The C-terminal regions of AKAP450 and pericentrin confer targeting to the centrosome. (A–F) Transfected COS cells expressing either GFP or RFP fused to the C-terminal regions of AKAP450 (C-terminal 266 residues, ‘AKAP’), pericentrin (241 residues, ‘peri’) or Drosophila CG6735 (226 residues, ‘Dros’), respectively. Cells were stained with antibodies against the indicated proteins, and nocodazole treatment was at 5 µM for 90 min. Scale bars, 5 µm.
Overexpression of the C-terminus of AKAP450 is sufficient to displace other centrosomal components
As described above, the fusion between GFP and the AKAP450 C-terminus colocalized with pericentrin in transfected cells. However, in cells expressing high levels of the AKAP fusion the centrosomal staining with antibodies against endogenous pericentrin was substantially reduced (Figure 2D). The focused GFP staining in Figure 2D, and the relative lack of an effect on γ-tubulin (data not shown), indicate that some centrosomal structure remained in these cells. This suggests that the C-terminal domain of AKAP450 was competing with the related domain in endogenous pericentrin for a binding site in the centrosome. Indeed a fusion of the C-terminal 241 amino acids of human pericentrin to red fluorescent protein (RFP) also accumulated in centrosomes, colocalizing with both anti-pericentrin antibodies and the GFP–AKAP450 chimera, and also reducing the staining of endogenous pericentrin when overexpressed (Figure 2E and data not shown). Taken together these results suggest that the C-terminal domains of AKAP450 and pericentrin share a binding site in the centrosome.
Investigation of the regions of AKAP450 required for centrosomal targeting
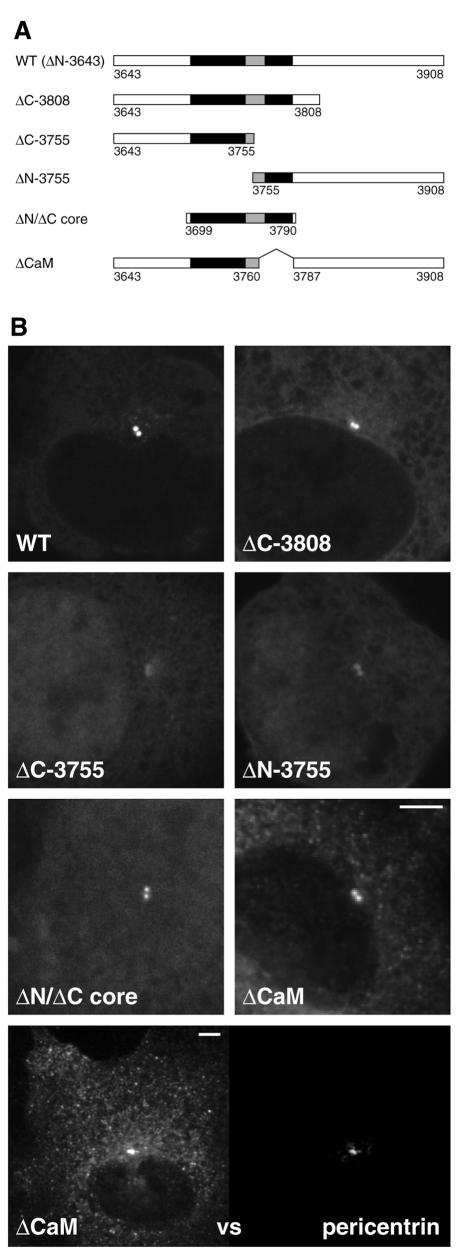
To examine the contribution of the conserved regions in the AKAP450 C-terminus to centrosomal targeting, truncated versions of this region were expressed in COS cells as fusions to GFP (Figure 3A). Protein blotting revealed bands with apparent sizes matching those predicted for these truncated versions (see below). Removal of the 100 amino acids C-terminal of the conserved region did not affect centrosomal localization (ΔC-3808, Figure 3B), indicating that the remaining conserved region contains the targeting activity. Indeed a 91 residue section covering this core conserved region is sufficient for centrosomal targeting (ΔN/ΔC core, Figure 3B). This region contains two highly conserved sections separated by a more variable stretch. When the C-terminus from AKAP450 was divided in its variable stretch both halves showed greatly reduced centrosomal targeting, but nonetheless centrosomal accumulation was not completely absent for either construct (ΔC-3755 and ΔN-3755, Figure 3B). This suggests that efficient recruitment of the domain to the centrosome does not involve binding through a single short motif, but rather requires an extended interaction of multiple motifs or a single large folded structure.
Fig. 3. At least two parts of the conserved region of the AKAP450 C-terminus contribute to centrosomal targeting. (A) The parts of the C-terminal region of AKAP450 fused to GFP in the indicated constructs. The residue numbers at the beginning and end of each construct are shown, with residue 3908 being the C-terminus of the whole protein. The two well conserved sections underlined in Figure 1B are shown in black. (B) COS cells expressing the six GFP fusions shown in (A). Cells were photographed using identical settings, and the perinuclear spots colocalized with endogenous pericentrin (not shown). In addition the lowest panel shows double labelling of endogenous pericentrin and ΔCaM at high levels of expression, the latter showing centrosomal staining and also a punctate pattern in the cytosol. Scale bars, 5 µm.
The C-terminus of AKAP450 binds to calmodulin
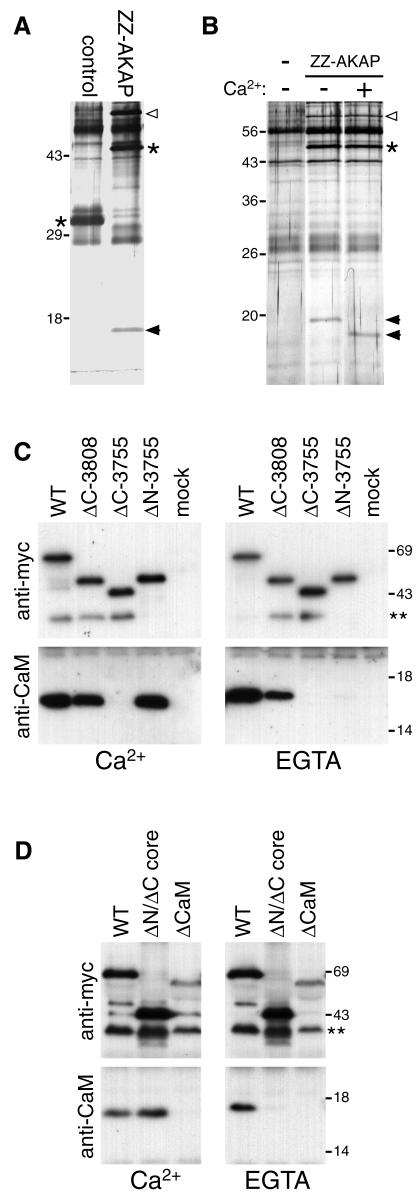
To identify proteins involved in the centrosomal targeting of AKAP450, the C-terminal domain was expressed in COS cells as a fusion to protein A [this chimera also localized to centrosomes (data not shown)]. Transfected cells were lysed, the ‘ZZ-AKAP’ fusion isolated on immunoglobulin beads, and an eluate from the beads examined for coprecipitating proteins. Figure 4A shows that the domain specifically coprecipitated with an ∼17 kDa protein. The isolation was repeated on a larger scale, and tryptic peptides analysed by mass spectrometry. The fragment masses gave a match to calmodulin, and this identification was confirmed in two ways. First, the mobility of the band upon electrophoresis increased in the presence of calcium, characteristic of calmodulin (Figure 4B), and secondly, anti-calmodulin antibodies detected a band of the appropriate size only in GFP–AKAP precipitates (see below).
Fig. 4. The C-terminal region of AKAP450 coprecipitates from cells in a complex with calmodulin. (A) Silver-stained gel of proteins isolated from COS cells (two 10 cm dishes per lane) expressing protein A fusions to either the C-terminal residues of AKAP450 or the C-terminal 134 residues of golgin-84 (control). Protein A fusions are indicated by asterisks, the coprecipitating bands by arrowheads. An additional band (open arrowhead) was seen at variable levels and identified as mitochondrial hsp60. We have found heat shock proteins associating with several protein A fusions, presumably reflecting binding to a partially folded proportion of overexpressed chimera. (B) As (A) except that the control was mock-transfected cells (–), and the samples in SDS buffer were divided in two and adjusted to 10 mM Ca2+ or 10 mM EGTA prior to electrophoresis. (C and D) Immunoblots of anti-GFP immunoprecipitates isolated from cells expressing GFP fused to parts of AKAP450 schematized in Figure 3A, or mock transfected (mock). Precipitation was in the presence of 10 mM Ca2+ or EGTA, and the blots were probed for calmodulin (CaM) or the Myc-tag present in the GFP fusion. A minor band corresponding to a smaller GFP fusion (**) was not visible in total cell samples and presumably reflects proteolysis post-lysis.
To investigate the requirements for this calmodulin interaction, the GFP–AKAP fusions described above were immunoprecipitated from transfected cells, and protein blots of the precipitated material probed for the fusion proteins and calmodulin. Figure 4C shows that although the different versions of the domain showed varying degrees of calmodulin binding, association of calmodulin with the complete C-terminal domain was calcium independent, and this association remained after removal of the poorly conserved C-terminal region (ΔC-3808). More surprisingly, removal of the first of the two conserved blocks resulted in the calmodulin binding becoming calcium dependent (ΔN-3755), although this first block did not bind calmodulin by itself. The core region binds calmodulin, but in a calcium sensitive fashion (ΔN/ΔC core, Figure 4D). A simple interpretation of these results is that, as with some other calmodulin binding proteins, there are at least two independent calmodulin-binding sites of distinct calcium sensitivity, and that these sites act synergistically to allow calmodulin binding that is insensitive to calcium levels (reviewed in Jurado et al., 1999). Indeed, the second of the two conserved regions resembles calcium-dependent calmodulin binding sites, being rich in basic and hydrophobic residues, and when this region was deleted calmodulin binding was abolished (ΔCaM, Figure 4D). Interestingly, this construct still showed centrosomal targeting, but at high levels of expression additional punctate staining was seen in the cytosol, rather than the diffuse staining seen with the other constructs, suggesting that its solubility had been compromised (ΔCaM, Figure 3B).
DISCUSSION
In this paper we report that two large coiled-coil proteins of the centrosome share a stretch of closely related sequence near their C-termini, and show that this region can confer a centrosomal location to heterologous proteins. Since coiled-coil proteins are likely to be dimeric a trivial explanation is that this putative domain is also involved in dimerization, and is dimerizing with the endogenous versions of AKAP450 or pericentrin. We believe that this is unlikely since overexpression of the AKAP450 C-terminus displaces endogenous pericentrin, indicating competition for a shared binding site. Thus, we propose that this C-terminal domain is in fact directly responsible for recruiting AKAP450 and pericentrin to the centrosome. We thus suggest that this region be called a PACT domain (pericentrin-AKAP450 centrosomal targeting). This domain is also present at the C-terminus of coiled-coil proteins from Drosophila and S. pombe, and that from the Drosophila protein is sufficient for targeting to the centrosome in mammalian cells. The function of these proteins is unknown but they seem good candidates for having a centrosomal or spindle pole body location.
We originally investigated AKAP450 because of a report that antisera against the protein also stained the Golgi apparatus in addition to the centrosome in some cell types (Takahashi et al., 1999). However, two other studies have found that a monoclonal antibody and a rabbit antiserum against AKAP450 gave only centrosomal staining in interphase cells (Schmidt et al., 1999; Witczak et al., 1999). Interestingly, a second rabbit antiserum was found to give some ‘Golgi-like’ staining, but on immunoblots this antiserum reacted with additional bands to those seen with the centrosomal-specific sera (Witczak et al., 1999). We believe that the simplest interpretation of these reports is that AKAP450 is not on the Golgi, but rather that antisera against coiled-coil regions can sometimes cross react with other coiled-coil proteins of which there are many on the Golgi apparatus. However, it is formally possible that there are versions of AKAP450 on the Golgi in some cells.
The specific association of the PACT domains from AKAP450 and pericentrin with the centrosome raises the question of what these domains are binding. When isolated from cells the C-terminus of AKAP450 is bound to calmodulin in a calcium-independent fashion. Deletions revealed the presence of a calcium-dependent calmodulin binding site, and indeed the second of the conserved regions is predicted to be such a site by the Ikura calmodulin binding site prediction server (http://calcium.oci.utoronto.ca/). A similar situation has been reported for some other calmodulin-binding proteins, such as phosphorylase kinase, which binds to calmodulin in a calcium-independent fashion through multiple sites, some of which are calcium-dependent in isolation (Dasgupta et al., 1989; Jurado et al., 1999).
As this manuscript was being prepared, Flory et al. (2000) reported that calmodulin binds to recombinant pericentrin in a gel-overlay assay, and that mutation of residues in the second of the conserved regions noted here abrogates binding, although the question of intracellular targeting was not addressed. This observation strengthens the conclusion that calmodulin binding could occur in vivo, but since calmodulin is widespread in the cell this conclusion, even if valid, does not help to explain centrosomal targeting. In yeasts a subset of calmodulin has been shown to be in the spindle pole body, but in mammalian cells a GFP–calmodulin fusion was found to be diffusely localized in interphase cells, and although in mitotic cells it was concentrated around spindle ends, it extended along the spindle, well beyond the centrosome (Stirling et al., 1994; Moser et al., 1997; Li et al., 1999). Indeed when the second of the conserved binding sites was removed (ΔCaM), calmodulin binding, but not centrosomal association, was affected. Thus, an alternative possibility is that calmodulin serves to chaperone the PACT domain in the cytoplasm, and could either remain bound upon centrosomal association, or be released to allow assembly with other proteins. A chaperone role has been proposed for yeast calmodulin, which binds to the C-terminus of the spindle pole protein Spc110p, although this binding site is not required for Spc110p to function (Geiser et al., 1993). Indeed the AKAP ΔCaM construct gave a coalesced rather than diffuse distribution at high expression levels, suggesting aggregation in the absence of calmodulin binding. The requirement for other regions of the domain for calmodulin binding to be calcium-insensitive suggests wrapping of an extended portion of the unassembled domain around calmodulin. Irrespective of whether the PACT domain binds calmodulin in the centrosome, or just in the cytosol, there must clearly be further interactions to explain the highly specific targeting reported here. The identification of the target of this domain may provide valuable insights into the organization of the centrosome.
Speculation
We would like to speculate that the reason that this PACT domain is well conserved in evolution is because it provides a critical link between a core of the centrosome and the components of the pericentriolar material such as pericentrin and AKAP450, which themselves serve to recruit further proteins to the centrosome.
METHODS
Plasmids. C-terminal regions of human AKAP450, pericentrin and fly CG6735 were PCR amplified from cDNA, and cloned into COS cell vectors containing a CMV promoter and GFP or RFP (Clontech) followed by a Myc epitope tag and the insertion site. Protein A fusions were expressed from similar vectors encoding two copies of the protein A ‘Z’ domain followed by a linker (VDANSGASENLYFQGS, where VD is the end of the second Z domain) and then the insertion site.
COS cell immunofluorescence. Cells transfected using Fugene (Roche) were fixed 30–48 h post-transfection with 4% paraformaldehyde (and permeabilized with 0.5% Triton X-100), or for pericentrin staining with methanol (5 min) and acetone (30 s). Rabbit anti-pericentrin (BabCo) or rat anti-tubulin (YL1/2) was detected with Alexa568 secondary antibodies (Molecular Probes) and an MRC-600 confocal microscope.
Protein analysis. COS cells in 10 cm dishes were transfected as above, were scraped into PBS 60 h later, pelleted (1 min at 2000 g), vortexed in lysis buffer (25 mM Tris–HCl pH 7.4, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride), debris removed by centrifugation (5 min at 14 000 g) and fusion proteins precipitated by the addition of IgG-Sepharose (Amersham Pharmacia Biotech) for protein A fusions, or rabbit anti-GFP (Rob Arkowitz, LMB Cambridge) and protein A–Sepharose for the GFP fusions. After binding for 2 h at 4°C, beads were washed five times in lysis buffer; for GFP the final wash was at 0.1% Triton X-100. Protein A fusions were eluted (0.5 M acetic acid pH 3.4), lyophilized and resuspended in SDS buffer. For protein identification, slices from a Coomassie-stained gel were digested with trypsin, and fragment masses determined by matrix-assisted laser desorption ionization mass spectrometry (Shevchenko et al., 1996). For immunoblotting mouse monoclonal antibodies against the Myc epitope tag (9E10) or calmodulin (Upstate Biotechnology) were detected with peroxidase-conjugated anti-mouse IgG and chemiluminescence (ECL; Amersham Pharmacia Biotech).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Sew Yeu Peak-Chew for mass spectrometric analysis, Nick Brown for fly cDNA, and John Kilmartin, Tim Levine and Hugh Pelham for comments on the manuscript.
REFERENCES
- Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta M., Honeycutt, T. and Blumenthal, D.K. (1989) The γ-subunit of skeletal muscle phosphorylase kinase contains two noncontiguous domains that act in concert to bind calmodulin. J. Biol. Chem., 264, 17156–17163. [PubMed] [Google Scholar]
- Dictenberg J.B., Zimmerman, W., Sparks, C.A., Young, A., Vidair, C., Zheng, Y., Carrington, W., Fay, F.S. and Doxsey, S.J. (1998) Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol., 141, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviani D., Langeberg, L.K., Doxsey, S.J. and Scott, J.D. (2000) Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr. Biol., 10, 417–420. [DOI] [PubMed] [Google Scholar]
- Doxsey S.J., Stein, P., Evans, L., Calarco, P.D. and Kirschner, M. (1994) Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell, 76, 639–650. [DOI] [PubMed] [Google Scholar]
- Flory M.R., Moser, M.J., Monnat, R.J. and Davis, T.N. (2000) Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin. Proc. Natl Acad. Sci. USA, 97, 5919–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser J.R., Sundberg, H.A., Chang, B.H., Muller, E.G. and Davis, T.N. (1993) The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado L.A., Chockalingam, P.S. and Jarrett, H.W. (1999) Apocalmodulin. Physiol. Rev., 79, 661–682. [DOI] [PubMed] [Google Scholar]
- Li C.J., Heim, R., Lu, P., Pu, Y., Tsien, R.Y. and Chang, D.C. (1999) Dynamic redistribution of calmodulin in HeLa cells during cell division as revealed by a GFP–calmodulin fusion protein technique. J. Cell Sci., 112, 1567–1577. [DOI] [PubMed] [Google Scholar]
- Moser M.J., Flory, M.R. and Davis, T.N. (1997) Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombe and performs an essential function in chromosome segregation. J. Cell Sci., 110, 1805–1812. [DOI] [PubMed] [Google Scholar]
- Munro S. and Nichols, B.J. (1999) The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol., 9, 377–380. [DOI] [PubMed] [Google Scholar]
- Paschal B.M., Holzbaur, E.L., Pfister, K.K., Clark, S., Meyer, D.I. and Vallee, R.B. (1993) Characterization of a 50-kDa polypeptide in cytoplasmic dynein preparations reveals a complex with p150Glued and a novel actin. J. Biol. Chem., 268, 15318–15323. [PubMed] [Google Scholar]
- Purohit A., Tynan, S.H., Vallee, R. and Doxsey, S.J. (1999) Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol., 147, 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel E. (2000) γ-tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol., 12, 113–118. [DOI] [PubMed] [Google Scholar]
- Schmidt P.H., Dransfield, D.T., Claudio, J.O., Hawley, R.G., Trotter, K.W., Milgram, S.L. and Goldenring, J.R. (1999) AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J. Biol. Chem., 274, 3055–3066. [DOI] [PubMed] [Google Scholar]
- Shevchenko A. et al. (1996) Linking genome and proteome by mass spectrometry—large-scale identification of yeast proteins from 2-dimensional gels. Proc. Natl Acad. Sci. USA, 93, 14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D.A., Welch, K.A. and Stark, M.J. (1994) Interaction with calmodulin is required for the function of Spc110p, an essential component of the yeast spindle pole body. EMBO J., 13, 4329–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Shibata, H., Shimakawa, M., Miyamoto, M., Mukai, H. and Ono, Y. (1999) Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the Golgi apparatus. J. Biol. Chem., 274, 17267–17274. [DOI] [PubMed] [Google Scholar]
- Witczak O., Skalhegg, B.S., Keryer, G., Bornens, M., Tasken, K., Jahnsen, T. and Orstavik, S. (1999) Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J., 18, 1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman W., Sparks, C.A. and Doxsey, S.J. (1999) Amorphous no longer: the centrosome comes into focus. Curr. Opin. Cell Biol., 11, 122–128. [DOI] [PubMed] [Google Scholar]