Abstract
In Escherichia coli cells, the origin of chromosomal replication is temporarily inactivated after initiation has occurred. Origin sequestration is the first line of defence against over-initiation, providing a time window during which the initiation potential can be reduced by: (i) titration of DnaA proteins to newly replicated chromosomal elements; (ii) regulation of the activity of the DnaA initiator protein; and (iii) sequestration of the dnaA gene promoter. This review represents the first attempt to consider together older and more recent data on such inactivation mechanisms in order to analyze their contributions to the overall tight replication control observed in vivo. All cells have developed mechanisms for origin inactivation, but those of other bacteria and eukaryotic cells are clearly distinct from those of E. coli. Possible differences and similarities are discussed.
Introduction
Any living cell must strike a balance between, on the one hand, promoting the reactions that are required for proliferation and, on the other hand, limiting such multiple reactions that may be deleterious to cell growth. This review describes the mechanisms known to limit DNA replication, with emphasis on the bacterium Escherichia coli.
When supplied with adequate nutrients and supplements, cells grow in size, replicate their chromosomes and divide into two daughter cells, in a tightly regulated manner. It is important that all chromosomes are replicated before cell division occurs, but it is also important to make sure that they are replicated only once. All cells that have been investigated, both prokaryotic and eukaryotic, seem to obey the once-and-only-once doctrine of DNA replication during normal, exponential growth (Boye, 1991a), although the doctrine can be deviated from under certain physiological situations. In the bacterium E. coli there are at least three different mechanisms blocking the occurrence of multiple replication events. The in vivo significance of each of the three mechanisms is not clear, but in their presence multiple replication events are extremely rare.
Chromosome replication in E. coli
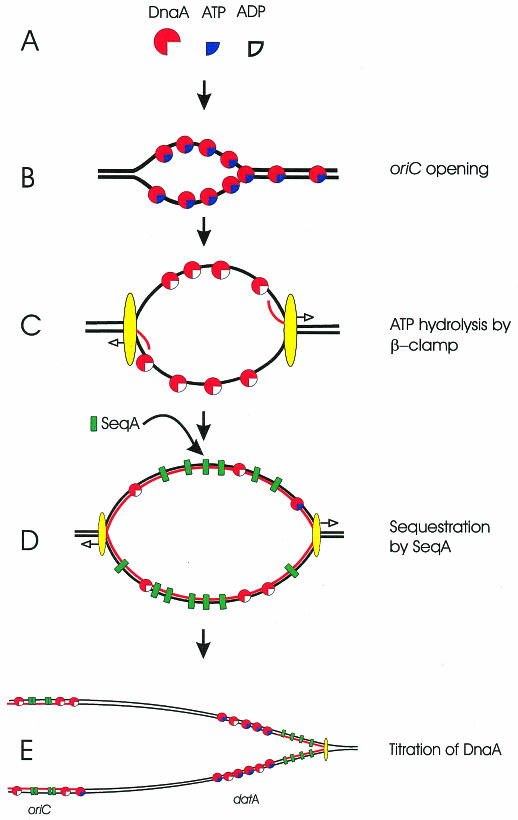
Escherichia coli has one circular chromosome which is replicated from a fixed point, called oriC. Initiation of replication occurs only once per cell division cycle, and simultaneously at all origins present in a cell (Skarstad et al., 1986). At initiation of replication, the initiator protein DnaA is bound to its five cognate binding sites within oriC and separates the two strands of the DNA helix in an AT-rich region of the origin (Figure 1B). The subsequent events include loading of the DnaB helicase into the open DNA structure, synthesis of an RNA primer and loading of the polymerase complex (Figure 1C) (Messer and Weigel, 1996). The intracellular availability of DnaA is the main regulator of replication initiation at oriC in vivo (Løbner-Olesen et al., 1989), strongly suggesting that the formation of an appropriate DnaA–oriC complex is the critical step in replication control. Once sufficient DnaA activity has accumulated and initiation has occurred, the main challenge for the cell is to prevent this activity from re-initiating one of the new origins. Below we describe three strategies that have been shown to contribute to the inactivation of a recently initiated origin of replication in E. coli.
Fig. 1. Schematic representation of mechanisms to limit initiation of chromosome replication in E. coli. The DnaA protein can bind ATP or ADP (A), bind to oriC and separate the DNA strands (B). After loading of the DnaB helicase, primase, and the elongation machinery the DnaA-bound ATP may be reduced to ADP by the β-clamp, which is part of the polymerase complex [yellow ellipse, (C)]. It should be noted that this ATP hydrolysis may continue as long as the β-clamp is loaded and not only shortly after initiation. The nascent DNA strands are unmethylated (red) and the SeqA protein binds hemimethylated DNA (D) and sequesters hemimethylated oriC. SeqA remains bound at the hemimethylated oriC long after the replication fork has passed datA, which titrates a large amount of DnaA (E). Note the difference in scale in the different panels.
Sequestration
The adenines of GATC sites in E. coli are methylated by Dam methyltransferase. The GATC sites on both strands are methylated but, during replication, unmethylated nucleotides are incorporated. Therefore, a region of hemimethylated DNA exists in the wake of the replication fork (Figure 1D and E). In general, Dam methyltransferase methylates these sites within less than a minute, but certain sites, e.g. oriC and the dnaA gene, remain hemimethylated for up to one-third of the cell cycle (Campbell and Kleckner, 1990). Hemimethylated origins are not initiated in vivo (Russell and Zinder, 1987), but are perfectly good substrates in vitro (Landoulsi et al., 1989; Boye, 1991b), arguing that some intracellular factor inactivates (sequesters) hemimethylated origins. This factor has been identified and named SeqA (Lu et al., 1994; von Freiesleben et al., 1994). The SeqA protein has a high affinity for fully methylated oriC and for hemimethylated DNA in general (Slater et al., 1995; Brendler and Austin, 1999), and it binds specifically to the same two sites in oriC, irrespective of whether the DNA is hemi- or fully methylated (Skarstad et al., 2000).
When oriC is replicated, it goes from being fully methylated to hemimethylated, a condition that is exploited by the cell as a kind of turnstile mechanism (Boye, 1991a): the origin cannot rapidly return to the fully methylated state because SeqA binds and sequesters the hemimethylated form (Figure 1D and E). However, this situation cannot persist, since oriC must eventually be remethylated and prepared for the next round of replication, but only after cell division has occurred. It is therefore important that the potential for initiation at oriC has been reduced by the time sequestration ends. The concentration of Dam methyltransferase influences both the duration of the hemimethylated state (Boye and Løbner-Olesen, 1990) and the minimum time between successive initiations of the same origin (von Freiesleben et al., 2000).
Sequestration is absolutely required for efficient inactivation of fired origins. In seqA (Lu et al., 1994; Boye et al., 1996) and dam (Boye and Løbner-Olesen, 1990) mutant cells, origin refiring occurs at a significant rate, even in the presence of the two additional inhibitory mechanisms below. Therefore, sequestration can be considered as the first line of defence against over-initiation, providing a time window (eclipse) during which the initiation potential can be reduced by other means.
Regulating the activity of DnaA
The DnaA protein binds the nucleotides ATP and ADP with high affinity (Figure 1A; Sekimizu et al., 1987). Both forms of the protein bind oriC and other sites with DnaA boxes, but ATP-DnaA is required to form an initiation complex that promotes strand separation and initiation of replication. The ATP form is therefore the active form of the DnaA protein, while the ADP form is inactive. Mutations in DnaA that affect binding or hydrolysis of ATP may cause the protein to be constitutively active, leading to over-initiation and even lethality (Katayama and Kornberg, 1994; Mizushima et al., 1997). Studies of one such mutant protein (DnaAcos), which over-initiates at high temperature, revealed that the over-initiation was due to a failure to inactivate DnaA, strongly suggesting that this inactivation is required in vivo. The mutant protein did not bind ATP and was therefore insensitive to normal inactivation by hydrolysis. The inactivating switch for DnaA turned out to be the β-subunit of the replication complex, together with a so far unidentified factor called IdaB (Katayama et al., 1998), and the two factors appear to work in concert. The β-subunit forms a multimeric, doughnut-shaped molecule, a so-called sliding clamp, which encircles the DNA and dramatically improves the processivity of DNA polymerase III. Efficient inactivation of wild type DnaA by hydrolysis of its bound ATP requires the β-subunit to be loaded onto DNA as a sliding clamp (Figure 1C), and the inactivation is further stimulated by ongoing DNA replication. By this mechanism the capacity for initiation of replication may be reduced as soon as one round of replication is underway. However, in the absence of sequestration and with excess DnaA activity the eclipse period is virtually non-existent (Bogan and Helmstetter, 1997), suggesting that the DnaA is not all inactivated, or is not inactivated fast enough, to prevent another round of replication from being started.
The lifetimes of ATP- and ADP-DnaA in vitro are ∼1 h. However, when DnaA is bound to DNA in the presence of anionic phospholipids the nucleotides can be released immediately and fresh nucleotide may bind DnaA (Sekimizu and Kornberg, 1988). Thus, in the presence of phospholipid and excess ATP, the inactive ADP form of DnaA can be rejuvenated to the active form.
A marked reduction of the intracellular ATP-DnaA level around the time of initiation further suggests that the ATP/ADP exchange has a biological function in vivo (Kurokawa et al., 1999). The DnaA protein is frequently found associated with the membrane (Newman and Crooke, 2000) and cells lacking anionic phospholipids cannot perform DnaA-dependent initiation at oriC, arguing, first, that the membrane is important for the function of DnaA in vivo and, secondly, that the DnaA-membrane interaction may facilitate rejuvenation of inactive DnaA.
Regulating the level of free DnaA
The datA locus. The affinity of DnaA for its cognate boxes is relatively high and the protein is mostly bound to DNA when DnaA boxes are available. The chromosome contains a hierarchy of ∼300 DnaA boxes with different affinities for the DnaA protein. Initiation of replication can be seen, at least in part, as a competition for active DnaA protein between the boxes in oriC and in other DnaA-binding regions. There are five regions of the chromosome which bind DnaA protein with particularly high affinity (Roth and Messer, 1998). The locus with the highest DnaA-binding capacity appears to be datA (Kitagawa et al., 1996), which can titrate 8-fold more DnaA protein than the region spanning both oriC and the neighbouring gene mioC, even though the two regions (datA and oriC/mioC) contain the same number of DnaA boxes. The reason for the difference in DnaA-binding capacity is not known, but may at least partly be explained by a competition between DnaA and SeqA for binding to oriC (von Freiesleben et al., 2000), whereas SeqA is not expected to bind much to datA, since it contains few GATC sites. Because datA is located close to oriC, it has been suggested to play a pivotal role in controlling replication initiation (Kitagawa et al., 1998). As the datA region is replicated it will titrate twice the amount of DnaA protein, thereby drastically reducing the intracellular concentration of free DnaA protein at this point in the cell cycle (Figure 1E).
There is direct evidence that datA has an important regulatory function in vivo. First, over-initiation is observed when datA is removed from the chromosome (Kitagawa et al., 1998). Secondly, introduction of additional intracellular copies of the datA locus is limiting for initiation (Kitagawa et al., 1998) and high datA copy numbers totally shut down intiation from oriC (our unpublished data). We conclude that the function of datA is required as a sink for DnaA protein for normal regulation of initiation. However, normal replication control does not require datA to be located close to oriC. This suggests that the timing of datA replication is not critical, as long as sequestration is active. It is not known how or whether datA replication can contribute to initiation control if its replication occurs after the sequestration period is over.
Sequestration of the dnaA gene. The dnaA gene is replicated shortly after oriC and, like oriC, becomes hemimethylated and is sequestered by a SeqA-dependent mechanism (Lu et al., 1994). This sequestration results in a transient suppression of dnaA gene transcription shortly after initiation (Theisen et al., 1993). Thus, following oriC initiation, there is a period of time when no new DnaA protein is being synthesized. Since the chromosome is replicated during this period, the ratio between DnaA protein and DnaA boxes is reduced, resulting in a reduction of the initiation capacity. This temporary repression of dnaA transcription is not important in the presence of the other mechanisms, though, since the gene can be expressed continuously, from a heterologous promoter, without any effect on replication control (Løbner-Olesen et al., 1989).
Gram-positive bacteria
Sequestration, as it is described here, is dependent upon methylation for discrimination between old and new replication origins. However, replication control is equally tight in Gram-positive bacteria that do not possess a Dam-like methylation system or a SeqA homologue, e.g. Bacillus subtilis (Seror et al., 1994). Presumably, these bacteria possess alternative, highly efficient regulatory mechanisms. In bacilli, oriC and dnaA are colocalized, representing a region with a high capacity to bind DnaA, particularly since SeqA is absent. Therefore, replication of this region will create a large sink for DnaA, which will effectively reduce the level of free DnaA. This mechanism may on its own represent the main form of replication control in Gram-positive bacteria. However, a separate regulatory mechanism that involves a strong interaction of oriC with the cell membrane may also be active (Firshein, 1989).
Eukaryotic cells
In eukaryotic cells, the major mechanism behind the restriction of chromosome replication to once per generation is termed licensing (Blow and Laskey, 1988). The term reflects the notion that each origin of replication is, in M/G1 phase, given a licence to replicate once, and the cells must pass through mitosis again to receive another licence. This mechanism is therefore distinct from sequestration. It involves assembly of the pre-replication complex, including loading of the minichromosome maintenance (MCM) proteins onto future replication origins, in late M or in G1, at a time when cdk/cyclin activity is low. Later in the replication cycle, a pre-replication complex cannot be assembled or re-assembled, for two reasons. First, the factor which allows MCM loading, the Cdc6 protein, is phosphorylated and degraded in G1 (Donovan et al., 1997; Tanaka et al., 1997). Secondly, some of the MCM proteins are phosphorylated in S phase (Coue et al., 1996) and, in the yeast Saccharomyces cerevisiae, this may cause the MCM proteins to be transported to the cytoplasm, thereby denying them access to the replication origins. Thus, a licence to assemble the pre-replication complex is issued only when the cells pass through mitosis, after the cyclins have been degraded and the cdk/cyclin activity is abolished.
Licensing may be sufficient to prevent re-initiation from occurring, but the ATP/ADP binding properties of some of the eukaryotic initiation proteins probably also have regulatory roles and may function in preventing re-initiation. The S. cerevisiae ORC and Cdc6 proteins, which are part of the pre-replication complex, both bind and hydrolyze ATP. Cdc6p requires ATP binding for proper replication complex assembly (Perkins and Diffley, 1998; Weinreich et al., 1999). In this situation, the assembly and disassembly of the pre-replication complex may well be controlled by ATP hydrolysis (Lee and Bell, 2000).
Conclusions
All cells investigated have built up a defense against replicating their chromosomes without purpose. However, the molecular details of the defense mechanisms vary between cell types. In E. coli, sequestration temporarily inactivates the origins and provides a time window during which other mechanisms can reduce the initiation potential. Titration of DnaA proteins by datA and reduction of DnaA activity by ATP/ADP exchange is operative during the sequestration period to reduce the initiation potential. Such mechanisms are important for optimal functioning and for genomic stability of individual cells, whether they are prokaryotic or eukaryotic.

Kirsten Skarstad

Anders Løbner-Olesen

Erik Boye
Acknowledgments
Acknowledgements
This work was supported by The Norwegian Cancer Society, the Norwegian Research Council and the Carlsberg Foundation.
References
- Blow J.J. and Laskey, R.A. (1988) A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature, 332, 546–548. [DOI] [PubMed] [Google Scholar]
- Bogan J.A. and Helmstetter, C.E. (1997) DNA sequestration and transcription in the oriC region of Escherichia coli. Mol. Microbiol., 26, 889–896. [DOI] [PubMed] [Google Scholar]
- Boye E. (1991a) A turnstile for initiation of DNA replication. Trends Cell Biol., 1, 107–109. [DOI] [PubMed] [Google Scholar]
- Boye E. (1991b) The hemimethylated replication origin of Escherichia coli can be initiated in vitro. J. Bacteriol., 173, 4537–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E. and Løbner-Olesen, A. (1990) The role of Dam methyltransferase in control of DNA replication in Escherichia coli. Cell, 62, 981–989. [DOI] [PubMed] [Google Scholar]
- Boye E., Stokke, T., Kleckner, N., and Skarstad, K. (1996) Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl Acad. Sci. USA, 93, 12206–12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T. and Austin, S. (1999) Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J., 18, 2304–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.L. and Kleckner, N. (1990) E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell, 62, 967–979. [DOI] [PubMed] [Google Scholar]
- Coue M., Kearsey, S.E., and Mechali, M. (1996) Chromotin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J., 15, 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Donovan S., Harwood, J. Drury, L.S., and Diffley, J.F.X. (1997) Cdc6-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firshein W. (1989) Role of the DNA/membrane complex in prokaryotic DNA replication. Annu. Rev. Microbiol., 43, 89–120. [DOI] [PubMed] [Google Scholar]
- Katayama T. and Kornberg, A. (1994) Hyperactive initiation of chromosomal replication in vivo and in vitro by a mutant initiator protein, DnaAcos, of Escherichia coli. J. Biol. Chem., 269, 12698–12703. [PubMed] [Google Scholar]
- Katayama T., Kubota, T., Kurokawa, K., Crooke, E., and Sekimizu, K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E.coli chromosomal replicase. Cell, 94, 61–71. [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Mitsuki, H., Okazaki, T., and Ogawa, T. (1996) A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol., 19, 1137–1147. [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Ozaki, T., Moriya, S., and Ogawa, T. (1998) Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev., 12, 3032–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Nishida, S., Emoto, A., Sekimizu, K., and Katayama, T. (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J., 18, 6642–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landoulsi A., Hughes, P., Kern, R., and Kohiyama, M. (1989) Dam methylation and the initiation of DNA replication on oriC plasmids. Mol. Gen. Genet., 216, 217–223. [DOI] [PubMed] [Google Scholar]
- Lee D.G. and Bell, S.P. (2000) ATPase switches controlling DNA replication initiation. Curr. Opin. Cell Biol., 12, 280–285. [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., Skarstad, K., Hansen, F.G., von Meyenburg, K., and Boye, E. (1989) The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell, 57, 881–889. [DOI] [PubMed] [Google Scholar]
- Lu M., Campbell, J.L., Boye, E., and Kleckner, N. (1994) SeqA: a negative modulator of replication initiation in E.coli. Cell, 77, 413–426. [DOI] [PubMed] [Google Scholar]
- Messer W. and Weigel, C. (1996) Initiation of chromosome replication. In Neidhardt, F.C. (ed.), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1579–1601.
- Mizushima T., Nishida, S., Kurokawa, K., Katayama, T., Miki, T., and Sekimizu, K. (1997) Negative control of DNA replication by hydrolysis of ATP bound to DnaA protein, the initiator of chromosomal DNA replication in Escherichia coli. EMBO J., 16, 3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman G. and Crooke, E. (2000) DnaA, the initiator of Escherichia coli chromosomal replication, is located at the cell membrane. J. Bacteriol., 182, 2604–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G. and Diffley, J. (1998) Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell, 2, 23–32. [DOI] [PubMed] [Google Scholar]
- Roth A. and Messer, W. (1998) High-affinity binding sites for the initiator protein DnaA on the chromosome of Escherichia coli. Mol. Microbiol., 28, 395–401. [DOI] [PubMed] [Google Scholar]
- Russell D.W. and Zinder, N.D. (1987) Hemimethylation prevents DNA replication in E.coli. Cell, 50, 1071–1079. [DOI] [PubMed] [Google Scholar]
- Sekimizu K. and Kornberg, A. (1988) Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem., 263, 7131–7135. [PubMed] [Google Scholar]
- Sekimizu K., Bramhill, D., and Kornberg, A. (1987) ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E.coli chromosome. Cell, 50, 259–265. [DOI] [PubMed] [Google Scholar]
- Seror S.J., Casaregola, S., Vannier, F., Zouari, N., Dahl, M., and Boye, E. (1994) A mutant cysteinyl-tRNA synthetase affecting timing of chromosomal replication initiation in B. subtilis and conferring resistance to a protein kinase C inhibitor. EMBO J., 13, 2472–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Boye, E., and Steen, H.B. (1986) Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J., 5, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K, Lueder, G, Lurz, R, Speck, C and Messer W. (2000) The Escherichia coli SeqA protein binds specifically and co-operatively to two sites in hemimethylated and fully methylated oriC. Mol. Microbiol., 36, 1319–1326 [DOI] [PubMed] [Google Scholar]
- Slater S., Wold, S., Lu, M., Boye, E., Skarstad, K., and Kleckner, N. (1995) E.coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell, 82, 927–936. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp, D., and Nasmyth, K. (1997) Loading of an Mcm protein onto DNA-replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Theisen P.W., Grimwade, J.E., Leonard, A.C., Bogan, J.A., and Helmstetter, C.E. (1993) Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol., 10, 575–584. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen, K.V., and Schaechter, M. (1994) SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol., 14, 763–772. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Krekling, M., Hansen, F.G., and Løbner-Olesen, A. (2000) The eclipse period of Escherichia coli. EMBO J., 19, 6240–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Liang, C., Stillman, B. (1999) The Cdc6p nucleotide-binding motif is required for loading MCM proteins onto chromatin. Proc. Natl Acad. Sci. USA, 96, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]