Abstract
Background:
The influence of cytokine in the reproductive system is becoming increasingly important. The polymorphisms of the transforming growth factor-β1 (TGF-β1) gene are involved in male infertility. This study aimed to demonstrate the association between TGF-β1 and infertility and to investigate its impact on semen quality.
Methods:
In this case-control study, serum TGF-β1 concentration was measured in 144 patients diagnosed with infertility and 40 fertile males by enzyme-linked immunosorbent assay (ELISA). The tetra-amplification refractory mutation system-PCR (T-ARMS-PCR) analysis was performed to detect the genotyping of the TGF-β1 (+869 C/T) (rs1800470) SNPs gene.
Results:
Serum concentration of TGF-β1 was less in infertile males compared to fertile ones. The detected and more effective genotypes and alleles of TGF-β1 gene polymorphic on male infertility were, in normozoospermic group, CT genotype, probability (p)= 0.45, relative risk (RR)= 1.56, confidence intervals (CI): 0.58–4.22, and T allele (p= 0.46, RR= 1.32, CI: 0.65—2.69), in oligozoospermic and azoospermic groups, CC genotype (p= 0.32, RR= 1.58, CI: 0.73–3.41), (p= 0.013, RR= 3.50, CI: 1.40–8.73), and allele C (p= 0.44, RR= 1.32, CI: 0.73–2.38), (p= 0.06, RR= 2.14, CI: 1.02–4.50), respectively. The recessive model (TT+CT) showed increased risk among normozoospermic group (p=0.44, RR=1.67, CI:0.60-4.62). The serum concentration of TGF-β1 with CT and TT genotypes was less than that of CC genotype. TGF-β1 C/T genotype correlated with low sperm number, high immotile sperm, and high abnormal sperm morphology.
Conclusions
Our study revealed that the TGF-β1(rs1800470) gene polymorphisms are associated negatively with semen quality.
Key Words: Asthenozoospermia, Non-obstructive azoospermia, Teratozoospermia, Oligozoospermia, TGF-β1 gene polymorphisms
Introduction
The family of transforming growth factors (TGF) is a class of cytokines responsible for a variety of essential body functions. The fundamental physiological processes comprise proliferation, differentiation, metabolism, and apoptosis (1). TGF-β family, in mammals, consists of 3 TGF-β1,2,3 isoforms (2).
The expression of TGF-β1 was found in normal Leydig cells, as well as sperm cells, and Sertoli cells (3). In the testis, it is expressed in many cells at various developmental stages. TGF-1 is mostly expressed in spermatogonia and Leydig cells during the neonatal period, but before puberty and in a variety of mature sperm cells, it is predominantly expressed in Sertoli cells (4). Studies on testicular spermatogenic epithelial cells illustrate that sperm express TGF-1 before they reach the epididymis. TGF-1 expression can be detected in the tight junctions separating Sertoli cells, which suggests that TGF-1 may be crucial for preserving the tiny tubules' structural integrity (5).
Researchers indicated that there are no differences in seminal plasma TGF-β1 concentrations between fertile and infertile subjects (6). Other studies revealed that the decline of TGF-β1 concentrations was correlated with poor semen parameters (7, 8). It has been reported that infertile males have high levels of seminal TGF-β1 concentration compared to the fertile group and suggested that increased seminal TGF-β1 levels are crucial risk factors that can lead to a number of morphological and functional modifications of spermatozoa (9). The normal spermatogenic activity of the body may be impacted by the testis' excessive TGF-1 expression (10). TGF-β1 is mostly expressed in Leydig cells and is detected in the testicular stroma in mature males because these cells are the principal site for the conversion of testosterone into oestrogen. Model rat testicular stroma was microinjected with a TGF-β1 receptor I (TβRI) blocker, which successfully prevented Leydig cells from expressing TGF-1 (11). Rat sperm density, sperm motility, and both rapid and slow forward movement (Grade A) can all be greatly increased by TβRI blockers. Under a high-powered microscope, it was found that TβRI blockers reduce the expression of TGF-β1, while increasing the expression of enzyme (CYP19) and sperm production in rat models. TβRI may raise serum sex hormone levels and enhance sperm quality (12).
The human TGF-β gene, which is existed on chromosome 19q13 (19q13.2–19q13.1), is responsible for encoding the TGF- β (13). Various single nucleotide polymorphisms (SNPs) have been discovered in the human TGF-1 exon 1 and exon 2 regions, and it has been hypothesized that these SNPs are connected to the cytokine's baseline levels (14). Two SNPs, the +869 C>T SNP (rs1800470) polymorphism, also known as c.+29C>T and Pro10Leu, and the +915 G>C SNP (rs1800471) polymorphism, also known as c.+74 G>C and Arg25Pro, are located in the signal peptide of exon 1. As well as, two unusual polymorphisms of rs201567874 and rs199758510 are discovered in exons 1 and 2, respectively. As a result, the researchers have found SNPs at the signal peptide of exon 1 (rs1800470 and rs1800471) that impact serum levels of this cytokine (15).
Materials and Methods
Study participants
The current study was implemented on 184 volunteers who attended Rizgary Teaching Hospital, Runahi IVF Center, and Shayi's private clinical laboratory in Erbil City from September 2021 to September 2022. The study was composed of two groups: Fertile group (control) : Included 40 (21.74%) fertile normozoospermic males (sperm concentration ≥ 15 million/ml) without any history of infertility problems or diseases, and their ages ranged between 21-47 years. Infertile group (patients) : Included 144 infertile males, categorized as normozoospermia [n=28 (15.22%)] oligozoospermia [n=76 (41.30%)], and azoospermia [n=40 (21.74%)], their ages ranged between 17-59 years.
The differences in the number of patients and controls are due to the sub-grouping of patients in addition to randomization and time specified for sample collection.
Exclusion criteria
Obstructive azoospermia, abnormal hormonal levels of (Luteinized hormone, Follicle stimulating hormone, and Testosterone), urogenital tract infections, cryptorchidism, and other chronic diseases were excluded from the study.
Ethical concerns
All the volunteers were fully informed regarding the objectives of the study and ensured that their specimens were used only for this particular study, also ensuring ethics remained a top priority throughout the study.
Seminal fluid analysis
After guiding the participants on how to collect the semen sample, the seminal fluid was incubated at 37 °C for 20 min. To categorize the seminal fluid of patient groups, the liquefied semen was assessed according to World Health Organization’s (WHO) guidelines (WHO, 2010).
Blood collection and storage
All volunteer participants donated around 7 ml of peripheral blood that was withdrawn with a disposable plastic syringe. In subsequent, they aliquot into a labelled EDTA tube (2 ml) and stored at -20 °C for genomic study. The remaining blood sample (5 ml) was collected in a gelatin tube, allowed to clot at room temperature then centrifuged at 3924 G for 5 minutes, the sera obtained were dispensed into a labeled and sterile Eppendorf tube and kept at -20 °C to determine the level of TGF-β1.
Serum TGF-β1 ELISA measurement
After thawing the frozen serum at room temperature, serum TGF-β1 concentration was quantified by a commercial ELISA kit (Sunlong Biotech Co., China, Lot No: 20220215, REF: SL1734Hu). Serum TGF-β1 concentration was calculated as pg/ml. TGF-β1 ELISA kit analysis was performed at the Biotechnology Laboratory, College of Education, Salahadden University.
DNA isolation and genotyping of TGF-β1 (+869C/T) SNPs
For the isolation of DNA, the frozen blood in an EDTA tube was allowed to thaw at room temperature, and BetaPrep genomic DNA extraction kit (Beta Bayern, German, Cat. No.: MDE 101) was used following manufacturer protocol. DNA purity and concentration were detected using nano-drop (Biometrics, OneDrop TOUCH Pro/Lite Micro-Volum Spectrophotometer, Wilmington, USA) by determining the absorbance at wavelengths 260 nm and 280 nm, which ranged from 10-97 ng, and the purity of all the genomic DNA samples was spotted in the range of 1.7 to 1.9.
The TGF-β1gene C and T alleles at positions +869 were genotyped using the T-ARMS-PCR reaction. Depending on the (16) source methodology, the designed primers to target the SNPs of the TGF-β1gene are provided in Table 1. The amplification protocol and the PCR reaction conditions for the T-ARMS technique were proceed using 2X Prime Taq Premix (Genet Bio, Korea, product code:35001) (Tables 2 and 3). After PCR cycles were performed, the amplified products were analyzed for their size and visualized using 2 % agarose gel electrophoresis which was carried out for 50min. (75 V/cm2 for 20 min. then 85 V/cm2 for 30 min.), stained with safe dye (Fig. 1). After electrophoresis was finished, the genotype was registered using the chart provided by the genotyping kit supplier (Table 1).
Table 1.
Designed primer sequences used in T-ARMS-PCR genotyping detection and interpretation.
| Gene polymorphism | Minor allele | Primer name | Primer sequence | allele | Amplicon size (bp) | Product size (bp) | Genotype |
|---|---|---|---|---|---|---|---|
| TGF-β1 (+869C/T)(rs1800470)(Gene code:7040) | T | FO | CAGCTTTCCCTCGAGGCCCTCCTACCTT | 255 | NH: 180, 255 | CC | |
| RO | TTCCGCTTCACCAGCTCCATGTCGATAG | 255 | HE: 123, 180, 255 | CT | |||
| FI | CTCCGGGCTGCGGCTGCTTCT | T | 123 | MH: 123, 255 | TT | ||
| RI | AGTAGCCACAGCAGCGGTAGCAGCATCG | C | 180 | ||||
NH: Normal homozygote; HE: Heterozygote; MH: Mutant homozygote.
Table 2.
DNA Amplification and PCR protocol for gene polymorphism detection.
| Amplification condition | ||
|---|---|---|
| Components | Volume (µl) | |
| Nuclease free water | 5 | |
| Taq master | 10 | |
| Forward primer | Outer | 1 |
| Inner | 1 | |
| Reverse primer | Outer | 1 |
| Inner | 1 | |
| DNA template | 1 | |
| Total reaction volume | 20 µl | |
Table 3.
PCR program used for the amplification.
| PCR program | |||
|---|---|---|---|
| PCR cycles | Temperature | Time | |
| Initial denaturation | 95 °C | 5 min | |
| PCR cycling (35 cycles) | Denaturation | 95 °C | 30 sec. |
| Annealing | 65 °C | 30 sec. | |
| Extension | 72 °C | 1 min. | |
| Final extension | 72°C | 10 min. | |
| Hold | 20°C | ∞ | |
Fig. 1.
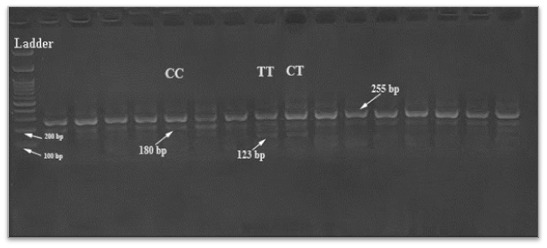
Agarose gel electrophoresis of the T-ARMS-PCR products for the TGF-β1 (+869C/T) SNPs.
Statistical analysis
Graph-Pad Prism version 9.0 was used to conduct all statistical analyses. Estimating the relative risk (RR) and 95% confidence intervals (CI) enabled the determination of the relationship between genotypes and infertility. Differences in the distributions of the diplotypes among the fertile and infertile groups were evaluated using the Hardy-Weinberg equilibrium (HWE). Statistical significance was defined as a P-value under 0.05.
Results
One hundred eighty-four volunteers enrolled in the current study. The mean age for control was 35.75±10.19, while for infertile patients as normozoospermia, oligozoospermia, and azoospermia were 35.93±9.10, 33.57±8.09, 33.35±8.16, respectively. No significant differences were found in the mean ages of the control and patient groups.
Evaluation of serum TGF-β1 and their correlation with seminogram
The TGF-β1 have been estimated in the serum of all participants and the mean values were as fertile males: TGF-β1=226.57±66.87 pg/ml; infertile normozoospermic: TGF-β1=206.41±72.19 pg/ml; infertile oligozoospermic: TGF-β1=180.05±52.40 pg/ml and infertile azoospermic: TGF-β1=122.55±36.33 pg/ml. The statistical analysis showed a declined level of serum TGF-β1 in infertile participants. Significant differences were observed relating to this factor between fertile and infertile oligozoospermic and azoospermic participants but not with infertile normozoospermic participants. Furthermore, Significant differences were observed among infertile participants, but not between normozoospermic and oligozoospermic infertile males (Fig. 2).
Fig. 2.
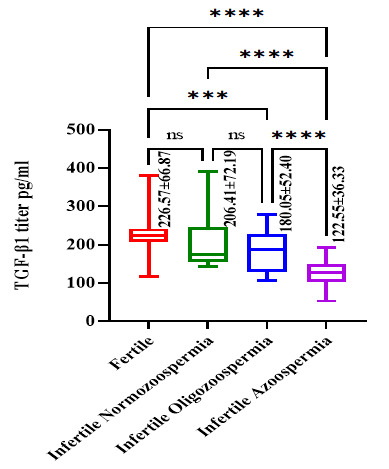
Level of serum TGF-β1 among volunteer participants.
Correlation analysis was carried out to assess the influence of serum TGF-β1 and semen parameters. In the oligozoospermic infertile males, the serum level of TGF-β1 positively associated with active progressive motile sperm (r= 0.346, p= 0.036) and slow progressive motile sperm (r=0.487, p=0.002), but negatively associated with immotile sperm (r=-0.470, p=0.003). In normozoospermic males, TGF-β1 negatively correlated with non-progressive motile sperm (r=-0.546, p= 0.040) (Table 4).
Table 4.
Correlation of TGF-β1 (+869 C/T) (rs1800470) with seminogram.
| Parameters | Infertile males’ categories | |||||
|---|---|---|---|---|---|---|
| Normozoospermia | Oligozoospermia | Azoospermia | ||||
| Pearson correlation (r) | p. value | Pearson correlation (r) | p. value | Pearson correlation (r) | p. value | |
| Volume | -0.347 | 0.133 | -0.049 | -0.276 | 0.385 | 0.194 |
| Concentration | -0.128 | 0.590 | -0.133 | 0.432 | NE | NE |
| Count | -0.266 | 0.258 | -0.078 | 0.607 | NE | NE |
| Active | -0.378 | 0.101 | 0.346 | 0.036* | NE | NE |
| Slow | -0.177 | 0.454 | 0.487 | 0.002** | NE | NE |
| Non-progressive | -0.546 | 0.040* | 0.155 | 0.361 | NE | NE |
| Immotile | 0.318 | 0.171 | -0.470 | 0.003** | NE | NE |
| Normal morphology | -0.146 | 0.539 | 0.236 | 0.159 | NE | NE |
| Abnormal morphology | 0.146 | 0.539 | -0.236 | 0.159 | NE | NE |
Correlation is significant at the 0.05 level (2-tailed).
Correlation is significant at the 0.01 level (2-tailed).
NE: Not estimated.
Associations of TGF-β1 (+869 C/T) genotypes distributions and alleles frequencies in infertile males
Our results show that the numbers of individuals of CC, CT, and TT genotypes of TGF-β1 (+869 C/T) (rs1800470) were 8 (28.57%); 19 (67.86%); 1 (3.57%) in the normozoospermic infertile group and 39 (51.32%); 35(46.05%); 2(2.63%) in the oligozoospermic infertile group and 28(70%); 10 (25%); 2(5%) in the azoospermic infertile group and 16 (40%); 23 (57.5%); 1(2.5%) in the fertile group, respectively. Moreover, the frequencies of the C and T alleles were 35 (62.5%); 21 (37.5%) in the normozoospermic infertile group and 113 (74.34%); 39 (25.66%) in the oligozoospermic infertile group and 66 (82.5%); 14 (17.5%) in the azoospermic infertile group and 55 (68.75%); 25 (31.25%) in the fertile group, respectively. Statistical analysis showed that there were differences but non-significant between the normozoospermic; oligozoospermic males and fertile males in the frequencies of TGF-β1 genotypes CC, CT, TT (p=0.41; p=0.45; p=1.00, and p=0.32; p=0.32; p=1.00, P < 0.05), respectively. However, statistically significant differences showed between the azoospermic males and fertile males in the distribution of TGF-β1 genotypes CC, and CT (p=0.013, p=0.006, P < 0.05), respectively, while non-significant differences in genotype TT (p=1.00, P < 0.05). Analysis of the allele frequencies for genotypes carrying the C allele at TGF-β1 (+869 C/T) compared to those carrying the T allele. There was a non-significant difference in the frequencies of C and T alleles for rs1800470 SNP in normozoospermic; oligozoospermic and azoospermic infertile participants as compared to the fertile group (p=0.46; p=0.44; p=0.06), respectively. The only recessive model (TT+CT) vs CC of the TGF-β1 (+869 C/T) showed an increased risk of male infertility among normozoospermic group (20 (71.43%) and the value was statistically non-significant (Table 5).
Table 5.
The genotypes and alleles frequencies of TGF-β1 +869 C/T in the fertile and infertile male categories.
| Seminogram categories | TGF- β1 | Infertile males’ frequency (N/%) | Fertile males’ frequency (N/%) | RR | Preventive Fraction | P | 95% CI |
|---|---|---|---|---|---|---|---|
| Normozoospermia | Genotype | ||||||
| CC | 8 (28.57%) | 16 (40%) | 0.60 | 0.6 | 0.41 | 0.22-1.66 | |
| CT | 19 (67.86%) | 23 (57.5%) | 1.56 | 0.24 | 0.45 | 0.58-4.22 | |
| TT | 1 (3.57%) | 1 (2.5%) | 1.44 | 0.01 | 1.00 | 0.09-23.09 | |
| (CC+CT) vs TT | 27 (96.43%) | 39 (97.5%) | 0.69 | 0.3 | 1.00 | 0.04-11.07 | |
| (TT+CT) vs CC | 20 (71.43%) | 24 (60.0%) | 1.67 | 0.28 | 0.44 | 0.60-4.62 | |
| Allele | |||||||
| C allele | 35 (62.5%) | 55 (68.75%) | 0.76 | 0.16 | 0.46 | 0.37-1.55 | |
| T allele | 21 (37.5%) | 25 (31.25%) | 1.32 | 0.09 | 0.46 | 0.65-2.69 | |
| Oligozoospermia | Genotype | ||||||
| CC | 39 (51.32%) | 16 (40%) | 1.58 | 0.18 | 0.32 | 0.73-3.41 | |
| CT | 35 (46.05%) | 23 (57.5%) | 0.63 | 0.21 | 0.32 | 0.29-1.35 | |
| TT | 2 (2.63%) | 1 (2.5%) | 1.05 | 0.001 | 1.00 | 0.10-11.69 | |
| (CC+CT) vs TT | 74 (97.37%) | 39 (97.5%) | 0.95 | 0.05 | 1.00 | 0.09-10.52 | |
| (TT+CT) vs CC | 37 (48.68%) | 24 (60.0%) | 0.63 | 0.22 | 0.32 | 0.29-1.36 | |
| Allele | |||||||
| C allele | 113 (74.34%) | 55 (68.75%) | 1.32 | 0.17 | 0.44 | 0.73-2.38 | |
| T allele | 39 (25.66%) | 25 (31.25%) | 0.76 | 0.07 | 0.44 | 0.42-1.37 | |
| Azoospermia | Genotype | ||||||
| CC | 28 (70%) | 16 (40%) | 3.50 | 0.5 | 0.013* | 1.40-8.73 | |
| CT | 10 (25%) | 23 (57.5%) | 0.25 | 0.43 | 0.006** | 0.10-0.63 | |
| TT | 2 (5%) | 1 (2.5%) | 2.05 | 0.02 | 1.00 | 0.18-22.88 | |
| (CC+CT) vs TT | 38 (95%) | 39 (97.5%) | 0.49 | 0.5 | 1.00 | 0.04-5.43 | |
| (TT+CT) vs CC | 12 (30%) | 24 (60.0%) | 0.29 | 0.42 | 0.013* | 0.11-0.71 | |
| Allele | |||||||
| C allele | 66 (82.5%) | 55 (68.75%) | 2.14 | 0.44 | 0.06 | 1.02-4.50 | |
| T allele | 14 (17.5%) | 25 (31.25%) | 0.47 | 0.16 | 0.06 | 0.22-0.98 | |
RR: Relative risk, CI: Confidence Intervals, Exact Fishers Probability (P).
The genotype frequencies of TGF-β1 among the categories of infertile males and fertile males were assessed by HWE calculation. The differences in frequency of heterozygous genotype (CT) between observed and expected were in normozoospermic group 19 (67.86%), 13.12 (46.86%), in oligozoosperic group 35 (46.05%), 28.99 (38.15%), in control 23 (57.5%), 17.19 (42.96%), respectively. While in azoospermic group the differences in frequencies of homozygous genotypes CC and TT between observed and expected were 28 (70%), 27.22 (68.05%) and 2 (5%), 1.22 (3.05%), respectively. The variance between the observed and expected values of genotype frequencies was statistically non-significant, indicating that the distribution of this cohort was under HWE (Table 6).
Table 6.
Hardy-Weinberg equilibrium (HWE) test for the genotypes and alleles distributions of TGF- β1 +869 C/T in the infertile and fertile male participants.
| Case Categories | TGF- β1 gene at position +869 C/T (rs1800470) | ||||||
|---|---|---|---|---|---|---|---|
| Genotypes | HWE p. value | Alleles | |||||
| CC | CT | TT | C | T | |||
| Normozoospermia | Observed | 8 (28.57%) | 19 (67.86%) | 1 (3.57%) | 0.06 | 35 (62.5%) | 21 (37.5%) |
| Expected | 10.94 (39.07%) | 13.12 (46.86%) | 3.94 (14.07%) | Not estimated | |||
| Oligozoospermia | Observed | 39 (51.32%) | 35 (46.05%) | 2 (2.63%) | 0.74 | 113 (74.34%) | 39 (25.66%) |
| Expected | 42 (55.27%) | 28.99 (38.15%) | 5 (6.58%) | Not estimated | |||
| Azoospermia | Observed | 28 (70%) | 10 (25%) | 2 (5%) | 0.69 | 66 (82.5%) | 14 (17.5%) |
| Expected | 27.22 (68.05%) | 11.55 (28.88%) | 1.22 (3.05%) | Not estimated | |||
| Control | Observed | 16 (40%) | 23 (57.5%) | 1 (2.5%) | 0.10 | 55 (68.75%) | 25 (31.25%) |
| Expected | 18.91(47.28%) | 17.19 (42.96%) | 3.91 (9.76%) | Not estimated | |||
Evaluation of serum TGF-β1 level with different TGF-β1 +869 C/T genotypes
We determined the correlations of serum TGF-β1concentrations with CC, CT, and TT genotypes in 184 participants to explore the relations between each genotype of the TGF-β1 (+869 C/T) polymorphisms and serum concentration of TGF-β1. The serum TGF-β1 concentration in infertile normozoospermic males was 291.2±77.96 pg/ml for the CC genotype, 174.1±30.40 pg/ml for the CT genotype, 142.5±0.00 pg/ml for the TT genotype, in infertile oligozoospermic males were 218.2±40.57 pg/ml for the CC genotype, 141.5±27.10 pg/ml for the CT genotype, 111.0±4.28 pg/ml for the TT genotype, in infertile azoospermic males, were 138.7±23.70 pg/ml for the CC genotype, 91.01±32.68 pg/ml for the CT genotype, 53.91±1.24 pg/ml for the TT genotype, in fertile males, were 244.5±87.56 pg/ml for the CC genotype, 218.9±174.1 pg/ml for the CT genotype, 116.2±0.00 pg/ml for the TT genotype. Thus, the serum TGF-β1 concentration was significantly lower in the CT (mutated heterozygous) and TT (mutated homozygous) genotypes compared with the CC genotype (non-mutated homozygous) among the infertile group, indicating that serum TGF-β1 level was significantly declined in mutated homozygous and mutated heterozygous compared to non-mutated homozygous infertile patients. In the fertile group, the serum TGF-β1level did not differ significantly between CC and CT genotypes, while CC and CT genotypes significantly differ from the TT genotype (Fig. 3).
Fig. 3.
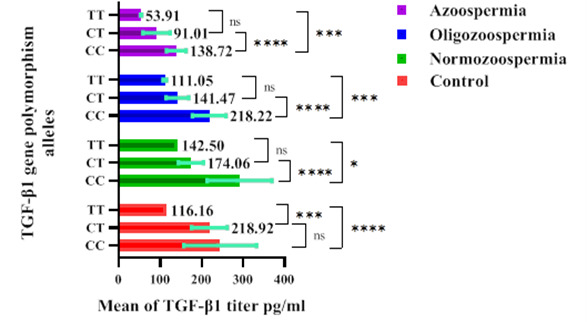
Serum TGF-β1 concentration according to TGF-β1 (+869 C/T) genotypes among volunteer participants.
Association of genotypes of TGF-β1 +869 C/T with semen quality
The results of association analyses between polymorphisms of the TGF-β1 +869 gene with the seminogram showed a considerable effect on sperm quality traits. The CT and TT genotypes produced lower sperm concentration compared to the CC genotype in infertile normozoospermic (31.53±20.41, 20.00±0.00, 44.50±27.53) and oligozoospermic (3.18±3.01, 0.62±0.53, 3.28±3.23) males, and the lower sperm count in infertile normozoospermic males (102.5±77.13, 40.00±0.00, 119.9±48.95) and oligozoospermic males (11.67±13.23, 2.00±2.12, 11.93±12.40) per ejaculation, respectively (Fig. 4A, B).
Fig. 4.
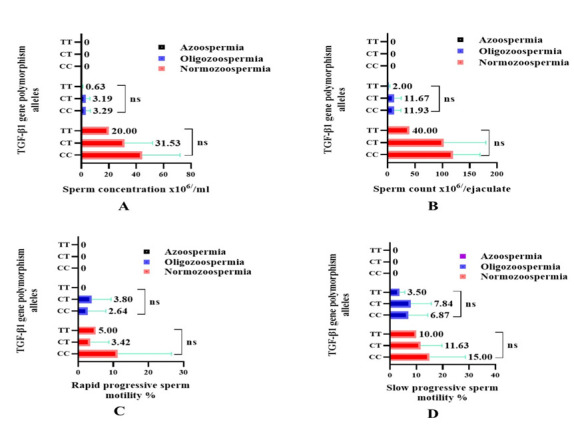
A. Sperm concentration, B. Sperm count, C. Rapid progressive sperm motility, D. Slow progressive sperm motility, according to TGF-β1 (+869 C/T) gene polymorphisms among infertile males.
Polymorphisms of TGF-β1 +869 gene with CT and TT genotypes in normozoospermic infertile males compared to CC genotype produced spermatozoa with a lower percentage of rapid (3.42±5.28, 5.00±0.00, 11.25±15.29); slow (11.63±8.09, 10.00±0.00, 15.00±13.63) progressive sperm motility and higher non-progressive sperm motility (4.47±2.83, 5.00±0.00, 2.50±2.67) and immotile sperm (80.47±14.92, 80.00±0.00, 71.25±30.33), respectively. In oligozoospermic infertile males, the percentage of progressive motility in CT and TT genotypes compared to CC genotype were rapid (3.80±5.56, 0.00±0.00, 2.64±5.20); slow (7.84±7.81, 3.50±2.12, 6.87±7.46); non-progressive (4.32±4.52, 3.00±2.82, 4.02±4.42) and immotile sperm (84.03±13.62, 93.50±4.95, 86.46±10.88), respectively (Fig. 4C, D and Fig. 5A, B).
Fig. 5.

A. Non-progressive sperm motility, B. Immotile sperm, C. Sperm normal morphology, D. Sperm abnormal morphology, according to TGF-β1 (+869 C/T) gene polymorphisms among infertile males.
The influence of the TGF-β1 +869 genes on sperm morphology was also detected. The CT and TT genotypes produced a lower percentage of normal sperm morphology compared to the CC genotype in infertile normozoospermic males (1.86±2.16, 0.50±0.00, 11.38±13.18) and oligozoospermic males (1.57±3.18, 0.75±0.35, 3.41±8.45), and the highest percentage of abnormal sperm morphology in infertile normozoospermic males (98.13±2.21, 99.50±0.00, 88.63±13.18) and oligozoospermic males (98.43±3.18, 99.25±0.35, 96.59±8.45), respectively (Fig. 5C, D).
Azoospermia is a condition characterized by the absence of sperm in the semen. That’s why the association between polymorphisms of the TGF-β1 +869 gene with the seminogram didn’t analyze to show in Figs. 4 and 5.
Discussion
The TGF-β plays a crucial role in regulating male pluripotency (proliferation and differentiation somatic and germline), testicular angiogenesis, and structure. Alterations in TGF-β may lead to testicular diseases and reduce fertility in males (17, 18). The expression of TGF-β1 diminished in several reproductive disorders including unexplained infertility, prostate cancer, and penile fibrosis (19)
To the best of our knowledge, there is a scarce study to define the correlation of TGF-β1 polymorphisms with male infertility and semen quality. After searching the literature of previous studies, no similar study relating TGF-β1 polymorphisms with semen parameters was specified. Subsequently, we based and compared our outcome of this study with the findings of some other diseases and reproductive disorders.
Regarding the impact of age factor on infertility among men, our study showed that this factor has no significant role and this came in agreement with previous studies (20, 21).
Our objective was to determine if there are differences in the serum level of TGF-β1 between fertile and infertile subjects and to determine its impact on semen quality. We found a high level of TGF-β1 in the serum of the fertile participants compared to the infertile participants, and this came in agreement with results reported by Al-Msaid and Al-Sallami (2018) (7).
In oligozoospermic infertile males, serum TGF-β1 concentration correlated positively with active and slow progressive motile sperm and negatively with immotile sperm. Concordant to our findings, previous research demonstrated that the decline of serum TGF-β1 concentrations influences sperm progressive motility (22).
Our findings showed that CT genotype was correlated to an increased risk of infertility in normozoospermic patients, while CC genotype was in oligozoospermic and azoospermic patients. Furthermore, the T allele was related to an increased risk of infertility in normozoospermic patients, while the risk of infertility increased in oligozoospermic and azoospermic patients with the C allele. Moreover, in normozoospermic patients, the recessive model (TT+CT) showed an increased risk of infertility when compared to CC genotype carriers. According to the study, TGF- β1 functional polymorphisms are correlating with a functional influence on TGF-β1 production. Even though the distribution of genotype CC and CT of TGF-1 +869 C/T in infertile men patients compared to the fertile participant did not meet the statistically significant criterion (14). Our results revealed that infertile males with the CC genotype and the C-allele also have a risk for infertility.
Polymorphisms can impact messenger RNA (mRNA) stability, microRNA target sequence, transportation of protein to the endoplasmic reticulum (ER) through signal peptides, gene expression, and alternative splicing changing protein function via amino acid alterations (23).
In our study, we revealed that in infertile males with the CC genotype serum TGF-β1 concentrations are higher than TGF-β1 concentrations in infertile males with the CT and TT genotypes. We assumed that the infertile males with the CT and TT genotypes their serum might be contained significantly different concentrations of TGF-β1 with proline protein than the CC genotype whose serum contained TGF-β1 with leucine protein. Our results are based on the facts obtained by previous studies that found in patients with the TGF-β1-CC genotype, the serum level of TGF-β1 was higher than those with the TGF-β1-CT and TT genotypes (24). Researchers described that the TGF-β1 gene polymorphisms had no significant relation with the TGF-β1 concentration (25). Other researchers mentioned that the TGF-β1 gene polymorphisms (C/T transition) result in the substitution of leucine for proline. The outcome of this conversion is increased levels of TGF-β1 and protein in individuals with the proline (minor allele) compared to those with the leucine (major allele) (26).
Spermatogonia undergo a series of divisions that differentiate into mature spermatozoa throughout the process of spermatogenesis (27, 28). Leucine plays a crucial role in male reproductive efficiency through promoting spermatogenesis, improving testes morphology, increased sperm viability, sperm count/ejaculation, and increase sperm motility (29-31). The mechanism by which leucine performs its functions through activating PI3K/mTOR/Akt signalling pathways (30, 32, 33). Accordingly, we proposed that the serum of the infertile men with TGF-β1-C/T genotype contain less amount of leucine and correlate with diminished sperm number, sperm motility, and sperm morphology.
The declining serum TGF-β1 concentration of infertile males provides evidence that TGF-β1 is significantly correlated with an increased risk of infertility. Our outcomes revealed that the existence of TGFβ1 +869 gene polymorphisms may result in male infertility. Moreover, the carriers of the CT genotype in normozoospermic and CC genotype in oligozoospermic and azoospermic infertile males were more prevalent among the infertile populations with poor semen quality. Furthermore, our study revealed a direct association between TGF-β1 gene polymorphisms and seminogram.
Ethical Approval
This project was approved by the Scientific Committee at Biology Department, College of Education, Salahaddin University.
Funding
No financial support was received.
Conflict of Interest
The authors state that they do not have any conflicts of interest.
Acknowledgements
The authors are thankful to the all-volunteer participants. As well as, Biotechnology Laboratory of Biology Department, College of Education, Salahaddin University for helping in conducting all laboratory experiments.
References
- 1.Rybska M, Knap S, Stefańska K, Jankowski M, Chamier-Gliszczyńska A, Popis M, et al. Transforming growth factor (TGF)–is it a key protein in mammalian reproductive biology? Med J Cell Biol. 2018;6(3):125–30. [Google Scholar]
- 2.Morikawa M, Derynck R, Miyazono K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb Perspect Biol. 2016;8(5):a021873. doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson L, Thompson DL Jr, Varner DD. Role of Sertoli cell number and function on regulation of spermatogenesis. Anim Reprod Sci. 2008;105(1-2):23–51. doi: 10.1016/j.anireprosci.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 4.Wang T, Zhang D, Song T, Sun M, Zhang J. Advances in research of TGF-Β1 in human testis. Food Sci Technol. 2022;42(e22521) [Google Scholar]
- 5.Stukenborg J-B, Schlatt S, Simoni M, Yeung C-H, Elhija MA, Luetjens CM, et al. New horizons for in vitro spermatogenesis? An update on novel three-dimensional culture systems as tools for meiotic and post-meiotic differentiation of testicular germ cells. Mol Hum Reprod. 2009;15(9):521–9. doi: 10.1093/molehr/gap052. [DOI] [PubMed] [Google Scholar]
- 6.Loras B, Vételé F, El Malki A, Rollet J, Soufir JC, Benahmed M. Seminal transforming growth factor-beta in normal and infertile men. Hum Reprod. 1999;14(6):1534–9. doi: 10.1093/humrep/14.6.1534. [DOI] [PubMed] [Google Scholar]
- 7.Al-Msaid HLF, Al-Sallami ASM. Study the level of cytokine in unexplained and idiopathic infertile men. J Pharm Sci & Rese. 2018;10(4):808–11. [Google Scholar]
- 8.von Wolff M, Nowak O, Pinheiro RM, Strowitzki T. Seminal plasma--immunomodulatory potential in men with normal and abnormal sperm count. Eur J Obstet Gynecol Reprod Biol. 2007;134(1):73–8. doi: 10.1016/j.ejogrb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Ben Ali H. Effect of Seminal Transforming Growth Factorβ1 (TGFβ1) and Glutathione on Apoptosis in Spermatozoa from Tunisian Infertile Men. Andrology (Los Angel). 2017;6(1):21.1000172. [Google Scholar]
- 10.Yao HH-C, Ungewitter E, Franco H, Capel B. Establishment of fetal Sertoli cells and their role in testis morphogenesis. Sertoli cell biology: Elsevier. 2015:57–79. [Google Scholar]
- 11.Oral O, Uchida I, Eto K, Nakayama Y, Nishimura O, Hirao Y. Promotion of spermatogonial proliferation by neuregulin 1 in newt (Cynops pyrrhogaster) testis. Mech Dev. 2008;125(9-10):906–17. doi: 10.1016/j.mod.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Nagaoka SI, Saitou M. Reconstitution of Female Germ Cell Fate Determination and Meiotic Initiation in Mammals. Cold Spring Harb Symp Quant Biol. 2017;82:213–222. doi: 10.1101/sqb.2017.82.033803. [DOI] [PubMed] [Google Scholar]
- 13.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor β in human disease. N Engl J Med. 2000;342(18):1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 14.Martelossi Cebinelli GC, Paiva Trugilo K, Badaró Garcia S, Brajão de Oliveira K. TGF-β1 functional polymorphisms: a review. Eur Cytokine Netw. 2016;27(4):81–89. doi: 10.1684/ecn.2016.0382. [DOI] [PubMed] [Google Scholar]
- 15.Khani M, Amani D, Taheripanah R, Sanadgol N, Feizollahzadeh S, Rahmani Z. Transforming growth factor beta-1 (TGF-β1) gene single nucleotide polymorphisms (SNPs) and susceptibility to pre-eclampsia in Iranian women: A case-control study. Pregnancy Hypertens. 2015;5(4):267–72. doi: 10.1016/j.preghy.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Eskandari E, Metanat M, Pahlevani E, Nakhzari-Khodakheir T. Association between TGFβ1 polymorphisms and chronic hepatitis B infection in an Iranian population. Rev Soc Bras Med Trop. 2017;50(3):301–308. doi: 10.1590/0037-8682-0266-2016. [DOI] [PubMed] [Google Scholar]
- 17.Young JC, Wakitani S, Loveland KL. TGF-β superfamily signaling in testis formation and early male germline development. Semin Cell Dev Biol. 2015;45:94–103. doi: 10.1016/j.semcdb.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 18.Kwiatkowski W, Gray PC, Choe S. Engineering TGF-β superfamily ligands for clinical applications. Trends Pharmacol Sci. 2014;35(12):648–57. doi: 10.1016/j.tips.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Kajdaniuk D, Marek B, Borgiel-Marek H, Kos-Kudła B. Transforming growth factor beta1 (TGFbeta1) in physiology and pathology. Endokrynol Pol. 2013;64(5):384–96. doi: 10.5603/EP.2013.0022. [DOI] [PubMed] [Google Scholar]
- 20.Ziaeemehr A, Sharebiani H, Taheri H, Fazeli B. Secondary Infertility: A Neglected Aspect of Buerger’s Disease. Rep Biochem Mol Biol. 2022;11(2):246. doi: 10.52547/rbmb.11.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torki A, Amirmozafari N, Talebi M, Talebi A. Using the PCR and Blood Agar in Diagnosis of Semen Bacterial Contamination of Fertile and Infertile Men. Rep Biochem Mol Biol. 2021;10(3):402. doi: 10.52547/rbmb.10.3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tongrueng S, Vongpralub T, Srimooltho W, Phasuk Y. Transforming growth factor beta1 in porcine seminal plasma on characteristic of sperm and reproductive efficiency in Sows. Sci Technol Asia. 2021:216–23. [Google Scholar]
- 23.Karamyshev AL, Tikhonova EB, Karamysheva ZN. Translational control of secretory proteins in health and disease. Int J Mol Sci. 2020;21(7):2538. doi: 10.3390/ijms21072538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunning AM, Ellis PD, McBride S, Kirschenlohr HL, Healey CS, Kemp PR, et al. A transforming growth factorβ1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003;63(10):2610–5. [PubMed] [Google Scholar]
- 25.Faria PC, Saba K, Neves AF, Cordeiro ER, Marangoni K, Freitas DG, et al. Transforming growth factor-beta 1 gene polymorphisms and expression in the blood of prostate cancer patients. CancerInvest. 2007;25(8):726–32. doi: 10.1080/07357900701600921. [DOI] [PubMed] [Google Scholar]
- 26.Mališić E, Petrović N, Brengues M, Azria D, Matić IZ, Srbljak Ćuk I, et al. Association of polymorphisms in TGFB1, XRCC1, XRCC3 genes and CD8 T-lymphocyte apoptosis with adverse effect of radiotherapy for prostate cancer. Sci Rep. 2022;12(1):21306. doi: 10.1038/s41598-022-25328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng CY, Mruk DD. The blood-testis barrier and its implications for male contraception. Pharmacol Rev. 2012;64(1):16–64. doi: 10.1124/pr.110.002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holstein A-F, Schulze W, Davidoff M. Understanding spermatogenesis is a prerequisite for treatment. Reprod Biol Endocrinol. 2003;1(1):1–16. doi: 10.1186/1477-7827-1-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolahian S, Sadri H, Larijani A, Hamidian G, Davasaz A. Supplementation of diabetic rats with leucine, zinc, and chromium: effects on function and histological structure of testes. Int J Vitam Nutr Res. 2015;85:311–21. doi: 10.1024/0300-9831/a000244. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Zhang X, Liu Y, Su Z, Dawar FU, Dan H, et al. Leucine mediates autophagosome-lysosome fusion and improves sperm motility by activating the PI3K/Akt pathway. Oncotarget. 2017;8(67):111807. doi: 10.18632/oncotarget.22910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y, Li J, Wang K, Fang Z, Che L, Xu S, et al. Effects of dietary L-leucine supplementation on testicular development and semen quality in boars. Front Vet Sci. 2022;9:904653. doi: 10.3389/fvets.2022.904653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vellai T. How the amino acid leucine activates the key cell-growth regulator mTOR. Nature. 2021;596(7871):192–194. doi: 10.1038/d41586-021-01943-7. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, Shen L, Chen X, Ding Y, He J, Zhu J, et al. mTOR/P70S6K promotes spermatogonia proliferation and spermatogenesis in Sprague Dawley rats. Reprod Biomed Online. 2016;32(2):207–17. doi: 10.1016/j.rbmo.2015.11.007. [DOI] [PubMed] [Google Scholar]


