Abstract
The nanodisc technology is increasingly used for structural studies on membrane proteins and drug delivery. The development of synthetic polymer nanodiscs and the recent discovery of non-ionic inulin-based polymers have significantly broadened the scope of nanodiscs. While the lipid exchange and size flexibility properties of the self-assembled polymer-based nanodiscs are valuable for various applications, the non-ionic polymer nanodiscs are remarkably unique in that they enable the reconstitution of any protein, protein-protein complexes, or drugs irrespective of their charge. However, the non-ionic nature of the belt could influence the stability and size homogeneity of inulin-based polymer nanodiscs. In this study, we investigate the size stability and homogeneity of nanodiscs formed by non-ionic lipid-solubilizing polymers using different biophysical methods. Polymer nanodiscs containing zwitterionic DMPC and different ratios of DMPC:DMPG lipids were made using anionic SMA-EA or non-ionic pentyl-inulin polymers. Non-ionic polymer nanodiscs made using zwitterionic DMPC lipids produced a very broad elution profile on SEC due to their instability in the column, thus affecting sample monodispersity which was confirmed by DLS experiments that showed multiple peaks. However, the inclusion of anionic DMPG lipids improved the stability as observed from SEC and DLS profiles, which was further confirmed by TEM images. Whereas, anionic SMA-EA-based DMPC-nanodiscs showed excellent stability and size homogeneity when solubilizing zwitterionic lipids. The stability of DMPC:DMPG non-ionic polymer nanodiscs is attributed to the inter-nanodisc repulsion by the anionic-DMPG that prevents the uncontrolled collision and fusion of nanodiscs. Thus, the reported results demonstrate the use of electrostatic interactions to tune the solubility, stability, and size homogeneity of non-ionic polymer nanodiscs which are important features for enabling functional and atomic-resolution structural studies of membrane proteins, other lipid-binding molecules, and water-soluble biomolecules including cytosolic proteins, nucleic acids and metabolites.
Keywords: Nanodisc, Polymer nanodisc, Charge, Lipids, Nanodisc stability, Nanodisc homogeneity
1. Introduction
Obtaining atomic-resolution structures of membrane proteins has been a long-standing challenge due to the hydrophobic nature of their transmembrane domains that require a suitable lipid-bilayer environment. The membrane mimetics that can be used to reconstitute various membrane proteins include micelles, bicelles, and lipid vesicles [1–7]. Each of these membrane mimetics has its own advantages and limitations. While the use of micelles has rendered high-resolution structural studies, the detergents used and the curvature are not desirable for some of the membrane proteins [2, 8–10]. Unlike detergent micelles, the planar lipid bilayer present in bicelles enables native-like folding of membrane proteins and, therefore, attractive for studies on the structure, dynamics, and membrane topology of membrane proteins by a variety of biophysical techniques, including solution NMR, solid-state NMR and X-ray crystallography [11–21]. However, the use of detergents (or detergent-like molecules) in bicelles could affect the stability of reconstituted membrane proteins. The use of liposomes is also limited by their lack of stability and the use of detergents is not desirable for the functional reconstitution of membrane proteins [22]. Therefore, long-term time-dependent functional and structural studies of membrane proteins may be difficult when using detergent-based membrane mimetics [9, 23, 24]. Also, a detergent-free and highly stable lipid-bilayer mimicking system is desirable for the functional reconstitution of membrane proteins, especially those that are intrinsically less-stable (e.g., GPCRs) [24, 25]. In particular, such stable lipid-bilayer reconstitution systems are useful for high-resolution NMR-based structural studies that require long data acquisition times lasting several days to weeks.
The introduction of nanodisc technology has been useful in overcoming some of the challenges mentioned above. A nanodisc is a lipid-bilayer membrane system surrounded by membrane scaffold proteins (MSP) [26–30], peptides [31–39], peptoids [40–42], amphipathic polymers [43–55], or DNA origami barrels [56]. While further studies are focused on the development of the different types of nanodiscs, the already reported nanodiscs are increasingly used for the reconstitution of membrane proteins for functional and high-resolution structural studies [57–60]. In the case of peptide- and polymer nanodiscs, the belt surrounding the lipid-bilayer is formed by a high-density of peptide or polymer molecules instead of a continuous belt as in protein-based nanodiscs. Hence, the peptide/polymer nanodiscs are highly dynamic, exhibiting an exchange of lipids between nearby nanodiscs [61–64]. Despite excellent lipid solubilizing capabilities, these tools do not affect lipid-bilayer properties as drastically as detergents [65]. Therefore, the lipid composition of the nanodiscs can be chosen to provide a native-like lipid membrane environment to characterize membrane proteins and other lipid-binding molecules using in vitro studies.
Synthetic amphipathic polymers that form nanodiscs have also been used for the detergent-free isolation of local lipid-membrane protein complexes directly from the cells where they are expressed [66–68]. However, despite the excellent advantages, most amphipathic polymers have some intrinsic limitations, such as low lipid-solubility, divalent metal-ion sensitivity, and non-specific interactions with target membrane proteins [68–71]. Additionally, all the above-mentioned nanodiscs-forming polymer molecules possess high-charge densities; hence there are Coulombic repulsions between nanodiscs, making them stable lipid-bilayer systems [72, 73]. But, how the charge of the lipids or polymer affects the nanodisc’s stability has not been studied so far as it is challenging to investigate due to the high charge density of amphipathic polymers.
Recently developed, non-ionic inulin-based polymers are on par with detergents in their ability to solubilize synthetic lipids and bacterial membranes [74, 75]. While the ability of these polymers to form nanodiscs has also been reported, purification of nanodiscs composed of different lipid compositions has not been carried out. Most studies on the reconstitution of detergent-purified membrane proteins into polymer nanodiscs use a single or a mixture of zwitterionic and charged lipids. Before use for reconstitution of any target membrane protein, polymer nanodisc samples are typically purified to remove excess free polymer, salt, and other impurities that come from polymer synthesis/functionalization by chemical methods. Since the inulin-based polymers are non-ionic, in this study, we took it as an advantage to investigate how the ionic nature of both polymer-belt and lipid-bilayer affect the stability and size homogeneity of nanodiscs.
This study used non-ionic pentyl-inulin and anionic SMA-EA polymers to solubilize lipids. The lipid composition of non-ionic polymer nanodiscs was varied using zwitterionic DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) and anionic DMPG (1,2-dimyristoyl-sn-glycero-3-phosphoglycerol) lipids, whereas only zwitterionic DMPC was used in anionic SMA-EA polymer nanodiscs (Fig. 1A). Size-exclusion chromatography (SEC) experiments were performed to examine the stability and size homogeneity of nanodiscs, followed by dynamic light scattering (DLS) to determine the particle size and distribution. Transmission electron microscopy (TEM) images were used to compare the size homogeneity of the nanodisc particles. In addition, 1H NMR experiments were performed to analyze the contents of the samples to ensure the presence of lipids and polymer, as well as to check for any impurities.
Fig. 1.
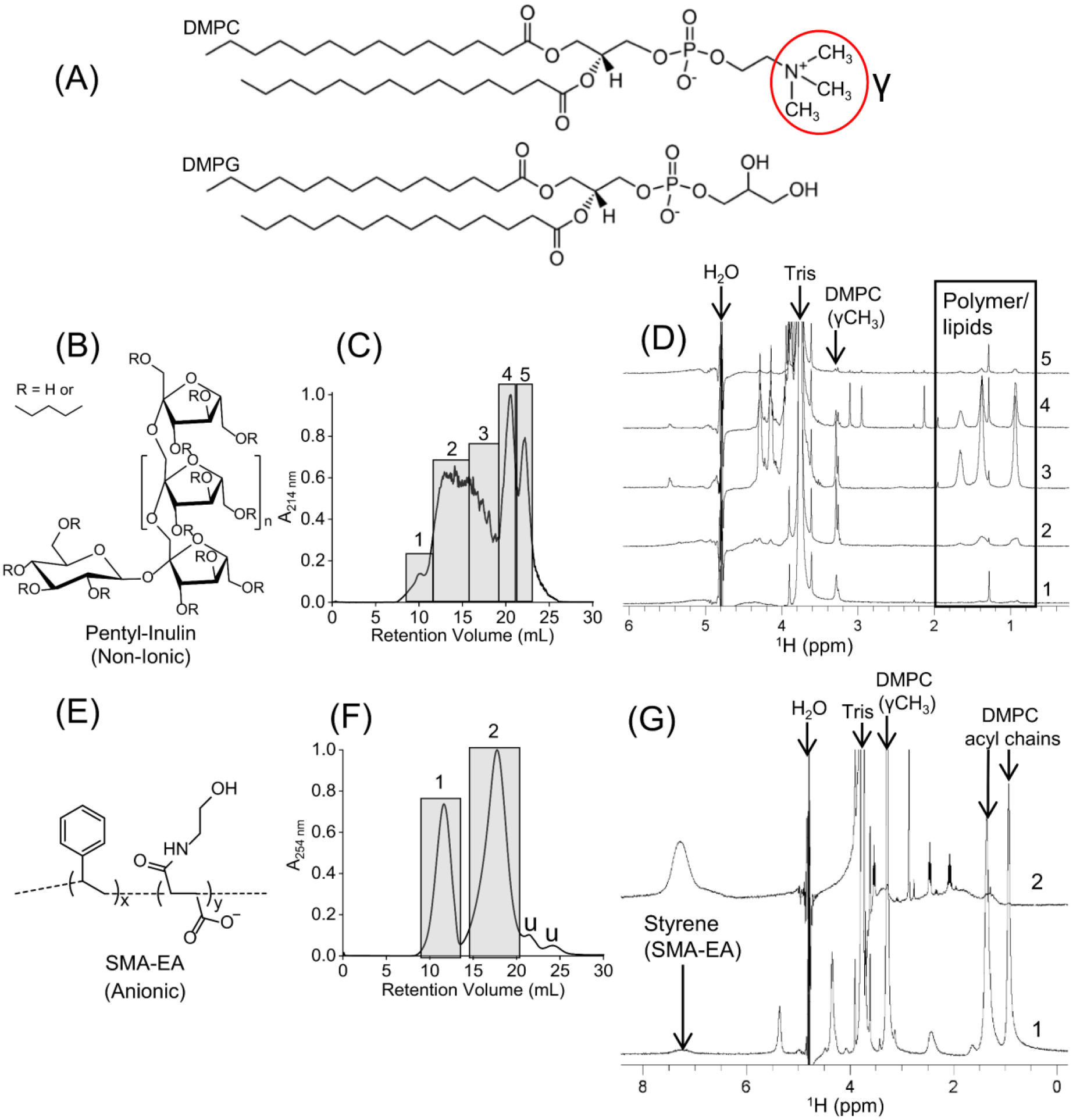
Stability of zwitterionic DMPC-nanodiscs prepared using two different polymers: pentyl-inulin (non-ionic) and SMA-EA (anionic). (A) Chemical structures of DMPC and DMPG. (B) Chemical structure of pentyl-inulin. (C) SEC chromatogram of pentyl-inulin DMPC mixture. The individual fractions from each rectangular box (labeled as 1–5) were analyzed. (D) 1H NMR spectra of different SEC fractions of pentyl-inulin DMPC mixture; NMR peaks from the pentyl-groups of inulin polymer and acyl chains of DMPC are boxed, and the peak from DMPC-γ-CH3 group is labeled. (E) Chemical structure of SMA-EA polymer. (F) SEC chromatogram of SMA-EA DMPC mixture. Peaks 1 and 2 in rectangular boxes correspond to nanodiscs and excess free polymer, respectively; ‘u’ represents the uncharacterized peaks. (G) 1H NMR spectra of SEC elution peaks 1 and 2 from (F). The characteristic peaks from DMPC and styrene (from SMA-EA) are labeled. All NMR spectra were recorded at 303 K.
2. Materials and methods
2.1. Lipids
DMPC and DMPG lipids were purchased from Avanti Polar Lipids, Inc (Alabaster, USA).
2.2. Polymer stock solution
Pentyl-inulin and SMA-EA were synthesized, purified, and characterized using published protocols [74, 76]. The lyophilized polymers were dissolved in a Tris buffer (10 mM Tris, 50 mM NaCl, pH 7.4) to a concentration of 100 mg/mL and checked the solution pH to ensure it was ~7.4 before using it for nanodisc preparation.
2.3. Preparation of liposomes
Liposomes were prepared by weighing the appropriate amounts of dry DMPC (6–10 mg) and DMPG (0–4 mg) into separate 1.5 mL centrifuge tubes. Each lipid sample was dissolved in 200 μL of 1:1 (v/v) methanol and chloroform. The dissolved lipids were then dried under nitrogen gas, followed by high vacuum drying for ~4 hours to obtain a lipid film and to remove any residual solvent. Next, the lipid film was resuspended in 900 μL of Tris buffer (10 mM Tris, 50 mM NaCl, pH 7.4) and subjected to three freeze-thaw cycles (liquid nitrogen to ~60 °C water). The solution was milky white throughout.
2.4. Preparation of polymer-lipid mixtures (polymer nanodiscs)
The liposomes were solubilized by adding the appropriate amount of polymer stock to obtain a 1:1 (w/w) lipid-to-polymer ratio. After mixing lipids and polymer, the pentyl-inulin sample was incubated overnight at 4 °C, while SMA-EA was incubated at 4 °C for 10 – 30 min, followed by overnight incubation at 30 °C.
2.5. Size-exclusion chromatography (SEC)
The SEC column (10x300 Superdex 200, GE Healthcare, Chicago, USA) was washed with ~2 column volumes of Milli-Q water followed by 2 column volumes of 10 mM Tris buffer with 50 mM of NaCl (pH 7.4). Nanodisc samples were loaded and run at a flow rate of 0.75 mL/min. Solutions used for SEC were filtered through a 0.2 μm Amicon membrane filter and degassed using an ultrasonic cleaner (Cole-Parmer, Chicago, USA) before use. A wavelength of 214 nm was used to detect pentyl-inulin lipid-nanodiscs, whereas a wavelength of 254 nm was used to detect SMA-EA lipid-nanodiscs.
2.6. Dynamic-light scattering (DLS)
DLS measurements were carried out using a Wyatt Technology® DynaPro® NanoStar® instrument equipped with a laser emitting at ~662 nm. All samples were measured in a 1 μL quartz MicroCuvette (Wyatt Technology, CA, USA). 1 μL of each SEC purified nanodisc sample was loaded into the cuvette. 10 scans were obtained with an acquisition time of 5 seconds and averaged to obtain the DLS profile. All experiments were recorded at 25 °C, allowing for at least 10 minutes for the loaded sample temperature to equilibrate. The spectra were obtained in mass-percentage mode.
2.7. 1H NMR spectroscopy
550 μL of the SEC purified nanodisc samples were mixed with 50 μL of 2H2O and loaded into 5 mm NMR tubes (Wilmad, NJ, USA). NMR spectra were then acquired using a 500 MHz Bruker NMR spectrometer (Billerica, MA, USA) operated at 303 K with 128 or 512 scans. The data were processed using Bruker Topspin (Version 3.6.2).
2.8. Transmission electron microscopy (TEM)
The polymer-lipid samples (10 mg/mL) without subjecting to any purification were used in TEM experiments. TEM images were obtained using a Technai® T-20® machine (FEI®, Netherlands) with an 80 kV operating voltage. 5 μL of each sample was loaded onto a carbon-coated copper grid and incubated for 5 min at room temperature. Then the solution was removed, and the grids were washed 3–4 times using Milli-Q water. Next, the sample-loaded grids were treated with 5 μL of 2% uranyl acetate for 2 min, followed by washing with Milli-Q water 3 times. Finally, all the grids were dried overnight at room temperature in a desiccator before collecting images.
3. Results and discussion
3.1. Charge of the polymer-belt modulates the stability and size homogeneity of polymer nanodiscs containing zwitterionic lipids
The stability and size homogeneity of zwitterionic DMPC-nanodiscs prepared using pentyl-inulin (Fig. 1B) were analyzed by SEC, 1H NMR, and DLS experiments. All polymer-lipid samples were prepared using a 1:1 (w/w) polymer:lipid ratio. The SEC chromatogram showed a broad non-symmetric peak between 9–20 mL (Fig. 1C). 1D 1H NMR spectra of SEC fractions from the broad peak showed resonances corresponding to both polymer and DMPC, indicating a broad elution of polymer-lipid assemblies in SEC (Fig. 1C and D (spectra 1–3)). DLS data showed the presence of particles with different hydrodynamic radii (Fig. S1). Large elution of pentyl-inulin-DMPC assemblies with different hydrodynamic radii suggests that polymer-lipid assemblies are highly heterogeneous in size. The peaks between 20–24 mL elution volume were due to excess free polymer in the solution (20–24 mL), as confirmed by 1H NMR (Fig. 1D (spectra 4,5)). A low-intensity DMPC peak in the free polymer samples is mostly due to the overlap of the free polymer peak (fractions 4, 5) with the polymer-DMPC peak (fractions 1–3) in SEC (Fig. 1C and D (spectrum 4)) and possibly due to the coexistence of a small amount of lipids with polymers. The width and intensity of NMR peaks are good indicators of the size of the nanodisc particle. Smaller the size of the nanodiscs, faster is the tumbling motion, and therefore higher peak intensity or narrower linewidth is observed. Comparing the 1H NMR spectra for the SEC fractions 1–3, the highest peak intensity for fraction-3 and the lowest peak intensity for fraction-1 reveal the size of nanodiscs/polymer-lipid assemblies to be fraction-1> fraction-2 >fraction-3, which is in agreement with the SEC elution profile. Interestingly, in addition to the narrowest and highest peak intensities, additional peaks are observed for fraction-3, further confirming the smallest size of the particles present in this fraction.
The SEC profile of anionic SMA-EA polymer nanodiscs showed a symmetric elution peak for nanodiscs (9–13 mL) (Fig. 1E and F), indicating a highly homogenous sample of nanodiscs compared to pentyl-inulin DMPC-nanodiscs (Fig. 1C). DLS experiments showed the presence of particles with a hydrodynamic radius of ~9 ±1 nm (Fig. S2). In addition, 1H NMR spectra confirmed the presence of both SMA-EA and DMPC in the nanodisc fraction, whereas free polymer and other impurities were present in excess free polymer fractions (14–20 mL) (Fig. 1G) [77]. While peaks from lipids are absent in the free polymer fraction (top trace in (G)), the nanodisc-bound polymer peaks (for example, the peak from styrene ~7.23 ppm) are broad (bottom trace in (G)). These observations indicate high stability and monodispersity of SMA-EA DMPC-nanodiscs as compared to pentyl-inulin DMPC-nanodiscs. The non-ionic nature of pentyl-inulin DMPC-nanodiscs allows unrestricted fusion of nearby nanodiscs, unlike the nanodiscs formed using the anionic SMA-EA, causing decreased stability of nanodiscs when passing through the SEC column and, therefore, polymer-lipid assemblies eluted in large elution volume as seen in Fig.1(C).
3.2. Charged lipids can stabilize and render homogeneous nanodisc particles for non-ionic polymers
The effect of inter-nanodiscs electrostatic interactions on the stability of nanodisc particles was tested by self-assembling nanodiscs samples containing non-ionic polymer and zwitterionic DMPC and anionic DMPG lipids. Nanodiscs with four different DMPC:DMPG concentration ratios were studied using a 1:1 (w/w) pentyl-inulin:lipid ratio. When DMPG was included in the polymer-lipid mixture at a 9:1 (w/w) DMPC:DMPG ratio, the SEC elution profile for the nanodiscs/polymer-lipid complexes was improved; hence the peaks arising from the excess free polymer were resolved better when compared to that of the sample containing only DMPC and pentyl-inulin (Figs. 2A, 1C, and S3). However, the SEC profiles obtained from different samples of 9:1 (w/w) DMPC:DMPG lipid ratio are not identical, indicating some variation between the samples (Fig. S3). 1H NMR and DLS experiments were used to analyze the SEC samples. 1H NMR spectra of the different SEC fractions (labeled 1–6 in Fig. 2A) showed peaks from both pentyl-inulin and lipids (Fig. 2B). Thus, the broader elution of polymer and lipids indicates the presence of polymer-lipid aggregates of various sizes but not stable nanodiscs on the column. NMR peak integration analysis was performed on the well-resolved peak from the DMPC-γ-CH3 group to estimate the DMPC lipid in all different SEC fractions. A large variation in the DMPC-γ-CH3 peak intensity was observed for the different fractions from SEC (Fig. 2A, C), further highlighting the size heterogeneity of varying polymer-lipid nanodisc particles. However, the DLS profile showed particles with a hydrodynamic radius between 8.5 to 10.4 nm from different SEC fractions (Fig. S3), indicating that the polymer and lipids reassembled into polymer nanodiscs in collection tubes after elution from the SEC column. Therefore, the non-identical SEC profiles obtained from different samples suggest a higher (>10%) concentration of anionic DMPG might be needed to stabilize the pentyl-inulin polymer nanodisc particles.
Fig. 2.
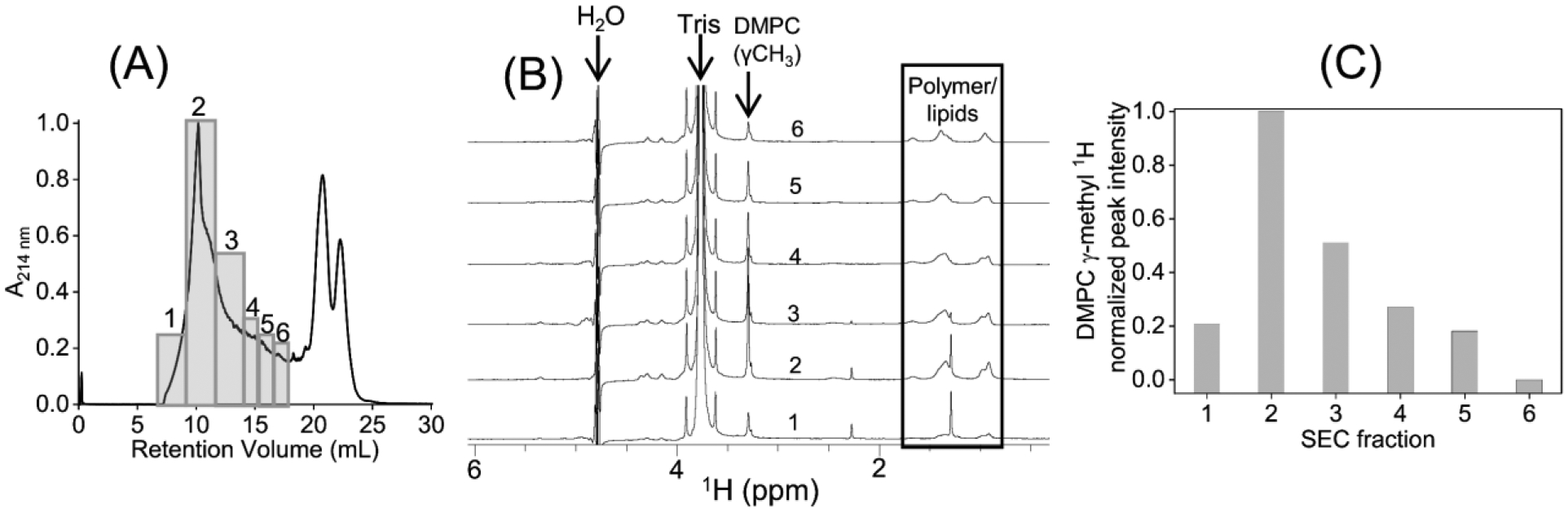
(A) SEC chromatogram of a solution mixture of pentyl-inulin polymer and 9:1 (w/w) DMPC:DMPG mixture. The fractions from SEC (boxed and labeled with 1–6) were separately analyzed using NMR and DLS experiments. (B) 1H NMR spectra of SEC fractions (1–6 from (A)). (C) The intensity of the 1H NMR peak from the DMPC-γ-CH3 group was measured from various SEC fractions (1–6 from (A)). Peak intensities are normalized to the intensity of DMPC γ-CH3 peak in spectrum-2 (see (B)).
In order to enhance the size stability of the nanodisc particles, we then prepared and tested pentyl-inulin nanodisc samples containing 8:2, 7:3, and 6:4 (w/w) DMPC:DMPG lipid ratios. Upon increasing DMPG concentration from 9:1 to 8:2 or 7:3 (w/w) DMPC:DMPG ratio, a more symmetric peak with a narrow elution volume was observed in SEC. The peak from excess free polymer was completely resolved (Fig. 3A, B). The fractions from nanodiscs were combined and analyzed together. DLS profiles showed a hydrodynamic radius between 9.5 and 11.4 nm (Figs. 3(C, D), S4, and S5), and 1H NMR spectra showed peaks from both pentyl-inulin and lipids (Fig. 3E), indicating the presence of polymer nanodisc particles. The symmetric shape of the elution peak and a narrow elution of nanodiscs indicate that the polymer nanodiscs were stable to unrestricted collision by the column pressure on the SEC column and suggest that the sample is highly homogeneous. The SEC profile for the 6:4 (w/w) DMPC:DMPG sample was similar to that of the 7:3 (w/w) DMPC:DMPG sample (Fig. S6). Overall, these results indicate the crucial role of charged lipids in stabilizing the non-ionic pentyl-inulin-nanodiscs.
Fig. 3.
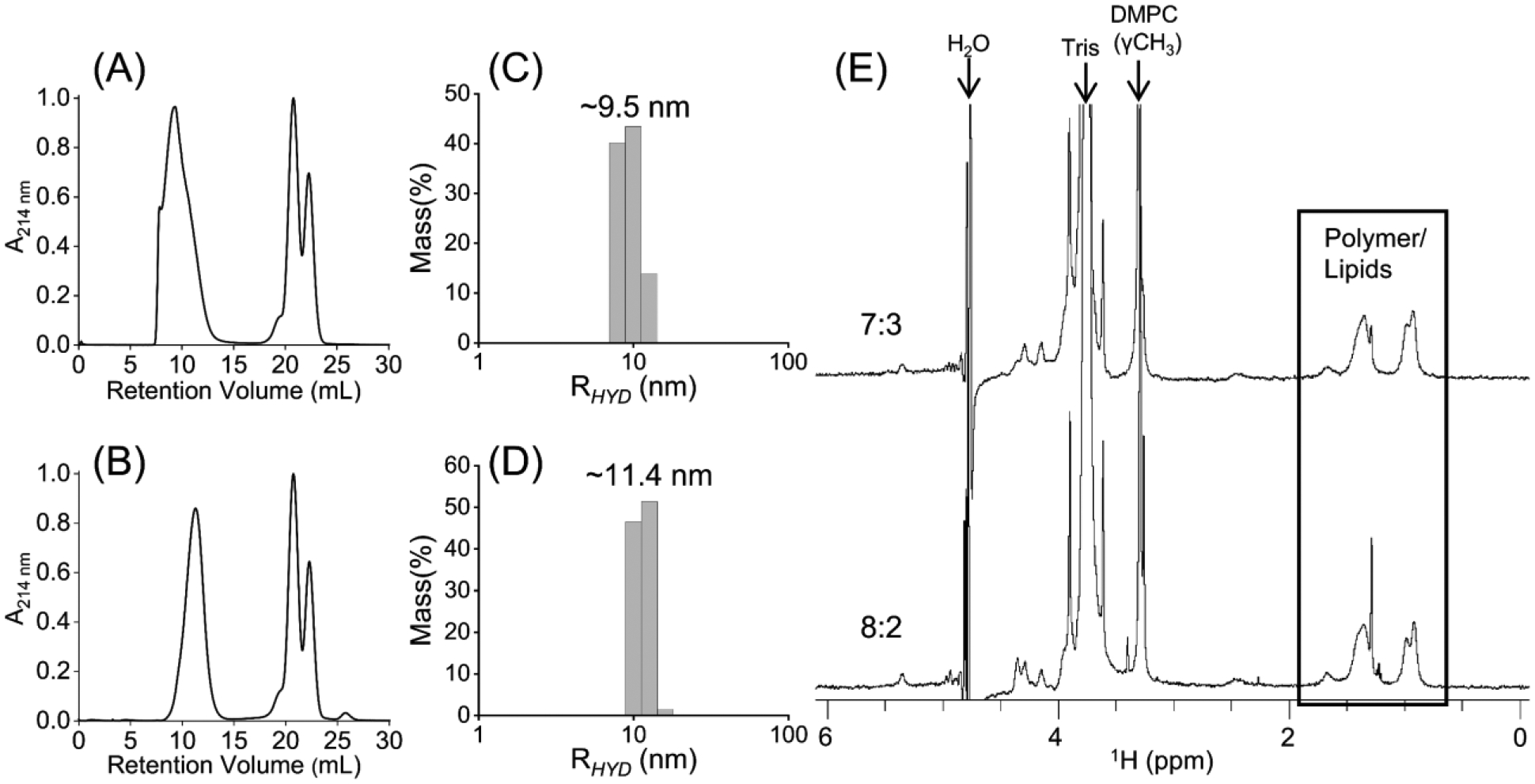
SEC chromatograms of polymer nanodiscs containing pentyl-inulin and (A) 8:2 or (B) 7:3 (w/w) DMPC:DMPG at 1:1 (w/w) polymer:lipid ratio. The SEC purified samples were analyzed by (C, D) DLS and (E) 1H NMR experiments. NMR peaks from polymer, lipids, buffer, and water are labeled. 8:2 and 7:3 labels in (E) indicate the w/w DMPC:DMPG ratios in pentyl inulin polymer nanodiscs.
The above observations are also reflected in the higher transparent solution exhibited by 7:3 and 8:2 (w/w) DMPC:DMPG polymer-lipid assemblies than by 9:1 (w/w) DMPC:DMPG and DMPC-only polymer-lipid assemblies at room temperature (Fig. 4A). The bluish color intensity decreased with increasing anionic DMPG lipids in the sample from the DMPC-only sample to the 7:3 (w/w) DMPC:DMPG sample, indicating a higher polydispersity of DMPC-only polymer-lipid assemblies as compared to DMPG-containing samples (Fig. 4A). A similar trend of temperature-dependent bluish color difference has been reported for DMPC containing bicelles [78]. Although the DMPC-only sample showed increased transparency at 4 °C, the solution transparency was improved upon the addition of DMPG, and it was complete in the samples containing 7:3 and 8:2 (w/w) DMPC:DMPG ratio of lipids (Fig. 4B). These observations indicate that the solubilization of lipids using non-ionic polymer is more efficient at low temperatures than at high temperatures. In other words, non-ionic polymer solubilizes the lipids below their phase-transition temperature which is ~24 °C for DMPC/DMPG lipids [79]. Therefore, the temperature condition could be an important parameter to consider for the solubilization of synthetic lipids as well as cell membranes to isolate a target membrane protein or any other non-ionic polymer-based nanodisc applications [80]. The increased lipid solubility at 4 °C may partly be due to the reduction in the collision and fusion of nanodiscs. In contrast, SMA-EA efficiently solubilized DMPC lipids at 30 °C, and therefore, it did not require any low-temperature treatment for nanodiscs formation. The solubility of the 7:3 (w/w) DMPC:DMPG mixture by different lipid:polymer ratios was also compared, and it was found that 1:1 w/w lipid:polymer is the minimum ratio that exhibited a maximum transparent solution at room temperature, as shown in Fig. 4C. Again, the gradual decrease of bluish color with increasing polymer concentration, indicates improved sample homogeneity when using polymer at 1:1 (w/w) ratio of polymer and lipids.
Fig. 4.
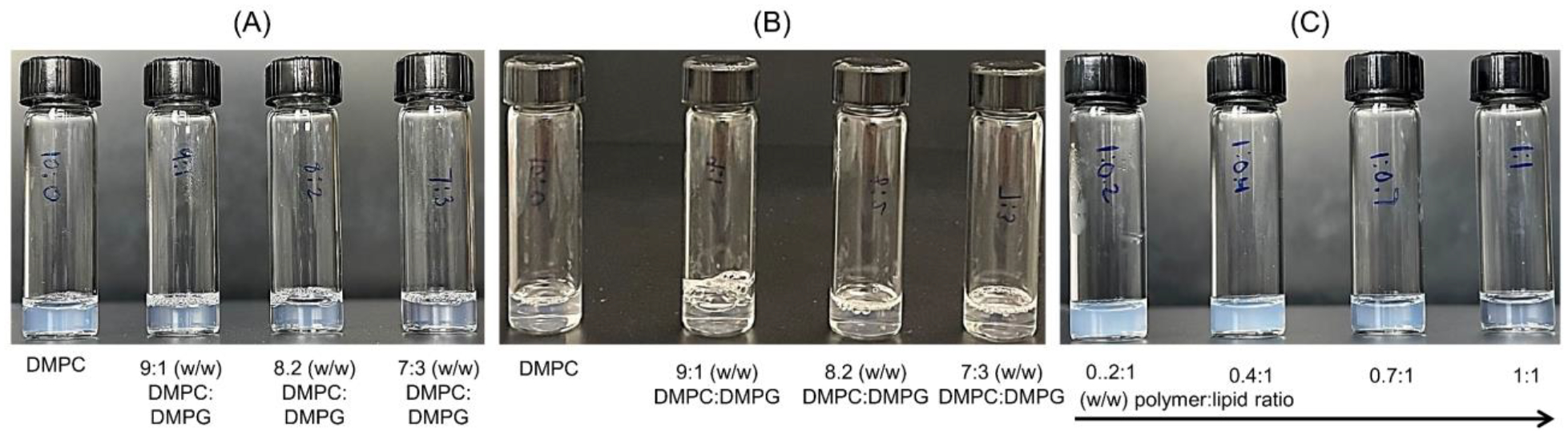
Solubilization of liposomes and nanodiscs formation in the presence of pentyl-inulin polymer. The photographs were taken 10 min after storing samples at room temperature (A) and 4 °C (B). All the samples were prepared using a 1:1 (w/w) ratio of polymer and lipids (10 mg/mL solution of 10 mg lipids and 10 mg polymer). (A and B; from left-to-right) zwitterionic DMPC alone, 9:1 (w/w) DMPC:DMPG, 8:2 (w/w) DMPC:DMPG, and 7:3 (w/w) DMPC:DMPG. (C) The solubilization of 7:3 (w/w) DMPC:DMPG lipid mixture in the presence of 0.2:1, 0.4:1, 0.7:1 and 1:1 (w/w) polymer:lipid ratio; 10 mg/mL lipids solution containing 2, 4, 7, and 10 mg/mL polymer (from left to right).
To further confirm the heterogeneity of the nanodisc particles, TEM images were obtained (Figs. 5 and S7). Although the images are of low resolution, the 7:3 (w/w) DMPC:DMPG nanodiscs sample exhibited more homogeneous nanodisc particles when compared to other samples with a low concentration of DMPG or without DMPG, as shown in Fig. 5. For the sample with DMPC alone, small micelle-like, fused, and circular nanodisc particles with a diameter of 30 ±5 nm were observed (Fig. 5A). For the 9:1 (w/w) DMPC:DMPG sample, micelle-like particles and a mixture of distorted and circular nanodisc particles with a diameter of 22 ±6 nm were observed (Fig. 5B). In the 8:2 (w/w) DMPC:DMPG sample, only a mixture of both distorted and circular nanodisc particles with a diameter of 23 ±5 nm were observed but did not find any substantial number of micelle-like particles (Fig. 5C). In comparison to the above mentioned three samples, the 7:3 (w/w) DMPC:DMPG sample showed more homogenous nanodisc particles with a diameter of 25 ±5 nm (Fig. 5D). Thus, our results suggest that a minimum of about 20% charged lipids are needed to obtain stable and homogenous polymer nanodiscs that can be purified by SEC before membrane protein reconstitution (Fig. 3A).
Fig. 5.
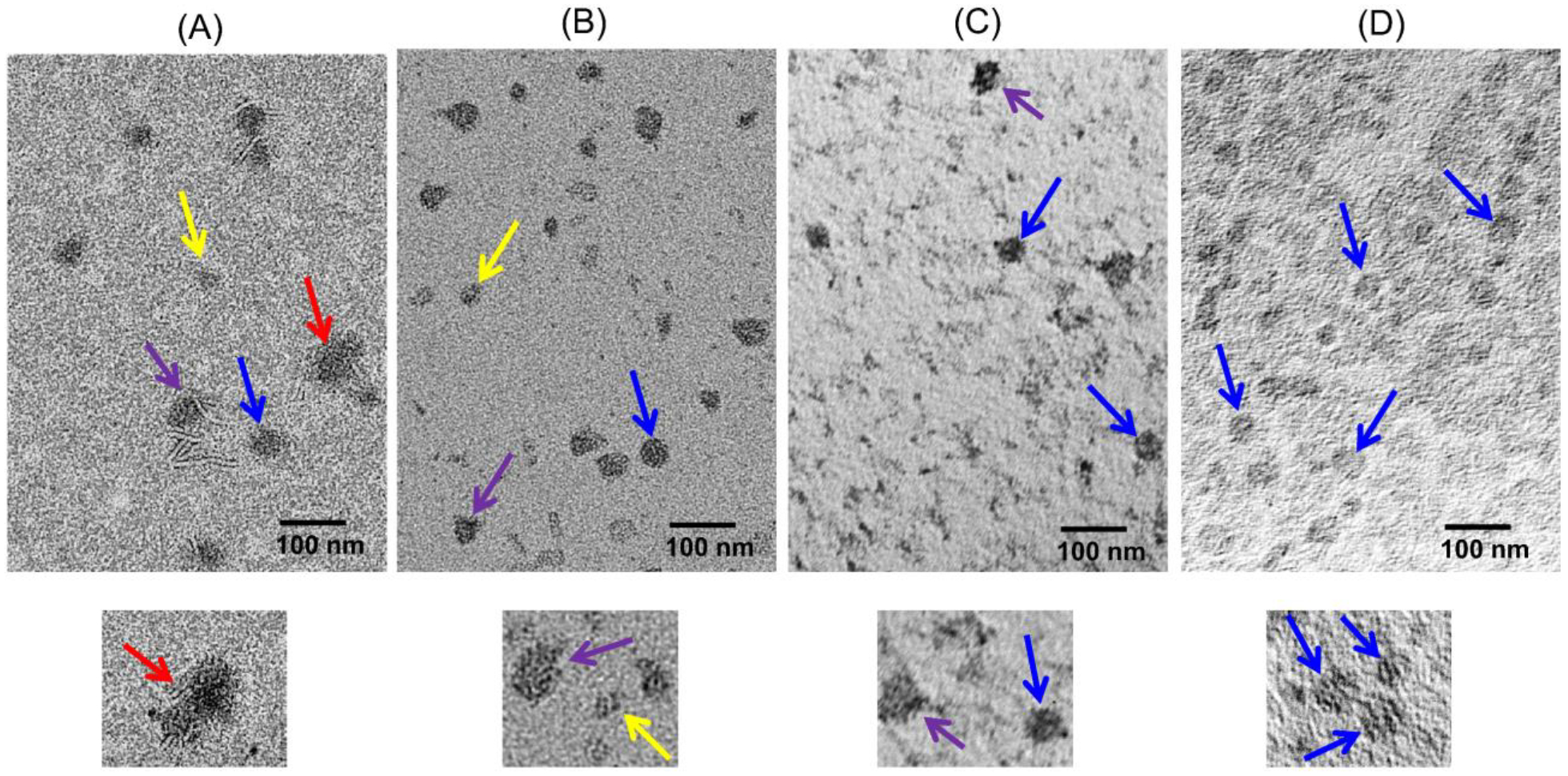
TEM images of nanodiscs composed of pentyl-inulin polymer and zwitterionic DMPC alone (A), 9:1 (w/w) DMPC:DMPG (B), 8:2 (w/w) DMPC:DMPG (C) and 7:3 (w/w) DMPC:DMPG (D). The bottom panel is an enlarged region of the image in the top panel. All the samples were made using 1:1 (w/w) lipid:polymer ratio. Nanodisc particles of different sizes/shapes are pointed with different colored arrows: small micelle-like particles (yellow), distorted assemblies (purple) and fused assemblies (red), and stable/circular nanodiscs (blue). Full images of these samples can be found in Fig. S7.
The main focus of this study is on the role of charge on non-ionic polymer-based nanodisc formation, and their stability and homogeneity. Amphipathic polymers (SMA and DIBMA) are great tools for making stable lipid-bilayered nanodiscs [72, 73, 81, 82] and also for isolating membrane proteins directly from cell membranes [67, 73]. However, similar to MSP [27, 83–86] and 4F peptides [87], these polymers are highly charged molecules; hence, they are not suitable for studying the role of charge on the nanodiscs’ stability and size homogeneity. Therefore, the non-ionic inulin-based polymer [74] is used to investigate the charge effect on nanodiscs stability by making nanodiscs with different lipid compositions and compared with the anionic SMA-EA polymer nanodiscs [74, 76]. The results reported here are helpful in quickly preparing non-ionic polymer nanodiscs with an optimized lipid composition to reconstitute any target membrane protein. Pentyl-inulin can solubilize both neutral (DMPC) and a mixture of neutral and charged lipids (DMPC:DMPG). But the broad elution of the polymer-DMPC complex is very intriguing when purified by SEC column under a column pressure of ~2.5 MPa. The broad SEC elution profile and multiple particles observed in TEM images and DLS profile reflects the instability and size heterogeneity of the pentyl-inulin/DMPC complexes. The sample heterogeneity is due to the uncontrolled fusion/fission of nanodiscs that leads to various sizes of polymer-lipid complexes (Fig. 6A).
Fig. 6.
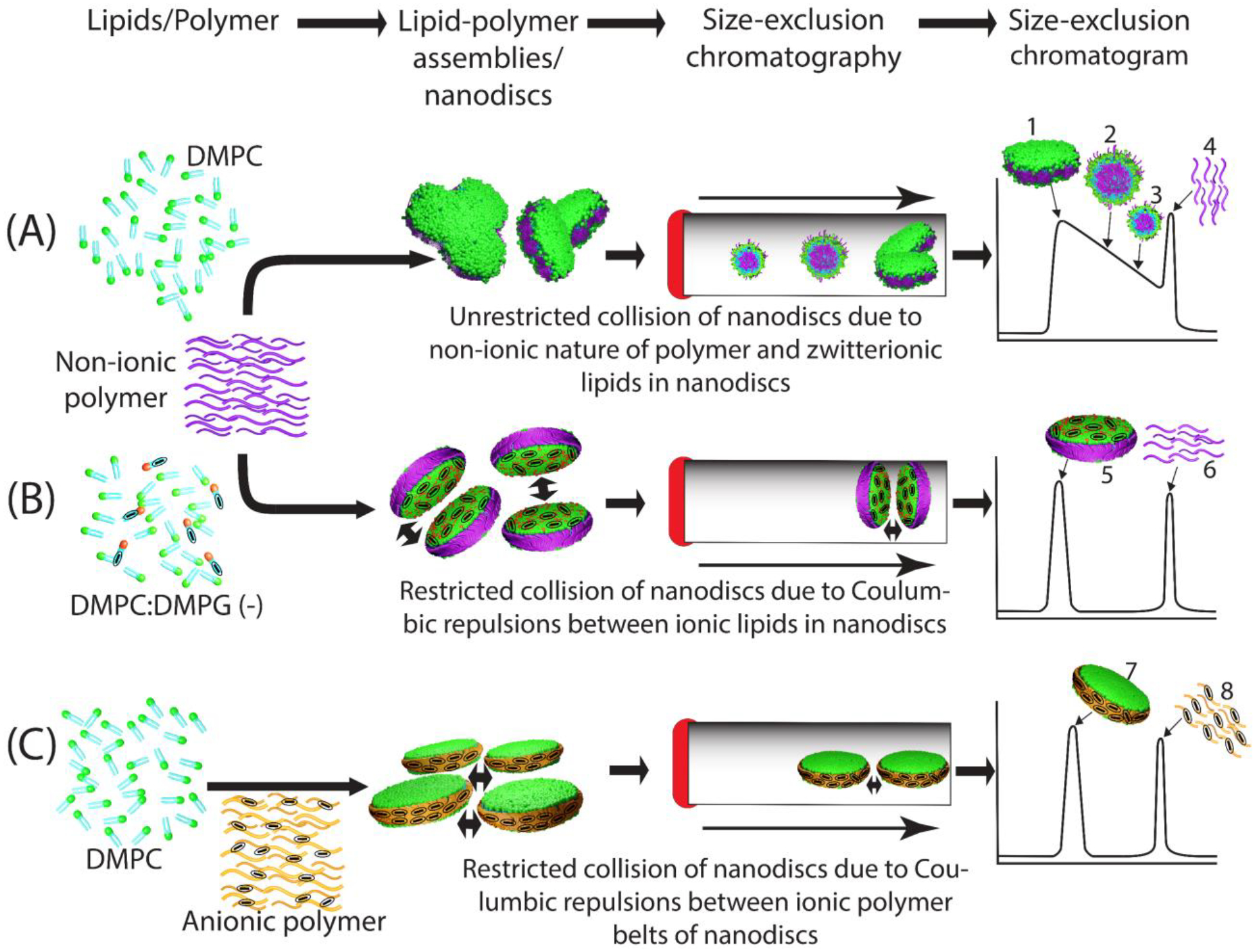
Schematic of charge-mediated stability and homogeneity of polymer nanodiscs. (A) non-ionic polymer-DMPC (zwitterionic lipid) nano assemblies/nanodiscs: Due to unrestricted collision, different size aggregates are formed on the SEC column under column pressure; hence they eluted with varying elution times as schematically shown in chromatogram: 1, nanodisc-like; 2–3, different size polymer-lipid aggregates; 4, excess free polymer. (B) non-ionic polymer-DMPC (zwitterionic) and DMPG (anionic) nanodiscs, and (C) anionic polymer-DMPC (zwitterionic) nanodiscs. The anionic nature of lipids/polymer is indicated with a minus symbol in oval circles. Charge-charge repulsions by anionic DMPG in non-ionic polymer nanodiscs (B) or by anionic SMA-EA belts in ionic polymer nanodiscs (C) resulted in a narrow elution volume, indicating high stability and homogeneity of nanodiscs. The two-headed arrows indicate ionic repulsions between charged nanodisc particles. In B and C, the nanodisc and free polymer peaks are labeled with 5/7 and 6/8, respectively.
On the other hand, the inclusion of the anionic DMPG produced an improved SEC profile, indicating a highly stable and homogeneous sample of pentyl-inulin polymer nanodiscs. The increased stability is because of the charge-charge repulsions between nearby nanodiscs containing negatively charged DMPG lipids that restrict their uncontrolled collision (Fig. 6B). These observations are similar to that reported for DMPG-containing bicelles where DMPG prevented the formation of vesicles due to the presence of strong Coulombic repulsion between lamellae at high temperatures [88]. Additionally, the Na+ ions from the buffer (50 mM NaCl) could form another charge layer by interacting with the negatively charged DMPG on the nanodisc surface. Thus, according to the Derjaguin-Landau-Verwey-Overbeek (DLVO) theory of repulsive electrostatic and attractive van der Waals forces, the electrical double layer formed by DMPG-Na+ may stabilize nanodiscs by creating electrostatic repulsive forces between nanodiscs as seen for charged bicelles complexed with Na+ [89, 90]. The charge repulsions between nanodiscs also make them resistant to the pressure effect on the SEC column, thus enabling easy purification of stable and homogenous nanodiscs. By contrast, the anionic SMA-EA polymer-DMPC nanodiscs produced a symmetric SEC elution profile. In this case, the negative charge of the SMA-EA polymer-belt inhibits any uncontrolled collision of nanodiscs (Fig. 6C).
Non-homogenous samples of nanodiscs might not be suitable for high-resolution structural studies of membrane proteins by techniques such as NMR, Cryo-EM, or other biophysical techniques that require highly homogenous samples. Thus, the study highlights the crucial role of ionic components (charged lipids in the current study) to obtain stable and homogeneous non-ionic polymer nanodisc samples for various functional and structural studies of membrane proteins or any other biomolecules of interest. Membrane-binding peptides (and antimicrobial peptides) with a net positive charge interact selectively with negatively charged bacterial membranes. Therefore, similar to bicelles, the negatively-charged lipid-containing non-ionic polymer nanodiscs could be suitable lipid-bilayer mimetics to study such membrane-binding peptides [91–93]. Furthermore, these polymer nanodiscs can also be applicable for the stable reconstitution and characterization of oppositely charged protein-protein complexes such as the redox cytochrome-P450 and its redox partner (P450 reductase or cytochrome-b5) [79] or other charged molecules including nucleic acids [94] and drugs [95]. While bicelles are well-studied membrane mimetics and used in various applications [96–98], their lipid:detergent molar ratio value has to be maintained above 1 to achieve a stable lipid bilayer [99]; lipid:detergent mixtures with q<1 form a micelle-like structure, which is less desirable for the functional reconstitution of membrane proteins [99, 100]. On the other hand, nanodiscs could be better lipid bilayer systems owing to their robustness, easy manipulation with different lipid compositions, and detergent-free environment for studying membrane proteins and other applications [101]. In addition, unlike nanodiscs, bicelles used for protein structural studies are not usually purified through SEC prior to protein reconstitution to remove any free detergent molecules or other impurities present due to their instability under column pressure. That said, polymer-based nanodiscs do have their own limitations. In general, polymers with a very high-charge density can be ineffective in solubilizing ionic lipids of the same charge due to Coulombic repulsions; this could be especially problematic when working with the detergent-free isolation of membrane proteins from anionic cell membranes [68, 80, 102].
Pentyl-inulin has been shown to solubilize lipids in the presence of high mM concentrations of divalent metal ions and a range of pH conditions [79]. However, how the metal ions in buffers with different pHs affect the stability of nanodiscs when purified by chromatography methods is unknown. Therefore, a systematic study on the effect of various metal ions on the stability of non-ionic polymer nanodiscs would be helpful for membrane protein reconstitution, especially for those that require/function in the presence of metal ions (for example, the Sarco-endoplasmic reticulum calcium ATPase) [103]. This information is also needed when using non-ionic polymer-based nanodiscs under magnetic-alignment conditions to determine high-resolution NMR structural parameters such as residual dipolar couplings for various water-soluble metal ion-binding biomolecules [104–106].
Studies have reported the development of bicelles for drug delivery applications [107–111]. Although bicelles are a promising tool for drug delivery studies, they are prone to interact with proteins and other molecules in plasma [112]. As a result, lipid bilayer destabilization and drug leakage may occur before reaching the drug target region. In contrast, the nonionic inulin-based polymer nanodiscs stabilized with suitable lipid composition could be useful in a broad range of applications, including drug delivery which requires pure, homogenous, and stable nanodiscs [113]. Furthermore, although inulins are not naturally present in humans, they occur in many plant species and have been shown to have health benefits [113]. Thus, the nanodiscs forming inulins may be suitable for the reconstitution and delivery of water-insoluble therapeutics through the gut or to deliver to the lungs, as shown with the MSP-based nanodiscs [114]. But, as mentioned above, highly stable nanodiscs with good size homogeneity and purity are needed. Therefore, the findings reported in this study, i.e., the preparation of stable and highly homogenous polymer nanodiscs using non-ionic inulin-based polymers and ionic lipids, would help to make a stable reconstitution system for drug delivery studies. Further studies characterizing the non-ionic polymer nanodiscs in the presence of plasma are needed to get insights into their interaction with different plasma proteins (protein corona) and to understand their lipid bilayer stability.
4. Conclusions
In conclusion, the charged lipids in nanodiscs play a significant role in the stability and size homogeneity of polymer nanodiscs. At least 20% charged lipids are needed to stabilize the non-ionic polymer nanodiscs. Without charged lipids, non-ionic polymers can still solubilize the zwitterionic lipids by hydrophobic-hydrophobic interactions. However, these soluble polymer-lipid assemblies are highly polydisperse when passed through an SEC column as they undergo unrestricted fusion/fission, forming polymer-lipid assemblies of different sizes (highly heterogeneous). But the inclusion of charged lipids can avoid the uncontrolled collision of nanodiscs in solution due to charge-charge repulsions between nearby nanodiscs; thus, obtaining a more homogenous nanodisc sample is charge-dependent. In addition to charged lipids, low-temperature treatment of polymer-lipid mixtures plays a role in non-ionic polymer-based lipid solubilization. When compared to the pentyl-inulin nanodiscs, SMA-EA nanodiscs are more homogeneous due to the high charge density of the polymer belt. Due to the non-ionic nature of pentyl-inulin, different types of charged lipids can be introduced to characterize their effects on various membrane-binding peptides/proteins and other biomolecules using non-ionic polymer nanodiscs. For high-resolution structural studies using static solid-state NMR spectroscopy on magnetically-aligned large-size nanodiscs, SEC-purified nanodiscs are recommended due to their high size homogeneity and, therefore, would likely result in narrow linewidths. Such purification would allow the unique advantages of the self-assembled non-ionic polymer nanodiscs to be utilized to successfully reconstitute large-size, dynamic integral membrane protein or protein-protein complexes.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (NIH) (R35 GM139572 to A.R.).
References
- [1].Sanders CR, Hare BJ, Howard KP, Prestegard JH, Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules, Prog. Nucl. Magn. Reson. Spectrosc 26 (1994) 421–444. [Google Scholar]
- [2].Dürr UHN, Gildenberg M, Ramamoorthy A, The magic of bicelles lights up membrane protein structure, Chem. Rev 112(11) (2012) 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pandey A, Shin K, Patterson RE, Liu X-Q, Rainey JK, Current strategies for protein production and purification enabling membrane protein structural biology, Biochem. Cell Biol 94(6) (2016) 507–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sgro GG, Costa TRD, Cryo-EM grid preparation of membrane protein samples for single particle analysis, Front Mol Biosci 5 (2018) 74–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sligar SG, Denisov IG, Nanodiscs: A toolkit for membrane protein science, Protein Sci 30(2) (2021) 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McCalpin SD, Ravula T, Ramamoorthy A, Saponins form nonionic lipid nanodiscs for protein structural studies by Nuclear Magnetic Resonance Spectroscopy, J. Phys. Chem. Lett 13(7) (2022) 1705–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mkam Tsengam IK, Omarova M, Kelley EG, McCormick A, Bothun GD, Raghavan SR, John VT, Transformation of lipid vesicles into micelles by adding nonionic surfactants: Elucidating the structural pathway and the intermediate structures, J. Phys. Chem. B 126(11) (2022) 2208–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Catoire LJ, Warnet XL, Warschawski DE, Micelles, bicelles, amphipols, nanodiscs, liposomes, or intact cells: The Hitchhiker’s guide to the study of membrane proteins by NMR, in: Mus-Veteau I (Ed.), Membrane Proteins Production for Structural Analysis, Springer; New York, New York, NY, 2014, pp. 315–345. [Google Scholar]
- [9].Yang Z, Wang C, Zhou Q, An J, Hildebrandt E, Aleksandrov LA, Kappes JC, DeLucas LJ, Riordan JR, Urbatsch IL, Hunt JF, Brouillette CG, Membrane protein stability can be compromised by detergent interactions with the extramembranous soluble domains, Protein Sci 23(6) (2014) 769–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Puthenveetil R, Vinogradova O, Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments, J. Biol. Chem 294(44) (2019) 15914–15931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].De Angelis AA, Opella SJ, Bicelle samples for solid-state NMR of membrane proteins, Nat. Protoc 2(10) (2007) 2332–8. [DOI] [PubMed] [Google Scholar]
- [12].Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, Bahl A, Kern D, Ha T, Henzler-Wildman KA, Antiparallel EmrE exports drugs by exchanging between asymmetric structures, Nature 481(7379) (2011) 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cho MK, Gayen A, Banigan JR, Leninger M, Traaseth NJ, Intrinsic conformational plasticity of native EmrE provides a pathway for multidrug resistance, J. Am. Chem. Soc 136(22) (2014) 8072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kobayashi H, Nagao S, Hirota S, Characterization of the cytochrome c membrane-binding site using cardiolipin-containing bicelles with NMR, Angewandte Chemie (International ed. in English) 55(45) (2016) 14019–14022. [DOI] [PubMed] [Google Scholar]
- [15].Koroloff SN, Tesch DM, Awosanya EO, Nevzorov AA, Sensitivity enhancement for membrane proteins reconstituted in parallel and perpendicular oriented bicelles obtained by using repetitive cross-polarization and membrane-incorporated free radicals, J. Biomol. NMR 67(2) (2017) 135–144. [DOI] [PubMed] [Google Scholar]
- [16].Nadezhdin KD, Goncharuk SA, Arseniev AS, Mineev KS, NMR structure of a full-length single-pass membrane protein NRADD, Proteins 87(9) (2019) 786–790. [DOI] [PubMed] [Google Scholar]
- [17].Aisenbrey C, Salnikov ES, Raya J, Michalek M, Bechinger B, Solid-state NMR approaches to study protein structure and protein-lipid interactions, Methods Mol. Biol 2003 (2019) 563–598. [DOI] [PubMed] [Google Scholar]
- [18].Hutchison JM, Shih K-C, Scheidt HA, Fantin SM, Parson KF, Pantelopulos GA, Harrington HR, Mittendorf KF, Qian S, Stein RA, Collier SE, Chambers MG, Katsaras J, Voehler MW, Ruotolo BT, Huster D, McFeeters RL, Straub JE, Nieh M-P, Sanders CR, Bicelles rich in both sphingolipids and cholesterol and their use in studies of membrane proteins, J. Am. Chem. Soc 142(29) (2020) 12715–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chiliveri SC, Robertson AJ, Shen Y, Torchia DA, Bax A, Advances in NMR spectroscopy of weakly aligned biomolecular systems, Chem. Rev 122(10) (2022) 9307–9330. [DOI] [PubMed] [Google Scholar]
- [20].Murugova TN, Ivankov OI, Ryzhykau YL, Soloviov DV, Kovalev KV, Skachkova DV, Round A, Baeken C, Ishchenko AV, Volkov OA, Rogachev AV, Vlasov AV, Kuklin AI, Gordeliy VI, Mechanisms of membrane protein crystallization in ‘bicelles’, Sci. Rep 12(1) (2022) 11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ujwal R, Bowie JU, Crystallizing membrane proteins using lipidic bicelles, Methods 55(4) (2011) 337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rigaud JL, Paternostre MT, Bluzat A, Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin, Biochemistry 27(8) (1988) 2677–88. [DOI] [PubMed] [Google Scholar]
- [23].Zhou HX, Cross TA, Influences of membrane mimetic environments on membrane protein structures, Annu. Rev. Biophys 42 (2013) 361–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].González Flecha FL, Kinetic stability of membrane proteins, Biophys. Rev 9(5) (2017) 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Heydenreich FM, Vuckovic Z, Matkovic M, Veprintsev DB, Stabilization of G protein-coupled receptors by point mutations, Frontiers in pharmacology 6 (2015) 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Denisov IG, Sligar SG, Nanodiscs in membrane biochemistry and biophysics, Chem. Rev 117(6) (2017) 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Plückthun A, Hagn F, Walz T, Hogle JM, Wagner G, Covalently circularized nanodiscs for studying membrane proteins and viral entry, Nat. Methods 14(1) (2017) 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nasr ML, Wagner G, Covalently circularized nanodiscs; challenges and applications, Curr. Opin. Struct 51 (2018) 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McLean MA, Denisov IG, Grinkova YV, Sligar SG, Dark, ultra-dark and ultra-bright nanodiscs for membrane protein investigations, Analytical Biochemistry 607 (2020) 113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang M, Gui M, Wang Z-F, Gorgulla C, Yu JJ, Wu H, Sun Z-YJ, Klenk C, Merklinger L, Morstein L, Hagn F, Plückthun A, Brown A, Nasr ML, Wagner G, Cryo-EM structure of an activated GPCR–G protein complex in lipid nanodiscs, Nat. Struct. Mol. Biol 28(3) (2021) 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Park SH, Berkamp S, Cook GA, Chan MK, Viadiu H, Opella SJ, Nanodiscs versus macrodiscs for NMR of membrane proteins, Biochemistry 50(42) (2011) 8983–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang M, Huang R, Ackermann R, Im S-C, Waskell L, Schwendeman A, Ramamoorthy A, Reconstitution of the Cytb5–CytP450 complex in nanodiscs for structural studies using NMR Spectroscopy, Angew. Chem. Int. Ed 55(14) (2016) 4497–4499. [DOI] [PubMed] [Google Scholar]
- [33].Salnikov ES, Anantharamaiah GM, Bechinger B, Supramolecular organization of apolipoprotein-A-I-derived peptides within disc-like arrangements, Biophys. J 115(3) (2018) 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im S-C, Anantharamaiah GM, Waskell L, Ramamoorthy A, Cytochrome-P450-induced ordering of microsomal membranes modulates affinity for drugs, Angew. Chem. Int. Ed 57(13) (2018) 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Barnaba C, Ramamoorthy A, Picturing the membrane-assisted choreography of cytochrome P450 with lipid nanodiscs, ChemPhysChem 19(20) (2018) 2603–2613. [DOI] [PubMed] [Google Scholar]
- [36].Salnikov ES, Aisenbrey C, Anantharamaiah GM, Bechinger B, Solid-state NMR structural investigations of peptide-based nanodiscs and of transmembrane helices in bicellar arrangements, Chem. Phys. Lipids 219 (2019) 58–71. [DOI] [PubMed] [Google Scholar]
- [37].Neumann B, Chao K, Chang CCY, Chang TY, Nanodisc scaffold peptide (NSP(r)) replaces detergent by reconstituting acyl-CoA:cholesterol acyltransferase 1 into peptidiscs, Arch. Biochem. Biophys 691 (2020) 108518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Anada C, Ikeda K, Egawa A, Fujiwara T, Nakao H, Nakano M, Temperature- and composition-dependent conformational transitions of amphipathic peptide–phospholipid nanodiscs, J. Colloid Interface Sci 588 (2021) 522–530. [DOI] [PubMed] [Google Scholar]
- [39].Anada C, Ikeda K, Nakao H, Nakano M, Improvement of thermal stability of amphipathic peptide–phospholipid nanodiscs via lateral association of α-helices by disulfide cross-linking, Langmuir 38(22) (2022) 6977–6983. [DOI] [PubMed] [Google Scholar]
- [40].Najafi H, Servoss SL, Altering the edge chemistry of bicelles with peptoids, Chem. Phys. Lipids 217 (2018) 43–50. [DOI] [PubMed] [Google Scholar]
- [41].Zhang Y, Heidari Z, Su Y, Yu T, Xuan S, Omarova M, Aydin Y, Dash S, Zhang D, John V, Amphiphilic polypeptoids rupture vesicle bilayers to form peptoid–lipid fragments effective in enhancing hydrophobic drug delivery, Langmuir 35(47) (2019) 15335–15343. [DOI] [PubMed] [Google Scholar]
- [42].Galiakhmetov AR, Davern CM, Esteves RJA, Awosanya EO, Guthrie QAE, Proulx C, Nevzorov AA, Aligned peptoid-based macrodiscs for structural studies of membrane proteins by oriented-sample NMR, Biophys. J 121(17) (2022) 3263–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cuevas Arenas R, Klingler J, Vargas C, Keller S, Influence of lipid bilayer properties on nanodisc formation mediated by styrene/maleic acid copolymers, Nanoscale 8(32) (2016) 15016–15026. [DOI] [PubMed] [Google Scholar]
- [44].Dominguez Pardo JJ, Dörr JM, Renne MF, Ould-Braham T, Koorengevel MC, van Steenbergen MJ, Killian JA, Thermotropic properties of phosphatidylcholine nanodiscs bounded by styrene-maleic acid copolymers, Chem. Phys. Lipids 208 (2017) 58–64. [DOI] [PubMed] [Google Scholar]
- [45].Dominguez Pardo JJ, Dörr JM, Iyer A, Cox RC, Scheidelaar S, Koorengevel MC, Subramaniam V, Killian JA, Solubilization of lipids and lipid phases by the styrene-maleic acid copolymer, Eur. Biophys 46(1) (2017) 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Radoicic J, Park SH, Opella SJ, Macrodiscs comprising SMALPs for oriented sample solid-state NMR spectroscopy of membrane proteins, Biophys. J 115(1) (2018) 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Overduin M, Esmaili M, Memtein: The fundamental unit of membrane-protein structure and function, Chem. Phys. Lipids 218 (2019) 73–84. [DOI] [PubMed] [Google Scholar]
- [48].Domínguez Pardo JJ, van Walree CA, Egmond MR, Koorengevel MC, Killian JA, Nanodiscs bounded by styrene-maleic acid allow trans-cis isomerization of enclosed photoswitches of azobenzene labeled lipids, Chem. Phys. Lipids 220 (2019) 1–5. [DOI] [PubMed] [Google Scholar]
- [49].Harding BD, Dixit G, Burridge KM, Sahu ID, Dabney-Smith C, Edelmann RE, Konkolewicz D, Lorigan GA, Characterizing the structure of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for membrane protein spectroscopic studies, Chem. Phys. Lipids 218 (2019) 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hall SCL, Clifton LA, Tognoloni C, Morrison KA, Knowles TJ, Kinane CJ, Dafforn TR, Edler KJ, Arnold T, Adsorption of a styrene maleic acid (SMA) copolymer-stabilized phospholipid nanodisc on a solid-supported planar lipid bilayer, J. Colloid Interface Sci 574 (2020) 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Burridge KM, Harding BD, Sahu ID, Kearns MM, Stowe RB, Dolan MT, Edelmann RE, Dabney-Smith C, Page RC, Konkolewicz D, Lorigan GA, Simple derivatization of RAFT-synthesized styrene-maleic anhydride copolymers for lipid disk formulations, Biomacromolecules 21(3) (2020) 1274–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Overduin M, Wille H, Westaway D, Multisite interactions of prions with membranes and native nanodiscs, Chem. Phys. Lipids 236 (2021) 105063. [DOI] [PubMed] [Google Scholar]
- [53].Brown CJ, Trieber C, Overduin M, Structural biology of endogenous membrane protein assemblies in native nanodiscs, Curr. Opin. Struct 69 (2021) 70–77. [DOI] [PubMed] [Google Scholar]
- [54].Ball LE, Riley LJ, Hadasha W, Pfukwa R, Smith CJI, Dafforn TR, Klumperman B, Influence of DIBMA polymer length on lipid nanodisc formation and membrane protein extraction, Biomacromolecules 22(2) (2021) 763–772. [DOI] [PubMed] [Google Scholar]
- [55].Hall SCL, Tognoloni C, Campbell RA, Richens J, O’Shea P, Terry AE, Price GJ, Dafforn TR, Edler KJ, Arnold T, The interaction of styrene maleic acid copolymers with phospholipids in Langmuir monolayers, vesicles and nanodiscs; a structural study, J. Colloid Interface Sci 625 (2022) 220–236. [DOI] [PubMed] [Google Scholar]
- [56].Zhao Z, Zhang M, Hogle JM, Shih WM, Wagner G, Nasr ML, DNA-corralled nanodiscs for the structural and functional characterization of membrane proteins and viral entry, J. Am. Chem. Soc 140(34) (2018) 10639–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Prade E, Mahajan M, Im S-C, Zhang M, Gentry KA, Anantharamaiah GM, Waskell L, Ramamoorthy A, A Minimal functional complex of cytochrome P450 and FBD of cytochrome P450 reductase in nanodiscs, Angew. Chem. Int. Ed 57(28) (2018) 8458–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gentry KA, Anantharamaiah GM, Ramamoorthy A, Probing protein–protein and protein–substrate interactions in the dynamic membrane-associated ternary complex of cytochromes P450, b5, and reductase, ChemComm 55(89) (2019) 13422–13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Krishnarjuna B, Yamazaki T, Anantharamaiah GM, Ramamoorthy A, Nanodisc reconstitution of flavin mononucleotide binding domain of cytochrome-P450-reductase enables high-resolution NMR probing, ChemComm 57(39) (2021) 4819–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Günsel U, Hagn F, Lipid nanodiscs for high-resolution NMR studies of membrane proteins, Chem. Rev 122(10) (2022) 9395–9421. [DOI] [PubMed] [Google Scholar]
- [61].Hazell G, Arnold T, Barker RD, Clifton LA, Steinke N-J, Tognoloni C, Edler KJ, Evidence of lipid exchange in styrene maleic acid lipid particle (SMALP) nanodisc systems, Langmuir 32(45) (2016) 11845–11853. [DOI] [PubMed] [Google Scholar]
- [62].Cuevas Arenas R, Danielczak B, Martel A, Porcar L, Breyton C, Ebel C, Keller S, Fast collisional lipid transfer among polymer-bounded nanodiscs, Sci. Rep 7 (2017) 45875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schmidt V, Sturgis JN, Modifying styrene-maleic acid co-polymer for studying lipid nanodiscs, BBA-Biomembranes 1860(3) (2018) 777–783. [DOI] [PubMed] [Google Scholar]
- [64].Ravula T, Ishikuro D, Kodera N, Ando T, Anantharamaiah GM, Ramamoorthy A, Real-time monitoring of lipid exchange via fusion of peptide based lipid-nanodiscs, Chem. Mater 30(10) (2018) 3204–3207. [Google Scholar]
- [65].Xue M, Cheng L, Faustino I, Guo W, Marrink SJ, Molecular mechanism of lipid nanodisk formation by styrene-maleic acid copolymers, Biophys. J 115(3) (2018) 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EAW, Dafforn TR, Baldus M, Killian JA, Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs, Proc. Natl. Acad. Sci. USA 111(52) (2014) 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR, A method for detergent-free isolation of membrane proteins in their local lipid environment, Nat. Protoc 11(7) (2016) 1149–62. [DOI] [PubMed] [Google Scholar]
- [68].Krishnarjuna B, Ravula T, Ramamoorthy A, Detergent-free extraction, reconstitution and characterization of membrane-anchored cytochrome-b5 in native lipids, ChemComm 56(48) (2020) 6511–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hall SCL, Tognoloni C, Charlton J, Bragginton EC, Rothnie AJ, Sridhar P, Wheatley M, Knowles TJ, Arnold T, Edler KJ, Dafforn TR, An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles, Nanoscale 10(22) (2018) 10609–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ravula T, Hardin NZ, Bai J, Im SC, Waskell L, Ramamoorthy A, Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs, ChemComm 54(69) (2018) 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gulamhussein AA, Uddin R, Tighe BJ, Poyner DR, Rothnie AJ, A comparison of SMA (styrene maleic acid) and DIBMA (di-isobutylene maleic acid) for membrane protein purification, BBA-Biomembranes 1862(7) (2020) 183281. [DOI] [PubMed] [Google Scholar]
- [72].Knowles TJ, Finka R, Smith C, Lin Y-P, Dafforn T, Overduin M, Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer, J. Am. Chem. Soc 131(22) (2009) 7484–7485. [DOI] [PubMed] [Google Scholar]
- [73].Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S, Solubilization of membrane proteins into functional lipid-bilayer nanodiscs using a diisobutylene/maleic acid copolymer, Angew. Chem. Int. Ed 56(7) (2017) 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ravula T, Ramamoorthy A, Synthesis, characterization, and nanodisc formation of non-ionic polymers, Angew. Chem. Int. Ed 60(31) (2021) 16885–16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Krishnarjuna B, Ravula T, Ramamoorthy A, Detergent-free isolation of CYP450-reductase’s FMN-binding domain in E.coli lipid-nanodiscs using a charge-free polymer, ChemComm 58(31) (2022) 4913–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A, Bioinspired, size-tunable self-assembly of polymer-lipid bilayer nanodiscs, Angew. Chem. Int. Ed 56(38) (2017) 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [77].Krishnarjuna B, Ravula T, Faison EM, Tonelli M, Zhang Q, Ramamoorthy A, Polymer-nanodiscs as a novel alignment medium for high-resolution NMR-based structural studies of nucleic acids, Biomolecules 12(11) (2022) 1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dargel C, Hannappel Y, Hellweg T, Heating-induced DMPC/glycyrrhizin bicelle-to-vesicle transition: A X-ray contrast variation study, Biophys. J 118(10) (2020) 2411–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Krishnarjuna B, Im SC, Ravula T, Marte J, Auchus RJ, Ramamoorthy A, Non-ionic inulin-based polymer nanodiscs enable functional reconstitution of a redox complex composed of oppositely charged CYP450 and CPR in a lipid bilayer membrane, Anal. Chem 94(34) (2022) 11908–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Krishnarjuna B, Ramamoorthy A, Detergent-free isolation of membrane proteins and strategies to study them in a near-native membrane environment, Biomolecules 12(8) (2022) 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Gröbner G, Watts A, Detergent-free formation and physicochemical characterization of nanosized lipid–polymer complexes: Lipodisq, Angew. Chem. Int. Ed 51(19) (2012) 4653–4657. [DOI] [PubMed] [Google Scholar]
- [82].Jamshad M, Grimard V, Idini I, Knowles TJ, Dowle MR, Schofield N, Sridhar P, Lin Y, Finka R, Wheatley M, Thomas ORT, Palmer RE, Overduin M, Govaerts C, Ruysschaert J-M, Edler KJ, Dafforn TR, Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins, Nano Research 8(3) (2015) 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bayburt TH, Grinkova YV, Sligar SG, Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins, Nano Lett 2(8) (2002) 853–856. [Google Scholar]
- [84].Denisov IG, Grinkova YV, Lazarides AA, Sligar SG, Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size, J. Am. Chem. Soc 126(11) (2004) 3477–3487. [DOI] [PubMed] [Google Scholar]
- [85].Denisov IG, Sligar SG, Nanodiscs for structural and functional studies of membrane proteins, Nat. Struct. Mol. Biol 23(6) (2016) 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hagn F, Nasr ML, Wagner G, Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR, Nat. Protoc 13(1) (2018) 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mishra VK, Anantharamaiah G, Segrest JP, Palgunachari MN, Chaddha M, Sham SS, Krishna N.R.J.J.o.B.C., Association of a model class A (apolipoprotein) amphipathic α helical peptide with lipid: High resolution NMR studies of peptide·lipid discoidal complexes, J. Biol. Chem 281(10) (2006) 6511–6519. [DOI] [PubMed] [Google Scholar]
- [88].Li M, Morales HH, Katsaras J, Kučerka N, Yang Y, Macdonald PM, Nieh M-P, Morphological characterization of DMPC/CHAPSO bicellar mixtures: A combined SANS and NMR study, Langmuir 29(51) (2013) 15943–15957. [DOI] [PubMed] [Google Scholar]
- [89].Leckband DE, Helm CA, Israelachvili J, Role of calcium in the adhesion and fusion of bilayers, Biochemistry 32(4) (1993) 1127–1140. [DOI] [PubMed] [Google Scholar]
- [90].Crowell KJ, Macdonald PM, Surface charge response of the phosphatidylcholine head group in bilayered micelles from phosphorus and deuterium nuclear magnetic resonance, BBA-Biomembranes 1416(1) (1999) 21–30. [DOI] [PubMed] [Google Scholar]
- [91].Struppe J, Whiles JA, Vold RR, Acidic phospholipid bicelles: A versatile model membrane system, Biophys. J 78(1) (2000) 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Baek S-B, Lim S-C, Lee H-J, Lee H-C, Kim C.J.B.o.t.K.C.S., An NMR study on the conformation of substance P in acidic bicelles, Bull. Korean Chem. Soc 32(10) (2011) 3702–3706. [Google Scholar]
- [93].Rohrbach TD, Shah N, Jackson WP, Feeney EV, Scanlon S, Gish R, Khodadadi R, Hyde SO, Hicks PH, Anderson JC, Jarboe JS, Willey CD, The effector domain of MARCKS is a nuclear localization signal that regulates cellular PIP2 levels and nuclear PIP2 localization, PLoS One 10(10) (2015) e0140870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Yang P-W, Lin T-L, Lin T-Y, Yang C-H, Hu Y, Jeng US, Packing DNA with disc-shaped bicelles, Soft Matter 9(48) (2013) 11542–11548. [Google Scholar]
- [95].Parker MA, King V, Howard KP, Nuclear magnetic resonance study of doxorubicin binding to cardiolipin containing magnetically oriented phospholipid bilayers, BBA-Biomembranes 1514(2) (2001) 206–216. [DOI] [PubMed] [Google Scholar]
- [96].Luchette PA, Vetman TN, Prosser RS, Hancock RE, Nieh MP, Glinka CJ, Krueger S, Katsaras J, Morphology of fast-tumbling bicelles: a small angle neutron scattering and NMR study, Biochim Biophys Acta 1513(2) (2001) 83–94. [DOI] [PubMed] [Google Scholar]
- [97].Hu A, Fan T-H, Katsaras J, Xia Y, Li M, Nieh M-P, Lipid-based nanodiscs as models for studying mesoscale coalescence – a transport limited case, Soft Matter 10(28) (2014) 5055–5060. [DOI] [PubMed] [Google Scholar]
- [98].Alahmadi I, Hoy D Jr., Tahmasbi Rad A, Patil S, Alahmadi A, Kinnun J, Scott HL, Katsaras J, Nieh M-P, Changes experienced by low-concentration lipid bicelles as a function of temperature, Langmuir 38(14) (2022) 4332–4340. [DOI] [PubMed] [Google Scholar]
- [99].Caldwell TA, Baoukina S, Brock AT, Oliver RC, Root KT, Krueger JK, Glover KJ, Tieleman DP, Columbus L, Low-q bicelles are mixed micelles, J. Phys. Chem. Lett 9(15) (2018) 4469–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gruenhagen TC, Ziarek JJ, Schlebach JP, Bicelle size modulates the rate of bacteriorhodopsin folding, Protein Sci 27(6) (2018) 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Dufourc EJ, Bicelles and nanodiscs for biophysical chemistry, BBA-Biomembranes 1863(1) (2021) 183478. [DOI] [PubMed] [Google Scholar]
- [102].Danielczak B, Rasche M, Lenz J, Pérez Patallo E, Weyrauch S, Mahler F, Agbadaola MT, Meister A, Babalola JO, Vargas C, Kolar C, Keller S, A bioinspired glycopolymer for capturing membrane proteins in native-like lipid-bilayer nanodiscs, Nanoscale 14(5) (2022) 1855–1867. [DOI] [PubMed] [Google Scholar]
- [103].Primeau JO, Armanious GP, Fisher MLE, Young HS, The sarcoendoplasmic reticulum calcium ATPase, in: Harris JR, Boekema EJ (Eds.), Membrane Protein Complexes: Structure and Function, Springer; Singapore, Singapore, 2018, pp. 229–258. [Google Scholar]
- [104].Maruyama T, Imai S, Kusakizako T, Hattori M, Ishitani R, Nureki O, Ito K, Maturana AD, Shimada I, Osawa M, Functional roles of Mg2+ binding sites in ion-dependent gating of a Mg2+ channel, MgtE, revealed by solution NMR, eLife 7 (2018) e31596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Krishnarjuna B, Sunanda P, Villegas–Moreno J, Csoti A, Morales RAV, Wai DCC, Panyi G, Prentis P, Norton RS, A disulfide-stabilised helical hairpin fold in acrorhagin I: An emerging structural motif in peptide toxins, J. Struct. Biol 213(2) (2021) 107692. [DOI] [PubMed] [Google Scholar]
- [106].Bej A, Ames JB, Chemical shift assignments of calmodulin under standard conditions at neutral pH, Biomolecular NMR Assignments (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Liu Y, Xia Y, Rad AT, Aresh W, Nieh MP, Stable discoidal bicelles: A platform of lipid nanocarriers for cellular delivery, Methods Mol. Biol 1522 (2017) 273–282. [DOI] [PubMed] [Google Scholar]
- [108].Uchida N, Nishizawa Horimoto N, Yamada K, Hikima T, Ishida Y, Kinetically stable bicelles with dilution tolerance, size tunability, and thermoresponsiveness for drug delivery applications, ChemBioChem 19(18) (2018) 1922–1926. [DOI] [PubMed] [Google Scholar]
- [109].Aramaki K, Adachi K, Maeda M, Mata J, Kamimoto-Kuroki J, Tsukamoto D, Konno Y, Formulation of bicelles based on lecithin-nonionic surfactant mixtures, Mater 13(14) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Choi S, Kang B, Taguchi S, Umakoshi H, Kim K, Kwak MK, Jung H-S, A simple method for continuous synthesis of bicelles in microfluidic systems, Langmuir 37(42) (2021) 12255–12262. [DOI] [PubMed] [Google Scholar]
- [111].Garcia CR, Rad AT, Saeedinejad F, Manojkumar A, Roy D, Rodrigo H, Chew SA, Rahman Z, Nieh MP, Roy U, Effect of drug-to-lipid ratio on nanodisc-based tenofovir drug delivery to the brain for HIV-1 infection, Nanomedicine 17(13) (2022) 959–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Chen H, Kim S, He W, Wang H, Low PS, Park K, Cheng J-X, Fast release of lipophilic agents from circulating PEG-PDLLA micelles revealed by in vivo förster resonance energy transfer imaging, Langmuir 24(10) (2008) 5213–5217. [DOI] [PubMed] [Google Scholar]
- [113].Hiel S, Bindels LB, Pachikian BD, Kalala G, Broers V, Zamariola G, Chang BPI, Kambashi B, Rodriguez J, Cani PD, Neyrinck AM, Thissen JP, Luminet O, Bindelle J, Delzenne NM, Effects of a diet based on inulin-rich vegetables on gut health and nutritional behavior in healthy humans, Am. J. Clin. Nutr 109(6) (2019) 1683–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Huda P, Binderup T, Pedersen MC, Midtgaard SR, Elema DR, Kjær A, Jensen M, Arleth L, PET/CT based in vivo evaluation of 64Cu labelled nanodiscs in tumor bearing mice, PLoS One 10(7) (2015) e0129310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


