Abstract
Changes in protein abundance and reversible protein phosphorylation (RPP) play important roles in regulating hypometabolism but have never been documented in overwintering frogs at high altitudes. To test the hypothesis that protein abundance and phosphorylation change in response to winter hibernation, we conducted a comprehensive and quantitative proteomic and phosphoproteomic analysis of the liver of the Xizang plateau frog, Nanorana parkeri, living on the Qinghai-Xizang Plateau. In total, 5170 proteins and 5695 phosphorylation sites in 1938 proteins were quantified. Based on proteomic analysis, 674 differentially expressed proteins (438 up-regulated, 236 down-regulated) were screened in hibernating N. parkeri versus summer individuals. Functional enrichment analysis revealed that higher expressed proteins in winter were significantly enriched in immune-related signaling pathways, whereas lower expressed proteins were mainly involved in metabolic processes. A total of 4251 modified sites (4147 up-regulated, 104 down-regulated) belonging to 1638 phosphoproteins (1555 up-regulated, 83 down-regulated) were significantly changed in the liver. During hibernation, RPP regulated a diverse array of proteins involved in multiple functions, including metabolic enzymatic activity, ion transport, protein turnover, signal transduction, and alternative splicing. These changes contribute to enhancing protection, suppressing energy-consuming processes, and inducing metabolic depression. Moreover, the activities of phosphofructokinase, glutamate dehydrogenase, and ATPase were all significantly lower in winter compared to summer. In conclusion, our results support the hypothesis and demonstrate the importance of RPP as a regulatory mechanism when animals transition into a hypometabolic state.
Keywords: Nanorana parkeri, Proteomic, Phosphoproteomic, Hibernation, Reversible protein phosphorylation, Metabolism
INTRODUCTION
Winter dormancy and hibernation are common survival strategies used by many species, including both endo- and ectotherms, to cope with prolonged cold exposure and poor food availability (Pinder et al., 1992; Tattersall & Ultsch, 2008). By entering dormancy or hibernation states, animals can reduce their metabolic rates, extend the duration for which stored food or endogenous energy reserves sustain survival, and retreat to safe locations conducive to protection. Depression of the metabolic rate involves the suppression of various biochemical (transcription, translation, cell cycle) and physiological processes (heart rate, respiratory rate) (Boutilier et al., 1997; Donohoe & Boutilier, 1998; Storey & Storey, 2004, 2007). Enhancement of defense mechanisms, such as the up-regulation of antifreeze proteins, antioxidant enzymes, and chaperone proteins, is also essential for long-term survival under hypometabolic conditions (Storey & Storey, 2007). This state allows for substantial energy conservation through targeted inhibition of various cellular and metabolic processes and decreased substrate utilization (Storey & Storey, 1990, 2004). Despite this strong suppression, essential metabolic functions must be retained to enable rapid restoration upon arousal (Storey & Storey, 1990, 2004). Thus, hypometabolism is generally characterized not by a significant reduction in metabolic capacity but by reversible control mechanisms (Storey & Storey, 2004).
The molecular mechanisms that underlie the hibernation phenotype have been elucidated in a variety of ectothermic vertebrates, including the Chinese alligator (Alligator sinensis) (Lin et al., 2020; Sun et al., 2018; Zhang et al., 2021), Australian central bearded dragon (Pogona vitticeps) (Capraro et al., 2019), Asiatic toad (Bufo gargarizans) (Jin et al., 2018), and wood frog (Rana sylvatica) (Kiss et al., 2011). In the case of A. sinensis, integrative multi-omics analysis revealed a down-regulation of pathways related to nutrient absorption and metabolism, muscle contraction, urinary excretion, and immunity function during hibernation (Lin et al., 2020). In addition, transcriptomic and proteomic studies of P. vitticeps revealed enrichment of protective mechanisms across various tissues during hibernation, including skeletal muscle atrophy resistance, neuroprotection in the brain, and cardiac hypertrophic processes (Capraro et al., 2019). Transcriptomic analysis of overwintering female toads found significant down-regulation of most genes in the liver during hibernation, involving metabolic depression and shifts in energy utilization (Jin et al., 2018). For R. sylvatica, hepatoproteomic analysis revealed significant up-regulation of proteins involved in cytoprotection (heat shock proteins and antioxidants) in the liver during hibernation, with significant down-regulation of proteins involved in cell proliferation, protein synthesis, and mitochondrial function (Kiss et al., 2011). Although the reorganization of specific proteomic components is crucial for survival under a hypometabolic state, the coordinated inhibition of non-essential or energy-intensive cellular functions is also crucial for successful overwintering (Storey & Storey, 2013). The extensive degradation of proteins and mRNA transcripts during dormant states would be counterproductive for organisms, as these components are immediately required upon arousal from torpor or rewarming. Therefore, mechanisms that can reversibly and comprehensively inhibit metabolic functions while differentially regulating various cellular processes are essential. One such efficient control mechanism is reversible protein phosphorylation (RPP). This fast-acting and easily reversible control mechanism necessitates minimal expenditure of adenosine triphosphate (ATP) and serves as a regulatory mechanism for fuel metabolism, ion channel inactivation, and protein synthesis inhibition during hypometabolic states (Storey & Storey, 2004, 2007). For example, various enzymes, such as glucose-6-phosphate dehydrogenase, catalase, creatine kinase, and hexokinase, are regulated by reversible phosphorylation in overwintering and/or frozen wood frogs (Dawson & Storey, 2016; Dieni & Storey, 2009, 2010, 2011). Although many studies have addressed the molecular mechanisms of overwintering in frog species inhabiting North America or high-latitude regions, the mechanisms governing metabolic depression in overwintering species residing in high-altitude regions remain unclear.
The Xizang plateau frog (Nanorana parkeri) (Anura: Dicroglossidae) inhabits the Qinghai-Xizang Plateau at elevations ranging from 2 850 to 5 100 m above sea level (a.s.l.), representing the highest known altitude of any amphibian species (Ma et al., 2009). As such, this species has been extensively used to explore adaptations to extreme high-altitude environments (Sun et al., 2015). Our previous studies on N. parkeri primarily focused on its physiological and biochemical mechanisms of adaptation to high altitude, as well as the physiological ecology of its winter hibernation (Niu et al., 2022a, 2022b). For example, N. parkeri individuals living at high elevations exhibit modified hematological parameters, such as higher blood hemoglobin content, increased hematocrit values, and higher red cell counts, in addition to stronger antioxidant defenses and reduced oxidative damage compared to low-elevation residing frogs (Niu et al., 2022b). Furthermore, during hibernation, N. parkeri demonstrates greater body mass, body mass index, liver index, liver glycogen concentration, and plasma bacteria-killing ability, as well as higher levels of oxidative stress but lower antioxidant defenses compared to summer-active individuals (Niu et al., 2018, 2022a). These frogs hibernate in shallow ponds within damp caves and display reduced freeze tolerance compared to R. sylvatica (Larson et al., 2014), although freeze exposure triggers substantial changes in liver metabolic profiles and tissue-specific activation of antioxidant systems (Niu et al., 2021a, 2021b). Moreover, acute warming leads to oxidative stress and a significant increase in total antioxidant capacity in most tissues in hibernating N. parkeri (Zhang et al., 2022a). Overwintering N. parkeri frogs also exhibit seasonal and temperature-independent metabolic depression, accompanied by marked changes in plasma metabolomic profiles (Niu et al., 2020, 2021b). Furthermore, integrated transcriptomic and metabolomic analyses have shown that adaptive regulation occurs in gene expression and metabolic processes of the liver in overwintering N. parkeri (Niu et al., 2023). However, information on the regulatory roles of protein abundance and RPP in hypometabolism in overwintering N. parkeri remains limited.
In the present study, we hypothesized that the adaptation of N. parkeri frogs to winter stress (such as low temperature, low oxygen, lack of food) also necessitates adjustments in protein abundance for maintenance of homeostasis, as well as reversible mechanisms (e.g., protein phosphorylation) for the differential regulation of metabolic processes in winter conditions. To test this hypothesis, we used quantitative proteomic and phosphoproteomic analyses to assess the seasonal variations in the abundance and phosphorylation status of proteins in the livers of N. parkeri. The liver was chosen as it is a key organ for the synthesis, metabolism, storage, and redistribution of carbohydrates, proteins, and lipids. The findings of this study provide insights into the macromolecular responses and regulatory roles of reversible phosphorylation that contribute to the winter survival of this species.
MATERIALS AND METHODS
Animals and sample collection
Adult male N. parkeri frogs (n=9 in each season) were captured and their liver tissues were collected, as reported previously (Niu et al., 2023). In brief, liver tissue samples were dissected and snap-frozen in liquid nitrogen, followed by storage at −80 °C in the laboratory. The mean snout-vent length (SVL) of frogs was 4.08±0.06 cm and mean body mass (BM) was 4.99±0.14 g in summer (n=9); comparable data were 4.06±0.06 cm and 5.14±0.16 g in winter. All animal procedures were approved by the Ethics Committee of Animal Experiments at Dezhou University (Approval No. DZXY2021003) and met the guidelines of the China Council on Animal Care.
Protein extraction and trypsin digestion
Protein extraction and trypsin digestion were performed as described by Lu et al. (2022) with minor modifications. Briefly, frozen samples (n=3 for each season) were fully powdered under liquid nitrogen and then sonicated in lysis buffer (8 mol/L urea (Sigma-Aldrich, USA), 1% protease inhibitor (Merck Millipore, USA), and 1% phosphatase inhibitor (Millipore, USA); w:v=1:4), followed by centrifugation at 12 000 ×g for 10 min at 4 °C. Supernatants were collected to determine protein concentration using a BCA kit (Beyotime Biotechnology, China). Supernatants were added to 20% (w:v) trichloroacetic acid (TCA) (Sigma-Aldrich, USA), followed by incubation at 4 °C for 2 h. After centrifugation at 4 500 ×g for 5 min at 4 °C, the precipitates were collected and washed three times with cooled acetone, then dissolved in 200 mmol/L tetraethylammonium bromide (TEAB) (Sigma-Aldrich, USA). For digestion, trypsin (Promega, USA) (w:w=1:50) was added and left overnight. Samples were then reduced with 5 mmol/L dithiothreitol (Sigma-Aldrich, USA) for 60 min at 37 °C and alkylated with 11 mmol/L iodoacetamide (Sigma-Aldrich, USA) for 45 min at room temperature in darkness. Finally, the peptides were desalted using a Strata X SPE column (Dr. Maisch GmbH, GER) and vacuum-dried.
Tandem mass tag (TMT) labeling, high-performance liquid chromatography (HPLC) fractionation, and affinity enrichment of phosphopeptides
Peptides from each sample were labeled with TMT reagent (Thermo Fisher Scientific, USA) and fractionated by high-pH reverse-phase HPLC (Thermo Fisher Scientific, USA), as reported in Lu et al. (2022) with minor modification. Briefly, the peptides were separated with a gradient of 8%–32% acetonitrile (Thermo Fisher Scientific, USA) in 10 mmol/L ammonium bicarbonate (Sigma-Aldrich, USA) (pH=9) over 60 min into 60 fractions, then combined into nine fractions and dried by vacuum centrifugation for liquid chromatography-tandem mass spectrometry (LC-MS-MS). Enrichment of phosphopeptides was carried out according to Lu et al. (2022).
LC-MS/MS analysis
LC-MS/MS analyses were performed as reported by Lu et al. (2022) with minor modification. Briefly, after dissolution in mobile phase A (0.1% formic acid, 2% acetonitrile in water, Thermo Fisher Scientific, USA), the peptides were separated using the EASY-nLC 1200 ultra-high-performance liquid system (Thermo Fisher Scientific, USA), followed by ionization in an NSI source and analysis using the Q ExactiveTM HF-X system (Thermo Fisher Scientific, USA). The mass spectrometry scan range was 350–1 600 m/z with a resolution of 120 000 and a second-order mass scan resolution of 30 000. The top 20 peptide parent ions with the highest signal intensity were used for higher-energy collisional dissociation (HCD) fragmentation at a normalized collision energy (NCE) of 28%. Mobile phase B contained 0.1% formic acid and 90% acetonitrile. For proteomic analysis, the elution settings were: 40 min, 6%–22% B; 14 min, 22%–32% B; 3 min, 32%–80% B; 3 min, 80% B, with a constant flow rate of 500 nL/min. For phosphoproteomic analysis, the gradient was as follows: 38 min, 5%–22% B; 15 min 22%–35% B; 4 min 35%–80% B; 3 min, 80% B, with a flow rate of 350 nL/min at 0–53 min and 600 nL/min at 53–60 min.
Proteomic and phosphoproteomic database search
The raw MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8), and tandem mass spectra were searched against the Nanorana_parkeri database (37 248 entries) concatenated with the reverse decoy database. The database search parameters were set as described in Zhang et al. (2022b). Briefly, trypsin/P was designated as a cleavage enzyme and missing cleavages of less than 2 were permitted. The mass error for precursor ions was set at 5 ppm in the main search and 20 ppm in the first search, and a value of 0.02 Da was used for mass error of fragment ions. Carbamidomethyl on Cys was specified as a fixed modification, and variable modifications included acetylation on protein N-terminal, oxidation on Met, deamidation (NQ), and phosphorylation on Ser, Thr, and Tyr. TMT-6plex was performed for quantification. The false discovery rate (FDR) was adjusted to <1% and the minimum score for peptides was set to >40.
Bioinformatics analysis for proteomics and phosphoproteomics
Gene Ontology (GO) annotation of proteomic and phosphoproteomic data was conducted using the UniProt-GOA database (http://www.ebi.ac.uk/GOA/) and included three categories: biological processes, cellular components, and molecular functions. Protein domains and pathways were annotated using the InterPro (http://www.ebi.ac.uk/interpro/) database and Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/), respectively. Subcellular localization was predicted using WoLF PSORT software (v.2.0), and sequence patterns around the phosphorylation modification site (spanning six amino acids both upstream and downstream) were identified using Soft motif-x (v.5.0.2). Two-tailed Fisher’s exact test was used to test functional enrichment of differentially expressed proteins with a critical corrected P-value of 0.05. Functional interaction network analysis of differentially expressed proteins and phosphorylated proteins was performed using STRING (v.11.0) and visualized using Cytoscape (v.3.9.1).
Enzymatic activity assays
All enzymatic activities were determined using an assay kit (Solarbio Science & Technology, China). Frozen liver tissues (n=6 for each season) were weighed and immediately homogenized on ice according to the manufacturer’s instructions. The resulting supernatant was immediately used to determine the activities of Na+/K+-ATPase, Ca2+-ATPase, phosphofructokinase (PFK; an important rate-controlling enzyme in the glycolytic pathway), and glutamate dehydrogenase (GDH). All enzymatic assays were conducted at 25 °C using an automatic microplate reader (BioTek Instruments, USA) and were run in triplicate.
Statistical analysis
All data were analyzed using SPSS v.20.0 (SPSS, USA) and expressed as mean±standard error of the mean (SEM). After verification of normality and homogeneity of variances to satisfy the assumptions for parametric tests, differences in enzyme activity between the two groups were evaluated using Student’s t-tests for independent samples. A threshold level of P<0.05 was used to establish statistical significance.
RESULTS
Summary of quantitative proteome and protein phosphoproteome analysis
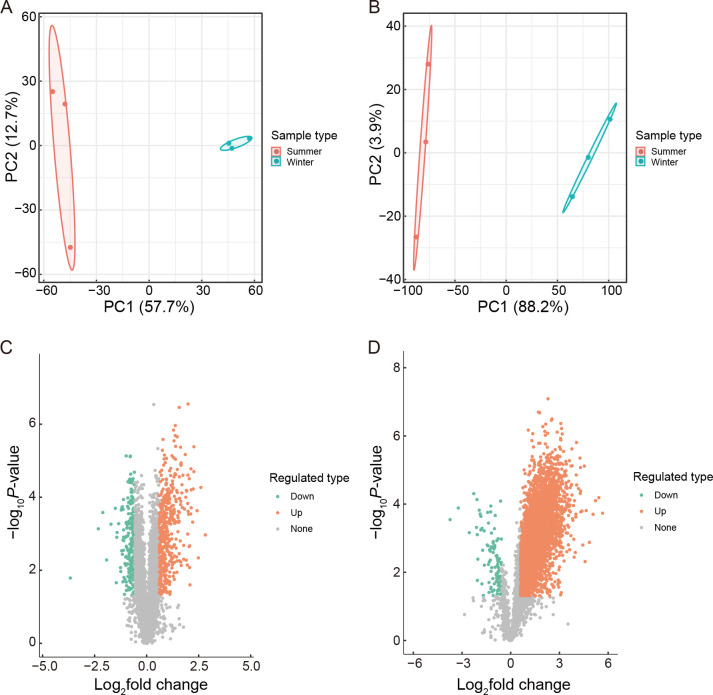
Principal component analysis (PCA) of the proteomic and phosphoproteomic datasets showed good clustering between biological replicates, as well as a clear summer and winter separation (Figure 1A, B). In total, 5 411 protein groups were identified, from which 5 170 proteins were quantified (Supplementary Table S1). Altogether, 12 299 phosphorylation sites in 4 119 protein groups were identified, among which 5 695 sites in 1 938 proteins were quantified (Supplementary Table S2).
Figure 1.
PCA and volcano plots showing differentially expressed proteins and phosphorylated proteins
A, C: Proteomic analysis. B, D: Phosphoproteomic analysis. Screening criteria were FC>1.5 (up-regulated, red) or <0.667 (down-regulated, green) and P<0.05.
Analysis of differentially expressed proteins and phosphorylated proteins
Based on established cut-off criteria (fold-change (FC)>1.5 (up-regulated) or <0.667 (down-regulated) and P<0.05), a total of 674 proteins were differentially expressed, among which 438 proteins (65%) showed higher abundance and 236 proteins (35%) showed lower abundance in winter compared to summer (Figure 1C; Supplementary Table S3). Phosphorylation levels changed significantly in 4 251 modified sites belonging to 1 638 phosphoproteins (FC>1.5 (up-regulated) or <0.667 (down-regulated) and P<0.05). Among them, 4 147 sites in 1 555 proteins were up-regulated and 104 sites in 83 proteins were down-regulated in the liver of hibernating N. parkeri compared to summer-active individuals (Figure 1D; Supplementary Table S4). The phosphorylation levels of ATPases showed a significant increase in overwintering N. parkeri, including plasma membrane calcium-transporting ATPase 1, v-type proton ATPase subunit d 1, v-type proton ATPase catalytic subunit A, cation-transporting ATPase, sarcoplasmic/endoplasmic reticulum calcium ATPase 1, sodium/potassium-transporting ATPase subunit alpha-1, and transitional endoplasmic reticulum ATPase. Based on motif-x analysis, 100 different phosphorylation motifs were identified from hibernation-regulated phosphopeptides (Supplementary Table S5). Among the top 10 phosphorylation motifs ranked by fold increase (Table 1), aspartic acid was conserved at the −1 position, while those motifs containing aspartic and glutamic acids at the +1, +2, or +3 positions of the central serine residue were highly enriched. Motifs containing arginine at the −3 position of the central serine and tyrosine residues were also strongly conserved.
Table 1. Motif-x analysis of significantly different phosphopeptides in Nanorana parkeri liver between summer and winter.
| Motif logo | Motif | Motif score | Foreground | Background | Fold increase | |||
| Matches | Size | Matches | Size | |||||

|
KxxxxD_S_ExExxx | 54.17 | 25 | 7 919 | 51 | 1 005 390 | 62.2 | |

|
xxxxxx_S_DDDxxx | 47.05 | 59 | 6 676 | 322 | 980 303 | 26.9 | |

|
xxxxxD_S_DxExxx | 48.00 | 117 | 9 266 | 493 | 1 024 378 | 26.2 | |

|
xxxxxx_T_PPxxxx | 32.00 | 167 | 1 375 | 3349 | 680 998 | 24.7 | |

|
xxxxxx_S_DDExxx | 44.42 | 66 | 8 344 | 352 | 1 012 577 | 22.8 | |

|
xxxxxD_S_DxDxxx | 40.76 | 35 | 5 657 | 282 | 952 528 | 20.9 | |

|
xRxRxx_S_xDxxxx | 39.02 | 40 | 8 971 | 223 | 1 020 862 | 20.4 | |

|
xxxRxx_S_PxPxxx | 48.00 | 76 | 10 662 | 366 | 1 041 899 | 20.3 | |

|
xxxRRx_T_xxxxxx | 32.00 | 68 | 1 030 | 2196 | 669 591 | 20.1 | |

|
xxxxxD_S_xEDxxx | 43.65 | 45 | 7 146 | 312 | 990 174 | 20.0 | |
Functional analysis of differentially quantified proteins
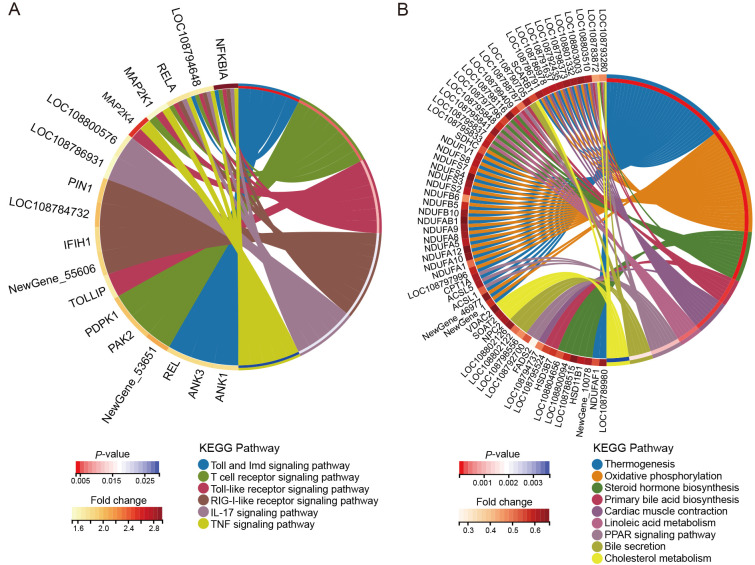
GO enrichment analysis indicated that higher expressed proteins in hibernating N. parkeri were enriched in cell-cell junction organization, regulation of cell morphogenesis, and cell junction assembly in the biological process category (Supplementary Figure S1A), double-stranded DNA binding, cytoskeletal protein binding, and DNA binding in the molecular function category (Supplementary Figure S1B), and in cell junction, anchoring junction, and adherens junction in the cellular component category (Supplementary Figure S1C). KEGG pathway enrichment analysis showed that higher expressed proteins in overwintering N. parkeri were mainly enriched in tight junction, vasopressin-regulated water reabsorption (Supplementary Figure S1D), and signaling pathways, including the Toll and Imd, T cell receptor, Toll-like receptor, RIG-I-like receptor, IL-17, and Ras signaling pathways (Figure 2A).
Figure 2.
Circle diagram showing KEGG enrichment analysis of differentially expressed proteins
A: Up-regulated proteins related to signaling pathways. B: Down-regulated proteins related to nutrient absorption and energy metabolism pathways.
Proteins with lower expression in the liver of hibernating N. parkeri were mainly enriched in oxidative phosphorylation, respiratory electron transport chain, cellular respiration, and ribose phosphate metabolic process in the biological process category (Supplementary Figure S2A), NADH dehydrogenase activity, oxidoreductase activity, and electron transfer activity in the molecular function category (Supplementary Figure S2B), and respirasome, respiratory chain complex, mitochondrial respirasome, and mitochondrial membrane in the cellular component category (Supplementary Figure S2C). KEGG pathway enrichment analysis showed that lower expressed proteins in overwintering N. parkeri were primarily enriched in metabolism-related pathways, including thermogenesis, oxidative phosphorylation, steroid hormone biosynthesis, retinol metabolism, primary bile acid biosynthesis, linoleic acid metabolism, cholesterol metabolism, and peroxisome proliferator-activated receptor (PPAR) signaling (Supplementary Figure S2D; Figure 2B).
Functional analysis of differentially phosphorylated proteins
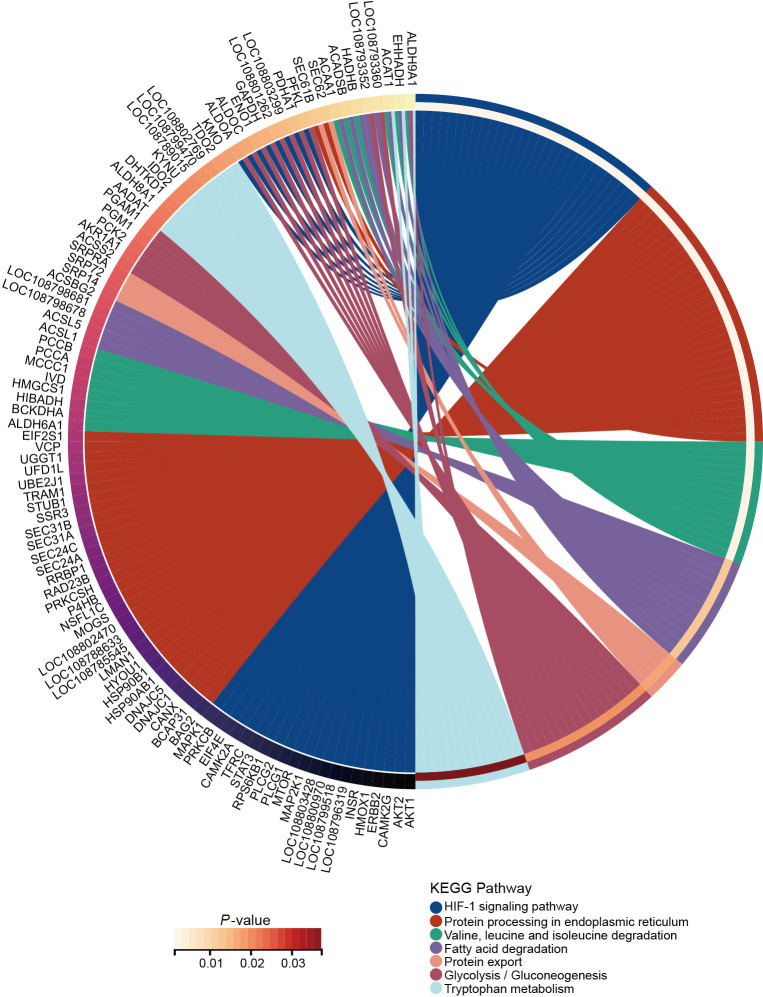
GO enrichment analysis showed that the higher abundances of phosphorylated proteins in hibernating N. parkeri were enriched in carboxylic acid metabolic process, carboxylic acid biosynthetic process, cellular amino acid metabolic process, Golgi vesicle transport, and lipid oxidation in the biological process category (Supplementary Figure S3A), oxidoreductase activity, mRNA binding, and cell adhesion molecule binding in the molecular function category (Supplementary Figure S3B), and mitochondrial matrix, intercalated disc, and endocytic vesicle in the cellular component category (Supplementary Figure S3C). For KEGG pathways analysis, a higher abundance of phosphorylated proteins in hibernating N. parkeri were enriched in metabolic pathways (such as valine, leucine and isoleucine degradation, fatty acid degradation, glycolysis/gluconeogenesis, and tryptophan metabolism), hypoxia-inducible factor-1 (HIF-1) signaling pathway, and protein processing in endoplasmic reticulum and protein export (Figure 3; Supplementary Figure S3D).
Figure 3.
Circle diagram showing KEGG enrichment analysis of up-regulated phosphorylated proteins related to signaling pathways, protein synthesis, and energy metabolism pathways in hibernating Nanorana parkeri
Proteins expressed at lower levels were mainly enriched in ribosome biogenesis, glucan metabolic processes, and energy reserve metabolic processes in the biological process category (Supplementary Figure S4A), rRNA methyltransferase activity, catalytic activity, acting on an rRNA, and poly(G) binding in the molecular function category (Supplementary Figure S4B), and mitotic spindle midzone and nephrocyte diaphragm in the cellular component category (Supplementary Figure S4C). KEGG pathway enrichment analysis showed that lower expressed proteins were enriched in starch and sucrose metabolism, metabolism of xenobiotics by cytochrome P450, and pathogenic Escherichia coli infection (Supplementary Figure S4D).
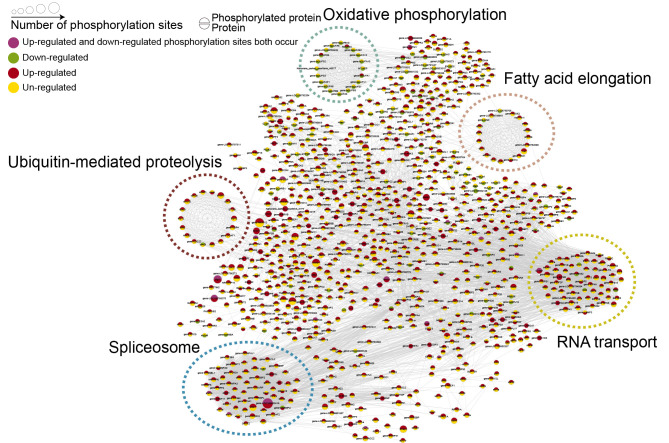
Protein-protein interaction (PPI) networks of differentially expressed proteins and phosphorylated proteins
PPI network analysis revealed significant enrichment in differentially expressed proteins and phosphorylated proteins in five major connected subnetworks, including oxidative phosphorylation, ubiquitin-mediated proteolysis, spliceosomes, fatty acid elongation, and RNA transport (Figure 4). In oxidative phosphorylation, the expression of most proteins was significantly decreased, but the phosphorylation levels of proteins did not change significantly. In contrast, for ubiquitin-mediated proteolysis, spliceosomes, fatty acid elongation, and RNA transport, phosphorylation levels increased substantially without apparent changes in protein expression, suggesting that protein phosphorylation plays an important regulatory role in these processes.
Figure 4.
PPI network of differentially regulated phosphopeptides and expressed proteins
Enriched pathways are surrounded by dotted circles. Green, red, and yellow in circles indicate down-, up-, or un-regulated phosphopeptides (upper half) and proteins (lower half). Purple in circle indicates that up-regulated and down-regulated phosphorylation sites occurred simultaneously.
Enzymatic activities
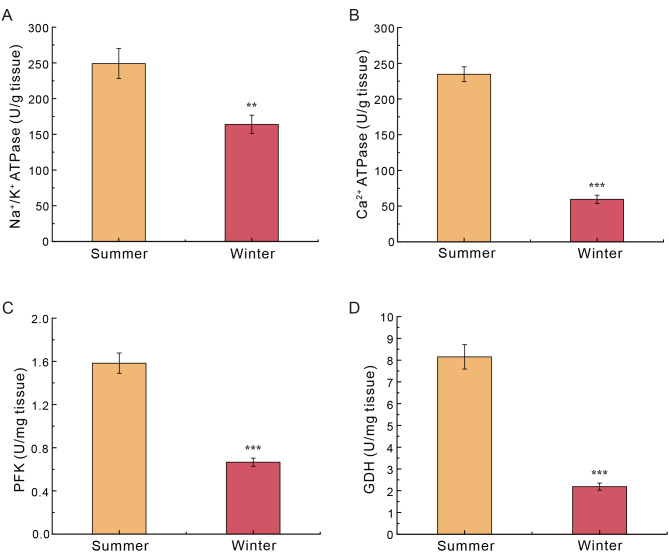
Overall, the activity of the four enzymes was significantly lower in winter compared to summer. Na+/K+ ATPase and Ca2+ ATPase activities were 34% and 75% lower, respectively, in the liver of hibernating N. parkeri frogs than those in summer-active individuals. Moreover, PFK and GDH activities decreased by 58% and 73%, respectively, in winter compared to summer (Figure 5).
Figure 5.
Enzymatic activities in liver of summer- and winter-collected Nanorana parkeri
A: Na+/K+ ATPase. B: Ca2+ ATPase. C: Phosphofructokinase (PFK). D: Glutamate dehydrogenase (GDH). Data are presented as mean±SEM (n=6 for each season). Asterisks (**: P<0.01; ***: P<0.001) indicate a significant difference between two seasons.
DISCUSSION
To the best of our knowledge, this study provides the first evidence of changes in the liver proteome and phosphorylated proteome during hibernation in a high-altitude frog (N. parkeri). Remarkably, in hibernating frogs, regulated changes were observed in both nutrient absorption and energy metabolism processes. These changes were affected though the down-regulation of protein levels as well as the utilization of RPP to modulate energy metabolism, ion motive ATPases, protein turnover, signal transduction cascades, and alternative splicing. In addition, the expression of proteins associated with protective mechanisms (a conserved response often seen in other hypometabolic states), such as chaperone proteins and antioxidant defense enzymes, was not extensively increased in hibernating N. parkeri (Hermes-Lima et al., 1998; Wu et al., 2018). These results highlight the commonality and specificity of regulatory mechanisms between hibernating N. parkeri and other frog species.
Nutrient absorption and energy metabolism
Nanorana parkeri frogs overwinter in shallow ponds within damp cold caves for up to 6 months without feeding (Niu et al., 2021a). Thus, not only is digestion in the stomach and intestines halted, but pathways associated with nutrient absorption and processing by the liver are also down-regulated. Indeed, our results showed that the expression levels of proteins involved in pathways related to primary bile acid biosynthesis and bile secretion were significantly down-regulated in hibernating N. parkeri. This observation aligns with our earlier transcriptomic and metabolomic data, showing a significant reduction in pathway-related genes, such as ACOT8, HSD3B7, and AMACR, and cholesterol (a precursor of bile acid synthesis), in winter (Niu et al., 2023). Similar results have also been found for the Chinese alligator (Alligator sinensis) (Lin et al., 2020) and Chinese soft-shelled turtle (Pelodiscus sinensis) (Huang et al., 2021). Cellular functions involving both ATP utilization and production are inhibited in all forms of natural hypometabolism, especially “optional” cellular functions (e.g., gene expression, protein turnover, cell proliferation, growth) (Storey & Storey, 1990, 2004, 2007). In the current study, proteins with reduced expression levels in winter were predominantly associated with the oxidative phosphorylation pathway. This is in line with our prior transcriptomic study showing a down-regulation of the ATP supply pathway (oxidative phosphorylation) in the liver of winter-collected frogs (Niu et al., 2023). Similar results have also been observed in overwintering A. sinensis, in which the expression levels of most genes involved in the oxidative phosphorylation pathway are significantly reduced in winter (Lin et al., 2020).
We also found that proteins involved in several metabolic pathways showed higher phosphorylation levels during hibernation, including the oxidative phosphorylation pathway, fatty acid degradation, glycolysis/gluconeogenesis, and valine, leucine, and isoleucine degradation. Increased phosphorylation and lower activity of glycolytic enzymes have also been reported in freeze-tolerant R. sylvatica after 24 h of freezing exposure (Hawkins et al., 2019). Our results also showed that the phosphorylation of PFK and GDH enzymes was up-regulated during winter, accompanied by a simultaneous decrease in their activities. These results suggest that RPP plays an important regulatory role in suppressing metabolic enzymatic activities by altering their conformation and charge state in winter. Our previous studies also showed a lower level of urea in winter (Niu et al., 2022a, 2021c), mainly attributed to suppressed GDH activity. Fatty acid oxidation is regulated by carnitine O-acetyltransferase (CPT1A, a rate-limiting enzyme) and PPARs (nuclear transcription factors) (Barger & Kelly, 2000; Britton et al., 1997). In this study, a lower level of CPT1A and elevated phosphorylation levels of the PPAR signaling pathway suggest suppressed fatty acid oxidation during winter. Moreover, the phosphorylation levels of acetyl-CoA acetyltransferase, which plays a crucial role in the degradation of fats and proteins, and acetyl-CoA carboxylase (ACC), which is the rate-limiting enzyme for fatty acid biosynthesis, were up-regulated in winter. These findings suggest that these ATP-consuming pathways were inhibited by RPP in hibernating N. parkeri, contributing to long-term survival during food-depleted winters.
Ion motive ATPases and protein turnover
Energy-consumptive processes are strongly inhibited in a hypometabolic state, among which ion-driven ATPases are particularly metabolically expensive (Clausen, 1986). In this study, increased phosphorylation and decreased activities of Na+/K+-ATPase were observed in overwintering N. parkeri frogs, suggesting that RPP plays a crucial role in depressing ion pump activity during hibernation. Inhibition of ion motive pumps is essential in the overall reduction of ATP turnover during hibernation. Similar results have also been reported in freeze-tolerant gall fly larvae (Eurosta solidaginis), with the activity of both Na+/K+-ATPase and Ca2+-ATPase strongly reduced by reversible phosphorylation in midwinter compared to early autumn (McMullen et al., 2010; McMullen & Storey, 2008).
As another major energy consumer in cells, protein turnover (both synthesis and degradation) is often suppressed when animals are subjected to stress or nutrient/energy limitations in hypometabolic systems (Degracia et al., 2002; Storey & Storey, 2004). In the present study, the phosphorylation levels of proteins associated with RNA transport, endoplasmic reticulum protein processing, and protein export were significantly up-regulated during winter, indicating that protein synthesis is inhibited by RPP. Previous research has indicated that RPP plays a pivotal role in regulating protein synthesis, particularly through ribosome initiation and elongation factor activities (Storey & Storey, 1990, 2004). Key sites for regulation include ribosome initiation factor 2 (eIF2), eIF4, and elongation factor 2 (eEF2). The relative content of phosphorylated eIF-2α is considered a sensitive indicator of protein synthesis activity in all eukaryotes (Storey & Storey, 2010). Here, the phosphorylation levels of many translation initiation factors and elongation factors, including eIF-3α, eIF-3f, eIF-2α, eIF-2β, eIF-2C, elongation factor 1 α/β/δ chain, and eEF2, were significantly increased in hibernating N. parkeri. Similarly, previous research has shown that the relative amount of phosphorylated eIF-2α is increased in the liver of aestivating desert frogs (Neobatrachus sutor) (Pakay et al., 2003). Protein degradation is also likely to be suppressed due to the strong inhibition of protein synthesis during dormancy, as reported in several hypometabolic systems (Storey & Storey, 2004). In this study, ubiquitin-mediated proteolysis exhibited increased phosphorylation during winter, suggesting that protein degradation is also inhibited by RPP in hibernating N. parkeri.
Cytoprotective mechanisms
Chaperone proteins, also known as heat shock proteins (HSPs), play important roles in the protection, renewal, and modification of the proteome to sustain cellular functions under hypometabolic states (Storey & Storey, 2004). Higher levels of HSPs have been observed in wood frogs during hibernation, facilitating the refolding and preventing the aggregation of denatured proteins (Kiss et al., 2011). In contrast, our results showed that molecular chaperones (stress-70 protein, HSP 90-β, DnaJ homolog subfamily B member 6) were significantly down-regulated in winter. Concordantly, our previous transcriptomic study showed a significant down-regulation of genes encoding HSPs, such as HSPD1, HSPE1, TRAP1, and HSPH1 (Niu et al., 2023). This reduced expression of HSPs during hibernation may be a consequence of metabolic inhibition. Similar findings have been reported in water frogs (Pelophylax ridibundus) (Feidantsis et al., 2012), with the lowest levels of HSP expression observed during hibernation. Enhancing antioxidant defenses is another prevalent feature of hypometabolism across phylogeny (Hermes-Lima et al., 1998; Storey & Storey, 2007), serving to mitigate oxidative damage to macromolecules during hibernation and recovery. We found that the phosphorylation levels of superoxide dismutase (SOD1), catalase, and glutathione S-transferase were significantly increased in winter. Although no significant changes in protein abundance levels were detected, enzymatic activities were markedly reduced, as also shown in our previous study (Niu et al., 2018). These results suggest that RPP plays a critical role in down-regulating the activity of antioxidant enzymes. Moreover, glutaredoxin 2, thioredoxin (TXN2), and glucose-6-phosphate dehydrogenase, which are implicated in antioxidant defense (Lillig et al., 2008; Lu & Holmgren, 2014), were significantly down-regulated in hibernating N. parkeri, providing evidence of reduced antioxidant defense. However, the genes encoding these antioxidants did not change significantly at the mRNA level. The inconsistency between the results obtained at the mRNA and protein level may be attributed to post-transcriptional regulation, which needs to be verified in future studies.
Evidence indicates that frogs exhibit reduced immune capabilities during winter, as mounting an effective immune response is energetically demanding (Zapata et al., 1992). In contrast, our previous study suggested an enhanced level of innate immunity in winter may be needed to protect frogs when sequestered in cold water hibernacula (Niu et al., 2022a). Notably, the up-regulated proteins identified in this study and up-regulated genes identified in our prior study (Niu et al., 2023) were significantly enriched in immune response-related pathways. Similar findings have been reported in juvenile Chinese soft-shelled turtles (Pelodiscus sinensis), in which cold-induced stress stimulates immune responses in the spleen and intestine, enhancing the complement pathway via up-regulation of the C1S, C3, and C6 genes (Zhang et al., 2018). Hence, our results are tentatively consistent with the winter immunity enhancement hypothesis (Nelson & Demas, 1996), although further studies of innate immune parameters, such as complement levels, phagocytes, pattern recognition receptors, natural killer cells, skin antimicrobial peptides, cytokines, chemokines, macrophages, and dendritic cells, are warranted.
Signal transduction pathways
Cell signaling is crucial for reorganizing multiple metabolic pathways and achieving a hypometabolic state (MacDonald, 2004; Storey & Storey, 2012). HIF-1 is the main transcriptional mediator of the hypoxic response in eukaryotic cells, activating expression of target genes involved in angiogenesis, oxygen transport, iron metabolism, glycolysis, glucose uptake, and apoptosis (Bárdos & Ashcroft, 2005; Semenza, 2001). Increasing evidence suggests that HIF-1α regulation occurs at the protein level and accumulates under hypoxic conditions (Kallio et al., 1997). In the present study, we observed higher levels of HIF-1α in overwintering frogs, suggesting potential hypoxia, consistent with our previous findings on the physiological ecology of hibernation in this species (Niu et al., 2022a). Moreover, higher phosphorylation levels of proteins were significantly enriched in the HIF-1 signaling pathway in the winter. HIF-1 plays an important role in inducing the expression of genes encoding glycolytic enzymes, vascular endothelial growth factor, and erythropoietin (Semenza, 2001). However, in our study, the expression of these genes showed no significant changes in hibernating N. parkeri. Therefore, we hypothesized that the regulation of HIF-1 by phosphorylation modification does not necessarily activate the expression of downstream targets. HIF-1 is also regulated by other factors, such as stabilization of the HIF-1α subunit, redox regulation, and interaction with HSP90 or selected coactivators (Semenza, 2001). The AMP-activated protein kinase (AMPK), a highly conserved serine/threonine protein kinase, is thought to be a sensor of cellular energy status and is activated by phosphorylation in response to low levels of intracellular ATP and/or high levels of AMP (Hardie et al., 2012). In the present study, AMPK was hyperphosphorylated in winter. The hyperphosphorylation of AMPK in winter has also been reported in hibernating leeches (Whitmania pigra) (Shi et al., 2020). To maintain energy homeostasis, AMPK phosphorylates downstream targets to turn on the ATP-producing pathway and turn off ATP-consuming processes such as lipid biosynthesis by phosphorylating ACC or protein synthesis by inactivating eEF2 (Hardie, 2007; Horman et al., 2002). Indeed, the phosphorylation levels of both ACC and eEF2 were up-regulated in hibernating N. parkeri, contributing to the down-regulation of lipid biosynthesis and inhibition of translational processes. Numerous studies have shown that AMPK is activated in a hypometabolic state (Bartrons et al., 2004; Rider, 2016). Our results also validate the role of AMPK in mediating metabolic depression through the phosphorylation of downstream targets.
Alternative splicing
Alternative splicing can generate multiple mRNA transcripts from a single mRNA precursor, greatly diversifying the transcriptome and proteome, and thereby altering protein activity, subcellular localization, and protein-protein interactions (Matlin et al., 2005). Alternative splicing also plays an important role in phenotypic plasticity in response to temperature at the cellular level. For instance, GO enrichment analyses have revealed that genes showing differential expression in fish under chronic low temperatures are enriched in spliceosomes (Healy & Schulte, 2019). In line with this, our results suggest that higher phosphorylation of spliceosomal proteins may facilitate an increase in alternative splicing events during cold shock in hibernating N. parkeri. Indeed, our previous transcriptomic study showed a significantly higher occurrence of alternative splicing events in winter compared to summer, including alternative 5’ first exon (transcription start site) the first exon splicing, alternative 3’ last exon (transcription terminal site) the last exon splicing, skipped exon single exon skipping, approximate SKIP single exon skipping (fuzzy boundary), approximate MSKIP multi-exon skipping (fuzzy boundary), and alternative exon ends (5’, 3’, or both) (Niu et al., 2023).
CONCLUSIONS
In summary, we performed comprehensive proteomic and phosphoproteomic profiling of the liver from summer- and winter-collected N. parkeri. Our study demonstrated significant down-regulation in various proteins that participate in active metabolic processes associated with warm-weather life (e.g., growth, reproduction, food acquisition, chaperones, and antioxidant defenses) as well as an up-regulation in proteins associated with the immune signaling pathways in hibernating N. parkeri. Moreover, we identified RPP as a crucial regulatory mechanism, which controlled the activities of metabolic enzymes and cellular signaling cascades, regulated transcription via mediation of alternative splicing, and inhibited energy-consuming processes, such as ion-motive ATPases and protein turnover, in hibernating N. parkeri. Collectively, our results provide insights into the genetic and epigenetic mechanisms underlying hibernation in a high-altitude frog species.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Y.G.N. and D.B.W. conceived and designed the project. Y.G.N., X.J.Z., T.S.X., X.Y.L., H.Y.Z., and Z.F.A. performed the experiments. Y.G.N. and X.J.Z. analyzed the data. T.S.X. and X.Y.L. collected the samples. Y.G.N. and X.J.Z. prepared the first draft of the manuscript. D.B.W., K.B.S., and Q.C. reviewed and edited the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We are grateful to Ruo-Yu Yang and Shi-Han Guo for their help in this study.
Funding Statement
This work was supported by the National Natural Science Foundation of China (32001110), Training Program for Cultivating High-level Talents by the China Scholarship Council (2021lxjjw01), and Open Project of State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University (2021-KF-004)
DATA AVAILABILITY
The mass spectrometry proteomic and phosphoproteomic data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD042165). The dataset is deposited in the Science Data Bank (https://www.scidb.cn/) under DOI: 10.57760/sciencedb.j00139.00079.
References
- Bárdos JI, Ashcroft M Negative and positive regulation of HIF-1: a complex network. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2005;1755(2):107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Barger PM, Kelly DP PPAR signaling in the control of cardiac energy metabolism. Trends in Cardiovascular Medicine. 2000;10(6):238–245. doi: 10.1016/S1050-1738(00)00077-3. [DOI] [PubMed] [Google Scholar]
- Bartrons M, Ortega E, Obach M, et al. 2004. Activation of AMP-dependent protein kinase by hypoxia and hypothermia in the liver of frog Rana perezi. Cryobiology, 49(2): 190–194.
- Boutilier RG, Donohoe PH, Tattersall GJ, et al Hypometabolic homeostasis in overwintering aquatic amphibians. Journal of Experimental Biology. 1997;200(2):387–400. doi: 10.1242/jeb.200.2.387. [DOI] [PubMed] [Google Scholar]
- Britton CH, Mackey DW, Esser V, et al Fine chromosome mapping of the genes for human liver and muscle carnitine palmitoyltransferase I (CPT1A and CPT1B) Genomics. 1997;40(1):209–211. doi: 10.1006/geno.1996.4539. [DOI] [PubMed] [Google Scholar]
- Capraro A, O’meally D, Waters SA, et al Waking the sleeping dragon: gene expression profiling reveals adaptive strategies of the hibernating reptile Pogona vitticeps. BMC Genomics. 2019;20(1):460. doi: 10.1186/s12864-019-5750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T Regulation of active Na+-K+ transport in skeletal muscle. Physiological Reviews. 1986;66(3):542–580. doi: 10.1152/physrev.1986.66.3.542. [DOI] [PubMed] [Google Scholar]
- Dawson NJ, Storey KB A hydrogen peroxide safety valve: the reversible phosphorylation of catalase from the freeze-tolerant North American wood frog. Rana sylvatica. Biochimica et Biophysica Acta (BBA)-General Subjects. 2016;1860(3):476–485. doi: 10.1016/j.bbagen.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Degracia DJ, Kumar R, Owen CR, et al Molecular pathways of protein synthesis inhibition during brain reperfusion: implications for neuronal survival or death. Journal of Cerebral Blood Flow & Metabolism. 2002;22(2):127–141. doi: 10.1097/00004647-200202000-00001. [DOI] [PubMed] [Google Scholar]
- Dieni CA, Storey KB Creatine kinase regulation by reversible phosphorylation in frog muscle. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology. 2009;152(4):405–412. doi: 10.1016/j.cbpb.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Dieni CA, Storey KB Regulation of glucose-6-phosphate dehydrogenase by reversible phosphorylation in liver of a freeze tolerant frog. Journal of Comparative Physiology B. 2010;180(8):1133–1142. doi: 10.1007/s00360-010-0487-5. [DOI] [PubMed] [Google Scholar]
- Dieni CA, Storey KB Regulation of hexokinase by reversible phosphorylation in skeletal muscle of a freeze-tolerant frog. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology. 2011;159(4):236–243. doi: 10.1016/j.cbpb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Donohoe PH, Boutilier RG The protective effects of metabolic rate depression in hypoxic cold submerged frogs. Respiration Physiology. 1998;111(3):325–336. doi: 10.1016/S0034-5687(97)00125-4. [DOI] [PubMed] [Google Scholar]
- Feidantsis K, Anestis A, Vasara E, et al Seasonal variations of cellular stress response in the heart and gastrocnemius muscle of the water frog (Pelophylax ridibundus) Comparative Biochemistry and Physiology Part A:Molecular & Integrative Physiology. 2012;162(4):331–339. doi: 10.1016/j.cbpa.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Hardie DG AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nature Reviews Molecular Cell Biology. 2007;8(10):774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Reviews Molecular Cell Biology. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins LJ, Wang MJ, Zhang BW, et al Glucose and urea metabolic enzymes are differentially phosphorylated during freezing, anoxia, and dehydration exposures in a freeze tolerant frog. Comparative Biochemistry and Physiology Part D:Genomics and Proteomics. 2019;30:1–13. doi: 10.1016/j.cbd.2019.01.009. [DOI] [PubMed] [Google Scholar]
- Healy TM, Schulte PM Patterns of alternative splicing in response to cold acclimation in fish. Journal of Experimental Biology. 2019;222(5):jeb193516. doi: 10.1242/jeb.193516. [DOI] [PubMed] [Google Scholar]
- Hermes-Lima M, Storey JM, Storey KB Antioxidant defenses and metabolic depression. The hypothesis of preparation for oxidative stress in land snails. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology. 1998;120(3):437–448. doi: 10.1016/S0305-0491(98)10053-6. [DOI] [PubMed] [Google Scholar]
- Horman S, Browne GJ, Krause U, et al Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Current Biology. 2002;12(16):1419–1423. doi: 10.1016/S0960-9822(02)01077-1. [DOI] [PubMed] [Google Scholar]
- Huang YF, Yang S, Bai XB, et al Molecular and cellular mechanisms of lipid droplet breakdown in the liver of Chinese soft-shelled turtle (Pelodiscus sinensis) Frontiers in Marine Science. 2021;8:633425. doi: 10.3389/fmars.2021.633425. [DOI] [Google Scholar]
- Jin L, Yu JP, Yang ZJ, et al Modulation of gene expression in liver of hibernating Asiatic toads (Bufo gargarizans) International Journal of Molecular Sciences. 2018;19(8):2363. doi: 10.3390/ijms19082363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio PJ, Pongratz I, Gradin K, et al Activation of hypoxia-inducible factor 1α: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(11):5667–5672. doi: 10.1073/pnas.94.11.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AJ, Muir TJ, Lee Jr RE, et al Seasonal variation in the hepatoproteome of the dehydration- and freeze-tolerant wood frog. Rana sylvatica. International Journal of Molecular Sciences. 2011;12(12):8406–8414. doi: 10.3390/ijms12128406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DJ, Middle L, Vu H, et al Wood frog adaptations to overwintering in Alaska: new limits to freezing tolerance. Journal of Experimental Biology. 2014;217(12):2193–2200. doi: 10.1242/jeb.101931. [DOI] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Holmgren A Glutaredoxin systems. Biochimica et Biophysica Acta (BBA)-General Subjects. 2008;1780(11):1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Lin JQ, Huang YY, Bian MY, et al A unique energy-saving strategy during hibernation revealed by multi-omics analysis in the Chinese alligator. iScience. 2020;23(6):101202. doi: 10.1016/j.isci.2020.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Holmgren A The thioredoxin antioxidant system. Free Radical Biology and Medicine. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- Lu Y, Li XY, Zhao K, et al Proteomic and phosphoproteomic profiling reveals the oncogenic role of protein kinase D family kinases in cholangiocarcinoma. Cells. 2022;11(19):3088. doi: 10.3390/cells11193088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Lu X, Merilä J Altitudinal decline of body size in a Tibetan frog. Journal of Zoology. 2009;279(4):364–371. doi: 10.1111/j.1469-7998.2009.00627.x. [DOI] [Google Scholar]
- MacDonald JA. 2004. Signal transduction pathways and the control of cellular responses to external stimuli. In: Storey KB. Functional Metabolism: Regulation and Adaptation. New York: Wiley-Liss, 87–123.
- Matlin AJ, Clark F, Smith CWJ Understanding alternative splicing: towards a cellular code. Nature Reviews Molecular Cell Biology. 2005;6(5):386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- McMullen DC, Ramnanan CJ, Storey KB In cold-hardy insects, seasonal, temperature, and reversible phosphorylation controls regulate sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) Physiological and Biochemical Zoology. 2010;83(4):677–686. doi: 10.1086/653489. [DOI] [PubMed] [Google Scholar]
- McMullen DC, Storey KB Suppression of Na+K+-ATPase activity by reversible phosphorylation over the winter in a freeze-tolerant insect. Journal of Insect Physiology. 2008;54(6):1023–1027. doi: 10.1016/j.jinsphys.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Demas GE Seasonal changes in immune function. The Quarterly Review of Biology. 1996;71(4):511–548. doi: 10.1086/419555. [DOI] [PubMed] [Google Scholar]
- Niu YG, Cao WJ, Storey KB, et al Metabolic characteristics of overwintering by the high-altitude dwelling Xizang plateau frog. Nanorana parkeri. Journal of Comparative Physiology B. 2020;190(4):433–444. doi: 10.1007/s00360-020-01275-4. [DOI] [PubMed] [Google Scholar]
- Niu YG, Cao WJ, Wang JZ, et al Freeze tolerance and the underlying metabolite responses in the Xizang plateau frog. Nanorana parkeri. Journal of Comparative Physiology B. 2021a;191(1):173–184. doi: 10.1007/s00360-020-01314-0. [DOI] [PubMed] [Google Scholar]
- Niu YG, Cao WJ, Zhao YF, et al. 2018. The levels of oxidative stress and antioxidant capacity in hibernating Nanorana parkeri. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 219–220: 19–27.
- Niu YG, Chen Q, Storey KB, et al. 2022a. Physiological ecology of winter hibernation by the high-altitude frog Nanorana parkeri. Physiological and Biochemical Zoology, 95(3): 201–211.
- Niu YG, Zhang XJ, Men SK, et al Integrated analysis of transcriptome and metabolome data reveals insights for molecular mechanisms in overwintering Tibetan frogs. Nanorana parkeri. Frontiers in Physiology. 2023;13:1104476. doi: 10.3389/fphys.2022.1104476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YG, Zhang XJ, Xu TS, et al Physiological and biochemical adaptations to high altitude in Tibetan frogs. Nanorana parkeri. Frontiers in Physiology. 2022b;13:942037. doi: 10.3389/fphys.2022.942037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YG, Zhang XJ, Zhang HY, et al Antioxidant and non-specific immune defenses in partially freeze-tolerant Xizang plateau frogs. Nanorana parkeri. Journal of Thermal Biology. 2021b;102:103132. doi: 10.1016/j.jtherbio.2021.103132. [DOI] [PubMed] [Google Scholar]
- Niu YG, Zhang XJ, Zhang HY, et al Metabolic responses of plasma to extreme environments in overwintering Tibetan frogs Nanorana parkeri: a metabolome integrated analysis. Frontiers in Zoology. 2021c;18(1):41. doi: 10.1186/s12983-021-00428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakay JL, Hobbs AA, Kimball SR, et al The role of eukaryotic initiation factor 2α during the metabolic depression associated with estivation. Journal of Experimental Biology. 2003;206(14):2363–2371. doi: 10.1242/jeb.00422. [DOI] [PubMed] [Google Scholar]
- Pinder AW, Storey KB, Ultsch GR. 1992. Estivation and hibernation. In: Feder ME, Burggren WW. Environmental Physiology of the Amphibians. Chicago: University of Chicago Press, 250–274.
- Rider MH Role of AMP-activated protein kinase in metabolic depression in animals. Journal of Comparative Physiology B. 2016;186(1):1–16. doi: 10.1007/s00360-015-0920-x. [DOI] [PubMed] [Google Scholar]
- Semenza GL HIF-1 and mechanisms of hypoxia sensing. Current Opinion in Cell Biology. 2001;13(2):167–171. doi: 10.1016/S0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- Shi HZ, Wang J, Liu F, et al Proteome and phosphoproteome profiling reveals the regulation mechanism of hibernation in a freshwater leech (Whitmania pigra) Journal of Proteomics. 2020;229:103866. doi: 10.1016/j.jprot.2020.103866. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM Metabolic rate depression and biochemical adaptation in anaerobiosis, hibernation and estivation. The Quarterly Review of Biology. 1990;65(2):145–174. doi: 10.1086/416717. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM Metabolic rate depression in animals: transcriptional and translational controls. Biological Reviews. 2004;79(1):207–233. doi: 10.1017/S1464793103006195. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM Tribute to P. L. Lutz: putting life on `pause'–molecular regulation of hypometabolism. Journal of Experimental Biology. 2007;210(10):1700–1714. doi: 10.1242/jeb.02716. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. 2010. Metabolic regulation and gene expression during aestivation. In: Navas CA, Carvalho JE. Aestivation. Berlin: Springer, 25–45.
- Storey KB, Storey JM Aestivation: signaling and hypometabolism. Journal of Experimental Biology. 2012;215(9):1425–1433. doi: 10.1242/jeb.054403. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM Molecular biology of freezing tolerance. Comprehensive Physiology. 2013;3(3):1283–1308. doi: 10.1002/cphy.c130007. [DOI] [PubMed] [Google Scholar]
- Sun HJ, Zuo XB, Sun L, et al Insights into the seasonal adaptive mechanisms of Chinese alligators (Alligator sinensis) from transcriptomic analyses. Australian Journal of Zoology. 2018;66(2):93–102. doi: 10.1071/ZO18005. [DOI] [Google Scholar]
- Sun YB, Xiong ZJ, Xiang XY, et al Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(11):E1257–E1262. doi: 10.1073/pnas.1501764112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall GJ, Ultsch GR Physiological ecology of aquatic overwintering in ranid frogs. Biological Reviews. 2008;83(2):119–140. doi: 10.1111/j.1469-185X.2008.00035.x. [DOI] [PubMed] [Google Scholar]
- Wu CW, Tessier SN, Storey KB. 2018. Stress-induced antioxidant defense and protein chaperone response in the freeze-tolerant wood frog Rana sylvatica. Cell Stress and Chaperones, 23(6): 1205–1217.
- Zapata AG, Varas A, Torroba M Seasonal variations in the immune system of lower vertebrates. Immunology Today. 1992;13(4):142–147. doi: 10.1016/0167-5699(92)90112-K. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Zhang XJ, Xu TS, et al Effects of acute heat exposure on oxidative stress and antioxidant defenses in overwintering frogs. Nanorana parkeri. Journal of Thermal Biology. 2022a;110:103355. doi: 10.1016/j.jtherbio.2022.103355. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Cai RQ, Liang JJ, et al Molecular mechanism of Chinese alligator (Alligator sinensis) adapting to hibernation. Journal of Experimental Zoology Part B:Molecular and Developmental Evolution. 2021;336(1):32–49. doi: 10.1002/jez.b.23013. [DOI] [PubMed] [Google Scholar]
- Zhang WY, Niu CJ, Chen BJ, et al Digital gene expression profiling reveals transcriptional responses to acute cold stress in Chinese soft-shelled turtle Pelodiscus sinensis juveniles. Cryobiology. 2018;81:43–56. doi: 10.1016/j.cryobiol.2018.02.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Q, Liang JQ et al Serum proteomic analysis of differentially expressed proteins and pathways involved in the mechanism of endemic osteoarthritis. Molecular Omics. 2022b;18(8):745–753. doi: 10.1039/D2MO00154C. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The mass spectrometry proteomic and phosphoproteomic data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD042165). The dataset is deposited in the Science Data Bank (https://www.scidb.cn/) under DOI: 10.57760/sciencedb.j00139.00079.