Abstract
The efficiency of translation termination is influenced by local contexts surrounding stop codons. In Saccharomyces cerevisiae, upstream and downstream sequences act synergistically to influence the translation termination efficiency. By analysing derivatives of a leaky stop codon context, we initially demonstrated that at least six nucleotides after the stop codon are a key determinant of readthrough efficiency in S. cerevisiae. We then developed a combinatorial-based strategy to identify poor 3′ termination contexts. By screening a degenerate oligonucleotide library, we identified a consensus sequence –CA(A/G)N(U/C/G)A–, which promotes >5% readthrough efficiency when located downstream of a UAG stop codon. Potential base pairing between this stimulatory motif and regions close to helix 18 and 44 of the 18S rRNA provides a model for the effect of the 3′ stop codon context on translation termination.
INTRODUCTION
Translation termination is determined by the three stop codons (UAA, UGA and UAG). The termination process is very efficient and errors occur at a level estimated at 0.3% in yeast (O. Namy, unpublished data). Readthrough occurs by the incorporation of an amino acid due to the decoding of the termination codon by a natural tRNA. One major determinant known to affect the efficiency of translation termination is the local sequence surrounding the termination codon. The effect of the upstream sequence is well documented in Escherichia coli and Saccharomyces cerevisiae (Mottagui-Tabar et al., 1994, 1998). In S. cerevisiae, the effect of the –2 codon has been correlated with the charge of the corresponding amino acid residue. In contrast, it has been suggested that the effect of the –1 codon is mediated by the identity of the peptidyl-tRNA (Mottagui-Tabar et al., 1998).
Numerous results indicate that the nucleotide immediately following the stop codon (defined as +4) is a crucial determinant of termination efficiency (Poole et al., 1995; Tate et al., 1996). A large-scale database analysis has shown a non-random distribution of this nucleotide, both in prokaryotes and eukaryotes (Dalphin et al., 1997), and crosslinking experiments have revealed that it interacts with the prokaryotic termination factor RF2 (Poole et al., 1998). These observations led to the proposal that the translation termination signal is constituted of at least 4 nucleotides (nt) (Brown et al., 1990). In yeast, the available data indicate that the three initial distal nucleotides play an important role in readthrough efficiency (Bonetti et al., 1995). Bedwell and co-workers identified the CAAUAGCAA readthrough motif as a leaky mutation in the STE6 gene and showed that upstream and downstream CAA act synergistically to promote a high readthrough level (Bonetti et al., 1995). This motif is also part of the tobacco mosaic virus (TMV) readthrough site, responsible for the synthesis of the replicase domain of the virus (Skuzeski et al., 1991).
We have analysed a readthrough sequence that contains the sequence CAAUAGCAA, but promotes a 10-fold lower readthrough efficiency than that of the STE6 and TMV sequences. To characterize the determinants responsible for this discrepancy, we performed a systematic analysis of the stop codon context involved in readthrough in yeast. We first demonstrated, by directed mutagenesis, that nucleotides +7, +8 and +9 (relative to the first base of the stop codon) are a key determinant of readthrough efficiency. We then developed a new in vivo reporter system based on the ADE2 gene, to easily identify recoding sequences in yeast. By screening a degenerated oligonucleotide library, we identified eight new sequences giving readthrough efficiencies >5%. This allowed us to define a consensus motif with some very biased positions. Potential base pairing between the most efficient 3′ sequence and two regions of the ribosomal 18S rRNA provides a model to explain the role of this stimulatory motif during the readthrough process.
RESULTS
The key determinant for readthrough is located at the 3′ side of the stop codon
In a screen aimed at finding genes harbouring leaky stop codons, we looked for the presence of the CAA STOP CAA motif in the yeast genome (O. Namy and J.-P. Rousset, submitted). Among several candidates, we identified the CAAp sequence bearing the CAAUAGCAA motif (see Table I). In order to precisely quantify the readthrough level driven by this sequence, it was sub-cloned into the pAC99 vector (Bidou et al., 2000). This vector carries a dual lacZ-luc reporter gene permitting accurate quantification of various recoding events. The readthrough level obtained with the pAC-CAAp was compared with that obtained with the pAC-TMV, which contains 9 nt from the TMV readthrough motif on each side of the stop codon (Stahl et al., 1995). As shown in Table I, the CAAp sequence directed a 10-fold lower readthrough level than the TMV sequence. This suggests that an extended context is required to achieve high readthrough in yeast.
Table I. The stimulatory sequence is not the –2 codon.
| Sequencea | Readthrough efficiency (%)b | Vector name | ||||||
|---|---|---|---|---|---|---|---|---|
| GGA | ACA | CAA | TAG | CAA | TTA | CAG | 23 | (pAC-TMV) |
| G | T | Q | * | Q | L | Q | ||
| AAA | CCA | CAA | TAG | CAA | GAA | TAT | 2.2 | (pAC-CAAp)c |
| K | P | Q | * | Q | E | Y | ||
| --- | TCA | --- | --- | --- | --- | --- | 2.4 | (pAC-CAAs) |
| --- | S | --- | --- | --- | --- | --- | ||
| --- | GCA | --- | --- | --- | --- | --- | 2.1 | (pAC-CAAa) |
| --- | A | --- | --- | --- | --- | --- | ||
| --- | ACA | --- | --- | --- | --- | --- | 2.2 | (pAC-CAAt) |
| --- | T | --- | --- | --- | --- | --- | ||
aSequences derived from the CAAp motif. The amino acids are presented below the nucleotide sequence. The modified nucleotide is indicated in bold, while – indicates unchanged nucleotides.
bThe assays were carried out using at least three independent transformants. The standard error of the mean of all data presented in this article is <10%.
cThis sequence has been identified in the PDE2 gene of S. cerevisiae (O. Namy, unpublished).
TMV and CAAp constructs bear different 5′ and 3′ contexts beyond the core CAAUAGCAA region (see Table I). One or several of these differences should thus be responsible for the high variation of readthrough efficiency observed. We began by the replacement of the first base of the –2 codon in the 5′ context of CAAp sequence by each of the other nucleotides. Replacing the CCA –2 codon by UCA, GCA or ACA had no effect on readthrough (Table I). This demonstrates that, in this case, changing the –2 codon and the –2 amino acid does not affect readthrough. It is interesting to note that the ACA codon, although found at the –2 position in the TMV sequence, also has no effect (Table I, compare lanes 1, 2 and 5).
The two motifs allowing >10% of readthrough in yeast, described by Bedwell and co-workers (Fearon et al., 1994; Bonetti et al., 1995), and the TMV motif share the presence of a glycine codon at the –3 position (see Table II). Since the nascent polypeptide is known to interfere with termination accuracy, we replaced the lysine (AAA) codon, present in CAAp, by the GGA glycine codon. As shown in Table II, this change had no effect on readthrough efficiency. This latter construct contains the same 5′ sequence as the TMV construct (Table II) but drives a readthrough efficiency equivalent to that promoted by the CAAp sequence. This result demonstrates that determinants of readthrough are not within three codons upstream of the stop codon.
Table II. Glycine at the –3 position does not influence readthrough.
| Sequencea | Readthrough efficiency (%) | Vector name | ||||||
|---|---|---|---|---|---|---|---|---|
| GGC | GGG | CAA | TAG | CAA | AGA | GT? | 10 | (pAC-STE6)b |
| G | G | Q | * | Q | R | V | ||
| GGA | TCT | CAA | TAG | CAA | GCA | AGC | 16 | (pAC-QXQ)b |
| G | S | Q | * | Q | A | S | ||
| GGA | ACA | CAA | TAG | CAA | TTA | CAG | 23 | (pAC-TMV) |
| G | T | Q | * | Q | L | Q | ||
| AAA | CCA | CAA | TAG | CAA | GAA | TAT | 2.2 | (pAC-CAAp) |
| K | P | Q | * | Q | E | Y | ||
| GGA | ACA | --- | --- | --- | --- | --- | 2.3 | (pAC-gga-2) |
| G | T | |||||||
aNucleotides changed are indicated in bold. ? indicates unknown nucleotide corresponding to cloning site.
bSTE6 and QXQ sequences have been described by Bedwell’s group (Fearon et al., 1994; Bonetti et al., 1995).
We then tested the influence of 3′ sequences on readthrough. The results, presented in Table III, show that an exchange UUA–GAA (+7, +8, +9 nt) reduced readthrough frequency 10-fold. This corresponds to the difference observed between TMV and CAAp sequences (Table I). Symmetrically, changing GAA to UUA in the CAAp sequence increased readthrough efficiency 10-fold (Table III, compare lanes 4 and 5). A CUU–UUA change in the CRc sequence, which carries a UAA stop codon and a 5′ context different from the TMV, also increased readthrough >10-fold (Table V). The stimulation mediated by the +7, +8 and +9 nt is thus independent of the nature of the stop codon and of the 5′ nucleotide context. The replacement of the CAG (+10, +11, +12 nt) sequence from TMV by the UAU sequence from CAAp also had a 2-fold effect on readthrough (Table III). Considered as a whole, these results demonstrate that nucleotides +7 or +8 are major determinants of translation termination efficiency in yeast, and that nucleotides up to position +12 also have a significant effect.
Table III. Nucleotides +7, +8 and +9 have a 10-fold effect on readthrough.
| Sequencea | Readthrough efficiency (%) | Vector name | ||||||
|---|---|---|---|---|---|---|---|---|
| +1 | +4 | +7 | +10 | |||||
| GCA | ACA | CAA | TAG | CAA | TTA | CAG | 30 | (pAC-TMV) |
| G | T | Q | * | Q | L | Q | ||
| --- | --- | --- | --- | --- | GAA | --- | 3 | (pAC-GAA) |
| E | ||||||||
| --- | --- | --- | --- | --- | --- | TAT | 14 | (pAC-TAT) |
| Y | ||||||||
| AAA | CCA | CAA | TAG | CAA | GAA | TAT | 2 | (pAC-CAAP) |
| K | P | Q | * | Q | E | Y | ||
| --- | --- | --- | --- | --- | TTA | --- | 22 | (pAC-TTA) |
| L | ||||||||
aNucleotide position (relative to the first base of the stop codon) is indicated.
Table V. The effect of the 3′ sequence is not correlated to properties of tRNAUAA.
| Sequence | Readthrough efficiency (%)a |
Vector name | |||||||
|---|---|---|---|---|---|---|---|---|---|
| –tRNAAAGLeu | +tRNAAAGLeu | ||||||||
| +1 | +4 | +7 | +10 | ||||||
| GGA | ACA | CAA | TAG | CAA | TTA | CAG | 30 | ND | (pAC-TMV) |
| G | T | Q | * | Q | L | Q | |||
| --- | --- | --- | --- | --- | CTT | --- | 3.5 | ND | (pAC-CTT) |
| L | |||||||||
| ACG | ACG | ATA | TAA | CAA | CTT | AAA | 0.8 | 0.9 | (pAC-CRc)b |
| T | T | I | * | Q | L | K | |||
| --- | --- | --- | --- | --- | TTA | --- | 9 | 9.1 | (pAC-CRctta) |
aThe UUA anticodon of tRNAUAALeu was modified to AAG. This new tRNAAAG is able to recognize the CUU codon and is functional in yeast (data not shown). The expression of tRNAAAG does not reproduce the effect of the replacement of CUU by the UUA triplet.
bThis sequence has been identified by O. Namy and J.-P. Rousset (unpublished).
Design and use of a combinatorial approach to isolate readthrough motifs
In order to identify 3′ readthrough motifs stimulating readthrough, we then developed a combinatorial approach. The principle was to introduce a library of oligonucleotides, centred on a stop codon and carrying a 3′ degenerated context, in a reporter gene whose activity could be monitored in living cells. We used the ADE2 gene, encoding the P-ribosyl-amino-imidazole-carboxylase (EC 4.1.1.21). Its inactivation results in the accumulation of a red pigment in the cell, and it has been widely used as a suppressible marker in yeast (Silhankova, 1972).
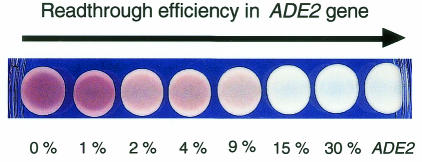
The ADE2 gene and its promoter were cloned in a centromeric URA3 vector (pFL38) (see Methods). To test the ability of the ADE2 system to report readthrough in vivo, we cloned different motifs promoting readthrough efficiencies ranging from 1 to 30%, in the unique HpaI restriction site located within the coding sequence of the ADE2 gene. Each construct was introduced into the FS1 strain. This strain is derived from Y349 and carries a frameshift point mutation in ADE2 (ade2-592), which completely abolishes Ade2p activity (Engebrecht and Roeder, 1990). The results, shown in Figure 1, defined three categories of clones: red clones, corresponding to targets that drive readthrough efficiencies lower than 2%; pink clones, bearing targets that drive between 2 and 10% of readthrough; and white clones, whose targets promote >15% readthrough efficiency. This demonstrates that the ADE2 reporter is suitable for screening large-scale libraries for readthrough motifs.
Fig. 1. Readthrough visualization. Different readthrough motifs, ranging from 1 to 30%, were introduced in the ADE2 gene cloned into a pFL38 plasmid. These plasmids were used to transform the FS1 strain. Exponentially growing cells were spotted on plates (1 × 106 cells/ml) and colour was checked after 3 days at 30°C.
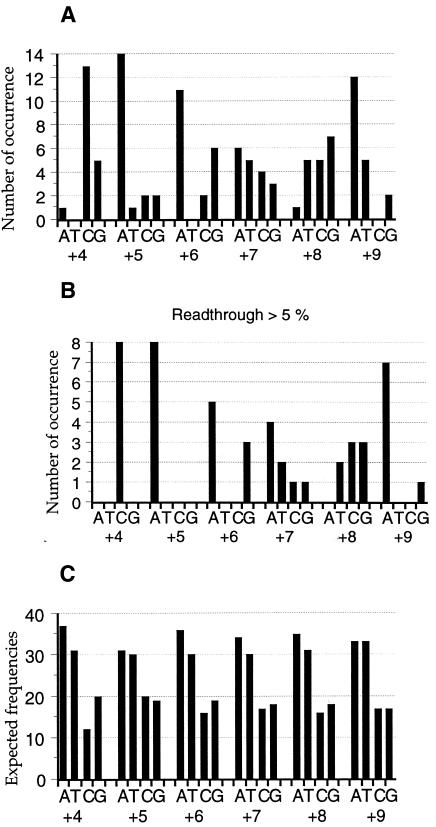
We cloned a library of degenerate sequences, concerning six positions following the stop codon, and derived from the oligonucleotide 5′-GGAACACAATAGNNNNNNCAG-3′. This library was introduced into the FS1 strain. After 3 days of growth at 30°C, 75 clones among 15 000 yeast transformants displayed a pink or white colour, indicating the presence of a readthrough motif. To confirm that the mutant ADE2 gene on the plasmid was responsible for the colour modification, each clone was grown on 5-FOA medium, which counter-selects cells carrying a wild-type URA3 gene and allows isolation of colonies having lost the vector. In each case, the loss of the plasmid reversed the colony colour, demonstrating that the phenotype was in fact due to the presence of the plasmid (data not shown). Each of the 75 plasmids was isolated and the stop codon region sequenced. Table IV exhibits the 3′ sequences identified and the number of vectors carrying each of these motifs. These sequences show an important bias at positions +4, +5, +6 and +9 (Figure 2A), while expected frequencies of nucleotides found at these positions in yeast do not show a similar bias (Figure 2C).
Table IV. Results of the genetic screening.
| Nucleotides identified at positions +4, +5, +6, +7, +8 and +9 | Number of occurrencea | Readthrough efficiency (%)b |
|---|---|---|
| CAATTA | 12 | 30 |
| CAATCA | 3 | 21 |
| CAGCTA | 6 | 12 |
| CAGACA | 3 | 11 |
| CAAAGA | 14 | 10 |
| CAAACA | 1 | 8 |
| CAGGGA | 3 | 6 |
| CAAAGG | 2 | 6 |
| CAGCCT | 1 | 4 |
| CAGGTT | 1 | 4 |
| GGATTA | 5 | 4 |
| CCACGA | 7 | 2.5 |
| CTAAAT | 1 | 2.5 |
| GAACGA | 9 | 2.5 |
| AAATTA | 1 | 2.5 |
| GGGTGT | 1 | 2.5 |
| GACTCA | 1 | 2 |
| CCAGCT | 1 | 1.5 |
| GACAGG | 1 | 1 |
aNumber of vectors carrying the same stop codon context.
bValues indicated are the mean of three independent transformants giving standard error <10%.
Fig. 2. Number of occurrences of each nucleotide in the 3′ sequence. (A) Results of the initial screening. The y-axis represents the number of occurrences at positions +4, +5, +6, +7, +8 and +9. (B) Only sequences giving readthrough values >5% were taken into account. (C) Expected frequencies of nucleotides at +4 to +9 positions from natural stop codons in the yeast genome. Results were obtained from the Transterm database (Jacobs et al., 2000).
To more precisely quantify the readthrough efficiencies directed by the selected sequences, each was cloned in the pAC99 vector. The results, shown in Table IV, demonstrate that the readthrough efficiencies varied from 1 to 30%. When we retained motifs yielding at least 5% readthrough (Figure 2B), we observed a more pronounced bias: only C and A were present at positions +4 and +5, respectively; A or G was present at position +6, while only A was found at position +9. No bias was observed at position +7, although A was the most frequent. Finally, A was completely absent from position +8. From these data we conclude that the CA(A/G)N(U/C/G)A consensus sequence located at the 3′ side of the stop codon is able to drive >5% of readthrough in yeast.
The effect of the 3′ sequence is not correlated with any property of tRNA or amino acids
The results presented above demonstrate that the 3′ stop codon nucleotide context is a key determinant for readthrough efficiency. The question they pose is how this context can modulate readthrough efficiency.
To test the hypothesis that the effect of the +2 triplet is mediated by the leucine tRNAUAA, possibly already in interaction with the UUA triplet, we made matched modifications in the +2 triplet and in the corresponding tRNA. We first changed the UUA triplet to the synonymous CUU triplet. As shown in Table V, this modification induced a 10-fold reduction of readthrough. This indicates that if the effect is due to the position of a tRNA, not all leucine tRNAs are able to stimulate stop codon readthrough.
To determine whether this effect is specific to tRNAUAALeu, we modified the anticodon loop, which recognizes the UUA codon, in order to obtain a tRNAAAGLeu capable of binding to the CUU codon. All the other characteristics of the tRNA were conserved, so that if this tRNA stimulates readthrough because of a specific conformation, the modified tRNA would guard that ability. No modification of readthrough efficiency was observed upon expression of this engineered tRNAAAGLeu, in a strain carrying a construct in which UUA is replaced by CUU (Table V). These data eliminate the hypothesis that the tRNA in interaction with the 3′ sequence mediates readthrough stimulation.
DISCUSSION
A consensus sequence drives high readthrough levels
This study describes the first combinatorial analysis of stop codon context. We have demonstrated that at least 6 nt following the stop codon are involved in high readthrough levels in yeast. An important positional effect of +4, +5, +6, +8 and +9 was observed on readthrough, while position +7 has no bias. The deduced consensus sequence, CAR NBA, is only slightly different from that identified in tobacco cells. Moreover, the most efficient readthrough motif identified by our study (CAAUUA) corresponds exactly to the 3′ sequence of the UAG leaky stop present in TMV.
The ADE2-based reporter system described is very powerful and easy to use. It will be very helpful in the development of the systematic combinatorial analysis of stop codon contexts whether they are 5′ or 3′ of the stop. It would be especially interesting to be able to identify 3′ readthrough motifs in various 5′ motifs and vice versa, since complex interactions are very probably involved in the recognition of the stop codon by the translational machinery (Bonetti et al., 1995; Cassan and Rousset, 2001). Preliminary results indicate that the readthrough stimulation directed by +7, +8 and +9 nt depends in fact on the identity of the +4, +5 and +6 nt (O. Namy and J.-P. Rousset, unpublished results). More generally, the combinatorial approach and the ADE2-based reporter system described herein can potentially be applied to numerous recoding events in which cis-acting sequences play a crucial role.
18S rRNA–mRNA interactions may be responsible for readthrough
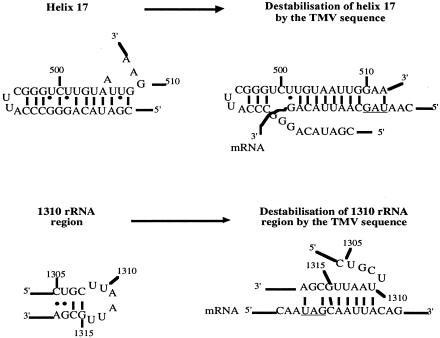
Given its small size, it is likely that the stimulatory context acts as a primary sequence rather than a secondary structure, and in fact no stable structure is predictable. Since rRNA–mRNA interactions are critical at different steps of translation, we sought rRNA regions potentially able to interact with the readthrough motif. Two such regions were identified (see Figure 3). The first (479–510) is located near helix 17, which has already been shown to control decoding accuracy in the prokaryotic A site (Van Ryk and Dahlberg, 1995). Furthermore, a sequence called ‘stimulator’, which highly stimulates +1 frameshifting at the Ty3 sequence, was recently identified 3′ to the frameshift site. This stimulatory element probably acts through interactions with helix 18 of the rRNA (Li et al., 2001). The second region (1305–1318) is absent from 16S rRNA of E. coli, and has never been implicated in translation termination, however it has been reported that a proximate region (helix 34 of the E. coli 16S rRNA) is involved in the binding of both RFs to the ribosome (Arkov et al., 2000). In S. cerevisiae the nucleotide located at position 1054 (corresponding to helix 34 in E. coli) is involved in translation termination accuracy (Chernoff et al., 1996). Finally, crosslinking data suggest that the prokaryotic 530 loop and helix 34 are located close to one another in the three-dimensional structure of the rRNA (Noller, 1991).
Fig. 3. Possible pairing between the 3′ readthrough motif and 18S rRNA. (A) Pairing with helix 17. (B) Pairing with the 1310 region. (•) Non-conventional base pairing.
Speculation
Both of the sequences complementary to the readthrough motif are engaged in imperfect secondary structures. We propose that perfect base pairing of these rRNA sequences with the mRNA may destabilize secondary structures in the ribosome. Such conformational changes would affect binding of release factors. Although the most efficient sequence identified in our screen is perfectly complementary to yeast 18S rRNA, there is one position without any bias (+7). It is striking that this nucleotide is supposed to pair with the only base not engaged in the secondary structure of helix 17 rRNA. The mRNA–rRNA base pairing remains to be experimentally tested, in concerning the identification of which of the two candidate interactions is in fact involved in readthrough. However, this model could explain how the 3′ motif promotes leaky termination.
METHODS
Yeast strains and media. The S. cerevisiae strains used for this work are Y349 (MATα lys2Δ201 leu2-3,112 his3Δ200 ura3-52) and its derivative FS1 (MATα ade2-592 lys2Δ201 leu2-3,112 his3Δ200 ura3-52). Both strains were grown in minimal media supplemented with the appropriate amino acids to allow maintenance of the different plasmids.
Yeast transformations were performed by the lithium acetate method (Ito et al., 1983). 5-FOA was added at a final concentration of 1 mg/ml to select for the loss of URA3 plasmids.
Colour screening was performed on plates containing a drop-out medium, CSM™, with all amino acids and 10 mg/l adenine. The colour intensity was checked after incubation for 3 days at 30°C.
Plasmids and molecular biology methods.
Sequencing. All constructs were verified by sequencing the region of interest with an ABI310 automatic sequencer.
Site-directed mutagenesis of tRNA. tRNAUAALeu was amplified from genomic DNA of the Y349 strain by PCR, using Pfu™ DNA polymerase and oligonucleotides tRNAUAA.w ACGTGTTGACAACACTTCGGGAG and tRNAUAA.c ATGGCTGCATAATGGAAAGGATC. This fragment was cloned into the pUC19 plasmid. tRNA site-directed mutagenesis was performed using the QuickChange™ Site-Directed Mutagenesis kit from Stratagene, and oligonucleotides tRNAAAG.w CTAAGGCGGCAGACTAAGGATCTGTTGGACGG and tRNAAAG.c CCGTCCAACAGATCCTTAGTCTGCCGCCTTAG. After sequencing, the tRNAUUGLeu fragment was subcloned into the yeast plasmid pFL44L.
Cloning of the wild-type allele of the ADE2 gene. The 2.1 kb fragment containing the ADE2 gene with promoter and terminator regions was amplified by Pfu™ Taq DNA polymerase using oligonucleotides ade2.216 AACACCAACATAACACTGACATC and ade2.2319 GGACACCTGTAAGCGTTGATTTC. The PCR fragment was purified using the Qiaquick Gel extraction kit™ (Qiagen), and cloned into the polylinker of the centromeric URA3 pFL38 vector. The unique HpaI site in the ADE2 gene was used to clone double-stranded degenerate oligonucleotides (GGAACACAATAGNNNNNNCAG).
Quantification of readthrough efficiency. pAC derivatives were constructed by cloning the fragment of interest in the unique MscI site of pAC99 (Bidou et al., 2000). Luciferase and β-galactosidase activities were assayed in the same crude extract as previously described (Stahl et al., 1995).
Acknowledgments
ACKNOWLEDGEMENTS
We thank L. Bidou for helpful discussions and M. Godon for technical help during this work. Part of this work was supported by the ‘Association pour la Recherche contre le Cancer’ (contract 9873 to J.-P.R.) and by the Association Française contre les Myopathies (contract 7623 to J.-P.R.).
REFERENCES
- Arkov A.L., Freistroffer, D.V., Pavlov, M.Y., Ehrenberg, M. and Murgola, E.J. (2000) Mutations in conserved regions of ribosomal RNAs decrease the productive association of peptide-chain release factors with the ribosome during translation termination. Biochimie, 82, 671–682. [DOI] [PubMed] [Google Scholar]
- Bidou L., Stahl, G., Hatin, I., Namy, O., Rousset, J.P. and Farabaugh, P.J. (2000) Nonsense-mediated decay mutants do not affect programmed –1 frameshifting. RNA, 6, 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti B., Fu, L.W., Moon, J. and Bedwell, D.M. (1995) The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol., 251, 334–345. [DOI] [PubMed] [Google Scholar]
- Brown C.M., Stockwell, P.A., Trotman, C.N. and Tate, W.P. (1990) Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res., 18, 6339–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan M. and Rousset, J.P. (2001) UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol., 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y.O., Newnam, G.P. and Liebman, S.W. (1996) The translational function of nucleotide C1054 in the small subunit rRNA is conserved throughout evolution: genetic evidence in yeast. Proc. Natl Acad. Sci. USA, 93, 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalphin M.E., Brown, C.M., Stockwell, P.A. and Tate, W.P. (1997) The translational signal database, TransTerm: more organisms, complete genomes. Nucleic Acids Res., 25, 246–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J. and Roeder, G.S. (1990) MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol. Cell. Biol., 10, 2379–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon K., McClendon, V., Bonetti, B. and Bedwell, D.M. (1994) Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J. Biol. Chem., 269, 17802–17808. [PubMed] [Google Scholar]
- Ito H., Fukuda, Y., Murata, K. and Kimura, A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs G.H., Stockwell, P.A., Schrieber, M.J., Tate, W.P. and Brown, C.M. (2000) Transterm: a database of messenger RNA components and signals. Nucleic Acids Res., 28, 293–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Stahl, G. and Farabaugh, P.J. (2001) Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA, 7, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Björnsson, A. and Isaksson, L.A. (1994) The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J., 13, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Tuite, M.F. and Isaksson, L.A. (1998) The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem., 257, 249–254. [DOI] [PubMed] [Google Scholar]
- Noller H.F. (1991) Ribosomal RNA and translation. Annu. Rev. Biochem., 60, 191–227. [DOI] [PubMed] [Google Scholar]
- Poole E.S., Brown, C.M. and Tate, W.P. (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E.S., Major, L.L., Mannering, S.A. and Tate, W.P. (1998) Translational termination in Escherichia coli: three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res., 26, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhankova L. (1972) Joined suppression of rough phenotype and of red colour of ade2-1 mutants in Saccharomyces cerevisiae. Folia Microbiol., 17, 479–489. [DOI] [PubMed] [Google Scholar]
- Skuzeski J.M., Nichols, L.M., Gesteland, R.F. and Atkins, J.F. (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol., 218, 365–373. [DOI] [PubMed] [Google Scholar]
- Stahl G., Bidou, L., Rousset, J.P. and Cassan, M. (1995) Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res., 23, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W.P., Poole, E.S., Dalphin, M.E., Major, L.L., Crawford, D.J. and Mannering, S.A. (1996) The translational stop signal: codon with a context, or extended factor recognition element? Biochimie, 78, 945–952. [DOI] [PubMed] [Google Scholar]
- Van Ryk D.I. and Dahlberg, A.E. (1995) Structural changes in the 530 loop of Escherichia coli 16S rRNA in mutants with impaired translational fidelity. Nucleic Acids Res., 23, 3563–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]