Abstract
Intrinsically disordered proteins and protein regions (IDRs) are abundant in eukaryotic proteomes and play a wide variety of essential roles. Instead of folding into a single stable structure, IDRs exist in an ensemble of interconverting conformations whose structure is biased by sequence-dependent interactions. The absence of a fixed 3D structure, combined with high solvent accessibility, means that IDR conformational biases are inherently sensitive to changes in their environment. Here we argue that IDRs are ideally poised to act as sensors and actuators of cellular physicochemistry. We review the physical principles that underlie IDR sensitivity, the molecular mechanisms that translate this sensitivity to function, and recent studies where environmental sensing by IDRs may play a key role in their downstream function.
Keywords: intrinsically disordered proteins, intrinsically disordered regions, biosensors, cellular environment, signaling
INTRINSICALLY DISORDERED REGIONS AND THEIR CONFORMATIONAL BIASES
Intrinsically disordered regions (IDRs) (see Glossary) make up around a third of most eukaryotic proteomes and play critical roles in various cellular functions [1]. Unlike folded domains, IDRs lack a fixed folded structure and instead exist in a set of interconverting conformations known as an ensemble (see Box 1, Fig. 1A). While IDRs are characterized as disordered, they are not “unstructured.” Instead, IDRs possess conformational biases (Box 1) that are dependent on their amino acid sequence [2–4]. These conformational biases may be driven by polar, hydrophobic, electrostatic, cation-pi, or pi-pi interactions between amino acid side chains that lead to attraction or repulsion between distal regions of an IDR [5–12]. Such interactions tune intramolecular distance distributions and ensemble-average global dimensions. As an example, long-range electrostatic interactions driven by clusters of oppositely charged residues can tune IDR global dimensions [13–15], as in the case of the cell cycle inhibitor protein p27Kip1 [14]. Alternatively, short-range transient secondary structure can manifest as specific conformational states that appear as distinct subpopulations within the overall ensemble [16], e.g., transient helicity within specific subregions of IDRs, as seen in the RNA binding protein TDP-43 or the transcription factor p53 [17,18]. For any given IDR, the emergent combination of sequence-encoded attractive and repulsive molecular interactions will dictate its conformational biases.
BOX 1: IDR CONFORMATIONAL ENSEMBLES and CONFORMATIONAL BIASES AND SENSING.
Conformational ensembles
An IDR’s conformational ensemble is the collection of accessible conformations assumed by the IDR in a solution. Although every protein actually exists somewhere on the continuum between rock-like rigidity and complete disorder, for simplicity the constantly-changing conformational ensembles of IDRs are commonly contrasted to the “native” structures of folded proteins.
Although highly susceptible to change, IDR conformational ensembles are far from random. Rather, every IDR conformational ensemble is influenced by conformational biases (see below) that depend on, among other things, the amino acid sequence of the IDR, the surrounding conditions (solution components, temperature, etc.), and interactions with folded domains to which an IDR may be tethered.
Information about the conformational biases of IDR conformational ensembles is accessible through measurements of ensemble-average properties such as average global dimensions. Ensemble-average global dimensions of an IDR in solution can be measured through methods such as Förster resonance energy transfer (FRET) and small-angle X-ray scattering (SAXS), and predicted through simulations and deep learning approaches.
Conformational biases and sensing
An IDR’s conformational biases (also called structural biases) are preferences in its conformational ensemble due to which certain conformations are observed more often than expected compared to an inert flexible polymer. This may include local structural biases (e.g., transient secondary structure), long-range intramolecular interactions, and biases in global dimensions. An IDR’s conformational biases can undergo pronounced changes as a result of changes in its physicochemical environment, and these changes can influence function.
Importantly, if two IDRs are chemically orthogonal, their conformational biases respond differently to a given physicochemical change in their surroundings. These differential responses give rise to the idea of an IDR being able to “sense” particular physicochemical changes. This perspective proposes that changes in IDR conformational ensembles offer a mechanism for intracellular sensing.
A major challenge in establishing IDRs as biological sensors is demonstrating that IDR conformational ensemble properties determine molecular function. Structural biology has benefited tremendously from the application of conservative separation-of-function point mutations motivated by 3D structures to infer structure-function relationships. For IDRs, the usability of these tools is diminished, because point mutations often have little effect on ensemble structure and function.
As a result, linking IDR ensemble to function, especially in a range of physicochemical conditions, requires novel approaches designed to test the ensemble-function relationship. Because of the resilience of ensemble structure to mutations, meaningful exploration of this relationship requires high- or medium-throughput approaches. Recent years have seen the expansion of computational and experimental approaches that help predict and test sequence-ensemble relationships. These include molecular simulations (coarse-grained and all-atom), high-throughput parallel reporter assays, solution-space scanning, and even direct prediction of ensemble properties from sequence.
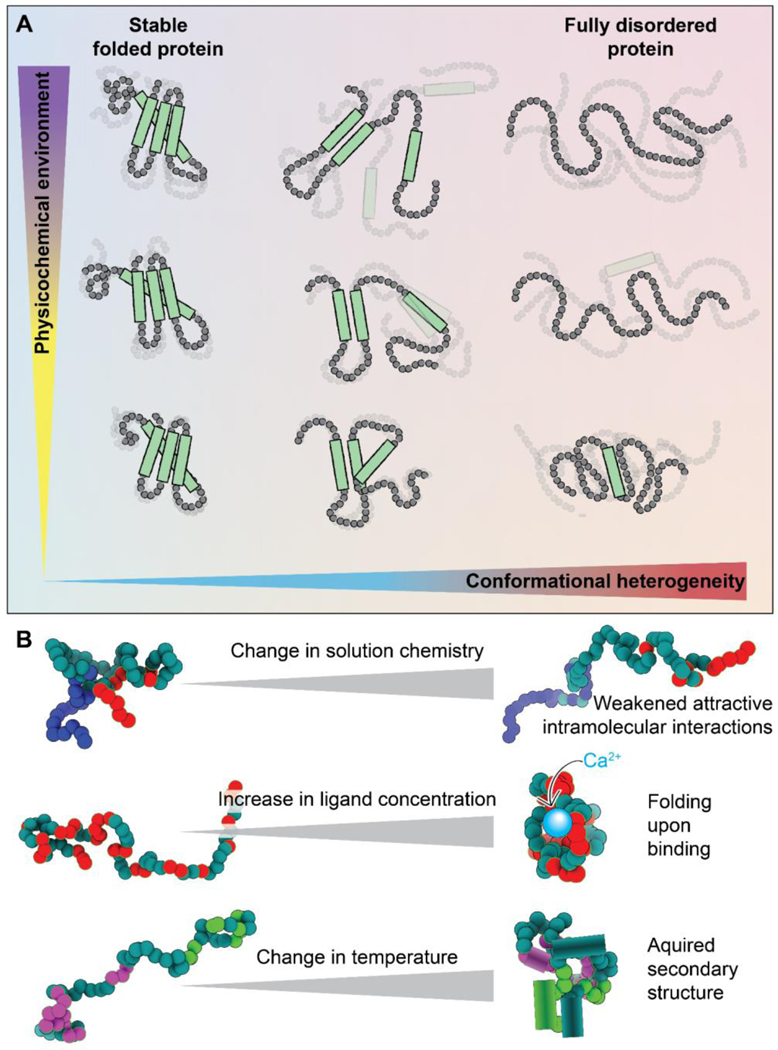
Figure 1. Disordered regions exist in an ensemble that is inherently sensitive to the physicochemical environment.
(A) Protein conformational heterogeneity (see Glossary) exists on a continuum, whereby well-folded domains are at one extreme and fully disordered regions with no strong conformational biases are at the other. Regions that are highly conformationally heterogeneous contain fewer intramolecular bonds and are more solvent-exposed, and hence, in general, are more sensitive to even modest changes in the physicochemical environment. Here, the x-axis represents conformational heterogeneity while the y-axis represents some change in solution chemistry (see Glossary). (B) Scheme showing some examples of how changes in the physicochemical environment can alter IDR conformational biases. Changes in solution chemistry (salt, osmolytes, pH) may weaken (or strengthen) intramolecular interactions leading to a decrease (or increase) in transient intramolecular interactions. The presence (or loss) of ligands, including specific ions, small molecules, second messengers, and other biomacromolecules, can lead to the gain (or loss) of structure upon binding (or unbinding). Changes in physical parameters such as temperature or pressure can lead to the enhancement (or suppression) of intramolecular interactions, which can drive the acquisition (or loss) of secondary or even tertiary structure. These are just a handful of examples of how changes in physicochemistry can be sensed by IDRs.
Besides amino acid sequence, another factor that influences IDR conformational biases, and therefore ensemble properties, is their physicochemical environment [19,20]. Folded domains benefit from a network of intramolecular non-covalent bonds that determine a consistent molecular topology. In IDRs, the lack of such a network has two implications. First, the designation of “buried” and “surface-exposed” residues commonly made in reference to folded proteins is not applicable (Fig. 1A). In general, all residues in an IDR will be at least transiently solvent-exposed. Thus, the entire sequence is in direct interaction with the solution and can sense any change in surrounding chemistry. A second implication is that the sparse interactions that exist in an IDR are often too weak to resist the push and pull of the chain’s interactions with its surrounding solution. For example, interactions with denaturants like urea can pull apart the non-covalent bonds that maintain a protein’s structure. However, denaturation of a folded protein often requires a high urea concentration (6–8 M as a standard) because a network of intramolecular bonds resists this pull. IDRs, on the other hand, can be dramatically extended even by urea concentrations that are almost an order of magnitude smaller (< 1 M) [19–22].
Why do IDR conformational biases matter? The sequence-ensemble-function paradigm posits that IDR function is at least partly dependent on an ensemble’s conformational biases [2,16]. Conformational biases can prime IDRs for molecular recognition that involves folding upon binding [18,23,24]. Alternatively, they can tune global dimensions or facilitate the formation of fuzzy complexes, where a bound structure lacks a defined 3D orientation [24–26]. Specific examples of the sequence-ensemble-function relationship include regions that form binding motifs when they exist as a transient helix [18], global dimensions tuning motif binding accessibility [27], and tuning of interactions by changing the overall volume occupied by the ensemble [28,29]. Additionally, IDRs can themselves play key functional roles without directly interacting with partners. For example, when two globular domains are tethered by an intervening IDR, the “effective concentration” of the two domains with respect to each other, and therefore the extent of their interactions, can be tuned by changing the end-to-end distance of the IDR tether [30,31]. Recent work has highlighted the importance of effective concentration to function by revealing that IDR dimensions – without conservation of a specific amino acid sequence – can be under evolutionary selection to ensure optimal linker lengths in a model termed “conformational buffering” [32]. In short, the relationship between sequence and ensemble can be critical for the biological function of IDRs.
The importance of conformational ensembles to IDR function, coupled to the inherent sensitivity of IDRs to their physicochemical environment, gives rise to the possibility of IDRs acting as molecular sensors of their surrounding physicochemical environment (Box 1). The broad palette of chemistry available through the twenty natural amino acids (plus their post-translational modifications) makes possible the evolution of chemically orthogonal IDRs that are differentially sensitive to a variety of distinct physicochemical changes [19,20,33,34]. Sensing based on IDR ensemble changes would bring obvious advantages to the cell. In contrast to, for example, kinase signaling, IDR ensemble changes require no expenditure of ATP. Also, given that IDRs undergo conformational rearrangement on timescales of 50–200 ns, sensing based on IDR ensemble changes could occur extremely rapidly [35]. These features position IDRs to be exceptionally efficient protein-based sensors.
This perspective focuses on the molecular mechanisms governing how IDRs sense and respond to changes in their physicochemical environment and on biomolecular systems where IDR sensitivity could be the mechanism underlying regulation and function. We will first discuss the conceptual and biophysical determinants of IDR sensing. Following this brief overview, we will consider how sensing can be measured, followed by examples where IDRs have been identified as playing a putative or demonstrable role in sensing their physicochemical environment.
THE MOLECULAR BASIS OF PHYSICOCHEMICAL SENSING
For an IDR to act as a physicochemical sensor, it must reproducibly respond to changes in its physicochemical environment (Fig. 1B). These responses may take the form of global changes in ensemble conformations or changes in local transient structure. Although these are often coupled, for simplicity we will consider them independently in our discussion below.
THE SOLUTION DEPENDENCE OF GLOBAL CONFORMATIONAL BIASES IN IDRS
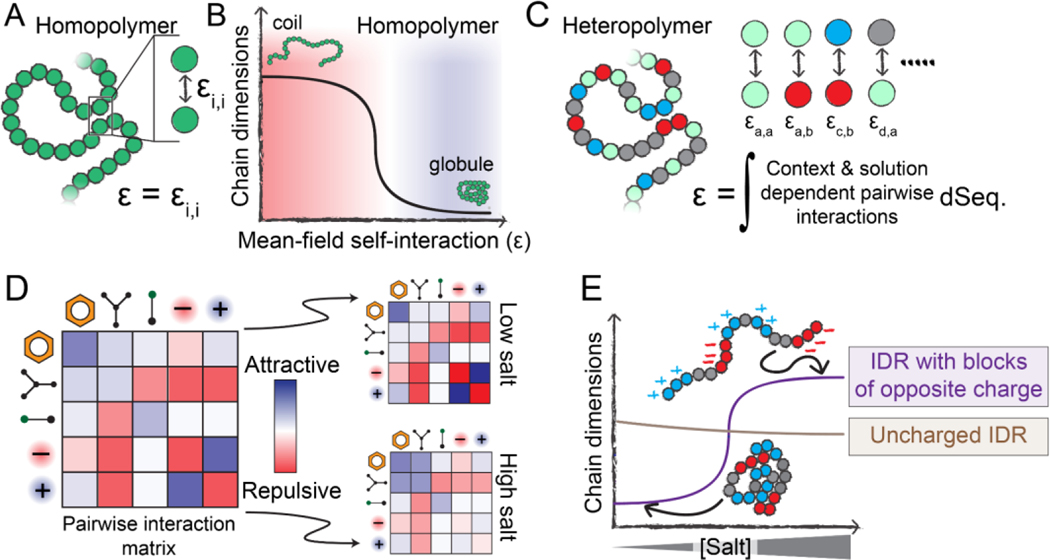
Changes in global IDR dimensions can be viewed through the lens of polymer physics [3,36]. If we represent an IDR as a homopolymer, its global dimensions depend on the balance between attractive and repulsive intramolecular interactions. This balance can be quantified as a single interaction energy that reflects the average overall attraction (or repulsion) of the polymer units (monomers) for one another, i.e., the mean-field self-interaction energy (ε) (Fig. 2A).
Figure 2. Physical principles that underlie sequence-specific IDR sensitivity to changes in physicochemistry.
(A) Homopolymers are defined by a single interaction strength between each polymer unit, which also defines the mean-field self-interaction energy (ε). (B) If ε is repulsive, a homopolymer behaves as an extended coil with large chain dimensions, whereas if ε is attractive, a homopolymer behaves as a compact globule. The mean-field interaction energy can be varied by changing the chemical identity of the polymer unit, but can also be varied by altering the physicochemical environment the polymer finds itself in (temperature, pH, solutes, etc.). (C) Unlike homopolymers, heteropolymers consist of many chemically distinct units. A complete description of a heteropolymer requires knowledge of how each unique inter-residue interaction behaves, and the mean-field self-interaction energy (ε) is now defined in terms of the composition-weighted and context-dependent integral over all possible interactions. (D) The various types of interactions that may occur between residues in a heteropolymer can be, to first order, described by an interaction matrix. The strengths of these interactions depend on solution conditions. (E) The response of a heteropolymer to changes in the solution environment depends on the heteropolymer’s chemistry. For example, a highly-charged IDR with blocks of oppositely-charged residues will be compact at low salt due to strong intramolecular electrostatic interactions. However, under high-salt conditions, those attractive interactions are screened, leading to an expanded ensemble driven by the substantial solvation free energies associated with charged groups. In contrast, a charge-depleted heteropolymer may be relatively salt-insensitive and is relatively compact compared to the blocky IDR in the high-salt limit.
The mean-field self-interaction energy is inherently dependent on the solution environment. In a solution of polymer and solvent, increasing solvent:monomer repulsion is equivalent, in a mean-field sense, to increasing monomer:monomer attraction. Moving from a solution in which the mean-field self-interaction is repulsive to one in which it is attractive can manifest as a coil-to-globule transition [36] (Fig. 2B). The sharpness of this transition depends on the chain length and the magnitude of the change in self-interaction energy.
For homopolymers, only a single type of monomer unit is present, so there exists only a single type of pairwise interaction energy (Fig. 2A). For heteropolymers (like IDRs), chemically distinct monomers give rise to a matrix of pairwise interaction strengths (Fig. 2C, 2D). A key concept in IDR sensitivity is that each of these individual pairwise interaction strengths may be modulated differently by changes in the physicochemical environment; that is, they may be chemically orthogonal (Fig. 2D). As a specific example, attractive pairwise interactions driven by electrostatics may be sensitive to salt, while attractive pairwise interactions driven by hydrogen bonding may not (Fig. 2D, 2E).
Two central conclusions emerge from this framework. First, IDR global dimensions must depend on amino acid sequence, as has been established by prior work [2,3,7,9,10,12,22,37]. Second, an IDR’s sensitivity – that is, how much global dimensions change as a function of the changing physicochemical environment – depends on (a) the underlying IDR sequence, i.e., where on the coil-to-globule curve an ensemble begins (Fig. 2E); and (b) how the intramolecular interactions encoded in this sequence respond to their environment, i.e., how much the overall mean-field pairwise interaction energy changes in response to physicochemical changes [19] (Fig. 3A).
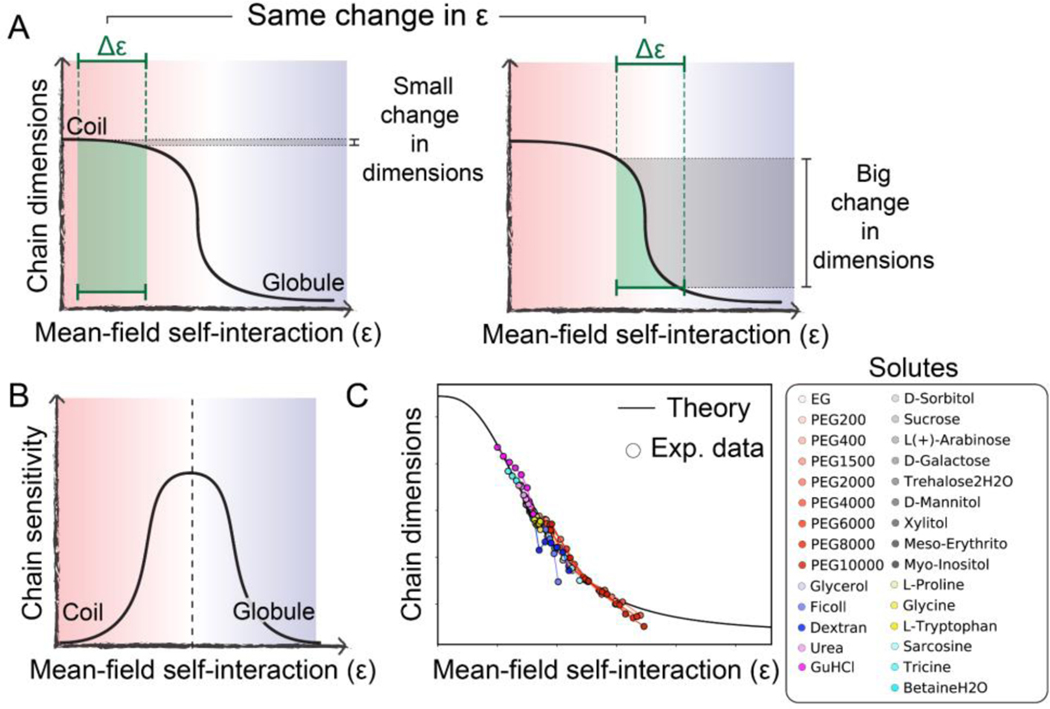
Figure 3. An IDR’s sensitivity to the physicochemical environment depends on its intrinsic conformational biases.
(A) The extent of change in chain dimensions (gray shaded region) in response to a change in mean-field self-interaction strength tuned by the physicochemical environment (green shaded region) depends on both the underlying sequence and the polymer’s behavior prior to the change. From equivalent changes in interaction strength, very different changes in polymer dimensions can emerge. (B) Chains at the midpoint of the coil-to-globule transition are most sensitive to changes in the solution environment. (C) Comparison of experimental data and analytical theory demonstrating the broad applicability of this framework. Adapted from [19].
For IDR ensembles that begin near one of the baselines (either coil or globule), large changes in the mean-field energy can have a relatively small impact on chain dimensions, making them less sensitive (Fig. 3A, left) [19]. Analogously, for ensembles that begin in the middle of the coil-to-globule transition, relatively small changes in the mean-field interaction energy drive large changes in global dimensions, making them more sensitive (Fig. 3A, right). One could consider folded domains to be at the globular extreme of this transition, illustrating their lack of solution sensitivity. The upshot of this is that chain sensitivity peaks at the midpoint of the coil-to-globule transition (Fig. 3B). Indeed, prior work has shown that this conceptual framework is able to quantitatively normalize the solution dependence of IDRs across a wide range of different cosolutes (Fig. 3C) [19].
In short, baseline conformational behavior and sensitivity to environmental change, both of which depend on sequence, combine to determine an IDR’s global dimensions (Fig. 2E). Together, these two features offer a quantitative framework through which IDR sensitivity can be interpreted and, looking forward, used as a design principle for the development of novel sensors.
THE SOLUTION DEPENDENCE OF LOCAL CONFORMATIONAL BIASES IN IDRS
Local conformational biases, such as the gain or loss of transient secondary structure (especially transient helicity), can also be tuned by the environment [38,39] (Fig. 4). Importantly, the ability of ensemble conformations to change locally and not just globally means that ensembles can have different structural features even though global dimensions are the same [12,40,41]. This poses an additional challenge to experiments which often measure only a single global dimension, as discussed below.
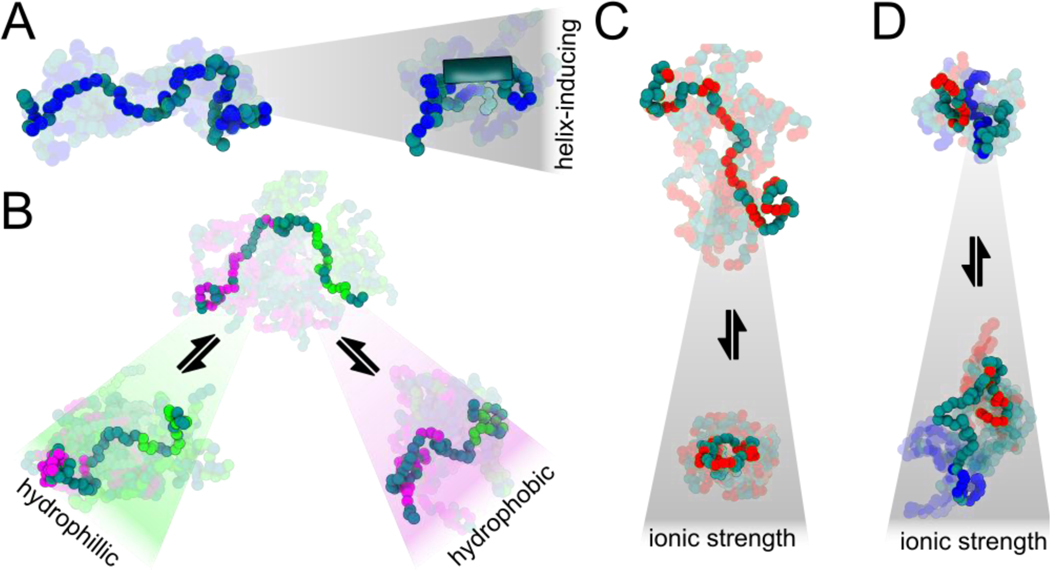
Figure 4. Examples of physicochemically-driven changes in IDR ensembles.
(A) Promotion of secondary structural elements such as residual helicity (shown by the tube on the right) can form or dissolve binding motifs, modulating binding affinities. By prepaying an entropic cost for binding, the effective concentration of binding motifs can be rapidly enhanced or suppressed without the need to alter protein copy number. (B) Amphipathic sequences with patches of hydrophobic or hydrophilic residues can compact or expand locally in different solutions, tuning accessibility of specific regions. (C) Sequences with high net charge (positive or negative) can compact when an increase in ionic strength screens out repulsive interactions. (D) Charged sequences with sequestered, opposite charges can expand at high ionic strength due to screening of attractive interactions.
CELLULAR SENSING THROUGH IDR-MEDIATED PHASE TRANSITIONS
IDRs can also contribute to the formation of biomolecular condensates through intracellular phase transitions, although we emphasize that IDRs are not necessary for phase transition to occur [10,42]. The same physical and chemical logic ascribed to IDR coil-to-globule transitions can and will tune phase behavior, meaning that changes in solution conditions can enhance or suppress biomolecular condensate formation [37,43–46].
Yoo et al. have made a compelling case that intracellular phase transitions offer a general class of sensing that benefits from the sharpness of a first-order phase transition [47]. Whereas coil-to-globule transitions of real chains are finitely-cooperative transitions with cooperativity dependent on sequence and solution (Fig. 2), first-order phase transitions are infinitely cooperative. This means phase transitions offer digital (on/off) sensors, while individual IDRs provide analog (dimmer switch) sensors. Depending on the scenario, digital or analog sensing may be preferable.
MEASURING IDR SENSITIVITY
In folded domains, chemical sensing can be facilitated through specific, evolutionarily optimized binding sites that enable picomolar-affinity binding to small molecules. IDRs, in contrast, cannot form well-defined binding pockets in their disordered state but instead can sense the chemical environment through changes in IDR:solvent interactions (Fig. 1). Because of this, sensing low concentrations (nM and below) of specific small molecules can be difficult to achieve unless IDRs engage in coupled folding-upon-binding with a ligand. Instead, IDRs sense the average physicochemical environment of their surroundings and respond with changes in ensemble structure – a holistic, integrated response to the environment that is difficult for well-folded proteins to achieve. Here we highlight experimental methods that probe how IDRs act as physicochemical sensors.
IN VITRO MEASUREMENTS OF IDR ENSEMBLE SENSITIVITY
Small-angle X-ray scattering (SAXS), single-molecule Förster Resonance Energy Transfer (FRET), nuclear magnetic resonance (NMR), and other biophysical methods provide a rich toolkit with which to study IDR ensembles in vitro. Such studies can measure the sensitivity of IDRs through changes in ensemble-average properties as a function of changes in solution physicochemistry [19,34,48].
Several groups have used changes in solution chemistry to elicit a structural response from IDRs. Perhaps the most well-studied examples are denaturants, which by weakening intramolecular interactions drive ensemble expansion [21,22,41,49]. To broadly explore the relationship between IDR sequence sensitivity and solution chemistry, recent work applied so-called solutionspace scanning to assess how different IDRs respond to a panel of different cosolutes. In this approach, an IDR of interest is sandwiched between a FRET pair, and the IDR’s dimensions as a function of osmolyte concentration and identity can be recorded [19]. This work showed that IDR sensitivity in vitro depends on the amino acid sequence and the specific changes in the physicochemical environment, strengthening the argument for IDRs as sensors and actuators of the cellular environment [19]. The importance of sequence to sensitivity was highlighted by the finding that an ensemble can be made more or less sensitive by scrambling its sequence while retaining the same amino acid composition, further supporting the idea that IDRs may be finely tuned to respond to environmental changes [34].
Solution-dependent changes in IDR dimensions have been examined in additional contexts. IDRs linked to desiccation tolerance undergo structural rearrangement upon dehydration and/or changes in solution composition [50–54]. IDR ensembles and their intermolecular interactions can also be tuned by ion concentration and identity [39,55,56]. Macromolecular crowding also modulates IDR ensembles, with the size and concentration of a crowding molecule key determinants of its ability to compact IDR ensembles [57,58]. In summary, in almost all in vitro systems examined, with a few notable exceptions, IDRs appear responsive to their environment, albeit to different degrees [12].
IN-CELL MEASUREMENTS OF IDR SENSITIVITY
In contrast to aqueous buffers, the cellular environment is constantly changing, causing spatial and temporal variations in crowding, pH, ion and osmolyte concentrations, and other solution properties [59]. These changes occur during routine cellular events such as cell cycle progression (e.g., entry into mitosis), due to external stress such as osmotic pressure or starvation, or due to pathology (e.g., the Warburg effect in most cancer cells) [60–63]. Given the sensitivity of IDRs, intracellular solution dynamics may be expected to alter IDR ensembles more than what is observed in vitro. To assess this, several methods, including NMR [64,65] and single-molecule FRET [48], have been used to study IDR ensembles in live cells. Despite some outliers that are more structured in the cell than in vitro [66], all IDRs studied to date remain disordered in the cell [34,65,67,68]. Despite the great differences between an aqueous buffer solution and the cellular environment, recent work has shown that conformational biases observed in vitro tend to persist in the cell [34,68].
Measuring IDR sensitivity in cells requires the ability to precisely and reproducibly change the cellular environment and measure the resulting response of the IDR ensemble. This makes in-cell experiments tricky to interpret: cells have well-established active mechanisms to maintain homeostasis and mitigate physicochemical perturbations. As a result, induced ensemble changes must be distinguished from changes driven by cellular pathways [69]. Rapid, laser-induced temperature jumps offer one route to produce high-speed perturbations, an approach that has been used to measure intracellular IDR dimensions as a function of temperature [70]. Several groups also have used rapid osmotic perturbations that occur in a matter of seconds. When studied using in-cell single-molecule FRET, the fully-disordered protein prothymosin alpha was found to compact under hyperosmotic stress, an effect quantitatively explained by changes in molecular crowding [48]. More recently, ensemble FRET measurements revealed that the osmotic sensitivity of IDRs inside the cell is often dramatically different from what is measured in vitro [34]. This highlights the ability to elicit different structural responses from the same IDR to the same perturbation depending on the IDR’s surroundings.
IDRS AS PHYSICOCHEMICAL AND BIOLOGICAL SENSORS
Recognition that proteins exist in a complex, spatially and temporally dynamic cellular environment has driven research into how cellular perturbations influence protein structure and function [71]. While folded domains are often robust to small physicochemical perturbations, IDRs can be much more sensitive to these changes [33,72]. The inherent flexibility of IDRs, together with the exposure of their residues to the surrounding environment, makes them ideal candidates to serve as sensors and actuators of physicochemical changes in the cellular environment [39,57,73]. However, whether this physicochemical sensing modality elicits a function can be hard to verify. An IDR is only a genuine biological sensor if it responds to a stimulus and then elicits a downstream biological response. Presented below and in Table 1 are examples of IDRs that mediate downstream function when exposed to physicochemical changes in the cellular environment. Despite clear links between IDR sensitivity and downstream function, whether this function is achieved through environmentally-mediated changes to ensemble or some other mechanism (e.g., post-translational modification or binding to other proteins) remains to be tested. Nonetheless, we expect that, at least for some of these examples, mechanistic studies will point to environmentally-mediated ensemble changes as underlying the production of downstream function.
Table 1.
IDRS SHOWING FUNCTIONAL RESPONSE TO PHYSICOCHEMICAL CHANGES
| Change | Protein | Function/Observation | Reference(s) |
|---|---|---|---|
| CO2 | Ptc2 phosphatase | Drives phenotypic (white-opaque) switching in C. albicans | [76] |
| Water availability/deficit | Hydrophilins, LEA proteins | Protection from water stress | [88][51][88] |
| LEA proteins | Sensing of osmotic stress | [33] | |
| CAHS D | Desiccation protection in tardigrades; acquires helical structure and forms hydrogels upon desiccation | [52–54,89] | |
| FLOE1 | Undergoes phase separation under hydration; signals A. thaliana to suppress germination in unfavorable environments | [90] | |
| SEUSS | Stress tolerance in A. thaliana; drives localization to condensates in response to hyperosmotic conditions | [91] | |
| Macromolecular crowding | YAP | Transcriptional coactivator in human cell growth that localizes in condensates in response to cell volume decrease and alters expression patterns | [121] |
| ASK3 | Kinase that forms condensates upon cell volume decrease and regulates volume recovery | [122] | |
| WNK1 | Kinase that forms condensates upon sensing crowding and regulates cell volume | [92] | |
| Redox state | CP12 | Regulates the Calvin-Benson-Bassham cycle | [95] |
| NPR1 | Regulates the ubiquitylation of stress response machinery through biomolecular condensates | [96] | |
| TMF | Gene control during flower development | [97] | |
| TDP-43 | Proposed to function as intracellular redox sensor | [98] | |
| Ataxin-2 | Proposed to function as intracellular redox sensor | [99] | |
| pH | Snf5 | Enables transcriptional rewiring in budding yeast | [80] |
| Sup35 | May tune local pKa values into the physiological range in yeast | [45] | |
| HSF1 | pH-responsive element involved in yeast stress response | [79] | |
| G3BP1 | Mammalian stress-granule formation | [86] | |
| Ions & metals | ASK3 | Na+ regulates the liquidity of ASK3 condensates | [123] |
| SK | Disordered regions that fold upon Ca2+ binding | [100] | |
| SilE | Disordered bacterial protein that folds upon silver binding | [102] | |
| PrPc | Binds copper via octa-repeat motifs in its disordered N-terminal IDR. This region may also bind other metals. | [103] | |
| Granulins | Small cysteine-rich disordered proteins that can sequester copper | [104] | |
| Mms6 | Bacterial protein whose C-terminal IDR coordinates iron in the context of magnetosome formation |
[105] |
|
| ProTα | Highly-charged IDR that can bind zinc; this binding has been proposed to act as an entropic switch. |
[106] |
|
| Insulin | SIRT1 | An insulin binding motif in the N-terminal IDR leads insulin-dependent structural acquisition | [124] |
| Temperature | Pab1 | Tunes stress granule assembly during heat stress in S. cerevisiae via the P-domain (however, P-domain is not required for condensation) | [44] |
| ELF3 | A polyglutamine tract in the ELF3 prion-like domain tunes temperature sensing in Arabidopsis | [117] | |
| FRIGIDA | C-terminal IDR contributes to cold-dependent condensate formation in Arabidopsis | [118] | |
| Phytochrome B | Encodes molecular timer to tune phytochrome revision in Arabidopsis | [120] |
CHEMICAL PERTURBATIONS
Early studies exploring the relationship between IDR:solvent interactions and IDR dimensions focused on the impact of denaturants [21,41,49]. Following this, a corpus of work showed that highly-charged IDRs could be extremely sensitive to changes in salt conditions, further illustrating how the solution environment can tune ensemble dimensions [7,56,57,74,75]. This work has paved the way for a growing appreciation that IDRs can and do respond to their chemical environment.
IDRS AS SENSORS OF CELLULAR CHEMISTRY
If IDRs can sense their environment, are there examples where this enables biological regulation? One such example is CO2 sensing enabled by a large IDR within the Ptc2 phosphatase in Candida albicans [76]. Here, a serine-rich IDR enables CO2 sensing by driving the formation of biomolecular condensates upon elevated CO2 levels, which in turn drives phenotypic (white-opaque) switching. Although CO2 sensing is conserved in functionally orthologous PP2C-family phosphatase IDRs, the primary sequence of the CO2-sensing IDR varies substantially across species. Conservation of function, even with poor sequence conservation, is gaining attention as a distinctive feature of IDRs, as highlighted in recent work across a range of organisms [32,77,78].
IDRs have also emerged as important participants in pH sensing [79,80]. Considering that pH changes can alter the ionization states of titratable side chains, and given the important role charged residues play in IDR ensembles, IDRs are well-poised to function as pH sensors [81,82]. In budding yeast, a glutamine-rich low-complexity domain in the transcriptional regulator Snf5 possesses a handful of histidine residues that enable large-scale pH-dependent transcriptional rewiring [80]. For Snf5, simulations predict that histidine protonation leads to an increase in IDR global dimensions. Similar IDR-based pH sensing in yeast has also been reported in HSF1 and Sup35, where the local sequence context may tune local pKa values into the physiological range [45,83–85]. These insights also offer a mechanistic explanation for the pH-dependent conformational rearrangement observed in G3BP1, a highly disordered mammalian protein essential for mammalian stress granule formation [86]. Taken together, pH-dependent conformational switching offers a mechanism through which IDRs can respond to intracellular changes or enable context-specific functionality.
Another situation in which large-scale intracellular physicochemical changes arise is desiccation. Responding to changing water availability is crucial, especially for sedentary or single-cell organisms. The precise physicochemical cues being sensed in response to changing water availability are unavoidably a convolution of many different factors (including dielectric constant, osmolyte concentrations, water potential, oxidative stress, etc.). However, organisms from all branches of the tree of life have evolved IDRs that can sense and/or protect them from water stress, highlighting the generality of this phenomenon [87]. The quintessential family of such protectants are hydrophilins – a family of largely disordered proteins that are accumulated under water deficit in bacteria, archaea, and eukaryotes [88]. Biophysical studies of sub-families, including the late-embryogenesis abundant (LEA) and the tardigrade-specific CAHS proteins, show that these proteins undergo ensemble-wide change upon exposure to water stress, often adopting a helical conformation [51–53,89]. This observation has enabled the design of novel water-sensing proteins, demonstrating the power of a biophysical understanding of IDR sensitivity [33]. Recent studies also highlight the role of condensate formation by IDRs under water stress. The highly disordered protein FLOE1 in Arabidopsis thaliana seeds reversibly undergoes phase separation under hydration, and the biophysical states of FLOE1 condensates signal the plant to suppress germination when the environment becomes unfavorable [90]. IDR-dependent condensation of another Arabidopsis protein, SEUSS, drives localization to condensates in response to hyperosmotic conditions and is indispensable for water stress tolerance [91]. Indeed, the ability of IDRs to sense osmotic challenges and respond through condensation has been highlighted as a common feature and observed in other systems [37,92,93].
Cellular redox state is another facet of the intracellular environment that can change substantially [94]. Sensing and responding to changes in redox states is vital for normal cellular function. However, like desiccation, changes in redox state often coincide with additional changes such that it may be impossible to deconvolve the relative contributions of related physicochemical changes. Nevertheless, recent studies have implicated IDRs as redox sensors in several different systems. In the context of CO2 assimilation in algae, redox-dependent conditional disorder in chloroplast protein of 12 kDa (CP12) regulates the Calvin-Benson-Bassham cycle in a redox- and light-dependent manner [95]. In plants, cell survival during pathogen response depends on three cysteine-containing redox-sensitive IDRs in NPR1, which regulate the ubiquitylation of stress response machinery through biomolecular condensates [96]. Similarly, in plant shoot apical meristems, the production of reactive oxygen species promotes phase separation of transcription factor Terminating Flower (TMF). Cysteine oxidation paired with the cooperation of N- and C-terminal IDRs in TMF enables redox-tunable transcriptional condensation and direct gene control during flower development in plants [97]. In humans, evolutionarily conserved methionine-rich IDRs in TAR DNA-binding protein 43 (TDP-43) and Ataxin-2 have been proposed to function as intracellular redox sensors [98,99]. Both TDP-43 and Ataxin-2 undergo self-assembly into redox-sensitive gel-like condensates, with implications for the dysregulation of redox homeostasis in human disease.
Ions and metals can influence IDR conformational biases in various ways. Changes in monovalent salt concentrations may screen attractive or repulsive electrostatic interactions, rewiring intramolecular and intermolecular interactions [22,74]. Beyond nonspecific electrostatic screening, direct binding of ions, metals, and even small organic molecules also represents a key mechanism by which IDRs can act as sensors [7,22,39]. As an example, a disorder-to-order transition upon Ca2+ binding may couple calcium sensing and ion channel opening in smallconductance calcium-activated potassium channels [100]. More generally, a growing number of IDRs appear to possess calcium-binding motifs, which may enable Ca2+ sequestration and Ca2+-dependent changes in ensemble behavior [101]. Beyond calcium, IDRs have been found to bind copper (PrPC, granulins), zinc (ProTα), silver (SilE), and ferric iron (Mms6) [102–106]. Ions can also tune IDR-mediated assembly with anion- and cation-specificity driven by charge density, ion solvation effects, and preferential interaction coefficients [107]. Regulatory logic involving ion-dependent changes in IDR properties (e.g., in developmental biology during calcium waves or neuronal action potentials) offers a potential mechanism for adaptive intracellular function. While the cellular consequences of metal binding are often unclear, these studies suggest that IDRs are poised to enable specific and tunable metal sensing.
OTHER INTRACELLULAR IDR SENSORS
In addition to sensing their chemical environment, IDRs can act as physicochemical sensors through other means. Sequence-dependent effects, namely sequence chemistry and length, can influence how IDRs sense or exert mechanical force [29,108]. Emerging data suggests IDRs may be central to sensing membrane curvature: mechanistically by negatively-charged IDRs electrostatically binding lipids, and entropically driven by preferential partitioning to convex surfaces of membranes [28,109,110].
IDRs may also function as sensors of intracellular crowding. Prior work combining experiments, theory, and simulations revealed that IDRs can show complex and sometimes unintuitive responses to crowders depending on IDR sequence and crowder size, shape, and chemistry [19,57,58,111,112]. Using single-molecule FRET, the impact of crowding on IDR function has also been examined as a potential means through which IDRs could – indirectly – enable sensing through crowding-dependent attenuation of molecular recognition [57,58,113]. Additionally, recent work on synthetic condensates indicates that crowding-induced condensate formation enables novel phosphorylation events to occur if those condensates recruit kinases [114]. The ability to encode mechanical-to-chemical signal transduction via intracellular phase transitions has broad implications for cell fate, human disease, and molecular evolution.
As a final note, IDRs are inherently temperature-sensitive, owing to the fact that the mean-field self-interaction energy has an unavoidable entropic component that comes from solvation effects [115,116]. Perhaps unsurprisingly, this temperature dependence depends on the solution environment, setting the stage for IDRs to act as tunable temperature sensors. As one example, the hydrophobic P domain of Pab1 in S. cerevisiae functions as a finely tuned temperature sensor that tunes stress granule assembly during heat stress [44]. Similar temperature-sensitive IDRs have been identified in plants [117,118], while rationally designed IDRs could enable novel thermosensors [119]. In addition to acting via self-assembly, IDRs have also been shown to encode short-lived molecular timers whose refractory period depends on the temperature, as is the case in the plant photoreceptor Phytochrome B [120].
Concluding remarks
IDRs are inherently sensitive to their physicochemical environment. Here we have considered physical bases of that sensitivity, biochemical explanations of how sensitivity manifests in IDR ensemble structure, and functional consequences of IDR-dependent sensing mechanisms. While this emerging paradigm of disordered sensors has the potential to enable new insights into cellular regulation, it also raises many questions, some of which are listed in the Outstanding Questions box.
A central challenge in studying IDRs in the context of cellular sensing is distinguishing between an effect that unavoidably happens versus one that reports on a bona fide sensor or actuator. We highlight this in our precise word choice, differentiating between a physicochemical sensor and a biological sensor (Box 1). Verifying biological responses as being due to physicochemically induced changes in IDRs remains a key challenge.
A second challenge is a corollary of the first. We reported here many examples in which IDRs have been shown to mediate cellular sensing, yet the actual mechanism through which this is achieved remains opaque in most – if not all – cases. Enabling molecular insights into how these IDRs actually work is an ongoing challenge, both in the context of IDR sensors and, more broadly, in understanding IDR function.
In summary, we propose that IDRs represent ubiquitous sensors of intracellular state, where they provide a means for complex integrative regulation. While studying IDRs is technically challenging, their biophysical plasticity and propensity for weak multivalent interactions make them ideal tunable cellular sensors. As we develop new methodologies to study links between sequence, environment, and function of IDRs, we expect the discovery of exciting new mechanisms by which they regulate biology.
OUTSTANDING QUESTIONS.
How do changes in IDR ensembles propagate to drive downstream effects?
How is chemical specificity for sensing encoded in IDR sequence?
Can IDR-based sensors decouple related chemical signals (e.g., desiccation and oxidative stress)?
Can IDR sensitivity cause malfunction in a dysregulated cellular environment (for example, the Warburg effect in cancer cells)?
HIGHLIGHTS:
Intrinsically disordered protein regions (IDRs) are ubiquitous across all kingdoms of life.
IDRs exist as a collection of interconverting conformations known as an ensemble.
Due to the absence of a fixed 3D structure, IDRs are – in general – more sensitive to their physicochemical surroundings than folded domains.
This sensitivity means IDRs are well suited to act as intracellular sensors, whereby sequence-encoded conformational biases are altered by varying salt, pH, temperature, metabolites, and other solution changes.
Here we discuss the biophysical and biochemical basis for IDR-mediated sensitivity, and highlight examples of this as a potential mechanism for biological regulation.
ACKNOWLEDGEMENTS
We thank members of the Sukenik and Holehouse labs for ongoing discussions, constructive and important feedback from our anonymous reviewers, as well as FNZ for feedback on topics pertaining to this work and beyond. Research reported in this publication was supported by the NIH under award R35GM137926 to SS, and by the NSF under award 2128067 to SS and 2128068 to ASH. DM is supported by a fellowship from NSF-CREST Center for Cellular and Biomolecular Machines (CCBM) at UC Merced, Grant No. NSF-HRD-1547848. GG is supported by a MilliporeSigma Fellowship. We thank members of the Water and Life Interface Institute (WALII), supported by NSF DBI grant # 2213983, for helpful discussions. We also thank the NSF for funding the Research Collaboration Network “Protein Folding Consortium (PFC)” (grant number 1516959), through which SS and ASH met in 2015 as trainees and established a long-term collaborative relationship.
GLOSSARY:
- Biological Sensor
in the context of IDRs, a physicochemical sensor that, by responding to a change in its physicochemical environment, elicits a downstream biological response
- Chemical Orthogonality
Modes of interaction that are driven by chemically distinct molecular mechanisms. In the context of IDRs, two chemically orthogonal IDRs may show divergent responses to the same change in solution chemistry. For example, one IDR may become compact in the presence of increased ionic strength but show no response to pH changes, while another could compact at elevated pH but show no response to elevated ionic strength
- Coil-to-globule Transition
A sigmoidal change in the global dimensions of a polymer from a maximally expanded state to a minimally expanded state (see Fig. 2B)
- Conformational Biases
Local and long-range intramolecular interactions (attractive or repulsive) that deviate from those expected for an inert, flexible polymer (see also Box 1)
- Conformational Ensembles
the collection of accessible conformations assumed by the IDR in a solution (see also Box 1)
- Conformational Heterogeneity
A measure of the range of different conformational states observed. Folded proteins have limited conformational heterogeneity, such that they are often well described by a single reference structure. Disordered regions have extensive conformational heterogeneity, necessitating their description in terms of average properties of a conformational ensemble
- Global Dimensions
Properties of an IDR’s conformational ensemble that relate to the overall volume being occupied by the ensemble. Typically reported in terms of the radius of gyration, the hydrodynamic radius, or the end-to-end distance. Global dimensions are the measured observable for most experimental methods (SAXS, ensemble FRET, smFRET, SEC, and others)
- Intrinsically Disordered Regions (IDRs)
Proteins or protein regions that are poorly described by a single three-dimensional structure and instead exist in a conformational ensemble
- IDR Sensitivity
The degree to which the conformational biases of an IDR change due to changes in the physicochemical environment. Different sequences can display sensitivity to different environments (see also chemical orthogonality)
- Physicochemical Environment
The physical and chemical environment surrounding an IDR, including but not limited to the solution chemistry, as well as physical parameters such as temperature and pressure
- Physicochemical Sensor
in the context of IDRs, an IDR that reproducibly changes its conformational ensemble (see Box 1) in response to a specific change in its physicochemical environment
- Solution Chemistry
The chemical identity and composition of a solution, including pH, water activity, and osmotic pressure, and the identity and concentration of solutes: ions, osmolytes, metabolites, other small solutes, and macromolecules (including other proteins)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.van der Lee R. et al. (2014) Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das RK et al. (2015) Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol 32, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao AH et al. (2013) Describing sequence–ensemble relationships for intrinsically disordered proteins. Biochem. J 449, 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forman-Kay JD and Mittag T. (2013) From sequence and forces to structure, function, and evolution of intrinsically disordered proteins. Structure 21, 1492–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukhopadhyay S. et al. (2007) A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proceedings of the National Academy of Sciences 104, 2649–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman MA et al. (2020) Properties of protein unfolded states suggest broad selection for expanded conformational ensembles. Proc. Natl. Acad. Sci. U. S. A 117, 23356–23364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller-Späth S. et al. (2010) Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A 107, 14609–14614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao AH et al. (2010) Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A 107, 8183–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marsh JA and Forman-Kay JD (2010) Sequence Determinants of Compaction in Intrinsically Disordered Proteins. Biophys. J 98, 2383–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin EW et al. (2020) Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S. et al. (2020) Comparative roles of charge, π, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A 117, 28795–28805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin EW et al. (2016) Sequence Determinants of the Conformational Properties of an Intrinsically Disordered Protein Prior to and upon Multisite Phosphorylation. J. Am. Chem. Soc 138, 15323–15335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das RK and Pappu RV (2013) Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U. S. A 110, 13392–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das RK et al. (2016) Cryptic sequence features within the disordered protein p27Kip1 regulate cell cycle signaling. Proc. Natl. Acad. Sci. U. S. A 113, 5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beveridge R. et al. (2019) Ion Mobility Mass Spectrometry Uncovers the Impact of the Patterning of Oppositely Charged Residues on the Conformational Distributions of Intrinsically Disordered Proteins. J. Am. Chem. Soc 141, 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davey NE (2019) The functional importance of structure in unstructured protein regions. Curr. Opin. Struct. Biol 56, 155–163 [DOI] [PubMed] [Google Scholar]
- 17.Conicella AE et al. (2016) ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure DOI: 10.1016/j.str.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borcherds W. et al. (2014) Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat. Chem. Biol 10, 1000–1002 [DOI] [PubMed] [Google Scholar]
- 19.Moses D. et al. (2020) Revealing the Hidden Sensitivity of Intrinsically Disordered Proteins to their Chemical Environment. J. Phys. Chem. Lett 11, 10131–10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holehouse AS and Sukenik S. (2020) Controlling Structural Bias in Intrinsically Disordered Proteins Using Solution Space Scanning. J. Chem. Theory Comput 16, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 21.Borgia A. et al. (2016) Consistent View of Polypeptide Chain Expansion in Chemical Denaturants from Multiple Experimental Methods. J. Am. Chem. Soc 138, 11714–11726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann H. et al. (2012) Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. U. S. A 109, 16155–16160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers JM et al. (2014) Interplay between partner and ligand facilitates the folding and binding of an intrinsically disordered protein. Proc. Natl. Acad. Sci. U. S. A 111, 15420–15425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staller MV et al. (2022) Directed mutational scanning reveals a balance between acidic and hydrophobic residues in strong human activation domains. Cell Syst 13, 334–345.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tompa P. and Fuxreiter M. (2008) Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci 33, 2–8 [DOI] [PubMed] [Google Scholar]
- 26.Tuttle LM et al. (2018) Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex. Cell Rep. 22, 3251–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateos B. et al. (2021) Hyperphosphorylation of Human Osteopontin and Its Impact on Structural Dynamics and Molecular Recognition. Biochemistry DOI: 10.1021/acs.biochem.1c00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeno WF et al. (2019) Molecular Mechanisms of Membrane Curvature Sensing by a Disordered Protein. J. Am. Chem. Soc 141, 10361–10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keul ND et al. (2018) The entropic force generated by intrinsically disordered segments tunes protein function. Nature 563, 584–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sørensen CS and Kjaergaard M. (2019) Effective concentrations enforced by intrinsically disordered linkers are governed by polymer physics. Proc. Natl. Acad. Sci. U. S. A 116, 23124–23131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyla M. and Kjaergaard M. (2020) Intrinsically disordered linkers control tethered kinases via effective concentration. Proc. Natl. Acad. Sci. U. S. A 117, 21413–21419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-Foutel NS et al. (2022) Conformational buffering underlies functional selection in intrinsically disordered protein regions. Nat. Struct. Mol. Biol 29, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuevas-Velazquez CL et al. (2021) Intrinsically disordered protein biosensor tracks the physical-chemical effects of osmotic stress on cells. Nat. Commun 12, 5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moses D. et al. (2022) Structural biases in disordered proteins are prevalent in the cellbioRxiv, 2021.11.24.469609 [Google Scholar]
- 35.Soranno A. et al. (2017) Integrated view of internal friction in unfolded proteins from single-molecule FRET, contact quenching, theory, and simulations. Proc. Natl. Acad. Sci. U. S. A 114, E1833–E1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soranno A. (2020) Physical basis of the disorder-order transition. Arch. Biochem. Biophys 685, 108305 [DOI] [PubMed] [Google Scholar]
- 37.Martin EW and Holehouse AS (2020) Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack thereof. Emerg Top Life Sci 4, 307–329 [DOI] [PubMed] [Google Scholar]
- 38.Elkjær S. et al. (2023) Evolutionary fine-tuning of residual helix structure in disordered proteins manifests in complex structure and lifetime. Commun Biol 6, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wicky BIM et al. (2017) Affinity of IDPs to their targets is modulated by ion-specific changes in kinetics and residual structure. Proc. Natl. Acad. Sci. U. S. A 114, 9882–9887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J. et al. (2017) Conformational Heterogeneity and FRET Data Interpretation for Dimensions of Unfolded Proteins. Biophys. J 113, 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuertes G. et al. (2017) Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proc. Natl. Acad. Sci. U. S. A 114, E6342–E6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banani SF et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nott TJ et al. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riback JA et al. (2017) Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168, 1028–1040.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franzmann TM et al. (2018) Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654 [DOI] [PubMed] [Google Scholar]
- 46.Patel A. et al. (2017) ATP as a biological hydrotrope. Science 356, 753–756 [DOI] [PubMed] [Google Scholar]
- 47.Yoo H. et al. (2019) Cellular sensing by phase separation: Using the process, not just the products. J. Biol. Chem 294, 7151–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuler B. et al. (2021) Impact of in-cell and in-vitro crowding on the conformations and dynamics of an intrinsically disordered protein. Angew. Chem. Int. Ed Engl DOI: 10.1002/anie.202016804 [DOI] [PubMed] [Google Scholar]
- 49.Zheng W. et al. (2016) Probing the action of chemical denaturant on an intrinsically disordered protein by simulation and experiment. J. Am. Chem. Soc 138, 11702–11713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuevas-Velazquez CL et al. (2016) The unstructured n-terminal region of Arabidopsis group 4 late embryogenesis abundant (LEA) proteins is required for folding and for chaperone-like activity under water deficit. J. Biol. Chem 291, 10893–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bremer A. et al. (2017) Folding of intrinsically disordered plant LEA proteins is driven by glycerol-induced crowding and the presence of membranes. FEBS J. 284, 919–936 [DOI] [PubMed] [Google Scholar]
- 52.Eicher JE et al. (2022) Secondary structure and stability of a gel-forming tardigrade desiccation-tolerance protein. Protein Sci. 31, e4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malki A. et al. (2022) Intrinsically Disordered Tardigrade Proteins Self-Assemble into Fibrous Gels in Response to Environmental Stress. Angew. Chem. Int. Ed Engl 61, e202109961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Martinez S. et al. (2023) Labile assembly of a tardigrade protein induces biostasisbioRxiv, 2023.06.30.547219 [Google Scholar]
- 55.Vancraenenbroeck R. et al. (2019) Polymer effects modulate binding affinities in disordered proteins. Proc. Natl. Acad. Sci. U. S. A 116, 19506–19512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maity H. et al. (2022) Salt-Induced Transitions in the Conformational Ensembles of Intrinsically Disordered Proteins. J. Phys. Chem. B 126, 5959–5971 [DOI] [PubMed] [Google Scholar]
- 57.Soranno A. et al. (2014) Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proc. Natl. Acad. Sci. U. S. A 111, 4874–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stringer MA et al. (2023) Excluded Volume and Weak Interactions in Crowded Solutions Modulate Conformations and RNA Binding of an Intrinsically Disordered Tail. J. Phys. Chem. B 127, 5837–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis CM et al. (2018) How does solvation in the cell affect protein folding and binding? Curr. Opin. Struct. Biol 48, 23–29 [DOI] [PubMed] [Google Scholar]
- 60.Schwartz L. et al. (2017) Out of Warburg effect: An effective cancer treatment targeting the tumor specific metabolism and dysregulated pH. Semin. Cancer Biol 43, 134–138 [DOI] [PubMed] [Google Scholar]
- 61.Thaker SK et al. (2019) Viral hijacking of cellular metabolism. BMC Biol. 17, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwartz L. et al. (2020) Cancer and Alzheimer’s disease: intracellular pH scales the metabolic disorders. Biogerontology 21, 683–694 [DOI] [PubMed] [Google Scholar]
- 63.Zhu J. and Thompson CB (2019) Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol 20, 436–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kosol S. et al. (2013) Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules 18, 10802–10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theillet F-X et al. (2016) Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature 530, 45–50 [DOI] [PubMed] [Google Scholar]
- 66.Dedmon MM et al. (2002) FlgM gains structure in living cells. Proc. Natl. Acad. Sci. U. S. A 99, 12681–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McNulty BC et al. (2006) Macromolecular crowding in the Escherichia coli periplasm maintains α-synuclein disorder. J. Mol. Biol 355, 893–897 [DOI] [PubMed] [Google Scholar]
- 68.König I. et al. (2015) Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat. Methods 12, 773–779 [DOI] [PubMed] [Google Scholar]
- 69.Sukenik S. et al. (2017) Weak protein-protein interactions in live cells are quantified by cell-volume modulation. Proc. Natl. Acad. Sci. U. S. A 114, 6776–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Büning S. et al. (2017) Conformational dynamics and self-association of intrinsically disordered Huntingtin exon 1 in cells. Phys. Chem. Chem. Phys 19, 10738–10747 [DOI] [PubMed] [Google Scholar]
- 71.Record MT, Jr et al. (1998) Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem 51, 281–353 [DOI] [PubMed] [Google Scholar]
- 72.Wang Y. et al. (2018) Cell Volume Controls Protein Stability and Compactness of the Unfolded State. J. Phys. Chem. B 122, 11762–11770 [DOI] [PubMed] [Google Scholar]
- 73.Wright PE and Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol 293, 321–331 [DOI] [PubMed] [Google Scholar]
- 74.Wohl S. et al. (2021) Salt-Dependent Conformational Changes of Intrinsically Disordered Proteins. J. Phys. Chem. Lett 12, 6684–6691 [DOI] [PubMed] [Google Scholar]
- 75.Huihui J. et al. (2018) Modulating charge patterning and ionic strength as a strategy to induce conformational changes in intrinsically disordered proteins. J. Chem. Phys 149, 085101 [DOI] [PubMed] [Google Scholar]
- 76.Zhang M. et al. (2022) The intrinsically disordered region from PP2C phosphatases functions as a conserved CO sensor. Nat. Cell Biol 24, 1029–1037 [DOI] [PubMed] [Google Scholar]
- 77.Zarin T. et al. (2021) Identifying molecular features that are associated with biological function of intrinsically disordered protein regions. Elife 10, e60220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langstein-Skora I. et al. (2022) Sequence- and chemical specificity define the functional landscape of intrinsically disordered regionsbioRxiv, 2022.02.10.480018 [Google Scholar]
- 79.Triandafillou CG et al. (2020) Transient intracellular acidification regulates the core transcriptional heat shock response. Elife 9, e54880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gutierrez JI et al. (2022) SWI/SNF senses carbon starvation with a pH-sensitive low-complexity sequence. Elife 11, e70344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dogra P. et al. (2019) Intermolecular Charge-Transfer Modulates Liquid-Liquid Phase Separation and Liquid-to-Solid Maturation of an Intrinsically Disordered pH-Responsive Domain. J. Am. Chem. Soc 141, 20380–20389 [DOI] [PubMed] [Google Scholar]
- 82.Baidya L. and Reddy G. (2022) pH Induced Switch in the Conformational Ensemble of Intrinsically Disordered Protein Prothymosin-α and Its Implications for Amyloid Fibril Formation. J. Phys. Chem. Lett 13, 9589–9598 [DOI] [PubMed] [Google Scholar]
- 83.Fossat MJ and Pappu RV (2019) q-Canonical Monte Carlo Sampling for Modeling the Linkage between Charge Regulation and Conformational Equilibria of Peptides. J. Phys. Chem. B 123, 6952–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fossat MJ et al. (2021) Quantifying charge state heterogeneity for proteins with multiple ionizable residues. Biophys. J 120, 5438–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fossat MJ et al. (2023) Uncovering the Contributions of Charge Regulation to the Stability of Single Alpha Helices. Chemphyschem 24, e202200746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guillén-Boixet J. et al. (2020) RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boothby TC and Pielak GJ (2017) Intrinsically Disordered Proteins and Desiccation Tolerance: Elucidating Functional and Mechanistic Underpinnings of Anhydrobiosis. Bioessays 39, 1700119 [DOI] [PubMed] [Google Scholar]
- 88.Battaglia M. et al. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boothby TC et al. (2017) Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol. Cell 65, 975–984.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dorone Y. et al. (2021) A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell 184, 4284–4298.e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang B. et al. (2022) Condensation of SEUSS promotes hyperosmotic stress tolerance in Arabidopsis. Nat. Chem. Biol 18, 1361–1369 [DOI] [PubMed] [Google Scholar]
- 92.Boyd-Shiwarski CR et al. (2022) WNK kinases sense molecular crowding and rescue cell volume via phase separation. Cell 185, 4488–4506.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jalihal AP et al. (2020) Multivalent Proteins Rapidly and Reversibly Phase-Separate upon Osmotic Cell Volume Change. Mol. Cell 79, 978–990.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamata H. and Hirata H. (1999) Redox regulation of cellular signalling. Cell. Signal 11, 1–14 [DOI] [PubMed] [Google Scholar]
- 95.Launay H. et al. (2019) Orchestration of algal metabolism by protein disorder. Arch. Biochem. Biophys 672, 108070 [DOI] [PubMed] [Google Scholar]
- 96.Zavaliev R. et al. (2020) Formation of NPR1 Condensates Promotes Cell Survival during the Plant Immune Response. Cell 182, 1093–1108.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang X. et al. (2021) ROS regulated reversible protein phase separation synchronizes plant flowering. Nat. Chem. Biol 17, 549–557 [DOI] [PubMed] [Google Scholar]
- 98.Lin Y. et al. (2020) Redox-mediated regulation of an evolutionarily conserved cross-β structure formed by the TDP43 low complexity domain. Proc. Natl. Acad. Sci. U. S. A 117, 28727–28734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kato M. et al. (2019) Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell 177, 711– 721.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang M. et al. (2013) Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation. Proceedings of the National Academy of Sciences 110, 4828–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Newcombe EA et al. (2021) Insight into Calcium-Binding Motifs of Intrinsically Disordered Proteins. Biomolecules 11, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Asiani KR et al. (2016) SilE is an intrinsically disordered periplasmic “molecular sponge” involved in bacterial silver resistance. Mol. Microbiol 101, 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Salzano G. et al. (2019) Structural Consequences of Copper Binding to the Prion Protein. Cells 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bhopatkar AA and Rangachari V. (2021) Are granulins copper sequestering proteins? Proteins 89, 450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rawlings AE et al. (2020) Investigating the ferric ion binding site of magnetite biomineralisation protein Mms6. PLoS One 15, e0228708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yi S. et al. (2007) Effects of zinc binding on the structure and dynamics of the intrinsically disordered protein prothymosin alpha: evidence for metalation as an entropic switch. Biochemistry 46, 13120–13130 [DOI] [PubMed] [Google Scholar]
- 107.Krainer G. et al. (2021) Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun 12, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu F. and Sukenik S. (2023) Structural Preferences Shape the Entropic Force of Disordered Protein Ensembles. J. Phys. Chem. B 127, 4235–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeno WF et al. (2018) Synergy between intrinsically disordered domains and structured proteins amplifies membrane curvature sensing. Nat. Commun 9, 4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Has C. et al. (2022) Insights into Membrane Curvature Sensing and Membrane Remodeling by Intrinsically Disordered Proteins and Protein Regions. J. Membr. Biol 255, 237–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.König I. et al. (2021) Impact of in- cell and in- vitro crowding on the conformations and dynamics of an intrinsically disordered protein. Angew. Chem. Weinheim Bergstr. Ger 133, 10819–10824 [DOI] [PubMed] [Google Scholar]
- 112.Banks A. et al. (2018) Intrinsically Disordered Protein Exhibits Both Compaction and Expansion under Macromolecular Crowding. Biophys. J 114, 1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zosel F. et al. (2020) Depletion interactions modulate the binding between disordered proteins in crowded environments. Proc. Natl. Acad. Sci. U. S. A 117, 13480–13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sang D. et al. (2022) Condensed-phase signaling can expand kinase specificity and respond to macromolecular crowding. Mol. Cell 82, 3693–3711.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wuttke R. et al. (2014) Temperature-dependent solvation modulates the dimensions of disordered proteins. Proc. Natl. Acad. Sci. U. S. A 111, 5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jephthah S. et al. (2019) Temperature Dependence of Intrinsically Disordered Proteins in Simulations: What are We Missing? J. Chem. Theory Comput 15, 2672–2683 [DOI] [PubMed] [Google Scholar]
- 117.Jung J-H et al. (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 [DOI] [PubMed] [Google Scholar]
- 118.Zhu P. et al. (2021) Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 599, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quiroz FG and Chilkoti A. (2015) Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater 14, 1164–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Burgie ES et al. (2021) Differing biophysical properties underpin the unique signaling potentials within the plant phytochrome photoreceptor families. Proc. Natl. Acad. Sci. U. S. A 118, e2105649118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cai D. et al. (2019) Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol 21, 1578–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watanabe K. et al. (2021) Cells recognize osmotic stress through liquid-liquid phase separation lubricated with poly(ADP-ribose). Nat. Commun 12, 1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morishita K. et al. (2023) Sodium ion influx regulates liquidity of biomolecular condensates in hyperosmotic stress response. Cell Rep. 42, 112315 [DOI] [PubMed] [Google Scholar]
- 124.Krzysiak TC et al. (2018) An Insulin-Responsive Sensor in the SIRT1 Disordered Region Binds DBC1 and PACS-2 to Control Enzyme Activity. Mol. Cell 72, 985–998.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]