Abstract
Transcriptional dysregulation is a hallmark of cancer and can be driven by altered enhancer landscapes. Recent studies in genome organization have revealed that multiple enhancers and promoters can spatially coalesce to form dynamic topological assemblies, known as promoter–enhancer hubs, that strongly correlate with elevated gene expression. In this review, we discuss the structure and complexity of promoter–enhancer hubs recently identified in multiple cancer types. We will further discuss underlying mechanisms driving dysregulation of promoter–enhancer hubs and speculate on their functional role in pathogenesis. Understanding the role of promoter–enhancer hubs in transcriptional dysregulation can provide insight into new therapeutic approaches to target these complex features of genome organization.
Keywords: promoter–enhancer hub, multiway interaction, cancer
Multiple enhancer and promoter elements can spatially coalesce
Spatial organization of mammalian genomes contributes to gene expression control and cell identity. The genome’s spatial organization is set at various length scales, ranging from chromosome territories (see Glossary) to megabase-scale transcriptionally active and inactive compartments, topologically associating domains (TADs), and fine-scale chromatin loops [1-4]. Compartments, TADs, and chromatin loops contribute to positioning of regulatory elements, including enhancers and promoters, influencing their activity and specificity (reviewed in [3]). Enhancers are non-protein coding, cis-regulatory sequences containing numerous binding sites for transcription factors (TFs). Upon binding to an enhancer, TFs facilitate increased chromatin accessibility and recruit coactivators and histone modifying enzymes [5,6]. Because TF binding depends on contextual information specified by cell lineage and/or signaling pathways, enhancers can integrate environmental and developmental cues to fine-tune gene expression [7].
Despite several proposed models (reviewed in [8]), it is not yet fully understood how distal enhancers exert their regulatory function across great genomic distances. Most models agree that proximity between the enhancer and promoter, even transiently, is essential for enhancer function [8,9]. Promoter–enhancer interactions could be mediated and/or stabilized by cohesin-mediated loop extrusion [10], transcriptional machinery and coactivator bridging [11], and TF/cofactor-mediated phase separation and oligomerization [12,13]. Adding to the complexity is the fact that the number of active genes in a cell is 2-3 times less than that of active enhancers [14]. Thus, it is often possible that the expression of a single gene is controlled by multiple enhancers, giving rise to complex gene expression control circuits [15,16]. In some cases, multiple enhancers in close genomic proximity cluster together, forming super-enhancers [17]. There are also cases where multiple gene promoters are controlled by the same set of enhancers, potentially allowing regulatory information to be relayed across multiple genes [18,19].
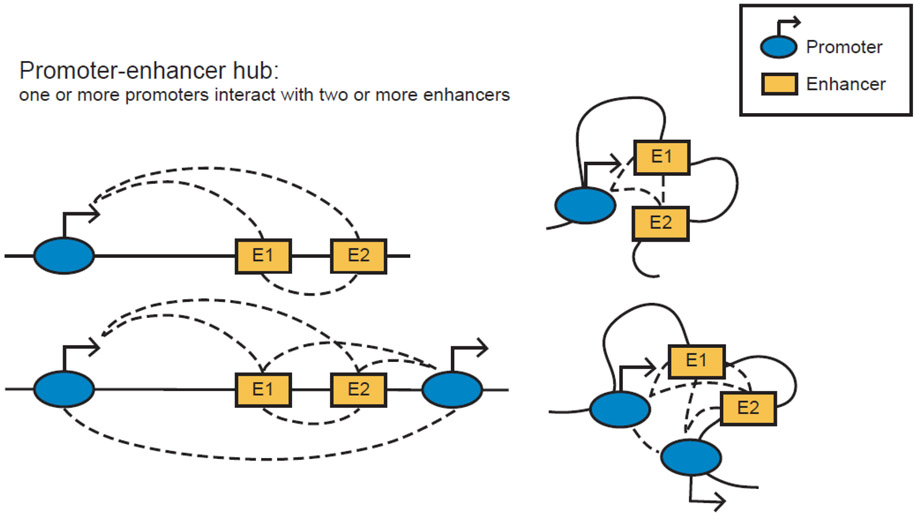
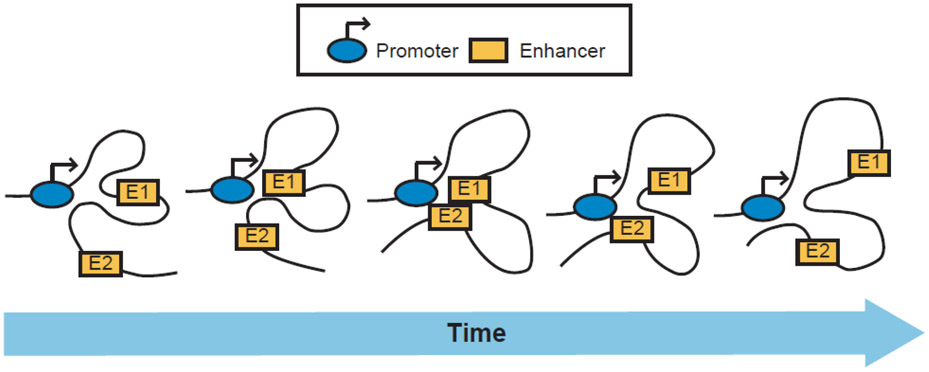
Technical advancements in chromatin conformation capture technologies have pointed to the existence of promoter–enhancer hubs or cliques, which we define here as dynamic topological assemblies with one or more promoters that interact, even transiently, amongst themselves and with two or more enhancers that may or may not be connected (Figure 1). Although most studies were limited to examining pairwise interactions between enhancers and promoters, recent work started to reveal that multiway interactions are more prevalent and complex than previously appreciated [20-24]. The frequency of multiway contacts is relatively low, suggesting that promoter–enhancer hubs are dynamic topological assemblies rather than rigid physical structures (Figure 2). Although the organizational principles, regulators, and functions of promoter–enhancer hubs remain largely unclear, detailed examination of multiple loci shows that spatial interactivity of enhancers with each other and cognate promoters strongly correlates with transcriptional activation and often regulate genes critical for cell identity [18,25-29]. Several reviews discussed the role of promoter–enhancer hubs as well as proposed mechanisms of their formation in normal differentiation and cell function [30-33] and hence will not be our focus here.
Figure 1: Graphical representation of promoter–enhancer hubs linearly and spatially.
Top: one promoter regulated by two enhancers. Bottom: two promoters that are regulated by two enhancers.
Figure 2: Evidence suggests that multiway interactions among enhancers and promoters are dynamic.
Hence, a promoter may be interacting with one, both, or none of its enhancers at a given time.
Recent evidence suggests that promoter–enhancer hubs can also play a role in regulating disease susceptibility and pathogenesis, including cancer. A key hallmark of cancer is aberrant gene expression control, which can be influenced by both genetic and epigenetic events that disrupt enhancer activity and nuclear positioning [34-41]. But how do cancer-specific promoter–enhancer hubs contribute to transcriptional dysregulation? This review will focus on recent evidence from diverse cancer types suggesting that coalescence or separation of promoter–enhancer hubs contribute to transcriptional regulation in cancer (Table 1, Key table). We will further speculate on their mechanisms of assembly and functional implication in pathogenesis based on available evidence from literature. We will start by describing new technological advancements that enable better detection of these dynamic topological assemblies.
Table 1. Key table.
Summary of loci/genes discussed in this review that potentially form a promoter–enhancer hub in different cancer types.
| Cancer type | Gene(s) affected |
Potential mechanism | Gene function |
Refs |
|---|---|---|---|---|
| Prostate Cancer | FOXO1 | Unknown | tumor suppressor | [71] |
| AR | Unknown; CTCF independent | oncogene | [72] | |
| Squamous cell lung carcinoma | FOXE1 | KLF5 binding | oncogene | [78] |
| TP63 | KLF5 binding | oncogene | [78] | |
| NUT carcinoma | MYC | BRD4-NUT fusion | oncogene | [83] |
| TP63 | BRD4-NUT fusion | oncogene | [83] | |
| SOX2 | BRD4-NUT fusion | oncogene | [83] | |
| Triple negative breast cancer | MYC | Notch Transcription Complex binding | oncogene | [24] |
| CCND1 | Notch Transcription Complex binding | oncogene | [24] | |
| TMPRSS2 | Notch Transcription Complex binding | oncogene | [24] | |
| RIPK4 | Notch Transcription Complex binding | oncogene | [24] | |
| Gastric cancer | ZFP36L2 | Tandem duplication | oncogene | [91] |
| Gastrointestinal stromal tumor | FGF3 | Disruption of insulation | oncogene | [93] |
| FGF4 | Disruption of insulation | oncogene | [93] | |
| KIT | Disruption of insulation | oncogene | [93] | |
| Colorectal cancer | CEACAM5 | KLF5 binding | oncogene | [78] |
| ETV4 | KLF5 binding | oncogene | [78] | |
| EPHA2 | Unknown | oncogene | [100] | |
| PDCD4 | Unknown | tumor suppressor | [100] | |
| Ph-like acute lymphoblastic leukemia | GATA3 | Single-nucleotide substitution rs3824662 | oncogene | [101] |
| CRLF2 | GATA3 binding | oncogene | [101] | |
| B-cell precursor acute lymphoblastic leukemia | FLT3 | Somatic 13q12.2 microdeletions | oncogene | [103] |
| Diffuse large B-cell lymphomas | Klf5 | H1 loss | oncogene | [105] |
| BCL6 | OCA-B-OCT2-MEF2B complex binding | oncogene | [106] | |
| Mantle cell lymphoma | MYC | Notch Transcription Complex binding | oncogene | [24] |
| LYN | Notch Transcription Complex binding | oncogene | [24] | |
| SH2B2 | Notch Transcription Complex binding | oncogene | [24] | |
| ORAI2 | Notch Transcription Complex binding | unknown | [24] | |
| LDLRAD4 | Notch Transcription Complex binding | unknown | [24] | |
| FAM210A | Notch Transcription Complex binding | unknown | [24] | |
| Chronic lymphocytic leukemia | EBF1 | Unknown | tumor suppressor | [110] |
| T-cell acute lymphoblastic leukemia | MYC | Disruption of insulation, Notch binding | oncogene | [113] |
| SOX4 | Unknown | oncogene | [114] | |
| HOXA genes | Fusion proteins such as SET-NUP214 | oncogene | [114] | |
| Acute Myeloid Leukemia | CDK5 | Chromosome 11 and 7 fusion | oncogene | [118] |
| HSF4 | inv(16) | unknown | [118] | |
| MYC | translocations | Oncogene | [118] | |
| CBL | translocations | Oncogene | [118] |
New technologies to study promoter–enhancer hubs
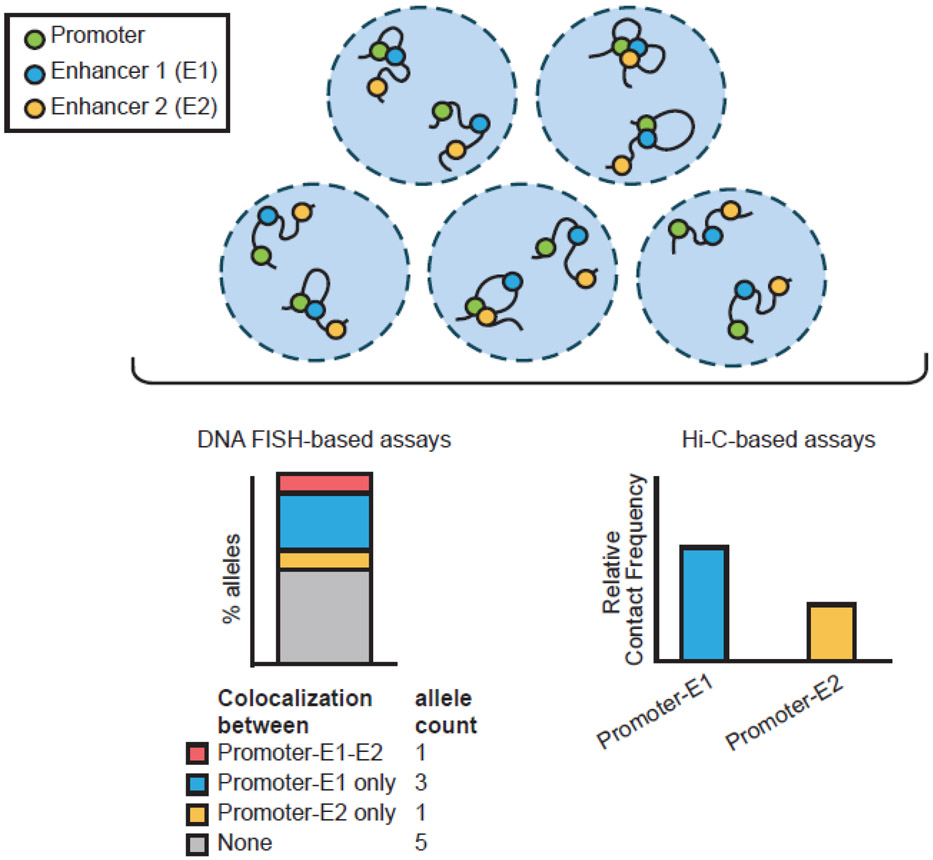
Proximity ligation-based chromatin conformation capture technologies such as Hi-C have enabled the genome-wide study of long-range enhancer–promoter interactions (Box 1). Yet, these assays can only measure pairwise interactions with high confidence. Further, results from chromatin conformation capture assays cannot distinguish between co-occurring interactions in the same cells and mutually exclusive interactions occurring in different cells (Figure 3). To overcome these limitations, variants of chromatin conformation capture protocols based on targeted (Tri-C, TM3C, COLA), long-read (MC-4C), and molecular barcoding (C-walks) technologies were developed [42-46]. Except for TM3C, these assays are not genome-wide. Further, the targeted-based methods cannot resolve beyond three-way interactions. Pore-C, a more recent approach, relies on long-read sequencing like MC-4C, but in addition enables genome-wide study of multiway contacts [47].
Box 1. Hypothesizing promoter–enhancer hubs’ existence from Hi-C-based assays.
Proximity ligation-based chromatin conformation capture technologies such as Hi-C enables the genome-wide study of genome folding, painting a more comprehensive picture of the organizational principles and function of DNA looping in health and disease [2,130,131]. Despite limited sequencing depth and resolution, earlier Hi-C-based studies confirmed the presence of lineage-restricted long-range pairwise enhancer–promoter, enhancer–enhancer, and promoter–promoter interactions [2,132]. Furthermore, these studies led to the hypothesis that promoter–enhancer hubs exist within mammalian genomes, formed as a result of long-range contacts among gene promoters and a number of distal enhancer elements [2,132,133].
To improve on the ability of Hi-C in detecting enhancer–promoter interactions, antibody or oligo-based enrichment steps can be added to the protocol. Antibody enrichment of chromatin factors including RNA Pol II, cohesin complex components, and active enhancer marks using HiChIP [134], ChIA-PET [135], or PLAC-seq [136] drastically improved the discovery rate of enhancer–promoter contacts by selectively enriching for the factors that are associated with these genomic elements. On the other hand, Promoter [137] and Enhancer [58] Capture Hi-C leverage oligo-based enrichment to enable higher resolution detection of either distal promoter-interacting or enhancer-interacting regions, respectively.
Figure 3: Various assays offer different perspectives of enhancer–promoter interactions.
Top: five nuclei depicting two alleles that contain a promoter (green), and two enhancers (blue and yellow). Bottom-left: DNA-FISH-based assays can be used to measure pairwise and multiway interactions at individual alleles of a given locus. Bottom-right: Hi-C-based assays cannot measure multiway interactions but can be used to find highly connected promoter–enhancer pairs genome-wide.
Approaches without proximity ligation have also been developed to study interactions among regulatory elements, including GAM and SPRITE [20,48,49]. SPRITE uniquely identifies fragment ends and overcomes resolution limitations of chromatin conformation capture-based techniques in identifying multiway interactions by repetitive pooling, splitting, and barcoding DNA fragments [20,48].
In addition to sequencing technologies, microscopy techniques have enabled the single-cell resolution study of the physical location and proximity of promoters and enhancers. For the past two decades, 3D DNA fluorescence in situ hybridization (FISH) has identified many features of chromatin architecture [50], but these studies were limited to probing a few genomic elements at a time (Figure 3). Advancements in oligopaint-based FISH probes combined with high-throughput imaging led to the development of chromosome-tracing technologies, including ORCA, Hi-M, MINA, DNA-MERFISH, and DNA-seqFISH+ [51-55]. The common principle of all these techniques is sequential cycling through adding and imaging a fluorophore that uniquely binds to a barcoded primary oligo corresponding to a distinct genomic locus, followed by removing and replacing it with another fluorophore that recognizes the adjacent barcoded primary oligo. Together, these chromosome-tracing technologies enabled the reconstruction of DNA polymer folding in individual cells. Furthermore, the single-cell resolution of these studies revealed that enhancer–promoter interactions are heterogeneous and occur in domain-like structures consisting of contacts among multiple regulatory elements [53-55]. More recently, these advanced imaging-based technologies also provided evidence of multiway contacts between multiple enhancers, promoters, and insulators [21,22,56]. Despite these intriguing findings, current chromatin tracing studies are limited to a few genomic loci. Future work using higher-throughput microscopy-based technologies can further elucidate the extent of enhancer cooperativity or competitiveness and the involvement of RNA and protein molecules in hub formation at single-cell resolution.
Promoter–enhancer hubs in health and disease
Recent investigations into cellular differentiation suggest that the dynamic rewiring of promoter–enhancer hubs may play a key role in cell-fate decision. In pluripotent stem cells, immune cells, and neurons, promoters exhibiting higher levels of chromatin interactivity are enriched for lineage-specific genes, enabling fine-tuning of transcription during differentiation [18,23,29,57]. For example, Enhancer Capture Hi-C conducted during differentiation of human mesenchymal stem cells into adipocytes and osteoblasts revealed that transcription of lineage-specific genes is regulated by the coordinated activation and convergence of multiple enhancers to promoters, forming promoter–enhancer hubs, amplifying TF occupancy, and increasing cofactor recruitment [58].
In addition to integrating spatiotemporal signals, enhancers within a promoter-enhancer hub may exhibit context–specific behavior to fine-tune transcription [29]. Prior studies of multi-enhancer gene regulation without long-range interaction measurement revealed cooperative, redundant, or repressive functions of enhancers [15,59-62]. Although it remains unclear whether enhancers and promoters examined in these studies participate in spatial hubs, recent work revealed that enhancers in spatial hub can behave additively, synergistically, or redundantly [24,63-65].
Given the role of promoter–enhancer hubs in controlling gene expression during normal development, it is unsurprising that the re-wiring of these topological assemblies contributes to aberrant transcription in various diseases. Disease-associated sequence variations have been linked to mis-regulation of promoter–enhancer hubs, increasing the risk of developing metabolic, immune, and brain disorders [19,23,66-68]. Additionally, accumulating evidence suggests that promoter–enhancer hub dysregulation coincides with aberrant oncogene and tumor suppressor expression in cancer. Next, we will describe the complexity, regulation, and function of promoter–enhancer hubs gleaned from recent studies on cancer genome organization. As this is a nascent topic in cancer biology, in some cases, we interpret and describe the data from the perspective of promoter–enhancer hubs even when the original study did not specifically examine these topological assemblies.
Promoter–enhancer hubs in cancer
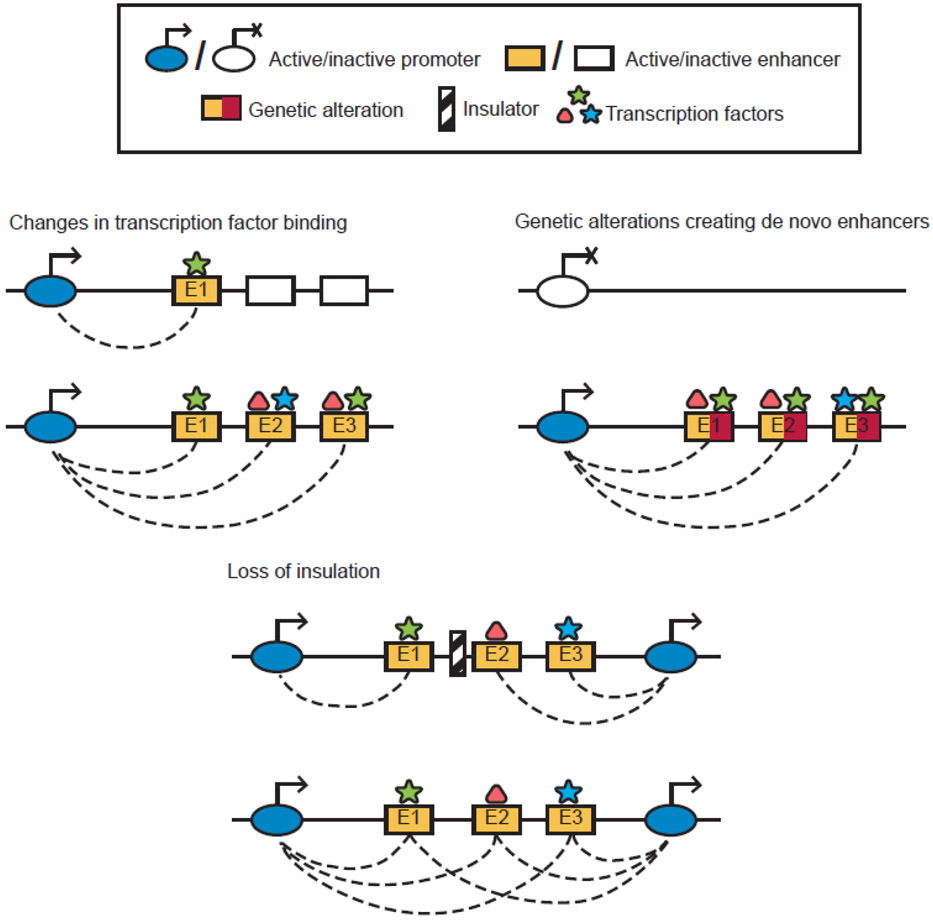
Promoter–enhancer hubs in cancer could be influenced by a plethora of genetic and epigenetic mechanisms (Figure 4). As described in the following sections, structural variants and deregulation of TFs and epigenetic regulators can alter enhancer function across different cancer types. The extent to which these events can mis-regulate gene expression often depends on a combination of enhancer activity and positioning relative to the target promoter [41,69]. Recent studies in various cancers uncovered cases where aberrant promoter–enhancer hubs dysregulate expression of oncogenes and tumor suppressors, contributing to malignant phenotypes.
Figure 4: Example of events influencing oncogenic promoter–enhancer hubs.
Top-left: changes in binding of one TF can activate previously inactive enhancers and recruit other partner TFs. Top-right: Genetic alterations can create novel binding sites for TFs and alter enhancer activity, inducing gene expression. Bottom: loss of insulation can lead to de novo contacts to enhancers.
Prostate Cancer
Prostate cancer is characterized by widespread transcriptional dysregulation [70]. To investigate the underlying mechanisms driving aberrant transcription in prostate cancer, several groups recently mapped the chromatin conformation in normal and malignant cells [71,72]. These studies identified major chromatin reorganization, including at the FOXO1 [71] and androgen receptor (AR) loci [72]. FOXO1, a tumor suppressor, is frequently downregulated in prostate cancer in the absence of genetic alterations [73]. Concomitant with FOXO1 downregulation, the promoter loses interactions with active compartments and connects to heterochromatic regions [71,74]. Although links between promoter–enhancer hubs and compartments remain unclear, this data shows the repression of multiple enhancers located at this locus, which can be interpreted as potential loss of the FOXO1 promoter–enhancer hub during tumorigenesis. In contrast, AR, whose upregulation is essential during tumorigenesis [75], acquires and interacts with oncogenic enhancers independent of CTCF binding changes [72]. Although the authors did not specifically define the AR locus as a hub, their observations suggest that AR participates in a cancer-restricted promoter–enhancer hub. Both studies showed that transcriptional changes correlate with alterations in enhancer activity and positioning in prostate cancer [71,72]. However, the chain of causality of these events in driving prostate cancer pathogenesis remains to be understood.
Squamous Cancers
Squamous cancer cells exhibit similar transcriptional programs and binding profiles of KLF5, a pro-proliferation TF, regardless of tissue of origin [76,77]. It was shown that KLF5 partners with lineage-specific TFs in squamous cancer cells to activate enhancers through CBP/EP300 methyltransferase complex and chromatin reader BRD4 recruitment [78]. KLF5-mediated histone acetylation promotes long-range interactions at loci with multiple enhancers, potentially forming promoter–enhancer hubs. This hypothesis is supported by the observation that several oncogenic enhancers synergize to promote Pol II elongation and activate transcription of oncogenes including FOXE1 and TP63. Similar to other TFs, direct inhibition of KLF5 is challenging. Liu et al. showed that degrading BRD4 can be an alternative strategy to selectively target KLF5-dependent cancer cells [78].
NUT carcinoma, a rare and aggressive squamous cell carcinoma subtype [79], is characterized by fusion between NUTM1 and a number of genes, mainly BRD4 (BRD4-NUT) [80,81]. BRD4-NUT drives the formation of mega-domains, or massive active genomic regions that contiguously bind BRD4-NUT as well as wild-type BRD4 and p300 [82]. Mega-domains are even larger than super-enhancers and coincide with key oncogenes, including MYC, TP63, and SOX2 (reviewed in [83]). The mega-domains form topological assemblies termed sub-compartment M that are characterized by high intradomain interactions across extremely large linear genomic distances. Sub-compartment M may potentiate an auto-regulatory feed-forward network to coregulate the expression of constituent genes [83,84]. Nevertheless, additional research is needed to investigate whether these topological assemblies can be selectively targeted in cancer.
Sarcoma
Ewing’s sarcoma is characterized by low mutation burden and the fusion between EWSR1 and ETS TFs, most commonly FLI1 [85]. The main oncogenic driver in this malignancy is EWS-FLI1, an aberrant fusion TF that drives epigenetic remodeling in permissive cells, resulting in their transformation [86]. Sanalkumar et al. demonstrated that EWS-FLI1 converts repetitive GGAA genomic sequences into highly inter-connected topological assemblies, connecting megabase-sized loci containing numerous enhancer–enhancer and enhancer–promoter interactions and promoting oncogenic expression [87]. Depletion of EWS-FLI1 induces chromatin reorganization in tumor cells and restores latent differentiation programs characteristic of the tumor precursor cells. Similar to NUT carcinomas, this study in Ewing’s sarcoma supports the idea that a single fusion protein can promote genome-wide nuclear reorganization and formation of oncogenic promoter–enhancer hubs.
Breast Cancer
Triple negative breast cancer (TNBC) accounts for more than 15% of breast cancer cases and exhibits an aggressive phenotype and higher relapse rate [88]. A recent study mapped promoter–enhancer interactions genome-wide in TNBC and found 140 hubs or cliques of enhancers and promoters with more than 100 interactions [24]. The spatial positioning of regulatory elements within cliques are preferentially regulated by oncogenic Notch, a TF that is crucial for TNBC tumorigenesis and its hyperactivation correlates with poor prognosis [88]. The authors found that many promoter–enhancer hubs form at key oncogenes, including MYC, CCND1, TMPRSS2, and RIPK4. The MYC promoter resides within a highly connected promoter–enhancer hub containing five super-enhancers [24]. The activity of the most distal and proximal super-enhancers in the MYC hub are the most Notch-dependent and behave synergistically. In contrast, the Notch-dependent enhancers within the CCND1 promoter–enhancer hub behave redundantly [24]. Notch can also coregulate expression of multiple genes by promoting looping within spatial hubs containing multiple promoters. For example, the TMPRSS2 / RIPK4 promoter–enhancer hub consists of highly interacting enhancers within a single super-enhancer. Comparison of the MYC and TMPRSS2 / RIPK4 loci suggests that some promoter–enhancer hubs closely align with aggregation of enhancer elements within a super-enhancer, while others involve multiple distally located super-enhancers. Overall, these results suggest that oncogenic TFs can markedly increase expression of key oncogenes by selectively targeting and positioning enhancers within highly connected promoter–enhancer hubs.
Gastrointestinal (GI) Cancers
Structural variations including tandem duplications can cause amplification of enhancer regions and activate proto-oncogenes [89]. ZFP36L2, an RNA-binding protein, behaves as an oncogene in pancreatic [90] and gastric cancers [91]. Xing et al. recently identified a novel tandem duplication hotspot in a 500 Kb region downstream of ZFP36L2, which is present in 10% of gastric cancers and associates with elevated ZFP36L2 [91]. The duplicated region contains up to eleven distal enhancer elements highly interacting with the ZFP36L2 promoter. The enhancers are organized hierarchically as deletion of one of the enhancers, but not the others, almost completely abrogates ZFP36L2 expression [91]. Although this study only reported enhancer–promoter but not enhancer–enhancer interactions, the data suggests the hypothesis that ZFP36L2 and its interacting enhancers form a promoter–enhancer hub. By contrast, ZFP36L2 is lowly expressed and functions as a tumor suppressor in esophageal squamous cell carcinoma, where unlike gastric cancer, the ZFP36L2 downstream region is hypermethylated and inactive [92]. These results illustrate that ZFP36L2 can exert opposite functions in different cancers.
Changes in TF specificity and DNA methylation are proposed epigenetic mechanisms altering promoter-enhancer hubs [78,93,94]. In colorectal cancer, KLF5, an essential TF, exhibits distinct binding patterns compared to squamous cell cancers [78]. Altered KLF5 binding promotes spatial hub formation at CEACAM5 and ETV4 loci in colorectal cancer, consistent with their aberrant upregulation [78]. CEACAM5 is overexpressed in 90% of colorectal cancer cases and correlates with poor prognosis [95], while ETV4 promotes proliferation and migration in this cancer type [96].
Succinate dehydrogenase (SDH) and isocitrate dehydrogenase (IDH) mutations are common initiating events in many tumor types [93]. In SDH and IDH-deficient tumors, the accumulation of succinate or 2-hydroxyglutarate, respectively, inhibits demethylases and causes DNA hypermethylation [97]. Increase in methylation disrupts CTCF binding [94], resulting in tumors with widespread insulator inactivation, local genome folding reorganization, and aberrant gene activation [93,98]. For example, in SDH-deficient GI stromal tumors, hypermethylation-mediated disruption of an insulator 3’ of oncogenes FGF3 and FGF4 leads to their upregulation and ectopic interactions between their promoters and multiple interconnected enhancers within a super-enhancer, potentially forming a hub consisting of two promoters and multiple enhancers [93]. This alteration is allele-specific and represents a stable epigenetic event, indicative of propagation through a malignant clone. SDH-deficiency similarly leads to the loss of an insulator 5’ of tyrosine kinase KIT in primary tumors, increasing looping between the KIT promoter and its upstream enhancers. Although KIT is already highly expressed in GI stromal tumors, loss of this insulator further upregulates KIT. Simultaneous FGF and KIT hyperactivation in these tumors increases crosstalk between the two signaling pathways, conferring resistance to KIT inhibitors [99].
Proto-oncogene EPHA2 encodes another receptor tyrosine kinase with an altered enhancer landscape in GI cancers. In colorectal tumors, one of the three enhancers in EPHA2’s promoter–enhancer hub gains activity and interactions with the promoter [100]. In the same samples, tumor suppressor PDCD4 is downregulated and less frequently interacts with one of its two distal enhancers located within a spatial hub [100]. Although changes in enhancer connectivity are observed across multiple colon tumor samples, the underlying mechanisms of these epigenetic changes remain unknown.
B Cell Malignancies
Cancer-associated mutations in non-coding regions are prevalent and growing evidence suggests that these mutations can impact chromatin organization. Inherited genetic variations are important determinants of susceptibility to acute lymphoblastic leukemia (ALL), but how these variants in non-coding sequences increase leukemia risk is less understood. Yang et al. showed that the rs3824662 variant at the GATA3 locus associates with susceptibility to pediatric Philadelphia chromosome-like (Ph)-like ALL [101]. This variant creates a de novo enhancer that loops to GATA3 and other putative GATA3 enhancers, forming a hub consisting of a single promoter and multiple enhancers. Alteration of the GATA3 promoter–enhancer hub directly increases GATA3 expression in lymphoblasts and is associated with higher GATA3 expression in multiple ALL subtypes [101]. Further, GATA3 upregulation associates with widespread genome reorganization in Ph-like ALL patients with rs3824662 variant. For example, increased GATA3 binding is associated with concomitant CRLF2 upregulation, formation of a new enhancer–promoter loop, and increased interactions with other enhancers within the CRLF2 promoter–enhancer hub [101]. CRLF2 mis-regulation is a hallmark of Ph-like ALL, mediates constitutive activation of the JAK-STAT pathway, and promotes leukemogenesis [102]. This study exemplifies how a single germline variation can increase cancer risk by altering TF binding and inducing genome reorganization, setting a key oncogenic transcriptional program.
Somatic 13q12.2 microdeletions at the FLT3 locus is one of the most common drivers of ALL [103]. In B-cell precursor ALL, the deletion disrupts insulation at the FLT3 locus, enabling the gene promoter to hijack and interact with an additional enhancer distal from the deletion breakpoints, leading to allele-specific FLT3 mis-regulation [103]. Although this study only showed enhancer–promoter and not enhancer–enhancer interactions, we hypothesized that changes in chromatin looping leads to gain of a promoter–enhancer hub comprised of FLT3 and its two interacting enhancers.
H1 proteins are linker histones that limit chromatin accessibility and act as transcriptional repressors [104]. Recurrent H1 mutations occur in approximately 30-40% of diffuse large B-cell lymphomas (DLBCL) [105]. Loss of H1 in germinal center (GC) B cells, the DLBCL cell-of-origin, decompacts and upregulates stem cell genes, including Klf5 [105]. Klf5 gains interactions with two distal enhancers that become accessible in H1-mutant GC B cells, consistent with Klf5 upregulation in H1-deficient lymphomas [105]. Enhanced self-renewal properties in lymphoma cells due to H1 loss may partly explain why H1-mutant DLBCL is highly aggressive. Further, the newly Klf5-interacting enhancers are enriched for OCT2 binding. Interestingly, another study on DLBCL found the enrichment of OCT2, OCA-B, and MEF2B ternary complexes as a key regulator of BCL6’s promoter–enhancer hub [106]. Sustained BCL6 expression is essential and regulated by a locus control region (LCR) containing 10 active enhancers highly interacting with the promoter in GC B and DLBCL cells [106-108]. Loss of one LCR enhancer reduces interaction frequency between the BCL6 promoter and non-targeted LCR enhancers, suggesting the LCR forms a cooperative promoter–enhancer hub with BCL6. A CRISPRi screen of the LCR revealed hierarchical organization of enhancers within the hub. Their individual essentiality in driving BCL6 expression and promoting cell survival is regulated by enrichment of the OCT2-MEF2B-OCA-B ternary complex and not enhancer activity [106]. Further studies in DLBCL are required to determine whether OCT2 and its partners can access closed chromatin to activate enhancers or if additional events like H1 loss are necessary to enhance chromatin accessibility.
Mutations affecting TFs can lead to their signal-independent activation and aberrant chromatin binding. For instance, activating NOTCH1 mutations can lead to ligand-independent release of NOTCH1 into the nucleus, recruiting transcriptional cofactors to chromatin [109]. In mantle cell lymphoma (MCL), aberrant Notch signaling upregulates crucial genes for cancer survival, including MYC, LYN, and SH2B2 [24]. In addition to TNBC, Notch signaling is also implicated in multi-gene coregulation in MCL by promoting hub formation at loci with multiple promoters, including the SH2B2 / ORAI2 and LDLRAD4 / FAM210A loci. Oncogenic NOTCH1 transcriptionally controls these genes by positioning their respective enhancers within promoter–enhancer hubs, even when the enhancer activity is NOTCH-independent. However, the factors determining why certain NOTCH-dependent promoter–enhancer hubs exhibit NOTCH-independent enhancer activity remain unclear.
Promoter–enhancer hubs can also be cancer-protective. For instance, EBF1 is a TF that is expressed and regulated by highly interconnected enhancers in naïve and mature B cells [110], which are thought to be the cells of origin for chronic lymphocytic leukemia (CLL) [111]. In CLL, most of the looping interactions and enhancer activity at the EBF1 locus are lost [110]. Loss of the promoter–enhancer hub results in EBF1 silencing, reducing levels of multiple B cell signaling factors and leading to anergy and low susceptibility to recognition by the host’s immune system, characteristics that are often observed in CLL patients [110].
T Cell Acute Lymphoblastic Leukemia (T-ALL)
T-ALL is an aggressive hematological malignancy resulting from transformed T cell progenitors [112]. Several groups mapped differential genome folding in primary T-ALL blasts and peripheral T cells to characterize chromatin reorganization during malignant transformation [113,114]. Widespread differences were observed in leukemic cells, including over 6000 differential loops [114].
NOTCH1 is mutated in more than 50% of T-ALL cases and contributes to leukemogenesis [112]. Like TNBC and MCL, MYC is a key Notch target in T-ALL [24,113-116]. Kloetgen et al. showed that loss of a MYC insulator element increases MYC expression and interactions with a NOTCH1-bound and a NOTCH1-unbound super-enhancer [113]. Although this study did not show interactions among the two MYC-interacting super-enhancers, another study found that the MYC promoter and its two super-enhancers are highly interconnected [115]. Zhou et al. further used Oligpaint DNA FISH and confirmed the formation of the MYC promoter–enhancer hub in individual T-ALL cells. Notch inhibition reduces MYC expression and NOTCH1-bound super-enhancer activity [113,115,116]. However, unlike TNBC and MCL [24], loss of Notch signaling has no impact on MYC promoter–enhancer looping in T-ALL. In contrast to Notch inhibition, CDK7 inhibition significantly reduces MYC promoter–enhancer interactions, suggesting that additional factors including CDK7 maintain enhancer positioning in a subset of T-ALL enhancers [113]. Further understanding of the factors maintaining or establishing genome folding could lead to new targets for T-ALL therapy.
Proto-oncogene SOX4 is another T-ALL associated TF. Yang et al. showed that SOX4 is derepressed in T-ALL and gains new loops to three distal enhancers, forming a T-ALL-restricted promoter–enhancer hub [114]. Differential looping is associated with changes in CTCF binding and H3K27ac loading, but not copy number alterations. In addition, HOXA genes are also commonly derepressed and correlates with poor prognosis [117]. Fusion proteins including SET-NUP214 can transactivate HOXA genes, increasing HOXA enhancer activity and promoter-interactions and credentialing the hypothesis that a spatial hub containing multiple promoters and enhancers forms at this locus. In line with sarcoma and NUT carcinomas, these observations in T-ALL provide another example where oncogenic fusion proteins can commission promoter–enhancer hubs.
Although Kloetgen et al. and Yang et al. reported widespread differential genome folding in T-ALL compared to healthy peripheral T cells [113,114], precaution must be taken when interpreting these results as many differences might be due to genome reorganization during normal T cell differentiation and not malignant transformation. Further studies using the corresponding cell-of-origin are needed to elucidate T-ALL-specific changes in genome organization.
Acute Myeloid Leukemia (AML)
To study genome organization changes in AML blasts compared to normal hematopoietic progenitors, Xu et al. integrated chromatin conformation, accessibility, enhancer activity, gene expression, and whole-genome sequencing data [118]. The authors identified several structural variations associated with enhancer hijacking events creating AML-specific enhancer–promoter loops, an observation also made in other cancer types [89,91,103]. For instance, a fusion between chromosomes 11 and 7 connects several enhancers to CDK5 and inv(16) creates new loops linking HSF4 to multiple enhancers [118]. Additionally, various translocation events in different patients can connect MYC and CBL to disjoint sets of enhancers [118]. Elevated CDK5, MYC and CBL are implicated in AML progression [119], but the clinical relevance of HSF4 hyperactivation is currently unclear. From this data, one could postulate that structural variants contribute to promoter–enhancer hub formation involving key oncogenes, a hypothesis that warrants further investigation.
Therapeutic opportunities
The role of promoter-enhancer hubs in regulating various oncogenes makes them an attractive target for treatment. One way to target hubs is by inhibiting their regulators. Because TF occupancy at one enhancer stabilizes the occupancy of TFs and cofactors at interacting enhancers [58], inhibiting TFs can specifically disrupt oncogenic promoter–enhancer hubs. For instance, Notch inhibition in multiple cancers is promising due to its strong anti-tumor effects and ability to disrupt Notch-dependent long-range interactions within promoter–enhancer hubs [24,115]. However, treatment resistance is still a challenge and could potentially be overcome with combination therapy [39,115,120,121]. Kloetgen et al. demonstrated that simultaneous inhibition of Notch and CDK7 disrupts Notch-insensitive enhancer–promoter loops [113]. Additionally, Notch-inhibitor-resistant T-ALL cells are more responsive to BRD4 inhibition [116], whose therapeutic value has been explored in other cancers [122].
Loss of insulation at oncogenic loci is prevalent [98,123-125]. Here, we provide specific examples from SDH-deficient GI stromal tumors, B-cell precursor ALL, and AML where insulator disruption could be interpreted as gain of enhancer–promoter hubs involving proto-oncogenes [93,103,118]. For instance, as we discussed, DNA hypermethylation coincide with altered enhancer–promoter interactions of key oncogenes. DNA methyltransferase inhibitors show promising anti-tumor effects and could disrupt AML-specific enhancer–promoter interactions [118], rationalizing for their use as potential therapeutic options. Yet, to fully leverage the potential of agents targeting promoter–enhancer hubs as part of combination therapies, several challenges need to be overcome, including toxicity and off-target effects as well as treatment resistance.
Concluding remarks
Our knowledge of enhancer biology and its contribution to cancer has drastically improved over the last few decades. Technological advancements continue to broaden our understanding of enhancer positioning and its functional importance in transcriptional regulation in cancer. Genome-wide and/or locus-focused examination of cancer genome folding show that promoter–enhancer hubs contribute to pathogenesis by preferentially promoting oncogenic expression. Some studies focused on loci with one promoter and multiple enhancers (Figure 1, top), while others examined loci with multiple promoters and enhancers (Figure 1, bottom).
Yet, a full understanding of enhancer-mediated gene expression control that can explain varying configurations of enhancers within the nuclear space remains enigmatic. Several long-standing questions remain (see Outstanding Questions). For instance, most studies discussed in this review identified promoter-enhancer hubs based on chromatin conformation capture data generated from populations of cells (Figure 3). But how many individual cancer cells do exhibit multiway interactions among enhancers and promoters? What is the functional importance of their formation? Do multiway contacts promote stronger gene expression than pairwise contacts? Further, while an association between changes in gene expression and alterations in promoter–enhancer hubs were observed, only a few studies dissected the details of these potential regulatory interactions. Hence, it remains unclear which multiway interactions are structurally competitive, cooperative, or independent, and what factors determine this context dependency. Moreover, despite recent evidence suggesting that dynamic rewiring of promoter–enhancer hubs may play a role in cell-fate decision, further systematic studies are needed to fully credential the hypothesis that these topological assemblies are lineage-restricted and contribute to cancer transformation and progression. To address these questions and develop better models of promoter–enhancer hub regulation and function, there is a need for systematic perturbations of their constituent components and testing their effects on gene expression.
Outstanding questions:
Do promoter–enhancer hubs form in individual cancer cells, and if they do, what is the extent of their formation and function?
What are the regulators of promoter–enhancer hubs?
Why does a subset of genes require promoter–enhancer hubs for their regulation?
What chromatin features distinguish the different types of multiway interactions within enhancer clusters?
To what extent do multiway enhancer interactions predict gene expression?
In addition, the precise identification of genome folding changes that happen during cancer transformation require comparison with the respective cells of origin, which is not known for many cancer types, or are not amenable to chromatin conformation capture assays [113,114]. Future studies using improved experimental setups and assays with lower input requirements can elucidate the underlying mechanisms driving chromatin architectural changes in cancer.
Further developments in chromatin-tracing methods and single-cell chromatin conformation capture assays allow the examination of chromatin folding heterogeneity at individual loci. Multi-modal extensions of these single-cell resolution methods, incorporating RNA and protein colocalization, will enable in-depth exploration of promoter–enhancer hub regulation and function. These techniques could also investigate links between promoter–enhancer hubs and nuclear bodies, including phase-separated condensates and PML nuclear bodies, which have been linked to super-enhancers [126-128]. On the other hand, super-resolution live-cell imaging help to elucidate the dynamics of promoter–enhancer hubs. However, genome-wide characterization of these topological assemblies in individual cells are still lacking and requires further technological advancements [129]. Overall, addressing these questions is an exciting avenue of research in genome architecture and may pave the way for reprogramming of promoter–enhancer hubs and exploiting the therapeutic potential of their disruption and/or formation in cancer treatment.
Highlights:
Genetic and epigenetic events can influence promoter–enhancer hubs in cancer.
Deregulation of promoter–enhancer hubs contributes to cancer pathogenesis through misregulation of oncogenes and tumor suppressors.
Promoter–enhancer hubs can also be cancer-protective when a tumor suppressor is activated.
Recent technological advancements have paved the way for in-depth study of the cause-and-effect relationships between transcriptional regulators, promoter–enhancer hubs, and gene expression control in cancer.
Glossary:
- Chromatin conformation capture
a class of experimental technique used to measure pairwise contact frequencies between two DNA sequences based on proximity ligation.
- Chromosome territory
the specific region of the nucleus occupied by different chromosomes.
- Compartments
domains found in Hi-C data formed by differential chromatin activity, and not CTCF binding.
- DNA binding transcription factors (TFs)
proteins with DNA binding domains that regulate transcription by binding to specific DNA sequences in regulatory elements.
- Insulator
a class of protein-binding DNA regulatory elements that protect genes from inappropriate signals from their surrounding environment.
- Oligopaint
a FISH method that labels DNA using short fluorescently labeled oligonucleotides for high-resolution imaging of chromatin.
- Promoter–enhancer hub
dynamic topological assemblies with one or more promoters that interact, even transiently, amongst themselves and with two or more enhancers that may or may not be connected.
- Topologically associating domains (TADs)
spatially self-associating intra-chromosomal regions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nuebler J. et al. (2018) Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A 115, E6697–E6706. 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao SS et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoenfelder S and Fraser P (2019) Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 20, 437–455. 10.1038/s41576-019-0128-0 [DOI] [PubMed] [Google Scholar]
- 4.Verschure PJ et al. (1999) Spatial relationship between transcription sites and chromosome territories. J Cell Biol 147, 13–24. 10.1083/jcb.147.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calo E and Wysocka J (2013) Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–837. 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W. et al. (2013) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520. 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spitz F and Furlong EE (2012) Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13, 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- 8.Karr JP et al. (2022) The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer-promoter communication. Genes Dev 36, 7–16. 10.1101/gad.349160.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuin J. et al. (2022) Nonlinear control of transcription through enhancer-promoter interactions. Nature 604, 571–577. 10.1038/s41586-022-04570-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vian L. et al. (2018) The Energetics and Physiological Impact of Cohesin Extrusion. Cell 175, 292–294. 10.1016/j.cell.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho WK et al. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma R. et al. (2021) Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization. Nat Commun 12, 5579. 10.1038/s41467-021-25761-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weintraub AS et al. (2017) YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588 e1528. 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grubert F. et al. (2015) Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell 162, 1051–1065. 10.1016/j.cell.2015.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel N. et al. (2010) Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493. 10.1038/nature09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J. et al. (2016) Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev Cell 36, 9–23. 10.1016/j.devcel.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hnisz D. et al. (2013) Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Giammartino DC et al. (2019) KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat Cell Biol 21, 1179–1190. 10.1038/s41556-019-0390-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miguel-Escalada I. et al. (2019) Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat Genet 51, 1137–1148. 10.1038/s41588-019-0457-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinodoz SA et al. (2018) Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 174, 744–757 e724. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki K and Fukaya T (2023) Functional coordination between transcription factor clustering and gene activity. Mol Cell 83, 1605–1622 e1609. 10.1016/j.molcel.2023.04.018 [DOI] [PubMed] [Google Scholar]
- 22.Chen LF et al. (2023) Structural elements promote architectural stripe formation and facilitate ultra-long-range gene regulation at a human disease locus. Mol Cell 83, 1446–1461 e1446. 10.1016/j.molcel.2023.03.009 [DOI] [PubMed] [Google Scholar]
- 23.Fasolino M. et al. (2020) Genetic Variation in Type 1 Diabetes Reconfigures the 3D Chromatin Organization of T Cells and Alters Gene Expression. Immunity 52, 257–274 e211. 10.1016/j.immuni.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrovic J. et al. (2019) Oncogenic Notch Promotes Long-Range Regulatory Interactions within Hyperconnected 3D Cliques. Mol Cell 73, 1174–1190 e1112. 10.1016/j.molcel.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng W. et al. (2014) Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158, 849–860. 10.1016/j.cell.2014.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu B. et al. (2018) Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359, 1050–1055. 10.1126/science.aao3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgs DR et al. (1990) A major positive regulatory region located far upstream of the human alpha-globin gene locus. Genes Dev 4, 1588–1601. 10.1101/gad.4.9.1588 [DOI] [PubMed] [Google Scholar]
- 28.Long HK et al. (2020) Loss of Extreme Long-Range Enhancers in Human Neural Crest Drives a Craniofacial Disorder. Cell Stem Cell 27, 765–783 e714. 10.1016/j.stem.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song M. et al. (2020) Cell-type-specific 3D epigenomes in the developing human cortex. Nature 587, 644–649. 10.1038/s41586-020-2825-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Giammartino DC et al. (2020) Transcription factors: building hubs in the 3D space. Cell Cycle 19, 2395–2410. 10.1080/15384101.2020.1805238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafner A and Boettiger A (2023) The spatial organization of transcriptional control. Nat Rev Genet 24, 53–68. 10.1038/s41576-022-00526-0 [DOI] [PubMed] [Google Scholar]
- 32.Lim B and Levine MS (2021) Enhancer-promoter communication: hubs or loops? Curr Opin Genet Dev 67, 5–9. 10.1016/j.gde.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uyehara CM and Apostolou E (2023) 3D enhancer-promoter interactions and multi-connected hubs: Organizational principles and functional roles. Cell Rep, 112068. 10.1016/j.celrep.2023.112068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H. et al. (2018) A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell 173, 386–399 e312. 10.1016/j.cell.2018.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 36.Sengupta S and George RE (2017) Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer 3, 269–281. 10.1016/j.trecan.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suva ML et al. (2013) Epigenetic reprogramming in cancer. Science 339, 1567–1570. 10.1126/science.1230184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhagwat AS et al. (2018) Enhancer dysfunction in leukemia. Blood 131, 1795–1804. 10.1182/blood-2017-11-737379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akdemir KC et al. (2020) Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat Genet 52, 294–305. 10.1038/s41588-019-0564-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X. et al. (2021) Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nat Methods 18, 661–668. 10.1038/s41592-021-01164-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z. et al. (2022) Structural variants drive context-dependent oncogene activation in cancer. Nature 612, 564–572. 10.1038/s41586-022-05504-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allahyar A. et al. (2018) Enhancer hubs and loop collisions identified from single-allele topologies. Nat Genet 50, 1151–1160. 10.1038/s41588-018-0161-5 [DOI] [PubMed] [Google Scholar]
- 43.Ay F. et al. (2015) Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics 16, 121. 10.1186/s12864-015-1236-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darrow EM et al. (2016) Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A 113, E4504–4512. 10.1073/pnas.1609643113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olivares-Chauvet P. et al. (2016) Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature 540, 296–300. 10.1038/nature20158 [DOI] [PubMed] [Google Scholar]
- 46.Oudelaar AM et al. (2018) Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat Genet 50, 1744–1751. 10.1038/s41588-018-0253-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshpande AS et al. (2022) Identifying synergistic high-order 3D chromatin conformations from genome-scale nanopore concatemer sequencing. Nat Biotechnol 40, 1488–1499. 10.1038/s41587-022-01289-z [DOI] [PubMed] [Google Scholar]
- 48.Vangala P. et al. (2020) High-Resolution Mapping of Multiway Enhancer-Promoter Interactions Regulating Pathogen Detection. Mol Cell 80, 359–373 e358. 10.1016/j.molcel.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beagrie RA et al. (2017) Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shopland LS et al. (2006) Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174, 27–38. 10.1083/jcb.200603083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardozo Gizzi AM et al. (2019) Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol Cell 74, 212–222 e215. 10.1016/j.molcel.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 52.Liu M. et al. (2020) Multiplexed imaging of nucleome architectures in single cells of mammalian tissue. Nat Commun 11, 2907. 10.1038/s41467-020-16732-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mateo LJ et al. (2019) Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su JH et al. (2020) Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell 182, 1641–1659 e1626. 10.1016/j.cell.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takei Y. et al. (2021) Integrated spatial genomics reveals global architecture of single nuclei. Nature 590, 344–350. 10.1038/s41586-020-03126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hafner A. et al. (2023) Loop stacking organizes genome folding from TADs to chromosomes. Mol Cell 83, 1377–1392 e1376. 10.1016/j.molcel.2023.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zelenka T. et al. (2022) The 3D enhancer network of the developing T cell genome is shaped by SATB1. Nat Commun 13, 6954. 10.1038/s41467-022-34345-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madsen JGS et al. (2020) Highly interconnected enhancer communities control lineage-determining genes in human mesenchymal stem cells. Nat Genet 52, 1227–1238. 10.1038/s41588-020-0709-z [DOI] [PubMed] [Google Scholar]
- 59.Dickel DE et al. (2018) Ultraconserved Enhancers Are Required for Normal Development. Cell 172, 491–499 e415. 10.1016/j.cell.2017.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunipace L. et al. (2019) Coacting enhancers can have complementary functions within gene regulatory networks and promote canalization. PLoS Genet 15, e1008525. 10.1371/journal.pgen.1008525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hay D. et al. (2016) Genetic dissection of the alpha-globin super-enhancer in vivo. Nat Genet 48, 895–903. 10.1038/ng.3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perry MW et al. (2011) Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A 108, 13570–13575. 10.1073/pnas.1109873108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai A. et al. (2019) Multi-enhancer transcriptional hubs confer phenotypic robustness. Elife 8. 10.7554/eLife.45325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phanstiel DH et al. (2017) Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development. Mol Cell 67, 1037–1048 e1036. 10.1016/j.molcel.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang J. et al. (2018) Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun 9, 943. 10.1038/s41467-018-03279-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sobreira DR et al. (2021) Extensive pleiotropism and allelic heterogeneity mediate metabolic effects of IRX3 and IRX5. Science 372, 1085–1091. 10.1126/science.abf1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mangnier L. et al. (2022) Cis-regulatory hubs: a new 3D model of complex disease genetics with an application to schizophrenia. Life Sci Alliance 5. 10.26508/lsa.202101156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chandra A. et al. (2023) Quantitative control of Ets1 dosage by a multi-enhancer hub promotes Th1 cell differentiation and protects from allergic inflammation. Immunity. 10.1016/j.immuni.2023.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fulco CP et al. (2019) Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet 51, 1664–1669. 10.1038/s41588-019-0538-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cancer Genome Atlas Research, N. (2015) The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025. 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y. et al. (2021) Systematic inference and comparison of multi-scale chromatin sub-compartments connects spatial organization to cell phenotypes. Nat Commun 12, 2439. 10.1038/s41467-021-22666-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhie SK et al. (2019) A high-resolution 3D epigenomic map reveals insights into the creation of the prostate cancer transcriptome. Nat Commun 10, 4154. 10.1038/s41467-019-12079-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez-Vega F. et al. (2018) Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173, 321–337 e310. 10.1016/j.cell.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo Y. et al. (2018) CRISPR-mediated deletion of prostate cancer risk-associated CTCF loop anchors identifies repressive chromatin loops. Genome Biol 19, 160. 10.1186/s13059-018-1531-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Formaggio N. et al. (2021) Loss and revival of androgen receptor signaling in advanced prostate cancer. Oncogene 40, 1205–1216. 10.1038/s41388-020-01598-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guan Y. et al. (2020) Unraveling cancer lineage drivers in squamous cell carcinomas. Pharmacol Ther 206, 107448. 10.1016/j.pharmthera.2019.107448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun R. et al. (2001) Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem 276, 6897–6900. 10.1074/jbc.C000870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y. et al. (2020) Chromatin Looping Shapes KLF5-Dependent Transcriptional Programs in Human Epithelial Cancers. Cancer Res 80, 5464–5477. 10.1158/0008-5472.CAN-20-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chau NG et al. (2020) An Anatomical Site and Genetic-Based Prognostic Model for Patients With Nuclear Protein in Testis (NUT) Midline Carcinoma: Analysis of 124 Patients. JNCI Cancer Spectr 4, pkz094. 10.1093/jncics/pkz094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.French CA et al. (2003) BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res 63, 304–307 [PubMed] [Google Scholar]
- 81.French CA et al. (2004) Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol 22, 4135–4139. 10.1200/JCO.2004.02.107 [DOI] [PubMed] [Google Scholar]
- 82.Alekseyenko AA et al. (2015) The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev 29, 1507–1523. 10.1101/gad.267583.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eagen KP and French CA (2021) Supercharging BRD4 with NUT in carcinoma. Oncogene 40, 1396–1408. 10.1038/s41388-020-01625-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosencrance CD et al. (2020) Chromatin Hyperacetylation Impacts Chromosome Folding by Forming a Nuclear Subcompartment. Mol Cell 78, 112–126 e112. 10.1016/j.molcel.2020.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Riggi N. et al. (2021) Ewing's Sarcoma. N Engl J Med 384, 154–164. 10.1056/NEJMra2028910 [DOI] [PubMed] [Google Scholar]
- 86.Riggi N. et al. (2014) EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 26, 668–681. 10.1016/j.ccell.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sanalkumar R. et al. (2023) Highly connected 3D chromatin networks established by an oncogenic fusion protein shape tumor cell identity. Sci Adv 9, eabo3789. 10.1126/sciadv.abo3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giuli MV et al. (2019) Notch Signaling Activation as a Hallmark for Triple-Negative Breast Cancer Subtype. J Oncol 2019, 8707053. 10.1155/2019/8707053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X. et al. (2016) Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 48, 176–182. 10.1038/ng.3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yonemori K. et al. (2017) ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci 108, 124–135. 10.1111/cas.13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xing R. et al. (2019) Whole-genome sequencing reveals novel tandem-duplication hotspots and a prognostic mutational signature in gastric cancer. Nat Commun 10, 2037. 10.1038/s41467-019-09644-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin DC et al. (2018) Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut 67, 1769–1779. 10.1136/gutjnl-2017-314607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Flavahan WA et al. (2019) Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 575, 229–233. 10.1038/s41586-019-1668-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tarjan DR et al. (2019) Epigenome editing strategies for the functional annotation of CTCF insulators. Nat Commun 10, 4258. 10.1038/s41467-019-12166-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou J. et al. (2015) Identification of CEACAM5 as a Biomarker for Prewarning and Prognosis in Gastric Cancer. J Histochem Cytochem 63, 922–930. 10.1369/0022155415609098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fonseca AS et al. (2021) ETV4 plays a role on the primary events during the adenoma-adenocarcinoma progression in colorectal cancer. BMC Cancer 21, 207. 10.1186/s12885-021-07857-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiao M. et al. (2012) Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 26, 1326–1338. 10.1101/gad.191056.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Flavahan WA et al. (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Javidi-Sharifi N. et al. (2015) Crosstalk between KIT and FGFR3 Promotes Gastrointestinal Stromal Tumor Cell Growth and Drug Resistance. Cancer Res 75, 880–891. 10.1158/0008-5472.CAN-14-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnstone SE et al. (2020) Large-Scale Topological Changes Restrain Malignant Progression in Colorectal Cancer. Cell 182, 1474–1489 e1423. 10.1016/j.cell.2020.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang H. et al. (2022) Noncoding genetic variation in GATA3 increases acute lymphoblastic leukemia risk through local and global changes in chromatin conformation. Nat Genet 54, 170–179. 10.1038/s41588-021-00993-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sasaki K. et al. (2022) Genome-wide CRISPR-Cas9 screen identifies rationally designed combination therapies for CRLF2-rearranged Ph-like ALL. Blood 139, 748–760. 10.1182/blood.2021012976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang M. et al. (2020) 13q12.2 deletions in acute lymphoblastic leukemia lead to upregulation of FLT3 through enhancer hijacking. Blood 136, 946–956. 10.1182/blood.2019004684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Willcockson MA et al. (2021) H1 histones control the epigenetic landscape by local chromatin compaction. Nature 589, 293–298. 10.1038/s41586-020-3032-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yusufova N. et al. (2021) Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 589, 299–305. 10.1038/s41586-020-3017-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chu CS et al. (2020) Unique Immune Cell Coactivators Specify Locus Control Region Function and Cell Stage. Mol Cell 80, 845–861 e810. 10.1016/j.molcel.2020.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Basso K and Dalla-Favera R (2015) Germinal centres and B cell lymphomagenesis. Nat Rev Immunol 15, 172–184. 10.1038/nri3814 [DOI] [PubMed] [Google Scholar]
- 108.Bunting KL et al. (2016) Multi-tiered Reorganization of the Genome during B Cell Affinity Maturation Anchored by a Germinal Center-Specific Locus Control Region. Immunity 45, 497–512. 10.1016/j.immuni.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aster JC et al. (2017) The Varied Roles of Notch in Cancer. Annu Rev Pathol 12, 245–275. 10.1146/annurev-pathol-052016-100127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vilarrasa-Blasi R. et al. (2021) Dynamics of genome architecture and chromatin function during human B cell differentiation and neoplastic transformation. Nat Commun 12, 651. 10.1038/s41467-020-20849-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Puente XS et al. (2018) Chronic lymphocytic leukemia and mantle cell lymphoma: crossroads of genetic and microenvironment interactions. Blood 131, 2283–2296. 10.1182/blood-2017-10-764373 [DOI] [PubMed] [Google Scholar]
- 112.Liu Y. et al. (2017) The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 49, 1211–1218. 10.1038/ng.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kloetgen A. et al. (2020) Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat Genet 52, 388–400. 10.1038/s41588-020-0602-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang L. et al. (2021) 3D genome alterations associated with dysregulated HOXA13 expression in high-risk T-lineage acute lymphoblastic leukemia. Nat Commun 12, 3708. 10.1038/s41467-021-24044-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Y. et al. (2022) EBF1 nuclear repositioning instructs chromatin refolding to promote therapy resistance in T leukemic cells. Mol Cell 82, 1003–1020 e1015. 10.1016/j.molcel.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yashiro-Ohtani Y. et al. (2014) Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci U S A 111, E4946–4953. 10.1073/pnas.1407079111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bond J. et al. (2016) An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. Haematologica 101, 732–740. 10.3324/haematol.2015.141218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu J. et al. (2022) Subtype-specific 3D genome alteration in acute myeloid leukaemia. Nature 611, 387–398. 10.1038/s41586-022-05365-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fernandes MS et al. (2010) Novel oncogenic mutations of CBL in human acute myeloid leukemia that activate growth and survival pathways depend on increased metabolism. J Biol Chem 285, 32596–32605. 10.1074/jbc.M110.106161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Achinger-Kawecka J. et al. (2020) Epigenetic reprogramming at estrogen-receptor binding sites alters 3D chromatin landscape in endocrine-resistant breast cancer. Nat Commun 11, 320. 10.1038/s41467-019-14098-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marine JC et al. (2020) Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 20, 743–756. 10.1038/s41568-020-00302-4 [DOI] [PubMed] [Google Scholar]
- 122.Jones PA et al. (2016) Targeting the cancer epigenome for therapy. Nat Rev Genet 17, 630–641. 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- 123.Hnisz D. et al. (2016) Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458. 10.1126/science.aad9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weischenfeldt J. et al. (2017) Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet 49, 65–74. 10.1038/ng.3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y. et al. (2018) A Pan-Cancer Compendium of Genes Deregulated by Somatic Genomic Rearrangement across More Than 1,400 Cases. Cell Rep 24, 515–527. 10.1016/j.celrep.2018.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361. 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ji X. et al. (2016) 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell 18, 262–275. 10.1016/j.stem.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Boija A. et al. (2021) Biomolecular Condensates and Cancer. Cancer Cell 39, 174–192. 10.1016/j.ccell.2020.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nagano T. et al. (2013) Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lieberman-Aiden E. et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu M and Ren B (2017) The Three-Dimensional Organization of Mammalian Genomes. Annu Rev Cell Dev Biol 33, 265–289. 10.1146/annurev-cellbio-100616-060531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jin F. et al. (2013) A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294. 10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sanyal A. et al. (2012) The long-range interaction landscape of gene promoters. Nature 489, 109–113. 10.1038/nature11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mumbach MR et al. (2017) Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet 49, 1602–1612. 10.1038/ng.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li G. et al. (2014) Chromatin Interaction Analysis with Paired-End Tag (ChIA-PET) sequencing technology and application. BMC Genomics 15 Suppl 12, S11. 10.1186/1471-2164-15-S12-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fang R. et al. (2016) Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res 26, 1345–1348. 10.1038/cr.2016.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schoenfelder S. et al. (2018) Promoter Capture Hi-C: High-resolution, Genome-wide Profiling of Promoter Interactions. J Vis Exp. 10.3791/57320 [DOI] [PMC free article] [PubMed] [Google Scholar]