Abstract
Recent statistics on lung cancer, including the steady decline of advanced diseases and the dramatically increasing detection of early-stage diseases and indeterminate pulmonary nodules, mark the significance of a comprehensive understanding of early lung carcinogenesis. Lung adenocarcinoma (ADC) is the most common histologic subtype of lung cancer, and atypical adenomatous hyperplasia is the only recognized preneoplasia to ADC, which may progress to adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA) and eventually to invasive ADC. Although molecular evolution during early lung carcinogenesis has been explored in recent years, the progress has been significantly hindered, largely due to insufficient materials from ADC precursors. Here, we employed state-of-the-art deep learning and artificial intelligence techniques to robustly segment and recognize cells on routinely used hematoxylin and eosin histopathology images and extracted 9 biology-relevant pathomic features to decode lung preneoplasia evolution. We analyzed 3 distinct cohorts (Japan, China, and United States) covering 98 patients, 162 slides, and 669 regions of interest, including 143 normal, 129 atypical adenomatous hyperplasia, 94 AIS, 98 MIA, and 205 ADC. Extracted pathomic features revealed progressive increase of atypical epithelial cells and progressive decrease of lymphocytic cells from normal to AAH, AIS, MIA, and ADC, consistent with the results from tissue-consuming and expensive molecular/immune profiling. Furthermore, pathomics analysis manifested progressively increasing cellular intratumor heterogeneity along with the evolution from normal lung to invasive ADC. These findings demonstrated the feasibility and substantial potential of pathomics in studying lung cancer carcinogenesis directly from the low-cost routine hematoxylin and eosin staining.
Keywords: artificial intelligence, computational pathology, deep learning, lung cancer, pathomic features, preneoplasia evolution, tumor heterogeneity
Introduction
Lung cancer is the leading cause of cancer-related mortality in the United States and worldwide, with approximately 350 deaths per day in 2022.1 Over the past 2 decades, the incidence of lung cancer, particularly advanced-stage non–small cell lung cancer (NSCLC), has declined steadily. Meanwhile, the proportion of localized NSCLC has drastically increased by 4.5% annually, from 17% in 2004 to 28% in 2018, largely due to the increasing implementation of low-dose computed tomography (LDCT)–guided lung cancer screening programs.2,3 These tendencies underscore the significance and urgency of a comprehensive understanding of early lung carcinogenesis to improve interception and treatment of lung cancer.
Adenocarcinoma (ADC) is the most common primary histologic subtype of lung cancer, accounting for approximately 30% of all cases and 40% of NSCLCs.4 In the past 2 decades, the proportion of lung ADC has increased in both sex in the United States, and a similar trend has been observed in Asian and European countries.5,6 Atypical adenomatous hyperplasia (AAH) is the only recognized preneoplasia of lung ADC, comprising a localized growth of premalignant and cuboidal cells lining the alveolar walls. AAH can progress to adenocarcinoma in situ (AIS), then minimally invasive adenocarcinoma (MIA), and finally invasive ADC. AAH, AIS, and MIA are deemed as precursors of invasive ADC.7–10 The widespread implementation of LDCT lung screening has contributed to the substantially increased detection of indeterminate pulmonary nodules (IPN), many of which are ADC precursors. Nevertheless, there is an insufficient understanding of IPN biology, and a definite diagnosis of IPN still bears a critical challenge for the management of patients with IPNs.11
Carcinogenesis is a complicated process, comprehensive understanding of which requires multiomics profiling. Using this approach, a series of studies from our group and others have explored the dynamic evolutionary progression of ADC precursors and demonstrated progressive accumulation of molecular alternations and progressive suppression of immune response from AAH to ADC.10,12–16 However, omics profiling needs a large number of tissue specimens, and most precancerous specimens are insufficient for comprehensive omics profiling. In addition, multiomics profiling is expensive, technically complicated, and time-consuming. These have been major challenges hindering the advance of our understanding during lung carcinogenesis.
Contrary to the tissue-demanding, time-consuming, and complex omics techniques, pathologic assessment by hematoxylin and eosin (H&E) staining is low-cost and robust and has been routinely used as the gold standard for cancer diagnosis.17–19 Plus, the technological upgrading of whole slide imaging has enabled high-quality and high-throughput slides digitization.20,21 This digital transformation has laid a foundation for implementing computer-aided pathology slide analysis, termed pathomics.22,23 The fast development of artificial intelligence (AI), in particular deep learning, has prompted numerous successful pathological applications, including but not limited to mitosis counting, tumor early detection and grading, patient prognostication, and treatment planning in subspecialties such as thyroid, bladder, kidney, and leukemia.19,24–32
To the best of our knowledge, no prior studies had yet investigated the cellular-level lung precancerous progression with pathomics, indicating the lagging behind in utilizing H&E staining to interrogate lung carcinogenesis. To fill this knowledge gap, we developed an AI-based pipeline utilizing tissue H&E slides to study the tumor and immune progressions and their coevolutions with molecular biomarkers from preneoplasia to ADC. Our results revealed that the trends of immune and molecular progression during early lung carcinogenesis are consistent with those inferred via omics profiling, highlighting the potential of pathomics in studying cancer biology, particularly in diseases with limited tissue specimens.
Materials and Methods
Patients and Study Design
This study was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center and conducted following the Declaration of Helsinki. We collected tissue slides from 98 patients, including 59, 21, and 18 patients from Japan, China, and United States, respectively. The sex, age, and tobacco usage of 3 cohort patients are summarized in Table 1. We only kept patients of White race in the US cohort. Patients originating from Japan and China were Asians.
Table 1.
Patient characteristics of the 3 study cohorts (Japan, China, and United States)
| Cohort | Japan (n = 59) | China (n = 21) | United States (n = 18) | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Sex | ||||||
| Female | 22 | 37.3% | 12 | 57.1% | 17 | 94.4% |
| Male | 37 | 62.7% | 9 | 42.9% | 1 | 5.6% |
| Age (y) | ||||||
| ≤65 | 12 | 20.3% | 19 | 90.5% | 3 | 16.7% |
| >65 | 47 | 79.7% | 2 | 9.5% | 15 | 83.3% |
| Tobacco | ||||||
| Never | 24 | 49.7% | 17 | 81.0% | 3 | 16.7% |
| Current/former | 35 | 59.3% | 4 | 19.0% | 15 | 83.3% |
The general pipeline of this study is shown in Figure 1. First, we curated and prepared lung neoplasia digitized slides, then annotated representative regions of interest (ROI), and further segmented and annotated cells into 3 subtypes, including atypical epithelial cells (AEC), lymphocytes (LYM), and other cells (OC). Next, cellular-based pathomic features were extracted to characterize each ROI, including proportion, density, spatial entropy, and embedded map textures. At last, we analyzed these pathomic features’ evolution trends along with pathological stage progression.
Figure 1.
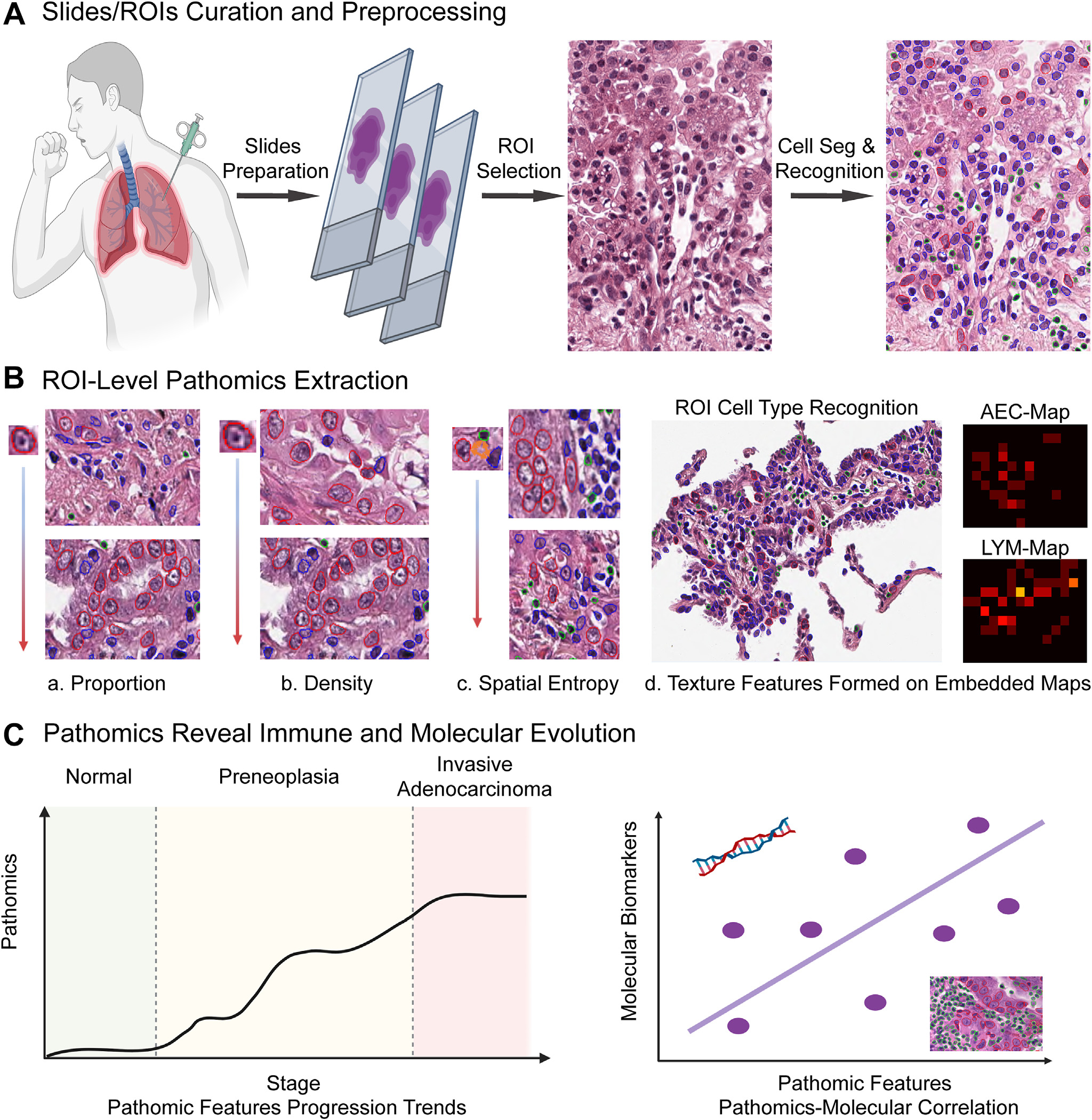
Pipeline illustrating the general study design. (A) Slide curation and preprocessing, region of interest (ROI) annotation, and cell segmentation (Seg) and recognition. (B) Extraction of 4 groups of ROI-level pathomic features, including cell proportions, cell densities, spatial entropy, and embedded map textures. (C) Investigation of extracted pathomic features’ evolution trends from normal to preneoplasia and eventually invasive adenocarcinoma, and the correlation between pathomic features with molecular biomarkers. AEC, atypical epithelial cell; LYM, lymphocyte.
Slides Acquisition and Preprocessing
H&E slides were digitized with an Aperio AT2 scanner (Leica Biosystems) at ×20 magnification (0.50 μm/pixel). One pathologist (F.R.R.) performed the quality check to exclude slides with evident artifacts, such as folding tissue, and 86, 30, and 46 slides remained from Japan, China, and US cohorts. Two pathologists (F.R.R. and L.M.S.S.) further confirmed the pathological diagnosis of each slide. Of note, as one patient can possess multiple slides with distinct diagnoses, we took the patient’s most severe diagnosis among all slides as the patient-level diagnosis.
As the pervasiveness of tissue heterogeneities, 1 slide might cover multiple pathological morphology patterns, for example, AAH and ADC can appear in different locations on the same slide. Although we conducted a slide-level quality check, the quality of certain regions inside the slides was still unsuitable for analysis. Besides, cellular-level analysis on whole slides is computationally expensive.33 With these considerations, we employed the ROI-based manner to analyze the tumor and immune progression for lung neoplasia. For each slide, diagnostically informative ROIs with high tissue quality were annotated by 1 pathologist (F.R.R.) and further checked by 3 more pathologists (A.S., J. Fujimoto., and L.M.S.S.). We only retained those ROIs with the consistently diagnosed pathological stages. We limited the minimum width and height of selected ROIs to 500 pixels (0.25 mm) to ensure a sufficient number of cells in each ROI, and the maximum ROI width and height to 5000 pixels (2.5 mm) to mitigate the computational burden. The numbers of patients, slides, and ROIs involved in the lung neoplasia progression analysis are shown in Supplementary Table S1.
To increase the number of studied ROIs, we occasionally annotated ROIs with a less severe pathological diagnosis, mostly normal, in certain tissue slides. The overall number of slides in each diagnosis and the number of annotated ROIs belonging to different diagnoses from each slide group with the same diagnosis are shown in Supplementary Table S2. Besides, we observed some evident stain color variations among ROIs delineated from different cohorts (Supplementary Fig. S1A). To ensure the robustness of the downstream intensity-related cellular features, we performed the Macenko34 normalization for all ROIs, and the color discrepancy among ROIs has been noticeably decreased after the stain normalization operation (Supplementary Fig. S1B).
Cellular Segmentation and Annotation From Tumor Microenvironment
We employed the state-of-the-art nuclei segmentation model, HoVer-Net35 pretrained on the PanNuke data set (covering nuclei curated from 19 different tissue types, including lungs), to segment nuclei on our experimented ROIs from 3 cohorts. HoVer-Net robustly separated and segmented nuclei, as qualitatively evaluated by pathologists (F.R.R., A.S., and L.M.S.S.). Exemplary nuclei segmentation overlaying ROIs are exhibited in Supplementary Figure S2. Since purplish blue regions denoting the ribosomes and chromatin within nuclei are major visible cell components under the H&E staining,36 cells were adopted to refer to segmented nuclei. Nonetheless, the cell classification outcomes from the HoVer-Net model, which classified cells into 6 categories, including neoplastic, inflammatory, connective, dead, nonneo-plastic, and others, were deviated from the acceptable performance.
Considering the biology of preneoplastic lesions, the major occurring cell phenotypes, and the technical feasibility of the machine learning-based pattern recognition, 3 cell subtypes, including AEC, LYM, and OC, were devised and annotated (Supplementary Fig. S3). AEC contained tumor epithelial cells and atypical pneumocytes. OC covered normal-appearing pneumocytes, fibroblasts, endothelial cells, neutrophils, macrophages, and normal epithelial bronchial cells. With the consensus of 2 pathologists (F.R.R. and A.S.), 306 cells (95 AEC, 88 LYM, and 123 OC) in 23 Japan ROIs and 576 cells (193 AEC, 190 LYM, and 193 OC) in 27 US ROIs were annotated for training cell classifiers. To guarantee that these cells cover a heterogeneous morphology spectrum, we selected ROIs from diverse stages and patients and annotated cells sparsely distributed within ROIs.
Inspired by H&E stained LYM’s morphology characteristics (small, dark, and round), we employed 3 cellular features, including cell area, mean intensity, and cell roundness, for machine recognition of AEC, LYM, and OC. The value distributions of 3 features based on annotated cells from Japan and United States were depicted by a violin plot (Supplementary Fig. S4). The cellular properties of the 2 cohorts were similar. We combined all annotated cells together to train the cell classifier using XGBoost37 and further applied the classifier to label segmented cells in 3 study cohorts.
We adopted 2 approaches to evaluate cell subtype recognition. On the cellular-level evaluation, we calculated the precision and sensitivity for each cell subtype and the overall accuracy for the whole cells. On the ROI-level visual evaluation, we used the precision to measure the recognition performance for each cell subtype, namely for all cells predicted as 1 particular cell type, divided by the true positive ones. Besides, we set 4 scales for ROI-level cell precision, with visual evaluation by pathologists (F.R.R. and A.S.), including outstanding (score = 100), good (score = 90 or 80), tolerable (score = 70 or 60), and poor (score < 60).
Pathomic Feature Extraction at the Region of Interest Level
Based on segmented and recognized cells inside each ROI, we extracted 4 groups of pathomic features: proportion, density, spatial entropy, and embedded map texture features.38 The cell proportion of each ROI was measured by dividing the number of 1-cell subtype by the total number of cells inside, which assessed the cells’ relative abundance. The cell density of each ROI was measured by dividing the number of 1-cell subtype by the occupied area, which evaluated the compactness of cells. Both cell proportion and density were independent measurements of the 1-cell subtype without consideration of the spatial arrangement among different cell subtypes. To quantify the interaction among different cell subtypes, we adopted Altieri’s spatial entropy,39,40 which accounts for the role of location and type of each cell inside the ROI to measure the heterogeneity. Besides, we embedded each ROI into smaller maps to summarize the cellular composition and architecture of each cell type.41,42 Here, we took LYMs to illustrate the generation process of embedded maps. We glided through the ROI from the image top-left to bottom-right with a window size of 50 × 50 pixels and a stride of 50 pixels. For each window, we counted the number of recognized LYMs as the value of the corresponding pixel of the down-sampled map. This map embedding procedure can be similarly applied to AECs and OCs. Several exemplary embedded maps are shown in Supplementary Figure S5. For each embedded map, we first calculated its gray-level co-occurrence matrix (GLCM). The contrast and energy of the GLCM matrix are then computed using the default scikit-image42 parameters to serve as a compact representation of the embedded maps. Supplementary Table S3 summarizes the 9 pathomic features extracted and used in our analysis.
Lung Carcinogenesis Decoded by Pathomics
With 9 pathomic features extracted from each ROI, we evaluated each feature’s mean value and distribution within each diagnosis stage to assess the progression trends from normal to AAH, AIS, MIA, and ultimately ADC. We also compared the trends across 3 different cohorts (Japan, China, and United States), 2different races (Asian and White), and different smoking statuses (never vs current/former).
Correlation of Pathomic Features and Molecular Markers
We assessed 3 molecular markers using a subset of Japan (21 patients; 23 slides) and China (9 patients; 12 slides) cohorts’ matched slides and conducted correlation analysis to evaluate the biological relevance of extracted pathomic features. These molecular markers comprised tumor mutational burden, copy number variation (CNV) burden, and allelic imbalance frequency. The molecular markers were measured based on the slide level, while multiple ROIs with distinct pathological stages were annotated on a single slide. To mitigate the gap, we only used the pathomic features from those ROIs with the same pathological stage as the slide to correlate with molecular measurements. We used Spearman’s rank correlation to evaluate the association of pathomics and molecular markers, with the false discovery rate method for the multiple testing adjustment of P values.43
Statistical Analysis
The 2-tailed Student’s t test was applied to assess the significant level of the difference between distinct pathological diagnoses or the difference between the 2 disparate cohorts with the same pathological diagnosis. All P values reported in this study were measured with 2-sided tests, and a P value of <.05 was considered statistically significant. We implemented all these statistical analyses using Python 3.8.3.
Results
Computational Pipeline With Robust Performance to Recognize Different Cell Subtypes as Confirmed by Thoracic Pathologists
We first explored 5 ways of cell classifier training and evaluation, including training on Japan test on United States, training on United States test on Japan, training and testing on Japan via cross-validation (CV), training and testing on United States via CV, and mix Japan and United States via CV. The confusion matrices of 5 experiments are shown in Figure 2. The accuracies of 3 CVs were 0.902, 0.894, and 0.897, on Japan, US, and Japan-US combined data, respectively (Fig. 2A–C). However, when evaluating the models across different data sets, the accuracy for the Japan cell classifier dropped to 0.892 and that for the US cell classifier dropped to 0.859 (Fig. 2D). This indicated the importance of encompassing diverse data to strengthen the cell classifier’s generalization and robustness. Although the Japan-US fusion model showed a similar accuracy to both Japan and US models, its SD (0.019) was smaller compared to that of the Japan (0.046) or US (0.037) models, demonstrating the robustness of the model built by fused data. We thus chose it to recognize the subtypes of all segmented cells in annotated ROIs. Additionally, the cell classifier using fused data showed high precision and sensitivity scores on both AEC and LYM (AEC precision = 0.931; AEC sensitivity = 0.902; LYM precision = 0.910; LYM sensitivity = 0.937) under the 5-fold CV settings, while an inferior recognition outcome is observed in the OC category (OC precision = 0.854; OC sensitivity = 0.857).
Figure 2.
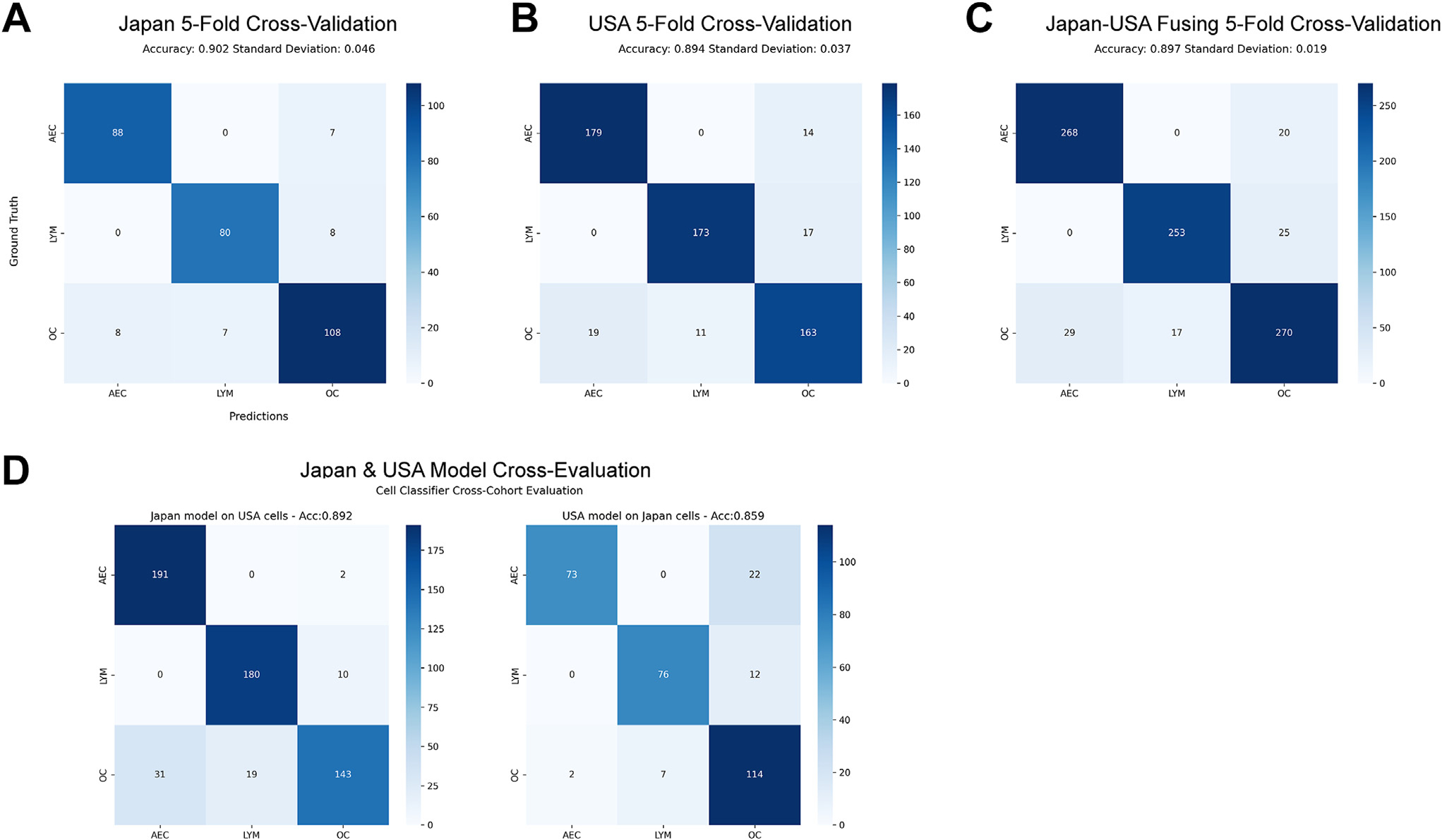
Cell recognition confusion matrices of 3 cell types, including atypical epithelial cell (AEC), lymphocyte (LYM), and other cell (OC). (A) Performance evaluation among 306 Japan annotated cells. (B) Performance valuation among 576 US annotated cells. (C) Performance valuation by fusing Japan and US annotated cells. (D) Cellular classifier cross-evaluations, with the left confusion matrix showing recognition of US cells evaluated on cellular classifier trained on Japan data, and the right showing the confusion matrix of Japan cells when evaluated on the US data trained cellular classifier.
To further validate the recognition performance of these 3 cell subtypes, dozens of ROIs (44 from the Japan cohort, 22 from the China cohort, and 22 from the US cohort) were randomly selected for pathologists to conduct a visual assessment for each cell type’s recognition precision, namely for those classified as a cell subtype category, and the percentage that belonged to that cell subtype category. Two pathologists (F.R.R. and A.S.) performed visual evaluations independently. As shown in Table 2, the ROI-level precisions on both AEC and LYM across 3 data sets by 2 pathologists were higher than 0.80 and mostly larger than 0.90, while OC’s evaluation was suboptimal, with precisions less than 0.80. In addition, consistency evaluation between 2 pathologists, with the denotations outstanding (score = 100), good (score = 90 or 80), tolerable (score = 70 or 60), and poor (score < 60) (Supplementary Fig. S6) showed interobserver variability, highlighting the challenges of histologic diagnosis of ADC precursors based on morphology. Most of the inconsistencies only happened between neighboring denotations. For instance, the inconsistencies between AEC and LYM mostly appeared within the outstanding and good regions.
Table 2.
Visual evaluations of region of interest—level cell recognition score distributions and precisions across 3 study cohorts of 2 pathologists, with their evaluation results on the 2 sides of “/,” respectively
| Recognition score | Japan (n = 44) |
China (n = 22) |
United States (n = 22) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| AEC | LYM | OC | AEC | LYM | OC | AEC | LYM | OC | |
|
| |||||||||
| 100 | 35/17 | 10/24 | 1/3 | 12/8 | 11/10 | 0/0 | 19/1 | 19/13 | 2/1 |
| 80–90 | 9/25 | 32/20 | 7/8 | 10/14 | 11/11 | 10/4 | 3/17 | 3/9 | 13/8 |
| 60–70 | 0/2 | 2/0 | 18/11 | 0/0 | 0/1 | 3/10 | 0/1 | 0/0 | 5/6 |
| <60 | 0/0 | 0/0 | 18/22 | 0/0 | 0/0 | 9/8 | 0/2 | 0/0 | 2/7 |
| Precision (%) | 97.3/91.8 | 90.7/95.5 | 60.9/55.7 | 94.5/93.2 | 95.0/92.7 | 65.0/59.5 | 98.6/81.4 | 98.6/95.5 | 78.6/65.5 |
AEC, atypical epithelial cell; LYM, lymphocyte; OC, other cell.
According to cellular- and ROI-level cell recognition evaluations, the recognition accuracy and precision of AEC and LYM were steady above 0.80, while OC was comparatively inferior. Thus, when quantifying cell proportions, cell densities, and texture features of embedded maps, we only considered the pathomic features from AEC and LYM.
Pathomic Features Characterize the Underlying Evolutionary Trends From Normal to Invasive Adenocarcinoma
The evolution trends of AEC proportion, AEC density, LYM proportion, and LYM density from normal to ADC are shown in Figure 3. Overall, the density and proportion of AEC gradually increased from normal to ADC, regardless of the cohort, ethnicity, or smoking status, reflecting the expansion of neoplastic cells along with the progression of ADC precursors. On the other hand, there was substantial variation between different lesions of the same stages and overlap between different stages, highlighting the profound heterogeneity between different patients even at the early stages of carcinogenesis. Additionally, the patterns of AEC density were more similar to each other between the Japan and China cohorts than with the US cohort, reflecting the potential racial and environmental differences. Interestingly, between never-smokers and current/former smokers, the difference in AEC proportion and density on both normal and ADC stages was minimal. However, current/former smokers exhibited a higher AEC proportion and density in AAH, AIS, and MIA stages.
Figure 3.
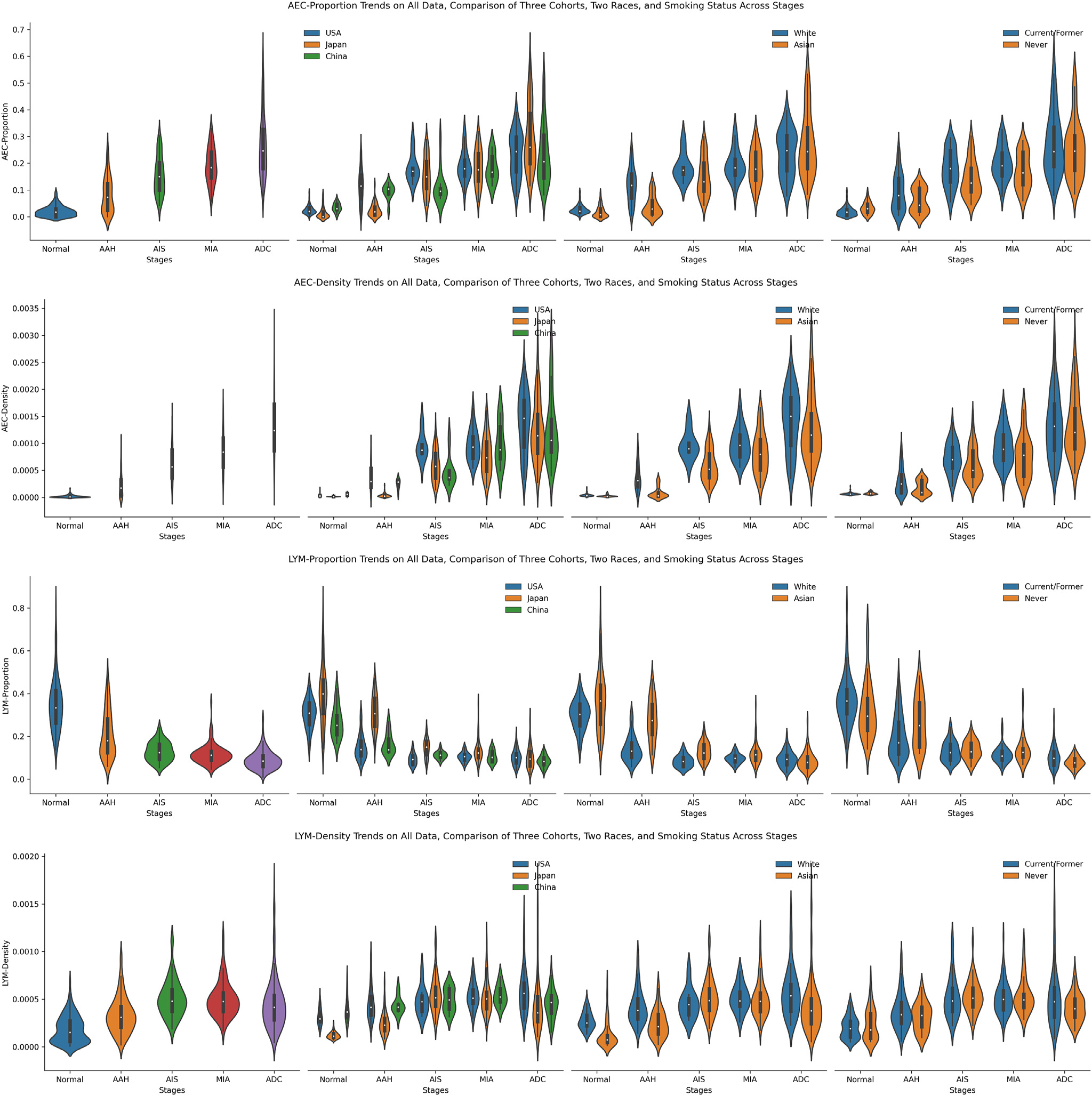
The trends of 4 pathomic features, including ratios and densities of both atypical epithelial cell (AEC) and lymphocyte (LYM). From top to bottom lie the AEC proportion, AEC density, LYM proportion, and LYM density. From left to right, present the trends of the fusion of all 3 data sets, the comparison of 3 data sets, the comparison of 2 races, and the comparison between never-smokers and former/current smokers. AAH, adenomatous hyperplasia; ADC, adenocarcinoma; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma.
Different from the AEC proportion, the LYM proportion first decreased from normal to AIS and then almost plateaued to ADC, which was in line with prior studies by molecular and immune profiling.44 Interestingly, although there were minor differences among AIS, MIA, and ADC, their values lay in a similar range compared to AAH and normal. Besides, the value range of the LYM proportion in these 3 stages was relatively narrow, mainly distributed between 0.05 and 0.20. Furthermore, the LYM density of all data steadily increased from normal to AIS and then slightly decreased until ADC, with a reverse trend compared to LYM proportion. The reverse evolution trend of LYM proportion and density also manifested when comparing subgroups (3 cohorts, 2 races, and different smoking statuses). Comparing between White and Asian races, the LYM proportion was more prominent in the Asian race in normal, AAH, and AIS stages, while the difference was minimal in MIA and ADC stages. The distinction of LYM proportion and density between never-smokers and current/former smokers was primarily slight.
Pathomics Analysis Reveals Increasing Cellular Heterogeneity Architecture Along With Progression of Lung Preneoplasia
Intratumor heterogeneity (ITH), a term referring to tumors being composed of cancer cells and stromal cells with distinct molecular and phenotypical features,45–51 is a fundamental phenomenon of malignancy with important biological and clinical impacts. A series of studies from our group and others have demonstrated ITH of lung cancer at different molecular levels, and increased molecular ITH was associated with impaired T cell response and increased risk of postsurgical recurrence.14,45,52–59 In these studies, ITH was assessed at the molecular level, which is often confounded by the composition of various cell types in the tumor. The H&E images provide a unique opportunity to delineate ITH of ADC and its precursors at the cellular level. We next applied Altieri entropy to depict the evolution of cellular ITH during progression of lung preneoplasia. The conventional Shannon entropy only considered the proportions of different cell types without considering their position information, which was one critical factor when analyzing ROIs’ characteristics at the cellular level. Here, we adopted the Altieri entropy, which considers both the proportions and relative positions of various cells, to quantify the spatial entropy of the cells inside ROIs. The spatial relationship was built by setting up multiple intervals to define the coexistence. As shown in Figure 4, the cellular ITH architecture was more complex in ADC and later-stage ADC precursors than in normal lung or AAH. It is thought-provoking to observe that the second law of thermodynamics, “entropy always increases,” was exquisitely held on the lung neoplasia progression process. When comparing 2 smoking statuses, the Altieri entropy was almost alike in the normal and AAH stages, while on AIS and ADC stages, the current/former smoker group presented higher entropy values. These results might reveal that the current/former smokers’ tumor microenvironment presented stronger ITH than never-smokers in advanced stages.
Figure 4.
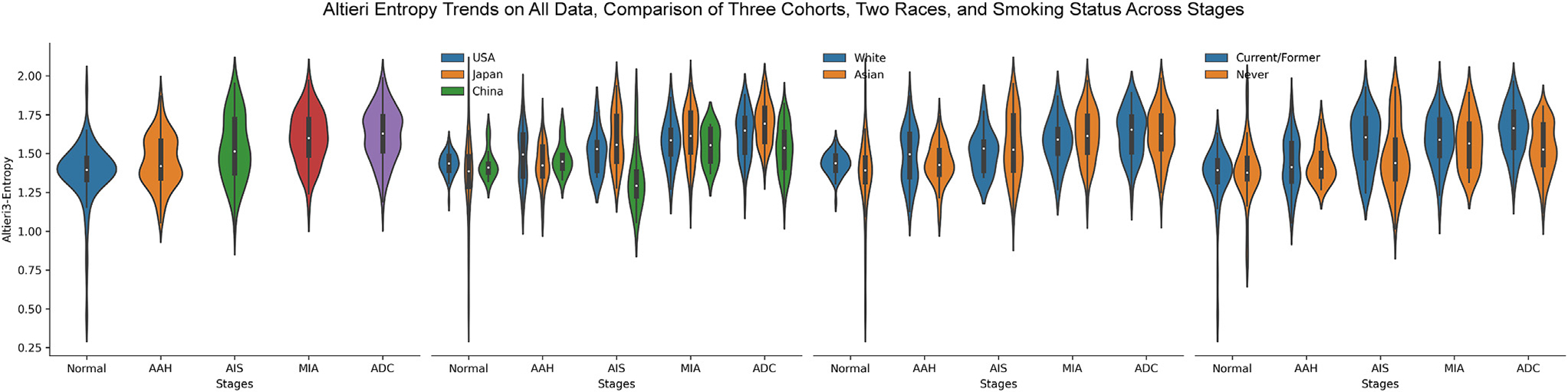
The trends of spatial entropies evolving from normal to invasive adenocarcinoma (ADC). The mean Altieri entropy steadily increases from normal to invasive ADC. AAH, adenomatous hyperplasia; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma.
We further encoded each ROI into 2 smaller embedded maps, AEC map and LYM map, to index the cellular spatial distribution from a textural perspective. For each embedded map, we explored the contrast and energy properties of embedded maps’ GLCM. The contrast represents the amount of local gray-level variations. A higher contrast value indicates the presence of edges or wrinkled structures inside. The energy measures the square root of uniformity of the gray-level distributions. A large energy value corresponds to a smaller number of gray levels, namely higher uniformity. The trends of 4 embedded map texture features are shown in Figure 5.
Figure 5.
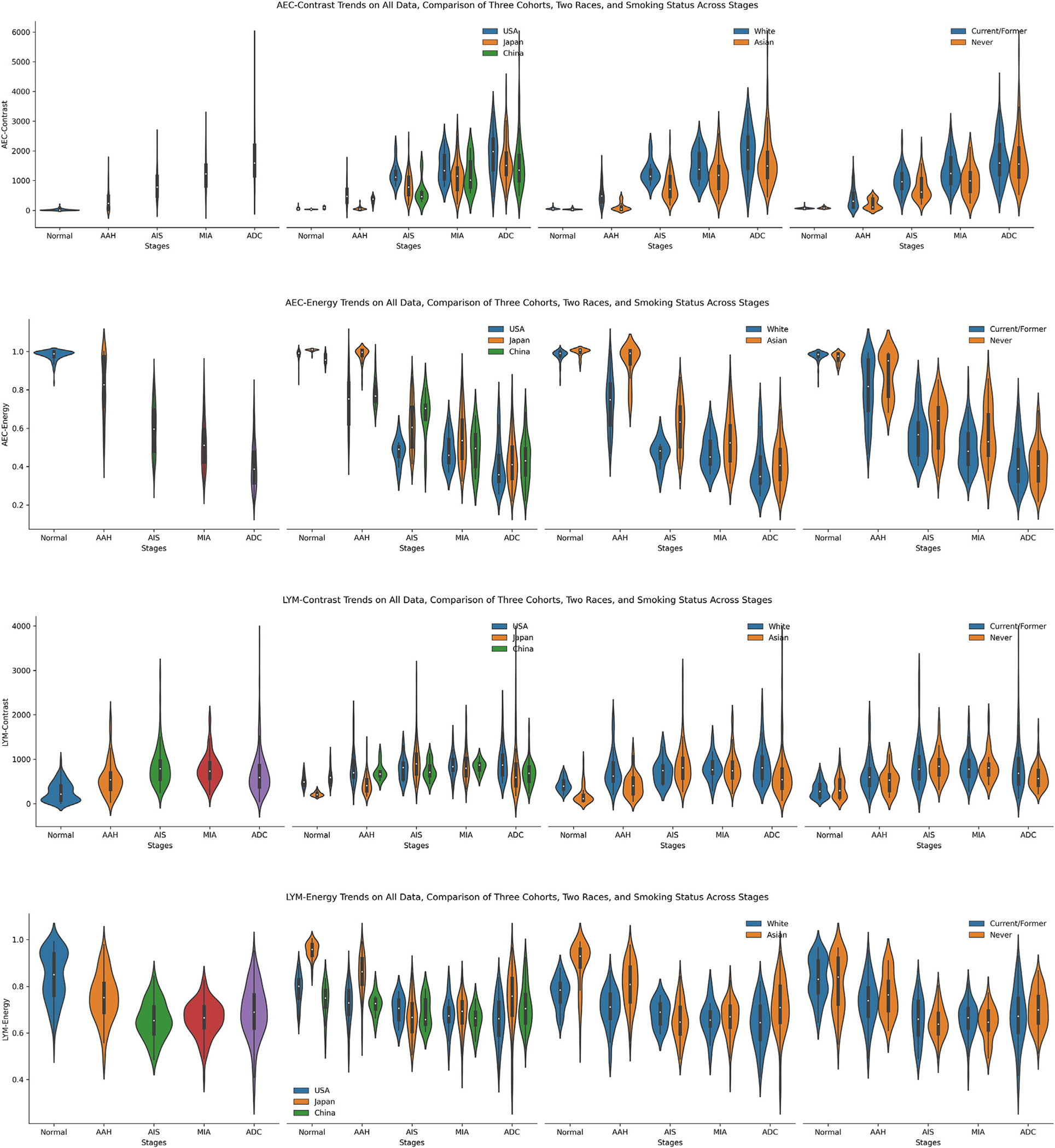
The trends of 4 embedded map texture features, including atypical epithelial cell (AEC) contrast, AEC energy, lymphocyte (LYM) contrast, and LYM energy presented from top to bottom. From left to right, present the trends of the fusion of all 3 data sets, the comparison of 3 data sets, the comparison of 2 races, and the comparison between never-smokers and former/current smokers. AAH, adenomatous hyperplasia; ADC, adenocarcinoma; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma.
In the AEC contrast, similar to the AEC proportion, it increased steadily from normal to ADC. From the biology perspective, as ROIs progressed to later stages, AEC were more widely scattered over ROIs, thus forming more edges or wrinkled structures inside the generated GLCMs, and accordingly accompanied with higher contrast values. However, the AEC energy exhibited a reverse trend compared to AEC contrast. Likewise, as the stages advance, the embedded map demonstrated more diverse distributions among local regions inside each ROI, namely lower uniformity accompanied by a lower energy value. The LYM contrast and LYM energy manifested more intricate trends than their AEC counter-parts, similar to the more subtle trends of LYM proportion and LYM density. The LYM contrast increased from normal to AIS, then slowly decreased on MIA, and further reduced in the ADC stage. The LYM energy manifested an opposite trend, with values steadily decreasing from normal to AIS and then continuously increasing from AIS to ADC. On smoking status subgroup analysis, the current/former smoker cohort exhibited higher AEC contrast and lower AEC energy in AAH, AIS, and MIA stages. Simultaneously, on LYM contrast, both never-smoker and current/former smoker cohorts manifested very close distribution on all 5 stages. In contrast, never-smokers showed higher LYM energy values in AAH and ADC stages.
Taken together, by different approaches, the pathomics analysis revealed a higher level of cellular ITH architecture in ADC and later-stage ADC precursors compared to early stages, marking the pattern of ITH evolution during early lung carcinogenesis. Just as for individual cell types, large variations between different lesions of the same stages and considerable overlap between different stages were observed regarding cellular ITH, once again high-lighting the profound heterogeneity among different patients.
Pathomic Features Reveal Distinct Associations With Molecular Markers
We next assessed whether pathomic features associated with genomics features from the gold standard whole exome sequencing in a subset of samples whose whole exome sequencing data were available.52 Here, we correlated 9 pathomic features with 3 important genomic features associated with the evolution of lung cancer, including total mutation burden, CNV burden, and allelic imbalance burden. To focus on those highly correlated pairs, we set a cutoff of 0.3 to mask those pairs with smaller absolute correlation values as 0.0.
As shown in Figure 6, the AEC proportion showed noticeable positive correlations with all 3 genomic features, in line with the facts that ADC and ADC later-stage precursors have more neoplastic cells, a high burden of genomic abnormalities, and a higher degree of ITH. On the other hand, the LYM proportion showed a negative correlation with CNV burden (rho =−0.328) consistent with previous knowledge that high CNV burden is associated with cold tumor immune microenvironment.44,55,60,61 On false discovery rate–adjusted P values, all pairs with absolute values larger than .3 presented significance (P ≤.05). Even some pairs with absolute values less than .3 also showed significance. These statistical significances further validated the potential associations between our extracted pathomic features with molecular markers. Additionally, the fitted regression of these pairs also manifested a relatively narrow CI (Fig. 6B).
Figure 6.
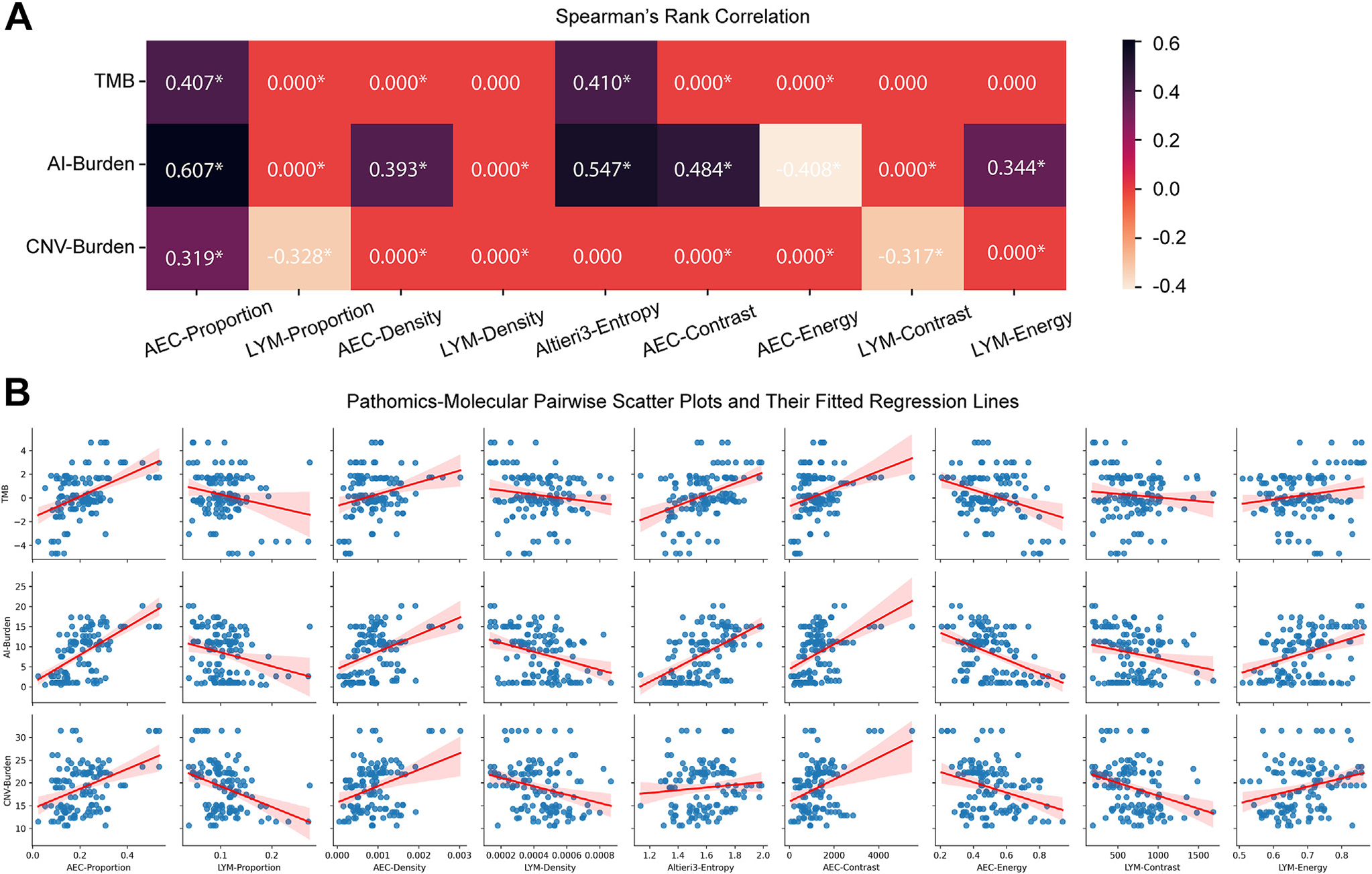
The correlations between 9 extracted pathomic features with 3 genomic markers. (A) Spearman’s rank correlation matrix. Absolute correlation coefficients less than 0.3 are masked as 0.000 to highlight those evident correlation pairs. Asterisks denote false discovery rate–adjusted P values less than .05. (B) Pairwise scatter plots and their fitted regression lines between correlation features. AEC, atypical epithelial cell; AI, allelic imbalance; CNV, copy number variation; LYM, lymphocyte; TMB, tumor mutational burden.
Discussion
Recent advances in pathology image digitization and AI have facilitated large-scale, objective, and low-cost pathomics studies. Although this approach has been applied to various malignancies in recent years,24–28 it has not been explored in the study on lung cancer precursors, for which image-based analysis such as pathomics is critically needed because of lack of study materials for multiomics analyses. In this study, we proposed a new pipeline using H&E slides to study lung ADC and its precursors. Using the H&E images from 3 countries, the proposed pipeline utilized the deep learning model HoVer-Net to segment cells inside annotated ROIs, constructed a robust cell classifier that can annotate segmented cells into 3 cell subtype categories (AEC, LYM, and OC), and extracted 9 pathomic features to characterize lung cancer evolution from normal to invasive ADC. The results from extracted pathomic features revealed progression of lung preneoplasia from different perspectives, including the proportion, density, textures of embedded maps of both AEC and LYM, and cell interaction and ITH architecture measured by Altieri entropy. Importantly, these trends obtained from pathomics are consistent with the findings from tissue-demanding, time-consuming, high-cost, and complicated molecular and immune profiling, which were further validated by correlations between pathomic features with molecular biomarkers. Taken together, these results demonstrated the substantial potential of H&E images and AI-based pathomics analysis in the study of diseases with limited materials for research, for example, lung preneoplasia.
Pathomics has also been broadly adopted to study a wide spectrum of lung diseases, including cancer, idiopathic pulmonary fibrosis, and COIVD-19, and further explored the utility of novel biomarkers for different clinical problems, including diagnosis, risk stratification, and treatment response prediction.62 To our knowledge, no prior studies have yet investigated the precancerous progression using the pathomics approach. With the increasing implementation of LDCT lung cancer screening, the drastically increased detection of IPNs, many of which are lung ADC precursors, demands an improved understanding of the biological features and more precise management of these IPNs. Lung carcinogenesis and its dynamic evolution have been explored by multiple different omics studies.10,44,52 However, it is well known that these omics techniques are expensive and require large tissue samples, which are often not amendable for ADC precursors. For these reasons, these omics studies can hardly be widely conducted, leading to lagging of our understanding of early lung carcinogenesis. In contrast, pathologic assessment by H&E staining has been introduced for more than 100 years, which is mature, robust, affordable, and widely available. Robust computational analytical tools to interrogate routinely used H&E tissue slides will be of great value to delineate the lung neoplasia progression.
As a study based on heterogeneous real-world cohorts, our work has several important limitations. First, although we have procured patients from 3 different countries, the cohort size is relatively small. Larger cohorts of ADC and its precursors of different studies are warranted to validate these intriguing findings before they can be widely applied. Second, we used a total of nearly 900 annotated cells to build the cell classifier and then used the cell classifier to annotate the remaining cells. We achieved accurate cell recognition performance on AEC and LYM. However, we had to group many other cell types important for the tumor microenvironment such as endothelial cells, fibroblasts, etc. into other cells. A more detailed cell annotation would further enhance cell recognition robustness and more comprehensively depict the tumor, precursor, and associated microenvironment. Moreover, due to computational cost, the current model did not incorporate the context information around the cells when conducting cellular recognition. A novel model that accounts for the pixels inside and outside the cellular contour and, meanwhile, is computationally efficient would further improve the pathomics pipelines. Additionally, a semisupervised or unsupervised learning strategy might be an option to make use of a large number of unannotated cells.63,64 Third, as a real-world study, the data quality from different institutions was very heterogeneous. Many factors may affect the quality and consistency of acquired images, for example, different protocols in glass slide preparations, varied tissue persevering periods/conditions, and different protocols for H&E stains. These factors can lead to inferior cell segmentation and classification and inaccurate quantified pathomics. Standardized tissue slide preparation, preservation, and digitalization can help mitigate confounding factors’ effects on the pathomics studies. On the other hand, these heterogeneous data from different institutions testified to the robustness of our pathomics pipeline and indicated its potential generalizability. Fourth, H&E images have inherited limitations. For example, boundaries of cell cytoplasm are difficult to mark on H&E images, which has limited our analysis to focusing only on nuclei. Moreover, H&E images are not able to provide more detailed phenotypic information on cell subtypes, so we could not recognize T LYM, B LYM, and natural killer cells within LYM. A deep understanding of these cell subtypes will have to depend on high-resolution immune staining images. However, a similar concept of machine learning can be applied to these immune staining images to improve the relevant image analyses. Finally, we have solely focused on the cell features such as the roundness, regularity, and size of cells in this study. However, the size of the lesion provides critical information for the diagnosis of ADC precursors. For example, among all 3 cohorts, AAH and AIS showed substantial disparity and overlap. One important reason is that lesion size, which was not captured in the current pipeline, is one of the major features to distinguish AAH from AIS. Future studies incorporating lesion size information from whole slide images or radiographic scans may further improve the performance of pathomics analysis.
In conclusion, we propose a new computational pipeline to study lung neoplasia progression using H&E images. Extracted pathomic features revealed progression trends in line with the results from molecular profiling studies. As a proof-for-concept study, our work proved the feasibility and laid a foundation for utilizing pathomics to investigate molecular and immune features of diseases with limited research tissues as exampled by the lung cancer precursors.
Supplementary Material
Funding
This work was supported by The University of Texas MD Anderson Lung Moon Shot Program, The University of Texas MD Anderson Cancer Center Core Grant P30 CA016672, the National Institutes of Health grants R00CA218667 and R01CA234629, the AACR–Johnson & Johnson Lung Cancer Innovation Science Grant (18–90-52-ZHAN), the Rexanna’s Foundation for Fighting Lung Cancer, Sabin Family Fund, Rydin Family Research Fund, and Permanent Health Fund. This work was supported by generous philanthropic contributions from Mrs Andrea Mugnaini and Dr Edward L. C. Smith.
Footnotes
Declaration of Competing Interest
J. Zhang reports grant from Merck, grants and personal fees from Johnson and Johnson and Novartis, and personal fees from Bristol Myers Squibb, AstraZeneca, GenePlus, Innovent, and Hengrui outside the submitted work. The other authors declare no potential conflicts of interest.
Ethics Approval and Consent to Participate
The study was approved by the Institutional Review Boards of MD Anderson Cancer Center, Nagasaki University Graduate School of Biomedical Sciences, and Zhejiang Cancer Hospital. Written informed consent was obtained from all patients.
Supplementary Material
The online version contains supplementary material available at https://doi.org/10.1016/j.modpat.2023.100326.
Data Availability
The experimental data that support the findings of this study have been deposited in Mendeley Data (https://doi.org/10.17632/7zc56ttd96.1). The code has been made publicly available at Zenodo (https://doi.org/10.5281/zenodo.8188290).
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2.National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oudkerk M, Devaraj A, Vliegenthart R, et al. European position statement on lung cancer screening. Lancet Oncol. 2017;18(12):e754–e766. 10.1016/S1470-2045(17)30861-6 [DOI] [PubMed] [Google Scholar]
- 4.Classification Zheng M. and pathology of lung cancer. Surg Oncol Clin N Am. 2016;25(3):447–468. 10.1016/j.soc.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 5.Bray F, Ferlay J, Laversanne M, et al. Cancer incidence in five continents: inclusion criteria, highlights from volume X and the global status of cancer registration. Int J Cancer. 2015;137(9):2060–2071. 10.1002/ijc.29670 [DOI] [PubMed] [Google Scholar]
- 6.Park K, Vansteenkiste J, Lee KH, et al. Pan-Asian adapted ESMO Clinical Practice Guidelines for the management of patients with locally advanced unresectable non-small-cell lung cancer: a KSMO-ESMO initiative endorsed by CSCO, ISMPO, JSMO, MOS, SSO and TOS. Ann Oncol. 2020;31(2):191–201. 10.1016/j.annonc.2019.10.026 [DOI] [PubMed] [Google Scholar]
- 7.Weichert W, Warth A. Early lung cancer with lepidic pattern: adenocarcinoma in situ, minimally invasive adenocarcinoma, and lepidic predominant adenocarcinoma. Curr Opin Pulm Med. 2014;20(4):309–316. 10.1097/MCP.0000000000000065 [DOI] [PubMed] [Google Scholar]
- 8.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Tang Y, Chen Y, et al. Joint use of the radiomics method and frozen sections should be considered in the prediction of the final classification of peripheral lung adenocarcinoma manifesting as ground-glass nodules. Lung Cancer. 2020;139:103–110. 10.1016/j.lungcan.2019.10.031 [DOI] [PubMed] [Google Scholar]
- 10.Nie M, Yao K, Zhu X, et al. Evolutionary metabolic landscape from preneoplasia to invasive lung adenocarcinoma. Nat Commun. 2021;12(1):6479. 10.1038/s41467-021-26685-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scafoglio CR, Villegas B, Abdelhady G, et al. Sodium-glucose transporter 2 is a diagnostic and therapeutic target for early-stage lung adenocarcinoma. Sci Transl Med. 2018;10(467), eaat5933. 10.1126/scitranslmed.aat5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Fan Y, Xiong Y, et al. Delineating the dynamic evolution from preneoplasia to invasive lung adenocarcinoma by integrating single-cell RNA sequencing and spatial transcriptomics. Exp Mol Med. 2022;54(11):2060–2076. 10.1038/s12276-022-00896-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Wang W, Xiong Y, et al. Evolution of lung adenocarcinoma from preneoplasia to invasive adenocarcinoma. Cancer Med. 2023;12(5):5545–5557. 10.1002/cam4.5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Estecio MR, Chen R, et al. Evolution of DNA methylome from precancerous lesions to invasive lung adenocarcinomas. Nat Commun. 2021;12(1):687. 10.1038/s41467-021-20907-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, Bai J, Reuben A, et al. Multiomics analysis reveals distinct immunogenomic features of lung cancer with ground-glass opacity. Am J Respir Crit Care Med. 2021;204(10):1180–1192. 10.1164/rccm.202101-0119OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Zhang J, Xu FP, et al. Genomic landscape and immune microenvironment features of preinvasive and early invasive lung adenocarcinoma. J Thorac Oncol. 2019;14(11):1912–1923. 10.1016/j.jtho.2019.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shamai G, Livne A, Polonia A, et al. Deep learning-based image analysis predicts PD-L1 status from H&E-stained histopathology images in breast cancer. Nat Commun. 2022;13(1):6753. 10.1038/s41467-022-34275-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naik N, Madani A, Esteva A, et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun. 2020;11(1):5727. 10.1038/s41467-020-19334-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br J Cancer. 2021;124(4):686–696. 10.1038/s41416-020-01122-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorstenson S, Molin J, Lundström C. Implementation of large-scale routine diagnostics using whole slide imaging in Sweden: digital pathology experiences 2006–2013. J Pathol Inform. 2014;5(1):14. 10.4103/2153-3539.129452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantanowitz L, Sharma A, Carter AB, Kurc T, Sussman A, Saltz J. Twenty years of digital pathology: an overview of the road travelled, what is on the horizon, and the emergence of vendor-neutral archives. J Pathol Inform. 2018;9:40. 10.4103/jpi.jpi_69_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta R, Kurc T, Sharma A, Almeida JS, Saltz J. The emergence of pathomics. Curr Pathobiol Rep. 2019;7(3):73–84. [Google Scholar]
- 23.Cui M, Zhang DY. Artificial intelligence and computational pathology. Lab Invest. 2021;101(4):412–422. 10.1038/s41374-020-00514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barisoni L, Lafata KJ, Hewitt SM, Madabhushi A, Balis UGJ. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. 2020;16(11):669–685. 10.1038/s41581-020-0321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Laak J, Litjens G, Ciompi F. Deep learning in histopathology: the path to the clinic. Nat Med. 2021;27(5):775–784. 10.1038/s41591-021-01343-4 [DOI] [PubMed] [Google Scholar]
- 26.Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer. 2022;3(9):1026–1038. 10.1038/s43018-022-00436-4 [DOI] [PubMed] [Google Scholar]
- 27.Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20(5):e253–e261. 10.1016/S1470-2045(19)30154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colling R, Pitman H, Oien K, et al. Artificial intelligence in digital pathology: a roadmap to routine use in clinical practice. J Pathol. 2019;249(2):143–150. 10.1002/path.5310 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Chen P, McGough M, et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat Mach Intell. 2019;1:236–245. 10.1038/s42256-019-0052-1 [DOI] [Google Scholar]
- 30.Li Y, Chen P, Li Z, Su H, Yang L, Zhong D. Rule-based automatic diagnosis of thyroid nodules from intraoperative frozen sections using deep learning. Artif Intell Med. 2020;108, 101918. 10.1016/j.artmed.2020.101918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P, Liang Y, Shi X, Yang L, Gader P. Automatic whole slide pathology image diagnosis framework via unit stochastic selection and attention fusion. Neurocomputing. 2021;453:312–325. 10.1016/j.neucom.2020.04.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Hussein S, Chen P, Medeiros LJ, et al. Artificial intelligence strategy integrating morphologic and architectural biomarkers provides robust diagnostic accuracy for disease progression in chronic lymphocytic leukemia. J Pathol. 2022;256(1):4–14. 10.1002/path.5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu W, Toss M, Dawood M, Rakha E, Rajpoot N, Minhas F. SlideGraph+: whole slide image level graphs to predict HER2 status in breast cancer. Med Image Anal. 2022;80, 102486. 10.1016/j.media.2022.102486 [DOI] [PubMed] [Google Scholar]
- 34.Macenko M, Niethammer M, Marron JS, et al. A method for normalizing histology slides for quantitative analysis. I S Biomed Imaging. 2009: 1107–1110. [Google Scholar]
- 35.Graham S, Vu QD, Raza SEA, et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal. 2019;58, 101563. 10.1016/j.media.2019.101563 [DOI] [PubMed] [Google Scholar]
- 36.Wersto RP, Chrest FJ, Leary JF, Morris C, Stetler-Stevenson MA, Gabrielson E. Doublet discrimination in DNA cell-cycle analysis. Cytometry. 2001;46(5):296–306. 10.1002/cyto.1171 [DOI] [PubMed] [Google Scholar]
- 37.Chen TQ, Guestrin C. XGBoost: a scalable tree boosting system. In: Kdd’16: Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining. Association for Computing Machinery; 2016:785–794. [Google Scholar]
- 38.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973:610–621. [Google Scholar]
- 39.Altieri L, Cocchi D, Roli G. A new approach to spatial entropy measures. Environ Ecol Stat. 2018;25(1):95–110. [Google Scholar]
- 40.Altieri L, Cocchi D, Roli G. Advances in spatial entropy measures. Stoch Environ Res Risk Assess. 2019;33(4–6):1223–1240. [Google Scholar]
- 41.Wang X, Barrera C, Bera K, et al. Spatial interplay patterns of cancer nuclei and tumor-infiltrating lymphocytes (TILs) predict clinical benefit for immune checkpoint inhibitors. Sci Adv. 2022;8(22), eabn3966. 10.1126/sciadv.abn3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen PJ, Saad MB, Rojas FR, et al. Cellular architecture on whole slide images allows the prediction of survival in lung adenocarcinoma. Lect Notes Comput Sc. 2022;13574:1–10. [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 44.Dejima H, Hu X, Chen R, et al. Immune evolution from preneoplasia to invasive lung adenocarcinomas and underlying molecular features. Nat Commun. 2021;12(1):2722. 10.1038/s41467-021-22890-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senosain MF, Massion PP. Intratumor heterogeneity in early lung adenocarcinoma. Front Oncol. 2020;10:349. 10.3389/fonc.2020.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marino FZ, Bianco R, Accardo M, et al. Molecular heterogeneity in lung cancer: from mechanisms of origin to clinical implications. Int J Med Sci. 2019;16(7):981–989. 10.7150/ijms.34739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non–small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 49.de Sousa VML, Carvalho L. Heterogeneity in lung cancer. Pathobiology. 2018;85(1–2):96–107. 10.1159/000487440 [DOI] [PubMed] [Google Scholar]
- 50.Ramon y Cajal S, Ses e M, Capdevila C, et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J Mol Med (Berl). 2020;98(2):161–177. 10.1007/s00109-020-01874-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;21(6):1258–1266. 10.1158/1078-0432.CCR-14-1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu X, Fujimoto J, Ying L, et al. Multi-region exome sequencing reveals genomic evolution from preneoplasia to lung adenocarcinoma. Nat Commun. 2019;10(1):2978. 10.1038/s41467-019-10877-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quek K, Li J, Estecio M, et al. DNA methylation intratumor heterogeneity in localized lung adenocarcinomas. Oncotarget. 2017;8(13):21994–22002. 10.18632/oncotarget.15777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee WC, Diao L, Wang J, et al. Multiregion gene expression profiling reveals heterogeneity in molecular subtypes and immunotherapy response signatures in lung cancer. Mod Pathol. 2018;31(6):947–955. 10.1038/s41379-018-0029-3 [DOI] [PubMed] [Google Scholar]
- 55.Lee WC, Reuben A, Hu X, et al. Multiomics profiling of primary lung cancers and distant metastases reveals immunosuppression as a common characteristic of tumor cells with metastatic plasticity. Genome Biol. 2020;21(1):271. 10.1186/s13059-020-02175-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nong J, Gong Y, Guan Y, et al. Circulating tumor DNA analysis depicts sub-clonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9(1):3114. 10.1038/s41467-018-05327-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin Cancer Res. 2018;24(24): 6195–6203. 10.1158/1078-0432.CCR-18-1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin Y, Bao H, Le X, et al. Distinct co-acquired alterations and genomic evolution during TKI treatment in non-small-cell lung cancer patients with or without acquired T790M mutation. Oncogene. 2020;39(9):1846–1859. 10.1038/s41388-019-1104-z [DOI] [PubMed] [Google Scholar]
- 59.Chen R, Lee WC, Fujimoto J, et al. Evolution of genomic and T-cell repertoire heterogeneity of malignant pleural mesothelioma under dasatinib treatment. Clin Cancer Res. 2020;26(20):5477–5486. 10.1158/1078-0432.CCR-20-1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M, Chen R, Jin Y, et al. Cold and heterogeneous T cell repertoire is associated with copy number aberrations and loss of immune genes in small-cell lung cancer. Nat Commun. 2021;12(1):6655. 10.1038/s41467-021-26821-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reuben A, Zhang J, Chiou SH, et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat Commun. 2020;11(1):603. 10.1038/s41467-019-14273-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viswanathan VS, Toro P, Corredor G, Mukhopadhyay S, Madabhushi A. The state of the art for artificial intelligence in lung digital pathology. J Pathol. 2022;257(4):413–429. 10.1002/path.5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen PJ, Aminu M, El Hussein S, Khoury JD, Wu J. Hierarchical phenotyping and graph modeling of spatial architecture in lymphoid neoplasms. Med Image Comput Comput Assist Interv. 2021;12908:164–174. [Google Scholar]
- 64.Chen PJ, El Hussein S, Xing FY, et al. Chronic lymphocytic leukemia progression diagnosis with intrinsic cellular patterns via unsupervised clustering. Cancers. 2022;14(10):2398. 10.3390/cancers14102398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental data that support the findings of this study have been deposited in Mendeley Data (https://doi.org/10.17632/7zc56ttd96.1). The code has been made publicly available at Zenodo (https://doi.org/10.5281/zenodo.8188290).


