Abstract
Although androgen deprivation treatment often effectively decreases prostate cancer, incurable metastatic castration-resistant prostate cancer (CRPC) eventually occurs. It is important to understand how CRPC metastasis progresses, which is not clearly defined. The loss of PTEN, a phosphatase to dephosphorylate phosphatidylinositol 3,4,5-trisphosphate in the PI3K pathway, occurs in up to 70~80% of CRPC. We generated a mouse androgen-independent prostate cancer cell line (PKO) from PTEN null and Hi-Myc transgenic mice in C57BL/6 background. We confirmed that this PKO cell line has an activated PI3K pathway and can metastasize into the femur and tibia of immunodeficient nude and immunocompetent C57BL/6 mice. In vitro, we found that androgen deprivation significantly enhanced PKO cell migration/invasion via the p110β isoform-depended PAK1-MAPK activation. Inhibition of the p110β-PAK1 axis significantly decreased prostate cancer cell migration/invasion. Of note, our analysis using clinical samples showed that PAK1 is more activated in CRPC than in advanced prostate cancer; high PAK1/phosphorylated-PAK1 levels are associated with decreased survival rates in CRPC patients. All the information suggests that this cell line reflects the characteristics of CRPC cells and can be applied to dissect the mechanism of CRPC initiation and progression. This study also shows that PAK1 is a potential target for CRPC treatment.
Introduction
Circulating androgen is critical for prostate development, and androgen deprivation is an effective method for prostate cancer treatment. Following androgen deprivation, a more severe refractory prostate cancer eventually occurs in almost all patients, independent of androgen availability. How this incurable Castration-Resistant Prostate Cancer (CRPC) progresses is largely unknown due to the lack of suitable models (1–5).
The phosphoinositide 3-kinase (PI3K) pathway is frequently dysregulated in most cancer types, including prostate cancer. As important regulators of tumor growth, proliferation, and survival, the Class 1A PI3Ks consist of a catalytic and a regulatory subunit. The p110α and p110β isoforms are the key catalytic subunits of PI3K in solid tissues. Activating mutations and amplifications of the PIK3CB gene encoding p110β are frequently found in the advanced CRPC (1). PTEN (phosphatase and tensin homolog deleted on chromosome 10), a dual phosphatase with both protein and lipid phosphatase activities, acts to dephosphorylate phosphatidylinositol 3,4,5-trisphosphate in the PI3K pathway. As a tumor suppressor, PTEN is inactivated in over 40% of primary and up to 70~80% of metastatic prostate tumors.
Our previous studies show that androgen deprivation accelerates the progression of High-Grade Prostatic Intraepithelial Neoplasia (HG-PIN) in PTEN null tumors to invasive CRPC (6), and p110β plays a key role in the initiation and development of PTEN null CRPC using genetically engineered mouse models (GEMMs) (7). Though our GEMMs study showed that p110β ablation diminished PTEN null CRPC invasion in the prostate lobes, whether and how p110β directly regulates the invasion/migration of CRPC cells is not clear. In the current study, we generated a murine PTEN null androgen-independent prostate cancer cell line to further dissect the molecular mechanisms of CRPC progression. We found that depletion/inhibition of p110β, but not p110α, reduced CRPC cell invasion/migration. Mechanistically we found that androgen deprivation up-regulates PAK1 expression and the mitogen-activated protein kinase (MAPK) activation to promote cell invasion. In addition, we found that PAK1-mediated MAPK activation depends on RAC1 and p110β in PTEN null prostate cancer cells. Consistently, clinical studies showed that the levels of PAK1/phosphorylated-PAK1/active MAPK are significantly increased in CRPC patients. Importantly, PAK1/phosphorylated-PAK1 negatively correlated with patient survival rates. Overall, our study using genetic and pharmacological methods showed that p110β regulates the RAC1-PAK1-MAPK axis to promote CRPC invasion, and androgen deprivation increases PAK1 expression and p110β/RAC1 interaction to further augment the invasion capability of CRPC cells. In addition, our murine cell line reflects the characteristics of human androgen-independent prostate cancer cells and can be applied for the mechanistic studies of PTEN null prostate cancer.
Materials and Methods
Antibodies:
The following antibodies were used in this study. PAK1 (Cell Signaling, 2602 for IB; Proteintech, 21401-1-AP for IHC), p-PAK1/2 (Cell Signaling, 2605 for IB; Biosynthesis biotechnology, bs-3315R for IHC), p110β (Cell Signaling, 3011), p110α (Cell Signaling, 4249), PTEN (Cell Signaling, 9552), p-AKT (Cell Signaling, 3787), AKT (Cell Signaling, 9272), p-ERK1/2 (Thr202/Tyr204) (Cell Signaling, 4370 for IB; Proteintech, 28733-1-AP for IHC), ERK (Cell Signaling, 9102), p-MEK1/2 (Cell Signaling, 9121), MEK1/2 (Cell Signaling, 9122), AR (N-20, SC-816), p-S6RP (Ser240/244) (Cell Signaling, 2215), p-S6RP (Ser235/236) (Cell Signaling, 2211), S6RP (Cell Signaling, 2217), Tubulin (Sigma-Aldrich, T5168), Vinculin (Sigma-Aldrich, clone V284), Actin (Cell Signaling, 4970), KRT5 (Covance, PRB-160P), KRT14 (Covance, PRB-155P), KRT18 (Epitomics, 1433-1), Rac1 (Millipore, clone 23A8), CD49f (BioLegend, 313611), Ep-CAM (BioLegend, 118217), Myc (9E10, Santa Cruz, sc-40), Myc-tag (9B11, Cell Signaling, 2276).
Plasmids and cells:
PC3 and HEK283T cells were originally purchased from ATCC. Though both lines had not been authenticated, none of these two lines is listed in the Database of Cross-Contaminated or Misidentified Cell Lines. Mycoplasma testing had not been conducted on these two lines. PKO cells were derived from PTEN null; Myc GEM mice housed at Dana-Farber Cancer Institute, Harvard Medical School. The Androgen receptor expression vector was a gift from Dr. Manav Korpal, Novartis, Boston. PAK1 expression plasmids and the shRNA vectors for PAK1 were purchased from Addgene. shRNA vector of AR was ordered from the shRNA core facility, Dana-Farber Cancer Institute. pBABE-hygro-2xMyc-Rac1-Q61L (addgene,128582), pWPXL-eGFP vector (addgene, 12257), pWPXL-c-Myc (addgene, 36980) were purchased from Addgene.
Mice used and caudal artery injection:
Adult (6~8 weeks old) athymic female nude mice (002019) and C57/B6 mice (female and male) (000664) were purchased from the Jackson Laboratory. Mouse experimental protocols were approved by the Institutional Animal Care and Use Committee of the Medical University of South Carolina. In brief, CS-PKO-luc cells (1X106) suspended in 200 μL PBS were injected into the caudal artery of mice using a 29 G syringe needle in a short time (<6 s). Bone metastasis was examined with bioluminescence imaging, which was acquired with IVIS Spectrum after intraperitoneal injection of D-luciferin (50 mg/kg). Mice were randomly grouped to receive either CS-PKO-luc cells or controls. The investigators were not blinded to the allocation of mice to mouse groups. The sample size was chosen based on the pilot experimental evidence from our laboratory, and no animal was excluded from the analysis.
Primary mouse prostate tumor epithelial cell cultures from PTEN null; Myc mice:
Prostate tissues from PTEN null; Myc mice were dissected and collected using a dissection microscope. Tumor tissues were minced into 2–3mm pieces with sterile scissors and transferred to a 10cm Petri dish containing cold, sterile PBS. The prostate tissues were digested with Collagenase/Hyaluronidase in DMEM/F-12 for 3hr, centrifuged at low speed for collection, and re-suspended in 0.05% Trypsin for an additional 1hr digestion on ice. Hanks’ solution supplemented with 2% FBS was added to neutralize the enzymatic reaction, and cells were centrifuged at low speed for 5 min for collection. Dispase and DNase were added to the cell pellet and pipetted softly for 1 min before neutralizing the reaction by adding Hanks’ solution supplemented with 2% FBS. The cell suspension was filtered through a 40μM cell strainer, centrifuged at low speed for collection, and re-suspended into prostate epithelial cell basal culture medium supplied with 10ng/ml EGF, bFGF, and 4μg/ml Heparin (StemCell, 05640). Culturing medium was refreshed every 3 days until cell colonies grew out. Cell colonies with typical epithelial morphology were carefully collected with sterile filter paper and were initially seeded on a 48-well culturing plate and later propagated in 10cm Petri dishes. The cells were adapted to culture in DMEM supplemented with 10% FBS or charcoal-stripped FBS (Sigma).
RT-PCR:
RNA RNeasy mini kit (Qiagen) was applied to extract total RNA from cells, followed by reverse-transcribing to cDNA using reverse transcriptase (Bio-Rad). qRT-PCR was performed in BioRad CFX96 Touch Real-Time PCR detection system with specific primers.
Lentivirus packaging and infection:
The shRNA-pLKO.1 expression plasmid containing the indicated sequences for shRNA generation was packaged in HEK293T cells. HEK293T cells were transfected with lipofectamine 2000. Lipofectamine 2000 was diluted in Opti-MEM medium, and pCMV-VSVG: pCMV-delta8.9: expression vector was mixed at a ratio of 1:5:10. Virus-containing medium was harvested 2 days after transfection. For virus infection, cells were seeded in 6 well plates and infected with 0.5ml lentivirus-containing medium for 24hr in the presence of 10μg/ml polybrene. Two days later, 1μg/ml puromycin was added for selection over 10 days for subsequent assays.
AKT1/2 knockdown:
shRNAs targeting mouse AKT1 (shAKT1-1: TRCN0000304683; shAKT1-2: TRCN0000304735) and AKT2 (shAKT2-1: TRCN0000055258; shAKT2-2: TRCN0000310882) were purchased from Millipore-Sigma. CS-PKO cells were first infected with the shAKT1-1 or shATK1-2 -encoding lentivirus followed by a five-day selection in the presence of 1 μg/ml puromycin. Then, these cells were infected with the shAKT2-1 or shAKT2-2 -encoding lentivirus, and infected CS-PKO cells were selected for one week in the presence of 2 μg/ml puromycin.
Immunofluorescence staining:
PKO cells were seeded on chamber slides at 30-50% confluence. The following day cells were washed with PBS twice, fixed with cold methanol for 15 min, then permeabilized with PBS/0.2% Triton X-100 for 15 min. PBS/2.5% BSA was added to chamber slides for 30 min for blocking. PKO cells were incubated with primary antibodies (1:100 dilutions in PBS/2.5% BSA) at 37°C for 1hr. After PBS washing, cells were incubated with secondary antibodies conjugated with Alexa-Fluor 568 dyes in PBS/2.5% BSA at room temperature for 1hr. Pictures were taken with a Zeiss Axio fluorescence microscope.
Immunoprecipitation and blotting:
Immunoprecipitation (IP) and blotting were performed as described before (6). Cell lysate (1mg) in IP lysis buffer was incubated with antibody (10ug) at 4°C overnight for immunoprecipitation. The next day, agarose beads (50ul) were added for an additional incubation for 2hr. After centrifuging at low speed, the immunoprecipitated complex was resuspended in 2 X SDS sample loading buffer (50ul) and denatured at 95°C for 10min. The denatured immunoprecipitated protein complex or cell lysates were separated in 10% or 15% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies for 3hr at room temperature or overnight at 4°C. The signal was detected and/or quantified with an Odyssey (LI-COR Biosciences).
Wound healing assay:
3X106 cells were seeded in a 10cm Petri dish overnight. The next day, multiple scratch lines were made with sterile 200ul tips. Pictures were taken at 0, 15, or 24hr post-scratching with a Nikon (Diaphot 200) microscope.
Invasion assay:
Modified Boyden chambers with diluted Matrigel (1:4) coated porous filters were applied for invasion assays. 4~5 X 105 cells were seeded in the upper chambers, and invaded cells were collected and counted after 24hr. Three repeats were performed for statistical analysis.
Human tissue samples:
The human specimens used in this study were approved by Sichuan Provincial People’s Hospital’s Committees for Ethical Review of Research Involving Human Subjects (2022377). Paraffin-embedded prostate cancer tissues were obtained from patients receiving radical prostatectomy or transurethral resection prostate at Sichuan Provincial People’s Hospital between Jan 1, 2017, and Dec 12, 2020. Tissues from patients showing a rising PSA level (PSA ≥ 4ng/ml) and/or imaging perspective tumor progression (CT or MRI showing new tumor lesions or bone metastasis) within 24 months after receiving radical prostatectomy or transurethral resection (serum testosterone <1.7nmol/L) were defined and analyzed as CRPC group. Tissues from patients with normal PSA levels and normal imaging after receiving radical prostatectomy or transurethral resection until our following were defined and analyzed as PCa group. All patients were followed up until August 31, 2022. All samples were diagnosed by two independent pathologists and urologists. TNM stage and Gleason score of samples were confirmed according to the guidelines.
Immunohistochemistry and scoring analyses:
The IHC was performed as described before (8). In brief, embedded tissues were deparaffinized with xylene and rehydrated with diluted ethanol. The tissue sections were processed for IHC staining. After antigen retrieval with sodium citrate buffer (pH6.0) at a sub-boiling temperature for 30 min, tissue sections were cooled down for an additional 30 min in sodium citrate buffer. The tissue sections were quenched with 3% H2O2 for 10 min and washed with ddH2O for 15 min. The sections were blocked with 5% normal goat serum. The sections were then incubated with primary antibody at room temperature for 2hr or 4°C overnight. The sections were incubated with biotinylated secondary antibodies (Servicebio, G1213) for 15 min, and Elite Peroxidase Kit (Servicebio, China) was applied to amplify staining signals. DAB (Servicebio, China) was used for enzymatic signal visualization. The nuclear counterstaining was performed with Mayer’s hematoxylin (Servicebio, China). Ultra-clean quick-drying sealant (Servicebio, China) was applied to the sections, and the sections were covered with coverslips. Images were visualized and scanned using a PANNORAMIC MIDI (3DHISTECH, Hungary) microscope and processed with Caseviewer software.
PAK1/p-PAK1 levels in the human samples were blind-quantified by two pathologists. The immunostaining intensity of each sample was divided into one of four categories: negative (0), weak (1), moderate (2), or strong (3). Subsequently, the proportion of positively stained cells for each slide was assessed and graded into <25% (1), 25–50% (2), 50–75% (3), and >75% (4). The overall score of each slide was calculated as the immunostaining intensity score multiplied by the proportion score (overall score = intensity X proportion score). The human samples were then classified as low (overall score ≤6) or high (overall score > 6) levels for PAK1 and p-PAK1.
RAC1/CDC42 activation assay:
RAC1/CDC42 activation levels in cells were measured by following the instruction of the Pull-Down Activation Assay Biochem Kit (Cytoskeleton, BK030). In brief, cells were seeded in 6-well plates and harvested on the next day in a lysate buffer. The GTP-bound form of RAC1/CDC42 was purified from cell lysates using the p21 Binding Domain of PAK1. The active RAC1/CDC42 was detected by immunoblotting using specific antibodies and normalized to input actin. For statistical comparison, three repeats were performed.
GSEA analysis:
GSEA analysis was performed using the GSEA tool v.4.2.3 (9), with the MSigDB v.7.1. Hallmarks gene sets collection and the ‘classic’ method for calculating enrichment scores.
Data availability:
Microarray data used to support the present study have been deposited in the Gene Expression Omnibus with an access number of GSE233306.
Statistical analyses:
Statistical results were represented as means ± SD or mean ± SEM. Analyses were performed with the two-tailed p-value or one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 and n.s., not significant.
Results
Generation of androgen-independent PTEN null cell lines
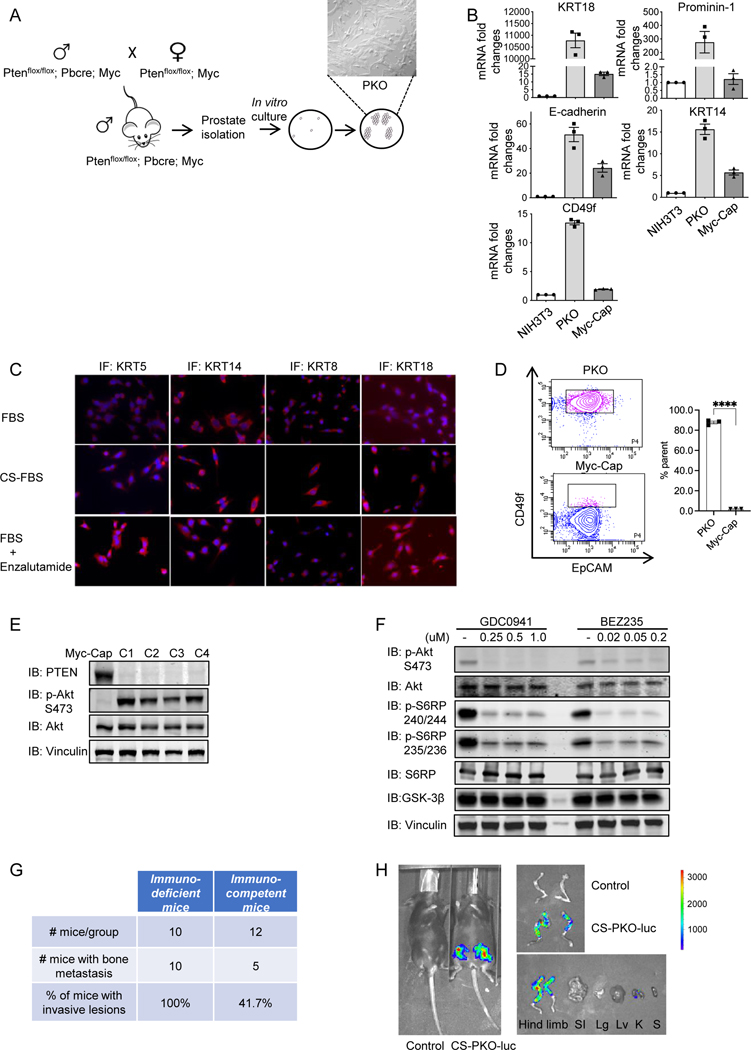
PTEN null GEMMs have been studied as models for androgen-sensitive and castration-resistant prostate tumorigenesis (10–12). As the prostate contains multiple cell types, including epithelial, stromal, and immune cells, it is difficult to use GEMMs to precisely define the mechanism of prostate cancer development (13). To investigate in detail the potential mechanisms through which PTEN null CRPC initiates, we utilized primary mouse cell cultures from our GEMMs of prostate cancer. Consistent with the unpublished findings of multiple laboratories that less aggressive prostate tumor models are difficult to culture, we failed to grow prostate epithelial cells in vitro from the HG-PIN that develops in the ventral lobes of the PTEN null model (6). c-Myc is one of the major drivers of the prostate tumorigenesis (14–17). Concurrent PTEN loss and Myc amplification markedly increase the risk of prostate cancer specific mortality, indicating the combination of these two genetic events may drive a more aggressive disease (16–18). We crossed the Hi-Myc transgene (19) into the PTEN null GEM (6), which both had been backcrossed to the C57BL/6 background, to generate an aggressive prostate cancer model (Supplementary Fig. S1). We successfully derived multiple clonal primary prostate cancer cells from the malignant tumors and established them as primary cultures (named PKO cells) (Fig. 1A). qRT-PCR (Fig. 1B) and/or immunofluorescent staining for cytokeratins and E-cadherin demonstrated the epithelial nature of these cells (Fig. 1C). The results of flow cytometry (Fig. 1D) and qRT-PCR (Fig. 1B) showed that PKO cells were mixed lineage cells (CD49fhighEpCAM+Keratin14highKeratin18high), which is different from Myc-Cap cells (CD49flowEpCAM+Keratin14lowKeratin18low), a control cell line derived from Hi-MYC mice. Immunoblotting and immunofluorescence results showed that PTEN was not expressed in PKO cells. In contrast, the PI3K pathway was activated due to PTEN deletion (Fig. 1E&Supplementary Fig. S2A). PI3K inhibitor treatment can decrease AKT and S6 ribosomal protein (S6RP) phosphorylation levels in PKO cells (Fig. 1F). Gene expression microarray analysis showed that PKO cells displayed compromised Androgen Receptor (AR) signaling under baseline conditions compared to the androgen-dependent murine cell line, Myc-Cap (Supplementary Fig. S2B, 2C&Supplementary Table 1). Gene Set Enrichment Analysis (GSEA) on the microarray data showed different pathways/processes between PKO and Myc-Cap cells (Supplementary Fig. S2D). The interferon-α response hallmark and the interferon-γ response hallmark were the top two enriched pathways/processes in PKO cells, in contrast, the E2F targets hallmark and the MYC targets hallmark were the top two pathways/processes highly expressed in Myc-Cap cells. To recapitulate CRPC progression in vitro, we cultured PKO cells in androgen-deprived media by supplying them with charcoal-stripped FBS (CS-PKO cells) or in the presence of Enzalutamide for a period of over a month. Immunofluorescence staining also showed the epithelial character (i.e., cytokeratin expression) in these androgen-deprived/AR-inhibited conditions (Fig. 1C). In addition, RT-PCR was performed to detect the mRNA levels of androgen response genes in PKO cells. The mRNA levels of KLF5 and NKX3.1 were down-regulated, FKBP5 was up-regulated, and PSCA did not significantly change under androgen deprivation conditions (Supplementary Fig. S2E).
Figure 1. Generation and characterization of androgen-independent prostate cancer cell lines.
1A. Schematic depiction of GEM model generation used to derive primary prostate cancer cell lines. PTEN−/−; Myc mice were generated by crossing PTEN−/− mice with Myc transgenic mice. Aged PTEN−/−; Myc mice (7 months old) with highly invasive tumors were utilized for prostate epithelial cell line generation.
1B. qRT-PCR analysis of epithelial markers expressed in PKO cells. Epithelial markers (E-cadherin, KRT14, KRT8, KRT18, KRT5) were positively expressed in PKO cells with a relatively higher expression level than Myc-Cap cells. mRNA from NIH3T3 cells, mouse fibroblasts, serves as the negative control, and mRNA from Myc-Cap cells (36) is the positive control for epithelial marker expression. mRNA levels of targets were normalized to that of GAPDH.
1C. Immunofluorescence (IF) staining of epithelial markers in PKO cells. The epithelial markers (KRT14. KRT8. KRT18. KRT5) showed positive staining in PKO cells cultured under either normal (supplied with 10% FBS) or androgen deprivation conditions (with 10% Charcoal Striped-FBS or 10% FBS+10μM enzalutamide (MDV3100), an androgen receptor inhibitor) DMEM medium.
1D. Flow cytometry analysis using CD49f and EpCAM staining showed that PKO cells were mixed populations of lineage, but not mixed populations with distinct lineages. Myc-Cap cells are controls for population analysis. PKO cells showed CD49fhighEpCAM+, and Myc-Cap cells showed CD49flow EpCAM+ lineages. n=3.
1E. PI3K pathway was activated in PKO cells. Immunoblots showed that PTEN was not detectable in PKO cell lines (C1 to C4) (IB: PTEN). Immunoblots of phosphorylated AKT at S473 (IB: p-Akt) showed PI3K pathway activation in PKO cell lines. Myc-Cap is a control mouse cell line with overexpressed Myc but a non-activated PI3K pathway.
1F. PI3K pan inhibitors repressed PI3K activity in PKO cells. To inspect the PI3K pathway in PKO cells, two pan-PI3K inhibitors (GDC0941 and BEZ235) were used to treat PKO cells. AKT activity and the PI3K/AKT downstream target S6 ribosomal protein (S6RP) were examined using phosphorylated AKT at S473 (IB: p-Akt S473), phosphorylated S6RP at 240/244 (IB: p-S6RP 240/244), 235/236 (IB: p-S6RP 235/236) as readouts. Vinculin is a loading control. DMSO is drug vehicle control.
1G&1H. Establishing murine CRPC bone metastasis models. Summary of bone metastases in immunodeficient and immunocompetent mice by injecting CS-PKO-Luc cells through the caudal artery (G). Whole-body bioluminescence imaging was applied to measure the metastatic lesions in bone after D-luciferin injection (H). The picture was taken on Day 14 post cell injection. SI stomach and intestine, Lg lung, Lv liver, K kidney, S spleen, PS prostate, and seminal vesicles.
Bone metastases develop in ~30% of patients within 2 years of castration resistance, and eventually in 80~90% of patients with CRPC (20,21). A recent study shows that intra-caudal arterial (CA) injection of prostate cancer cells can achieve bone metastasis, which is more efficient than the intracardiac injection (22). As the testosterone levels in female mice are less than one-tenth of that in males, female mice are often applied in the CRPC study (23–25). To test whether CS-PKO cells have the potential to develop bone metastases in vivo, we injected luciferase-expressing CS-PKO cells (PKO-luc) into immunodeficient and immunocompetent female mice through CA. Whole-body bioluminescence imaging (BLI) was applied to measure the metastatic lesions in bone after intraperitoneal injection of D-luciferin. Within 3 weeks, PKO-luc developed bone metastasis in 5 out of 12 immunocompetent mice and 10 out of 10 immunodeficient nude mice (Fig. 1G). Luciferase signals were also measured in dissected mouse tissues to confirm the imaging findings. Only the femur and tibia of the hindlimbs, but not other organs, showed enhanced luciferase signals (Fig. 1H). All these results indicate that this cell line is an epithelial cell line, which inherits the characteristics of PTEN null prostate cancer, and can be applied to the study of PTEN null prostate cancer progression.
Androgen deprivation upregulates the PAK-MEK axis to increase invasion
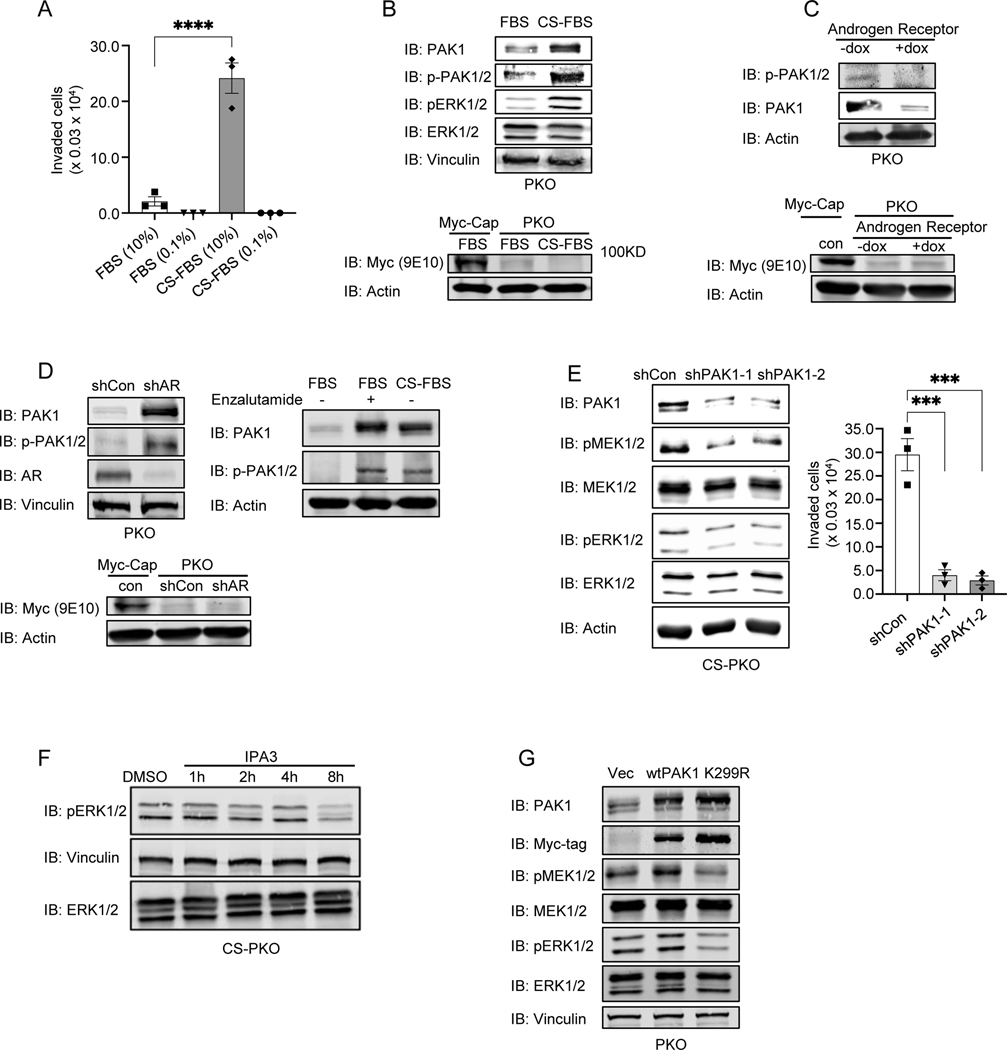
Our previous study showed that either surgical or pharmaceutical inhibition of AR in PTEN null mice induces the invasive CRPC (6). We measured the invasion capability of CS-PKO cells with a period of culturing under androgen deprivation. We found that the invasion rate of CS-PKO cells was dramatically enhanced (Fig. 2A&Supplementary Fig. S2F). Our previous study showed that the mRNA level of p21-activated kinase 1 (PAK1) is significantly increased in mouse tissues of CRPC compared to HG-PIN (7). PAKs are major effectors downstream of the Rho/RAC family of GTPases, and activated PAKs play important roles in promoting cell invasion and metastasis (26–28). It has been reported that PAK1 is highly expressed in invasive prostate cancer cells compared to non-invasive prostate cancer cells (29). Whether androgen deprivation upregulates PAK1 expression is not known. Immunoblotting showed that the expression and phosphorylation levels of PAK1 were significantly enhanced in CS-PKO cells (Fig. 2B). RT-PCR using PAK1-specific primers showed that mRNA level of PAK1 was also upregulated in androgen deprivation condition (Supplementary Fig. S3A). Androgen deprivation suppresses the AR activity (30,31). To further examine whether the AR pathway could regulate PAK1 expression, we induced AR expression using a doxycycline-regulated promoter in stably transfected PKO cells. A significant decrease in PAK1 expression was observed in PKO cells as exogenous AR expression increased (Fig. 2C). Conversely, a dramatic increase of PAK1 expression was seen in PKO cells when AR expression was knocked down with an shRNA (Fig. 2D, left) or AR activity was inhibited by Enzalutamide (Fig. 2D, right). These results indicate that enhanced PAK1 expression is an outcome occasioned by a reduction of AR activity in PTEN null CRPC. To examine whether PAK1 phosphorylation was regulated, we overexpressed, and immunoprecipitated exogenous PAK1 from the lysates of PKO and CS-PKO cells, and then immunoblotted IP-ed PAK1 with phosphorylated PAK1 antibodies. Our results showed that PAK1 phosphorylation was upregulated in androgen deprivation conditions (Supplementary Fig. S3B). Thus, we concluded that PAK1 expression and phosphorylation are both increased in androgen-deprived PTEN null cancer cells.
Figure 2. PAK1 expression/activity is up-regulated in PKO cells in androgen-deprived conditions to activate the MAPK pathway.
2A. Androgen deprivation increased the invasion capability of PKO cells. The invasion assays were performed in modified Boyden chambers. PKO/CS-PKO cells were starved with 0.1% FBS (FBS) or charcoal-stripped FBS (CS-FBS) overnight and seeded in the upper chambers. 10% FBS or CS-FBS was applied as attractants in the bottom chambers. The invaded cells were countered after 24hr.
2B. The expression and phosphorylation levels of PAK1 were enhanced in PKO cells cultured in androgen deprivation conditions. The expression and phosphorylation levels of PAK1 in cell lysates of PKO cells cultured in DMEM with FBS (FBS) or charcoal-stripped FBS (CS-FBS) were determined by immunoblotting with an antibody against PAK1 (IB: PAK1) or phosphorylated PAK1/2 (IB: p-PAK1/2). Immunoblots of vinculin (IB: Vinculin) and actin (IB: Actin) serve as loading controls.
2C. Inducible AR expression in PKO cells decreased PAK1 expression and phosphorylation. AR expression was rendered inducible with doxycycline in PKO cells using a pLKO-TREX-HA-Neo construct.
2D. The expression and phosphorylation levels of PAK1 were increased in PKO cells when AR was knocked down (Left) or inhibited by enzalutamide (10uM) (Right). AR expression was knocked down with shRNA in PKO cells and measured with AR antibody blotting (IB: AR).
In 2B-2D, Myc antibodies (9E10) was applied to detect transgenic MYC expression in PKO cells. Myc-Cap is a control for transgenic MYC expression.
2E. PAK1 knockdown decreased MAPK activity (Left) and invasion (Right) in CS-PKO cells. PAK1 expression was knocked down with two individual shRNA (shPAK1–1 and shPAK1–2). MAPK activity was measured with phosphorylated MEK1/2 (IB: pMEK1/2) or ERK1/2 (IB: pERK1/2) antibodies. The invasion capabilities of PAK1 knockdown CS-PKO cells were measured with modified Boyden chambers.
2F. Inhibition of PAK1 decreased MAPK phosphorylation in CS-PKO cells. MAPK activity was measured via ERK1/2 phosphorylation (IB: pERK1/2) in CS-PKO cells treated with the PAK1 inhibitor IPA-3. CS-PKO cells were cultured in androgen-depleted DMEM and starved in 0.5% CS-FBS DMEM overnight before adding inhibitors. Total protein levels of ERK1/2 were measured as a loading control (IB: ERK1/2).
2G. PAK1 overexpression increased the activity of the MAPK pathway. MAPK activity was measured with antibodies to phosphorylated MEK1/2 (IB: pMEK1/2) or phosphorylated ERK1/2 (IB: pERK1/2). PAK1 levels were examined with PAK1-specific antibodies (IB: PAK1). Expression of exogenous PAK1 was detected with a Myc-tag antibody (IB: Myc-tag). Vinculin (IB: Vinculin) is a loading control. Wt is wild-type PAK1. Vec is empty vector control. K299R is a PAK1 kinase-dead mutant.
Our work and others report that the MAPK pathway plays a critical role in CRPC development and is significantly elevated in CRPC in vivo (6,32,33). Our in vitro study showed that androgen deprivation, indeed, enhanced the MAPK pathway, as evidenced by phosphorylation of MEK-ERK in CS-PKO cells (Fig. 2B). Meanwhile, PAK1 knockdown with specific shRNAs significantly decreased the activity of MAPK pathway in CS-PKO cells (Fig. 2E, left). In addition, IPA3, a selective allosteric PAK1 inhibitor, decreased ERK phosphorylation in CS-PKO cells (Fig. 2F). Overexpression wild-type PAK1, but not a kinase-dead mutant (K299R-PAK1), increased the phosphorylation of MEK-ERK (Fig. 2G). PAK1 knockdown significantly decreased the invasion capability of CS-PKO cells (Fig. 2E, right). These findings suggest that PAK1 regulates MEK-ERK activation and cell invasion.
PAK1-mediated MAPK pathway relies on p110β to regulate CRPC cell migration/invasion
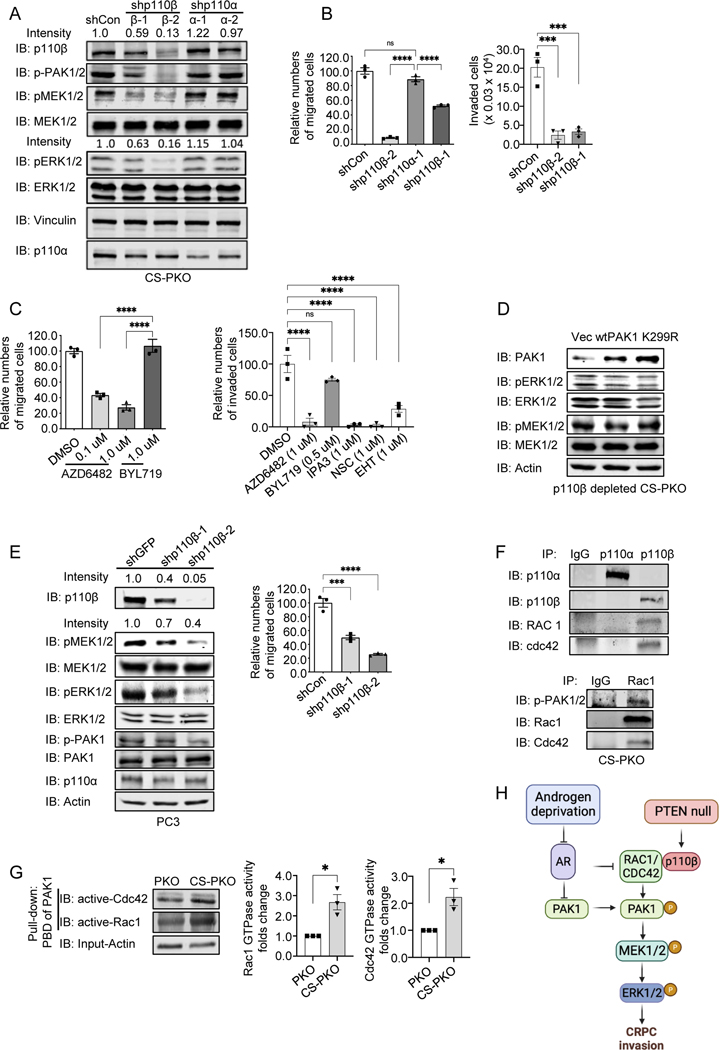
Our previous study showed that the p110β isoform of PI3K plays a critical role in CRPC initiation via unknown mechanisms (7). To investigate the biological role of p110β in CRPC, we knocked down p110β with a specific shRNA in CS-PKO cells. PAK1-MEK-ERK phosphorylation, as well as the migration/invasion capability, in p110β knockdown CS-PKO cells was decreased, which was not observed in p110α knockdown cells (Fig. 3A, 3B&Supplementary Fig. S3C). Treatment with the p110β inhibitor, AZD6482, also decreased CS-PKO cell migration/invasion, while the p110α inhibitor, BYL719 was ineffective (Fig. 3C&Supplementary Fig. S3D). The invasion capability of CS-PKO cells was significantly decreased when PAK1 was inhibited with IPA3 (Fig. 3C, right). Overexpressing PAK1 in PKO cells increased the phosphorylation of MEK-ERK (Fig. 2G), while overexpressing PAK1 in p110β knockdown CS-PKO cells did not increase the phosphorylation of MEK-ERK, indicating that PAK1-mediated MAPK activation depends on p110β (Fig. 3D).
Figure 3. p110β is required for PAK1-MEK-dependent migration.
3A. Knockdown of p110β but not p110α decreased the phosphorylation levels of PAK1 and MAPK activity in CS-PKO cells. p110 isoforms were stably knocked down with shRNA in CS-PKO cells cultured in androgen-depleted DMEM. The knockdown efficiencies in p110s were examined with either p110β (IB: p110β) or p110α (IB: p110α) antibodies. Phosphorylation levels of ERK1/2 (IB: pERK1/2) and MEK1/2 (pMEK1/2) were detected with individual antibodies. Expression levels of total ERK (IB: ERK1/2) and MEK1/2 (IB: MEK1/2) were examined with specific antibodies.
3B&3C. Left: Migration quantification of CS-PKO cells with p110 isoform knocked down (3B, left) or with p110 isoform inhibitor treatment (3C, left) at 24hr post-scratching in scratch assays. Cell number was counted and normalized to controls in random regions in scratched areas. The numbers were counted in three regions for statistical analysis.
Rights: Invasion quantification of CS-PKO cells with p110β knocked down (3B, right) or under the treatment of inhibitors for p110 isoform (AZD6482 for p110β, BYL719 for p110α), PAK1 (IPA3) or RAC1 (NSC23766, EHT1864) (3C, right). Modified Boyden chambers were applied for migration assays. The migrated cells were countered after 24hr.
3D. Compared to Figure 2G, overexpressing wild-type PAK1 in p110β knockdown CS-PKO cells no longer increased the phosphorylation of MAPK. PAK1 levels were examined with PAK1-specific antibodies (IB: PAK1). Actin (IB: Actin) is a loading control.
3E. p110β knockdown in PC3 cells decreases p-PAK/MAPK activity and cell migration capability. p110β KD in PC3 cells was induced by adding doxycycline to cells expressing a tet regulated shRNA construct (left). Knockdown efficiency was examined with a p110β-specific antibody (IB: p110β). p-PAK/MAPK activities were detected using antibodies against phosphorylated MEK1/2, ERK1/2 and p-PAK1. Right: Quantification of migration of PC3 cells with p110β knocked down at 72hr post-scratching in scratch assays.
3F. Co-immunoprecipitation of p110β with RAC1 and CDC42 in CS-PKO cells cultured in androgen deprivation media. p110α/β (IP/IB: p110α or p110β) was immunoprecipitated and immunoblotted from whole cell lysate of CS-PKO cells. The presence of either RAC1 or CDC42 associated with p110s was measured by blotting with RAC1 (IB: RAC1) or CDC42 (IB: Cdc42) antibodies.
3G. Immunoblotting using RAC1 or CDC42 specific antibodies of the GTP-bound form of RAC1/CDC42 pulled down from cell lysate using the p21 Binding Domain of PAK1 (left). Actin was the loading control for input proteins for the pull-down assay. The Middle and Right panels are Statistical analyses of three pull-down assays.
3H. The schematic figure depicts the presented study model. Created with BioRender.com.
To test whether our observations are also true in human prostate cancer cells, we investigated the pathway in a human prostate cancer cell line, PC3, which is a PTEN null, androgen-independent cell line. An inducible shRNA was utilized to knock down p110β in PC3 cells. Both the phosphorylation levels of MAPK and cell migration capability were decreased in PC3 cells upon p110β knockdown (Fig. 3E&Supplementary Fig. S3E). The treatment of p110β inhibitor (AZD6482), but not p110α inhibitor (BYL719), also decreased PC3 cell invasion (Supplementary Fig. S3F).
The PI3K isoform p110β is responsible for RAC activation and enhanced mobility in PTEN null hematopoietic tumors (34). Whether p110β mediates RAC activation in solid tumors is not known. We tested the interaction of PI3K isoforms with RAC1/CDC42 in CS-PKO cells. Co-immunoprecipitation results showed that the p110β isoform, but not p110α, interacts with RAC1/CDC42 (Fig. 3F). RAC1/CDC42 Pull-Down Activation Assay using the p21 Binding Domain of PAK1 showed that more GTP-bound form of RAC1/CDC42 (active form) was founded in CS-PKO cells than that in PKO cells (Fig. 3G). Co-immunoprecipitation results showed that the interaction of p110β and RAC1 was increased under androgen deprivation conditions (Supplementary Fig. S3G). Further, the enhanced RAC1 activity in CS-PKO cells (Fig. 3G) was diminished in p110β depleted CS-PKO cells, indicating that p110β expression is critical for RAC1 activity in CS-PKO cells (Supplementary Fig. S3H). RAC1/CDC42 is known to increase PAK1 phosphorylation and activate PAK1 (35). Overexpressing the constitutively active mutant, RAC1-Q61L, in p110β-depleted CS-PKO cells rescued and increased p-PAK/p-MEK/p-ERK (Supplementary Fig. S3I). Furthermore, the treatment of RAC1/CDC42 inhibitors, NSC23766 or EHT1864, decreased CS-PKO cell invasion (Fig. 3C, right). Our study suggests that p110β interacts with RAC1/CDC42 to enhance PAK1 activation, but not PAK1 expression, and androgen deprivation increases this regulation (PAK1 expression and p110β-RAC1 interaction) to augment PTEN null prostate cancer cell invasion (Fig. 3H).
In PTEN null tumors, there is a negative feedback loop between AKT and AR through the regulation of PHLPP (12). To test whether the AKT pathway is involved in RAC1-PAK1 activation under androgen deprivation conditions, we applied pharmacological and genetic approaches to inactivate AKT. A pan-Akt inhibitor, Capivasertib (AZD5363), was applied to inhibit AKT activity in CS-PKO cells. In parallel, AKT1 and AKT2 were sequentially knocked down in CS-PKO cells. Compared to the vehicle-treated/control cells, the expression and phosphorylation levels of PAK1 in AKT-inhibited/depleted-CS-PKO cells were not decreased. Meanwhile, the RAC1 activity was not significantly affected by AKT knockdown. S6RP and GSK3β are two downstream targets of AKT. In this study, the phosphorylation levels of S6RP and GSK3β were decreased, which are the positive controls for AKT inhibition/depletion treatment. Thus, we concluded that the regulation of RAC1 and PAK1 is independent of the AKT pathway (Supplementary Fig. S3J).
Immunoblotting using MYC antibodies (9E10) (19,36) showed that the transgenic MYC expression was significantly low in PKO cells compared to Myc-Cap cells, a control cell line derived from Hi-MYC mice (Fig. 2B). Consistently, GSEA of the microarray array showed that the MYC targets hallmark was highly enriched in Myc-Cap cells compared to PKO cells (Supplementary Fig. S2E). Since PKO cells were derived from the combo PTEN null;Hi-Myc mice, the transgenic MYC (ARR2/probasin-Myc) expression in PKO cells depends on the presence of AR. AR pathway manipulation by androgen deprivation, or AR knockdown, slightly decreased the transgenic MYC expression in PKO cells (Fig. 2B & 2D). Induced AR expression in PKO cells marginally increased the transgenic MYC expression (Fig. 2C). Although AR manipulation barely changed the transgenic MYC expression, it is important to test whether exogenous MYC expression affects the PAK/MEK/ERK signaling in PKO cells. Immunoblotting results showed that exogenous MYC expression did not significantly modulate the levels of PAK1/p-PAK/p-MEK/p-ERK in CS-PKO cells (Supplementary Fig. S3K). These results suggest that the alteration of PAK/MEK/ERK signaling in PKO cells under AR inhibition is not through MYC regulation.
Clinical data analysis showed that PAK1 expression and phosphorylation are increased in CRPC patients
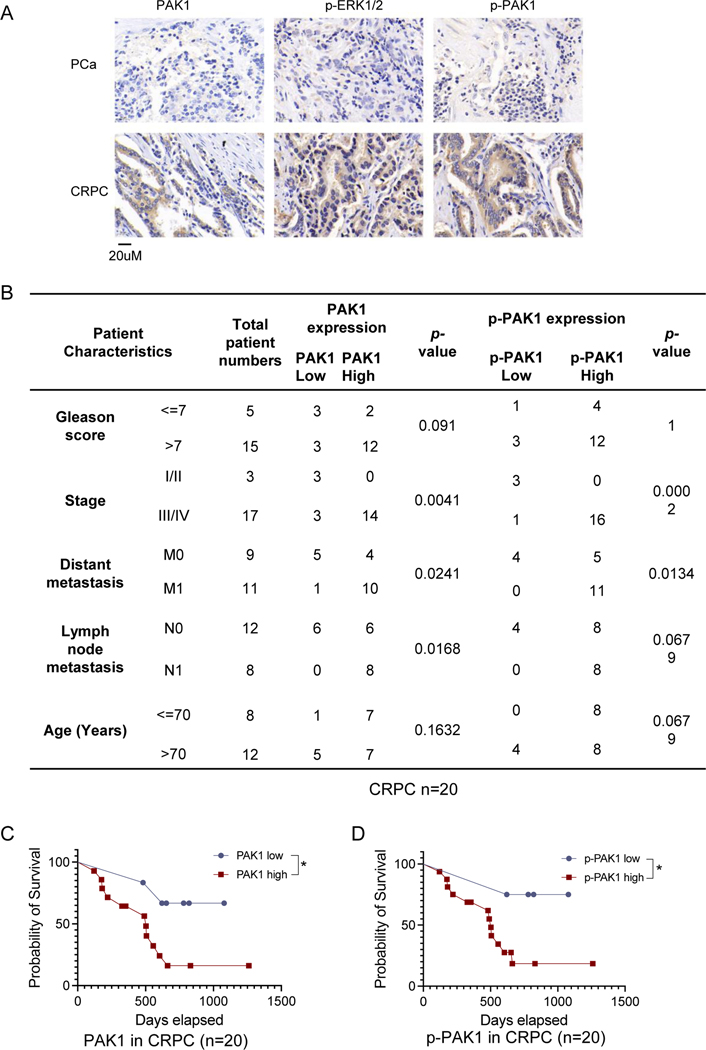
To investigate whether androgen deprivation increases PAK1 expression in a more clinical setting, we performed IHC with validated antibodies to analyze the PAK1/p-PAK1 levels in CRPC patient samples (Supplementary Fig. S4A). IHC staining showed that PAK1 is highly expressed in CRPC, accompanied by enhanced levels of phosphorylated PAK1 and MAPK (Fig. 4A). Statistical analyses showed that in CRPC patient samples, PAK1/p-PAK1 levels were positively correlated with the advanced tumor stage and metastasis status (Fig. 4B). In PCa, PAK1/p-PAK1 levels were positively associated with a high Gleason score and the advanced tumor stage but not other factors (Supplementary Fig. S4B). CRPC patients, as expected, show decreased survival rates compared to PCa patients (Supplementary Fig. S5A). Notably, survival rates of CRPC patients, but not of PCa patients, with high PAK1/p-PAK1 staining were significantly decreased compared with those of low staining (Fig. 4C, 4D&Supplementary Fig. S5B).
Figure 4. Clinical analysis of the expression and phosphorylation levels of PAK1 in prostate cancer patients.
4A. IHC staining shows the upregulation of PAK1 expression/phosphorylation and MAPK phosphorylation in CRPC tissues. The scale bar is 20uM.
4B. PAK1/p-PAK1 expression is statistically associated with advanced tumor stage, and metastasis in CRPC patients (n=20).
4C&4D. High PAK1/p-PAK1 expression is associated with decreased survival rates in CRPC patients (n=20).
In summary, our study established a PTEN null androgen-independent prostate cancer cell line and shows that androgen deprivation increased CRPC cell invasion accompanied by enhanced PAK1/p-PAK1/MAPK signaling. Inhibition of the p110β/RAC/PAK1 pathway decreased CRPC invasion, indicating a critical role of this pathway in CRPC initiation and treatment. The levels of PAK1/p-PAK1 may have prognostic value, as they negatively correlate with survival rates in CRPC patients.
Discussion
The most commonly used human prostate cancer cell lines are not ideal for studying the initiation/development of CRPC, as they are originally derived from metastatic lesions, such as lymph nodes (LNCap) (37), bones (PC3) (38), or brain (DU145) (39). As primary human prostate cancer often has morphologically and topographically distinct tumor foci, it is also not ideal for cells to be cultured in vitro for mechanistic studies (40). Thus, we turned to a primary culture from GEM mice. Primary culture of PTEN null prostate cells is useful but challenging, mainly due to the low malignancy of transgenic mouse models (as only HG-PIN is developed in our C57BL/6 background GEM models (6)). It has been reported that primary prostate cancer cells can be cultured from metastatic murine PTEN null models with a 129/Balb/c and C57BL/6 mixed background (41,42). We didn’t succeed in growing primary cells from PTEN null GEM mice with a C57BL/6 background (6), instead, we generated primary cells (PKO) from the combo PTEN null;Hi-MYC GEM mice with a C57BL/6 background. Interestingly, PKO cells exhibited an activated PI3K pathway as the consequence of PTEN deletion, and also displayed low expression of the transgenic ARR2/probasin-Myc, which contains two additional androgen response elements besides probasin. We reasoned that the low expression of ARR2/probasin-Myc in PKO cells may be due to the reciprocal feedback regulation of PI3K and androgen receptor signaling (12). Androgen deprivation could reduce transgenic Myc expression since it is probasin-driven, which may be a potential limitation of our model though we found that Myc overexpression did not affect the PAK-MAPK signaling. In addition, it would be worthy to use RNA-seq to detect AR regulated transcriptome in this model. PKO cells showed androgen-independent growth, which is different from the androgen-dependent Myc-Cap cells derived from Hi-MYC mice (36). PKO cells gained enhanced RAC1-PAK1-MAPK signaling and invasive capability under androgen deprivation conditions, which is consistent with our previous observation about castrated PTEN null mouse models showing invasive lesions and an enhanced MAPK pathway signaling (6). All the information suggests that PKO cells exhibit the characteristics of PTEN null prostate cancer cells, and can be applied to investigate PTEN null CRPC initiation/progression.
Our previous study using GEMMs shows that p110β plays a critical role in PTEN null CRPC initiation (7). Our current study using this newly established cell line mechanistically demonstrated that p110β regulated RAC1-PAK1-MAPK to mediate cell invasion. We first showed that AR deprivation/inhibition upregulated the p110β-RAC1 interaction and RAC1 activity. Meanwhile, AR deprivation/inhibition also increased PAK1 expression and phosphorylation to increase MAPK signaling and cell invasion/migration (Fig. 2A–2D). We then found that p110β played a critical role in regulating PAK1 phosphorylation, and its downstream MAPK signaling pathway (Fig. 3A). Our study showed that RAC1 played a critical role as an active mutant of RAC1 can rescue the PAK1-MAPK signaling pathway without changing PAK1 expression in p110β depleted CS-PKO cells (Supplementary Fig. S3I). In addition, PAK1 overexpression alone in p110β depleted CS-PKO cells did not rescue the MAPK signaling pathway (Fig. 3D), which further suggests that p110β-RAC1/CDC42 mediated PAK1 phosphorylation is critical in regulating MAPK pathway and this regulation is independent of AR-regulated PAK1 transcription. We previously reported the upregulation of MEK/ERK signaling in our murine CRPC model and human CRPC (6,7). The data presented here suggest that the p110β/RAC/PAK axis may account for the activation of MEK/ERK in CRPC. Of note, PTEN-deficient tumors rely more on p110β (43,44), so it is possible that the lack of effect of p110α inhibition is due to the lack of p110α activity in PKO cells. In PTEN-competent tumors, the pathway described here could exhibit p110α dependence.
PAK1 has been suggested as a potential anti-tumor target in the breast and squamous NSCLCs (45,46). Our previous study also showed that the inhibition of both p110β and PAK1 significantly increased apoptosis of CRPC organoids (7). Our current study demonstrated a mechanism for how androgen deprivation induces PAK1-MAPK activation and cell invasion. It is critical to use patient-derived CRPC organoids from localized sites to further examine this p110β-RAC1-PAK1-MAPK pathway in the future. It is worth testing whether targeting PAK1 can inhibit CRPC metastasis. Our analysis using clinical samples shows that PAK1 is more activated in CRPC than in advanced prostate cancer. However, the cohort size of the immuno-staining numbers of human samples is relatively small. With a large sample number, it would be helpful to know whether PAK1/p-PAK1 staining is associated with CRPC independent of other clinical variables. It would also be good to know whether PAK1/p-PAK1 staining correlates with Gleason Grade/Stage/biochemical relapse.
Supplementary Material
Implications:
This study uses a newly generated PTEN null prostate cancer cell line to define a critical functional role of p110β-PAK1 in CRPC migration/invasion. This study also shows that the p110β-PAK1 axis can potentially be a therapeutic target in CRPC metastasis.
Acknowledgments:
We thank Thomas M. Roberts, Dana-Farber Cancer Institute, Harvard Medical School, for providing reagents and tremendous support of this study. We thank all members of X.G. laboratories for helpful discussion and technical help. We apologize for not citing other peers’ primary studies due to space constraints. This work was supported by the National Institutes of Health (R37CA251165), the Bristol-Myers Squibb Melanoma Research Alliance (MRA) Young Investigator Award (821901) to H.W.; the National Natural Science Foundation of China (81970825), the Department of Science and Technology of Sichuan Province (2022JDJQ0062) to Y. Z.; the American Cancer Society Institutional Research Grant (IRG-19-137-20), the SC COBRE in Oxidants, Redox Balance and Stress Signaling Pilot Projects Program (1P30CM140964) to X. G.
Abbreviations
- CRPC
Castration-resistant prostate cancer
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- PTEN
Phosphatase and tensin homolog
- AR
Androgen receptor
- MAPK
Mitogen-activated protein kinase
- PAK1:
P21 protein (Cdc42/Rac)-activated kinase 1
Footnotes
Disclosure of Potential Conflicts of Interest: The authors disclosed no potential conflicts of interest.
References:
- 1.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–28 doi S0092–8674(15)00548–6 [pii] 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487(7406):239–43 doi nature11125 [pii] 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18(1):11–22 doi 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong L, Zieren RC, Xue W, de Reijke TM, Pienta KJ. Metastatic prostate cancer remains incurable, why? Asian J Urol 2019;6(1):26–41 doi 10.1016/j.ajur.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010;24(18):1967–2000 doi 24/18/1967 [pii] 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia S, Gao X, Lee SH, Maira SM, Wu X, Stack EC, et al. Opposing effects of androgen deprivation and targeted therapy on prostate cancer prevention. Cancer Discov 2013;3(1):44–51 doi 2159–8290.CD-12–0262 [pii] 10.1158/2159-8290.CD-12-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X, Wang Y, Ribeiro CF, Manokaran C, Chang H, Von T, et al. Blocking PI3K p110beta Attenuates Development of PTEN-Deficient Castration-Resistant Prostate Cancer. Mol Cancer Res 2022. doi 10.1158/1541-7786.MCR-21-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X, Qin S, Wu Y, Chu C, Jiang B, Johnson RH, et al. Nuclear PFKP promotes CXCR4-dependent infiltration by T cell acute lymphoblastic leukemia. J Clin Invest 2021;131(16) doi 10.1172/JCI143119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunardi A, Ala U, Epping MT, Salmena L, Clohessy JG, Webster KA, et al. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat Genet 2013;45(7):747–55 doi 10.1038/ng.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011;19(6):792–804 doi S1535–6108(11)00164–4 [pii] 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011;19(5):575–86 doi S1535–6108(11)00155–3 [pii] 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song H, Weinstein HNW, Allegakoen P, Wadsworth MH, 2nd, Xie J, Yang H, et al. Single-cell analysis of human primary prostate cancer reveals the heterogeneity of tumor-associated epithelial cell states. Nat Commun 2022;13(1):141 doi 10.1038/s41467-021-27322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu X, Boufaied N, Hallal T, Feit A, de Polo A, Luoma AM, et al. MYC drives aggressive prostate cancer by disrupting transcriptional pause release at androgen receptor targets. Nat Commun 2022;13(1):2559 doi 10.1038/s41467-022-30257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH. Myc confers androgen-independent prostate cancer cell growth. J Clin Invest 2003;112(11):1724–31 doi 10.1172/JCI19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard GK, Mutton LN, Khalili M, McMullin RP, Hicks JL, Bianchi-Frias D, et al. Combined MYC Activation and Pten Loss Are Sufficient to Create Genomic Instability and Lethal Metastatic Prostate Cancer. Cancer Res 2016;76(2):283–92 doi 10.1158/0008-5472.CAN-14-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Eltoum IE, Roh M, Wang J, Abdulkadir SA. Interactions between cells with distinct mutations in c-MYC and Pten in prostate cancer. PLoS Genet 2009;5(7):e1000542 doi 10.1371/journal.pgen.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Xie CC, Thomas CY, Kim ST, Lindberg J, Egevad L, et al. Genetic markers associated with early cancer-specific mortality following prostatectomy. Cancer 2013;119(13):2405–12 doi 10.1002/cncr.27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 2003;4(3):223–38 doi S1535610803001971 [pii]. [DOI] [PubMed] [Google Scholar]
- 20.Wirth M, Tammela T, Cicalese V, Gomez Veiga F, Delaere K, Miller K, et al. Prevention of bone metastases in patients with high-risk nonmetastatic prostate cancer treated with zoledronic acid: efficacy and safety results of the Zometa European Study (ZEUS). Eur Urol 2015;67(3):482–91 doi 10.1016/j.eururo.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer 2002;2(8):584–93 doi 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 22.Kuchimaru T, Kataoka N, Nakagawa K, Isozaki T, Miyabara H, Minegishi M, et al. A reliable murine model of bone metastasis by injecting cancer cells through caudal arteries. Nat Commun 2018;9(1):2981 doi 10.1038/s41467-018-05366-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michiel Sedelaar JP, Dalrymple SS, Isaacs JT. Of mice and men--warning: intact versus castrated adult male mice as xenograft hosts are equivalent to hypogonadal versus abiraterone treated aging human males, respectively. Prostate 2013;73(12):1316–25 doi 10.1002/pros.22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura Y, Uchida N, Hayata I, Yamaguchi K, Okamoto S, Narita N, et al. Effect of serial passage in female nude athymic mice on androgen dependency of Shionogi carcinoma 115. Cancer Res 1980;40(12):4781–5. [PubMed] [Google Scholar]
- 25.Nordeen SK, Su LJ, Osborne GA, Hayman PM, Orlicky DJ, Wessells VM, et al. Titration of Androgen Signaling: How Basic Studies Have Informed Clinical Trials Using High-Dose Testosterone Therapy in Castrate-Resistant Prostate Cancer. Life (Basel) 2021;11(9) doi 10.3390/life11090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Najahi-Missaoui W, Quach ND, Jenkins A, Dabke I, Somanath PR, Cummings BS. Effect of P21-activated kinase 1 (PAK-1) inhibition on cancer cell growth, migration, and invasion. Pharmacol Res Perspect 2019;7(5):e00518 doi 10.1002/prp2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Bi S, Hou J, Zhao Z, Wang C, Xie S. Targeting p21-activated kinase 1 inhibits growth and metastasis via Raf1/MEK1/ERK signaling in esophageal squamous cell carcinoma cells. Cell Commun Signal 2019;17(1):31 doi 10.1186/s12964-019-0343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. Nat Rev Cancer 2014;14(1):13–25 doi 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goc A, Al-Azayzih A, Abdalla M, Al-Husein B, Kavuri S, Lee J, et al. P21 activated kinase-1 (Pak1) promotes prostate tumor growth and microinvasion via inhibition of transforming growth factor beta expression and enhanced matrix metalloproteinase 9 secretion. J Biol Chem 2013;288(5):3025–35 doi 10.1074/jbc.M112.424770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011;20(4):457–71 doi 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol 2009;6(2):76–85 doi 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nickols NG, Nazarian R, Zhao SG, Tan V, Uzunangelov V, Xia Z, et al. MEK-ERK signaling is a therapeutic target in metastatic castration resistant prostate cancer. Prostate Cancer Prostatic Dis 2019;22(4):531–8 doi 10.1038/s41391-019-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res 2012;72(7):1878–89 doi 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuzugullu H, Baitsch L, Von T, Steiner A, Tong H, Ni J, et al. A PI3K p110beta-Rac signalling loop mediates Pten-loss-induced perturbation of haematopoiesis and leukaemogenesis. Nat Commun 2015;6:8501 doi ncomms9501 [pii] 10.1038/ncomms9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parrini MC, Lei M, Harrison SC, Mayer BJ. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell 2002;9(1):73–83 doi 10.1016/s1097-2765(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 36.Watson PA, Ellwood-Yen K, King JC, Wongvipat J, Lebeau MM, Sawyers CL. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer Res 2005;65(24):11565–71 doi 10.1158/0008-5472.CAN-05-3441. [DOI] [PubMed] [Google Scholar]
- 37.Horoszewicz JS, Leong SS, Chu TM, Wajsman ZL, Friedman M, Papsidero L, et al. The LNCaP cell line--a new model for studies on human prostatic carcinoma. Prog Clin Biol Res 1980;37:115–32. [PubMed] [Google Scholar]
- 38.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol 1979;17(1):16–23. [PubMed] [Google Scholar]
- 39.Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145). Int J Cancer 1978;21(3):274–81 doi 10.1002/ijc.2910210305. [DOI] [PubMed] [Google Scholar]
- 40.Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol 2021;18(2):79–92 doi 10.1038/s41585-020-00400-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiao J, Wang S, Qiao R, Vivanco I, Watson PA, Sawyers CL, et al. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res 2007;67(13):6083–91 doi 67/13/6083 [pii] 10.1158/0008-5472.CAN-06-4202. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003;4(3):209–21 doi S1535610803002150 [pii]. [DOI] [PubMed] [Google Scholar]
- 43.Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A 2008;105(35):13057–62 doi 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Gao X, Schmit F, Adelmant G, Eck MJ, Marto JA, et al. CRKL Mediates p110beta-Dependent PI3K Signaling in PTEN-Deficient Cancer Cells. Cell Rep 2017;20(3):549–57 doi 10.1016/j.celrep.2017.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ong CC, Jubb AM, Haverty PM, Zhou W, Tran V, Truong T, et al. Targeting p21-activated kinase 1 (PAK1) to induce apoptosis of tumor cells. Proc Natl Acad Sci U S A 2011;108(17):7177–82 doi 10.1073/pnas.1103350108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrestha Y, Schafer EJ, Boehm JS, Thomas SR, He F, Du J, et al. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene 2012;31(29):3397–408 doi onc2011515 [pii] 10.1038/onc.2011.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data used to support the present study have been deposited in the Gene Expression Omnibus with an access number of GSE233306.