Abstract
We have identified previously a repressor element in the transcription start site region of the cyclin E1 promoter that periodically associates with an atypical, high molecular weight E2F complex, termed CERC. Purification of native CERC reveals the presence of the type II arginine methyltransferase PRMT5, which can mono- or symetrically dimethylate arginine residues in proteins. Chromatin immunoprecipitations (ChIPs) show that PRMT5 is associated specifically with the transcription start site region of the cyclin E1 promoter. ChIP analyses also show that this correlates with the presence on the same promoter region of arginine-methylated proteins including histone H4, an in vitro substrate of PRMT5. Consistent with its presence within the repressor complex, forced expression of PRMT5 negatively affects cyclin E1 promoter activity and cellular proliferation, effects that require its methyltransferase activity. These data provide the first direct experimental evidence that a type II arginine methylase is involved in the control of transcription and proliferation.
INTRODUCTION
Cyclin E1 gene transcription is undetectable in G0 and G1 phases of the cell cycle but then rises sharply for a brief period prior to each entry into S phase. Biochemical and genetic evidence suggests that E2F- and E2F/pocket protein-associated complexes regulate its activation and repression, respectively (Sardet et al., 1997). Consistent with this, the cyclin E1 promoter contains several E2F binding sites that are occupied in vivo (Geng et al., 1996; Le Cam et al., 1999). Complexes bound to these sites in G0 and G1 recruit enzymatic activities that induce dynamic changes in chromatin, including members of the SNF2-like helicase family (Zhang et al., 2000), type I histone deacetylases (Brehm et al., 1998; Zhang et al., 2000) and the lysine methyltransferase SUVARH1 (Nielsen et al., 2001). Consistent with this, repression of the cyclin E1 gene correlates with an E2F/pocket protein-dependent lysine deacetylation (histone H4/H3) and methylation [H3 lysine(K9)] of a single nucleosome positioned at the transcriptional start site (Nielsen et al., 2001; Morrison et al., 2002). Interestingly, the promoter region within this nucleosome includes an E2F-bound DNA element, the Cyclin E1 Repressor Module (CERM), which participate in downregulation of the cyclin E1 promoter in G0 and G1. CERM functions through both a variant binding site for E2F and a contiguous upstream AT-rich sequence that associate periodically with an unusual high molecular weight E2F-pocket protein complex, CERC (Cyclin E1 Repressor Complex). CERC differs in both binding behaviour and size from previously characterised E2F complexes (Le Cam et al., 1999; Polanowska et al., 2001). Consistent with a role in chromatin remodelling, CERC contains a TSA-sensitive histone deacetylase activity and RbAp48/46, a component of several distinct nucleosome-modifying complexes (Polanowska et al., 2001). In this report we demonstrate that native CERC also contains PRMT5, a type II arginine methyltransferase.
Arginine can be mono- or di-methylated, with the latter in symmetrical or asymmetrical configuration. This is catalysed by two types of protein arginine methyltransferase (PRMT). Type I enzymes (PRMT1–4, 6) generate NG-monomethylarginine and asymmetrical NG,NG-dimethylarginine residues, while type II (PRMT5) leads to NG-monomethylarginine and symmetrical NG,N′G-dimethylarginine (Gary and Clarke, 1998; Zhang and Reinberg, 2001). The type I PRMT1 and PRMT4 (also termed CARM1) were proposed to function as transcriptional coactivators for nuclear receptors (Chen et al., 1999; Strahl et al., 2001). Accordingly, CARM1 and PRMT1 lead to arginine-specific methylations of histone H3 and H4 tails that coincide with transcriptional activation (Ma et al., 2001; Strahl et al., 2001; Wang et al., 2001; Bauer et al., 2002). In addition, the function of the transcriptional activators stat1 and CBP/p300 are regulated by type I PRMT-mediated methylation (Mowen et al., 2001; Xu et al., 2001). In contrast, the involvment of a type II arginine methyltransferase in transcription had not yet been firmly established. PRMT5 (Pollack et al., 1999; Branscombe et al., 2001; Frankel et al., 2001) is the homologue of the Schizosaccharomyces pombe Shk1 kinase-binding protein 1, Skb1 (Gilbreth et al., 1998) and of the Saccharomyces cerevisiae HSL7p protein (Histone Synthetic Lethal 7) (Ma et al., 1996). These type II enzymes are involved in several cellular functions. PRMT5 (also termed JBP1), was identified initially in mammals as a Janus kinase 2-binding protein, implying a role in cytokine-activated transduction pathways (Pollack et al., 1999). It is also an essential component of the ‘Methylosome’, a 20S complex involved in the assembly of snRNP core particles (Friesen et al., 2001). Moreover, HSL7p and Skb1 negatively regulate Swe1 and Ste20/Shk1 kinases, with null mutants exhibiting cell cycle abnormalities and phenotypic alterations characteristic of mitotic inhibitors (Gilbreth et al., 1998; Fujita et al., 1999).
We show here that PRMT5 is present at the cyclin E1 promoter in vivo, where it functions as a corepressor of cyclin E1 transcription.
RESULTS AND DISCUSSION
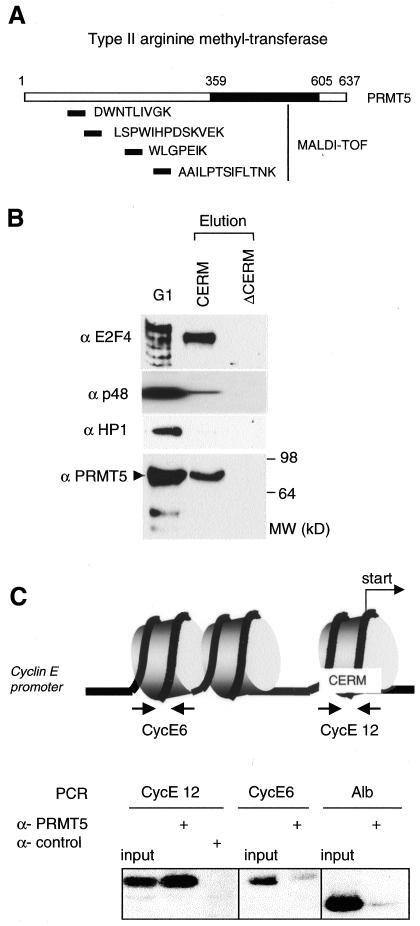
CERC was affinity purified from nuclear extracts prepared from 25 adult rat livers. Extracts were fractionated successively by gel filtration, ion exchange, WGA affinity and finally DNA affinity chromatography using an oligonucleotide column bearing either the wild-type (WT) CERM sequence or its AT-mutated version (ΔCERM). Components specifically present in the WT CERM eluate were analysed by MALDI-MS. Notably, this analysis identified four peptides corresponding to the type II arginine methyltransferase PRMT5 (Figure 1A).
Fig. 1. The type II arginine methyltransferase, PRMT5, is present in CERC. (A) PRMT5 is a component of the native CERC complex; MALDI-MS analysis of affinity purified CERC. Monoisotopic peptides corresponding to PRMT5 are indicated. (B) Western blot analyses of proteins eluted from the CERM affinity column or from the control column (ΔCERM) confirm the presence of PRMT5 in native CERC. Membranes were probed with various antisera as indicated. (C) ChIPs show that PRMT5 is present on the cyclin E1 promoter in vivo. Formaldehyde crosslinked chromatin from G0-arrested NIH 3T3 cells was immunoprecipitated with PRMT5 antisera or control antibody (anti-Flag epitope). One-tenth of the input chromatin and the immunoprecipitated chromatin were analysed by PCR for mouse cyclin E1 promoter fragments (CE12, centred around the transcription start site region and encompassing the CERM element: –40, +46; CE6, centred 500 bp upstream in the promoter region: –432, –526) or the mouse albumin gene. These cyclin E1 promoter DNA fragments are present in two distinct nucleosomes.
Subsequent western blot analyses revealed the presence of PRMT5 in CERC purified from various cells (NIH 3T3, Swiss 3T3 and MEF), as shown in Figure 1B with CERC prepared from a G1 subpopulation of K562 human cells (Polanowska et al., 2001). As expected, we detected neither PRMT5 nor other components of CERC, such as Rbp48 and E2F4, in eluates from the ΔCERM column (Figure 1B). Notably, other proteins implicated in chromatin remodelling of the cyclin E1 promoter region, such as HP1, SUVARH1 or BRG1/hbrm (Zhang et al., 2000; Nielsen et al., 2001), were not detected in purified CERC (Figure 1B; Polanowska et al., 2001), suggesting the presence of several distinct remodelling complexes on this promoter.
To confirm that PRMT5 is recruited to the cyclin E1 promoter in vivo, anti-PRMT5 antibodies were used in chromatin immunoprecipitation (ChIP) assays on cross-linked mononucleosomal-size chromatin fragments prepared from G0-arrested NIH 3T3 cells (Morrison et al., 2002). We assayed for DNA fragments corresponding to two distinct nucleosomal regions of the Cyclin E1 promoter by PCR (Morrison et al., 2002). One target sequence (CE12) encompasses the transcription initiation site and contains the CERM element. The other (CE6) is centred 500 bp upstream in the promoter region. This high resolution relied on previous studies establishing that these cyclin E1 promoter DNA fragments are present in two distinct nucleosomes (Nielsen et al., 2001; Morrison et al., 2002). The CE12 fragment, but neither the CE6 fragment nor an unrelated DNA fragment (Alb), could be detected in anti-PRMT5 immunoprecipitates (Figure 1C), indicating that PRMT5 is associated selectively with the transcription start site region of the cyclin E1 promoter in cells arrested in G0. This result, together with its purification as a component of CERC, suggested strongly that PRMT5 could be involved in the transcriptional control of Cyclin E1 expression. To test this hypothesis, we analysed the effect of forced expression of PRMT5 on cyclin E1 promoter activity.
Luciferase-reporter plasmids driven by either the mouse cyclin E1 promoter, the viral thymidine kinase minimal promoter or a minimal promoter with upstream E2F binding sites were transfected, along with expression vectors for PRMT5 and the cyclin E1 activators, E2F1 and DP1 (Geng et al., 1996) (Figure 2A). PRMT5 overexpression inhibited both basal and stimulated transcription of the cyclin E1 reporter, unlike the vTK reporter (Figure 2A) or the E2F-responsive synthetic promoter (data not shown) used as controls. Significantly, PRMT5 mutated in the S-adenosyl-methionine-binding domain, which impairs its ability to methylate histone H4 or MBP in vitro (Pollack et al., 1999), did not block cyclin E1 promoter activity (Figure 2A). These results suggest strongly that the methyltransferase activity of PRMT5 is important for repression. Notably, similar mutations in CARM1 and PRMT1 impaired their activity as transcriptional coactivators (Chen et al., 1999; Wang et al., 2001; Xu et al., 2001).
Fig. 2. PRMT5 has an inhibitory effect on cyclin E1 gene transcription and cell proliferation. (A) NIH 3T3 cells were transfected with luciferase reporter genes driven by either the mouse cyclin E1 promoter (pCEluc) or the HSV TK promoter (pvTKluc), together with combinations of expression vectors encoding E2F1, DP1, PRMT5 or an enzymatically inactive PRMT5mut, as indicated. All were co-transfected with a CMV–β-gal reporter construct. Results are expressed in relative luciferase units (RLU), normalised to β-gal activity. (B) Xenopus oocytes were injected with pCEluc or pvTKluc, together with expression vectors encoding either PRMT5 or PRMT5mut, as indicated. Luciferase activities were normalised to β-gal values. (C) NIH 3T3 cells grown on coverslips were transfected with PRMT5 or PRMT5mut expression vectors together with a CMV-driven GFP expression plasmid used to identify transfected cells. Twenty-four hours later, cells were analysed for endogenous cyclin E1 expression by immunofluorescence. Panels show Hoechst staining and cyclin E1 immunodetection in GFP-positive cells. (D) Quantification of several experiments performed as described in (C). Results are expressed as the percentage of transfected cells (GFP positive) that expressed endogenous cyclin E1 protein. (E) Forced expression of PRMT5 blocks cells in G1. PRMT5/GFP co-transfectants were synchronised in G0 by serum depletion and, after serum restimulation, individual cells were monitored for S phase entry by immunodetection of BrdU incorporation during the 24 h period after stimulation. Results are expressed as the percentage of GFP-positive cells incorporating BrdU. (F) Cells overexpressing both PRMT5 and cyclin E1 resume DNA synthesis. NIH 3T3 cells were co-transfected with expression plasmids encoding CD20 marker and PRMT5 together with cyclin E or an empty control vector. DNA profiles of CD20-positive cells were obtained using bivariate flow cytometry. The y-axis shows the increase in percentage of cells in the G0/G1 phase upon expression of the indicated proteins. The baseline represents the percentage of G0/G1 cells in mock-transfected cells.
In non-dividing Xenopus oocytes, PRMT5 overexpression also repressed cyclin E reporter constructs, an effect not observed with either mutant PRMT5 or the control reporter gene (Figure 2B). Thus, PRMT5’s negative effect on cyclin E1 promoter activity is not due to blockage of the cell cycle, but rather is direct and dependent on its methyltransferase activity.
Significantly, most NIH 3T3 cells overexpressing PRMT5 did not express the endogenous cyclin E1 protein (Figure 2C and D). These PRMT5 transfectants were synchronised in G0 and serum stimulated, and we monitored the ability of individual cells to re-enter S phase by BrdU incorporation. NIH 3T3 cells expressing PRMT5, but not the mutant protein, failed to incorporate BrdU, suggesting that they were blocked in G1 (Figure 2E). Moreover, proliferating NIH 3T3 cells ectopically expressing WT PRMT5 accumulated in the G0–G1 compartment, an effect that was reverted partially by overexpression of cyclin E1, suggesting that the cyclin E1 gene is a key mediator of this PRMT5-dependent cell cycle arrest (Figure 2F).
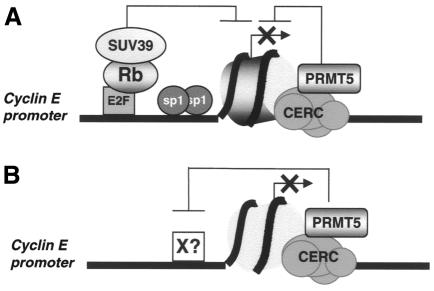
Using xChIP, we investigated whether any potential PRMT5 substrates might be present on the cyclin E1 promoter in vivo. PRMT5 methylates a variety of different proteins in vitro, in particular histones H2A and H4 (Pollack et al., 1999), although no genuine substrate has been identified in a cellular context. Therefore, we initially performed xChIP with a monoclonal antibody that recognises monomethylarginine and dimethylarginine residues in many proteins, including STAT1 (Mowen et al., 2001) and several histone species (E. Fabbrizio et al., manuscript in preparation). The region (CE12) of the cyclin E1 promoter where CERC/PRMT5 binds is associated with a methylated protein recognised by this antibody, unlike the upstream region of the same promoter (CE6) or a control gene (Figure 3A). This methylated protein could be a transcriptional regulator (Figure 4B), since stat1 and CBP/p300 are regulated by PRMTs-mediated methylation (Mowen et al., 2001; Xu et al., 2001). Alternatively, it could be a protein component(s) of the chromatin that surrounds the transcription start site. Indeed, the CERC/PRMT5 complex displays several features of a chromatin remodelling complex since it copurifies with a TSA-sensitive histone deacetylase activity and contains Rbp46/48, a component of several distinct nucleosome-modifying complexes (Polanowska et al., 2001). Moreover, PRMT5 can methylate histones H2A and H4 in vitro (Pollack et al., 1999). It is also noteworthy that the yeast homologue of PRMT5, HSL7, was identified in a genetic screen scoring for genes whose mutation is lethal in combination with deletions of histone tails (Ma et al., 1996). Finally, histone H4 is methylated in vivo at R3, a modification that interferes with the acetylation of surrounding lysine residues, therefore suggesting a link between arginine methylation of histone H4, chromatin remodelling and gene expression (Strahl et al., 2001; Wang et al., 2001). Therefore, we repeated our xChIP analyses with an antibody that recognises specifically this methylated form of histone H4 (Me-R3-H4) (Strahl et al., 2001; Wang et al., 2001). It efficiently precipitated only the chromatin around the transcription start site/CERM region (CE12) (Figure 3B). Thus, R3 methylation of H4 occurs in vivo on the nucleosome that surrounds the regulatory element associated with PRMT5 but not the upstream promoter region. This indicates that E2F/CERC binding to the CERM element recruits PRMT5 at the cyclin E1 promoter in quiescent cells, where it might induce directly or undirectly methylation of histone H4 on a single nucleosome located on the start site of transcription (Figure 4A). While attractive, this model awaits further proof, especially in light of recent reports showing that another arginine methyltransferase, the type I PRMT1, is the most prominent enzyme targeting R3 on histone H4 in vivo (Strahl et al., 2001; Wang et al., 2001). Potential antagonistic effects between PRMT1 and PRMT5 will be addressed by future studies. In light of the ‘histone code’ hypothesis (Jenuwein and Allis, 2001), it is interesting that the same nucleosome on the cyclin E1 promoter was reported to contain other modifications associated with repression in arrested cells. It is specifically hypoacetylated on both histones H3 and H4 (Morrison et al., 2002), and methylated on K9 of histone H3 (Nielsen et al., 2001) (Figure 4A).
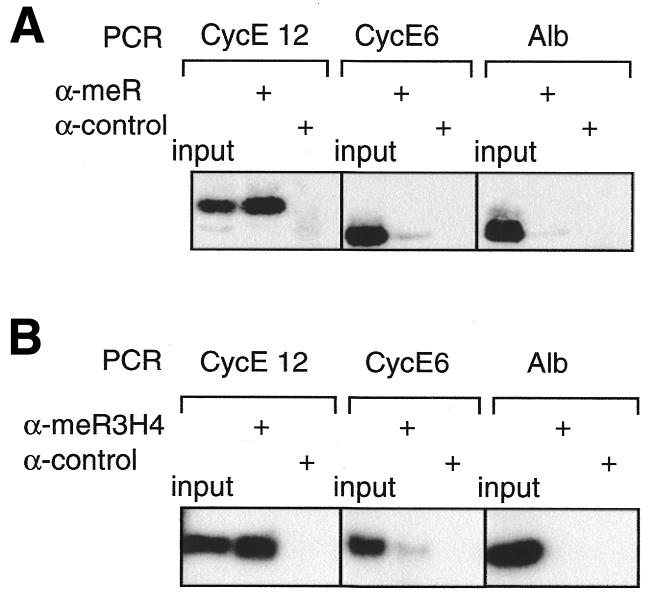
Fig. 3. ChIP analyses reveal the association of proteins methylated on arginine residues with the transcription start site region of the cyclin E1 promoter in vivo. (A) Crosslinked chromatin from G0-arrested NIH 3T3 fibroblasts was immunoprecipitated with an anti-methylarginine monoclonal antibody or a control antibody (anti-Flag epitope). One-tenth of the input chromatin and the immunoprecipitated chromatin were analysed by quantitative PCR for the presence of mouse cyclin E1 promoter fragments CE12 or CE6, or for fragments corresponding to the mouse albumin gene (Alb). (B) ChIPs performed as in (A) but using an antiserum directed against methylated histone H4 (me-R3-H4).
Fig. 4. Role of the type II arginine methyltransferase, PRMT5, in the repressor function of CERC. (A) Rb- and CERC-associated deacetylase, lysine and arginine methyltransferase activities maintain repression via a single nucleosome located near the transcription start site of the cyclin E1 promoter. (B) Alternatively, the target of CERC-associated methyltransferase activity is a non-nucleosomal regulatory protein located in the same region of the cyclin E1 promoter.
Collectively, these data suggest strongly that multiple histone modifying activities are recruited near a single nucleosome associated with the start site of the cyclin E1 gene, working in concert to silence the promoter in arrested cells. Further studies will elucidate the interplay between these activities and their respective roles in controlling cyclin E1 transcription.
METHODS
Affinity purification of CERC and MALDI/TOF analysis.
Nuclear extracts prepared from 25 adult rat livers were fractionated successively by gel filtration, ion exchange, WGA affinity and finally DNA affinity chromatography using an oligonucleotide column bearing either the WT CERM sequence (AGCTCGATGACGCTGGGATTTTTAAATGTCCCGCTCGAAGTCATC) or its AT-mutated version (CAGCTCGATGACGCTGGGATTCTGCTATGTCCCGCTCGAAGTCATC) (mutated bases are underlined). Fractions were tested for CERC and E2F activity by EMSA (see Supplementary data available at EMBO reports Online). Components of eluates from the WT and mutant CERM columns were then separated by two-dimensional gel electrophoresis and compared. Silver-stained spots specifically present in the WT CERM eluate were analysed by MALDI-MS (see Supplementary data available at EMBO reports Online).
Cell culture, transfection, oocytes injection, reporter and proliferation assays.
NIH 3T3 fibroblasts were grown, synchronised and transfected with normalised combinations and amounts of plasmids as indicated in the figure legends using a calcium phosphate precipitation technique as described previously (Fajas et al., 2001). Reporter plasmids pCE-Luc, pTKluc, pCH110 and expression vectors for E2F (pCMVE2F1), DP1 (pCMVDP1) and PRMT5 (pcDEF3PRMT5) have been described previously (Geng et al., 1996; Le Cam et al., 1999; Pollack et al., 1999). Measurements of luciferase and β-galactosidase activities were performed on 1 × 105 cells as in Le Cam et al. (1999) and proliferation assays as in Fajas et al. (2001). The data presented in Figure 2 are representative of a series of at least three independent transfection experiments.
Thirty freshly isolated stage VI Xenopus laevis oocytes were microinjected into the nucleus with combinations of plasmids as indicated; 18 h later they were lysed using the lysis buffer provided with the dual luciferase assay kit (Promega). After two rounds of centrifugation at 14 000 r.p.m., supernatants were processed for luciferase and β-galactosidase activities as above. Western blot analysis revealed that the levels of expression of PRMT5 and PRMT5(RXS) in oocyte extracts were similar (data not shown).
Site-directed mutagenesis.
Mutations in the catalytic site of PRMT5 were performed according to Pollack et al. (1999) using the QuikChange™ site-directed mutagenesis kit (Stratagene) and oligonucleotides mut1-PRMT5 (GGGAGCAGGATCCGGACCCCTGGTGAACGC) and mut2-PRMT5 (GTTCACCAGGGGTCCGGATCCTGCTCCCAGCACCATC).
Western blots and immunofluorescence studies.
Immunofluoresence analyses and western blot detections were performed as described previously (Fajas et al., 2001; Polanowska et al., 2001).
ChIP assay.
NIH 3T3 cells were used for ChIP and processed essentially as described previously (Fajas et al., 2001; Morrison et al., 2002). Cyclin E1 primers CE6 (–432, –526, +1/start site) and CE12 (–47, +46) were as described in Morrison et al. (2002). Anti-methylarginine (7E6), anti-diMeR3H4 and anti-skb1Hs (PRMT5) antibodies were purchased from Abcam, Euromedex and Transduction Laboratories, respectively.
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We would like to express special thanks to B. Hipskind, J.M. Blanchard and G. Almouzni for critical reading of the manuscript. This work was supported by grants to C.S. from the CNRS, l’ARC, La Ligue Contre le Cancer and from the Human Frontier in Science Program and by grants to S.P. from the US Public Health Services Grants RO1-CA46465 from the NCI, RO1 AI36450, RO1-AI43369 and 2T32AI07403 from the National Institute of Allergy and Infectious Diseases, and an award from the Milstein Family Foundation to S.P. S.E. is supported by a fellowship from La Ligue Contre le Cancer.
REFERENCES
- Bauer U.M., Daujat, S., Nielsen, S., Nightingale, K. and Kouzarides, T. (2002) Methylation at arg 17 of histone H3 is linked to gene activation. EMBO rep., 3, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscombe T.L., Frankel, A., Lee, J.H., Cook, J.R., Yang, Z., Pestka, S. and Clarke, S. (2001) PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem., 276, 32971–32976. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska, E.A., McCance, D., Reid, J.L., Bannister, A. and Kouzarides, T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Chen D., Ma, H., Hong, H., Koh, S., Huang, S.M., Schurter, B.T., Aswad, D.W. and Stallcup, M.R. (1999) Regulation of transcription by a protein methyltransferase. Science, 284, 2174–2177. [DOI] [PubMed] [Google Scholar]
- Fajas L., Paul, C., Vie, A., Estrach, S., Medema, R., Blanchard, J.M., Sardet, C. and Vignais, M.L. (2001) CyclinA is a mediator of p120(E4F)-dependent cell cycle arrest in G1. Mol. Cell. Biol., 21, 2956–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen W.J., Paushkin, S., Wyce, A., Massenet, S., Pesiridis, G., Van Duyne, G., Rappsilber, J., Mann, M. and Dreyfuss, G. (2001) The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol., 21, 8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Tonouchi, A., Hiroko, T., Inose, F., Nagashima, T., Satoh, R. and Tanaka, S. (1999) Hsl7p, a negative regulator of Ste20p protein kinase in the S. cerevisiae filamentous growth-signaling pathway. Proc. Natl Acad. Sci. USA, 96, 8522–8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary J.D. and Clarke, S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol., 61, 65–131. [DOI] [PubMed] [Google Scholar]
- Geng Y., Eaton, E., Picon, M., Roberts, J.M., Lundberg, A.S., Gifford, A., Sardet, C. and Weinberg, R.A. (1996) Regulation of the cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene, 12, 1173–1180. [PubMed] [Google Scholar]
- Gilbreth M., Yang, P., Bartholomeusz, G., Pimental, R.A., Kansra, S., Gadiraju, R. and Marcus, S. (1998) Negative regulation of mitosis in fission yeast by the shk1 interacting protein skb1 and its human homolog, Skb1Hs. Proc. Natl Acad. Sci. USA, 95, 14781–14786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis, C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Le Cam L., Polanowska, J., Fabbrizio, E., Olivier, M., Philips, A., Ng Eaton, E., Classon, M., Geng, Y. and Sardet, C. (1999) Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J., 18, 1878–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.J., Lu, Q. and Grunstein, M. (1996) A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in S. cerevisiae. Genes Dev., 10, 1327–1340. [DOI] [PubMed] [Google Scholar]
- Ma H. et al. (2001) Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol., 11, 1981–1985. [DOI] [PubMed] [Google Scholar]
- Morrison A.J., Sardet, C. and Herrera, R. (2002) Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol. Cell. Biol., 22, 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen K.A., Tang, J., Zhu, W., Schurter, B.T., Shuai, K., Herschman, H.R. and David, M. (2001) Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell, 104, 731–741. [DOI] [PubMed] [Google Scholar]
- Nielsen S.J. et al. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature, 412, 561–565. [DOI] [PubMed] [Google Scholar]
- Polanowska J., Fabbrizio, E., Le Cam, L., Trouche, D., Emiliani, S., Herrera, R. and Sardet, C. (2001) The periodic down regulation of Cyclin E1 gene expression from exit of mitosis to end of G1 is controlled by a deacetylase- and E2F-associated bipartite repressor element. Oncogene, 20, 4115–4127. [DOI] [PubMed] [Google Scholar]
- Pollack B.P., Kotenko, S.V., He, W., Izotova, L.S., Barnoski, B.L. and Pestka, S. (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem., 274, 31531–31542. [DOI] [PubMed] [Google Scholar]
- Sardet C., Le Cam, L., Fabbrizio, E. and Vidal, M. (1997) E2Fs and the retinoblastoma protein family. In Ghysdael, J. and Yaniv, M. (eds), Progress in Gene Expression. Oncogenes as Transcriptional regulators, Volume 2. Birkhauser, Basel, Switzerland, pp. 1–62.
- Strahl B.D. et al. (2001) Methylation of histone H4 at arg 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol., 11, 996–1000. [DOI] [PubMed] [Google Scholar]
- Wang H. et al. (2001) Methylation of histone H4 at arg3 facilitating transcriptional activation by nuclear hormone receptor. Science, 293, 853–857. [DOI] [PubMed] [Google Scholar]
- Xu W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B.M., Montminy, M. and Evans, R.M. (2001) A transcriptional switch mediated by cofactor methylation. Science, 8, 8–11. [DOI] [PubMed] [Google Scholar]
- Zhang H.S., Gavin, M., Dahiya, A., Postigo, A., Ma, D., Luo, R.X., Harbour, J.W. and Dean, D.C. (2000) Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing. HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell, 101, 79–89. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Reinberg, D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev., 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.