Abstract
E2F transcription factors play an important role in regulating mammalian cell proliferation. E2F6, the most recently identified E2F family member, is a transcriptional repressor. In an effort to ascertain the in vivo biological function of E2F6, we have generated an E2f6 mutant mouse strain. Mice lacking E2F6 are viable and healthy. Surprisingly, E2f6–/– embryonic fibroblasts proliferate normally. However, E2f6–/– animals display overt homeotic transformations of the axial skeleton that are strikingly similar to the skeletal transformations observed in polycomb mutant mice. This observation is compatible with the recent finding that endogenous E2F6 and one or more mammalian polycomb proteins are components of the same multiprotein complex. The accumulated evidence suggests that, during development, E2F6 participates in the recruitment of polycomb proteins to specific target promoters.
INTRODUCTION
E2F family members have been intensively studied for their ability to control cellular proliferation. E2F-responsive elements have been identified in genes that play key roles in cell cycle progression, DNA replication and apoptosis (Trimarchi and Lees, 2002). E2F forms a heterodimeric complex containing an E2F and a DP subunit. In mammals, six E2F proteins (E2F1–E2F6) and two DP proteins (DP-1 and DP-2) have been identified. E2F1–E2F5 function as transcriptional activators. Their transcriptional activity is negatively regulated by binding to pRB, the product of the retinoblastoma tumor suppressor protein, and to two related ‘pocket proteins’, p107 and p130 (Dyson, 1998).
Except for a closely related DNA-binding and dimerization domain, E2F6, the most recently identified E2F family member, is only weakly homologous to the other E2F proteins. It does not bind to pocket proteins, lacks a transactivation domain and is a transcriptional repressor (Morkel et al., 1997; Cartwright et al., 1998; Gaubatz et al., 1998; Trimarchi et al., 1998). Ectopic overproduction of E2F6 can result in S-phase accumulation and, in quiescent cells, it can delay re-entry into the cell cycle (Cartwright et al., 1998; Gaubatz et al., 1998). Recently, a novel complex that contains E2F6, polycomb proteins and chromatin modifiers was shown to occupy target promoters in G0 (Ogawa et al., 2002). Thus, it was suggested that one function of E2F6 is to inactivate E2F-dependent genes in quiescent cells.
In another study, consistent with a role for E2F6 in mediating transcriptional silencing, E2F6 was found to associate with a number of mammalian polycomb group (Pc-G) proteins, including Bmi-1, Mel-18, Mph1 and Ring1 (Trimarchi et al., 2001). Pc-G proteins form large multimeric complexes needed to maintain stable transcriptional repression of Hox genes (Gould, 1997; Schumacher and Magnuson, 1997). Hox genes encode a family of regulatory proteins that are expressed along the antero-posterior axis in the vertebrate embryo (Cillo et al., 2001). According to the ‘Hox code’ model, the particular combination of Hox genes expressed in an individual embryonic segment determines the identity of individual vertebrae (Kessel and Gruss, 1991). In Pc-G mutant mice, the expression boundaries of certain Hox genes are shifted anteriorly, resulting in posterior transformations of the axial skeleton. In addition to their role in Hox gene regulation, Pc-G proteins also display other activities. For example, Bmi-1 is a critical regulator of proliferation, senescence and apoptosis (Jacobs et al., 1999).
Since most polycomb proteins cannot bind directly to DNA, it is not well understood how they are targeted to specific promoters (Pirrotta, 1999). Because E2F6 exists in a complex with certain polycomb proteins, one wonders whether E2F6 functions to recruit Pc-G complexes to target promoters. If so, one might speculate that E2F6 contributes to the specific biological functions of certain Pc-G proteins, e.g. the regulation of developmental patterning, proliferation and/or cellular senescence. However, to date, there have been no biological data to support this model.
We have begun to probe the function of E2F6 in vivo by introducing a null mutation into the E2f6 gene in murine embryonic stem (ES) cells. Mice unable to synthesize the E2F6 transcription factor have now been generated and analyzed. These animals are viable and healthy. However, they appear to be defective in spermatogenesis, and they display homeotic transformations of the axial skeleton. The latter are strikingly similar to the skeletal transformations observed in polycomb knockout mice. Our observations support a model that foresees E2F6 mediating the recruitment of polycomb complexes to specific target promoters during development.
RESULTS
E2F6 is not required for mouse viability
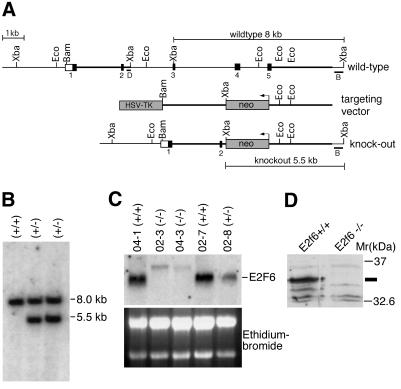
We inactivated the E2f6 gene in mice using a targeting strategy that replaced exons 3–5 of E2f6 with a neomycin resistance gene. This deletes the dimerization and DNA-binding domains of E2F6 and is, thus, expected to result in a null mutation (Figure 1). Interbreeding of heterozygous mutant animals yielded E2f6 null (E2f6–/–) animals, from which E2F6 transcripts and protein were absent.
Fig. 1. Targeting strategy and germline transmission. (A) Top: genomic organization of the E2f6 gene. Middle: targeting vector. Bottom: targeted allele. Exons are shown by boxes and introns by lines. The open box represents the non-coding region of exon 1. (B) Southern blot of genomic DNA isolated from different ES cell lines digested with XbaI and hybridized with probe B [see (A)]. Correct targeting of the left flank was confirmed by hybridizing EcoRI-digested genomic ES cell DNA with probe D (not shown). Genotypes are indicated. (C) Northern blot analysis to confirm the absence of E2F6 mRNA in MEFs derived from E2f6–/– animals. Genotypes are indicated. (D) Western blot analysis of lysates from E2f6+/+ and E2f6–/– MEFs to confirm the absence of the E2F6 protein in animals.
E2f6 null animals were born with a normal Mendelian frequency, showed no obvious external defects and appeared to be healthy for >12 months (not shown). The average growth of E2f6+/+ and E2f6–/– animals over time from birth to 1 year of age was indistinguishable (not shown). Female and male E2f6 nullizygous animals were fertile and gave rise to healthy offspring. The appearance and size of their internal organs were indistinguishable from those of wild-type (wt) animals. Furthermore, there were no significant differences in the abundance of the various classes of circulating hematopoietic cells between E2f6–/– and wt animals (not shown).
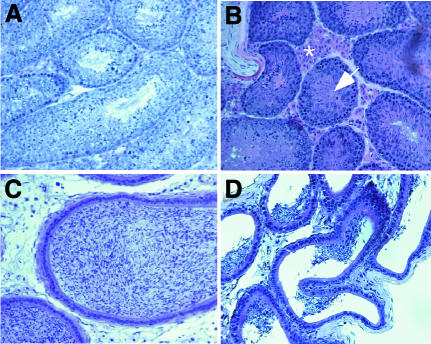
A detailed histological analysis of the organs of several 2- to 3-month-old male E2f6–/– animals revealed no abnormalities except in the testes (Figure 2; data not shown). The testes of these animals showed a paucity of mature spermatocytes and unusually abundant Leydig cells. In addition, there was an incomplete filling of the ductus epididymidis with spermatocytes (Figure 2). Taken together, these data suggest a defect in spermatocyte development in E2f6–/– animals. Notably, these abnormalities failed to affect fertility. Interestingly, in E2f1–/– animals, there was also an increase in interstitial Leydig cells and an incomplete filling of tubuli with spermatocytes, and they, too, are fertile (Yamasaki et al., 1996). Presumably, the extent of the defect is insufficient to affect fertility. In this regard, others have shown that rodent spermatogenesis must be reduced by at least 90% to affect fertility (Russell et al., 1990).
Fig. 2. Testicular abnormalities in E2f6–/– animals. Histological sections (hematoxylin and eosin) of testes (A and B) and ductus epididymidis (C and D) of wt (A and C) and E2f6–/– (B and D) animals. An increase of interstitial Leydig cells (asterisk) and of eosinophilic material (residual bodies) in the seminiferous tubuli (arrow) is evident in E2f6–/– testes (B). Incomplete filling of the ductus epididymidis with spermatocytes in E2f6–/– animals (D). In (C), a wt ductus epididymidis is shown at the same magnification as (D).
Homeotic transformations of the axial skeleton in E2f6–/– mice
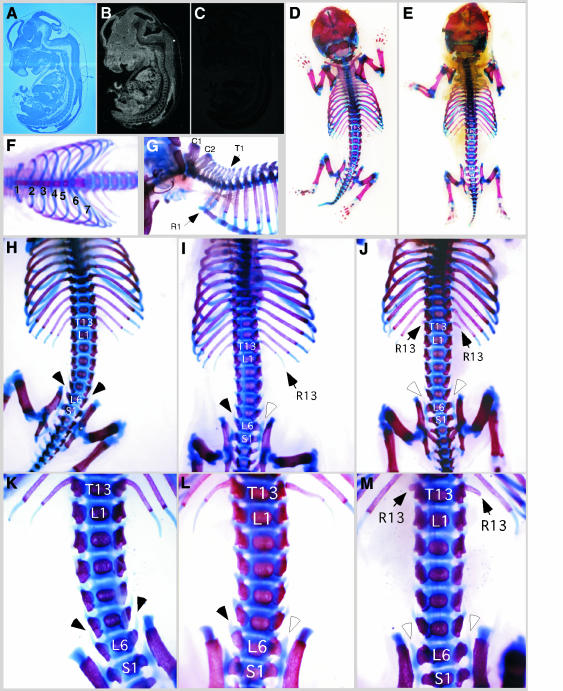
Mice deficient in certain Pc-G genes display homeotic transformations of the axial skeleton. For example, posterior transformations of the axial skeleton have been observed in mice deficient for the Bmi-1 polycomb protein, a protein known to associate with E2F6 (van der Lugt et al., 1994). Like Bmi-1 and other polycomb group genes, E2F6 is ubiquitously expressed at low levels in mouse embryos, as shown by in situ hybridization on sections from wt 14.5 d.p.c. embryos (Figure 3A–C). We were interested in the skeletal development of E2f6–/– animals and compared skeletal preparations of E2f6–/– embryos and newborn mice with those of wt animals (Figure 3D–M). These analyses revealed abnormalities along the antero-posterior skeletal axis of the knockout mice. Specifically, we observed a transformation of the sixth lumbar vertebra (L6) to the first sacral vertebra (S1), indicated by abnormally located sacro-iliac joints at L6 (Figure 3I, J and L, M). There was also a transformation of the thirteenth thoracic vertebra (T13) to the first lumbar vertebra (L1), as demonstrated by degeneration of the ribs that are normally associated with T13 (Figure 3J and M). The aforementioned changes can be interpreted as posterior transformations, because the T13 and L6 vertebrae acquire morphological characteristics of adjacent posterior vertebrae. In contrast to Bmi-1-deficient mice, which reveal more widespread skeletal transformations involving the atlas and multiple ribs (van der Lugt et al., 1994), the E2f6–/– transformations were restricted to the lumbar and sacral regions (Figure 3F and G). No other gross abnormalities of the skeletons were observed (Figure 3D and E). As indicated in Table I, in some E2f6–/– animals, there were asymmetric skeletal transformations, which presented on only one side of the body. Also, the penetrance of the transformations within litters was incomplete, similar to what was observed for many polycomb nullizygous mice, e.g. bmi-1–/– mice (van der Lugt et al., 1994). Skeletal transformations were also noted in heterozygous E2f6+/– animals, albeit at a lower frequency (see Table I).
Fig. 3. Skeletal transformations of E2f6–/– animals. (A–C) In situ hybridization of E2F6 in 14.5-day-old mouse embryos: (A) Bright field; (B) E2F6 antisense probe; (C) E2F6 sense probe (control). (D–M) Alcian Blue (cartilage) and Alizarin Red (bone) staining of cleared newborn animals. Overview of the skeletons of wt (D) and E2f6–/– (E) skeletons. Ventral view of the thoracic region (F) and lateral view of the cervical region (G) of E2f6–/– animals. Note the normal number and attachment of the ribs, and normal size and appearance of the C1 and C2 vertebrae. Dorsal view of the thoraco-lumbo-sacral region of wt (H) and E2f6–/– (I and J) animals. In (J), the thirteenth ribs are degenerated (arrows). Moreover, the sixth lumbar vertebra (L6) has been transformed into the first sacral vertebra (S1) on one side (I and L) or two sides (J and M) (white arrowheads). Black arrowheads point to normal L6 vertebral elements. The skeletal transformations of E2f6–/– animals can also be seen in the ventral view of the lumbo-sacral region (L and M). The formations of sacro-iliac joints at L6 are evident in (L) and (M). Compare with the lumbo-sacral region of a wt animal (K).
Table I. Skeletal abnormalities in E2f6–/– animals.
| Phenotype |
|
Penetrance of skeletal abnormalities (%) |
||
|---|---|---|---|---|
| E2f6+/+ | E2f6+/– | E2f6–/– | ||
| (n = 16) | (n = 5) | (n = 16) | ||
| T13 > L1a | One side | 0 | 0 | 19 |
| Both sides | 0 | 0 | 6 | |
| L6 > S1 | One side | 0 | 0 | 44 |
| Both sides | 0 | 40 | 31 | |
aDemonstrated by rudimentary ribs.
Proliferation of E2f6 mutant and wt fibroblasts
In addition to their role in Hox gene regulation during development, polycomb proteins are also involved in the control of cell cycle proliferation and senescence. The proliferation defects of bmi-1–/– cells result, at least in part, from the upregulation of the p16INK4a and p19ARF tumor suppressor genes (Jacobs et al., 1999), suggesting that transcriptional repression of these genes is normally mediated by Bmi-1-containing polycomb complexes. Since E2F proteins have also been implicated in the regulation of p19ARF expression (DeGregori et al., 1997; Bates et al., 1998), it was speculated that E2F6 could mediate the polycomb-dependent repression of the INK4a locus (Trimarchi and Lees, 2002). Moreover, E2F6, when overproduced, can restrain exit from quiescence (G0) in mouse embryonic fibroblasts (MEFs) (Gaubatz et al., 1998), and, in human cells, a complex that contains E2F6 and chromatin modifiers was recently shown to occupy target promoters in G0 (Ogawa et al., 2002).
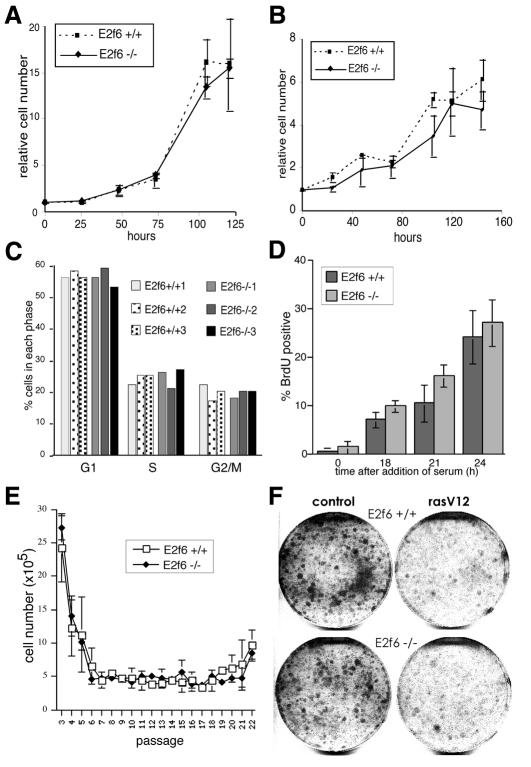
To directly assess the role of endogenous E2F6 in cell proliferation, we cultured MEFs from E2f6 null and littermate wt embryos. Surprisingly, proliferation of both sets of MEFs was indistinguishable both in normal and low serum-containing medium (Figure 4A and B), and, by FACS analysis, neither cell strain revealed a significant cell cycle abnormality (Figure 4C). Moreover, the nullizygous cells entered and exited G0 normally (Figure 4D). We conclude that E2F6 is neither required for growth arrest following serum removal nor for timely re-entry into the cell cycle from G0.
Fig. 4. Properties of E2f6 null fibroblasts. (A) Proliferation of wt and E2f6–/– MEFs in medium with 10% FCS. Growth values represent the average of three independently isolated wt MEF cultures and three different E2f6–/– MEF cultures isolated from littermate embryos. Error bars represent the standard deviation. (B) Growth of E2f6–/– and E2f6+/+ MEFs under low serum conditions [for details see (A)]. (C) Cell cycle distribution of independently isolated asynchronously growing E2f6–/– MEF lines. (D) Serum starvation and re-entry into the cell cycle of E2f6+/+ and E2f6–/– MEFs was analyzed by BrdU incorporation. (E) Wild-type and E2f6–/– MEFs were passaged according to a standard 3T3 protocol. Cell numbers were determined at each passage. (F) Premature senescence of E2f6+/+ and E2f6–/– MEFs: E2f6+/+ and E2f6–/– MEFs were infected with either a control retrovirus or a RasV12-expressing retrovirus together with a puromycin resistance gene. Dishes were stained with Crystal Violet following 10 days of selection with puromycin.
Senescence of E2f6–/– and wt fibroblasts
Bmi-1 knockout fibroblasts undergo premature senescence stimulated by the upregulation of p16INK4a and p19ARF (Jacobs et al., 1999). To determine whether E2f6 nullizygous MEFs have a similar phenotype, we analyzed their longevity in culture. Like wt MEFs, proliferation of the mutant cells began to slow after passage 5 and, eventually, most cells senesced. Moreover, spontaneous immortalization was observed after passage 20, as was the case for the wt cells. This demonstrates that loss of E2F6 is not sufficient to result in premature senescence (Figure 4E). In addition, retrovirus-mediated expression of oncogenic Ras (RasV12) resulted in senescence of both wt and E2f6–/– MEFs (Figure 4F), demonstrating that E2F6 is also not required for the RasV12-induced premature senescence process.
DISCUSSION
We have generated E2F6-deficient mice to establish the role of E2F6 in cellular proliferation and mouse development. Surprisingly, we found that E2F6 is dispensable for proliferation and quiescence of MEFs. This was unexpected as it was recently shown that an E2F6 complex that contains chromatin modifiers occupies promoters of cell cycle genes in G0 cells (Ogawa et al., 2002). Although our findings seem to contradict the conclusion that E2F6 is important for quiescence, there are several possible explanations for the seeming discrepancy between these two studies. First, it is likely that E2F6 is normally involved in quiescence, but that in its absence its function is provided by at least one other protein. In this regard, the E2F6 complex purified by Ogawa et al. (2000) contains several distinct DNA-binding activities in addition to E2F6. Thus, in the absence of E2F6, the complex might be targeted to certain cell cycle promoters through its Mga/Max DNA-binding subunit. Therefore, it will be critical to determine whether the complex is able to bind to cell cycle promoters in E2f6–/– cells and, if so, how it is targeted to the relevant promoters. Secondly, it is possible that in the absence of E2F6, E2F–pocket protein complexes compensate for its function in G0. For example, previous experiments have shown that E2F4–p130 complexes occupy promoters of cell cycle genes in quiescence (Takahashi et al., 2000; Wells et al., 2000). Thus, in future, it will be important to characterize E2F–pocket protein complexes in E2F6-deficient cells in detail. Finally, effects on target gene expression in E2f6–/– cells may be too subtle to abolish entry into or exit from quiescence. It will be interesting to carefully analyze expression of cell cycle genes in E2F6-deficient cells.
Although E2F6 is not essential for quiescence of mouse fibroblasts, we found that it is critical for developmental patterning. E2f6–/– mice are born with posterior homeotic transformations of the axial skeleton. The skeletal transformations in E2f6–/– mice are strikingly similar to those observed in mice deficient in the Bmi-1, Mel-18, Rae28 and M33 polycomb proteins (van der Lugt et al., 1994; Akasaka et al., 1996; Core et al., 1997; Takihara et al., 1997). However, while these polycomb knockout animals present with transformations along the complete antero-posterior axis of the skeleton, homeotic transformations in E2f6–/– mice are restricted to the lumbar and sacral regions of the vertebral column. Posterior transformations can arise when Hox genes are mis-expressed in an anterior position where they are normally transcriptionally repressed by polycomb proteins. Therefore, together with the previous observations that E2F6 is a component of the polycomb complex (Trimarchi et al., 2001; Ogawa et al., 2002), our findings suggest that E2F6 is normally required for the polycomb-dependent transcriptional repression of a subset of Hox genes. However, we can not exclude a more indirect role for E2F6 in the regulation of Hox function. Further work will be necessary to define the role of E2F6 in the regulation of the spatial and temporal expression of selected Hox genes.
In conclusion, our data demonstrate that the function of E2F6 is distinct from that of other E2F family members. We propose that E2F6 acts as a sequence-specific DNA-binding subunit of certain Pc-G complexes that mediate Hox gene regulation during development.
METHODS
Gene targeting and generation of E2f6 mutant animals. An XbaI–HindIII restriction fragment from mouse E2F6 cDNA was used to screen a Lambda Fix II library derived from 129Sv genomic DNA. E2f6 exons were identified by Southern blotting and sequence analysis. A 3 kb BamHI–XbaI fragment containing exon 1 and intron 1 sequences and a 3 kb HpaI–NotI fragment containing intron 4 sequences were inserted into the targeting vector, pPNT, which was electroporated into 129/Sv J1 ES cells. Cells were selected in medium containing 300 µg/ml G418 and 2 µM gancyclovir. Two correctly targeted clones were obtained from 150 selected ES cell clones. They were injected into C57BL/6 and Balb/c blastocysts to generate chimeric animals that passed the mutant E2f6 allele to their offspring, as determined by PCR on DNA isolated from tail biopsies. Sequences of primers used for PCR genotyping of E2f6 animals are available upon request.
Primary fibroblasts. MEFs were isolated from 13.5 d.p.c. embryos. Growth was analyzed by plating MEFs in triplicate in 24-well plates. At the indicated time points, cells were fixed and stained with Crystal Violet. The dye was extracted with 10% acetic acid and the optical density was determined. Proliferation of three independently isolated wt and three E2f6 null fibroblast lines was compared in media containing 10 and 2% serum. For serum-starvation/release experiments, MEFs were growth arrested by incubation in medium containing 0.2% fetal bovine serum for 60 h. Cells were then stimulated with 10% fetal bovine serum and pulsed with 10 µM bromodeoxyuridine (BrdU). BrdU incorporation was analyzed by immunostaining. For retroviral expression, MEFs were infected with a RasV12-expressing retrovirus or with a control virus. Two days after infection, MEFs were selected with 1.5 µg/ml puromycin. After 10 days, cells were fixed and stained with Crystal Violet. For immunoblotting, MEFs were lysed, and aliquots of these lysates (500 µg) were separated by SDS–gel electrophoresis and transferred to Immobilon-P. E2F6 was detected with affinity-purified polyclonal antiserum (C10) raised in rabbits against the keyhole limpet hemocyanin-coupled peptide C-10 (CPEKEDEPPQ).
Histology, skeletal preparations and in situ hybridization. Spleen, thymus, pancreas, stomach, duodenum, colon, kidney, lung, heart, skeletal muscle, cerebellum and testis were harvested from 3-month-old male animals and fixed in 60% ethanol, 30% chloroform, 10% acetic acid. Paraffin sections (6 µm) were prepared and stained with hematoxylin and eosin. Skeletal analysis was performed with E2f6 mice that were bred for 3–4 generations to the C57/Bl6 strain. E16.5 embryos and newborn pups were stained with cartilage-specific Alcian Blue and bone-specific Alizarin Red. Soft tissue was cleared with alkali. For in situ hybridization, 14.5-day-old embryos were embedded in Tissue-Tek, frozen on the surface of a liquid nitrogen pool. Sectioning, post-fixation, preparation of a single-stranded DNA probe and hybridization were performed as described previously (Millauer et al., 1993). Slides were coated with Kodak NTB2 film emulsion and exposed for ∼7 days.
Acknowledgments
ACKNOWLEDGEMENTS
We thank S. Hauser, M. Eilers and our laboratory colleagues for their advice. We thank W. Sperling, G. Lindeman, H. Göllner, A. Sharp, L. Du, J. Wood, G. Schemken, M. Zouzarte, J. Dick, U. Herz and V. Kräling for assistance in generating and analyzing E2f6 mice. This work was supported by grants from the National Cancer Institute to D.M.L. and from The Leukemia and Lymphoma Society and the VolkswagenStiftung to S.G.
REFERENCES
- Akasaka T., Kanno, M., Balling, R., Mieza, M.A., Taniguchi, M. and Koseki, H. (1996) A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development, 122, 1513–1522. [DOI] [PubMed] [Google Scholar]
- Bates S., Phillips, A.C., Clark, P.A., Stott, F., Peters, G., Ludwig, R.L. and Vousden, K.H. (1998) p14ARF links the tumour suppressors RB and p53. Nature, 395, 124–125. [DOI] [PubMed] [Google Scholar]
- Cartwright P., Muller, H., Wagener, C., Holm, K. and Helin, K. (1998) E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene, 17, 611–623. [DOI] [PubMed] [Google Scholar]
- Cillo C., Cantile, M., Faiella, A. and Boncinelli, E. (2001) Homeobox genes in normal and malignant cells. J. Cell Physiol., 188, 161–169. [DOI] [PubMed] [Google Scholar]
- Core N., Bel, S., Gaunt, S.J., Aurrand-Lions, M., Pearce, J., Fisher, A. and Djabali, M. (1997) Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development, 124, 721–729. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Leone, G., Miron, A., Jakoi, L. and Nevins, J.R. (1997) Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl Acad. Sci. USA, 94, 7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Gaubatz S., Wood, J.G. and Livingston, D.M. (1998) Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl Acad. Sci. USA, 95, 9190–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould A. (1997) Functions of mammalian Polycomb group and trithorax group related genes. Curr. Opin. Genet. Dev., 7, 488–494. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom, K., Marino, S., DePinho, R.A. and van Lohuizen, M. (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature, 397, 164–168. [DOI] [PubMed] [Google Scholar]
- Kessel M. and Gruss, P. (1991) Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell, 67, 89–104. [DOI] [PubMed] [Google Scholar]
- Millauer B., Wizigmann-Voos, S., Schnurch, H., Martinez, R., Moller, N.P., Risau, W. and Ullrich, A. (1993) High affinity VEGF binding and developmental expression suggest Flk-1 as a major regulator of vasculogenesis and angiogenesis. Cell, 72, 835–846. [DOI] [PubMed] [Google Scholar]
- Morkel M., Wenkel, J., Bannister, A., Kouzarides, J.T. and Hagemeier, C. (1997) An E2F-like repressor of transcription. Nature, 390, 567–568. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro, K., Gaubatz, S., Livingston, D.M. and Nakatani, Y. (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science, 296, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1999) Polycomb silencing and the maintenance of stable chromatin states. Results Probl. Cell Differ., 25, 205–228. [DOI] [PubMed] [Google Scholar]
- Russell L.D., Ettlin, R.A., SinahiHikim, A.P. and Clegg, E.D. (1990) Histological and Histopathological Evaluation of the Testis. Cache River Press, Clearwater, FL.
- Schumacher A. and Magnuson, T. (1997) Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet., 13, 167–170. [PubMed] [Google Scholar]
- Takahashi Y., Rayman, J.B. and Dynlacht, B.D. (2000) Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev., 14, 804–816. [PMC free article] [PubMed] [Google Scholar]
- Takihara Y. et al. (1997) Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development, 124, 3673–3682. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees, J.A. (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol., 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild, B., Verona, R., Moberg, K., Andon, N. and Lees, J.A. (1998) E2F-6, a member of the E2F family that can behave as transcriptional repressor. Proc. Natl Acad. Sci. USA, 95, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild, B., Wen, J. and Lees, J.A. (2001) The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl Acad. Sci. USA, 98, 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lugt N.M. et al. (1994) Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev., 8, 757–769. [DOI] [PubMed] [Google Scholar]
- Wells J., Boyd, K.E., Fry, C.J., Bartley, S.M. and Farnham, P.J. (2000) Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol., 20, 5797–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki L., Jacks, T., Bromson, R., Goillot, E., Harlow, E. and Dyson, N.J. (1996) Tumor induction and tissue atrophy in mice lacking E2F-1. Cell, 85, 537–548. [DOI] [PubMed] [Google Scholar]