Abstract
The Escherichia coli chaperonin machine is composed of two members, GroEL and GroES. The GroEL chaperonin can bind 10–15% of E. coli’s unfolded proteins in one of its central cavities and help them fold in cooperation with the GroES cochaperonin. Both proteins are absolutely essential for bacterial growth. Several large, lytic bacteriophages, such as T4 and RB49, use the host-encoded GroEL in conjunction with their own bacteriophage-encoded cochaperonin for the correct assembly of their major capsid protein, suggesting a cochaperonin specificity for the in vivo folding of certain substrates. Here, we demonstrate that, when the cochaperonin of either bacteriophage T4 (Gp31) or RB49 (CocO) is expressed in E. coli, the otherwise essential groES gene can be deleted. Thus, it appears that, despite very little sequence identity with groES, the bacteriophage-encoded Gp31 and CocO proteins are capable of replacing GroES in the folding of E. coli’s essential, housekeeping proteins.
INTRODUCTION
Molecular chaperones are present in all cells to prevent aggregation and thus help many of the polypeptides to achieve their native state (reviewed in Ellis, 1994). In Escherichia coli, the GroES/GroEL chaperonin machine is encoded by two genes, groES and groEL, which form an operon. These genes were first identified as host factors required for bacteriophage morphogenesis (reviewed in Georgopoulos and Welch, 1993). Subsequent work established that the GroES and GroEL proteins were essential for the correct assembly of λ proheads and T5 tails (Georgopoulos et al., 1973; Zweig and Cummings, 1973). In addition, both genes are indispensable for E. coli growth at all temperatures (Fayet et al., 1989).
The groEL gene, but not groES, is required for bacteriophage T4 propagation, since T4 is specifically unable to propagate on certain groEL mutants (reviewed in Georgopoulos and Welch, 1993). T4-encoded compensatory mutations can be readily isolated as plaque formers on the restrictive groEL mutants and are shown to map in gene 31 (Georgopoulos and Welch, 1993; Ang et al., 2000). The gene 31 product, Gp31, acts at an early stage of prohead assembly in the T4 life cycle, as it is needed for the correct folding of Gp23, the major capsid protein of T4. In the absence of functional Gp31, the capsid protein aggregates into amorphous ‘lumps’ on the bacterial membrane (Laemmli, 1970). Genetic and biochemical evidence has revealed that the Gp31 and GroEL proteins interact in vivo and in vitro. Gene 31 can partially complement the growth defect of groES temperature-sensitive mutations at the non-permissive temperature (van der Vies et al., 1994). In addition, Gp31 can substitute for GroES in vitro in the refolding of prokaryotic Rubisco protein and citrate synthase (van der Vies et al., 1994; Richardson et al., 1999). This result was somewhat surprising, considering that Gp31 and GroES possess only 14% identity at the sequence level (Keppel et al., 1990; Koonin and van der Vies, 1995).
The structures of GroES and Gp31 have been solved by a combination of X-ray crystallography and NMR experiments (Landry et al., 1993; Hunt et al., 1996, 1997). In addition to an overall similar core structure, both cochaperonins possess a mobile loop, which interacts directly with GroEL and which has been shown to play a central regulatory role in the mechanism of action of the GroES/GroEL chaperonin machine (Landry et al., 1993, 1996; Richardson et al., 1999).
Recently, Ang et al. (2001) isolated a 31-like gene from coliphage RB49, distantly related to T-even bacteriophages (Repoila et al., 1994). The gene 31 homolog has been named cocO. Although the CocO protein is distantly related to GroES, it is 35% identical at the amino acid sequence level to Gp31 and can functionally substitute for it in the folding of either citrate synthase or T4-encoded Gp23 capsid protein (Richardson et al., 1999; Ang et al., 2001).
In view of the similarity between GroES and Gp31 at the structural and functional levels, we asked whether the T4 and RB49 cochaperonins could totally substitute for E. coli’s GroES in vivo. We found that either Gp31 or CocO can replace E. coli’s GroES for bacterial growth and the growth of bacteriophages λ and T5.
RESULTS AND DISCUSSION
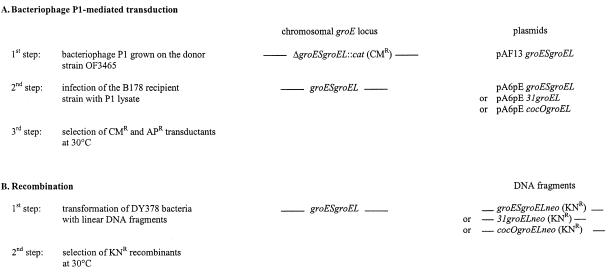
We employed two different strategies to determine whether gene 31 of bacteriophage T4 or gene cocO of bacteriophage RB49 could replace the otherwise essential groES gene of E. coli. The first method was based on bacteriophage P1-mediated transduction. In the donor strain, OF3465, the chromosomal copy of the groESgroEL (groE) operon is deleted and replaced by an Ω-cassette containing the chloramphenicol resistance gene cat (Prentki and Krisch, 1984). Since groES and groEL are essential genes (Fayet et al., 1989), the donor strain is maintained alive by a plasmid expressing the wild-type groE operon. A bacteriophage P1 lysate was grown on this donor strain and used to transduce B178, the recipient wild-type strain, carrying one of the various cochaperonin-groEL operon constructs shown in Figure 1A. Chloramphenicol-resistant (CMR) transductants were readily obtained at 30°C from strains carrying either the groES, 31 or cocO gene constructs, but, as expected, none were obtained with B178 recipient bacteria harboring vector sequences only. Thus, it appears that either Gp31 or CocO can completely replace GroES in E. coli growth when expressed from a multicopy plasmid.
Fig. 1. Genetic strategies employed in gene replacements.
To see whether a single copy of the bacteriophage-encoded cochaperonin is sufficient for bacterial growth, the E. coli groES gene was replaced on the chromosome by transformation with linear DNA fragments encoding gene 31 or gene cocO. We used strain DY378, which contains a defective λ prophage, whose transient induction simultaneously inhibits the RecBCD nuclease and enhances recombinogenic activity by employing the bacteriophage λ-encoded Exo and Beta recombination proteins (Yu et al., 2000). DY378 bacteria were electroporated with a series of DNA fragment constructs encoding the GroES, Gp31 or CocO cochaperonins, the groEL gene and the neo gene encoding for kanamycin resistance (KNR), inserted immediately downstream of groEL (Figure 1B). The integration of the DNA fragments into the host chromosome was efficient, as judged by the fact that KNR transformants were obtained at a frequency of 10–4 to 10–5 per surviving bacterium at 30°C.
The KNR recombinants obtained with strain DY378 cannot grow at high temperature because of the induction of the kil gene present in the defective λ prophage (Yu et al., 2000). To further analyze the growth properties of these recombinants at higher temperatures, the KNR-encoding neo gene and the recombined groE operon were transferred into our B178 wild-type strain by bacteriophage P1-mediated transduction. As expected, the frequency of cotransduction between the cochaperonin and the neo marker (KNR) was ∼100%, due to the fact that groEL and the inserted neo gene are immediately adjacent on the bacterial chromosome (data not shown).
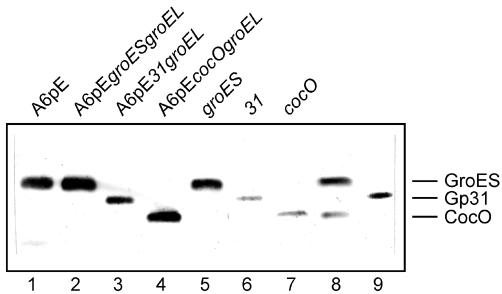
In order to biochemically verify that all recombinants expressed GroES, Gp31 or CocO, total protein samples were prepared from the various B178 multicopy or single-copy constructs. Extracts of each strain were separated by SDS–PAGE and analyzed by western immunoblot technology with a mixture of antibodies against GroES, Gp31 and CocO. The results are shown in Figure 2. The first lane is a control displaying the endogenous level of GroES in the B178 parental strain. Only one cochaperonin is detected in each experimental lane (lanes 2–7), demonstrating that groES has been replaced by either the 31 or the cocO gene. The level of cochaperonin is very similar in the multicopy constructs (lanes 2–4), whereas Gp31 and CocO are relatively less expressed in the corresponding single-copy, chromosomally encoded strains (lanes 5–7).
Fig. 2. Western blot analysis of cochaperonin proteins expressed in our various bacterial constructs. Cellular extracts were separated by SDS–PAGE and immunoblotted as described in Methods. Lane 1 represents an extract from the wild-type B178(pA6pE) parental strain. Lanes 2–4 represent extracts from multicopy constructs and lanes 5–7 extracts from single-copy constructs. Lanes 8 and 9 contain 100 ng of each purified protein.
The physical presence or absence of the groES gene was also tested in the variously constructed strains. The Southern blot analysis shown in Figure 3 establishes that groES sequences are absent in all transductants that carry either the 31 or cocO gene on a plasmid (lanes 3 and 4) and in single-copy constructs, where the groES gene has been replaced by either 31 or cocO on the chromosome (lanes 6 and 7). As expected, the EcoRI–PvuII DNA fragment that hybridizes to the groES probe is shortened in lanes 2 and 5, where an extra EcoRI site has been introduced in front of groES. These results, coupled with the western blot experiments, amply demonstrate the appropriate replacement of groES by the corresponding bacteriophage-encoded cochaperonin orthologs.
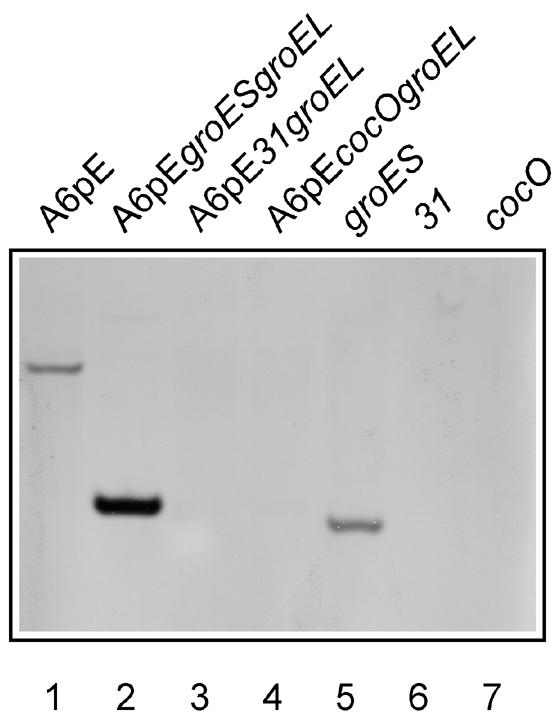
Fig. 3. Southern blot analysis of groES-encoding sequences in the various bacterial constructs. Total DNA was digested, blotted and hybridized with a groES-specific DNA probe. Lane 1 represents total DNA from the wild-type B178(pA6pE) parental strain. Lanes 2–4 contain DNA from multicopy constructs and lanes 5–7 DNA from single-copy constructs. Four times less DNA has been loaded on lane 2 so that the band that corresponds to the groES DNA present on the plasmid is not excessively over-represented.
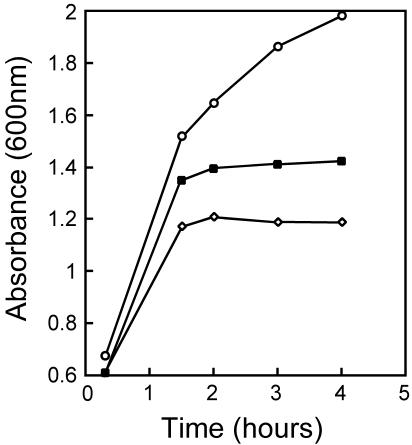
Since GroES and GroEL belong to the so-called heat shock protein family, whose intracellular levels progressively rise with increasing temperature, we asked whether our various bacterial constructs are able to grow at higher temperatures. The data in Table IA show that there is no detectable difference in bacterial growth between 15 and 42°C when the transduced cells carry either the groES, 31 or cocO gene on the pA6pE plasmid. In contrast, when either 31 or cocO have replaced groES on the chromosome, bacterial growth is impaired above 40°C (Table IB). The growth curves shown in Figure 4 indicate that cellular growth ceases after one or two cellular divisions at 42°C in bacteria encoding either 31 or cocO instead of groES. The growth defect is bactericidal after overnight incubation at 42°C, as judged by the progressive loss of colony formation (data not shown). All of these observations, taken together, indicate that the bacteriophage-encoded cochaperonins can fold all of E. coli’s essential proteins at all temperatures, provided that they are not in limiting amounts. We do not know the exact reason(s) for the inability of the single-copy 31 and cocO genes to sustain E. coli’s viability at 42°C. The chromosomally encoded 31 and cocO genes were constructed so as to possess the exact Shine–Dalgarno ribosomal binding sequence as that of groES. Nevertheless, semi-quantitative western immunoblotting analysis indicated that the intracellular levels of Gp31 and CocO are substantially reduced to ∼15–40% of the levels of GroES (Figure 2; data not shown). Thus, it is likely that the temperature sensitivity of the E. coli strains carrying the chromosomally encoded 31 or cocO gene is due to the relatively low levels of Gp31 or CocO. We do not know whether the relatively low levels of Gp31 or CocO are due to lower levels of synthesis and/or intracellular instability of the bacteriophage-encoded cochaperonins.
Table I. Colony-forming ability of various E. coli constructs at four different temperatures.
| 15°C | 30°C | 37°C | 42°C | |
|---|---|---|---|---|
| A. Replacement of the groE operon by a plasmid-encoded bacteriophage gene and groEL | ||||
| B178(pA6pE) | 1 | 1 | 1 | 1 |
| B178(pA6pE groESgroEL) | 1 | 1 | 1 | 1 |
| B178(pA6pE 31groEL) | 1 | 1 | 1 | 1 |
| B178(pA6pE cocOgroEL) | 1 | 1 | 1 | 1 |
| B. Replacement of the chromosomal groES copy by a bacteriophage-encoded gene | ||||
| B178(groES) | 1 | 1 | 1 | 1 |
| B178(31) | ∼10–1 | 1 | 1 | <10–5 |
| B178(cocO) | 1 | 1 | 1 | <10–5 |
See Methods for details of the experimental procedures.
∼10–1 indicates the formation of very small colonies compared with the parental strain B178(pA6pE) at an approximate efficiency of 10–1.
<10–5 indicates no visible colony formation at an efficiency of <10–5.
Fig. 4. Growth curves of bacteria encoding various cochaperonins on the chromosome. Bacterial cultures were shifted from 30 to 42°C at time 0 and light scattering was measured at 600 nm every 30 min. Circles, bacteria encoding groES; squares, bacteria encoding 31; diamonds, bacteria encoding cocO.
Finally, we examined the plating of bacteriophages on our various bacterial constructs. For simplicity, we omitted in Table II the data obtained with bacteriophages T5, T4 and RB49, since they all propagate equally well on all of our E. coli constructs at either 30 or 40°C. As shown in Table IIA, the strains that express either Gp31 or CocO from a multicopy plasmid can fully complement an amber mutation in gene 31 or cocO, whereas, as already known, GroES cannot substitute for either Gp31 or CocO in bacteriophage growth. The growth of bacteriophage λ is impaired at 40°C, especially when groES is replaced by cocO. This result suggests that the λB and λE proteins, whose folding is rate limiting in E. coli groE mutant strains (Georgopoulos et al., 1973), cannot be folded readily by CocO at 40°C. With the single-copy constructs, we observed that cross-complementation between Gp31 and CocO is only partial at 30°C (Table IIB), due most likely to the lower levels of expression of Gp31 and CocO. This result highlights the fact that, at least under limiting conditions, Gp31 and CocO somehow exhibit a preference for their own capsid protein. The growth of bacteriophage λ is also impaired on the single-copy constructs (Table IIB). However, as seen above, the effect on bacteriophage λ plating is more severe at higher temperatures, especially in the presence of CocO. Indeed, Gp31 seems to work better for the assembly of λ proheads compared with CocO, whereas both bacteriophage-encoded cochaperonins help equally well in the assembly of T5 tail components and E. coli’s essential proteins. Taken together, these last observations suggest that the cochaperonins of groEL may play an important role in the specificity of the substrate being folded by the GroEL machine. We do not know yet why, at relatively low intracellular concentrations, the bacteriophage-encoded Gp31 and CocO cochaperonins exhibit a preference for their corresponding capsid protein. This preference could be due to either a general effect on the efficiency or the kinetics of the GroEL reaction cycle. Alternatively, but not mutually exclusively, the preference could reflect a specific recognition of the cochaperonin for its own capsid protein.
Table II. Plaque-forming ability of three different bacteriophages on various E. coli constructs.
| λ cI | T431am | RB49cocOam | ||||
|---|---|---|---|---|---|---|
| 30°C | 40°C | 30°C | 40°C | 30°C | 40°C | |
| A. Replacement of the groE operon by a plasmid-encoded bacteriophage gene and groEL | ||||||
| B178(pA6pE) | 1 | 1 | <10–5 | <10–5 | <10–5 | <10–5 |
| B178(pA6pE groESgroEL) | 1 | 1 | <10–5 | <10–5 | <10–5 | <10–5 |
| B178(pA6pE 31groEL) | 1 | 1 | 1 | 1 | 1 | 1 |
| B178(pA6pE cocOgroEL) | ∼10–1 | ∼10–2 | 1 | 1 | 1 | 1 |
| B. Replacement of the chromosomal groES copy by a bacteriophage-encoded gene | ||||||
| B178(groES) | 1 | 1 | <10–5 | <10–5 | <10–5 | <10–5 |
| B178(31) | ∼10–1 | <10–5 | 1 | 1 | ∼10–3 | 1 |
| B178(cocO) | ∼10–3 | <10–5 | ∼10–3 | 1 | 1 | 1 |
See Methods for details of the experimental procedures.
∼10–1, ∼10–2 and ∼10–3 indicate progressively smaller plaques compared to those on the parental strain B178(pA6pE) at an approximate efficiency of 10–1, 10–2 or 10–3, respectively.
<10–5 indicates no detectable plaque formation at an efficiency of <10–5.
METHODS
Strains. The various bacterial strains, bacteriophages and plasmids used in this study are listed in Table III.
Table III. Bacterial strains, bacteriophages and plasmids.
| Relevant genotype | Source | |
|---|---|---|
| Bacterial strains | ||
| OF3465 | ΔgroESgroEL :: cat (CMR) (pAF13, APR, groESgroEL) | O. Fayet and M.-P. Castanié |
| B178 | W3110 galE sup+ | Our collection |
| DY378 | W3110 λcI857 Δ(cro-bio) | Yu et al. (2000) |
| Bacteriophages | ||
| P1L4 | P1 clear-plaque former | Our collection |
| λcI | λ clear-plaque former | Our collection |
| T4Do | Wild type | Our collection |
| T5 | Wild type | Our collection |
| T431amNG71 | Amber mutation in gene 31 | Epstein et al. (1963) |
| RB49 | Wild type | Repoila et al. (1994) |
| RB49cocOamE45 | Amber mutation in cocO | Ang et al. (2001) |
| RB43 | Wild type | Repoila et al. (1994) |
| Plasmids | ||
| pMPM-A6 | p15Aori, APR, pBAD | Mayer (1995) |
| pA6pE | p15Aori, APR, pgroE (=pE) | This study |
| pA6pEgroESgroEL | groES+groEL+ | This study |
| pA6pE31groEL | 31+groEL+ | This study |
| pA6pEcocOgroEL | cocO+groEL+ | This study |
| pLS1 | pBR322, APR, groES+ groEL+ | Chandrasekhar et al. (1986) |
ori, origin of replication; APR and CMR, resistance to ampicillin and chloramphenicol, respectively, encoded by the bla and cat genes, respectively.
Plasmids. Various derivatives of the pBAD vector pMPM-A6 (Mayer, 1995) have been constructed. The DNA region comprising the promoter of the arabinose operon was deleted and replaced by a 150 bp BamHI–EcoRI DNA fragment (obtained by PCR amplification) carrying the wild-type heat shock promoter of the E. coli groE operon; the resulting plasmid is referred to as pA6pE. The groES, 31 (Richardson et al., 1999) or cocO (Ang et al., 2001) genes were cloned at the 3′-end of the EcoRI site in pA6pE, starting at the first ATG of the coding sequence. Escherichia coli groEL was subsequently added downstream of the groES, 31 or cocO genes. Plasmid pLS1 (Chandrasekhar et al., 1986) was used as a source of the 8 kb E. coli DNA fragment carrying the groE operon. An EcoRI site was introduced by site-directed mutagenesis in front of groES in order to replace it by its bacteriophage-encoded orthologs.
Transduction experiments. Bacteriophage P1-mediated transduction experiments were conducted essentially as described previously (Weber et al., 1998). A bacteriophage P1 lysate was grown on the donor strain OF3465 (a kind gift from Drs O. Fayet and M.-P. Castanié, Laboratoire de microbiologie et génétique moléculaire, Toulouse, France) and used to transduce B178, our wild-type recipient strain. The transductants were selected on LB plates supplemented with 50 µg/ml ampicillin, 15 µg/ml chloramphenicol and 10 mM sodium citrate at 30°C.
Recombination experiments. Recombination experiments in E. coli were performed essentially according to the method of Yu et al. (2000). DY378 cells were induced at 42°C for 15 min and made electrocompetent. They were subsequently electroporated with linear 5.8 kb DraI-derived DNA fragments. Recombined cells were selected at 30°C on LB plates supplemented with 40 µg/ml kanamycin and then assayed for complementing the growth of either T431amNG71 or RB49cocOamE45 mutant bacteriophages. A bacteriophage P1 lysate was grown on the recombined bacteria, which carry either groES, 31 or cocO, and used to transduce the wild-type strain B178 to KNR. Transductants were selected on LB plates supplemented with 40 µg/ml kanamycin and 10 mM sodium citrate at 30°C and assayed for complementation as described above. A 500 bp DNA fragment containing the appropriate cochaperonin gene and the flanking regions homologous to E. coli DNA were amplified by PCR and then sequenced to prove the occurence of the recombination event.
Protein analysis. Whole-cell extracts were prepared from the B178 parental strain and its various isogenic constructs following a 30 min incubation at 42°C, to enhance the expression of the groE heat shock operon. The proteins were then separated on 12.5% SDS–polyacrylamide gels (Laemmli, 1970) and subsequently transfered to a nitrocellulose filter (Schleicher and Schuell). The presence of the individual cochaperonins was detected by the western immunoblotting technique using a mixture of three polyclonal rabbit antibodies directed against GroES, Gp31 or CocO (1:5000 dilution; our collection), followed by development using a goat anti-rabbit IgG AP-conjugated secondary antibody from Bio-Rad (1:3000). Purified GroES, Gp31 and CocO proteins were used to standardize their relative intracellular levels in semi-quantitative western immunoblotting experiments.
Southern analysis. Total DNA was extracted from the B178 parental strain and its various isogenic constructs. Two micrograms of total DNA were digested with the EcoRI and PvuII restriction enzymes and separated on agarose gels. The DNA fragments were transferred to Hybond membrane (Amersham) using standard methodology (Southern, 1975). The filters were subsequently hybridized with a 295 bp groES-specific probe generated by PCR, using primers internal to groES, and labeled with the DIG-High-Prime reaction from Boehringer. Finally, groES sequences were detected immunologically using the anti-DIG-AP conjugated antibody from Boehringer (1:10 000), essentially according to the manufacturer’s instructions.
Bacterial and bacteriophage growth analysis. Bacteria were grown in LB broth and then diluted serially (20-fold dilution steps). Five microliters of each dilution were spot-tested on LB plates supplemented with the appropriate antibiotics and incubated at various temperatures. Bacterial colony formation was monitored following overnight incubation.
Bacterial growth curves in liquid media were carried out as follows: overnight cultures were diluted 100-fold in LB and grown at 30°C for 2 h. The cultures were subsequently shifted to 42°C, and optical density at 600 µm was monitored at 30 min intervals.
The growth of bacteriophages was similarly scored by spot-testing 5 µl of serial dilutions on lawns of the various bacterial constructs on LB plates supplemented with the appropriate antibiotic. The plates were incubated at various temperatures, and bacteriophage plaque-forming ability was monitored following overnight incubation.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Marie-Pierre Castanié and Olivier Fayet for the generous gift of their OF3465 strain. We thank Pierre Genevaux and Kyle Tanner for help with the illustrations and Luli Billecchi-Mestre for cheerful editorial assistance. This work was supported by a grant from the Swiss National Science Foundation (FN 31-065403.01) and the Canton of Geneva.
REFERENCES
- Ang D., Keppel, F., Klein, G., Richardson, A. and Georgopoulos, C. (2000) Genetic analysis of bacteriophage-encoded cochaperonins. Annu. Rev. Genet., 34, 439–456. [DOI] [PubMed] [Google Scholar]
- Ang D., Richardson, A., Mayer, M.P., Keppel, F., Krisch, H. and Georgopoulos, C. (2001) Pseudo-T-even bacteriophage RB49 encodes CocO, a cochaperonin for GroEL, which can substitute for Escherichia coli’s GroES and bacteriophage T4’s Gp31. J. Biol. Chem., 276, 8720–8726. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar G.N., Woolford, C., Hendrix, R. and Georgopoulos, C. (1986) Purification and properties of the GroES morphogenetic protein of Escherichia coli. J. Biol. Chem., 261, 12414–12419. [PubMed] [Google Scholar]
- Ellis R.J. (1994) Roles of molecular chaperones in protein folding. Curr. Opin. Struct. Biol., 4, 117–122. [Google Scholar]
- Epstein R.H., Bolle, A., Steinberg, C.M., Kellenberger, E., Boy de la Tour, E., Chevallay, R., Edgar, R.S., Susman, M., Denhardt, G.H. and Lielausis, A. (1963) Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp. Quant. Biol., 28, 375–384. [Google Scholar]
- Fayet O., Ziegelhoffer, T. and Georgopoulos, C. (1989) The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol., 171, 1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. and Welch, W.J. (1993) Role of major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol., 9, 601–635. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C., Hendrix, R.W., Casjens, S.R. and Kaiser, A.D. (1973) Host participation in bacteriophage λ head assembly. J. Mol. Biol., 76, 45–60. [DOI] [PubMed] [Google Scholar]
- Hunt J.F., Weaver, A.J., Landry, S.J., Gierasch, L. and Diesenhofer, J. (1996) The crystal structure of the GroES cochaperonin at 2.8 Å resolution. Nature, 379, 37–45. [DOI] [PubMed] [Google Scholar]
- Hunt J.F., van der Vies, S.M., Henry, L. and Deisenhofer, J. (1997) Structural adaptations in the specialized bacteriophage T4 co-chaperonin Gp31 expand the size of the Anfinsen cage. Cell, 90, 361–371. [DOI] [PubMed] [Google Scholar]
- Keppel F., Lipinska, B., Ang, D. and Georgopoulos, C. (1990) Mutational analysis of the phage T4 morphogenetic gene 31, whose product interacts with the E. coli GroEL protein. Gene, 90, 19–25. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. and van der Vies, S.M. (1995) Conserved sequence motifs in bacterial and bacteriophage chaperonins. Trends Biochem. Sci., 20, 14–15. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Landry S.J., Zeilstra-Ryalls, J., Fayet, O., Georgopoulos, C. and Gierasch, L.M. (1993) Characterization of a functionally important mobile domain of GroES. Nature, 364, 255–258. [DOI] [PubMed] [Google Scholar]
- Landry S.J., Taher, A., Georgopoulos, C. and van der Vies, S. (1996) Interplay of structure and disorder in bacteriophage T4 and Escherichia coli chaperonin-10 mobile loops. Proc. Natl Acad. Sci. USA, 93, 11622–11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P. (1995) A new set of useful cloning and expression vectors derived from pBlueScript. Gene, 163, 41–46. [DOI] [PubMed] [Google Scholar]
- Prentki P. and Krisch, H.M. (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene, 29, 303–313. [DOI] [PubMed] [Google Scholar]
- Repoila F., Tétart, F., Bouet, J.-Y. and Krisch, H.M. (1994) Genomic polymorphism in the T-even bacteriophages. EMBO J., 13, 4181–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A., van der Vies, S., Keppel, F., Taher, A., Landry, S.J. and Georgopoulos, C. (1999) Compensatory changes in GroEL/Gp31 affinity as a mechanism for allele-specific genetic interaction. J. Biol. Chem., 274, 52–58. [DOI] [PubMed] [Google Scholar]
- Southern E.M. (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol., 98, 503–517. [DOI] [PubMed] [Google Scholar]
- van der Vies S.M., Gatenby, A.A. and Georgopoulos, C. (1994) Bacteriophage T4 encodes a cochaperonin that can substitute for Escherichia coli GroES in protein folding. Nature, 368, 654–656. [DOI] [PubMed] [Google Scholar]
- Weber F., Keppel, F., Georgopoulos, C., Hayer-Hartl, M.K. and Hartl, F.U. (1998) The oligomeric structure of GroEL/GroES is required for biologically significant chaperonin function in protein folding. Nat. Struct. Biol., 5, 977–985. [DOI] [PubMed] [Google Scholar]
- Yu D., Ellis, H.M., Lee, E.C., Jenkins, N.A., Copeland, N.G. and Court, D.L. (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweig M. and Cummings, D.J. (1973) Cleavage of head and tails proteins during bacteriophage T5 assembly: selective host involvement in the cleavage of a tail protein. J. Mol. Biol., 80, 505–518. [DOI] [PubMed] [Google Scholar]