Conspectus:
Higher-order or supramolecular protein assemblies, usually regulated by enzymatic reactions, are ubiquitous and essential for cellular functions. This evolutionary fact has provided a rigorous scientific foundation, as well as an inspiring blueprint, for exploring supramolecular assemblies of man-made molecules that are responsive to biological cues as a novel class of therapeutics for biomedicine. Among the emerging man-made supramolecular structures, peptide assemblies, formed by enzyme reactions or other stimuli, have received most of the research attention and advanced most rapidly.
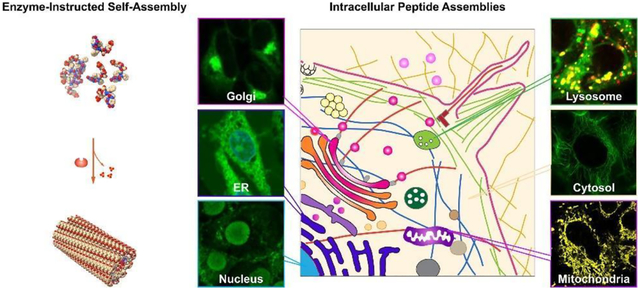
In this article, we will review works that apply enzyme-instructed self-assembly (EISA) to generate intracellular peptide assemblies for developing a new kind of biomedicine, especially in the field of novel cancer nanomedicines and modulating cell morphogenesis. As a versatile and cell-compatible approach, EISA can generate non-diffusive peptide assemblies locally; thus, it provides a unique approach to target subcellular organelles with exceptional cell selectivity. We have arranged this article in the following way: after introducing the concept, simplicity, and uniqueness of EISA, we discuss the EISA-formed intracellular peptide assemblies, including as artificial filaments, in the cell cytosol. Then, we describe the representative examples on targeting subcellular organelles, such as mitochondria, endoplasmic reticulum, Golgi apparatus, lysosomes, and the nucleus, by enzyme-instructed intracellular peptide assemblies for potential cancer therapeutics. After that, we highlight the recent exploration of the transcytosis of peptide assemblies for controlling cell morphogenesis. Finally, we provide a brief outlook of enzyme-instructed intracellular peptide assemblies. This article aims to illustrate the promise of EISA-generated intracellular peptide assemblies in understanding diseases, controlling cell behaviors, and developing new therapeutics from a class of less explored molecular entities, which are substrates of enzymes and become building blocks of self-assembly after the enzymatic reactions.
Graphical Abstract:
1. Introduction
1.1. Biogenesis of Endogenous Supramolecular Assemblies
Supramolecular assemblies, which are molecular complexes held together by noncovalent interactions, play a pivotal role in the fundamental processes of life. For example, lipid membranes, composed of amphipathic molecules, form the basis of cellular boundaries, providing compartmentalization essential for maintaining the delicate balance of intracellular and extracellular environments.5 On the other hand, higher-order protein structures, such as cytoskeletons6–8 and signalosomes9–10, are crucial for carrying out specific biochemical reactions and cellular functions. These assemblies allow for dynamic yet precise regulation, substrate recognition, and catalytic efficiency, ensuring vital cell processes. These facts highlight the significance of supramolecular assemblies as indispensable features in the complex and intricate machinery of life.
The studies of endogenous supramolecular assemblies reveal that their biogenesis relies on two fundamental molecular processes, that is, enzymatic reaction and self-assembly.11 For example, the dynamic nature of endoplasmic reticulum membrane requires enzymatic reactions to regulate network formation with proteins and phospholipids.12–13 Inflammasomes, which act as supramolecular organization centers for host defense within cells, are formed through the self-assembly of NLRP3, ASC, and caspase-1 proteins, which are controlled by enzymatic posttranslational modifications.14 Actin functions as an enzyme and its substrate is ATP, making it an ATPase. The crucial reaction for preserving actin filaments is the hydrolysis of ATP on actin, which is pivotal for maintaining their structure and function.6 Like actin, tubulin, acting as a GTPase, catalyzes GTP to GDP hydrolysis, maintaining microtubule’s asymmetric growth.8 These insights underscore the importance of enzymatic reactions in controlling noncovalent interactions for self-assembly to generate supramolecular assemblies that perform biological functions.
1.2. The Concept of Enzyme-Instructed Self-Assembly (EISA)
The ubiquitous and essential integration of enzyme reactions and self-assembly in cells has bestowed a solid scientific foundation while also serving as an inspiring blueprint, for harnessing these fundamental processes to create assemblies of various other molecules, particularly those of synthetic origin. Inspired by the biogenesis of supramolecular protein structures, we integrated enzymatic reactions and self-assembly for generating soft materials, such as hydrogels,15 and termed such an integrated processes as enzyme-instructed self-assembly (EISA).16 Figure 1 shows the concept of EISA, a multiple step process that consists of enzymatic reactions and self-assembly. In EISA, enzyme plays a crucial role in initiating self-assembly by transforming non-self-assembling precursors (also substrates of the enzyme) into self-assembling molecules through bond cleavage. This self-assembly process typically leads to the formation of supramolecular nanoscale assemblies, such as nanofibers, in water. When the concentration of these nanoscale assemblies surpasses a specific threshold, they entangle to create a network, leading to hydrogelation in most cases.15, 17
Figure 1.
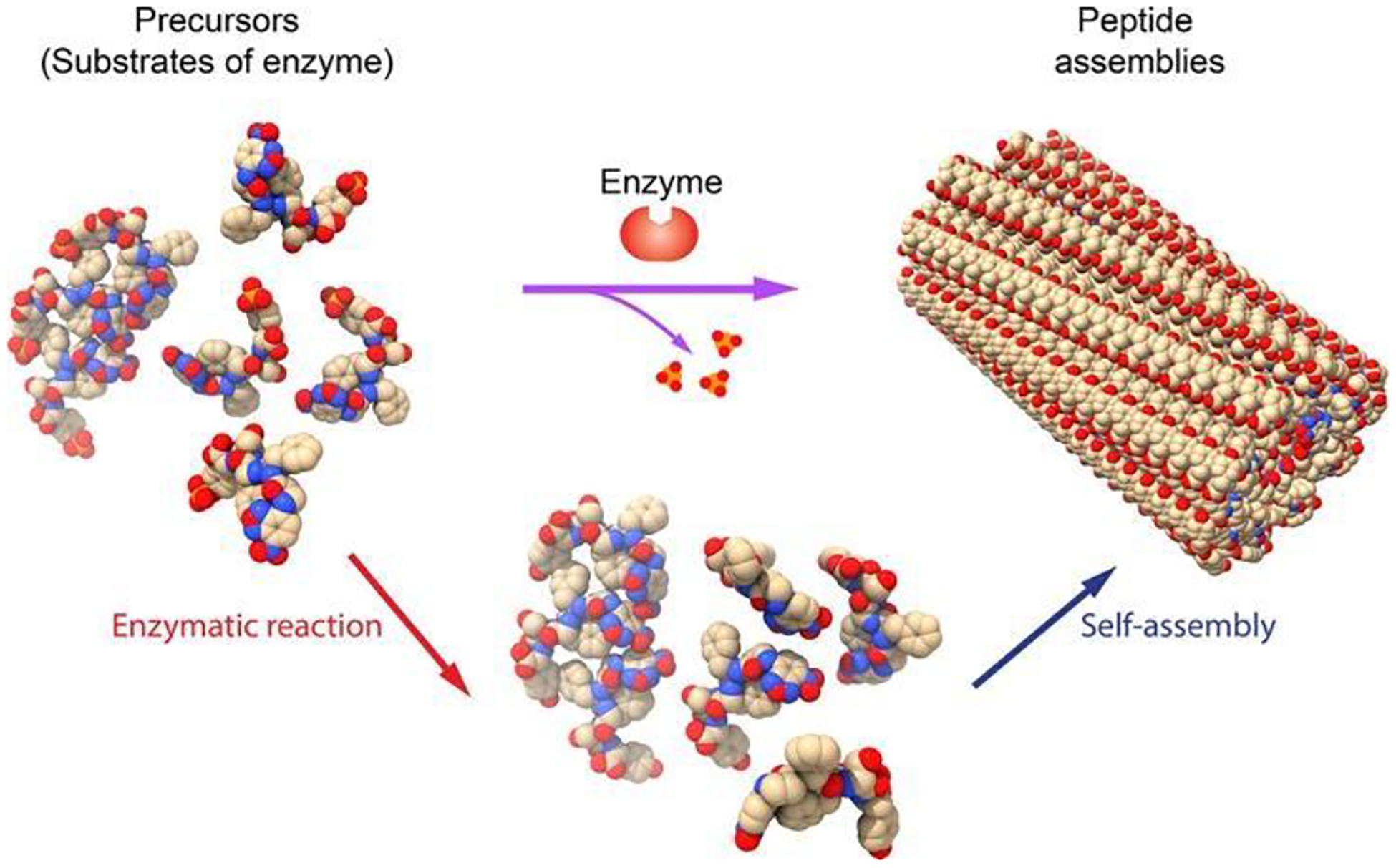
Illustration of enzyme-instructed self-assembly (EISA), which integrates enzymatic reaction and self-assembly, for generating peptide assemblies.
Building upon these principles, we reported the first example of EISA of man-made molecules.1 In that study, Fmoc-phosphotyrosine (Fmoc-pY) acts as the substrate of alkaline phosphatase (ALP), and ALP, a readily available enzyme with high catalytic efficiency, serves as the instructive agent for molecular self-assembly. Dephosphorylation of Fmoc-phosphotyrosine by ALP generates Fmoc-tyrosine (Fmoc-Y), which self-assembles into nanofibers/hydrogel.1 It is also feasible to employ enzymes to form new bonds between two substrates, thereby generating self-assembling molecules for EISA.18
An overlooked, unique feature of EISA is that it involves a form of self-assembly under far-from-equilibrium conditions because the concentration of the self-assembling building blocks is influenced by enzymatic reactions and scarcely remains at equilibrium (though it may reach a steady state) during the self-assembly process. While the detailed kinetics remain to be formulated in a more rigorous manner, EISA can generate non-diffusive assemblies, even continuously, allowing the localization of the supramolecular assemblies in cellular environment. This local control of aggregates opens a new way to use assemblies of small molecules for controlling cell fate, as discussed in the following sections.
2. Intracellular Peptide Assemblies Formed by EISA
2.1. Inhibiting Bacteria Growth
Among supramolecular assemblies of small molecules, peptide assemblies have received considerable research attention over the last few decades.19–34 Therefore, it is not surprising that peptide assemblies formed by EISA35 have become the most explored molecular platform for various applications since the demonstration of the EISA of small molecules.1 The first example of EISA in a living organism is the use of EISA of peptides to inhibit the growth of bacteria.2 The EISA precursor is a phosphotripeptide (Nap-FFpY (1), Figure 2A). ALP catalytically dephosphorylates 1 to generate Nap-FFY (2), which self-assembles in water to form nanofibers (Figure 2B) and induce hydrogelation. Incubating 1 with the phosphatase (PhoA) overexpressing E. coli leads to the accumulation of 2 inside the E. coli, reaching about seven times the incubation concentration. EISA catalyzed by PhoA can generate enough 2 inside the E. coli to form nanofibers (Figure 2C) and inhibit E. coli growth. This study demonstrates a new methodology—enzyme-regulated intracellular self-assembly of small molecules for creating artificial nanostructures (Figure 2D)—for controlling the fate of a cell.
Figure 2.
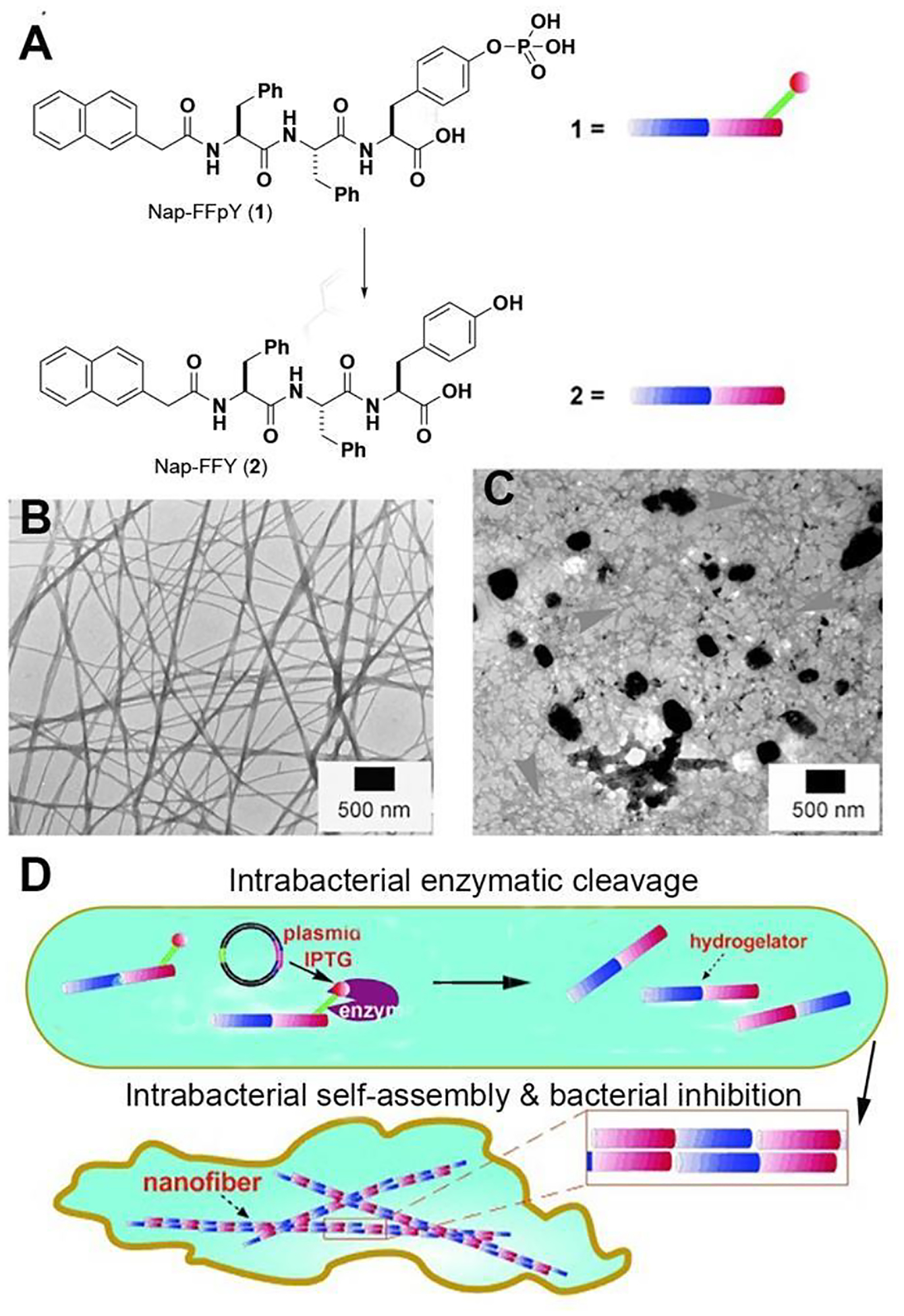
(A) Structures of 1 and 2. (B) Nanofibers formed by 2 upon the addition of ALP in the PBS solution of 1. (C) Transmission electron microscope (TEM) images of the nanofibers of 2 formed inside the bacteria. (D) Illustration of intracellular nanofiber for hydrogelation and the inhibition of bacterial growth. Adapted from Ref.2 with permission. Copyright Wiley 2007.
2.2. Selectively Inhibiting Cancer Cells
The ability to create self-assembled nanostructures by EISA within cells opens the opportunity for in vivo or in situ self-assembly, thus providing a new way to generate peptide assemblies for selectively inhibiting cancer cells, as demonstrated in the treatment of HeLa (a cervical cancer cell) and NIH3T3 (a mouse fibroblast cell) with a peptide substrate (Nap-FF-es-suc (3), Figure 3A) of esterase.30 Esterase converts 3 to 4, leading form nanofibers and inducing hydrogelation (Figure 3B). MTT assay (Figure 3C) shows that the IC50 of 3 against HeLa cells is around 320 μM, but the IC50 of 3 against NIH3T3 cells is above 1.28 mM. TEM of the cell lysate of HeLa cells also reveals nanofibers of 4, indicating that the formation of nanofibers and subsequent hydrogelation within the HeLa cells to results in cell death. This observation is consistent with the high-level expression of esterase in HeLa cell and low-level of esterase expression in NIH3T3 cells. This impressive selectivity against cancer cells stems from variations in enzyme expression and molecular self-assembly, thus demonstrating the feasibility of employing EISA of peptide assemblies for cancer therapy.
Figure 3.
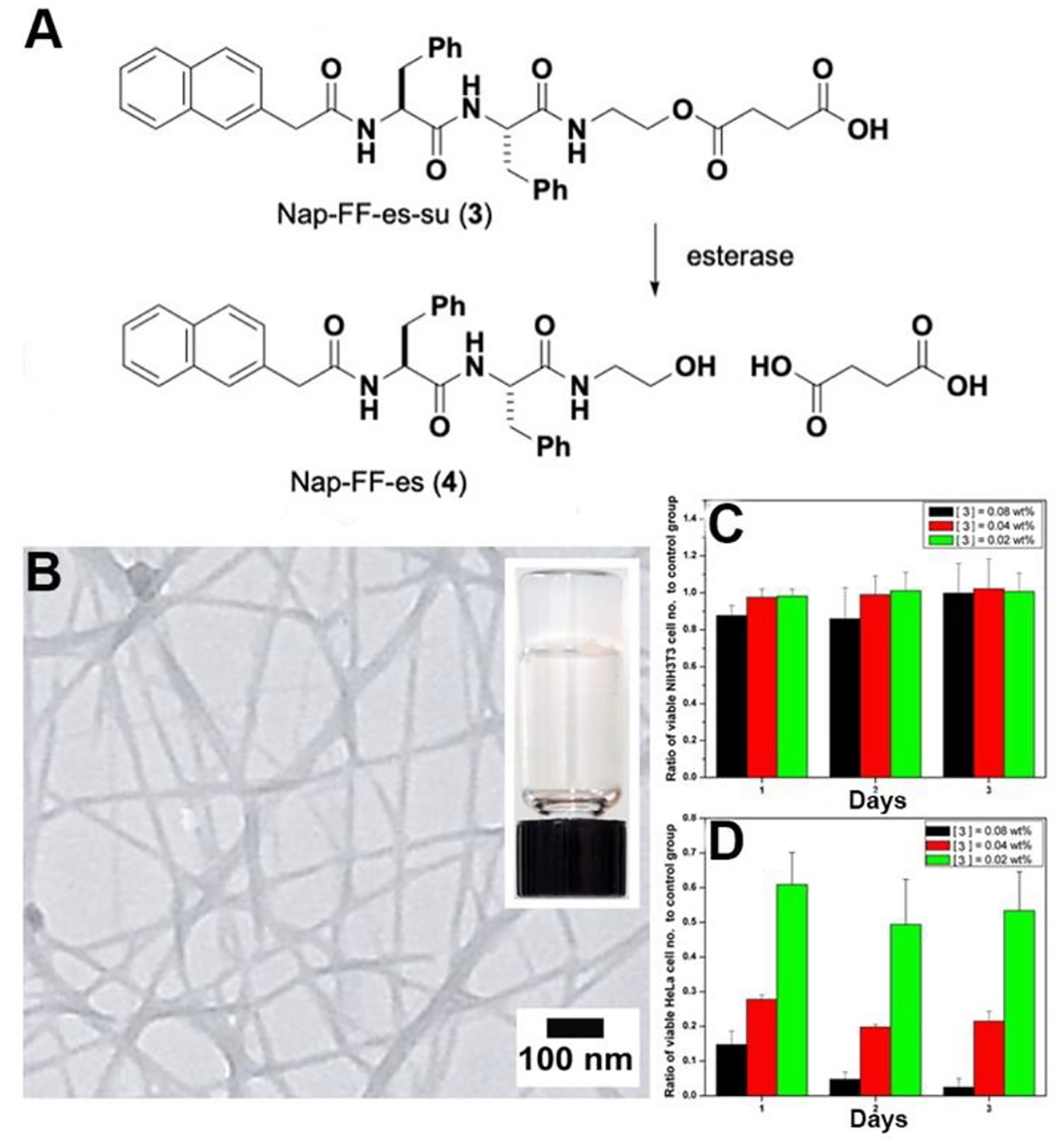
(A) Structures of 3 and 4. (B) Nanofibers formed by 4 upon the addition of esterase in the PBS solution of 3 (0.5 wt%). MTT assays of (C) NIH3T3 cells and (D) HeLa cells treated with 3 at concentrations of 1280, 640, and 320 μM. Adapted from Ref.30 with permission. Copyright Wiley 2007
2.3. Inhibits Immunosuppressive Bone Tumor
The choice of ALP, which is a highly efficient hydrolase36, for EISA was initially random, but in hindsight, it turned out to be a fortunate decision due to ALP’s metabolic inertness in serum37 and its overexpression in certain cancer cells, leading to immunosuppression in solid tumors38. Given its crucial roles in embryogenesis, bone metabolism, and neuron functions,39–40 ALP is “undruggable” by inhibitors. In contrast, EISA transforms enzyme substrates to produce self-assembling peptides, making it an ideal strategy to target tumors that overexpress ALP (Figure 4A).4 Specifically, the precursor (5) comprises a self-assembling peptide backbone (Nap-ff), a dephosphorylation site (py), and an esterase detoxification module (eMe2). It transforms into the self-assembling molecule (Nap-ffyeMe2 (6)), forming uniform nanofibers (Figure 4B). While 5 exhibits potent inhibition of Saos-2 cells (IC50 = 4 μM), its IC50 on hepatocyte cells (HepG2) surpasses Saos-2’s by over two orders of magnitude (Figure 4C). This outcome is particularly significant for potential application of 5 in clinical setting as it demonstrates that 5 selectively inhibits osteosarcoma cells without harming liver cells. Treatment of tumor-bearing mice with 5 or saline for four weeks results in a 25-fold reduction in tumor volume for the group treated with 5 (Figure 4D). These results further validate the efficient tumor growth inhibition in osteosarcoma with 5. Furthermore, this treatment significantly extends the survival time of osteosarcoma-bearing nude mice (Figure 4E), confirming the benefits of EISA using 5. As the first example of EISA of peptides targeting immunosuppressive tumors in vivo, this work introduces a novel approach to develop cancer therapeutics that counter immunosuppression in the tumor microenvironment.
Figure 4.
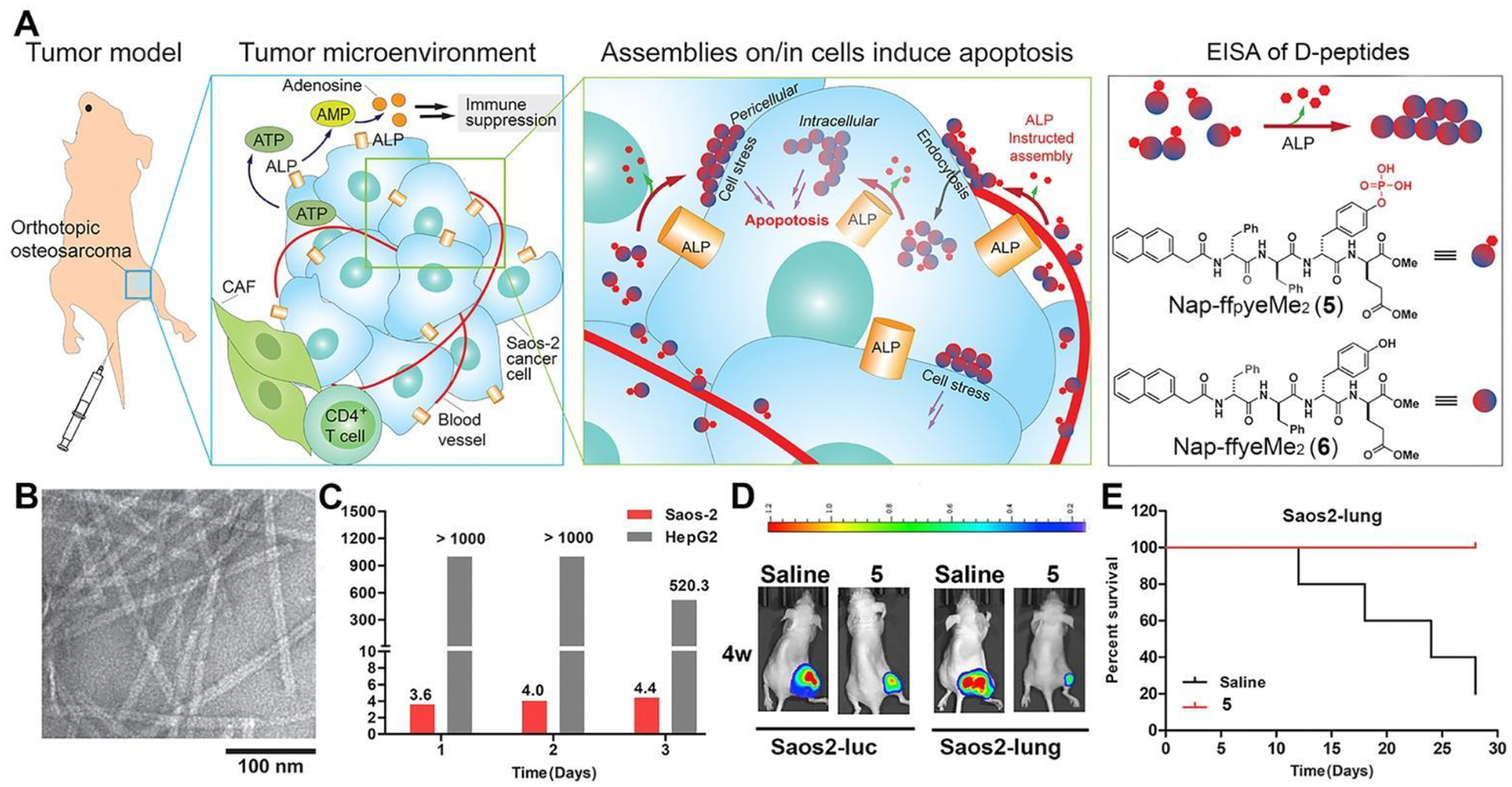
(A) Illustration of ALP-instructed assembly for inhibiting metastatic, immunosuppressive osteosarcoma in an orthotopic mice model and structures of 5 and 6. (B) TEM images of nanofibers of 6. (C) IC50 values of 5 against Saos-2 or HepG2 cells. (D) Images of osteosarcoma tumor growth for orthotopic osteosarcoma models established by Saos2-luc and Saos2-lung cells at week 4 after 5 or saline treatment. (E) Kaplan-Meier survival curves for orthotopic osteosarcoma nude mice (n = 5) treated with 5. Data are presented as mean ± standard deviation (S.D.). NS, not significant, ***P < 0.001. Adapted from Ref.4 with permission. Copyright Elsevier 2019.
2.4. Artificial Intracellular Filaments
Despite the profound implications of man-made supramolecular nanostructures in cells,11, 41 the direct and unambiguous confirmation of intracellular nanostructures made of small molecules was unexpectedly difficult. This difficulty arises due to the crowded intracellular space and the low contrast between synthetic nanostructures and endogenous biomacromolecules. Although it is feasible to use fluorescent imaging to establish intracellular nanostructures made by EISA,3 biorthogonal reaction,42 and redox reaction43, two key issues, namely the atomistic structures of the peptide assemblies and their intracellular architectures, remain unresolved until a recent study of bundles of peptide filaments made of a peptide bearing trimethylated lysine residue.44
In that particular study, a complex precursor (NBD-ffpyKMe3 (7)) was utilized, consisting of several components: a polarity-sensitive fluorescent dye known as nitrobenzoxadiazole (NBD), a self-assembling D-peptide backbone (ff),25 a phosphatase cleavage site (D-phosphotyrosine (py)), and a C-terminal trimethyl-L-lysine (KMe3). This design facilitated the conversion of precursor 7 to a new self-assembly building block (NBD-ffyKMe3 (8)) (Figure 5A). TEM shows that 7 self-assembled into nanoparticles. Subsequent ALP-catalyzed dephosphorylation of 7 resulted in the formation of filaments of 8, characterized by monodispersed diameters of approximately 6±1 nm. Surprisingly, the introduction of trimethylation on lysine enhanced the interfilamental interactions, leading to the formation of bundles of peptide filaments composed of 8.
Figure 5.
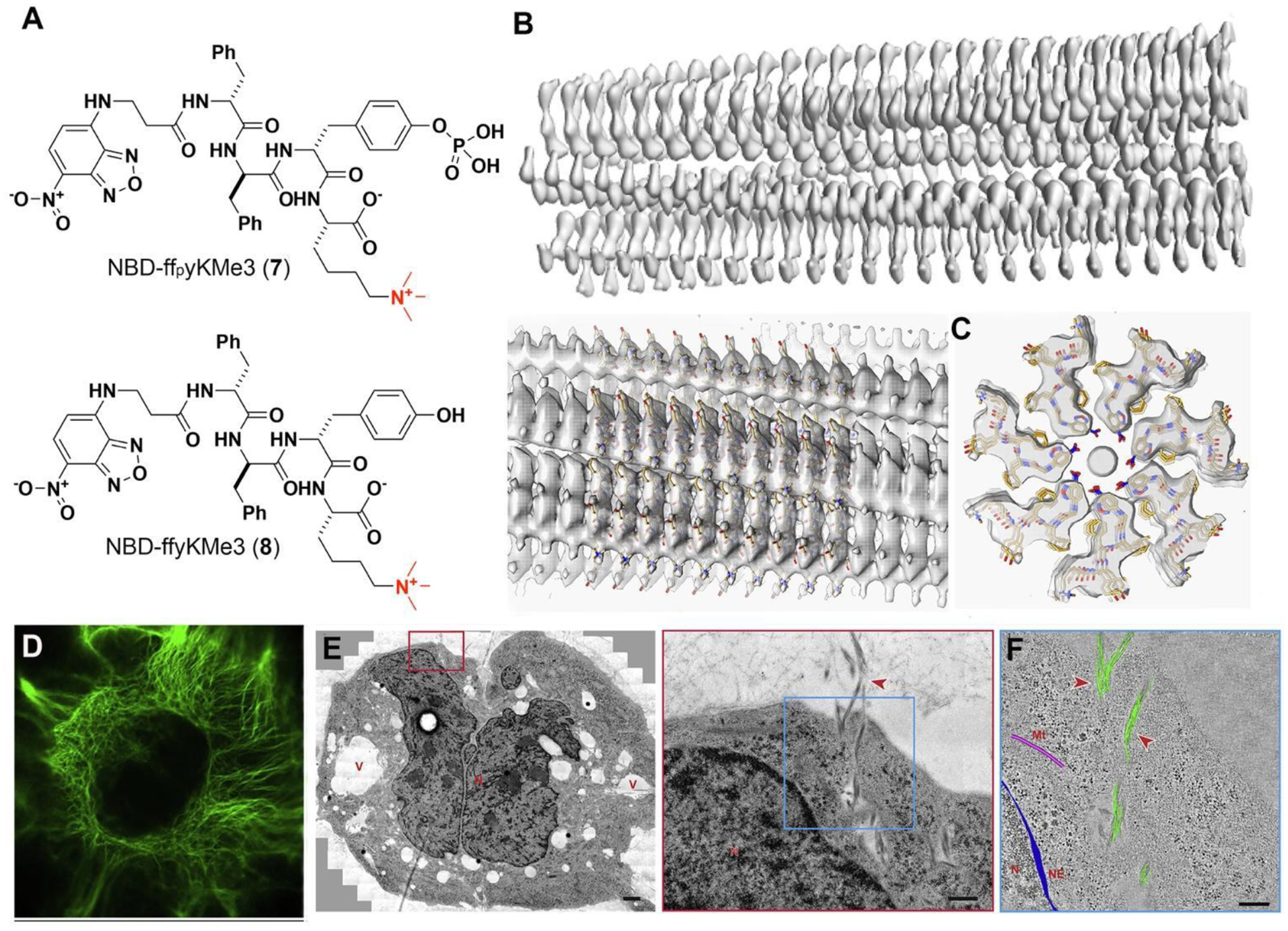
(A) Structures of 7 and 8. (B) 3D cryo-EM reconstruction of the filaments of 8. (C) Top views of the cross-section of the EM density of the filament and the stick representation of the peptides. (D) CLSM images of Saos-2 cells treated with 7. Scale bars, 10 mm. (E) TEM image treated Saos-2 cell (7, 200 μM, 24 h) and higher-magnification electron micrograph of the red boxed area. (F) 3D reconstruction models of the filament bundles (green), microtubules (pink), and nuclear envelope (blue) on an electron tomographic image of the blue boxed area in (E). Adapted from Ref.44. Copyright of the authors 2020 CC BY-NC-ND.
Cryo-EM reveals that the peptide self-assembles into unique cross-β structures exhibiting C7 symmetry, as shown in Figure 5B,C. Within the cellular context, the peptide filaments demonstrated consistent diameters and organized into twist bundles, extending from the plasma membrane to the nuclear membrane, as depicted in Figure 5D. Notably, these filaments exhibited an orthogonal arrangement relative to endogenous cytoskeletons and showed minimal interactions with other cellular components, but appearing as cytoskeleton-like structures.
Moreover, electron tomography (ET) demonstrated the formation of artificial filaments inside cells, further substantiating their extensive presence within cellular environments, as illustrated in Figure 5E. 3D ET-reconstruction (Figure 5F) confirmed that these bundles consisted of clusters of intertwining filaments, corroborating the in vitro TEM findings. Beyond establishing the existence of artificial intracellular peptide filaments through EISA, this research emphasized the essential role of diverse techniques, such as cryo-EM, fluorescent microscopy, electron tomography, molecular dynamics, and molecular engineering, in comprehending and developing artificial nanostructures within live cells.
3. Subcellular Localization of Peptide Assemblies
In eukaryotic cells, organelles function as essential hubs for cellular activities, and the communication between them plays a pivotal role in critical cellular processes such as programmed cell death. For example, nuclear DNA damage sets off a chain reaction, including the induction of mitochondrial membrane permeabilization (MMP). Similarly, endoplasmic reticulum (ER) stress and the permeabilization of lysosomes also induce cell death that is executed via mitochondria. These events collectively integrate pro-apoptotic signaling pathways and underscore that targeting subcellular organelles holds great promise for advancing therapeutic development and enhancing our comprehension of various diseases.45 However, the task of selectively targeting the organelles within specific cells presents a significant challenge. In this context, EISA emerges as a unique solution to address this challenge, as will be expounded upon in the subsequent sections.
3.1. Targeting Mitochondria
Mitochondria are complex organelles that play a central role in key cellular processes, and are becoming one of the most important drug targets for treating a wide range of diseases, including cancer, cardiovascular, inflammation, and neurological disorders. While most of the reported mitochondria targeting molecules are lipophilic and cationic, which may become cytotoxic with accumulation,46 EISA of branched peptides that carry negative charges, unexpectedly, can selectively target mitochondria of cancer cells.47 Conjugating a well-established protein tag (i.e., FLAG-tag)48 to self-assembling motifs49–50 yields a precursor (9) as the substrate of enterokinase (ENTK) (Figure 6A).47 TEM shows that 9 self-assembles to form micelles, which then transform into the nanofibers of 10 after ENTK clips the branch off 9 (Figure 6B). After being taken up by cells, the micelles of 9, upon the action of intracellular ENTK, turn into nanofibers, mainly located at mitochondria (Figure 6C).47 The micelles of the precursors can deliver cargos (such as plasmids) into cells for transfection of proteins inside mitochondria (Figure 6D,E).51 As the first-in-kind studies of using EISA for targeting mitochondria and delivering cargos to mitochondria, these works illustrate EISA as a fundamentally new way to target subcellular organelles for biomedicine.
Figure 6.
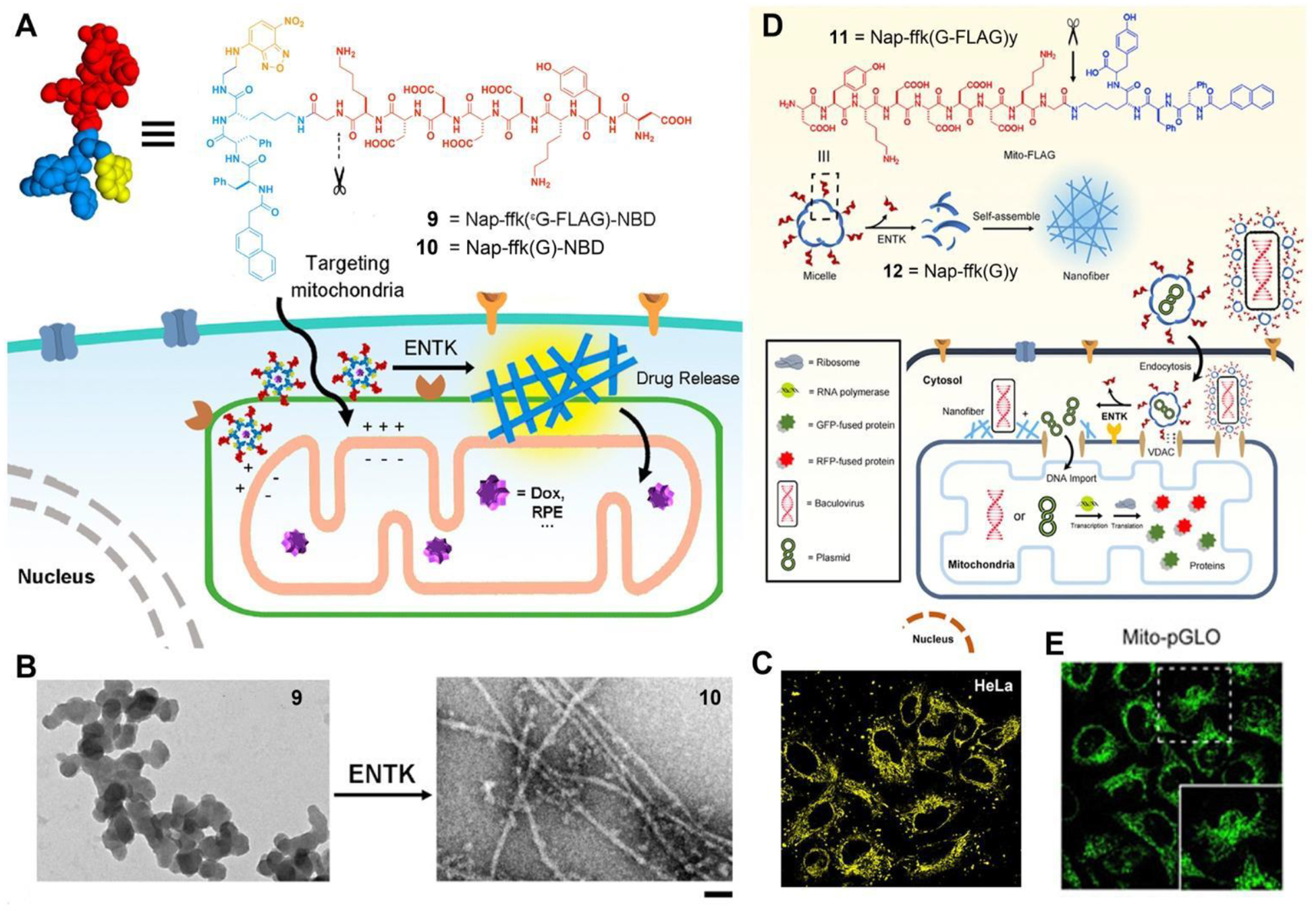
(A) Structures of 9 and 10. (B) TEM of 9 before (left) and after (right) adding ENTK, bar = 100 nm. (C) Fluorescent images of HeLa cells incubated with 9 for 2 h. (D) Structure of 11 and illustration of the proteolysis (ENTK cleaving off the Flag-tag) of 11 to result in supramolecular assemblies of Nap-ffk(G)y (12) and the consequential phase/morphology transition on mitochondria to facilitate the mitochondrial genetic engineering. (E) Fluorescent images of the HeLa cells incubated with Mito-pGLO plasmid in the presence of 11. Adapted from Ref.47 Copyright ACS 2018 and Ref.51.
3.2. Targeting Endoplasmic Reticulum
Endoplasmic reticulum (ER) is involved in various crucial cellular processes, making it a potential target for cancer therapy. However, current strategies for selectively targeting the ER of cancer cells are limited. We have shown that the peptide assemblies interact with cell membranes, leading to membrane disruption, generating ER stress, and ultimately inducing cancer cell death.52 Specifically, the precursor for EISA is a phosphotetrapeptide (13), which, upon dephosphorylation by ALP, transforms into a tetrapeptide derivative (14) (Figure 7A). Peptide 14 self-assembles into crescent-shaped aggregates (Figure 7B). It appears to be crucial to attach an L-amino acid at the C-terminal of a D-tripeptide for obtaining the crescent-shaped morphology of the assemblies. The peptide crescent-shaped assemblies on the cancer cell surface and interact with the lipid membrane, causing membrane integrity impairment and leakage. Once internalized by the cancer cells, these assemblies accumulate in the ER, leading to ER stress (Figure 7C) and activating the caspase signaling cascade, resulting in cancer cell death (Figure 7D). Fluorescent imaging also indicates an even dispersion of nanoparticles within the cytoplasm, likely due to the endosomal escape of these nanoparticles, while the details of this process remain to be elucidated. This unique shape was essential for selectively inhibiting cancer cells, as it facilitated efficient membrane disruption and leakage, leading to cancer cell death. The work demonstrates a reaction-based process that allows controlled disruption of membranes in a spatiotemporally controlled manner and offers a new concept in controlling cell fates via EISA.
Figure 7.
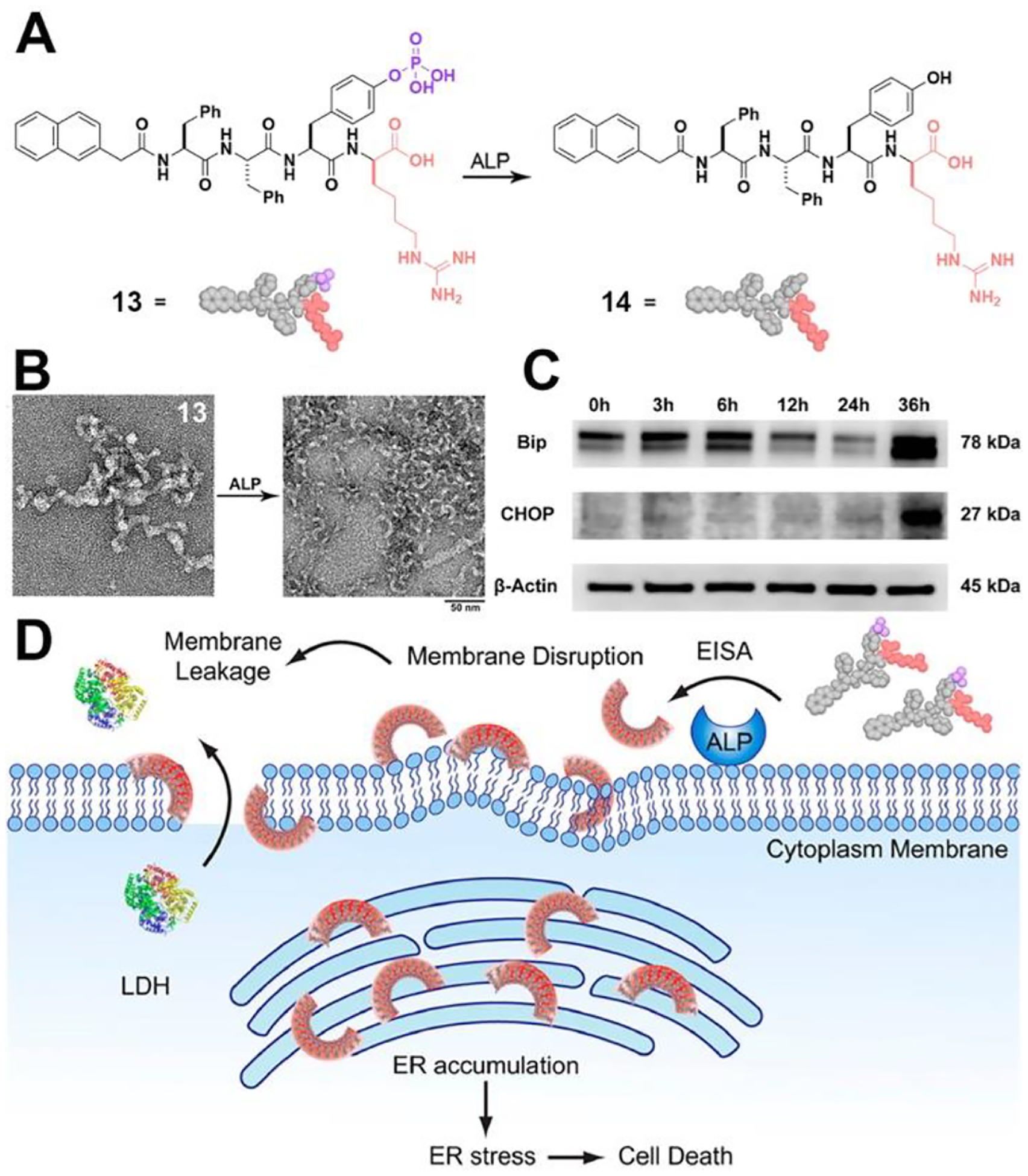
(A) Structures of 13 and 14. (B) HRTEM image of nanostructures formed before and after adding ALP to the solution of 13. (C) Western blot analysis of ER-stress marker (Bip, CHOP) after treating HeLa cells with 13 (50 μM) at different time (i.e., 0, 3, 6, 12, 24 or 36 h). (D) Illustration of EISA assemblies to disrupt cell membrane and to target ER and molecular structure of the EISA precursor (13). Adapted from Ref.52 Copyright ACS 2018.
3.3. Targeting Golgi Apparatus
The Golgi apparatus (GA) is a crucial signaling hub in cells and a promising target for cancer therapy. However, there are scarce approaches for specifically targeting Golgi and selectively killing cancer cells. We found that by replacing an oxygen atom with a sulfur atom in phosphopeptides, they could instantaneously target the Golgi apparatus and selectively eliminate cancer cells through EISA.53 A thiophosphopeptide (15, Figure 8A) serves as a substrate for ALP, which catalyzes rapid dephosphorylation, resulting in the formation of a thiopeptide (16) that self-assembles. Remarkably, the thiophosphopeptide is efficiently internalized into cells through caveolin-mediated endocytosis and macropinocytosis, rapidly accumulating within the Golgi apparatus (Figure 8B) due to dephosphorylation and plausible disulfide bond formation with Golgi proteins. Moreover, replacing NBD in 15 produce another thiophosphopeptide (17), which exhibits potent and selective inhibition of cancer cells, such as HeLa cells, with an IC50 (half-maximal inhibitory concentration) of approximately 3 μM. This potency is significantly higher than that of the parent phosphopeptide. This work represents the first use of thiopeptide for targeting the Golgi and demonstrates enzyme and redox responsive molecules that can target specific subcellular compartments for therapeutic purposes.
Figure 8.
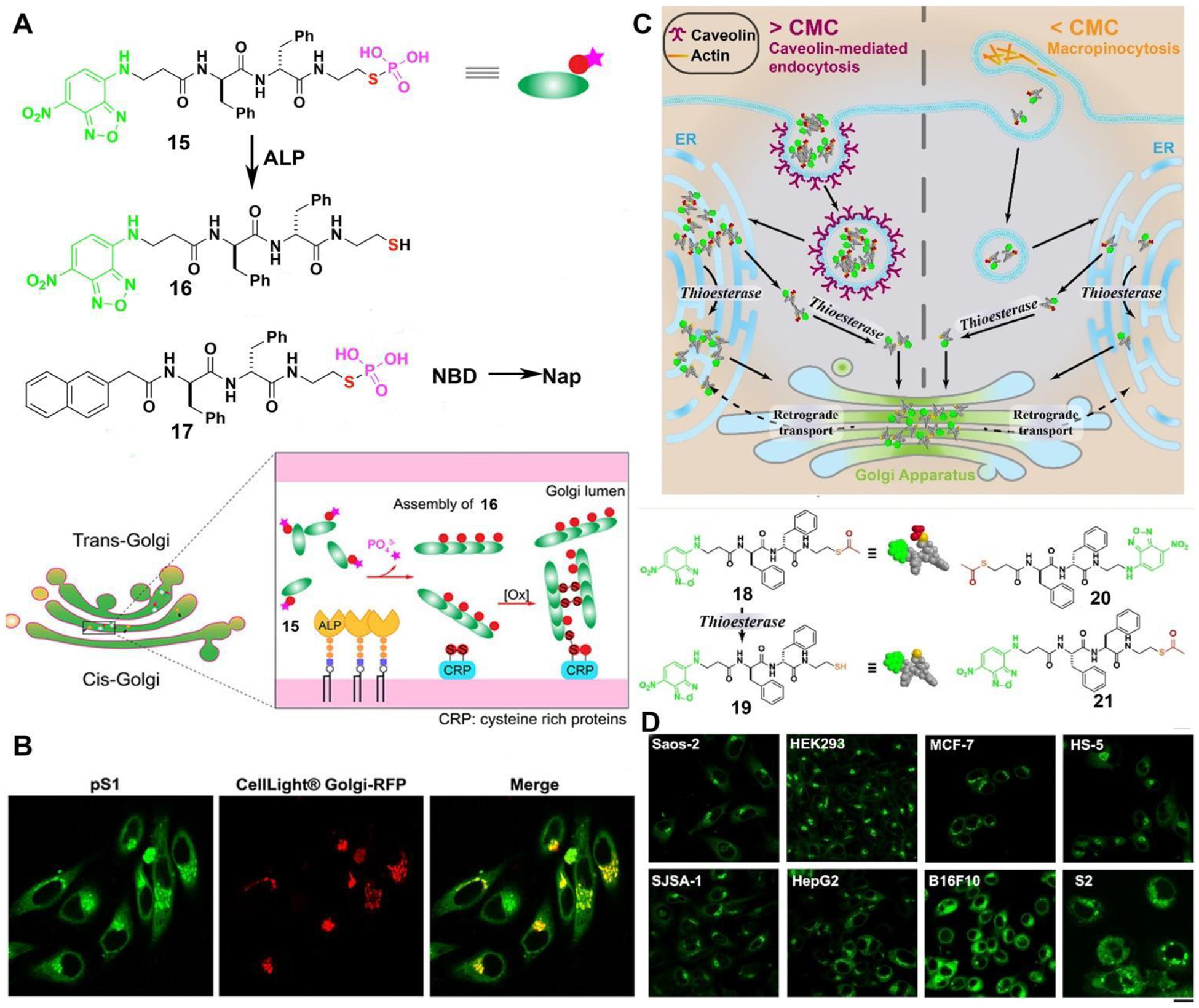
(A) Illustration of thiophosphopeptides instantly targeting the GA by enzymatic assembling and forming disulfide bonds. (B) CLSM images of HeLa cells stained with CellLight® Golgi-RFP after treating with 15 for 8 minutes. (C) Enzyme-responsive peptide thioesters targeting GA. (D) Different cells treated with 18 for 8 min ([18] = 10 μM, scale bar 20 μm). Adapted from Ref.53. Copyright Wiley 2021, and from Ref.58 Copyright ACS 2022.
In a subsequent investigation focusing on the control molecules of 15, we uncovered a novel type of peptide thioesters (18, 20, 21).54 These thioesters have the remarkable capability of instantaneously targeting the GA within cells. The peptide thioesters, whether above or below their critical micelle concentrations, can effectively enter cells through two distinct mechanisms: caveolin-mediated endocytosis and micropinocytosis (Figure 8C). Once inside the cells, these peptide thioesters are hydrolyzed by GA-associated thioesterases (e.g., PPT1,55 LYPLA1,56 or LYPLA2,57), leading to the formation of thiopeptide dimers that accumulate in the GA. Eventually, the GA becomes saturated, causing the thiopeptides to concentrate in the endoplasmic reticulum (ER). These peptide thioesters can specifically target the GA in various types of cells, including human, murine, and Drosophila cells (Figure 8D). Moreover, the accumulation of the thiopeptides at GA disrupts protein trafficking (such as oncoprotein NRAS in MCF-7 cell), triggering cell death through multiple pathways, and yet still selectively inhibit cancer cells without harming hepatocytes.58 This study demonstrates an novel molecular platform that is both thioesterase-responsive and redox-active, providing a promising means to target the GA and control cell fates effectively.
3.4. Targeting Lysosome
Lysosomes, containing more than 60 different enzymes and over 50 membrane proteins, serve as crucial sites for biomolecular degradation within cells. Lysosomes also play essential roles in various physiological functions.59 Thus, selectively targeting lysosome promises a new way to modulate cellular functions. In a recent report,60 Wang et al. introduced an innovative design of an EISA substrate (22) to achieve precise targeting lysosome of cells (Figure 9A). This substrate utilized a protected tyrosine phosphate to prevent undesired dephosphorylation at the cell membrane and cytoplasm (Figure 9B), while facilitating acid phosphatase (ACP) catalyzed dephosphorylation at lysosomes (Figure 9C). At pH 5.0, 22 turns into 23. Subsequently, ACP in lysosomes catalyzed the dephosphorylation of 23 at pH 5.0, yielding 24, which further self-assembled into nanofibers. TEM revealed that 22 displayed minimal observable nanostructures at pH 7.4, but it formed short and discrete nanofibers of approximately 3.7 nm in diameter. Over 24 hours (or longer) of ACP addition at pH 5.0, thicker nanofibers with a diameter of 6.7 nm were formed (Figure 9D). Using fluorescent analogs, Wang et al. confirmed that 22 localized to lysosomes (Figure 9E), while 23 led to pericellular localization. The study also found that protecting tyrosine phosphate in 22 reduced the cytotoxicity of the EISA precursors, highlighting the potential of this approach for spatiotemporal control of peptide assemblies. Interestingly it is also feasible to achieve lysosomal targeting by using tryptophane to replace the phenylalanine in 1.61 Considering the diversity of enzymes and their substrates in lysosomes, these works illustrate that the combination of EISA and molecular engineering promise precise targeting lysosomes with cell selectivity.
Figure 9.
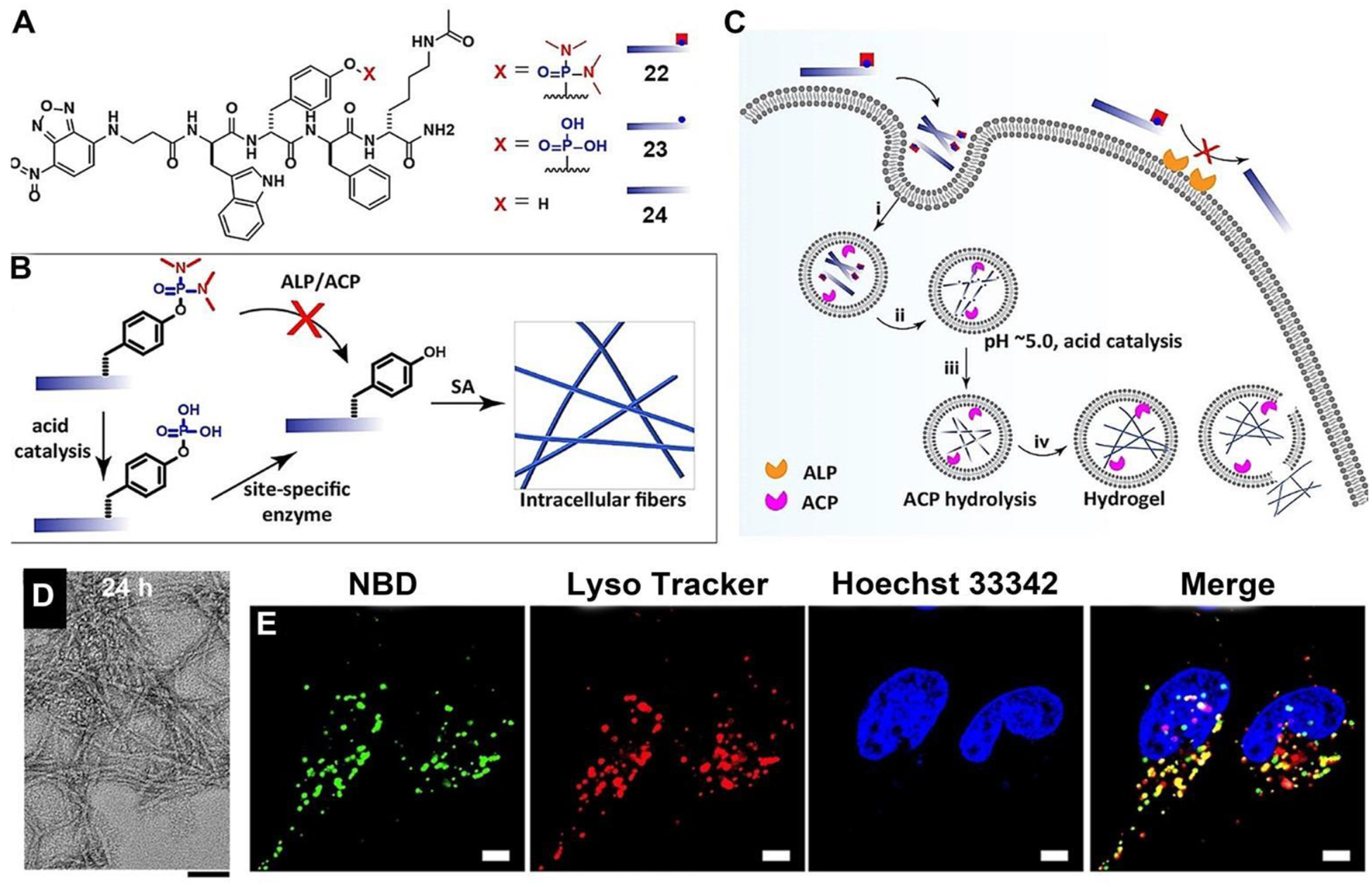
(A) Structures and (B) illustration of multistage chemical reactions in vitro. (C) Illustration of the site-specific construction of nanofibers in living cells through multistage processes for targeting lysosomes. (D) Cryo-EM images of 22 incubated with ACP at pH 5.0 for 24 h. Bar = 50 nm. (E) CLSM images of Saos-2 cells incubated with 22 (100 μM, 4h). Bar = 5 μm. Adapted from Ref.60 Copyright Wiley 2022
3.5. Targeting Nucleus
The nucleus is the largest and probably the most important membrane-bound organelle in a eukaryotic cell. Thus, targeting the nucleus is of pivotal importance. While there are nuclear localization sequences (NLS), incorporating NLS into synthetic molecules has produced mixed results due to NLS being subject to proteolysis. Surprisingly, EISA of peptides can provide an effective way to target the nucleus of cells overexpressing ALP.62–63 An L-phosphopentapeptide (25), upon dephosphorylation catalyzed by ALP, turns into the self-assembling peptide (26) (Figure 10A). TEM reveals that at 400 μM and in PBS, 25 self-assembles to form short nanofibers with a diameter of 9 ± 2 nm and a few nanoparticles. ALP turns the nanoparticles into nanoribbons with widths of 74 ± 13 nm (Figure 10B). The morphology of the pentapeptide assemblies of 26 is dependent on the concentration of ALP. High expression level (800 U/L) of ALP leads to nanoribbon formation, while normal expression level (100 U/L) results in nanofibers, depending on the incubation time.
Figure 10.
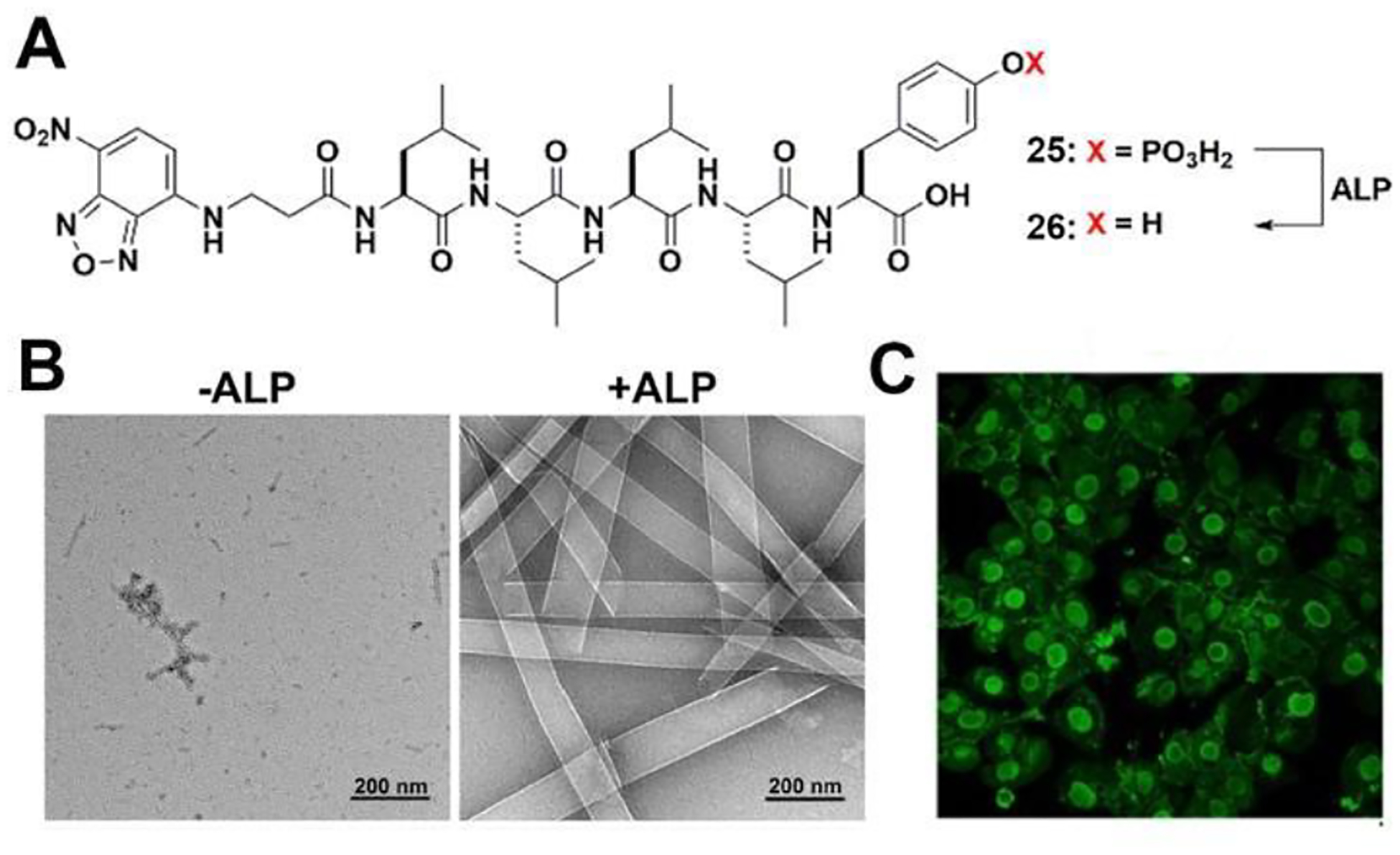
(A) Structures of 25 and 26. (B) TEM imaging of 25 (400 μM, PBS) and the corresponding 26 formed by adding ALP (0.5 U/mL) for 24 h, (C) CLSM images of iPS cells after being treated by 25 (400 μM) for 2 h. Adapted from Ref.62 Copyright ACS 2022.
Incubating 25 with human induced pluripotent stem cells (iPSCs) results in their rapid and selective killing (within 2 hours) as the pentapeptide assemblies accumulate in the iPSC nuclei (Figure 10C). Importantly, these phosphopentapeptides are innocuous to normal cells such as HEK293 and hematopoietic progenitor cells (HPC). Inhibiting ALP abolishes the formation of intranuclear assemblies. This work represents the first instance of intranuclear assemblies of peptides and highlights the application of EISA for selectively targeting the nuclei of cells.63
4. Transcytosis of Peptide Assemblies
A perplexing and probably one of the most important features of high-ordered protein structures is their context dependent functions. EISA of peptide assemblies is able to recapitulate such a context dependency, as shown in a dynamic continuum of peptide assemblies that can kill osteosarcoma cells, but induce cell spheroids of fibroblast cells.64 Although previous studies have reported several examples of incorporating bioactive molecules (vancomycin64–65 and biotin66) and EISA of short peptides for generating cell spheroids from monolayer of cells, the mechanism of the ability of EISA to induce cell morphogenesis remains elusive until the recent study67 to reveal that the transcytosis of peptide assemblies created by EISA.
Specifically, a protease-resistant D-peptide, NBD-ffspy (27) undergoes transcytosis (endocytosis and exocytosis) and intracellular dephosphorylation during cellular trafficking to generate intercellular hydrogels that colocalize with fibronectin to enable cell spheroids (Figure 11A). In this process, 27 undergoes partial dephosphorylation to generate NBD-ffsy (28) (Figure 11B), which results in morphological transformation to form nanofibers (Figure 11C) and enzymatic hydrogelation in PBS (Figure 11D). Cryo-EM reveals polymorphism among the filaments of 28. Both classes of filaments display parallel cross-β packing; however, they differ in the arrangement of 28 molecules. 10 copies of 28 pack in class 1 filaments, while the class 2 filament possesses C3 symmetry, and each asymmetrical unit contains 6 copies of 28 (Figure 11E–F). On the other hand, 30 packs into homogenous cross-β filaments with C3 symmetry (Figure 11G).
Figure 11.
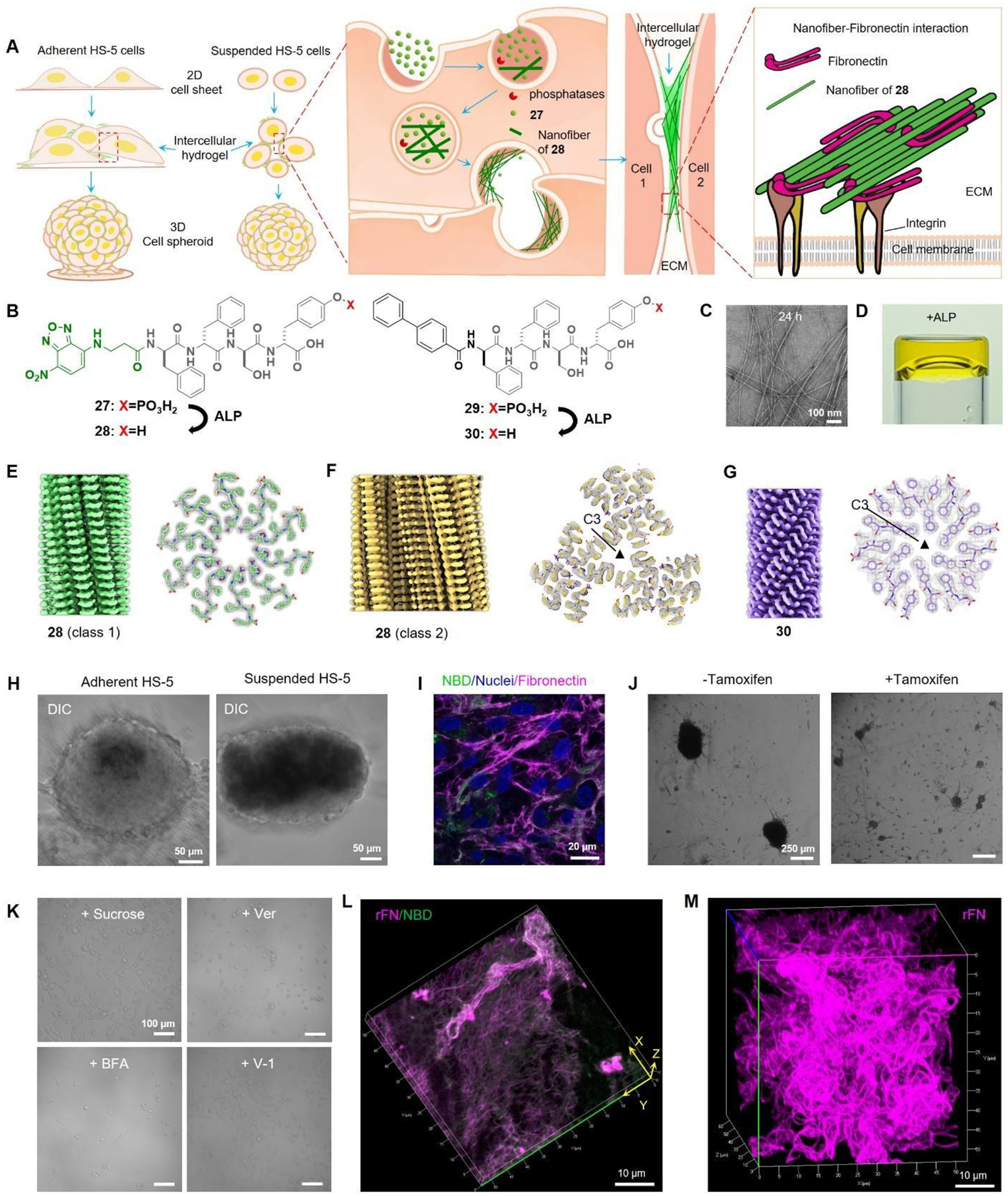
(A) Illustration of the transcytotic dephosphorylation of 27 forming intercellular hydrogels that colocalize with fibronectin to enable spheroids. (B) Structures of 27, 28 and 29. (C) TEM images of 27. (D) ALP converts 27 to gel. 3D reconstruction and cross section of (E) 28 (class 1), (F) 28 (class 2), and (G) 29 filaments. (H) Adherent or suspended HS-5 cells treated with 27 to form spheroids. (I) Immunofluorescence staining of fibronectin in HS-5 cells treated with 27. (J) Suspended TKO cells with or without tamoxifen knockout incubated with 27. (K) Suspended HS-5 cells under different conditions (27 at 200 μM): + Sucrose (sucrose pre-treated cells + 27); + Ver (27 + 50 μM Verapamil); + BFA (27 + 300 nM Brefeldin A); + V-1 (27 + 10 μM Vacuolin-1). (L) 3D rendering of 27 treated with ALP and then incubated with rhodamine-fibronectin (rFN). (M) 3D rendering of 29 treated with ALP and then incubated with rFN. Adapted from Ref.67 Copyright Springer 2023.
Incubating with HS-5 cells, 27 turns both suspended and adherent cells into spheroids within 24 hours (Figure 11H), with the intercellular assemblies colocalization with fibronectin (Figure 11I). With biphenyl, 29 exhibits more potent spheroid-inducing effects (Figure 11B). Minimizing endocytosis by tamoxifen-induced dynamin knockout (Figure 11J) and inhibiting exocytosis via distinct inhibitors undermine the spheroids generation (Figure 11K), suggesting the crucial role of transcytosis in this process. Moreover, incubating 27 with ALP generates filaments of 28. These filaments transform the globular fibronectin into a fibrillar morphology, accompanied by colocalization between the filaments and fibronectin (Figure 11L). In the case of the more potent analog, 29, the filaments induced by ALP (30) lead to the formation of curly fibronectin fibers that entangle into a three-dimensional network (Figure 11M). This study illustrates the dynamic nature of EISA of peptide assemblies, which enables this subtle transcytotic process. This study also confirms the fibers formed by the assemblies of a D-peptide are able to induce the fibrillation of fibronectins.
5. Summary and Outlook
In the last two decades, intracellular peptide assemblies have emerged as a promising class of molecules with potential applications in developing biomedicine, especially in anticancer therapeutics. As shown in this review, enzymatic reactions offer an endogenous control to generate intracellular, yet exogeneous, peptide assemblies in vivo. The concepts and promises illustrated by these examples should be applicable for generating intracellular supramolecular assemblies by other means, such as chemical reactions and physical stimuli.41 Despite the rapid advancements, there are several limitations that need to be addressed to fully capitalize the potential of enzyme-instructed intracellular peptide assemblies. These limitations include issues related to efficiency, atomistic structures, and mechanisms. Regarding efficiency, recent efforts have identified peptide assemblies, such as assemblies of 6 (formed in-situ by EISA), that exhibit significant inhibitory effects against osteosarcoma tumors in murine models. However, further molecular engineering is necessary to develop EISA precursors that are responsive to a broad range of enzymes and exhibit nanomolar-level inhibitory concentrations. Achieving this goal requires an atomic-level structural understanding of the peptide assemblies. While the resolution revolution of cryo-electron microscopy (cryo-EM) has shown promise in resolving the structures of peptide nanofibers, an approach to determine the structures of non-helical peptide assemblies would be more useful and remains to be developed. Utilizing cryo-electron tomography (cryo-ET) could be the holy grail in gaining a deeper understanding of how these intracellular assemblies interact with other biomacromolecules, particularly inside cells. The dynamic nature of intracellular peptide assemblies presents a challenge in understanding their cellular mechanisms. For example, there is a need to elucidate how the intracellular peptide assemblies pleiotropically modulate cellular signaling pathway for controlling the cell fates and what are the molecular interactions between the intracellular peptide assemblies and protein assemblies of cells. These unresolved issues are crucial for developing enzyme-instructed intracellular peptide assemblies to modulate cell behaviors.
The above challenges also provide important goals and exciting opportunities to explore enzyme-instructed intracellular peptide assemblies. Prior studies, both in cell cultures and animal models, have already demonstrated the success and versatility of EISA in generating supramolecular nanostructures in living organisms. EISA is emerging as an approach that has shown great potential in various biomedical applications, such as targeted drug delivery, regenerative medicine, and diagnostics. Further applying this simple concept at the intersection of chemistry and cell biology would be fruitful and likely lead to bountiful discoveries. The key to future success and discovery lies in engineering molecules for EISA inside cells or in animals. The potential to fine-tune and customize these assemblies for specific purposes makes EISA a highly attractive field of study with numerous practical applications in chemistry, biology, medicine, and other disciplines.
Acknowledgement:
This work was partially supported by NIH (CA142746 and CA262920) and NSF (DMR-2011846).
Biographies
Zhiyu Liu received his BS and MS degrees from the school of biomedical engineering at Southeast University in 2020 and 2022, respectively. He is currently in his second year as a Ph. D student majoring in chemistry at Brandeis University, supervised by Prof. Bing Xu. His research focuses on enzyme-instructed peptide assemblies and their applications in biomaterials.
Jiaqi Guo received her B.S. degree in chemical biology from Xiamen University in 2018. She joined the laboratory of Prof. Bing Xu at Brandeis University in the same year. Her research focuses on stimuli-responsive peptide assemblies and their emergent properties in cells.
Yuchen Qiao obtained her BS degree from the school of biomedical engineering at Southeast University, China. She is currently in her third year as a graduate student in chemistry supervised by Professor Bing Xu at Brandeis University. Her current research interest lies in the design of self-assembling materials for biological applications.
Bing Xu received his BS and MS degrees from Nanjing University in 1987 and 1990, respectively. He obtained his PhD in 1996 from the University of Pennsylvania. Prior to beginning his independent research at the Hong Kong University of Science and Technology (HKUST) in 2000, he worked as an NIH postdoctoral fellow at Harvard University. He was tenured as an associate professor in January 2006 and became a full professor in July 2008 at HKUST. Currently, he is a professor at the Department of Chemistry, Brandeis University. His research focuses on the applications of enzymatic noncovalent synthesis in materials, biology, and medicine.
References
- 1.Yang Z; Gu H; Fu D; Gao P; Lam JK; Xu B, Enzymatic Formation of Supramolecular Hydrogels. Adv Mater 2004, 16 (16), 1440–1444. [Google Scholar]; This is the first study that introduces enzymatic reactions triggering self-assembly of synthetic molecules in water, forming the basis of enzyme-instructed self-assembly (EISA), showcasing enzyme-responsive material, and demonstrating non-equilibrium sol-to-gel phase transition.
- 2.Yang Z; Liang G; Guo Z; Guo Z; Xu B, Intracellular hydrogelation of small molecules inhibits bacterial growth. Angew Chem Int Ed 2007, 46 (43), 8216–9. [DOI] [PubMed] [Google Scholar]; The first report of using an enzymatic reaction to trigger the self-assembly of man-made molecules inside living organisms, thus establishing EISA as a first-in-kind, intracellular, supramolecular chemistry methodology to control cell fate.
- 3.Gao Y; Shi J; Yuan D; Xu B, Imaging enzyme-triggered self-assembly of small molecules inside live cells. Nat Commun 2012, 3, 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work reports imaging enzyme-triggered self-assembly of small molecules inside live cells, illustrating a simple and versatile way to explore supramolecular chemistry inside cells and at the interface of chemistry and biology.
- 4.Feng Z; Han X; Wang H; Tang T; Xu B, Enzyme-Instructed Peptide Assemblies Selectively Inhibit Bone Tumors. Chem 2019, 5 (9), 2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work reports on enzyme-instructed assembly of peptides for selectively inhibiting tumors that overexpress ALP, an “undruggable” target that results in immunosuppression in solid tumors, establishing the use of enzyme substrates to generate intracellular peptide assemblies as potential anticancer therapeutics.
- 5.Van Meer G; Voelker DR; Feigenson GW, Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Bio 2008, 9 (2), 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugyi B; Carlier MF, Control of actin filament treadmilling in cell motility. In Annu Rev Biophys, 2010; Vol. 39, pp 449–470. [DOI] [PubMed] [Google Scholar]
- 7.Lazarides E, Intermediate filaments as mechanical integrators of cellular space. Nature 1980, 283 (5744), 249–255. [DOI] [PubMed] [Google Scholar]
- 8.Al-Bassam J; Chang F, Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol 2011, 21 (10), 604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H; Fuxreiter M, The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell 2016, 165 (5), 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du M; Chen ZJ, DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361 (6403), 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He H; Tan W; Guo J; Yi M; Shy AN; Xu B, Enzymatic Noncovalent Synthesis. Chem Rev 2020, 120 (18), 9994–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powers RE; Wang S; Liu TY; Rapoport TA, Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 2017, 543, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J; Prinz WA; Rapoport TA, Weaving the web of ER tubules. Cell 2011, 147 (6), 1226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y; Wang H; Kouadir M; Song H; Shi F, Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death & Disease 2019, 10 (2), 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z; Liang G; Xu B, Enzymatic hydrogelation of small molecules. Acc Chem Res 2008, 41 (2), 315–26. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J; Xu B, Enzyme-instructed self-assembly: a multistep process for potential cancer therapy. Bioconjug Chem 2015, 26 (6), 987–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y; Yang Z; Kuang Y; Ma M-L; Li J; Zhao F; Xu B, Enzyme-instructed self-assembly of peptide derivatives to form nanofibers and hydrogels. Biopolymers 2010, 94 (1), 19–31. [DOI] [PubMed] [Google Scholar]
- 18.Toledano S; Williams RJ; Jayawarna V; Ulijn RV, Enzyme-Triggered Self-Assembly of Peptide Hydrogels via Reversed Hydrolysis. Journal of the American Chemical Society 2006, 128 (4), 1070–1071. [DOI] [PubMed] [Google Scholar]
- 19.Ghadiri MR; Granja JR; Milligan RA; McRee DE; Khazanovich N, Self-assembling organic nanotubes based on a cyclic peptide architecture. Nature 1993, 366 (6453), 324–327. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S; Holmes T; Lockshin C; Rich A, Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proceedings of the National Academy of Sciences of the United States of America 1993, 90 (8), 3334–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berndt P; Fields GB; Tirrell M, Synthetic Lipidation of Peptides and Amino Acids: Monolayer Structure and Properties. J Am Chem Soc 1995, 117 (37), 9515–9522. [Google Scholar]
- 22.Cha X; Ariga K; Onda M; Kunitake T, Molecular Recognition of Aqueous Dipeptides by Noncovalently Aligned Oligoglycine Units at the Air/Water Interface. J Am Chem Soc 1995, 117 (48), 11833–11838. [Google Scholar]
- 23.Aggeli A; Nyrkova IA; Bell M; Harding R; Carrick L; McLeish TCB; Semenov AN; Boden N, Hierarchical self-assembly of chiral rod-like molecules as a model for peptide β-sheet tapes, ribbons, fibrils, and fibers. Proceedings of the National Academy of Sciences of the United States of America 2001, 98 (21), 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartgerink JD; Beniash E; Stupp SI, Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 2001, 294 (5547), 1684–1688. [DOI] [PubMed] [Google Scholar]
- 25.Reches M; Gazit E, Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300 (5619), 625–627. [DOI] [PubMed] [Google Scholar]
- 26.Xing B; Yu CW; Chow KH; Ho PL; Fu D; Xu B, Hydrophobic interaction and hydrogen bonding cooperatively confer a vancomycin hydrogel: a potential candidate for biomaterials. J Am Chem Soc 2002, 124 (50), 14846–7. [DOI] [PubMed] [Google Scholar]
- 27.Silva GA; Czeisler C; Niece KL; Beniash E; Harrington DA; Kessler JA; Stupp SI, Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science 2004, 303 (5662), 1352–5. [DOI] [PubMed] [Google Scholar]
- 28.Schneider JP; Pochan DJ; Ozbas B; Rajagopal K; Pakstis L; Kretsinger J, Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc 2002, 124 (50), 15030–7. [DOI] [PubMed] [Google Scholar]
- 29.Majumder P; Singh A; Wang Z; Dutta K; Pahwa R; Liang C; Andrews C; Patel NL; Shi J; de Val N; Walsh STR; Jeon AB; Karim B; Hoang CD; Schneider JP, Surface-fill hydrogel attenuates the oncogenic signature of complex anatomical surface cancer in a single application. Nat Nanotechnol 2021, 16 (11), 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang ZM; Xu KM; Guo ZF; Guo ZH; Xu B, Intracellular Enzymatic Formation of Nanofibers Results in Hydrogelation and Regulated Cell Death. Adv Mater 2007, 19 (20), 3152–3156. [Google Scholar]
- 31.Jiang Q; Zhan W; Liu X; Bai L; Wang M; Xu Y; Liang G, Assembly drives regioselective azide-alkyne cycloaddition reaction. Nat Commun 2023, 14 (1), 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall LJ; Wallace M; Mahmoudi N; Ciccone G; Wilson C; Vassalli M; Adams DJ, Hierarchical Composite Self-Sorted Supramolecular Gel Noodles. Adv Mater 2023, 35 (17), e2211277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita K; Nishimura K; Yamamoto S; Shimizu N; Yashiro T; Kawabata R; Aoi T; Tamura A; Maruyama T, In Situ Synthesis of an Anticancer Peptide Amphiphile Using Tyrosine Kinase Overexpressed in Cancer Cells. JACS Au 2022, 2 (9), 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F; Huang Q; Su H; Sun M; Wang Z; Chen Z; Zheng M; Chakroun RW; Monroe MK; Chen D; Wang Z; Gorelick N; Serra R; Wang H; Guan Y; Suk JS; Tyler B; Brem H; Hanes J; Cui H, Self-assembling paclitaxel-mediated stimulation of tumor-associated macrophages for postoperative treatment of glioblastoma. Proc Natl Acad Sci U S A 2023, 120 (18), e2204621120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z; Liang G; Wang L; Xu B, Using a kinase/phosphatase switch to regulate a supramolecular hydrogel and forming the supramolecular hydrogel in vivo. J Am Chem Soc 2006, 128 (9), 3038–43. [DOI] [PubMed] [Google Scholar]
- 36.Simopoulos TT; Jencks WP, Alkaline phosphatase is an almost perfect enzyme. Biochemistry 1994, 33 (34), 10375–80. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan MM, Alkaline phosphatase. N Engl J Med 1972, 286 (4), 200–2. [DOI] [PubMed] [Google Scholar]
- 38.Vijayan D; Young A; Teng MWL; Smyth MJ, Targeting immunosuppressive adenosine in cancer. Nature Reviews Cancer 2017, 17 (12), 709–724. [DOI] [PubMed] [Google Scholar]
- 39.Fonta C; Négyessy L, Neuronal Tissue-Nonspecific Alkaline Phosphatase (TNAP). Springer, 2015. [Google Scholar]
- 40.Millán JL, Mammalian Alkaline Phosphatases: From Biology to Applications in Mdicine and Biotechnology. John Wiley & Sons: 2006. [Google Scholar]
- 41.Chagri S; Ng DYW; Weil T, Designing bioresponsive nanomaterials for intracellular self-assembly. Nat Rev Chem 2022, 6 (5), 320–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang G; Ren H; Rao J, A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat Chem 2010, 2 (1), 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pieszka M; Han S; Volkmann C; Graf R; Lieberwirth I; Landfester K; Ng DYW; Weil T, Controlled Supramolecular Assembly Inside Living Cells by Sequential Multistaged Chemical Reactions. J Am Chem Soc 2020, 142 (37), 15780–15789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Z; Wang H; Wang F; Oh Y; Berciu C; Cui Q; Egelman EH; Xu B, Artificial Intracellular Filaments. Cell Rep Phys Sci 2020, 1 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferri KF; Kroemer G, Organelle-specific initiation of cell death pathways. Nat Cell Biol 2001, 3 (11), E255–63. [DOI] [PubMed] [Google Scholar]
- 46.Murphy MP, Selective targeting of bioactive compounds to mitochondria. Trends Biotechnol 1997, 15 (8), 326–330. [DOI] [PubMed] [Google Scholar]
- 47.He H; Wang J; Wang H; Zhou N; Yang D; Green DR; Xu B, Enzymatic Cleavage of Branched Peptides for Targeting Mitochondria. J Am Chem Soc 2018, 140 (4), 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopp TP; Prickett KS; Price VL; Libby RT; March CJ; Cerretti DP; Urdal DL; Conlon PJ, A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat. Biotechnol 1988, 6 (10), 1204–1210. [Google Scholar]
- 49.Feng Z; Wang H; Chen X; Xu B, Self-Assembling Ability Determines the Activity of Enzyme-Instructed Self-Assembly for Inhibiting Cancer Cells. J Am Chem Soc 2017, 139 (43), 15377–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y; Kuang Y; Gao Y; Xu B, Versatile small-molecule motifs for self-assembly in water and the formation of biofunctional supramolecular hydrogels. Langmuir 2010, 27 (2), 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He H; Lin X; Wu D; Wang J; Guo J; Green DR; Zhang H; Xu B, Enzymatic Noncovalent Synthesis for Mitochondrial Genetic Engineering of Cancer Cells. Cell Rep Phys Sci 2020, 1 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Z; Wang H; Wang S; Zhang Q; Zhang X; Rodal AA; Xu B, Enzymatic Assemblies Disrupt the Membrane and Target Endoplasmic Reticulum for Selective Cancer Cell Death. J Am Chem Soc 2018, 140 (30), 9566–9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan W; Zhang Q; Wang J; Yi M; He H; Xu B, Enzymatic Assemblies of Thiophosphopeptides Instantly Target Golgi Apparatus and Selectively Kill Cancer Cells*. Angew Chem Int Ed 2021, 60 (23), 12796–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi M; Wang F; Tan W; Hsieh JT; Egelman EH; Xu B, Enzyme Responsive Rigid-Rod Aromatics Target “Undruggable” Phosphatases to Kill Cancer Cells in a Mimetic Bone Microenvironment. J Am Chem Soc 2022, 144 (29), 13055–13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Camp LA; Verkruyse LA; Afendis SJ; Slaughter CA; Hofmann SL, Molecular cloning and expression of palmitoyl-protein thioesterase. J Biol Chem 1994, 269 (37), 23212–23219. [PubMed] [Google Scholar]
- 56.Duncan JA; Gilman AG, A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21RAS. J Biol Chem 1998, 273 (25), 15830–15837. [DOI] [PubMed] [Google Scholar]
- 57.Toyoda T; Sugimoto H; Yamashita S, Sequence, expression in Escherichia coli, and characterization of lysophospholipase II. Biochim. Biophys. Acta. Mol. Cell. Biol. Lipids 1999, 1437 (2), 182–193. [DOI] [PubMed] [Google Scholar]
- 58.Tan W; Zhang Q; Quinones-Frias MC; Hsu AY; Zhang Y; Rodal A; Hong P; Luo HR; Xu B, Enzyme-Responsive Peptide Thioesters for Targeting Golgi Apparatus. J Am Chem Soc 2022, 144 (15), 6709–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saftig P; Klumperman J, Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Bio 2009, 10 (9), 623–635. [DOI] [PubMed] [Google Scholar]
- 60.Yang X; Lu H; Tao Y; Zhou L; Wang H, Spatiotemporal Control over Chemical Assembly in Living Cells by Integration of Acid-Catalyzed Hydrolysis and Enzymatic Reactions. Angew Chem Int Ed 2021, 60 (44), 23797–23804. [DOI] [PubMed] [Google Scholar]
- 61.Yang D; Kim BJ; He H; Xu B, Enzymatically Forming Cell Compatible Supramolecular Assemblies of Tryptophan-Rich Short Peptides. Pept Sci (Hoboken) 2021, 113 (2), e24173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu S; Zhang Q; Shy AN; Yi M; He H; Lu S; Xu B, Enzymatically Forming Intranuclear Peptide Assemblies for Selectively Killing Human Induced Pluripotent Stem Cells. J Am Chem Soc 2021, 143 (38), 15852–15862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu S; Zhang Q; He H; Yi M; Tan W; Guo J; Xu B, Intranuclear Nanoribbons for Selective Killing of Osteosarcoma Cells. Angew Chem Int Ed 2022, 61 (44), e202210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H; Feng Z; Xu B, Instructed Assembly as Context-Dependent Signaling for the Death and Morphogenesis of Cells. Angew Chem Int Ed 2019, 58 (17), 5567–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H; Shi J; Feng Z; Zhou R; Wang S; Rodal AA; Xu B, An in situ Dynamic Continuum of Supramolecular Phosphoglycopeptides Enables Formation of 3D Cell Spheroids. Angew Chem Int Ed Engl 2017, 56 (51), 16297–16301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H; Feng Z; Xu B, Intercellular Instructed-Assembly Mimics Protein Dynamics To Induce Cell Spheroids. J Am Chem Soc 2019, 141 (18), 7271–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo J; Wang F; Huang Y; He H; Tan W; Yi M; Egelman EH; Xu B, Cell Spheroid Creation by Transcytotic Intercellular Gelation. Nat Nanotechnol 2023, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]


