Abstract
The aberrant accumulation of tau protein is implicated as a pathogenic factor in many neurodegenerative diseases. Tau seeding may underlie its predictable spread in these diseases. Molecular chaperones can modulate tau pathology, but their effects have mainly been studied in isolation. This study employed a semi-high throughput assay to identify molecular chaperones influencing tau seeding using Tau RD P301S FRET Biosensor cells, which express a portion of tau containing the frontotemporal dementia-related P301S tau mutation fused to a FRET biosensor. Approximately fifty chaperones from five major families were screened using live cell imaging to monitor FRET-positive tau seeding. Among the tested chaperones, five exhibited significant effects on tau in the primary screen. Notably, three of these were from the DnaJ family. In subsequent studies, overexpression of DnaJA2, DnaJB1, and DnaJB6b resulted in significant reductions in tau levels. Knockdown experiments by shRNA revealed an inverse correlation between DnaJB1 and DnaJB6b with tau levels. DnaJB6b overexpression, specifically, reduced total tau levels in a cellular model with a pre-existing pool of tau, partially through enhanced proteasomal degradation. Further, DnaJB6b interacted with tau complexes. These findings highlight the potent chaperone activity within the DnaJ family, particularly DnaJB6b, towards tau.
Keywords: molecular chaperone, tau, DnaJ
1. Introduction
Tau pathology accumulation is the major driver for more than 20 neurodegenerative diseases, including Alzheimer’s disease (AD) [1–5]. Mutations in the MAPT gene, which encodes the tau protein, can lead to the development of frontotemporal dementia (FTD) [6, 7]. Tau pathology is the most common pathological entity observed in diseased brains, which develops in a distinct and predictable manner [8–10]. Recent research suggests this expected pattern is likely due to the propagation of tau deposits through a prion-like mechanism known as “tau seeding” [11–15]. However, the local accumulation of normal tau by pathogenic tau has been shown to limit this process [16, 17]. In mouse models of tauopathy, tau reduction prevents neuronal loss and mitigates tau accumulation and seeding [18–22]. Given the close association between tau pathology and disease progression, as well as the detectability of tau seeding prior to significant neuronal loss and notable pathology, there is a pressing need to identify regulators of tau seeding and accumulation [15, 23–25]. Such discoveries could pave the way for the development of novel therapeutic targets and strategies to effectively manage tau-related neurodegenerative diseases.
Molecular chaperones are a diverse set of proteins that play a crucial role in maintaining homeostasis and regulating protein quality control [26–28]. In the context of tau, discrete molecular chaperones are involved in the processing of normal and pathological tau in the brain [29–34]. However, in aging and neurodegenerative diseases, like AD, the balance of chaperone levels becomes dysregulated, potentially contributing to the initiation and progression of tau pathogenesis [35–37]. Despite the extensive network of chaperones and overlap in roles, there is still a high degree of functional diversity between and within many chaperone families[38–45]. Work from our group and others has shed light on the distinct interactions between tau and molecular chaperones from several major families, including heat shock protein (Hsp) 90s and Hsp90 cochaperones, Hsp70s, Hsp40s (commonly DnaJs), FK506-binding proteins (FKBPs), and small Hsps (sHsps). Among these, both Hsp90 and Hsp70, two highly abundant chaperones involved in protein triage and folding, are essential for tau processing and stability [30, 46–51]. Although the Hsp70/Hsp90 machinery often works in conjunction with cochaperones to recognize misfolded clients, such as tau, Hsp70 and Hsp90 can also act independently [52]. For example, Hsp90 can regulate tau alone or with cochaperones, such as Carboxy-terminus of Hsc70-interacting protein (CHIP) and the activator of Hsp90 ATPase homolog 1 (Aha1), which alter how Hsp90 modulates tau function and stability [40, 48, 50, 53–57]. While FKBP51 coordinates with Hsp90 to regulate tau, other FKBPs, like FKBP52 and FKBP12, regulate tau independently [33, 58, 59]. Hsp70 typically relies on DnaJ cochaperones for its function but can also affect tau turnover and phosphorylation in an isoform- and ATP-dependent manner [60–63]. The DnaJs, the largest chaperone family, can mitigate tau pathology through refolding, disaggregation, and degradation [29, 30, 32, 64–66]. DnaJC5, also known as cysteine string protein α (CSPα), can promote tau secretion [30, 67]. Lastly, the ATP-independent sHsps, including Hsp22 and Hsp27, act as “first responders” to protein aggregation and have been shown to decrease tau hyperphosphorylation, aggregation, and improve synaptic plasticity in vivo [38, 68–72].
While many chaperones have been previously studied for their effects on tau, individual members and their respective families have been predominantly examined in isolation, with limited comparative analysis across chaperone families. Moreover, the role of molecular chaperones in tau seeding is only beginning to be understood. The Tau RD P301S FRET Biosensor cell model has been used by several groups to study tau seeding in vitro [11, 14, 73, 74], which includes four studies using molecular chaperones [29, 45, 66, 75]. However, these studies are limited to exploring the effects of silenced molecular chaperones, using recombinant tau fibrils formed in the presence of molecular chaperones, or overexpression within a single chaperone family. The effects of high intracellular expression of chaperones from different families on tau seeding has not been investigated.
The present study aimed to directly compare the impact of a wide range of molecular chaperones on tau seeding using a semi-high throughput live-cell quantitative assay. Therefore, Tau RD P301S FRET Biosensor cells were used to assess the effect of chaperones from five major families on tau seeding in vitro. One family in particular was found to be enriched with members that can regulate tau seeding. Follow-up overexpression and knockdown studies were performed to confirm these hits and understand more about their effects on tau. Overall, a new semi-high throughput quantitative method to screen tau regulators was characterized, which led to the identification of novel targets that should be further examined in future work.
2. Results
2.1. Discrete molecular chaperones target tau seeding in vitro
Tau RD P301S FRET Biosensor cells have been widely used as an in vitro model of tau seeding [14, 15, 73, 76]. This HEK293T cell-based model stably expresses the tau repeat domain containing the disease associated mutation P301S, fused to either a cyan or yellow fluorescent protein (CFP/YFP). Under normal cellular conditions these tau proteins remain soluble with low background signal, however when templated by exogenous tau aggregates, the Tau RD-CFP and Tau RD-YFP proteins multimerize and create a fluorescence resonance energy transfer (FRET) that can be measured. This model has demonstrated high sensitivity to low amounts of tau aggregates, which is increased by utilizing lipofectamine to promote cellular uptake of preformed tau seeds [11, 66]. Tau RD P301S FRET Biosensor cells also have high selectivity to tau as neither tau monomers nor seeds from other pathological proteins result in significant FRET signal [11, 66, 77]. While there are caveats of this model [78], correlations have been made to in vivo studies [11]. Therefore, the Tau RD P301S FRET Biosensor model was selected to screen molecular chaperone regulators of tau seeding. Initially, a robust quantitative screening method was established in a 96-well plate format in which Tau RD P301S FRET Biosensor cells yielded a significant FRET signal when seeded with sonicated recombinant P301L tau fibrils, but not when exposed to vehicle alone (Fig. 1A–C). Imaging over time allows dynamic analysis of tau seeding in live cells. This is advantageous to the typical readout employed for this cell line, flow cytometry, which only allows for static timepoint measurements [11, 66, 79–81].
Figure 1. Schematic of semi-high throughput chaperone screen.
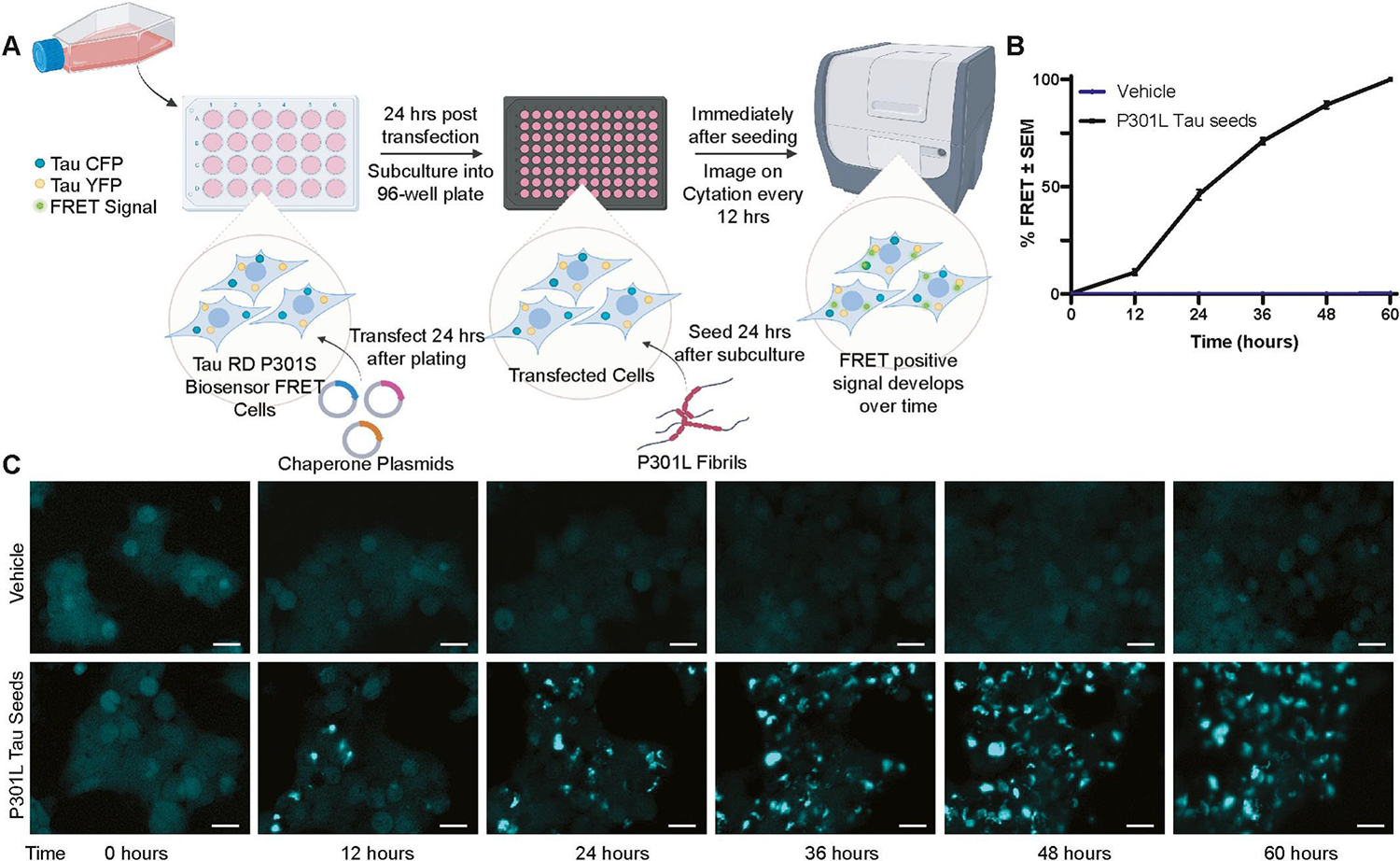
(A) Workflow of the semi-high throughput assay developed to identify molecular chaperones that alter tau seeding. In short, Tau RD P301S FRET Biosensor cells were transfected with chaperone plasmids, subcultured into 96-well plates with 3 wells per chaperone and seeded with sonicated recombinant P301L tau fibrils or vehicle control (100 mM sodium acetate pH 7.0) 48 hours after transfection. (B) Cells were imaged every 12 hours for 60 hours with 4 non-overlapping images per well. Threshold masking was used to measure the FRET intensity within the total cell area at each time point and then normalized to the final time point to calculate %FRET signal. (C) Representative images of FRET signal at each time point are shown from cells treated with vehicle or P301L tau seed. Scale bar = 20 μm. Image created with Biorender.com
Upon validation of the assay, Tau RD P301S FRET Biosensor cells were then transfected with molecular chaperones representing five distinct families and subcultured into 96-well plates, a strategy which aided in cell viability over the course of the assay. After 24 hours, the cells were seeded with sonicated recombinant P301L tau fibrils, and FRET signal was imaged every 12 hours for a total of 60 hours (Fig. S1). Total FRET signal within the cell for each timepoint was analyzed for each chaperone family by repeated measures ANOVA with a Greenhouse-Geisser correction and Dunnett’s post hoc test. A summary of statistical analysis can be found in Table S1. Across different publications, the percentage of FRET-positive cells can vary widely from 5–100% of the cells. This variation is driven by experimental conditions, such as the lipofectamine amount, seed concentration, type of seed, timing, cell density, and whether the readout is based on imaging methods or flow cytometry [79, 82]. In this chaperone screen the percentage of FRET-positive cells at 60 hours was approximately 18±4% across the entire screen. Similar to other screening protocols and for ease of comparison [83], the percentage of FRET-positive cells without chaperone was normalized to 100%. DnaJA2, DnaJB1, DnaJB6b, Hsp90α, and FKBP19 significantly reduced tau seeding (Fig. 2). This was evident from the notable decrease in FRET signal compared to their respective empty vector (EV) controls. Among the chaperone families tested, the DnaJs had the most significant overall impact on reducing tau seeding in vitro (Fig. 2A), while other chaperone families had only single members that reached significance, such as Hsp90α (Fig. 2B) and FKBP19 (Fig. 2C). None of the members from the Hsp70 (Fig. 2D) or sHsp (Fig. 2E) chaperone families had a significant effect on tau seeding in this assay.
Figure 2. Discrete molecular chaperones alter tau seeding in vitro.
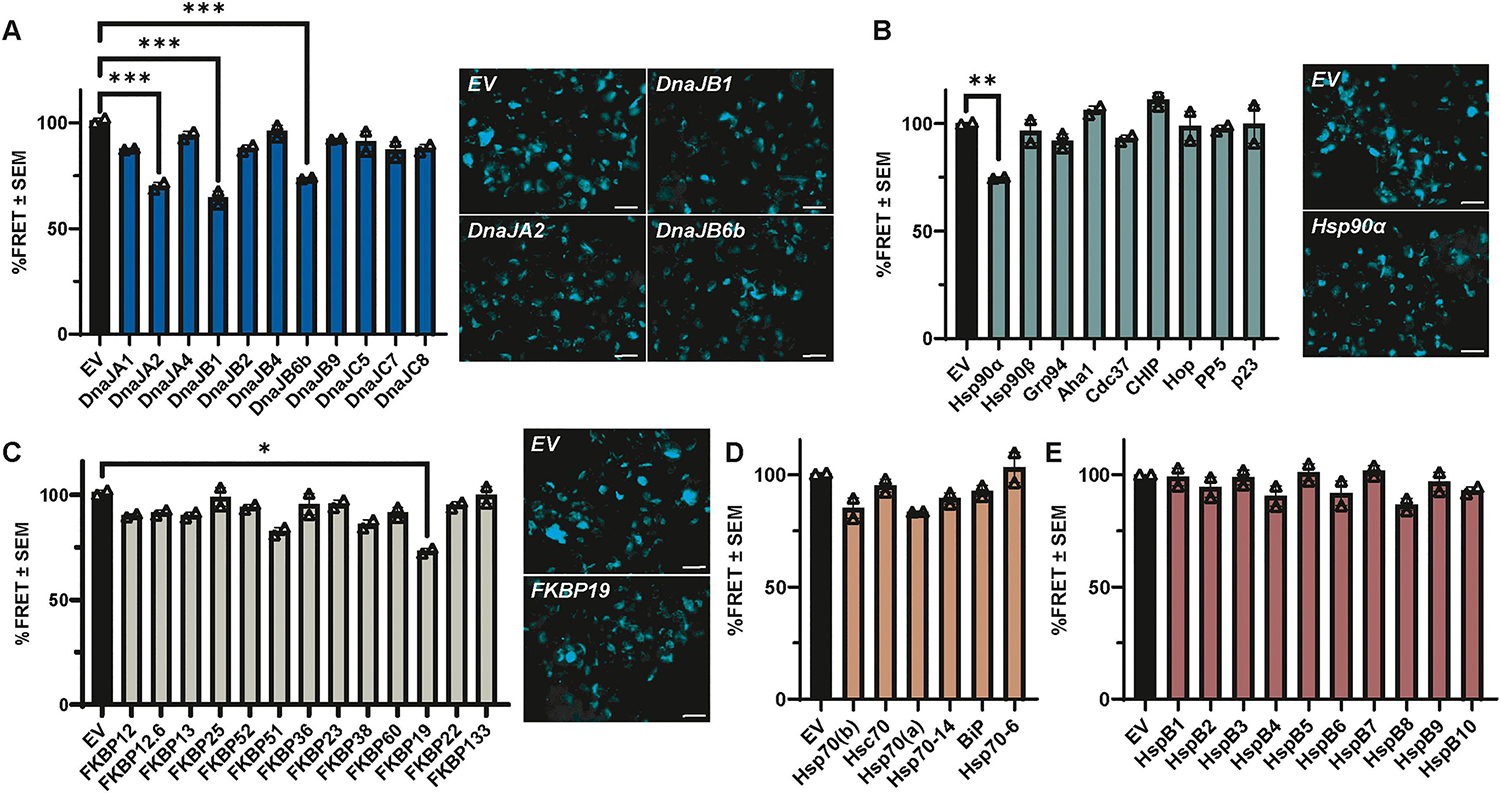
Using the semi-high throughput Tau RD P301S FRET biosensor assay, 49 molecular chaperones from five chaperone families were screened for their effects on tau seeding. For each chaperone, FRET intensity within the total cell area at 60 hours was normalized to EV control to calculate the relative %FRET signal. Bar graphs from the 60-hour timepoint show the average of 2 independent experiments as %FRET ± S.E.M. for chaperone members of the (A) DnaJ family, (B) Hsp90 and Hsp90 cochaperone families, (C) FKBP family, (D) Hsp70 family, and (E) sHsp family compared to their EV control, respectively. Data analysis was performed by repeated measures ANOVA with a Greenhouse-Geisser correction over the course of the experiment (Fig. S1) across each family of chaperones, except for Hsp90 and Hsp90 cochaperones, which were combined. The 60-hour timepoint from Fig. S1 is displayed here for simplicity. Significance indicated as follows: *p<0.05, **p<0.01, ***p<0.001. Representative images of select chaperones found to be significant in the post hoc analysis are shown. Scale bar = 20 μm.
2.2. DnaJB1 and DnaJB6b inversely correlate with tau levels
To investigate whether changes in tau-seeded aggregation could be attributed to alterations in overall tau levels, tau overexpression in HEK293T cells was utilized to measure tau protein under non-aggregation-inducing conditions in the presence or absence of the chaperones that significantly impacted tau seeding in the FRET screen. Specifically, P301L tau was co-transfected with DnaJA2, DnaJB1, DnaJB6b, Hsp90α, and FKBP19. Following 48 hours of co-expression, cells were harvested and assessed for changes in total tau levels by western blot. Remarkably, overexpression of all tested DnaJ chaperones resulted in a robust decrease in total tau, by one-way ANOVA with Dunnett’s post hoc (Fig. 3A–B). Conversely, overexpression of Hsp90α and FKBP19 did not yield any significant changes (Fig. 3A–B). These findings suggest that DnaJ chaperones may be enriched in members that potently reduce tau protein levels, even under the condition of tau overexpression, which can contribute to the formation of soluble, pathogenic forms of tau [84].
Figure 3. Overexpression of DnaJs significantly affect intracellular tau levels.
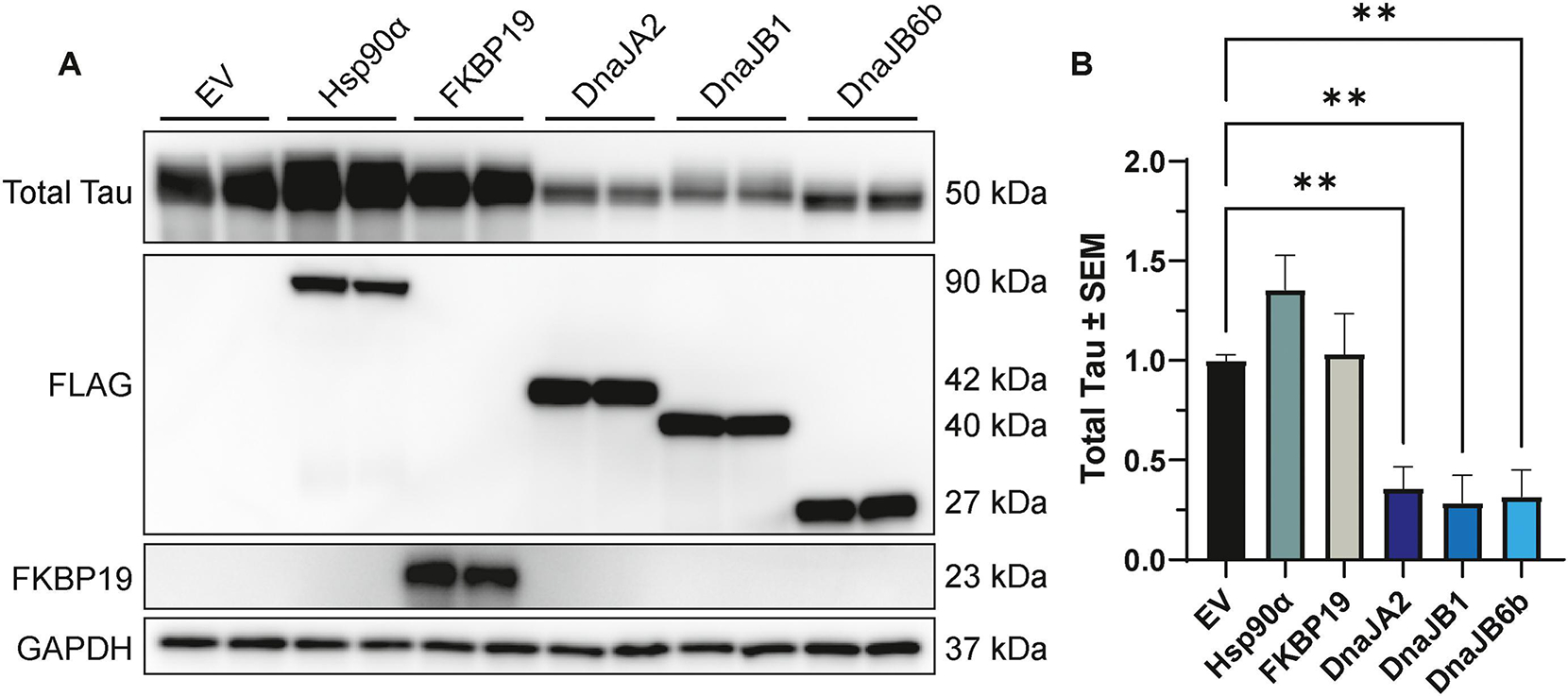
(A) Representative western blot images of cell lysates from HEK293T cells co-transfected with P301L tau and Hsp90α, FKBP19, DnaJA2, DnaJB1, DnaJB6b, or EV control for 48 hours, prior to harvesting for western blot analysis. (B) Quantification of total tau relative to GAPDH normalized to EV control is shown from three independent experiments ± S.E.M. A one-way ANOVA with Dunnett’s post hoc test was used to identify significance. Significance indicated as follows: **p<0.01.
To further corroborate the role of DnaJA2, DnaJB1, and DnaJB6b as regulators of tau, knockdown experiments were conducted to examine whether reducing the expression of these molecular chaperones would increase tau levels. HEK293T cells were transfected with either DnaJA2, DnaJB1, DnaJB6b, or GFP-targeting (control) shRNA and subsequently transfected with P301L tau the next day. Cells were harvested 48 hours after tau transfection and assessed for changes in total tau levels by western blot. Analysis using one-way ANOVA with Dunnett’s post hoc revealed that the ablation of DnaJB1 and DnaJB6b resulted in a significant increase in total tau. In contrast, DnaJA2 knockdown did not exhibit a significant effect (Fig. 4A–B). Importantly, these observations were not influenced by the efficiency of knockdown among these chaperones (Fig. 4C–E). Collectively, these findings provide compelling evidence supporting an inverse correlation between the levels of DnaJB1 and DnaJB6b with tau levels in vitro.
Figure 4. Knockdown of DnaJB1 or DnaJB6b significantly increases total tau levels.
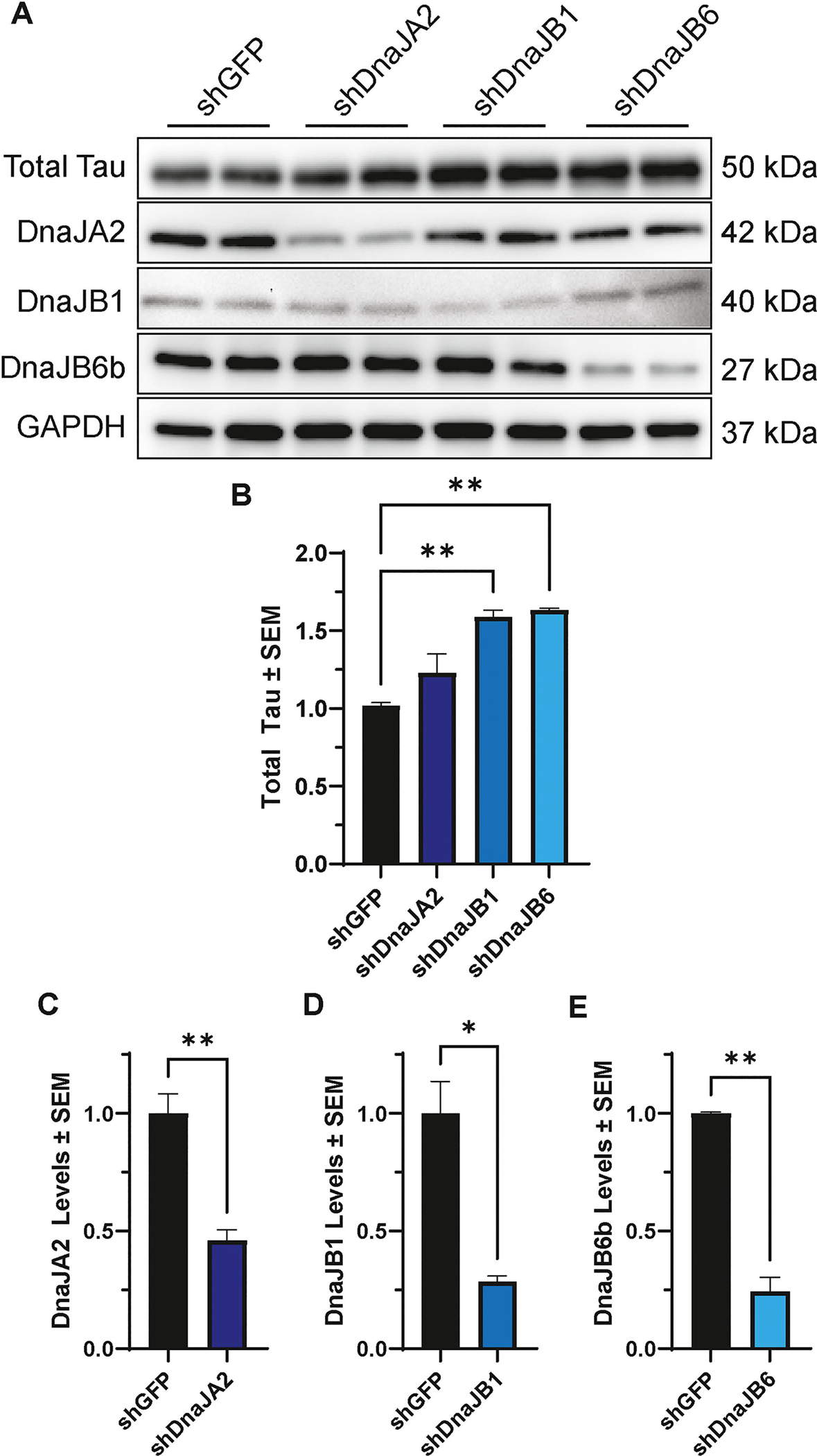
(A) Representative western blot of cell lysates from HEK293T cells transfected with P301L tau and shDnaJA2, shDnaJB1, shDnaJB6, or shGFP control, as indicated. (B) Quantification of total tau relative to GAPDH normalized to shGFP control is shown from two independent experiments ± S.E.M. A one-way ANOVA with Dunnett’s post hoc test was used to identify significance. Quantification of knockdown was confirmed for (C) DnaJA2, (D) DnaJB1 and (E) DnaJB6b by unpaired t-test. Significance indicated as follows: *p<0.05, **p<0.01.
2.3. DnaJB6b mitigates tau accumulation in aggressive cell models
To gain further insights into the effects of DnaJB1 and DnaJB6b on tau, a more aggressive model of tau accumulation was selected that uses a tetracycline-inducible system to express wild-type (WT) tau or tau containing the FTD-associated mutations, P301L and ΔK280, in HEK293 (iHEK) cells [34, 85, 86]. In these assays, tau expression was induced for 48 hours to create a preexisting pool of tau with a subset of aggregated tau. Then, cells were transfected with DnaJB1, DnaJB6b, or EV control. Cell lysates were collected 48 hours after transfection for western blot analysis. Notably, the overexpression of DnaJB6b, but not DnaJB1, led to a significant reduction in total tau levels in all three iHEK cell lines—iHEK P301L tau (Fig. 5A–B), iHEK WT tau (Fig. 5C–D), and iHEK ΔK280 tau (Fig. 5E–F). DnaJB6b, but not DnaJB1, also reduced the total tau recovered in the low-speed pellet fraction (Fig. S2). To start to understand how DnaJB6b lowers tau levels, the iHEK P301L tau model described above was treated with a 26S proteasome inhibitor, Peptide aldehyde (PSI), a 20S proteasome inhibitor, bortezomib (BTZ), or the lysosomal inhibitor, leupeptin (Leup). This assay revealed that proteasome inhibition counteracted the tau reductions by DnaJB6b (Fig. 6). Overall, these results suggest that DnaJB6b exerts a regulatory effect on tau accumulation across different tau species likely through proteasomal degradation.
Figure 5. DnaJB6b overexpression mitigates tau levels in iHEK P301L, WT, and ΔK280 tau cells.
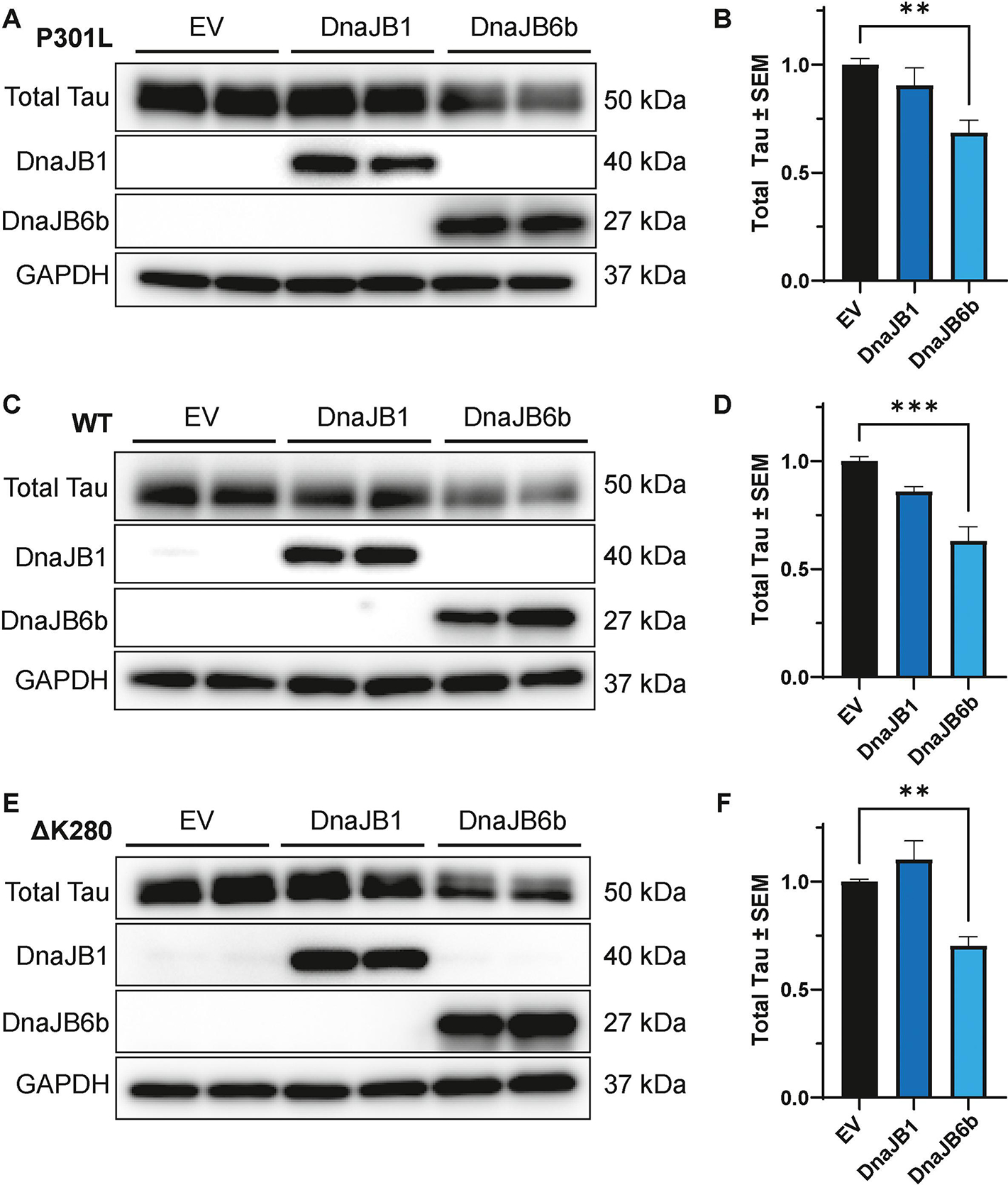
Tau expression was induced by tetracycline for 48 hours in iHEK P301L, WT, and ΔK280 tau cells, followed by transfection of DnaJB1, DnaJB6b, or EV for 48 hours, prior to harvesting for western blot analysis. Representative western blot and corresponding quantification of cell lysates from iHEK (A-B) P301L, (C-D) WT and (D-E) ΔK280 tau cells expressing DnaJB1, DnaJB6b, or EV control. Quantification was performed for total tau relative to GAPDH normalized to EV control from two independent experiments ± S.E.M. One-way ANOVA with Dunnett’s post hoc test was used to identify significance. Significance indicated as follows: **p<0.01, ***p<0.001.
Figure 6. Inhibition of the proteasome counteracts the reduction of tau by DnaJB6b.
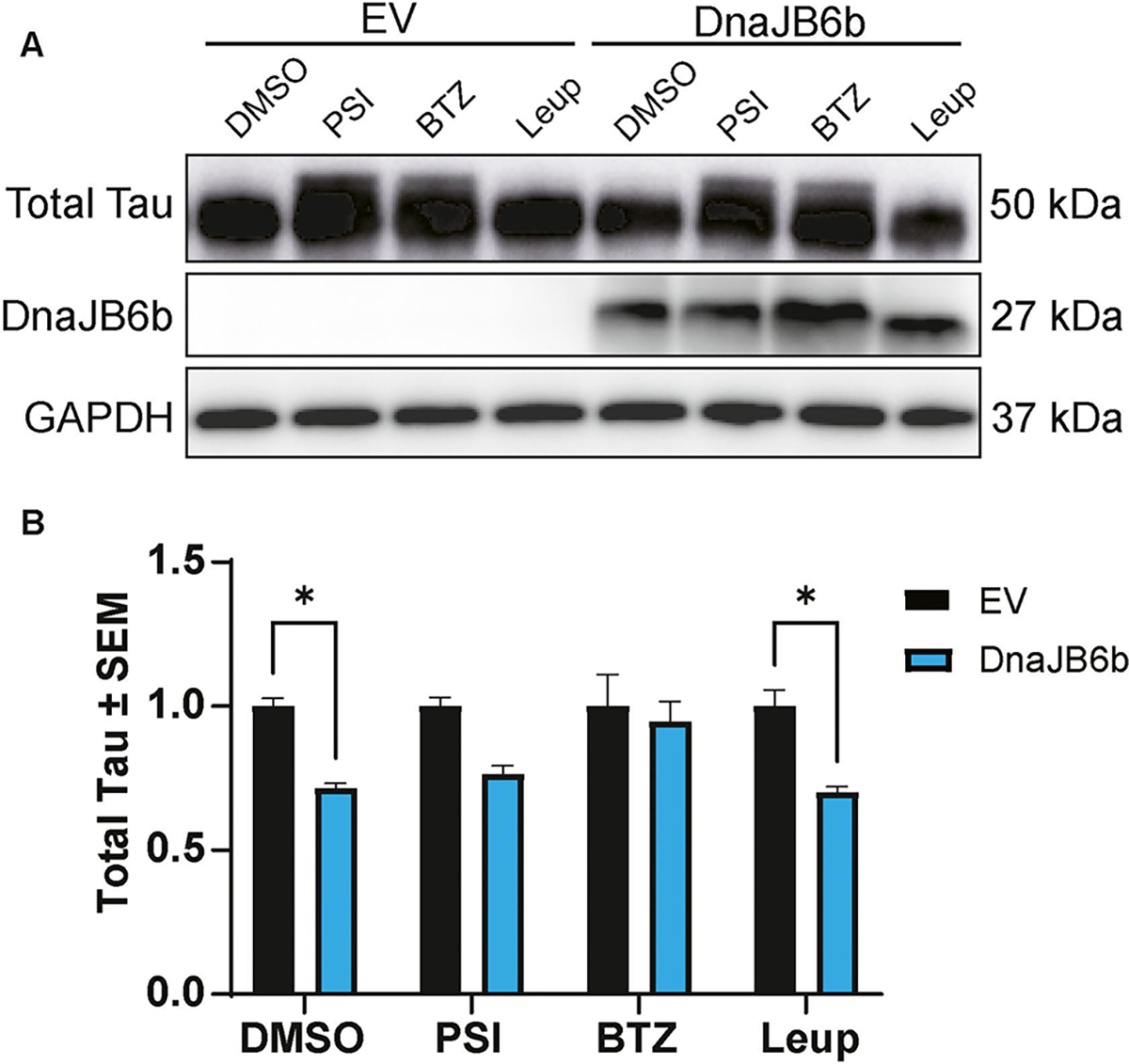
(A) Representative western blot from iHEK P301L tau cells induced by tetracycline for 48 hours, followed by transfection of DnaJB6b or EV. Twenty-four hours after transfection, cells were treated with DMSO control, PSI, Bortezomib (BTZ), or Leupeptin (Leup) for 16 hours prior to harvesting cells for western blot analysis. (B) Quantification of total tau relative to GAPDH normalized to respective EV treatment controls is shown from two independent experiments ± S.E.M. Two-way ANOVA with Šídák’s post hoc test was used to identify significance. Significance indicated as follows: *p<0.05.
2.4. DnaJB6b complexes with tau
The interaction between DnaJB6b and tau has not yet been described, so we examined whether these proteins can form a complex in a cellular environment. Specifically, iHEK P301L tau cells were treated with tetracycline for 48 hours, followed by transfection with FLAG-tagged DnaJB6b or EV. Co-immunoprecipitation for FLAG was performed 48 hours after transfection to assess the association between tau and DnaJB6b. The results revealed the formation of complexes between tau and DnaJB6b (Fig. 7), indicating either a direct protein-protein interaction or a stable multi-protein complex. This suggests DnaJB6b may play a crucial role in directly regulating tau accumulation.
Figure 7. DnaJB6b complexes with P301L tau.
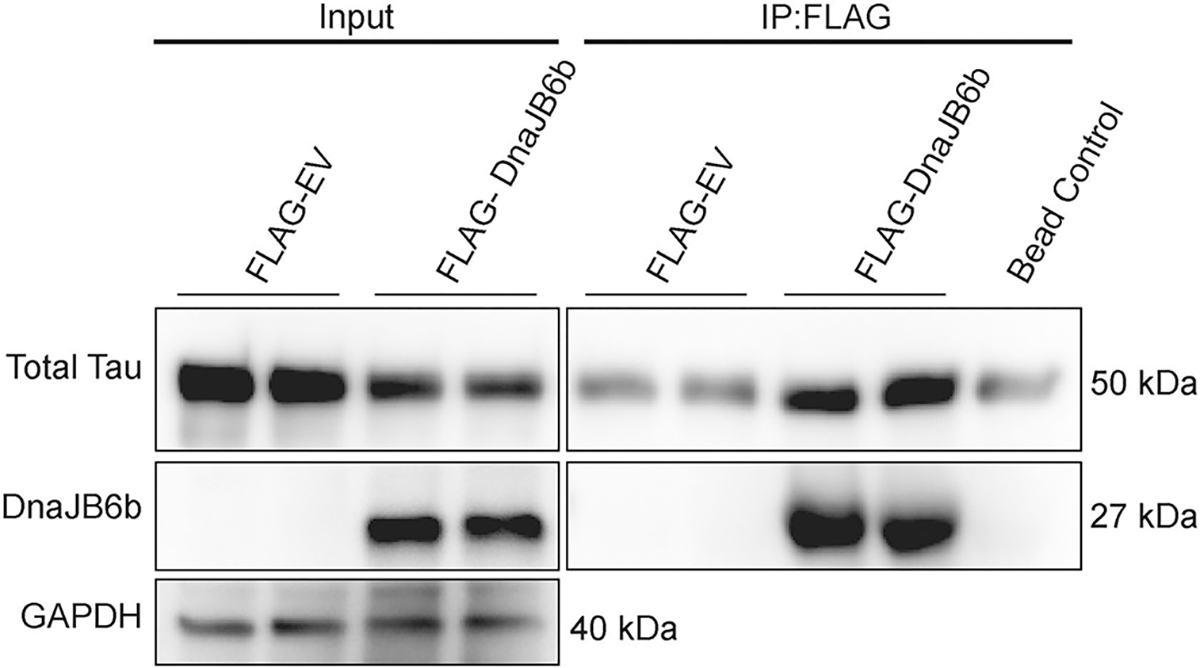
Tau expression was induced by tetracycline for 48 hours in iHEK P301L tau cells, followed by transfection of FLAG-DnaJB6b, FLAG-EV, or EV for 48 hours, prior to harvesting. DnaJB6b and DnaJB6b interacting proteins were immunoprecipitated using FLAG-tag conjugated beads and analyzed by western blot as indicated. Representative blot represents results from two independent experiments.
3. Discussion
This study investigated the impact of chaperone overexpression on tau seeding using a semi-high throughput quantitative assay. Through the systematic screening of molecular chaperones across five major families, our findings revealed significant effects on tau seeding by five select chaperones, three of which belong to the DnaJs. In follow up studies, the overexpression of DnaJA2, DnaJB1, and DnaJB6b significantly reduced tau. However, only the knockdown of DnaJB1 and DnaJB6b significantly increased tau levels. This suggests an inverse correlation between the expression of DnaJB1 and DnaJB6b with tau. When tested in a more aggressive tau model, the overexpression of DnaJB6b, but not DnaJB1, led to a significant reduction in total tau. The effect of DnaJB6b on tau levels may rely on proteasomal degradation. Importantly, tau co-immunoprecipitated with DnaJB6b, which suggests DnaJB6b may affect tau through direct interactions. Overall, this work highlights DnaJB6b as a potent tau regulator.
It is important to acknowledge that the use of Tau RD P301S FRET Biosensor cells, like many models of tau pathology, has its limitations and caveats. Here, liposomes were utilized to enhance the seed uptake efficiency, but it is important to note that tau seeds can be endocytosed in the absence of liposomes by micropinocytosis in an heparan sulfate proteoglycan-dependent manner [87]. This is similar to the uptake mechanisms of other aggregating proteins like SOD1, which has been shown to enter cells through micropinocytosis or endocytosis [88, 89]. FRET signal is not a direct readout of uptake but rather a downstream effect, which has been shown to correlate with the amount of cargo or seed concentration contained within the liposomes [11]. A recent study suggests that the observed FRET signal does not directly reflect templated aggregation of tau, but rather an alternative pathology [78]. However, it is worth noting that this model is still able to capture some important elements of the pathological templating and accumulation of tau [11, 79]. Additionally, our data using this model demonstrates that effects in this FRET-based model can be recapitulated in other tau models, supporting Tau RD P301S FRET Biosensor cells as a useful tool. While used in this study, it is also important to acknowledge that overexpression can lead to atypical protein-protein interactions. However, chaperone overexpression studies were complemented by knockdown studies, which provide further evidence for a connection between DnaJB1 and DnaJB6b with tau. Lastly, a limitation of cellular models is that they do not replicate all aspects of protein aggregation observed in disease, and while there are methods for identifying disease-related pathological forms of tau, complementary in vivo studies are needed to validate in vitro observations.
Our screen identified Hsp90α, FKBP19, DnaJA2, DnaJB1, and DnaJB6b as regulators of tau seeding. While Hsp90α overexpression reduced tau seeding, no changes were measured in co-expression assays. Prior work has implicated Hsp90α as a promoter of tau fibrillization [40, 48, 90]. Taken together, this suggests the action of Hsp90α on tau may be different based on the state of tau. Further studies to fully comprehend the role of Hsp90α in tau pathology are warranted but this is challenging given the numerous Hsp90α interacting proteins, which may confound interpretation [26, 91]. Like Hsp90α, FKBP19 reduced tau seeding without effects on tau accumulation. This transmembrane chaperone has high expression in secretory tissues [92], but has yet to be investigated in the context of tau or other aggregating proteins. While evidence suggests that FKBP19 may have chaperoning activity outside of its conserved enzymatic function, more studies are required to further understand its effect on tau [93].
Identifying an enrichment of tau regulators within the DnaJ family is not completely surprising, since these data complement prior work supporting an association between DnaJs and tau [94]. One of these studies reported a twofold decrease in the amount of DnaJB1 and DnaJB6b bound to tau in brain lysate from tau transgenic PS19 mice over time, while DnaJA2 showed no changes [65]. However, DnaJA2 was shown to co-localize with pathological tau in neurons and AD brain tissue [29, 32]. Recombinantly, DnaJA2 and DnaJB1 have been shown to reduce tau aggregation [29, 32], while studies using recombinant DnaJB6b in the context of tau have not yet been reported. In a FRET Tau Biosensor cell line, similar to the one used in this study, recombinant tau fibrils pre-incubated with recombinant DnaJA2 had reduced tau seeding, whereas tau fibrils pre-incubated with recombinant DnaJB1 did not [29]. Other members of the DnaJ family have been implicated in tau regulation, including DnaJC5, which is involved in tau secretion [30], and DnaJC7, which binds multiple forms of pathological and WT tau with high affinity [65, 66]. In the same study, Tau RD P301S Biosensor cells lacking DnaJC7 or DnaJB6 exhibited a significant increase in seeding [65, 66]. However, no changes in tau seeding were found in our study with the overexpression of DnaJC5 or DnaJC7 [65, 66]. Previous research has also indicated that other DnaJs, like DnaJA2 and DnaJB1, can directly interact with tau [29, 32]. A recent review highlights differences in the J-domain of DnaJ proteins that alters the coordination with Hsp70 and may drive the varied interactions with tau between the classes of DnaJ chaperones [94]. While both DnaJA and DnaJB proteins bind to many clients, Class B DnaJs have been shown to bind tau fibrils and seeds, while Class A DnaJ proteins can bind to monomeric tau as a substrate [31, 64]. In addition, Class B DnaJs have been shown to be capable of promoting tau disaggregation as part of the Hsp70 disaggregation machinery, while Class A DnaJs do not promote disaggregation [31]. Remarkably the most significant regulator of tau accumulation in this study, DnaJB6b, has only recently been described to have a potential role in regulating tau, where knockdown of DnaJB6 increased tau seeding in a cell model similar to the one we used here [66].
DnaJB6b has been reported to counteract the aggregation of several amyloidogenic proteins, including polyQ Huntingtin, polyQ ataxin, amyloid-β, α-synuclein, and TDP-43 [95–98]. Our results show DnaJB6b forms complexes with tau, suggesting that like other DnaJ family members, DnaJB6b may also engage in direct interactions with tau. This is in line with a recent report showing DnaJB6 as a tau aggregate interactor [66] and is further complemented by prior work demonstrating DnaJB6b can directly bind other neurodegenerative disease-related proteins, including amyloid-β [97]. Moreover, recent evidence indicates an inverse correlation between DnaJB6 expression and late-onset AD [37], but further studies are needed to confirm the direct interaction between DnaJB6b and tau to understand its role in neurodegenerative disease. Our results indicate that DnaJB6b may promote tau turnover through the proteasome. These data are consistent with DnaJB6b’s effect on polyQ degradation, where turnover of pathogenic proteins depends on proteasomal degradation through Hsp70 [99]. DnaJB6b may not solely rely on the proteasome to mitigate the accumulation of client proteins. In regards to seeding, questions still remain as to how DnaJB6b can reduce tau aggregation after the addition of exogenous seeds. Does DnaJB6b protect against cytoplasmic mislocalization of endogenous tau and the early signs of apoptosis similar to SH-SY5Y cells induced with SOD-1 oligomers [89]? Does DnaJB6b use the Hsp70 chaperone machinery to disaggregate tau similar to other Class B DnaJs [31]? Does DnaJB6b somehow reduce the uptake or release of seeds? It is possible DnaJB6b is the most potent regulator of tau because it synergistically uses multiple mechanisms of action. Additional studies are warranted to identify the complementary pathways DnaJB6b uses to reduce tau.
4. Conclusion
Overall, our study demonstrates the significant impact of molecular chaperones, particularly members of the DnaJ family, on tau seeding and accumulation. DnaJA2, DnaJB1, DnaJB6b, Hsp90α, and FKBP19 were identified as regulators of tau seeding. Unlike Hsp90α and FKBP19, the DnaJ proteins were found to affect intracellular tau levels, with DnaJB6b having the most potent effects. However, further in vivo studies are necessary to confirm these findings and validate DnaJB6b as a viable target for tauopathies.
5. Materials and Method
5.1. Molecular Cloning
FLAG-Hsp90α and FLAG- Hsp90β plasmids were provided by Dr. Leonard Neckers (National Institutes of Health, Rockville, MD). BiP/Grp78 pCMV-Myc-KDEL-wt was a gift from Ron Prywes (Addgene plasmid #27164; http://n2t.net/addgene:27164; RRID:Addgene_27164). The plasmids encoded in the pcDNA4/FRT/TO/V5 backbone include HSPA14 (Addgene plasmid #19515; http://n2t.net/addgene:19515; RRID:Addgene_19515, HSPA6 (Addgene plasmid #19513; http://n2t.net/addgene:19513; RRID:Addgene_19513), HSPA1A (Addgene plasmid #19510; http://n2t.net/addgene:19510; RRID:Addgene_19510), DNAJA2 (Addgene plasmid #19519; http://n2t.net/addgene:19519; RRID:Addgene_19519), DNAJA4 (Addgene plasmid #19494; http://n2t.net/addgene:19494; RRID:Addgene_19494, DNAJB1 (Addgene plasmid #19522; http://n2t.net/addgene:19522; RRID:Addgene_19522), DNAJB2A (Addgene plasmid # 19523; http://n2t.net/addgene:19523; RRID:Addgene_19523), DNAJB4 (Addgene plasmid #19526; http://n2t.net/addgene:19526; RRID:Addgene_19526), DNAJB6b (Addgene plasmid #19528; http://n2t.net/addgene:19528; RRID:Addgene_19528), DNAJB9 (Addgene plasmid #19532; http://n2t.net/addgene:19532; RRID:Addgene_19532) and were gifts from Harm Kampinga. cDNA of DnaJA2, DnaJA4, DnaJB2a, DnaJB6b, and DnaJB9 were subcloned into pCMV6 vector with an N-terminal FLAG. The plasmids encoded in the pcDNA4/FRT/TO backbone include HspB1 (Addgene plasmid #63092; http://n2t.net/addgene:63092; RRID:Addgene_63092), HspB2 (Addgene plasmid #63093; http://n2t.net/addgene:63093; RRID:Addgene_63093), HspB3 (Addgene plasmid #63094; http://n2t.net/addgene:63094; RRID:Addgene_63094), HspB4 (Addgene plasmid #63095; http://n2t.net/addgene:63095; RRID:Addgene_63095), HspB5 (Addgene plasmid #63096; http://n2t.net/addgene:63096; RRID:Addgene_63096), HspB6 (Addgene plasmid #63097; http://n2t.net/addgene:63097; RRID:Addgene_63097), HspB7 (Addgene plasmid #63098; http://n2t.net/addgene:63098; RRID:Addgene_63098), HspB8 (Addgene plasmid #63099; http://n2t.net/addgene:63099; RRID:Addgene_63099), HspB9 (Addgene plasmid #63100; http://n2t.net/addgene:63100; RRID:Addgene_63100), HspB10 (Addgene plasmid #63101; http://n2t.net/addgene:63101; RRID:Addgene_63101) and were additional gifts from Harm Kampinga. DnaJC7 and DnaJC8 pcDNA4/FRT/TO/V5 were also purchased from Addgene. Cdc37 was PCR-amplified from a human cDNA library (Invitrogen) and cloned into the pCMV6 vector. FKBP12.6 (HsCD00022455), FKBP13 (HsCD00002722), FKBP19 (HsCD00618224), FKBP22 (HsCD00616399), FKBP23 (HsCD00640901), FKBP25 (HsCD00002303), FKBP36 (HsCD00076429), FKBP133 (HsCD00294942) were subcloned from DNASU purchased donor plasmid into the pCMV6 vector. pCMV6 (Origene, #RC223397), pCMV6 with an N-terminal FLAG tag (Origene, #PCMV6XL6) and pCDNA5 FRT/TO (Invitrogen #V6520–20) empty vectors were purchased from verified sources. Hsp70 (HSPA1B), Hsc70, DnaJA1, DnaJC5, FKBP12, FKBP52, FKBP51, FKBP38, FKBP60, Grp94, CHIP, Aha1, p23, PP5C and HOP were generated by our lab. Pet28-a(+)-TEV and pCMV6 P301L 4R0N plasmids were also generated in our lab. All plasmids were propagated in DH5α competent cells (Fisher Scientific #18265017) and verified by DNA sequencing (Eurofins Genomics). A summary of these plasmids can be found in Table S2.
5.2. Generation of recombinant P301L tau seeds
Recombinant P301L tau was produced and purified, as we have previously done in our lab [45]. In summary, a glycerol stock of 4R0N P301L tau in a pET-28a(+)-TEV vector was used to inoculate a 10 mL starter culture of LB broth with kanamycin and was incubated overnight at 37°C. Cultures were grown to an OD600 of 0.7–0.8 then induced with a final concentration of 1 mM IPTG, grown for 3 hours, and subsequently harvested. Cells were spun down at 3,500 × g for 20 min, supernatant discarded, and each pellet resuspended in 35 mL of running buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 10 mM Imidazole) supplemented with 1x EDTA-free protease inhibitor cocktail III (Bimake #B14002) and 1 mM PMSF (Sigma Aldrich #P7626). Resuspended pellets were frozen and stored at −80°C.
For protein purification, each cell pellet was thawed, sonicated on ice using a Sonic Dismembrator (Fisher Scientific Model 120) for 60 pulses at 85% amplitude for 5 seconds on/10 seconds off, and spun at 50,000 × g for 30 min at 4°C to isolate supernatant. The supernatant was loaded onto a standard gravity nickel column packed with Nickel Sepharose high performance resin (Cytiva #17–5268-02), washed with 35 mL of running buffer, and eluted with 25 mL of elution buffer (20 mM Tris base pH 8.0, 500 mM NaCl, 250 mM Imidazole). TEV protease (~2–4 mg produced in-house) was added directly to the elution fraction to remove N-terminal 6x His tag, which was placed into a 3,500 MW cutoff dialysis bag for 4 hours at room temperature in a 1 L beaker of TEV cleavage buffer (50 mM Tris base pH 8.0, 250 mM NaCl). Post cleavage, the solution was then dialyzed back into running buffer overnight at 4°C and another nickel column purification was performed to remove TEV protease and uncleaved protein. Finally, using a HiLoad 16/600 Superdex 200 pg column, P301L tau was separated by size exclusion chromatography in 20 mM Tris base pH 7.6, 500 mM NaCl, 0.5 mM EDTA, and 0.5 mM DTT. Protein purity was confirmed by Coomassie stained SDS-PAGE gel to be >90% pure and protein concentration was measured by BCA (Fisher Scientific #PI23225). P301L tau was diluted to 2 mg/mL, aliquoted, and flash-frozen in liquid nitrogen for storage at −80°C.
To generate recombinant P301L tau fibrils for seeding assays, 10 μM recombinant P301L tau, previously dialyzed into 100 mM sodium acetate pH 7.0 buffer overnight, was mixed with 2.5 μM low-molecular-weight heparin (MP Biomedicals #194114) and 2 mM DTT in 100 mM sodium acetate pH 7.0 buffer supplemented with 10 μM Thioflavin T (ThT). Subsequently, the mixture was pipetted into a 96-well, black, clear-bottom nonbinding surface plate (Corning #3651) at 200 μL per well, sealed, and incubated at 37°C without agitation for 120 hours. Formation of P301L tau fibrils was confirmed by an increase in ThT fluorescence. Following ThT confirmation, aggregated P301L tau from all wells was combined into a single tube, aliquoted, flash-frozen, and stored at −80°C. Prior to use in the FRET biosensor screening assay, recombinant P301L tau fibrils were sonicated using a Sonic Dismembrator for 3 pulses at 25% amplitude for 5 seconds on/5 seconds off.
5.3. Tau FRET Biosensor Cell Screening Assay
HEK293T cells stably expressing Tau RD P301S-CFP and Tau RD P301S-YFP (ATCC #CRL-3275) were subcultured into 24-well poly-L-Lysine (Sigma-Aldrich #P4707) coated plates (0.5 × 106 cells per well) and co-transfected using 2.5 μL Lipofectamine 2000 (Invitrogen #11668019) per well with mKate (Evrogen #pmKate2-C) and chaperone plasmids at a 1:4 ratio of mKate to chaperone (0.2 μg mKate:0.8 μg chaperone). Twenty-four hours after transfection, cells were subcultured into 96-well, black, clear-bottom plates (Corning #3601) at a density of 0.4 × 105 cells per well and allowed to recover for 24 hours, reaching about 50–60% confluency, before seeding with a final concentration of 300 nM sonicated recombinant P301L tau fibrils in 100 mM sodium acetate pH 7.0 per well using 0.25 μL Lipofectamine 2000 per well. Cells were then housed in a BioSpa8 and imaged on a Cytation 3 using brightfield, CFP-YFP FRET (excitation 445 nm/emission 542 nm—BioTek #1225110), and Texas Red (excitation 486 nm/ emission 647 nm—BioTek #1225102) LED cubes using a 10x objective (Plan Fluorite WD 2.7 NA 0.6) every 12 hours for 60 hours. Each condition was measured in triplicate, and 4 images per well were used for analysis with Gen5 Image Prime Software plus Spot Counting. Relative FRET positive intensity was calculated as the FRET positive aggregate area within total cell area (using the brightfield image) and then normalized to the EV control. mKate (Texas red) was used to confirm successful transfection.
5.4. Follow-up cell-based assays
For co-overexpression studies, HEK293T cells (ATCC #CRL-3216) were subcultured into 12-well plates (Thermo Scientific #130185) coated with poly-L-Lysine and co-transfected using 2.5 μL Lipofectamine 2000 with P301L tau and chaperone plasmids at a ratio of 1:10, tau:chaperone (0.1 μg tau: 1 μg chaperone for 12-well plates). Forty-eight hours after transfection, cells were washed with PBS and harvested for western blots.
For shRNA studies, HEK293T cells were subcultured into 12-well plates coated with poly-L-Lysine and co-transfected using with chaperone shRNA (1 μg) using 2.5 μL Lipofectamine 2000, and subsequently transfected with P301L tau (0.1 μg) twenty-four hours later. Forty-eight hours after tau transfection, cells were washed with PBS and harvested for western blots.
For the iHEK cell lines (Invitrogen #R71007), iHEK P301L, WT, or ΔK280 tau cells were subcultured into 12-well plates coated with poly-L-Lysine with complete media containing 1 μg/mL tetracycline (Sigma-Aldrich #T7660–5G) to induce tau expression. After 48 hours, media was changed to complete media without tetracycline and transfected with DnaJ chaperone and control plasmids (1 μg) using 2.5 μL Lipofectamine 2000. Forty-eight hours after transfection, cells were washed with PBS and harvested for western blots.
For proteasome and lysosome inhibition assays, iHEK P301L tau cells were subcultured into 12-well plates and treated with tetracycline, as described above. After 48 hours, the cells were transfected with DnaJ chaperone and control plasmids (1 μg). Media was replaced 24 hours after transfection with complete media containing Bortezomib (50 nM) (Selleck Chemicals #S10103), Peptide aldehyde (PSI) (10 μM) (APExBio #A1900), Leupeptin (200 nM) (Millipore #E18), or vehicle control (1% DMSO). Cells were harvested 16 hours after treatment for western blot analysis.
For co-immunoprecipitation studies, iHEK P301L tau cells were subcultured into 100 mm dishes (Sarstedt #83–3902) coated with poly-L-Lysine, with complete media containing 1 μg/mL tetracycline to induce tau expression. After 48 hours, media was changed to complete media without tetracycline and transfected with either N-terminal FLAG-tagged DnaJB6b (FLAG-DnaJB6b), N-terminal FLAG-tagged EV (FLAG-EV), or EV plasmids (12 μg) using 12.5 μL Lipofectamine 2000. Forty-eight hours following transfection, cells were harvested in IP Lysis Buffer (Thermo Scientific Pierce #PI87787) and co-immunoprecipitated with Anti-DYDDDDK Magnetic Agarose beads (Thermo Scientific Pierce #PIA36797), according to the manufacturer’s protocol, and processed by western blot.
5.5. Western blotting and antibodies
All samples, diluted into a final concentration of 1× sodium dodecyl sulfate (SDS) sample buffer (Bio-Rad #1610737) and heated at 100°C for 10 min, were loaded into Any kD SDS-PAGE gels (Bio-Rad #4569034) and transferred to PVDF membranes (MilliporeSigma #IPVH00010) followed by blocking for 1 hour in 7% milk. Rabbit anti-tau polyclonal antibody (Dako/Agilent #A0024) at 1:10,000 was used to detect total tau protein and mouse anti-GAPDH monoclonal antibody (Proteintech #1E6D9) was used at 1:1,000 to detect loading control. Mouse anti-FLAG M2 (Sigma-Aldrich #F1804) and rabbit anti-FKBP11/FKBP19 (Sigma-Aldrich #HPA041709) were used to detect overexpressed chaperones in the initial co-expression assays. Rabbit anti-DnaJA2 antibody (Thermo Scientific # PA5–63874), rabbit anti-DnaJB1 antibody (Cell Signaling #4868), and mouse anti-DnaJB6 antibody (Proteintech #66587–1) were used at 1:1,000 to detect respective DnaJs. HRP-conjugated secondary antibodies at 1:1,000 for mouse (Southern Biotech #1030–05) and rabbit (Southern Biotech #4010–05) were used for ECL visualization by the FluorChem System E (ProteinSimple # 92–14860-00).
5.6. Statistical analysis
Data distributions were first tested using the Kolmogorov–Smirnov test for normality and Bartlett’s test for homogeneity of variance, and all groups were found to have no violations. Time-course imaging data were analyzed by repeated measures ANOVA with the Greenhouse-Geisser correction and Dunnett’s post hoc tests using SPSS Statistics 28 (IBM Corp, Version 28.0. Armonk, NY) to compare the effect of chaperone overexpression within chaperone families on tau seeding, as described in the figure legends and in Table S1. In groups containing more than one control vector, which include DnaJ, Hsp90, and Hsp70 families, a repeated measures ANOVA determined the control vector data did not statistically differ across all time points, so the control vector data was grouped together to allow whole family comparisons. Statistical analysis for cell-based assays analyzed by western blot were done through one-way ANOVA with respective post hoc tests, as described in figure legends and in Table S1, using GraphPad 9 (Prism. La Jolla, CA). All graphs were generated using GraphPad 9. Significant differences of the mean values were determined at P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001).
Supplementary Material
Highlights.
Molecular chaperones across five distinct families were assessed to determine their effect on tau seeding.
Hsp90α, FKBP19, DnaJA2, DnaJB1, and DnaJB6b significantly reduced tau seeding.
DnaJA2, DnaJB1, and DnaJB6b reduce intracellular tau levels.
DnaJB1 and DnaJB6b knockdown increases tau levels.
DnaJB6b forms complexes with tau.
Acknowledgements
We would like to thank Dr. Danielle Gulick for her guidance and advice on statistical analysis.
Funding
This research was funded by the Ed and Ethel Moore Alzheimer’s Disease Research Program (Grant Number: 21A24) through the Florida Department of Health. Research reported in this publication was supported in part by Merit Review Award [I01BX004626] from the United States (U.S.) Department of Veterans Affairs Biomedical Laboratory Research and Development Service. The views expressed in this article are those of the authors and do not necessarily reflect the position, policy or views of the Department of Veterans Affairs or the United States Government. Research reported in this publication was supported in part by the National Institute On Aging of the National Institutes of Health under Award Number F31AG082505 and R01NS073899, which is co-funded by the National Institute of Neurological Disorders and Stroke and the National Institute On Aging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Credit authorship contribution statement
Abigail R. Esquivel: Methodology, Validation, Formal analysis, Investigation, Visualization, Writing - Original Draft, Writing - Review & Editing; Shannon E. Hill: Methodology, Investigation, Writing - Review & Editing; Laura J. Blair: Conceptualization, Methodology, Resources, Writing - Original Draft, Writing - Review & Editing, Supervision, Project administration, Funding acquisition
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Narasimhan S, Guo JL, Changolkar L, Stieber A, McBride JD, Silva LV, He Z, Zhang B, Gathagan RJ, Trojanowski JQ, Lee VMY, Pathological Tau Strains from Human Brains Recapitulate the Diversity of Tauopathies in Nontransgenic Mouse Brain, J. Neurosci. 37(47) (2017) 11406–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].He Z, McBride JD, Xu H, Changolkar L, Kim S.-j., Zhang B, Narasimhan S, Gibbons GS, Guo JL, Kozak M, Schellenberg GD, Trojanowski JQ, Lee VMY, Transmission of tauopathy strains is independent of their isoform composition, Nature Communications 11(1) (2020) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lopes S, Vaz-Silva J, Pinto V, Dalla C, Kokras N, Bedenk B, Mack N, Czisch M, Almeida OF, Sousa N, Sotiropoulos I, Tau protein is essential for stress-induced brain pathology, Proc. Natl. Acad. Sci. U. S. A. 113(26) (2016) E3755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kaufman SK, Sanders DW, Thomas TL, Ruchinskas AJ, Vaquer-Alicea J, Sharma AM, Miller TM, Diamond MI, Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability In Vivo, Neuron 92(4) (2016) 796–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Irwin DJ, Tauopathies as Clinicopathological Entities, Parkinsonism Relat. Disord. 22(0 1) (2016) S29–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tacik P, Sanchez-Contreras M, Rademakers R, Dickson DW, Wszolek ZK, Genetic Disorders with Tau Pathology: A Review of the Literature and Report of Two Patients with Tauopathy and Positive Family Histories, NDD 16(1–2) (2016) 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hardy J, Orr H, The genetics of neurodegenerative diseases, J. Neurochem. 97(6) (2006) 1690–9. [DOI] [PubMed] [Google Scholar]
- [8].Adams JN, Maass A, Harrison TM, Baker SL, Jagust WJ, Cortical tau deposition follows patterns of entorhinal functional connectivity in aging, Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].La Joie R, Visani AV, Baker SL, Brown JA, Bourakova V, Cha J, Chaudhary K, Edwards L, Iaccarino L, Janabi M, Lesman-Segev OH, Miller ZA, Perry DC, O’Neil JP, Pham J, Rojas JC, Rosen HJ, Seeley WW, Tsai RM, Miller BL, Jagust WJ, Rabinovici GD, Prospective longitudinal atrophy in Alzheimer’s disease correlates with the intensity and topography of baseline tau-PET, Sci. Transl. Med. 12(524) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Spillantini MG, Goedert M, Tau pathology and neurodegeneration, Lancet Neurol. 12(6) (2013) 609–22. [DOI] [PubMed] [Google Scholar]
- [11].Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, Belaygorod L, Cairns NJ, Holtzman DM, Diamond MI, Proteopathic tau seeding predicts tauopathy in vivo, Proc. Natl. Acad. Sci. U. S. A. 111(41) (2014) E4376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vogel JW, Iturria-Medina Y, Strandberg OT, Smith R, Levitis E, Evans AC, Hansson O, Alzheimer’s Disease Neuroimaging I, Swedish BioFinder S, Spread of pathological tau proteins through communicating neurons in human Alzheimer’s disease, Nat Commun 11(1) (2020) 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brunello CA, Merezhko M, Uronen RL, Huttunen HJ, Mechanisms of secretion and spreading of pathological tau protein, Cell. Mol. Life Sci. 77(9) (2020) 1721–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].DeVos SL, Corjuc BT, Oakley DH, Nobuhara CK, Bannon RN, Chase A, Commins C, Gonzalez JA, Dooley PM, Frosch MP, Hyman BT, Synaptic Tau Seeding Precedes Tau Pathology in Human Alzheimer’s Disease Brain, Front. Neurosci. 12 (2018) 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mudher A, Colin M, Dujardin S, Medina M, Dewachter I, Alavi Naini SM, Mandelkow EM, Mandelkow E, Buee L, Goedert M, Brion JP, What is the evidence that tau pathology spreads through prion-like propagation?, Acta Neuropathol Commun 5(1) (2017) 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Franzmeier N, Neitzel J, Rubinski A, Smith R, Strandberg O, Ossenkoppele R, Hansson O, Ewers M, Alzheimer’s Disease Neuroimaging I, Functional brain architecture is associated with the rate of tau accumulation in Alzheimer’s disease, Nat Commun 11(1) (2020) 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meisl G, Hidari E, Allinson K, Rittman T, DeVos SL, Sanchez JS, Xu CK, Duff KE, Johnson KA, Rowe JB, Hyman BT, Knowles TPJ, Klenerman D, In vivo rate-determining steps of tau seed accumulation in Alzheimer’s disease, Sci Adv 7(44) (2021) eabh1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Criado-Marrero M, Sabbagh JJ, Jones MR, Chaput D, Dickey CA, Blair LJ, Hippocampal Neurogenesis Is Enhanced in Adult Tau Deficient Mice, Cells 9(1) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].DeVos SL, Corjuc BT, Commins C, Dujardin S, Bannon RN, Corjuc D, Moore BD, Bennett RE, Jorfi M, Gonzales JA, Dooley PM, Roe AD, Pitstick R, Irimia D, Frosch MP, Carlson GA, Hyman BT, Tau reduction in the presence of amyloid-beta prevents tau pathology and neuronal death in vivo, Brain 141(7) (2018) 2194–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].DeVos SL, Miller RL, Schoch KM, Holmes BB, Kebodeaux CS, Wegener AJ, Chen G, Shen T, Tran H, Nichols B, Zanardi TA, Kordasiewicz HB, Swayze EE, Bennett CF, Diamond MI, Miller TM, Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy, Sci. Transl. Med. 9(374) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nicholls SB, DeVos SL, Commins C, Nobuhara C, Bennett RE, Corjuc DL, Maury E, Eftekharzadeh B, Akingbade O, Fan Z, Roe AD, Takeda S, Wegmann S, Hyman BT, Characterization of TauC3 antibody and demonstration of its potential to block tau propagation, PLoS One 12(5) (2017) e0177914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ittner A, Bertz J, Suh LS, Stevens CH, Gotz J, Ittner LM, Tau-targeting passive immunization modulates aspects of pathology in tau transgenic mice, J Neurochem 132(1) (2015) 135–45. [DOI] [PubMed] [Google Scholar]
- [23].Stancu IC, Vasconcelos B, Ris L, Wang P, Villers A, Peeraer E, Buist A, Terwel D, Baatsen P, Oyelami T, Pierrot N, Casteels C, Bormans G, Kienlen-Campard P, Octave JN, Moechars D, Dewachter I, Templated misfolding of Tau by prion-like seeding along neuronal connections impairs neuronal network function and associated behavioral outcomes in Tau transgenic mice, Acta Neuropathol. 129(6) (2015) 875–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Han ZZ, Kang SG, Arce L, Westaway D, Prion-like strain effects in tauopathies, Cell Tissue Res (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pickett EK, Henstridge CM, Allison E, Pitstick R, Pooler A, Wegmann S, Carlson G, Hyman BT, Spires-Jones TL, Spread of tau down neural circuits precedes synapse and neuronal loss in the rTgTauEC mouse model of early Alzheimer’s disease, Synapse 71(6) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schopf FH, Biebl MM, Buchner J, The HSP90 chaperone machinery, Nat. Rev. Mol. Cell Biol. 18(6) (2017) 345–360. [DOI] [PubMed] [Google Scholar]
- [27].Sahasrabudhe P, Rohrberg J, Biebl MM, Rutz DA, Buchner J, The Plasticity of the Hsp90 Co-chaperone System, Mol. Cell 67(6) (2017) 947–961 e5. [DOI] [PubMed] [Google Scholar]
- [28].Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B, The Hsp70 chaperone network, Nature Reviews Molecular Cell Biology 20(11) (2019) 665–680. [DOI] [PubMed] [Google Scholar]
- [29].Mok SA, Condello C, Freilich R, Gillies A, Arhar T, Oroz J, Kadavath H, Julien O, Assimon VA, Rauch JN, Dunyak BM, Lee J, Tsai FTF, Wilson MR, Zweckstetter M, Dickey CA, Gestwicki JE, Mapping interactions with the chaperone network reveals factors that protect against tau aggregation, Nat. Struct. Mol. Biol. 25(5) (2018) 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fontaine SN, Zheng D, Sabbagh JJ, Martin MD, Chaput D, Darling A, Trotter JH, Stothert AR, Nordhues BA, Lussier A, Baker J, Shelton L, Kahn M, Blair LJ, Stevens SM Jr., Dickey CA, DnaJ/Hsc70 chaperone complexes control the extracellular release of neurodegenerative-associated proteins, EMBO J. 35(14) (2016) 1537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nachman E, Wentink AS, Madiona K, Bousset L, Katsinelos T, Allinson K, Kampinga H, McEwan WA, Jahn TR, Melki R, Mogk A, Bukau B, Nussbaum-Krammer C, Disassembly of Tau fibrils by the human Hsp70 disaggregation machinery generates small seeding-competent species, J Biol Chem 295(28) (2020) 9676–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Irwin R, Faust O, Petrovic I, Wolf SG, Hofmann H, Rosenzweig R, Hsp40s play complementary roles in the prevention of tau amyloid formation, Elife 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Criado-Marrero M, Gebru NT, Blazier DM, Gould LA, Baker JD, Beaulieu-Abdelahad D, Blair LJ, Hsp90 co-chaperones, FKBP52 and Aha1, promote tau pathogenesis in aged wild-type mice, Acta Neuropathol Commun 9(1) (2021) 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Baker JD, Shelton LB, Zheng D, Favretto F, Nordhues BA, Darling A, Sullivan LE, Sun Z, Solanki PK, Martin MD, Suntharalingam A, Sabbagh JJ, Becker S, Mandelkow E, Uversky VN, Zweckstetter M, Dickey CA, Koren J 3rd, Blair LJ, Human cyclophilin 40 unravels neurotoxic amyloids, PLoS Biol. 15(6) (2017) e2001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yu A, Fox SG, Cavallini A, Kerridge C, O’Neill MJ, Wolak J, Bose S, Morimoto RI, Tau protein aggregates inhibit the protein-folding and vesicular trafficking arms of the cellular proteostasis network, J. Biol. Chem. 294(19) (2019) 7917–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fu H, Possenti A, Freer R, Nakano Y, Hernandez Villegas NC, Tang M, Cauhy PVM, Lassus BA, Chen S, Fowler SL, Figueroa HY, Huey ED, Johnson GVW, Vendruscolo M, Duff KE, A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology, Nat. Neurosci. 22(1) (2019) 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ayoub CA, Wagner CS, Kuret J, Identification of gene networks mediating regional resistance to tauopathy in late-onset Alzheimer’s disease, PLoS Genet 19(3) (2023) e1010681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Abisambra JF, Blair LJ, Hill SE, Jones JR, Kraft C, Rogers J, Koren J 3rd, Jinwal UK, Lawson L, Johnson AG, Wilcock D, O’Leary JC, Jansen-West K, Muschol M, Golde TE, Weeber EJ, Banko J, Dickey CA, Phosphorylation dynamics regulate Hsp27-mediated rescue of neuronal plasticity deficits in tau transgenic mice, J. Neurosci. 30(46) (2010) 15374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Brehme M, Voisine C, Rolland T, Wachi S, Soper JH, Zhu Y, Orton K, Villella A, Garza D, Vidal M, Ge H, Morimoto RI, A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease, Cell Rep 9(3) (2014) 1135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shelton LB, Baker JD, Zheng D, Sullivan LE, Solanki PK, Webster JM, Sun Z, Sabbagh JJ, Nordhues BA, Koren J 3rd, Ghosh S, Blagg BSJ, Blair LJ, Dickey CA, Hsp90 activator Aha1 drives production of pathological tau aggregates, Proc. Natl. Acad. Sci. U. S. A. 114(36) (2017) 9707–9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shemesh N, Jubran J, Dror S, Simonovsky E, Basha O, Argov C, Hekselman I, Abu-Qarn M, Vinogradov E, Mauer O, Tiago T, Carra S, Ben-Zvi A, Yeger-Lotem E, The landscape of molecular chaperones across human tissues reveals a layered architecture of core and variable chaperones, Nat Commun 12(1) (2021) 2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Serlidaki D, van Waarde M, Rohland L, Wentink AS, Dekker SL, Kamphuis MJ, Boertien JM, Brunsting JF, Nillegoda NB, Bukau B, Mayer MP, Kampinga HH, Bergink S, Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones, J Biol Chem 295(21) (2020) 7301–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qiu XB, Shao YM, Miao S, Wang L, The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones, Cell. Mol. Life Sci. 63(22) (2006) 2560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Biebl MM, Riedl M, Buchner J, Hsp90 Co-chaperones Form Plastic Genetic Networks Adapted to Client Maturation, Cell Rep 32(8) (2020) 108063. [DOI] [PubMed] [Google Scholar]
- [45].Hill SE, Esquivel AR, Ospina SR, Rahal LM, Dickey CA, Blair LJ, Chaperoning activity of the cyclophilin family prevents tau aggregation, Protein Sci. 31(11) (2022) e4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kundel F, De S, Flagmeier P, Horrocks MH, Kjaergaard M, Shammas SL, Jackson SE, Dobson CM, Klenerman D, Hsp70 Inhibits the Nucleation and Elongation of Tau and Sequesters Tau Aggregates with High Affinity, ACS Chem. Biol. 13(3) (2018) 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jinwal UK, O’Leary JC 3rd, Borysov SI, Jones JR, Li Q, Koren J 3rd, Abisambra JF, Vestal GD, Lawson LY, Johnson AG, Blair LJ, Jin Y, Miyata Y, Gestwicki JE, Dickey CA, Hsc70 rapidly engages tau after microtubule destabilization, J. Biol. Chem. 285(22) (2010) 16798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Weickert S, Wawrzyniuk M, John LH, Rudiger SGD, Drescher M, The mechanism of Hsp90-induced oligomerizaton of Tau, Sci Adv 6(11) (2020) eaax6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Moll A, Ramirez LM, Ninov M, Schwarz J, Urlaub H, Zweckstetter M, Hsp multichaperone complex buffers pathologically modified Tau, Nat Commun 13(1) (2022) 3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Oroz J, Chang BJ, Wysoczanski P, Lee C-T, Pérez-Lara Á, Chakraborty P, Hofele RV, Baker JD, Blair LJ, Biernat J, Urlaub H, Mandelkow E, Dickey CA, Zweckstetter M, Structure and pro-toxic mechanism of the human Hsp90/PPIase/Tau complex, Nature Communications 9(1) (2018) 4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Luengo TM, Kityk R, Mayer MP, Rüdiger SGD, Hsp90 Breaks the Deadlock of the Hsp70 Chaperone System, Mol. Cell 70(3) (2018) 545–552.e9. [DOI] [PubMed] [Google Scholar]
- [52].Rutledge BS, Choy WY, Duennwald ML, Folding or holding?-Hsp70 and Hsp90 chaperoning of misfolded proteins in neurodegenerative disease, J. Biol. Chem. 298(5) (2022) 101905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gracia L, Lora G, Blair LJ, Jinwal UK, Therapeutic Potential of the Hsp90/Cdc37 Interaction in Neurodegenerative Diseases, Front. Neurosci. 13 (2019) 1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zgajnar NR, De Leo SA, Lotufo CM, Erlejman AG, Piwien-Pilipuk G, Galigniana MD, Biological Actions of the Hsp90-binding Immunophilins FKBP51 and FKBP52, Biomolecules 9(2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Giustiniani J, Chambraud B, Sardin E, Dounane O, Guillemeau K, Nakatani H, Paquet D, Kamah A, Landrieu I, Lippens G, Baulieu EE, Tawk M, Immunophilin FKBP52 induces Tau-P301L filamentous assembly in vitro and modulates its activity in a model of tauopathy, Proc. Natl. Acad. Sci. U. S. A. 111(12) (2014) 4584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lee K, Thwin AC, Nadel CM, Tse E, Gates SN, Gestwicki JE, Southworth DR, The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state, Mol Cell 81(17) (2021) 3496–3508 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nadel CM, Thwin AC, Callahan M, Lee K, Connelly E, Craik CS, Southworth DR, Gestwicki JE, The E3 Ubiquitin Ligase, CHIP/STUB1, Inhibits Aggregation of Phosphorylated Proteoforms of Microtubule-associated Protein Tau (MAPT), J. Mol. Biol. (2023) 168026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Blair LJ, Baker JD, Sabbagh JJ, Dickey CA, The emerging role of peptidyl-prolyl isomerase chaperones in tau oligomerization, amyloid processing, and Alzheimer’s disease, J. Neurochem. 133(1) (2015) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Jiang L, Chakraborty P, Zhang L, Wong M, Hill SE, Webber CJ, Libera J, Blair LJ, Wolozin B, Zweckstetter M, Chaperoning of specific tau structure by immunophilin FKBP12 regulates the neuronal resilience to extracellular stress, Science Advances 9(5) (2023) eadd9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Young ZT, Rauch JN, Assimon VA, Jinwal U, Ahn M, Li X, Dunyak BM, Ahmad A, Carlson G, Srinivasan SR, Zuiderweg ERP, Dickey CA, Gestwicki JE, Stabilizing the Hsp70-Tau Complex Promotes Turnover in Models of Tauopathy, Cell Chemical Biology 23(8) (2016) 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jinwal UK, Akoury E, Abisambra JF, O’Leary JC 3rd, Thompson AD, Blair LJ, Jin Y, Bacon J, Nordhues BA, Cockman M, Zhang J, Li P, Zhang B, Borysov S, Uversky VN, Biernat J, Mandelkow E, Gestwicki JE, Zweckstetter M, Dickey CA, Imbalance of Hsp70 family variants fosters tau accumulation, FASEB journal : official publication of the Federation of American Societies for Experimental Biology 27(4) (2013) 1450–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fontaine SN, Martin MD, Akoury E, Assimon VA, Borysov S, Nordhues BA, Sabbagh JJ, Cockman M, Gestwicki JE, Zweckstetter M, Dickey CA, The active Hsc70/tau complex can be exploited to enhance tau turnover without damaging microtubule dynamics, Human Molecular Genetics 24(14) (2015) 3971–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Taylor IR, Ahmad A, Wu T, Nordhues BA, Bhullar A, Gestwicki JE, Zuiderweg ERP, The disorderly conduct of Hsc70 and its interaction with the Alzheimer’s-related Tau protein, J Biol Chem 293(27) (2018) 10796–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Abisambra JF, Jinwal UK, Suntharalingam A, Arulselvam K, Brady S, Cockman M, Jin Y, Zhang B, Dickey CA, DnaJA1 antagonizes constitutive Hsp70-mediated stabilization of tau, J. Mol. Biol. 421(4–5) (2012) 653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hou Z, Wydorski PM, Perez VA, Mendoza-Oliva A, Ryder BD, Mirbaha H, Kashmer O, Joachimiak LA, DnaJC7 binds natively folded structural elements in tau to inhibit amyloid formation, Nat Commun 12(1) (2021) 5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Perez VA, Sanders DW, Mendoza-Oliva A, Stopschinski BE, Mullapudi V, White CL, Joachimiak LA, Diamond MI, DnaJC7 specifically regulates tau seeding, Elife 12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xu Y, Cui L, Dibello A, Wang L, Lee J, Saidi L, Lee JG, Ye Y, DNAJC5 facilitates USP19-dependent unconventional secretion of misfolded cytosolic proteins, Cell Discov 4 (2018) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Baughman HER, Pham TT, Adams CS, Nath A, Klevit RE, Release of a disordered domain enhances HspB1 chaperone activity toward tau, Proc. Natl. Acad. Sci. U. S. A. 117(6) (2020) 2923–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Baughman HER, Clouser AF, Klevit RE, Nath A, HspB1 and Hsc70 chaperones engage distinct tau species and have different inhibitory effects on amyloid formation, J. Biol. Chem. 293(8) (2018) 2687–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Janowska MK, Baughman HER, Woods CN, Klevit RE, Mechanisms of Small Heat Shock Proteins, Cold Spring Harb Perspect Biol 11(10) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rodriguez Ospina S, Blazier DM, Criado-Marrero M, Gould LA, Gebru NT, Beaulieu-Abdelahad D, Wang X, Remily-Wood E, Chaput D, Stevens S, Uversky VN, Bickford PC, Dickey CA, Blair LJ, Small Heat Shock Protein 22 Improves Cognition and Learning in the Tauopathic Brain, Int. J. Mol. Sci. 23(2) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Webster JM, Darling AL, Sanders TA, Blazier DM, Vidal-Aguiar Y, Beaulieu-Abdelahad D, Plemmons DG, Hill SE, Uversky VN, Bickford PC, Dickey CA, Blair LJ, Hsp22 with an N-Terminal Domain Truncation Mediates a Reduction in Tau Protein Levels, Int. J. Mol. Sci. 21(15) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Furman JL, Holmes BB, Diamond MI, Sensitive Detection of Proteopathic Seeding Activity with FRET Flow Cytometry, J Vis Exp (106) (2015) e53205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kaufman SK, Thomas TL, Del Tredici K, Braak H, Diamond MI, Characterization of tau prion seeding activity and strains from formaldehyde-fixed tissue, Acta Neuropathol Commun 5(1) (2017) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Chen JJ, Nathaniel DL, Raghavan P, Nelson M, Tian R, Tse E, Hong JY, See SK, Mok SA, Hein MY, Southworth DR, Grinberg LT, Gestwicki JE, Leonetti MD, Kampmann M, Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation, J. Biol. Chem. 294(50) (2019) 18952–18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Tak H, Haque MM, Kim MJ, Lee JH, Baik JH, Kim Y, Kim DJ, Grailhe R, Kim YK, Bimolecular fluorescence complementation; lighting-up tau-tau interaction in living cells, PLoS One 8(12) (2013) e81682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sala-Jarque J, Zimkowska K, Avila J, Ferrer I, Del Rio JA, Towards a Mechanistic Model of Tau-Mediated Pathology in Tauopathies: What Can We Learn from Cell-Based In Vitro Assays?, Int. J. Mol. Sci. 23(19) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kaniyappan S, Tepper K, Biernat J, Chandupatla RR, Hubschmann S, Irsen S, Bicher S, Klatt C, Mandelkow EM, Mandelkow E, FRET-based Tau seeding assay does not represent prion-like templated assembly of Tau filaments, Mol Neurodegener 15(1) (2020) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hitt BD, Vaquer-Alicea J, Manon VA, Beaver JD, Kashmer OM, Garcia JN, Diamond MI, Ultrasensitive tau biosensor cells detect no seeding in Alzheimer’s disease CSF, Acta Neuropathol Commun 9(1) (2021) 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Polanco JC, Akimov Y, Fernandes A, Briner A, Hand GR, van Roijen M, Balistreri G, Gotz J, CRISPRi screening reveals regulators of tau pathology shared between exosomal and vesicle-free tau, Life Sci Alliance 6(1) (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhu J, Pittman S, Dhavale D, French R, Patterson JN, Kaleelurrrahuman MS, Sun Y, Vaquer-Alicea J, Maggiore G, Clemen CS, Buscher WJ, Bieschke J, Kotzbauer P, Ayala Y, Diamond MI, Davis AA, Weihl C, VCP suppresses proteopathic seeding in neurons, Mol Neurodegener 17(1) (2022) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chen D, Drombosky KW, Hou Z, Sari L, Kashmer OM, Ryder BD, Perez VA, Woodard DR, Lin MM, Diamond MI, Joachimiak LA, Tau local structure shields an amyloid-forming motif and controls aggregation propensity, Nat Commun 10(1) (2019) 2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Dujardin S, Commins C, Lathuiliere A, Beerepoot P, Fernandes AR, Kamath TV, De Los Santos MB, Klickstein N, Corjuc DL, Corjuc BT, Dooley PM, Viode A, Oakley DH, Moore BD, Mullin K, Jean-Gilles D, Clark R, Atchison K, Moore R, Chibnik LB, Tanzi RE, Frosch MP, Serrano-Pozo A, Elwood F, Steen JA, Kennedy ME, Hyman BT, Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease, Nat. Med. 26(8) (2020) 1256–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kopeikina KJ, Hyman BT, Spires-Jones TL, Soluble forms of tau are toxic in Alzheimer’s disease, Transl. Neurosci. 3(3) (2012) 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Abisambra JF, Jinwal UK, Blair LJ, O’Leary JC 3rd, Li Q, Brady S, Wang L, Guidi CE, Zhang B, Nordhues BA, Cockman M, Suntharalingham A, Li P, Jin Y, Atkins CA, Dickey CA, Tau accumulation activates the unfolded protein response by impairing endoplasmic reticulum-associated degradation, J. Neurosci. 33(22) (2013) 9498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rosso SM, Donker Kaat L, Baks T, Joosse M, de Koning I, Pijnenburg Y, de Jong D, Dooijes D, Kamphorst W, Ravid R, Niermeijer MF, Verheij F, Kremer HP, Scheltens P, van Duijn CM, Heutink P, van Swieten JC, Frontotemporal dementia in The Netherlands: patient characteristics and prevalence estimates from a population-based study, Brain 126(Pt 9) (2003) 2016–22. [DOI] [PubMed] [Google Scholar]
- [87].Holmes BB, DeVos SL, Kfoury N, Li M, Jacks R, Yanamandra K, Ouidja MO, Brodsky FM, Marasa J, Bagchi DP, Kotzbauer PT, Miller TM, Papy-Garcia D, Diamond MI, Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds, Proc. Natl. Acad. Sci. U. S. A. 110(33) (2013) E3138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Xu WC, Liang JZ, Li C, He ZX, Yuan HY, Huang BY, Liu XL, Tang B, Pang DW, Du HN, Yang Y, Chen J, Wang L, Zhang M, Liang Y, Pathological hydrogen peroxide triggers the fibrillization of wild-type SOD1 via sulfenic acid modification of Cys-111, Cell Death Dis. 9(2) (2018) 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Munch C, O’Brien J, Bertolotti A, Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells, Proc. Natl. Acad. Sci. U. S. A. 108(9) (2011) 3548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Woo JA, Liu T, Zhao X, Trotter C, Yrigoin K, Cazzaro S, Narvaez E, Khan H, Witas R, Bukhari A, Makati K, Wang X, Dickey C, Kang DE, Enhanced tau pathology via RanBP9 and Hsp90/Hsc70 chaperone complexes, Hum. Mol. Genet. 26(20) (2017) 3973–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Echeverria PC, Bernthaler A, Dupuis P, Mayer B, Picard D, An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine, PLoS One 6(10) (2011) e26044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rulten SL, Kinloch RA, Tateossian H, Robinson C, Gettins L, Kay JE, The human FK506-binding proteins: characterization of human FKBP19, Mamm. Genome 17(4) (2006) 322–31. [DOI] [PubMed] [Google Scholar]
- [93].Ruer-Laventie J, Simoni L, Schickel JN, Soley A, Duval M, Knapp AM, Marcellin L, Lamon D, Korganow AS, Martin T, Pasquali JL, Soulas-Sprauel P, Overexpression of Fkbp11, a feature of lupus B cells, leads to B cell tolerance breakdown and initiates plasma cell differentiation, Immun Inflamm Dis 3(3) (2015) 265–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ryder BD, Wydorski PM, Hou Z, Joachimiak LA, Chaperoning shape-shifting tau in disease, Trends Biochem. Sci. 47(4) (2022) 301–313. [DOI] [PubMed] [Google Scholar]
- [95].Deshayes N, Arkan S, Hansen C, The Molecular Chaperone DNAJB6, but Not DNAJB1, Suppresses the Seeded Aggregation of Alpha-Synuclein in Cells, Int J Mol Sci 20(18) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Gillis J, Schipper-Krom S, Juenemann K, Gruber A, Coolen S, van den Nieuwendijk R, van Veen H, Overkleeft H, Goedhart J, Kampinga HH, Reits EA, The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides, J Biol Chem 288(24) (2013) 17225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Mansson C, Arosio P, Hussein R, Kampinga HH, Hashem RM, Boelens WC, Dobson CM, Knowles TP, Linse S, Emanuelsson C, Interaction of the molecular chaperone DNAJB6 with growing amyloid-beta 42 (Abeta42) aggregates leads to sub-stoichiometric inhibition of amyloid formation, J Biol Chem 289(45) (2014) 31066–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rodriguez-Gonzalez C, Lin S, Arkan S, Hansen C, Co-chaperones DNAJA1 and DNAJB6 are critical for regulation of polyglutamine aggregation, Sci Rep 10(1) (2020) 8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hageman J, Rujano MA, van Waarde MA, Kakkar V, Dirks RP, Govorukhina N, Oosterveld-Hut HM, Lubsen NH, Kampinga HH, A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation, Mol Cell 37(3) (2010) 355–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


