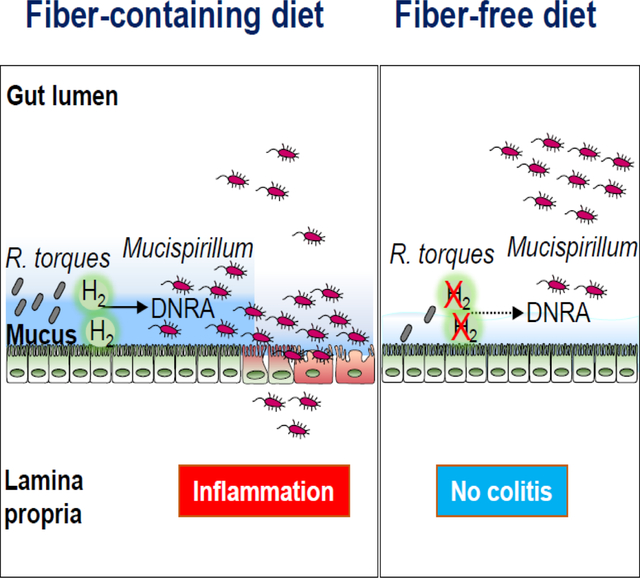
SUMMARY
Exclusive enteral nutrition (EEN) with fiber-free diets is an effective steroid-sparing treatment to induce clinical remission in children with Crohn’s disease (CD). However, the mechanism underlying the beneficial effects of EEN remains obscure. Using a model of microbiota-dependent colitis with the hallmarks of CD, we find that administration of a fiber-free diet prevents the development of colitis and inhibits intestinal inflammation in colitic animals. Remarkably, fiber-free diet alters the intestinal localization of Mucispirillum schaedleri, a mucus-dwelling pathobiont, which is required for triggering disease. Mechanistically, the absence of dietary fiber reduces nutrient availability and impairs the DNRA metabolic pathway of M. schaedleri, leading to exclusion from the mucus layer and disease remission. Thus, appropriate localization of the specific pathobiont in the mucus layer is critical for disease development, which is disrupted by fiber exclusion. These results suggest strategies to treat CD by targeting the intestinal niche and metabolism of disease-causing microbes.
Keywords: Crohn’s disease, exclusive enteral nutrition, fiber-free diet, mucus-dwelling pathobiont, dissimilatory nitrate reduction to ammonia, Ruminococcus torques, Mucispirillum schaedleri
Graphical Abstract
eTOC Blurb
Kuffa et al. demonstrate that fiber-free diet prevents and inhibits disease in a Crohn’s disease-like colitis model. Fiber exclusion reduces the abundance of M. schaedleri, a mucus-dwelling pathobiont required for triggering disease. Mechanistically, fiber deprivation reduces nutrient availability and impairs the DNRA metabolic pathway in M. schaedleri to promote re-localization.
INTRODUCTION
Crohn’s disease (CD), one of the major forms of inflammatory bowel disease (IBD), is a chronic inflammatory disorder of the gastrointestinal tract that affects both children and adults 1,2. While the incidence of adult-onset IBD has plateaued in many westernized regions 3,4, the IBD incidence in children including very early onset IBD (VEOIBD) has increased in the Western world over the past two decades, with a more pronounced increase in CD 5,6. Although the pathogenesis of CD remains poorly understood, accumulating evidence suggests that CD arises from a confluence of genetic and environmental factors that alter gut homeostasis 1,7. Genetic studies have identified mutations in more than 100 genes that are associated with CD, many of which regulate microbial recognition, killing, and immune responses to microbes 8,9. Furthermore, increased genetic burden has been associated with earlier age of onset for CD including VEOIBD 10. Both VEOIBD and adult CD are associated with mutations in NOD2, a gene that regulates bacterial recognition, and genes that encode components of the phagocyte NADPH oxidase complex including CYBB that are critical for bacterial killing 7,11,12. Notably, NOD2 deficiency together with deficiency in CYBB triggers spontaneous early-onset CD-like colitis in mice exposed to a specific microbiota 13. Thus, specific interactions between host genes and the microbiota are critical for the development of disease.
Alterations in the gut microbiota or dysbiosis are commonly found in patients with IBD including CD 7. For instance, a decreased community diversity and a shift in certain bacterial taxa is a common feature of the disease in both adults and children with CD 7,14. However, such microbial alterations seen in CD patients may be secondary to inflammation. Although direct evidence for a causal role for specific microbes in CD is lacking, the imbalance in the gut microbial community towards a dysbiotic state is more pronounced in mucosal samples than in the stool of adult CD and treatment-naïve pediatric CD patients 14,15. Thus, it is conceivable that mucus-dwelling microbes, due to their proximity to the intestinal epithelium, are more likely to trigger abnormal mucosal immune responses in genetically susceptible individuals. Thus, strategies that target disease-causing microbes living near the epithelium may be beneficial in patients with CD.
Given the potential importance of the gut microbiota in triggering CD, the development of therapeutic approaches that target causal intestinal microbes may provide a unique approach to treatment of this disease. Diet is one of the available tools that can be used to modify the intestinal microbiota 7. As of now, there are no clear evidence-based recommendations for diet as therapy for CD, except for EEN. EEN is a complete liquid formula diet that contains all nutritional requirements but is typically free of or low in plant fiber 16,17. EEN is a first line therapy for mild to moderately active pediatric CD, providing complete nutrition while simultaneously inducing remission in 60–85% of cases 18–20. EEN is also effective in select adult patients with CD 21. However, EEN is a very restrictive diet, which has limited its acceptability and clinical use, particularly in adults with CD. The effectiveness of EEN in the treatment of pediatric CD is somewhat unexpected, given that the administration of low fiber diets in murine models is associated with thinning of the mucus layer and increased mucus permeability 22–24. The mechanism by which EEN induces clinical remission in children with CD remains unclear, although it has been suggested that EEN may act by reducing the luminal load of dietary antigens or altering the composition of the gut microbiota 25,26. Using a spontaneous model of CD, we found that a fiber-free diet ameliorates colitis by altering the intestinal localization of a specific gut pathobiont that is required to trigger CD-like disease in genetically susceptible mice. Mechanistically, fiber-free diet inhibited the ability of mucin-degrading microbes to supply critical molecules that are required for the metabolism and maintenance of the disease-causing pathobiont in the mucus layer.
RESULTS
Exclusion of dietary fiber inhibits spontaneous CD-like colitis
To investigate the role of dietary fiber in intestinal inflammation, we utilized a spontaneous model of CD-like colitis. In this model, mice doubly deficient in Nod2 and phagocyte NADPH oxidase/Cybb (DKO), two genes associated with human CD, develop a spontaneous early-onset Th1-type CD-like colitis when exposed to Taconic microbiota (Tac) 13. To determine whether dietary fiber prevents the development of colitis, littermate Tac-DKO mice were separated at weaning before the development of colitis and fed ad libitum a fiber-containing mouse chow, referred to as regular chow (RC), or a fiber-deficient diet (FD) for up to 9 weeks (Figure 1A). The macronutrient composition of FD resembles some low fiber/no fiber EEN diets clinically used in pediatric CD patients for up to 12 weeks to induce disease remission 17,18,25. First, we monitored diet consumption and found no difference between the RC and the FD (Figure S1A). Tac-DKO mice maintained on the RC spontaneously developed CD-like colitis, as verified by increased levels of fecal lipocalin-2 (Lcn-2) (Figure 1B), a marker of intestinal inflammation 27, and by histological examination (Figures 1C and 1D). Consistent with our previous study 13, gut inflammation in RC-fed Tac DKO mice was characterized by crypt architectural distortion, transmural inflammatory cell infiltrates, and ulcers (Figure 1C), all hallmarks of CD in both adults and children 1,28. In contrast, Tac-DKO animals maintained on FD were protected from colitis (Figures 1B–D). Consistent with these observations, transcript levels of pro-inflammatory cytokines, such as tumor necrosis factor α (TNFα), interferon γ (IFNγ), interleukin-6 (IL-6), but not IL-17A, were decreased in colonic explants of Tac-DKO mice on FD as compared to the animals maintained on RC (Figure 1E).
Figure 1. Fiber-deficient diet inhibits intestinal inflammation in Tac-DKO mice.
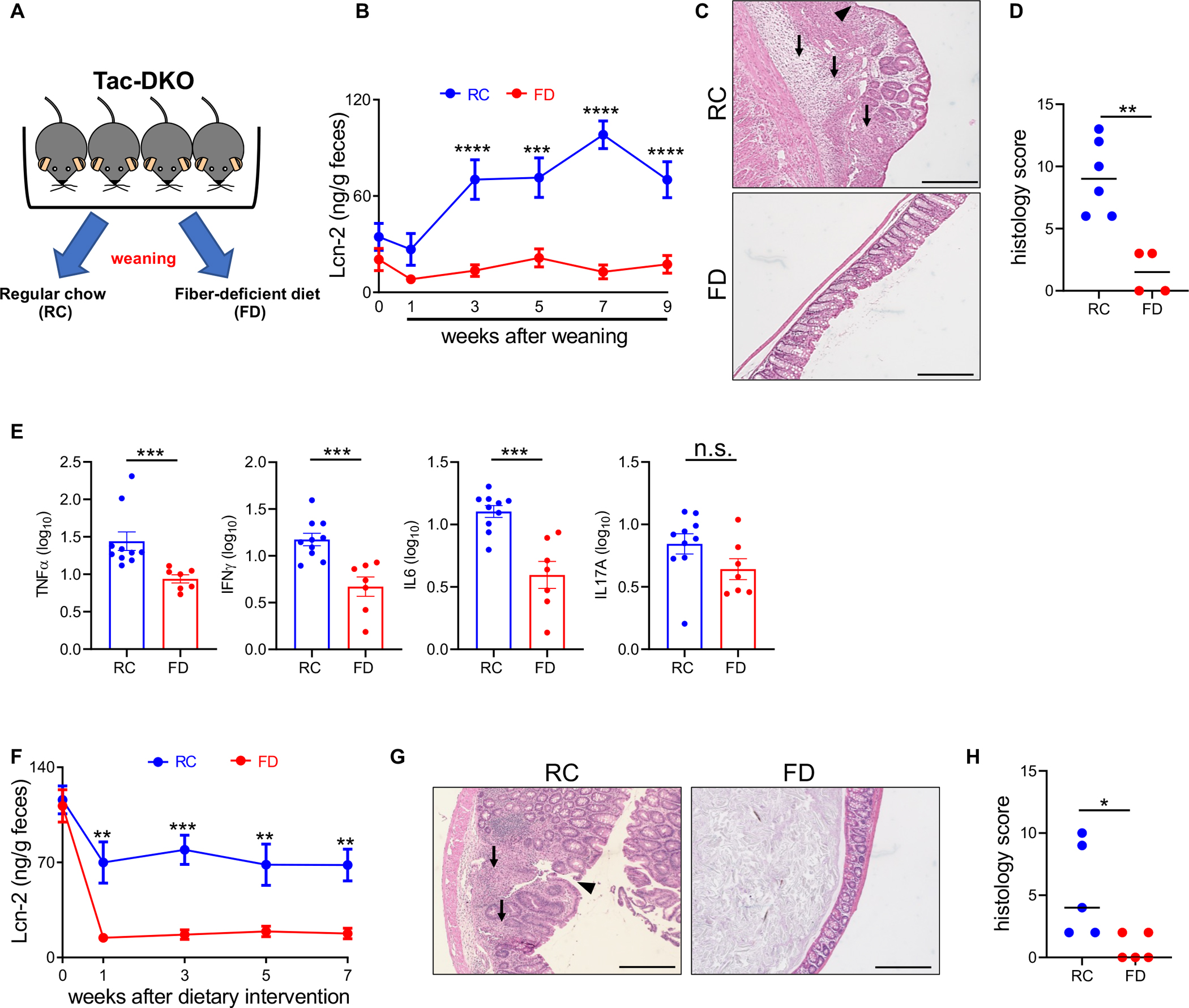
(A) Schematic representation of the experimental design for feeding strategies. Fecal Lcn-2 levels in Tac-DKO mice fed either the RC or the FD during preventive (B), and therapeutic approach (F). Representative hematoxylin and eosin (H&E)-stained colonic sections from RC- and FD-fed Tac-DKO mice during preventive (C) and therapeutic (G) approach. Scale bar, 200 μm. Arrowhead shows ulcer. Black arrows indicate colonic transmural inflammation. Histology scores of colonic tissue from RC-fed Tac-DKO and FD-fed Tac-DKO mice during preventive (D) and therapeutic (H) approach. (E) Gene expression in colonic tissue of RC-fed and FD-fed Tac-DKO mice was normalized to GAPDH expression.
Each symbol represents one mouse. Data are mean ± SEM (B, E and F), or median (D and H); and representative of at least two independent experiments, n= 4–12 per group. **p<0.01; ***p<0.001; ****p<0.0001; n.s. not significant by two-way repeated measures ANOVA followed by Sidak’s post-test (B and F), by two-tailed unpaired t-test (E) or by two-tailed Mann-Whitney U test (**p=0.0095, D; **p=0.0476, H). See also Figure S1; Tables S1 and S7.
The RC and FD diets differ substantially in several aspects of their macronutrient composition in addition to fiber. To specifically test the influence of fiber on disease severity, we utilized a custom-made control mouse diet (CTR), in which roughly 50% of the simple sugar FD contains (glucose) was removed and replaced by several natural fiber sources to match the percentage of neutral detergent fiber present in RC (Table S1). In a preventive protocol, littermate Tac-DKO mice were fed either the CTR or the FD, before the development of colitis (at 21 days of age), up to 9 weeks (Figure S1B). While no difference in food consumption was noted between the two groups (Figure S1C), restoring dietary fiber using the CTR diet to Tac-DKO mice promoted the development of colitis, as confirmed by Lcn-2 measurements, histological examination, and pro-inflammatory cytokine profiles (Figures S1D–G).
Next, we examined the therapeutic impact of fiber exclusion on mice that had already developed gut inflammation. In these studies, littermate colitic Tac-DKO mice (4–5 weeks of age) were separated and fed either the RC or the FD for up to 7 weeks. Notably, Tac-DKO mice on FD exhibited decreased Lcn-2 levels and histology compared to mice maintained on RC (Figures 1F–H). Collectively, these results indicate that exclusion of fiber from the diet inhibits intestinal inflammation in our model of CD-like colitis using both preventive and therapeutic protocols.
Fiber-deficient diet reduces the abundance of pathobiont Mucispirillum in the mucus layer
Administration of a FD results in thinning of the intestinal mucus layer, which is associated with increased mucin-degrading activity of mucus-dwelling symbionts 22,23,29. Consistent with these observations, FD-fed Tac-DKO mice displayed increased permeability to FITC-dextran (Figure S2A) and reduced mucus thickness compared to their RC-fed (Figures 2A–C) or CTR-fed (Figure S2C) animal counterparts. Similar thinning of the colonic mucus layer was observed in colitic Tac DKO mice fed the FD in contrast to RC-fed littermates (Figure S2B). As expected, degradation of the mucus layer during periods of fiber exclusion brought luminal microbes closer to the gut epithelium (Figures 2A and 2B, right panels). Next, we examined whether mucus production was altered in Tac-DKO mice fed the FD. To this end, we determined the abundance of transcripts encoding several key proteins involved in synthesis and regulation of the mucus layer, such as Muc2, Muc5ac, Tff1, Tff3, and Klf3 30. Our results show no differences in transcript levels of any genes examined between FD and RC groups (Figure 2D). Consistent with these findings, qualitative visualization by Alcian blue and Alcian blue/Periodic acid-Schiff (PAS) staining showed that the colonic tissue of FD- and RC-fed Tac DKO mice contained similar numbers of goblet cells (Figures 2A and 2B, first two panels from the left). Despite displaying reduced mucus layer thickness (Figure 2C), the FD-fed Tac-DKO showed less inflammation compared to the RC group (Figures 1B–D).
Figure 2. Fiber-deficient diet leads to altered localization of gut pathobiont Mucispirillum in Tac-DKO mice.
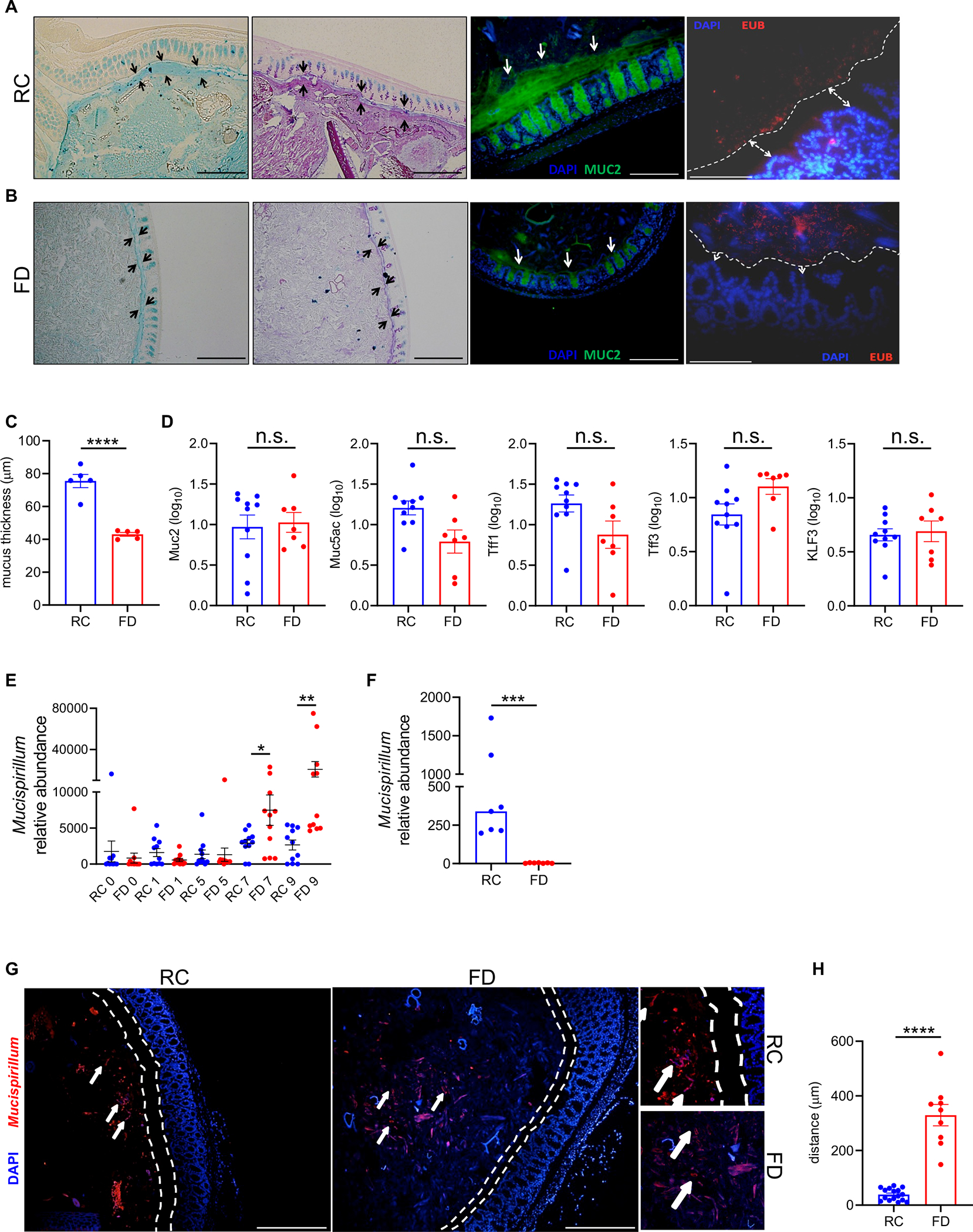
Alcian blue-stained (first panel from the left) and Alcian blue/PAS-stained (second panel from the left) colonic sections from RC-fed (A) and FD-fed (B) Tac-DKO mice. Opposing black arrows delineate the mucus layer. Scale bars, 200 μm. Immunofluorescence images of colonic sections (third panel from the left) from RC-fed (A) and FD-fed (B) Tac-DKO mice stained with anti-Muc2 antibody (green) and DAPI (blue). White arrows indicate the mucus layer. Scale bars, 200 μm. Fluorescence in situ hybridization (FISH) (right panel) of colonic sections from RC-fed (A) and FD-fed (B) Tac-DKO mice (red, EUB338 probe; blue, DAPI). Dashed white line and white arrows delineate mucus layer. Scale bars, 100 μm. (C) Colonic mucus layer measurements in RC-fed and FD-fed Tac-DKO mice. (D) Gene expression in colonic tissue of RC-fed and FD-fed Tac-DKO mice was normalized to GAPDH expression. (E) Abundance of Mucispirillum in fecal DNA from RC-fed and FD-fed Tac-DKO mice was normalized to the universal 16S rRNA gene. (F) Abundance of Mucispirillum in colonic mucus scrapings harvested from RC-fed and FD-fed Tac-DKO mice was normalized to the universal 16S rRNA gene. (G) FISH analysis using Mucispirillum-specific probe (red) and DAPI (blue) of colonic sections from RC-fed (left panel) and FD-fed (right panel) Tac-DKO mice. Dashed white lines delineate mucus layer. White arrows indicate Mucispirillum. Scale bars, 200 μm. Insets: shows a higher magnification of Mucispirillum in proximity to host epithelium in RC-fed animals (top inset), and in the lumen in FD-fed mice (lower inset). (H) Distance from bacterium Mucispirillum to the inner mucus layer in RC- or FD-fed Tac-DKO mice.
Each symbol represents one mouse. Data are mean ± SEM (C, D, E and H), or median (F), and representative of at least two independent experiments, n= 5–11 per group. *p< 0.05; **p<0.01; ***p<0.001; ****p<0.0001; n.s. not significant by two-tailed unpaired t-test (C, D and H), by one-way ANOVA followed by Tukey’s post-test (E), or by two-tailed Mann-Whitney U test (***p=0.0006, F). See also Figure S2; Table S7.
Colitis in Tac-DKO mice is triggered by intestinal accumulation and penetration of the pathobiont Mucispirillum schaedleri (referred here as Mucispirillum) 13. We thus examined the intestinal abundance of Mucispirillum during dietary intervention. Surprisingly, time-course analysis revealed increased luminal Mucispirillum abundance in fecal samples obtained from FD-fed Tac-DKO mice compared to RC-fed animals at later time points (i.e., 7 and 9 weeks of dietary intervention) (Figure 2E). These results were somewhat unexpected given that FD-fed Tac-DKO were protected from the development of colitis (Figure 1). Because Mucispirillum primarily localizes to the mucus layer 31, we set out to quantify Mucispirillum abundance in the mucus layer during dietary interventions. Remarkably, we found that unlike the RC-fed Tac-DKO mice whose mucus layer was enriched with Mucispirillum, FD-fed animals exhibited a marked reduction of Mucispirillum abundance in the mucus layer (Figure 2F), despite having similar bacterial genome copies (16S rRNA) and host genomic copy numbers (ActB) in both luminal and mucosal compartments (Figures S2D and S2E). In line with these findings, Mucispirillum accumulated in the proximity of the gut epithelium in RC-fed mice, whereas the bacterium in FD-fed animals primarily localized in the intestinal lumen in FD-fed animals, as shown by fluorescence in situ hybridization (Figures 2G and 2H). Taken together, these results indicate that the absence of dietary fiber leads to altered localization of the disease-causing Mucispirillum in the intestine.
The mucus layer is critical for Mucispirillum intestinal colonization and colitis induction
Mucispirillum was barely detectable in the mucus layer of FD-fed Tac-DKO mice (Figure 2F), which were protected from disease development (Figures 1B–D). Thus, we hypothesized that appropriate localization of Mucispirillum in the mucus layer is critical for its ability to locally invade the intestinal tissue and to trigger inflammation. To test this hypothesis, adult littermate DKO mice harboring Mucispirillum-free microbiota (microbiota from Jackson Laboratory (Jax) mice) 13, were fed either the RC or the FD for two weeks prior to bacterial challenge (Figure 3A). While remaining on either the RC or the FD, Jax-DKO mice were pre-treated with vancomycin and then orally gavaged with Mucispirillum (Figure 3A). As expected, the absence of dietary fiber resulted in reduced mucus thickness in Jax-DKO mice (Figure 3B and Figure S3A). Notably, Mucispirillum colonized RC-fed Jax-DKO mice over time after gavage, while its intestinal colonization was markedly reduced in FD-fed animals (Figure 3C). In line with these observations, oral challenge with Mucispirillum resulted in weight loss (Figure 3D) and colitis as determined by increased levels of fecal Lcn-2 in RC-fed mice, but not in FD-fed DKO animals (Figure 3E). Histological examination of Mucispirillum-colonized mice confirmed the development of colitis in RC-fed DKO mice, which was inhibited in FD-fed animals (Figures 3F and 3G). To further test the role of the mucus layer in Mucispirillum colonization, we utilized Muc2−/− mice that are deficient in Muc2, the dominant colonic mucin glycoprotein. Consistent with previous evidence 32, Muc2−/− mice show a very thin colonic mucus layer (Figure S3B and S3C). Littermate mutant Muc2−/− and heterozygous Muc2+/−mice were pre-treated with vancomycin and then orally gavaged with Mucispirillum (Figure 3H). Because Muc2−/− mice can develop colitis over time 33, we used young mice and performed a short bacterial challenge in Muc2−/− animals to avoid confounding effects of inflammation on Muscispirillum colonization (Figure 3H). The Muc2−/− and Muc2+/− mice were euthanized 1 day after bacterial gavage, when levels of fecal Lcn-2 were low and comparable between both groups of mice (Figure 3I). At this time point, Muc2−/−mice exhibited a very thin mucus layer, and a loss of Alcian-blue-positive goblet cells, compared to Muc2+/− animals (Figure 3J and S3D). Notably, Mucispirillum colonization was markedly reduced in Muc2−/−mice compared to their heterozygous littermates (Figure 3K). Collectively, these results indicate an intact mucus layer is important for Mucispirillum intestinal colonization.
Figure 3. The intestinal mucus layer is critical for Mucispirillum intestinal colonization.
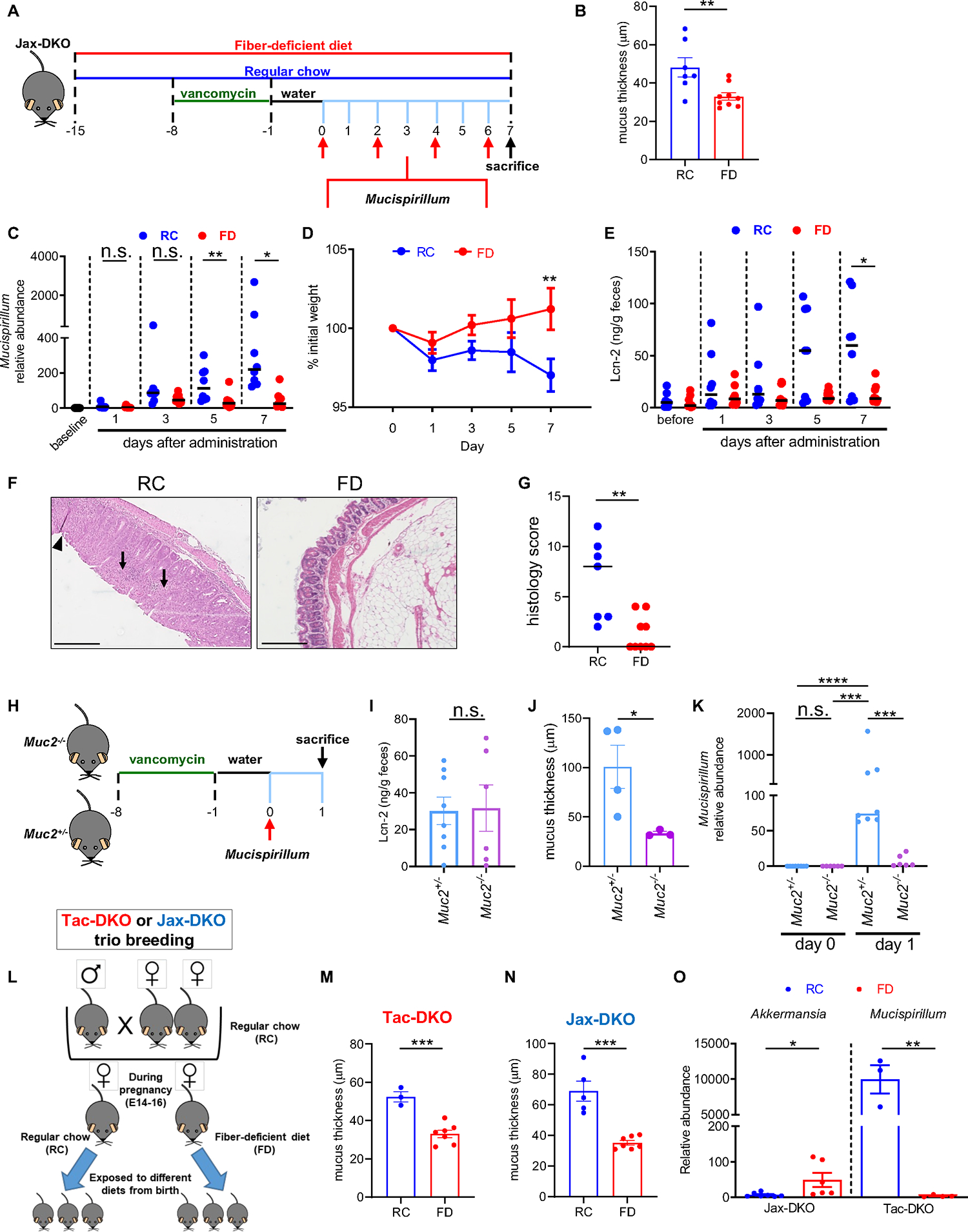
(A) Schematic representation of Mucispirillum challenge in Jax-DKO mice. (B) Colonic mucus layer measurements in RC-fed and FD-fed Jax-DKO mice. (C) Luminal abundance of Mucispirillum prior (baseline) and every other day up to 7 days after bacterial administration in RC-fed and FD-fed Jax-DKO mice was normalized to the universal 16S rRNA gene. (D) Body weight during the 7-day bacterial challenge in RC-fed and FD-fed Jax-DKO mice. (E) Fecal Lcn-2 levels before (day 0) and 7 days after the first Mucispirillum oral gavage. (F) Representative H&E-stained colonic sections from Mucispirillum-infected Jax-DKO mice fed either RC or the FD. Scale bar, 200 μm. Black arrows indicate colonic inflammatory infiltrate and crypt architectural distortion. (G) Histology scores of colonic tissues from Mucispirillum-infected Jax-DKO fed either RC or the FD. (H) Schematic representation of Mucispirillum challenge in Muc2−/− mice. (I) Fecal Lcn-2 levels 1 day after the oral gavage in Muc2+/− and Muc2−/− mice. (J) Colonic mucus thickness measurements in Mucispirillum-infected Muc2+/− and Muc2−/− mice. (K) Luminal Abundance of Mucispirillum in in Muc2+/− and Muc2−/− mice prior (day 0), and 1 day after bacterial administration was normalized to the universal 16S rRNA gene. (L) Schematic representation of the experimental design for trio breeding experiments. Colonic mucus thickness measurements in Tac-DKO (M) and Jax-DKO (N) pups fed either RC or the FD from birth. (O) Abundance of A. muciniphila (Akkermansia) and Mucispirillum in Jax-DKO and Tac-DKO pups, respectively, was normalized to the universal 16S rRNA gene.
Each symbol represents one mouse. Data are mean ± SEM (B, D, I-J, M-O), or median (C, E and K), and representative of at least two independent experiments, n= 3–9 per group. *p< 0.05; **p<0.01; ***p<0.001; ****p<0.0001; n.s. not significant by Kruskal-Wallis test followed by Dunn’s post-test (C, E and K), by two-way ANOVA followed by Sidak’s post-test (D), by two-tailed Mann-Whitney U test (**p=0.0053, G), or by two-tailed unpaired t-test (**p=0.0081, B; n.s., I; *p=0.0498, J; ***p=0.0006, M; ***p=0.0002, N; **p=0.0424 Jax-DKO, O; **p=0.0018 Tac-DKO, O). See also Figure S3; Table S7.
We also sought to assess the role of FD and the mucus layer in maternal transmission of Mucispirillum. To this end, we modified our protocol to perform dietary interventions during the neonatal period by feeding pregnant Tac-DKO dams either the RC or the FD, which allows pups to be exposed to different diets from birth (Figure 3L). Jax-DKO dams fed either the RC or the FD were included as controls (Figure 3L). As expected, fiber exclusion resulted in thinning of the mucus layer in both Tac-DKO (Figure 3M) and Jax-DKO pups (Figure 3N). In line with previous evidence 22, switching to the FD induced a microbiota shift in favor of the mucin-degrading species, such as Akkermansia muciniphila, in Jax-DKO pups (Figure 3O). In contrast, the mucus-dwelling Mucispirillum was depleted in Tac-DKO pups in response to low fiber administration compared to RC-fed pups (Figure 3O). Consistent with the inability of Mucispirillum to colonize the mucus layer in the absence of dietary fiber, FD-fed Tac-DKO pups did not develop spontaneous colitis (Figures S3C–E). In contrast, Mucispirillum-colonized RC-fed Tac-DKO pups spontaneously developed intestinal inflammation, as assessed by fecal Lcn-2 levels and histological examination (Figures S3E–G). As expected, Jax-DKO pups, whose microbiota is devoid of Mucispirillum 13, did not develop intestinal inflammation regardless of the diet (Figures S3H–J). These results indicate that diet-induced alteration of the mucus layer affects vertical transmission of Mucispirillum. Collectively, these results indicate that appropriate localization of Mucispirillum in the mucus layer is critical for its ability to trigger intestinal inflammation.
R. torques promotes Mucispirillum growth
Since our data indicate that the mucus layer plays a crucial role in Mucispirillum colonization (Figure 3), we next assessed the ability of Mucispirillum to grow in BHI medium in the presence of different sugars using a custom carbohydrate array that contains most of the common mono- and polysaccharides present in plant and animal tissue, including purified mucin O-glycans 22. Unlike Escherichia coli, which exhibited optimal growth on monosaccharides (Figure 4A and Table S2), Mucispirillum grew poorly in BHI medium supplemented with either mono- or polysaccharides (Figure 4A and Table S2), while the bacterium grew in a dose-dependent manner in the presence of either amino acids or glycerol (Figure S4A). Taken together, our results suggest that Mucispirillum is not a primary degrader of dietary fiber and/or host-derived polysaccharides. Thus, we reasoned that Mucispirillum may utilize breakdown products produced by mucus-degrading microbes. To test this hypothesis, we utilized specific-pathogen free (SPF) and germ-free (GF) C57BL/6 mice and fed them either the RC or the FD for 2 weeks. Sterile supernatants were prepared anaerobically from luminal contents and mucus isolated from the ceca of RC- and FD-fed SPF and GF C57BL/6 mice and used as a medium for either Mucispirillum or E. coli growth. E. coli bloomed in the presence of GF-derived, but not SPF-derived, supernatants regardless of the diet (Figure S4B), suggesting that E. coli and intestinal microbes compete for nutrients needed for growth. In contrast, Mucispirillum growth was slightly supported by GF-derived supernatants, but markedly enhanced by SPF-derived supernatants, particularly by the cecal supernatant from SPF mice fed the FD (Figure 4B). Taken together, these results indicate that Mucispirillum growth is promoted by nutrients provided by intestinal microbes.
Figure 4. Mucin-degrading microbes promote Mucispirillum growth.
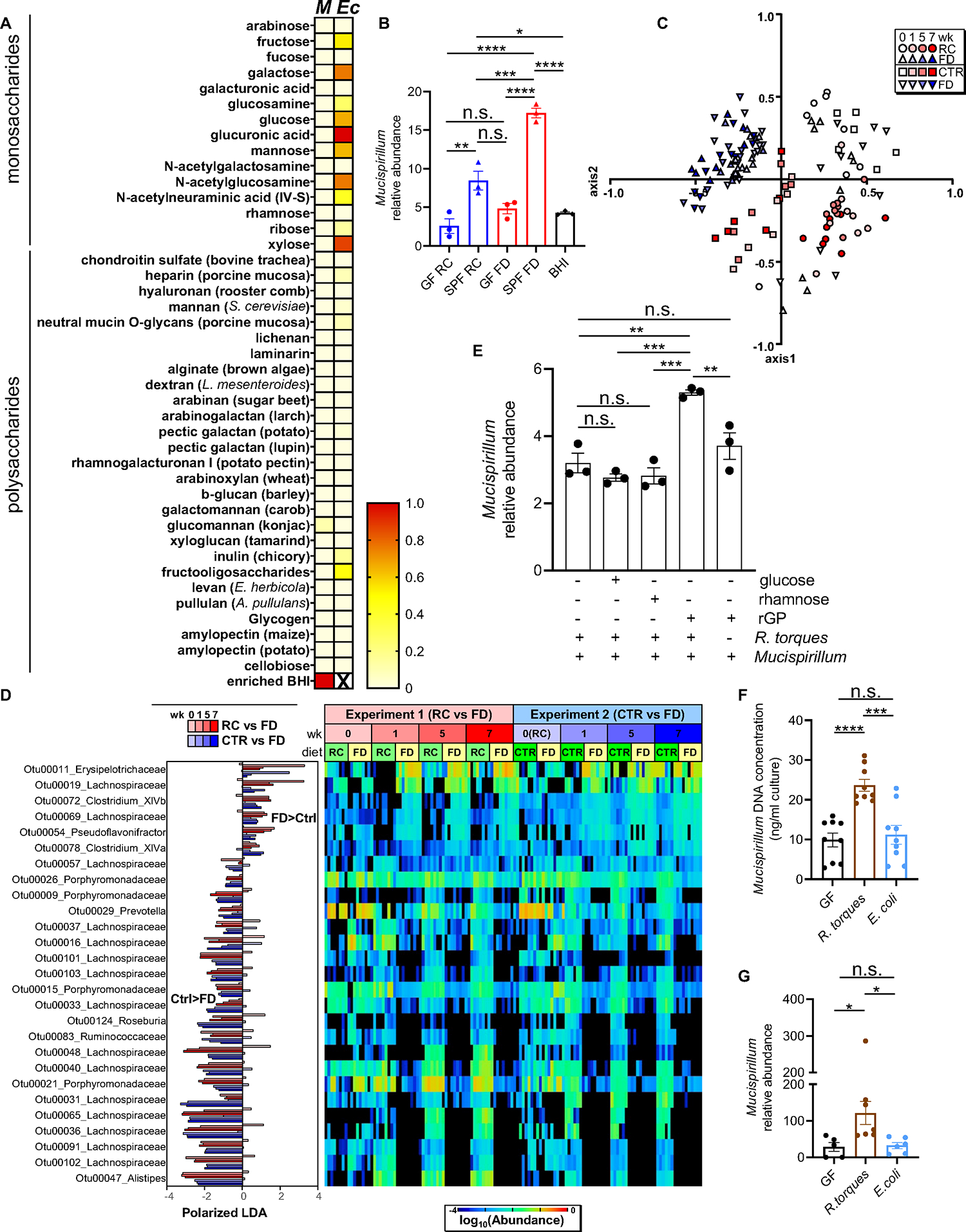
(A) Heat map showing normalized growth values of Mucispirillum (M) or E. coli (Ec) (enriched BHI, positive control for Mucispirillum). (B) Mucispirillum growth in the presence of either GF- or SPF-derived supernatants (enriched BHI (BHI), positive control). Mucispirillum abundance was normalized to the universal 16S rRNA gene. (C) NMDS plot of θYC β-diversity indexes of fecal microbiota from Tac-DKO mice fed either RC (circles), CTR (squares) or FD (triangles) for up to 7 weeks. (D) OTUs differentially abundant in fecal microbiota of Tac-DKO mice fed the FD or the control diets (RC or CTR) shown with LDA values of LEfSe with p<0.05, false discovery rate <0.05 and maximal abundance cutoff <1%. (E) Mucispirillum and R. torques co-culture in custom chopped meat broth in the presence of rectal glycoproteins (rGP), glucose or rhamnose. Mucispirillum abundance was normalized to the universal 16S rRNA gene. (F) Mucispirillum growth in the presence of supernatants derived from GF, R. torques-monocolonized and E. coli-monocolonized mice. (G) Abundance of Mucispirillum in GF, R. torques-monocolonized and E. coli-monocolonized mice was normalized to the universal 16S rRNA gene.
Each symbol represents one mouse, except for panel B, in which each dot represents data pooled from 3 individual mice. Data are mean ± SEM, representative of at least two independent experiments, n= 5–9 per group. *p< 0.05; **p<0.01; ***p<0.001; ****p<0.0001; n.s. not significant by one-way ANOVA followed by Tukey’s post-test. See also Figures S4 and S5; Tables S2, S3 and S7.
To examine the bacterial species that are affected by the exclusion of dietary fiber, we analyzed the composition of the gut microbiota in Tac-DKO mice over time. The α diversity in the fecal microbiota of RC-fed and the FD-fed Tac-DKO mice remained comparable, except for week 7 after dietary intervention (Figure S4C). Likewise, no α diversity differences were noted in the fecal microbiota of CTR- vs FD-fed Tac-DKO mice, except for week 3 (Figure S4D). Importantly, fecal microbiota composition significantly differed among the three diet groups (Figure 4C). As expected, a decrease in the abundance of fiber-degrading species (i.e., Porphyromonadaceae) was observed in the absence of fiber (Figure 4D and Table S3). Moreover, the abundance of several species in the Lachnospiraceae family varied markedly between mice fed fiber-containing diets (either RC or CTR) and a FD diet as determined by linear discriminant analysis effect size (LEfSe) analysis (p < 0.05) (Figure 4D and Table S3). While specific Lachnospiraceae species were reduced in the FD group, other Lachnospiraceae species were increased in the absence of dietary fiber (Figure 4D and Table S3). Furthermore, 16S rRNA sequence alignment homology analysis revealed that the Lachnospiraceae species whose abundance decreased after fiber deprivation (Figure 4D), were similar to that of Ruminococcus torques (Figure S5A), a well-characterized member of the Lachnospiraceae family 34. We thus assessed whether the growth of Mucispirillum is promoted by R. torques. Consistent with previous evidence 35, we found that R. torques was able to grow on purified rectal glycoproteins which was not affected by the presence of Mucispirillum (Figure S5B). To assess whether mucin-degrading R. torques regulates the growth of Mucispirillum, we co-cultured R. torques and Mucispirillum in the presence of purified rectal glycoproteins or monosaccharides (i.e., glucose and rhamnose). We found that R. torques enhanced Mucispirillum growth in the presence of mucin-containing, but not in mucin-free, medium (Figure 4E). Additionally, GF C57BL/6 mice monocolonized with R. torques or non-mucin degrading E. coli as a control were fed a RC (Figure S5C and S5D). Monocolonization with R. torques did not affect the intestinal mucus thickness (Figure S5E). We found that supernatants derived from R torques-monocolonized mice enhanced the growth of Mucispirillum in vitro compared to supernatants derived from either GF or E. coli-monocolonized animals (Figure 4F). Likewise, the intestinal colonization of Mucispirillum was increased after gavage into R. torques-monocolonized mice when compared to gavage of the bacterium into GF or E. coli-monocolonized mice (Figure 4G). Thus, dietary fiber exclusion alters the abundance of specific members of the Lachnospiraceae family that can promote the growth of Mucispirillum.
Exclusion of dietary fiber affects the DNRA metabolic pathway in Mucispirillum
The exclusion of fiber from the diet led to a relocation of Mucispirillum from the mucus layer to the luminal compartment (Figures 2E and 2F). We reasoned that reduced abundance of Mucispirillum in the mucus layer may be the result of altered metabolism of the bacterium at that site after administration of a fiber-free diet. To assess this, we performed metatranscriptomic analysis to identify Mucispirillum genes differentially expressed in the intestine of mice maintained on RC compared to animals fed the FD. Our transcriptomic analysis revealed 8 Mucispirillum genes that were more abundant and 2 genes less abundant in RC-fed mice compared to FD-fed animals (Figure 5A and Table S4). Of these genes, only 2 genes, nroB and nroD, have been functionally annotated in an operon we named nro (nitrate reductase operon) in the Mucispirillum genome and predicted to act in the metabolic pathway of dissimilatory nitrate reduction to ammonia (DNRA) (Figure S6), a part of anaerobic respiration 36. Mucispirillum can utilize nitrate, which is abundant in the inflamed gut 37, as an electron acceptor, and hydrogen (H2) as an electron donor for anaerobic respiration 38. To test whether Mucispirillum was capable of DNRA, we incubated the bacterium in enriched brain heart infusion (BHI) with different combinations of nitrate and/or H2 under anaerobic conditions. Nitrate increased Mucispirillum growth in the presence of H2 (Figure 5B), which was associated with H2 consumption and ammonium production (Figures 5C and 5D), indicating that Mucispirillum was indeed capable of DNRA under our growth conditions. Further transcriptomic analysis revealed reduced expression of all genes belonging to the unique DNRA-associated nitrate reductase operon (nroA, nroB, nroC, nroD, nroE and nroF) in the mucus layer of FD-fed Tac-DKO mice compared to RC-fed animals (Figure 5E and 5F, and Table S4). Taken together, these results suggest that the exclusion of dietary fiber decreases within the mucus layer the function of DNRA, a metabolic pathway that promotes Mucispirillum growth.
Figure 5. Dietary fiber affects dissimilatory nitrate reduction to ammonium in Mucispirillum.
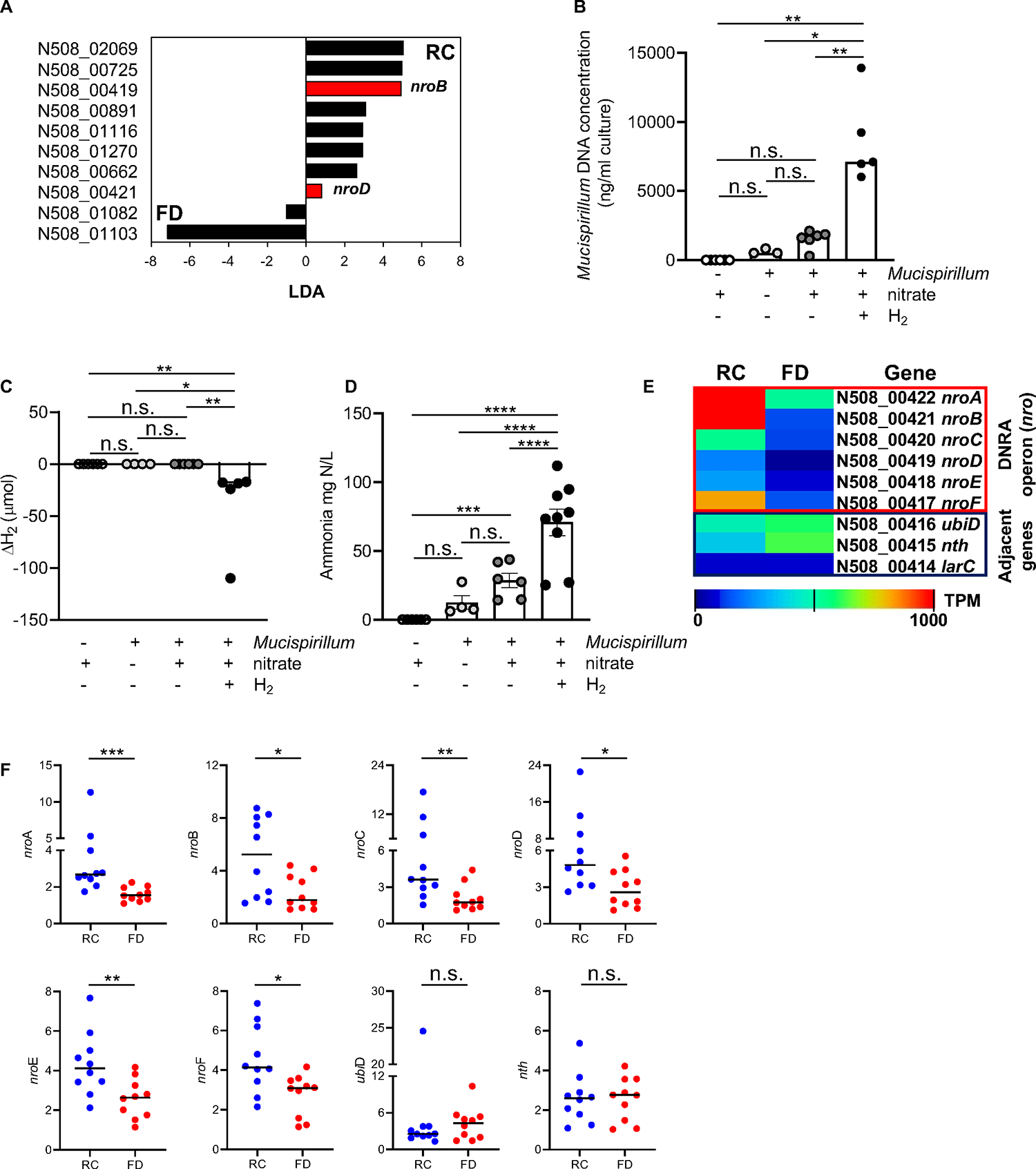
(A) Mucispirillum genes differentially expressed in the two groups of diets (RC vs FD) shown with LDA values by LEfSe with p<0.05, maximum transcript per million (TPM)>10, and >5-fold difference. Red bars indicate nroB and nroD genes that encode putative α and δ subunits of the nitrate reductase, respectively. Black bars indicate genes encoding for uncharacterized proteins. Mucispirillum was grown in the presence/absence of nitrate (10mM) and H2 (80%) in enriched BHI and then Mucispirillum growth (B), H2 consumption (C) and ammonium measurements (D) were assessed after three days. (E) Heat map showing the DNRA operon (nro) (loci N508_00422-N508_00417) in mucus isolates from RC- or FD-fed Tac-DKO mice (adjacent genes, control). Average transcript levels (log10 TPM) in RC- and FD-fed mice. p=not significant. (F) Gene expression in mucus isolates from RC-fed or FD-fed Tac-DKO animals normalized to gap expression.
Each symbol represents one mouse. Data are mean ± SEM (D), or median (B, C and F), and representative of at least two independent experiments, n= 4–10 per group. *p< 0.05; **p<0.01; ***p<0.001; ****p<0.0001; n.s. not significant by Kruskal-Wallis test followed by Dunn’s post-test (B-C), by one-way ANOVA followed by Tukey’s post-test (D), by two-tailed unpaired t-test (E), or by two-tailed Mann-Whitney U test (***p=0.0002, nroA; *p=0.0355, nroB; **p=0.0089, nroC; *p=0.0232, nroD; **p=0.0068, nroE; *p=0.0185, nroF; n.s. not significant, nth and ubiD, F). See also Figure S6; Tables S4 and S7.
Fiber exclusion alters fermentative H2 metabolism and interspecies H2 transfer
H2 fuels Mucispirillum growth in the presence of nitrate (Figure 5B), but Mucispirillum lacks hydrogenases that are required for H2 production 38. Thus, we hypothesized that H2 is provided to Mucispirillum by fermentative microbes, which may be impaired in mice fed a FD. We thus surveyed the distribution of hydrogenase-encoding genes that leads to the production of diffusible H2 in Tac-DKO mice to determine the dominant mechanisms of H2 cycling during dietary fiber exclusion. Two distinct classes of hydrogenases, FeFe Groups A1 and B and NiFe Group 4a, are involved in hydrogenogenic fermentation 39. Transcriptomic analysis revealed a reduction in the total transcript levels of FeFe Groups A1 and B hydrogenase homologues in the mucus layer of Tac-DKO mice that were fed the FD compared to mice maintained on RC (Figure 6A and Table S5). In contrast, no changes in the transcript levels of these hydrogenases were found in the luminal compartment (Figure 6A and Table S5). These results suggest that H2 production is diminished in the mucus layer after dietary fiber exclusion. Furthermore, we found that distinct members of the Lachnospiraceae family harboring [FeFe]-hydrogenase genes were differentially abundant in each diet group (i.e., RC vs FD) and in each intestinal compartment (i.e., mucus vs lumen) (Figure 6B and Table S5). We also assessed H2 metabolism in the human gut during fiber exclusion by analyzing available metagenomics data from pediatric CD patients who received EEN 26. A marginal increase in the number of microbes (relative abundance) encoding FeFe Groups A1 and B hydrogenases was observed in fecal samples isolated from EEN-treated CD children (1 week of EEN) compared to samples of patients before starting EEN (Figure 6C and Table S6). More importantly, consistent with our mouse transcriptomic data (Figure 6B), we found that distinct members of the Lachnospiraceae family harboring [FeFe]-hydrogenase genes were differentially abundant in CD children before starting EEN (baseline) and after 1 week of nutritional therapy (Figure 6D and Table S6). Thus, our findings indicate that the exclusion of dietary fiber alters the levels of Fe-Fe hydrogenases-expressing Lachnospiraceae both in mice and in humans with CD.
Figure 6. Fiber exclusion alters fermentative H2 metabolism and interspecies H2 transfer.
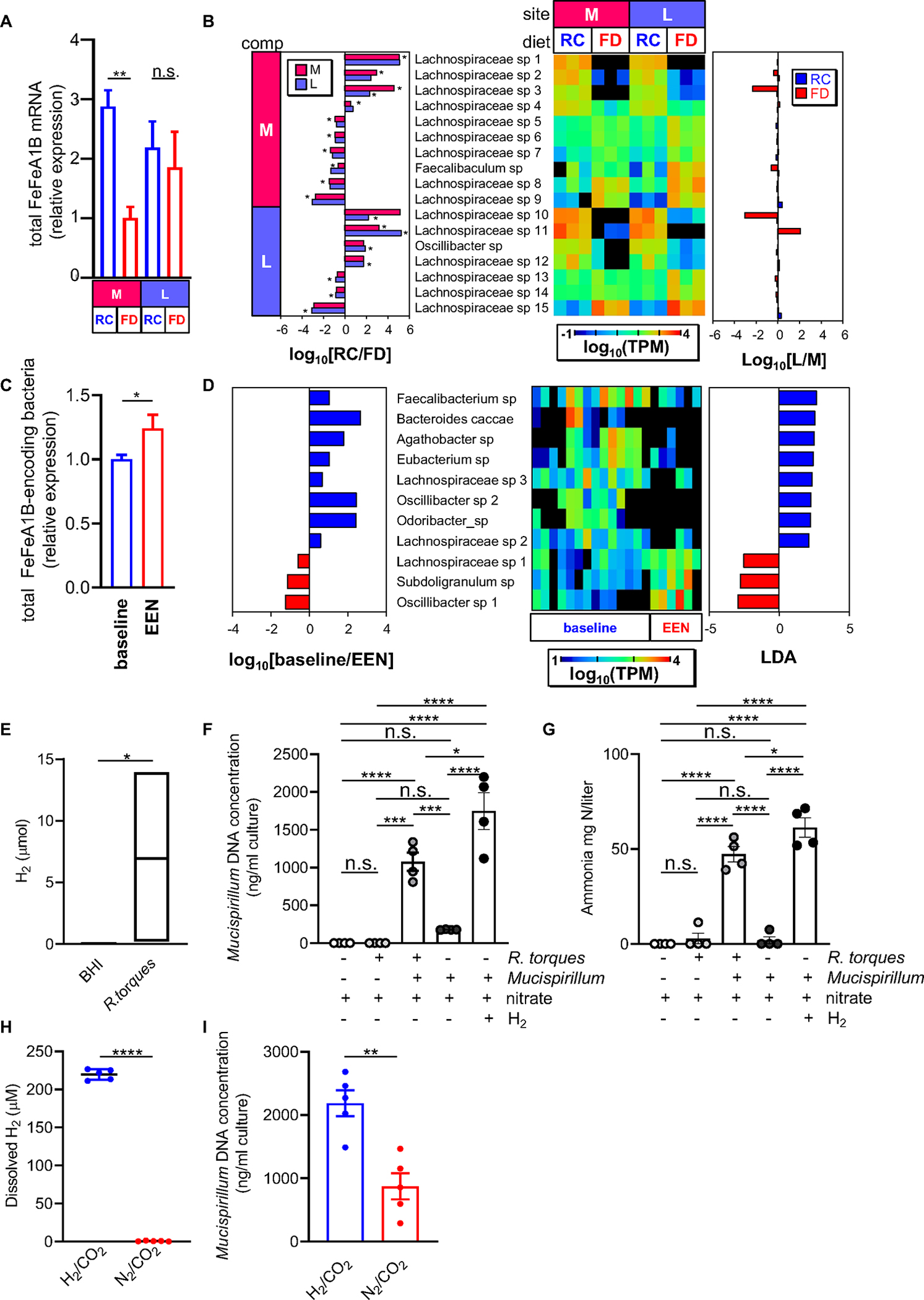
(A) Relative expression level of total FeFe hydrogenase groups A1 and B in mucus (M) and luminal (L) compartments of RC-fed and FD-fed Tac-DKO mice. FeFe Groups A1 and B expression was normalized to 20 housekeeping genes. (B) Genes were selected by comparison between RC and FD at L and M compartments. Microbial species that were differentially abundant in RC vs FD group, and in L vs M are shown as log10 RC/FD (left panel), log10 TPM (central heat map), and log10 L/M (right panel). (C) Abundance of total bacterial FeFe hydrogenase group A1 and B homologues normalized by the abundance of 20 housekeeping genes in the data obtained from CD patients (Original raw data SRP057027 retrieved from 26). (D) Specific FeFe A1/B homologues-harboring species with differential abundance between baseline and 1-week EEN treatment (right panel) are shown with heat map (central panel, log10 TPM) and LDA values of LEfSe with p<0.05 (left panel). (E) H2 content in R. torques cultures (enriched BHI medium, control). Mucispirillum was grown in the presence/absence of R. torques, nitrate (10mM) and H2 (80%) in enriched BHI. Mucispirillum growth (F) and ammonium measurements (G) were assessed after three days. Co-cultures of R. torques and Mucispirillum in bioreactor vessels sparged with a mixed gas containing 20% CO2 and either 80% N2 or 80% H2. (H) Dissolved H2 concentration at 26.5 hours post-inoculation. (I) Mucispirillum growth at day 3 post-inoculation.
Each symbol represents one mouse. Data are mean ± SEM (A, C, F-I), or floating bars show max and min values and line at median (E), and representative of at least two independent experiments, n= 4–5 per group. **p<0.01; ***p<0.001; ****p<0.0001; n.s. not significant by one-way ANOVA followed by Tukey’s post-test (A, F-G), by two-tailed Mann-Whitney U test (*p=0.02, C; *p=0.0286, E), or by two-tailed unpaired t-test (****p<0.0001, H; **p=0.0020, I). See also Figure S7; Table S5, S6 and S7.
We tested whether Mucispirillum utilizes H2 produced by Lachnospiraceae species to promote its growth. In these experiments we utilized R. torques, which can enhance Mucispirillum growth (Figures 4E and 4F), and tested if R. torques can promote Mucispirillum growth by transferring H2. To this end, we first confirmed that R. torques grew (Figure S7A) and produced H2 in nitrate-supplemented BHI medium (Figure 6E). Next, we co-cultured Mucispirillum with R. torques in the absence of exogenous H2 in nitrate-supplemented BHI. Notably, R. torques promoted Mucispirillum growth in the absence of H2 to the same extent as it was observed in the presence of exogenous H2 (Figure 6F). The enhancement of Mucispirillum growth by R. torques was associated with ammonium production (Figure 6G), suggesting that Mucispirillum uses anaerobic respiration through DNRA in the presence of R. torques. We next examined whether R. torques promotes Mucispirillum growth through H2 cross-feeding. To this end, we utilized a nitrogen sparging technique that has been shown to decrease the dissolved H2 concentration in a fermentative H2-producing reactor 40. In these experiments co-cultures of R. torques and Mucispirillum were grown in bioreactor vessels and sparged with a mixed gas containing 20% carbon dioxide (CO2) and either 80% nitrogen (N2) or 80% H2 (Figure S7B). N2 sparging resulted in a marked reduction of dissolved H2 in the bacterial co-cultures (Figure 6H) that was stable over a 3-day culture period (Figure S7C). N2 sparging resulted in a marked reduction of Mucispirillum growth compared to the H2-sparged cultures (Figure 6I), which is consistent with a reduced H2 concentration (Figure 6H and Figure S7C). Taken together, these results suggest that Lachnospiraceae species play an important role in H2 metabolism, both in the mouse and human colon, during periods of fiber exclusion. Furthermore, Lachnospiraceae species, such as R. torques, can support the growth of disease-causing Mucispirillum via interspecies H2 transfer.
DISCUSSION
The gut microbiota is thought to be a crucial determinant of host susceptibility to several diseases, including CD 41. Although dietary fiber is typically considered beneficial in humans 42, fiber-free (or fiber-low) diet therapies such as EEN markedly improve disease outcomes in children with CD 18–20. Consistent with the human data, our findings indicate that the administration of a fiber-free diet prevents and/or ameliorates the CD-like colitis in our animal model that is based on mutations associated with human disease. The mechanism by which EEN is effective in inducing clinical remission in pediatric CD remains unknown, but it has been proposed that EEN may act by reducing the amounts of dietary antigens or changing the composition of the gut microbiota 25,26. Our results reveal a distinct and unique mechanism through which the absence of dietary fiber inhibits intestinal inflammation by targeting the localization and metabolism of the pathobiont Mucispirillum that is required for triggering colitis in our model 13. Mucus-inhabiting microbes such as Mucispirillum live near the intestinal epithelium and thus are well positioned to locally penetrate into the lamina propria to trigger intestinal inflammation 35,43. Beside the proximity to the gut epithelium, Mucispirillum harbors some virulence traits, including a type VI secretion system and putative effector proteins 38. Such bacterial features may lower the threshold for local penetration increasing the ability of Mucispirillum to trigger colitis when host protective antimicrobial mechanisms are compromised by IBD-associated mutations, such as NOD2 and CYBB.
Our results show that localization in the mucus layer, and not abundance in the intestinal lumen, is critical for Mucispirillum to trigger inflammation. Although these findings provide a plausible mechanism to account for the efficacy of EEN in pediatric CD, further studies are needed to determine whether the beneficial effect of EEN is mediated via a similar mechanism in human CD. Although EEN remains the most validated nutritional recommendation for inducing remission in CD, there are some challenges to the use of EEN, especially in adults, due to its poor palatability. In this context, the use of a fiber-free food-based diet, with similar composition to EEN 44, might represent a more acceptable dietary treatment particularly in adults in whom EEN compliance is low, and may raise the prospect of long-term dietary maintenance therapy in CD.
Members of the Lachnospiraceae family, a subgroup of Clostridia cluster XIVa 34, were most responsive to our dietary intervention with fiber-free diet. Although Lachnospiraceae species can utilize plant pectin, some Lachnospiraceae species can degrade and utilize mucin for growth 34. It is, therefore, not surprising that some Lachnospiraceae species were abundant in mice fed a fiber-containing diet, while the abundance of other species of the same family were more abundant in animals fed a fiber-free diet. Although Lachnospiraceae are among the main producers of short-chain fatty acids, such as butyrate, which promote intestinal barrier function, different taxa of Lachnospiraceae are also associated with intestinal disease. For instance, mucin-degrading R. torques and R. gnavus are two Lachnospiraceae species particularly abundant in CD patients 43,45. Furthermore, R torques is also more frequently found in the gut of relatives of patients with CD that harbor a dysbiotic microbiota 46. Most importantly, a recent prospective cohort study of individuals at risk of developing CD revealed that mucolytic R. torques is the top important taxon associated with the future onset of CD, suggesting that R. torques can be an important contributor to CD development 47. Indeed, we show that mucolytic R. torques enables the pathobiont Mucispirillum to acquire host-derived nutrients required for its growth. R. torques produce H2, a fermentation byproduct, which, in turn, supports Mucispirillum growth. Our findings indicate that fiber exclusion reduces the expression of hydrogenases in the mucus layer and alters the levels of hydrogenase-expressing Lachnospiraceae. Thus, the absence of dietary fiber affects the activity of fermentative bacteria (hydrogenogens), which, in turn, limit nutrient availability necessary for Mucispirillum colonization in the mucus layer. Notably, our findings indicate that fiber-free EEN alters H2 metabolism in CD children, suggesting that one potential beneficial effect of fiber-free therapy in CD might be linked to reduced H2 availability to gut pathobionts such as Mucispirillum that depend on mucolytic microbes for survival at the mucus layer. Overall, our studies provide the concept that diet can control the proximity of the pathobionts to the host epithelium through mucolytic microbes that promote their metabolism and growth at the mucus layer.
Our transcriptomic data reveal a unique metabolic pathway, DNRA, utilized by Mucispirillum that was reduced in the mucus layer by dietary fiber exclusion. Mucispirillum can grow in the presence of nitrate and H2, as it can utilize nitrate as an electron acceptor and H2 as an electron donor in the context of the DRNA pathway. Importantly, our findings could be broadly applied to other gut pathobionts that use anaerobic respiration. In line with this, Proteobacteria take advantage of more abundant electron acceptors allowing respiration, such as nitrate, produced in the inflamed gut to bloom during periods of active IBD 37. Consequently, targeting the molybdenum cofactor-dependent enzymes that are required by some nitrate-utilizing pathogenic bacteria for anaerobic respiration protects mice from colitis by blunting the expansion of pathogenic Enterobacteriaceae 48. Our studies further highlight the importance of identifying metabolic pathways utilized by pathobionts to promote their colonization and demonstrate the feasibility of a dietary approach to target these metabolic pathways as a therapeutic strategy for the treatment of CD.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Roberta Caruso (rocaruso@med.umich.edu).
Materials availability
Any newly generated items in this work will be shared on request.
Data and code availability
Raw data files for 16S and metatranscriptomics sequencing are freely available at BioProject database, ID number: PRJNA984252. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals and diet treatments
All animal experiments followed protocols approved by the University of Michigan, Institutional Animal Care and Use Committee (IACUC).
Three- to eight-week-old female and male SPF mice were used. The SFP C57BL/6J animals were purchased from the Jackson Laboratory (JAX: 000664). SPF doubly-deficient Nod2/Cybb mice harboring microbiota from either Taconic Biosciences or Jackson Laboratory previously described 13 were housed in the Animal Facility at the University of Michigan. SPF Muc2−/− on a C57BL/6J background were provided by E. C. Martens (University of Michigan) and originally obtained from Dr. Leonard Augenlicht (Albert Einstein College of Medicine) 49. Eight- to twelve-week-old female and male GF C57BL/6NTac mice (from Taconic Biosciences, B6-F; B6-M) were housed in flexible film isolators at the University of Michigan Germ-Free Mouse Facility. The absence of microbes in GF mice was verified weekly by aerobic and anaerobic microbial culture and microscopic analysis of stained cecal contents. Mice were housed in groups as appropriate for gender, litter and diet requirements, and provided ad libitum with autoclaved distilled water and the diets described below.
The fiber-deficient (FD) diet (TD.130343) was manufactured by Teklad/Envigo (Indianapolis, IN) and, as previously described 22, consisted of a modified version of Harlan TD.08810, in which starch and maldodextrin were replaced with glucose. The custom-made control diet (CTR) (TD.190428, Teklad/Envigo, Indianapolis, IN) was designed in collaboration with a nutritionist at Envigo. It is a version of the FD diet from which roughly 50% of the simple sugar content (glucose) was removed and replaced by several natural fiber sources, including wheat middlings, alfalfa meal, wheat germ, beet pulps and oats. Additionally, to account for protein and fat coming from natural ingredients, casein and corn oil were reduced in the CTR diet to keep protein at 23.8% (26.3% kcal) and fat at 15.3% by weight (38.0% kcal), as in the FD diet (Table S1). FD and CTR diets were sterilized by gamma irradiation and regular chow (RC) (LabDiet 5L0D, laboratory rodent diet, LabDiet, St. Louis, MO) was sterilized by autoclaving. GF mice were fed regular chow (LabDiet 5013, rodent breeder diet, LabDiet, St. Louis, MO) that had also been sterilized by autoclaving. Daily food consumption amount per mouse in each cage was calculated. Mice of similar age and similar weight were randomly assigned to experimental groups. None of the animal experiments were blinded except for the pathology assessment and mucus thickness measurement.
Trio breeding strategy
Eight- to twelve-week-old doubly-deficient Nod2/Cybb mice, harboring microbiota from either Taconic Biosciences or Jackson Laboratory, were used to set up trio breeding cages, consisting of one male and two females maintained on RC. At embryonic day 14–16, pregnant dams were separated, single-housed and either switched to FD diet or maintained on RC for the duration of the experiments.
Bacteria
Mucispirillum schaedleri isolated from the mouse cecum was a gift from Jani L. O’Rourke (The University of New South Wales, Sydney, Australia) 31. E. coli K12 strain BW25113 was originally received from Yale Coli Genetic Stock Center (https://cgsc.biology.yale.edu/). Ruminococcus torques strain VIII was provided by E. C. Martens (University of Michigan), and originally isolated and characterized by L.C. Hoskins 50.
METHOD DETAILS
Bacterial cultures
Mucispirillum was grown anaerobically in enriched Brain Heart Infusion (BHI) (plate and/or broth), which contained 37 g/l BHI (BD Difco, Franklin Lakes, NJ), 5 g/l yeast extract (Sigma-Aldrich, St. Louis, MO), 0.5 g/l l-cysteine (Sigma, St. Louis, MO), and added 2ml vitamin K3 (menadione, 1mg/ml stock concentration, Sigma, St. Louis, MO), 4 ml hematin/l-histidine solution (1.9mM hematin/0.2M l-histidine stock concentration, both from Sigma, St. Louis, MO), 16 g/l agar (only added to the plates) (BD Difco, Franklin Lakes, NJ), 5% new born calf serum (Gibco, Thermo Fisher Scientific, Waltham, MA), 5% horse serum (Gibco, Thermo Fisher Scientific, Waltham, MA) and 5% sheep serum (Sigma, St. Louis, MO). For Mucispirillum carbohydrate growth array, we used a BHI broth, which was made as described above except for the absence of glucose (we used a dextrose-free BHI (MyBiosource, San Diego, CA)) and for the absence of the three sera. E. coli was grown overnight in LB medium (10 g/l peptone, 5 g/l yeast extract, 10 g/l sodium chloride, Invitrogen, Waltham, MA) with shaking at 37°. R. torques was grown anaerobically for two days at 37°C in custom chopped meat broth 13. The custom chopped meat broth was made as follows: 10g/l beef extract, 30g/l pancreatic digest of casein, 5g/l yeast extract, 5g/l dipotassium phosphate 1mg/l cysteine, 1g/l glucose, 1ml of vitamin K3 (menadione, 1mg/ml stock concentration), 4 ml of hematin/l-histidine solution (1.9mM hematin/0.2M l-histidine stock concentration), 1ml of vitamin B12 (Cyanocobalamin, 0.01mg/ml stock concentration), 10ml of Balch’s Vitamins (5mg/l p-aminobenzoic acid, 2mg/l folic acid, 2mg/l biotin, 5mg/l nicotinic acid, 5mg/l calcium pantothenate, 5mg/l riboflavin, 5mg/l thiamine HCl, 10mg/l pyridoxine HCl, 0.1mg/l cyanocobalamin, 5mg/l thioctic acid), 10 ml of Trace Mineral Solution (0.5g/l EDTA, 3g/l magnesium sulfate heptahydrate, 0.5g/l manganese (II) sulfate monohydrate, 1g/l sodium chloride, 0.1g/l iron(II) sulfate heptahydrate, 0.1g/l calcium chloride, 0.1g/l zinc sulfate heptahydrate, 0.01g/l copper (II) sulfate pentahydrate, 0.01g/l boric acid , 0.01g/l sodium molybdate dihydrate, 0.02g/l nickel (II) chloride hexahydrate), 10 ml of Purine/Pyrimidine Solution [adenine, guanine, thymine, cytosine, uracil (all at 200mg/l concentration)] and 10 ml of Amino Acid Solution [alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, valine (all at 250mg/l concentration)]. All the reagents were purchased from Sigma, St. Louis, MO, except for the beef extract (BD Difco, Franklin Lakes, NJ).
Intestinal histopathology and disease evaluation
SPF mice were sacrificed at various time points. The cecum and colon were harvested and immediately preserved in freshly made Carnoy’s fixative [(anhydrous methanol:chloroform:glacial acetic acid, ratio 60:30:10 v/v, (methanol and chloroform are from Sigma, St. Louis, MO, glacial acetic acid is from Thermo Fisher Scientific, Waltham, MA)] using a protocol described previously with some modifications 51. The intestinal samples were fixed in Carnoy’s solution for 3 h, then transferred to fresh Carnoy’s solution for additional 24 h. The samples were then washed in dry methanol and kept at 4 °C until further use. An additional set of mice from all diet experiments was used to prepare H&E-stained sections. The cecum and colon from this set were flushed with Phosphate buffered saline (PBS, Gibco, Thermo Fisher Scientific, Waltham, MA), fixed in 10% (v/v) formalin (Thermo Fisher Scientific, Waltham, MA) and then processed for H&E staining. Histologic evaluation was performed in a blinded fashion, using a scoring system described previously 13.
Thickness measurements of the colonic mucus layer
Methanol-Carnoy-fixed intestinal samples were embedded in paraffin, cut, and stained with Alcian blue and Alcian blue/PAS (both from Sigma, St. Louis, MO). The staining was performed by the School of Dentistry Histology Core (University of Michigan). The thickness of the colonic inner mucus layer was measured as previously described 22 using partially overlapping photographs, taken from nearly the entire length of each colon based on the Alcian blue stained slides after cross-validation using anti-Muc2 staining. The thickness of the mucus in colonic sections was measured in a blinded fashion using Aperio ImageScope v 12.1.0.5029 (Aperio Technologies, Inc., Leica Microsystems, Wetzlar, Germany). Only regions in which the mucus layer was sandwiched between epithelium on one side and fecal material on the other were used.
Intestinal permeability assay
Intestinal permeability assay was performed using fluorescein isothiocyanate (FITC)-dextran, as described previously 52. Briefly, mice were deprived of food for 4 h and then gavaged with 0.6 mg/g body weight 4 kDa FITC–dextran (FD4, Sigma-Aldrich St. Louis, MO). Blood was collected 3 h later. The concentration of FITC in serum was determined by spectrophotofluorometry at an excitation of 485 nm and an emission wavelength of 520 nm. FITC-dextran concentrations were determined using a standard curve generated by the serial dilutions of FITC-dextran.
Dextran Sodium Sulfate colitis
Mice were administrated 3% DSS (36–50kDa, MP Biomedicals, Solon, OH) in autoclaved drinking water for 5 days to induce colitis.
ELISA for fecal Lipocalin-2
Frozen fecal samples (−20°C) were used to measure the levels of fecal Lipocalin-2 (Lcn-2) (Duoset murine Lcn-2 ELISA kit, R&D Systems, Minneapolis, MN). Blocking was performed in 1% bovine serum albumin (Thermo Fisher Scientific, Waltham, MA) in PBS, according to manufacturer’s instructions. Feces were resuspended in sterile PBS (100 mg/ml), vortexed for 5 min and then centrifuged for 10 min at 13,000 rpm at 4°C. Next, the supernatants were carefully recovered and stored at −20°C until analysis.
Immunofluorescence staining
Slides were deparaffinized twice by submerging in xylene (Thermo Fisher Scientific, Waltham, MA) for five minutes each. The tissue sections were then washed twice in 100% ethanol for five minutes each, followed by rehydration in Milli-Q water for 5 minutes. Antigen retrieval was performed in 10 mM sodium citrate solution (pH 6.0) (Sigma, St. Louis, MO). The submerged sections were heated to 89°C for 10 minutes and then allowed to cool down for 20 minutes at room temperature. Slides were then washed with Milli-Q water and submerged in a blocking buffer [1:10 goat serum (Thermo Fisher Scientific, Waltham, MA) in Tris-buffered Saline (TBS; 500 mM sodium chloride (Sigma, St. Louis, MO), 50 mM Tris (Thermo Fisher Scientific, Waltham, MA), pH 7.4,] for an hour at room temperature. pH of TBS was adjusted with hydrochloric acid (Thermo Fisher Scientific, Waltham, MA). Sections were then incubated with a 1:80 dilution of Mucin 2 antibody (C3) (GeneTex, Irvine, CA) in a blocking buffer for two hours at room temperature. The slides were rinsed three times in TBS, five minutes each. The secondary antibody staining was performed with a 1:200 dilution of Alexa Fluor 488 conjugated goat anti-rabbit IgG antibody (Thermo Fisher Scientific, Waltham, MA) in a blocking buffer for one hour at room temperature in the dark. Finally, sections were washed twice in TBS for 5 minutes, blotted, and covered with ProLong Gold Antifade reagent with DAPI (Invitrogen, Waltham, MA) and cover slips. The slides were kept overnight at room temperature in the dark, and then kept at 4°C until imaging. The mucus layer in the sections was visualized using an Olympus IX71 inverted fluorescent microscope (Olympus, Center Valley, PA).
Fluorescent In-Situ Hybridization
Methanol-Carnoy-fixed intestinal samples were embedded in paraffin and then cut. FISH was performed on 6μm paraffin-embedded sections as previously described with some modifications 51. Paraffin sections were dewaxed and washed in 99.5% ethanol. The tissue sections were incubated with 5 μg of Alexa Fluor 555-conjugated EUB (5′-GCTGCCTCCCGTAGGAGT-3′) (bp 337–354 in bacteria EU622773) 51 or a double 3′-and 5′-labelled 16S rRNA targeting probe specific for Mucispirillum (Msc487_correct-2xCy3 (CAG TCA CTC CGA ACA ACG CT) 53 in 500 μl of hybridization buffer [20mM Tris-HCl (pH 7.4), 0.1% (w/v) SDS (both from Thermo Fisher Scientific, Waltham, MA), 0.9M NaCl (Sigma, St. Louis, MO)] at 50°C overnight. The sections were rinsed in a wash buffer [20 mM Tris-HCl (pH 7.4), 0.9 M sodium chloride], washed at 50°C for 20 min and counterstained with DAPI (Invitrogen, Waltham, MA). To reduce autofluorescence, the sections were treated with an autofluorescence quenching kit (Vector Laboratories, Burlingame, CA), according to the manufacturer’s instruction. Images were recorded using an Olympus IX71 inverted fluorescent microscope (Olympus, Center Valley, PA) within 24 h.
Cecal homogenate preparation
SPF and GF C57BL/6 mice (fed RC or FD for 2 weeks) were euthanized, and the cecum was removed inside an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). The cecum was opened, contents squeezed into 2 ml of pre-reduced ultrapure water, and the mucus layer was scraped off using coverslips and added to the conical tube. The mixture was then homogenized by vortexing and pipetting, passed through a 0.45μm centrifuge tube filter (Costar, Corning Inc., Corning, NY) to remove large debris, and finally through a 0.22 μm centrifuge tube filter (Costar, Corning Inc., Corning, NY). The resulting cecal homogenates were then dispensed as 200 μl aliquots into a 96-well round bottom plate (Costar, Corning Inc., Corning, NY). Mucispirillum was grown on enriched BHI plates for 3 days at 37°C under anaerobic conditions (10% H2, 5% CO2 and 85% N2). Mucispirillum was then harvested from the plates with a loop and 1ml of enriched BHI broth, transferred into 4ml of enriched BHI broth and grown anaerobically overnight at 37°C. E. coli was grown aerobically in LB broth with shaking overnight at 37°C. For in vitro studies, either 5 μl of E. coli culture or 10 μl of Mucispirillum culture was added to the 96-well round bottom plate (containing the homogenates) and incubated anaerobically at 37°C for up to 3 days. E. coli CFUs were enumerated by serial dilution and spot plating on LB plates (LB agar, Invitrogen, Waltham, MA) after 24 h. Mucispirillum concentration was quantified by qPCR after 3 days. For ex-vivo studies, the cecal homogenates were prepared as described above from GF, E. coli-, and R. torques-monocolonized mice. Once prepared, the cecal homogenates were dispensed as 200 μl aliquots into a 96-well round bottom plate, mixed with 10 μl of Mucispirillum culture and incubated anaerobically at 37°C for 3 days. Mucispirillum concentration was quantified by qPCR.
Extraction of nucleic acids and quantitative Real-Time PCR
RNA was extracted from both cecal luminal and mucus contents isolated from Tac-DKO mice maintained on RC or FD (for 8 weeks) using a phenol:chloroform:isoamyl alcohol method with bead beating as previously described 22 with a few modifications. Briefly, after the cecal contents and mucus scrapings were freshly harvested, garnet beads (PowerBead tubes, Qiagen, Hilden, Germany) were added to the samples instead of acid-washed glass beads. Next, 500 μl of Buffer A [200 mM NaCl (Sigma, St. Louis, MO), 20 mM EDTA (EDTA solution from Thermo Fisher Scientific, Waltham, MA)], 210 μl of 20% SDS (filter sterilized) (Thermo Fisher Scientific, Waltham, MA) and 500 μl of phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.5; Invitrogen, Waltham, MA) were added to the samples. The mixture was then bead beaten (mini beadbeater, Biospec products, Bartlesville, OK) for 2 min and centrifuged at 4 °C (3 min at 13000 rpm). The aqueous phase was recovered and mixed with 500 μl of the aforementioned phenol:chloroform:isoamyl alcohol solution. Afterwards, the mixture was centrifuged at 4°C (3 min at 13,000 rpm) and the aqueous phase was recovered. 1/10 volume of the 3M sodium acetate solution (pH: 5.2, Sigma, St. Louis, MO) and 1 volume of −20°C chilled 100% ethanol (Thermo Fisher Scientific, Waltham, MA) were added to the aqueous phase. The resulting solution was then mixed by inversion and incubated for 20 min on ice. Afterwards, the mixture was centrifuged at 4°C for 20 min at 13,000 rpm. The pellet was recovered and washed in 500 μl of cold 70% ethanol. The mixture was centrifuged at 4°C (5 min at 13,000 rpm) and the RNA pellet was recovered, air dried and then resuspended in 100μl nuclease-free water. The RNA extracts were then further purified using an RNeasy Mini kit (Qiagen, Hilden, Germany) for luminal contents and an Rneasy Micro kit (Qiagen, Hilden, Germany) for mucus samples. 10μl of 2-Mercaptoethanol (Sigma, St. Louis, MO) was added to the provided extraction buffer according to the manufacturer’s protocol. Total RNA was submitted to the Advanced Genomics Core at the University of Michigan for sequencing.
RNA was extracted from colonic tissue using the E.Z.N.A. Total RNA Kit (Omega Biotek, Norcross, GA) according to the manufacturer’s instructions. RNA was reverse transcribed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Waltham, MA) and the resulting cDNA was utilized for quantitative real time RT-PCR (qPCR). Genomic DNA was extracted from fecal pellets using the E.Z.N.A stool DNA kit (Omega Bio-Tek, Norcross, GA) according to the manufacturer’s protocol. Genomic DNA was extracted from colonic mucus scrapings using the E.Z.N.A stool DNA kit (Omega Bio-Tek, Norcross, GA) following incubation with 1mg lysozyme (Sigma, St. Louis, MO) for 15 min at room temperature. DNA was then used for qPCR.
qPCR was performed using a SYBR green PCR master mix (Alkali Scientific, Fort Lauderdale, FL) and StepOne Real-time PCR system (Applied Biosystems, Waltham, MA). The primer sequences used for the detection of each gene were summarized separately (Table S7) 13,54–59. Relative mRNA expression was calculated by the ΔΔCt method and normalized to the expression of either GAPDH or to the universal 16S rRNA gene (for in vivo studies). Analysis was performed in duplicate sample sets. Specificity of each primer was validated by the following approaches: 1) by qPCR against target species genome and melting curve analysis (for a single peak), 2) by qPCR for each primer set against a non-target template. Genomic DNA of Mucispirillum and R. torques were used to generate standard curves to determine target concentrations (for in vitro studies). Copy numbers of host DNA and bacterial 16S rRNA gene sequences were determined by comparison with known quantities of mouse and E. coli genomic DNA. Bacterial genome copies were estimated by qPCR using universal v4 16S rRNA primers and normalized by total DNA. Host genomic copy numbers were determined by qPCR using ACTB primers and normalized by total DNA.
Microbiota analyses
For the 16S rRNA sequencing, PCR and library preparation were performed by the Microbiome Core by the University of Michigan. Amplicons of the V4 region within the 16S rRNA gene were generated and both ends of the fragments were sequenced using an Illumina MiSeq instrument. The paired end sequences were curated using Mothur (v.1.40.5) as previously described 60,61. Briefly, the paired end sequences were assembled into ≈ 250 bp contigs, aligned to the SILVA 16S rRNA database release 13862, made free of possible chimeric sequences by UCHIME63, binned into Operational taxonomic units (OTUs) at >97 % identity level and taxonomically assigned with 16S rRNA gene training set version 16 of Ribosomal Database Project. α-diversity (Shannon index) and NMDS plot of β-diversity values, LEfSe Linear discriminant analysis (LDA) values of OTUs were all determined using Mothur. To identify significant differentially abundant OTUs, bacterial taxa were screened with pairwise LEfSe analysis, and further verified by multiple hypothesis testing using the false discovery rate (FDR) of signal-to-noise ratio with Morpheus (https://software.broadinstitute.org/morpheus/, permutations=9999, markers=0). Heat maps of OTUs were visualized by MeV. To label individual OTUs, we chose the lowest taxa that were not identified as “unclassified”. All the samples that had zero Mucispirillum reads during all time points of the dietary interventions were removed. The phylogenetic tree was calculated by Phylogeny.fr 64 using the most abundant sequences of indicated OTUs that were extracted by Mothur and sort/uniq Linux commands. R. torques VIII-239, derived from PRJNA227979, was included in the phylogenetic analysis. The figure (Figure S5A) was drawn by TreeGraph2.
Microbial RNA-Sequencing analysis
Library preparation and sequencing of the RNA-seq libraries were performed by the Advanced Genomics Core at the University of Michigan. Briefly, RNA was assessed for quality using the TapeStation (Agilent, Santa Clara, CA). Samples were prepared using the New England BioLabs (NEB)Next Ultra II Directional RNA Library Prep Kit for Illumina, the Ribo depletion Module NEBNext rRNA (Human/Mouse/Rat), the NEBNext rRNA Depletion Kit (Bacteria), and the NEBNext Multiplex Oligos for Illumina Unique dual (all from NEB, Ipswitch, MA) where 115ng of total RNA was ribosomal depleted using the rRNA Depletion modules. The rRNA-depleted RNA is then fragmented 15 minutes determined by RIN (RNA Integrity Number) of input RNA as per protocol and copied into first strand cDNA using reverse transcriptase and dUTP mix. Samples underwent end repair and dA-Tailing step, followed by ligation of NEBNext adapters. The products were purified and enriched by 13 cycles of PCR to create the final cDNA library, which were checked for quality and quantity by Qubit hsDNA (Thermo Fisher Scientific, Waltham, MA) and LabChip (PerkinElmer, Waltham, MA). The samples were pooled and sequenced on the Illumina NovaSeq S4 Paired-end 150bp, according to manufacturer’s recommended protocols (Illumina, San Diego, CA). bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files. Contaminated host sequences were removed by Bowtie2 65.
Analysis of Mucispirillum gene transcripts
The transcript levels of individual genes in Mucispirillum schaedleri ASF457 (GCA_000487995) were mapped, counted and annotated by Salmon 66. For a more detailed annotation, closest protein sequences and species were identified by Diamond using Uniref100 of UniProt database (https://www.uniprot.org/).
Analysis of hydrogenase transcripts
To identify hydrogenase transcripts and metagenome-assembled genomes (MAGs), core sequences with homology were obtained by Diamond (e-value < 10−5) 67 from either mouse (mucus and luminal) or human pediatric (stool) IBD sequences (SRP057027,26 respectively, and assembled by metaSPAdes 68. Finally, pooled sequences were verified by Diamond using reference sequences downloaded from HydDB database 69 Transcript reads were further obtained by four-time repetitive expansion of transcript sizes (detected by Bowtie2) and de novo assembly by metaSPAdes. Metagenomic reads were directly counted after removing low read samples (<5000 target reads). For putative H2-generating hydrogenases, we used all HydDB reference sequences from NiFe group 4a and FeFe groups A1 and B. Hydrogenases (from FeFe groups A1 and B) were further screened by two criteria, >70 % identity to the references and higher homology to FeFe group A1/B rather than non-A1/B FeFe hydrogenases. For a more detailed annotation, closest protein sequences and species were identified by Diamond using Uniref100 of UniProt database. Microbial species that were identical to Lachnospiraceae unclassified, Lachnospiraceae [Ruminococcus] (not Ruminococcaceae Ruminococcus), and the species with higher homology (but not identical) to Lachnospiraceae spp. were classified as Lachnospiraceae spp (Figure 6B). The reads were counted by Salmon and a pairwise comparison of TPM (individual transcript per million of hydrogenases) was performed by the “marker selection” function of Morpheus (https://software.broadinstitute.org/morpheus/). Log10[TPM] values were assigned (visualized by MeV), 1/2 being the minimal TPM, to all transcripts as the detection threshold for error suppression of log calculation.
Bacterial growth assays in a custom carbohydrate array
The custom carbohydrate array was formulated and performed as previously described 22. The carbon sources (all at 10mg/ml, except those specified below) included in the array were: arabinose, fructose, fucose, galactose, galacturonic acid, glucosamine, glucose, glucuronic acid, mannose, N-acetylgalactosamine, N-acetylglucosamine, N-acetylneuraminic acid (IV-S), rhamnose, ribose, xylose, inulin (chicory), amylopectin (maize), amylopectin (potato), cellobiose, chondroitin sulfate, heparin hyaluronan, glycogen mannan (S. cerevisiae), laminarin, alginate (brown algae), dextran (L. mesenteroides), levan (E. herbicola), pullulan (A. pullulans) (all from Sigma, St. Louis, MO), arabinan (sugar beet), arabinogalactan (larch), pectic galactan (potato), pectic galactan (lupin), rhamnogalacturonan I (potato pectin) (20mg/ml), arabinoxylan (wheat), β-glucan (barley), galactomannan (carob), glucomannan (koniac), xyloglucan (tamarind), lichenan (all from Megazyme-Neogen, Lansing, MI) fructooligosaccharides (commercial food supply), mucin O-glycans [provided by Dr. E.C. Martens, (20mg/ml)]. Additional carbon sources, such as acid casein peptone (1% and 10% w/v) and glycerol (1% and 10% v/v) (both from Thermo Fisher Scientific, Waltham, MA) were tested. Growth assays for all carbohydrates were carried out in non-adjacent duplicates and contained two non-adjacent water only samples (negative controls) and two non-adjacent enriched BHI (with glucose and serum) samples (positive controls). Mucispirillum was grown on enriched BHI plates for 3 days at 37°C under anaerobic conditions (10% H2, 5% CO2 and 85% N2). Mucispirillum was then harvested from the plates with a loop and 1ml of enriched BHI broth, transferred into 4ml of enriched BHI broth and grown anaerobically overnight at 37°C. E.coli was grown aerobically overnight at 37°C in LB broth with shaking. For growth profiling in a custom carbohydrate array, 1 ml of each culture was anaerobically centrifugated. The pellet was recovered and washed 2 times in 2x concentrated BHI medium without any carbohydrates and serum for Mucispirillum, and 2x concentrated carbohydrate-free minimal medium (M9 with the addition of 4mM magnesium sulfate, 200μM calcium chloride, 2μM ferrous sulfate, all from Sigma, St. Louis, MO) for E.coli to remove carried over carbohydrates from the culture media. The pellets were finally resuspended in 1 ml of 2x concentrated BHI broth and M9 minimal medium without any carbohydrates and serum, respectively. This 1 ml culture was used to inoculate 50 ml of 2x concentrated medium (either BHI broth or M9 minimal medium without carbohydrates and serum) at a 1:50 ratio, respectively. 100 μl of the resulting cultures were then added to the individual wells of the carbohydrate solutions in the 96-well plates, resulting in 200 μl of final volume.
Absorbance values were measured at 600 nm (A600) at an interval of 10 min over 72 h. To construct a heat map containing relative growth values (Figure 4A), absorbance data of all bacterial species were normalized as follows: only carbohydrate growth assays for which both replicate cultures produced an increase in absorbance of more than 0.1 were scored as positive (all other values were set to 0), the starting absorbance on each substrate was subtracted. Next, the maximum change in absorbance was normalized within each individual species by setting its best growth to 1.0 and normalizing all other positive growths to this maximum value (normalized values were thus between 0 and 1.0). Both raw and normalized values are provided in Table S2. Each positive growth curve was manually inspected to validate the presence of an exponential growth profile.
Additionally, 50 μl of the 1 ml Mucispirillum culture (resuspended in BHI medium without carbohydrates and serum) were used to inoculate 5 ml of BHI medium (without carbohydrates and serum) in the presence of either acid casein peptone or glycerol and grown in borosilicate glass culture tubes (Thermo Fisher Scientific, Waltham, MA) for three days. Mucispirillum growth was then analyzed by qPCR. BHI medium only (negative) and carbohydrate-free BHI medium with serum (positive) were included as controls.
Purification of porcine colonic mucin oligosaccharides and bacterial growth
Highly sulfated porcine colonic mucin oligosaccharides (referred as rectal glycoproteins, rGP) were purified from the distal porcine colon and rectum using a protocol described previously 70. Briefly, distal porcine colon and rectum were opened, and the fecal contents were removed. The mucosa was scraped off, and mucus was extracted by homogenizing the tissue in at least 5 volumes of extraction buffer (6 M guanidine chloride, 5 mM EDTA, 10 mM sodium phosphate monobasic, pH 6.5, all from Sigma, St. Louis, MO). The samples were gently stirred at 4 °C for 16 h. The solution was then spun down at 15,000 r.p.m. for 30 min at 10 °C, and the supernatant was discarded. Pellets were resuspended in the extraction buffer, and the process was repeated until the supernatant was clear for at least two extractions. After extraction, the mucins were solubilized by disulfide bonds reduction. Pellets were resuspended in the fresh reduction buffer [6 M guanidine chloride, 5 mM EDTA (both from Sigma, St. Louis, MO), 0.1 M Tris (Thermo Fisher Scientific, Waltham, MA) pH 8.0] containing 25 mM of 1,4-dithiothreitol (DL-Dithiothreitol from Sigma, St. Louis, MO) and slowly stirred at 37 °C for 5 h. After this incubation, 62.5 mM iodoacetamide (Sigma, St. Louis, MO) was added and the solution was stirred slowly in the dark at room temperature for 16 h. The solution was then centrifuged at 10,000 r.p.m. at 4 °C for 30 min, and the supernatant containing the solubilized mucins was extensively dialysed into water. Samples were dissolved into 100 mM Tris-HCl, pH 8.0 (Thermo Fisher Scientific, Waltham, MA), containing 1 mg ml–1 trypsin (Sigma-Aldrich, St. Louis, MO), and incubated with slow stirring at 37 °C for 16 h. Additionally, after trypsin treatment the sample was dialyzed against water and lyophilized. Finally, rGP solution (5 mg/ml concentration in PBS) was autoclaved for 5 minutes (sterilization cycle). 100 μl of rGP (5 mg/ml concentration in PBS), glucose (10 mg/ml concentration, sterile filtered in water) and rhamnose (10 mg/ml concentration, sterile filtered in water) solutions were added to flat bottom 96-well plates (Costar, Corning Inc., Corning, NY). The plates were then transferred to an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) and allowed to equilibrate in the anaerobic atmosphere for ~3–4 h. Growth assays were carried out in triplicates and contained three medium only samples (negative controls). Mucispirillum was grown on enriched BHI plates for 3 days at 37°C under anaerobic conditions. Mucispirillum was then recovered from the plates with a loop and 1 ml of enriched BHI broth, transferred into 5ml of enriched BHI broth and grown anaerobically overnight at 37°C. R. torques was grown in pre-reduced custom chopped meat broth for 2 days at 37°C under anaerobic conditions. For growth profiling in the presence of rGP, glucose and rhamnose, 1 ml of each culture was anaerobically centrifugated. The pellet was recovered and washed 2 times in 2x concentrated chopped meat broth without any carbohydrates to remove carried over carbohydrates from the culture media. The pellet was then resuspended in 1 ml of 2x concentrated chopped meat broth without any carbohydrates. This 1 ml culture was used to inoculate 50 ml of 2x concentrated chopped meat broth without any carbohydrates at a 1:50 ratio. 100 μl (for single R. torques or Mucispirillum cultures) or 50 μl (for R. torques and Mucispirillum co-cultures) of the resulting cultures were then added to the individual wells of the rGP, glucose and rhamnose solutions in the 96-well plates, resulting in 200 μl final volumes. The absorbance measurements were performed as described above, and bacterial growth was quantified by qPCR.
Bacterial oral challenge
For in vivo inoculations, Mucispirillum was grown anaerobically on enriched BHI plates. A ratio of one plate/mouse was used for in vivo experiments. Three-week-old (both male and female) Jax DKO mice were co-housed as appropriate for gender, right after weaning, for 5 weeks. Jax-DKO mice were then randomly assigned to groups and fed either the RC or the FD diet (for the duration of the experiment). One week later, the animals were treated with vancomycin (250 mg/l in drinking water, Xellia Pharmaceuticals, Buffalo Grove, IL) for 7 days and finally switched to regular water. Among antibiotics usually used to deplete microbiota in SPF, vancomycin is one that mainly targets Gram-positive microbe, and it was used as it doesn’t not affect colonization of Gram-negative Mucispirillum and rather enhances it in SPF mice (based on our preliminary experiments). Mice were then colonized (oral gavage) with 0.2 ml of BHI broth containing approximately 1 × 108 CFU of Mucispirillum every other day for 7 days. Mouse survival and body weights were monitored for 7 days after the first oral administration. RC-fed male and female littermate Muc2−/− and Muc2+/− mice were separated at five-week of age (after 2 weeks of cohousing), then treated with vancomycin (250 mg/l in drinking water) for 7 days and finally switched to regular water. Mice were then infected (oral gavage) with 0.2 ml of BHI broth containing approximately 1 × 108 CFU of Mucispirillum and sacrificed one day after bacterial administration. To determine bacterial loads and to monitor the development of intestinal inflammation, fecal samples were collected before and every other day for 7 days in Jax-DKO mice, and until day 1 in Muc2−/− and Muc2+/− mice, following the first bacterial administration. The abundance of Mucispirillum was quantified by qPCR and level of intestinal inflammation was determined using lipocalin-2 ELISA. Jax-DKO mice were sacrificed 7 days after the first infection, while Muc2−/− and Muc2+/− mice were sacrificed on day 1. Cecum and colon tissue was harvested and processed for mucus thickness measurement and histological examination.
GF C57BL/6 mice were inoculated with either E. coli K12 BW25113 or R. torques VIII (0.2 ml of bacterial suspension/mouse, ~109 CFU). Mono-colonization with either E.coli or R. torques was confirmed by qPCR. Mono-colonization with R. torques VIII was achieved by treatment with 100mM succinate (disodium succinate, Sigma, St. Louis, MO) for 7 days prior the bacterial gavage, a strategy which is used to reduce the oxygen concentration in the gut of GF mice and allow colonization with strict anaerobes 71. Succinate treatment was administrated only to R. torques group and stopped two weeks prior Mucispirillum administration to avoid it could have affected it. For ex-vivo studies, GF, E. coli- and R. torques-monocolonized mice were euthanized 2 weeks after reconstitution and used to prepared cecal homogenates (see above). For in vivo studies, GF, E. coli- and R. torques monocolonized C57BL/6 mice (2 weeks after reconstitution) received 0.2 ml of BHI broth (oral gavage) containing approximately 1 × 108 CFU of Mucispirillum. To determine bacterial loads, fecal samples were collected before and every week for up to 2 weeks following Mucispirillum administration. The abundance of Mucispirillum was quantified by qPCR. In additional in vivo experiments, GF and R. torques monocolonized C57BL/6 mice were sacrificed 2 weeks after reconstitution. Large intestine was harvested, and the mucus thickness was assessed as described above.
Bacterial co-culture in absence/presence of exogenous H2
Culture medium tubes were prepared by adding 5 ml nitrate-supplemented enriched BHI [(BHI with 10mM sodium nitrate (Sigma, St. Louis, MO)] to sterilized Balch tubes that were subsequently sealed with sterilized butyl rubber stoppers. The culture medium (nitrate-supplemented BHI) was then rendered anaerobic by the vacuum-vortex technique of Wolfe and Metcalf 72 with six cycles, using a mixture of either 80% N2-20% CO2 or 80% H2-20% CO2 (Cryogenic Gases, Detroit, MI). Medium tubes were incubated in a water bath at 50°C until the resazurin indicator (Sigma, St. Louis, MO) was observed to become colorless. Medium tubes were inoculated with 50ul of Mucispirillum starter culture in the presence of absence of R. torques (50ul starter culture) and incubated with shaking at 37°C for 3 days. Mucispirillum and R. torques concentrations were quantified by qPCR.
Bioreactors
Co-cultures of R. torques and Mucispirillum species were grown in bioreactor vessels (Wyse Glass, Midland, MI) operated in batch mode. The bioreactors were filled with 150 ml of nitrate-supplemented enriched BHI and inoculated with 1.5 ml each of R. torques and Mucispirillum species starter cultures. The cultures were kept at 37°C using a recirculating water jacket, stirred, and sparged with a mixed gas containing 20% CO2 and either 80% N2 (Cryogenic Gases, Detroit, MI) or 80% H2 (Cryogenic Gases, Detroit, MI) for 3 days. Mucispirillum concentration was quantified by qPCR.
H2 measurements
Dissolved H2 was quantified using a method derived from Kraemer and Bagley 40. Samples of 0.5 mL culture liquid were immediately added to 1 ml 3M sulfuric acid (Sigma, St. Louis, MO) in sealed Hungate tubes. The tubes were vortexed vigorously and incubated for >2 hrs at room temperature to extract dissolved H2 into the gas phase. The pressure in the tube headspaces was then adjusted to 10 psig with pure N2 and 2 ml samples were withdrawn for analysis of H2 content.
H2 content was measured using a Peak Performer 1 gas chromatograph (Cat#910-105) with a reducing compound photometer (RCP) detector and post-column diluter (Peak Laboratories, Mountain View, CA) calibrated using a 10 ppm H2 standard (GASCO 105L-H2N-10, Cal Gas Direct Incorporate, Huntington Beach, CA). Ultra-high purity N2 was used as the carrier gas.
To quantify H2 in culture headspaces, 0.5 ml of headspace gas was transferred from each culture into a sealed Balch tube containing air. 10 ml of air was then added to each tube to facilitate sample withdrawal and pumped in and out several times to mix the gases. 2 ml samples were withdrawn from the Balch tubes, further diluted using syringes fitted with stopcocks if necessary, and analyzed for H2 content using the Peak Performer 1 gas chromatograph (Peak Laboratories, Mountain View, CA).
Ammonia assay
The concentration of ammonium in culture supernatants was measured in triplicates using a Synergy H1 Multi-Mode Microplate Reader (BioTek Instruments, Winooski, VT) using Gen5™ software (BioTek Instruments, Winooski, VT) following the manual phenol hypochlorite method (detection limit 1μg N l−1) 73. Briefly, 500μl of culture supernatant was mixed with 20ul of phenol solution [(11% w/vol phenol (Thermo Fisher Scientific, Waltham, MA) dissolved in ethanol)]. Next, 20 μl of sodium nitroprusside solution [(0.5% w/vol sodium nitroprusside (Fisher Scientific, Waltham, MA) dissolved in distilled deionized water)] was added to the mixture and the solution was thoroughly mixed. Then 50 μl of oxidizing solution [(16% w/vol sodium citrate (Thermo Fisher Scientific, Waltham, MA), 0.8% w/vol sodium hydroxide (Thermo Fisher Scientific, Waltham, MA), and 4% sodium hypochlorite dissolved in distilled deionized water] was added and mixed thoroughly. The mixture was incubated for 4 h at room temperature, then 200 μl of the mixture was transferred to an assay plate and absorbance was measured at 640 nm. Ammonia/ammonium concentration was determined according to a standard curve [prepared using ammonium chloride (Fisher Scientific, Waltham, MA)].
Quantification and Statistical analysis
Unless otherwise stated in individual method sections above, all statistical analyses were performed using Prism 9.1.0 (GraphPad Software, San Diego, CA). Sample sizes were chosen to reach statistical significance (p < 0.05) for a pre-determined effect based on the sample variation observed in previous studies. Sample sizes for microbiota analysis were chosen based on sample sizes published in previous studies. Differences between two groups were evaluated using two-tailed unpaired t-test (parametric) or Mann–Whitney U test (nonparametric). Comparisons of more than two groups were performed with one-way ANOVA (parametric) or two-way repeated measures ANOVA (parametric) or Kruskal–Wallis test (non-parametric) followed by Tukey’s, Sidak’s or Dunn’s multiple comparisons test. Differences of p < 0.05 were considered significant in all statistical analyses. Numbers of animals (n) used in individual experiments, details of the statistical tests used and pooled values for several biological replicates are indicated in the respective figure legends.
Supplementary Material
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mucin 2 antibody (C3) | GeneTex | Cat# GTX100664 RRID: AB_1950958 |
| Alexa Fluor 488 goat anti-rabbit IgG | Invitrogen | Cat# A11008; RRID: AB_143165 |
| Alexa Fluor 555-conjugated EUB338 probe | Invitrogen | Johansson et al.51 |
| Msc487_correct-2xCy3-conjugated Mucispirillum probe | Sigma-Aldrich | Herp et al.53 |
| Bacterial and virus strains | ||
| Mucispirillum schaedleri | Dr. Jani L. O’Rourke | Robertson et al.31 |
| Escherichia coli K12 strain BW25113 | Yale Coli Genetic Stock Center | https://cgsc.biology.yale.edu/ |
| Ruminococcus torques strain VIII | Dr. Eric C. Martens | Hoskins et al.50 |
| Chemicals, peptides, and recombinant proteins | ||
| Acid casein peptone | Thermo Fisher Scientific | Cat# BP1424-100 |
| Acetic acid, glacial | Thermo Fisher Scientific | Cat# A38S |
| Adenine | Sigma-Aldrich | Cat# A2786 |
| Alanine | Sigma-Aldrich | Cat# A7627 |
| Alcian blue | Sigma-Aldrich | Cat# A5268 |
| Alginate | Sigma-Aldrich | Cat# 180947 |
| Agar | BD Difco | Cat# 214010 |
| Ammonium Chloride | Fisher Scientific | Cat# A661-500 |
| Amylopectin from maize | Sigma-Aldrich | Cat# 10120 |
| Arabinose | Sigma-Aldrich | Cat# A3131 |
| Arginine | Sigma-Aldrich | Cat# A8094 |
| Asparagine | Sigma-Aldrich | Cat# A4159 |
| Aspartic Acid | Sigma-Aldrich | Cat# A93100 |
| Barley β-glucan | Megazyme | Cat# P-BGBL |
| Beef extract | BD Difco | Cat# 212303 |
| Biotin | Sigma-Aldrich | Cat# B4501 |
| Boric acid | Sigma-Aldrich | Cat# B6768 |
| Bovine Serum Albumin | Thermo Fisher Scientific | Cat# BP9703-100 |
| Brain Heart Infusion | BD Difco | Cat# 237500 |
| Brain Heart Infusion (dextrose-free) | MyBiosource | Cat# MBS652861 |
| Calcium chloride | Sigma-Aldrich | Cat# C5670 |
| Calcium pantothenate | Sigma-Aldrich | Cat# P2250 |
| Cellobiose | Sigma-Aldrich | Cat# C7252 |
| Chloroform | Sigma-Aldrich | Cat# 288306 |
| Chondroitin sulfate | Sigma Aldrich | Cat# C9819 |
| Citrate buffer (pH 6.0) | Sigma Aldrich | Cat# C9999 |
| Copper (II) sulfate pentahydrate | Sigma Aldrich | Cat# 209198 |
| Cyanocobalamin | Sigma Aldrich | Cat# C3607 |
| Cysteine | Sigma Aldrich | Cat# C7352 |
| Cytosine | Sigma Aldrich | Cat# C3506 |
| DAPI (ProLong Gold Antifade reagent) | Invitrogen | Cat# P36931 |
| Dextran | Sigma Aldrich | Cat# D5251 |
| Dextran Sodium Sulfate | MP Biomedical | Cat# 0216011025 |
| Dipotassium phosphate | Sigma Aldrich | Cat# P3786 |
| Disodium succinate | Sigma Aldrich | Cat# W327700 |
| DL-Dithiothreitol | Sigma Aldrich | Cat# D9779 |
| EDTA | Sigma Aldrich | Cat# ED4SS |
| EDTA solution (pH 8.0) | Thermo Fisher Scientific | Cat# BMA51201 |
| Ethanol | Thermo Fisher Scientific | Cat# BP2818-4 |
| Ferrous sulfate | Sigma Aldrich | Cat# F8048 |
| FITC-dextran | Sigma Aldrich | FD4 |
| Folic acid | Sigma Aldrich | Cat# F7876 |
| Formalin Phosphate (10% buffered) | Thermo Fisher Scientific | Cat# SF100-20 |
| Fructooligosaccharides | Commercial food supply | N/A |
| Fructose | Sigma Aldrich | Cat# F0127 |
| Fucose | Sigma Aldrich | Cat# F2252 |
| Galactomannan | Megazyme | Cat# P-GALML |
| Galactose | Sigma Aldrich | Cat# G0750 |
| Galacturonic acid | Sigma Aldrich | Cat# 73960 |
| Glucomannan | Megazyme | Cat# P-GLCML |
| Glucosamine | Sigma Aldrich | Cat# G4875 |
| Glucose | Sigma Aldrich | Cat# G8270 |
| Glucuronic acid | Sigma Aldrich | Cat# G8645 |
| Glutamic Acid | Sigma Aldrich | Cat# G1501 |
| Glutamine | Sigma Aldrich | Cat# G8540 |
| Glycerol | Thermo Fisher Scientific | Cat# BP229-4 |
| Glycine | Sigma Aldrich | Cat# G8898 |
| Glycogen | Sigma Aldrich | Cat# G0885 |
| Goat serum | Thermo Fisher Scientific | Cat# 16210-064; Lot# 2226570 |
| Guanine | Sigma Aldrich | Cat# G11950 |
| Guanidine hydrochloride | Sigma Aldrich | Cat# 50950 |
| Hematin | Sigma Aldrich | Cat# H3281 |
| Heparin | Sigma Aldrich | Cat# H0777 |
| Histidine | Sigma Aldrich | Cat# H8000 |
| Horse serum | Thermo Fisher Scientific | Cat# 16050-122; Lot# 2540511 |
| Hydrochloric Acid | Thermo Fisher Scientific | Cat# A144-212 |
| Hyaluronic acid | Sigma Aldrich | Cat# H5388 |
| Inulin | Sigma Aldrich | Cat# I2255 |
| Iodoacetamide | Sigma Aldrich | Cat# I6125 |
| Iron(II) sulfate heptahydrate | Sigma Aldrich | Cat# 215422 |
| Isoleucine | Sigma Aldrich | Cat# W527602 |
| Lamarin | Sigma Aldrich | Cat# L9634 |
| Larch arabinogalactan | Megazyme | Cat# P-ARGAL |
| LB agar | Invitrogen | Cat# 22700-041 |
| LB broth | Invitrogen | Cat# L3022 |
| Leucine | Sigma Aldrich | Cat# L8000 |
| Levan | Sigma Aldrich | Cat# L8647 |
| Lichenan | Megazyme | Cat# P-LICHN |
| Lupin pectic galactan | Megazyme | Cat# P-PGALU |
| Lysine | Sigma Aldrich | Cat# L5501 |
| Lysozyme | Sigma Aldrich | Cat# L6876 |
| Magnesium sulfate | Sigma Aldrich | Cat# MX0075 |
| Magnesium sulfate heptahydrate | Sigma Aldrich | Cat# M1880 |
| Manganese (II) sulfate monohydrate | Sigma Aldrich | Cat# M8179 |
| Mannan | Sigma Aldrich | Cat# M7504 |
| Mannose | Sigma Aldrich | Cat# M4625 |
| Menadione | Sigma Aldrich | Cat# M5625 |
| Methanol | Sigma Aldrich | Cat# 322415 |
| Methionine | Sigma Aldrich | Cat# M9625 |
| Mucin O-glycans (porcine gastric mucosa) | Dr. Eric C. Martens | N/A |
| M9 minimal salt | Sigma Aldrich | Cat# M6030 |
| N-acetyl galactosamine | Sigma Aldrich | Cat# A2795 |
| N-acetyl glucosamine | Sigma Aldrich | Cat# A3286 |
| N-acetyl neuraminic acid | Sigma Aldrich | Cat# A0812 |
| Newborn Calf serum | Thermo Fisher Scientific | Cat# 16010-159; Lot#2394148 |
| Nicotinic acid | Sigma Aldrich | Cat# N4126 |
| Nickel (II) chloride hexahydrate | Sigma Aldrich | Cat# 654507 |
| p-Aminobenzoic acid | Sigma-Aldrich | Cat# A9878 |
| Pancreatic digest of casein | Sigma Aldrich | Cat# 70169 |
| PAS staining kit | Sigma-Aldrich | Cat# 95B-1KT |
| PBS | Thermo Fisher Scientific | Cat# 10010-023 |
| Phenol solution | Thermo Fisher Scientific | Cat# A931L |
| Phenol:Chloroform:Isoamyl Alcohol (ph: 4.5) | Thermo Fisher Scientific | Cat# AM9722 |
| Phenylalanine | Sigma Aldrich | Cat# P2126 |
| Potato amylopectin | Sigma Aldrich | Cat# 10118 |
| Potato pectic galactan | Megazyme | Cat# P-GAPT |
| Proline | Sigma Aldrich | Cat# P5607 |
| Pullulan | Sigma Aldrich | Cat# P4516 |
| Pyridoxine HCl | Sigma Aldrich | Cat# P9755 |
| Resazurin | Sigma Aldrich | Cat# R7017 |
| rhamnose | Sigma Aldrich | Cat# R3875 |
| Rhamnogalacturonan I | Megazyme | Cat# P-RHAM1 |
| Riboflavin | Sigma Aldrich | Cat# R7649 |
| Ribose | Sigma Aldrich | Cat# R7500 |
| SDS | Thermo Fisher Scientific | Cat# BP166-500 |
| Serine | Sigma Aldrich | Cat# 84959 |
| Sheep serum | Sigma-Aldrich | Cat# S2263; Lot# 21H266 |
| Sodium acetate buffer solution (pH 5.2) | Sigma Aldrich | Cat# S7899 |
| Sodium chloride | Sigma Aldrich | Cat# S7653 |
| Sodium citrate dihydrate | Thermo Fisher Scientific | Cat# S279-500 |
| Sodium hydroxide | Thermo Fisher Scientific | Cat# S318-100 |
| Sodium hypochlorite | Clorox | Cat# US001237 |
| Sodium molybdate dihydrate | Sigma Aldrich | Cat# M1651 |
| Sodium nitrate | Sigma Aldrich | Cat# S5506 |
| Sodium nitroprusside dihydrate | Fisher Scientific | Cat# ICN15206125 |
| Sodium phosphate monobasic | Sigma Aldrich | Cat# S8282 |
| Sulfuric acid | Sigma Aldrich | Cat# 84736 |
| Sugar beet arabinan | Megazyme | Cat# P-ARAB |
| Tamarind xyloglucan | Megazyme | Cat# P-XYGLN |
| Thiamine HCl | Sigma Aldrich | Cat# T4625 |
| Thioctic acid | Sigma Aldrich | Cat# T5625 |
| Threonine | Sigma Aldrich | Cat# T8625 |
| Thymine | Sigma Aldrich | Cat# T0895 |
| Tris | Thermo Fisher Scientific | Cat# BP152-10 |
| Tris-HCL (pH8) | Thermo Fisher Scientific | Cat# BP1758-500 |
| Trypsin from porcine pancreas | Sigma Aldrich | Cat# T4799 |
| Tryptophan | Sigma Aldrich | Cat# T0254 |
| Tyrosine | Sigma Aldrich | Cat# T3754 |
| Uracil | Sigma Aldrich | Cat# U1128 |
| Valine | Sigma Aldrich | Cat# V0500 |
| Vancomycin | Xellia Pharmaceuticals | NDC#70594-048-01 |
| Wheat arabinoxylan | Megazyme | Cat# P-WAXYL |
| Xylenes | Thermo Fisher Scientific | Cat# X3S-4 |
| Xylose | Sigma Aldrich | Cat# X3877 |
| Yeast extract | Sigma Aldrich | Cat# 92144 |
| Zinc sulfate heptahydrate | Sigma Aldrich | Cat# Z0251 |
| 2-Mercaptoethanol | Sigma Aldrich | Cat# M3148 |
| Critical commercial assays | ||
| E.Z.N.A.Total RNA Kit I | OMEGA | Cat# R6834-02 |
| E.Z.N.A. Stool DNA Kit | OMEGA | Cat# D4015-02 |
| TrueVIEW® Autofluorescence | Vector Laboratories | Cat# SP-8400 |
| High-Capacity RNA-to-cDNA Kit | Thermo Fisher Scientific | Cat# 4387406 |
| Radiant™ SYBR Green qPCR Kit | Alkali Scientific | Cat# QS1050 |
| RNeasy Mini Kit | QIAGEN | Cat# 74134 |
| RNeasy Micro Kit | QIAGEN | Cat# 74004 |
| Mouse Lipocalin-2/NGAL DuoSet ELISA kit | R & D Biosystems | Cat# DY1857 |
| NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina | New England BioLabs | Cat# E7760L |
| Ribo depletion Module NEBNext rRNA Human/Mouse/Rat | New England BioLabs | Cat# E6310X |
| NEBNext rRNA Depletion Kit Bacteria | New England BioLabs | Cat# E7850X |
| NEBNext Multiplex Oligos for Illumina Unique dual | New England BioLabs | Cat# E6440L |
| Deposited data | ||
| 16S rRNA and RNA sequencing data | This study | BioProject: PRJNA984252 |
| Other | ||
| PowerBead Tubes, garnet (0.70 mm) | QIAGEN | Cat# 13123-50 |
| Fiber-Free diet | Envigo | Cat# TD.130343 |
| Custom CTR diet | Envigo | Cat# TD.190428 |
| Laboratory Autoclavable Rodent Diet - 5LOD | LabDiet | Cat# 3005659-220 |
| Laboratory Autoclavable Rodent Breeder Diet - 5013 | LabDiet | Cat# 0006529 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J: WT | The Jackson Laboratory | JAX: 000664 |
| Mouse: C57BL/6NTac: WT | Taconic Biosciences | B6-F; B6-M |
| Mouse: Nod2−/−-Cybb−/− | Caruso et al.13 | |
| Mouse: Muc2−/− | Dr. Eric C. Martens | Velcich et al.49 |
| Oligonucleotides | ||
| See Table S7 | ||
| Software and algorithms | ||
| Bowtie2 | Langmead et al.65 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Morpheus | N/A | https://software.broadinstitute.org/morpheus/ |
| Aperio ImageScope v 12.1.0.5029 | Aperio Technologies, Inc. | https://www2.leicabiosystems.com/ |
| Mothur (v.1.40.5) | Schloss et al.61 | https://mothur.org/ |
| UCHIME | Edgar et al.63 | https://drive5.com/usearch/manual/uchime_algo.html |
| SILVA 16S rRNA database release 138 | Pruesse et al.62 | https://www.arb-silva.de/ |
| Phylogeny.fr | Dereeper et al.64 | https://www.phylogeny.fr/simple_phylogeny.cgi |
| Prism 9.1.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| TreeGraph2 | N/A | http://treegraph.bioinfweb.info/ |
| Salmon | Patro et al.66 | https://salmon.readthedocs.io/en/latest/salmon.html |
| bcl2fastq2 Conversion Software | Illumina | https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2-20.html |
| UniProt database | N/A | https://www.uniprot.org/ |
| Diamond | Buchfink et al.67 | https://gensoft.pasteur.fr/docs/diamond/0.8.22/diamond_manual.pdf |
| metaSPAdes | Nurk et al.68 | https://cab.spbu.ru/files/release3.12.0/manual.html |
| HydDB database | Sondergaard et al.69 | https://services.birc.au.dk/hyddb/ |
| Gen5™ | BioTek | https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imagersoftware/ |
Highlights.
Dietary fiber exclusion is beneficial in a spontaneous Crohn’s disease-like colitis model
Fiber exclusion alters the localization of a mucus-dwelling pathobiont, M. schaedleri
Fiber deprivation impairs a critical M. schaedleri metabolic pathway
Pathobiont localization in the intestinal mucus layer is critical for causing disease
ACKNOWLEDGMENTS
The authors thank Kelsey Martin and Amr Aboud for animal husbandry, Jani L. O’Rourke for bacterial strain, Grace Chen for critical reading of the manuscript, Nicholas A. Pudlo for technical assistance, and the Microbiome Core, the Germ-Free Mouse Facility, the Advanced Genomics Core, the School of Dentistry Histology Core, the Digital Pathology (Slide Scanning service) at the University of Michigan for support. This work was supported by Senior Research Award from the Crohn’s and Colitis Foundation (1005572) (R.C), Pilot Feasibility Project through the University of Michigan Center for Gastrointestinal Research (NIH P30 DK034933) (R.C.), and NIH grant 5R01 DK121504 (G.N.).
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Xavier RJ, and Podolsky DK (2007). Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434. 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Peloquin JM, Goel G, Villablanca EJ, and Xavier RJ (2016). Mechanisms of Pediatric Inflammatory Bowel Disease. Annu Rev Immunol 34, 31–64. 10.1146/annurev-immunol-032414-112151. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. (2017). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390, 2769–2778. 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 4.Windsor JW, and Kaplan GG (2019). Evolving Epidemiology of IBD. Curr Gastroenterol Rep 21, 40. 10.1007/s11894-019-0705-6. [DOI] [PubMed] [Google Scholar]
- 5.Benchimol EI, Mack DR, Nguyen GC, Snapper SB, Li W, Mojaverian N, Quach P, and Muise AM (2014). Incidence, outcomes, and health services burden of very early onset inflammatory bowel disease. Gastroenterology 147, 803–813 e807; quiz e814–805. 10.1053/j.gastro.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Benchimol EI, Fortinsky KJ, Gozdyra P, Van den Heuvel M, Van Limbergen J, and Griffiths AM (2011). Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis 17, 423–439. 10.1002/ibd.21349. [DOI] [PubMed] [Google Scholar]
- 7.Caruso R, Lo BC, and Nunez G (2020). Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol 20, 411–426. 10.1038/s41577-019-0268-7. [DOI] [PubMed] [Google Scholar]
- 8.McGovern DP, Kugathasan S, and Cho JH (2015). Genetics of Inflammatory Bowel Diseases. Gastroenterology 149, 1163–1176 e1162. 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 47, 979–986. 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ananthakrishnan AN, Huang H, Nguyen DD, Sauk J, Yajnik V, and Xavier RJ (2014). Differential effect of genetic burden on disease phenotypes in Crohn’s disease and ulcerative colitis: analysis of a North American cohort. Am J Gastroenterol 109, 395–400. 10.1038/ajg.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Lee CH, Peters LA, Mastropaolo LA, Thoeni C, Elkadri A, Schwerd T, Zhu J, Zhang B, Zhao Y, et al. (2016). Variants in TRIM22 That Affect NOD2 Signaling Are Associated With Very-Early-Onset Inflammatory Bowel Disease. Gastroenterology 150, 1196–1207. 10.1053/j.gastro.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muise AM, Xu W, Guo CH, Walters TD, Wolters VM, Fattouh R, Lam GY, Hu P, Murchie R, Sherlock M, et al. (2012). NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut 61, 1028–1035. 10.1136/gutjnl-2011-300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caruso R, Mathes T, Martens EC, Kamada N, Nusrat A, Inohara N, and Nunez G (2019). A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol 4. 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cipcic Paljetak H, Baresic A, Panek M, Peric M, Matijasic M, Lojkic I, Barisic A, Vranesic Bender D, Ljubas Kelecic D, Brinar M, et al. (2022). Gut microbiota in mucosa and feces of newly diagnosed, treatment-naive adult inflammatory bowel disease and irritable bowel syndrome patients. Gut Microbes 14, 2083419. 10.1080/19490976.2022.2083419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limketkai BN, Shah ND, Sheikh GN, and Allen K (2019). Classifying Enteral Nutrition: Tailored for Clinical Practice. Curr Gastroenterol Rep 21, 47. 10.1007/s11894-019-0708-3. [DOI] [PubMed] [Google Scholar]
- 17.Logan M, Gkikas K, Svolos V, Nichols B, Milling S, Gaya DR, Seenan JP, Macdonald J, Hansen R, Ijaz UZ, et al. (2020). Analysis of 61 exclusive enteral nutrition formulas used in the management of active Crohn’s disease-new insights into dietary disease triggers. Aliment Pharmacol Ther 51, 935–947. 10.1111/apt.15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Rheenen PF, Aloi M, Assa A, Bronsky J, Escher JC, Fagerberg UL, Gasparetto M, Gerasimidis K, Griffiths A, Henderson P, et al. (2020). The Medical Management of Paediatric Crohn’s Disease: an ECCO-ESPGHAN Guideline Update. J Crohns Colitis. 10.1093/ecco-jcc/jjaa161. [DOI] [PubMed] [Google Scholar]
- 19.Lee D, Baldassano RN, Otley AR, Albenberg L, Griffiths AM, Compher C, Chen EZ, Li H, Gilroy E, Nessel L, et al. (2015). Comparative Effectiveness of Nutritional and Biological Therapy in North American Children with Active Crohn’s Disease. Inflamm Bowel Dis 21, 1786–1793. 10.1097/MIB.0000000000000426. [DOI] [PubMed] [Google Scholar]
- 20.Cohen-Dolev N, Sladek M, Hussey S, Turner D, Veres G, Koletzko S, Martin de Carpi J, Staiano A, Shaoul R, Lionetti P, et al. (2018). Differences in Outcomes Over Time With Exclusive Enteral Nutrition Compared With Steroids in Children With Mild to Moderate Crohn’s Disease: Results From the GROWTH CD Study. J Crohns Colitis 12, 306–312. 10.1093/ecco-jcc/jjx150. [DOI] [PubMed] [Google Scholar]
- 21.Damas OM, Garces L, and Abreu MT (2019). Diet as Adjunctive Treatment for Inflammatory Bowel Disease: Review and Update of the Latest Literature. Curr Treat Options Gastroenterol 17, 313–325. 10.1007/s11938-019-00231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353 e1321. 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, and Sonnenburg JL (2015). Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe 18, 478–488. 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, and Backhed F (2018). Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe 23, 27–40 e27. 10.1016/j.chom.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D, Albenberg L, Compher C, Baldassano R, Piccoli D, Lewis JD, and Wu GD (2015). Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology 148, 1087–1106. 10.1053/j.gastro.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, et al. (2015). Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 18, 489–500. 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, and Vijay-Kumar M (2012). Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One 7, e44328. 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad MA, Carreon CK, Dawany N, Russo P, and Kelsen JR (2019). Distinct Histopathological Features at Diagnosis of Very Early Onset Inflammatory Bowel Disease. J Crohns Colitis 13, 615–625. 10.1093/ecco-jcc/jjy212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish A, Boudaud M, Grant ET, Willieme S, Neumann M, Wolter M, Craig SZ, De Sciscio A, Cosma A, Hunewald O, et al. (2023). Akkermansia muciniphila exacerbates food allergy in fibre-deprived mice. Nat Microbiol. 10.1038/s41564-023-01464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aihara E, Engevik KA, and Montrose MH (2017). Trefoil Factor Peptides and Gastrointestinal Function. Annu Rev Physiol 79, 357–380. 10.1146/annurev-physiol-021115-105447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SLW, Fox JG, and Lee A (2005). Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 55, 1199–1204. 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 32.Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, and Phillipson M (2011). Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol 300, G327–333. 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, and Hansson GC (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105, 15064–15069. 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, and De Angelis M (2020). The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 8. 10.3390/microorganisms8040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, and Florin TH (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105, 2420–2428. 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 36.Zumft WG (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61, 533–616. 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loy A, Pfann C, Steinberger M, Hanson B, Herp S, Brugiroux S, Gomes Neto JC, Boekschoten MV, Schwab C, Urich T, et al. (2017). Lifestyle and Horizontal Gene Transfer-Mediated Evolution of Mucispirillum schaedleri, a Core Member of the Murine Gut Microbiota. mSystems 2. 10.1128/mSystems.00171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf PG, Biswas A, Morales SE, Greening C, and Gaskins HR (2016). H2 metabolism is widespread and diverse among human colonic microbes. Gut Microbes 7, 235–245. 10.1080/19490976.2016.1182288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraemer JT, and Bagley DM (2006). Supersaturation of dissolved H(2) and CO (2) during fermentative hydrogen production with N(2) sparging. Biotechnol Lett 28, 1485–1491. 10.1007/s10529-006-9114-7. [DOI] [PubMed] [Google Scholar]
- 41.Durack J, and Lynch SV (2019). The gut microbiome: Relationships with disease and opportunities for therapy. J Exp Med 216, 20–40. 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burkitt DP, Walker AR, and Painter NS (1972). Effect of dietary fibre on stools and the transit-times, and its role in the causation of disease. Lancet 2, 1408–1412. 10.1016/s0140-6736(72)92974-1. [DOI] [PubMed] [Google Scholar]
- 43.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. (2017). A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 9, 103. 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svolos V, Hansen R, Nichols B, Quince C, Ijaz UZ, Papadopoulou RT, Edwards CA, Watson D, Alghamdi A, Brejnrod A, et al. (2019). Treatment of Active Crohn’s Disease With an Ordinary Food-based Diet That Replicates Exclusive Enteral Nutrition. Gastroenterology 156, 1354–1367 e1356. 10.1053/j.gastro.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, and Vermeire S (2011). Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637. 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 47.Raygoza Garay JA, Turpin W, Lee SH, Smith MI, Goethel A, Griffiths AM, Moayyedi P, Espin-Garcia O, Abreu M, Aumais GL, et al. (2023). Gut Microbiome Composition Is Associated With Future Onset of Crohn’s Disease in Healthy First-Degree Relatives. Gastroenterology 165, 670–681. 10.1053/j.gastro.2023.05.032. [DOI] [PubMed] [Google Scholar]
- 48.Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Buttner L, de Lima Romao E, Behrendt CL, Lopez CA, et al. (2018). Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211. 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, and Augenlicht L (2002). Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729. 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 50.Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, and Niedermeyer G (1985). Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest 75, 944–953. 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johansson ME, and Hansson GC (2012). Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol 842, 229–235. 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 52.Chassaing B, Aitken JD, Malleshappa M, and Vijay-Kumar M (2014). Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol 104, 15 25 11–15 25 14. 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herp S, Brugiroux S, Garzetti D, Ring D, Jochum LM, Beutler M, Eberl C, Hussain S, Walter S, Gerlach RG, et al. (2019). Mucispirillum schaedleri Antagonizes Salmonella Virulence to Protect Mice against Colitis. Cell Host Microbe 25, 681–694 e688. 10.1016/j.chom.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 54.Bergstrom A, Kristensen MB, Bahl MI, Metzdorff SB, Fink LN, Frokiaer H, and Licht TR (2012). Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res Notes 5, 402. 10.1186/1756-0500-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J, Li P, Wang L, Li M, Ge Z, Noordam L, Lieshout R, Verstegen MMA, Ma B, Su J, et al. (2021). Cancer-Associated Fibroblasts Provide a Stromal Niche for Liver Cancer Organoids That Confers Trophic Effects and Therapy Resistance. Cell Mol Gastroenterol Hepatol 11, 407–431. 10.1016/j.jcmgh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L, Zhai Y, Wang Y, Fearon ER, Nunez G, Inohara N, and Cho KR (2021). Altering the Microbiome Inhibits Tumorigenesis in a Mouse Model of Oviductal High-Grade Serous Carcinoma. Cancer Res 81, 3309–3318. 10.1158/0008-5472.CAN-21-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gomes-Neto JC, Mantz S, Held K, Sinha R, Segura Munoz RR, Schmaltz R, Benson AK, Walter J, and Ramer-Tait AE (2017). A real-time PCR assay for accurate quantification of the individual members of the Altered Schaedler Flora microbiota in gnotobiotic mice. J Microbiol Methods 135, 52–62. 10.1016/j.mimet.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG 3rd, Hayashi A, Imai J, Sugihara K, Miyoshi M, Brazil JC, Kuffa P, et al. (2020). The Intermucosal Connection between the Mouth and Gut in Commensal Pathobiont-Driven Colitis. Cell 182, 447–462 e414. 10.1016/j.cell.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, Baxter NT, Zhao L, Schloss PD, Martens EC, et al. (2017). NLRP6 Protects Il10(−/−) Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell Rep 19, 733–745. 10.1016/j.celrep.2017.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caruso R, Ono M, Bunker ME, Nunez G, and Inohara N (2019). Dynamic and Asymmetric Changes of the Microbial Communities after Cohousing in Laboratory Mice. Cell Rep 27, 3401–3412 e3403. 10.1016/j.celrep.2019.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, and Glockner FO (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35, 7188–7196. 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar RC, Haas BJ, Clemente JC, Quince C, and Knight R (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36, W465–469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchfink B, Xie C, and Huson DH (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12, 59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 68.Nurk S, Meleshko D, Korobeynikov A, and Pevzner PA (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res 27, 824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sondergaard D, Pedersen CN, and Greening C (2016). HydDB: A web tool for hydrogenase classification and analysis. Sci Rep 6, 34212. 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luis AS, Jin C, Pereira GV, Glowacki RWP, Gugel SR, Singh S, Byrne DP, Pudlo NA, London JA, Basle A, et al. (2021). A single sulfatase is required to access colonic mucin by a gut bacterium. Nature 598, 332–337. 10.1038/s41586-021-03967-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG 3rd, Pudlo NA, Hoostal M, Li X, Wang TD, Feehley T, et al. (2017). Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319. 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolfe RS, and Metcalf WW (2010). A vacuum-vortex technique for preparation of anoxic solutions or liquid culture media in small volumes for cultivating methanogens or other strict anaerobes. Anaerobe 16, 216–219. 10.1016/j.anaerobe.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 73.Bronk DA, Roberts QN, Sanderson MP, Canuel EA, Hatcher PG, Mesfioui R, Filippino KC, Mulholland MR, and Love NG (2010). Effluent organic nitrogen (EON): bioavailability and photochemical and salinity-mediated release. Environ Sci Technol 44, 5830–5835. 10.1021/es101115g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data files for 16S and metatranscriptomics sequencing are freely available at BioProject database, ID number: PRJNA984252. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


