Abstract
Transgenic mice have been used to explore the role of chromosomal translocations in the genesis of tumors. But none of these efforts has actually involved induction of a translocation in vivo. Here we report the use of Cre recombinase to replicate in vivo the t(8;21) translocation found in human acute myeloid leukemia (AML). As in the human tumors, the murine translocation fuses the genes AML1 and ETO. We used homologous recombination to place loxP sites at loci that were syntenic with the break points for the human translocation. Cre activity was provided in mice by a transgene under the control of the Nestin promoter, or in cultured B cells by infecting with a retroviral vector encoding Cre. In both instances, Cre activity mediated interchromosomal translocations that fused the AML1 and ETO genes. Thus, reciprocal chromosomal translocations that closely resemble rearrangements found in human cancers can be achieved in mice.
INTRODUCTION
Animal models are important tools for studying the mechanisms of tumorigenesis and for the preclinical testing of new therapeutic agents (Bedell et al., 1997). The mouse is a popular model organism because of its relatively short reproductive cycle and because its genome can be readily manipulated by molecular means.
Chromosomal translocations appear to be involved in the genesis of many types of human tumors, often by forming fusion proteins that are pathogenic (Mitelman et al., 1997). Mouse models for several human leukemias have been established by the tissue-specific expression of transgenes encoding fusion proteins (Look, 1997). But these models often possess features that detract from their utility.
A drawback of many transgenic models is that the expression profile of the transgene frequently differs from the endogenous fusion genes generated by the translocation. Better models are provided by the gene ‘knock-in’ approach that expresses a fusion protein under the control of the appropriate endogenous promoter (Corral et al., 1996). However, this approach also fails to recapitulate exactly the situation found in human cancers with chromosomal translocations. The fusion gene is present from the inception of embryogenesis, whereas in humans the chromosomal translocation is believed to occur during a later stage of development. The timing of the translocation might be an important factor involved in the development of cancer. For example, the ‘knock-in’ approach for an AML/ETO mouse model resulted in early lethality of the developing embryo (Yergeau et al., 1997; Okuda et al., 1998), which prevented the investigation of the role of AML/ETO in leukemogenesis.
Another drawback of the ‘knock-in’ approach is that only one fusion protein is generated, whereas in balanced chromosomal translocations two fusion proteins can be produced. It has been shown that both fusion proteins can play a role in some leukemias (Pollock et al., 1999). Systems that allow exact recapitulation of genetic events observed in human cancers are therefore critical to model the disease as closely as possible.
We set out to recapitulate events that are observed in patients with the M2 subtype of acute myeloid leukemia (AML) with reciprocal translocation t(8;21) (Nucifora and Rowley, 1994). Our plan was to induce a translocation between mouse chromosomes 4 and 16, which harbor the syntenic regions of human chromosomes 8 and 21. We targeted introns that were frequently found to be the point of translocation in many human AMLs. We used homologous recombination to place loxP sites into intron sequences of the murine AML1 and ETO genes. Translocation between AML1 and ETO was then elicited with Cre recombinase, driven by the transcriptional control element for the Nestin gene (Zimmerman et al., 1994). We used a line of Nestin–Cre mice (Nes–cre1), which apparently do not express Cre early in embryogenesis (Trumpp et al., 1999), a feature that allowed us to circumvent the possible embryonic lethality encountered when AML1/ETO was introduced as a transgene by the ‘knock-in’ technique (Yergeau et al., 1997; Okuda et al., 1998). We also tested the ability to achieve chromosomal translocations in primary B cells, delivering Cre recombinase with a retrovirus in vitro.
We found that interchromosomal recombination at the loxP sites in AML1 and ETO could be induced by Cre. To our knowledge this is the first example that elicits chromosomal translocations at loxP sites in the mouse and in somatic cells ex vivo.
RESULTS
Generation of mice carrying a loxP site in introns of the AML1 and ETO genes
To evaluate the utility of the Cre/loxP system for inducible chromosomal translocations, we first had to insert loxP sites into the AML1 and ETO genes (Figure 1). We isolated genomic fragments from the murine AML1 and ETO genes corresponding to the breakpoint regions described in human leukemias and placed a neomycin cassette flanked by loxP sites (floxed neomycin) 1496 bp downstream of AML1 exon 5 and 893 bp upstream of ETO exon 2 (Figure 2A and B). ES cells were electroporated with these constructs and correctly targeted clones were identified for both AML1 and ETO (Figure 2C and D). Chimeric mice were generated by blastocyst injection and each of three chimeras transmitted the targeted allele through the germline.
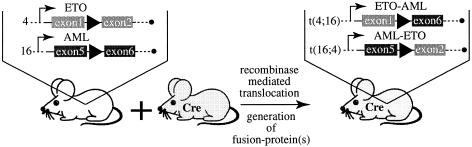
Fig. 1. Scheme for induction of chromosomal translocation by Cre. Mice containing a loxP site in intron 1 of the ETO gene on chromosome 4 and a loxP site in intron 5 of the AML1-gene on chromosome 16 were crossed to mice expressing Cre recombinase. Interchromosomal site-specific recombination at the loxP sites resulted in Cre-mediated chromosomal translocation [t(4;16), t(16;4)] in the offspring. Filled triangles depict loxP sites.
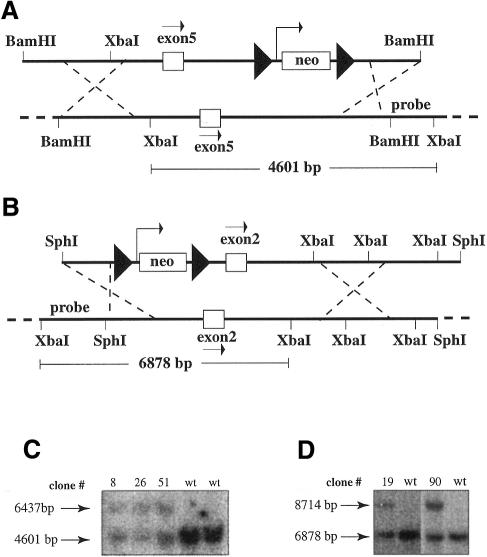
Fig. 2. Targeting of AML1 and ETO in mouse ES cells. The targeting constructs and the genomic regions for the AML1 gene (A) and the ETO gene (B) are shown. Relevant restriction sites and the probes used in Southern blot analysis in (C) and in (D) are presented. loxP sites are depicted as filled triangles flanking the neomycin resistance gene (neo). Exon 5 of AML1 and exon 2 of ETO are presented as white boxes and the direction of transcription is depicted by an arrow. The XbaI fragment size of the wild-type genomic fragment is shown for the AML1 (4601 bp) and ETO (6878 bp) genes. The floxed neomycin gene adds 1836 bp to each fragment. The increased fragment size can be detected in DNA from correctly targeted ES cells for AML1 (C) (6437 bp) and ETO (D) (8714 bp). Positive clones #8, 26, 51 for AML1 and clones #19 and 90 for ETO are shown. bp, base pairs; wt, wild type.
No obvious phenotype was observed in mice carrying the floxed neomycin cassette in the AML1 intron (designated AMLX) or in the ETO intron (ETOX). Blood counts from these mice appeared normal (data not shown). Mice that had been bred to homozygosity for either AMLX or ETOX were also normal (data not shown).
Identification of t(16;4) in primary hematopoetic cells infected with a Cre-expressing retrovirus
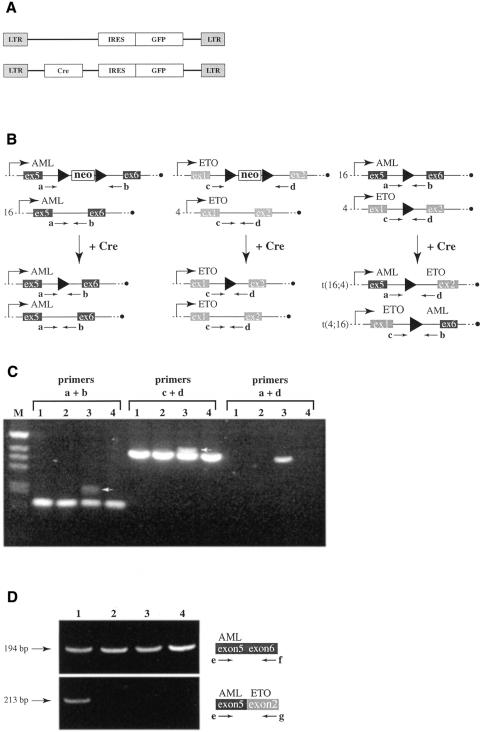
We obtained AMLX/ETOX mice carrying the floxed neomycin cassette in both the AML1 and ETO genes by breeding. Naive B cells from AMLX/ETOX and from wild-type mice were isolated, activated and infected with a retrovirus expressing either Cre recombinase in addition to GFP (pMIG–Cre) or GFP alone (pMIG) (Figure 3A). Initial infection rates were ∼15% as measured by flow cytometry, assaying for GFP positive cells (data not shown). After 10 days of culture, genomic DNA was prepared from each pool of infected cells and analyzed by PCR. AMLX and ETOX cells infected with pMIG–Cre should excise the neomycin cassette, leaving behind the additional sequence of a loxP site (34 bp) in the intron (Figure 3B). Indeed, the extra 34 bp were detected in a PCR analysis with primers annealing 5′ and 3′ of the loxP sites (Figure 3C, primers a + b and primers c + d, lane 3).
Fig. 3. Cre-mediated chromosomal translocation in primary B cells. A schematic presentation of the pMIG and pMIG–Cre vectors used to generate retroviruses is shown in (A). LTR, long terminal repeat; IRES, internal ribosomal entry site; GFP, green fluorescent protein; Cre, Cre recombinase. Predicted events of Cre-mediated site-specific recombination are presented in (B). Primers a, b, c and d are shown at their position of anealing within the genomic locus as arrows. Neo, neomycin resistance cassette; ex, exon. An agarose gel analyzing the PCRs on DNA isolated from primary B cells is shown in (C). Lanes 1: wild-type cells infected with pMIG–Cre; lanes 2: wild-type cells infected with pMIG; lanes 3: AMLX/ETOX cells infected with pMIG–Cre; lanes 4: AMLX/ETOX cells infected with pMIG. Primer pairs used in the PCR are shown above the lanes. M, marker. The band specific for the targeted allele that has recombined out the neomycin cassette is highlighted by a white arrow. RT–PCR analysis on RNA isolated from B cells infected with different retroviruses is shown in (D). Lanes 1: AMLX/ETOX cells infected with pMIG–Cre; lanes 2: AMLX/ETOX cells infected with pMIG; lanes 3: wild-type cells infected with pMIG–Cre; lanes 4: wild-type cells infected with pMIG. The upper panel shows the PCR products obtained with primers e and f. The lower panel shows the PCR products obtained with primers e and g. A schematic presentation of the PCRs is shown to the right and the size of the PCR fragment is presented on the left. bp, base pairs.
To investigate whether Cre-mediated interchromosomal recombination between two loxP sites on chromosomes 4 and 16 had taken place we performed PCRs with primers annealing to AML1 and ETO (Figure 3B, primers a + d). A 382 bp band was obtained when DNA isolated from AMLX/ETOX B cells infected with pMIG–Cre was used as the template, but not when DNA isolated from wild-type cells infected with the same virus or DNA isolated from cells infected with pMIG was used (Figure 3C). Sequencing of the 382 bp band confirmed that interchromosomal recombination had occurred, suggesting that the chromosomal translocation of chromosomes 4 and 16 had been promoted by Cre-mediated recombination.
To test whether the fusion transcript could be detected, we performed RT–PCR analysis on RNA isolated from virus infected B cells (Figure 3D). Expression of AML1 was detectable with primers annealing to exon 5 (primer e) and exon 6 (primer f) in all samples examined, indicating that AML1 was expressed in the activated B cells. RT–PCRs with primers annealing to exon 5 of AML1 (primer e) and exon 2 of ETO (primer g) revealed that the fusion gene was detectable only in AMLX/ETOX B cells infected with pMIG–Cre, consistent with the detection of the translocation product in PCRs on genomic DNA.
Detection of t(16;4) in Nestin–Cre mice
To test whether Cre-mediated interchromosomal recombination could be achieved in mice, we crossed an AMLX mouse also carrying the Nestin-Cre transgene (Nes–cre1), which contains a Cre gene under the control of the rat Nestin promoter and intron 2 enhancer (Trumpp et al., 1999), with an ETOX mouse and analyzed tail DNA obtained from the offspring by PCR.
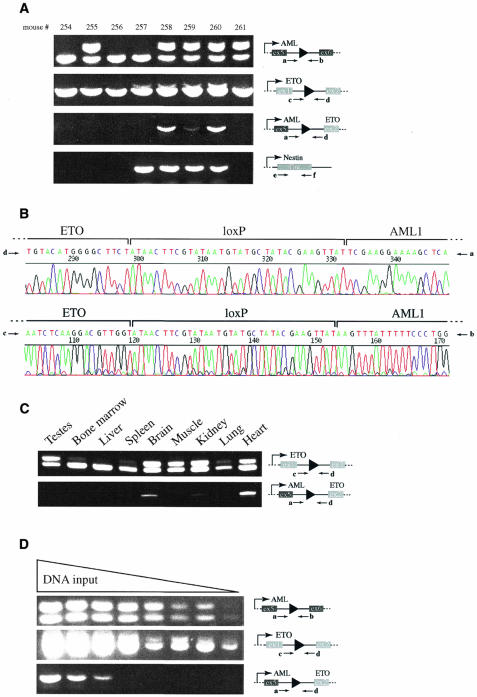
Three mice tested positive for Cre, the deleted neomycin cassette in AML1, and the deleted neomycin cassette in ETO (Figure 4, mouse #258, 259 and 260). These were the only mice that also tested positive in the PCR designed to detect the chromosomal translocation of chromosomes 4 and 16 using primers a and d. Primer pair b and c appeared to function less efficiently in the detection of the translocation under tested PCR conditions. Only trace amounts of the translocation PCR product were detected with these primers, using tail DNA from mouse #258 and #260 (data not shown). However, sequencing of re-amplified fragments with primers b and c confirmed that these primers can detect the translocation (see Figure 4B, primers b and c). We conclude that Cre-mediated translocation between chromosomes 4 and 16 had taken place in these mice.
Fig. 4. Cre-mediated chromosomal translocation in the mouse. Agarose gels analyzing the PCRs on DNA isolated from mouse tails are illustrated in (A). PCR results from mouse #254, 255, 256, 257, 258, 259, 260 and 261 are presented. Schematic presentations of relevant loci and utilized primers (a, b, c and d) are depicted to the right of each panel. Ex, exon. Sequencing reactions obtained from PCR products with primers a + d, and c + d, respectively, are presented in (B). The loxP sites and the respective genomic intron sequences from AML1 and ETO are indicated. PCR analysis on DNA isolated from mouse #260 and indicated tissues is presented in (C). Results with primer pairs c + d, and a + d, respectively are shown. Serial dilution PCR analysis on heart DNA isolated from mouse #260 is presented in (D). Schematic representations of relevant loci and utilized primers (a, b, c and d) are shown on the right.
Mouse #255 did not inherit the Cre transgene and therefore tested negative in the PCR designed to detect Cre (Figure 4). However, the PCR examining the AML1 locus showed that the neomycin cassette had been deleted in this mouse, indicating that Cre-mediated deletion of the neomycin cassette had taken place in the germ cell before meiosis. Mouse #257 tested positive for Cre and the deleted neomycin cassette in ETO but not for the deleted neomycin cassette in AML1. Therefore, this mouse did not carry a loxP site in the AML1 gene, which prevented Cre-mediated translocations of chromosomes 4 and 16.
Mouse #260 was sacrificed at the age of four months and genomic DNA from various tissues was isolated and analyzed. PCR analysis with primers c and d allowed the estimation of Cre-mediated recombination in different tissues. The ratio of the slower migrating band containing the loxP site versus the faster migrating band is a reflection of how many cells had excised the neomycin cassette from their genome. We observed strong recombination in testes, brain, muscle, kidney and heart (Figure 4C). These results are consistent with previous experiments utilizing the Nes–Cre1 mouse, where recombination of floxed alleles has been demonstrated in various tissues including the spinal cord, muscle and germ cells (Bates et al., 1999; Trumpp et al., 1999).
We readily detected the translocation-specific PCR product with primers a and d in brain, kidney and heart (Figure 4C), indicating that cells carrying the translocated chromosomes are present in these tissues. We were unable to detect the translocation in DNA isolated from bone marrow, muscle and lung. However, trace amounts of the translocation-specific PCR product were also obtained when DNA isolated from testes, liver and spleen was used (data not shown), indicating that these tissues also contain cells carrying the translocation.
To estimate the frequency of cells carrying the translocated chromosomes we performed serial dilution PCR experiments with DNA isolated from brain, kidney and heart (Figure 4D and data not shown). Based on these experiments we estimate that approximately one in 104 to one in 106 cells carry the translocated chromosomes in these tissues.
DISCUSSION
The Cre/loxP system is a powerful tool for genome manipulations in the mouse (Rajewsky et al., 1996; Nagy, 2000). Chromosome engineering resulting in large deletions has been reported utilizing the Cre/loxP system (Ramirez-Solis et al., 1995). Cre-mediated recombination between homologous chromosomes through the reconstitution of a selectable marker gene has been shown in mouse ES cells (Smith et al., 1995; Van Deursen et al., 1995). Trans-allelic targeted mitotic recombination between homologous chromosomes during the first mitotic division has also been demonstrated in the mouse (Hérault et al., 1998). However, previous efforts to achieve recombination between loxP sites positioned on different chromosomes in somatic cells have been unsuccessful (Hérault et al., 1998). We show that chromosomal translocations utilizing the Cre/loxP system can be achieved in the mouse and in primary B cells in culture. This finding makes the Cre/loxP system an ideal tool to generate predefined chromosomal translocations in the mouse, thus allowing recapitulation of genetic events observed in human cancers.
The application of this system might allow the precise dissection of molecular details occurring during the development of cancer. For instance, the significance of timing of the translocation can be addressed by the choice of Cre-expressing mouse lines. The importance of timing is illustrated by the present work. By using a promoter that is apparently not active early in embryogenesis, we were able to circumvent the early lethality encountered when AML1/ETO was introduced into the germ line by the ‘knock-in’ strategy (Yergeau et al., 1997; Okuda et al., 1998). We have not yet detected malignancies in AMLX/ETOX/Nes–cre1 mice at the age of five months or younger, and we do not expect the AMLX/ETOX/Nes–cre1 mice to develop leukemia because the promoter is not known to be active in haematopoietic cells (Zimmerman et al., 1994). But having demonstrated the feasibility of inducing translocations with Cre, we are now attempting to express the enzyme specifically in haematopietic cells.
Mice that allow inducible expression of Cre recombinase might prove to be valuable in future experiments (Schwenk et al., 1998; Holzenberger et al., 2000). By this means the translocation could be induced at different time points during development or in adult animals, and consequences monitored over time. In addition, molecular dissection of functions of the generated fusion gene and the molecular effects of the newly synthesized fusion protein can be investigated in vivo and ex vivo.
Chromosomal translocations can also be induced in different cell types using mice expressing Cre from tissue-specific promoters. This would allow the study of in vivo mechanisms by which the generated fusion protein may transform only certain cell types.
The frequency of Cre-mediated chromosomal translocations in AMLX/ETOX/Nes–cre1 mice appears to be relatively low, when compared with the reported efficiency of deletion of floxed alleles in mice (Rajewsky et al., 1996). However, it may not be necessary to achieve efficient interchromosomal recombination in translocation mouse models, because the translocations found in human cancers are thought to be rare events.
In summary, the proof that chromosomal translocations between non-homologous chromosomes can be achieved in mice and ex vivo in somatic cells utilizing the Cre/loxP system opens new doors in the field of advanced genome engineering. Establishing mouse lines that make use of this system should result in improved mouse models of human cancers that involve chromosomal translocations.
MATERIALS AND METHODS
Gene targeting. Targeting constructs were generated by inserting a neomycin/kanamycin cassette flanked by loxP sites 1496 bp downstream of exon 5 of AML1 and 893 bp upstream of exon 2 of ETO using homologous recombination in E. coli (ET cloning) (Zhang et al., 1998) (Figure 2). Feederless E14 ES cells (2 × 107) were electroporated with 20 µg of each targeting construct and clones were selected at 150 µg/ml Geneticin (Gibco-BRL). Two correctly targeted ES cell clones for AML1 and ETO were used for blastocyst injections and three chimeras each transmitted the targeted allele through the germline. Mice positive for targeted AML1 and ETO genes were obtained by breeding.
Culture and infection of primary B cells. Naive B cells were isolated and purified from pooled spleen and lymph nodes and grown in RPMI medium, supplemented with 10% heat inactivated fetal calf serum, penicillin/streptomycin, l-glutamine, Na-pyruvate, 2-β-ME, HEPES and non-essential amino acids. Cells were activated with 1 µg/ml anti-CD40 antibody (Pharmingen), and infected twice at 24 h intervals with virus-containing supernatant produced in BOSC 23 cells, as previously described (Refaeli et al., 1998). The viruses used were either pMIG or pMIG–Cre (Van Parijs et al., 1999). Initial infection efficiency was monitored 3 days after cell isolation by flow cytometry.
PCR analysis. PCRs were performed with 0.6 µg of genomic DNA (isolated from either mouse tails or primary B cells) and 40 cycles of 95°C for 1 min; 62°C for 1 min; 72°C for 0.5 min. The dilution factor for each reaction in serial dilution PCR experiments was 10. Frangments were resolved on 2% MetaPhor agarose (FMC BioProducts). Tail DNA was isolated from 21-day-old mice. RT–PCR analysis was performed with the Titan™ one tube RT–PCR system from Roche on RNA isolated with the RNeasy Mini Kit from Qiagen. The RT–PCR program was as follows: 50°C for 30 min; 94°C for 2 min; 10× (94°C for 30 s; 60°C for 30 s; 68°C for 10 s); 30× (94°C for 30 s; 61°C for 20 s; 68°C for 20 s); 68°C for 5 min. The following primers were used in PCRs as shown in the figures:
primer a: 5′-TTCCCTACCAAATGTTGA
primer b: 5′-AGGTTTCCTGGGCTGTGTGAG
primer c: 5′-TTTCAGCGAGAATCTACTGCCTTG
primer d: 5′-ATTCCTGGCCTGTCCTGTTAGTTA
primer e: 5′-TTACAAATCCGCCACAAGTT
primer f: 5′-GGCTGGGTGGTGCGGGCTGAC
primer g: 5′-ATGTCGTTGGCGTAAATGAG
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the Bishop laboratory for critical experimental advice and useful discussions and Luk Van Parijs (Caltech, Pasadena, CA) for the kind gift of pMIG–Cre. We are grateful to the transgenics core facility at UCSF for excellent service. F.B. is a Fellow of the Leukemia and Lymphoma Society. Y.R. is a Merck fellow of the Life Sciences Research Foundation. This work was supported by funds from the National Institutes of Health (Grant CA 44338) and the G.W. Hooper Research Foundation.
REFERENCES
- Bates B., Rios, M., Trumpp, A., Chen, C., Fan, G., Bishop, J.M. and Jaenisch, R. (1999) Neurotrophin-3 is required for proper cerebellar development. Nature Neurosci., 2, 115–117. [DOI] [PubMed] [Google Scholar]
- Bedell M.A., Largaespada, D.A., Jenkins, N.A. and Copeland, N.G. (1997) Mouse models of human disease. Part II: recent progress and future directions. Genes Dev., 11, 11–43. [DOI] [PubMed] [Google Scholar]
- Corral J. et al. (1996) An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell, 85, 853–861. [DOI] [PubMed] [Google Scholar]
- Hérault Y., Rassoulzadegan, M., Cuzin, F. and Duboule, D. (1998) Engineering chromosomes in mice through targeted meiotic recombination (TAMERE). Nature Genet., 20, 381–384. [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Zaoui, R., Leneuve, P., Hamard, G. and Le Bouc, Y. (2000) Ubiquitous postnatal LoxP recombination using a doxycycline auto-inducible Cre transgene (DAI-Cre). Genesis, 26, 157–159. [PubMed] [Google Scholar]
- Look A.T. (1997) Oncogenic transcription factors in the human acute leukemias. Science, 278, 1059–1064. [DOI] [PubMed] [Google Scholar]
- Mitelman F., Mertens, F. and Johansson, B. (1997) A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nature Genet., 15, 417–474. [DOI] [PubMed] [Google Scholar]
- Nagy A. (2000) Cre recombinase: the universal reagent for genome tailoring. Genesis, 26, 99–109. [PubMed] [Google Scholar]
- Nucifora G. and Rowley, J.D. (1994) The AML1 and ETO genes in acute myeloid leukemia with a t(8;21). Leuk. Lymphoma, 14, 353–362. [DOI] [PubMed] [Google Scholar]
- Okuda T., Cai, Z., Yang, S., Lenny, N., Lyu, C.J., van Deursen, J.M., Harada, H. and Downing, J.R. (1998) Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood, 91, 3134–3143. [PubMed] [Google Scholar]
- Pollock J.L., Westervelt, P., Kurichety, A.K., Pelicci, P.G., Grisolano, J.L. and Ley, T.J. (1999) A bcr-3 isoform of RARα-PML potentiates the development of PML-RARα-driven acute promyelocytic leukemia. Proc. Natl Acad. Sci. USA, 96, 15103–15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K., Gu, H., Kuhn, R., Betz, U.A., Muller, W., Roes, J. and Schwenk, F. (1996) Conditional gene targeting. J. Clin. Invest., 98, 600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R., Liu, P. and Bradley, A. (1995) Chromosome engineering in mice. Nature, 378, 720–724. [DOI] [PubMed] [Google Scholar]
- Refaeli Y., Van Parijs, L., London, C.A., Tschopp, J. and Abbas, A.K. (1998) Biochemical mechanisms of IL-2-regulated Fas-mediated T cell apoptosis. Immunity, 8, 615–623. [DOI] [PubMed] [Google Scholar]
- Schwenk F., Kuhn, R., Angrand, P.O., Rajewsky, K. and Stewart, A.F. (1998) Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res., 26, 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J., De Sousa, M.A., Kwabi-Addo, B., Heppell-Parton, A., Impey, H. and Rabbitts, P. (1995) A site-directed chromosomal translocation induced in embryonic stem cells by Cre-loxP recombination [published erratum appears in Nature Genet. (1996), 12, 110]. Nature Genet., 9, 376–385. [DOI] [PubMed] [Google Scholar]
- Trumpp A., Depew, M.J., Rubenstein, J.L., Bishop, J.M. and Martin, G.R. (1999) Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev., 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deursen J., Fornerod, M., Van Rees, B. and Grosveld, G. (1995) Cre-mediated site-specific translocation between non-homologous mouse chromosomes. Proc. Natl Acad. Sci. USA, 92, 7376–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Parijs L., Refaeli, Y., Lord, J.D., Nelson, B.H., Abbas, A.K. and Baltimore, D. (1999) Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity, 11, 281–288. [DOI] [PubMed] [Google Scholar]
- Yergeau D.A. et al. (1997) Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nature Genet., 15, 303–306. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Buchholz, F., Muyrers, J.P. and Stewart, A.F. (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet., 20, 123–128. [DOI] [PubMed] [Google Scholar]
- Zimmerman L., Parr, B., Lendahl, U., Cunningham, M., McKay, R., Gavin, B., Mann, J., Vassileva, G. and McMahon, A. (1994) Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors [published erratum appears in Neuron (1994), 12, following pp. 1388]. Neuron, 12, 11–24. [DOI] [PubMed] [Google Scholar]