The site-specific recombinases (SSRs), Cre and FLP, have added a variety of possibilities to the field of genome engineering. Two articles in this issue of EMBO Reports (Buchholz et al., 2000; Collins et al., 2000) now extend the repertoire of SSR-driven genomic alterations in the mouse by adding possibly the most difficult task ever asked of these enzymes, namely translocation between non-homologous chromosomes. Aiming at establishing better mouse models of human leukemias, the two groups introduced loxP sites into the mouse at the relevant positions on two chromosomes and show that, upon Cre expression, it is possible to detect the heterologous translocation at reasonable frequencies. Although FLP-mediated translocation between homologous chromosomes in Drosophila was one of the first applications of site-specific recombination in genome engineering (Golic, 1991), the findings reported in this issue were by no means an expected advance along a linear pathway. Scepticism had lingered over the possibility that an SSR-driven non-homologous translocation could be achieved in the mouse at a detectable frequency. It was clear from other (‘easier’) genome engineering exercises that efficiency dropped as a function of the distance between SSR target-sites (Ramirez-Solis et al., 1995; Ringrose et al., 1999; Zheng et al., 2000). Additionally, evidence indicating that reasonable efficiencies of SSR translocations require the pairing of homologous chromosomes achieved during mitosis (Golic and Golic, 1996) or meiosis (Herault et al., 1998) was obtained in both flies and mice. A further source of doubt arose with the emergence of the chromosomal territory model of nuclear organization (Figure 1) (Lamond and Earnshaw, 1998; Zink and Cremer, 1998). How would the two SSR target sites, buried in distinct and possibly distant chromosomal territories, ever manage to find each other for the crucial handshake?
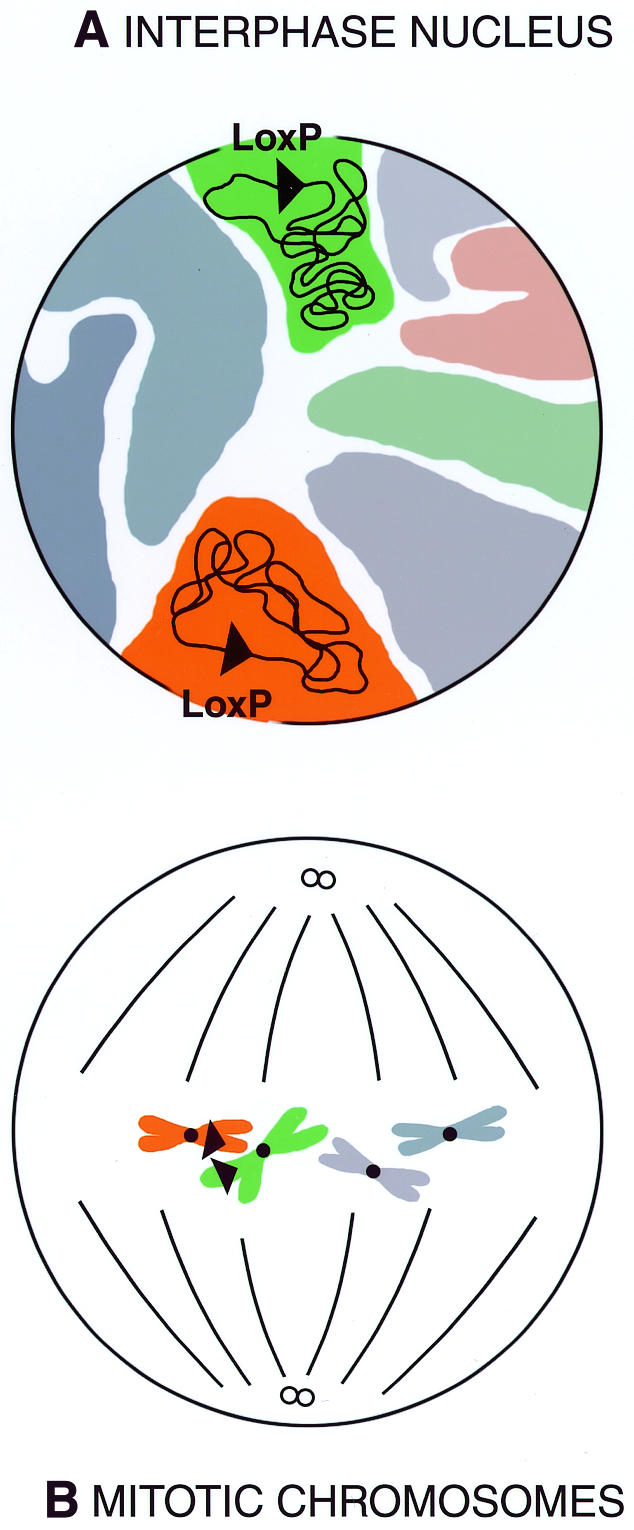
Fig. 1. (A) According to the chromosome territory model, during interphase each chromosome resides in a unique region within the nucleus, thus separating the two LoxP sites into different chromosomal territories (colored sectors). (B) During chromosomal resortment at mitotic metaphase, the two LoxP sites can stochastically come in close contact for Cre-mediated translocation to occur.
Hints for optimism, but also pessimism, came from pioneering experiments with Cre-mediated translocations between non-homologous chromosomes in mouse embryonic stem cells (Smith et al., 1995; Van Deursen et al., 1995). In spite of the success at achieving translocations, the very low efficiencies observed and the fact that these experiments were limited to cells in culture gave rise to legitimate doubts as to whether the result could be recapitulated in a living mouse. The work of Collins et al. (2000) and Buchholz et al. (2000) now dismisses these doubts. At the same time, their accomplishments prompt questions regarding how the two SSR target sites come together given the territorial organization of the nucleus. More work is needed to address this issue, but it is possible that global reshuffling of chromosomal positions may occur during mitosis, thus enabling pairing between two loxP sites. This scenario predicts that the rate of non-homologous SSR translocations will be higher among actively cycling cell populations. Alternatively, if recombination happens in interphase or resting cells, its frequency would be predicted to mirror the relative orientation of the two chromosomal territories in a given cell population.
Beyond the substantial technical achievement and inherent interest, what motivated these brave but risky enterprises considering that technically easier ways to make mouse models are available? As is well illustrated by the path taken by Rabbitts and co-workers, the goal is to create the best possible mouse model. Previously this group had created a mouse model of the same leukemia by a knock-in strategy to the Mll (mixed-lineage leukemia) locus (Corral et al., 1996; Dobson et al., 1999). Although a good model was obtained, a knock-in strategy does not include the balanced translocation and does not imitate the cell-type-specific, somatic aspects of a tumorigenic translocation. The possibility that both expression products of a translocation can alter the disease phenotype is well illustrated by work on mouse models of the PML-RARα (promyelocytic leukemia-retinoic acid receptor α) leukemogenic translocation. Using conventional transgenesis, it was shown that expression of the non-leukemogenic RARα-PML translocation product affects the phenotype of PML-RARα leukemias (Pollock et al., 1999). This important observation emphasizes the need to create the most authentic models possible. Similarly, the drawbacks of creating a model where all cells carry the tumorigenic translocation product from the start of development are illustrated by work with the AML–ETO (acute myeloid leukemia–eight twenty-one) fusion product. When introduced as a fusion knock-in into the AML locus, the AML–ETO fusion protein is embryonically lethal (Yergeau et al., 1997; Okuda et al., 1998), thus precluding the creation of a simple model and underlying the loxP translocation efforts of Buchholz et al. (2000). Furthermore, loxP translocation strategies should permit cell-type-specific and temporal inductions of balanced translocations when combined with appropriate Cre-expressing mouse lines. This will provide an unprecedented opportunity to investigate leukemogenic fusion protein function at different stages throughout the development of the hematopoietic lineages.
The results of Collins et al. (2000) and Buchholz et al. (2000) represent another chapter in the remarkable story of SSRs in genome engineering. While we wait for the development of the leukemias as final proof of the applicability of this approach to disease modeling, other questions can be contemplated. Amongst these are how translocation frequencies relate to the levels of Cre expression, the relative positions of the targeted chromosomal territories and the rate of target cell proliferation.
References
- Buchholz F., Refaeli, Y., Trumpp, A. and Bishop, J.M. (2000) Inducible chromosomal translocation of AML and ETO genes through Cre-loxP-mediated recombination in the mouse. EMBO Rep., 1, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins E.C., Pannell, R., Simpson, E.M., Forster, A. and Rabbitts, T.H. (2000) Inter-chromosomal recombination of Mll and Af9 genes mediated by cre-loxP in mouse development. EMBO Rep., 1, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral J. et al. (1996) An Mll–AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell, 85, 853–861. [DOI] [PubMed] [Google Scholar]
- Dobson C.L., Warren, A.J., Pannell, R., Forster, A., Lavenir, I., Corral, J., Smith, A.J. and Rabbitts, T.H. (1999) The mll–AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J., 18, 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic K.G. (1991) Site-specific recombination between homologous chromosomes in Drosophila. Science, 252, 958–961. [DOI] [PubMed] [Google Scholar]
- Golic K.G. and Golic, M.M. (1996) Engineering the Drosophila genome: chromosome rearrangements by design. Genetics, 144, 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herault Y., Rassoulzadegan, M., Cuzin, F. and Duboule, D. (1998) Engineering chromosomes in mice through targeted meiotic recombination (TAMERE). Nature Genet., 20, 381–384. [DOI] [PubMed] [Google Scholar]
- Lamond A.I. and Earnshaw, W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Okuda T., Cai, Z., Yang, S., Lenny, N., Lyu, C.J., van Deursen, J.M., Harada, H. and Downing, J.R. (1998) Expression of a knocked-in AML1–ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood, 91, 3134–3143. [PubMed] [Google Scholar]
- Pollock J.L., Westervelt, P., Kurichety, A.K., Pelicci, P.G., Grisolano, J.L. and Ley, T.J. (1999) A bcr-3 isoform of RARα-PML potentiates the development of PML-RARα-driven acute promyelocytic leukemia. Proc. Natl Acad. Sci. USA, 96, 15103–15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R., Liu, P. and Bradley, A. (1995) Chromosome engineering in mice. Nature, 378, 720–724. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Chabanis, S., Angrand, P.O., Woodroofe, C. and Stewart, A.F. (1999) Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J., 18, 6630–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J., De Sousa, M.A., Kwabi-Addo, B., Heppell-Parton, A., Impey, H. and Rabbitts, P. (1995) A site-directed chromosomal translocation induced in embryonic stem cells by Cre-loxP recombination [published erratum appears in Nature Genet. (1996) 12, 110]. Nature Genet., 9, 376–385. [DOI] [PubMed] [Google Scholar]
- Van Deursen J., Fornerod, M., Van Rees, B. and Grosveld, G. (1995) Cre-mediated site-specific translocation between nonhomologous mouse chromosomes. Proc. Natl Acad. Sci. USA, 92, 7376–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau D.A. et al. (1997) Embryonic lethality and impairment of hematopoiesis in mice heterozygous for an AML1–ETO fusion gene. Nature Genet., 3, 303–306. [DOI] [PubMed] [Google Scholar]
- Zheng B., Sage, M., Sheppeard, E.A., Jurecic, V. and Bradley, A. (2000) Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol. Cell. Biol., 20, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D. and Cremer, T. (1998) Cell nucleus: chromosome dynamics in nuclei of living cells. Curr. Biol., 8, R321–R324. [DOI] [PubMed] [Google Scholar]


