Abstract
FAT10 is a small ubiquitin-like modifier that is encoded in the major histocompatibility complex and is synergistically inducible by tumor necrosis factor alpha and gamma interferon. It is composed of two ubiquitin-like domains and possesses a free C-terminal diglycine motif that is required for the formation of FAT10 conjugates. Here we show that unconjugated FAT10 and a FAT10 conjugate were rapidly degraded by the proteasome at a similar rate. Fusion of FAT10 to the N terminus of very long-lived proteins enhanced their degradation rate as potently as fusion with ubiquitin did. FAT10-green fluorescent protein fusion proteins were not cleaved but entirely degraded, suggesting that FAT10-specific deconjugating enzymes were not present in the analyzed cell lines. Interestingly, the prevention of ubiquitylation of FAT10 by mutation of all lysines or by expression in ubiquitylation-deficient cells did not affect FAT10 degradation. Thus, conjugation with FAT10 is an alternative and ubiquitin-independent targeting mechanism for degradation by the proteasome, which, in contrast to polyubiquitylation, is cytokine inducible and irreversible.
The ubiquitin (Ub)-proteasome pathway is the main system for the targeted degradation of intracellular proteins (37). Depending on the metabolic and functional requirement of a cell, regulatory proteins like cell cycle regulators, transcription factors, or key enzymes can be specifically selected for degradation by the 26S proteasome. The basis for selectivity does not lie in the protease itself but rather in the selective covalent modification of target proteins with Lys48-linked polyubiquitin chains. Polyubiquitylation is achieved by an enzymatic cascade of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). The specificity of substrate recognition is afforded by the numerous ubiquitin ligases which selectively bind a substrate as well as an E2 enzyme, thus facilitating the formation of isopeptide bonds between the ɛ-amino group of lysines in a substrate protein and the diglycine motif at the carboxy terminus of ubiquitin (11). Monomeric ubiquitin is processed from precursor proteins through ubiquitin-specific proteases which recognize the C-terminal diglycine motif along with the ubiquitin domain and which cleave after the diglycine motif irrespective of whether it is isopeptide linked or linked through a conventional peptide bond (38). Before degradation, the ubiquitin chains are removed from the substrate and disassembled into monomeric ubiquitin which can be reused. Ubiquitin levels are hence kept at a steady-state level, and the ubiquitin protein itself is long lived (9, 26).
A growing number of proteins which contain domains with significant homology to ubiquitin have been discovered over the past 5 years. These ubiquitin-like proteins can be assigned either to the group of ubiquitin domain proteins which contain a ubiquitin homology domain but which do not become covalently linked to target proteins or to the group of ubiquitin-like modifiers (UBLs) which become isopeptide linked to target proteins (14). Prominent members of the UBL family, such as SUMO, APG12, and ISG15, have been investigated in detail, and specific E1, E2, and/or E3 enzymes have been identified. However, none of these modifiers target proteins for proteasomal degradation.
FAT10 is a fairly new member of the UBL family. It was recognized as a ubiquitin-like protein after chromosomal sequencing of the human major histocompatibility complex class I locus (6). FAT10 is an 18-kDa protein that consists of two ubiquitin-like domains with 29% and 36% identity to ubiquitin in its N- and C-terminal parts, respectively. Unlike other UBLs, FAT10 is synthesized with a free diglycine motif at its C terminus, which implies that it can become conjugated immediately after translation and folding. FAT10 is constitutively expressed in mature dendritic cells and B cells (1, 6), but it is also inducible by the proinflammatory cytokines gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) in cells of various tissue origins (23, 28). The ectopic expression of murine FAT10 led to the induction of caspase-dependent apoptosis, which would be consistent with a role of FAT10 in the TNF-α-mediated induction of apoptosis. Evidence that FAT10 becomes covalently linked to target proteins via its C terminus was obtained in inducible FAT10 transfectants because, in addition to monomeric FAT10, a prominent band of about 35 kDa appeared upon FAT10 induction that was detected with FAT10-specific antibodies and resisted boiling in sodium dodecyl sulfate (SDS) under reducing conditions. This band was not observed when the diglycine motif of FAT10 was mutated, thus strongly suggesting that it represented a covalent FAT10 conjugate (29).
The functional consequences of FAT10 conjugation have so far not been thoroughly investigated, but it is noteworthy that the inhibition of proteasome activity lead to an accumulation of FAT10 (23, 29). Moreover, we have recently identified a noncovalent interaction partner of FAT10 named NEDD8 ultimate buster 1L (NUB-1L) that bound to the proteasome and markedly accelerated FAT10 degradation (12). In this study we show that monomeric FAT10 as well as its conjugates is rapidly degraded by the proteasome in a ubiquitin-independent manner. Interestingly, ubiquitin and FAT10 turned out to be equally efficient at targeting long-lived proteins for degradation, thus indicating that FAT10 is the first ubiquitin-like modifier which, like polyubiquitylation, functions as a protein signal for rapid degradation of substrate proteins through the proteasome.
MATERIALS AND METHODS
Cell lines, tissue culture, and transfectants.
The tetracycline-inducible mouse FAT10 transfectant TB1N has been described previously (29). TB1N cells were cultured in Iscove's modified Dulbecco's medium (Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal calf serum (Invitrogen), 100 U/ml penicillin/streptomycin (Sigma, Taufkirchen, Germany), 400 μg/ml hygromycin (Calbiochem, San Diego, CA), 5 μg/ml puromycin (Calbiochem), and 1 μg/ml tetracycline (Sigma). The epitheloid cervix carcinoma cell line HeLa and the human embryonic kidney line HEK293T were obtained from American Type Culture Collection (Manassas, VA). Stimulation with 100 U/ml recombinant human IFN-γ (Roche) and 100 U/ml recombinant human TNF-α (Roche) was performed for at least 12 h. The E1 thermosensitive cell line E36-ts20 and E36-ts20 cells retransfected with a wild-type E1 (E36-ts20/E1) were kindly provided by M. Piechaczyk and have been described previously (2, 10, 18). E36-ts20 and E36-ts20/E1 cells were cultured in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 20% fetal calf serum (Invitrogen) and 100 U/ml penicillin/streptomycin. Cells were kept at 32°C (permissive temperature for the ts20 cell line). For studies at the restrictive temperature, cells were incubated for 120 min at 42°C (to inactivate the E1 in the ts20 cells) and then switched to 39.5°C for up to 5 h. For transfection experiments, HEK293T or E36-ts20 cells were grown in 100-mm dishes to 30 to 40% confluence before transfection with 5 μg cDNA/dish, using the Fugene reagent (Roche, Mannheim, Germany) according to the manufacturer's protocol.
Chemicals and antibodies.
Lactacystin (PI-104) was purchased from Biomol (Plymouth Meeting, PA), LLnL (calpain inhibitor I) was purchased from Roche, iodacetamide and N-ethyl-maleimide was purchased from Sigma, and ubiquitin-aldehyde was purchased from Calbiochem (Darmstadt, Germany). The following antibodies were used: monoclonal anti-hemagglutinin (anti-HA), clone HA-7 (Sigma), monoclonal anti-β-galactosidase (anti-β-Gal), clone GAL-13 (Sigma), monoclonal anti-green fluorescent protein (anti-GFP) antibodies, clones 7.1 and 13.1 (Roche), monoclonal anti-His6 (clone BMG-His-1 [Roche]), polyclonal antiubiquitin, code no. Z0458 (DAKO, Glostrup, Denmark), and monoclonal anti-HA high-affinity matrix (Roche). Horseradish peroxidase coupled secondary antibodies were purchased from DAKO.
Generation of FAT10 polyclonal antibodies.
A FAT10-specific polyclonal antibody was raised in rabbits by immunization with a GST-FAT10 recombinant protein. For production of the GST-FAT10 fusion protein, the FAT10 cDNA was retrotranscribed from mRNA of JY B cells and amplified, using sense primer 5′-CCATGGATCCATGGCTCCCAATGCTTCCTGCCTC-3′ and antisense primer 5′-CCGTCTCGAGTCTCACCCTCCAATACAATAAGATGC-3′ and cloned via BamHI and XhoI sites into the expression vector pGEX-4T-3 (Amersham Biosciences). BL21 cells were transformed with this construct, and the GST-FAT10 fusion protein was induced with isopropyl-β-d-1-thiogalactopyranoside (IPTG) and purified by glutathione-Sepharose 4B column chromatography (Amersham). The GST-FAT10 fusion protein (5 mg) was used for immunization of four different rabbits. One of the raised antibodies was very specific for detection of human and mouse FAT10 in Western analysis and immunoprecipitation experiments.
Pulse-chase experiments and immunoprecipitation.
Pulse-chase experiments and immunoprecipitations were performed as previously described (12). In immunoprecipitation experiments (see Fig. 4 and 5), we included 50 mg/ml ubiquitin aldehyde, 25 mM iodoacetamide, 25 mM N-ethyl-maleimide, and 5 mM EDTA in the lysis and washing buffers in order to inhibit ubiquitin-specific proteases.
FIG. 4.
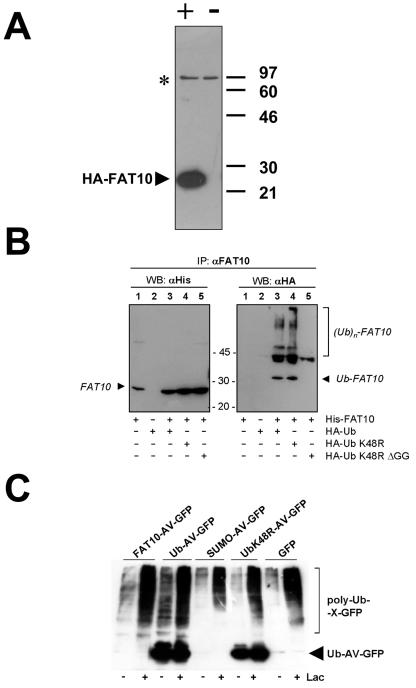
Ubiquitylation status of FAT10. (A) Characterization of a polyclonal anti-human FAT10 antibody. HEK293T cells were transiently transfected with an HA-FAT10 expression construct (+) or a vector control (−). A Western blot with a polyclonal antibody raised in rabbits against a GST-FAT10 fusion protein was then performed. The HA-FAT10 protein is labeled with an arrowhead, and an unspecific band also present in untransfected cells is indicated by an asterisk. (B) HEK293T cells were transiently transfected with His6-FAT10, HA-Ub, HA-UbK48R, or HA-UbK48R/ΔGG. Prior to lysis, cells were incubated with 100 μM of the proteasome inhibitor LLnL for 6 h. After an immunoprecipitation (IP) with the anti-FAT10 antibody characterized in panel A, the precipitates were analyzed by Western blotting (WB) with either anti-His6 (left panel) or anti-HA (right panel) antibodies. (C) HEK293T cells were transiently transfected with constructs encoding FAT10-AV-GFP, ubiquitin-AV-GFP, SUMO-AV-GFP, UbK48R-AV-GFP, or GFP. Four hours before lysis, cells were treated with 50 μM of the proteasome inhibitor lactacystin (Lac) where indicated. Lysates were immunoprecipitated with anti-GFP antibody, and immunoprecipitates were analyzed by Western blotting with an antiubiquitin antibody. The arrowhead indicates the signal for Ub-AV-GFP and UbK48-AV-GFP, and polyubiquitin conjugates of the respective proteins are labeled poly-Ub-X-GFP.
FIG. 5.
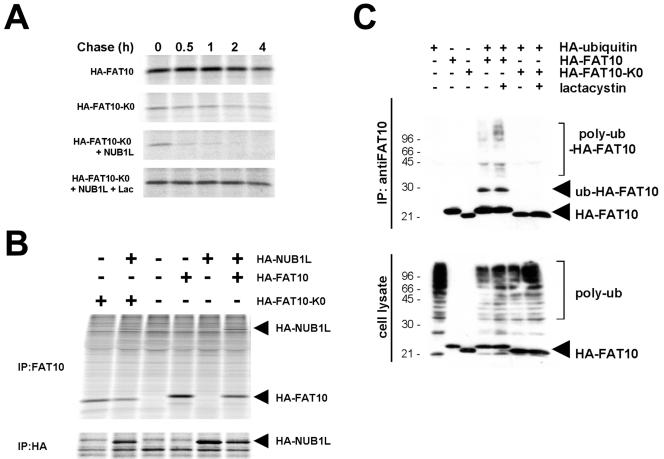
Ubiquitylation of FAT10 is not necessary for FAT10 degradation. (A) FAT10, a lysineless FAT10 mutant (FAT10-K0), and NUB1L were transiently expressed as HA-tagged proteins in HEK293T cells and treated with 50 μM of lactacystin (Lac) during the labeling and the chase where indicated. The cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to HA-specific immunoprecipitation, SDS/PAGE, and autoradiography. (B) HEK293T cells were transfected with HA-FAT10, HA-FAT10-K0, and HA-NUB1L as indicated on the top. After labeling, lysates were immunoprecipitated (IP) with anti-FAT10 or anti-HA antibodies as indicated and analyzed by SDS/PAGE and autoradiography. (C) HEK293T cells were transfected with HA-FAT10, HA-FAT10-K0, and HA-ubiquitin as indicated above. Four hours before lysis, cells were treated with 50 μM of lactacystin where indicated. As shown in the upper panel, lysates were immunoprecipitated with anti-FAT10 antibody, and immunoprecipitates were analyzed by Western blotting with anti-HA antibody. The lower panel shows an anti-HA Western blot of total lysates.
Plasmids and generation of expression constructs.
The plasmids pcDNA3.1/HA-FAT10, pcDNA3.1/His6-FAT10, pcDNA3.1/HA-Ub, pcDNA3.1/HA-UbK48R, and pcDNA3.1/HA-UbK48RΔGG have been described previously (12, 29). The vector pEGFP N1 was purchased from Clontech (Heidelberg, Germany). The generation of the vectors encoding HA-FAT10-GG-GFP, HA-FAT10-AV-GFP, HA-ubiquitin-AV-GFP, HA-Sumo-AV-GFP, and HA-NUB1long pCDNA3.1 has been described elsewhere (12). The vector containing DHFR-HA-UBK48R was a kind gift from F. Levi (21). HA-Fat10-AV-DHFR, and HA-ubiquitin-AV-DHFR were generated by PCR amplification of dihydrofolate reductase (DHFR) and replacement of the GFP gene in the corresponding vectors by the DHFR gene using conventional cloning methods. The vector expressing HA-Ubiquitin K48R was generated by replacing ubiquitin with PCR-amplified ubiquitin K48R using pcDNA3.1/HA-UbK48R as template. The mutant of FAT10 in which all lysines were replaced by arginines (HA-FAT10-K0) was generated by consecutive site-directed mutagenesis using PCR; the sequences of the 18 primers used for this purpose will be made available by us upon request. All sequences were verified by dideoxy sequencing.
RESULTS
FAT10 and its endogenous conjugates are rapidly degraded by the proteasome.
It has been shown previously that the inhibition of proteasome activity resulted in an accumulation of FAT10 suggesting that FAT10 is rapdily degraded by the proteasome (23, 29). To determine the degradation rate of FAT10, we performed a pulse-chase experiment with the inducible FAT10 transfectant TB1N. The induction of FAT10 in this cell line leads to the appearance of an additional major band at 35 kDa which we have previously shown to be a covalent FAT10 conjugate (29). FAT10 was induced for 24 h by tetracycline removal, and the cells were then labeled for 1 hour and chased for the indicated time periods (Fig. 1 A). Quantification of the FAT10 monomer band on a radioimager (Fig. 1B) demonstrated that 50% of FAT10 was degraded within 0.9 h and that degradation was strongly attenuated when the cells were chased in the presence of 80 μM of the proteasome inhibitor lactacystin (Fig. 1C and D). Interestingly, the 35-kDa FAT10 conjugate which was apparent in the induced but not in the uninduced cells, had a similar degradation rate as FAT10 itself and was similarly stabilized by lactacystin treatment. Since our quantifications provided no evidence for the release of FAT10 from the conjugate over time, the data strongly suggest that both FAT10 and its target protein were degraded by the proteasome.
FIG. 1.
FAT10 and a FAT10 conjugate are rapidly degraded in a proteasome-dependent manner. The tetracycline-inducible FAT10 transfectant TB1N was labeled with [35S]Met-Cys and chased for the indicated time periods in the absence (A) or presence (C) of the proteasome inhibitor lactacystin (80 μM) followed by immunoprecipitation against HA-FAT10. Lanes 1 in panels A and C represent noninduced TB1N cells (n.i.). The bands shown in panels A and C were quantified on a radioimager and plotted in panels B and D, respectively, as percent radioactivity based on values obtained after the pulse.
FAT10 and ubiquitin are equally potent at reducing the half-lives of long-lived proteins when fused to their N termini.
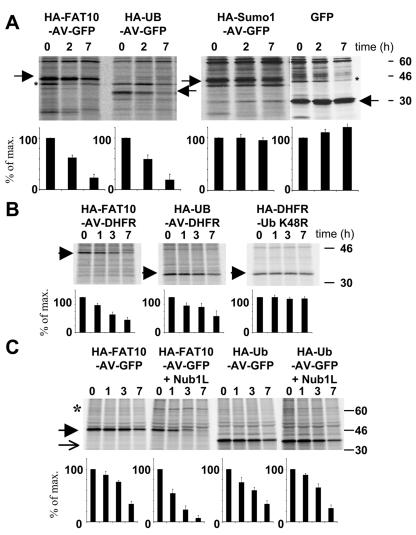
Since FAT10 and its conjugate were degraded by the proteasome at similar rates, we decided to test whether FAT10 may function as a degradation signal. The N-terminal fusion of ubiquitin to long-lived proteins is known to markedly reduce their half-lives (15). This requires, however, that the two C-terminal glycine residues be replaced by different amino acids, because otherwise ubiquitin-specific proteases will rapidly cleave off the ubiquitin moiety. Hence, we created expression constructs for fusion proteins between the long-lived GFP at the C terminus and either HA-tagged FAT10, ubiquitin, or SUMO-1 at the N terminus. In these constructs the diglycine motif was replaced with Ala and Val, and for SUMO-1 the C-terminal extension was removed. The constructs were transiently transfected into HeLa cells, and the degradation of the three fusion proteins as well as GFP alone was monitored in pulse-chase experiments (Fig. 2A, top panel). As expected, GFP was not degraded within 7 h, and also the HA-SUMO-AV-GFP fusion protein, which served as our negative control, remained stable. Interestingly, quantification of the bands with a radioimager revealed that FAT10 and ubiquitin were equally potent in targeting GFP for fast degradation (Fig. 2A, bottom panel). This demonstrates that FAT10, like ubiquitin, but in contrast to SUMO-1 and other UBLs, functions as a degradation signal.
FIG. 2.
FAT10 and ubiquitin are equally potent in targeting for degradation. (A) FAT10, SUMO, and ubiquitin were transiently expressed as uncleavable AV/GG mutated and HA-tagged GFP fusion proteins in HeLa cells. The cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to HA-specific immunoprecipitation, SDS-polyacrylamide gel electrophoresis (PAGE), autoradiography, and quantification on a radioimager. As a control, GFP was expressed and immunoprecipitated with an anti-GFP antibody. The arrowheads denote the different GFP fusion proteins and GFP as indicated above; an asterisk indicates an unspecific band. (B) Fusion proteins of DHFR with HA-tagged FAT10 and ubiquitin as well as a DHFR-HA-UbK48R control were expressed in HeLa cells, and a pulse-chase experiment was performed as shown for panel A. The arrowheads denote the respective DHFR fusion proteins as indicated above the panels. (C) HA-FAT10-GFP and HA-ubiquitin-GFP were transiently expressed in HeLa cells without or together with HA-NUB1L. A pulse-chase analysis was performed as shown for panels A and B. An arrow denotes HA-Ub-AV-GFP, and an arrowhead denotes HA-FAT10-AV-GFP. All bands were quantified on a radioimager and plotted below each lane as percent radioactivity based on values obtained after the pulse. The data represent means of results from three independent experiments ± standard errors of the means.
Subsequently, we tested whether this property of FAT10 is also observed with another long-lived protein and chose DHFR for this purpose. We generated HA-FAT10-AV-DHFR expression constructs and compared the half-life of the encoded fusion protein to that of HA-Ub-AV-DHFR as a positive control. As a negative control we used a DHFR-HA-UbK48R construct because it has been shown that the attachment of a single ubiquitin to the C terminus of DHFR does not lead to accelerated degradation (21, 36). A quantitative evaluation of the experiment shown in Fig. 2B and two additional experiments indicated that the N-terminal fusion of FAT10 and ubiquitin similarly reduced the half-life of DHFR.
NUB1L targets a FAT10-AV-GFP fusion protein but not Ubiquitin-AV-GFP for accelerated degradation.
Previously, we have shown that the coexpression of NUB1L caused a marked acceleration of FAT10 degradation (12). In order to examine whether fusion proteins containing FAT10 were likewise degraded in an accelerated manner, we performed a pulse-chase experiment similar to that described above, but this time HeLa cells were transiently transfected with a FAT10-AV-GFP plasmid either alone or together with an expression construct for HA-NUB1L (12). Interestingly, the coexpression of NUB1L markedly accelerated the degradation of FAT10-AV-GFP as revealed by the quantitative analysis of three independent experiments (Fig. 2C). Moreover, a similar NUB1L-dependent acceleration of FAT10-AV-GFP degradation was observed when the same transfection experiments were performed with HEK293T cells (data not shown). Evidently, NUB1L also accelerated the degradation of FAT10 when it was covalently linked to another protein. This function appeared to be FAT10 specific, as NUB1L coexpression had virtually no effect on the proteolysis of a Ub-AV-GFP fusion protein (Fig. 2C).
Lack of evidence for a FAT10-specific protease.
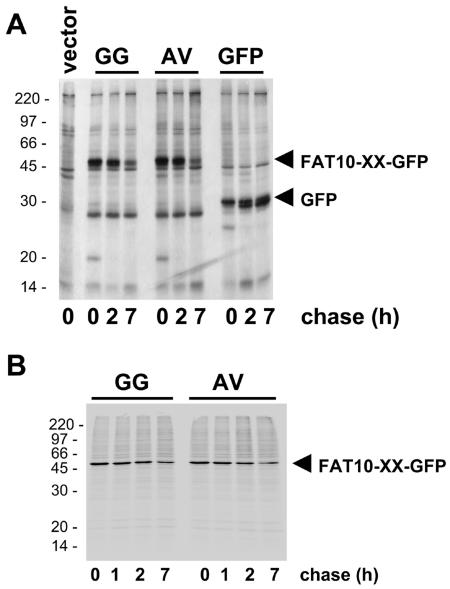
In order to investigate whether a FAT10-specific protease exists, we compared two FAT10-GFP fusion proteins, one with the regular diglycine C terminus of FAT10 and another one where the diglycine motif was replaced by Ala and Val. This mutation can abolish the cleavage at the C termini of most other UBLs and ubiquitin itself (38). HeLa cells were transiently transfected with GFP, and the two FAT10-GFP fusion proteins and the fate of these proteins were analyzed in pulse-chase experiments (Fig. 3A). Both FAT10-GFP fusion proteins were degraded with the same kinetics, and neither GFP nor FAT10 was released from the HA-FAT10-GG-GFP fusion protein. Moreover, repetition of the same experiments in IFN-γ-treated HeLa cells (data not shown) or in HEK293T cells (Fig. 3B) also showed degradation but no processing of either HA-FAT10-GG-GFP or HA-FAT10-AV-GFP. This result indicates that a FAT10-specific processing protease which ought to remove FAT10 from isopeptide-linked or N-terminally fused target proteins was not active in either HeLa or HEK293T cells.
FIG. 3.
Lack of evidence for a FAT10-specific processing protease. GFP and FAT10-GFP fusion proteins with either wild-type FAT10 or with a GG-to-AV mutation of the FAT10 C terminus were transiently expressed in (A) HeLa or (B) HEK293T cells. The cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to GFP-specific immunoprecipitation, SDS/PAGE, and autoradiography. The leftmost lane in panel A shows a vector control.
Determination of the ubiquitylation status of FAT10.
Next we investigated whether targeting to the proteasome is mediated by the two ubiquitin-like domains of FAT10 itself or whether FAT10 needs to be polyubiquitylated for degradation by the 26S proteasome. For these and other experiments we generated a polyclonal antibody by immunizing rabbits with a GST-FAT10 fusion protein. This antibody prominently detected HA-FAT10 in immunoblots of HEK293T cells that were transiently transfected with an HA-FAT10 expression construct but not in untransfected cells (Fig. 4A). To determine the ubiquitylation status of FAT10, we transiently expressed human His6-FAT10 in HEK293T cells in the absence or presence of HA-tagged ubiquitin in its wild-type form or in a K48R mutant or a K48R/ΔGG double-mutant form. After treatment with the proteasome inhibitor LLnL, FAT10 was immunoprecipitated, and the amount of FAT10 was determined by anti-His6 Western blotting. In addition, ubiquitin-FAT10 conjugates were visualized by anti-HA Western blotting (Fig. 4B). From the anti-His6 Western blot it was apparent that roughly the same amount of FAT10 was immunoprecipitated in all transfection experiments. The anti-HA Western blot revealed that bands that correspond to one, two, and three ubiquitin molecules linked to FAT10 became visible, but high-molecular-weight polyubiquitin conjugates were not very prominent. Since none of the ubiquitin conjugates were visible when HA-UbK48R/ΔGG was coexpressed, we can be confident that the apparent bands do not represent ubiquitin molecules that have themselves been modified by FAT10 conjugation.
The low level of polyubiquitylated FAT10 shown in Fig. 4A and B cast some doubt on whether FAT10 needs to be polyubiquitylated for proteasomal degradation. Moreover, when we examined the degree of polyubiquitylation of GFP fusion proteins immunoprecipitated from HEK293T cells transiently transfected with expression constructs for FAT10-AV-GFP, Ub-AV-GFP, SUMO-AV-GFP, UbK48R-AV-GFP, and GFP (Fig. 4C), we noted that the degrees of polyubiquitylation of the respective fusion proteins did not correlate with their rates of degradation (Fig. 2A). Hence, we decided to investigate whether the degradation of FAT10 could be ubiquitin independent.
Ubiquitylation of FAT10 is not necessary for FAT10 degradation.
An expression construct in which all 17 lysines of human FAT10 were replaced by arginines was generated (designated HA-FAT10-K0). HA-FAT10 and HA-FAT10-KO were transiently expressed in HEK293T cells, and their half-lives were determined in a pulse-chase experiment (Fig. 5A). A quantitative evaluation of the results revealed that the degradation rates of HA-FAT10 and HA-FAT10-K0 were identical. Moreover, the coexpression of NUB1L accelerated HA-FAT10-K0 degradation to the same extent as we had shown before for wild-type FAT10 (12) and the FAT10-AV-GFP fusion protein (Fig. 2C). The degradation of HA-FAT10-K0 also appeared to be proteasome dependent, as the protein could be stabilized with the proteasome inhibitor lactacystin even in the presence of NUB1L (Fig. 5A, bottom).
A concern with these experiments could be that HA-FAT10-K0 may not be folded properly. Several observations argue against this possibility. First, we could efficiently immunoprecipitate HA-FAT10-K0 with our polyclonal anti-FAT10 antibody from transiently transfected HEK293T cells (Fig. 5B, first lane from the left). Second, NUB1L was as efficiently coimmunoprecipitated with HA-FAT10 as with HA-FAT10-K0 from transiently transfected HEK293T cells (Fig. 5B, second and sixth lanes from the left). Third, NUB1L accelerated HA-FAT10-K0 degradation as efficiently as that of HA-FAT10 (Fig. 5A) (12). It is hence unlikely that the rapid degradation of HA-FAT10-K0 is caused by misfolding.
In general, lysine residues are required for the conjugation of substrate proteins with ubiquitin. In some proteins, however, the amino termini were found to be ubiquitylated (4). To test whether the HA-FAT10-K0 protein can be ubiquitylated, we expressed HA-FAT10 or HA-FAT10-K0 alone or together with HA-ubiquitin in transiently transfected HEK293T cells. FAT10 was immunoprecipitated with an anti-FAT10 antibody, and the immunoprecipitates were analyzed by immunoblotting with an anti-HA antibody. Although similar amounts of the FAT10 proteins were expressed, and although polyubiquitin conjugates were readily detected in the lysates of the transfected cells (Fig. 5C, bottom), we detected mono- and polyubiquitylation only for HA-FAT10 but not for HA-FAT10-K0 even after prolonged exposure (Fig. 5C, top). This indicates that ubiquitin conjugation of FAT10 is not required for its proteasome-dependent degradation.
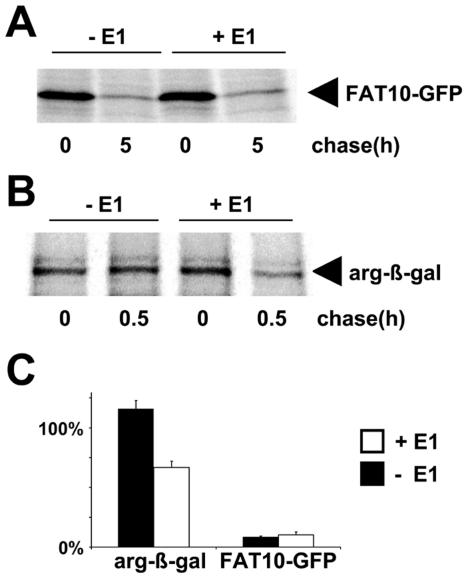
FAT10 degradation occurs normally in E1 temperature-sensitive mutants.
A prerequisite for ubiquitin conjugation is the ATP-dependent activation of the carboxy terminus of ubiquitin by the ubiquitin-activating enzyme (E1). An extensively characterized mutant cell line which expresses a temperature-sensitive form of E1 is the Chinese hamster ovary line E36-ts20 (18). We used this mutant cell line and E36-ts20/E1 cells that are reconstituted with a transfected wild-type E1 enzyme to investigate whether the degradation of FAT10 or FAT10-GFP fusion proteins is in fact independent of ubiquitin conjugation to FAT10. For this purpose, E36-ts20 and E36-ts20/E1 cells were transiently transfected with a FAT10-AV-GFP expression construct, and a pulse-chase experiment was performed at the restrictive temperature (Fig. 6A) and quantitatively evaluated (Fig. 6C). Interestingly, the FAT10-AV-GFP protein was degraded at the same rate in both cell types, indicating that FAT10-AV-GFP catabolism occurred even when ubiquitin conjugation was defective (10). To confirm that the E36-ts20 line was defective in ubiquitylation, we transfected E36-ts20 and E36-ts20/E1 cells with an HA-DHFR-ubiquitin-Arg-β-galactosidase expression plasmid. This construct encodes a fusion protein which is cleaved by ubiquitin-specific proteases into a stable N-terminal HA-DHFR-ubiquitin part that was used as a transfection control (data not shown) and a C-terminal Arg-β-galactosidase (Arg-β-Gal) part which is an N-end rule substrate and therefore rapidly degraded in a ubiquitin-dependent manner (21). As shown in Fig. 6B and C, the Arg-β-Gal protein was stable in the E36-ts20 mutant while it was degraded in the E36-ts20/E1 transfectant. Taken together, these results agree with the data obtained with the lysine-deficient FAT10 mutant (Fig. 5) in that they establish FAT10 as a ubiquitin-like modifier that can target proteins for degradation by the proteasome without the need for polyubiquitylation.
FIG. 6.
Degradation of FAT10 is ubiquitin independent. (A) The E1 thermosensitive cell line E36-ts20 (−E1) and E36-ts20 cells retransfected with a wild-type E1 (+E1) were transiently transfected with HA-tagged FAT10-AV-GFP. After inactivation of the thermolabile E1 by temperature shifting, the cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to HA-specific immunoprecipitation, SDS/PAGE, and autoradiography. (B) Similar experiments were performed transfecting a short-lived HA-DHFR-ubiquitin-Arg-β-Galactosidase construct. The immunoprecipitation was performed with an anti-β-Gal antibody. (C) The bands were quantified on a radioimager and plotted as percent radioactivity based on values obtained directly after the pulse. The means of results from three independent experiments are shown, and error bars indicate the standard errors of the means.
DISCUSSION
When UBLs are described it is usually emphasized that they form isopeptide linkages with target proteins just like ubiquitin but that they do not serve proteolytic functions. In this work we show that FAT10 is the first UBL which, like ubiquitin, targets conjugated proteins for degradation through the proteasome. Similar to ubiquitin, FAT10 served as a degradation signal when it was fused to the N termini of two long-lived proteins, and it is remarkable that FAT10 was as potent as ubiquitin in accelerating their proteolysis (Fig. 2A). Since proteasomal targeting by FAT10 did not rely on the ubiquitin conjugation system, modification with a single FAT10 molecule appears to be an independent alternative to polyubiquitylation.
When we initially observed that FAT10 can act as a transferable degradation tag, two scenarios seemed possible: firstly, FAT10 could, like other degrons, bind to an E3 ubiquitin ligase and by that means initiate the assembly of polyubiquitin chains, and, secondly, FAT10 itself may act like polyubiquitin in functioning as a proteasome-targeting signal. First we addressed the question of whether FAT10 is ubiquitylated in cells, and we observed that FAT10 was primarily modified with only one to three copies of ubiquitin that do not mediate efficient targeting to the 26S proteasome (35) (Fig. 4B). This was an incentive for us to further investigate the ubiquitin dependence of FAT10 degradation.
Subsequent experiments revealed that FAT10 is rapidly degraded by the proteasome without requiring the ubiquitylation system in general and polyubiquitylation of FAT10 itself in particular. In Fig. 6, the degradation rate of FAT10-AV-GFP in E36-ts20 cells that express temperature-sensitive ubiquitin-activating enzyme E1 is compared with the degradation rate in the reconstituted E1 transfectant E36-ts20/E1. Consistently, we observed no differences in FAT10-AV-GFP degradation between E36-ts20 and E36-ts20/E1 cells. It should be emphasized though, that ubiquitin conjugation in E36-ts20 is largely deficient at the restrictive temperature but that some 20% of polyubiquitin conjugate formation remains that, nevertheless, did not suffice to maintain viability or to degrade the bulk of short-lived proteins (8, 10, 18). Since FAT10-AV-GFP degradation occurred at a normal pace in these mutants, it is fair to conclude that FAT10 degradation does not rely on polyubiquitylation to an extent similar to that of other short-lived proteins, like p53 (3) or FOS (32). We cannot, however, rule out the unlikely scenario that the 20% polyubiquitylation capacity remaining at the restrictive temperature contributed to FAT10 degradation.
To further investigate this issue, we generated a FAT10 mutant in which all 17 lysines were replaced by arginines. This mutant protein, designated HA-FAT10-K0, bound normally to anti-FAT10 polyclonal antibodies and to the FAT10-interacting protein NUB1L (Fig. 5B), indicating that HA-FAT10-K0 was properly folded. Since HA-FAT10-K0 was degraded at the same rate as HA-FAT10, in both the presence and the absence of NUB1L, we conclude that isopeptide linkage to ubiquitin at lysine residues was not required for HA-FAT10-K0 degradation. In a few cases, however, the α-amino group at the N terminus of proteins was shown to be linked to the C terminus of ubiquitin by formation of a conventional peptide bond (4). Lysineless variants of proteins have been shown to be polyubiquitylated at the N terminus and degraded, although in some of these proteins the degradation was significantly slowed down when all lysines were mutated. We found that wild-type HA-FAT10 was ubiquitylated, but no ubiquitylation was detected for the lysineless variant HA-FAT10-K0 either in the absence or in the presence of proteasome inhibitors (Fig. 5C). One could argue that a very minor portion of HA-FAT10-K0 that escaped our detection was ubiquitylated, but if this was the case, such trace amounts are very unlikely to mediate HA-FAT10-K0 degradation at the same efficiency as lysine-proficient HA-FAT10. Moreover, the HA-FAT10-K0 protein bears an HA tag at the N terminus which most likely is acetylated since it bears an alanine at position 2 that, after cleavage of the N-terminal methionine by methionine aminopeptidases, is predicted to become acetylated by the N-terminal acetyltransferase type A (27). N-terminal acetylation interferes with N-terminal ubiquitylation and hence is another strong argument against N-terminal ubiquitylation of HA-FAT10-K0. Taken together, the data shown in Fig. 5 and 6 indicate that FAT10 and FAT10-conjugated proteins can be efficiently degraded in a ubiquitin-independent manner.
These results raise the question of how FAT10 can mediate degradation by the proteasome. Ubiquitin-independent degradation by the proteasome has been described previously for ornithine decarboxylase (25), which is targeted to the 26S proteasome by a polyamine-induced protein called antizyme 1 (39). Also, the cell cycle inhibitor p21 appears to be degraded by the proteasome without the need for ubiquitylation (31), as unmodified p21 can be efficiently degraded by the 20S and 26S proteasomes in vitro (22).
Ubiquitin-like domains can mediate binding to the 26S proteasome as has been demonstrated for the ubiquitin domain proteins Rad23, Dsk2, and BAG-1 (5, 7, 24, 30). Rad23 and Dsk2 may act as adaptor proteins in that they bind to the Rpn1 subunit of the 26S proteasome through their ubiquitin-like domain and to polyubiquitin chains through their ubiquitin-associated domains. In this respect it is intriguing that we recently identified NUB1L as a noncovalent interaction partner of FAT10 (12). NUB1L contains three ubiquitin-associated domains in its C-terminal part and a ubiquitin-like domain at the N terminus (12, 34). In addition, NUB1L was found to be associated with the 26S proteasome and to bind to the 19S regulator subunit Rpn10 in vitro (16). Given that NUB1L, which is also IFN-γ inducible (17), markedly accelerated the degradation of FAT10 (12) and FAT10 fusion proteins (Fig. 2C), it is attractive to hypothesize that NUB1L functions as an adaptor that ties FAT10 and FAT10-conjugated proteins to the proteasome. A recent study demonstrated that proteins that were targeted to the proteasome with an artificial tagging system were rapidly degraded by the proteasome (13). Proximity to the proteasome may therefore be sufficient for degradation, and polyubiquitin chains may not be the only signal for proteasomal degradation.
A striking difference between ubiquitin and FAT10 is in their rates of turnover. Although investigations on the half-life of ubiquitin yielded quite different results ranging from 320 h (26) to 28 h (9), it is clear that ubiquitin is much more stable than FAT10, which was degraded by 50% within approximately 1 hour in human and murine cells. Since FAT10 conjugates are degraded at the same speed as monomeric FAT10 and since both are similarly stabilized through lactacystin, we consider it very likely that FAT10 is being degraded along with its substrate. This notion is supported by the observation that monomeric FAT10 did not accumulate during our pulse-chase experiments (Fig. 1A), thus indicating that FAT10 was not liberated from FAT10 conjugates. Liberation of FAT10 from its conjugates would require the existence of FAT10-specific proteases. Using FAT10-GFP fusion proteins, we have examined this possibility, but no evidence for cleavage of the fusion protein could be obtained with HeLa cells (Fig. 3A) or HEK293T cells (Fig. 3B). HeLa cells express FAT10 upon induction with IFN-γ (28), but even in the presence of IFN-γ we failed to obtain any evidence for a FAT10-specific protease (data not shown). We are aware that our experiments cannot rule out the expression of a FAT10-specific protease in cells which we have not yet tested, but given that FAT10 is expressed in HeLa cells, we find this possibility unlikely. A lack of FAT10-specific processing proteases would be in striking contrast to ubiquitin, NEDD8, SUMO-1, or ISG15, for which specific processing proteases have been described. This feature would, however, be in accordance with another unique trait of FAT10, i.e., the direct biosynthesis with a free diglycine motif at the C terminus which does not need further processing before conjugation. Interestingly, in mutants that lack the proteasome-associated deubiquitinating enzymes Ubp6 and Doa4, the half-life of ubiquitin is dramatically shortened; this finding was attributed to a failure to remove ubiquitin from polyubiquitylated substrates before they are degraded by the 26S proteasome (20, 33). In these mutants ubiquitin appears to have roughly the same low persistence and fate as FAT10.
The rapid proteolysis of FAT10 conjugates may also explain why it has been difficult to detect these conjugates. Liu et al. reported that using a FAT10-specific antibody, they did not detect proteins that could represent FAT10 conjugates (23). The first time we could detect a FAT10 conjugate with the molecular mass of about 35 kDa was after the tetracycline induced overexpression of HA-FAT10 in mouse fibroblasts (29). Recently, Lee et al. published a study showing that FAT10 expression was markedly upregulated in 90% of hepatocellular carcinoma tissue as well as several tumors of the gastrointestinal tract and female reproductive organs (19). Interestingly, the anti-FAT10 antibody generated in that study also detected a 35-kDa band in addition to monomeric FAT10 in NIH 3T3 fibroblasts. It will now be crucial to identify FAT10-conjugated target proteins in order to learn more about the biological functions of FAT10 and to understand why these proteins can be rapidly and irreversibly targeted for proteasomal degradation without the involvement of the ubiquitin system.
Acknowledgments
We thank E. Naidoo for excellent technical support and C. M. Pickart for valuable advice. We acknowledge M. Piechaczyk for contributing ts20 cells, F. Levy for DHFR constructs, and M. Basler for HA-ubiquitin plasmids.
We declare that we have no financial conflict of interest. This work was funded by the Swiss National Science Foundation (grant 31-63387.00) in its initial phase and by the German Research Foundation (grants GR 1517/-2-1 and GR 1517/3-1) in its later phase.
REFERENCES
- 1.Bates, E. F. M., O. Ravel, M. C. Dieu, S. Ho, C. Guret, J. M. Bridon, S. Ait-Yahia, F. Briere, C. Caux, J. Banchereau, and S. Lebecque. 1997. Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells. Eur. J. Immunol. 27:2471-2477. [DOI] [PubMed] [Google Scholar]
- 2.Bossis, G., P. Ferrara, C. Acquaviva, I. Jariel-Encontre, and M. Piechaczyk. 2003. c-Fos proto-oncoprotein is degraded by the proteasome independently of its own ubiquitinylation in vivo. Mol. Cell. Biol. 23:7425-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdary, D. R., J. J. Dermody, K. K. Jha, and H. L. Ozer. 1994. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciechanover, A., and R. Ben-Saadon. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14:103-106. [DOI] [PubMed] [Google Scholar]
- 5.Elsasser, S., R. R. Gali, M. Schwickart, C. N. Larsen, D. S. Leggett, B. Muller, M. T. Feng, F. Tubing, G. A. G. Dittmar, and D. Finley. 2002. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4:725-730. [DOI] [PubMed] [Google Scholar]
- 6.Fan, W., W. Cai, S. Parimoo, G. G. Lennon, and S. M. Weissman. 1996. Identification of seven new human MHC class I region genes around the HLA-F locus. Immunogenetics 44:97-103. [DOI] [PubMed] [Google Scholar]
- 7.Funakoshi, M., T. Sasaki, T. Nishimoto, and H. Kobayashi. 2002. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 99:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gropper, R., R. A. Brandt, S. Elias, C. F. Bean, A. Mayer, A. L. Schwartz, and A. Ciechanover. 1991. The ubiquitin-activating enzyme, E1, is required for stress induced lysosomal degradation of cellular proteins. J. Biol. Chem. 266:3602-3610. [PubMed] [Google Scholar]
- 9.Haas, A. L., and P. M. Bright. 1987. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J. Biol. Chem. 262:345-351. [PubMed] [Google Scholar]
- 10.Handley-Gearhart, P. M., J. S. Trausch-Azar, A. Ciechanover, and A. L. Schwartz. 1994. Rescue of the complex temperature-sensitive phenotype of Chinese hamster ovary E36ts20 cells by expression of the human ubiquitin-activating enzyme cDNA. Biochem. J. 304:1015-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 12.Hipp, M. S., S. Raasi, M. Groettrup, and G. Schmidtke. 2004. NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation. J. Biol. Chem. 279:16503-16510. [DOI] [PubMed] [Google Scholar]
- 13.Janse, D. M., B. Crosas, D. Finley, and G. M. Church. 2004. Localization to the proteasome is sufficient for degradation. J. Biol. Chem. 279:21415-21420. [DOI] [PubMed] [Google Scholar]
- 14.Jentsch, S., and G. Pyrowolakis. 2000. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10:335-342. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, E. S., B. Bartel, W. Seufert, and A. Varshavsky. 1992. Ubiquitin as a degradation signal. EMBO J. 11:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamitani, T., K. Kito, T. Fukuda-Kamitani, and E. T. H. Yeh. 2001. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem. 276:46655-46660. [DOI] [PubMed] [Google Scholar]
- 17.Kito, K., E. T. H. Yeh, and T. Kamitani. 2001. NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J. Biol. Chem. 276:20603-20609. [DOI] [PubMed] [Google Scholar]
- 18.Kulka, R. G., B. Raboy, R. Schuster, H. A. Parag, G. Diamond, A. Ciechanover, and M. Marcus. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 263:15726-15731. [PubMed] [Google Scholar]
- 19.Lee, C. G., J. Ren, I. S. Cheong, K. H. Ban, L. L. Ooi, S. Yong Tan, A. Kan, I. Nuchprayoon, R. Jin, K. H. Lee, M. Choti, and L. A. Lee. 2003. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene 22:2592-2603. [DOI] [PubMed] [Google Scholar]
- 20.Leggett, D. S., J. Hanna, A. Borodovsky, B. Crosas, M. Schmidt, R. T. Baker, T. Walz, H. Ploegh, and D. Finley. 2002. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10:495-507. [DOI] [PubMed] [Google Scholar]
- 21.Levy, F., N. Johnsson, T. Rümenapf, and A. Varshavsky. 1996. Using ubiquitin to follow the metabolic fate of a protein. Proc. Natl. Acad. Sci. USA 93:4907-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, C. W., M. J. Corboy, G. N. DeMartino, and P. J. Thomas. 2003. Endoproteolytic activity of the proteasome. Science 299:408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Y., J. Pan, C. Zhang, W. Fan, M. Collinge, J. R. Bender, and S. M. Weissman. 1999. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc. Natl. Acad. Sci. USA 96:4313-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luders, J., J. Demand, and J. Hohfeld. 2000. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275:4613-4617. [DOI] [PubMed] [Google Scholar]
- 25.Murakami, Y., S. Matsufuji, T. Kameji, S. Hayashi, K. Igarashi, T. Tamura, K. Tanaka, and A. Ichihara. 1992. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360:597-599. [DOI] [PubMed] [Google Scholar]
- 26.Neff, N. T., L. Bourret, P. Miao, and J. F. Dice. 1981. Degradation of proteins microinjected into IMR-90 human diploid fibroblasts. J. Cell Biol. 91:184-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polevoda, B., and F. Sherman. 2003. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325:595-622. [DOI] [PubMed] [Google Scholar]
- 28.Raasi, S., G. Schmidtke, R. D. Giuli, and M. Groettrup. 1999. A ubiquitin-like protein which is synergistically inducible by interferon-γ and tumor necrosis factor-α. Eur. J. Immunol. 29:4030-4036. [DOI] [PubMed] [Google Scholar]
- 29.Raasi, S., G. Schmidtke, and M. Groettrup. 2001. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J. Biol. Chem. 276:35334-35343. [DOI] [PubMed] [Google Scholar]
- 30.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, D. Lambertson, W. Potts, and K. Madura. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715-718. [DOI] [PubMed] [Google Scholar]
- 31.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5:403-410. [DOI] [PubMed] [Google Scholar]
- 32.Stancovski, I., H. Gonen, A. Orian, A. L. Schwartz, and A. Ciechanover. 1995. Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol. Cell. Biol. 15:7106-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swaminathan, S., A. Y. Amerik, and M. Hochstrasser. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10:2583-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka, T., H. Kawashima, E. T. H. Yeh, and T. Kamitani. 2003. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J. Biol. Chem. 278:32905-32913. [DOI] [PubMed] [Google Scholar]
- 35.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner, G. C., and A. Varshavsky. 2000. Detecting and measuring cotranslational protein degradation in vivo. Science 289:2117-2120. [DOI] [PubMed] [Google Scholar]
- 37.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson, K. D. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141-148. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, M. S., C. M. Pickart, and P. Coffino. 2003. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 22:1488-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]