Abstract
Chromium(VI) is a toxic and carcinogenic metal that causes the formation of DNA phosphate-based adducts. Cr-DNA adducts are genotoxic in human cells, although they do not block replication in vitro. Here, we report that induction of cytotoxicity in Cr(VI)-treated human colon cells and mouse embryonic fibroblasts requires the presence of all major mismatch repair (MMR) proteins. Cr-DNA adducts lost their ability to block replication of Cr-modified plasmids in human colon cells lacking MLH1 protein. The presence of functional mismatch repair caused induction of p53-independent apoptosis associated with activation of caspases 2 and 7. Processing of Cr-DNA damage by mismatch repair resulted in the extensive formation of γ-H2AX foci in G2 phase, indicating generation of double-stranded breaks as secondary toxic lesions. Induction of γ-H2AX foci was observed at 6 to 12 h postexposure, which was followed by activation of apoptosis in the absence of significant G2 arrest. Our results demonstrate that mismatch repair system triggers toxic responses to Cr-DNA backbone modifications through stress mechanisms that are significantly different from those for other forms of DNA damage. Selection for Cr(VI) resistant, MMR-deficient cells may explain the very high frequency of lung cancers with microsatellite instability among chromate workers.
Mismatch repair (MMR) is the major mechanism for correction of replication errors in all organisms. MMR recognizes and repairs single-base mispairs, as well as small insertions or deletion loops (26, 29). MMR proteins are also involved in control of fidelity of homologous recombination. Inactivation of MMR leads to an increase in the frequency of homeologous recombination that involves imperfectly matched DNA sequences (28). Mismatch recognition in human cells is largely performed by a heterodimeric MutSα complex composed of MSH2 and MSH6 proteins (1, 15). This complex recognizes and binds single-base mismatches and loops of one to three nucleotides (15). A less-abundant MutSβ heterodimer, which is composed of MSH2 and MSH3 proteins, primarily corrects DNA loops containing two or more bases (1, 15). Following the recognition step, both MutS dimers recruit another heterodimeric complex, MutLα, which is composed of MLH1 and PMS2. The formation of MutS-MutL complex activates the excision of up to 1 kb of the newly synthesized DNA strand containing the mismatch (26, 28, 29). The single-stranded gap is filled in by a DNA polymerase, and DNA ligase restores the integrity of the DNA strand. Germ line mutations in MMR proteins have been identified as the cause of the majority of cases of hereditary nonpolyposis colorectal cancer (HNPCC) (53, 60). Inactivation of MSH2 and MLH1 genes is the most frequent genetic change in HNPCC patients. HNPCC tumors exhibit highly elevated rates of mutagenesis in simple repetitive sequences (microsatellite instability) (26, 28, 29). Several other forms of cancer can also exhibit microsatellite instability.
In addition to their functions in repair of mismatches, MMR proteins also play an important role in cellular responses to some forms of DNA damage, such as those induced by Sn1-methylating agents N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and N-methyl-N-nitrosourea (MNU) (3, 5, 20, 49) and by a commonly used chemotherapeutic drug, cisplatin (2, 33, 39). Cells deficient in the main MMR proteins are highly resistant to toxic effects of O6-methylguanine, which is the principal cytotoxic lesion in MNNG- and MNU-treated cells (19, 38). An increase in the resistance of MMR-deficient cells to cisplatin is more modest (39). A full complement of MMR proteins is required for both cell cycle arrest and induction of apoptosis by DNA-methylating damage and cisplatin (9, 33, 59). The specific mechanisms by which MMR proteins activate cytotoxic responses have not yet been identified. One hypothesis, the futile repair model, suggests that MMR proteins recognize misincorporated nucleotide or damaged base sites as mismatches and attempts to repair them (27, 49). During this futile repair, DNA breaks and gaps are continuously produced, and it is these secondary lesions that are responsible for the activation of cell cycle arrest and apoptosis. A second theory suggests that the MMR proteins can directly initiate stress signaling, leading to the induction of toxic responses (14). Support for this model was initially provided by the discovery that MSH2, MLH1, and MSH6 are components of the BRCA1-associated genome surveillance complex damage-signaling complex (57). Recent reports of the isolation of MSH2 and MSH6 mutants displaying normal sensitivity to MNNG and cisplatin but unable to correct DNA mismatches strongly indicated that repair and activation of toxicity are two separate functions of MMR proteins (12, 33, 61). In addition to its role in stress responses to DNA damage, the MMR system has also been found to be responsible for a high mutagenic activity of the carcinogenic metal cadmium in yeast (10, 25). Cadmium strongly inhibited both MutSα- and MutSβ-dependent repair activities causing extreme hypermutability, due to the inability of yeast cells to repair replication errors. Suppression of MMR activity resulted from the ability of cadmium to interfere with binding of MutS complexes to mismatched DNA and to inhibit ATP hydrolysis by Msh6 (10).
Hexavalent chromium [Cr(VI)] is a recognized human carcinogen causing exposure in several dozens of professional groups and major public health concerns, due to frequent environmental contamination (32). Intracellular reduction of Cr(VI) to Cr(III) leads to the extensive formation of Cr-DNA adducts. The major form of Cr-DNA adducts in mammalian cells are Cr(III)-mediated DNA cross-links with glutathione, cysteine, or ascorbate (42, 65). The formation of these ternary adducts proceeds through the attachment of binary Cr(III)-ligand complexes to DNA phosphates (42, 63, 64). Nucleotide-level mapping of major Cr adducts detected no apparent base specificity in Cr-DNA binding (55). Shuttle-vector experiments have shown that Cr-DNA adducts containing cross-linked ligands were mutagenic and inhibited replication of Cr-modified vectors in human cells (43, 55, 63). Formation of Cr-DNA adducts is also a major cause of Cr(VI)-induced toxicity, because human cells unable to remove these lesions due to inactive nucleotide excision repair were much more sensitive to apoptosis and clonogenic lethality (44). In contrast to replication experiments in cells, studies with purified polymerases in vitro have shown that ternary Cr-DNA adducts have very little if any blocking potential (36, 37), which is consistent with the lack of direct Cr coordination to hydrogen-bonding groups of DNA bases. Thus, it appears that genotoxic activity of Cr-DNA adducts is not caused by direct interference with replicative polymerases but likely results from the interactions with other cellular proteins or processes.
In this work, we examined the possibility that MMR proteins function as activators of genotoxic responses to a major form of DNA damage generated by Cr(VI) in mammalian cells. Our initial interest in exploring a role of MMR was based on the fact that, similarly to base mismatches (22), DNA phosphate-based adducts facilitate DNA bending and kinking, due to neutralization of the negative charge (50). We found that MMR proteins are required for the induction of toxicity and replication inhibition by Cr-DNA adducts. Activation of toxicity in apoptotically proficient cells by this MMR-dependent mechanism proceeded through the accumulation of DNA double-strand breaks (DSB) in G2 phase and rapid activation of cell death without a significant G2 arrest. Our results extend the role of MMR proteins as activators of genotoxic responses to the DNA backbone damage and demonstrate a lesion specificity in downstream stress signaling.
MATERIALS AND METHODS
Cell cultures and drug treatments.
Human lung epithelial A549 and colon HCT116 (MLH1−/−) and DLD1 (MSH6−/−) cell lines were purchased from the American Type Culture Collection. HCT116+ch3 (MLH1+) and DLD1+ch2 (MSH6+) cell lines were generous gifts from T. Kunkel. The cells were cultured in Dulbecco's minimum essential medium-Hanks' F-12 medium supplemented with 10% serum and penicillin-streptomycin. Growth media for chromosome-complemented cell lines additionally contained 400 μg/ml Geneticin. Mlh1+/+, Mlh1−/−, Pms2+/+, and Pms2−/− mouse embryonic fibroblasts (MEFs) (gifts from M. Nguyen and P. Glazer) were cultured in Dulbecco's minimum essential medium containing 10% serum and penicillin-streptomycin. Cells were exposed to K2CrO4, H2O2, or formaldehyde (HCHO) for 3 h and to mitomycin C (MMC) for 4 h in serum-free medium. In selected experiments, cells were preincubated for 90 min with dehydroascorbic acid in Krebs-HEPES buffer (30 mM HEPES [pH 7.5], 130 mM NaCl, 4 mM KH2PO4, 1 mM MgSO4, 1 mM CaCl2, 0.5 mM glucose) prior to treatment with Cr(VI). This incubation with dehydroascorbic acid led to cellular accumulation of 0.9 ± 0.1 mM ascorbate, as determined by high-performance liquid chromatography analysis of cellular extracts (42). MNNG exposure was carried out as described previously (49). Briefly, cells were pretreated for 2 h with 10 μM O6-benzylguanine prior to the addition of 0.2 μM MNNG.
Clonogenic survival.
A total of 500 or 1,000 cells were seeded onto 100-mm dishes and allowed to attach overnight. Three dishes were used for each concentration of a drug. Cells were treated with Cr(VI), H2O2, HCHO, or MMC as described above. Ten to 14 days after exposure, colonies were stained with Giemsa solution and counted. Clonogenic experiments were repeated at least three times.
Western blotting.
Cells were collected by scraping, washed with cold phosphate-buffered saline (PBS), and resuspended by being vortexed in whole-cell lysis buffer (50 mM Tris [pH 8.0], 250 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 5 mM EDTA, 2 mM Na3VO4, 10 mM Na2P2O7, 10 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 0.5 μg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride). Lysates were incubated on ice for 10 min, and then cell debris was spun down at a speed of 10,000 × g for 10 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to an ImmunoBlot PVDF membrane (Bio-Rad). Primary antibodies used were anti-poly(ADP-ribose) polymerase (anti-PARP), anti-cleaved PARP, anti-caspase 3, anti-caspase 7, and anti-phospho-p53 (Ser-15) 16G8 monoclonal antibody (Cell Signaling Technology), anti-p53 (DO-1) (Santa Cruz Biotechnology), anti-caspase 2 (Pharmingen), and anti-γ-tubulin clone GTU-88 (Sigma). Protein bands were visualized using horseradish peroxidase-conjugated secondary antibodies (Upstate) and an enhanced chemiluminescence kit (Amersham).
Flow cytometry.
Both attached and floating cells were collected and washed with cold PBS. For cell cycle analysis, cells were fixed for at least 25 min in cold 70% ethanol. Cells were washed once with 3% fetal bovine serum-PBS solution and stained in 300 μl of propidium iodide (PI) (0.9 mg/ml) and 100 μl of RNase A (1 mg/ml) for 40 min at 37°C, and then samples were analyzed on a Becton Dickinson FACSCalibur. For annexin V staining, freshly collected cells were resuspended in 1× binding buffer, stained with annexin V and 7-aminoactinomycin D (7-AAD) (BD Biosciences) following the manufacturer's protocol, and analyzed immediately on a Becton Dickinson FACSCalibur.
Stable siRNA knockdown of p53.
Stable knockdown of p53 levels was achieved by expression of hairpin small interfering RNA (siRNA) from a retroviral vector. The sequence for p53-targeting siRNA was taken from Brummelkamp et al. (6). The control green fluorescent protein-targeting sequence was 5′-GCAAGCTGACCCTGAAGTT-3′. Double-stranded oligonucleotides were ligated into the pSUPER retroviral vector that was linearized with BamHI and HindIII. Retroviral infections and selection of puromycin-resistant cells were performed as recently described (44).
In vivo plasmid replication assay.
Ternary ascorbate-Cr-DNA adducts were formed as previously described (42). Briefly, pSP189 DNA was modified with 50 μM Cr(VI) in the presence of 1 mM ascorbate and purified by Bio-Gel P-30 chromatography and ethanol precipitation. Control and Cr-modified plasmids were transfected into MLH1−/− (HCT116) and MLH1+ (HCT116+ch3) cells and allowed to replicate for 40 h. The replicative progeny of the plasmids was isolated, and the yield of replicated molecules was determined by a bacterial transformation assay (63).
Immunofluorescence.
Cells were grown on Superfrost Plus slides and exposed to Cr(VI) for 3 h and then returned to complete medium. Bromodeoxyuridine (BrdU) labeling was performed by incubation with a cell proliferation labeling reagent (Amersham) in complete medium for 1 h in the dark. At the indicated times, cells were washed twice with PBS and fixed with 2% paraformaldehyde in PBS for 15 min. Cells were then permeabilized for 15 min with 1% Triton X-100, followed by incubation with 2% fetal bovine serum in PBS for 30 min. Double labeling was performed by simultaneous incubation of primary antibodies for γ-H2AX at 1:150 dilution (Upstate), anti-BrdU at 1:100 dilution (Pharmingen), or anti-cyclin B1 at 1:200 dilution (Santa Cruz) for 2 h at 37° in a humidified chamber. The secondary antibodies, Alexa Fluor 488-conjugated anti-mouse immunoglobulin G and Alexa Fluor 594-conjugated anti-rabbit immunoglobulin G (Molecular Probes), were incubated for 1 h at room temperature. All antibodies were diluted in PBS containing 1% bovine serum albumin and 0.5% Tween 20, with the exception of anti-BrdU staining solution, which additionally contained 1 mM MgCl2 and 125 U/ml Benzonase nuclease (Novagen). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and mounted with Vectashield hard set mounting medium (Vector Laboratories). Fluorescence images were recorded with a Zeiss Axiovert 100 confocal microscope and analyzed by Renaissance 410 software. All experiments were repeated at least four times. At least 100 cells were analyzed in each experiment.
p53 reporter assay.
Cells were transfected with p53-Luc (Stratagene) and pRL-TK (Promega) plasmids, and they were exposed the next day to 0 to 30 μM Cr(VI) for 3 h. Cell lysates were obtained 24 h after Cr treatment. As a positive control, cells were treated with 1 μg/ml doxorubicin for 24 h in complete medium. Luciferase activity was measured on a Tecan Spectrafluor Plus using a Dual-Luciferase Reporter Assay Kit (Promega). The expression of p53-driven firefly luciferase was normalized using the activity of constitutively expressed Renilla luciferase.
Determination of Cr-DNA adducts.
Control and Cr(VI)-exposed cells were collected by trypsinization and washed with cold PBS, and then DNA was isolated by an RNase-phenol-chloroform-proteinase K procedure. The amount of DNA-bound Cr was determined by graphite furnace atomic absorption spectroscopy using Zeeman background correction (42). Measurements were done using a model 41002L Perkin-Elmer GF-AAS instrument. The detection limit was 0.4 pmol of chromium.
RESULTS
Increased clonogenic survival of MMR-deficient cells after Cr(VI) damage.
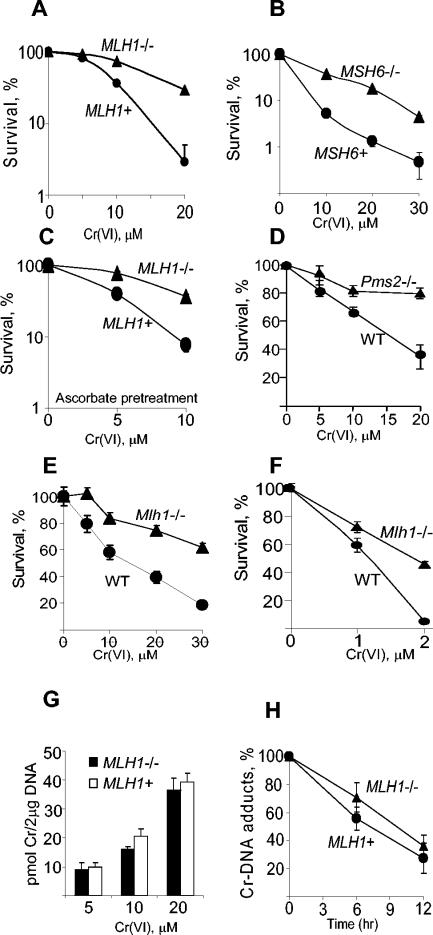
To determine if MMR participates in cellular responses to Cr(VI), we first examined clonogenic survival of MMR-proficient (MMR+) and MMR-deficient (MMR−) cells. Human colon cells deficient for MLH1 (HCT116) and MSH6 (DLD1) MMR proteins showed much greater survival than their chromosome-complemented MMR+ counterparts (Fig. 1A and B). HCT116 cells stably transfected with a MLH1-expressing plasmid also had much higher sensitivity to Cr(VI) than the empty vector control (data not shown). MLH1 deficiency was also associated with greater survival after Cr(VI) treatment of HCT116 cells that were preloaded with physiological levels of ascorbate (Fig. 1C), which is an important reducer of Cr(VI) in the target tissues for this genotoxin (51). Ascorbate is either absent or present at very low levels in HCT116 and other cultured human cells (42). Increased tolerance of Cr-DNA damage was not limited to MMR− human colon cells, as Pms2−/− and Mlh1−/− knockout MEFs also showed much higher survival than wild-type cells (Fig. 1D and E). MMR deficiency in Mlh1−/− MEFs was also associated with greater clonogenic survival after chronic exposure to low doses of Cr(VI) (Fig. 1F). Control experiments showed that MMR status had no significant effect on the formation of Cr-DNA damage, as indicated by similar levels of Cr-DNA adducts after exposure to different concentrations of Cr(VI) (Fig. 1G). As MMR proteins have been found in some cases to influence repair of DNA lesions (35), we also examined rates of removal of Cr-DNA adducts in MLH1+ and MLH1−/− cells (Fig. 1H). Both cell lines were highly proficient in repair of Cr-DNA adducts without significant differences in rates of repair (t1/2 = 6.4 ± 1.8 for MLH1+ cells and 8.1 ± 1.5 h for MLH1−/− cells). These values are within the range of repair rates recently determined for four other MMR+ human cell lines (t1/2 = 6.7 to 8.2 h) (39). Thus, diminished sensitivity of MLH1−/− HCT116 cells to Cr(VI) did not result from changes in the formation of Cr-DNA adducts or their rates of repair. Collectively, data on increased tolerance of Cr(VI) by cells deficient in three different MMR proteins (MLH1, PMS2, and MSH6) strongly indicate that the entire MMR machinery is required for the manifestation of cytotoxicity in response to Cr-DNA damage.
FIG. 1.
Clonogenic survival and DNA repair in MMR-deficient and normal cells. Cells were treated with Cr(VI) for 3 h (panels A to E and G) or 48 h (F). Clonogenic data are means ± standard deviation (SD) from three to six independent experiments each, with three dishes per dose. Where not seen, error bars were smaller than the symbol. (A) Survival of human HCT116 (MLH1−/−) and HCT116+ch3 (MLH1+) cells exposed to Cr(VI) under standard culture conditions (six experiments). (B) Survival of human DLD1 (MSH6−/−) and DLD1+ch2 (MSH6+) cells (three experiments). (C) Clonogenic survival of MLH1−/− and MLH1+ HCT116 cells treated with Cr(VI) after preloading with 0.9 mM ascorbate (three experiments). Survival of isogenic pairs of Pms2−/− and WT (D) and Mlh1−/− and WT (E) MEFs (three experiments). (F) Survival of Mlh1−/− and Mlh1+/+ MEFs continuously exposed to Cr(VI) for 48 h (three experiments). (G) Formation of Cr-DNA adducts in HCT116 and HCT116+ch3 cells. DNA was isolated immediately after exposure to Cr(VI) for 3 h. (H) Time course of removal of Cr-DNA adducts in HCT116 and HCT116+ch3 cells. Cells were treated with 10 μM Cr(VI) for 3 h, and DNA was isolated either immediately or 6 and 12 h later. Data are means ± SD from three independent DNA preparations analyzed twice for Cr content.
MMR− cells exhibit deficient activation of apoptosis in response to Cr-DNA damage.
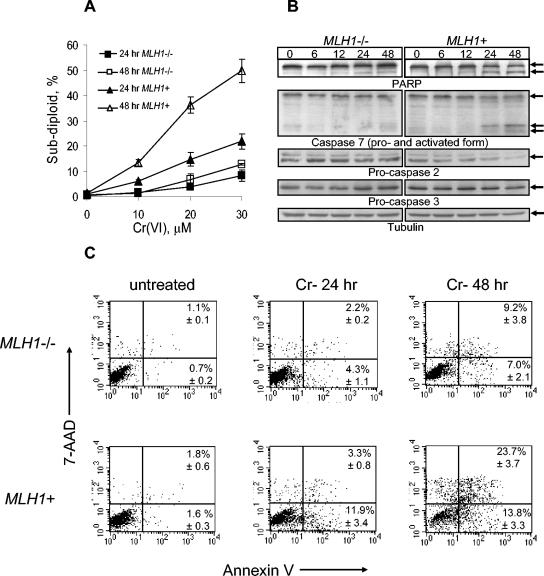
Next, we sought to determine whether differential survival of MMR+ and MMR− cells was related to their apoptotic responses to Cr damage. To establish the presence of apoptosis, we looked at several markers associated with this form of cell death. First, we analyzed PI-stained cells by fluorescence-activated cell sorter (FACS) to determine the appearance of cellular populations with subdiploid amount of DNA. MLH1+ cells exhibited a very strong, dose-dependent increase in subdiploid cells at both 24 and 48 h postexposure (Fig. 2A). The presence of subdiploid populations in Cr(VI)-treated MLH1−/− cells was also detected but to a much smaller extent than in MLH1+ cells. The percentage of apoptotic cells was, on average, three to five times higher in MLH1+ cells. To verify these findings, MLH1+ and MLH1−/− cells were stained with annexin V and 7-AAD and analyzed by flow cytometry. Annexin V binds to phosphatidylserine on the outer leaflet of apoptotic cells, and 7-AAD stains DNA. Early apoptotic cells are annexin V positive only; DNA was not stained because the membrane was intact. Late-apoptotic cells are positive for both annexin V and 7-AAD because the cell membrane becomes permeable to this dye. Consistent with the results seen with PI-stained cells, the amount of apoptotic, annexin V-positive cells was substantially smaller for the MLH1−/− cell line than for its MMR+ counterpart at both 24 and 48 h postexposure (Fig. 2C). The presence of functional MLH1 protein was required for Cr(VI)-dependent activation of executioner caspases 2 and 7, as well as for apoptotic cleavage of PARP (Fig. 2B). Caspase 3, another executioner caspase, underwent only modest activating cleavage in MLH1+ cells, but it remained completely intact in MLH1−/− cells.
FIG. 2.
Different levels of apoptosis in MLH1−/− and MLH1+ HCT116 cells treated with Cr(VI). Human HCT116 and HCT116+ch3 cells were treated with Cr(VI) for 3 h as described in Materials and Methods. (A) Percentage of subdiploid cells determined by FACS analysis of PI-stained samples. Data are means ± SD from four independent experiments. If not seen, error bars were smaller than the data point. (B) Western blots for intact and cleaved PARP; caspases 2, 3, and 7; and a γ-tubulin loading control. Cells were treated with 30 μM Cr(VI) for 3 h and collected at indicated times. (C) Detection of apoptosis by FACS analysis of cells stained with annexin V-phycoerythrin and 7-AAD. Cells were treated with 0 or 30 μM Cr(VI) for 3 h, collected at 24 and 48 h postexposure, stained, and immediately subjected to cell sorting. Representative FACS profiles are shown. Numbers in the right quadrants are means ± SD from four independent experiments.
Loss of replication-blocking potential of Cr-DNA adducts in MMR-deficient human cells.
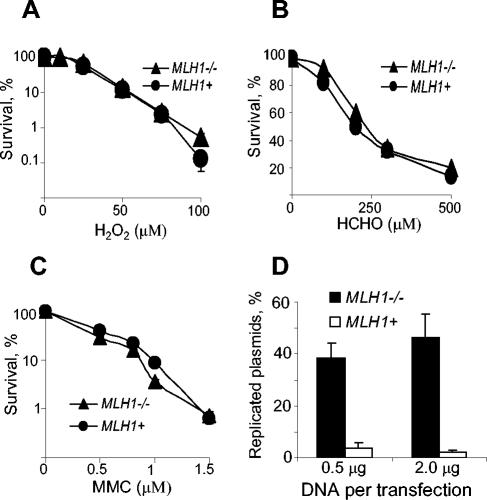
Cellular metabolism of Cr(VI) generates ternary Cr-DNA adducts as the most abundant form of DNA damage (65), but small amounts of other lesions, such as DNA-protein and interstrand cross-links, are also formed (34, 37, 52). Exposure to high doses of Cr(VI) leads to the induction of oxidative stress (31, 54). To identify DNA lesions whose genotoxic potential is enhanced by the presence of MMR proteins, we treated MLH1+ and MLH1−/− cells with DNA-damaging agents that primarily cause specific types of lesions. We found that toxicity of oxidative damage (hydrogen peroxide exposure), DNA-protein cross-links (formaldehyde exposure), and DNA interstrand cross-links (mitomycin C exposure) was the same for both MMR+ and MMR− human HCT116 cells (Fig. 3A to C). Similar results were found with Mlh1+/+ and Mlh1−/− MEFs (data not shown). These data suggested that MMR proteins probably potentiate genotoxicity of Cr-DNA adducts. To test this possibility more directly, we examined the dependence of replication-blocking activity of Cr-DNA adducts on MLH1 status. We formed ternary ascorbate-Cr-DNA adducts in vitro and then transfected control and Cr-modified plasmids into MLH1+ and MLH1−/− cells. Plasmids were propagated for 40 h, their replicated progeny was isolated, and the number of replicated molecules was scored by bacterial transformation assay. We found that the presence of functional MLH1 protein severely (10 to 20 times) inhibited the yield of replicated plasmids from Cr-modified templates, identifying Cr-DNA adducts as the MMR-dependent form of Cr(VI)-induced DNA damage (Fig. 3D). Preliminary studies also indicated that MMR proteins were capable of specific binding to ascorbate-Cr-DNA adducts in vitro.
FIG. 3.
Effect of MLH1 status on toxicity of various forms of DNA damage. (A to C) Clonogenic survival of human HCT116 (MLH1−/−) and HCT116+ch3 (MLH1+) cells treated with H2O2, HCHO, or MMC, respectively. Data are means ±SD from three independent clonogenic experiments. For many data points, error bars were smaller than the symbols. (D) Yield of replicated progeny from Cr adduct-containing plasmids propagated in HCT116 (MLH1−/−) and HCT116+ch3 (MLH1+) cells. The pSP189 plasmids were modified with 50 μM Cr(VI) in the presence of 1 mM ascorbate, then transfected into cells, and allowed to replicate for 40 h. Replicated plasmids were isolated, and their amounts were determined by bacterial transformation assay. Data are means ± SD from four independent DNA transfections.
Tumor suppressor p53 is not required for activation of cell death by Cr(VI).
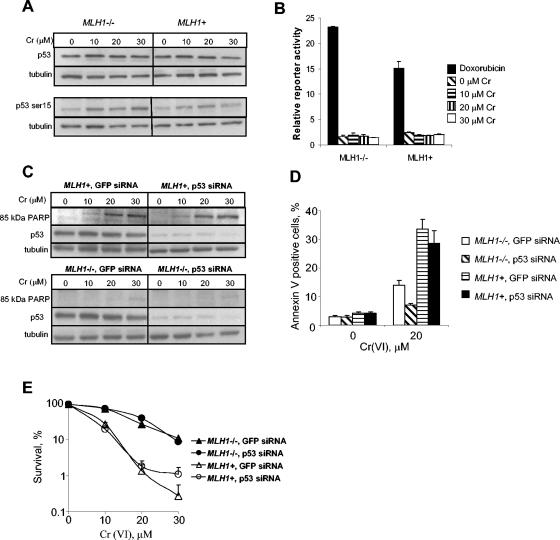
Previous studies have pointed to a potential significance of p53 protein in the induction of apoptosis in human cells treated with high doses of Cr(VI) (8, 56). Therefore, we decided to examine whether MMR transmits stress signaling to apoptotic machinery via activation of a p53 transcriptional factor. We first looked at p53 protein levels and the extent of phosphorylation at Ser-15, which are important indicators of p53 activation following genotoxic stress (7). We did not find any significant changes in p53 protein levels in either MLH1−/− or MLH1+ cells that were treated with Cr(VI) for 3 h and analyzed 48 h postexposure during the maximal induction of apoptosis (Fig. 4A). Protein levels of p53 were also unchanged at earlier postexposure times, varying from 1 to 24 h (data not shown). At 48 h postexposure, we did observe a modest increase in the amount of phosphorylation at Ser-15 of p53 in both MLH1−/− and MLH1+ cells (Fig. 4A). Consistent with previous reports (8, 56), a much more toxic exposure regimen (<0.1% survival by clonogenic assay) involving continuous treatment with Cr(VI) for 24 h led to a modest but significant elevation of p53 levels and a strong increase in Ser-15 phosphorylation; however, both responses were again independent of the MLH1 status (data not shown). To determine whether Cr-DNA damage stimulated transactivation activity of p53, we used a reporter plasmid containing a firefly luciferase gene linked to a promoter with multiple copies of the p53 response element. Our positive control, doxorubicin-treated cells, showed an approximately 20-fold increase in p53-dependent transcription of the luciferase activity over untreated cells. However, increasing doses of Cr gave no response over the untreated control (Fig. 4B).
FIG. 4.
Cr(VI)-induced cell death is independent of p53. (A) Western blots for p53 protein and phosphorylated Ser-15 of p53. Cells were treated with Cr(VI) for 3 h, and protein extracts were prepared 48 h later. (B) Reporter assay for p53 transactivation. Cells were treated with 0 to 30 μΜ Cr(VI) for 3 h, and cell lysates were prepared 24 h later. As a positive control, cells were treated with 1 μg/ml doxorubicin for 24 h. Relative p53 reporter activity was calculated by dividing firefly luciferase activity by the internal control, Renilla luciferase activity. Results are means ± SD of duplicate determinations from two dishes. Similar results were obtained in two more independent experiments. (C) Western blotting with antibodies against a caspase-cleaved, 85-kDa fragment of PARP and anti-p53 and anti-γ-tubulin antibodies. Cells were treated with Cr(VI) for 3 h, and protein extracts were obtained 48 h later. (D) Effect of p53 levels on the amounts of apoptotic cells following Cr(VI) treatment. Cells were treated with 0 or 20 μM Cr(VI) for 3 h, incubated for additional 48 h in normal medium, and then stained with annexin-phycoerythrin. Annexin V-positive, apoptotic cells were quantified by FACS. Shown are means ± SD from four independent experiments. (E) Clonogenic survival of cells with normal and stably downregulated levels of p53 protein. Results are means ± SD from three independent clonogenic experiments.
To test a potential role of p53 in Cr-induced apoptosis more directly, we downregulated levels of this protein in HCT116 and HCT116+ch3 cells by stable expression of hairpin siRNA from a retroviral vector. Western blots detected more than a 10-fold reduction in p53 levels in both cell lines (Fig. 4C). Control experiments showed that this decrease in p53 amounts was sufficient to suppress apoptosis and clonogenic death by the DNA-breaking drug bleomycin (data not shown). The levels of Cr(VI)-induced apoptosis measured by the appearance of caspase-cleaved PARP fragment (Fig. 4C) and FACS analysis of annexin V-positive cells (Fig. 4D) were not significantly different in MLH1+ cells with normal and downregulated levels of p53. Knockdown of p53 protein in MLH1−/− cells led to a some decrease in the percentage of annexin V-positive cells, indicating that MMR-independent apoptosis is partially p53 dependent. Because the MMR-potentiated fraction accounts for >80% of all apoptosis, we conclude that Cr-induced apoptosis is primarily p53 independent. Knockdown of p53 had no effect on clonogenicity of cells treated with low and moderate Cr(VI) concentrations, but it slightly increased the number of colonies at the highest dose causing <1% clonogenic survival (Fig. 4E). Thus, p53 protein appears to be dispensable for Cr(VI)-induced apoptosis and plays only a minor role in clonogenic death at highly toxic Cr(VI) doses that likely induced nonspecific oxidative stress.
Increased formation of DNA double-strand breaks in MMR-proficient cells.
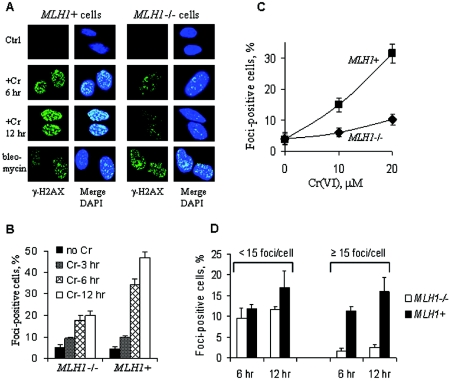
To explore potential mechanisms by which MMR proteins potentiate toxicity of Cr(VI), we tested the possibility that processing of Cr-DNA damage leads to the formation of DNA DSB as more toxic secondary lesions. H2AX is a histone protein that has been shown to be specifically phosphorylated (γ-H2AX) and forms foci at the sites of DSB (45). Indirect immunofluorescence for γ-H2AX following Cr(VI) exposure showed higher focus formation in MLH1+ than in MLH1−/− cells (Fig. 5A). The presence of functional MLH1 protein was also associated with a much greater overall number of focus-positive, DSB-containing cells at all time points examined (Fig. 5B). The differences in the number of focus-containing cells were even more striking at mildly and moderately toxic doses of Cr(VI) (Fig. 5C). The presence of MLH1 was specifically associated with the appearance of multifoci cells that were very rare in MLH1-deficient populations (Fig. 5D). Increased formation of γ-H2AX in Cr(VI)-treated MLH1+ HCT116 and Mlh1+/+ MEFs relative to their isogenic MMR-deficient controls was also detected by Western blotting (not shown). The absence of MLH1 did not lead to a generalized defect in the ability to phosphorylate H2AX because MLH1+ and MLH1−/− cells treated with bleomycin showed similar levels of γ-H2AX foci per cell (Fig. 5A) and nearly identical percentages of focus-positive cells (75.1% ± 5.6% for MLH1+ cells and 71.3% ± 3.5% for MLH1−/− cells).
FIG. 5.
Increased formation of DNA double-strand breaks in MMR-proficient cells following Cr(VI) exposure. (A) Confocal images of control, Cr(VI)- and bleomycin-treated HCT116 (MLH1−/−), and HCT116+ch3 (MLH1+) cells stained with anti-γ-H2AX antibodies and DAPI. Cells were treated with 30 μM Cr(VI) for 3 h and then fixed with paraformaldehyde at the indicated times. Bleomycin-treated cells (50 μg/ml for 1 h in serum-free medium) were analyzed 4 h postexposure. The formation of γ-H2AX foci represents the biochemical marker of DNA double-strand breaks, while counterstaining with DAPI was used to detect nuclei. (B) Time-dependent formation of γ-H2AX foci after treatment of cells with 30 μM Cr(VI). Shown are means ± SD from six independent experiments. (C) Dose-dependent induction of γ-H2AX focus-containing cells by Cr(VI). Focus-positive cells were counted at 12 h postexposure. Data are means ± SD from three independent experiments. (D) Distribution of cells with low and high numbers of γ-H2AX foci in MLH1−/− and MLH1+ HCT116 cells. Cells were treated with 20 μM Cr(VI) for 3 h and processed for indirect immunofluorescence at 6 and 12 h postexposure. Values are means ± SD from four independent experiments.
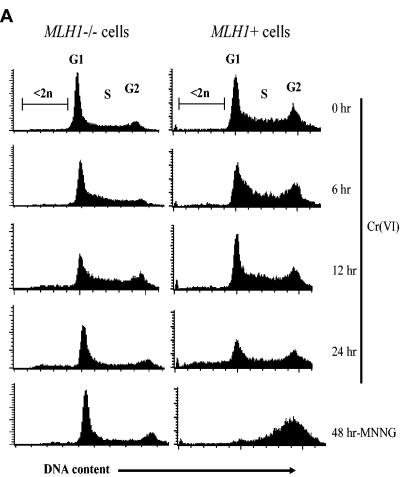
Accumulation of γ-H2AX foci in G2 phase and cell cycle changes.
Since functional MMR is known to be most active in S and G2 phases, we wanted to determine if foci formation was cell cycle dependent. Using BrdU incorporation as a marker of S phase cells, we found only modest colocalization of γ-H2AX foci with BrdU-positive cells analyzed 12 h after treatment with 20 μM Cr(VI) (Fig. 6A and C). The frequency of BrdU and γ-H2AX double-positive cells was similar in MLH1+ and MLH1−/− cells. We next examined the presence of DSB in G2 cells using cyclin B1 expression as a marker (24, 46). At 12 h after exposure to 20 μM Cr(VI), approximately 80% of γ-H2AX focus-containing MLH1+ cells were positive for cyclin B1 staining, indicating a predominant production of DSB in G2 phase (Fig. 6B and C). The percentage of γ-H2AX-positive G2 cells at an earlier, 6-h postexposure time was also very high (69.3% ± 4.7% for three experiments). The presence of MLH1 protein led to about five-times-higher overall frequency of cyclin B1 and γ-H2AX double-positive cells. A preferential accumulation of γ-H2AX in cyclin B1-positive cells was not specific to the selected dose of Cr(VI), because it was also found after treatment with a lower, 10 μM dose of Cr(VI): 69.3% ± 5.5% at 6 h postexposure (three experiments) and 67.7 ± 5.5% of γ-H2AX-containing MLH1+ cells at 12 h postexposure (three experiments) were in G2 phase. A slightly smaller colocalization of cyclin B1 and γ-H2AX at lower doses and earlier times probably reflected a relatively higher contribution of background γ-H2AX-containing cells that are typically present in S phase. The induction of γ-H2AX foci was not a result of apoptotic DNA fragmentation, because none of the apoptotic markers examined showed initiation of apoptotic events at 12 h postexposure (Fig. 2). A more detailed time course of caspase-mediated PARP cleavage and sub-G1 peak formation determined that in 30 μM Cr(VI)-treated cells apoptosis was initiated at 18 h postexposure, whereas at a low 10 μM dose, apoptosis was detected only at 24 h postexposure (not shown). Thus, induction of γ-H2AX preceded apoptotic events by 12 to 18 h. Similar distributions of low-focus and high-focus cells at 6 and 12 h postexposure also argues against any significant contribution of apoptotic DNA fragmentation, which usually causes extremely high levels of γ-H2AX staining and coincides with caspase activation (23).
FIG. 6.
G2-specific induction of γ-H2AX foci by Cr(VI). HCT116 (MLH1−/−) and HCT116+ch3 (MLH1+) cells were treated with 0 or 20 μM Cr(VI) for 3 h and fixed 12 h later with paraformaldehyde. (A) Representative images of cells stained with anti-γ-H2AX antibodies, anti-BrdU antibodies (marker of S phase), and DAPI. (B) Images of cells stained with anti-γ-H2AX antibodies, anti-cyclin B1 antibodies (marker of G2 phase), and DAPI. Cyclin B1 staining was most commonly cytoplasmic, but in a small fraction of positive cells, it was found in both the cytoplasm and nucleus. (C) Quantitation of colocalization of γ-H2AX foci with markers of S (BrdU incorporation) and G2 (cyclin B1 staining) phases in MLH1−/− and MLH1+ cells. Results are means ± SD from three independent experiments in each of which at least 100 cells were analyzed.
The high production of double-strand breaks by Cr(VI) in MLH1+ cells prompted us to investigate whether this response was associated with cell cycle arrest in G2 phase. We did not detect significant changes in tetraploid populations by FACS analysis of propidium iodide-stained cells collected at 6, 12, or 24 h postexposure (Fig. 7A). To ensure that our cells contained an intact G2 checkpoint, we examined cell cycle distribution of these cells following DNA methylating damage by MNNG. In agreement with a recently published study (49), we detected a massive accumulation of MLH1-expressing cells in G2 phase. To verify our FACS data, we scored G2 cells by immunostaining for cyclin B1 expression (Fig. 7B and C). With the exception of a marginal increase in MLH1+ cells at 12 h postexposure, treatment with 20 μM Cr(VI) did not cause any significant changes in the size of G2 populations in either MLH1+ or MLH1−/− cells. Treatment of cells with lower doses of Cr(VI) also failed to cause significant G2 accumulation (data not shown). Control experiments showed that both MLH1−/− and MLH1+ cells were equally proficient in the induction of G2 arrest after DNA damage by bleomycin (50 μg/ml for 1 h, staining for cyclin B1 at 5 h postexposure). The number of G2 cells increased from 30.6% ± 5% to 66.7% ± 7.9% in MLH1−/− (four experiments) and from 31.7% ± 5.6% to 63.6% ± 7.7% in MLH1+ (four experiments) populations.
FIG. 7.
Cell cycle changes following DNA damage induced by Cr(VI) and MNNG. (A) Representative FACS profiles of propidium iodide-stained HCT116 (MLH1−/−) and HCT116+ch3 (MLH1+) cells. Treatment with 20 μM Cr(VI) was for 3 h, and cells were collected at 0 to 24 h postexposure. Cells were treated with 0.2 μM MNNG continuously for 48 h. The numbers of cyclin B1-positive cells at 6 and 12 h (B) and 24 h (C) after exposure to 20 μM Cr(VI) are shown. Results are means ± SD from three independent experiments (D) G2 arrest by Cr(VI) in apoptosis-resistant human A549 cells. Cells were treated with 50 μM Cr(VI) for 3 h, collected 24 h later, stained with propidium iodide, and analyzed by FACS. Shown are means ± SD from four independent experiments.
Unlike MNNG, Cr(VI) induced a more rapid onset of apoptosis, evident by a significant fraction of subdiploid cells at 24 h postexposure (Fig. 7A). To test whether apoptotic deficiency can result in G2 arrest, we examined cell cycle changes in human A549 cells that are resistant to the induction of apoptosis by Cr(VI). The formation of Cr-DNA damage in A549 cells did cause the accumulation of a large number of G2/M-arrested cells at 24 h posttreatment (Fig. 7D). There was no detectable increase in subdiploid populations at this and later time points (at 48 h, 0.7% subdiploid cells in control and 0.7% in Cr-treated group), demonstrating a high apoptotic resistance of A549 cells. Thus, despite such an important similarity as G2-specific induction of double-strand breaks, MMR proteins activate different stress signaling in response to the presence of O6-methylguanine and Cr-DNA adducts in human colon HCT116+ch3 cells.
DISCUSSION
In this work, we have found that MMR proteins strongly modulate cellular responses to an important environmental carcinogen, hexavalent Cr. The use of four cell lines deficient in three different MMR proteins clearly showed that loss of either MSH6, a component of the MutSα dimer, or components of the MutL dimer (MLH1 or PMS2) leads to greatly increased survival of Cr(VI)-treated human or mouse cells. The need for multiple MMR proteins to induce toxicity implies activation of the entire MMR complex by Cr-DNA damage. MMR apparently recognizes Cr-DNA adducts because the toxicity of oxidative damage, interstrand, and DNA-protein cross-links was not influenced by the absence of MLH1 protein in human or mouse cells. This conclusion was further supported by results on the loss of replication inhibition of Cr-adducted plasmids in cells lacking MLH1 protein.
Poor clonogenic survival of Cr-damaged MMR+ cells reflected their higher apoptotic response than MMR− cells. Several markers of apoptosis, including DNA fragmentation, annexin V staining, activation of caspases, and apoptotic cleavage of PARP, have shown that MMR+ cells undergo more extensive and rapid apoptosis after Cr(VI) treatment. Many DNA-damaging agents have been shown to induce apoptosis through p53-dependent mechanisms. It has also been reported that p53 protein levels and phosphorylation at Ser-15 increase following Cr(VI) damage (17, 56). Levels of apoptosis have been found to be lower in p53−/− mouse dermal fibroblasts exposed to high Cr(VI) concentrations (8). However, we did not detect strong phosphorylation of p53 at Ser-15 or overall stabilization of p53 at doses that induced extensive apoptosis and led up to 99% lethality by the clonogenic assay. In addition, there was no significant increase in p53 transactivation by Cr-DNA damage. Importantly, stable knockdown of p53 protein levels showed no significant effect on clonogenic survival or apoptosis by low and moderate doses of Cr(VI). Previous studies differ from our work in two ways. First, our study used lower doses of Cr(VI). At the highest dose we used, 30 μM Cr(VI), we did observe some improvement in clonogenic survival in MMR+ cells expressing p53-targeting siRNA. This result probably reflected the contribution of nonspecific oxidative stress to toxicity under the conditions of a very high Cr load. Consistent with this suggestion, a study that used high doses of Cr(VI) to detect p53-dependent apoptosis also showed significant oxidative damage (62). Therefore, it appears that Cr-DNA adducts activate apoptosis by a p53-independent mechanism, whereas oxidative damage induced by very high levels of Cr(VI) engages a p53-dependent apoptotic pathway. At doses of Cr(VI) that caused up to 95% clonogenic lethality, the contribution of p53 to activation of apoptosis was negligible, as indicated by identical survival of cells with normal and 10- to 20-times-lower levels of p53. A decrease in Cr(VI)-induced apoptosis observed in cells expressing human papillomavirus type 16 E6 protein (8) might not have necessarily reflected downregulated levels of p53 because E6 has other cellular targets including the p53 homolog, p73 (40), which is important for apoptosis by some DNA-damaging agents (16, 47).
The mechanism of MMR-dependent sensitization is not well understood even for the forms of DNA damage whose toxicity has long been known to require the presence of MMR proteins. Currently there are two models, futile repair (27, 49) and direct signaling (14). In the futile repair model, the product of DNA methylation by MNNG or MNU, O6-methylguanine, mispairs with T during replication, which leads to the activation of MMR at this site (13). Since MMR removes the newly synthesized strand, O6-methylguanine is not eliminated from DNA, which can lead to multiple cycles of attempted repair and ultimately to the formation of secondary lesions and toxicity. In support of this theory, G2 arrest and apoptosis occur in MMR+ cells only after the second S phase following methylating damage (49, 59). During that time, there is a massive induction of γ-H2AX foci, indicating the presence of DNA double-strand breaks that are thought to arise from collapsed replication forks at the site of single-strand breaks produced after the first S phase (49). Recent studies provided a strong set of experimental evidence for separate roles of MutSα complex in MMR and induction of cell death. Three groups have shown that in both yeast and mouse species, mutations in the ATPase domain of Msh2 or Msh6 led to uncoupling of MMR and cytotoxicity in response to cisplatin and MNNG (12, 33, 61). The mutated Msh2 and Msh6 proteins were still able to form heterodimers and bind DNA with the same affinity, but they were unable to repair mismatches. Although MMR was nonfunctional, the mutant protein retained the ability to sensitize cells to Pt-DNA and O6-methylguanine (33). These findings pointed to the possibility that MMR proteins may be directly involved in the initiation or potentiation of signaling responses. This suggestion is supported by direct association of major MMR proteins with the components of stress signaling, such as ATM and CHK2 (6), ATR (58), and proapoptotic p73 (41).
In contrast to the Sn1-type methylating agents, the induction of apoptosis in Cr(VI)-treated cells started much earlier, at 18 to 24 h postexposure, indicating a different mechanism of toxicity. The initiation of apoptosis in Cr-damaged MMR+ cells was preceded by extensive accumulation of γ-H2AX foci, indicating the formation of highly toxic DNA double-strand breaks. A major increase in the number of the γ-H2AX focus-containing MMR+ cells was observed at 6 h postexposure, which suggests that DNA breakage was not caused directly by Cr(VI) but resulted from processing of the damaged DNA. The majority of γ-H2AX-containing cells were positive for cyclin B1, a protein that is highly expressed in G2 phase (24, 46). These results point to the possibility that the formation of double-strand breaks requires passage of cells through S phase. This suggestion is supported by recent findings that only treatment of cycling but not growth-arrested human fibroblasts with Cr(VI) led to the production of γ-H2AX (18).
A more rapid onset of double-strand breakage in Cr(VI)-treated cells could reflect a stronger replication blockage by MMR complexes bound to Cr-DNA adducts. Extensive formation of Cr-DNA adducts in Cr(VI)-treated cells (reference 44 and this work) provides the opportunity for frequent stalling of replication complexes in relatively short stretches of DNA, leading to a higher probability of complete arrest and collapse of replication forks. Every collapsed replication fork ultimately results in the formation of a double-strand break (11, 48). Studies of toxicity and the production of γ-H2AX foci by MNNG have typically been performed under conditions of completely inactive repair of O6-methylguanine, which probably leads to the induction of cell death by a lower number of persistent lesions and may account for a somewhat different activation of toxicity. In all our studies, we used cells proficient in nucleotide excision repair, the main mechanism for removal of Cr-DNA adducts in human cells (44). MMR-dependent apoptotic signaling in Cr(VI)- and MNNG-treated cells also appears to be different, as judged by the much more prominent activation of caspase 3 by O6-methylguanine (19, 38) but not by Cr-DNA damage (this study). A lack of apparent G2 arrest in Cr(VI)-treated MLH1-complemented HCT116 cells containing a large number of double-strand breaks probably reflects a strong and rapid activation of apoptotic processes. Human A549 cells that are much more resistant to the induction of Cr(VI)-dependent apoptosis than HCT116+ch3 cells did show G2 arrest in response to Cr-DNA damage.
MMR-dependent toxicity and Cr(VI) carcinogenesis.
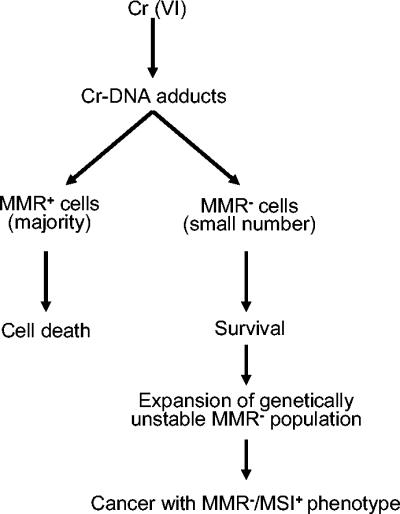
Our findings have potentially important implications for understanding the mechanisms of Cr(VI) carcinogenesis. In any given organ system in the body, there is a small population of cells that are deficient in MMR, due to spontaneous mutagenesis and epigenetic changes. When the lung tissue sustains high levels of Cr-DNA damage, normal lung cells will undergo apoptosis, but MMR− variants will survive. Thus, chronic exposure to Cr(VI) may actually select for MMR− cells through multiple cycles of cell death of sensitive cells and proliferation of resistant clones. In addition to mutations directly induced by Cr-DNA adducts, MMR− cells would have a very high spontaneous mutation rate and exhibit microsatellite instability (4), making them highly prone to malignant transformation (41). Cr(VI)-induced death of normal cells provides the opportunity for MMR− cells to expand and acquire additional mutations necessary for tumorigenesis. This model of carcinogenesis would predict that Cr(VI)-induced lung tumors should be deficient in one or more MMR proteins and exhibit microsatellite instability (Fig. 8). A recent molecular epidemiological study has found that 80% of lung tumors from chromate-exposed workers had microsatellite instability compared to only 15% in the control population (21). Interestingly, only a small fraction of chromate-associated cancers contained mutated p53 protein (30). This suggests that Cr(VI) carcinogenesis does not exert a strong selective pressure to inactivate p53, which could reflect its lack of involvement in toxic effects of Cr(VI) and, consequently, survival and outgrowth of resistant clones.
FIG. 8.
Model of Cr(VI) carcinogenesis through selection of mismatch repair-deficient cells.
Acknowledgments
We are grateful to T. Kunkel, M. Nguyen, and P. Glazer for their generous gifts of cells.
This work was supported by grants 2R01 ES008786, 1R01 ES012915, and 5T32 ES007272 from the National Institute of Environmental Health Sciences.
REFERENCES
- 1.Acharya, S., T. Wilson, S. Gradia, M. F. Kane, S. Guerrette, G. T. Marsischky, R. Kolodner, and R. Fishel. 1996. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. USA 93:13629-13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, S., B. Kurdi-Haidar, R. Gordon, B. Cenni, H. Zheng, D. Fink, R. D. Christen, C. R. Boland, M. Koi, R. Fishel, and S. B. Howell. 1996. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 56:3087-3090. [PubMed] [Google Scholar]
- 3.Aquilina, G., M. Crescenzi, and M. Bignami. 1999. Mismatch repair, G2/M cell cycle arrest and lethality after DNA damage. Carcinogenesis 20:2317-2326. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, J. C., A. Umar, J. I. Risinger, J. R. Lipford, M. Kane, S. Yin, J. C. Barrett, R. D. Kolodner, and T. A. Kunkel. 1995. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res. 55:6063-6070. [PubMed] [Google Scholar]
- 5.Branch, P., G. Aquilina, M. Bignami, and P. Karran. 1993. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature 362:652-654. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 7.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 8.Carlisle, D. L., D. E. Pritchard, J. Singh, and S. R. Patierno. 2000. Chromium(VI) induces p53-dependent apoptosis in diploid human lung and mouse dermal fibroblasts. Mol. Carcinog. 28:111-118. [DOI] [PubMed] [Google Scholar]
- 9.Cejka, P., L. Stojic, N. Mojas, A. M. Russell, K. Heinimann, E. Cannavo, M. di Pietro, G. Marra, and J. Jiricny. 2003. Methylation-induced G2/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 22:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, A. B., and T. A. Kunkel. 2004. Cadmium inhibits the functions of eukaryotic MutS complexes. J. Biol. Chem. 279:53903-53906. [DOI] [PubMed] [Google Scholar]
- 11.Courcelle, J., J. R. Donaldson, K. H. Chow, and C. T. Courcelle. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299:1064-1067. [DOI] [PubMed] [Google Scholar]
- 12.Drotschmann, K., R. P. Topping, J. E. Clodfelter, and F. R. Salsbury. 2004. Mutations in the nucleotide-binding domain of MutS homologs uncouple cell death from cell survival. DNA Repair (Amsterdam) 3:729-742. [DOI] [PubMed] [Google Scholar]
- 13.Duckett, D. R., J. T. Drummond, A. I. Murchie, J. T. Reardon, A. Sancar, D. M. Lilley, and P. Modrich. 1996. Human MutSα recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. USA 93:6443-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fishel, R. 1998. Mismatch repair, molecular switches, and signal transduction. Genes Dev. 12:2096-2101. [DOI] [PubMed] [Google Scholar]
- 15.Genschel, J., S. J. Littman, J. T. Drummond, and P. Modrich. 1998. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 273:19895-19901. [DOI] [PubMed] [Google Scholar]
- 16.Gong, J. G., A. Costanzo, H. Q. Yang, G. Melino, W. G. Kaelin, Jr., M. Levrero, and J. Y. Wang. 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399:806-809. [DOI] [PubMed] [Google Scholar]
- 17.Ha, L., S. Ceryak, and S. R. Patierno. 2003. Chromium (VI) activates ataxia telangiectasia mutated (ATM) protein. Requirement of ATM for both apoptosis and recovery from terminal growth arrest. J. Biol. Chem. 278:17885-17894. [DOI] [PubMed] [Google Scholar]
- 18.Ha, L., S. Ceryak, and S. R. Patierno. 2004. Generation of S phase-dependent DNA double strand breaks by Cr(VI) exposure: involvement of ATM in Cr(VI) induction of γ-H2AX. Carcinogenesis 25:2265-2274. [DOI] [PubMed] [Google Scholar]
- 19.Hickman, M. J., and L. D. Samson. 2004. Apoptotic signaling in response to a single type of DNA lesion, O(6)-methylguanine. Mol. Cell 14:105-116. [DOI] [PubMed] [Google Scholar]
- 20.Hickman, M. J., and L. D. Samson. 1999. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl. Acad. Sci. USA 96:10764-10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirose, T., K. Kondo, Y. Takahashi, H. Ishikura, H. Fujino, M. Tsuyuguchi, M. Hashimoto, T. Yokose, K. Mukai, T. Kodama, and Y. Monden. 2002. Frequent microsatellite instability in lung cancer from chromate-exposed workers. Mol. Carcinogenesis. 33:172-180. [DOI] [PubMed] [Google Scholar]
- 22.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 23.Huang, X., M. Okafuji, F. Traganos, E. Luther, E. Holden, and Z. Darzynkiewicz. 2004. Assessment of histone H2AX phosphorylation induced by DNA topoisomerase I and II inhibitors topotecan and mitoxantrone and by the DNA cross-linking agent cisplatin. Cytometry A 58A:99-110. [DOI] [PubMed] [Google Scholar]
- 24.Hwang, A., A. Maity, W. G. McKenna, and R. J. Muschel. 1995. Cell cycle-dependent regulation of the cyclin B1 promoter. J. Biol. Chem. 270:28419-28424. [DOI] [PubMed] [Google Scholar]
- 25.Jin, Y. H., A. B. Clark, R. J. Slebos, H. Al-Refai, J. A. Taylor, T. A. Kunkel, M. A. Resnick, and D. A. Gordenin. 2003. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34:326-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiricny, J., and M. Nystrom-Lahti. 2000. Mismatch repair defects in cancer. Curr. Opin. Genet. Dev. 10:157-161. [DOI] [PubMed] [Google Scholar]
- 27.Karran, P. 2001. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis 22:1931-1937. [DOI] [PubMed] [Google Scholar]
- 28.Kolodner, R. D. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 29.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 30.Kondo, K., N. Hino, M. Sasa, Y. Kamamura, S. Sakiyama, M. Tsuyuguchi, M. Hashimoto, T. Uyama, and Y. Monden. 1997. Mutations of the p53 gene in human lung cancer from chromate-exposed workers. Biochem. Biophys. Res. Commun. 239:95-100. [DOI] [PubMed] [Google Scholar]
- 31.Kortenkamp, A., M. Casadevall, and P. Da Cruz Fresco. 1996. The reductive conversion of the carcinogen chromium (VI) and its role in the formation of DNA lesions. Ann. Clin. Lab. Sci. 26:160-175. [PubMed] [Google Scholar]
- 32.Langard, S. 1990. One hundred years of chromium and cancer: a review of epidemiological evidence and selected case reports. Am. J. Ind. Med. 17:189-215. [DOI] [PubMed] [Google Scholar]
- 33.Lin, D. P., Y. Wang, S. J. Scherer, A. B. Clark, K. Yang, E. Avdievich, B. Jin, U. Werling, T. Parris, N. Kurihara, A. Umar, R. Kucherlapati, M. Lipkin, T. A. Kunkel, and W. Edelmann. 2004. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 64:517-522. [DOI] [PubMed] [Google Scholar]
- 34.Mattagajasingh, S. N., and H. P. Misra. 1999. Analysis of EDTA-chelatable proteins from DNA-protein crosslinks induced by a carcinogenic chromium(VI) in cultured intact human cells. Mol. Cell. Biochem. 199:149-162. [DOI] [PubMed] [Google Scholar]
- 35.Mellon, I., D. K. Rajpal, M. Koi, C. R. Boland, and G. N. Champe. 1996. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science 272:557-560. [DOI] [PubMed] [Google Scholar]
- 36.O'Brien, T., H. G. Mandel, D. E. Pritchard, and S. R. Patierno. 2002. Critical role of chromium (Cr)-DNA interactions in the formation of Cr-induced polymerase arresting lesions. Biochemistry 41:12529-12537. [DOI] [PubMed] [Google Scholar]
- 37.O'Brien, T., J. Xu, and S. R. Patierno. 2001. Effects of glutathione on chromium-induced DNA crosslinking and DNA polymerase arrest. Mol. Cell. Biochem. 222:173-182. [PubMed] [Google Scholar]
- 38.Ochs, K., and B. Kaina. 2000. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 60:5815-5824. [PubMed] [Google Scholar]
- 39.Papouli, E., P. Cejka, and J. Jiricny. 2004. Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair status of human cells. Cancer Res. 64:3391-3394. [DOI] [PubMed] [Google Scholar]
- 40.Park, J. S., E. J. Kim, J. Y. Lee, H. S. Sin, S. E. Namkoong, and S. J. Um. 2001. Functional inactivation of p73, a homolog of p53 tumor suppressor protein, by human papillomavirus E6 proteins. Int. J. Cancer 91:822-827. [DOI] [PubMed] [Google Scholar]
- 41.Prolla, T. A., S. M. Baker, A. C. Harris, J. L. Tsao, X. Yao, C. E. Bronner, B. Zheng, M. Gordon, J. Reneker, N. Arnheim, D. Shibata, A. Bradley, and R. M. Liskay. 1998. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat. Genet. 18:276-279. [DOI] [PubMed] [Google Scholar]
- 42.Quievryn, G., J. Messer, and A. Zhitkovich. 2002. Carcinogenic chromium(VI) induces cross-linking of vitamin C to DNA in vitro and in human lung A549 cells. Biochemistry 41:3156-3167. [DOI] [PubMed] [Google Scholar]
- 43.Quievryn, G., E. Peterson, J. Messer, and A. Zhitkovich. 2003. Genotoxicity and mutagenicity of chromium(VI)/ascorbate-generated DNA adducts in human and bacterial cells. Biochemistry 42:1062-1070. [DOI] [PubMed] [Google Scholar]
- 44.Reynolds, M., E. Peterson, G. Quievryn, and A. Zhitkovich. 2004. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J. Biol. Chem. 279:30419-30424. [DOI] [PubMed] [Google Scholar]
- 45.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherwood, S. W., D. F. Rush, A. L. Kung, and R. T. Schimke. 1994. Cyclin B1 expression in HeLa S3 cells studied by flow cytometry. Exp. Cell Res. 211:275-281. [DOI] [PubMed] [Google Scholar]
- 47.Shimodaira, H., A. Yoshioka-Yamashita, R. D. Kolodner, and J. Y. Wang. 2003. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc. Natl. Acad. Sci. USA 100:2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 49.Stojic, L., N. Mojas, P. Cejka, M. Di Pietro, S. Ferrari, G. Marra, and J. Jiricny. 2004. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 18:1331-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strauss-Soukup, J. K., M. M. Vaghefi, R. I. Hogrefe, and L. J. Maher, 3rd. 1997. Effects of neutralization pattern and stereochemistry on DNA bending by methylphosphonate substitutions. Biochemistry 36:8692-8698. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki, Y., and K. Fukuda. 1990. Reduction of hexavalent chromium by ascorbic acid and glutathione with special reference to the rat lung. Arch. Toxicol. 64:169-176. [DOI] [PubMed] [Google Scholar]
- 52.Taioli, E., A. Zhitkovich, P. Kinney, I. Udasin, P. Toniolo, and M. Costa. 1995. Increased DNA-protein crosslinks in lymphocytes of residents living in chromium-contaminated areas. Biol. Trace Elem. Res. 50:175-180. [DOI] [PubMed] [Google Scholar]
- 53.Tsao, J. L., Y. Yatabe, R. Salovaara, H. J. Jarvinen, J. P. Mecklin, L. A. Aaltonen, S. Tavare, and D. Shibata. 2000. Genetic reconstruction of individual colorectal tumor histories. Proc. Natl. Acad. Sci. USA 97:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsou, T. C., H. J. Lai, and J. L. Yang. 1999. Effects of mannitol or catalase on the generation of reactive oxygen species leading to DNA damage by chromium(VI) reduction with ascorbate. Chem. Res. Toxicol. 12:1002-1009. [DOI] [PubMed] [Google Scholar]
- 55.Voitkun, V., A. Zhitkovich, and M. Costa. 1998. Cr(III)-mediated crosslinks of glutathione or amino acids to the DNA phosphate backbone are mutagenic in human cells. Nucleic Acids Res. 26:2024-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, S., and X. Shi. 2001. Mechanisms of Cr(VI)-induced p53 activation: the role of phosphorylation, mdm2 and ERK. Carcinogenesis 22:757-762. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Y., and J. Qin. 2003. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. USA 100:15387-15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe, Y., A. Haugen-Strano, A. Umar, K. Yamada, H. Hemmi, Y. Kikuchi, S. Takano, Y. Shibata, J. C. Barrett, T. A. Kunkel, and M. Koi. 2000. Complementation of an hMSH2 defect in human colorectal carcinoma cells by human chromosome 2 transfer. Mol. Carcinog. 29:37-49. [PubMed] [Google Scholar]
- 60.Wheeler, J. M., W. F. Bodmer, and N. J. Mortensen. 2000. DNA mismatch repair genes and colorectal cancer. Gut 47:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, G., S. J. Scherer, S. S. Shell, K. Yang, M. Kim, M. Lipkin, R. Kucherlapati, R. D. Kolodner, and W. Edelmann. 2004. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell 6:139-150. [DOI] [PubMed] [Google Scholar]
- 62.Ye, J., S. Wang, S. S. Leonard, Y. Sun, L. Butterworth, J. Antonini, M. Ding, Y. Rojanasakul, V. Vallyathan, V. Castranova, and X. Shi. 1999. Role of reactive oxygen species and p53 in chromium(VI)-induced apoptosis. J. Biol. Chem. 274:34974-34980. [DOI] [PubMed] [Google Scholar]
- 63.Zhitkovich, A., Y. Song, G. Quievryn, and V. Voitkun. 2001. Non-oxidative mechanisms are responsible for the induction of mutagenesis by reduction of Cr(VI) with cysteine: role of ternary DNA adducts in Cr(III)-dependent mutagenesis. Biochemistry 40:549-560. [DOI] [PubMed] [Google Scholar]
- 64.Zhitkovich, A., V. Voitkun, and M. Costa. 1996. Formation of the amino acid-DNA complexes by hexavalent and trivalent chromium in vitro: importance of trivalent chromium and the phosphate group. Biochemistry 35:7275-7282. [DOI] [PubMed] [Google Scholar]
- 65.Zhitkovich, A., V. Voitkun, and M. Costa. 1995. Glutathione and free amino acids form stable complexes with DNA following exposure of intact mammalian cells to chromate. Carcinogenesis 16:907-913. [DOI] [PubMed] [Google Scholar]