Abstract
Type II DNA topoisomerases have been classified into two families, Topo IIA and Topo IIB, based on structural and mechanistic dissimilarities. Topo IIA is the target of many important antibiotics and antitumoural drugs, most of them being inactive on Topo IIB. The effects and mode of action of Topo IIA inhibitors in vitro and in vivo have been extensively studied for the last twenty-five years. In contrast, studies of Topo IIB inhibitors were lacking. To document this field, we have studied two Hsp90 inhibitors (radicicol and geldanamycin), known to interact with the ATP-binding site of Hsp90 (the Bergerat fold), which is also present in Topo IIB. Here, we report that radicicol inhibits the decatenation and relaxation activities of Sulfolobus shibatae DNA topoisomerase VI (a Topo IIB) while geldanamycin does not. In addition, radicicol has no effect on the Topo IIA Escherichia coli DNA gyrase. In agreement with their different effects on DNA topoisomerase VI, we found that radicicol can theoretically fit in the ATP-binding pocket of the DNA topoisomerase VI ‘Bergerat fold’, whereas geldanamycin cannot. Radicicol inhibited growths of Sulfolobus acidocaldarius (a crenarchaeon) and of Haloferax volcanii (a euryarchaeon) at the same doses that inhibited DNA topoisomerase VI in vitro. In contrast, the bacteria E.coli was resistant to this drug. Radicicol thus appears to be a very promising compound to study the mechanism of Topo IIB in vitro, as well as the biological roles of these enzymes in vivo.
INTRODUCTION
Type II DNA topoisomerases (Topo II) are ubiquitous enzymes that catalyse the ATP-dependent crossing of two DNA duplexes through each other via transient double-strand breaks (DSBs) (1,2). They are implicated in major biological processes such as replication, recombination and transcription. In particular, the decatenation activity of Topo II is essential in all organisms to separate daughter chromosomes at the end of one replication round. In bacteria and archaea, Topo II are also the only enzymes that can remove positive superturns induced by the movement of RNA and DNA polymerases, since prokaryotic type I DNA topoisomerases (Topo I) can only relax negative superturns.
Topo II has been classified into two evolutionarily distinct protein families: Topo IIA and Topo IIB [(3), for review see (4)]. The Topo IIA family is represented by the ‘classical’ eukaryotic Topo II, the bacterial DNA gyrase (also present in some archaea), the DNA topoisomerase IV and several viral DNA topoisomerases. The prototype of the Topo IIB family is the archaeal DNA topoisomerase VI. Most archaea only contain DNA topoisomerase VI, but a few of them also contain a bacterial-like DNA gyrase (4). Furthermore recently, putative DNA topoisomerase VI turned out to be also present in plants beside the classical Topo IIA (5,6).
The two families of Topo II resemble each other in terms of enzymatic activities and organization of their functional domains. In both cases, an ATP-binding module, which triggers conformational change in the protein, is associated with a DNA nicking-closing module that controls the formation of transient DSBs. Both Topo IIA and Topo IIB use a tyrosine to cleave DNA, leading to the formation of a transient 5′ phospho-tyrosine linkage which is used to store the energy required for the subsequent ligation step. The ATP-binding modules of Topo IIA and IIB are homologues, since they exhibit both sequence similarities in their N-terminal regions and a common three-dimensional structure known as the ‘Bergerat fold’ (2,7–9). In contrast, the nicking-closing modules of Topo IIA and Topo IIB are non-homologous, being structurally dissimilar (10). Furthermore, the production of DSB by Topo IIA is ATP independent and generates single-stranded extension of 4 bp, whereas DSB formation by Topo IIB is ATP-dependent and generates single-stranded extension of 2 bp (11). These data indicate that the type II DNA topoisomerase activity has been invented twice in the course of evolution by the independent recruitment of two different nicking-closing modules to work with the same type of ATP-binding module (4).
Topo IIA have been studied extensively, in particular because they are important cellular targets for anti-cancer agents and antibiotics [for recent reviews, see (12–14)]. Two main classes of drugs have been described that affect different steps of the catalytic cycle: drugs of the first class stabilizes the covalent complexes formed between the DNA and the Topo II linked to the DSB (known as the cleavable complex) converting the enzyme into a physiological poison and drugs of the second class, sometimes referred as ‘catalytic inhibitors’, which are those acting at any other steps of the cycle. Most antitumoural drugs (such as epidophylotoxins, ellipticins and anthracyclins) as well as antibiotics of the quinolone and fluoroquinolone families (like nalidixic acid or perfloxacin) belong to the first class, whereas a few antitumoural drugs and antibiotics of the coumarin family belong to the second one. These inhibitors might either interfere with the binding between DNA and the enzyme (i.e. the anthracyclin aclarubicin), stabilize the non-covalent complex between the DNA and the Topo II (i.e. merbarone, ICRF-193 and its bisdioxopiperazine derivatives) or else inhibit ATP binding (coumarins). In parallel with structural studies, the variety of inhibitors available to study Topo IIA has been essential to describe in detail the catalytic cycle of these fascinating enzymes and to analyse their numerous cellular functions.
In contrast to the wealth of studies performed with Topo IIA inhibitors, very little information is available on inhibitors of Topo IIB. We have previously shown that the unknotting activity of Sulfolobus shibatae DNA topoisomerase VI was inhibited by several antitumoural drugs known to be DNA intercalants (ellipticin, m-AMSA, donorubicin and doxorubicin) and by VP16, a DNA topoisomerase II poison which inhibits the resealing of DNA breaks created by the enzyme, at concentrations similar to those used to inhibit eukaryotic Topo IIA (15). In contrast, S.shibatae DNA topoisomerase VI was not sensitive to compounds which have no DNA-binding properties, such the bacterial Topo IIA inhibitors (novobiocin, coumermycin and nalidixic acid).
In order to look for new drugs active against Topo IIB, we have tested the effect of two inhibitors of the heat-shock protein Hsp90, radicicol and geldanamycin, on S.shibatae DNA topoisomerase VI. These two drugs are known to interact with the Bergerat fold of Hsp90 (16), suggesting that they could also interact with the Bergerat fold of DNA topoisomerase VI. We also tested the effect of radicicol and geldanamycin on the growth of the archaea, Haloferax volcanii and Sulfolobus acidocaldarius, which are representatives of the two known archaeal phyla, euryarchaeota and crenarchaeota, respectively. Finally, we performed an in silico analysis of the complexes between radicicol, geldanamycin and the archaeal DNA topoisomerase VI. Our results show that radicicol, but not geldanamycin, inhibits the archaeal DNA topoisomerase VI in vitro and the archaeal growth in vivo. Radicicol thus appears to be a very promising compound to study the mechanism of Topo IIB in vitro, as well as the biological roles of these enzymes in vivo.
MATERIALS AND METHODS
Drugs were purchased from Sigma Aldrich. Concentrated stock solutions (100 mM) were prepared in dimethyl sulfoxide (DMSO), except novobiocin (H20). Stock solutions were aliquoted and stored at −20°C in the dark. For in vitro tests, the drugs were diluted in DMSO. S.shibatae DNA topoisomerase VI was purified as a heterotetramer after co-expression and overproduction of the two subunits, Top6A and Top6B in Escherichia coli, as previously described (11). E.coli DNA gyrase was purchased from TopoGEN. The enzymes were tested using as substrates kDNA for decatenation assay, negatively supercoiled pBR322 plasmids for relaxation assay, and relaxed pBR322 plasmids for supercoiling assay. kDNA and plasmids were purchased from Promega, TopoGEN or invitrogen.
In vitro enzymatic assays
DNA topoisomerase VI assays
The enzyme activities were carried out in a final volume of 20 μl containing 35 mM HEPES (pH 7.5), 40 mM KCl, 10 mM MgCl2, 0.5 mM ATP, 2 mM DTT, 1 mM spermidine, 0.1 mM EDTA and either 0.2 μg of kDNA (for decatenation assays) or 0.2 μg of pBR322 plasmids (for relaxation and supercoiling assays). Reactions were incubated with 2 U of enzyme for 4 or 6 min at 74°C (1 U of enzyme being defined as the amount of enzyme required to completely decatenate 0.2 μg of kDNA in 6 min at 74°C or relaxed 0.2 μg of pBR322 in 4 min at 74°C) and with various concentrations of drugs dissolved in DMSO (or H2O for novobiocin), ranging from 25 to 1000 μM. The reactions were terminated by cooling to 0°C, and immediately after the addition of 0.1 volume of loading dye (50% glycerol and 0.025% bromophenol blue). Samples were loaded and run at 35 mV (for relaxation assays) or 50 mV (for decatenation assays) directly onto a 1% agarose gel with or without ethidium bromide (EtBr). Gels were stained with 0.5 μg/ml of EtBr for 20 min and photographed. The stability of the drugs at the DNA topoisomerase VI incubation temperature (74°C) were tested by preincubation of these drugs during 2–30 min.
The kDNA assay was done using a catenated DNA substrate prepared from the kinetoplast of the insect trypanosome Crithidia fasciculata. kDNA is an aggregate of interlocked DNA minicircles (mostly 2.5 kb) that form extremely large networks of high molecular weight. As a result, these networks fail to enter an agarose gel. Upon incubation with a type II DNA topoisomerase, which engages DNA in a double-stranded breaking and reunion cycle, minicircular DNAs are released (decatenated). The decatenated minicircles move rapidly into the gel owing to their small size.
DNA gyrase assays
Assays with DNA gyrase were carried out at 37°C in the same way as DNA topoisomerase VI, but with its specific buffer and for 30 min (see TopoGEN protocols: http://www.topogen.com).
In vivo drugs treatments
S.acidocaldarius (strain DSM639), H.volcanii (strain DS2) and E.coli K12 C600 were grown in liquid shaken cultures (200 r.p.m.) at 78, 45 and 37°C, respectively. The growth media were as described by Lopez-Garcia and Forterre (17) for S.acidocaldarius, by Allers et al. (18) for H.volcanii (medium AHv-YPC). LB classical medium was used for E.coli. To test the effect of drugs on cell growth, stationary phase cultures were diluted into fresh medium and grown up to an optical density in the range of 0.05–0.2 OD. The cultures were then divided into several samples and supplemented with drugs at various concentrations. The growth of the three strains was insensitive to DMSO at the concentrations used.
The different cultures were cultivated up to 24 h in the presence of drugs and regularly sampled for absorbance monitoring (at 600 nm) and microscopic examination (with a Nikon eclipse E600 microscope ×100). Control cultures without drugs were grown in parallel under the same conditions. To test the reversibility of the drug effects, some 24 h cultures were centrifuged at 10 000 r.p.m. in a Sorvall centrifuge, the pellets were washed with fresh media, resuspended in the initial volume of growth medium, and grown for an additional 24 h. To test the stability of the radicicol at the condition employed for Sulfolobus growth, five flasks with 10 ml of Sulfolobus culture medium were incubated at 74°C, one for control without drug, and the four others with 100 μM of radicicol. The Sulfolobus cells (1 ml at optical density of 0.62) were added at time 0, 2, 4 or 6 h after radicicol. The absorbance of the different cultures after the inoculation was checked every 2 h for growth curves.
RESULTS
In vitro assays
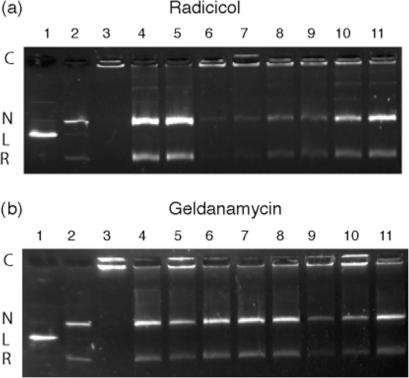
We first tested the effect of various concentrations of radicicol and geldanamycin on the decatenation activity of S.shibatae DNA topoisomerase VI. As shown in Figure 1a, DNA topoisomerase VI was strongly inhibited by radicicol in a dose-dependent manner. Some inhibition was visible above 50–100 μM of drug and nearly complete inhibition was observed at radicicol concentrations of 200 μM. In contrast, geldanamycin was inactive in the decatenation assay at concentrations up to 250 μM (Figure 1b). Radicicol also inhibited strongly relaxation by S.shibatae DNA topoisomerase VI in a dose-dependent manner and in the same range of concentrations. As shown in Figure 2, inhibition of relaxation was already clearly visible at 50–100 μM and complete at 125 μM. To be sure that radicicol did not induce the cleavable complex, we have also performed the classical experiment used to reveal such complex (addition of SDS and proteinase K before electrophoresis). No linear form appeared after such treatment (data not shown). As for decatenation, relaxation by S.shibatae DNA topoisomerase VI was not sensitive to geldanamycin (data not shown).
Figure 1.
Agarose gel electrophoresis showing the effect of radicicol (a) and geldanamycin (b) on decatenation of kDNA by the DNA topoisomerase VI. Lane 1, linear kDNA marker (L); lane 2, decatenated kDNA marker: nicked, open circular (N) and relaxed DNA (R); lane 3, catenated kDNA (C); lane 4, kDNA with Topo VI; lane 5, kDNA with Topo VI and DMSO; and lanes 6–11, kDNA with Topo VI in the presence of radicicol or geldanamycin (250, 200, 125, 100, 50 and 25 μM, respectively).
Figure 2.
Agarose gel electrophoresis showing the effect of radicicol on relaxation of the plasmid pBR322. Lane 1, supercoiled pBR322; lane 2, supercoiled pBR322 in the presence of radicicol (250 μM); lane 3, supercoiled pBR322 with Topo VI; lane 4, supercoiled pBR322 with Topo VI and DMSO; and lanes 5–10, supercoiled pBR322 with Topo VI in the presence of radicicol (25, 50, 100, 125, 200 and 250 μM, respectively). SC: negatively supercoiled pBR322 DNA; R: relaxed DNA; OC: open circular (nicked) pBR322 DNA.
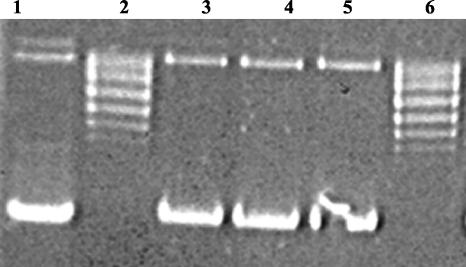
Both Topo IIA and Topo IIB bear a Bergerat fold, so radicicol could have been a general inhibitor of type II DNA topoisomerases. To check this possibility, we tested the effect of radicicol on the supercoiling activity of E.coli DNA gyrase (a Topo IIA). As shown in Figure 3, supercoiling of E.coli DNA gyrase was not inhibited by radicicol (up to 500 μM), indicating that radicicol exhibits specificity towards different type II DNA topoisomerases. Novobiocin was used as a positive control against gyrase activity.
Figure 3.
Agarose gel electrophoresis showing the effect of radicicol on supercoiling of the plasmid pBR322 by the DNA gyrase. Lane 1, control pBR322 negatively supercoiled; lane 2, relaxed pBR322 and 500 μM radicicol without gyrase; lane 3, relaxed pBR322 with gyrase; lanes 4 and 5, relaxed pBR322 with gyrase in the presence of 100 or 500 μM radicicol; and lane 6, relaxed pBR322 with gyrase in the presence of 100 μM novobiocin.
In vivo assays
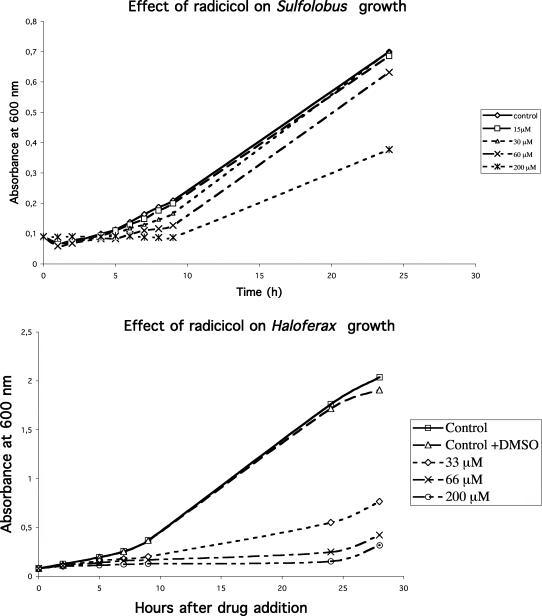
We have tested the effect of radicicol and geldanamycin in vivo on the growth of two archaea harbouring a DNA topoisomerase VI, the crenarchaeon S.acidocaldarius (an extreme thermophile and acidophile) and the euryarchaeon H.volcanii (an extreme halophile). As illustrated in Figure 4, both Archaea were sensitive to radicicol in the same concentration range than Sulfolobus DNA topoisomerase VI in vitro. The MIC for radicicol in vivo with Haloferax was below 12 μg/ml, and was between 36 and 73 with Sulfolobus. In agreement with in vitro data, the growth of both S.acidocaldarius and H.volcanii were in contrast fully resistant to geldanamycin (data not shown).
Figure 4.
In vivo inhibition of radicicol on Haloferax and Sulfolobus growths. Effect of different drug concentrations of radicicol measured by monitoring the cultures' absorbances at 600 nm.
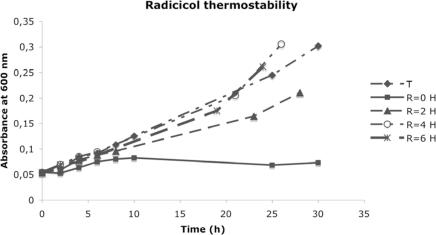
As shown in Figure 4, Sulfolobus growth resumed after 10 h of exposure to radicicol, suggesting that this drug was unstable at high temperature in the acidic medium used to cultivate Sulfolobus. Figure 5 shows that preincubation of this drug in the culture medium for >6 h completely suppressed the radicicol inhibition. In another experiment (data not shown), addition of a second radicicol dose (after centrifugation and resuspension of the culture), 2 or 4 h after the first one produced a complete arrest of Sulfolobus growth.
Figure 5.
Stability of radicicol at 74°C in the Sulfolobus medium. Five flasks with 10 ml of Sulfolobus culture medium were incubated at 74°C, one for control without drug (flask T), and the four others with 100 μM of radicicol (flasks R:0, R:2, R:4 and R:6). At time = 0 h, 1 ml of a Sulfolobus culture (OD of 0.62) was added to flasks T and R:0, at time = 2, 4 and 6 h, 1 ml of the same Sulfolobus culture (OD of 0.62) was added, respectively, to flasks R:2, 4 and 6.
In contrast, Haloferax growth only resumed after 25 h of exposure, indicating that this drug was more stable in the medium and culture conditions used to cultivate halophilic archaea.
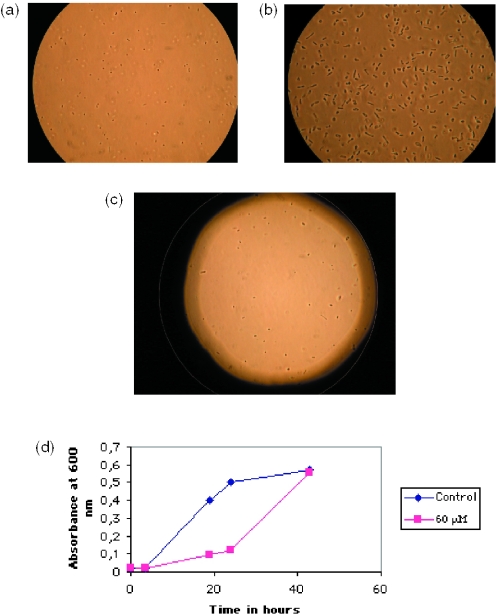
The morphology of S.acidocaldarius cells was not affected by the addition of radicicol (data not shown) whereas Haloferax cells morphology (Figure 6a) was drastically altered, with the appearance of large rod-shaped cells (Figure 6b). The effect of radicicol on H.volcanii was reversible, since growth resumed (Figure 6d) and cells regained their initial morphology after drug removal (Figure 6c).
Figure 6.
Effect of radicicol treatment (60 μM) on the morphology of H.volcanii. Microscopic observations ×100. (a) Cells before radicicol addition; (b) cells at time 24 h after radicicol addition (the culture has been concentrated by centrifugation for microscopic observation); (c) cells at time 24 h after removal of radicicol and resuspension; and (d) growth curves before and after removal of radicicol.
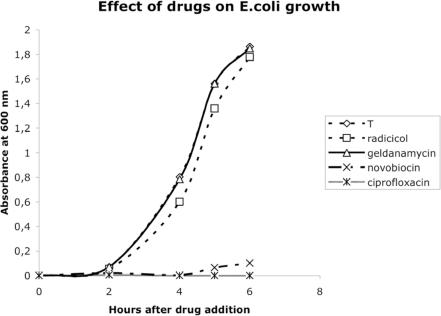
In order to identify possible alternative in vivo targets of radicicol in S.acidocaldarius and H.volcanii, we looked for all Bf proteins present in H.volcanii, Sulfolobus solfataricus and Sulfolobus tokodai by screening the complete set of proteins encoded by their genomes for conserved amino acid signatures specific for the Bergerat fold (data not shown). DNA topoisomerase VI turned out to be the only Bf protein that we could detect in the genomes of the two Sulfolobus, strengthening the idea that this enzyme is the intracellular target of radicicol in vivo in S.acidocaldarius. In contrast, we could detect 16 Bf proteins encoded by the H.volcanii genome, including DNA topoisomerase VI, DNA gyrase, 13 histidine kinases and a MutL homologue. As a control, we thus tested the effect of radicicol on the growth of E.coli which similarly encodes MutL and up to 29 histidines kinases (Table 1) but only contain members of the Topo IIA family. As shown in Figure 7, E.coli was not sensitive to radicicol and geldanamycin, but fully inhibited by two specific Topo IIA inhibitors, novobiocin and ciprofloxacin.
Table 1.
Potential targets of radicicol (containing the ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase: http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/index.html)
Figure 7.
Effect of different drugs on E.coli growth measured by monitoring the cultures' absorbances at 600 nm.
Modelization of radicicol–DNA topoisomerase VI complex
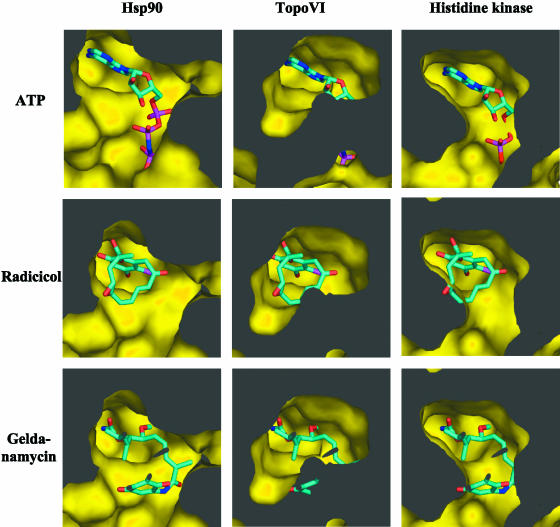
Both radicicol and geldanamycin inhibit Hsp90 by interacting with its Bergerat fold (16). Our data suggested that only radicicol could interact with the Bergerat fold of DNA topoisomerase VI or that interaction with geldanamycin did not produce inhibition. To get insight into this question, we have modelled the putative interactions of these two drugs and ATP with DNA topoisomerase VI, Hsp90 and the histidine kinase using coordinates obtained from the crystal structure of the B subunit of S.shibatae DNA topoisomerase VI [(9), PDB code:1MX0], the Saccharomyces cerevisiae Hsp90 radicicol complex [(16), PDB code: 1BGQ], and the Thermotoga maritima histidine kinase CheA [(19), PDB code: 1I59]. The superimposition of these structures (root mean square deviation of 1.5 Å) shows that radicicol can be fitted in the ATP-binding pocket of DNA topoisomerase VI without steric clashes. On the contrary, a similar model built from the structure of the geldanamycin-Hsp90 ATP-binding domain [(19), PDB code 1a4h], shows that geldanamycin is too large to be accommodated without drastic conformational changes in the ATP-binding pocket of DNA topoisomerase VI (Figure 8). As controls, we compare our models with the Hsp90-radicicol and geldanamycin complexes and model their interaction with the histidine kinase CheA (Figure 8). As previously shown (16), both radicicol and geldanamycin fit perfectly in the ATP-binding site of Hsp90. In contrast, radicicol could not fit in the ATP-binding site of CheA. The latter observation could explain why the presence of histidine kinase does not seem to be correlated to the sensitivity to radicicol. These in silico data could nicely explain the differences observed between the effects of radicicol and geldanamycin on archaeal DNA topoisomerase VI.
Figure 8.
Sliced surface representation of the S.shibatae DNA topoisomerase VI, S.cerivisiae Hsp90 and T.maritima histidine kinase ATP-binding sites complexed with ATP, radicicol and geldanamycin. The ligands are shown in sticks. Opaque regions correspond to the interior of the enzymes molecules.
DISCUSSION
Radicicol was isolated 50 years ago from the fungus Monospirium bonorden (20) and was first described as a tyrosine kinase inhibitor. Later on, it was shown to inhibit signal transduction of oncogene products and to exhibit potent in vitro antiproliferative activity against a wide variety of tumour cell lines (21). Finally, the actual intracellular target of radicicol in mammalian cells turned out to be the Hsp90 protein (16,22). Hsp90, which is one of the most abundant chaperones in eukaryotes, participates in folding and stabilization of signal-transducing molecules, including steroid hormone receptors and protein kinases. Radicicol induces destabilization of Hsp90-dependent client proteins by binding with high affinities to the N-terminal domain of the chaperone (16,22,23). This binding prevents the correct maturation of the client proteins, leading to their degradation by the proteasome.
The three-dimensional structure of the complex between radicicol and Hsp90 has shown that the drug specifically binds to the ATP-binding site of Hsp90 (24). This site corresponds to a recently described ATP-binding module known as the Bergerat fold (8). This fold is also present in the B subunits of type II DNA topoisomerases (both Topo IIA and Topo IIB), in the mismatch repair protein MutL and in various histidine and serine kinases (2,8,25,26). The specific binding of radicicol and geldanamycin to the Hsp90 Bergerat fold suggested that other proteins with a Bergerat fold (Bf proteins) could be sensitive to these drugs. This turned out to be the case for branched-chain alpha-keto acid dehydrogenase kinase (BCKDHK) and Sln1 yeast histidine kinase (27).
Here, we have shown that radicicol inhibits in vitro the activity of another Bf protein, S.shibatae DNA topoisomerase VI (the prototype of the Topo IIB family). In contrast, radicicol has no effect on a Bf protein representative of the Topo IIA family, E.coli DNA gyrase. The resistance of bacterial DNA gyrase to radicicol indicate that this drug can target specifically some families of Bf proteins, since bacterial Topo IIA (both DNA gyrase and Topo IV) also bear a Bergerat fold. This is reminiscent of data previously obtained with another Bf interacting drug, novobiocin (28,29), since this classical inhibitor of Topo IIA has no effect on S.shibatae DNA topoisomerase VI (15).
In agreement with our in vitro observations, we found that radicicol inhibits the growth of two archaea that contain DNA topoisomerase VI, S.acidocaldarius and H.volcanii but has no effect on E.coli that only contains Topo IIA (DNA gyrase and DNA topoisomerase IV). Although the definitive proof will require genetic experiments, the good correlation between our in vitro and in vivo data strongly suggests that archaeal DNA topoisomerase VI are the targets of radicicol in vivo. In agreement with this conclusion, the inhibition of H.volcanii and S.acidocaldarius by radicicol in vivo occurs at doses that are in the concentration range of those effective on S.shibatae Topo VI in vitro. Furthermore, DNA topoisomerase VI is the only Bf protein that can be detected in the genome of Sulfolobus.
In the case of H.volcanii, one cannot formally exclude that radicicol inhibits in vivo other Bf proteins encoded by the genome of this archaeon (either MutL and/or histidine kinases) but our modelization prediction with the histidine kinase CheA suggests that this enzyme should not be inhibited by radicicol, strengthens our point that Topo VI is also probably the target of radicicol in Haloferax.
The inhibition of both S.acidocaldarius and H.volcanii by radicicol suggests that this drug is a general inhibitor for archaeal DNA topoisomerase VI and possibly for their homologues in plants.
In contrast to radicicol, the drug geldanamycin, another Hsp90 inhibitor that also interacts with the Bergerat fold, has no effect on the activity of S.shibatae DNA topoisomerase VI in vitro and did not inhibit the growth of the two archaea tested. This result fits well with modelling experiments showing that radicicol can be located in the ATP-binding pocket located in the Bergerat fold of DNA topoisomerase VI, whereas geldanamycin cannot (Figure 8). This favours the idea that, as in the case of Hsp90, radicicol inhibits DNA topoisomerase VI by interacting with the Bergerat fold of its B subunit.
If we assume that the in vivo target of radicicol in S.acidocaldarius and H.volcanii is DNA topoisomerase VI, our in vivo results point to an essential role for this enzyme in these two archaea. This is not surprising in the case of S.acidocaldarius, since DNA topoisomerase VI is the only type II DNA topoisomerase present in the two Sulfolobus whose genomes have been completely sequenced. Furthermore, the three other DNA topoisomerases (type I) present in Sulfolobus species (two reverse gyrases and a relaxing enzyme) belong to a superfamily (Topo IA) whose members cannot relax positive supercoils and cannot decatenate intact DNA duplexes. Accordingly, DNA topoisomerase VI should be essential in Sulfolobus both for the decatenation of daughter chromosomes at the end of one DNA replication round, and to remove positive supercoils that would otherwise accumulate in front of DNA replication forks and transcribing RNA polymerases.
In the case of H.volcanii, the inhibition by radicicol was not predictable, since this archaeon also contains a DNA gyrase that could theoretically replace the missing activities of DNA topoisomerase VI in radicicol-treated cells. It was already known that DNA gyrase is essential for halophilic archaea, since their growth is inhibited by novobiocin (30,31). The inhibition of Haloarchaea by radicicol reported here thus indicates that both DNA gyrase and DNA topoisomerase VI are essential in these archaea. These two enzymes have possibly specialized roles, as in the case of DNA gyrase and DNA topoisomerase IV in bacteria. DNA gyrase is probably essential in Haloarchaea because some critical steps in DNA function, such as initiation of DNA replication and/or transcription at some promoters, are dependent on negative supercoiling. Indeed, negatively supercoiled plasmid of Halobacterium GRB became positively supercoiled after novobiocin treatment in vivo (31). It is not so clear why DNA topoisomerase VI is essential in H.volcanii. One possibility is that DNA topoisomerase VI specifically interacts with other proteins for proper chromosome segregation and cell division. This would be reminiscent of the specific interaction of DNA topoisomerase IV with the cell division protein FtsK in E.coli (32). A strong involvement of DNA topoisomerase VI in cell division would explain why addition of radicicol to cultures of H.volcanii triggers cell filamentation (Figure 6).
We anticipate that radicicol will be a very useful tool to further study the mechanism of action of DNA topoisomerase VI in vitro, and to analyse its role in vivo in archaea. An appealing property of radicicol is its ability to discriminate between Topo IIA and Topo IIB. The discovery of a new archaeal inhibitor should also help for the development of the genetic tools in archaea by providing the possibility to design selectable markers based on the top6B gene. The discovery that radicicol inhibits DNA topoisomerase VI can also have implication for studies in eukaryotic systems. For instance, DNA topoisomerase VI appears to be essential for endo-reduplication in plant, a process that triggers cell polyploidy and controls cellular and plant size (33,34). It has already been shown that addition of radicicol has a profound effect on plant morphology and this effect has been attributed to Hsp90 (33). However, some of the observed effects could have been also mediated via DNA topoisomerase VI inhibition. Finally, our experimental observations combined with modelization confirm that distinct families of Bf proteins can be sensitive to radicicol while some others cannot.
Acknowledgments
The work on archaeal topoisomerase in our laboratory is supported by the Association pour la Recherche sur la Cancer (ARC) and M.G. is a fellowship of ARC. Funding to pay the Open Access publication charges for this article was provided by the Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J.J. DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. Review. [DOI] [PubMed] [Google Scholar]
- 3.Bergerat A., de Massy B., Gadelle D., Varoutas P.C., Nicolas A., Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 4.Gadelle D., Filee J., Buhler C., Forterre P. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003;25:232–242. doi: 10.1002/bies.10245. [DOI] [PubMed] [Google Scholar]
- 5.Hartung F., Puchta H. Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene. 2001;271:81–86. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- 6.Yin Y., Cheong H., Friedrichsen D., Zhao Y., Hu J., Mora-Garcia S., Chory J.A. Crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc. Natl Acad. Sci., USA. 2002;99:10191–10196. doi: 10.1073/pnas.152337599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mushegian A.R., Bassett D.E., Jr, Boguski M.S., Bork P., Koonin E.V. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc. Natl Acad. Sci., USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutta R., Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 9.Corbett K.D., Berger J.M. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 2003;22:151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols M.D., DeAngelis K., Keck J.L., Berger J.M. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buhler C., Lebbink J.H., Bocs C., Ladenstein R., Forterre P. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J. Biol. Chem. 2001;276:37215–37222. doi: 10.1074/jbc.M101823200. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell A. DNA gyrase as a drug target. Trends Microbiol. 1997;5:102–109. doi: 10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 13.Andoh T., Ishida R. Catalytic inhibitors of DNA topoisomerase II. Biochim. Biophys. Acta. 1998;1400:155–171. doi: 10.1016/s0167-4781(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 14.Larsen A.K., Escargueil A.E., Skladanowski A. Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol. Ther. 2003;99:167–181. doi: 10.1016/s0163-7258(03)00058-5. [DOI] [PubMed] [Google Scholar]
- 15.Bergerat A., Gadelle D., Forterre P. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 1994;269:27663–27669. [PubMed] [Google Scholar]
- 16.Roe S.M., Prodromou C., O'Brien R., Ladbury J.E., Piper P.W., Pearl L.H. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Garcia P., Forterre P. DNA topology in hyperthermophilic archaea: reference states and their variation with growth phase, growth temperature, and temperature stresses. Mol. Microbiol. 1997;23:1267–1279. doi: 10.1046/j.1365-2958.1997.3051668.x. [DOI] [PubMed] [Google Scholar]
- 18.Allers T., Ngo H., Mevarech M., Lloyd R.G. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilwes A.M., Quezada C.M., Croal L.R., Crane B.R., Simon M.I. Nucleotide binding by the histidine kinase CheA. Nature Struct. Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- 20.Delmotte P., Delmotte-Plaque J. A new antifungal substance of fungal origin. Nature. 1953;171:344. doi: 10.1038/171344a0. [DOI] [PubMed] [Google Scholar]
- 21.Soga S., Neckers L.M., Schulte T.W., Shiotsu Y., Akasaka K., Narumi H., Agatsuma T., Ikuina Y., Murakata C., Tamaoki T., Akinaga S. KF25706, a novel oxime derivative of radicicol, exhibits in vivo antitumor activity via selective depletion of Hsp90 binding signaling molecules. Cancer Res. 1999;59:2931–2938. [PubMed] [Google Scholar]
- 22.Schulte T.W., Akinaga S., Soga S., Sullivan W., Stensgard B., Toft D., Neckers L.M. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones. 1998;3:100–108. doi: 10.1379/1466-1268(1998)003<0100:arbttn>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stebbins C.E., Russo A.A., Schneider C., Rosen N., Hartl F.U., Pavletich N.P. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 24.Prodromou C., Roe S.M., O'Brien R., Ladbury J.E., Piper P.W., Pearl L.H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 25.Yang W. Histidine kinases: extended relationships with GHL ATPases. In: Masayori Inouye, Rinku Dutta, editors. Histidine Kinases in Signal Transduction. New York: Academic Press; 2002. pp. 219–236. [Google Scholar]
- 26.Hu X., Machius M., Yang W. Monovalent cation dependence and preference of GHKL ATPases and kinases. FEBS Lett. 2003;544:268–273. doi: 10.1016/s0014-5793(03)00519-2. [DOI] [PubMed] [Google Scholar]
- 27.Besant P.G., Lasker M.V., Bui C.D., Turck C.W. Inhibition of branched-chain alpha-keto acid dehydrogenase kinase and Sln1 yeast histidine kinase by the antifungal antibiotic radicicol. Mol. Pharmacol. 2002;62:289–296. doi: 10.1124/mol.62.2.289. [DOI] [PubMed] [Google Scholar]
- 28.Lamour V., Hoermann L., Jeltsch J.M., Oudet P., Moras D. An open conformation of the Thermus thermophilus gyrase B ATP-binding domain. J. Biol. Chem. 2002;277:18947–18953. doi: 10.1074/jbc.M111740200. [DOI] [PubMed] [Google Scholar]
- 29.Lewis R.J., Singh O.M., Smith C.V., Skarzynski T., Maxwell A., Wonacott A.J., Wigley D.B. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 1996;15:1412–1420. [PMC free article] [PubMed] [Google Scholar]
- 30.Sioud M., Baldacci G., de Recondo A.M., Forterre P. Inhibitors of DNA topoisomerase II induce topological changes in an archaebacterial plasmid in vivo. Biochem. Pharmacol. 1988;37:1879–1880. doi: 10.1016/0006-2952(88)90492-3. [DOI] [PubMed] [Google Scholar]
- 31.Holmes M.L., Dyall-Smith M.L. Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. J. Bacteriol. 1991;173:642–648. doi: 10.1128/jb.173.2.642-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espeli O., Lee C., Marians K.J. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 2003;278:44639–44644. doi: 10.1074/jbc.M308926200. [DOI] [PubMed] [Google Scholar]
- 33.Hartung F., Angelis K.J., Meister A., Schubert I., Melzer M., Puchta H. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 2002;12:1787–1791. doi: 10.1016/s0960-9822(02)01218-6. [DOI] [PubMed] [Google Scholar]
- 34.Sugimoto-Shirasu K., Stacey N.J., Corsar J., Roberts K., McCann M.C. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 2002;12:1782–1786. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]