Abstract
Background:
Evidence is accumulating that growth hormone (GH) protects against the development of steatosis and progression of non-alcoholic fatty liver disease (NAFLD). GH may control steatosis indirectly by altering systemic insulin sensitivity and substrate delivery to the liver and/or by the direct actions of GH on hepatocyte function.
Approach:
To better define the hepatocyte-specific role of GH receptor (GHR) signaling on regulating steatosis, we used a mouse model with adult-onset, hepatocyte-specific GHR knockdown (aHepGHRkd). To prevent the reduction in circulating insulin-like growth factor 1 (IGF1) and the subsequent increase in GH observed after aHepGHRkd, subsets of aHepGHRkd mice were treated with adeno-associated viral vectors (AAV) driving hepatocyte-specific expression of IGF1 or a constitutively active form of STAT5b (STAT5bCA). The impact of hepatocyte-specific modulation of GHR, IGF1 and STAT5b on carbohydrate and lipid metabolism was studied across multiple nutritional states and in the context of hyperinsulinemic:euglycemic clamps.
Results:
Chow-fed male aHepGHRkd mice developed steatosis associated with an increase in hepatic glucokinase (GCK) and ketohexokinase (KHK) expression and de novo lipogenesis (DNL) rate, in the post-absorptive state and in response to refeeding after an overnight fast. The aHepGHRkd-associated increase in hepatic KHK, but not GCK and steatosis, was dependent on hepatocyte expression of carbohydrate response element binding protein (ChREBP), in re-fed mice. Interestingly, under clamp conditions, aHepGHRkd also increased the rate of DNL and expression of GCK and KHK, but impaired insulin-mediated suppression of hepatic glucose production, without altering plasma NEFA levels. These effects were normalized with AAV-mediated hepatocyte expression of IGF1 or STAT5bCA. Comparison of the impact of AAV-mediated hepatocyte IGF1 versus STAT5bCA in aHepGHRkd mice across multiple nutritional states, indicated the restorative actions of IGF1 are indirect, by improving systemic insulin sensitivity, independent of changes in the liver transcriptome. In contrast, the actions of STAT5b are due to the combined effects of raising IGF1 and direct alterations in the hepatocyte gene program that may involve suppression of BCL6 and FOXO1 activity. However, the direct and IGF1-dependent actions of STAT5b cannot fully account for enhanced GCK activity and lipogenic gene expression observed after aHepGHRkd, suggesting other GHR-mediated signals are involved.
Conclusion:
These studies demonstrate hepatocyte GHR-signaling controls hepatic glycolysis, DNL, steatosis and hepatic insulin sensitivity indirectly (via IGF1) and directly (via STAT5b). The relative contribution of these indirect and direct actions of GH on hepatocytes is modified by insulin and nutrient availability. These results improve our understanding of the physiologic actions of GH on regulating adult metabolism to protect against NAFLD progression.
Keywords: Growth hormone (GH), STAT5b, Insulin like growth factor 1 (IGF1), Liver, Glucokinase, de novo lipogenesis (DNL)
1. Introduction
Growth hormone (GH) is negatively associated with steatosis and the progression of non-alcoholic fatty liver disease (NAFLD). Patients with primary GH deficiency (GHD) have a higher prevalence of NAFLD, while those with elevated GH levels due to GH-producing pituitary tumors (acromegaly) have reduced liver fat [1]. Obesity is also associated with reduced GH levels, where up to 70 % of patients with obesity present with NAFLD [1–3]. In fact, patients with NAFLD demonstrate a reduced capacity to secrete GH in response to a GH-secretagogue challenge [1]. Low GH levels appear to contribute to excess hepatic fat accumulation based on studies showing GH treatment reduces steatosis in patients with GHD [1] and dramatically reduces hepatic triglyceride content in wild type mice made obese by high-fat diet feeding [4]. Both clinical and animal studies indicate that GH may protect against steatosis by acting directly on hepatocytes via GH receptor (GHR) signaling or indirectly via GH’s effects to augment whole body energy expenditure, alter other peripheral endocrine signals and/or change immune cell function [1,5]. Some of these non-hepatocyte effects may be due to GH-mediated insulin-like growth factor 1 (IGF1) [1–3]. However, high concentrations of GH can antagonize the actions of insulin leading to insulin resistance, which is also associated with NAFLD [5–8].
To tease apart the complex interplay between the systemic metabolic actions of GH and IGF1 from the direct actions of GH on hepatocyte metabolism, our laboratory has generated a mouse model of adult-onset, hepatocyte-specific, GHR knockdown (aHepGHRkd) [9]. Chow-fed, male aHepGHRkd mice develop steatosis within one week of GHR knockdown. Steatosis is associated with increased de novo lipogenesis (DNL), assessed by incorporation of deuterated water into newly formed fatty acids (FA) associated with triglycerides (TG). Of note, the elevated hepatic DNL was observed in the post-absorptive state, that was associated with increased expression of genes important for glycolysis and DNL [9,10]. The steatosis and increased rate of DNL observed in aHepGHRkd mice was not accompanied by glucose intolerance and could not be explained by altered hepatic VLDL release or by increases in white adipose tissue (WAT) lipolysis or hepatic FA uptake [9,10]. Interestingly, female aHepGHRkd mice were protected from steatosis and this protection was lost after ovariectomy and restored with estrogen replacement [9,11]. These changes in liver phenotype persist with age, independent of IGF1, with males developing mild non-alcoholic steatohepatitis, even when maintained on a standard chow diet [11].
The goal of the current study was to determine the key mechanisms driving DNL and steatosis after aHepGHRkd, and if these are regulated directly by GHR signaling in hepatocytes, or due to systemic alterations in metabolic function evoked by the loss of hepatocyte GHR expression. Short term studies were conducted to minimize compensatory mechanisms that may evolve over time that could mask the early events that drive alterations in liver metabolism after GHR loss. We previously demonstrated that aHepGHRkd dramatically reduces circulating IGF1 levels, which removes negative feedback from pituitary somatotrope and increases circulating GH levels [9,11]. Since this reciprocal shift in IGF1 and GH alters systemic metabolic function, which could indirectly impact liver metabolism via changes in systemic insulin sensitivity and production [5], we sought to maintain IGF1 expression in hepatocytes of aHepGHRkd mice by treatment with an adeno-associated viral vector driving expression of a rat IGF1 transgene under the control of a hepatocyte-specific, thyroxine-binding globulin promoter (AAV-TBGp-rIGF1; [11]). Many of the direct hepatocyte actions of GHR signaling are mediated by JAK2-dependent, STAT5b activation, including GHR-dependent IGF1 gene expression [12]. Therefore, to determine if GHR-mediated STAT5b is a key signal transduction pathway that alters liver metabolic function, aHepGHRkd mice were treated with an AAV-TBGp that expresses a constitutively active mouse STAT5b (STAT5bCA), a construct we recently showed can maintain STAT5b activity and IGF1 in the absence of hepatocyte GHR signaling [13]. Comparison of the relative impact of maintaining hepatocyte expression of IGF1 versus STAT5b activity in aHepGHRkd mice provides insight into the IGF1-dependent versus IGF1-independent actions of hepatocyte STAT5b on liver metabolism.
2. Materials and methods
2.1. Generation of aHepGHRkd mice without or with maintenance of hepatocyte IGF1 or STAT5b activity
Mouse studies were approved by the Institutional Animal Care and Use Committees of the Jesse Brown VA Medical Center, University of Illinois at Chicago and Vanderbilt University (hyperinsulinemic:euglycemic clamps). Mice were housed in a specific pathogen-free barrier facility maintained at 22–24 °C with a 12 h light-dark cycle (lights on 0600 h) on a standard chow diet (Teklad LM-485, Envigo, Madison, WI), with water provided ad libitum. As previously described [9,11], 10–12 week-old GHRfl/fl mice [14] received a single injection (lateral tail vein or retro-orbital venous plexus) of 1.5 × 1011 genome copies (GC) of AAV-TBGp expressing Cre recombinase (AAV8-TBGp-Cre; AAV.TBG.PI. Cre.rBG, Addgene, Watertown, MA). Subsets of mice were co-injected with AAV8-TBGp-rIGF1 (2.0 × 1011 GC; AAV8.TBG.PI.ratIGF1.WPRE. bGH; [11]) or AAV8-TBGp-STAT5bCA (0.75 × 1011 GC; AAV8.TBG.PI.N-FLAG-mSTAT5bCA.WPRE.bGH; [15]). Injection of an AAV8-TBGp-Null (pAAV8.TBG.PI.Null.bGH, Addgene) was used to generate GHR-intact control mice and to equalize the total amount of AAV8 injected in each mouse (total 3.5 × 1011 GC per mouse). Tissue (liver) and cell type (hepatocyte) specificity of the AAV8-TBGp vector was reported previously [9,16].
Consistent with the ARRIVE guidelines, within each nutritional state tested (post-absorptive, overnight fasting without or with 6 h refeeding), the GHRfl/fl mice used were born within a 2 weeks’ time frame and housed with littermates (3–4 mice/per cage within sex). Within cage, each mouse randomly received a different AAV. All mice within each cage were euthanized sequentially, and all mice within each experiment were euthanized within a 2 h window. Within each experiment, all samples of each tissue type (liver, pituitary, plasma) were processed at the same time, blinded to experimental group, and analytical assays used to assess the various endpoints described below.
2.2. Hormones and metabolic parameters
Plasma GH (MilliporeSigma, Burlington, MA), insulin (Mercodia, Uppsala, Sweden), IGF1 (ALPCO Diagnostics, Salem, NH), NEFA, 3-hydroxybutyrate and TG (FUJIFILM Wako Diagnostics, Mountain View, CA), and hepatic glycogen (BioVision, Milpitas, CA) were measured following the manufacturer’s instructions. Blood glucose was assessed by glucometer (Accu-Chek® Aviva Plus; Roche, Basel, Switzerland). Tissue-specific FA uptake was determined by incorporation of boron-dipyrromethene labeled palmitate (BODIPY-C16) [10]. The rate of hepatic VLDL-TG secretion was assessed after tyloxapol injection as previously described [9].
2.3. Lipids and DNL analysis
Hepatic neutral lipids were extracted as previously described [9,10], to determine hepatic TG content (FUJFILM Wako Diagnostics). Hepatic DNL was assessed using the deuterated water (2H2O; Sigma-Aldrich, Madison, WI) method, as measured by the 2H-labeled FA, and newly made TG were assessed by TG bound 2H-labeled glycerol as previously described [9] by the Metabolic Phenotyping Mass Spectrometry Core, Dr. Michelle Puchowicz Director (UTHSC).
2.4. Gene expression and protein analysis
Pituitary gland and liver gene expression was assessed by quantitative PCR (qPCR) after reverse transcription of RNA to cDNA, where Actb, Ppia and Hprt were used as reference genes to calculate a normalization factor, as previously reported [11,17]. The specific sequences of qPCR primers are provided in Table S1. For protein analysis, 50 μg of liver protein extract from each sample was separated by SDS-PAGE gels (Bio-Rad, Hercules, CA), transferred to nitrocellulose membranes, and incubated with the respective primary and secondary antibodies (Table S2), as previously described [11]. Blots were developed by ECL (Clarity ECL, Bio-Rad).
2.5. RNA-seq analysis
Total liver RNA was extracted using RNeasy Plus Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions. RNA-seq library preparation and sequencing were performed by Novogene Corporation Inc. (Sacramento, CA) as previously described [11]. Data analysis was performed using an in-house custom RNA-seq pipeline as described [15], with sequence reads aligned to mouse genome build mm9 (NCBI 37) using STAR aligner and FeatureCounts used to count sequence reads mapping to the union of exonic regions in all isoforms of a given gene. Counting was based on an mm9 Gene Transfer Format file comprised of 75,798 mouse genes, including 20,884 RefSeq protein coding genes, 48,360 mouse liver-expressed lncRNAs genes described elsewhere [18] and 6554 other non-coding RNA genes [15]. Raw sequencing files and processed data files are available at GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE222513. Differential gene expression across four comparisons (Kd/Control, rIGF1/Kd, STAT5bCA/Kd, and STAT5bCA/rIGF1) was analyzed using edgeR (ver. 3.28), which yielded a total of 1504 genes meeting the threshold values of |fold-change| > 2 and false discovery rate < 0.05 in one or more of the four comparisons. Expression values (FPKM units, fragments per kilobase RNA per million sequence reads) for the 1504 genes were used to create a tSNE (t-distributed stochastic neighbor embedding) dimensionality reduction plot (Rtsne R package, ver. 0.15) with the following parameters: perplexity = 2, ndims = 3, theta = 0.5. The tSNE plot is a non-deterministic, unsupervised non-linear dimensionality reduction and data visualization technique that tries to preserve the local structure (cluster) of the data. Venn diagrams were used to compare the gene overlaps across the four comparisons. Pathway enrichment analysis was implemented using ClueGO from Cytoscape [19].
2.6. Metabolomic analysis
For metabolomic analysis, liver samples were homogenized at a ratio of 20 μL/mg tissue in 80 % cold methanol. Then, 200 μL of the homogenate were mixed with 800 μL of 80 % cold methanol and incubated overnight at − 80 °C. Samples were then centrifuged at 20,000g, for 30 min, at 4 °C. Supernatants were transferred to the Metabolomics Core Facility of the Northwestern University (Chicago, IL) and liquid chromatography-mass spectrometry was used to assess hydrophilic metabolites. Pellets were resuspended in 8 M urea buffer, quantified using Pierce™ BCA protein assay kit (Thermo Fisher Scientific) and total protein levels used to normalize metabolomic data. Results were analyzed using MetaboAnalyst 5.0 [20].
2.7. Hyperinsulinemic:euglycemic clamp
Mice were shipped to Vanderbilt University, Mouse Metabolic Phenotyping Center (Associate Director, Dr. Owen McGuiness). Mice were implanted with jugular vein and carotid artery catheters as previously described [21] and injected iv with AAVs 7d prior to clamp study. On the day of the clamp, food was withdrawn at 0600 h and 3 h later primed, then followed by a continuous infusion of [6,6-2H2]-glucose and 2H20, for 2 h. Ten minutes before starting the insulin clamp, a basal blood sample (t = −10) was drawn. At time t0 the constant [6,6-2H2]-glucose was stopped and a variable glucose infusion containing, [6,6-2H2]-glucose and 2H20 along with constant infusions of insulin (2 mU/kg/min) and saline-washed erythrocytes (5.5 μL/min) were initiated. Blood was drawn at t80, t90, t110, t120 and t145 min after the start of the insulin infusion. At t120 min, 2-deoxy-[14C]-glucose bolus was given and arterial blood samples were taken over the subsequent 25 min period and mice were then sacrificed under pentobarbital anesthesia. Blood and tissues were collected, weighed and snap-frozen. Assessment of isotope labeled glucose metabolites was as previously reported [22]. Newly formed TG, with newly formed palmitate (DNL) was assessed in liver samples, as well in plasma samples taken at t-10 and t145. Plasma NEFA was assessed in lipase-inhibitor treated samples at t10 and t145.
2.8. Statistical analysis
Except for RNA-seq and metabolomics data, values are represented as mean ± SEM and individual data points shown. Grubbs’ test was used to identify outliers, which were excluded from the analysis and graphs. One-way or two-way ANOVA was used, followed by Bonferroni post hoc test or Kruskal-Wallis followed by a Dunn’s multiple comparison test if the data set did not pass the normality test. Statistical analysis was performed using GraphPad Prism 8 (San Diego, CA). p-Values <0.05 were considered significant.
3. Results
3.1. In the post-absorptive state, maintenance of hepatocyte IGF1 expression or STAT5b activity prevents the aHepGHRkd-mediated increase in GH levels, but does not normalize hepatic steatosis, DNL rate, or glycolytic and lipogenic gene expression
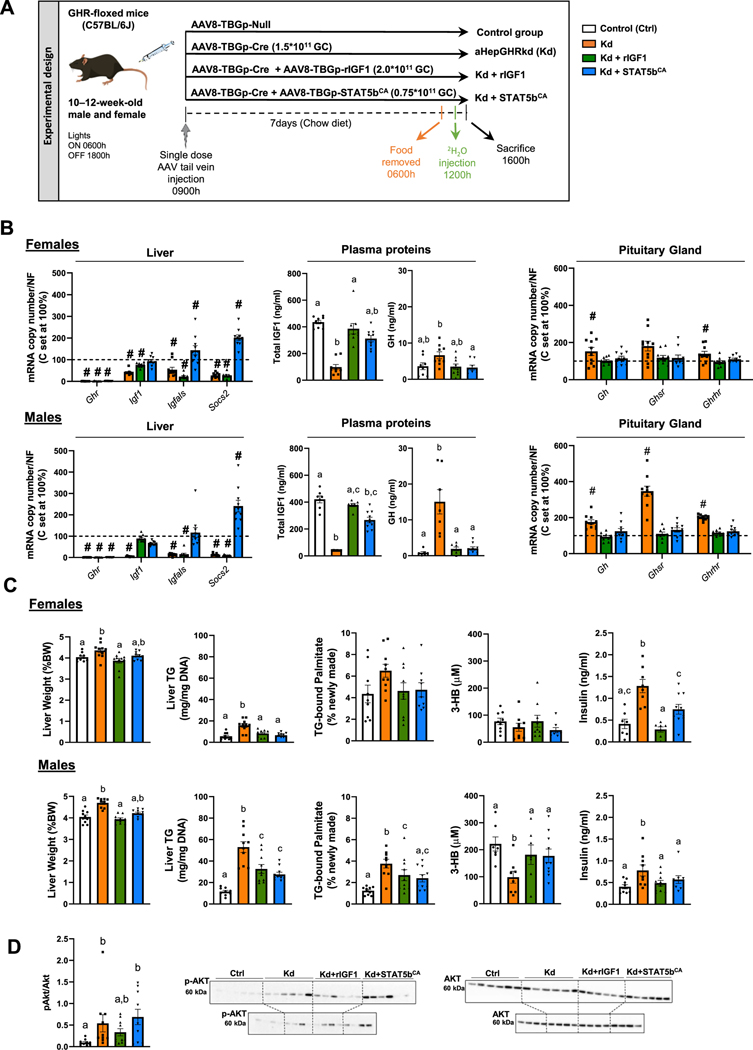
Schematic representation of the study design is illustrated in Fig. 1A. Loss of the hepatocyte GHR led to a reduction in hepatic Igf1 mRNA and circulating IGF1 levels, in both female and male mice (Fig. 1B). This was associated with an increase in pituitary Gh mRNA and circulating GH levels, as well as an increase in pituitary expression of GH stimulatory receptors, Ghsr and Ghrhr. When hepatocyte Igf1 expression was maintained in aHepGHRkd mice, plasma IGF1 levels were normalized. It should be noted that plasma IGF1 in this context would be considered largely in the free form (bioactive), due to the fact that GHR is also required to maintain the expression of the binding protein, IGF1 acid label subunit (Igfals) [23]. The increase in IGF1 normalized circulating GH levels and pituitary expression of Gh, Ghsr and Ghrhr. When hepatocyte-specific STAT5b activity was sustained in the absence of GHR, hepatic Igf1 and Igfals mRNA and circulating IGF1 levels were normalized, which was also sufficient to restore negative feedback to the pituitary, thereby reducing circulating GH levels. The activity of STAT5bCA was further confirmed by a significant increase in hepatic expression of Socs2, a direct STAT5b target that suppresses GHR signaling via JAK2 [24].
Fig. 1.
Impact of aHepGHRkd without and with IGF1 or STAT5bCA on GH-axis function and liver metabolic phenotype, assessed in the post-absorptive state. (A) Experimental design: 10 to 12-week-old GHR-floxed male and female mice were maintained on a standard chow diet and treated with a single dose of different types of AAVs to generate four experimental groups (n = 10 mice/group). Mice were studied in the post-absorptive state, 7 days after the AAV injection. Specifically, food was removed at 0600 h. At 1200 h, mice were injected with deuterated water and tissues and blood collected at 1600 h. (B) Female (top panel) and male (bottom panel), liver: Ghr, Igf1, Igfals and Socs2 mRNA expression levels, assessed by qPCR; plasma proteins: total IGF1 and GH assessed by ELISA; pituitary gland: Gh, Ghsr and Ghrhr mRNA expression levels, assessed by qPCR. #, indicates differences in liver and pituitary gene expression levels, compared to GHR-intact controls set at 100 %, p < 0.05. (C) Female (top panel) and male (bottom panel) relative liver weight, liver triglyceride (TG) content, plasma β-hydroxybutyrate (3-HB), percentage of hepatic newly formed palmitate and plasma insulin. (D) Male only hepatic pAKT/AKT protein quantification and western blot. It should be noted that two separate gels/blots were run (top panel n = 6/group and bottom panel n = 4/group) representing all biological replicates within each group (n = 10/group). Values are represented as mean ± SEM, with individual data points included. For plasma IGF1, GH and insulin levels and all other endpoints shown in panels C and D, values that do not share a common letter (a, b, c) statistically differ (p < 0.05).
In the postabsorptive state, liver weights were significantly increased in female and male aHepGHRkd mice, associated with an increase in liver triglyceride (TG) content, which was greater in males compared to females (Fig. 1C). In male (but not female) aHepGHRkd livers, the rate of hepatic DNL significantly increased, as indicated by an increase in the percent of newly formed palmitate (Fig. 1C), but not stearate or oleate (Fig. S1A). In addition, in male (but not female) aHepGHRkd mice, plasma β-hydroxybutyrate levels were significantly reduced (Fig. 1C). Taken together these results suggest enhanced DNL and reduced FA oxidation contribute to the steatosis observed in male aHepGHRkd mice.
Female mice are relatively protected from diet-induced steatosis, compared to males, when housed at standard temperatures (22–24 °C), but this protection is lost when mice are housed at thermoneutrality (30 °C), which reduces the requirement for basal energy expenditure to maintain core body temperature [25,26]. When aHepGHRkd mice were maintained at 30 °C, males developed steatosis, as well as elevated plasma TG (Fig. S2). However, females remained relatively protected (Fig. S2), suggesting alterations in basal energy expenditure do not play a major role in the sexually dimorphic impact of aHepGHRkd on steatosis.
Under standard housing temperatures (22–24 °C), maintaining hepatocyte-specific IGF1 expression or increasing STAT5b activity in aHepGHRkd male mice reduced, but did not normalize, hepatic TG and DNL, while plasma β-hydroxybutyrate was increased to the levels observed in GHR-intact controls (Fig. 1C). These changes were not associated with significant alterations in circulating glucose, non-esterified FA (NEFA) or TG levels (Table S3), or differences in hepatic VLDL release (Fig. S1B). However, aHepGHRkd increased insulin levels in both female and male mice, which were reduced by both IGF1 and STAT5bCA (Fig. 1C). In males, IGF1, but not STAT5bCA, fully normalized insulin levels and this pattern of insulin was reflected by hepatic pAKT levels (Fig. 1D). Although elevated insulin levels may contribute to the increased steatosis and DNL observed in aHepGHRkd male mice, the fact that restoring IGF1 fully controlled insulin levels, but failed to normalize DNL and steatosis, suggest GHR can act independently of IGF1 to control liver metabolism.
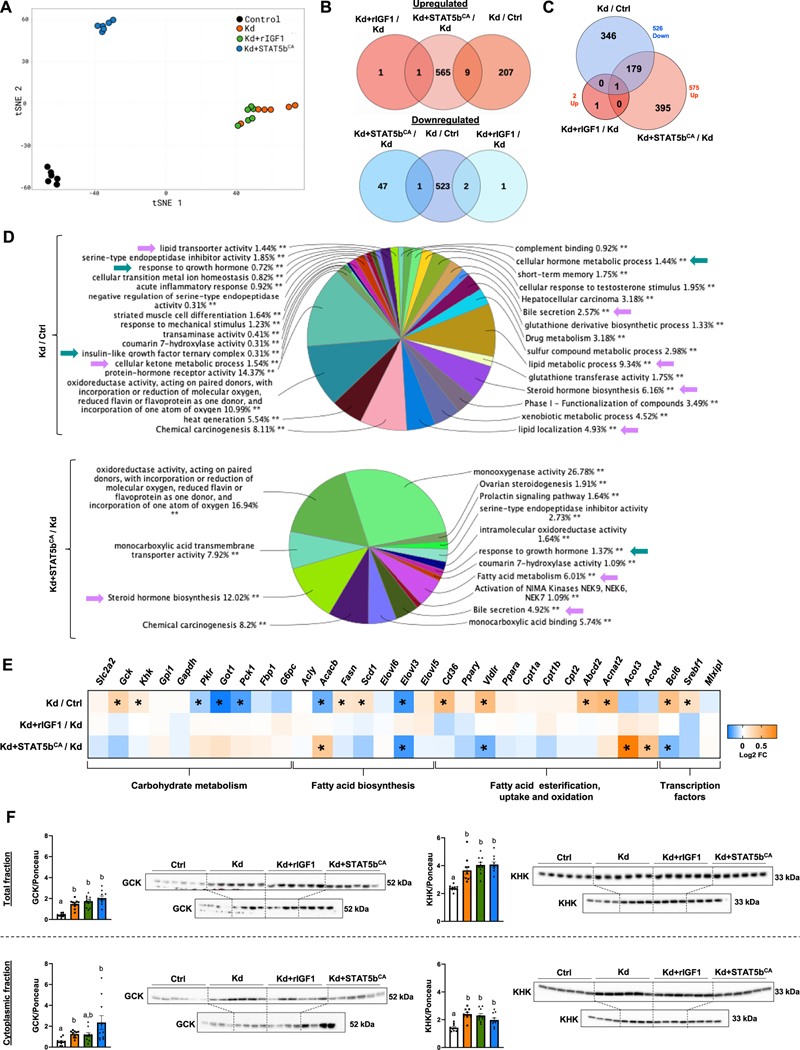
Given the more dramatic impact of aHepGHRkd seen in male compared to female mice, RNA-seq analysis was performed on a subset of male liver samples (n = 6/group) to identify key genes and pathways altered by aHepGHRkd that may contribute to enhanced DNL and steatosis (Fig. 2). The differentially expressed genes (DEGs) identified are shown in Supplemental Excel file S1. Analysis of the DEGs using a tSNE dimensionality reduction plot (Fig. 2A) revealed that loss of hepatocyte GHR caused a dramatic shift in the liver transcriptome, compared to GHR-intact controls, with 216 upregulated genes and 526 downregulated genes (Fig. 2B). Maintenance of hepatocyte IGF1 expression, in the absence of the GHR, had almost no effect on global hepatic gene expression, as previously observed in long-term aHepGHRkd livers ([11]; GEO, GSM4667026). Since the IGF1 receptor is not expressed by adult hepatocytes [27], the effect of IGF1 on hepatic steatosis and DNL in aHepGHRkd mice, is likely due to reduced insulin input to the liver or to enhanced systemic insulin-like effects of IGF1 that could alter nutrient flux to the liver. Nonetheless, there were no differences in circulating glucose or NEFA levels across all groups (Table S3). In contrast, increasing the activity of STAT5b by expressing STAT5bCA in male aHepGHRkd livers partially reversed the downregulation of gene expression in the absence of liver GHR, with 180 of the aHepGHRkd-repressed genes being upregulated by STAT5bCA, while IGF1 had virtually no effect (Fig. 2C). Furthermore, STAT5bCA expression shifted the transcriptome away from that observed in male GHR intact controls (Fig. 2A), as shown by the increased expression of 575 genes and decreased expression of 48 other genes (Fig. 2B). This is explained in part by the ability of STAT5b, when activated persistently, as in the case of the constitutively active STAT5bCA used in our experiments, to feminize the liver transcriptome (see Supplemental Excel file S1), as we recently reported [15]. Thus, STAT5bCA mimics the effects of the more continuous patterns of pituitary GH release in females (compared to pulsatile GH release in males) in sustaining STAT5b activity in the liver and in establishing a female pattern of hepatic gene expression [28].
Fig. 2.
Impact of aHepGHRkd without and with IGF1 or STAT5bCA on hepatic transcriptome and glucokinase (GCK) and ketohexokinase (KHK) protein levels, assessed in the post-absorptive state.
(A–E) Data derived from hepatic RNA-Seq analysis in control (Ctrl), aHepGHRkd (Kd), Kd + rIGF1 and Kd + STAT5bCA (n = 6 mice/group, subset of samples shown in Fig. 1A). (A) tSNE plot based on all 1504 genes differentially expressed in at least one of the four RNA-seq comparisons, where each datapoint represents an individual biological replicate. (B) Venn diagrams showing intersection of up-regulated genes (top panel, red) and down-regulated genes (bottom panel, blue) from the Kd/Crtl, rIGF1 + Kd/Kd, and Kd + Stat5bCA/Kd comparisons. (C) Venn Diagram showing the intersection of down-regulated genes (blue) from the Kd/Ctrl comparison with the up-regulated genes (red) from the Kd + rIGF1/Kd and Kd + STAT5CA/Kd comparisons. (D) Enrichment analysis of DEG in Kd/Ctrl and Kd + STAT5bCA/Kd using ClueGO from Cytoscape. Results are illustrated as a functionally grouped pie chart of terms including Gene Ontology (Biological Processes, Molecular Functions and Cellular Components) and KEGG and Reactome pathways. Percentages indicate the number of DEG included in each group with respect to total. Green arrows indicate processes related with GH signaling and purple arrows indicate processes mainly related to lipid metabolism. (E) Heat-map showing relative expression levels (Log2FC) of selected genes related to carbohydrate metabolism, fatty acid biosynthesis, fatty acid esterification, fatty acid uptake and oxidation, and transcription factors (n = 6 mice/group). (F) Total (top panel) and cytoplasmic (bottom panel) of glucokinase (GCK) and ketohexokinase (KHK) hepatic protein levels assessed by Western Blot. It should be noted that for Westerns two separate gels/blots were run and shown for each protein/fraction (top panel n = 6/group and bottom panel n = 4/group) representing all biological replicates within each group (n = 10/group). Protein levels are normalized by Ponceau S (Ponceau S images are shown in Supplemental Fig. S3). Values in graphs are represented as mean ± SEM, with individual data points included. Values that do not share a common letter (a, b, c) are statistically different, p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Pathway analysis confirmed the altered expression of genes important in the GH response and IGF1 ternary complex in aHepGHRkd livers, compared to GHR-intact controls (Fig. 2D). Pathways specifically related to alterations in carbohydrate and lipid metabolism included lipid transport activity, cellular ketone metabolism, lipid metabolism and lipid localization. Expression of STAT5bCA, in the context of aHepGHRkd, significantly altered pathways important in monocarboxylic acid activity and FA metabolism. The specific DEGs in each of the pathways illustrated by the pie charts in Fig. 2D are provided in Supplemental Excel file S2.
The impact of aHepGHRkd without and with IGF1 or STAT5bCA on the relative expression of specific genes related to glycolysis, gluconeogenesis, glycogenesis, lipogenesis, lipid processing/transport and FA oxidation is summarized in Fig. 2E. A subset of the DEG identified by RNA-seq, was confirmed by qPCR using the full sample set (n = 10; Fig. S1C). In the context of the post-absorptive state, aHepGHRkd increased mRNA levels for glucokinase (Gck), ketohexokinase (Khk), as well as genes important for lipogenesis (Fasn, Scd1) and lipid transport (Cd36, Vldlr). With respect to lipid oxidation, there were no differences in expression of Cpt1/2, which is important for mitochondrial lipid oxidation. However, genes associated with peroxisomal lipid oxidation were differentially impacted, with Abcd2 and Acnat2 upregulation; and Acot3 and Acot4 downregulation. In addition, aHepGHRkd increased the expression of the transcription factor Bcl6, which suppresses PPARα-mediated lipid oxidation [29], and increased Srebf1 expression, which helps maintain lipogenic gene expression [30]. Conversely, expression of genes important for gluconeogenesis were reduced (Got1, Pck1). Curiously, the expression of pyruvate kinase (Pklr), which catalyzes the final step in glycolysis, was also reduced.
Hepatocyte expression of STAT5bCA reversed aHepGHRkd-mediated upregulation of Bcl6 (Figs. 2E, S1C). This response was associated with an increase in some (Acot3/4), but not all (Abcd2, Acnat2, Acacb) PPARα target genes [29]. In addition, STAT5bCA further suppressed the expression of Elovl3, a known male-specific gene, previously shown to be suppressed by sustained STAT5b activity [15]. However, the relative changes in Gck, Khk, Fasn, Cd36 and Srebf1 observed in aHepGHRkd livers, were not completely corrected by STAT5bCA ( Figs. 2E, S1C). This suggest that loss of hepatocyte GHR, independent of STAT5b, plays a role in altering the hepatic transcriptome to favor glycolysis, DNL and steatosis. Of note, CD36, important in fatty acid transport [31], is elevated in mice with congenital defects in hepatocyte GHR-signaling and is considered a major driver of steatosis in these models [32,33]. However, the physiologic significance of the rise in Cd36 gene expression observed in aHepGHRkd livers and its contribution to steatosis in this acute model may be limited, in that the increase in Cd36 mRNA did not translate to an increase in CD36 protein levels (Fig. S1D) or enhanced hepatic fatty acid uptake, as measured by BODIPY-labeled palmitate (Fig. S1E). However, it should be noted CD36 protein levels were significantly increased with STAT5bCA treatment (Fig. S1D).
In contrast to the disconnect between Cd36 mRNA and protein levels, the increase in Gck and Khk gene expression observed in aHepGHRkd livers is translated into an increase in total protein, as well as cytoplasmic (bioactive) protein levels (Fig. 2F, Ponceau images used for data normalization are shown in Fig. S3). In fact, an increase in cytoplasmic GCK protein levels was observed as early as 3d post aHepGHRkd, and is associated with an increase in hepatic TG and glycogen content (Fig. S1F). The increase in GCK and KHK, observed after 7d aHepGHRkd, was not normalized by IGF1 or STAT5bCA (Fig. 2F).
3.2. Hepatic expression of GCK is elevated in aHepGHRkd livers in response to refeeding after an overnight fast, associated with steatosis that is independent of ChREBP, and normalized by STAT5bCA, but not IGF1 alone
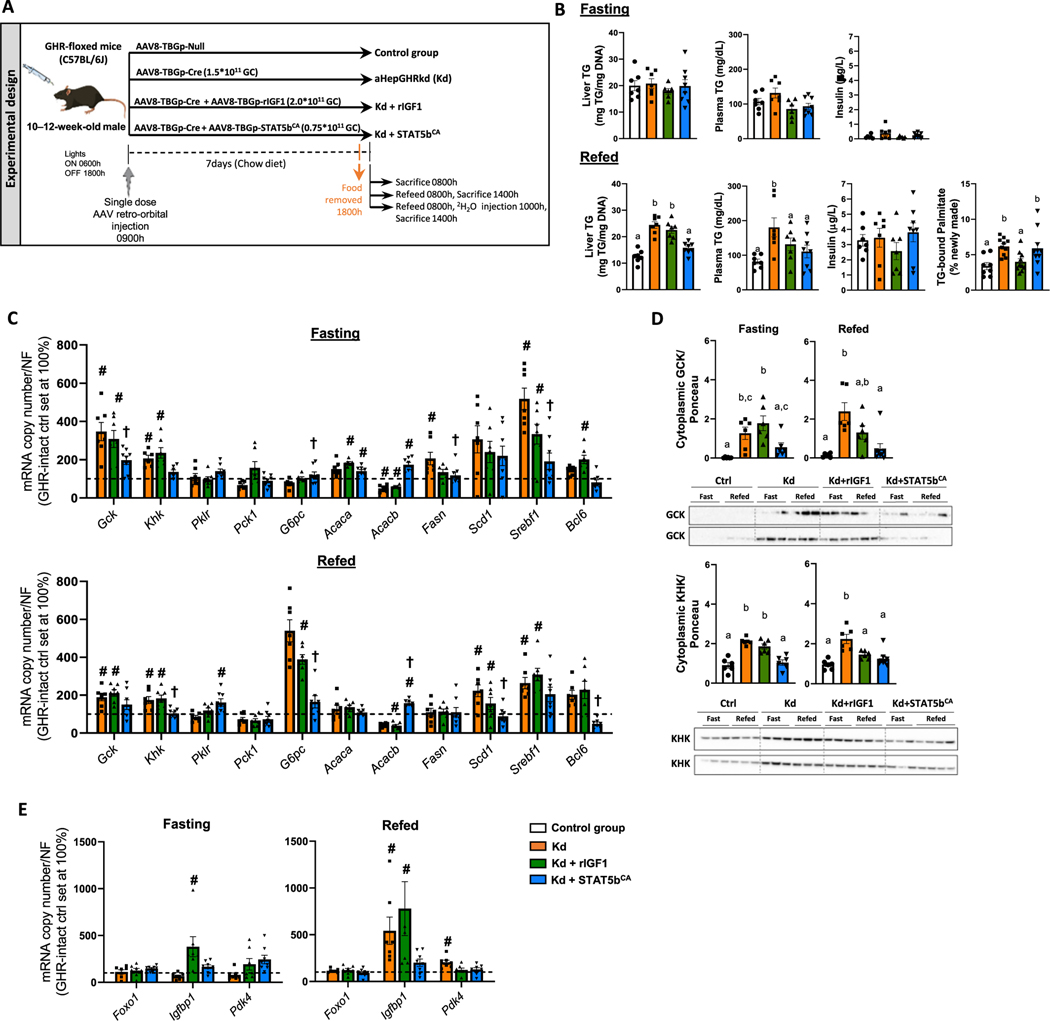
Given glycolysis and lipogenesis (de novo, as well as complex lipids formed from dietary fatty acids) is greatest in the absorptive state (following a meal), due to elevated insulin and dietary nutrient supply, we sought to determine if enhanced DNL and steatosis in aHepGHRkd male livers would persist in this context. To this end, we examined the response of aHepGHRkd male mice without or with IGF1 or Stat5bCA treatment, to refeeding (6 h standard chow) after an overnight fast (Fig. 3A).
Fig. 3.
Impact of aHepGHRkd without and with IGF1 or STAT5bCA on liver metabolic phenotype in response to fasting and refeeding. (A) Experimental design: 10 to 12-week-old GHR-floxed male mice were treated with a single dose of different types of AAVs to generate four experimental groups (n = 6–7 mice/group). 7 days after the injection, food was removed at 1600 h and tissues and blood collected the following morning at 0800 h or at 1400 h after 6 h of refeeding a standard chow diet. A subset of refed mice was injected with 2H20 at 1000 h. (B) Liver and plasma TG, plasma insulin (fasting and refeeding) and percentage of hepatic newly formed palmitate after refeeding. (C) Expression of glycolytic, gluconeogenic and fatty acid synthesis-related genes (assessed by qPCR, controls were set at 100 %). (D) Cytoplasmic glucokinase (GCK) and ketohexokinase (KHK) hepatic protein levels in fasted and refed mice assessed by Western, where two separate gels/blots were run and shown for each protein (top and bottom panels contains n = 3 fasted and 3–4 refed/group) representing all biological replicates within each group (n = 6 fasted and 6–8 refed/group). Protein levels are normalized by Ponceau S (Ponceau S images are shown in Supplemental Fig. S3). (E) Expression of Foxo1 and Foxo1 target genes Igfbp1 and Pdk4, assessed by qPCR with controls were set at 100 %. Graphical values are represented as mean ± SEM, with individual data points included (n = 6–8 mice/group). #, in panels C and E, indicates differences in hepatic gene expression levels compared to GHR intact controls, where † indicates a significant effect of STAT5bCA, compared to aHepGHRkd alone, p < 0.05. In panels B and D, values that do not share a common letter (a, b, c) are statistically different, p < 0.05.
In mice subjected to overnight fasting, FAs derived from white adipose tissue (WAT) lipolysis fuels gluconeogenesis, however excess FA are re-esterified, leading to steatosis [34]. In this context aHepGHRkd, without or with IGF1 or STAT5bCA, did not alter hepatic TG content or circulating TG, insulin, glucose, NEFA or β-hydroxybutyrate (Fig. 3B; Table S3). Surprisingly, elevation in expression of GCK and KHK (mRNA and cytoplasmic protein) persisted in the fasted aHepGHRkd liver, associated with increased expression of Fasn and Srebf1, where STAT5bCA, but not IGF1 alone, consistently normalized these endpoints (Fig. 3C, D).
With refeeding, aHepGHRkd without or with IGF1 or STAT5bCA did not alter total food intake or plasma glucose, β-hydroxybutyrate or plasma insulin (Table S3, Fig. 3B). However, plasma NEFA levels, although low, were significantly greater than GHR-intact controls (Table S3). Liver TG content and circulating TG levels were greater in refed aHepGHRkd mice, compared to GHR-intact controls (Fig. 3B), that could not be accounted for by an increase in mature SREBP1c protein levels, despite the increase in Sreb1f mRNA levels ( Figs. 3C, S4B). However, refed aHepGHRkd mice did exhibit an increase in hepatic GCK, KHK, and rate of DNL ( Figs. 3B–D, S4A), reflected by an increase in hepatic tricarboxylic acid cycle metabolites, assessed by unbiased metabolomics analysis (Fig. S5; Supplemental Excel file S3).
Since aHepGHRkd livers consistently exhibit an increase in cytoplasmic GCK protein levels, and GCK activity and downstream glucose metabolites have been shown to activate the lipogenic transcription factor, carbohydrate response element binding protein (ChREBP), we sought to determine if ChREBP contributes to the alterations in hepatic gene expression and steatosis observed after aHepGHRkd. To this end we generated mice with adult-onset, hepatocyte-specific knockdown of both GHR and ChREBP (aHepGHR-ChREBPkd; Fig. S4C). Again, refeeding after an overnight fast increased Gck and Srebf1 mRNA and protein levels and steatosis in aHepGHR-ChREBPkd mice, compared to GHR/ChREBP-intact controls, although the expression level of known ChREBP target genes (Khk, Tkfc, Fasn, Scd1, Elovl6, and Pklr) was blunted. Of note, aHepChREBPkd alone also increased Gck mRNA and cytoplasmic GCK protein levels (Fig. S4D), but SREBP1c expression and protein levels tended to be reduced, and steatosis did not develop. These results indicate ChREBP activation may promote, but is not essential, for the steatosis observed after aHepGHRkd.
In the context of refeeding, IGF1 normalized DNL in aHepGHRkd livers ( Figs. 3B, S4A), but this was not reflected by normalization of GCK, KHK or lipogenic gene expression (Fig. 3C, D). In contrast, STAT5bCA suppressed the expression of GCK, KHK and lipogenic genes, as well as expression of Bcl6, but failed to normalize DNL (Fig. 3B–D). These results emphasize that the actions of STAT5bCA on liver metabolism are not solely due to the rise in IGF1. In addition, these results coupled with those observed in the post-absorptive state demonstrate the ultimate actions of GHR-mediated STAT5b – IGF1 regulation on DNL and steatosis is highly dependent on the nutritional state.
3.3. FOXO1 target genes are elevated in refed aHepGHRkd mice
Curiously, G6pc mRNA levels were elevated in refed aHepGHRkd livers, which was normalized by STAT5bCA, but not IGF1 (Fig. 3C). Elevated G6pc expression was also observed in refed aHepGHR-ChREBPkd mice, compared to intact-controls (Fig. S4C), despite the fact that G6pc is a known ChREBP target gene [35]. Since G6pc expression is also stimulated by FOXO1, where FOXO1 activity should be suppressed by insulin in response to refeeding, this suggests that in the refed aHepGHRkd liver, the gluco-regulatory arm of hepatic insulin-signaling is resistant. This is further supported by the observation that the expression of Igfbp1, which is also a FOXO1 target, is elevated in aHepGHRkd liver after refeeding ( Figs. 3E, S4C), and this increase is normalized by STAT5bCA, but not IGF1 (Fig. 3E).
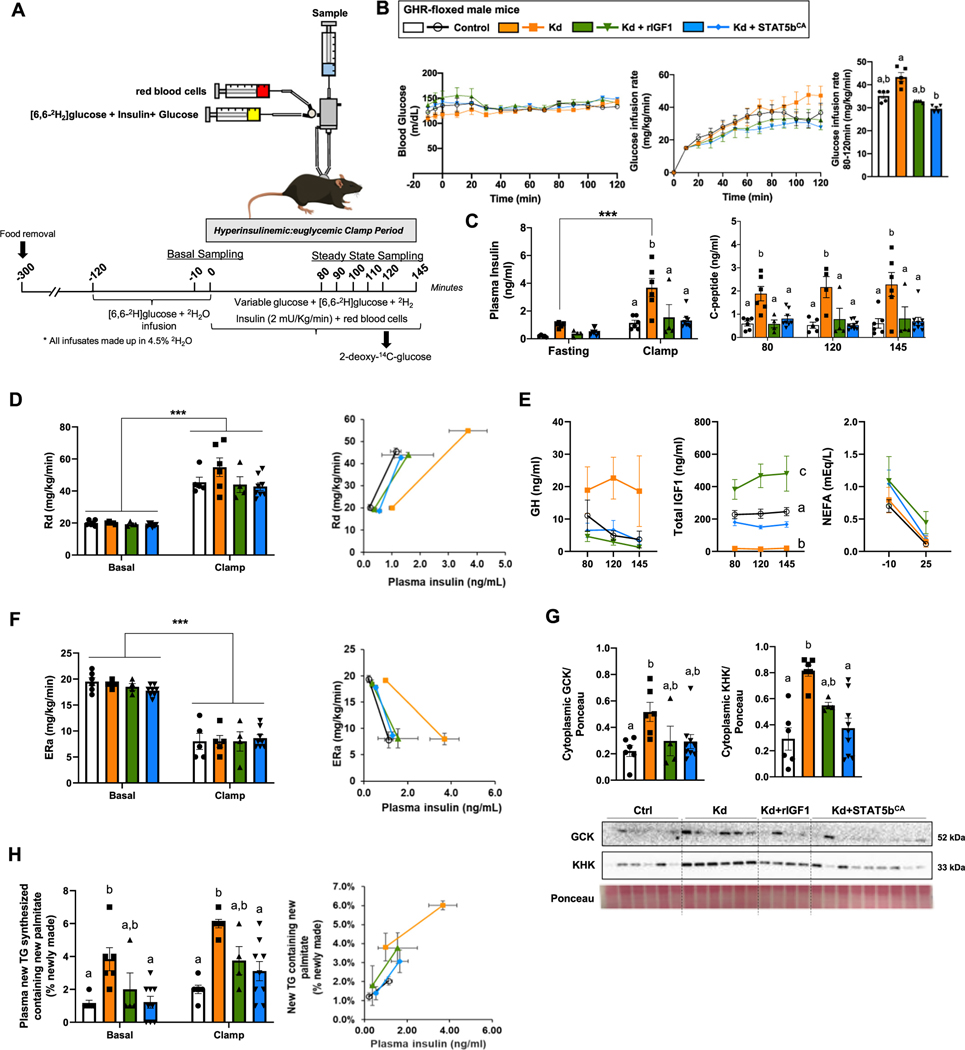
3.4. aHepGHRkd impairs systemic insulin sensitivity and insulin-mediated hepatic glucose production, while augmenting hepatic DNL rate, which can be corrected by both IGF1 and STAT5bCA
To better evaluate the impact of aHepGHRkd, without and with IGF1 or STAT5bCA on tissue-specific insulin sensitivity, male mice were subjected to hyperinsulinemic:euglycemic clamps with stable isotope tracing (Fig. 4A–B). aHepGHRkd mice exhibited basal and clamped hyperinsulinemia (Fig. 4C). In this context, both IGF1 and STAT5CA normalized insulin levels. Since C-peptide levels mimicked that of insulin under clamped conditions (Fig. 4C), the rise in insulin observed in aHepGHRkd mice is primarily due to enhanced insulin secretion and not impairment in insulin clearance. During the clamp, the glucose infusion rate was modestly elevated in aHepGHRkd mice, that was significantly suppressed with STAT5bCA (Fig. 4B). Under these conditions there were no differences between groups related to rate of glucose disappearance (Fig. 4D). Also, across groups there were no differences in glucose uptake in adipose tissue, heart and brain, however Stat5bCA actually reduced glucose uptake in the soleus and gastrocnemius relative to GHR-intact controls (Fig. S6A). As illustrated in Fig. 4D (right panel), the fact that more insulin is required to achieve the same level of glucose disappearance indicates aHepGHRkd mice have systemic insulin resistance, that is corrected by maintaining hepatocyte IGF1, as well as STAT5b activity. Impaired systemic insulin sensitivity could be attributed to the reciprocal shift in IGF1 and GH (Fig. 4E), where it is commonly accepted that IGF1 improves insulin sensitivity, while GH antagonizes the action of insulin [5].
Fig. 4.
Hyperinsulinemic:euglycemic clamps in aHepGHRkd male mice without or with IGF1 or STAT5bCA maintenance. (A) Schematic representation of the hyperinsulinemic:euglycemic clamp design. All mice were injected with AAVs 7 days before the experiment to generate the four experimental groups (Control, aHepGHRkd [Kd], aHepGHRkd [Kd] + rIGF1 and aHepGHRkd [Kd] + STAT5bCA). (B) Blood glucose and glucose infusion rate during hyperinsulinemic:euglycemic clamps. (C) Plasma insulin levels before and after the clamps and C-peptide levels at different time points during the clamps. (D) Rate of glucose disappearance (Rd) and correlation between Rd and plasma insulin levels before and after the clamps. (E) Plasma GH and IGF1 levels at different time points during the clamps and circulating NEFA levels before and after the clamps. (F) Endogenous glucose production (ERa) and correlation between ERa and plasma insulin levels before and after the clamps. (G) Cytoplasmic GCK and KHK protein levels at steady state. (H) Percentage of plasma new glyceride with new palmitate and correlation between newly synthesized glyceride containing new palmitate and plasma insulin levels at baseline and during the clamps. Values are represented as mean ± SEM, with individual points included (n = 4–9 mice/group). Values that do not share a common letter (a, b, c) are statistically different (p < 0.05). ***p < 0.001.
However, loss of the hepatocyte GHR had an unexpected impact on hepatic insulin sensitivity. Insulin suppression of endogenous glucose production (Fig. 4F), as well as hepatic gluconeogenesis and glycogenolysis (Fig. S6B), was unaltered in aHepGHRkd mice, but required more insulin tone to achieve the same effect (Fig. 4F, right panel). These data demonstrate the gluco-regulatory arm of hepatic insulin signaling is resistant in the absence of the hepatocyte GHR. This observation is also supported by preliminary studies (Fig. S7), showing aHepGHRkd mice produce more glucose in response to a pyruvate challenge, compared to GHR-intact controls and this was reduced by STAT5bCA. How this extreme hyperinsulinemia and hepatic insulin resistance develops in this acute model remains to be determined. Nonetheless, this effect cannot be clearly attributed to an increase in fatty acid flux to the liver, shown in other conditions to contribute to hepatic insulin resistance [36], since NEFA levels did not differ between groups under basal or clamped conditions (Fig. 4E).
Like that observed in the refed state, under clamped conditions cytoplasmic GCK and KHK levels were increased in aHepGHRkd mice, above GHR-intact controls, and this increase was normalized by STAT5bCA (Fig. 4G). In addition, aHepGHRkd dramatically increased the percent of esterified newly synthesized palmitate in the plasma and in the liver (Fig. S6C, D), as well as the percent newly synthesized palmitate, esterified to newly synthesized glyceride in the plasma (Fig. 4H). The finding that these changes also track with elevated insulin (Fig. 4H, right panel), indicates insulin-mediated DNL remains intact.
4. Discussion
This series of studies rigorously examined the impact of aHepGHRkd without and with IGF1 and STAT5bCA, in multiple nutritional states (post-absorptive [end of day] and overnight fasted without or with refeeding), as well as in the controlled experimental condition of a hyperinsulinemic:euglycemic clamp. Overall, the results reveal that GH, specifically in males, regulates liver metabolism at multiple levels (direct and indirect) dependent on the context studied.
In both the post-absorptive and refed state, aHepGHRkd increases hepatic GCK mRNA and cytoplasmic (active) protein levels, expression of lipogenic genes, DNL and steatosis. Some of these changes can be attributed to an increase in insulin tone, which may be driven in part by the reciprocal shift in circulating IGF1 and GH levels, that promotes systemic insulin resistance. This was dramatically illustrated using hyperinsulinemic:euglycemic clamp conditions, where AAV-mediated hepatocyte expression of both IGF1 and STAT5bCA, normalized the elevated levels of plasma insulin and hepatic DNL observed in aHepGHRkd mice. However, maintaining IGF1 alone, in the absence of the hepatocyte GHR, failed to prevent steatosis in the post-absorptive and refed states, and did not prevent the increase in the expression of Gck and lipogenic genes observed across the three nutritional states tested. These results support the conclusion that hepatocyte GHR signaling also acts independent of the systemic actions of IGF1, to control glycolytic and lipogenic programs. In fact, cytoplasmic GCK protein levels observed after an overnight fast, in aHepGHRkd livers, without or with IGF1, were even higher than that observed in refed GHR-intact controls. The sustained activity of hepatic GCK observed in aHepGHRkd may serve to prime the liver to promote DNL when the insulin and nutrient milieu is optimal. This is consistent with reports showing overexpression of hepatic Gck increases DNL in humans and mice [37,38].
GHR signals through multiple signal transduction pathways to regulate cellular function [39]. In the context of the liver, the GHR-mediated increase in STAT5b activity is thought to be a major factor in preventing steatosis insofar as congenital liver-specific deletion of the GHR, JAK2 or STAT5 exhibit fatty liver as adults, in association with impaired systemic metabolic function [33,40–43]. In the current report, sustaining hepatocyte STAT5b activity, but not sustaining IGF1, in the absence of the GHR, normalized the glycolytic and lipogenic transcriptional programs in the overnight fasted and refed states, but failed to fully correct these endpoints in the post-absorptive state, where DNL and steatosis remained greater than GHR-intact controls. These observations indicate some of the anti-steatotic actions of STAT5b are independent of IGF1’s effects on peripheral tissue. However, the actions of STAT5b cannot be fully accounted for a direct regulation of the glycolytic and lipogenic gene expression.
A STAT5b-dependent pathway that may account for some of the anti-steatotic effects of STAT5bCA is direct regulation of BCL6 [44], a transcriptional repressor of PPARα target genes, important in lipid oxidation [29]. Hepatic BCL6 is expressed at higher levels in males than females [45], and its expression is reduced in fasting and increased with refeeding [29]. Differential expression of hepatic Bcl6 can explain in part the increased susceptibility of males, as compared to females, to diet-induced steatosis [46,47]. In the context of aHepGHRkd, hepatic expression of Bcl6 was further increased in male livers, most dramatically in the post-absorptive state, and was reduced by STAT5bCA. Elevated Bcl6 would be predicted to contribute to the steatosis by repression of PPARα-mediated lipid oxidation, where RNA-seq revealed this may be in part be due to Bcl6 transcriptional regulation of a subset of genes important in peroxisomal lipid oxidation (Acot3/Acot4) [48]. STAT5b may also alter liver metabolism by impacting the FOXO1 transcriptional regulation, based on the observation that expression of classic FOXO1 target genes (G6pc and Igfbb1) were inappropriately elevated in aHepGHRkd after refeeding, that could be suppressed by STAT5bCA, but not IGF1. Specifically, it was previously reported adenoviral expression of STAT5b in the livers of hypophysectomized rats suppressed expression of both G6pc and Igfbp1, and in vitro STAT5b directly suppressed FOXO1-mediated increase in Igfbp1 expression without altering FOXO1 nuclear localization or DNA binding [49]. Sustained hepatic FOXO1 activity in aHepGHRkd mice may contribute to the relative insulin resistance of the gluco-regulatory arm of hepatic insulin signaling revealed by hyperinsulinemic:euglycemic clamps. Although these observations are intriguing, additional studies are required to understand the relationship between GHR/STAT5b-mediated signaling with BCL6 and FOXO1 activity, in controlling liver metabolism across nutritional states.
GH has been shown to antagonize the actions of insulin [5–8], consistent with the observation that aHepGHRkd mice with elevated endogenous GH levels exhibit a relative impairment in insulin-mediated glucose uptake observed in basal and clamped conditions (i.e. more insulin is required to achieve the same effect) that was normalized by IGF1 and STAT5bCA. However, at the level of the liver, loss of GHR from hepatocytes resulted in a relative impairment in insulin-mediated glucose suppression. Our group has previously reported a similar tissue-specific impact of GH on insulin-mediated glucose homeostasis. Specifically, endogenous GH levels were negatively associated with insulin-mediated glucose uptake, but positively associated with insulin-mediated hepatic glucose suppression in mice with adult-onset isolated GH deficiency and mice with elevated GH levels due to loss of pituitary insulin and IGF1 negative feedback [17]. Taken together, these results confirm GH acts systemically to suppress the actions of insulin, while at the level of the hepatocyte GH serves to maintain insulin-mediated suppression of hepatic glucose production. Although under basal and clamped conditions both IGF1 alone and STAT5bCA corrected the hyperinsulinemia observed, based on our findings across multiple nutritional states, we might speculate that the actions of free IGF1 were primarily due to improving systemic insulin sensitivity, while the actions of STAT5b are due to the combined effects of raising IGF1 and direct alterations in the hepatocyte gene program, as discussed above.
Despite the relative resistance of the gluco-regulatory arm of hepatic insulin signaling, basal and clamp data suggest the insulin-mediated stimulation of hepatic glycolysis and DNL is sustained, suggesting loss of hepatocyte GHR signaling leads to pathway selective insulin resistance. Pathway selective insulin resistance has been reported in multiple mouse models with diet-induced obesity and type II diabetes, where under these conditions the primary driver may not be impaired hepatic insulin-signaling per se, but altered substrate delivery to the liver, due to excess dietary intake and/or systemic insulin resistance promoting WAT lipolysis and impaired glucose disposal, as well as the repartitioning of hepatic nutrient handling [36,50]. However, in the current study aHepGHRkd mice were maintained on a standard chow diet and under basal and clamp conditions the hyperinsulinemia observed is sufficient to normalize systemic glucose uptake and suppress WAT lipolysis, based on normal circulating NEFA levels. In addition, in all nutrient conditions tested, circulating glucose and NEFA levels did not appreciably differ across groups. Therefore, the dramatic increase in DNL observed under clamp conditions is unlikely due to an increase in substrate delivery to the liver. These results suggest that hepatocyte GHR signaling plays an important role in partitioning of hepatic nutrients in response to insulin.
This series of studies reveals novel actions of hepatocyte GHR signaling that control hepatic carbohydrate and lipid metabolism. The major findings include: 1) loss of hepatocyte GHR signaling increases glucokinase activity, independent of IGF1 and nutritional state; 2) loss of hepatocyte GHR signaling enhances DNL and steatosis, dependent on the nutritional state; 3) sustaining STAT5b activity in the absence of the GHR can normalize liver metabolism, in some but not all nutritional states; and 4) loss of hepatocyte GHR signaling sustains DNL, but impairs insulin suppression of HGP, which cannot be explained by enhanced substrate delivery to the liver. These results provide a foundation to design future studies to better understand the role GH plays in regulation of adult metabolism, which may explain the increase in NAFLD prevalence observed in patients with primary GHD, or relative GHD observed in obese patients.
Funding
This work was funded by NIH R01DK114326, VA Merit BX001114 and VA RCS BX005382 to RDK, NIH U2C DK059637, S10OD025199 and P30 DK020593 (to OPM) and NIH R01 DK121998 (to DJW),
Supplementary Material
Acknowledgements
The authors would wish to thank Dr. John J Kopchick (Ohio University, Athens, OH) for providing the original breeding colony of GHRfl/fl mice and Dr. Terry Unterman (University of Illinois at Chicago, Chicago, IL) for reviewing the final draft of the manuscript and helpful discussions.
Footnotes
CRediT authorship contribution statement
MCVB and MRM contributed equally to this work and were involved in the study design, tissue preparation and analysis, figure preparation, and all stages of manuscript preparation; MP: performed RNA-seq data analysis; ASC, MM, NP and MW participated in mouse management and tissue preparation. JCC: involved in early studies of the mouse model and current study design; DJW: performed RNA-seq data analysis; MAP: performed analysis of hepatic DNL in livers from the nutritional states tested; OPM: performed the hyperinsulinemic:euglycemic clamp with stable isotope tracers; RDK: designed and supervised the study, involved in all stages of manuscript preparation. RDK, MCVB, MRM, JCC, DJW, MAP and OPM were involved in extensive discussion regarding data interpretation. All authors reviewed the manuscript and approved the final version.
Declaration of competing interest
Authors have nothing to disclose.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.metabol.2023.155589.
References
- [1].Dichtel LE, Cordoba-Chacon J, Kineman RD. Growth hormone and insulin-like growth factor I regulation of nonalcoholic fatty liver disease. J Clin Endocrinol Metab 2022. 10.1210/clinem/dgac088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Quek J, Chan KE, Wong ZY, Tan C, Tan B, Lim WH, et al. Global prevalence of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in the overweight and obese population: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2023;8:20–30. 10.1016/S2468-1253(22)00317-X. [DOI] [PubMed] [Google Scholar]
- [3].Dichtel LE, Corey KE, Haines MS, Chicote ML, Kimball A, Colling C, et al. The GH/IGF-1 axis is associated with intrahepatic lipid content and hepatocellular damage in overweight/obesity. J Clin Endocrinol Metab 2022;107:e3624–32. 10.1210/clinem/dgac405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].List EO, Palmer AJ, Berryman DE, Bower B, Kelder B, Kopchick JJ. Growth hormone improves body composition, fasting blood glucose, glucose tolerance and liver triacylglycerol in a mouse model of diet-induced obesity and type 2 diabetes. Diabetologia 2009;52:1647–55. 10.1007/s00125-009-1402-z. [DOI] [PubMed] [Google Scholar]
- [5].Vazquez-Borrego MC, Del Rio-Moreno M, Kineman RD. Towards understanding the direct and indirect actions of growth hormone in controlling hepatocyte carbohydrate and lipid metabolism. Cells 2021;10. 10.3390/cells10102532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sharma R, Kopchick JJ, Puri V, Sharma VM. Effect of growth hormone on insulin signaling. Mol Cell Endocrinol 2020;518:111038. 10.1016/j.mce.2020.111038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kopchick JJ, Berryman DE, Puri V, Lee KY, Jorgensen JOL. The effects of growth hormone on adipose tissue: old observations, new mechanisms. Nat Rev Endocrinol 2020;16:135–46. 10.1038/s41574-019-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Targher G, Corey KE, Byrne CD, Roden M. The complex link between NAFLD and type 2 diabetes mellitus - mechanisms and treatments. Nat Rev Gastroenterol Hepatol 2021;18:599–612. 10.1038/s41575-021-00448-y. [DOI] [PubMed] [Google Scholar]
- [9].Cordoba-Chacon J, Majumdar N, List EO, Diaz-Ruiz A, Frank SJ, Manzano A, et al. Growth hormone inhibits hepatic de novo lipogenesis in adult mice. Diabetes 2015; 64:3093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cordoba-Chacon J, Sarmento-Cabral A, Del Rio-Moreno M, Diaz-Ruiz A, Subbaiah PV, Kineman RD. Adult-onset hepatocyte GH resistance promotes NASH in male mice, without severe systemic metabolic dysfunction. Endocrinology 2018; 159:3761–74. 10.1210/en.2018-00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sarmento-Cabral A, Del Rio-Moreno M, Vazquez-Borrego MC, Mahmood M, Gutierrez-Casado E, Pelke N, et al. GH directly inhibits steatosis and liver injury in a sex-dependent and IGF1-independent manner. J Endocrinol 2021;248:31–44. 10.1530/JOE-20-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Davey HW, Xie T, McLachlan MJ, Wilkins RJ, Waxman DJ, Grattan DR. STAT5b is required for GH-induced liver IGF-I gene expression. Endocrinology 2001;142: 3836–41. 10.1210/endo.142.9.8400. [DOI] [PubMed] [Google Scholar]
- [13].Lau-Corona D, Ma H, Vergato C, Sarmento-Cabral A, Del Rio-Moreno M, Kineman RD, et al. Constitutively active STAT5b feminizes mouse liver gene expression. Endocrinology 2022;163. 10.1210/endocr/bqac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].List EO, Berryman DE, Funk K, Gosney ES, Jara A, Kelder B, et al. The role of GH in adipose tissue: lessons from adipose-specific GH receptor gene-disrupted mice. Mol Endocrinol 2013;27:524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lau-Corona D, Ma H, Vergato C, Sarmento-Cabral A, del Rio-Moreno M, Kineman RD, et al. Constitutively active STAT5 b feminizes mouse liver gene expression. bioRxiv 2022:2022.2002.2014.480424. 10.1101/2022.02.14.480424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wolf Greenstein A, Majumdar N, Yang P, Subbaiah PV, Kineman RD, Cordoba-Chacon J. Hepatocyte-specific, PPARgamma-regulated mechanisms to promote steatosis in adult mice. J Endocrinol 2017;231:107–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cordoba-Chacon J, Gahete MD, McGuinness OP, Kineman RD. Differential impact of selective GH deficiency and endogenous GH excess on insulin-mediated actions in muscle and liver of male mice. Am J Physiol Endocrinol Metab 2014;307: E928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Goldfarb CN, Karri K, Pyatkov M, Waxman DJ. Interplay between GH-regulated, sex-biased liver transcriptome and hepatic zonation revealed by single-nucleus RNA sequencing. Endocrinology 2022;163. 10.1210/endocr/bqac059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pang Z, Zhou G, Ewald J, Chang L, Hacariz O, Basu N, et al. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat Protoc 2022;17:1735–61. 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- [21].Ayala JE, Bracy DP, Malabanan C, James FD, Ansari T, Fueger PT, et al. Hyperinsulinemic-euglycemic clamps in conscious, unrestrained mice. J Vis Exp 2011. 10.3791/3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hasenour CM, Wall ML, Ridley DE, Hughey CC, James FD, Wasserman DH, et al. Mass spectrometry-based microassay of (2)H and (13)C plasma glucose labeling to quantify liver metabolic fluxes in vivo. Am J Physiol Endocrinol Metab 2015;309: E191–203. 10.1152/ajpendo.00003.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Domene S, Domene HM. The role of acid-labile subunit (ALS) in the modulation of GH-IGF-I action. Mol Cell Endocrinol 2020;518:111006. 10.1016/j.mce.2020.111006. [DOI] [PubMed] [Google Scholar]
- [24].Davey HW, McLachlan MJ, Wilkins RJ, Hilton DJ, Adams TE. STAT5b mediates the GH-induced expression of SOCS-2 and SOCS-3 mRNA in the liver. Mol Cell Endocrinol 1999;158:111–6. 10.1016/s0303-7207(99)00175-6. [DOI] [PubMed] [Google Scholar]
- [25].Sadler DG, Treas L, Sikes JD, Porter C. A modest change in housing temperature alters whole body energy expenditure and adipocyte thermogenic capacity in mice. Am J Physiol Endocrinol Metab 2022;323:E517–28. 10.1152/ajpendo.00079.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Giles DA, Moreno-Fernandez ME, Stankiewicz TE, Graspeuntner S, Cappelletti M, Wu D, et al. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat Med 2017;23:829–38. 10.1038/nm.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Waraky A, Aleem E, Larsson O. Downregulation of IGF-1 receptor occurs after hepatic linage commitment during hepatocyte differentiation from human embryonic stem cells. Biochem Biophys Res Commun 2016;478:1575–81. 10.1016/j.bbrc.2016.08.157. [DOI] [PubMed] [Google Scholar]
- [28].Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol 2006;20:2613–29. 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- [29].Sommars MA, Ramachandran K, Senagolage MD, Futtner CR, Germain DM, Allred AL, et al. Dynamic repression by BCL6 controls the genome-wide liver response to fasting and steatosis. Elife 2019;8. 10.7554/eLife.43922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ferre P, Phan F, Foufelle F. SREBP-1c and lipogenesis in the liver: an update1. Biochem J 2021;478:3723–39. 10.1042/BCJ20210071. [DOI] [PubMed] [Google Scholar]
- [31].Rada P, Gonzalez-Rodriguez A, Garcia-Monzon C, Valverde AM. Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death Dis 2020;11: 802. 10.1038/s41419-020-03003-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hosui A, Tatsumi T, Hikita H, Saito Y, Hiramatsu N, Tsujii M, et al. Signal transducer and activator of transcription 5 plays a crucial role in hepatic lipid metabolism through regulation of CD36 expression. Hepatol Res 2017;47:813–25. 10.1111/hepr.12816. [DOI] [PubMed] [Google Scholar]
- [33].Sos BC, Harris C, Nordstrom SM, Tran JL, Balazs M, Caplazi P, et al. Abrogation of growth hormone secretion rescues fatty liver in mice with hepatocyte-specific deletion of JAK2. J Clin Invest 2011;121:1412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guan HP, Goldstein JL, Brown MS, Liang G. Accelerated fatty acid oxidation in muscle averts fasting-induced hepatic steatosis in SJL/J mice. J Biol Chem 2009; 284:24644–52. 10.1074/jbc.M109.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xu X, So JS, Park JG, Lee AH. Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP. Semin Liver Dis 2013;33:301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Petersen MC, Shulman GI. Mechanisms of insulin action and insulin resistance. Physiol Rev 2018;98:2133–223. 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Peter A, Stefan N, Cegan A, Walenta M, Wagner S, Konigsrainer A, et al. Hepatic glucokinase expression is associated with lipogenesis and fatty liver in humans. J Clin Endocrinol Metab 2011;96:E1126–30. [DOI] [PubMed] [Google Scholar]
- [38].Ferre T, Riu E, Franckhauser S, Agudo J, Bosch F. Long-term overexpression of glucokinase in the liver of transgenic mice leads to insulin resistance. Diabetologia 2003;46:1662–8. [DOI] [PubMed] [Google Scholar]
- [39].Chhabra Y, Lee CMM, Muller AF, Brooks AJ. GHR signalling: receptor activation and degradation mechanisms. Mol Cell Endocrinol 2021;520:111075. 10.1016/j.mce.2020.111075. [DOI] [PubMed] [Google Scholar]
- [40].Cui Y, Hosui A, Sun R, Shen K, Gavrilova O, Chen W, et al. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration. Hepatology 2007;46:504–13. [DOI] [PubMed] [Google Scholar]
- [41].Fan Y, Menon RK, Cohen P, Hwang D, Clemens T, DiGirolamo DJ, et al. Liver-specific deletion of the growth hormone receptor reveals essential role of growth hormone signaling in hepatic lipid metabolism. J Biol Chem 2009;284:19937–44. 10.1074/jbc.M109.014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].List EO, Berryman DE, Funk K, Jara A, Kelder B, Wang F, et al. Liver-specific GH receptor gene-disrupted (LiGHRKO) mice have decreased endocrine IGF-I, increased local IGF-I, and altered body size, body composition, and adipokine profiles. Endocrinology 2014;155:1793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mueller KM, Kornfeld JW, Friedbichler K, Blaas L, Egger G, Esterbauer H, et al. Impairment of hepatic growth hormone and glucocorticoid receptor signaling causes steatosis and hepatocellular carcinoma in mice. Hepatology 2011;54: 1398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang Y, Laz EV, Waxman DJ. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol Cell Biol 2012;32:880–96. 10.1128/mcb.06312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Meyer RD, Laz EV, Su T, Waxman DJ. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol Endocrinol 2009;23:1914–26. 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nikkanen J, Leong YA, Krause WC, Dermadi D, Maschek JA, Van Ry T, et al. An evolutionary trade-off between host immunity and metabolism drives fatty liver in male mice. Science 2022;378:290–5. 10.1126/science.abn9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Waxman DJ, Kineman RD. Sex matters in liver fat regulation. Science 2022;378: 252–3. 10.1126/science.ade7614. [DOI] [PubMed] [Google Scholar]
- [48].Tillander V, Alexson SEH, Cohen DE. Deactivating fatty acids: acyl-CoA thioesterase-mediated control of lipid metabolism. Trends Endocrinol Metab 2017; 28:473–84. 10.1016/j.tem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ono M, Chia DJ, Merino-Martinez R, Flores-Morales A, Unterman TG, Rotwein P. Signal transducer and activator of transcription (Stat) 5b-mediated inhibition of insulin-like growth factor binding protein-1 gene transcription: a mechanism for repression of gene expression by growth hormone. Mol Endocrinol 2007;21: 1443–57. [DOI] [PubMed] [Google Scholar]
- [50].Otero YF, Stafford JM, McGuinness OP. Pathway-selective insulin resistance and metabolic disease: the importance of nutrient flux. J Biol Chem 2014;289:20462–9. 10.1074/jbc.R114.576355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.