Abstract
T follicular helper (TFH) cell lymphomas (TFHLs) are characterized by TFH-like properties and accompanied by substantial immune-cell infiltration into tumor tissues. Nevertheless, the comprehensive understanding of tumor-cell heterogeneity and immune profiles of TFHL remains elusive. To address this, we conducted single-cell transcriptomic analysis on 9 lymph node (LN) and 16 peripheral blood (PB) samples from TFHL patients. Tumor cells were divided into 5 distinct subclusters, with significant heterogeneity observed in the expression levels of TFH markers. Copy number variation (CNV) and trajectory analyses indicated that the accumulation of CNVs, together with gene mutations, may drive the clonal evolution of tumor cells towards TFH-like and cell proliferation phenotypes. Additionally, we identified a novel tumor-cell-specific marker, PLS3. Notably, we found a significant increase in exhausted CD8+ T cells with oligoclonal expansion in TFHL LNs and PB, along with distinctive immune evasion characteristics exhibited by infiltrating regulatory T, myeloid, B, and natural killer cells. Finally, in-silico and spatial cell-cell interaction analyses revealed complex networking between tumor and immune cells, driving the formation of an immunosuppressive microenvironment. These findings highlight the remarkable tumor-cell heterogeneity and immunoevasion in TFHL beyond previous expectations, suggesting potential roles in treatment resistance.
Subject terms: T-cell lymphoma, Cancer microenvironment, Cancer therapeutic resistance, Cancer genomics
Introduction
Nodal T follicular helper cell (TFH) lymphomas (TFHLs) represent a distinct subtype of peripheral T-cell lymphomas (PTCLs), characterized by tumor cells exhibiting TFH-like properties [1–3]. Physiologically, TFH cells interact with B cells through CD40 ligand (CD40LG)-CD40 signaling, as well as by secreting chemokines and cytokines, such as CXCL13, interleukin (IL)-21, and IL-4, to support B cell proliferation and differentiation in the germinal center of follicles [4]. In TFHL, malignant T cells also express CXCL13 and CD40LG [1, 5], and patients show high serum levels of cytokines, including IL-4, IL-6, and IL-21 [6, 7]. Although this release of cytokines and chemokines may lead to the prominent infiltration into TFHL tissue by immune cells [1], the details are unknown.
TFHLs carry a poor prognosis, with a 5-year overall survival of 30–40% and chemotherapy resistance [8]. While tumor-cell heterogeneity contributes to therapeutic resistance in various cancers [9], it remains incompletely understood in TFHL. Molecularly, genomic analysis has revealed that the RHOA G17V mutation (G17V) controls TFH cell lineage differentiation, promoting the development of TFHL [10–12]. Moreover, G17V mutants acquire VAV1 binding capability, leading to activation of the T-cell receptor (TCR) signaling [10]. However, factors driving further tumor evolution and heterogeneity after tumor initiation remain poorly elucidated.
Generally, the tumor microenvironment (TME) controls immune evasion to support growth and metastasis [13], with high expression levels of immunosuppressive signatures within the TME associated with a poor TFHL prognosis [6, 7, 14, 15]. Although mounting evidence of this nodal interface indicates a primary role in tumor growth control, comprehensive immune profiling of the TFHL microenvironment remains incomplete.
Therefore, we aimed to profile the cell types and tumor cell characteristics involved in immune evasion and therapeutic resistance in TFHL by single-cell transcriptomic analysis.
Methods
Detailed methods are described in Supplementary Methods.
Human samples
This study was approved by the Institutional Review Board of the University of Tsukuba Hospital (Tsukuba, Japan) and other participating institutions and conducted in accordance with the Declaration of Helsinki. Written, informed consent was obtained from all participating patients. Prospectively, nine lymph node (LN; six from newly diagnosed [ND] patients and three from relapsed or refractory [RR] patients) and 16 peripheral blood (PB; including sequential samples from two patients) samples were collected from 14 TFHL patients (Fig. 1A; Table S1). Diagnoses were performed by expert hematopathologists at each institution. Seven homeostatic LNs (HLNs) from patients with non-hematologic cancers were collected for comparison.
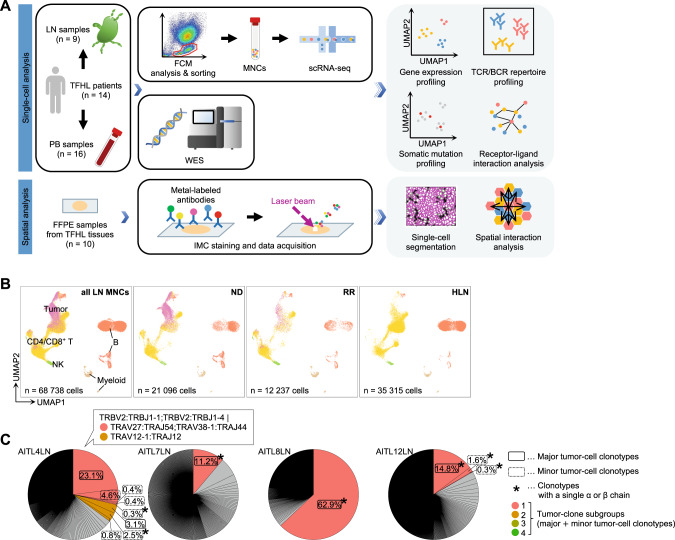
Fig. 1. Transcriptomic profile of T follicular helper cell lymphoma (TFHL) samples.
A Study overview and workflow of analysis. BCR B cell receptor, scRNA-seq, single-cell RNA sequencing, FCM flow cytometry, FFPE formalin-fixed paraffin-embedded, IMC imaging mass cytometry, LN lymph node, MNC mononuclear cell, PB peripheral blood, TCR T-cell receptor, UMAP Uniform Manifold Approximation and Projection for Dimension Reduction, WES whole-exome sequencing, B UMAP plots of all MNCs from nine TFHL LNs and seven homeostatic lymph nodes (HLNs), colored by cell type (left). Cells are shown separately for each clinical status (middle two and right). B, B cell; CD4/CD8+ T, CD4+ and CD8+ T cell; Myeloid, myeloid cell; NK, natural killer cell; ND newly diagnosed, TFHL; RR relapsed or refractory TFHL, Tumor, tumor cell. C Representative pie charts of TCR clonotypes for each sample. Tumor clones are colored.
Flow cytometry (FCM) and single-cell library construction
Mononuclear cells (MNCs) from LN and PB samples were sorted for 5′ single-cell RNA (scRNA)- and T-/B-cell receptor (scTCR/BCR)-sequencing (seq) (Fig. S1A). Additionally, T cell-enriched scRNA/TCR-seq libraries were constructed from TFHL LN CD4/CD8+ T cells when sufficient cells were available (Fig. S1B; Table S1). Single-cell libraries were constructed using the Chromium system (10x Genomics, Pleasanton, CA, USA). Publicly available 5′ scRNA/TCR/BCR-seq data of 5 PB samples from healthy donors [16] were used as controls. HLN samples were verified as malignancy-free by pan-cytokeratin negativity (Fig. S1C). TFHL tumor cells were detected as a PD1bright CD4+ T-cell population by FCM, as previously reported [17] (Fig. S1D).
Genomic mutation detection using whole-exome sequencing (WES)
WES was performed on genomic DNA extracted from LNs, PB, and buccal swabs of the 14 TFHL patients (Table S1). Somatic mutations were described by the Genomon2 pipeline (v.2.6.2, https://github.com/Genomon-Project) as previously described [18, 19] with minor modifications. Germline mutations were excluded by buccal swab comparison. Somatic copy number variations (CNVs) were called using the Genome Analysis Tool Kit (v4.2) [20].
Spatial analysis using imaging mass cytometry (IMC)
Spatial interaction analysis was performed on 10 formalin-fixed, paraffin-embedded samples from TFHL LNs using the Hyperion Imaging System (Standard Bio Tools Inc., South San Francisco, CA, USA) (Table S2). Cell-cell interactions were evaluated from cell-type spatial distributions [21].
Statistical analysis
All statistical analyses were performed using R (v3.6.0, v3.6.2, or v4.0.2) on RStudio (v1.2.1578 or v1.2.5019). The Wilcoxon rank-sum test or MAST (for single-cell data only, v1.12.0) [22] was used to analyze differences between the two groups. Two-tailed statistical tests were performed, and P < 0.05 was considered statistically significant.
Results
Single-cell transcriptomic and TCR repertoire profiles of TFHL
We first analyzed scRNA-seq data of 68,738 LN and 98,891 PB cells collected from 14 TFHL patients and controls (Fig. 1A; Table S1). Unsupervised clustering and annotation by canonical markers revealed four main cell types: CD4/CD8+ T, natural killer (NK; NK/gamma-delta [γδ] T for PB), B, and myeloid cells (Fig. 1B, Fig. S2A).
Subsequently, we analyzed LN scTCR-seq data to identify tumor cells and found unique TCR clonotypes in 93.7% of T cells (Fig. S2B, C). Major and minor tumor-cell clonotypes were defined based on clonality (Fig. 1C, Fig. S2D; Table S3). The median proportion of tumor cells in all LN MNCs was 14.1% (range 5.7–83.8%; Table S4). In 7 of 9 TFHL LNs, tumor cells were clones within a patient, suggesting single-TCR clonal expansion (Fig. 1C, Fig. S2D). Contrastingly, in AITL4, two clones with identical TCRβ chains and differing TCRα chains expanded (Fig. 1C; Table S3). Notably, 31.3% (2652 cells) of tumor cells expressed only one productive α or β chain, contrasting dogma that αβ T cells physiologically express a pair of productive α and β chains (Fig. 1C, Fig. S2D). In three TFHLs (AITL7, AITL8, and AITL12), the single-TCR clonotype was detected in most tumor cells (Fig. S2E). Moreover, in these TFHLs, FCM analysis revealed the expansion of abnormal CD4+ PD1high T-cell populations lacking cell-surface expression of TCRα and β chain-interacting CD3ε [23], which was not observed in TFHLs with physiological paired-chain TCRs (Fig. S2F, G, and data not shown). Tumor cell infiltration was also identified in 12 of 16 PB samples (0.24–74.0% in PB MNCs; Table S4), five of which showed expansion of single-chain TCR tumor cells with CD3ε- CD4+ PD1high T-cell populations by FCM, consistent with the LN data (Fig. S2F and S3A–C).
Genomic characterization of TFHL
We next analyzed somatic mutations using bulk WES data to construct genomic profiles of TFHL, finding 397 total somatic mutations in 14 TFHL patients. This included recurrent TET2 mutations in 11 (78.6%), G17V in 6 (42.9%), DNMT3A in 6 (42.9%), and IDH2 R172 in 2 (14.3%), comparable to previous reports [24–27] (Fig. S4A; Table S5). Furthermore, we analyzed CNVs and found that gain of chromosome (chr) 5 was most frequently observed (35.7%; 5/14), and gains of chr7/7q, 19, 21, or 22q were also detected in ≥2 of 14 cases (14.3%) with co-occurrence of chr5 and 21 gain in 1 case (7.1%), consistent with previous reports [28] (Fig. S4B).
Inter-patient and intra-tumor TFHL heterogeneity
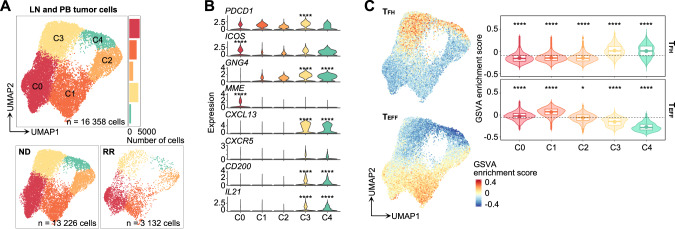
To investigate tumor cell characteristics and heterogeneity, we sorted 16,358 tumor cells from scRNA-seq data into five clusters by unsupervised clustering (C0–C4; Fig. 2A, Fig. S5A), finding consistent inter-patient heterogeneity (Fig. S5B). Notably, TFH marker [4] expressions were highly heterogeneous between clusters (Fig. 2B, Fig. S5C; Table S6). PDCD1 and ICOS were globally expressed, while MME (encoding CD10) and other TFH markers were predominantly expressed in C0 and C3–4, respectively (Fig. 2B). Subsequently, we conducted gene set variation analysis (GSVA) [29] on each cell using TFH and non-TFH effector T cell (TEFF) signatures derived from bulk RNA-seq data (GSE58596 [30]). This revealed enrichment of the TFH signature in C3–4 and the TEFF signature in C0–2 (Fig. 2C). Moreover, cell-cycle scores revealed that C0, C1, and C3 were primarily in G1 stage, but C2 and C4 were exclusively in G2M or S stages, indicating active cell proliferation in C2 and C4 (Fig. S5D).
Fig. 2. Single-cell analysis of tumor cells from TFHL LN and PB samples.
A UMAP plots of LN and PB tumor cell subclusters (top). Cells are shown separately for each clinical status (bottom). B The expression levels of known T follicular helper (TFH) cell markers for each cluster. Adjusted P values are calculated by the Wilcoxon rank-sum test with Bonferroni correction across all genes expressed by tumor cells and shown only when there is a significant difference. C Feature (left panel) and violin (right panel) plots of gene set variation analysis (GSVA) enrichment score for TFH (top) or CD4+ effector T cell (TEFF, bottom) signatures in tumor cells. In the violin plots, the dotted lines represent mean enrichment scores across all clusters. The boxplots show the median (center line), mean (center dot), interquartile range (box limits), and minimum to max values (whiskers) for each group. Adjusted P values are calculated by the pairwise Wilcoxon rank-sum test for each cluster against the mean value of all clusters. *P < 5.0 × 10−2, ****P < 1.0 × 10−4.
Next, we compared LN and PB tumor cells, revealing that C3–4 were predominant in LNs while C1–2 were more abundant in PB (Fig. S5E). Moreover, differentially expressed gene (DEG) analysis and GSVA revealed that LN tumor cells exhibited relatively higher expression of TFH markers whereas PB tumor cells showed high expression of cytotoxicity-associated genes [31] (Fig. S5F, G). FCM analysis confirmed significantly higher PD1 expression in LN tumor cells versus those of PB (Fig. S5H, I). Subsequent trajectory analysis using Monocle3 [32] inferred that LN tumor cells terminally differentiated starting from C0 through C3 to C4, whereas PB tumor cells differentiated from C0 to C2 via C1 (Fig. S5J).
We next performed DEG and gene ontology (GO) analyses between tumor cells with single-chain and paired-chain TCRs (Fig. S5K). Unexpectedly, both types had activated TCR and cytokine signaling pathways (Fig. S5L). Notably, genomic mutations known to activate TCR pathways (G17V, CTNNB1), or those in TCR signaling-related genes (LCK, KRAS) [33], were detected in 4 of 5 cases with expanded single-chain TCR clonotypes (Table S5).
Estimation of tumor evolution at single-cell resolution
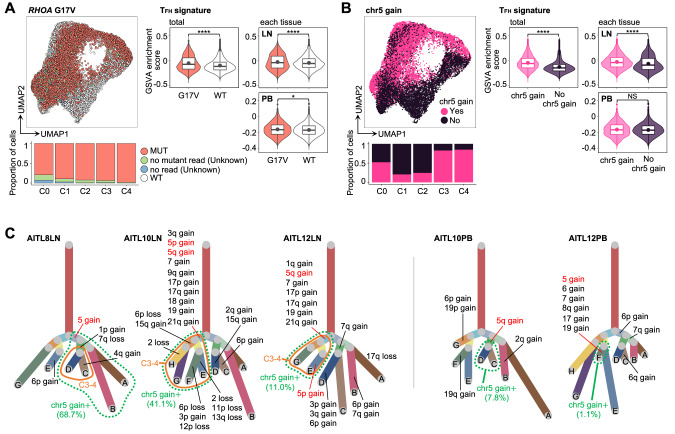
To assess mutational evolution using scRNA-seq tumor cell data, we reanalyzed the initially detected 11 somatic mutations and focused on mutations with a total read coverage ≥100×, identifying seven mutations present in over 20 cells, of which only G17V was detected in multiple patients (Fig. S6A–C; Table S7). In TFHL patients where G17V was initially identified by WES, G17V was detected in 86.7% of tumor cells while 9.4% of the cells lacked mutant reads and 3.9% had no coverage (Fig. 3A). Considering the frequent allelic dropout in scRNA-seq [34], cells without mutant reads or no coverage status were categorized as “unknown”. Conversely, cells from TFHL patients without G17V mutations by WES were dubbed “G17V wild-type (WT)” (Fig. 3A). GSVA, DEG, and GO analyses revealed enrichment of TFH signatures and pathways related to the adaptive immune system and B-cell activation, including CD40LG, in G17V mutant cells compared to G17V WT cells, consistent with our previous report [18] (Fig. 3A, Fig. S6D, E). In AITL4LN/PB, with two distinctive tumor clones featuring identical TCRβ chains and different TCRα, LRRC41 mutations were detected only in tumor-clone 1 (Fig. S6F). Based on UMAP visualization, these cells with different clones exhibited different gene expression profiles, indicating clonal evolution across divergent genetic/transcriptomic directions (Fig. S6G).
Fig. 3. Estimation of genomic alternations at the single-cell level.
A Distribution of RHOA G17V (G17V) mutant cells (left panel) and comparison of GSVA enrichment score for the TFH signature between G17V mutant (red) and wild-type (WT, white) cells (right panel) in LN and PB tumor cells. In the violin plots of the right panel, adjusted P values are calculated for all tumor cells (left) and each tissue (right). “Unknown” cells had no mutant reads or no coverage. “WT” cells were from TFHL patients without G17V mutations by WES. MUT, mutant cells. B Distribution of tumor cells with chromosome (chr)5 gain (left panel) and comparison of GSVA enrichment score for the TFH signature between tumor cells with chr5 gain (pink) and those without (blue) (right panel). In the violin plots of the right panel, adjusted P values are calculated for all tumor cells (left) and each tissue (right). NS, not significant. C Phylogenetic trees based on copy number variation (CNV) patterns identified by inferCNV [35] in TFHL LN (left panel) and PB (right panel) samples with partial chr5 gain. All boxplots show the median (center line), mean (center dot), interquartile range (box limits), and minimum to max values (whiskers) for each group. *P < 5.0 × 10−2, ****P < 1.0 × 10−4.
Next, we conducted single-cell level estimation of CNVs in LN tumor cells and classified them into subgroups based on their CNV patterns using inferCNV [35]. The estimated CNVs exhibited a high degree of consistency with paired bulk WES results (Fig. S4B and S7A). Similar to WES, the most frequently observed CNV in LN tumor cells was chr5 gain (6/9; 66.7%), comprising four samples detected by both scRNA-seq and WES, and two samples exclusively identified through scRNA-seq (AITL10LN and AITL12LN; Fig. S7A). In these two samples and AITL8LN, chr5 gain was identified only in a portion of tumor cells with the identical TCR clones, suggesting that it was acquired after TCR clonal expansion (Fig. S7A). Notably, chr5 gain was most prevalent in C3–4, showing enriched TFH phenotypes (C3–4) and active cell proliferation (C4), followed by C0 (Fig. 3B). Consistently, DEG analysis revealed that TFH-related genes, including CXCR5, CXCL13, IL21, and CD40LG, were significantly upregulated in chr5-gain tumor cells compared to those without and GSVA showed that the TFH signature enrichment was observed in total and LN tumor cells while being insignificant in PB tumor cells (Fig. 3B, Fig. S7B). Furthermore, the phylogenetic trees of partial chr5-gain samples diverged into two major branches based on the presence or absence of chr5 gain, plus the subsets with chr5 gain accompanied by other CNVs frequently detected in TFHL [28] (e.g., gains of chr7, 19, and 21) that evolved into C3–4 (Fig. 3C). In these samples, the TFH signature was significantly enriched in chr5-gain positive cells compared to non-gain (Fig. S7C). Contrastingly, in AITL10PB and AITL12PB, where partial chr5 gain was detected in LN tumor cells, fewer chr5-gain cells were detected in PB tumor cells compared to those of LNs (41.1 versus 7.8% and 11.0 versus 1.1%), and most of the tumor cells belonged to C0–2, suggesting that C0–2 without chr5 gain and TFH signatures favored infiltration into PB versus C3–4 (Fig. 3C, Fig. S7D). Indeed, CNV scores [36] were significantly higher in C3–4 and LN tumor cells than those in C0–2 and PB tumor cells, respectively, indicating the genomic complexity of C3–4 and LN tumor cells (Fig. S7E).
In summary, genomic aberrations, particularly G17V and chr5 gain, were closely correlated with distinct transcriptomic profiles in TFHL tumor cells at the single-cell resolution.
PLS3 as a novel, tumor-specific marker
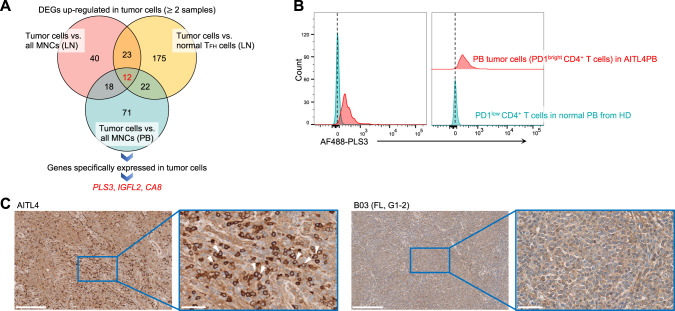
To discover novel tumor-specific markers, we conducted DEG analysis using scRNA-seq data between LN/PB tumor cells and non-malignant MNCs or normal TFH cells (for LNs only). DEGs with significantly higher expression in tumor cells across ≥2 samples, as well as those upregulated in both LN and PB tumor cells, were extracted (Fig. 4A; Table S8). Finally, PLS3, IGFL2, and CA8 were identified as candidate genes (Fig. S8A; Table S9). FCM was performed to validate PLS3, revealing high expression in both LN and PB tumor cells versus non-malignant T cells (Fig. 4B). Furthermore, immunohistochemical staining revealed that PLS3 was strongly expressed on the tumor cell-surface in 17 of 35 PTCLs (10 of 20 angioimmunoblastic T-cell lymphomas, 2 of 5 nodal PTCLs with TFH phenotypes, and 5 of 10 PTCLs not otherwise specified), accounting for 48.6% PTCL samples examined, while significant expression was observed in only one of 168 (0.6%) mature B-cell lymphomas (Fig. 4C; Table S10). These findings suggest that PLS3 is a promising, tumor-specific marker in PTCL.
Fig. 4. Identification of novel tumor-specific cell markers.
A Overview of tumor-specific cell marker identification. For both LN (n = 9) and tumor cell-bearing PB (n = 12) samples, differentially expressed gene (DEG) analysis was performed to select genes whose expression was significantly elevated in tumor cells of two or more samples versus (vs.) all MNCs or normal TFH cells (for only LN tumor cells) for all samples. Next, genes specifically expressed in tumor cells were selected by manual inspection using UMAP plots. B Representative FCM plots of PLS3 expression in PD1bright CD4+ cells from PB of TFHL (AITL4, red) and PD1dim CD4+ cells from PB of a healthy donor (HD, blue). C Representative images of PLS3 expression by immunohistochemical staining of FFPE samples from TFHL (AITL4, left) and B-cell lymphoma (B03, right). White triangles indicate tumor cells expressing PLS3 on the cell membrane surface. Images were scanned using the NanoZoomer (2.0HT, Hamamatsu Photonics, Shizuoka, Japan) and acquired at ×10 and ×40 with the NDP.view.2 software (v2.9.29). FL, follicular lymphoma; G, grade of FL according to the 4th edition of the World Health Organization classification [1]; Scale bar, 250 μm (left) and 50 μm (right).
Dysfunctional CD8+ and regulatory T-cell expansion
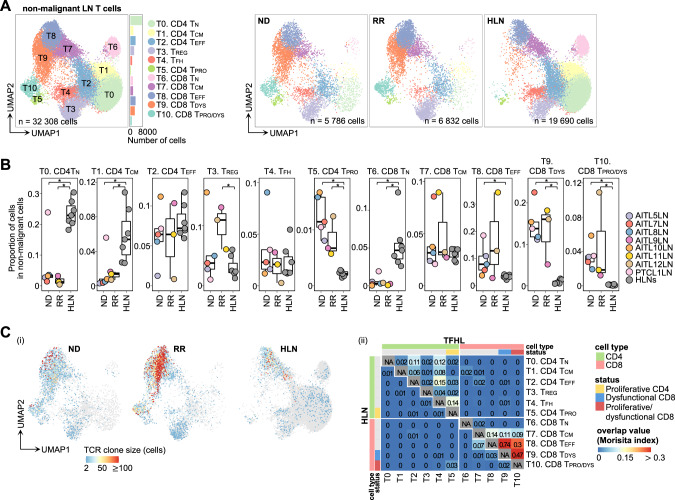
Subsequently, subclustering of non-malignant LN T cells highlighted 11 clusters (T0–10). Based on canonical markers, these clusters were classified as conventional CD4+ or CD8+ T, encompassing naïve (TN, T0 and T6), central memory (TCM, T1 and T7), TEFF (T2 and T8), regulatory T (TREG, T3), TFH (T4), proliferating CD4+ T (CD4 TPRO, T5), dysfunctional CD8+ T (CD8 TDYS, T9), and proliferating dysfunctional CD8+ T (CD8 TPRO/DYS, T10) (Fig. 5A, Fig. S9A–E). T9 and T10 expressed “exhaustion” or “dysfunctional” markers such as PDCD1, HAVCR2 (encoding TIM-3), TIGIT, and LAG3 [37, 38] (Fig. S9C, D). T5 was subclassified into two FOXP3+ IL2RA+ clusters (T5 FOXP3-1 and T5 FOXP3-2) and one cluster positive for the TFH marker (T5 PDCD1) (Fig. S9F), suggesting that T5 was a mixture of proliferating TREG and TFH. These manual annotations were further validated using SingleR [39, 40] (Fig. S9G).
Fig. 5. Subclustering of non-malignant T cells from LNs.
A UMAP plots of non-malignant LN T cell subclusters. Cells are shown separately for each clinical status (right panels). TCM, central memory T cell; TDYS, dysfunctional T cell; TEFF, effector T cell; TN, naïve T cell; TPRO, proliferative T cell; TPRO/DYS, proliferative dysfunctional T cell; TREG, regulatory T cell. B Comparison of proportions of each cluster in non-malignant MNCs per sample. The boxplots show the median (center line), interquartile range (box limits), minimum to max values (whiskers), and samples (dots) for each group. P values are shown only for significant differences. *P < 5.0 × 10−2. C Clone size (i) and overlap analysis (ii) of TCRs in non-malignant LN T cells. The number of cells expressing each clonotype was defined as clone size and illustrated for each cell. The “repOverlap” function of Immunarch measured the TCR sharing between each cluster, analyzed and illustrated for TFHL LNs (upper right) and HLNs (lower left), respectively.
The proportions of CD4 TPRO and CD8 TDYS/TPRO/DYS significantly increased in TFHL LNs versus HLNs, whereas those of CD4/CD8 TN and CD4 TCM decreased as previously reported [41] (Fig. 5B). Additionally, the proportion of TREG was higher in RR TFHL LNs than HLNs (Fig. 5B, Fig. S9H). Moreover, compared with HLNs, TREG in RR TFHL LNs exhibited higher expressions of genes associated with TREG inhibitory functions, such as FOXP3, IL2RA, TNFRSF4, TNFRSF9, and LAG3 [42], suggesting TREG activation in RR TFHL (Fig. S9I; Table S11). Additionally, non-malignant PB T cell subclustering demonstrated the expansion of CD8 TDYS/TPRO/DYS and upregulation of activated markers in TREG from TFHL (Fig. S10A–F; Supplementary Note 1).
We next analyzed the TCR clonality of non-malignant LN T cells and identified oligo-clonal expansion of TCRs in CD8 TEFF, TDYS, and TPRO/DYS of TFHL (Fig. 5C(i), Fig. S10G). Indicative of a common cellular origin, these cells partly shared identical TCRs between clusters, an overlap not observed in HLNs (Fig. 5C(ii)). Moreover, trajectory analysis revealed that a portion of CD8 TEFF diverged and differentiated into CD8 TDYS and TPRO/DYS in TFHL, consistent with TCR analysis (Fig. S10H). Notably, TCR commonality was observed between LN and PB CD8 TPRO/DYS of TFHL, suggesting that enrichment of immunoevasive phenotypes in PB potentially reflects the LN immune environment (Fig. S10I).
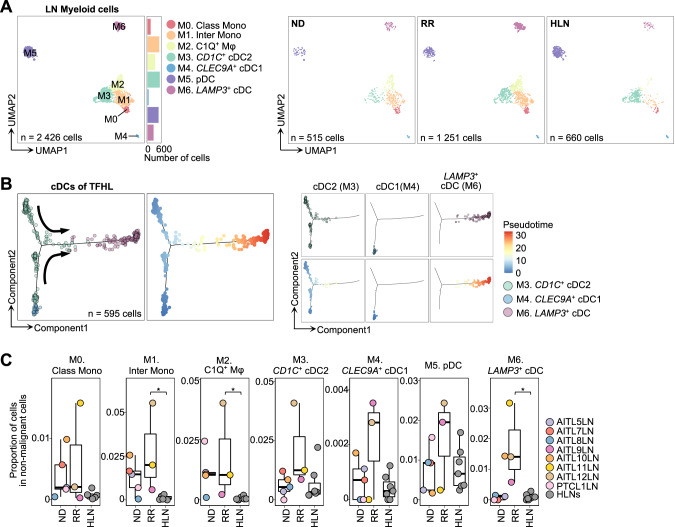
Increased immunosuppressive myeloid cells
Subclustering LN myeloid cells distinguished seven clusters (M0–6; Fig. 6A). Employing canonical markers and SingleR analysis [43, 44], we annotated each cluster as classical and intermediate monocytes (Class Mono, M0; Inter Mono, M1, respectively), complement component 1Q (C1Q)-positive macrophages (C1Q+ Mφ, M2), CD1C-positive type 2 conventional dendritic cells (CD1C+ cDC2, M3), CLEC9A-positive type 1 cDCs (CLEC9A+ cDC1, M4), plasmacytoid DCs (pDC, M5), and LAMP3-positive cDCs (LAMP3+ cDC, M6) (Fig. 6A, Fig. S11A–D).
Fig. 6. Subclustering of myeloid cells from LNs.
A UMAP plots of LN myeloid cell subclusters. Cells are shown separately for each clinical status (right panels). C1Q+ Mφ, complement component C1q positive macrophage; CD1C+ cDC2, CD1C-positive type2 conventional dendritic cell; Class Mono, classical monocyte; CLEC9A+ cDC1, CLEC9A-positive type1 cDC; Inter Mono, intermediate monocyte; LAMP3+ cDC, LAMP3-positive cDC; pDC, plasmacytoid DC. B Trajectory inference by Monocle2 for cDCs of TFHL, color-coded by cluster (left of the left panel) and pseudo-time (right of the left panel). Cells are shown separately for each cluster (right panel). C Comparison of proportions of each cluster in non-malignant MNCs per sample. The boxplots show the median (center line), interquartile range (box limits), minimum to max values (whiskers), and samples (dots) for each group. P values are shown only when there is a significant difference. *P < 5.0 × 10−2.
Notably, C1Q+ Mφ exhibited high C1Q-family gene expression (C1QA, C1QB, and C1QC), CD163 expression, and ligands for T-cell immune checkpoints, including LGALS9 (binds to TIM-3) and NECTIN2 (binds to TIGIT) (Fig. S11C), resembling tumor-associated macrophages with immunosuppressive signatures reported in multiple cancers [43, 45]. Trajectory inference showed a continuous spectrum from Class/Inter Mono to C1Q+ Mφ (Fig. S11E). LAMP3+ cDCs were characterized by high immune regulatory gene expression, including CD274 (encoding PD-L1) and PDCD1LG2 (encoding PD-L2) [46], consistent with “mregDCs,” a subset of DCs co-expressing maturation and immunoregulatory genes identified in various cancers [43, 46] (Fig. S11B, C and F). LAMP3+ cDCs have been proposed to develop from either cDC1s or cDC2s [43, 46]. Using a previously reported scoring system [43, 46], we inferred that most LAMP3+ cDCs in TFHL originated from cDC2s, unlike many cancers [43, 46] (Fig. S11G). Trajectory analyses further supported this, revealing a continuous connection from cDC2 to LAMP3+ cDCs (Fig. 6B). Next, we compared the proportions of each cluster and observed a significant increase in Inter Mono, C1Q+ Mφ, and LAMP3+ cDCs in RR TFHL LNs than in HLNs (Fig. 6C). Finally, DEG analysis showed the expression level of CCL17, a ligand for CCR4 [47], was significantly higher in LAMP3+ cDCs of RR TFHL LNs than HLNs (Fig. S11H, I).
Taken together, a distinct increase in immunoregulatory myeloid cells, particularly LAMP3+ cDCs, was observed in RR TFHL, which could support an immune-evasive TME.
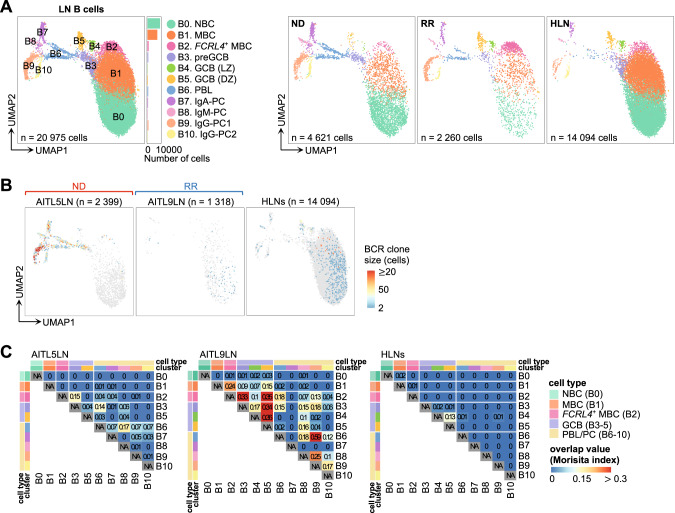
Exhausted phenotype and clonal expansion of B cells
Subclustering analysis of LN B cells revealed 11 clusters: naïve B cell (NBC, B0), memory B cell (MBC, B1), FCRL4-positive MBC (FCRL4+ MBC, B2), pre-germinal center B cell (preGCB, B3), light-zone GCB (GCB [LZ], B4), dark-zone GCB (GCB [DZ], B5), plasmablast (PBL, B6), and plasma cell (PC, B7–10) (Fig. 7A, Fig. S12A–C). FCRL4+ MBCs displayed “exhausted” or “atypical” MBC phenotypes, as observed in chronic inflammatory or immunodeficiency conditions [48–50], distinguished by high expression of TBX21 (encoding T-bet), B cell inhibitory receptor genes (FCRL4 and SIGLEC6 [49]), and homing receptor genes (CXCR3 and ITGAX [48, 49]) as well as low expression of CR2 (encoding CD21) and CD27 (Fig. S12D; Table S12). Compared to HLNs, the proportions of each cluster varied between TFHL patients (Fig. S12A). While statistically insignificant, the proportion of FCRL4+ MBCs was markedly increased (>20% of B cells) in 2 (AITL9LN and AITL12LN) out of 3 RR TFHLs (Fig. S12A). Moreover, DEG analysis highlighted the greater upregulation of genes related to exhausted MBC phenotypes [48–51] in FCRL4+ MBCs from RR TFHL LNs versus HLNs, although not significantly in ND TFHL (Fig. S12E; Table S13). Additionally, GO analysis and gene set enrichment analysis revealed the extensive enrichment of GCB- and CD40-related and AITL-specific GCB signatures [18] in broad B-cell clusters of TFHL, consistent with our previous findings [18] (Fig. S12F, G; Table S14).
Fig. 7. Subclustering of B cells from LNs.
A UMAP plots of LN B cell subclusters. Cells are shown separately for each clinical status (right panels). FCRL4+ MBC, FCRL4-positive memory B cell; GCB (DZ), germinal center B cell in the dark zone; GCB (LZ), GCB in the light zone; MBC, memory B cell; NBC, naïve B cell; PBL, plasmablast; PC, plasma cell; preGCB, pre-germinal center B cell. B Clone size of BCRs in B cells from AITL5LN, AITL9LN, and HLNs. The number of cells expressing each clonotype was defined as clone size and calculated for each sample. HLN samples are shown as integrated (right). C BCR overlap analysis of B cells from AITL5LN, AITL9LN, and HLNs. AITL5LN and AITL9LN were analyzed individually while HLN samples were analyzed after integration.
Repertoire analysis was performed for five TFHL LNs (AITL5LN and AITL7–10LN), as well as all HLNs, revealing clonally expanded BCRs of GCBs and PBL/PCs in AITL5LN and AITL9LN (Fig. 7B, Fig. S12H–I). The BCRs of GCBs and PBL/PCs from AITL5LN and AITL9LN overlapped with FCRL4+ MBCs (Fig. 7C). Particularly in AITL9LN, trajectory analysis showed continuity of trajectories from GCBs to FCRL4+ MBCs and PBL/PCs among the cells sharing BCRs (Fig. S12J).
NK-cell activation and exhaustion in RR TFHL
LN NK cell subclustering identified three clusters: proliferative NK (NKPRO, NK0), XCL1-positive tissue-resident NK [52] (XCL1+ NK, NK1), and FCGR3A (encoding CD16)-positive activated NK [53] (FCGR3A+ NK, NK2) (Fig. S13A–D). We found increases in the proportions of NKPRO and FCGR3A+ NKs in TFHL LNs compared to all TFHL and HLNs (Fig. S13E). Moreover, DEG and GO analyses revealed proinflammatory cytokine pathway enhancement (e.g., interferon-γ) as well as reduced expression of a receptor-activating adapter molecule, TYROBP [54] (encoding DAP12), specifically in XCL1+ NK of TFHL LNs versus HLNs (Fig. S13F; Table S15, 16). Additionally, higher expressions of inhibitory genes, such as HAVCR2 and KLRC1 (encoding NKG2A) [53], were observed in XCL1+ NKs of RR TFHL but not in those of ND TFHL (Fig. S13G; Table S15, 16). These data suggest sustained activation of NK cells in TFHL LNs and subsequent elevation of inhibitory signaling, particularly in RR TFHL.
In-silico and spatial intercellular interactions and formation of the TFHL immune-evasive microenvironment
To investigate the cellular interactions within the TME of TFHL LNs, we conducted in silico cell-cell network analysis using sc-RNAseq data through CellphoneDB [55] and NicheNetR [56] pipelines. The CellphoneDB analysis identified significant crosstalk among various cell types related to immunoregulation, including TREG, CD8 TDYS, FCRL4+ MBCs, C1Q+ Mφ, and tumor cells, which was more prominent in RR than ND TFHL (Fig. S14A). Moreover, the NicheNetR analysis revealed the potential ligands expressed by C3–4 of LN tumor cells that drive exhausted signatures of CD8 TDYS in RR TFHL, including IL21, TNF, TGFB1, CD80, and CD86 (Fig. S14B–D).
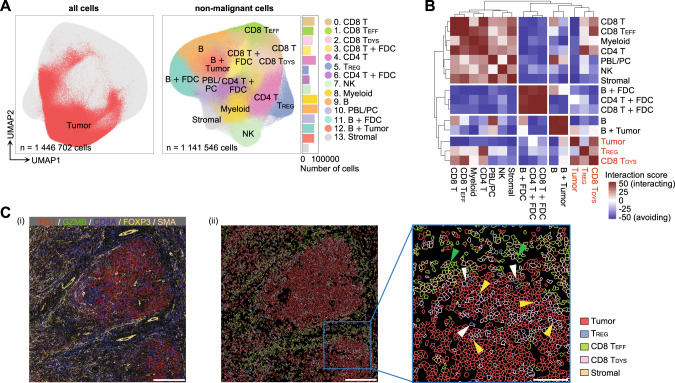
To further validate the results of scRNA-seq data, we performed single-cell spatial analysis by IMC using 27 antibodies (Table S17) for samples from 10 TFHL patients (Table S2). Overall, 1,446,702 cells were identified by the single-cell segmentation process. Unsupervised clustering revealed expansion of a TFH-like tumor cell cluster (Fig. 8A, Fig. S15A, B). Subsequent clustering of non-malignant cells identified 14 subclusters, including GZMBhigh PD1low CD8 TEFF (cluster 1), GZMBlow PD1high CD8 TDYS (cluster 2), and TREG (cluster 5) (Fig. 8A, Fig. S15C, D; Table S18; Supplementary Note 2).
Fig. 8. Cell-cell interaction analysis between tumor and immune cells from TFHL tissues.
A UMAP plots of all (left) and non-malignant (right) cells detected by single-cell segmentation of IMC data from 10 FFPE samples of TFHL tissues. The tumor-cell cluster, identified by unsupervised clustering of all cells, is colored in red (left). The non-malignant cells are color-coded by subclusters identified by unsupervised clustering without the tumor-cell cluster (right). FDC, follicular dendritic cell; Stromal, stromal cell; +, a mixture of two cell types. B Spatial interaction analysis for each cell type using IMC data of TFHL tissues. Higher interaction scores indicate closer distance and more interaction between cells, whereas lower scores indicate farther distance and less interaction. C Representative IMC images (P5) color-coded by (i) expression levels of markers for CD8 TDYS (PD1), CD8 TEFF (GZMB), TREG (FOXP3), and stromal cells (SMA) and (ii) cell types identified by clustering at the single-cell resolution. In panel (ii), only tumor cells, stromal cells, TREG (white arrowheads), CD8 TEFF (green arrowheads), and CD8 TDYS (yellow arrowheads) are shown. Scale bar, 300 μm ((i) and left of (ii)) and 100 μm (right of (ii)).
To assess cell-cell interactions [21], intercellular distances were calculated from the spatial distribution of each cell (Table S19). Tumor cells were significantly adjacent to CD8 TDYS but distant from CD8 TEFF and TREG were adjacent to CD8 TDYS, suggesting potential cell-cell interactions within the TFHL TME (Fig. 8B–C).
Discussion
Integration of single-cell and spatial analyses allowed us to comprehensively elucidate tumor cell profiles, as well as immunosuppressive and dysfunctional immune cells, revealing synergy in establishing an immunoevasive TME that co-evolves with disease progression.
Although increased exhausted CD8+ T cells and TREGs have been reported in TFHL [41], as well as solid cancers [37, 38], we found oligoclonal expansion and TCR commonality of CD8 TDYS between LNs and PB for the first time in TFHL. Persistent antigenic stimulation and TREG infiltration might lead to clonal expansion and augment exhaustion phenotypes of CD8+ T cells [38]. Clonal proliferation of B-lineage cells, as well as T-lineage tumor cells, is well reported for TFHL [1–3]. We previously reported that abnormal GCBs clonally expand under direct interaction with tumor cells [18]. In this study, we further identified that exhausted MBCs, resembling MBC subsets that regulate inappropriate immune responses in chronic infections [48, 49], also expanded with common clones with GCBs and PCs in TFHL. By employing single-cell analysis, we identified immunosuppressive LAMP3+ cDCs, likely promoting TREG migration into tumor tissues through the CCL17-CCR4 axis [47], in addition to increased CD163-positive macrophages previously associated with adverse TFHL prognoses [7, 14].
Tumor cells may acquire TFH-like and cell proliferation phenotypes during clonal evolution through CNV accumulation, including chr5 gain, consistent with the previously reported increased expression of T-cell activation-associated genes, including IL4, in cases with chr5 gain [28]. Acquisition of TFH-like properties by tumor cells may modulate the TME to impart a self-survival advantage, driving self-activation and proliferation through interactions with immune cells [4]. As a limitation of tumor cell analysis in this study, AITL4PB accounts for the majority of PB tumor cells, which may lead to biased characteristics of PB tumor cells.
Intriguingly, the absence of CD3 surface expression in tumor cells, a known hallmark of TFHL [57], was associated with aberrant single-chain TCRs. Despite lacking a functional TCR-CD3 complex, these tumor cells exhibited upregulated TCR signaling pathways essential for T-cell development and survival [58] and frequent TCR-related gene mutations, indicating the acquisition of autonomous TCR signaling capacity during tumor progression without external antigen stimulation.
We identified PLS3 as a tumor-specific marker through scRNA-seq and further confirmed its protein expression on the PTCL tumor cell-surface. PLS3, an actin-bundling protein, is physiologically expressed in cells within solid tissues such as fibroblasts and endothelial cells [59]. Notably, PLS3 is ectopically expressed in Sézary syndrome tumor cells, a leukemic variant of cutaneous T-cell lymphomas [60], but remains uninvestigated in other T-cell lymphomas. In our cohort of nodal PTCLs, approximately half expressed PLS3, with or without TFH phenotypes. Moreover, we found that the expression profiles of known TFH markers were very heterogeneous even within the same TFHL sample. PD1, a broadly positive marker for TFHL tumor cells, is also detectable in normal TFH cells; combining PD1 with PLS3 may thus enhance diagnostic accuracy.
Overall, our study provides valuable insights into tumor-immune interactions and tumor cell characteristics mediated by genomic alterations which contribute to an immunoevasive environment in TFHL. This study may lead to the development of innovative therapeutic strategies targeting tumor immunity, ultimately addressing the challenge of therapeutic resistance in TFHL.
Supplementary information
Acknowledgements
We thank the staff at the associated institutions and departments for helping with human sample collection, as well as Y. Sakashita and E. Matsuzawa for technical assistance. The authors would like to thank Dr. Bryan J. Mathis of the International Medical Center (University of Tsukuba Hospital, Tsukuba, Japan) and Editage (www.editage.com) for language editing. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI: JP23K15317 [SS], JP20J20851 [YA], JP23K15316 [KH], JP23K15293 [KM], JP21K16261 [TS], and JP22K19451 and JP21H02945 [MS-Y]) from the Ministry of Education, Culture, Sports, and Science of Japan; AMED (JP22ck0106544, JP23ck0106797, the Moonshot Research and Development Program [JP22zf0127009] [MS-Y]); Japan Leukemia Research Fund, SENSHIN Medical Research Foundation, SGH Foundation, the Yasuda Medical Foundation, and Daiichi Sankyo Foundation of Life Science [YA]; and Kobayashi Foundation for Cancer Research, Kobayashi Foundation, and the Chemo-Sero Therapeutic Research Institute [MS-Y].
Author contributions
Contribution: SS collected the human samples, performed the experiments and computational analyses, and generated all figures and tables; YS built the environment of data analysis using a supercomputer and supported the experiments and computational analyses; MS-Y and MF developed the experimental and analytical scRNA-seq systems; YA, KH, KM, TS, AS, HB, DK, TS, KK, KA, KM and SC contributed to human sample collection; JC and NN performed IHC staining and judged positivity of TFH markers and PLS3; AS and YS contributed to scRNA-seq analysis; KI performed IMC staining and acquisition of MCD data; HS contributed to spatial analysis; MS-Y conceived the study; SS and MS-Y designed the project and wrote the manuscript; all authors participated in the discussion and interpretation of the data and results.
Data availability
The scRNA-seq data are deposited in the European Genome-phenome Archive and available upon request. All other data are available from the corresponding author upon reasonable request (sakatama@md.tsukuba.ac.jp [MS-Y]).
Competing interests
KI and HS are employees of Eisai Co., Ltd., and this research was funded by Eisai Co., Ltd. Other authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-023-02093-7.
References
- 1.Swerdlow SH, Campo E, Pileri SA, Lee Harris N, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campo E, Jaffe ES, Cook JR, Quintanilla-Martinez L, Swerdlow SH, Anderson KC, et al. The international consensus classification of mature lymphoid neoplasms: a report from the clinical advisory committee. Blood. 2022;140:1229–53. doi: 10.1182/blood.2022015851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36:1720–48. doi: 10.1038/s41375-022-01620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–48. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Leval L, Rickman DS, Thielen C, De Reynies A, Huang YL, Delsol G, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109:4952–63. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 6.Yi JH, Ryu KJ, Ko YH, Kim WS, Kim SJ. Profiles of serum cytokines and their clinical implications in patients with peripheral T-cell lymphoma. Cytokine. 2019;113:371–9. doi: 10.1016/j.cyto.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Ham JS, Park HY, Ryu KJ, Ko YH, Kim WS, Kim SJ. Elevated serum interleukin-10 level and M2 macrophage infiltration are associated with poor survival in angioimmunoblastic T-cell lymphoma. Oncotarget. 2017;8:76231–40. doi: 10.18632/oncotarget.19301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vose JM, Neumann M, Harris ME. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes international T-cell lymphoma project. J Clin Oncol. 2008;26:4124–30. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- 9.Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–53. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujisawa M, Sakata-Yanagimoto M, Nishizawa S, Komori D, Gershon P, Kiryu M, et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia. 2018;32:694–702. doi: 10.1038/leu.2017.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes JR, Ambesi-Impiombato A, Couronné L, Quinn SA, Kim CS, da Silva Almeida AC, et al. RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018;33:259–273.e7. doi: 10.1016/j.ccell.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng SY, Brown L, Stevenson K, DeSouza T, Aster JC, Louissaint A, et al. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood. 2018;132:935–47. doi: 10.1182/blood-2017-11-818617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niino D, Komohara Y, Murayama T, Aoki R, Kimura Y, Hashikawa K, et al. Ratio of M2 macrophage expression is closely associated with poor prognosis for Angioimmunoblastic T-cell lymphoma (AITL) Pathol Int. 2010;60:278–83. doi: 10.1111/j.1440-1827.2010.02514.x. [DOI] [PubMed] [Google Scholar]
- 15.Iqbal J, Weisenburger DD, Greiner TC, Vose JM, McKeithan T, Kucuk C, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010;115:1026–36. doi: 10.1182/blood-2009-06-227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yabe M, Gao Q, Ozkaya N, Huet S, Lewis N, Pichardo JD, et al. Bright PD-1 expression by flow cytometry is a powerful tool for diagnosis and monitoring of angioimmunoblastic T-cell lymphoma. Blood Cancer J. 2020;10:32. doi: 10.1038/s41408-020-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujisawa M, Nguyen TB, Abe Y, Suehara Y, Fukumoto K, Suma S, et al. Clonal germinal center B cells function as a niche for T-cell lymphoma. Blood. 2022;140:1937–50. doi: 10.1182/blood.2022015451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe Y, Sakata-Yanagimoto M, Fujisawa M, Miyoshi H, Suehara Y, Hattori K, et al. A single-cell atlas of non-haematopoietic cells in human lymph nodes and lymphoma reveals a landscape of stromal remodelling. Nat Cell Biol. 2022;24:565–78. doi: 10.1038/s41556-022-00866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14:873–6. doi: 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong D, Zheng L, Lin J, Zhang B, Zhu Y, Li N, et al. Structural basis of assembly of the human T cell receptor-CD3 complex. Nature. 2019;573:546–52. doi: 10.1038/s41586-019-1537-0. [DOI] [PubMed] [Google Scholar]
- 24.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–5. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 25.Cairns RA, Iqbal J, Lemonnier F, Kucuk C, De Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–3. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–6. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 27.Lemonnier F, Couronné L, Parrens M, Jaïs JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with T FH-like features and adverse clinical parameters. Blood. 2012;120:1466–9. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 28.Heavican TB, Bouska A, Yu J, Lone W, Amador C, Gong Q, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019;133:1664–76. doi: 10.1182/blood-2018-09-872549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstein JS, Lezon-Geyda K, Maksimova Y, Craft S, Zhang Y, Su M, et al. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood. 2014;124:3719–29. doi: 10.1182/blood-2014-06-582700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978–85. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 32.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallois D, Dobay MPD, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128:1490–502. doi: 10.1182/blood-2016-02-698977. [DOI] [PubMed] [Google Scholar]
- 34.Petti AA, Williams SR, Miller CA, Fiddes IT, Srivatsan SN, Chen DY, et al. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun. 2019;10:3660. doi: 10.1038/s41467-019-11591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–38. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–99. [DOI] [PMC free article] [PubMed]
- 38.van der Leun AM, Thommen DS, Schumacher TN. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20:218–32. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–72. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–72. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 41.Pritchett JC, Yang Z-Z, Kim HJ, Villasboas JC, Tang X, Jalali S, et al. High-dimensional and single-cell transcriptome analysis of the tumor microenvironment in angioimmunoblastic T cell lymphoma (AITL) Leukemia. 2022;36:165–76. doi: 10.1038/s41375-021-01321-2. [DOI] [PubMed] [Google Scholar]
- 42.Sakaguchi S, Mikami N, Wing JB, Tanaka A, Ichiyama K, Ohkura N. Regulatory T Cells and Human Disease. Annu Rev Immunol. 2020;38:541–66. doi: 10.1146/annurev-immunol-042718-041717. [DOI] [PubMed] [Google Scholar]
- 43.Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.e23. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. [DOI] [PMC free article] [PubMed]
- 45.Obradovic A, Chowdhury N, Haake SM, Ager C, Wang V, Vlahos L, et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell. 2021;184:2988–3005.e16. doi: 10.1016/j.cell.2021.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 2020;580:257–62. doi: 10.1038/s41586-020-2134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishida T, Ueda R. CCR4 as a novel molecular target for immunotherapy of cancer. Cancer Sci. 2006;97:1139–46. doi: 10.1111/j.1349-7006.2006.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O’Shea MA, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008;205:1797–805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kardava L, Moir S, Wang W, Ho J, Buckner CM, Posada JG, et al. Attenuation of HIV-associated human B cell exhaustion by siRNA downregulation of inhibitory receptors. J Clin Invest. 2011;121:2614–24. doi: 10.1172/JCI45685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warnatz K, Wehr C, Dräger R, Schmidt S, Eibel H, Schlesier M, et al. Expansion of CD19(hi)CD21(lo/neg) B cells in common variable immunodeficiency (CVID) patients with autoimmune cytopenia. Immunobiology. 2002;206:502–13. doi: 10.1078/0171-2985-00198. [DOI] [PubMed] [Google Scholar]
- 51.Ehrhardt GRA, Hijikata A, Kitamura H, Ohara O, Wang J-Y, Cooper MD. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med. 2008;205:1807–17. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subedi N, Verhagen LP, Bosman EM, van Roessel I, Tel J. Understanding natural killer cell biology from a single cell perspective. Cell Immunol. 2022;373:104497. doi: 10.1016/j.cellimm.2022.104497. [DOI] [PubMed] [Google Scholar]
- 53.Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19:200–18. doi: 10.1038/s41573-019-0052-1. [DOI] [PubMed] [Google Scholar]
- 54.Coudert JD, Zimmer J, Tomasello E, Cebecauer M, Colonna M, Vivier E, et al. Altered NKG2D function in NK cells induced by chronic exposure to NKG2D ligand-expressing tumor cells. Blood. 2005;106:1711–7. doi: 10.1182/blood-2005-03-0918. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Alonso L, Handfield L-F, Roberts K, Nikolakopoulou K, Fernando RC, Gardner L, et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat Genet. 2021;53:1698–711. doi: 10.1038/s41588-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17:159–62. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 57.Loghavi S, Wang SA, Jeffrey Medeiros L, Jorgensen JL, Li X, Xu-Monette ZY, et al. Immunophenotypic and diagnostic characterization of angioimmunoblastic T-cell lymphoma by advanced flow cytometric technology. Leuk Lymphoma. 2016;57:2804–12. doi: 10.3109/10428194.2016.1170827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hwang J-R, Byeon Y, Kim D, Park S-G. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp Mol Med. 2020;52:750–61. doi: 10.1038/s12276-020-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin CS, Park T, Chen ZP, Leavitt J. Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J Biol Chem. 1993;268:2781–92. doi: 10.1016/S0021-9258(18)53842-4. [DOI] [PubMed] [Google Scholar]
- 60.Kari L, Loboda A, Nebozhyn M, Rook AH, Vonderheid EC, Nichols C, et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med. 2003;197:1477–88. doi: 10.1084/jem.20021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data are deposited in the European Genome-phenome Archive and available upon request. All other data are available from the corresponding author upon reasonable request (sakatama@md.tsukuba.ac.jp [MS-Y]).