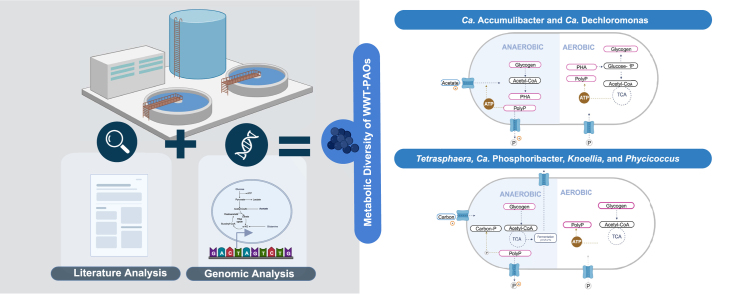
Abstract
Currently, the most cost-effective and efficient method for phosphorus (P) removal from wastewater is enhanced biological P removal (EPBR) via polyphosphate-accumulating organisms (PAOs). This study integrates a literature review with genomic analysis to uncover the phylogenetic and metabolic diversity of the relevant PAOs for wastewater treatment. The findings highlight significant differences in the metabolic capabilities of PAOs relevant to wastewater treatment. Notably, Candidatus Dechloromonas and Candidatus Accumulibacter can synthesize polyhydroxyalkanoates, possess specific enzymes for ATP production from polyphosphate, and have electrochemical transporters for acetate and C4-dicarboxylates. In contrast, Tetrasphaera, Candidatus Phosphoribacter, Knoellia, and Phycicoccus possess PolyP-glucokinase and electrochemical transporters for sugars/amino acids. Additionally, this review explores various detection methods for polyphosphate and PAOs in activated sludge wastewater treatment plants. Notably, FISH-Raman spectroscopy emerges as one of the most advanced detection techniques. Overall, this review provides critical insights into PAO research, underscoring the need for enhanced strategies in biological phosphorus removal.
Keywords: Polyphosphate-accumulating organisms (PAOs), Enhanced biological phosphorus removal (EBPR), Tetrasphaera, Ca. accumulibacter, Ca. phosphoribacter, Knoellia, Phycicoccus
Graphical abstract
Highlights
-
•
We integrate literature review and genomic analysis to investigate WWT-relevant PAOs.
-
•
PAOs can harvest energy from PolyP via ADP (pap) or sugar phosphorylation (ppgk).
-
•
Ca. Dechloromonas & Ca. Accumulibacter produce ADP from PolyP.
-
•
Tetrasphaera, Ca. Phosphoribacter, Knoellia, & Phycicoccus use sugar phosphorylation.
-
•
WWT-relevant PAOs exhibit metabolic variance in PHA, glycogen & denitrification.
1. Introduction
Before the Industrial Revolution, erosion was the main source of phosphorus (P) in water bodies. However, as time progressed, wastewater and agricultural run-off have become the primary source of phosphorus pollution due to the disposal of fertilizers, detergents, and human waste [1]. Unfortunately, this has led to eutrophication, where dissolved P stimulates the growth of microorganisms and aquatic plant life, reducing dissolved oxygen (DO) and biodiversity [2]. To prevent eutrophication, regulatory limits have been established for the amount of P present in the effluent of wastewater treatment plants (WWTPs) discharged into water bodies. For instance, the European Environment Agency requires an 80% reduction in P release or a limit of 1–2 mg L−1 in the effluent of WWTPs [3]. Meanwhile, the U.S. Environmental Protection Agency mandates an effluent P level of less than 1 mg L−1 in WWTPs [4]. To achieve these standards, various chemical and biological methods have been developed and implemented for the removal of P from wastewater [5].
Currently, the most cost-effective and efficient method for P removal from wastewater is enhanced biological P removal (EPBR) [6,7]. In the EBPR process, bacteria uptake P from their environment for growth and metabolism [8]. This process, known as P assimilation, contributes to nearly 50% of the total P removal in WWTPs [8]. However, to achieve higher removal of P, the presence of bacteria possessing the ability to not only assimilate P but also to uptake an excess amount of P beyond their assimilatory demand is essential. In this regard, polyphosphate-accumulating organisms (PAOs) play a critical role in P removal due to their capacity to store P in the form of polyphosphate (PolyP). Traditionally, the activity of PAOs encompasses P uptake under aerobic conditions to synthesize PolyP, and in the anaerobic phase, PolyP is hydrolyzed, and P is released for energy generation and substrate uptake [9].
The relevance of PAOs in EBPR has led to extensive research focusing on uncovering new PAOs and gaining a deeper understanding of their metabolic capacities and phylogenetic relationships [[10], [11], [12], [13], [14], [15]]. Furthermore, several review articles have provided valuable insights into various aspects of PAOs, including their distribution in diverse ecosystems [1], the influence of operational conditions on their performance [16], the mechanisms by which they derive reducing power in anaerobic metabolism [17], and a general overview of the metabolic capabilities of Ca. Accumulibacter, Ca. Dechloromonas, and Tetrasphaera [16]. Additionally, other reviews have explored techniques for enriching and isolating PAOs [18], methods for detecting PolyP in microbial cells [19], and methodologies for characterizing P species in natural environments [20]. However, existing reviews do not cover the discovery of novel PAOs, such as Ca. Phosphoribacter, nor do they properly discuss the reclassification of Tetrasphaera into Ca. Phosphoribacter, Knoellia, and Phycicoccus. Furthermore, there is a noticeable lack of emphasis on the existing methodologies for detecting and targeting PAOs in sludge from WWTPs.
Thus, it is crucial to have an updated review that thoroughly covers the phylogenetic and metabolic diversity of all existing wastewater treatment (WWT)-relevant PAOs, along with the latest detection methods. To address these gaps, this review integrates for the first time existing literature with metagenome-assembled genomes (MAGs) analysis of WWT-relevant PAOs, including Ca. Accumulibacter, Ca. Dechloromonas, Tetrasphaera, Ca. Phosphoribacter, Knoellia, and Phycicoccus. This approach allows for exploring functional capabilities not explicitly described in previous reviews and facilitates comparisons of relevant metabolisms across different species and genera. Moreover, the study provides a comprehensive discussion of past and recent detection methods for PAOs, including a detailed list of probes designed for targeting PAOs, PolyP detection methods, and methodologies for identifying PAO activity in complex samples. The key motivation behind this review is to fill the knowledge gaps in WWT-relevant PAOs by comprehensively exploring their phylogenetic diversity, metabolic capabilities, and the latest detection methods.
2. Phylogenetic diversity of PAOs
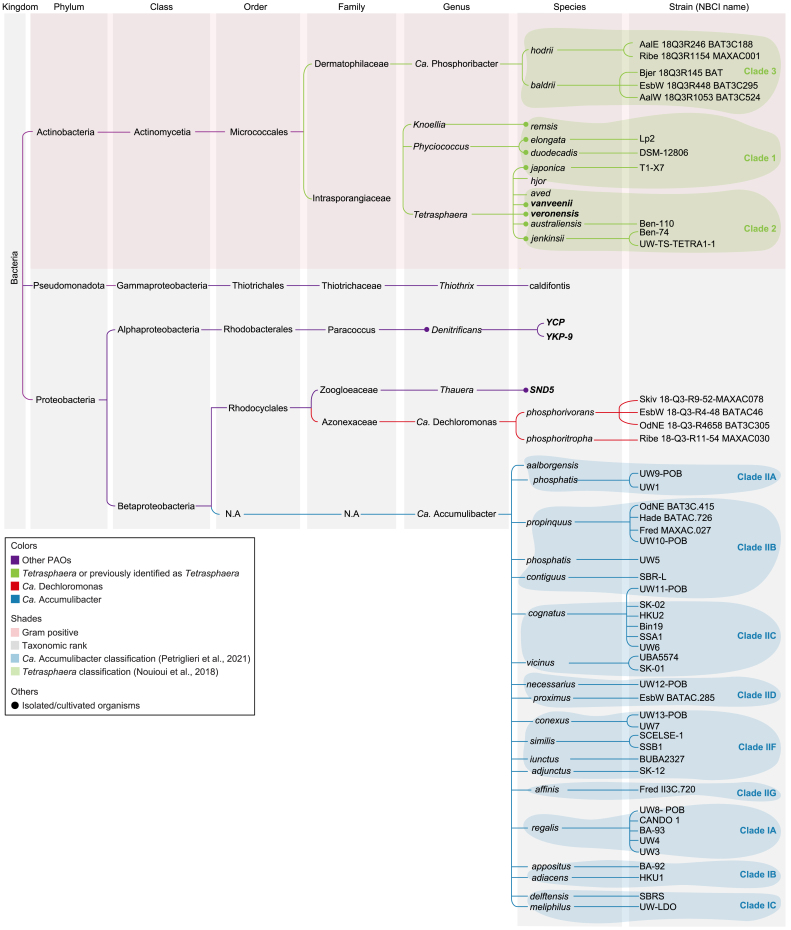
PAOs have a remarkable ability to store unusually large quantities of PolyP, making them crucial for efficiently removing P in EBPR WWTPs. Among all reported PAOs (Fig. 1), Ca. Accumulibacter, Ca. Dechloromonas, and Tetrasphaera were identified as the most significant PAOs, primarily due to their high frequencies and abundance in a global sampling of WWTPs [21].
Fig. 1.
This tree visually represents the taxonomic classification of organisms displaying PAO activity (nine genera). This classification is based on information from the NCBI taxonomy database. Isolated and cultured organisms are marked with dots. Additionally, most genomes are accessible on NCBI, except for those highlighted in bold text, which only have a partial 16S rRNA gene available.
Ca. Accumulibacter was the first PAO discovered, and subsequently, this genus was further classified into type I or II, with clades ranging from A to C in type I and A to G in type II (Fig. 1), based on the ppk1 gene [22,23]. Later, in 2023, the novel Ca. Accumulibacter genomes were classified as Type III [24]. Although these organisms have been reported exclusively in lab-scale bioreactors and thus are currently not considered in the analysis of WWT-relevant PAOs, it is essential to highlight their remarkable ability to utilize dimethyl sulfoxide for energy production [24]. Consequently, there is a potential to intentionally introduce or naturally enrich Type III Ca. Accumulibacter organisms in full-scale WWTPs that manage high concentrations of both dimethyl sulfoxide and P [24].
In 2000, Maszenan and colleagues discovered three Actinobacteria isolates capable of accumulating PolyP, giving Tetrasphaera as their genus name [23]. Tetrasphaera was initially composed of eight species: australiensis (strains Ben 109 and Ben 110), japonica (strain T1-X7) [23], elongata (strain Lp2 and ASP12) [25,26], jenkinsii, veronensis, vanveenii [27], duodecadis [23], and remsis [28]. Later, during the study of WWTPs, novel species of Tetrasphaera were described, and as a result, this genus was classified into three clades [29]. Clades 1 and 2 encompassed the eight species mentioned above, while clade 3 comprised only environmental sequences [29].
However, recent phylogenetic analyses resulted in the reclassification of the different species, with Tetrasphaera elongata and Tetrasphaera duodecadis being moved into the genus Phyciococcus, and Tetrasphaera remsis is now classified as Knoellia remsis [30,31]. Furthermore, the Tetrasphaera classified as clade 3, now belong to two distinct genera: Ca. Phosphoribacter (Ca. Phosphoribacter baldrii and Ca Phosphoribacter hodrii) and Ca. Lutibacillus [14]. Lastly, two new species were introduced to clades 1 and 2, with Tetrasphaera hjor (Tetrasphaera midas_s_299) being integrated into clade 1, and Tetrasphaera aved (Tetrasphaera midas_s_1404) becoming part of clade 2 [14]. Fig. 1 illustrates the most recent categorization of these organisms; where green lines represent all the taxa previously categorized as Tetrasphaera.
Finally, the ability of some members of the genus Dechloromonas to accumulate PolyP was first reported in 2005 [32]. Recently, strains belonging to two species of Dechloromonas are well-identified as PAOs; these include Ca. Dechloromonas phosphoritropha and Ca. Dechloromonas phosphorivorans [12]. Notably, most WWT-relevant PAOs are classified as Candidatus since they have not been cultured yet, encompassing Ca. Accumulibacter, Ca. Phosphoribacter, and the two Dechloromonas species (Ca. Dechloromonas phosphoritropha and Ca. Dechloromonas phosphorivorans) [12,14].
3. Metabolic diversity of PAOs
In this section, we gathered publicly available amino acid sequences of MAGs from WWT-relevant PAOs. These sequences were collected from the National Center for Biotechnology Information (NCBI) database as of May 2023 (Table S1). Subsequently, functional gene annotation for each MAG was performed on these sequences using BlastKoala [33], which assigned Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) identifiers based on sequence similarity. Finally, all MAGs were analyzed to assess the presence or absence of a specifically targeted KEGG ID in each PAO. However, it is important to know that such analyses may be affected by various biological and bioinformatic factors that could lead to conflicting information. Examples of biological errors include DNA loss due to sample collection, storage, and DNA extraction procedures. While bioinformatic errors can result from sequencing, assembly, binning, and annotation [34]. Moreover, it is worth noting that our analysis aligns with the findings reported in existing literature, where researchers have examined the presence and absence of some studied genes. This literature includes Ca. Accumulibacter [13], Ca. Dechloromonas [12], Tetrasphaera [14], Ca. Phosphoribacter [14], and Phycicoccus [14].
In terms of general metabolism, Ca. Accumulibacter, Ca. Dechloromonas, and Ca. Phosphoribacter possess the capacity for Glycolysis through the Embden-Meyerhof pathway. In contrast, Tetrasphaera, Knoellia, and Phycicoccus lack the 6-phosphofructokinase or ADP-dependent phosphofructokinase enzyme that is essential for phosphorylating d-Fructose-6P to d-Fructose-1,6P in the Embden-Meyerhof pathway. The fact that Tetrasphaera, Knoellia, and Phycicoccus lack this enzyme underscores the necessity for these organisms to evolve alternative pathways or mechanisms to compensate for this deficiency in carbon metabolism.
Furthermore, none of the PAOs can undergo assimilatory nitrate or nitrite reduction (Table S2), meaning they cannot incorporate the nitrogen from these molecules into cellular components for growth and maintenance. However, all PAOs possess the metabolic capacity for pyruvate oxidation, citrate cycle (TCA cycle or Krebs cycle), pentose phosphate pathway, and ribose-P ryrophosphokinase (PRPP) biosynthesis. Moreover, the following section will provide a more comprehensive analysis of the relevant metabolism for PAOs, encompassing PolyP synthesis and hydrolysis, transport capabilities, glycogen metabolism, PHA (Polyhydroxyalkanoates) metabolism, fermentation, and denitrification capability.
3.1. PolyP synthesis and hydrolysis, and P transport
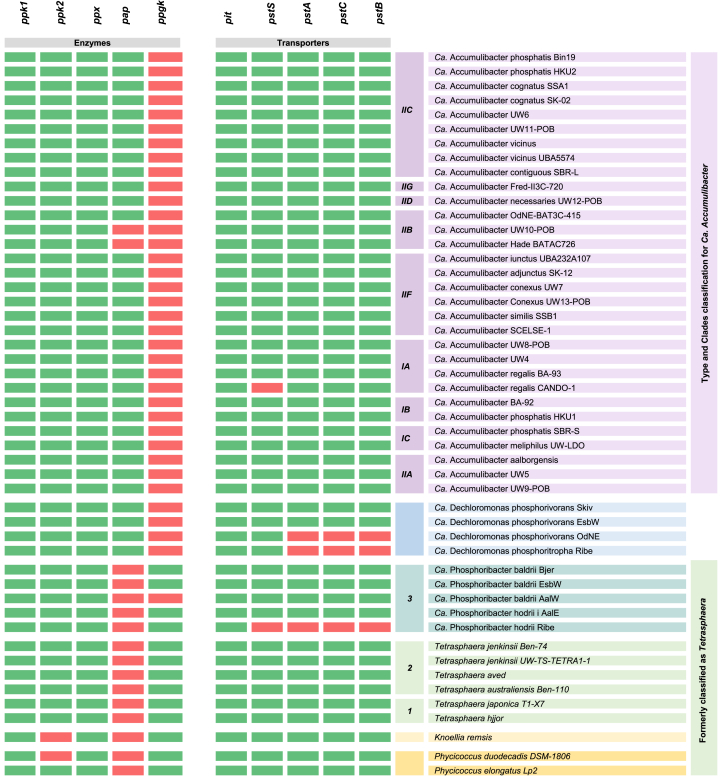
The PolyP metabolic process involves a set of genes responsible for PolyP synthesis, hydrolysis, and P transport (Fig. S1 and Table S3). PolyP synthesis predominates in aerobic or denitrifying conditions and is catalyzed by the polyphosphate kinases PPK1 (gene ppk1) and PPK2 (gene ppk2) [35]. PPK1 and PPK2 differ in function, structure, and equilibrium preference. PPK1 is a soluble enzyme found in various bacteria and eukaryotic organisms, while PPK2 is a membrane-bound enzyme found primarily in bacteria. Although both enzyme families can catalyze PolyP synthesis, PPK1 preferentially synthesizes PolyP from nucleoside triphosphates, and PPK2 preferentially uses PolyP to phosphorylate nucleoside mono- or di-phosphates, such as adenosine monophosphate (AMP), dimethyl phosphate (DMP), and guanosine diphosphate (GDP) [36,37].
PolyP hydrolysis in PAOs occurs under anaerobic conditions and can be catalyzed by various enzymes, including PPKs, exopolyphosphatases, and endopolyphosphatases [38,39]. As previously stated, the PPKs reverse reaction contributes to the synthesis of adenosine triphosphate (ATP) and guanosine triphosphate (GTP) from PolyP [39]. Besides, exopolyphosphatases (gene ppx) hydrolyze PolyP from polymer ends, releasing free phosphate [40]. Lastly, endopolyphosphatases break down the PolyP chain within the polymer, releasing shorter PolyP chains or PPi molecules [39]. As observed in Table 1, genes encoding for polyphosphate kinases (genes ppk1 and ppk2) and exopolyphosphatase (gene ppx) are well conserved among all genera. However, endopolyphosphatase was not found in any organism using KEGG. The absence of endopolyphosphatase in the studied PAOs, as indicated by the KEGG database, could be attributed to two potential reasons: (1) the endopolyphosphatase in these PAOs may differ from the annotated endopolyphosphatase in the KEGG database, leading to its non-recognition in genomic analyses, and (2) this enzyme is actually absent in WWT-relevant PAOs.
Table 1.
Genes involved in PolyP metabolism in WWT-relevant PAOs. Green indicates the presence of a gene, while red denotes its absence. Complete gene names and KEGG nomenclature are available in Supplementary Materials (Table S8); Microorganism accession numbers (NCBI) are available in Supplemental Materials (Table S1).
Additionally, PolyP degradation can occur by transfer of the terminal P groups to an acceptor molecule, facilitated by a phosphotransferase. This process conserves the energy of the energy-rich phosphoanhydride bond in the newly formed phosphoanhydride or phosphoester bond at the acceptor molecule [39]. For example, a PolyP-AMP phosphotransferase (gene pap) transfers the P group from PolyP to AMP, while a PolyP glucokinase (gene ppgk) phosphorylates glucose using PolyP [7]. Although PPGK primarily targets glucose as its substrate, it has also demonstrated the potential to phosphorylate other sugars [41]. According to Table 1, phosphotransferase in Ca. Accumulibacter and Ca. Dechloromonas occurs via PolyP-producing ADP, whereas in Tetrasphaera, Ca. Phosphoribacter, Knoellia, and Phycicoccus, it is achieved through the phosphorylation of a sugar.
P transporters play a crucial role in regulating the uptake and utilization of P within cells. There are different P transporters in bacteria, including low-affinity transporters (genes pit) and high-affinity transporters (genes pstSCAB), where PSTSCAB is likely to perform better in low P concentrations than PIT. The PSTSCAB proteins comprise a periplasmic P-binding protein (gene pstS), integral membrane proteins (genes pstC and pstA), and an ATP-binding protein (gene pstB) [42]. In contrast, the P transporter encoded by gene pit is a periplasmic single-component symporter that facilitates the transport of P or sulfate using either Na+ or H+ ions [43]. In anaerobic environments, PAOs employ the PIT to excrete P and cations simultaneously, generating an electrochemical gradient that facilitates carbon uptake [44]. For example, in organisms like Ca. Accumulibacter and Ca. Dechloromonas, the ACTP symporter uses an electrochemical gradient to drive acetate uptake [44]. As observed in Table 1, the pit gene is present in all PAOs, whereas the pstSABC cluster is not found in all species but exhibits high conservation within PAOs. Notably, the pit gene is not exclusive to PAOs and can also be found in organisms that do not show PAO activity.
3.2. Organic substrate transport
When examining the ABC transporters (Table 2), all genera show the potential for uptaking branched-chain amino acids (genes livKHMGF), polar amino acids (genes paSPA), suggesting a conserved capacity for uptaking different nitrogen sources. Additionally, Ca. Accumulibacter, Ca. Dechloromonas, and Ca. Phosphoribacter, possess the genes for lipopolysaccharide synthesis (genes rfbAB). However, it should be noted that Ca. Phosphoribacter lacks the potential for exporting lipopolysaccharides (genes lptFGB). On the other hand, all PAOs, except for Ca. Accumulibacter and Ca. Dechloromonas, possess genes responsible for the transport of nucleosides (genes nupABC and bmpABC), simple sugars (gene ssSPA), and multiple sugars (genes gguAB, msSPA, chvE, and sugC). These findings support previous reports on the ability of Tetrasphaera to utilize a diverse range of carbon sources, including yeast extract, peptone, glucose, and soluble starch [26,45]. Likewise, it is in agreement with the potential of Ca. Accumulibacter to utilize and uptake glucose [45].
Table 2.
Genes for transporting various carbon substrates in WWT-relevant PAOs. Green indicates the presence of a gene, while red denotes its absence. Complete gene names and KEGG nomenclature are available in Supplementary Materials (Table S8); Microorganism accession numbers (NCBI) are available in Supplemental Materials (Table S1).
Moreover, all PAOs possess electrochemical potential-driven transporters, such as the cation/acetate (gene actP) and the malonate-proton symporter (gene mdcF), which allow these organisms to efficiently transport carbon compounds without requiring ATP. The presence of acetate transporters in all PAOs is consistent with the documented uptake of VFAs (acetate and propionate) by Ca. Accumulibacter and Ca. Dechloromonas during the anaerobic phase [46]. Similarly, a sludge enriched with Tetrasphaera and Ca. Phosphoribacter effectively removed acetate, though the specific genera accountable for this removal remains unidentified [45]. On the other hand, certain PAO species, such as Tetrasphaera australiensis, Tetrasphaera japonica, Phyciococcus elongata, and Tetrasphaera jenkinsii, do not uptake acetate despite possessing the necessary transporters [47]. It has been hypothesized that the inability of these species to uptake acetate could be explained by the lack of the phaC gene and glyoxylate cycle genes required for processing acetate for PHA synthesis [29,44,45]. However, evidence suggests that ActP transporters have a broader range of substrates beyond acetate, such as glycolate [44]. Therefore, these species may utilize the ActP transporters to transport other molecules than acetate.
Moreover, Ca. Accumulibacter and Ca. Dechloromonas species feature an enrichment of electrochemical potential-driven transporters responsible for C4-dicarboxylate uptake (genes dcuAB, dctA, and dctPQM). This aligns with the reported ability of Ca. Accumulibacter to utilize fermentation products, such as succinate, as a carbon source [48]. In contrast, the Ca. Dechloromonas genus exclusively possesses the glutamate-sodium symporter (gene gltS), incorporating glutamate into its central carbon metabolism via glutamate dehydrogenase (gene gdhA) [12]. Conversely, Tetrasphaera exhibits a diverse array of specific electrochemical gradient transporters, including putrescine importer (gene puuP), the transport of sugars such as maltose/maltodextrin (gene malY), and the fucose-proton symporter (gene fucP), as well as amino acid transporters, such as l-asparagine permease (gene ansP) and glycine betaine transporter (gene opuD or betL). This agrees with the reported preference of these genera for consuming sugars and accumulating amino acids [49].
Certain transporters are exclusive to a particular species. For example, Phycicoccus duodecadis possesses the proline-proton symporter (gene proP), Knoellia remsis has the cytosine permease (gene codB) and the benzoate-proton symporter (gene benK) as specific transporters, Tetrasphaera japonica has the tartrate transporter (gene ttuB), galactose-proton symporter (gene galP), and lactate-proton symport (gene jen), Tetrasphaera australiensis Ben-110 has the ethanolamine permease (gene eat) and the oxalate-formate antiporter (gene oxlT).
3.3. Fermentation
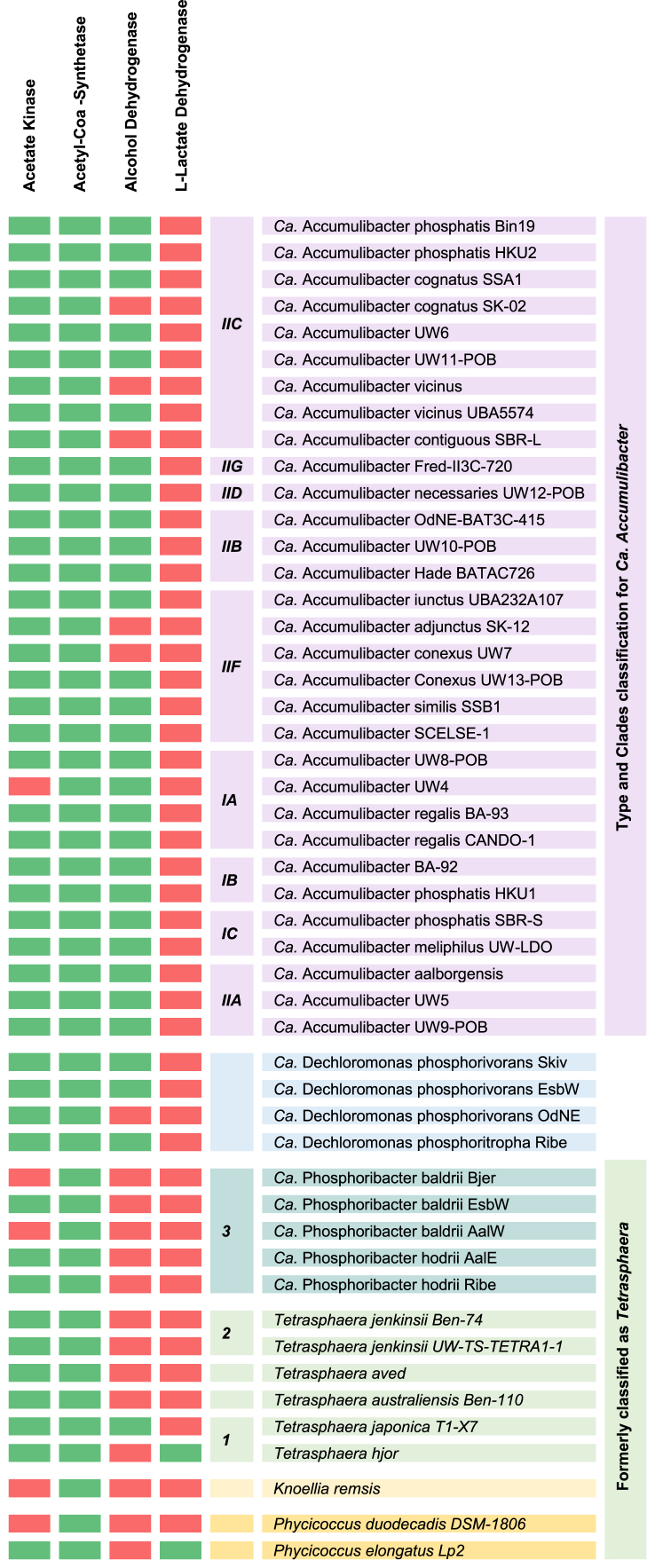
Fermentation capabilities have not been reported in Ca. Accumulibacter and Ca. Dechloromonas. However, Tetrasphaera has been shown to have the capacity for anaerobic P release; it is also capable of anaerobic P uptake using the energy generated from the fermentation of various organic compounds [50]. It has been reported that Tetrasphaera can ferment glucose and amino acids to produce acetate, succinate, lactate, and ethanol [[51], [52], [53]]. Additionally, Ca. Phosphoribacter can ferment alanine to pyruvate, while Ca. Phosphoribacter possesses the complete fermentation pathway to convert pyruvate to acetate [14]. Likewise, it has been hypothesized that Phyciococcus elongata can produce lactate as the end product of glucose fermentation.
The capability of these organisms to produce succinate and acetate is supported by the presence of the TCA cycle and the acetate kinase (Table 3). However, it is crucial to note that a specific gene alone cannot conclusively predict acetate fermentation, as indicated by its occurrence in non-fermenting genomes. Lactate production in Tetrasphaera and Phycicoccus is substantiated by the data presented in Table 3. This table shows that Phycicoccus elongata and Tetrasphaera Hjor contain the gene encoding lactate dehydrogenase. Moreover, Tetrasphaera japonica possesses alcohol dehydrogenase (Table 3), confirming the potential of this genus for alcohol production.
Table 3.
Genes involved in fermentation in WWT-relevant PAOs. Green indicates the presence of a gene, while red denotes its absence; Microorganism accession numbers (NCBI) are available in Supplemental Materials (Table S1).
3.4. Glycogen
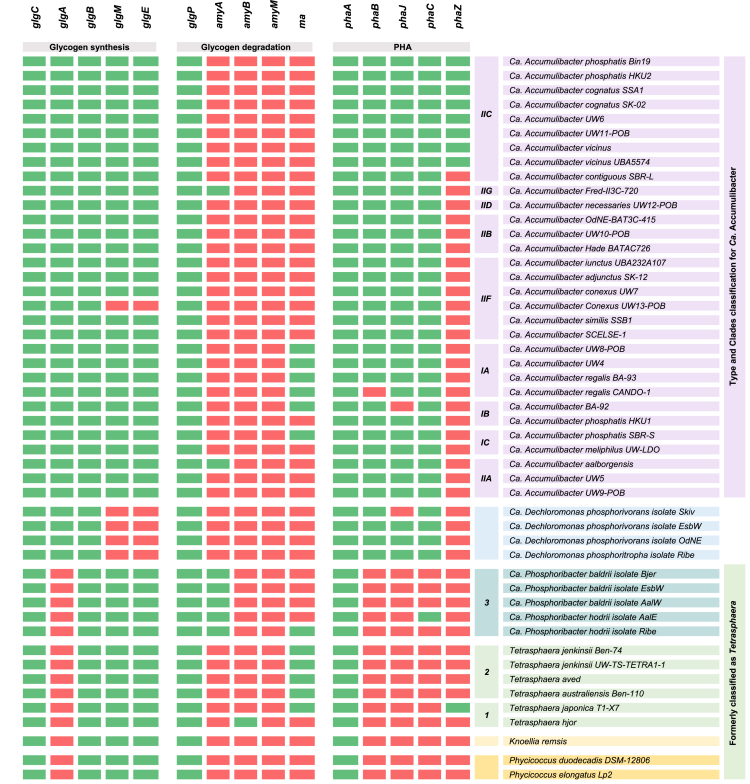
All WWT-relevant PAOs possess the capacity for glycogen synthesis but differ in the enzymes they use (Table 4). The classical biosynthetic pathway of glycogen involves three key enzymes: glucose-1-phosphate adenylyltransferase (gene glgC), ADP-glucose-specific glycogen synthase (gene glgA), and branching enzyme (gene glgB) [54]. GLGC is crucial in converting glucose-1-phosphate to ADP-glucose, serving as a substrate for GLGA to add glucosyl units to the growing glycogen chain, while GLGB further enhances glycogen complexity by cleaving portions of the chain and forming α-1,6 glycosidic bonds [54]. Ca. Accumulibacter and Ca. Dechloromonas exhibit the capability for glycogen production through the glgCAB pathway, as indicated by the data presented in Table 4 and corroborated by the current literature [12,16]. In contrast, Tetrasphaera, Ca. Phosphoribacter, Knoellia, and Phycioccocus lack the GLGA enzyme required for glycogen elongation. Although, research has shown that the alpha-maltose-1-phosphate synthase (gene glgM) can employ the GLGC product to generate maltose, while the maltosyl-transferase (gene glgE) can synthesize glycogen through maltose elongation [55]. Given these insights, it can be inferred that Tetrasphaera, Ca. Phosphoribacter, Knoellia, and Phycioccocus possess the potential to synthesize glycogen employing maltose as the monomer (Table 4).
Table 4.
Genes involved in PHA synthesis, PHA hydrolysis, glycogen synthesis, and glycogen degradation in WWT-relevant PAOs. Green indicates the presence of a gene, while red denotes its absence. Complete gene names and KEGG nomenclature are available in Supplementary Materials (Table S8); Microorganism accession numbers (NCBI) are available in Supplemental Materials (Table S1).
On the other hand, during glycogen catabolism, two essential enzymes come into play: glycogen phosphorylase (gene glgP) and the debranching enzyme (gene amy) [56]. Glycogen phosphorylase helps remove glucose units from glycogen through phosphorolysis, generating glucose-1-phosphate. The debranching enzyme removes branch points in the glycogen chain, making it more accessible for further breakdown [56]. The presence of conserved GLGP enzymes across all genera indicates that all PAOs have the capacity to utilize glycogen as a carbon source (Table 4). Besides, Ca. Phosphoribacter is the only genus that possesses the alpha-amylase gene (amyA), which indicates its capacity to efficiently break down glycogen into shorter oligosaccharides, such as maltose, maltotriose, and maltotetraose (Table 4) [57]. Additionally, beta-amylase, responsible for breaking down glycogen into maltose, is exclusively found in Tetrasphaera hjor [58].
3.5. Polyhydroxyalkanoates (PHA) metabolism
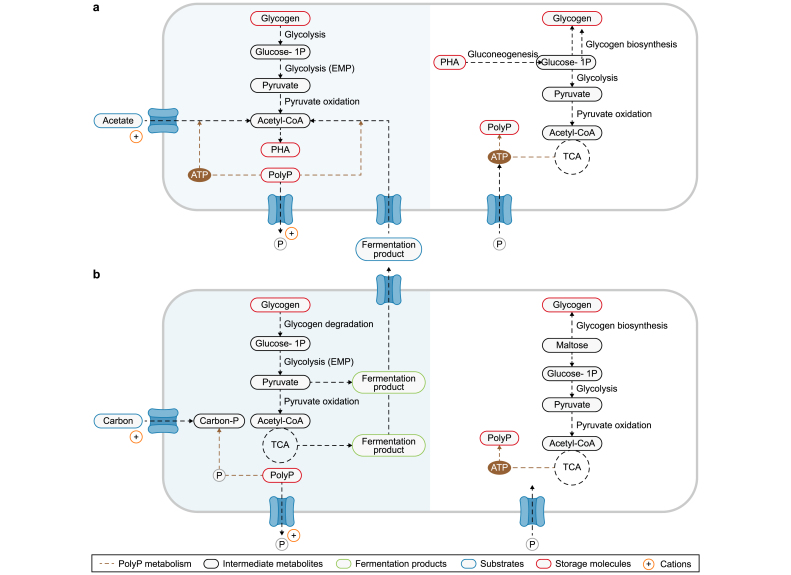
PHA production occurs during periods of abundant carbon and energy, often in the presence of PolyP, in the anaerobic phase. Subsequently, PHA can be hydrolyzed to serve as a carbon substrate under aerobic conditions for PAOs and anoxic conditions for DPAOs (Fig. 2) [36,59]. In PAOs, it has been demonstrated that both acetate and glycogen contribute to anaerobic PHA synthesis through their conversion to acetyl-CoA. The PHA generated through this process is subsequently utilized aerobically for glycogen production [60]. Furthermore, glycogen breakdown via the Entner-Doudoroff (ED) pathway allows it to serve as a considerable source of reducing power for PHA production. Regarding the proteins involved in PHA synthesis, they are encoded by the PHA gene cluster, which is composed of a set of genes that may differ based on the microorganism in question. The PHA gene cluster typically includes phaC, phaA, phaB, phaG, and phaJ [61]. The genes phaA, phaB, phaG, and phaJ encode for a group of enzymes that are essential for producing PHA monomers from different carbon sources, including acetyl-CoA (genes phaA and phaB), fatty acids (gene phaG), and lipids (gene phaJ) [62]. Then, these monomers are polymerized into PHA by the PHA synthase (gene phaC). As a result, phaC is regarded as a marker for determining microorganisms' potential for PHA synthesis [63].
Fig. 2.
Metabolism of WWT-relevant PAOs under anaerobic (shade) and aerobic conditions: a, Metabolic capabilities of Ca. Accumulibacter and Ca. Dechloromonas, encompassing substrate uptake, polyphosphate (PolyP), glycogen, and polyhydroxyalkanoates (PHA) synthesis. b, Metabolic characteristics of Ca. Phosphoribacter, Tetrasphaera, Knoellia, and Phycicoccus, specifically addressing carbon uptake, storage, and PolyP and glycogen synthesis.
Table 4 demonstrates that both Ca. Accumulibacter and Ca. Dechloromonas contain phaA, phaB, phaJ, and phaC genes, indicating they can potentially use acetate and lipids as substrates for PHA synthesis. This is consistent with studies showing that Ca. Accumulibacter and Ca. Dechloromonas can synthesize PHA using VFAs [12,46]. In contrast, Tetrasphaera, Ca. Phosphoribacter, Knoellia, and Phycicoccus have been found to lack the phaB, phaJ, and phaC genes. This genetic profile aligns with the reported inability of Tetrasphaera to produce PHA [47]. Interestingly, Ca. Phosphoribacter hodrii showed the potential for synthesizing PHA due to the presence of the phaC gene (Table 4). Moreover, it has been established that the synthesis of different PHAs (Table S4) depends on the types of VFA present. For example, polyhydroxy butyrate (PHB) can be synthesized from acetate only, polyhydroxy-2-methyl valerate (PH2MV) from propionate only, polyhydroxy valerate (PHV) and polyhydroxy-2-methyl butyrate (PH2MB) from acetate and propionate [64].
The enzyme PHA depolymerase, encoded by the phaZ gene, plays a vital role in hydrolyzing water-insoluble PHAs into soluble 3-hydroxyalkanoic acids [65]. These enzymes lack common sequences and substrate preferences [65,66]. Still, they share two consistent structural elements: the presence of the α/β-hydrolase fold and an active-site catalytic triad (composed of serine, histidine, and aspartic acid) [65,66]. Among the various phaZ genes reported in KEGG, only the poly(3-hydroxyoctanoate) depolymerase was present in the WWT-relevant PAOs. Furthermore, this enzyme was found exclusively in species within the Ca. Accumulibacter clade IIC (Table 4). The absence of the phaZ gene in Ca. Dechloromonas is consistent with previous reports [12]. However, it is essential to note that PHA degradation has been reported in these genera. Therefore, it is possible that other species of Ca. Accumulibacter and Ca. Dechloromonas may possess PHA depolymerases with significant differences in enzyme structure compared to those reported in KEGG, or they might utilize alternative pathways for PHA degradation.
3.6. Denitrification capabilities of PAOs
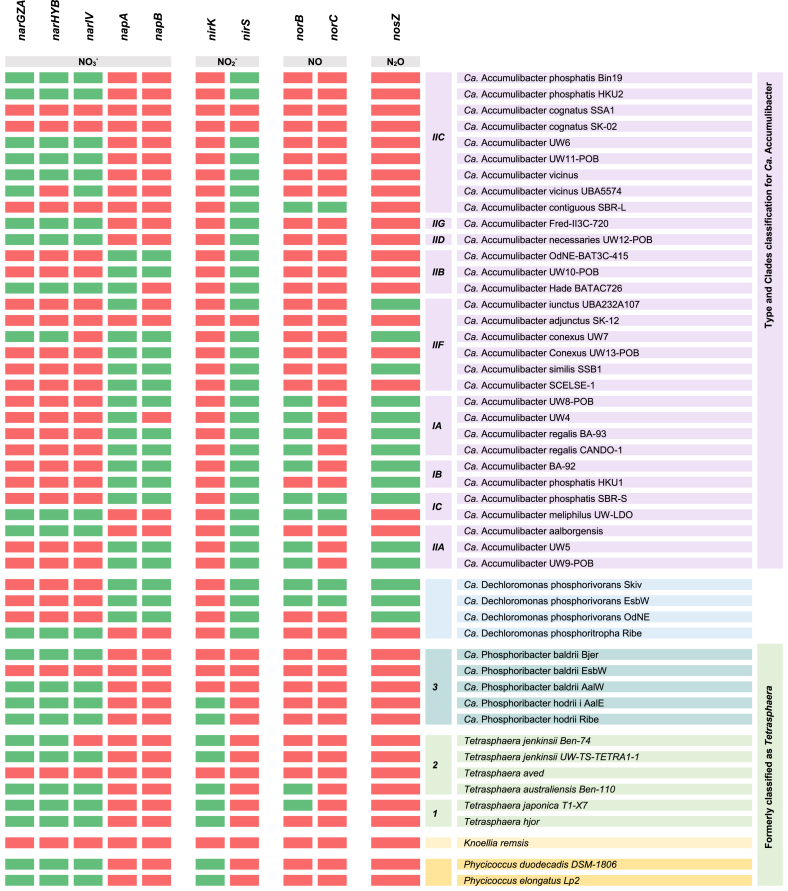
Denitrification is a respiratory process in which nitrogen compounds, such as nitrate or nitrite, can be used as electron acceptors [67]. The denitrification process involves several steps and enzymes. The first step involves reducing nitrate (NO3−) to nitrite (NO2−) via nitrate reductases. There are three types of known nitrate reductases: periplasmic nitrate reductase (gene nap), respiratory nitrate reductase (gene nar), and assimilatory nitrate reductase (gene nas) [67]. After nitrate reduction, nitrite reductase (gene nir) catalyzes the conversion of NO2− into nitric oxide (NO) and plays an essential role in transforming N from liquid to gas. Nitrite reduction in denitrification could be done by two distinct nitrite reductase enzymes encoded by the nirK and nirS genes. The nirK gene encodes a copper-containing enzyme found in low-oxygen environments, whereas nirS encodes a cytochrome cd1-containing enzyme found in high-oxygen environments [68,69]. Next, nitric oxide reductase (gene nor) catalyzes the reduction of NO to nitrous oxide (N2O) [70]. In the final step, the nitrous oxide reductase (gene nos) reduces N2O to N2 and completes denitrification [71].
PAOs that demonstrated complete denitrification capability (i.e., reducing NO3− to N2) are called DPAOs [72]. Consequently, if present, DPAOs can remove P, hydrolyze PHA, and grow in anoxic conditions [71]. However, it is important to highlight that their ability to remove P in anoxic conditions is approximately half as fast and less efficient than aerobic conditions [73]. So far, identified DPAOs from WWTPs include Paracoccus denitrificans YCP [74], Paracoccus sp. YKP-9 [67], Thauera sp. SND5 [68] (Table S5). Besides the already identified DPAOs, certain species from WWT-relevant PAOs have shown the potential for denitrification, and as a result, this section will focus on discussing their denitrification capabilities.
As observed in Table 5, several PAO species, such as Knoellia remsis, Tetrasphaera aved, Ca. Accumulibacter adjunctus, and Ca. Accumulibacter cognatus, were found to lack the genes necessary for denitrification. However, most PAOs possess the necessary genes for reducing nitrate and nitrite, indicating their potential to utilize these compounds as electron acceptors, while interesting distinctions exist among different genera regarding their enzyme capabilities for nitrate and nitrite reduction. For instance, Ca. Accumulibacter and Ca. Dechloromonas have the ability to utilize either the NAP or NAR enzyme for nitrate reduction, whereas the other genera exclusively possess the NAR enzyme. On the other hand, when it comes to nitrite reduction, the nirS gene is found in Ca. Dechloromonas and Ca. Accumulibacter species, whereas the nirK gene is present in Tetrasphaera, Phycicoccus, and Ca. Phosphoribacter. This distinction is interesting because the presence of the nirK gene suggests that these species may be more adept at denitrification in low-oxygen environments [75].
Table 5.
Genes involved in denitrification in WWT-relevant PAOs. Green indicates the presence of a gene, while red denotes its absence. Complete gene names and KEGG nomenclature are available in Supplementary Materials (Table S8); Microorganism accession numbers (NCBI) are available in Supplemental Materials (Table S1).
On the other hand, most PAO species lack the genes responsible for reducing nitric oxide (gene nor) and nitrous oxide (gene nos). The absence of these enzymes, which act as electron sinks and are crucial for reducing toxic intermediate products (i.e., NO), results in a diminished ability to utilize NO3− and NO2− as terminal electron acceptors [64]. Furthermore, it is worth noting that Ca. Accumulibacter meliphilus UW-LDO, Ca. Accumulibacter contiguous SBR-L, Tetrasphaera australiensis, and Tetrasphaera japonica, possess the nor gene and lack the nos gene. This genetic configuration could potentially produce nitrous oxide, a potent greenhouse gas, by these PAOs [69]. Nevertheless, in WWTPs, PAOs coexist with a diverse microbial community that can utilize PAOs' byproducts, effectively mitigating the aforementioned issues [70].
Lastly, certain PAOs, such as Ca. Accumulibacter (including BA-92, phosphatis SBR-S, UW5, UW9-POB, and all species in cluster IA) and Ca. Dechloromonas phosphorivorans Skiv and EsbW, possess all the necessary genes for complete denitrification. Interestingly, it has been observed that PAOs possessing all genes required for denitrification possess the NAP enzyme for nitrate reduction, which is considered more efficient than nar enzymes [64]. These findings align with the reported synergistic role of the NAP-NIR enzyme in generating a proton motive force to support denitrification, even in the absence of the NAR enzyme [64]. Furthermore, the complete denitrification ability of species from clade IA has been experimentally demonstrated [71]. However, as the study utilized fluorescence in situ hybridization (FISH) probes targeting clade IA, it lacks the ability to differentiate whether the observed activity is shared by all species within this clade or limited to a specific subset. As a result, there is uncertainty about which particular species are prominently exhibiting DPAO activity.
4. Methods for the detection and quantification of PolyP and PAOs
4.1. Staining and visualization techniques for PolyP detection in cells
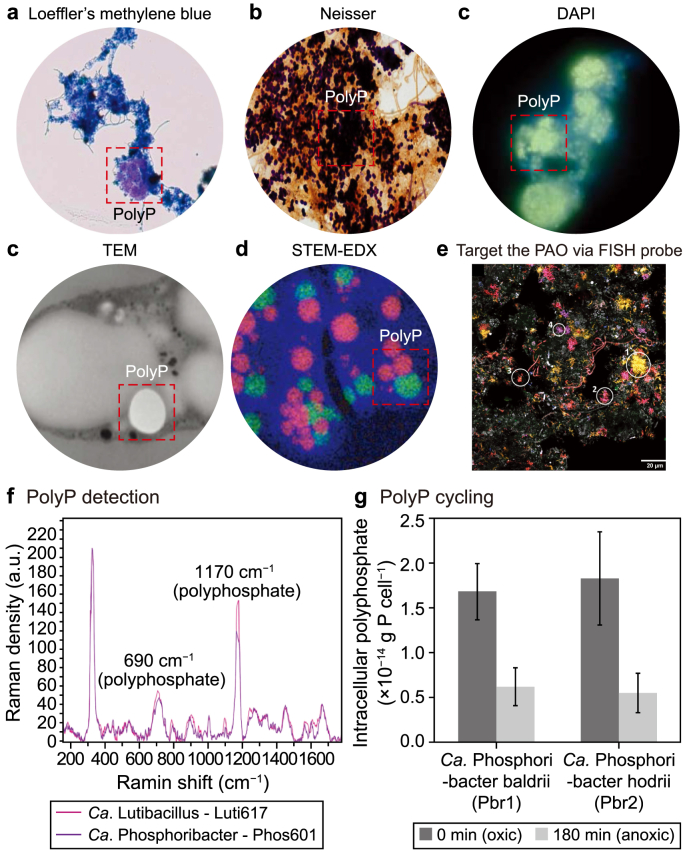
PolyP can be observed as an electron-dense material using transmission electron microscopy (TEM) and scanning electron microscopy (SEM). However, electron microscopy may confuse PolyP with another molecule (Fig. 3) [76]. To provide a more accurate identification of PolyP clusters, a combination of electron microscopy and PolyP staining techniques can be used [77]. The most common PolyP stains are Loeffler's Methylene Blue, Neisser, and 4′,6-diamidino-2-phenylindole (DAPI) [78] (Fig. 3). When using Loeffler's staining method, PolyP appears to be pink violet on a blue cell background, whereas Neisser's staining method colors the PolyP in purple-black in a yellowish-brown background of counterstained cells. The latter procedure is more effective than other light microscopic staining techniques since it yields the highest contrast between PolyP granules and cells [19,26]. DAPI staining also stains PolyP granules with a yellow fluorescence when used at high concentrations. However, DAPI is not specific because it also stains other polymeric ions, such as DNA and lipids, where DAPI-DNA fluorescence is blue, whereas both DAPI-PolyP and DAPI-lipids show a yellow fluorescence [78]. Additionally, PolyP can be visualized using scanning transmission electron microscopy (STEM) combined with energy-dispersive X-ray spectroscopy (STEM-EDX), enabling the detection of the X-ray emission spectrum of P [79]. On the other hand, Saito et al. developed a novel technique for PolyP detection using a recombinant PolyP binding domain (PPBD) from an exopolyphosphatase enzyme found in Escherichia coli [80]. The PPBD has a high affinity for long-chain PolyP and a low affinity for short-chain PolyP, which allows for the specific detection and isolation of long-chain PolyP molecules in a sample [80].
Fig. 3.
Techniques for detecting PolyP and PAOs. a–c, The commonly used staining methods include Loeffler's Methylene Blue [102] (a), Neisser [103] (b), and 4′,6-diamidino-2-phenylindole (DAPI) [104] (c). d–e, Staining-independent visualization techniques include Transmission electron microscopy (TEM) (d) [104] and scanning transmission electron microscopy (STEM) coupled with X-ray spectroscopy (STEM-EDX) (e) [79]. f–h, The methodology for identifying PAOs [14] includes FISH probes targeting PAOs (f), Raman for detecting the presence of PolyP in the targeted cell (g), and Raman for measuring the PolyP cycling in anaerobic and aerobic conditions (h).
PolyP can also be identified and quantified within cells through flow cytometry, nuclear magnetic resonance (NMR) spectroscopy, and Raman spectroscopy [77]. Flow cytometry can be used to detect PolyP molecules by incorporating a staining technique and modifying the laser's excitation wavelength to detect the used dye. For example, in the case of DAPI-stained PolyP molecules, the fluorescence emitted can be detected at approximately 550 nm [76]. It is important to emphasize that achieving accurate separation of cluster cells into single cells for flow cytometry analysis requires crucial steps of homogenization, sonication, and filtration of sludge samples, especially for microorganisms like Ca. Accumulibacter that tend to grow in clusters [81]. On the other hand, Raman spectroscopy and NMR can achieve quantification without requiring the dyeing of PolyP, as they focus on the absorption/emission spectra or mass-to-charge ratio of P and PolyP [82]. In NMR, interferences may arise due to the similarity of linkages in PolyP and other molecules containing phosphoranhydride bonds, such as nucleotides, resulting in limited accuracy in determining PolyP concentration [83]. Raman spectroscopy accurately identifies PolyP based on its unique spectral signature, though determining PolyP length is challenging due to the overlapping band [84,85]. Therefore, Raman spectroscopy is the simplest and most widely used method for accurately detecting PolyP in non-cultivated PAOs, particularly in complex samples [14,49].
4.2. Molecular-based techniques for identifying PAOs
Initially, the taxonomic classification of PAOs relied on two criteria: DNA GC content and chemotaxonomic properties [22,30,86,87]. However, with the advent of molecular biology tools, the DNA sequencing of three genes (16S rRNA, PPK1, and 23S rRNA) has been used to reflect the phylogenetic relationships among different genera of PAOs [12,14,88]. For example, the full 16S rRNA gene sequence was used to classify PAOs from the genus Dechloromonas (Ca. Dechloromonas phosphoritropha and Ca. Dechloromonas phosphorivorans) [12]. Furthermore, the full-length 16S rRNA gene divides the Tetrasphaera genus into three clades [29]. Nevertheless, the utilization of the 23S rRNA gene has been demonstrated to be more effective in revealing the phylogenetic classification within this genus, preventing incorrect assignment of genera such as Ca. Phosphoribbacter, Phycicoccus, and Knoellia [14]. Finally, the 16S rRNA was used to identify Ca. Accumulibacter. However, to reveal finer-scale population structure in Ca. Accumulibacter, McMahon and coworkers used polyphosphate kinase 1 (gene ppk1) [89]. The ppk1 gene encodes for PPK1 enzyme that participates in PolyP synthesis and is typically only present in one copy in Ca. Accumulibacter genome [89]. The Ca. Accumulibacter ppk1 gene classification revealed the presence of two genotypes (types I and II) and five clades (one clade within type I and four within type II). The ppk1 primers used to categorize type I and type II are shown in Table S6, along with improved primers for Ca. Accumulibacter type and clades identification [71,88,90]. The only consistent difference observed in the classification of clades is that clade IA exhibits the capacity to couple nitrate reduction with P uptake, whereas clade IIA does not possess this capability [71]. However, in addition to this distinction, relying solely on clade description is insufficient for accurately characterizing a specific metabolic capability [91,92].
The most used method for identifying PAOs in wastewater samples is sequencing the variable regions of the 16S rRNA gene and comparing the sequences to publicly available databases of 16S rRNA, such as MiDAS. The MiDAS guide is a well-known database for identifying PAOs in WWTPs, and it is based on the V1–V3 and V4 variable regions of the 16S rRNA gene. Of these regions, the V1–V3 region provides the most reliable taxonomic resolution for identifying PAOs in WWTPs [21]. However, it has a poor species-level resolution for PAOs. For instance, Ca. Accumulibacter affinis, Ca. Accumulibacter proximus, Ca. Accumulibacter propinquus, and Ca. Accumulibacter phosphatis will all be classified as Ca. Accumulibacter phosphatis despite being a different species [13]. Therefore, this limits MiDAS's ability to provide accurate species-level identification of PAOs in WWTPs.
Another commonly used method to identify known PAOs in WWTPs is FISH with 16S/23S rRNA-targeted oligonucleotide probes. Using FISH allows for identifying target organisms and provides a better understanding of the distribution and morphology of the targeted PAOs [93]. Out of all the FISH probes designed to target the 16S rRNA of Ca. Accumulibacter, the PAOmix (a combination of PAO462, PAO651, and PAO846) was considered the most sensitive and specific for targeting Ca. Accumulibacter [22,86]. However, subsequent research revealed that the traditionally used PAOmix also targets Propionivibrio GAO [94]. Consequently, it is now recommended to employ PAO651 alone to ensure accurate targeting of Ca. Accumulibacter [94]. Furthermore, a set of new species-level FISH probes (Table S7) were designed in 2022 using the full-length 16S rRNA, resulting in a more accurate phylogenetic classification [13].
The FISH probes for Tetrasphaera were initially discovered using probes HGC69a and HGC236, which are specific to Actinobacteria [95,96]. In 2000, Maszenan and colleagues discovered three Actinobacteria isolates capable of accumulating PolyP, giving Tetrasphaera as their genus name. These isolates were classified into two species: Tetrasphaera japonica sp. (T1-X7T) and Tetrasphaera australiensis sp. (Ben 109 and Ben 110) [23]. In 2001, the FISH probe Actino-1011 was used to target Tetrasphaera japonica [97]. Tetrasphaera elongata was discovered in 2002, but no FISH probes were developed despite sequencing its 16S rRNA gene [25,26]. In 2005, two FISH probes, Actino-221 and Actino-658, were designed to specifically target new, uncultivated Tetrasphaera species (still unnamed) that exhibited PolyP accumulation, with Actino-658 targeting short and Actino-221 targeting tetrad-arranged cocci [98]. Actino-221 and Actino-658 do not target clade 1 and a significant portion of clade 2, while HGC69a and HGC236 fail to target branched rods and thin filaments, resulting in an underestimation of the total abundance of Tetrasphaera [29]. In 2006, McKenzie and coworkers proposed that Ca. Nostocoida limicola (NLIMII175 probe) should be re-classified as three novel species belonging to the genus Tetrasphaera: Tetrasphaera jenkinsii, Tetrasphaera veronensis, and Tetrasphaera vanveeni [27,99]. Later, FISH probes were designed to target the different clades in Tetrasphaera: Tet1-266 for clade 1, Tet2-892 and Tet2-174 for clade 2, and Tet3-654 for clade 3 [29]. However, in this classification, Tetrasphaera japonica does not belong to the described clades [95,96,98].
With the discovery of other genera within the organisms initially classified as Tetrasphaera, new FISH probes were developed to improve accuracy and avoid unintentional targeting of these additional genera. Therefore, Tetra67 specifically targeted clade 1 Tetrasphaera species, such as Tetrasphaera elongate, Tetrasphaera japonica, and Tetrasphaera Hjor. Tetra732, on the other hand, is designed to target certain Tetrasphaera organisms from clade 2 [14]. Furthermore, Tetra183 has been developed to target a wide range of Tetrasphaera-related organisms, covering both clade 1 and clade 2. Specific FISH probes have also been developed to target the recently discovered Ca. Phosphoribacter genera, with Phos1260-23S-Pbr1 designed for Ca. Phosphoribacter baldrii and Phos601 for Ca. Phosphoribacter hodrii [14].
In 2007, the BET135 probe became the first FISH probe targeting Dechloromonas with the capability for PolyP accumulation [97]. Subsequently, those FISH probes were coupled with Raman spectroscopy to detect the P uptake and release cycles, elucidating the PAO activity in Ca. Dechloromonas phosphoritropha and Ca. Dechloromonas phosphorivorans [12]. However, these FISH probes are not species-specific and may hybridize with other Dechloromonas species not associated with PAO activity.
As mentioned above, WWT-relevant PAOs typically reside within complex communities and cannot be readily isolated. For identifying known PAOs, the prevailing methods include utilizing specialized FISH probes to target the desired PAO(s) and subsequently detecting PolyP, mainly through Raman microspectroscopy [49]. In addition, metagenomics has proven valuable in discovering novel PAOs within complex microbial communities present in WWTPs. The procedure involves using high-quality MAGs to design FISH probes that target microorganisms carrying genes related to PolyP metabolism and P transport [12]. Then, Raman microspectroscopy is employed to verify the dynamic levels of PolyP during anaerobic-aerobic conditions in the cells targeted by the novel FISH probes (Fig. 3) [12].
Furthermore, novel approaches have been proposed for identifying novel PAOs. One suggestion involves employing a trait-based comparative omics approach, integrating genomics, transcriptomics, and proteomics data [100]. This method aims to identify gene expression patterns associated with specific traits, such as P-cycling. However, the specific trait accurately reflecting the PAO activity is still uncertain [100]. Moreover, recent analysis suggests that proteomics may offer a potential avenue for identifying and measuring the abundance of PAOs [101].
To summarize, various methods exist for PAO detection, and the choice of the most suitable method depends on the specific objective. For the detection of known PAOs, sequencing methods enable their classification and abundance quantification. In this context, metagenomic analysis offers improved phylogenetic resolution, whereas 16S rRNA amplicon sequencing provides a faster and more cost-effective genus identification [12,13,21]. For visualizing the distribution of PAOs within sludge, it is recommended to utilize FISH probes designed for the desired genera in conjunction with Raman spectroscopy to confirm the presence of PolyP [12,49]. Lastly, the FISH-Raman method is recommended for identifying novel PAOs, as it enables simultaneous detection and quantification of PolyP and specific PAO genera [12,14,49].
5. Conclusions
Considering the significance of PAOs in WWT, this article reviews the literature and analyzes the MAG annotations from WWT-relevant PAOs to provide valuable insights into their metabolic capabilities. The main conclusions derived from this review are.
-
•
Ca. Accumulibacter and Ca. Dechloromonas exhibit similar metabolic capabilities. Under anaerobic conditions, they can utilize PolyP to generate an electrochemical gradient for VFAs and C4-dicarboxylates uptake; likewise, they can use PolyP and glycogen for PHA synthesis. Under oxic conditions, they can use PHA for glycogen synthesis.
-
•
Tetrasphaera, Ca. Phosphoribacter, Knoellia, and Phycicoccus share similar metabolic capabilities. Under anaerobic conditions, they utilize PolyP to generate an electrochemical gradient, enabling the uptake of sugars and amino acids through electrochemical potential-driven transporters. Additionally, they can use PolyP for sugar phosphorylation. Furthermore, these genera have been reported to possess fermentation capacity, with Tetrasphaera exhibiting the ability to couple this process with P removal capabilities. Under aerobic conditions, they have the potential to produce glycogen via maltose. However, further research is essential to understand energy production mechanisms and unveil the yet unknown carbon storage molecules in these genera.
-
•
This review describes the most relevant FISH probes developed for PAO detection and highlights the limitations of current methods in accurately reflecting the diversity of PAO subgenera and distinguishing PAOs from non-PAOs. It emphasizes the importance of developing comprehensive and reliable markers to better understand their diversity and identify PAO indicators to accurately forecast their activity.
CRediT authorship contribution statement
LucÍa Ruiz Haddad: Conceptualization, Writing - Original Draft, Methodology, Formal Analysis. Muhammad Ali: Conceptualization, Supervision, Writing - Review & Editing. Mario Pronk: Writing - Review & Editing, Supervision. Mark C.M. Van Loosdrecht: Writing - Review & Editing, Supervision. Pascal E. Saikaly: Conceptualization, Project Administration, Supervision, Writing - Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by King Abdullah University of Science and Technology (KAUST).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2024.100387.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Zheng Y., Wan Y., Zhang Y., Huang J., Yang Y., Tsang D.C.W., Wang H., Chen H., Gao B. Recovery of phosphorus from wastewater: a review based on current phosphorous removal technologies. Crit. Rev. Environ. Sci. Technol. 2023;53:1148–1172. doi: 10.1080/10643389.2022.2128194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blöch H. EU policy on nutrients emissions: legislation and implementation. Water Sci. Technol. 2001;44:1–6. [PubMed] [Google Scholar]
- 3.European Environment Agency . 2019. Phosphorus and Freshwater Eutrophication Pressure Narrative. [Google Scholar]
- 4.Elser J.J. Phosphorus: a limiting nutrient for humanity. Curr. Opin. Biotechnol. 2012;23:833–838. doi: 10.1016/j.copbio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Bunce J.T., Ndam E., Ofiteru I.D., Moore A., Graham D.W. A review of phosphorus removal technologies and their applicability to small-scale domestic wastewater treatment systems. Front. Environ. Sci. 2018;6:8. doi: 10.3389/fenvs.2018.00008. [DOI] [Google Scholar]
- 6.Kulakovskaya T.V., Vagabov V.M., Kulaev I.S. Inorganic polyphosphate in industry, agriculture and medicine: modern state and outlook. Process Biochem. 2012;47:1–10. doi: 10.1016/j.procbio.2011.10.028. [DOI] [Google Scholar]
- 7.Pepin C.A., Wood H.G. Polyphosphate glucokinase from Propionibacterium shermanii kinetics and demonstration that the mechanism involves both processive and nonprocessive type reactions. J. Biol. Chem. 1986;261:4476–4480. doi: 10.1016/S0021-9258(17)38524-1. [DOI] [PubMed] [Google Scholar]
- 8.Cyprowski M., Stobnicka-Kupiec A., Ławniczek-Wałczyk A., Bakal-Kijek A., Gołofit-Szymczak M., Górny R.L. Anaerobic bacteria in wastewater treatment plant. Int. Arch. Occup. Environ. Health. 2018;91:571–579. doi: 10.1007/s00420-018-1307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernando E.Y., McIlroy S.J., Nierychlo M., Herbst F.-A., Petriglieri F., Schmid M.C., Wagner M., Nielsen J.L., Nielsen P.H. Resolving the individual contribution of key microbial populations to enhanced biological phosphorus removal with Raman–FISH. ISME J. 2019;13:1933–1946. doi: 10.1038/s41396-019-0399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izadi P., Izadi P., Eldyasti A. Understanding microbial shift of enhanced biological phosphorus removal process (EBPR) under different dissolved oxygen (DO) concentrations and hydraulic retention time (HRTs) Biochem. Eng. J. 2021;166 doi: 10.1016/j.bej.2020.107833. [DOI] [Google Scholar]
- 11.Oehmen A., Zeng R.J., Yuan Z., Keller J. Anaerobic metabolism of propionate by polyphosphate accumulating organisms in enhanced biological phosphorus removal systems. Biotechnol. Bioeng. 2005;91:43–53. doi: 10.1002/bit.20480. [DOI] [PubMed] [Google Scholar]
- 12.Petriglieri F., Singleton C., Peces M., Petersen J.F., Nierychlo M., Nielsen P.H. Candidatus Dechloromonas phosphoritropha and Ca. D. Phosphorivorans, novel polyphosphate accumulating organisms abundant in wastewater treatment systems. ISME J. 2021;15:3605–3614. doi: 10.1038/s41396-021-01029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petriglieri F., Singleton C.M., Kondrotaite Z., Dueholm M.K.D., McDaniel E.A., McMahon K.D., Nielsen P.H. Reevaluation of the phylogenetic diversity and global distribution of the genus Candidatus accumulibacter. mSystems. 2022;7 doi: 10.1128/msystems.00016-22. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singleton C.M., Petriglieri F., Wasmund K., Nierychlo M., Kondrotaite Z., Petersen J.F., Peces M., Dueholm M.S., Wagner M., Nielsen P.H. The novel genus, Candidatus Phosphoribacter, previously identified as Tetrasphaera, is the dominant polyphosphate accumulating lineage in EBPR wastewater treatment plants worldwide. ISME J. 2022;16:1605–1616. doi: 10.1038/s41396-022-01212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rollemberg S.L.D.S., De Oliveira L.Q., Barros A.R.M., Melo V.M.M., Firmino P.I.M., Dos Santos A.B. Effects of carbon source on the formation, stability, bioactivity and biodiversity of the aerobic granule sludge. Bioresour. Technol. 2019;278:195–204. doi: 10.1016/j.biortech.2019.01.071. [DOI] [PubMed] [Google Scholar]
- 16.Zhao W., Bi X., Peng Y., Bai M. Research advances of the phosphorus-accumulating organisms of Candidatus accumulibacter, Dechloromonas and Tetrasphaera: metabolic mechanisms, applications and influencing factors. Chemosphere. 2022;307 doi: 10.1016/j.chemosphere.2022.135675. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y., Pijuan M., Oehmen A., Yuan Z. The source of reducing power in the anaerobic metabolism of polyphosphate accumulating organisms (PAOs) – a mini-review. Water Sci. Technol. 2010;61:1653–1662. doi: 10.2166/wst.2010.983. [DOI] [PubMed] [Google Scholar]
- 18.Tarayre C., Nguyen H.-T., Brognaux A., Delepierre A., De Clercq L., Charlier R., Michels E., Meers E., Delvigne F. Characterisation of phosphate accumulating organisms. Sensors. 2016;16:797. doi: 10.3390/s16060797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serafim L.S., Lemos P.C., Levantesi C., Tandoi V., Santos H., Reis M.A.M. Methods for detection and visualization of intracellular polymers stored by polyphosphate accumulating microorganisms. J. Microbiol. Methods. 2002;51:1–18. doi: 10.1016/S0167-7012(02)00056-8. [DOI] [PubMed] [Google Scholar]
- 20.Alam M.M., Srinivasan V., Mueller A.V., Gu A.Z. Status and advances in technologies for phosphorus species detection and characterization in natural environment- A comprehensive review. Talanta. 2021;233 doi: 10.1016/j.talanta.2021.122458. [DOI] [PubMed] [Google Scholar]
- 21.Dueholm M.K.D., Nierychlo M., Andersen K.S., Rudkjøbing V., Knutsson S., MiDAS Global Consortium, Albertsen M., Nielsen P.H. MiDAS 4: a global catalogue of full-length 16S rRna gene sequences and taxonomy for studies of bacterial communities in wastewater treatment plants. Nat. Commun. 2022;13:1908. doi: 10.1038/s41467-022-29438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hesselmann R.P.X., Werlen C., Hahn D., Van Der Meer J.R., Zehnder A.J.B. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 23.Maszenan A.M., Seviour R.J., Patel B.K., Schumann P., Burghardt J., Tokiwa Y., Stratton H.M. Three isolates of novel polyphosphate-accumulating gram-positive cocci, obtained from activated sludge, belong to a new genus, Tetrasphaera gen. Nov., and description of two new species, Tetrasphaera japonica sp. nov. And Tetrasphaera australiensis sp. nov. Int. J. Syst. Evol. Microbiol. 2000;50:593–603. doi: 10.1099/00207713-50-2-593. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Song W., Zhang X., Zheng M., Li H., Yu K., Guo F. Expanding the diversity of accumulibacter with a novel type and deciphering the transcriptional and morphological features among Co-occurring strains. Appl. Environ. Microbiol. 2023;89 doi: 10.1128/aem.00771-23. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanada S. Tetrasphaera elongata sp. nov., a polyphosphate accumulating bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2002;52:883–887. doi: 10.1099/ijs.0.01990-0. [DOI] [PubMed] [Google Scholar]
- 26.Onda S., Takii S. Isolation and characterization of a gram-positive polyphosphate accumulating bacterium. J. Gen. Appl. Microbiol. 2002;48:125–133. doi: 10.2323/jgam.48.125. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie C.M., Seviour E.M., Schumann P., Maszenan A.M., Liu J.-R., Webb R.I., Monis P., Saint C.P., Steiner U., Seviour R.J. Isolates of Candidatus Nostocoida limicola Blackall et al. 2000 Should be Described as Three Novel Species of the Genus Tetrasphaera, as Tetrasphaera jenkinsii sp. nov., Tetrasphaera vanveenii sp. nov. and Tetrasphaera veronensis sp. nov. Int. J. Syst. Evol. Microbiol. 2006;56:2279–2290. doi: 10.1099/ijs.0.63978-0. [DOI] [PubMed] [Google Scholar]
- 28.Osman S., Moissl C., Hosoya N., Briegel A., Mayilraj S., Satomi M., Venkateswaran K. Tetrasphaera remsis sp. nov., isolated from the regenerative enclosed life support module simulator (REMS) air system. Int. J. Syst. Evol. Microbiol. 2007;57:2749–2753. doi: 10.1099/ijs.0.65137-0. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen H.T.T., Le V.Q., Hansen A.A., Nielsen J.L., Nielsen P.H. High diversity and abundance of putative polyphosphate accumulating Tetrasphaera related bacteria in activated sludge systems. FEMS Microbiol. Ecol. 2011;76:256–267. doi: 10.1111/j.1574-6941.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- 30.Nouioui I., Carro L., García-López M., Meier-Kolthoff J.P., Woyke T., Kyrpides N.C., Pukall R., Klenk H.-P., Goodfellow M., Göker M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2007;9(2018) doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oren A., Garrity G.M. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2020;70:1–5. doi: 10.1099/ijsem.0.003881. [DOI] [PubMed] [Google Scholar]
- 32.Goel R.K., Sanhueza P., Noguera D.R. Evidence of Dechloromonas sp. participating in enhanced biological phosphorus removal (EBPR) in a bench-scale aerated-anoxic reactor. Proc. Water Environ. Fed. 2005;2005:3864–3871. doi: 10.2175/193864705783866261. [DOI] [Google Scholar]
- 33.Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du H., Yang L., Wu J., Xiao L., Wang X., Jiang L. Simultaneous removal of phosphorus and nitrogen in a sequencing batch biofilm reactor with transgenic bacteria expressing polyphosphate kinase. Appl. Microbiol. Biotechnol. 2012;96:265–272. doi: 10.1007/s00253-011-3839-5. [DOI] [PubMed] [Google Scholar]
- 36.García H., Ivanova N., Kunin V., Warnecke F., Barry K.W., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., Dalin E., Putnam N.H., Shapiro H.J., Pangilinan J.L., Rigoutsos I., Kyrpides N.C., Blackall L.L., McMahon K.D., Hugenholtz P. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- 37.Neville N., Roberge N., Jia Z. Polyphosphate kinase 2 (PPK2) enzymes: structure, function, and roles in bacterial physiology and virulence. Int. J. Mol. Sci. 2022;23:670. doi: 10.3390/ijms23020670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller W.E.G., Schröder H.C., Wang X. Inorganic polyphosphates as storage for and generator of metabolic energy in the extracellular matrix. Chem. Rev. 2019;119:12337–12374. doi: 10.1021/acs.chemrev.9b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong C., Fu J., Jiang T., Zhang C., Cao G. Polyphosphate metabolic gene expression analyses reveal mechanisms of phosphorus accumulation and release in Microlunatus phosphovorus strain JN459. FEMS Microbiol. Lett. 2018;365 doi: 10.1093/femsle/fny034. [DOI] [PubMed] [Google Scholar]
- 40.Albi T., Serrano A. Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J. Microbiol. Biotechnol. 2016;32:27. doi: 10.1007/s11274-015-1983-2. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka S., Lee S.-O., Hamaoka K., Kato J., Takiguchi N., Nakamura K., Ohtake H., Kuroda A. Strictly polyphosphate-dependent glucokinase in a polyphosphate-accumulating bacterium Microlunatus phosphovorus. J. Bacteriol. 2003;185:5654–5656. doi: 10.1128/JB.185.18.5654-5656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan Z.-C., Zaheer R., Finan T.M. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 2006;188:1089–1102. doi: 10.1128/JB.188.3.1089-1102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hauska G., Thauer R.K., editors. The Molecular Basis of Bacterial Metabolism. Springer Berlin Heidelberg; Berlin, Heidelberg: 1990. [DOI] [Google Scholar]
- 44.Gimenez R., Nuñez M.F., Badia J., Aguilar J., Baldoma L. The gene yjcG cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 2003;185:6448–6455. doi: 10.1128/JB.185.21.6448-6455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziliani A., Bovio-Winkler P., Cabezas A., Etchebehere C., Garcia H.A., López-Vázquez C.M., Brdjanovic D., Van Loosdrecht M.C.M., Rubio-Rincón F.J. Putative metabolism of Ca. Accumulibacter via the utilization of glucose. Water Res. 2023;229 doi: 10.1016/j.watres.2022.119446. [DOI] [PubMed] [Google Scholar]
- 46.He S., McMahon K.D. Microbiology of Candidatus accumulibacter in activated sludge. Microb. Biotechnol. 2011;4:603–619. doi: 10.1111/j.1751-7915.2011.00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kristiansen R., Nguyen H.T.T., Saunders A.M., Nielsen J.L., Wimmer R., Le V.Q., McIlroy S.J., Petrovski S., Seviour R.J., Calteau A., Nielsen K.L., Nielsen P.H. A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal. ISME J. 2013;7:543–554. doi: 10.1038/ismej.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L., Wei G., Zhang Y., Wang K., Wang C., Deng X., Li Y., Xie X., Chen J., Huang F., Chen H., Zhang B., Wei C., Qiu G. Candidatus accumulibacter use fermentation products for enhanced biological phosphorus removal. Water Res. 2023;246 doi: 10.1016/j.watres.2023.120713. [DOI] [PubMed] [Google Scholar]
- 49.Close K., Marques R., Carvalho V.C.F., Freitas E.B., Reis M.A.M., Carvalho G., Oehmen A. The storage compounds associated with Tetrasphaera PAO metabolism and the relationship between diversity and P removal. Water Res. 2021;204 doi: 10.1016/j.watres.2021.117621. [DOI] [PubMed] [Google Scholar]
- 50.Marques R., Ribera-Guardia A., Santos J., Carvalho G., Reis M.A.M., Pijuan M., Oehmen A. Denitrifying capabilities of Tetrasphaera and their contribution towards nitrous oxide production in enhanced biological phosphorus removal processes. Water Res. 2018;137:262–272. doi: 10.1016/j.watres.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Otieno J., Kowal P., Mąkinia J. The occurrence and role of Tetrasphaera in enhanced biological phosphorus removal systems. Water. 2022;14:3428. doi: 10.3390/w14213428. [DOI] [Google Scholar]
- 52.Kong Y., Xia Y., Nielsen P.H. Activity and identity of fermenting microorganisms in full-scale biological nutrient removing wastewater treatment plants. Environ. Microbiol. 2008;10:2008–2019. doi: 10.1111/j.1462-2920.2008.01617.x. [DOI] [PubMed] [Google Scholar]
- 53.Herbst Dueholm, Wimmer Nielsen. The proteome of Tetrasphaera elongata is adapted to changing conditions in wastewater treatment plants. Proteomes. 2019;7:16. doi: 10.3390/proteomes7020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J.-T., Shim J.-H., Tran P.L., Hong I.-H., Yong H.-U., Oktavina E.F., Nguyen H.D., Kim J.-W., Lee T.S., Park S.-H., Boos W., Park K.-H. Role of maltose enzymes in glycogen synthesis by Escherichia coli. J. Bacteriol. 2011;193:2517–2526. doi: 10.1128/JB.01238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elbein A.D., Pastuszak I., Tackett A.J., Wilson T., Pan Y.T. Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J. Biol. Chem. 2010;285:9803–9812. doi: 10.1074/jbc.M109.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson W.A., Roach P.J., Montero M., Baroja-Fernández E., Muñoz F.J., Eydallin G., Viale A.M., Pozueta-Romero J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 2010;34:952–985. doi: 10.1111/j.1574-6976.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monteiro De Souza P.M. Application of microbial - amylase in industry, braz. J. Microbiol. 2010;4:850–861. doi: 10.1590/S1517-83822010000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fox G. Starch Food. Elsevier; 2018. Starch in brewing applications; pp. 633–659. [DOI] [Google Scholar]
- 59.Oehmen A., Lemos P., Carvalho G., Yuan Z., Keller J., Blackall L., Reis M. Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res. 2007;41:2271–2300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 60.Seviour R.J., McIlroy S. The microbiology of phosphorus removal in activated sludge processes: the current state of play. J. Microbiol. 2008;46:115–124. doi: 10.1007/s12275-008-0051-0. [DOI] [PubMed] [Google Scholar]
- 61.Zou H., Shi M., Zhang T., Li L., Li L., Xian M. Natural and engineered polyhydroxyalkanoate (PHA) synthase: key enzyme in biopolyester production. Appl. Microbiol. Biotechnol. 2017;101:7417–7426. doi: 10.1007/s00253-017-8485-0. [DOI] [PubMed] [Google Scholar]
- 62.Uribe Acosta M., Villa Restrepo A.F. In silico analysis of phaG-like protein in ralstonia eutropha h16, potentially involved in polyhydroxyalkanoates synthesis. Rev. Politécnica. 2019;15:55–64. doi: 10.33571/rpolitec.v15n29a5. [DOI] [Google Scholar]
- 63.Zher Neoh S., Fey Chek M., Tiang Tan H., Linares-Pastén J.A., Nandakumar A., Hakoshima T., Sudesh K. Polyhydroxyalkanoate synthase (PhaC): the key enzyme for biopolyester synthesis. Curr. Res. Biotechnol. 2022;4:87–101. doi: 10.1016/j.crbiot.2022.01.002. [DOI] [Google Scholar]
- 64.Skennerton C.T., Barr J.J., Slater F.R., Bond P.L., Tyson G.W. Expanding our view of genomic diversity in Candidatus accumulibacter clades. Environ. Microbiol. 2015;17:1574–1585. doi: 10.1111/1462-2920.12582. [DOI] [PubMed] [Google Scholar]
- 65.Knoll M., Hamm T.M., Wagner F., Martinez V., Pleiss J. The PHA depolymerase engineering database: a systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinf. 2009;10:89. doi: 10.1186/1471-2105-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morohoshi T., Oi T., Suzuki T., Sato S. Identification and characterization of a novel extracellular polyhydroxyalkanoate depolymerase in the complete genome sequence of undibacterium sp. KW1 and YM2 strains. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han-Wong L., Keun-Park Y. Characterizations of denitrifying polyphosphate-accumulating bacterium Paracoccus sp. strain YKP-9. J. Microbiol. Biotechnol. 2008;2:1958–1965. [PubMed] [Google Scholar]
- 68.Wang Q., He J. Complete nitrogen removal via simultaneous nitrification and denitrification by a novel phosphate accumulating Thauera sp. strain SND5. Water Res. 2020;185 doi: 10.1016/j.watres.2020.116300. [DOI] [PubMed] [Google Scholar]
- 69.Miao Z., Li D., Guo S., Zhao Z., Fang X., Wen X., Wan J., Li A. Effect of free nitrous acid on nitrous oxide production and denitrifying phosphorus removal by polyphosphorus-accumulating organisms in wastewater treatment. BioMed Res. Int. 2018;2018:1–10. doi: 10.1155/2018/9192607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen G., Huang J., Tian X., Chu Q., Zhao Y., Zhao H. Effects of influent loads on performance and microbial community dynamics of aerobic granular sludge treating piggery wastewater. J. Chem. Technol. Biotechnol. 2018;93:1443–1452. doi: 10.1002/jctb.5512. [DOI] [Google Scholar]
- 71.Flowers J.J., He S., Yilmaz S., Noguera D.R., McMahon K.D. Denitrification capabilities of two biological phosphorus removal sludges dominated by different Candidatus accumulibacter clades. Environ. Microbiol. Rep. 2009;1:583–588. doi: 10.1111/j.1758-2229.2009.00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang D., Huang Q., Wang C., Ma M., Wang Z. The effects of different electron donors on anaerobic nitrogen transformations and denitrification processes in lake taihu sediments. Hydrobiologia. 2007;581:71–77. doi: 10.1007/s10750-006-0499-z. [DOI] [Google Scholar]
- 73.Peng Y., Wang X., Li B. Anoxic biological phosphorus uptake and the effect of excessive aeration on biological phosphorus removal in the A2O process. Desalination. 2006;189:155–164. doi: 10.1016/j.desal.2005.06.023. [DOI] [Google Scholar]
- 74.Liu H., Sun Y., Jia X., Li J., Zhou K., Qu X., Tao X., Chen Y. Identification and metabolic mechanism of non-fermentative short cut denitrifying phosphorus removing bacteria. Chin. J. Chem. Eng. 2013;21:332–340. doi: 10.1016/S1004-9541(13)60465-6. [DOI] [Google Scholar]
- 75.Zhang Y., Kinyua M.N. Identification and classification of the Tetrasphaera genus in enhanced biological phosphorus removal process: a review. Rev. Environ. Sci. Biotechnol. 2020;19:699–715. doi: 10.1007/s11157-020-09549-7. [DOI] [Google Scholar]
- 76.Günther S., Trutnau M., Kleinsteuber S., Hause G., Bley T., Röske I., Harms H., Müller S. Dynamics of polyphosphate-accumulating bacteria in wastewater treatment plant microbial communities detected via DAPI (4′,6′-diamidino-2-phenylindole) and tetracycline labeling. Appl. Environ. Microbiol. 2009;75:2111–2121. doi: 10.1128/AEM.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Majed N., Li Y., Gu A.Z. Advances in techniques for phosphorus analysis in biological sources. Curr. Opin. Biotechnol. 2012;23:852–859. doi: 10.1016/j.copbio.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 78.Hupfer M., Glöss S., Schmieder P., Grossart H.-P. Methods for detection and quantification of polyphosphate and polyphosphate accumulating microorganisms in aquatic sediments. Int. Rev. Hydrobiol. 2008;93:1–30. doi: 10.1002/iroh.200610935. [DOI] [Google Scholar]
- 79.Li J., Margaret Oliver I., Cam N., Boudier T., Blondeau M., Leroy E., Cosmidis J., Skouri-Panet F., Guigner J.-M., Férard C., Poinsot M., Moreira D., Lopez-Garcia P., Cassier-Chauvat C., Chauvat F., Benzerara K. Biomineralization patterns of intracellular carbonatogenesis in cyanobacteria: molecular hypotheses. Minerals. 2016;6:10. doi: 10.3390/min6010010. [DOI] [Google Scholar]
- 80.Saito K., Ohtomo R., Kuga-Uetake Y., Aono T., Saito Masanori. Direct labeling of polyphosphate at the ultrastructural level in Saccharomyces cerevisiae by using the affinity of the polyphosphate binding domain of Escherichia coli exopolyphosphatase. Appl. Environ. Microbiol. 2005;10:5692–5701. doi: 10.1128/AEM.71.10.5692-5701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim J.M., Lee H.J., Kim S.Y., Song J.J., Park W., Jeon C.O. Analysis of the fine-scale population structure of Candidatus accumulibacter phosphatis in enhanced biological phosphorus removal sludge, using fluorescence in situ hybridization and flow cytometric sorting. Appl. Environ. Microbiol. 2010;76:3825–3835. doi: 10.1128/AEM.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Espina R., Yu L., Wang J., Tong Z., Vashishtha S., Talaat R., Scatina J., Mutlib A. Nuclear magnetic resonance spectroscopy as a quantitative tool to determine the concentrations of biologically produced metabolites: implications in metabolites in safety testing. Chem. Res. Toxicol. 2009;22:299–310. doi: 10.1021/tx800251p. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H., Lo V.K., Thompson J.R., Koch F.A., Liao P.H., Lobanov S., Mavinic D.S., Atwater J.W. Recovery of phosphorus from dairy manure: a pilot-scale study. Environ. Technol. 2015;36:1398–1404. doi: 10.1080/09593330.2014.991354. [DOI] [PubMed] [Google Scholar]
- 84.De Jager H.-J., Heyns A.M. Study of the hydrolysis of sodium polyphosphate in water using Raman spectro. Appl. Spectrosc. 1998;52:808–814. doi: 10.1366/0003702981944535. [DOI] [Google Scholar]
- 85.Schuster K.C., Urlaub E., Gapes J.R. Single cell analysis of bacteria by Raman microscopy: spectral information on the chemical composition of cells and on the heterogeneity in a culture. J. Microbiol. Methods. 2000;42:29–38. doi: 10.1016/S0167-7012(00)00169-X. [DOI] [PubMed] [Google Scholar]
- 86.Crocetti G.R., Hugenholtz P., Bond P.L., Schuler A., Keller J., Jenkins D., Blackall L.L. Identification of polyphosphate accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 2000;66:1175–1182. doi: 10.1128/AEM.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vandamme P., Pot B., Gillis M. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60 doi: 10.1128/mr.60.2.407-438.1996. 07–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McMahon K.D., Yilmaz S., He S., Gall D.L., Jenkins D., Keasling J.D. Polyphosphate kinase genes from full scale activated sludge plants. Appl. Microbiol. Biotechnol. 2007;77:167–173. doi: 10.1007/s00253-007-1122-6. [DOI] [PubMed] [Google Scholar]
- 89.McMahon K.D., Dojka M.A., Pace N.R., Jenkins D., Keasling J.D. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl. Environ. Microbiol. 2002;68:4971–4978. doi: 10.1128/AEM.68.10.4971-4978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He S., Gall D.L., McMahon K.D. Candidatus accumulibacter population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl. Environ. Microbiol. 2007;73:5865–5874. doi: 10.1128/AEM.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Camejo P.Y., Oyserman B.O., McMahon K.D., Noguera D.R. Integrated omic analyses provide evidence that a Candidatus accumulibacter phosphatis strain performs denitrification under microaerobic conditions. mSystems. 2019;4 doi: 10.1128/mSystems.00193-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rubio-Rincón F.J., Weissbrodt D.G., Lopez-Vazquez C.M., Welles L., Abbas B., Albertsen M., Nielsen P.H., Van Loosdrecht M.C.M., Brdjanovic D. Candidatus accumulibacter delftensis: a clade IC novel polyphosphate accumulating organism without denitrifying activity on nitrate. Water Res. 2019;161:136–151. doi: 10.1016/j.watres.2019.03.053. [DOI] [PubMed] [Google Scholar]
- 93.Zhou Y., Pijuan M., Zeng R.J., Lu H., Yuan Z. Could polyphosphate-accumulating organisms (PAOs) Be glycogen-accumulating organisms (GAOs)? Water Res. 2008;42:2361–2368. doi: 10.1016/j.watres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 94.Albertsen M., McIlroy S.J., Stokholm-Bjerregaard M., Karst S.M., Nielsen P.H. Candidatus Propionivibrio aalborgensis: a novel glycogen accumulating organism abundant in full-scale enhanced biological phosphorus removal plants. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roller C., Wagner M., Amann R., Ludwig W., Schleifer K.-H. In situ probing of gram-positive bacteria with high DNA G + C content using 23s rRNA-targeted oligonucleotides. Microbiol. Soc. 1994;140 doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 96.Erhart R., Bradford D., Seviour R.J., Amann R., Blackall L.L. Development and use of fluorescent in situ hybridization probes for the detection and identification of Microthrix parvicella in activated sludge. Syst. Appl. Microbiol. 1997;20:310–318. doi: 10.1016/S0723-2020(97)80078-1. [DOI] [Google Scholar]
- 97.Kong Y., Xia Y., Nielsen J.L., Nielsen P.H. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology. 2007;153:4061–4073. doi: 10.1099/mic.0.2007/007245-0. [DOI] [PubMed] [Google Scholar]
- 98.Kong Y., Nielsen J.L., Nielsen P.H. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 2005;71:4076–4085. doi: 10.1128/AEM.71.7.4076-4085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blackall L.L., Seviour E.M., Bradford D., Rossetti S., Tandoi V., Seviour R.J. Candidatus nostocoida limicola a filamentous bacterium from activated sludge. Int. J. Syst. Evol. Microbiol. 2000;50:703–709. doi: 10.1099/00207713-50-2-703. [DOI] [PubMed] [Google Scholar]
- 100.McDaniel E.A., Van Steenbrugge J.J.M., Noguera D.R., McMahon K.D., Raaijmakers J.M., Medema M.H., Oyserman B.O. TbasCO: trait-based comparative omics identifies ecosystem-level and niche-1 differentiating adaptations of an engineered microbiome. ISME Commun. 2022;2:111. doi: 10.1038/s43705-022-00189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tomás-Martínez S., Zwolsman E.J., Merlier F., Pabst M., Lin Y., Van Loosdrecht M.C.M., Weissbrodt D.G. Turnover of the extracellular polymeric matrix of granules performing biological phosphate removal. Appl. Microbiol. Biotechnol. 2023;107:1997–2009. doi: 10.1007/s00253-023-12421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.da Fonseca A.P. A digital image based method to quantify intracellular polyphosphates in microbial aggregates. Universidade do Minho. 2018;29 http://hdl.handle.net/1822/66151 [Google Scholar]
- 103.Tu Y. Univ. N. M.; 2012. The Effects of Acetate Concentrations on Competition between Microbial Populations in Enhanced Biological Phosphorus Removal from Wastewater.https://digitalrepository.unm.edu/ce_etds/10 [Google Scholar]
- 104.Ahn J., McIlroy S., Schroeder S., Seviour R. Biomass granulation in an aerobic:anaerobic-enhanced biological phosphorus removal process in a sequencing batch reactor with varying pH. J. Ind. Microbiol. Biotechnol. 2009;36:885–893. doi: 10.1007/s10295-009-0566-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.