Abstract
Objective
This study aimed to assess the current status of carcinoembryonic antigen (CEA) detection. We evaluated the correlation, consistency, and comparability of CEA results among six automated immunoassays, and combined with the results of CEA trueness verification of the Beijing Center for Clinical Laboratories (BCCL) for further analysis.
Methods
Abbott Architect i2000, Beckman DxI800, Roche Cobas E601, Diasorin Liaison XL, Maccura IS1200, and Autolumo A2000 were used to detect 40 individual serum CEA samples. Taking the optimal analytical quality specifications calculated from data on biological variation as the evaluation criterion. Passing-Bablok regression and Bland-Altman analysis were performed between each assay and all-assays median values to evaluate the correlation and relative difference. The concordance correlation coefficient (CCC) was used for consistency analysis. Additionally, the trueness verification program used samples at three concentration levels to assess the bias, coefficient of variation (CV), and total error (TE) between the average measured values and the target value.
Results
The Spearman's rank correlation coefficient (rs) was ≥0.996 and the CCC ranged between 0.9448 and 0.9990 for each assay vs. all-assays median. Considering the all-assays median value of each sample as a reference, there were proportional and systematic differences according to the Passing-bablok regression analysis. The relative difference of the four assays (Abbott Architect i2000, Autolumo A2000, Diasorin Liaison XL, and Maccura IS1200) met the optimal analytical quality specifications. On the other hand, Beckman DxI800 (13.2 %) and Roche Cobas E601 (−9.0 %) were only able to fulfill the desirable analytical quality specifications. The average pass rates for bias, CV, and TE of the trueness verification program were 80 %, 98 %, and 96 %, respectively.
Conclusions
The six automated immunoassays vs. all-assays median have a good correlation in CEA detection. However, there is a lack of comparability of CEA results. Further improvements are needed in harmonization among CEA detections.
Keywords: Carcinoembryonic antigen, Immunoassay, Assay comparison, Trueness verification program
Highlights
-
•
Six assays were compared with all-assays median to evaluate the comparability of CEA.
-
•
45 laboratories participated in trueness verification to evaluate the status of CEA.
-
•
Rs and CCC were used to evaluate the correlation and consistency between assays.
-
•
The harmonization of CEA results needs to be further improved.
1. Introduction
Carcinoembryonic antigen (CEA) is an acidic glycoprotein with a molecular weight of approximately 200 kDa, produced in colorectal cancer tissue. It displays characteristics with the human embryonic antigen and is closely related to cell adhesion. As an antigen, CEA can induce an immune response in patients and is commonly used as a marker for the early diagnosis of colon and rectal cancer. Serum levels of CEA are also elevated in patients with lung cancer, gastric cancer and other malignant tumors [1]. Mild elevations of CEA is associated with benign diseases, such as colitis, pancreatitis, liver cirrhosis, hepatitis, renal insufficiency, and heavy smokers. Therefore, CEA functions as a broad-spectrum tumor marker. Although being unsuitable as a specific marker for diagnosing certain malignant tumors, it still plays an important role in the differential diagnosis of malignant tumors, disease monitoring, and evaluation of therapeutic efficacy, and therefore has clinical significance [2]. However, the CEA request suffers from the general concept that "CEA is a broad-spectrum tumor marker". Thus, overuse and misuse of the marker determination generally occur [3].
At present, the detection of CEA mainly includes enzyme-linked immunosorbent assay (ELISA), radioimmunoassay (RIA), and chemiluminescence immunoassay (CLIA) [[4], [5], [6]]. RIA and ELISA have been largely replaced by fully automated CLIA assays. In addition, there are burgeoning technologies for CEA detection in recent decades, such as a signal-on/split-type photoelectrochemical sensing approach, an enhanced pressure-based immunoassay and a novel homogeneous visual fluorescence sensing system [[7], [8], [9], [10]]. The reason to compare assays is to assure that patients with a disease will be equally classified by all clinically validated assays and that monitoring may be recommended performed using the same assays, or independently from the assays if results are interchangeable. However, in practice, CEA assay results vary from laboratory to laboratory due to the differences in assay principles, detection instruments and reagents, epitopes, and antibody specificities, which directly affect clinical diagnosis and treatment. We assessed the comparability of the results from six different automated immunoassays using 40 individual samples. The trueness verification program was performed in 45 laboratories to assess the consistency of the CEA assays and to compare their performance with analytical quality specifications. This study can provide the current status of CEA detection for manufacturers and clinical laboratories to promote the harmonization of CEA immunoassays and the improvement of clinical guidelines.
2. Materials and methods
2.1. Materials
2.1.1. Reagents and instruments
The six immunoassays and reagents used in this study were as follows: Abbott Architect i2000 (Abbott Diagnostics); Beckman DxI800 (Beckman Coulter Inc.); Roche Cobas E601 (Roche Diagnostics GmbH); Diasorin Liaison XL (DiaSorin S.p.A); Maccura IS1200 (Maccura Biology Co., Ltd.); and Autolumo A2000 (Autobio Diagnostics). Performance characteristics of the six automated CEA immunoassay analyzers based on information provided by the manufacturers are summarized in Supplemental Table 1.
2.1.2. Individual serum samples
A total of 40 individual serum samples containing different levels of CEA (low, medium, and high) were collected from the laboratory department of Beijing Chaoyang Hospital in 2019, and the CEA concentrations ranged from 2.41 to 888.57 ng/mL (as measured by the Roche Cobas E601). Each sample (at least 2.5 mL) was aliquoted into six tubes and stored at −80 °C until analysis. The use of leftover patient samples was approved by the Ethics Committee of the Beijing Center for Clinical Laboratories, and the study procedure adhered to the tenets of the Declaration of Helsinki. Detailed patient information was not needed and the data were analyzed anonymously; therefore, participants did not provide written informed consent.
2.1.3. Samples of the trueness verification program
According to the EQA program of the BCCL, CEA samples of high (designated 01), medium (02), and low (03) concentrations were delivered to 45 laboratories for low-temperature transportation, with 3 sets of 9 samples in one laboratory.
2.2. Methods
2.2.1. Imprecision
The six assays were evaluated for imprecision using low-, medium-, and high-level quality controls. The Standard Deviation (SD) and Coefficient of Variation (CV) were calculated for each level of quality controls.
2.2.2. Comparability analysis
The Passing-Bablok regression and the Bland-Altman analysis were performed on the 40 individual serum samples. The slope, intercept, and Spearman's rank correlation coefficient (rs) were calculated to evaluate the correlation and to identify proportional and systematic bias. The relative difference plots between the six assays and all-assays median were performed [11]. The consistency between the six assays measured values and all-assays median value was evaluated using the concordance correlation coefficient (CCC): CCC >0.99, almost perfect; 0.99–0.95, substantial; 0.95–0.90, moderate; <0.90, poor consistency [12].
2.2.3. EQA program
The CEA samples of three concentration levels were distributed to 45 laboratories. Each laboratory was requested to measure one aliquot of each concentration level on three consecutive days, and each aliquot was tested for repeatability thrice time. Therefore, a total of 9 results were obtained for each concentration level, and the average of these 9 results was considered as the laboratory tested value. The target values were assigned as 806.49 ng/mL, 87.57 ng/mL and 15.38 ng/mL [13]. The bias, CV, and total error (TE) between the average measured value and the target value were used to evaluate measurement performance, and the pass rates of bias, CV, and TE were calculated with the minimal analytical quality specifications calculated from data on biological variation as the evaluation criterion separately.
2.3. Statistical analysis
Considering the median value of each sample for all assays as a reference, Passing-Bablok regression analysis and relative difference plots were performed. Average absolute bias (the difference between the measured value of the assay and all-assays median value) and average relative bias (ratio of average absolute bias to all-assays median value) were calculated. Bias, CV, and TE evaluation criterion were calculated from data on biological variation (https://www.westgard.com/biodatabase1.htm) to evaluate the results of the trueness verification program. Intralaboratory CV was calculated from the 9 repeated results, and TE was calculated as the bias + 2CV. Accordingly, the optimal analytical quality specifications of the measurements were 7.13 %, 3.18 % and 12.37 % for CEA, respectively; the desirable analytical quality specifications of the measurements were 14.26 %, 6.35 % and 24.74 % for CEA, respectively; the minimal analytical quality specifications of the measurements were 21.39 %, 9.53 %, and 37.10 % for CEA, respectively. Excel and MedCalc were used for statistical data analysis.
3. Results
3.1. Imprecision
The CVs of the six assays ranged between 1.23 % and 3.15 % at low concentration, which met the requirement of imprecision in optimal analytical quality specifications (3.18 %). 2 %–6.03 % for medium concentration, and 0.6 %–3.58 % for high concentration of CEA. The CVs of the six assays met the requirement of imprecision in desirable analytical quality specifications (6.35 %) for medium and high concentrations (Supplemental Table 2). Among them, only the CV of Roche Cobas E601 and Abbott Architect i2000 met the optimal analytical quality specifications at three concentration levels.
3.2. Assay comparison of six CEA immunoassays
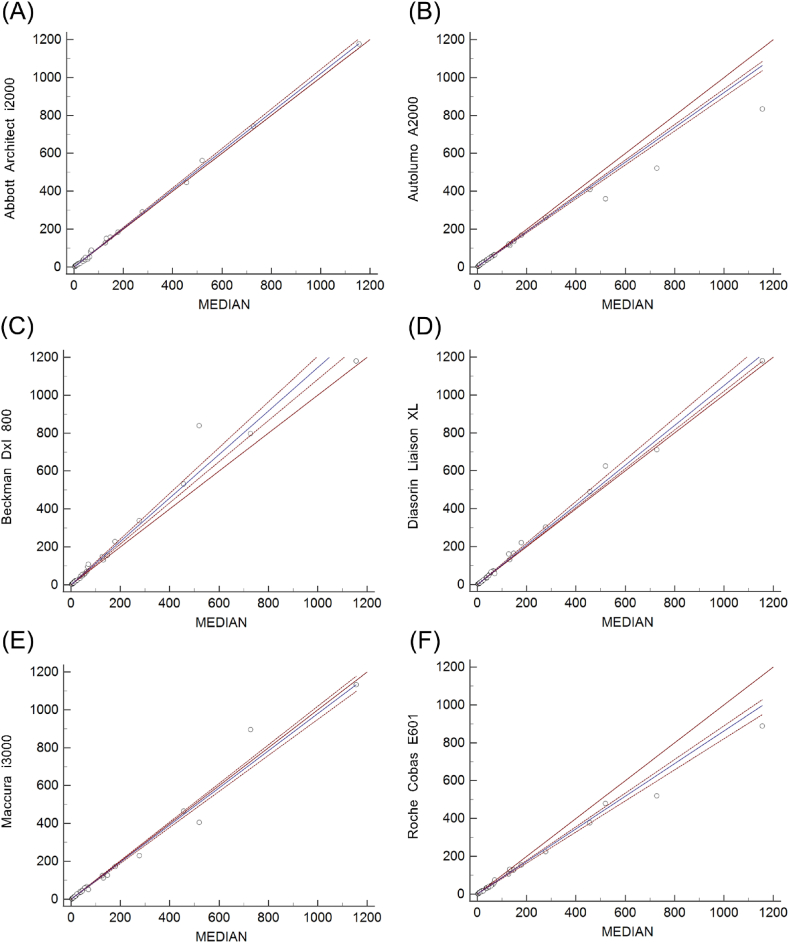
The measured values of each assay were compared to the all-assays median value, and the regression parameters, average bias estimates, and consistency parameters are shown in Table 1. For the individual serum CEA samples, each assay vs. all-assays median exhibited strong correlations, with all Spearman's rank correlation coefficient (rs) ≥ 0.96 (p < 0.0001). The CCC, ranging from 0.9448 to 0.9990, indicated a substantial concordance for each assay vs. all-assays median. The Cusum test showed no significant deviation from linearity for each assay vs. all-assays median (p > 0.05). Additionally, the regression slope ranged from 0.8617 to 1.1462 with the 95 % confidence interval of the slope excluding 1, except for Abbott Architect i2000/Maccura IS1200 vs. all-assays median. The 95 % confidence interval of the intercept excluded 0 for Autolumo A2000/Roche Cobas E601 vs. all-assays median. Thus, there are proportional and systematic differences between the assays and all-assays median [14].
Table 1.
Comparison data of six assays vs. all-assays median values of CEA results for 40 individual serum samples.
| Measuring system | Regression parameters |
||||||
|---|---|---|---|---|---|---|---|
| Slope (95 % CI) | Intercept (95 %CI) | Cusum test | rs (95 %CI) | Average bias, mg/L (95 % CI) | Average bias, % (95 % CI)a |
CCC (95 %CI) | |
| Abbott Architect i2000 |
1.0201 (1.000–1.0422) | 0.0913 (−0.1325 - 0.4250) | P = 0.53 | 0.996 (0.993–0.998) | 2.97 (−0.29 - 6.23) | 1.67 (−1.78 - 5.12) | 0.9990 (0.9982–0.9994) |
| Autolumo A2000 |
0.9184 (0.8954–0.9363) | 2.3815 (1.8409–3.2455) | P = 0.15 | 0.998 (0.997–0.999) | −19.30 (−39.74 - 1.15) | 6.16 (0.46–11.86) | 0.9448 (0.9293–0.9570) |
| Beckman DxI800 |
1.1462 (1.0845–1.2054) | −0.4720 (−1.2793 - 0.3001) | P = 0.97 | 0.998 (0.997–0.999) | 18.97 (2.10–35.84) | 13.18 (8.47–17.89) | 0.9737 (0.9556–0.9844) |
| Diasorin Liaison XL |
1.0508 (1.0209–1.0984) | −08629 (−1.3395 to −0.4254) | P = 0.15 | 0.997 (0.995–0.999) | 7.26 (0.80–13.73) | 1.50 (−1.79 - 4.78) | 0.9958 (0.9925–0.9976) |
| Maccura IS1200 |
0.9804 (0.9504–1.0189) | −0.3010 (−0.7041 - 0.0813) | P = 0.15 | 0.997 (0.994–0.998) | −1.72 (−12.64 - 9.21) | −4.17 (−7.34 to −1.00) | 0.9891 (0.9797–0.9942) |
| Roche Cobas E601 |
0.8617 (0.8208–0.8883) | 0.7981 (0.4616–1.2332) | P = 0.53 | 0.999 (0.997–0.999) | −19.72 (−36.83 to −2.61) | −9.00 (−11.86 to −6.13) | 0.9614 (0.9489–0.9709) |
Assays not fulfilling optimal bias analytical quality specifications are in bold. CCC, concordance correlation coefficient.
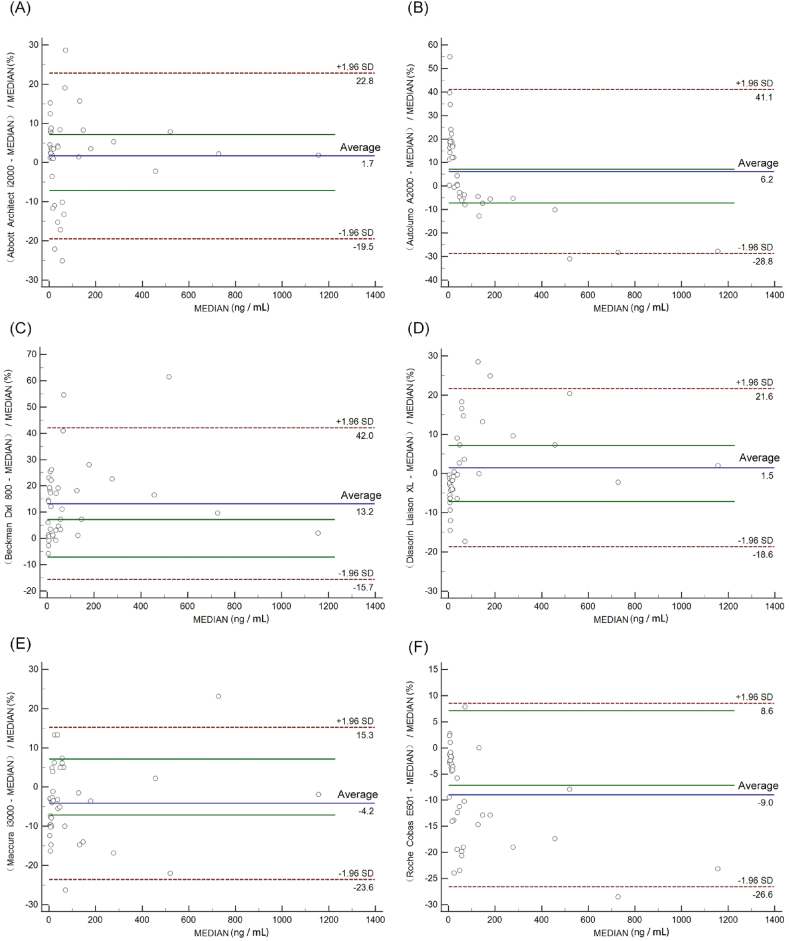
Fig. 1 A-F depicts the Passing-Bablok regression graphs, and relative difference plots of six assays vs. all-assays median are shown in Fig. 2 A-F. The average bias between four assays (Abbott Architect i2000, Autolumo A2000, Diasorin Liaison XL and Maccura IS1200) and all-assays median met optimal analytical quality specifications, while Beckman DxI800 and Roche Cobas E601 were only able to fulfill desirable analytical quality specifications (Table1).
Fig. 1.
Comparison of the six immunoassays for CEA detection by Passing-Bablok regression analysis. MEDIAN indicates all-assays median. Results are in ng/mL. The blue solid lines indicate the Passing-Bablok regression analysis results, while the red dashed lines around the regression lines describe 95 % confidence interval areas. The red solid lines indicate x = y.
Fig. 2.
Comparison of six CEA immunoassays by Bland-Altman plots in 40 serum samples. MEDIAN indicates all-assays median. The blue solid line indicates the percent difference from the average of the two assays. The red dashed lines indicate the upper and lower 95 % confidence limits of the percent difference between the two assays. The green solid lines are the accepted bias limits of 7.13 % optimal analytical quality specifications.
3.3. Analysis of the trueness verification program results
The pass rates of laboratories that met the bias, CV, and TE limits was calculated from minimal analytical quality specifications, as shown in Table 2. The total pass rates of bias varied from 69 % to 87 %. The intralaboratory CV pass rates were 98 % at three concentrations with only one laboratory failing to meet the requirement. The pass rates for TE, ranging from 93 % to 98 %, were approximately equal to the CV. Among them, the Roche system achieved the highest pass rates (100 %) for bias, CV and TE. The pass rates of bias, CV and TE for all other systems except for the Roche system differed greatly.
Table 2.
Results of the trueness verification program.
| 01 |
02 |
03 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Bias | CV | TE | Bias | CV | TE | Bias | CV | TE | |
| Total | 87 % (39/45) | 98 % (44/45) | 98 % (44/45) | 69 % (31/45) | 98 % (44/45) | 93 % (42/45) | 84 % (38/45) | 98 % (44/45) | 98 % (44/45) |
| Abbott Architect i2000 | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) | 17 % (1/6) | 100 % (6/6) | 100 % (6/6) | 83 % (5/6) | 100 % (6/6) | 100 % (6/6) |
| Abbott AxSym | 100 % (3/3) | 100 % (3/3) | 100 % (3/3) | 33 % (1/3) | 100 % (3/3) | 100 % (3/3) | 67 % (2/3) | 100 % (3/3) | 100 % (3/3) |
| Beckman Access | 67 % (2/3) | 100 % (3/3) | 100 % (3/3) | 33 % (1/3) | 100 % (3/3) | 100 % (3/3) | 33 % (1/3) | 100 % (3/3) | 100 % (3/3) |
| Beckman DxI800 | 50 % (3/6) | 100 % (6/6) | 100 % (6/6) | 50 % (3/6) | 100 % (6/6) | 67 % (4/6) | 50 % (3/6) | 100 % (6/6) | 100 % (6/6) |
| Roche Cobas E601 | 100 % (13/13) | 100 % (13/13) | 100 % (13/13) | 100 % (13/13) | 100 % (13/13) | 100 % (13/13) | 100 % (13/13) | 100 % (13/13) | 100 % (13/13) |
| Roche Elecsys E411 | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) | 100 % (6/6) |
| Roche ES 300 | 100 % (2/2) | 100 % (2/2) | 100 % (2/2) | 100 % (2/2) | 100 % (2/2) | 100 % (2/2) | 100 % (2/2) | 100 % (2/2) | 100 % (2/2) |
| Siemens ADVIA Centaur XP | 50 % (2/4) | 100 % (4/4) | 100 % (4/4) | 50 % (2/4) | 100 % (4/4) | 100 % (4/4) | 100 % (4/4) | 100 % (4/4) | 100 % (4/4) |
| Bioscience Peteck 96-I | 100 % (1/1) | 100 % (1/1) | 100 % (1/1) | 100 % (1/1) | 100 % (1/1) | 100 % (1/1) | 100 % (1/1) | 100 % (1/1) | 100 % (1/1) |
| Snibe Maglumi 2000plus | 100 % (1/1) | 0 % (0/1) | 0 % (0/1) | 100 % (1/1) | 0 % (0/1) | 0 % (0/1) | 100 % (1/1) | 0 % (0/1) | 0 % (0/1) |
The percentage represents the laboratories meeting the minimal bias, CV, and TE analytical quality specifications, and the fraction in bracket was the number of labs meeting criterion/total number of labs of this type. CV, coefficient of avriation; TE, total error.
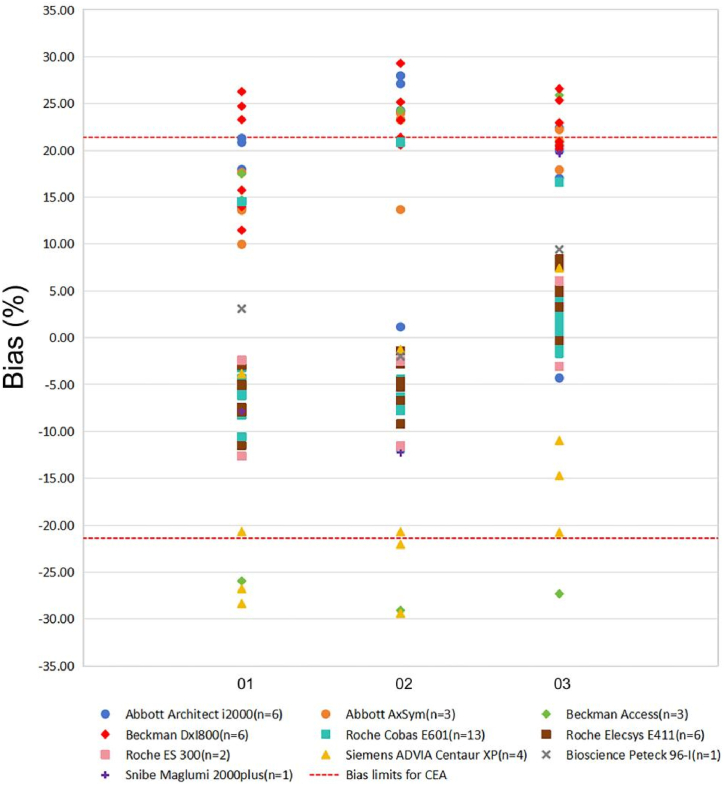
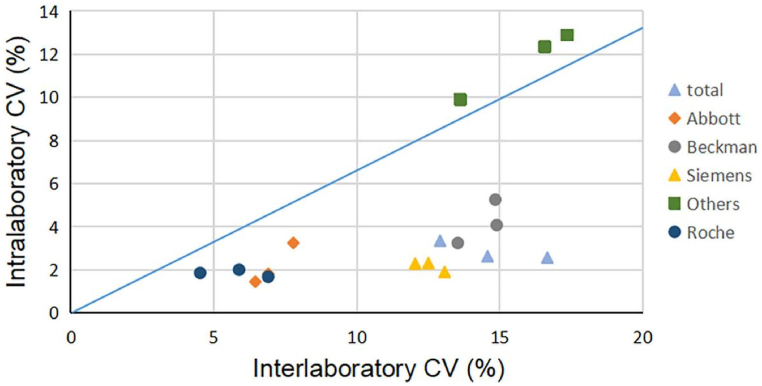
As shown in Fig. 3, the ranges of bias were highly dispersed. The bias at the low concentration was higher than those at the high concentration for the Abbot system and Beckman system. Beckman DxI800 and Abbott system showed positive bias was predominant for three concentrations, and Beckman DxI800 have the biggest positive mean bias. Meanwhile, the Siemens system exhibited the largest negative bias, and the bias at low concentration was higher than high concentration. The biases of these systems were large and only the Roche system met bias limits at three concentrations. For most systems, the ratio of interlaboratory/intralaboratory CVs was >2.0 except for two minority systems (Bioscience Peteck 96-I and Snibe Maglumi 2000plus) (Fig. 4).
Fig. 3.
Percent difference in different assays against bias limits for CEA at three concentrations. The X-axis represents the sample codes of the three concentrations of CEA, the Y-axis represents the relative bias for peer groups. The red dashed line represents the minimal bias limits for CEA derived from the biological variability.
Fig. 4.
Potential to improve equivalence with standardization. The X-axis displays the interlaboratory CVs of different systems, and the Y-axis displays the mean intralaboratory CVs of each system. The red and blue solid lines display interlaboratory/intralaboratory CVs ratios of 2.0 and 1.5, respectively.
4. Discussion
Automated systems are able to process numerous test samples with smaller amounts of reagents, resulting in cost savings and improved efficiency. However, the results obtained by different automated assays have not always been satisfactory [15]. Our results indicated a variation in the CEA levels determined by different assays, which is consistent with previous studies. A study by Joonhong Park et al. [6] evaluated the agreement among automated immunoassays for CEA in four different medical laboratories, including ADVIA Centaur XP, ARCHITECT i2000sr, Elecsys E170, and Unicel DxI800, and the results showed considerable variability and significant differences. Another study by Kuo Zhang et al. [16], individual serum CEA samples were measured using four different assays: Abbott Architect i2000SR, Beckman Access DXI800, Roche Cobas E601, and Siemens ADVIA Centaur XP. They discovered that there was no perfect correlation between any two assays, and the results of four assays varied.
The results of Passing-Bablok regression analysis indicated that there were proportional and systematic differences between the assays and all-assays median. It was presumed that the variation of results obtained from six different assays could be attributed to the current CEA detection primarily relying on the immunoassays, whichuse the principle of specific binding between antigens and antibodies. Difference may be attributed to the use of different monoclonal antibodies in the assays and the specificity of the specificity of the antigen-antibody binding site. Besides, interference such as non-specific cross-reactions, heterophilic antibodies, and paraproteins may be inevitable [17]. A previous study found that 83 % of monoclonal antibodies targeting CEA could be categorized into five epitope groups denoted as GOLD 1–5. These groups are non-interacting and these antibodies have been categorized into 7 groups (groups A-G) based on the domain structure of the CEA molecule [18,19]. According to the manufacturer's instructions, the Roche system's detection kit monoclonal antibodies reacted with the 2 and 5 antigenic determinants. In contrast, the Diasorin system employs three different highly specific monoclonal antibodies, for the coating and tracer of solid phase (magnetic particles), respectively. The tracer recognizes the peptide epitopes specific to CEA belong to category 5. The lack of standardized methods for the production and purification monoclonal antibodies may lead to differences in the detection of the same group of epitopes [16]. Additionally, differences in the structure of the carbohydrate chains present in the CEA molecule may result in varying specificity of anti-CEA antibodies. Therefore, selecting monoclonal antibodies that remain unaffected by variations in carbohydrate chains and forgo cross-reactivity with CEA-related antigens is pivotal in improving the accuracy of CEA assays [20].
In the Bland-Altman analysis, the average bias of Maccura IS1200, Diasorin Liaison XL, Autolumo A2000, and Abbott Architect i2000 vs. all-assays median met the requirement for optimal analytical quality specifications, while those of Roche Cobas E601 and Beckman DxI800 vs. all-assays median were only able to fulfill the desirable analytical quality specifications. This difference could be related to the traceability of calibration products. Among the six assays, only the calibrations of Roche Cobas E601, Maccura IS1200, and Diasorin Liaison XL could be calibrated to WHO 73/601. The use of a common standard may decrease the variation among different CEA immunoassays.
To achieve a comprehensive understanding of the current state of CEA detection, a trueness verification program was conducted with the following aims: to assess individual laboratories and systems for various factors such as bias and reproducibility, stability, calibration traceability, and consistency across different laboratories and systems. The pass rates of imprecision (CV), trueness (bias), and inaccuracy (TE) of each system's CEA detection were evaluated using minimal analytical quality specifications. The results showed that the Roche system achieved the highest pass rates. Furthermore, an average of 98 % of laboratories met the requirement for intralaboratory CV, whereas the pass rates for bias were lower compared to CV and TE. These findings indicated that bias was the primary factor limiting the accuracy and comparability of CEA results.
The intralaboratory CVs serve as an indicator of how reproducible and stable the assay is. Factors such as instrument imprecision, consistency of reagents and calibrators, and the stability of the reagents all contributed significantly to the stability of assays [21,22]. The pass rates of laboratories meeting the bias were approximately equal to the TE. The bias of the systems, except for the Roche system, was large, indicating obvious system bias in these systems. The ability of the participating laboratory is assessed by comparing the bias of each system and the acceptable criterion. The poor results with a large dispersion and a large deviation from the target value, indicate a significant issue of the trueness. This may be due to differences in the principle of detection methods, incorrect calibration, inappropriate use of standards and quality control products, lack of operator skills or other reasons caused by systematic errors. The interlaboratory CVs of results using the same system were used to illustrate the degree of bias and imprecision, and are also one of the indicators used to measure the overall detection quality of these laboratories. The greater the difference between interlaboratory and intralaboratory CVs, the higher the contribution of bias to the dispersion between laboratory results and the greater the effect of standardization on the improvement of equivalence. In our study, interlaboratory CVs were much larger than intralaboratory CVs (for most systems, the ratio of interlaboratory/intralaboratory CVs was over 2.0), indicating significant differences and unsatisfactory comparability among laboratories. Efforts need to be made to achieve better equivalence through standardization or harmonization of CEA [21]. The trueness verification program also had some limitations, as only a few specific concentrations of samples were available, making it difficult to achieve assay comparisons across the entire range of assays [23].
In brief, the clinical measurement of CEA is based on immunological techniques, however, the absence of internationally accepted reference methods and high levels of accuracy is the primary impediments to the standardisation and consistency of measurement. Therefore, each laboratory needs to establish its reference range to accurately reflect the situation of a specific population. The laboratory's report to the clinician should detail the characteristics of the reagent used. If the test reagent used changes during patient monitoring with the same assay is required until comparability of CEA detection values is achieved. The results of CEA should be evaulated comprehensively in conjunction with the patient's medical history, clinical examination, and other clinical information. It is also necessary to select the most suitable antigen-antibody combination to improve the accuracy of the detection assay. Moreover, in the absence of internationally recognized reference methods, good laboratory internal quality control is an important guarantee to ensure the quality of testing [8].
5. Conclusions
Our study showed that there were quite difference in CEA results of the six common chemiluminescence automated immunoassays, and the harmonization of CEA needs to be further improved. However, this study also had certain limitations, such as only six assays were included, and only 40 individual serum samples were tested at the same time. The sample size was relatively small, and the number of laboratories participating in the EQA program was small. Further studies should consider additional assays and a large number of patient serum samples.
Research funding
This study was supported by Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support, China (ZYLX202137) and Beijing Chaoyang Hospital Science and Technology Innovation Fund (grant numbers 22kcjjzd-7)
Data availability
Data will be made available on request.
Employment or leadership: Not applicable.
Honorarium
Not applicable.
Ethical approval
The study involved use of leftover patient serum samples. The leftover patient samples were all de-identified during the collection. The use of patient samples in the present study was reviewed by the Ethics Committee of Beijing Chaoyang Hospital. The ethical approval number was 2022-ke-47. Detailed patient information was not needed, and the data were analyzed anonymously; therefore, participants did not provide written informed consent.
CRediT authorship contribution statement
Wenxuan Fu: Writing – original draft, Visualization, Resources, Methodology, Investigation, Data curation, Conceptualization. Yuhong Yue: Visualization, Validation, Resources, Methodology, Conceptualization. Yichuan Song: Methodology, Conceptualization. Shunli Zhang: Methodology. Jie Shi: Methodology. Rui Zhao: Methodology. Qingtao Wang: Supervision. Rui Zhang: Writing – review & editing, Validation, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e25158.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Zheng T.S., Chen X.F. Higher Education Press; Beijing: 2012. Clinical Biochemical Testing; p. 203p. [Google Scholar]
- 2.Zhang T.Z., Xu G.W. Tianjin Science Press; 1996. Oncology. Tianjin; pp. 546–547p. [Google Scholar]
- 3.Ferraro S., Mozzi R., Panteghini M. Tumor marker ordering: do not lose control: a prospective clinical trial. Am. J. Clin. Pathol. 2015 Oct;144(4):649–658. doi: 10.1309/AJCPNZAPJRB3T6KK. 10.1309/AJCPNZAPJRB3T6KK. PMID: 26386087. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J., Diao X., Wang S., Yao Y. Diagnosis value of combined detection of serum SF, CEA and CRP in non-small cell lung cancer. Cancer Manag. Res. 2020 Sep 22;12:8813–8819. doi: 10.2147/CMAR.S268565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elessawi D.F., Alkady M.M., Ibrahim I.M. Diagnostic and prognostic value of serum IL-23 in colorectal cancer. Arab J Gastroenterol. 2019 Jun;20(2):65–68. doi: 10.1016/j.ajg.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Park J., Lee S., Kim Y., Choi A., Lee H., Lim J., et al. Comparison of four automated carcinoembryonic antigen immunoassays: ADVIA Centaur XP, ARCHITECT I2000sr, Elecsys E170, and Unicel Dxi800. J Ann Lab Med. 2018;38:355–361. doi: 10.3343/alm.2018.38.4.355. PMID: 29611386; PMCID: PMC5895865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X., Lin Q., Lu L., Li M., Tang D. In2O3/CdIn2S4 heterojunction-based photoelectrochemical immunoassay of carcinoembryonic antigen with enzymatic biocatalytic precipitation for signal amplification. Anal. Chim. Acta. 2022 Oct 2;1228 doi: 10.1016/j.aca.2022.340358. [DOI] [PubMed] [Google Scholar]
- 8.Qiu Z., Shu J., Tang D. Bioresponsive release system for visual fluorescence detection of carcinoembryonic antigen from mesoporous silica nanocontainers mediated optical color on quantum dot-enzyme-impregnated paper. Anal. Chem. 2017 May 2;89(9):5152–5160. doi: 10.1021/acs.analchem.7b00989. [DOI] [PubMed] [Google Scholar]
- 9.Huang L., Zeng Y., Liu X., Tang D. Pressure-based immunoassays with versatile electronic sensors for carcinoembryonic antigen detection. ACS Appl. Mater. Interfaces. 2021 Oct 6;13(39):46440–46450. doi: 10.1021/acsami.1c16514. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Kangyao, Lv Shuzhen, Zhou Qian, Tang Dianping. CoOOH nanosheets-coated g-C3N4/CuInS2 nanohybrids for photoelectrochemical biosensor of carcinoembryonic antigen coupling hybridization chain reaction with etching reaction. Sensors and Actuators B-chemical. 2019 Dec;307 doi: 10.1016/j.snb.2019.127631. [DOI] [Google Scholar]
- 11.Ferraro S., Bussetti M., Rizzardi S., Braga F., Panteghini M. Verification of harmonization of serum total and free prostate-specific antigen (PSA) measurements and implications for medical decisions. Clin. Chem. 2021 Mar 1;67(3):543–553. doi: 10.1093/clinchem/hvaa268. [DOI] [PubMed] [Google Scholar]
- 12.Mcbride G.B. 2005. A proposal for strength-of-agreement criteria for Lin’s concordance correlation coefficient; pp. 1–10. NIWA Client Rep. HAM2005-062. [Google Scholar]
- 13.Zhang R., Xu Z., Zhao R., Fu W., Song Y., Wang Q., Yue Y. Accurate method for value assignment of carcinoembryonic antigen reference materials. J. Clin. Lab. Anal. 2023 Jun;37(11–12) doi: 10.1002/jcla.24936. Epub 2023 Jul 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zidan M., Thomas R.L., Slovis T.L. What you need to know about statistics, part II: reliability of diagnostic and screening tests. Pediatr. Radiol. 2015 Mar;45(3):317–328. doi: 10.1007/s00247-014-2944-x. Epub 2015 Mar 1. PMID: 25726014. [DOI] [PubMed] [Google Scholar]
- 15.Du L., Zhang Q.X., Tang J.X. Current situation of serum protein tumor biomarkers standardization. J Chin J Lab Med. 2019;42(1):10–13. doi: 10.3760/cma.j.issn.1009-9158.2019.01.003. [DOI] [Google Scholar]
- 16.Zhang K., Huo H., Lin G., Yue Y., Wang Q., Li J. A long way to go for the harmonization of four immunoassays for carcinoembryonic antigen. Clin. Chim. Acta. 2016 Feb 15;454:15–19. doi: 10.1016/j.cca.2015.12.029. Epub 2015 Dec 23. PMID: 26721316. [DOI] [PubMed] [Google Scholar]
- 17.Wauthier L., Plebani M., Favresse J. Interferences in immunoassays: review and practical algorithm. Clin. Chem. Lab. Med. 2022 Mar 18;60(6):808–820. doi: 10.1515/cclm-2021-1288. PMID: 35304841. [DOI] [PubMed] [Google Scholar]
- 18.Rousserie G., Grinevich R., Brazhnik K., Even-Desrumeaux K., Reveil B., Tabary T., Chames P., Baty D., Cohen J.H., Nabiev I., Sukhanova A. Detection of carcinoembryonic antigen using single-domain or full-size antibodies stained with quantum dot conjugates. Anal. Biochem. 2015 Jun 1;478:26–32. doi: 10.1016/j.ab.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 19.Kuroki M., Arakawa F., Haruno M., Murakami M., Wakisaka M., Higuchi H., Oikawa S., Nakazato H., Matsuoka Y. Biochemical characterization of 25 distinct carcinoembryonic antigen (CEA) epitopes recognized by 57 monoclonal antibodies and categorized into seven groups in terms of domain structure of the CEA molecule. Hybridoma. 1992 Aug;11(4):391–407. doi: 10.1089/hyb.1992.11.391. 10.1089/hyb.1992.11.391. PMID: 1383122. [DOI] [PubMed] [Google Scholar]
- 20.Wojtalewicz N., Vierbaum L., Kaufmann A., Schellenberg I., Holdenrieder S. Longitudinal evaluation of AFP and CEA external proficiency testing reveals need for method harmonization. Diagnostics. 2019;13(12) doi: 10.3390/diagnostics13122019. 2023 Jun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge M., Zhao H., Yan Y., Zhang T., Zeng J., Zhou W., Wang Y., Meng Q., Zhang C. Performance of electrolyte measurements assessed by a trueness verification program. Clin. Chem. Lab. Med. 2016 Aug 1;54(8):1319–1327. doi: 10.1515/cclm-2015-1110. PMID: 27010777. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Wang J., Zhao H., Zhang J., Zhang T., Zeng J., Zhou W., Zhang C. Assessment of enzyme measurement procedures in China through a trueness verification program. Clin. Chim. Acta. 2016 Oct 1;461:98–102. doi: 10.1016/j.cca.2016.07.008. Epub 2016 Jul 15. PMID: 27425848. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T.J., Pu Y.G., Zhou H.J., Ma R., Zhang J.T., Wang D.G., et al. Application of IFCC reference assay in the evaluation of five glycosylated hemoglobin detection assays. J Chin J Lab Med. 2018;41(11):821–826. doi: 10.3760/cma.j.issn.1009-9158.2018.11.007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.
Employment or leadership: Not applicable.