Abstract
We have analyzed DNA of Euglena gracilis for the presence of the unusual minor base β-d-glucosyl-hydroxymethyluracil or J, thus far only found in kinetoplastid flagellates and in Diplonema. Using antibodies specific for J and post-labeling of DNA digests followed by two-dimensional thin-layer chromatography of labeled nucleotides, we show that ~0.2 mole percent of Euglena DNA consists of J, an amount similar to that found in DNA of Trypanosoma brucei. By staining permeabilized Euglena cells with anti-J antibodies, we show that J is rather uniformly distributed in the Euglena nucleus, and does not co-localize to a substantial extent with (GGGTTA)n repeats, the putative telomeric repeats of Euglena. Hence, most of J in Euglena appears to be non-telomeric. Our results add to the existing evidence for a close phylogenetic relation between kinetoplastids and euglenids.
INTRODUCTION
β-d-glucosyl-hydroxymethyluracil or J is a DNA modification discovered in the DNA of the African trypanosome, Trypanosoma brucei, where it replaces 0.5–1.0% of all thymines (1,2). J has only been found in non-transcribed, and partially transcribed, repetitive sequences of T.brucei (3) and a substantial fraction of J is present in both strands of the telomeric hexamer (GGGTTA)n repeats of this organism (4). We have speculated that J may have an analogous function in trypanosomes as 5-methylcytosine (5-MeC) in plants and animals, and that it is involved in transcriptional repression and/or suppression of recombination between homeologous sequences (2,3,5).
To screen other organisms than T.brucei for the presence of J, we have generated antibodies that can detect J in DNA with high specificity (4). These antibodies can be used to detect J on DNA blots or by immunoprecipitation. The immunoprecipitated DNA can be analyzed by combined 32P-postlabeling and two-dimensional thin-layer chromatography (2D-TLC) experiments (6,7) to verify that J is present. Using these methods we have shown that J is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres (5). J was not detected in the animals, plants, or fungi tested, nor in a range of other simple eukaryotes, such as Plasmodium, Toxoplasma, Entamoeba, Trichomonas and Giardia (5). Outside the Kinetoplastida, J was only found in Diplonema, a small phagotrophic marine flagellate, in which we also identified 5-MeC. Fractionation of Diplonema DNA showed that the two modifications are present in a common genome compartment supporting the idea that they may have a similar function (5).
In our initial survey (5) we did not test Euglena DNA. Yet this is important for understanding the origin and distribution of J. On the basis of shared cytological traits, kinetoplastids and euglenids are traditionally placed in a common phylum, Euglenozoa (8,9). This common ancestry is supported by molecular phylogenetic analyses of nuclear-encoded genes such as ribosomal RNA (10,11), tubulin (12), glyceraldehyde-3-phosphate dehydrogenase (13), the ER-specific protein calreticulin (14), mitochondrial hsp60 (15) and the mitochondrial-encoded coxI gene (11,15,16). The major difference between the two groups is that the euglenids possess plastids whereas the kinetoplastids (trypanosomatids and bodonids) do not. However, this difference may not be fundamental, as Euglena appears to have acquired its plastids through engulfment of a eukaryotic unicellular green alga, a chlorophyte (17). Phylogenetic analyses of completely sequenced chloroplast genomes have lent overwhelming support to this view (18,19). Accordingly, Euglena can be viewed as a descendant of an ancient member of the non-photosynthetic kinetoplastid stem that engulfed a unicellular green alga and retained the plastid of its eukaryotic symbiont and its photosynthetic lifestyle.
We have now tested Euglena gracilis DNA and find that it contains J.
MATERIALS AND METHODS
Isolation of E.gracilis DNA
Cultures of E.gracilis strain SAG 1224-5/25 were grown as described (13) under a 14:10 light:dark regime aerated with 1.5% CO2. Ten grams of 4-day-old Euglena cells were harvested by centrifugation. These were ground in liquid nitrogen and gently suspended for 1 h in 100 ml of 100 mM NaCl, 50 mM EDTA, 50 mM Tris–HCl, 1% v/v sodium N-laurylsarkosin, 7 mM 2-mercaptoethanol, 100 mM sodium diethyldithiocarbaminate (Merck, Darmstadt, Germany), pH 7.5, followed by gentle phenol extraction. Nucleic acids from the aqueous phase were collected by ethanol precipitation and dissolved in 20 ml of 10 mM Tris–HCl, 1 mM EDTA, pH 8.0. RNA was precipitated by addition of LiCl to a final concentration of 2 M. DNA from the supernatant was collected by isopropanol precipitation, dissolved in 30 ml of 10 mM Tris–HCl, 1 mM EDTA, pH 8.0 containing 1 µg DNase-free RNase A (Sigma, St Louis, MO) per milliliter and purified by two rounds of CsCl centrifugation in a Ti 70.1 rotor. The collected DNA was diluted 10-fold with 10 mM Tris–HCl, 1 mM EDTA, pH 8.0, gently extracted with isoamyl alcohol, and precipitated twice with 0.6 vol of isopropanol. The final precipitate was dissolved in 10 mM Tris–HCl, 1 mM EDTA, pH 8.0.
Analysis of J
Quantitation of J with anti-J antibodies on DNA blotted on nitrocellulose was done as described (5), using the polyclonal rabbit antibody 538αJ (3). A pig horseradish peroxidase (HRP) conjugated anti-rabbit antibody (DAKO) was used to detect the rabbit antibody bound to J-DNA by enhanced chemiluminescence. The chemical analysis of J by post-labeling and 2D-TLC was done as described (7). Post-labeling of synthesized standards has shown that the labeling efficiency of J can be somewhat variable and is only ~50% (7).
In some experiments, sonicated DNA was immunoprecipitated with anti-J antibody and the precipitate used for chemical analysis by post-labeling and 2D-TLC, as described (5).
Immunofluorescense assays
Probe labeling, cell fixation, in situ hybridization and immunocytochemical detection were done essentially as described (5,20) with some adaptations: E.gracilis cells were subjected to a pepsin treatment of 10 min instead of 5 min. For Figure 2 streptavidin-Texas Red (Vector Laboratories, Burlingame, CA) was replaced by streptavidin-Alexa 594 (Molecular Probes Inc., Eugene, OR). For Figure 3, the GGGTTA repeat probe was labeled with digoxigenin-11-2′-deoxy-uridine-5′-triphosphate (DIG-11-dUTP) by nick translation instead of Biotin-16-dUTP. For the detection of the digoxigenin labeled probe, cells were incubated with mouse anti-digoxigenin (Roche, Mannheim, Germany) and as a second layer with goat anti-mouse-Alexa488 (Molecular Probes) also containing the dye TO-PRO-3 (Molecular Probes) to counterstain the nucleus. Cells were mounted in Vectashield (Vector Laboratories). Confocal fluorescence images were obtained on a Leica TCS NT (Leica Microsystems, Heidelberg, Germany) confocal system, equipped with an Ar/Kr laser. Images were taken using a 100× NA 1.4 objective. A standard filter combination for Alexa488/TO-PRO-3 and Kalman averaging was used. Processing of images for presentation was done on a PC using the software packages Photoshop (Adobe Systems Inc., Mountain View, CA) and Freelance Graphics (Lotus Development Corp., Cambridge, MA).
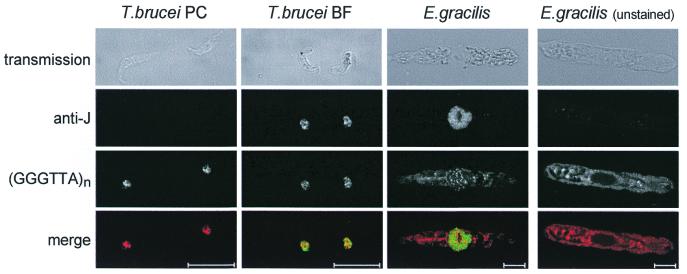
Figure 2.
Immunofluorescence assay of cells with anti-J antiserum combined with fluorescence in situ hybridisation with a telomeric repeat probe. The preparations were analysed with a Leica Confocal Laser Scanning Microscope. The scale bars represents 10 µm. From left to right: T.brucei procyclic form (PC), T.brucei bloodstream form (BF), E.gracilis, and unstained E.gracilis (as negative control for the cytoplasmic red autofluorescence). From top to bottom: transmission image, detection of J with anti-J antibodies, detection of (GGGTTA)n repeats with a (GGGTTA)n probe, and merge of the J and (GGGTTA)n images (green and red, respectively). Cells incubated with the pre-immune serum (or another rabbit antiserum) showed no staining in the case of T.brucei and only red autofluorescence in the case of E.gracilis (not shown).
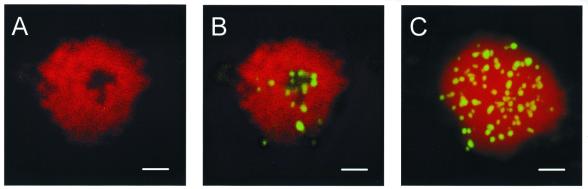
Figure 3.
Fluorescent images of a E.gracilis nucleus hybridized with the GGGTTA repeat probe (green) and counterstained with TO-PRO-3 (red). (A) CLSM section of a nucleus showing the understained region (the putative nucleolus). (B) Same section as (A) but showing hybridization with the GGGTTA probe around the putative nucleolus. (C) The superposition of all sections of a different Euglena nucleus, in which we count a total of 93 spots. Note that the edge of the nucleus counterstains weakly and that all spots are in the nucleus. The scale bars represent 2 µm. See Materials and Methods for details.
Sequence analysis of putative hexamer repeats from E.gracilis
For the single-stranded PCR (32 cycles at 52°C annealing temperature) we used 10 ng of genomic E.gracilis DNA, 50 pmol of primer (5′-CGGAATTCGGTTAGGGTTAGGGTTAG-3′; underlined is an EcoRI site with two extra nucleotides) and 1.5 U Taq DNA polymerase (Gibco BRL, Life Technologies, Inc., Rockville, MD). The product gave a smear on gel up to 600 bp and the 300–600 bp region of the gel was isolated and purified using the Qiaex II Gel Extraction Kit (Qiagen Inc., Valencia, CA).
Twenty-five and fifty nanograms of single-stranded PCR fragments were used for cycle-sequencing using 7.5 pmol of the 5′-CGGGATCCCTAACCCTAACCCTAACC-3′ primer (underlined is a BamHI site with two extra nucleotides). Cycle sequencing was done with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Foster City, CA) on a GeneAmp PCR System 9600 (PE Applied Biosystems). The extension products were purified by EtOH precipitation and run on an ABI Prism 377 sequenator.
RESULTS
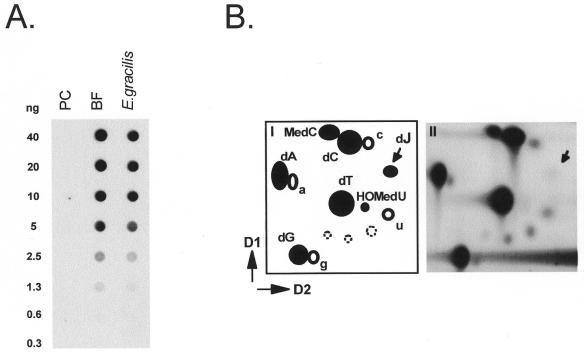
To test whether E.gracilis DNA contains J, we spotted a dilution series of E.gracilis DNA on nitrocellulose filters and analyzed the amounts of J with polyclonal antibodies against J. For comparison a dilution series of equal amounts of T.brucei bloodstream form (BF) DNA (contains J) and procyclic (PC) insect form DNA [contains no J (2)] was spotted. Figure 1A shows that the amount of J present in Euglena DNA is nearly as high as in T.brucei DNA. Two independent Euglena DNA preparations gave similar results, i.e. a value between 0.1 and 0.2 mole percent of J, compared to 0.2% in T.brucei DNA (7).
Figure 1.
Detection of J in E.gracilis. (A) Detection of J-containing DNA on filters with polyclonal anti-J antibodies. The dot-blot with serial dilutions of total genomic DNA was incubated with rabbit antiserum 538αJ and bound antibodies were detected as described in Materials and Methods. DNA loading was checked by ethidium bromide staining of the DNA samples. From left to right: procyclic (insect form) T.brucei (PC), bloodstream form T.brucei (BF) and E.gracilis DNA. PC trypanosomes do not contain J. (B) Analysis of E.gracilis DNA by 32P-nucleotide post-labeling combined with 2D-TLC (D1 and D2 are indicated), as described in Materials and Methods. The positions of the labeled 5′-deoxynucleosidemonophosphates are indicated by filled circles, 5′-ribonucleoside monophosphates are indicated by open circles, and background spots (open circles with dotted lines) are explained in panel I. Panel II: labeling of total DNA of E.gracilis. The position of J is indicated by an arrow.
To verify that J is a constituent of Euglena DNA we did 32P-postlabeling experiments. Panel II of Figure 1B shows that a weak J spot is indeed present on the 2D-TLC of post-labeled Euglena DNA. This spot was increased in intensity in DNA fragments immunoprecipitated with anti-J antibodies (not shown). In the total E.gracilis DNA sample (panel II, Fig. 1B) a spot can be observed next to dC. This spot is absent in T.brucei DNA (6,7,21) and migrates at the position of 5-methyl-dCMP. Whether the extra spot is really 5-methyl-dCMP remains to be verified.
To determine the intra-nuclear distribution of J in E.gracilis we used the anti-J antibodies in an immunofluorescence assay on fixed and permeabilized cells. For comparison, we analyzed DNA from T.brucei insect form (PC, no J) and BF (contains J) trypanosomes. Figure 2 shows that the nuclei of E.gracilis stain intensely with the anti-J antiserum with the exception of a roundish structure. This is probably the nucleolus, but we have not verified this. In T.brucei we have recently found, however, that J is absent from the ribosomal repeat DNA present in the nucleolus (unpublished results). There was no detectable staining in the cytoplasm of E.gracilis suggesting that the organelle DNAs, mitochondrial and chloroplast DNA, do not contain high amounts of J. This is in line with results obtained with T.brucei, as no J was detected in kinetoplast DNA, the mitochondrial DNA of trypanosomatids (6).
The intense nuclear staining in Euglena compared to T.brucei is not surprising, since Euglena diploid nuclei contain 3.0 pg DNA (22) and T.brucei nuclei only 0.1 pg (23). In T.brucei, a substantial fraction of J is in the telomeric (GGGTTA)n repeats (24) and this is also evident from the pictures in Figure 2 that show that part of the punctate J staining coincides with the punctate staining of the clustered telomeres of T.brucei. To test whether E.gracilis DNA contains arrays of GGGTTA that could represent the telomeric repeats, we sequenced single-stranded DNA generated with a (GGGTTA)3 primer (Materials and Methods). This yielded (TAACCC)n arrays with n up to 23. We assume that these arrays of GGGTTA repeats are present in the telomeric repeats of E.gracilis DNA. On the basis of this assumption, we hybridized the Euglena cells with a (GGGTTA)n probe labeled with biotin. The probe was detected with fluorescently-labeled streptavidin. Figure 2 shows the discrete punctate pattern obtained, which contrasts with the intense global staining of the nucleus obtained with anti-J antibodies. We conclude that only a small fraction of E.gracilis J is in telomeric repeats, contrary to the situation in Kinetoplastida.
The number of chromosomes in Euglena is controversial, estimates varying between four (25) and 45 (26). If the arrays of GGGTTA repeats that we detect by in situ hybridization indeed represent the telomeric repeats, the number of hybridizing spots would provide information on the number of chromosomes present in Euglena nuclei. Our initial attempts to quantitate spots by eye gave around 30 spots per nucleus. A more precise analysis by Confocal Laser Scanning Microscopy (CLSM) (Materials and Methods) gave a much higher number. For 32 nuclei we found 84 ± 9 (mean ± SD) spots with a range of 66–107. The understained roundish structure, the putative nucleolus, when visible usually did not contain spots, but often spots were arranged around it, resulting in rather striking halos (Fig. 3B).
DISCUSSION
Our results show that nuclear DNA of E.gracilis contains J. The amount is substantial, ~0.2% of total nucleotides. This is similar to the level found in T.brucei and higher than in most other kinetoplastids (5). The presence of J in Euglena is in agreement with molecular phylogeny, which places the kinetoplastids and euglenids in a common phylum (Introduction).
We find long arrays of GGGTTA repeats in Euglena and we assume that these are the telomeric repeats, as in Kinetoplastida. If all spots hybridizing with the GGGTTA probe represent telomeric repeat arrays, rather than chromosome-internal arrays, an average of 84 spots per nucleus suggests the presence of at least 42 linear chromosomes, very close to the 45 reported more than 40 years ago by Leedale (26,27) using light microscopy of mitotic cells. Why O’Donnell found only four haploid chromosomes by electron microscopy (25) is unclear.
As T.brucei has 30-fold less DNA per nucleus and about 125 chromosomes rather than 45, it is not surprising that only a small fraction of J immunofluorescence in Euglena nuclei coincides with telomeric repeats. In T.brucei, J is prominent in telomeric repeats (4), but also present in sub-telomeric repeats (3) and in intra-chromosomal repeats, such as the spliced leader repeats and 5S RNA encoding repeats (unpublished results). Hence, it seems likely that Euglena has much longer chromosomes than T.brucei and that most of the J is intra-chromosomal in this organism.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mike Cross, Herlinde Gerrits, Rainer Mußmann, Robert Sabatini, Sebastian Ulbert (this laboratory) and Fred van Leeuwen (Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Washington, DC) for critical reading of this manuscript. We thank Roeland Dirks (University of Leiden) for useful discussions and for providing us with the possibility to label the probes in his laboratory. We thank Lauran Oomen (The Netherlands Cancer Institute, Amsterdam) for his advice and technical assistance with the microscope. This work was supported by a grant form the Gulbenkian PhD Program in Biology and Medicine (Portugal) to I.C. and by The Netherlands Foundation for Chemical Research (SON), with financial aid from the Netherlands Organisation for Scientific Research (NWO), to P.B.
REFERENCES
- 1.Gommers-Ampt J.H., Van Leeuwen,F., De Beer,A.L.J., Vliegenthart,F.G., Dizdaroglu,M., Kowalak,J.A., Crain,P.F. and Borst,P. (1993) Cell, 75, 1129–1136. [DOI] [PubMed] [Google Scholar]
- 2.Borst P. and Van Leeuwen,F. (1997) Mol. Biochem. Parasitol., 90, 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Van Leeuwen F., Wijsman,E.R., Kieft,R., van der Marel,G.A., Van Boom,J.H. and Borst,P. (1997) Genes Dev., 11, 3232–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Leeuwen F., Wijsman,E.R., Kuyl-Yeheskiely,E., van der Marel,G.A., Van Boom,J.H. and Borst,P. (1996) Nucleic Acids Res., 24, 2476–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Leeuwen F., Taylor,M.C., Mondragon,A., Moreau,H., Gibson,W., Kieft,R. and Borst,P. (1998) Proc. Natl Acad. Sci. USA, 95, 2366–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gommers-Ampt J., Lutgerink,J. and Borst,P. (1991) Nucleic Acids Res., 19, 1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Leeuwen F., De Kort,M., van der Marel,G.A., Van Boom,J.H. and Borst,P. (1998) Anal. Biochem., 258, 223–229. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T. (1993) Microbiol. Rev., 57, 953–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corliss J. (1994) Acta Protozool., 33, 1–51. [Google Scholar]
- 10.Sogin M., Gunderson,J., Elwood,H., Alonso,R. and Peattie,D. (1989) Science, 243, 75–77. [DOI] [PubMed] [Google Scholar]
- 11.Maslov D.A., Yasuhira,S. and Simpson,L. (1999) Protist, 150, 33–42. [DOI] [PubMed] [Google Scholar]
- 12.Levasseur P.J., Meng,Q. and Bouck,B. (1994) J. Eukaryot. Microbiol., 41, 468–477. [DOI] [PubMed] [Google Scholar]
- 13.Henze K., Badr,A., Wettern,M., Cerff,R. and Martin,W. (1995) Proc. Natl Acad. Sci. USA, 92, 9122–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navazio L., Nardi,C., Baldan,B., Dainese,P., Fitchette,A.C., Martin,W. and Mariani,P. (1998) J. Eukaryot. Microbiol., 45, 307–313. [DOI] [PubMed] [Google Scholar]
- 15.Yasuhira S. and Simpson,L. (1997) J. Mol. Evol., 44, 341–347. [DOI] [PubMed] [Google Scholar]
- 16.Tessier L.H., Van der Speck,H., Gualberto,J.M. and Grienenberger,J.M. (1997) Curr. Genet., 31, 208–213. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs S.P. (1978) Can. J. Bot., 56, 2883–2889. [Google Scholar]
- 18.Martin W., Stoebe,B., Goremykin,V., Hansmann,S., Hasegawa,M. and Kowallik,K.V. (1998) Nature, 393, 162–165. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart P.J., Howe,C.J., Barbrook,A.C., Larkum,A.W.D. and Penny,D. (1999) Mol. Biol. Evol., 16, 573–576. [Google Scholar]
- 20.Chaves I., Zomerdijk,J., Dirks-Mulder,A., Dirks,R.W., Raap,A.K. and Borst,P. (1998) Proc. Natl Acad. Sci. USA, 95, 12328–12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Leeuwen F., Kieft,R., Cross,M. and Borst,P. (1998) Mol. Cell. Biol., 10, 5643–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawson J.R., Eckenrode,V.K., Boerma,C.L. and Curtis,S. (1979) Biochim. Biophys. Acta, 563, 1–16. [DOI] [PubMed] [Google Scholar]
- 23.Borst P., van der Ploeg,M., van Hoek,J.F., Tas,J. and James,J. (1982) Mol. Biochem. Parasitol., 6, 13–23. [DOI] [PubMed] [Google Scholar]
- 24.Van Leeuwen F., Dirks-Mulder,A., Dirks,R.W., Borst,P. and Gibson,W. (1998) Mol. Biochem. Parasitol., 94, 127–130. [DOI] [PubMed] [Google Scholar]
- 25.O’Donnell E.H.J. (1965) Cytologia, 30, 118–154. [Google Scholar]
- 26.Leedale G.F. (1958) Nature, 181, 502–503.13517196 [Google Scholar]
- 27.Leedale G.F. (1958) Arch. Mikrobiol., 32, 32–64. [DOI] [PubMed] [Google Scholar]