Abstract
microbeMASST, a taxonomically informed mass spectrometry (MS) search tool, tackles limited microbial metabolite annotation in untargeted metabolomics experiments. Leveraging a curated database of >60,000 microbial monocultures, users can search known and unknown MS/MS spectra and link them to their respective microbial producers via MS/MS fragmentation patterns. Identification of microbe-derived metabolites and relative producers without a priori knowledge will vastly enhance the understanding of microorganisms’ role in ecology and human health.
Subject terms: Microbiology, Computational biology and bioinformatics, Metabolomics
microbeMASST is a tool to associate known and unknown metabolites to microbial producers leveraging untargeted metabolomics data.
Main
Microorganisms drive the global carbon cycle1 and can establish symbiotic relationships with host organisms, influencing their health, aging and behaviour2–6. Microbial populations interact with different ecosystems through the alteration of available metabolite pools and the production of specialized small molecules7,8. The vast genetic potential of these communities is exemplified by human-associated microorganisms, which encode ~100 times more genes than the human genome9,10. However, this metabolic potential remains unreflected in modern untargeted metabolomics experiments, where typically <1% of the annotated molecules can be classified as microbial. This problem particularly affects mass spectrometry (MS)-based untargeted metabolomics, a common technique to investigate molecules produced or modified by microorganisms11, which famously struggles with spectral annotation of complex biological samples. This is because most spectral reference libraries are biased towards commercially available or otherwise accessible standards of primary metabolites, drugs or industrial chemicals. Even when metabolites are annotated, extensive literature searches are required to understand whether these molecules have microbial origins and to identify the respective microbial producers. Public databases, such as KEGG12, MiMeDB13, NPAtlas14 and LOTUS15, can assist in this interpretation, but they are mostly limited to well-established, largely genome-inferred metabolic models or to fully characterized and published molecular structures. In addition, while targeted metabolomics efforts aimed at interrogating the gut microbiome mechanistically have been developed16, these focus only on relatively few commercially available microbial molecules. Hence, the majority of the microbial chemical space remains unknown despite the continuous expansion of MS reference libraries. To fill this gap, we have developed microbeMASST (https://masst.gnps2.org/microbemasst/), a search tool that leverages public MS repository data to identify the microbial origin of known and unknown metabolites and map them to their microbial producers.
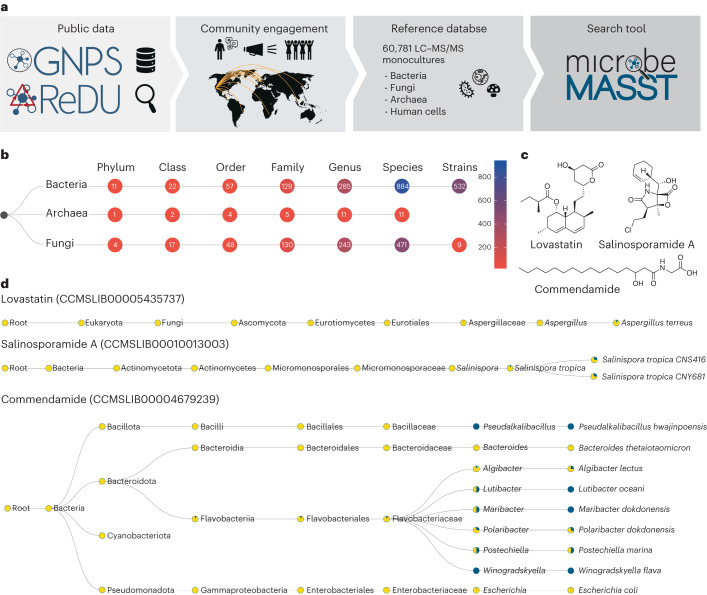
microbeMASST is a community-sourced tool that works within the GNPS ecosystem17. Users can search tandem MS (MS/MS) spectra obtained from their experiments against the GNPS/MassIVE repository and retrieve matching samples exclusively acquired from extracts of bacterial, fungal or archaeal monocultures. No other available resource or tool allows linking uncharacterized MS/MS spectra to characterized microorganisms. The microbeMASST reference database of microbial monocultures has been generated through years of community contributions and metadata curation, and it contains microorganisms isolated from plants, soils, oceans, lakes, fish, terrestrial animals and humans (Fig. 1a). All available microorganisms have been categorized according to the NCBI taxonomy18 at different taxonomic resolutions (that is, species, genus, family and so on) or mapped to the closest taxonomically accurate level, if no NCBI ID was available at the time of database creation. As of September 2023, microbeMASST includes 60,781 liquid chromatography (LC)–MS/MS files comprising >100 million MS/MS spectra mapped to 541 strains, 1,336 species, 539 genera, 264 families, 109 orders, 41 classes and 16 phyla from the three domains of life: Bacteria, Archaea and Eukaryota (Fig. 1b). Different from MASST19, which uses a precomputed network of ~110 million MS/MS spectra to enable spectral searching, microbeMASST is based on the recently introduced Fast Search Tool (https://fasst.gnps2.org/fastsearch/)20. This tool, originally designed for proteomics, drastically improves search speed by several orders of magnitude by indexing all the MS/MS spectra present in GNPS/MassIVE and restricting the search space to the user input parameters. Because of this, search results are returned within seconds as opposed to 20 min per search or 24–48 h for modification tolerant searches in the original implementation of MASST. In addition, microbeMASST leverages pre-curated file-associated metadata to aggregate results into easy-to-interpret taxonomic trees. This represents a major enhancement over MASST, where users have to manually inspect results tables and contextualize them, making interpretations tedious. Finally, users can leverage microbeMASST Python code to perform batch searches of thousands of MS/MS spectra by providing either a formatted MS/MS file (.mgf) or a list of Universal Spectrum Identifiers (USIs)21, which represent paths to spectra in public datasets22. This is particularly useful for creating integrated data analysis pipelines using the standard outputs (.mgf) of already established data processing tools, such as MZmine23.
Fig. 1. The microbeMASST search tool and reference database.
a, Community contributions of data and knowledge to GNPS17, ReDU57 and MassIVE from 2014 to 2022 were used to generate the microbeMASST reference database. In addition, a public invitation to deposit data in June 2022 resulted in the further deposition of LC–MS/MS files from 25 different laboratories from 15 different countries across the globe, leading to the curation of a total of 60,781 LC–MS/MS files of microbial monoculture extracts. b, microbeMASST comprises 1,858 unique lineages across three different domains of life mapped to 541 unique strains, 1,336 species, 539 genera, 264 families, 109 orders, 41 classes and 16 phyla. c, Examples of medically relevant small molecules known to be produced by bacteria or fungi. Lovastatin, a cholesterol-lowering drug originally isolated from Aspergillus genus25; salinosporamide A, a Phase III candidate to treat glioblastoma produced by Salinispora tropica27; and commendamide, a human G-protein-coupled receptor agonist28. d, microbeMASST search outputs of the three different molecules of interest confirm that they were exclusively found in monocultures of the only known producers. Pie charts display the proportion of MS/MS matches found in the deposited reference database. Blue indicates a match with a monoculture, while yellow represents a non-match. Searches were performed using MS/MS spectra deposited in the GNPS reference library: lovastatin (CCMSLIB00005435737), salinosporamide A (CCMSLIB00010013003) and commendamide (CCMSLIB00004679239). GNPS, ReDU and microbeMASST logos reproduced under a Creative Commons license CC BY 4.0.
In the microbeMASST web app (https://masst.gnps2.org/microbemasst/), users can search single MS/MS spectra and obtain matching results from the reference database of microbeMASST, providing either a USI or a precursor ion mass and its spectral fragmentation pattern (Supplementary Fig. 1). Analogue search can also be enabled to discover molecules related to the MS/MS spectrum of interest across the taxonomic tree17,19,24. The microbeMASST web app displays query results in interactive taxonomic trees, which can be downloaded as HTML files. Nodes in the trees represent specific taxa and display rich information, such as taxon scientific name, NCBI taxonomic ID, number of deposited sample data files, number of sample data files containing a match to the queried spectrum, within the user search criteria, and a proportion of the number of sample data files matching the queried spectrum to the number of total available sample data files for that specific taxon in the reference database of microbeMASST. This proportion is also visualized through pie charts. Information for an MS/MS match in a particular taxon is propagated upstream through its lineage. The reactive interface of microbeMASST enables filtering of the tree to specific taxonomic levels or to a minimum number of matches observed per taxon. In addition, three data tables are generated, linking the search job to other resources in the GNPS/MassIVE ecosystem. For example, each MS/MS query is also searched against the public MS/MS reference library of GNPS (587,213 MS/MS spectra, September 2023) to provide spectra annotations when available. The annotations to reference compounds are listed under the ‘Library matches’ tab (Supplementary Fig. 2a). The ‘Datasets matches’ tab contains information on the matching scans, displaying scientific name, NCBI taxonomic ID and taxonomic rank, number of matching fragment ions and modified cosine score together with a link to a mirror plot visualization (Supplementary Fig. 2b). Finally, the ‘Taxa matches’ tab informs on how many matches were found per taxon and the number of samples available for that taxon (Supplementary Fig. 2c). Quality controls (QCs) and blank samples (n = 2,902) present in the reference datasets of microbeMASST have been retained to provide information on possible contaminants and media components. In addition, data from human cell line cultures (n = 1,199) have been included to enable assessment of whether molecules can be produced by both human hosts and microorganisms. It is important to point out that microbeMASST allows linking of both partly annotated, through MS/MS match to reference library spectra, and fully uncharacterized spectra to possible microbial producers but that technical limitations inherent to mass spectrometry or the experiment itself are present. For example, the absence of a matching spectrum in a specific taxon does not necessarily indicate that it is not capable of producing the searched molecule but rather that the methodology used to acquire the data did not allow its detection. These and other limitations are described in Methods. Despite these limitations, microbeMASST can uniquely enable the discovery of links between uncharacterized MS/MS spectra and defined microorganisms, providing valuable information for future mechanistic studies.
Search results for lovastatin, salinosporamide A and commendamide MS/MS spectra highlight how microbeMASST can correctly connect microbial molecules to their known producers (Fig. 1c). In the case of lovastatin, a clinically used cholesterol-lowering drug originally isolated from Aspergillus terreus25, spectral matches were unique to the genus Aspergillus (Fig. 1d). The MS/MS spectrum for salinosporamide A, a Phase III candidate to treat glioblastoma26, only matched two strains of Salinispora tropica (Fig. 1d), the only known producer27. Commendamide, first observed in cultures of Bacteroides vulgatus (recently reclassified as Phocaeicola vulgatus), is a G-protein-coupled receptor agonist28. Surprisingly it had many matches to several bacterial cultures, including in Flavobacteriaceae (Algibacter, Lutibacter, Maribacter, Polaribacter, Postechiella and Winogradskyella) and Bacteroides cultures (Fig. 1d). Additional examples include searches of mevastatin, arylomycin A4, yersiniabactin, promicroferrioxamine, and the microbial bile acid conjugates29–31 glutamate-cholic acid (Glu-CA) and glutamate-deoxycholic acid (Glu-DCA) (Supplementary Fig. 3). Mevastatin, another cholesterol-lowering drug originally isolated from Penicillium citrinum32, was only found in samples classified as fungi. The antibiotic arylomycin A4 was observed in different Streptomyces species, and it was originally isolated from Streptomyces sp. Tue 6075 in 200233. Yersiniabactin, a siderophore originally isolated from Yersinia pestis34 whose monoculture is not yet present in the reference database of microbeMASST, was observed in Escherichia coli and Klebsiella species, consistent with previous observations35,36. Promicroferrioxamine, another siderophore, was observed to match Micromonospora chokoriensis and Streptomyces species. This molecule was originally isolated from an uncharacterized Promicromonosporaceae isolate37. The MS/MS spectrum of the gut microbiota-derived Glu-CA, an amidated tri-hydroxylated bile acid, was most frequently observed in cultures of Bifidobacterium species, while Glu-DCA was found only in one Bifidobacterium strain but also in two Enterococcus and Clostridium species. None of the molecules were found in cultured human cell lines, highlighting the ability of microbeMASST to distinguish MS/MS spectra of molecules that can be exclusively produced by either bacteria or fungi. It is important to acknowledge that MS/MS data generally do not differentiate stereoisomers, but it can nevertheless provide crucial information on molecular families.
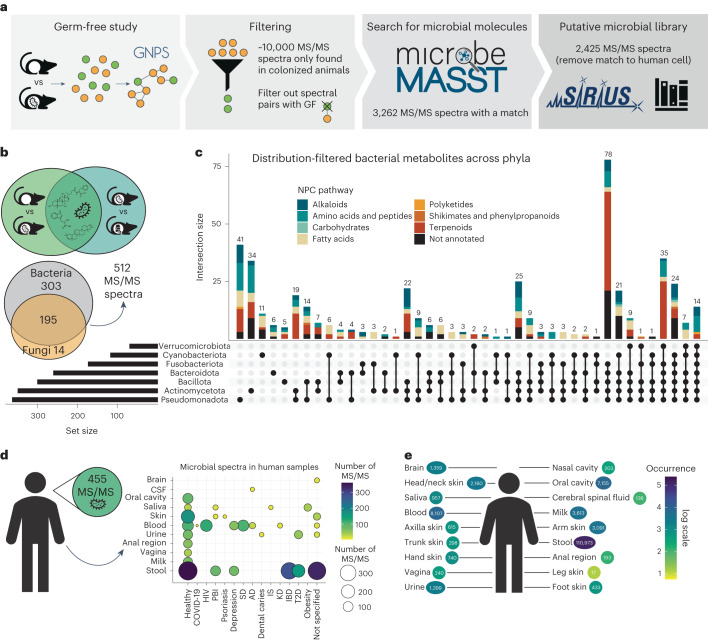
microbeMASST can be also used to extract microbial information from mass spectrometry-based metabolomics studies without any a priori knowledge. To illustrate this, we reprocessed an untargeted metabolomics study with data acquired from 29 different organs and biofluids comprising tissues including brain, heart, liver, blood and stool of germ-free (GF) mice and mice harbouring microbial communities, also known as specific pathogen-free (SPF) mice30 (Fig. 2a). We extracted 10,047 consensus MS/MS spectra uniquely present in SPF mice and queried them with microbeMASST. A total of 3,262 MS/MS spectra were found to have a microbial match to the microbeMASST reference database. Of these, 837 were also found in human cell lines and for this reason were removed from further analysis. Among the remaining 2,425 MS/MS spectra, 1,673 were exclusively found in bacteria, 95 in fungi and 657 in both (Supplementary Fig. 4). These MS/MS spectra were then processed with SIRIUS38 and CANOPUS39 to tentatively annotate the metabolites and identify their chemical classes. A file containing all these spectra of interest can be explored and downloaded in .mgf format from GNPS (see Methods). To further validate the microbial origin of these MS/MS spectra, we assessed their overlap with data acquired from a different study comparing SPF mice treated with a cocktail of antibiotics to untreated controls40. Interestingly, 621 MS/MS spectra were also found in this second dataset and 512 were only present in animals not treated with antibiotics (Fig. 2b). The distribution of these spectra and their putative classes across bacterial phyla was visualized using an UpSet plot41 (Fig. 2c). Notably, most of the spectra classified as terpenoids were commonly observed across phyla, while amino acids and peptides appeared to be more phylum specific. Of these 512 spectra, 23% had a level 2 putative annotation according to the 2007 Metabolomics Standards Initiative42, matching the GNPS reference library (Supplementary Table 1). A level 2 annotation within the user-specified search criteria might result in MS/MS matches between molecules belonging to related families as opposed to unique molecules. Annotations included the recently described amidated microbial bile acids19,29–31,43–48, free bile acids originating from the hydrolysis of host-derived taurine bile acid conjugates49, keto bile acids formed via microbial oxidation of alcohols30, N-acyl-lipids belonging to a similar class of metabolites as commendamide28 (a microbial N-acyl lipid), di- and tri- peptides seen in microbial digestion of proteins50, and soyasapogenol, a by-product of the microbial digestion of complex saccharides from dietary soyasaponins30. Part of the remaining unannotated spectra can be identified as chemical modifications of the above annotated microbial metabolites through spectral similarity obtained from molecular networking (Supplementary Fig. 5). This list of annotated MS/MS spectra included metabolites that are not yet widely considered to be of microbial origins, such as the di- and tri-hydroxylated bile acids and the glycine-conjugated bile acids43. One interpretation of these findings is that microorganisms are capable of producing metabolites previously described to be only of mammalian origins. Notable examples of metabolites that have been established to be produced by both the mammalian host and bacteria include serotonin51, γ-aminobutyric acid (GABA)52 and most recently, glycocholic acid43,53–55. In addition, an alternative hypothesis is that microorganisms can also selectively stimulate the production of host metabolites. Other limitations regarding annotations are discussed in Methods. To assess whether the observations from the mouse models translate to humans, we searched and found that 455 out of the 512 MS/MS spectra of interest matched public human data (Fig. 2d). Interestingly, these spectra were found in both healthy individuals and individuals affected by different diseases, including type II diabetes, inflammatory bowel disease, Alzheimer’s disease and other conditions. These spectra were most commonly found in stool samples (n = 110,973 MS/MS matches), followed by blood, breast milk and the oral cavity, as well as other organs including the brain, skin, vagina and biofluids (for example, cerebrospinal fluid and urine) (Fig. 2e). These findings support the concept that a substantial number of microbial metabolites reach and influence distant organs in the human body56.
Fig. 2. microbeMASST can identify microbial MS/MS spectra within mouse and human datasets.
a, Workflow to extract microbial MS/MS spectra from biochemical profiles of 29 different tissues and biofluids of SPF mice that are not observed in GF mice30. The MS/MS spectra unique to SPF mice (10,047) were searched with microbeMASST. A total of 3,262 MS/MS spectra had a match; those MS/MS also matching human cell lines were removed, leaving a total of 2,425 putative microbial MS/MS spectra (see Methods to download .mgf file). b, The presence of the 2,425 MS/MS spectra was evaluated in an additional animal study looking at antibiotic usage40. A total of 512 MS/MS spectra, out of the 621 overlapping, were exclusively found in animals not receiving antibiotics. c, UpSet plot of the distribution of the detected MS/MS spectra (512) across bacterial phyla. Terpenoids were more commonly observed across phyla, while amino acids and peptides appeared to be more phylum specific. d, The 512 MS/MS spectra were searched in human datasets and 455 were found to have a match. These MS/MS spectra were present in both healthy individuals and individuals affected by different diseases. e, Most of the MS/MS spectra (n = 411) matched faecal samples (n = 110,973 matches), followed by blood, oral cavity, breast milk, urine and several other organs. CSF, cerebral spinal fluid; COVID-19, coronavirus disease 2019; HIV, human immunodeficiency virus; PBI, primary bacterial infectious disease; SD, sleep disorder; AD, Alzheimer’s disease; IS, ischaemic stroke; KD, Kawasaki disease; IBD, inflammatory bowel disease; T2D, type II diabetes. GNPS and and microbeMASST logos reproduced under a Creative Commons license CC BY 4.0; SIRIUS logo reproduced under a Creative Commons license CC BY 4.0-ND.
We anticipate that microbeMASST will be a key resource to enhance understanding of the role of microbial metabolites across a wide range of ecosystems, including oceans, plants, soils, insects, animals and humans. This expanding resource will enable the scientific community to gain valuable taxonomic and functional insights into diverse microbial populations. The mass spectrometry community will play a key role in the evolution of this tool in the future through the continued deposition of data associated with microbial monocultures and the expansion of spectral reference libraries. Moreover, microbeMASST holds potential for various applications ranging from aquaculture and agriculture to biotechnology and the study of microbe-mediated human health conditions. By harnessing the power of public data, we can unlock opportunities for advancements in multiple fields and deepen our understanding of the intricate relationships between microorganisms and their ecosystems.
Methods
Data collection and harmonization
Data deposited in GNPS/MassIVE were investigated manually and systematically using ReDU57 (https://redu.ucsd.edu/) to extract all publicly available MS/MS files (.mzML or .mzXML formats) acquired from monocultures of bacteria, fungi, archaea and human cell lines. Only monocultures were included in the reference database of this search tool to unequivocally associate the production of the detected metabolites to each specific taxon. A total of 60,781 files from 537 different GNPS/MassIVE datasets were selected to be used as the reference database of microbeMASST (Supplementary Table 2). These include files deposited in response to our call to the scientific community. Between May and July 2022, 25 different research groups deposited 65 distinct datasets in GNPS/MassIVE, comprising a total of 3,142 unique LC–MS/MS files. This represented a 5.45% increase in publicly available MS/MS data acquired from monocultures in just 2 months. To qualify as a contributor and be credited as one of the authors, researchers had to deposit high-resolution LC–MS/MS data acquired either in positive or negative ionization modes from monocultures of either bacteria, fungi or achaea. Harmonization of the acquired data and metadata represented a challenge. The NCBI taxonomic database is constantly expanding and evolving, and the ReDU latest update (December 2021) does not accommodate the latest deposited taxa. For this reason, an additional metadata file (microbeMASST_metadata_massiveID) was generated specifically for the microbeMASST project and uploaded to the respective GNPS/MassIVE datasets deposited by the collaborators if the ReDU workflow failed. All the collected information was finally aggregated in a single .csv file (microbe_masst_table.csv) that can be found on GitHub, which contains: (1) full MassIVE path of each sample, (2) file name of each sample reported as its MassIVE ID/file name to avoid the presence of duplicated names, (3) MassIVE ID, (4) taxonomic name of the isolate as reported by the author submitting the associated metadata, (5) alternative taxonomic name if the provided taxonomic name was incorrect or not present in NCBI, (6) associated NCBI ID to the taxonomic name or the alternative taxonomic name, when present, (7) definition if the taxonomic ID was automatically assigned or manually curated, and information if (8) ReDU metadata are available for that specific file and if the file correspond to a (9) blank or (10) QC rather than a unique biological sample.
Unique taxonomic names and NCBI IDs were extracted from the metadata associated with the selected samples. When metadata were not available and multiple species of bacteria or fungi were present in the same dataset, samples were generically classified as bacteria or fungi. Concordance between taxonomic names and NCBI IDs was checked by blasting taxonomic names against NCBI (https://www.ncbi.nlm.nih.gov/Taxonomy/TaxIdentifier/tax_identifier.cgi) to obtain respective NCBI IDs and updated taxonomic names. Results were manually investigated and missing IDs were recovered using the NCBI browser (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi). If the taxonomic name was not found in NCBI, most probably because it was not deposited yet, the NCBI of the closest taxon was retrieved and used. For example, the strain Staphylococcus aureus CM05 was unavailable in NCBI and was curated to the species Staphylococcus aureus instead.
Taxonomic tree generation
The microbeMASST taxonomic tree was generated using both R 4.2.2 and Python 3.10. In R, the microbeMASST table was filtered and only unique NCBI IDs were retained (n = 1,834). The classification function of the ‘taxize’ package (v.0.9.100) was used to retrieve the full lineage of each NCBI ID58. Main taxonomic ranks (kingdom to strain) plus subgenus, subspecies and varieties were kept to obtain taxonomic trees with a similar number of nodes per lineage. The list of NCBI IDs of all lineages was then imported to Python, where the ETE3 toolkit was used to generate a taxonomic tree on the basis of the provided NCBI IDs59. The generated Newick tree was then converted into JSON format and information such as taxonomic rank and number of available samples per taxon was added. In addition, children nodes for blanks and QCs were created to be visualized in the same tree.
MASST query
The microbeMASST web application was built using Dash and Flask open-source libraries for Python (https://github.com/mwang87/GNPS_MASST/blob/master/dash_microbemasst.py). The web app can receive as inputs either a USI or an MS/MS spectrum (fragment ions and their intensities). In addition, batch searches can be performed using a customizable Python script that can read either a .tsv file containing a list of USIs or a single .mgf file (https://github.com/robinschmid/microbe_masst). Through the manuscript, we showcase how we were able to search for more than 10,000 MS/MS spectra contained in a single .mgf file (~2 h run time). After receiving input information, microbeMASST leverages the Fast Search Tool (https://fasst.gnps2.org/fastsearch/) API and the sample-specific associated metadata to generate taxonomic trees. Fast searches are based on indexing all the MS/MS spectra present in GNPS/MassIVE according to the mass and intensity of their precursor ions and then restricting the search to only a set of relevant spectra that have a greater chance of achieving a high spectral similarity (modified cosine score) to the MS/MS of interest. Searches are customizable and default settings are the following: precursor and fragment ion mass tolerances, 0.05; minimum cosine score threshold, 0.7; minimum number of matching fragment ions, 3; and analogue search, False. Users can modify these parameters on the basis of their data and research questions. Once matches are obtained, it is good practice to inspect the associated mirror plots for confirmation. To create the final taxonomic tree, the JSON file of the complete microbeMASST taxonomic tree is filtered according to the results and converted into a D3 JavaScript object that can be visualized as an HTML file.
Applications
We envision microbeMASST to have several applications. First, we showcase how researchers can investigate single MS/MS spectra using the web interface and obtain matching results if their known or unknown MS/MS spectrum was previously observed in any of the microbial monocultures present in the microbeMASST database. Nine small molecules of interest were investigated using MS/MS spectra already deposited in the GNPS reference library (see ‘Data availability’ and ‘Code availability’). Second, we show how microbeMASST can be leveraged to mine for known or unknown microbial metabolites in MS studies. To test this hypothesis, we reprocessed an LC–MS/MS dataset acquired from 29 different organs and biofluids of GF and SPF mice30. A comprehensive molecular network was generated (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=893fd89b52dc4c07a292485404f97723). From the obtained job, the qiime2 artefact (qiime2_table.qza), the .mgf file (METABOLOMICS-SNETS-V2-893fd89b-download_clustered_spectra-main.mgf) containing all the captured MS/MS spectra, and the annotation table (METABOLOMICS-SNETS-V2-893fd89b-view_all_annotations_DB-main.tsv) were extracted. The .qza file was first converted into a .biom file and then a .tsv file using QIIME2 (ref. 60) to extract the feature table. This was then imported to R where only spectra present in tissues and biofluids of SPF animals were retained (n = 10,047). To add an extra layer of filtering, all MS/MS spectra that had an edge (cosine similarity >0.7) and a delta parent ion mass of ±0.02 Da with MS/MS spectra present in GF animals were removed. Spectral pairs information was contained in a networkedges_selfloop file. All the MS/MS spectra were then run in batch using a custom Python script of microbeMASST (processing time: ~2 h, Apple M2 Max, 64 GB RAM) to obtain microbial matches. Matching and filtered MS/MS spectra (n = 2,425) were aggregated into a single .mgf file that can be downloaded from GNPS (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=aecd30b9febd43bd8f57b88598a05553). The compound class of each MS/MS spectrum with parent ion mass <850 Da was predicted using SIRIUS38 and CANOPUS39. The 2,425 MS/MS spectra were then searched against the MSV000080918 dataset containing mice treated or not with antibiotics40. Matching and filtered MS/MS spectra (n = 512) were aggregated into a single .mgf file that can be downloaded from GNPS (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c33855fc32c948049331e9730189d5c1). A list of the spectra with putative chemical class classification is available in Supplementary Table 1. Venn diagrams and UpSet plots were generated in R using VennDiagram 1.7.3, UpSetR 1.4.0 and ComplexUpset 1.3.3. Finally, the 512 MS/MS spectra were searched in batch against the GNPS public repository to observe whether they were detected in human datasets (Supplementary Table 3). ReDU metadata information associated with the human datasets was then used to observe the distribution of the MS/MS spectra across different diseases and body parts.
Technical limitations
Analysis of the results should be considered with the following limitations in mind. Molecule detection in microbeMASST is dependent on the availability of specific substrates in the reference monocultures. If the culture lacks the necessary substrates (or any other culture condition) to produce a certain molecule, this molecule will not be detected. Nevertheless, if related substrates are present, then a different but related molecule may be produced instead, which can be detected using the analogue search function. To address this problem, it is crucial for the community to continue to gather data from as many diverse experimental conditions as possible to capture the full range of metabolites produced by different microorganisms. This will help in building the most comprehensive reference database that encompasses diverse microbial metabolic profiles. In addition, isomers and stereoisomers, which are molecules with the same molecular formula but different structural arrangements, often exhibit similar MS/MS spectra. This means that their fragmentation patterns may not provide enough information to distinguish them. Finally, differences in extraction conditions and instrument settings can lead to variations in the obtained MS/MS spectra. For example, the intensity of precursor ions used for fragmentation can impact the resulting spectra. If the precursor ion intensity is low, the fragmented spectrum may lack ions that are present in spectra obtained from high-intensity precursor ions. This may result in ‘data leakage’ as the MS/MS spectrum may be missing ions, leading to the two molecules not being recognized as the same molecule. To partially overcome this, more permissive settings can be created. The majority of the data deposited in public repositories, GNPS included, and used in microbeMASST were acquired using positive electrospray ionization mode, which limits the observation of molecules that cannot be ionized in positive mode. This means that certain metabolites may be underrepresented or not detected at all. The continuous curation of the microbeMASST reference database involves adding more diverse data in terms of ionization modes to improve the coverage of metabolites. The taxonomic tree was generated using associated NCBI IDs provided by the community. Specimen assignment before metabolomic analysis cannot be checked by microbeMASST. The majority of the deposited data do not have associated genetic information and even if available, it was not used to build the taxonomic tree. Thus, specimen misidentification cannot be excluded. By addressing these challenges and continuously curating the reference database with more comprehensive and diverse data, microbeMASST coverage can be expanded to provide valuable insights into the role of microbiota and to facilitate our understanding of microbial metabolism in diverse ecosystems.
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Figs. 1–5.
Supplementary Tables 1–3.
Acknowledgements
This work was carried out through the collaborative microbial metabolite centre, which is supported by the National Institutes of Health (NIH) grant U24DK133658, the Alzheimer’s gut project U19AG063744 and BBSRC-NSF award 2152526. We thank T. Adkins and L. McCormick from the USDA ARS Culture Collection for assistance in selecting and providing microbial strains used in this research. This project was supported in part by the US Department of Agriculture, Agricultural Research Service. A.M.C.-R. and H.M. were supported by NIH grant 1DP2GM137413. Research reported in this publication was supported in part by the National Center for Complementary and Integrative Health of the NIH under award number F32AT011475 to N.E.A. K.B.K. was supported by National Research Foundation of Korea (NRF) grants funded by the Korean Government (MSIT) (NRF-2020R1C1C1004046, 2022R1A5A2021216 and 2022M3H9A2096191). B.S. was supported by the Austrian Science Fund (FWF) P31915 and K.H. was financed by the FWF research group programme (grant FG3). D.P. was supported by the German Research Foundation (DFG) CMFI Cluster of Excellence (EXC 2124) and Collaborative Research Center CellMap (TRR 261). A.B., E.A.M., P.C.J., L.V.C.-L. and N.P.L. were supported by The São Paulo Research Foundation (FAPESP #2018/24865-4, #2019/03008-9, #2020/06430-0, #2022/12654-4, #2015/17177-6, #2020/02207-5, #2021/10603-0) and CNPq. C.L.-C. received financial support from StrainBiotech and the FEMSA Biotechnology Center from Tecnológico de Monterrey. L.R.-O. received a scholarship from the Mexican National Council of Science and Technology (CONACYT). W.H.G. was supported by NIH R01 GM107550. N.G. was supported by the NSF CAREER Award #2047235. N.J.B., S.D. and J.S.S.D. were supported by ERC Horizon 2020 (grant agreement no. 694569) and by a Spinoza award (to J.S.S.D. from NWO). B.C.W. was supported by CMFI Cluster of Excellence (EXC 2124). M.B. was supported by DZIF (Grant no. TTU 09.722). A.V.P.d.L., S.L.L.R. and P.B.P. were supported by ERA-Net Cofund project BlueBio (grant agreement no. 311913), Research Council of Norway (300846). H.M.R., M.F.L. and G.L.B. were supported by Novo Nordisk Foundation (grant NNF19OC0056246; PRIMA—toward Personalized dietary Recommendations based on the Interaction between diet, Microbiome and Abiotic conditions in the gut). H.M.R. was supported by the Independent Research Fund Denmark (MOTILITY; grant no. 0171-00006B). E.C.O.N. was supported by a Nottingham Research Fellowship. A.I.P.-L. was supported by FPU (FPU19/00289). M.F.T. and R.d.C.P. were supported by R35GM12889. C.M.-S. was supported by Juan de la Cierva-Incorporación (IJC2018-036923-I) and Proyectos dirigidos por jóvenes investigadores de la Universidad de Málaga (B1-2021_21). D.R. was supported by Plan Nacional de I+D+i of the Ministerio de Ciencia e Innovación (PID2019-107724GB-I00) and Junta de Andalucía (P20_00479). X.L.G. was supported by a Nanyang Assistant Professorship. K.-S.J. was supported by NIH R01 GM137135. A.S. was supported by EMBO fellowship ALTF 996-2021. S.P. was supported by ETH Zurich Career Seed Fellowship. R.G. was supported by the Simons Foundation Postdoctoral Fellowship in Marine Microbial Ecology. H.C. was supported by NIH R01AI167860 and CIFAR. M.C.T. was supported by T32 DK007202 (NIDDK), the National Academies of Sciences, Engineering and Medicine through the Predoctoral Fellowship of the Ford Foundation, and the Howard Hughes Medical Institute (HHMI) Graduate Fellowships grant GT15123. E.E.-L. was supported by VI-Universidad de Costa Rica, Grants numbers C1604 and C0469. P.C. was supported by VI-Universidad de Costa Rica, Grants numbers C1604 and C0469; US National Science Foundation DEB-1638976. D.A.-V. and G.T.-C. were supported by VI-Universidad de Costa Rica, Grants numbers C1604 and C0469. K.F. was supported by CMFI Cluster of Excellence (EXC 2124). H.H.F.K. and G.F.d.S. were supported by Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM). D.B.S. was supported by Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul - FUNDECT (process number: 71/032.390/2022, FUNDECT number: 311/2022). K.L.M. was supported by NIH/1R01GM132649. P.-M.A. was supported by a swissuniversities Open Research Data grant. M.C.M.K. and S.L.J. were supported by NIH R35GM142938. A.D.P. was supported by NIH U01 DK119702 and S10 OD021750. A.T.A. was supported by the Betty and Gordon Moore Foundation. M.L. and P.B. were supported by the Max Planck Society. Omnia Group Ltd. is duly acknowledged for microbial cultures. B.S.P. and N.B. were partially supported by NIH 1R01LM013115 and NSF ABI 1759980. R.K. was supported by NIH DP1AT010885. L.P.N. was supported by Omnia Group Ltd. We thank Shimadzu South Africa Ltd. for analytical support, and J. MacRae, head of The Metabolomics STP at the Francis Crick Institute, for guidance. J.-L.W. is supported by the Swiss National Science Foundation (SNSF) Bridge – Discovery 40B2-0_211759 for studies on fungal metabolomics.
Author contributions
S.Z., R.S., A.B., M.W. and P.C.D. conceptualized the method. R.S., S.Z. and M.W. developed microbeMASST. P.C.D., S.Z., R.S., A.B., P.W.P.G., A.M.C.-R., Y.E.A., A.T.A., E.C.G., J. Zemlin, M.J.M., N.E.A., R.H.C., E.B., M.C.T., C.-Y.H., R.O., A.V.A., J. Zhao, H.C., M.C.M.K., S.L.J., F.T., L.P.N., N.E.M., I.A.D., E.A.M., L.V.C.-L., N.P.L., P.R.-T., P.C.J., B.R., A.D.P., M.F.T., R.d.C.P., G.T.-C., P.C., E.E.-L., D.A.-V., L.-M.Q.-G., J.-L.W., A.S., S.P., J.J., W.H.G., K.G., J.M.-C., P.-M.A., B.C.W., K.F., D.P., N.A., N.G., M.L., P.B., K.B.K., H.G., L.P.S.d.C., M.S.d.S., A.I.P.-L., C.M.-S., D.R., R.F., M.B., A.V.P.d.L., P.B.P., S.L.L.R., G.L.B., M.F.L., H.M.R., A.R., B.S., F.H., A.J.B., U.P., C.L.-C., L.R.-O., E.R., F.H., G.K., H.S., K.H., L.P., R.G., E.C.O.N., E.T.R., J.O., N.J.B., S.D., J.S.S.D., X.L.G., J.J.C., K.-S.J., D.B.S., F.M.R.S., G.F.d.S., H.H.F.K., C.G., J.A.C., H.M., K.B., K.L.M., S.E.O.-S., C.M.R., D.M. and R.K. contributed data and curated metadata. S.Z. generated the taxonomic tree and performed analyses. R.S. developed the tree visualizer for enriched ontologies and output summaries. B.S.P. and N.B. developed the FASST algorithm. M.W. developed the Fast Search Tool API. S.Z., R.S. and P.C.D. tested microbeMASST. S.Z., R.S. and P.C.D. wrote the manuscript. All authors reviewed the manuscript.
Peer review
Peer review information
Nature Microbiology thanks Matej Orešič and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Data used to generate the reference database of microbeMASST are publicly available at GNPS/MassIVE (https://massive.ucsd.edu/). A list of all the accession numbers (MassIVE IDs) of the studies used to generate the reference database of this tool is available in Supplementary Table 2. Interactive examples of the MS/MS queries illustrated in Fig. 1d and Supplementary Fig. 3 can be visualized at the microbeMASST website (https://masst.gnps2.org/microbemasst/). A video tutorial on how to use microbeMASST is available on YouTube. Known molecules already present in the GNPS library (https://library.gnps2.org/) were used to facilitate interpretation and confirm that specific bacterial and fungal molecules of interest were exclusively observed in the respective monocultures.
Lovastatin - CCMSLIB00005435737
Salinosporamide A - CCMSLIB00010013003
Commendamide - CCMSLIB00004679239
Mevastatin - CCMSLIB00005435644
Arylomycin A4 - CCMSLIB00000075066
Yersiniabactin - CCMSLIB00005435750
Promicroferrioxamine - CCMSLIB00005716848
Glutamate-cholic acid (Glu-CA) - CCMSLIB00006582258
Glutamate-deoxycholic acid (Glu-DCA) - CCMSLIB00006582092
Data used to extract MS/MS spectra exclusively present in colonized (SPF) mice are publicly available in GNPS/MassIVE under the accession number MSV000079949. Data used to validate and assess antibiotics effect on microbial MS/MS spectra of interest are available under the accession number MSV000080918. A list of datasets with data acquired from human biosamples that presented matches to the putative microbial MS/MS spectra of interest is available in Supplementary Table 3.
Code availability
The microbeMASST code to query spectra, create interactive trees and analyse results is available under an open-source license on GitHub (https://github.com/robinschmid/microbe_masst). This repository also contains code to run batch searches of thousands of MS/MS spectra by providing either a .tsv file containing a list of USIs or a .mgf file generated for example through the MZmine data processing pipeline. Code used to generate the microbeMASST web interface can be accessed on GitHub (https://github.com/mwang87/GNPS_MASST). Code used to perform the analysis and generate the figures presented in the manuscript can be downloaded from GitHub (https://github.com/simonezuffa/Manuscript_microbeMASST).
Competing interests
P.C.D. is an advisor to Cybele, consulted for MSD animal health in 2023 and is a co-founder and scientific advisor for Ometa Labs, Arome and Enveda, with previous approval from the University of California, San Diego. M.W. is a co-founder of Ometa labs. There are no known conflicts of interest in this work by the USDA, Agricultural Research Service, National Center for Agricultural Utilization Research, Mycotoxin Prevention and Applied Microbiology Research Unit. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. R.K. is a scientific advisory board member and consultant for BiomeSense, Inc., where he has equity and receives income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Simone Zuffa, Robin Schmid, Anelize Bauermeister.
Supplementary information
The online version contains supplementary material available at 10.1038/s41564-023-01575-9.
References
- 1.Jansson JK, Hofmockel KS. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020;18:35–46. doi: 10.1038/s41579-019-0265-7. [DOI] [PubMed] [Google Scholar]
- 2.Malard F, Dore J, Gaugler B, Mohty M. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol. 2021;14:547–554. doi: 10.1038/s41385-020-00365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 4.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023;186:243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Radjabzadeh D, et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 2022;13:7128. doi: 10.1038/s41467-022-34502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morais LH, Schreiber HLIV, Mazmanian SK. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021;19:241–255. doi: 10.1038/s41579-020-00460-0. [DOI] [PubMed] [Google Scholar]
- 7.Milshteyn A, Colosimo DA, Brady SF. Accessing bioactive natural products from the human microbiome. Cell Host Microbe. 2018;23:725–736. doi: 10.1016/j.chom.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paoli L, et al. Biosynthetic potential of the global ocean microbiome. Nature. 2022;607:111–118. doi: 10.1038/s41586-022-04862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grice EA, Segre JA. The human microbiome: our second genome. Annu. Rev. Genomics Hum. Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tierney BT, et al. The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe. 2019;26:283–295.e8. doi: 10.1016/j.chom.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK, Dorrestein PC. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 2022;20:143–160. doi: 10.1038/s41579-021-00621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS, et al. MiMeDB: the Human Microbial Metabolome Database. Nucleic Acids Res. 2023;51:D611–D620. doi: 10.1093/nar/gkac868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Santen JA, et al. The Natural Products Atlas 2.0: a database of microbially-derived natural products. Nucleic Acids Res. 2022;50:D1317–D1323. doi: 10.1093/nar/gkab941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutz A, et al. The LOTUS initiative for open knowledge management in natural products research. eLife. 2022;11:e70780. doi: 10.7554/eLife.70780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han S, et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature. 2021;595:415–420. doi: 10.1038/s41586-021-03707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoch, C. L. et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database2020, baaa062 (2020). [DOI] [PMC free article] [PubMed]
- 19.Wang M, et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 2020;38:23–26. doi: 10.1038/s41587-019-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batsoyol, N., Pullman, B., Wang, M., Bandeira, N. & Swanson, S. P-massive: a real-time search engine for a multi-terabyte mass spectrometry database. In Proc. International Conference on High Performance Computing, Networking, Storage and Analysis 1–15 (IEEE, 2022).
- 21.Deutsch EW, et al. Universal Spectrum Identifier for mass spectra. Nat. Methods. 2021;18:768–770. doi: 10.1038/s41592-021-01184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bittremieux, W. et al. Universal MS/MS visualization and retrieval with the Metabolomics Spectrum Resolver Web Service. Preprint at bioRxiv10.1101/2020.05.09.086066 (2020).
- 23.Schmid R, et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 2023;41:447–449. doi: 10.1038/s41587-023-01690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl Acad. Sci. USA. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberts AW, et al. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc. Natl Acad. Sci. USA. 1980;77:3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A phase III trial of with marizomib in patients with newly diagnosed glioblastoma (MIRAGE). CTG Labs (NCBI)https://beta.clinicaltrials.gov/study/NCT03345095?distance=50&intr=NPI-0052&rank=9 (2023).
- 27.Fenical W, et al. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorg. Med. Chem. 2009;17:2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen LJ, et al. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl Acad. Sci. USA. 2015;112:E4825–E4834. doi: 10.1073/pnas.1508737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentry, E. C. et al. Reverse metabolomics for the discovery of chemical structures from humans. Nature10.1038/s41586-023-06906-8 (2023). [DOI] [PMC free article] [PubMed]
- 30.Quinn RA, et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579:123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson, A. et al. Bile acids are substrates for amine N-acyl transferase activity by bile salt hydrolase. Preprint at Res. Square10.21203/rs.3.rs-2050120/v1 (2022).
- 32.Endo A. The origin of the statins. Atheroscler. Suppl. 2004;5:125–130. doi: 10.1016/j.atherosclerosissup.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 33.Schimana, J. et al. Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by Streptomyces sp. Tü 6075. J. Antibiot.55, 565–570 (2002). [DOI] [PubMed]
- 34.Drechsel H, et al. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann. Org. Bioorg. Chem. 1995;1995:1727–1733. [Google Scholar]
- 35.Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 2002;70:5335–5337. doi: 10.1128/IAI.70.9.5335-5337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawlor MS, O’connor C, Miller VL. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 2007;75:1463–1472. doi: 10.1128/IAI.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y-L, et al. Connecting chemotypes and phenotypes of cultured marine microbial assemblages by imaging mass spectrometry. Angew. Chem. Int. Ed. Engl. 2011;50:5839–5842. doi: 10.1002/anie.201101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dührkop K, et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods. 2019;16:299–302. doi: 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- 39.Dührkop K, et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021;39:462–471. doi: 10.1038/s41587-020-0740-8. [DOI] [PubMed] [Google Scholar]
- 40.Shalapour S, et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature. 2017;551:340–345. doi: 10.1038/nature24302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucas, L. N. et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. mSystems6, e0080521 (2021). [DOI] [PubMed]
- 44.Hoffmann MA, et al. High-confidence structural annotation of metabolites absent from spectral libraries. Nat. Biotechnol. 2022;40:411–421. doi: 10.1038/s41587-021-01045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guzior, D. et al. Bile salt hydrolase/aminoacyltransferase shapes the microbiome. Preprint at Res. Square10.21203/rs.3.rs-2050406/v1 (2022).
- 46.Foley MH, et al. Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut. Nat. Microbiol. 2023;8:611–628. doi: 10.1038/s41564-023-01337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Folz J, et al. Human metabolome variation along the upper intestinal tract. Nat. Metab. 2023;5:777–788. doi: 10.1038/s42255-023-00777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shalon D, et al. Profiling the human intestinal environment under physiological conditions. Nature. 2023;617:581–591. doi: 10.1038/s41586-023-05989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao L, et al. A selective gut bacterial bile salt hydrolase alters host metabolism. eLife. 2018;7:e37182. doi: 10.7554/eLife.37182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartlett, A. & Kleiner, M. Dietary protein and the intestinal microbiota: an understudied relationship. iScience25, 105313 (2022). [DOI] [PMC free article] [PubMed]
- 51.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strandwitz P, et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019;4:396–403. doi: 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maneerat S, Nitoda T, Kanzaki H, Kawai F. Bile acids are new products of a marine bacterium, Myroides sp. strain SM1. Appl. Microbiol. Biotechnol. 2005;67:679–683. doi: 10.1007/s00253-004-1777-1. [DOI] [PubMed] [Google Scholar]
- 54.Kim D, et al. Biosynthesis of bile acids in a variety of marine bacterial taxa. J. Microbiol. Biotechnol. 2007;17:403–407. [PubMed] [Google Scholar]
- 55.Ohashi K, Miyagawa Y, Nakamura Y, Shibuya H. Bioproduction of bile acids and the glycine conjugates by Penicillium fungus. J. Nat. Med. 2008;62:83–86. doi: 10.1007/s11418-007-0190-3. [DOI] [PubMed] [Google Scholar]
- 56.Lai Y, et al. High-coverage metabolomics uncovers microbiota-driven biochemical landscape of interorgan transport and gut–brain communication in mice. Nat. Commun. 2021;12:6000. doi: 10.1038/s41467-021-26209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jarmusch AK, et al. ReDU: a framework to find and reanalyze public mass spectrometry data. Nat. Methods. 2020;17:901–904. doi: 10.1038/s41592-020-0916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chamberlain SA, Szöcs E. taxize: taxonomic search and retrieval in R. F1000Research. 2013;2:191. doi: 10.12688/f1000research.2-191.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–5.
Supplementary Tables 1–3.
Data Availability Statement
Data used to generate the reference database of microbeMASST are publicly available at GNPS/MassIVE (https://massive.ucsd.edu/). A list of all the accession numbers (MassIVE IDs) of the studies used to generate the reference database of this tool is available in Supplementary Table 2. Interactive examples of the MS/MS queries illustrated in Fig. 1d and Supplementary Fig. 3 can be visualized at the microbeMASST website (https://masst.gnps2.org/microbemasst/). A video tutorial on how to use microbeMASST is available on YouTube. Known molecules already present in the GNPS library (https://library.gnps2.org/) were used to facilitate interpretation and confirm that specific bacterial and fungal molecules of interest were exclusively observed in the respective monocultures.
Lovastatin - CCMSLIB00005435737
Salinosporamide A - CCMSLIB00010013003
Commendamide - CCMSLIB00004679239
Mevastatin - CCMSLIB00005435644
Arylomycin A4 - CCMSLIB00000075066
Yersiniabactin - CCMSLIB00005435750
Promicroferrioxamine - CCMSLIB00005716848
Glutamate-cholic acid (Glu-CA) - CCMSLIB00006582258
Glutamate-deoxycholic acid (Glu-DCA) - CCMSLIB00006582092
Data used to extract MS/MS spectra exclusively present in colonized (SPF) mice are publicly available in GNPS/MassIVE under the accession number MSV000079949. Data used to validate and assess antibiotics effect on microbial MS/MS spectra of interest are available under the accession number MSV000080918. A list of datasets with data acquired from human biosamples that presented matches to the putative microbial MS/MS spectra of interest is available in Supplementary Table 3.
The microbeMASST code to query spectra, create interactive trees and analyse results is available under an open-source license on GitHub (https://github.com/robinschmid/microbe_masst). This repository also contains code to run batch searches of thousands of MS/MS spectra by providing either a .tsv file containing a list of USIs or a .mgf file generated for example through the MZmine data processing pipeline. Code used to generate the microbeMASST web interface can be accessed on GitHub (https://github.com/mwang87/GNPS_MASST). Code used to perform the analysis and generate the figures presented in the manuscript can be downloaded from GitHub (https://github.com/simonezuffa/Manuscript_microbeMASST).