Abstract
Background and Aims
Breast cancer is one of the deadliest diseases affecting women in Bangladesh, and its prevalence is increasing year by year. Although several IL‐6 single nucleotide polymorphisms have been implicated in BC susceptibility and prognosis in various studies, no research has been done to investigate the relationship between breast cancer and IL‐6 in Bangladeshi women. This investigation aimed to explore the linkage between the rs1800797 variant of IL‐6 and the susceptibility to breast carcinoma among women in Bangladesh.
Methods
The IL‐6 rs1800797 variant was genotyped in 218 subjects (110 cases and 108 controls) using the tetra‐primer ARMS‐PCR method. The statistical analysis was applied utilizing the SPSS software version 24.0. UALCAN database was used for IL‐6 mRNA analysis, and genotype‐based gene expression was retrieved from GTEx Portal.
Results
This study found a significant link between IL‐6 rs1800797 variants and increased chance of breast cancer across different genetic inheritance models, including additive model 1 (AG vs. GG: OR = 2.16, p = 0.035); dominant model (AG + AA vs. GG: OR = 2.26, p < 0.05); overdominant model (AG vs. GG + AA: OR = 2.08, p < 0.05); and allelic model (A vs. G: OR = 2.15, p < 0.05). However, an insignificant association of breast cancer was found in both additive model 2 (AA vs. GG: OR = 2.91, p > 0.05) and the recessive model (AA vs. GG + AG: OR = 2.52, p > 0.05). Under the analysis of the probability of false positive reports, no significant values were found in different models when the OR was 1.5, and the prior probability was 0.25.
Conclusions
A significant relationship was found between the IL‐6 rs1800797 genetic variant and the risk of breast cancer. However, the findings of the study should be further investigated with a larger sample size to validate the correlation.
Keywords: ARMS‐PCR, breast cancer, IL‐6 gene, polymorphisms
1. INTRODUCTION
Breast cancer (BC) is one of the most frequently diagnosed malignancies across the world. 1 It usually begins in the breast lobules, tubes, or connective tissue. 2 A recent report from the International Agency for Research on Cancer (IARC) identified 2.3 million new BC cases (11.7%) and a death rate of 6.9% across 185 countries. 3 Notably, about 42,000 women die from BC each year, making it the sixth leading cause of death. 4 The rising incidence of BC has led to it becoming a global public health crisis 5 that affects women of all ethnicities and income levels. 6 The development of BC is influenced by both environmental and genetic factors. 7 While only a small percentage of BC is genetically predisposed, mutation‐induced BC can be more lethal, with mutations in cancer susceptibility genes causing 8%–10% of BC. 8 Inflammation is one of the many factors that contribute to the pathogenesis of BC, as it can cause cellular events that lead to malignant cell transformation and carcinogenesis. 9
Interleukin‐6 (IL‐6), a pro‐inflammatory cytokine, is located on chromosome 7p21. 10 When the body is infected or injured, innate immune cells like macrophages and monocytes produce IL‐6. This stimulates hematopoiesis, immunological responses, and acute phase reactions to help protect the host. 11 However, if this regulation is disrupted, IL‐6 is produced excessively and persistently, triggering the onset of several diseases. 12
Various cells in the tumor microenvironment express IL‐6, including adipocytes, myeloid‐derived suppressor cells (MDSCs), fibroblasts, and lymphatic endothelial tissues. Many studies have found evidence linking IL‐6 release from various cell types to BC growth over time. 13 Increased IL‐6 production has been linked to the initiation and promotion of tumor growth following the malignant conversion, invasion, and metastasis of cancer. Polymorphisms in the IL‐6 promoter region were found to be significantly related to cancer susceptibility and prognosis. 10 Individuals with breast cancer have a poor prognosis and a low chance of survival when their blood IL‐6 levels are elevated. 14
If tumor‐specific mutations function as metastasis‐promoting factors, the gene expression characteristics of tumor tissue are critical. 15 Several mutations have been discovered in the genes encoding IL‐6, members of its family, and its receptors in humans. 16 Despite significant variation across studies, candidate gene studies and genome‐wide association studies (GWAS) demonstrate the significance of these variants in BC risk. 17 Several single nucleotide polymorphisms (SNPs) of IL‐6 have been found to be linked with BC risk and prognosis in various studies. One of the SNPs, rs1800797, which is located in the 50 flanking regions of the IL‐6 gene promoter, can impact the expression of IL‐6. A study conducted on postmenopausal women in the southwestern United States revealed that those with the AA genotype of rs1800797 SNP had a higher risk of developing BC. Another study conducted in Sweden showed that patients with GG genotype of rs1800797 SNP had shorter disease‐free survival (DFS) times compared with those with AA or GA genotypes and estrogen receptor (ER)‐positive tumors. 18 Moreover, IL‐6 rs1800797 AG or AA genotypes were linked to a lower DFS, and the A allele was linked to hormone receptor‐positive BC in Iranian women. 19 IL‐6 rs1800797, an allele, was also linked to human epidermal growth factor receptor 2 (HER2) negativity. In a separate study, researchers found that functional SNPs in genes linked to angiogenesis and inflammation were connected to the prognosis of early‐stage breast cancer in the Lithuanian population. 17 However, no research has been done to investigate the relationship between BC and IL‐6 in Bangladeshi women. Using the tetra‐primer ARMS‐PCR technique, this study aims to investigate the link between BC and IL‐6 1800797 polymorphism in Bangladeshi women based on preliminary findings.
2. MATERIALS AND METHODS
2.1. Study population
This case‐control investigation involved 218 women from the Bangladeshi population, out of which 110 were breast cancer patients and 108 were healthy individuals. The study was performed at the Laboratory of Pharmacogenomics and Molecular Biology situated at Noakhali Science and Technology University in Noakhali‐3814, Bangladesh. It was acknowledged by the respective ethical committee of the National Institute of Cancer Research and Hospital (NICRH/Ethics/2019/447). Patients' demographic data was collected through written consent and a detailed questionnaire. For breast carcinoma patients, we collected data on age, primary tumor location and tumor size from their medical records with the help of a physician. People under the age of 18 who were unable to provide the necessary information and those with cancer or other diseases such as kidney, hepatic, or pulmonary disease were excluded from this research. The investigation was completed following the Helsinki Declaration's principle of researching human subjects, as amended (World Medical Association Declaration of Helsinki‐Seoul revision: Ethical principles for medical research involving human subjects 2010).
2.2. Sample preparation
A blood sample (3 mL) was obtained using a sterile plastic syringe and put into plastic tubes containing EDTA‐Na2. All patients selected for this case‐control research provided approximately 3 mL of blood, which was extracted with the assistance of an experienced nurse. After collecting blood samples, they were stored at −80°C until the DNA extraction. Genomic DNA was then isolated from the entire peripheral blood sample that is typically used in our laboratory. 20 We determined the purity and concentration of the DNA by transferring 2 µL of the extracted DNA into a micro‐volume spectrophotometer (Genova Nano, Jenway), with the absorbance ratio set at 260 and 280 nm. We found an acceptable range for the absorbance ratio, which helped determine the purity of genomic DNA. The primer sequences were designed using the online software Primer1 (http://primer1.soton.ac.uk/primer1help.html#allele2) (Supporting Information: Table S1).
2.3. Genotyping
To genotype IL‐6 rs1800797, we used the tetra‐primer amplification refractory mutation system (ARMS) technique based on polymerase chain reaction (PCR). We designed four primers, including forward outer primer, reverse outer primer, forward inner primer, and reverse inner primer (Supporting Information: Table S1), to amplify the desired allele. To create the PCR premix, we combined EmeraldAmp GT PCR Master Mix, nuclease‐free water, MgCl2, and primers at the appropriate concentrations. For preparing a 120 µL PCR master mix solution (for 12 samples at 10 µL per reaction), we used 1.5 µL of each outer primer (forward outer and reverse outer), with a concentration of 0.25 µM in 1x solution. To prepare for the PCR, 2 µL (0.33 µM in 1x solution) of each inner primer (forward inner and reverse inner) was added to the mixture. Subsequently, the DNA sample was combined with the premix (10 µL) at the desired concentration. Finally, the PCR products were examined through gel electrophoresis (1% agarose gel) to verify the presence of allele‐specific DNA bands that were stained with ethidium bromide. The target area for rs1800797 had a 315 bp nonallele specific segment, and pieces of 210 and 161 bp were discovered for the G allele and the A allele, respectively (Figure 1). Supporting Information: Table S2 shows the PCR conditions, fragment sizes, and features of the rs1800797 SNP. To validate the findings, a 20% sample of hetero and homozygous mutant samples was checked again. There was no use of Sanger sequencing or any other sequencing techniques.
Figure 1.
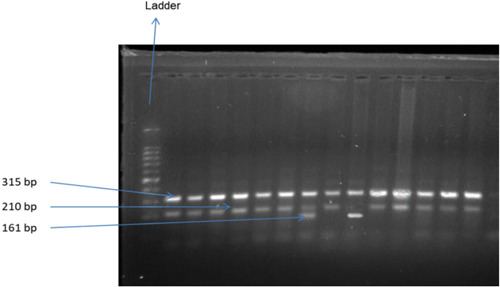
Agarose gel electrophoresis of (1% w/v agarose) of genomic DNA. From the side of the ladder, lanes 1,2,3,4,5,6,8,10,11,12,13,14 contain GG; lane 7 contains AG; and lane 9 contains AA genotype.
2.4. False‐positive report probability (FPRP)
The FPRP was calculated to assess the findings' reliability. 21 , 22 The threshold for FPRP was set at 0.2, and prior probabilities of 0.25, 0.1, 0.01, 0.001, and 0.0001 were specified to find an odds ratio (OR) of 1.5 related to cancer risk. Findings were considered important if their FPRP values were below 0.20.
2.5. In silico gene expression analysis
The UALCAN database (http://ualcan.path.uab.edu/) was used for IL‐6 mRNA analysis and genotype‐based gene expression was retrieved for rs1800797 polymorphism from the Genotype‐Tissue Expression (GTEx) Portal.
2.6. Statistical analysis
To calculate Hardy‐Weinberg Equilibrium (HWE) and BC risk, we used the Chi‐square (□ 2) test and the OR at the corresponding 95% confidence interval (CI). We presented the percentages of genotype and allelic frequencies. The likelihood of developing cancer was determined using the OR calculator MedCalc (https://www.medcalc.org/calc/odds_ratio.php). The risk was evaluated using the OR and its 95% CI. We considered results statistically significant when p values were 0.05 or less and nonsignificant when p > 0.05. 23
3. RESULTS
3.1. Characteristics of individuals based on demographic and clinicopathological data
Table 1 displays demographic and clinicopathological data for BC patients and controls. 37.27% of patients were below the age of 45, 50.91% were between 45 and 60, and 11.82% were over 60 years. The average age of cases was 48.11 years, with an age range of 26–72 years. The age range of control volunteers was 23–67 years, with an average age of 37.97. The average BMI in the patient group was 27.59 kg/m2, while the control group had an average BMI of 21.62 kg/m2. Almost 92% of the cases and 85% of the controls were married. 53.33% of the patients had invasive duct cell carcinoma, while 20% had infiltrated duct cell carcinoma. 58.54% of the patients had grade II cancer. Biopsy, computerized tomography (CT), Fine Needle Aspiration Cytology (FNAC), echocardiogram, ultrasound sonography test (USG), and X‐ray were used to diagnose approximately 67.78%, 18.89%, 27.78%, 18.89%, 66.67%, and 31.11% of patients, respectively. The majority of patients (45.68%, 43.21%, and 22.22%, respectively) had surgery, CT scan, and RT as their previous treatments. Around 57.45% of patients received 4–8 cycles of chemotherapy, while 36.17% received 1–3 cycles. Approximately 40% of patients were ER (+) and 52% were ER (‐). Patients with PR (+) and PR (‐) were 56% and 52%, respectively. 53.33% of patients had HER2 (‐), while 32% had HER2 (+). Approximately 2.67% of patients had triple‐negative breast cancer. According to the history of drugs taken by patients, the majority of them (61.82%, 50.91%, and 45.45%, respectively) took cyclophosphamide, doxorubicin, and paclitaxel for BC. Others were given dexamethasone, ranitidine, normal saline, and the other medications listed in Table 2.
Table 1.
Distribution of demographic variables of BC patients and controls.
| Variables | Cases (N = 110) N (%) | Controls (n = 108) N (%) | p Value |
|---|---|---|---|
| Age (years) | |||
| <45 | 41 (37.27) | 55 (50.93) | 0.151 |
| 45–60 | 56 (50.91) | 43 (39.81) | |
| >60 | 13 (11.82) | 10 (9.26) | |
| 45–60 + >60 | 69 (62.73) | 53 (49.07) | |
| Mean age (years) | |||
| Minimum age | 26 | 23 | 0.023 |
| Maximum age | 72 | 67 | |
| Average ± SD | 48.11 ± 3.57 | 37.97 ± 2.84 | – |
| BMI (kg/m2) | |||
| Average ± SD | 27.59 ± 1.55 | 21.61 ± 1.36 | – |
| Marital status | |||
| Married | 101 (91.82) | 92 (85.19) | 0.124 |
| Unmarried | 9 (8.18) | 16 (14.81) | |
| Type of BC | |||
| Atypical ductal hyperplasia | 5/90 (5.55) | N/A | |
| Duct cell carcinoma | 7/90 (7.78) | N/A | |
| Infiltrating duct cell carcinoma | 18/90 (20.00) | N/A | |
| Intraductal carcinoma | 1/90 (1.11) | N/A | |
| Invasive duct cell carcinoma | 48/90 (53.33) | N/A | |
| Medullary carcinoma | 4/90 ((4.44) | N/A | |
| Metastatic duct cell carcinoma | 4/90 (4.44) | N/A | |
| Triple negative BC | 3/90 (3.33) | N/A | |
| No data | 20/110 (18.18) | N/A | |
| Grade of BC | |||
| Ⅰ | 7/41 (17.07) | N/A | |
| Ⅱ | 24/41 (58.54) | N/A | |
| Ⅲ | 9/41 (21.95) | N/A | |
| No data | 69/110 (62.73) | N/A | |
| Diagnosis | |||
| Biopsy | 61/90 (67.78) | N/A | |
| CA 15‐3 | 2/90 (2.22) | N/A | |
| CT | 17/90 (18.89) | N/A | |
| CXR | 3/90 (3.33) | N/A | |
| Echocardiogram | 17/90 (18.89) | N/A | |
| FNAC | 25/90 (27.78) | N/A | |
| Lumpectomy | 4/90 (4.44) | N/A | |
| Mastectomy | 11/90 (12.22) | N/A | |
| RT | 2/90 (2.22) | N/A | |
| USG | 60/90 (66.67) | N/A | |
| R‐Ray | 28/90 (31.11) | N/A | |
| No data | 20/110 (18.18) | N/A | |
| Current treatment | |||
| Chemotherapy | 38/69 (55.07) | N/A | |
| CT | 56/69 (81.16) | N/A | |
| Mastectomy | 5/69 (7.25) | N/A | |
| MRM | 5/69 (7.25) | N/A | |
| RT | 6/69 (8.69) | N/A | |
| Surgery | 13/69 (18.84) | N/A | |
| No data | 41/110 (37.27) | N/A | |
| Previous treatment | |||
| CT | 35/81 (43.21) | N/A | |
| Hormone therapy | 14/81 (17.28) | N/A | |
| Lumpectomy | 2/81 (2.46) | N/A | |
| Mastectomy | 8/81 (9.88) | N/A | |
| MRM | 1/81 (1.23) | N/A | |
| RT | 18/81 (22.22) | N/A | |
| Surgery | 37/81 (45.68) | N/A | |
| No data | 29/110 (26.36) | N/A | |
| Chemotherapy cycle | |||
| 1–3 | 17/47 (36.17) | N/A | |
| 4–8 | 27/47 (57.45) | N/A | |
| 9–12 | 3/47 (6.38) | N/A | |
| No data | 63/110 (57.27) | N/A | |
| ER, PR, HER2 status | |||
| ER (+) | 30/75 (40.00) | N/A | |
| ER (‐) | 39/75 (52.00) | N/A | |
| PR (+) | 42/75 (56.00) | N/A | |
| PR (‐) | 39/75 (52.00) | N/A | |
| HER2 (+) | 24/75 (32.00) | N/A | |
| HER2 (‐) | 40/75 (53.33) | N/A | |
| Triple negative | 2/75 (2.67) | N/A | |
| No data | 35/110 (31.82) | N/A | |
| Drug name | |||
| 5‐FU | 10/110 (9.09) | N/A | |
| 5% DNS | 6/110 (5.45) | N/A | |
| Avil | 10/110 (9.09) | N/A | |
| Carboplatin | 5/110 (4.54) | N/A | |
| Cisplatin | 14/110 (12.73) | N/A | |
| Cyclophosphamide | 68/110 (61.82) | N/A | |
| Dexamethasone | 28/110 (25.45) | N/A | |
| Docetaxel | 2/110 (1.82) | N/A | |
| Doxorubicin | 56/110 (50.91) | N/A | |
| Emeset | 3/110 (2.73) | N/A | |
| Epirubicin | 10/110 (9.09) | N/A | |
| Filgrastim | 10/110 (9.09) | N/A | |
| Gemcitabine | 2/110 (1.82) | N/A | |
| Herceptin | 5/110 (4.54) | N/A | |
| Ranitidine | 35/110 (31.82) | N/A | |
| Normal saline | 26/110 (23.64) | N/A | |
| Paclitaxel | 50/110 (45.45) | N/A | |
| Tamoxifen | 7/110 (6.36) | N/A | |
| Zoledronic acid | 8/110 (7.27) | N/A | |
Note: p < 0.05 was considered a statistically significant.
Abbreviation: N/A, not applicable.
Table 2.
Genotypic association of IL6 rs1800797 variants with breast cancer.
| Genetic Models | Genotype/Allele | Cases (n = 110) | HWE | Controls (n = 108) | HWE | Risk analysis | |||
|---|---|---|---|---|---|---|---|---|---|
| χ 2 | p Value | χ 2 | p Value | OR(95% Cl) | p Value | ||||
| GG | 79 (71.82%) | 2.05 | 0.152 | 92 (85.19%) | 2.48 | 0.115 | 1 | ||
| Additive model 1 (AG vs. GG) | AG | 26 (23.64%) | 14 (12.96%) | 2.16 (1.06–4.43) | 0.035 | ||||
| Additive model 2 (AA vs. GG) | AA | 5 (4.55%) | 2 (1.85%) | 2.91 (0.55–15.42) | 0.209 | ||||
| Dominant model (AG + AA vs. GG) | GG | 79 (71.82%) | 92 (85.19%) | 1 | |||||
| AG + AA | 31 (28.18) | 16 (14.81) | 2.26 (1.15–4.43) | 0.018 | |||||
| Recessive model (AA vs. GG + AG) | GG + AG | 105 (95.45) | 106 (98.15) | 1 | |||||
| AA | 5 (4.55%) | 2 (1.85%) | 2.52 (0.48–13.30) | 0.275 | |||||
| Overdominant model (AG vs. GG + AA) | GG + AA | 84 (76.36) | 94 (87.04) | 1 | |||||
| AG | 26 (23.64%) | 14 (12.96%) | 2.08 (1.02–4.24) | 0.044 | |||||
| Allele (A vs. G) | G | 184 (83.64) | 198 (91.67) | 1 | |||||
| A | 36 (16.36) | 18 (8.33) | 2.15 (1.18–3.92) | 0.012 | |||||
Note: p < 0.05 was considered a statistically significant. Bold values indicate statistically significant.
Abbreviation: OR, odds ratio.
3.2. Frequency distribution of IL‐6 rs1800797 polymorphism
The genotypes and allele frequencies of the IL‐6 rs1800797 polymorphisms in breast cancer patients and healthy individuals are revealed in Table 2. For cases, the frequency distribution of GG, AG, and AA genotypes was 71.82%, 23.64%, and 4.55%, respectively. This compared with 85.19%, 12.96%, and 1.85% for controls, respectively. Furthermore, the major G allele was prevalent in both cases (83.64%) and controls (91.67%). In addition, the genotype distribution of IL‐6 rs1800797 variants was consistent with the HWE in both cases and controls.
3.3. Genotypic association of IL‐6 rs1800797 variants with breast cancer
Six genetic models were used to calculate the risk of the IL‐6 rs1800797 polymorphisms in BC patients (Table 2). The ORs of each genetic model were used to calculate the risk of BC. Our study found a correlation between IL‐6 rs1800797 variants and an elevated risk of breast cancer in all genetic inheritance models. Specifically, the study revealed a significant genetic link between the IL‐6 rs1800797 variants and an increased risk of breast cancer in different genetic inheritance models, including additive model 1, dominant model, overdominant model, and allelic model. However, both the additive model 2 and the recessive model revealed an insignificant association of breast cancer risk. In the additive model 1 for BC patients, heterozygote AG carriers had a 2.16‐fold higher BC risk (OR = 2.16, CI = 1.06–4.43, p < 0.05; AG vs. GG). When comparing homozygous recessive AA carriers to reference homozygous dominant GG carriers, additive model 2 revealed a 2.91‐fold higher risk (AA vs. GG: OR = 2.91, CI = 0.55–15.42, p > 0.05). The dominating model indicated that there was a 2.26‐fold more significant risk in the BC patients compared with the control group (AG + AA vs. GG: OR = 2.26, CI = 1.15–4.43, p < 0.05). In comparison to the control group, the recessive model likewise indicated a 2.52‐fold higher risk in cancer patients (AA vs. GG + AG: OR = 2.52, CI = 0.48–13.30, p > 0.05). This outcome lacked statistical significance. The overdominant model revealed a 2.08‐fold significant risk in BC patients (AG vs. GG + AA: OR = 2.08, CI = 1.02–4.24, p < 0.05). When compared with wild‐type G allele carriers, mutant allele A carriers had a substantially greater risk (A vs. G: OR = 2.15, CI = 1.18–3.92, p < 0.05) according to the allele frequency analysis. Thus, IL‐6 rs1800797 polymorphism showed a significantly strong association with BC risk in Bangladeshi women.
3.4. FPRP test
The FPRP values with statistical power for findings with the IL‐6 rs1800797 polymorphism are shown in Table 3. In accordance with the FPRP analysis, no values in different models were significant when the OR was 1.5, and the prior probability was 0.25. IL‐6 rs1800797 polymorphism was nonsignificantly (p < 0.05) linked to the increased BC risk with statistical powers of 16.0%, 10.0%, 18.4%, and 12.0% when the prior probability value was 0.25.
Table 3.
False‐positive report probability values for the association between rs1800797 and breast cancer risk.
| Genetic Models | OR (95% Cl) | p Value | Statistical powera | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| AG versus GG | 2.16 (1.06–4.43) | 0.035 | 0.160 | 0.401 | 0.667 | 0.957 | 0.996 | 1.000 |
| AA versus GG | 2.91 (0.55–15.42) | 0.209 | 0.218 | 0.742 | 0.896 | 0.990 | 0.999 | 1.000 |
| AG + AA versus GG | 2.26 (1.15–4.43) | 0.018 | 0.100 | 0.312 | 0.576 | 0.937 | 0.993 | 0.999 |
| AA versus GG + AG | 2.52 (0.48–13.30) | 0.275 | 0.271 | 0.754 | 0.902 | 0.990 | 0.999 | 1.000 |
| AG versus GG + AA | 2.08 (1.02–4.24) | 0.044 | 0.184 | 0.417 | 0.682 | 0.959 | 0.996 | 1.000 |
| A versus G | 2.15 (1.18–3.92) | 0.012 | 0.120 | 0.238 | 0.484 | 0.912 | 0.990 | 0.999 |
Note: The results in false‐positive report probability analysis were in bold if the prior probability <0.2.
Statistical power was calculated using the number of observations in the subgroup and the OR and p values in this table.
3.5. In silico gene expression profile
While in silico mRNA expression indicates a significant difference in expression data in the breast cancer tissues and healthy tissues, genotype‐based gene expression in cultured fibroblasts suggests there may be a significant difference in the gene expression of the variant allele carrier gene compared with the wild allele carrier gene (Figures 2 and 3). The violin plots illustrate the allelic effect of rs1800797 on normalized IL‐6 gene expression levels. The major and minor allele types, A and G, respectively, are represented along with the number of subjects for each genotype. The sample density distribution is depicted in each genotype. The median value of gene expression for each genotype is represented by a white stripe in the black box plot.
Figure 2.
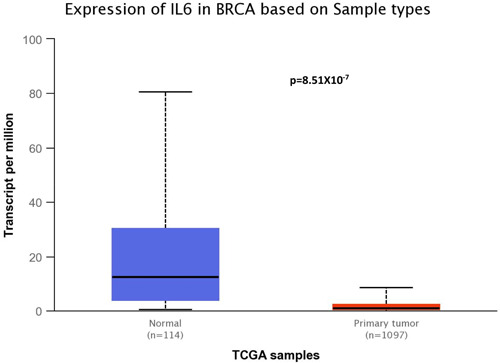
Box plots of IL‐6 gene expression between tumor tissue sample (n = 1097) and normal tissue sample (n = 114) using the UALCAN database (http://ualcan.path.uab.edu/).
Figure 3.
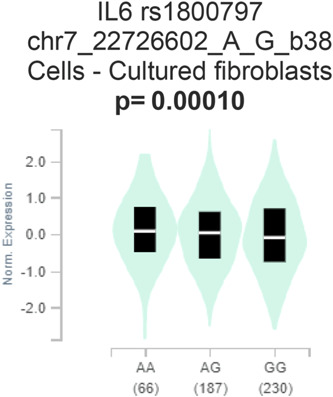
Genotype‐Tissue Expression (GTEx) of IL6 rs1800797 polymorphism in cultured fibroblast indicates a significant difference in the expression between the mutant genotypes compared with the wild genotypes.
4. DISCUSSION
According to the findings of this study, the allele frequency of rs1800797 may be linked to the development of BC. In a meta‐analysis of IL‐6 promoter genetic variants in cancer risk and prognosis, rs1800797 was found to have a significant association in BC (OR = 1.14). 10 Another study discovered that postmenopausal women with the rs1800797 AA genotype were prone to develop BC, even if they had not been exposed to hormones. In addition, the genotyping model revealed a remarkable correlation between IL‐6 rs1800797 genotypes and ER (p < 0.05) and PR (p < 0.05) status in a single‐locus analysis. Based on the allelic model, the A allele of this SNP is highly linked with positive ER status (OR 2.23). Patients who carried the IL‐6 rs1800797 A allele had a greater risk of developing HER2‐negative breast cancer (OR 2.21; p < 0.05). The IL‐6 rs1800797 gene genotypes AG or AA were also linked to a lower DFS rate. 24
For IL‐6 rs1800797 polymorphisms, we found that the risk of BC was 2.16 times higher for patients carrying the mutant heterozygote genotype (AG) than for the patients carrying homozygous dominant genotype (GG) (AG vs. GG: OR = 2.16; p = 0.035). However, the risk was not statistically remarkable (p < 0.05). Again, the AA homozygous recessive genotype was found to have a 2.91‐fold higher risk of developing breast carcinoma than the GG homozygous dominant genotype. However, this difference was not statistically significant (AA vs. GG: OR = 2.91; p > 0.05). On the other hand, when compared with the GG genotype, the AG + AA combined genotype showed a 2.26 times higher risk of breast cancer in the dominant model (AG + AA vs. GG: OR = 2.26; p < 0.05). In the recessive model (AA vs. GG + AG), the AA genotype had a 2.52 times higher risk of breast carcinoma, which was statistically insignificant (OR = 2.52; p > 0.05). Finally, the overdominant model (AG vs. GG + AA) revealed a statistically significant 2.08‐fold increase in risk. When minor allele A was compared with major allele G in the allele model, it showed 2.15 times increased association with BC risk, which is statistically significant (A vs. G: OR = 2.15; p < 0.05). As a result, the wild allele (G) demonstrated a protective function against the development of breast neoplasia, whereas the variant allele (A) demonstrated the opposite, implying that the A allele is a risk factor for developing breast carcinoma for this SNP. However, we did not find any evidence to support our findings. In a systematic review and meta‐analysis, IL‐6 promoter genetic variants were found to be strongly connected with BC. 25
Moreover, the IL‐6 (rs1800797) gene expression was significantly overexpressed in breast carcinoma tissues compared with normal tissues. Expression Quantitative Trait Loci (eQLT) analysis of SNP rs1800797 in cultured fibroblasts represents that there may be a significant difference in the gene expression of the variant allele carrier gene compared with the wild allele carrier gene.
4.1. Limitations
There are several potential limitations to this case‐control study that must be explained. To begin, if we had included some other SNPs of this gene that have an impact on breast cancer, we would have obtained a broader and more precise link between IL‐6 rs1800797 variant and breast carcinoma. Second, the small sample size might not portray the entire image of all patients with breast cancer in Bangladesh. Finally, demographic characteristics were insufficient to accurately represent the real‐world scenario of breast cancer patients and their relationship with the genotype distribution of the respective gene. As a result, larger sample size studies with additional clarified environmental exposure data are essential to validate the prognostic and therapeutic value of this research work.
5. CONCLUSION
This study has discovered a strong connection between the IL‐6 rs1800797 genetic variant and breast cancer susceptibility in Bangladeshi women. It provides a basis for further investigation into genes directly associated with breast cancer. The findings should be confirmed through a large‐scale study to provide substantial evidence.
AUTHOR CONTRIBUTIONS
Mohima Khanom: Investigation; methodology; validation; writing—original draft. Md. Shafiul Hossen: Formal analysis; investigation; software; writing—original draft; writing—review and editing. Md. Abdul Barek: Formal analysis; methodology; project administration; software; writing—original draft; writing—review and editing. Md. Shuvo Ahamed: Methodology; validation; visualization; writing—original draft. Md. Sohanur Alam: Data curation; investigation; methodology; validation; visualization. Khokon Kanti Bhowmik: Methodology; software; validation; writing—original draft; writing—review and editing. Sarah Jafrin: Formal analysis; project administration; resources; software; validation; writing—original draft. Md. Abdul Aziz: Data curation; methodology; resources; writing—original draft; writing—review and editing. Mohammad Safiqul Islam: Conceptualization; formal analysis; funding acquisition; resources; software; supervision; writing—original draft; writing—review and editing.
CONFLICT OF INTEREST STATEMENT
Dr. Mohammad Safiqul Islam, a coauthor of this article, is a member of the Editorial Board of Health Science Reports. However, to minimize bias, he was excluded from any editorial decision‐making related to the acceptance of this article for publication. The remaining authors declare no conflict of interest.
TRANSPARENCY STATEMENT
The lead author Mohammad Safiqul Islam affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
This research project was partially funded by the University Grants Commission, Bangladesh (ID: 37.01.0000.073.04.019.22.713).
Khanom M, Hossen MS, Barek MA, et al. The linkage between IL‐6 rs1800797 variant and breast cancer susceptibility in Bangladeshi women: A case‐control study. Health Sci Rep. 2024;7:e1875. 10.1002/hsr2.1875
DATA AVAILABILITY STATEMENT
The data supporting this study's findings are available from the corresponding author upon reasonable request. The corresponding author takes complete responsibility for the integrity of the data and the accuracy of the data analysis.
REFERENCES
- 1. Sahaba SA, Rashid MA, Islam MS, et al. The link of ERCC2 rs13181 and ERCC4 rs2276466 polymorphisms with breast cancer in the Bangladeshi population. Mol Biol Rep. 2022;49:1847‐1856. [DOI] [PubMed] [Google Scholar]
- 2. Park M, Kim D, Ko S, Kim A, Mo K, Yoon H. Breast cancer metastasis: mechanisms and therapeutic implications. Int J Mol Sci. 2022;23(12):6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kashyap D, Pal D, Sharma R, et al. Global increase in breast cancer incidence: risk factors and preventive measures. BioMed Res Int. 2022;2022:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4. Akter T, Aziz MA, Islam MS, Sarwar MS. Association of MMP1 gene polymorphisms with breast cancer risk: a narrative review. Health Sci Rep. 2023;6:e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yedjou CG, Sims JN, Miele L, et al. Health and racial disparity in breast cancer. Adv Exp Med Biol. 2019;1152:31‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banerjee DK, Seijo Lebrón A, Baksi K. Glycotherapy: a new paradigm in breast cancer research. Biomolecules. 2022;12:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rudolph A, Chang‐Claude J, Schmidt MK. Gene–environment interaction and risk of breast cancer. Br J Cancer. 2016;114:125‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klassen CL, Gilman E, Kaur A, Lester SP, Pruthi S. Breast cancer risk evaluation for the primary care physician. Cleve Clin J Med. 2022;89:139‐146. [DOI] [PubMed] [Google Scholar]
- 9. Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peng X, Shi J, Sun W, et al. Genetic polymorphisms of IL‐6 promoter in cancer susceptibility and prognosis: a meta‐analysis. Oncotarget. 2018;9:12351‐12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka T, Narazaki M, Kishimoto T. IL‐6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect Biol. 2014;6:a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka T, Narazaki M, Kishimoto T. Interleukin (IL‐6) immunotherapy. Cold Spring Harbor Perspect Biol. 2017;10:a028456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Felcher CM, Bogni ES, Kordon EC. IL‐6 cytokine family: a putative target for breast cancer prevention and treatment. Int J Mol Sci. 2022;23:1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aziz MA, Jafrin S, Islam MS, Kabir Y. Interleukins in the development and progression of breast cancer. In: Interdisciplinary Cancer Research. Springer; 2022:1‐22. [Google Scholar]
- 15. Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8:110635‐110649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang S, Narazaki M, Metwally H, Kishimoto T. Historical overview of the interleukin‐6 family cytokine. J Exp Med. 2020;217:e20190347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korobeinikova E, Ugenskiene R, Insodaite R, et al. Association of angiogenesis and inflammation‐related gene functional polymorphisms with early‐stage breast cancer prognosis. Oncol Lett. 2020;19:3687‐3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Markkula A, Simonsson M, Ingvar C, Rose C, Jernström H. IL6 genotype, tumour ER‐status, and treatment predicted disease‐free survival in a prospective breast cancer cohort. BMC Cancer. 2014;14:759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masjedi A, Hashemi V, Hojjat‐Farsangi M, et al. The significant role of interleukin‐6 and its signaling pathway in the immunopathogenesis and treatment of breast cancer. Biomed Pharmacother. 2018;108:1415‐1424. [DOI] [PubMed] [Google Scholar]
- 20. Islam MS, Ahmed MU, Bin Sayeed MS, et al. Lung cancer risk in relation to nicotinic acetylcholine receptor, CYP2A6 and CYP1A1 genotypes in the Bangladeshi population. Clin Chim Acta. 2013;416:11‐19. [DOI] [PubMed] [Google Scholar]
- 21. Wacholder S, Chanock S, Garcia‐Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Islam MR, Aziz MA, Shahriar M, Islam MS. Polymorphisms in IL‐17A gene and susceptibility of colorectal cancer in Bangladeshi population: a case‐control analysis. Cancer Control. 2022;29:107327482211438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morgan JF. p Value fetishism and use of the Bonferroni adjustment. Evid Based Ment Health. 2007;10:34‐35. [DOI] [PubMed] [Google Scholar]
- 24. Sa‐nguanraksa D, Suntiparpluacha M, Kulprom A, et al. Association of estrogen receptor alpha and interleukin 6 polymorphisms with lymphovascular invasion, extranodal extension, and lower disease‐free survival in thai breast cancer patients. Asian Pac J Cancer Prev. 2016;17:2935‐2940. [PubMed] [Google Scholar]
- 25. Barek MA, Begum M, Noor F, Aziz MA, Islam MS. The link between IL‐6 rs2069840 SNP and cancer risk: evidence from a systematic review and meta‐analysis. Meta Gene. 2021;30:100972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data supporting this study's findings are available from the corresponding author upon reasonable request. The corresponding author takes complete responsibility for the integrity of the data and the accuracy of the data analysis.


