The construction of clean and unmarked mutations in bacteria, where a gene is replaced by an in vitro-modified allele is a fundamental approach to the understanding of pathogenicity at a molecular level, the definition of structure-function relationships, and the production of vaccine candidates. With the increasing availability of complete bacterial genome sequences, the potential for such mutagenesis has grown exponentially. However, to date a great number of open reading frames (ORFs) remain unannotated since they present no homology with sequences already present in the databases (9, 13). The precise function of some of these unknown genes will probably be deduced through in silico predictions, by comparing different genomes, or by the use of modern genetic strategies such as serial analysis of gene expression, in vivo expression technology, representational difference analysis, and signature tagged mutagenesis that help in extraction of information without a priori knowledge of the sequence (9, 17). Nevertheless, reverse genetic analysis, one of the logical approaches to be undertaken, will be necessary to identify a phenotype and to attribute a precise function to many undefined ORFs. Reverse genetics is a powerful approach for the identification of gene function, in which the gene of interest is mutated or inactivated to study the resulting effects on the microorganism (17).
Although allelic exchange is easy to perform with many bacteria, it remains very difficult or impractical with others. The classical method of using a suicide plasmid that is unable to replicate in the studied strain to deliver an inactivated allele of the gene in the chromosome is often not efficient because the frequency of double crossover events may be low and because illegitimate recombination may occur (30). Consequently, allelic exchange mutants may represent only a small fraction of the transformants and may be difficult to isolate. Counterselectable markers are often instrumental for the construction of such mutants, especially in microorganisms for which the genetics schemes is poorly developed. Under appropriate growth conditions, a counterselectable gene promotes the death of the microorganisms harboring it (49). Hence, transformants which have integrated a suicide vector containing a counterselectable marker, either by a single event of homologous or illegitimate recombination, retain a copy of the counterselectable marker in the chromosome and are therefore eliminated in the presence of the counterselective compound.
Consequently, counterselectable markers have been used for the positive selection of mutants that have undergone defined genetic alterations leading to the loss of the marker. In different studies, applications such as the construction of mutants, the isolation of insertion sequence (IS) elements, and the curing of plasmids have been described. The most-used counterselectable markers are the genes that confer sucrose, streptomycin, or fusaric acid sensitivity. They have been used to construct mutants or vaccine strains in Mycobacterium tuberculosis, Helicobacter pylori, Bordetella pertussis, and many other bacteria.
Here we provide a short review of the situations in which the use of a counterselectable marker has proven to be particularly advantageous.
COUNTERSELECTABLE MARKERS
The goal of this review is not to give an exhaustive list of the available counterselectable markers but rather to describe those that have proved to be the most useful. Three of the most popular ones are described in some detail below (for a comprehensive list, see Table 1 and reference 2).
TABLE 1.
Some counterselectable markers and their mechanisms of toxicity
| Counterselectable marker | Description |
|---|---|
| sacB | B. subtilis gene encoding levansucrase that converts sucrose to levans, which is harmful to the bacteria (14) |
| rpsL(strA) | Encodes the ribosomal subunit protein (S12) target of streptomycin (10) |
| tetAR | Confers resistance to tetracycline but sensitivity to lipophilic compounds (fusaric and quinalic acids) (27) |
| pheS | Encodes the α subunits of Phe-tRNA synthetase, which renders bacteria sensitive to p-chlorophenylalanine, a phenylalanine analog (21) |
| thyA | Encodes thymidilate synthetase, which confers sensitivity to trimethoprim and related compounds (47) |
| lacY | Encodes lactose permease, which renders bacteria sensitive to t-o-nitrophenyl-β-d-galactopyranoside (32) |
| gata-1 | Encodes a zinc finger DNA-binding protein which inhibits the initiation of bacterial replication (53) |
| ccdB | Encodes a cell-killing protein which is a potent poison of bacterial gyrase (3) |
The fusaric acid sensitivity system.
The first counterselectable marker described was tetAR, a gene coding for tetracycline resistance or sensitivity. Expression of tetAR results in alteration of the host cell membrane which interferes with tetracycline permeation and thereby renders the cell resistant to tetracycline. These alterations render the bacteria hypersensitive to lipophilic chelating agents such as fusaric or quinalic acids (5, 27). It is therefore possible to select clones which have lost the tetAR gene by virtue of their resistance to fusaric acid. This system is, to the best of our knowledge, effective only in Escherichia coli, in which the threshold of fusaric acid sensitivity is strongly dependent on the host strain. As a consequence, this marker was mostly used for abrogating tetracycline resistance in E. coli that had been previously mutagenized with a transposon harboring a tetracycline cassette (27). One problem encountered with this system is the generation of out-of-frame deletions with polar effects on downstream genes, which obscure the interpretation of the phenotype.
The streptomycin sensitivity system.
The streptomycin sensitivity system takes advantage of the fact that the S12 ribosomal protein is the target of streptomycin, a widely used antibiotic. Mutations in the rpsL gene which encodes this protein are responsible for resistance to high concentrations of streptomycin. However, resistance is recessive in a merodiploid strain (24). When both wild-type and mutant alleles of rpsL are expressed in the same strain, the strain is sensitive to streptomycin, possibly because of a general inhibition of translation by the wild-type ribosome. Consequently, it is possible to select mutants that have lost the wild-type allele encoding streptomycin sensitivity by plating the culture on streptomycin (10). Positive selection based on streptomycin sensitivity functions in different bacteria, such as E. coli (10), B. pertussis (50), or Mycobacterium smegmatis (45). A prerequisite for this technology to be effective is a streptomycin-resistant strain. Therefore, the use of rpsL as a counterselectable marker leads to mutants resistant to streptomycin, an antibiotic which is used in some chemotherapeutic regimens (56). This is a severe limitation, particularly in the construction of attenuated strains meant to be used as live vaccine candidates.
The sucrose sensitivity system.
The Bacillus subtilis sacB gene coding for levansucrase is perhaps the most popular counterselectable marker. In its natural gram-positive environment its expression is harmless to the bacterium. However, cloning of sacB in E. coli and other gram-negative bacteria (15) leads to the death of the transformed bacteria when they are plated in the presence of sucrose (48). The mechanism of this toxicity is not completely understood, but it has been proposed that the accumulation of levans (high molecular-weight fructose polymers synthesized by the levansucrase) in the periplasm of gram-negative bacteria might be toxic (48). The effect of the sacB gene in the presence of sucrose has been demonstrated in most of the gram-negative bacteria in which it has been tested, including Erwinia chrysanthemi, Legionella pneumophila, Anabaena sp., Yersinia sp., Rhizobium sp., Xanthomonas sp., Pseudomonas aeruginosa, Klebsiella pneumoniae, H. pylori, and Agrobacterium tumefaciens. The same mechanism of toxicity has been invoked for Corynebacterium glutamicum (19) and Mycobacterium sp. (36), the only gram-positive bacteria for which sacB was shown to be functional as a counterselectable marker.
CREATION OF CHROMOSOMAL MUTATIONS
Allelic exchange mutagenesis and site-directed mutagenesis.
Any gene to be studied can be interrupted by the insertion of an antibiotic cassette, but this strategy itself may cause a mutant phenotype. Indeed, bacterial genes involved in a particular pathway tend to be grouped in cotranscribed clusters or operons. Insertional frameshift, nonsense or antisense disruption of an ORF within an operon, can therefore affect upstream or downstream gene expression in addition to the targeted gene. The so-called polar effects, also due to RNA instability, could confuse the assignment of a mutant phenotype to the disrupted gene. In fact, when a Kmr gene was inserted into the E. coli hdeA gene at two different locations and in two orientations, both essential and nonessential phenotypes were seen (26). In order to minimize the polar effect, Link and coworkers adapted a method for use with yeast, called “pop-out” mutagenesis, which allows the introduction of in-frame deletions (26). This method demonstrated unambiguously that the hdeA gene was nonessential and that the apparent lethal effect of the Kmr insertion was certainly due to an effect on neighboring genes. However, it should not be considered that in-frame deletions are never polar since polar effects can be observed when cis-acting element are deleted. Nevertheless, the use of similar in-frame deletion systems could overcome these regulation problems and help to unravel the multilayered networks that control gene expression.
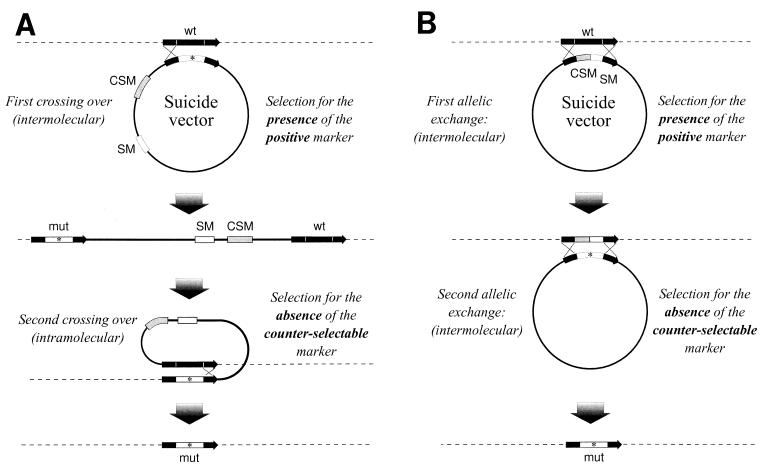
The gene to be studied can also be mutagenized in a specific subregion in order to study the function of a particular domain of the protein. This possibility of constructing unmarked mutations may be used to introduce several mutations into a single gene or into a single strain independently of the number of selectable markers available. This methodology permits disruption of a protein’s function without affecting its antigenic properties, thereby enabling construction of attenuated strains with vaccine potential. In this case counterselectable markers are essential. Two strategies known as single or double selection have been described (Fig. 1).
FIG. 1.
(A) Double-selection strategy. Positive selection of allelic exchange mutants in a two-step selection strategy, using a counterselectable marker. (B) Single-selection strategy. CSM, counterselectable marker; SM, selectable marker; WT, wild-type allele; mut, mutated allele.
Double selection is a more general strategy since there is no prerequisite of constructing a recipient strain carrying the counterselectable marker inside the gene to be studied (20, 37, 38). A double selection in two steps allows the construction of defined unmarked mutants with changes that are usually undetectable, such as point mutations or in-frame deletions. In the first step, clones that have integrated the vector by a single crossover are selected owing to an antibiotic resistance gene and are propagated to allow for a second crossover. Clones that have lost the vector during a second crossover are selected by plating on counterselective medium. When the selectable marker is inserted inside the target gene, mutants can be selected directly when selection and counterselection are applied simultaneously; however, a two-step selection is often more efficient (Fig. 1A). This two-step method has been successfully applied for construction of an eae (effacing and attaching effect) deletion mutant of enteropathogenic E. coli by using a vector with the counterselectable property of sacB (11). Without a counterselectable marker, such an event is detected by the laborious screening of tens of thousands of colonies for sensitivity to the antibiotic originally used in selection.
Single selection consists in the introduction of the counterselectable marker together with a selectable marker into the gene to be mutated (43). Allelic exchange results in a strain in which the target gene has been interrupted by the selectable-counterselectable cassette. This strain is used subsequently as a host for introducing different mutations into the selected gene. Allelic exchange within the mutated gene results in the loss of the cassette (Fig. 1B).
Another method which combines the selective properties of a counterselectable marker and a temperature-sensitive replicon has been developed. The strain is first transformed at a permissive temperature and selected for the integration of the plasmid by growing the transformants at a nonpermissive temperature. In the second step, by plating in the presence of the counterselective compound it is possible to positively select for both plasmid excision and curing. This protocol permitted highly efficient gene exchange in both E. coli (4) and M. tuberculosis (35).
Transposon mutagenesis.
Despite the similarities between allelic exchange and transposon mutagenesis, counterselectable markers have rarely been used for transposon mutagenesis (12, 25, 35). Clearly, all the above-described double-selection strategies can be used for transposon mutagenesis if the vector used to deliver the transposon is lost, or in other words, when the mobile element transposes in a conservative fashion, or when the cointegrate is resolved by a specific resolvase or by homologous recombination.
Study of mutant alleles of essential genes.
Genes essential for bacterial life cannot be disrupted unless a complementing wild-type copy of the gene is present in the cell or conditionally lethal mutants with a thermosensitive phenotype are obtained. Mutational analysis is however complicated by the fact that these mutants may retain some function and are consequently not null alleles. An elegant method to overcome these problems is to bracket the complementing copy of the studied gene by two counterselectable markers (32). The efficiency of a double counterselection (with both counterselectable markers) is very high and permits events with a frequency of 10−9 to 10−10 to be recovered. Thus, any cell surviving the two selections must either have lost the plasmid or have suffered a deletion which simultaneously deleted both genes (encoding the counterselectable markers) plus the studied gene. In either case, the selection would generate cells that had lost the studied gene. Extremely rare plasmidfree mutants with bypass suppressors of a null missense allele or intragenic pseudorevertants can be selected simply by plating on double-counterselection medium. This approach could also be used to test the ability of defined mutant alleles to complement an essential function.
Chromosomal deletions.
Transposons carrying a counterselectable marker can be used to assess genomic stability. Mutants in which the transposon has integrated into the chromosome can be propagated in liquid culture, and clones arising from the spontaneous deletion of the transposon can be selected on counterselective medium. The frequency at which these mutants appear reflects the genetic stability of the region in which the transposon has integrated. A transposon containing sacB was used to demonstrate that Xanthomonas campestris pv. campestris, a xanthan gum-producing bacterium which was thought to be genetically unstable, was as stable as E. coli (29).
A novel approach, termed island probing, has recently been described for Shigella flexneri (41). The she gene was insertionally disrupted by allelic exchange with the use of a Tn10-derived tetAR cassette. Then, to detect loss of the she locus, the tetracycline-resistant derivative was plated onto fusaric medium to select for the tetracycline-sensitive revertant, which was observed to arise at a frequency of 10−5 to 10−6. This new method provides a useful tool for studying the stability of pathogenicity islands in general (41).
Identification of positive regulatory regions.
The use of counterselectable markers are of interest for analysis of the expression of defined genes, as demonstrated with Rhodobacter sphaeroides for analysis of the regulation of the puc operon, which encodes proteins involved in photosynthesis. A promoterless sacB gene was fused to the puc genes and expressed under the control of their expression signals (44). Clones with reduced levels of levansucrase expression were positively selected by virtue of their ability to survive in the presence of sucrose. Both cis and trans mutations have been detected; cis mutations, which were located in the regulatory region, hindered the transcription of the operon or the interaction with a positive regulator, whereas trans mutations were located elsewhere in the chromosome and corresponded to the disruption of regulatory genes. This finding led to the identification of the mgpS locus, which controls the expression of the puc operon (44).
SELECTION OF PLASMID ALTERATIONS
Curing or deleting plasmids.
Plasmids can encode functions that are very important for pathogenicity, as in the case of Shigella or Salmonella spp., and also for symbiosis, as in Rhizobium spp. Plasmid-containing strains from the wild rarely segregate plasmidfree derivatives. It is therefore often necessary to cure the plasmid from its host bacterium to prove that it encodes a gene responsible for a given phenotype. Curing of some stably maintained plasmids may be difficult to achieve. Moreover, most curing treatments are also mutagenic, so that a plasmidfree strain might not be isogenic to the initial strain, rendering the phenotypic analysis uncertain. An elegant solution is to use a transposon carrying a counterselectable marker. Tn5-derived mini-transposons, containing rpsL or sacB, were used to cure plasmids from Salmonella (51) and Rhizobium (18) strains in two-step selection protocols. First, a mutant in which the mini-transposon has integrated into the plasmid to be cured is selected either by DNA-DNA hybridization with the mini-transposon as a probe or by exconjugation. Then, the mutant is propagated in liquid culture and plated on counterselective medium, streptomycin, or sucrose. Normally, only a few mutants able to survive on counterselective medium need to be screened in order to isolate plasmid-cured strains.
Counterselectable markers can also be used for the selection of mutants harboring deletions or inversions within a plasmid that can result from the recombination between two palindromic sequences (55) and also from the intramolecular transposition of mobile elements such as Tn5 or γδ. Based on this methodology, a counterselectable cosmid, pDUAL, representing a “deletion factory,” was constructed. pDUAL, with an origin of replication within a mini-γδ-transposon, contains a multiple cloning site where DNA inserts can be cloned and bracketed by two selectable and two counterselectable markers (54). Upon in vivo random intramolecular transposition, two daughter molecules, each containing a part of the original insert and one of the two counterselectable markers, are formed. It is easy to recover, on an appropriate counterselective medium, a set of nested deletions extending from the side that contains the counterselectable marker that is not selected against. pDUAL is thus a powerful tool for genome sequencing projects which require time- and cost-effective methods. Both commonly used in vitro methods for sequencing large DNA regions, shotgun subcloning and primer walking, have drawbacks in terms of cost, efficiency, ease of gap closure, and aligning of repeated sequences. On the contrary, sets of nested deletions generated in a “deletion factory” can be easily sequenced with the use of a unique pair of primers that are just interior to the transposon ends (54).
Isolation of mobile elements.
Insertion sequences (IS) play an important role in the plasticity and evolution of bacterial genomes. Moreover, IS are useful tools for monitoring the epidemiology of pathogenic strains by restriction fragment length polymorphism techniques and also for random mutagenesis after it has been engineered to contain an antibiotic resistance cassette. The detection of an IS that possesses no detectable genes which could allow its positive selection is very tedious and often fortuitous. Classically, ISs have been detected by their capacity to induce mutations or by the detection of plasmids or phages with increased molecular weights. The simplest solution is to use a transposon trap consisting of a replicative vector harboring a counterselectable marker (14). The vector is propagated in the bacterium for a period of time during which resistant mutants, in which an IS has transposed into the counterselectable marker, may appear. These mutants are positively selected by plating onto counterselective medium (14, 46). The power of this technology is particularly well illustrated by the study of Simon et al., who reported the isolation of 20 different ISs from various gram-negative bacteria (46).
Selection of recombinant vectors.
In DNA cloning experiments, nonrecombinant transformants are often an annoying problem, especially when the amount of insert DNA is small or when the percentage of recombinants is suboptimal, as in the construction of gene libraries. Although phosphatase treatment of the vector ends may reduce the proportion of nonrecombinants by preventing the vector from religating, dephosphorylation presents the penalty of reduced ligation frequencies. However, if the vector’s cloning sites are within a counterselectable marker or within its promoter, the counterselectable marker will be disrupted by DNA inserted, and transformants will be able to grow on counterselective medium.
Most of the early-described positive selection vectors were severely limited because they required specialized strains and presented a limited range of available cloning sites, limitations which made the recovery of the inserted fragments difficult (6). Nevertheless, some recently described vectors are more suitable because they meet all the requirements for an ideal E. coli cloning vector. They (i) are small amplifiable plasmids, (ii) carry a counterselectable marker, and (iii) contain several restriction sites suitable for cloning. One such vector, pKSS, a 4.1-kbp derivative of pBluescript, contains the pheS gene (21). pKSS gives reproducibly high levels of recombinants (more than 99%) and is therefore an interesting alternative to classical cloning vectors, especially in constructing DNA libraries. A positive selection cosmid, pAd10-SacBII, which uses the bacteriophage P1 system and sacB as a counterselectable marker (39), has also been described. The P1 system allows in vitro packaging of foreign DNA inserts as large as 95 kbp. This cosmid vector is particularly useful because it also contains rare restriction sites bracketing the insert and thus allows the characterization of its size by pulsed-field gel electrophoresis (39).
USE OF COUNTERSELECTABLE MARKERS TO DECIPHER MECHANISMS OF PATHOGENICITY AND TO CONSTRUCT VACCINES
B. pertussis.
A double-selection strategy has been used to construct a B. pertussis strain that is presently used to produce an acellular vaccine against whooping cough. This strain produces a genetically detoxified form of the pertussis toxin (PT-9K/129G), an AB toxin composed of five subunits (40). The wild-type molecule, a major virulence factor of B. pertussis, could not be used as such in vaccines because of its high toxicity. In order to render the molecule nontoxic, two approaches have been used: chemical detoxification of the purified molecule by addition of formaldehyde, glutaraldehyde, hydrogen peroxide or other chemicals and in vitro mutagenesis of the toxin gene to eliminate the enzymatic activity (31).
To construct a strain producing a nontoxic form of the toxin, the five genes coding for the PT operon were cloned in E. coli and unmarked mutations were introduced to knock-out the ADP-ribosylating activity responsible for the toxicity. Two amino acid substitutions in the S1 subunit, Arg-9→Lys and Glu-129→Gly, were finally selected because they were able to eliminate the enzymatic activity without altering the structure or stability of the molecule. The in vitro-mutagenized PT operon was then reintroduced into the B. pertussis chromosome in a two-step approach. First, the wild-type gene was deleted from the chromosome and replaced by a kanamycin resistance cassette. This replacement was performed by allelic exchange with a suicide vector which was unable to replicate in B. pertussis that contained a selectable marker gene (gentamicin resistance cassette) and a counterselectable marker (rpsL) (50). Following conjugation into B. pertussis and selection for gentamicin and kanamycin resistance, integrons of the suicide plasmid into the chromosome were isolated. The plasmid was then eliminated from the chromosome by plating onto streptomycin and selecting for Strr Gms. During the second step of allelic replacement, the kanamycin resistance cassette was replaced with a nontoxic allele of the PT gene harboring the two amino acid substitutions. The strain obtained is used in the industrial production of a genetically detoxified toxin included in the acellular B. pertussis vaccine. This molecule has been shown to be more immunogenic than the chemically detoxified PT and to induce excellent protection from the disease (16, 42).
H. pylori.
H. pylori is a gram-negative microaerophilic bacterium involved in the development of acute gastritis, peptic ulcers, and gastric adenocarcinomas in humans (33). In the pregenomic area a number of virulence factors, such as a urease, various adhesins, a vacuolating cytotoxin (VacA), and the cag region, have been identified. The recently completed sequence of the genome (52) has revealed some additional virulence factors such as other putative adhesins, enzymes involved in lipopolysaccharide biosynthesis, pilin, and proteins involved in the scavenging of iron. Careful analysis of the nucleotide sequence has also showed stretches of dinucleotide repeats as well as poly(C) or poly(G) tracts, suggesting the possibility of antigenic variation (52). Due to their importance in H. pylori pathogenesis, these genes are priority targets for reverse genetic analysis.
A sucrose-based allelic exchange system has been reported for H. pylori (8), allowing the creation of unmarked mutations that are helpful in deciphering the role of some virulence genes in the pathogenesis of this bacterium. A cassette, consisting of sacB and kanamycin resistance genes under the control of the H. pylori flagellin promoter, was introduced by homologous recombination into the vacA gene. The resulting strain was sucrose sensitive and kanamycin resistant. In this regard, this strategy is identical to the strategy described by Ried and Collmer (43). Following transformation with a mutated allele, growth on sucrose-containing medium allowed the selection of strains that had lost the kan-sacB module and had integrated the unmarked allele. This strategy was used to perform a site-directed modification of two histidine residues of the vacA gene (8) but can be used to modify any cloned H. pylori gene. Consecutive rounds of mutagenesis can be performed to create strains with the desired genetic background, without introducing multiple antibiotic-resistant markers. Furthermore, this gene replacement system can be used to create nonpolar mutations or deletions in putative virulence factors, to modify various promoter sequences, and to introduce high-affinity antibody tags facilitating protein purification and immunohistochemical analysis.
M. tuberculosis.
More than three million people die annually from tuberculosis. The number of deaths result in part from the low efficacies of the antituberculous vaccines and the requirement of a long-term treatment. In this regard, the complete sequence of the M. tuberculosis genome, that is soon expected to be completed, promises to be of invaluable interest. Mycobacterial molecular genetics has lagged behind the genomic advances of other microorganisms because of the difficulty of generating mutants (30). The recent demonstration of the use of counterselectable markers in Mycobacterium sp. will greatly facilitate the genetic analysis of this important pathogen.
Two of the most popular counterselectable markers, rpsL and sacB, have been successfully adapted to mycobacteria for the positive selection of allelic exchange events. Double-selection methods with rpsL or sacB as a counterselectable marker are very efficient with M. smegmatis (34, 37, 45). Two-step selection with sacB (37) or pyrF, which confers susceptibility to 5-fluoroorotic acid (22), enabled the construction of mycobacterial allelic exchange mutants bearing unmarked mutations. Although the above-described markers should be usable for the mutagenesis of mycobacteria from the M. tuberculosis complex, sacB is, to the best of our knowledge, the only one for which it has been demonstrated (1, 38).
The above methodologies, using suicide delivery vectors, have however two severe limitations. They are very dependent on the target gene and are less efficient with M. tuberculosis. The detection of rare allelic exchange events in the tubercle bacillus is indeed hindered by the low transformation efficiencies observed when suicide vectors are used and by the high frequencies of illegitimate recombination. To circumvent these problems a series of replicative thermosensitive delivery vectors harboring the sacB gene was recently constructed (35). By allowing the delivery vector to replicate, allelic exchange mutants can accumulate and problems arising from low transformation efficiencies are overcome. By plating on counterselective medium at nonpermissive temperature, allelic exchange mutants can be directly selected. When these vectors were used in M. tuberculosis to deliver a mutated allele of purC, a gene from the purine biosynthetic pathway that was refractory to mutagenesis by previously available methods, all the selected clones were allelic exchange mutants. Interestingly, it was recently demonstrated that these mutants displayed an attenuated virulence both in vitro and in mice and could therefore have potential as vaccine candidates. Moreover, this ts-sacB system was also used to deliver a transposon into the chromosome of M. tuberculosis, permitting the construction of the first insertion mutant library described for this species (35).
Unmarked, attenuated mutants as live vaccines candidates.
Live attenuated vaccines have in several cases, notably shigellosis and tuberculosis, been shown to be more effective at eliciting a protective immune response than killed bacteria. An advantage of live vaccines is that an immune response is induced against multiple antigens, some of which may be expressed only in vivo. Another reason for using live bacteria is that when no obvious virulence factor is known, the design of subunit vaccines remains impractical. Moreover, attenuated strains expressing heterologous antigens might be used as multivalent vaccines. Originally, attenuated strains were obtained by serial passage in vitro, as is the case for the vaccines against tuberculosis (Mycobacterium bovis BCG), polio, and smallpox. The main limitation of this strategy is that the mutations that attenuate the virulence are very difficult to characterize; therefore, this method is no longer acceptable to regulatory agencies for vaccine purposes.
A more rational solution is to identify virulence factors or dispensable metabolic pathways and to impair their function by producing a well-defined mutation which renders the bacteria avirulent (40). Both vaccine development and the understanding of pathogenesis benefit from the construction and the study of these mutants. Usual targets for this approach are genes encoding enzymes of the purine biosynthesis pathway. Purine auxotrophy results in reduction of the virulence of intracellular bacteria such as Salmonella, Listeria, Shigella, and Mycobacterium because of the low availability of purines in the eukaryotic cell. Unmarked attenuated strains can be constructed by homologous recombination or by using a transposon mutagenesis system. However, in addition to carrying antibiotic-resistant genes, transposons may also excise or induce genetic rearrangements within the strain’s genome; therefore, these strains cannot be used as vaccines in human trials. The transposon has to be cured on a specific counterselectable medium, allowing the selection of unmarked mutants that are more suitable for vaccine development.
Currently, a vaccine candidate based on an icsA mutant of S. flexneri, the etiologic agent of dysenteric syndrome, is undergoing clinical trials at Fort Detrick, Md. Interruption of the icsA gene, which is required for the actin-based spreading of the bacterium from cell to cell, significantly reduced the survival capacity of S. flexneri in vivo. This icsA mutation was combined with a deletion of the genes encoding aerobactin production (siderophore) that was produced by curing of a Tn10 insertion located in the iucABCD operon. This icsA Δiuc mutant is well tolerated and induces significant protective immunity in humans (45a). It is interesting that the sacB technology was not efficient for the mutagenesis of the icsA gene, which is present on a virulence plasmid, because the frequency with which the virulence plasmid was lost was too high.
Attenuated unmarked mutants are also used as veterinary vaccines. A vaccine against turkey colibacillosis has been obtained by introducing a mutation in the carAB gene, which resulted in arginine and pyrimidine auxotrophies (23). The carAB mutant was obtained by transposon mutagenesis with a Tn10-based system and subsequent curing of the transposon on Bochner’s medium (5). The resulting E. coli strain, shown to be tetracycline sensitive and to require arginine and uracil for growth, was avirulent in turkeys. Four-week-old turkey poults vaccinated orally with the carAB mutant were completely protected against a challenge with the virulent wild-type strain (23). It has been suggested that the low level of pyrimidines in animal tissues may cause the induction of the carAB operon and that failure to produce carbamoylphosphate synthetase may result in the observed attenuation.
Mutations in the genes required for synthesis of aromatic amino acids have also been shown to result in the attenuation of the virulence. For example, an aroA mutant of Aeromonas salmonicida, the etiologic agent of furonculosis in rainbow trout, has been constructed and is being analyzed for its vaccine potential. This mutant was obtained by integration of a suicide vector harboring an internal deletion of the aroA gene (ΔaroA) (28). This mutation was achieved via a single crossover, followed by the search for clones that had excised the plasmid during a second recombination which resulted in allele replacement. These kinds of events are very rare and were therefore very tedious to detect. The construction of this unmarked mutant could have greatly benefited from the use of a counterselectable marker.
Counterselectable markers have also been used to construct strains expressing heterologous antigens. Heterologous vaccination takes advantage of the adjuvant immunogenicity of the antigen carrier to develop long-term immunity. Attenuated mutants of Vibrio cholerae or Salmonella are particularly effective organisms for the delivery of heterologous antigens in order to stimulate a common mucosal immune response. For example, stxB, the gene encoding the B subunit of Shiga-like toxin I, was cloned under the transcriptional control of the iron-regulated irgA promoter and introduced into an attenuated strain of V. cholerae by gene exchange with the use of a suicide vector containing the sacB gene (7). The production of StxB was shown to be tightly iron regulated and greater than in the reference strain S. dysenteriae 60R. This method can be used to express other antigens of interest in V. cholerae under control of the irgA promoter. Heterologous antigens may also be expressed under the control of a variety of other promoters, either constitutive or responsive to other environmental signals, in order to obtain the optimal expression system in humans.
CONCLUDING REMARKS
The number of counterselectable markers already in existence and their broad host range make it very likely that an effective marker will be found for every bacterial genus (2). As described above, counterselectable markers are useful tools for basic genetics, for the study of pathogenesis, and for genome analysis. Indeed, this simple methodology allows the positive selection of recombinant vectors, the isolation of IS elements, the selection of plasmid-cured strains, the study of mutant alleles of essential genes, the discovery of positive regulatory regions, and the construction of transposon delivery vectors. Hence, counterselectable markers represent a straightforward way to construct unmarked mutants, which are a prerequisite for assigning functions to the numerous unannotated ORFs in the genomic databases, for understanding the mechanisms of pathogenicity and for constructing live attenuated strains for vaccination. Therefore, with the development of genomics (9), counterselectable markers are likely to become favorites in the geneticist’s tool chest.
ACKNOWLEDGMENTS
J.-M. Reyrat and V. Pelicic contributed equally to this work.
We gratefully acknowledge J. L. Telford, A. Covacci, and C. Mallia for critical reading of the manuscript and helpful suggestions.
REFERENCES
- 1.Azad A K, Sirakova T D, Fernandes N D, Kolattukudy P E. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J Biol Chem. 1997;272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- 2.Berg C M, Berg D E. Transposable element tools for microbial genetics. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 2588–2612. [Google Scholar]
- 3.Bernard P, Gabant P, Bahassi E M, Couturier M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature sensitive pSC101 replicon. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 5.Bochner B R. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns D M, Beacham I R. Positive selection vectors: a small plasmid vector useful for the direct selection of Sau3A-generated overlapping fragments. Gene. 1984;27:323–325. doi: 10.1016/0378-1119(84)90077-5. [DOI] [PubMed] [Google Scholar]
- 7.Butterton J R, Boyko S A, Calderwood S B. Use of the Vibrio cholerae irgA gene as a locus for insertion and expression of heterologous antigens in cholera vaccine strain. Vaccine. 1993;11:1327–1335. doi: 10.1016/0264-410x(93)90103-5. [DOI] [PubMed] [Google Scholar]
- 8.Copass M, Grandi G, Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun. 1997;65:1949–1952. doi: 10.1128/iai.65.5.1949-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Covacci A, Kennedy G C, Cormack B, Rappuoli R, Falkow S. From microbial genomics to meta-genomics. Drug Dev Res. 1998;41:180–192. [Google Scholar]
- 10.Dean D. A plasmid cloning vector for the direct selection of strains carrying recombinant plasmids. Gene. 1981;15:99–102. doi: 10.1016/0378-1119(81)90108-6. [DOI] [PubMed] [Google Scholar]
- 11.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumais Pope C, Dhand L, Cianciotto N P. Random mutagenesis of Legionella pneumophila with mini-Tn10. FEMS Microbiol Lett. 1994;124:107–112. doi: 10.1111/j.1574-6968.1994.tb07269.x. [DOI] [PubMed] [Google Scholar]
- 13.Gabor-Miklos G L, Rubin G M. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 14.Gay P, Lecoq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gay P, Lecoq D, Steinmetz M, Ferrari E, Hoch J A. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greco D, Salmaso S, Mastrantonio P, Giuliano M, Tozzi A E, Anemona A, Ciofi degli Atti M L, Giammanco A, Panei P, Blackwelder W C, Klein D L, Wassilak S G. A controlled trial of two acellular vaccines and one whole-cell vaccine against pertussis. N Engl J Med. 1996;334:341–348. doi: 10.1056/NEJM199602083340601. [DOI] [PubMed] [Google Scholar]
- 17.Hensel M, Holden D W. Molecular genetic approaches for the study of virulence in both pathogenic bacteria and fungi. Microbiology. 1996;142:1049–1058. doi: 10.1099/13500872-142-5-1049. [DOI] [PubMed] [Google Scholar]
- 18.Hynes M F, Quandt J, O’Connell M P, Pühler A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene. 1989;78:111–120. doi: 10.1016/0378-1119(89)90319-3. [DOI] [PubMed] [Google Scholar]
- 19.Jäger W, Schäfer A, Pühler A, Labes G, Wohlleben W. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J Bacteriol. 1992;174:5462–5465. doi: 10.1128/jb.174.16.5462-5465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 21.Kast P. PKSS—a second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene. 1994;138:109–114. doi: 10.1016/0378-1119(94)90790-0. [DOI] [PubMed] [Google Scholar]
- 22.Knipfer N E, Shrader T E. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–383. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- 23.Kwaga J K P, Allan B J, van den Hurk J V, Seida H, Potter A A. A carAB mutant of avian pathogenic Escherichia coli serogroup O2 is attenuated and effective as a live oral vaccine against collibacillosis in turkeys. Infect Immun. 1994;62:3766–3772. doi: 10.1128/iai.62.9.3766-3772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lederberg J. Streptomycin resistance: a genetically recessive mutation. J Bacteriol. 1951;61:549–550. doi: 10.1128/jb.61.5.549-550.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsden M J, Vaughan L M, Foster T J, Secombes C J. A live (ΔaroA) Aeromonas salmonicida vaccine for furunculosis preferentially stimulates T-cell responses relative to B-cell responses in rainbow trout (Oncorhinchus mykiss) Infect Immun. 1996;64:3863–3869. doi: 10.1128/iai.64.9.3863-3869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Salazar J M, Palacios A N, Sánchez R, Caro A, Soberón-Chavez G. Genetic stability and xanthan gum production in Xanthomonas campestris pv. campestris NRRL B1459. Mol Microbiol. 1993;8:1053–1061. doi: 10.1111/j.1365-2958.1993.tb01650.x. [DOI] [PubMed] [Google Scholar]
- 30.McFadden J. Recombination in mycobacteria. Mol Microbiol. 1996;21:205–211. doi: 10.1046/j.1365-2958.1996.6271345.x. [DOI] [PubMed] [Google Scholar]
- 31.Moxon R, Rappuoli R. Haemophilus influenzae infections and whooping cough. Lancet. 1990;i:1324–1329. doi: 10.1016/0140-6736(90)91200-t. [DOI] [PubMed] [Google Scholar]
- 32.Murphy C K, Stewart E J, Beckwith J. A double counter-selection system for the study of null alleles of essential genes in Escherichia coli. Gene. 1995;155:1–7. doi: 10.1016/0378-1119(94)00920-n. [DOI] [PubMed] [Google Scholar]
- 33.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 34.Pavelka M S, Jr, Jacobs W R., Jr Biosynthesis of diaminopimelate (DAP), the precursor of lysine and a component of the peptidoglycan, is an essential function of Mycobacterium smegmatis. J Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelicic V, Jackson M, Reyrat J-M, Jacobs W R, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelicic V, Reyrat J-M, Gicquel B. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J Bacteriol. 1996;178:1197–1199. doi: 10.1128/jb.178.4.1197-1199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelicic V, Reyrat J-M, Gicquel B. Generation of unmarked directed mutations in mycobacteria, using sucrose counterselectable suicide vectors. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 38.Pelicic V, Reyrat J-M, Gicquel B. Positive selection of allelic exchange mutants in Mycobacterium bovis BCG. FEMS Microbiol Lett. 1996;144:161–166. doi: 10.1111/j.1574-6968.1996.tb08524.x. [DOI] [PubMed] [Google Scholar]
- 39.Pierce J C, Sauer B, Sternberg N. A positive selection vector for cloning high molecular weight DNA by the bacteriophage P1 system: improved cloning efficacy. Proc Natl Acad Sci USA. 1992;89:2056–2060. doi: 10.1073/pnas.89.6.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizza M, Covacci A, Bartoloni A, Perugini M, Nencioni L, de Magistris M T, Villa L, Nucci D, Manetti R, Bugnoli M, Giovannoni F, Olivieri R, Barbieri J T, Sato H, Rappuoli R. Mutants of pertussis toxin suitable for vaccine development. Science. 1989;246:497–500. doi: 10.1126/science.2683073. [DOI] [PubMed] [Google Scholar]
- 41.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappuoli R. Rational design of vaccines. The recombinant pertussis vaccine induces early and long-lasting protection. Nat Med. 1997;3:374–376. doi: 10.1038/nm0497-374. [DOI] [PubMed] [Google Scholar]
- 43.Ried J L, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 44.Sabaty M, Kaplan S. mgpS, a complex regulatory locus involved in the transcriptional control of the puc and puf operons in Rhodobacter sphaeroides. J Bacteriol. 1996;178:35–45. doi: 10.1128/jb.178.1.35-45.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sander P, Meier A, Böttger E C. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol Microbiol. 1995;16:991–1000. doi: 10.1111/j.1365-2958.1995.tb02324.x. [DOI] [PubMed] [Google Scholar]
- 45a.Sansonetti, P. J. Personal communication.
- 46.Simon R, Hötte B, Klauke B, Kosier B. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J Bacteriol. 1991;173:1502–1508. doi: 10.1128/jb.173.4.1502-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stacey K A, Simson E. Improved method for the isolation of thymine-requiring mutants of Escherichia coli. J Bacteriol. 1965;90:554–555. doi: 10.1128/jb.90.2.554-555.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinmetz M, Lecoq D, Ben Djemia H, Gay P. Analyse génétique de sacB, gène de structure d’une enzyme secrétée, la lévane-saccharase de Bacillus subtilis Marburg. Mol Gen Genet. 1983;191:138–144. doi: 10.1007/BF00330901. [DOI] [PubMed] [Google Scholar]
- 49.Stibitz S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 1994;235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 50.Stibitz S, Black W, Falkow S. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene. 1986;50:133–140. doi: 10.1016/0378-1119(86)90318-5. [DOI] [PubMed] [Google Scholar]
- 51.Stojiljkovic I, Trgovcevic Z, Salaj-Smic E. Tn5-rpsL: a new derivative of transposon Tn5 useful in plasmide curing. Gene. 1991;99:101–104. doi: 10.1016/0378-1119(91)90039-e. [DOI] [PubMed] [Google Scholar]
- 52.Tomb J-F, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 53.Trudel P, Provost S, Massie B, Chartrand P, Wall L. pGATA: a positive selection vector based on the toxicity of the transcription factor GATA-1 to bacteria. BioTechniques. 1996;20:684–693. doi: 10.2144/19962004684. [DOI] [PubMed] [Google Scholar]
- 54.Wang G, Blakesley R W, Berg D E, Berg C M. pDUAL: a transposon-based cosmid cloning vector for generating nested deletions and DNA sequencing templates in vivo. Proc Natl Acad Sci USA. 1993;90:7874–7878. doi: 10.1073/pnas.90.16.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weston-Hafer K, Berg D E. Specificity of deletion events in pBR322. Plasmid. 1989;21:251–253. doi: 10.1016/0147-619x(89)90050-4. [DOI] [PubMed] [Google Scholar]
- 56.World Health Organization. Drugs used in mycobacterial infections. Geneva, Switzerland: World Health Organization; 1991. [Google Scholar]