Abstract
Attention-deficit hyperactivity disorder (ADHD) and autism spectrum disorder (ASD) are highly heritable neurodevelopmental conditions with a considerable overlap in their genetic etiology. We dissected their shared and distinct genetic etiology by cross-disorder analyses of large datasets. We identified seven loci shared by the disorders and five loci differentiating them. All five differentiating loci showed opposite allelic directions in the two disorders and significant associations with other traits, e.g., educational attainment, neuroticism and regional brain volume. Integration with brain transcriptome data identified and prioritized several significantly associated genes. The shared genomic fraction contributing to both disorders was strongly correlated with other psychiatric phenotypes, while the differentiating portion correlated most strongly with cognitive traits. Additional analyses revealed that individuals diagnosed with both ASD and ADHD are double-loaded with genetic predisposition for both disorders and show distinctive patterns of genetic association with other traits when compared to the ASD-only and ADHD-only subgroups. The results provide novel insights into the biological foundation for developing just one or both conditions and for driving the psychopathology discriminatively towards either ADHD or ASD.
Attention-deficit hyperactivity disorder (ADHD) and autism spectrum disorder (ASD) are among the most common neurodevelopmental disorders in children and often persist throughout adulthood1. ADHD and ASD are both highly heritable (60–93%)2–4 and the mode of their inheritance is complex and polygenic. Despite high family-based heritability estimates, genome-wide association studies (GWAS) have only recently identified common variants robustly associated with each disorder5–7. Although differing from one another with regard to core clinical symptoms, genetic studies have demonstrated significant overlap between the two disorders, with a genetic correlation (rG) from common variation of 0.365,8 and substantial sharing of rare genetic risk variants such as large copy number variants9 and protein-truncating variants10. These findings are consistent with clinical and epidemiological evidence showing overlap in phenotypic features11, high comorbidity rates between ASD and ADHD12,13 in both females and males14, and familial co-aggregation of the disorders with increased risk of ADHD among relatives of ASD probands (odds ratios monozygotic twins: 17.8; dizygotic twins: 4.3; full-siblings: 4.6; full cousins: 1.6)15. Identification of the genetic components that are shared or distinct for the disorders may provide insights into the underlying biology and potentially inform on sub-classification, course and treatment.
Here we utilize large collections of genotyped samples of ADHD and ASD from the Psychiatric Genomics Consortium (PGC) and the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) to address two questions: (1) What specific variants and genes are shared by, or differentiate, ASD and ADHD? (2) Are there distinct genetic signatures in terms of polygenic burden for subgroups within these disorders such as cases diagnosed with both disorders (comorbid cases) or with just one of them (ASD-only, ADHD-only cases)?
Results
Shared genetic liability to ADHD and ASD.
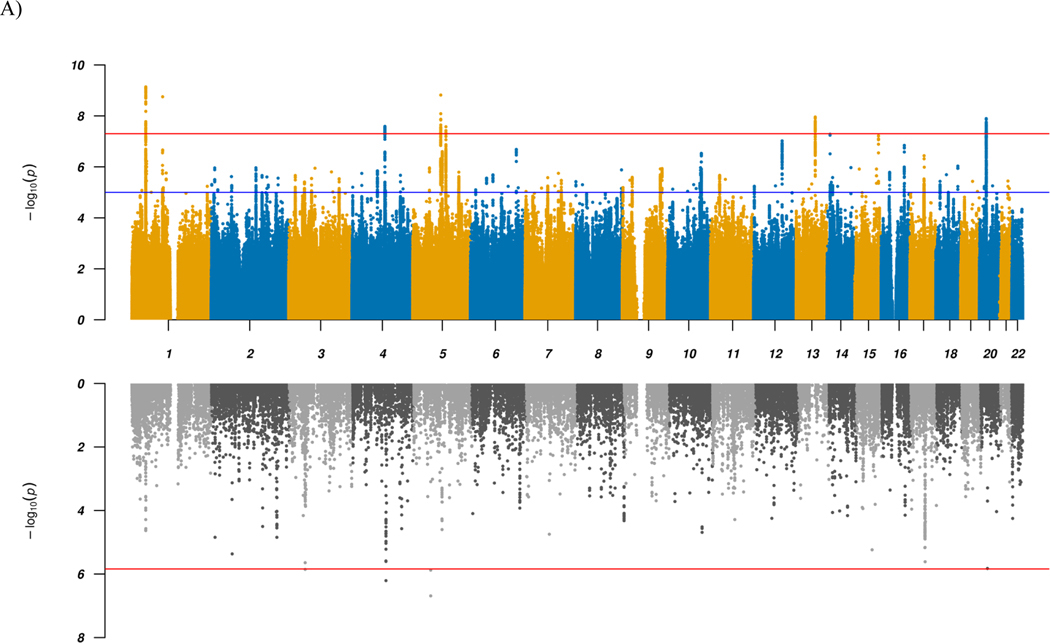
We performed a GWAS of diagnosed ADHD and/or ASD combined into a single phenotype (‘combined GWAS’), totaling 34,462 cases and 41,201 controls on 8.9 million SNP allele dosages imputed from 1000 Genomes phase 316. Using LD score regression (LDSC)17, we found evidence for a strong polygenic signal with an intercept of 1.0134 (ratio = 0.0558) and calculated the liability scale SNP-heritability to be 0.128 (for an assumed population prevalence of 0.055). We identified 263 genome-wide significant SNPs in seven distinct loci (Table 1, Fig. 1, and Supplementary Fig. 1). All these loci showed associations with both of the disorders separately at P-values below 1 × 10−4 except one, which is genome-wide significant in ADHD and has a P-value of 0.009 in ASD. Overall, the findings corroborate previous results8,18, but two loci have not been identified before as shared between ADHD and ASD. The novel shared associations are located in a highly pleiotropic multigene locus on chromosome 1 (rs7538463) and on chromosome 4 (rs227293) in the gene encoding mannosidase beta (MANBA). Mutations in MANBA are associated with beta-mannosidosis, a lysosomal storage disease that has a wide spectrum of neurological phenotypes, including intellectual disability, hearing loss and speech impairment19. More details on the seven loci can be found in Table 1, and results from lookups in the open GWAS project database (https://gwas.mrcieu.ac.uk/about/) and comparisons with previous cross-disorder studies are available in the Supplementary Note, Supplementary Data 1 and 2, and as PheWAS plots in Supplementary Fig. 2.
Table 1 |.
Results of combined GWAS (ADHD or ASD)
| META | ASD | ADHD | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP (#CS) | CHR | BP | A1 | A2 | FRQca | FRQco | OR | P | OR | P | OR | P | GENES | OTHER |
|
|
|
|
|
|
|
|||||||||
| rs7538463 (2/2/2) | 1 | 44196416 | A | T | 0.707 | 0.721 | 0.928 | 7.26 • 10−10 | 0.961 | 0.0091 | 0.914 | 1.00 • 10−9 | PTPRF, KDM4A, ST3GAL3, MIR6079 | ADHDa, Manyb |
| rs4916723 (5/5/5) | 5 | 87854395 | A | C | 0.558 | 0.573 | 0.935 | 1.52 • 10−9 | 0.935 | 1.92 • 10−6 | 0.925 | 1.81 • 10−8 | MIR9–2 (58.3) | ALCc, Neuroticismd, ADHDa, ADHD-CDGe, CDGf, sexual partnersg. CDGh |
| rs2391769 (2/2/2) | 1 | 96978961 | A | G | 0.351 | 0.364 | 0.934 | 1.77 • 10−9 | 0.926 | 1.14 • 10−7 | 0.928 | 1.04 • 10−7 | - | ADHD-CDGe, CDGf, CDGh |
| rs9530773 (0/0/0) | 13 | 78852243 | T | G | 0.674 | 0.689 | 0.935 | 1.14 • 10−8 | 0.938 | 1.76 • 10−5 | 0.933 | 1.78 • 10−6 | - | ADHDa, CDGh |
| rs13869664 5 (4/4/4) | 20 | 21154234 | A | AAAG | 0.644 | 0.659 | 0.937 | 1.27 • 10−8 | 0.926 | 1.22 • 10−7 | 0.940 | 1.11 • 10−5 | PLK1S1, KIZ, XRN2 | CDGf, CDGh, Manyi |
| rs227293 (0/0/0) | 4 | 103623491 | T | C | 0.689 | 0.672 | 1.061 | 2.57 • 10−8 | 1.061 | 7.02 • 10−5 | 1.080 | 1.08 • 10−7 | MANBA | ADHD-CDGj, Bloodk |
| rs325506 (23/27/24) | 5 | 104012303 | C | G | 0.441 | 0.428 | 1.064 | 2.66 • 10−8 | 1.074 | 3.50 • 10−7 | 1.070 | 8.40 • 10−7 | - | ASD-CDGl, ADHDa, ADHD-CDGe, CDGf, CDGh, Manym |
Results shown in the table are for three different GWAS: META refers to our combined ADHD or ASD GWAS described in the main text body of this manuscript, ADHD refers to results from the previously published GWAS on ADHD (PMID 30478444), and ASD refers to results from the previously published GWAS on ASD (PMID 30804558). Results from lookups in the open GWAS project database (https://gwas.mrcieu.ac.uk/about/, accessed 14 October 2020) are available in Supplementary Data 1 and as PheWAS plots in Supplementary Fig. 2. SNP (#CS), marker name and number of reported GWASs where this marker is in the 95% credible set in (FINEMAP/PAINTOR/CAVIARBF) according to http://mulinlab.org/causaldb/; please note that SNPs do not need to be genome-wide significant in those reported GWASs to be in the list of credible SNPs. SNPs representing novel shared loci for ASD and ADHD are highlighted in bold. CHR, chromosome; BP, base pair position on the chromosome; A1, effect allele; A2, other allele; FRQca, frequency in the cases; FRQco, frequency in the controls; OR, odds ratio based on effect allele; P, P value for association results (two-sided from logistic regression); GENES, protein-coding genes and/or microRNAs in a LD region around lead SNP (r2 = 0.6), or in cases where no protein coding gene or microRNA is present in the region the nearest protein-coding gene or microRNA within a 100-kb window around the LD region is provided together with the distance in kb (if there is no gene present a ‘-’ will be shown); OTHER, previously reported associations with the lead SNP (underlined letters) or other SNPs (italic letters) in LD with the lead SNP (r2 = 0.6), reported P values needed to be genome-wide significant to be listed. In case of the ASD and ADHD P values, these are the P values in the original GWAS. Please note that the OR and the P values reported for the ADHD and ASD GWASs both times include the comorbid cases (i.e. in each of the two GWASs) as well as related individuals across studies. Markers highlighted in bold letters indicate previously unidentified associations with either of the two disorders (ADHD/ASD).
ADHD (PMID 30478444);
Cross Disorder GWAS in the PGC (PMID 31835028), Educational attainment (years of education, PMID 30038396), Intelligence (MTAG, PMID 29326435), Adventurousness (PMID 30643258), Feeling worry (neuroticism item; 29500382), Household income (PMID 31844048), Balding type 1 (PMID 30595370), Number of sexual partners (PMID 30643258);
Alcohol consumption (PMIDs 30643258, 31358974, 30643251);
Neuroticism (PMID 29942085), Worry (neuroticism item; PMID 29942085);
Attention deficit hyperactivity disorder or cannabis use (PMID 30610198);
Cross Disorder GWAS in the PGC (PMID 31835028);
Number of sexual partners (PMID 30643258);
Cross disorder GWAS for TS-ADHD-ASD (PMID 33714545);
Fat-free mass (PMID 30593698), Appendicular lean mass (PMID 31761296), Height (PMIDs 30595370 and 25282103);
Asthma and attention-deficit hyperactivity disorder (PMID 31619474);
Blood protein levels (PMID 29875488);
Autism and major depressive disorder (MTAG, PMID 30804558);
Educational attainment (PMID 30038396), Life satisfaction (PMID 30643256), Well-being spectrum (multivariate analysis, PMIDs 30643256 and 29292387), Depressive symptoms (PMIDs 30643256 and 29292387), Neuroticism (PMID 29292387), Positive affect (PMID 30643256), Loneliness (PMID 31518406), Asthma and attention deficit hyperactivity disorder (PMID 31619474), Asthma and major depressive disorder (PMID 31619474), Insomnia (PMIDs 30804566 and 30804565), Risk-taking tendency (4-domain principal component model, PMID 30643258), BMI (PMIDs 31669095, 30595370, and 30239722), Highest math class taken (PMID 30038396), Hand grip strength (PMID 29691431), Predicted visceral adipose tissue (PMID 31501611).
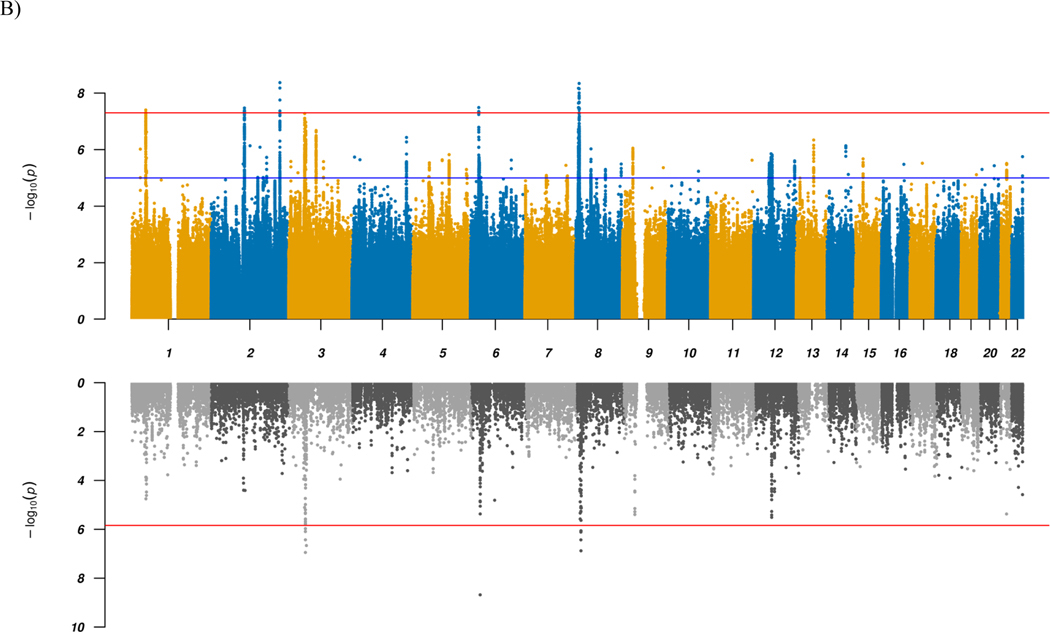
Figure 1 |. Manhattan plots for GWAS and TWAS results.
a,b, Results for GWAS (top panels) and TWAS for DLPFC transcripts (bottom panels) for combined (a) and ADHD vs. ASD (b) analyses. In the top panel, blue line in the Manhattan plot indicates a P-value of 1 × 10−5, and red line a P-value of 5 × 10−8 (genome-wide significance). Each dot represents a tested SNP. In the bottom panel, genes are represented by both imputed gene expression and isoform expression (features, represented by the dots); two-tailed P-values are derived from the z-scores (Wald statistic) of the gene-trait association. Red line indicates Bonferroni-corrected genome-wide significance within analyses (combined or ADHD vs. ASD; P < 1.44 × 10−6; corresponding to Bonferroni correction of all the 34,646 features). We implement an imputation R2 filter (pred_perf_r2) of 0.01 in this study, which means that at least 10% of the variance in expression of each gene can be explained by cis-heritability. Please also refer to the results in Supplementary Data 4.
To identify and prioritize putative causal shared genes, we performed a transcriptome-wide association study (TWAS), imputing the genetically regulated gene expression using EpiXcan20 and expression data from the PsychENCODE Consortium21 for genes as well as isoforms detected in 924 samples from the dorsolateral prefrontal cortex (DLPFC). Applying a conservative significance threshold (P < 1.44 × 10−6; corresponding to Bonferroni correction of all 34,646 genes and isoforms tested), we identified five genes/isoforms showing significant differential expression between the combined case group and controls, and 177 genes/isoforms significant at a false discovery rate (FDR) < 0.05 (Fig. 1 and Supplementary Data 4). One of the five Bonferroni significant transcripts, KRT8P46–201, is located in the identified chromosome 4 GWAS locus in an intron of MANBA, which is among the genes with an FDR < 0.05 (Supplementary Fig. 3a). The four other top findings are the two genes MOCS2 and CCDC71 or their isoforms, which are not located in any of the identified GWAS loci and thus represent additional novel candidate genes for shared ADHD and ASD risk.
Gene-based analysis using MAGMA v1.0822,23 largely corroborated the results from the GWAS and TWAS, highlighting, e.g., MANBA (Supplementary Fig. 4a and Supplementary Data 5). Furthermore, two of the significant genes—sortilin related VPS10 domain containing receptor 3 (SORCS3) and dual specificity phosphatase 6 (DUSP6)—are located in regions that were not identified in the GWAS, suggesting these as additional shared loci.
Differentiating genetic liability to ADHD and ASD.
To identify loci with divergent effects on ADHD and ASD, we performed an association analysis comparing 11,964 ADHD-only cases with 9,315 ASD-only cases from the iPSYCH cohort, excluding all 2,304 comorbid cases (‘ADHDvsASD GWAS’). Using LDSC17, we found an intercept of 0.9863 and a SNP-heritability of 0.4468 on the observed scale, the latter indicating that a substantial part of the variance in the phenotypic representation differentiating the two case groups can be explained by common variants (see Supplementary Note for more details). Five genome-wide significant loci were identified, three of which have not previously been identified in GWAS of either of the two disorders separately (although one has been reported as an ADHD-ASD differentiating locus24). All loci have been reported in related disorders and, remarkably, all but one are associated with cognitive abilities and/or neuroticism or neuroticism sub-items (Table 2, Fig. 1, and Supplementary Data 2 and 7). The lead variants all show opposite directions of effects in the two disorders.
Table 2 |.
Results of differentiating GWAS (ADHD vs. ASD)
| ADHDvsASD | ASD | ADHD | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP (#CS) | CHR | BP | A1 | A2 | FRQadhd | FRQasd | OR | P | OR | P | OR | P | GENES | OTHER |
|
|
|
|
|
|
|
|||||||||
| rs13023832 (NA/NA/NA) | 2 | 215219808 | A | G | 0.121 | 0.102 | 1.207 | 4.28 • 10−9 | 0.956 | 0.0484 | 1.122 | 9.33 • 10−8 | SPAG16 | ADHDb, CDGc |
| rs7821914 (3/5/5) | 8 | 10805015 | T | C | 0.584 | 0.556 | 1.127 | 4.58 • 10−9 | 0.935 | 1.86 • 10−6 | 1.022 | 0.1113 | XKR6 | Neuroticismd, Manye |
| rs147420422 (16/17/17) | 2 | 104139422 | CAT | C | 0.529 | 0.502 | 1.118 | 3.37 • 10−8 | 0.947 | 6.89 • 10−5 | 1.036 | 0.0092 | - | Neuroticismf, Manyg |
| rs3791033n (6/7/6) | 1 | 44134077 | T | C | 0.681 | 0.656 | 1.124 | 3.98 • 10−8 | 0.979 | 0.1407 | 1.095 | 2.76 • 10−10 | PTPRF, KDM4A, ST3GAL3, MIR6079 | EAh, ADHDi, Manyj |
| rs9379833 (58/59/58) | 6 | 26207175 | A | C | 0.251 | 0.275 | 0.884 | 4.51 • 10−8 | 1.041 | 0.0102 | 0.949 | 0.0007 | HIST1a | EAh, Neuroticismk, Heightl, Manym |
Results shown in the table are for three different GWAS: ADHDvsASD refers to our ADHD vs. ASD GWAS described in the main text body of this manuscript, ADHD refers to results from the previously published GWAS on ADHD (PMID 30478444), and ASD refers to results from the previously published GWAS on ASD (PMID 30804558). Results from lookups in the open GWAS project database (https://gwas.mrcieu.ac.uk/about/, accessed 14 October 2020) are available in Supplementary Data 7 and as PheWAS plots in Supplementary Fig. 11. SNP (#CS), marker name and number of reported GWASs where this marker is in the 95% credible set in (FINEMAP/PAINTOR/CAVIARBF) according to http://mulinlab.org/causaldb/; please note that SNPs do not need to be genome-wide significant in those reported GWASs to be in the list of credible SNPs. If instead of a number ‘NA’ appears, this means the SNP has not been reported in a credible set before. SNPs highlighted in bold have not been identified in GWASs of ADHD and ASD before. CHR, chromosome; BP, base pair position on the chromosome; A1, effect allele; A2, other allele; FRQADHD, frequency in the iPSYCH ADHD-only cases; FRQASD, frequency in the iPSYCH ASD-only cases; OR, odds ratio based on effect allele; P, P value for association result (two-sided from logistic regression); GENES, protein coding-genes and/or microRNAs in an LD region around lead SNP (r2 = 0.6), or in cases where no protein coding gene or microRNA is present in the region the nearest protein-coding gene or microRNA within a 100-kb window around the LD region is provided together with the distance in kb (if there is no gene present a ‘-’ will be shown); OTHER, previously reported associations with the lead SNP or other SNPs in LD with the lead SNP (r2 = 0.6), reported P values needed to be genome-wide significant to be listed. For ASD and ADHD, P values these are the P values in the original GWAS. Please note that the OR and the P values reported for the ADHD and ASD GWAS both times include the ADHD/ASD comorbid cases (i.e. in each of the two GWASs) as well as related individuals across studies.
Genes in the HIST1 region (PMID 12408966): HIST1H1E, HIST1H2BD, HIST1H2BE, HIST1H4D, HIST1H3D, HIST1H2AD, HIST1H2BF, HIST1H4E, HIST1H2BG, HIST1H2AE, HIST1H3E, HIST1H1D, HIST1H4F, HIST1H4G, HIST1H3F, HIST1H2BH.
ADHD GWAS (PMID 30478444);
Cross disorder GWAS (PMID 31835028);
General factor of Neuroticism (PMID 30867560), Neuroticism (PMIDs 29255261 and 30643256);
Remission after SSRI treatment in MDD or neuroticism (PMID 29559929), Gene alcohol interaction for blood pressure (PMID 29912962), White matter microstructure (PMID 31666681), Estimated glomerular filtration rate (PMID 31152163);
Worry (neuroticism item; PMID 29942085), Feeling nervous (neuroticism item; PMID 29500382), Anxiety/tension (special factor of neuroticism; PMID 30867560);
Smoking related phenotypes (PMIDs 30617275, 30643251, 30643258, 30595370, 30679032), Number of sexual partners (PMID 30643258), Age at first sexual intercourse (PMID 27089180), Reaction time (PMID 29844566), Risk-taking tendency (4-domain principal component model, PMID 30643258), General risk tolerance (MTAG, PMID 30643258), BMI (PMID 30239722), Pneumonia (PMID 28928442), Photic sneeze reflex (PMID 27182965);
Educational Attainment (PMID 30038396);
ADHD GWAS (PMID 30478444), Attention deficit hyperactivity disorder or cannabis use (PMID 30610198);
Highest math class taken (PMID 30038396), Self-reported math ability (PMID 30038396), Cognitive ability, years of educational attainment or schizophrenia (pleiotropy, PMID 31374203), Intelligence (PMIDs 29326435 and 29942086), Educational attainment (years of education, PMID 27225129), General cognitive ability (PMIDs 29844566 and 29186694), Smoking related phenotypes (PMID 30643251), Household income (MTAG, PMID 31844048), C-reactive protein levels (PMID 31900758), Menarche (age at onset, PMID 30595370), Red blood cell count (PMID 30595370), Height (PMID 30595370);
Worry too long after an embarrassing experience (neuroticism item; PMID 29500382);
Height (PMID 31562340);
Brain region volumes (PMID 31676860), Smoking related phenotypes (PMID 30643251), Strenuous sports or other exercises (PMID 29899525), Height (PMIDs 28552196, 28270201, 23563607, 20881960, 25282103, 25429064, 18391950, 18391951, 19343178, 31217584), Body fat percentage (PMID 30593698), Predicted visceral adipose tissue (PMID 31501611), Hip circumference adjusted for BMI (PMID 25673412), Hip circumference (PMID 25673412), Waist circumference (PMID 25673412), Waist circumference adjusted for BMI (joint analysis main effects and physical activity interaction; PMID 28448500), Waist circumference adjusted for body mass (PMID 28448500), Body fat distribution (leg fat ratio, PMID 30664634), Birth weight (PMIDs 27680694 and 31043758);
rs7538463(A) allele from Table 1 is correlated with rs3791033(C) allele in this table, r2 = 0.1687, D’= 0.8989 (LDpair Tool at LDlink website, EUR reference).
Two of the five lead SNPs have previously been found associated with educational attainment25. For the first SNP (rs3791033 on chromosome 1; P = 4.65 × 10−23), the C allele confers an increased risk for ASD and increased cognitive performance while the ADHD risk allele (T) is associated with decreased performance. Similarly, for the second SNP (rs9379833 on chromosome 6; P = 2.26 × 10−8), the A allele confers an increased risk for ASD and increased cognitive performance while the ADHD risk allele (C) is associated with decreased performance. Notably, this SNP (rs9379833) is located in the large histone gene cluster HIST126 and has also been associated with regional brain volume, specifically of the left globus pallidus27 (P = 2.95 × 10−8; the C allele confers an increased risk for ADHD and a decreased volume while the ASD risk allele (A) is associated with an increased volume). It is also of note that the lead SNP on chromosome 8 (rs7821914) is associated with neuroticism28 (P = 9.46 × 10−21). For this SNP, the effect allele (C) in the neuroticism GWAS leads to an increased risk for ASD and a decreased risk for ADHD. Two additional lead SNPs are in LD (r2 > 0.6) with SNPs that have previously been identified in neuroticism or one of its subdimensions (rs147420422 and rs9379833; see Table 2). Results from additional lookups in the open GWAS project database (https://gwas.mrcieu.ac.uk/about/) are available in Supplementary Data 7 and as PheWAS plots in Supplementary Fig. 6.
TWAS using EpiXcan identified 11 Bonferroni significant genes/isoforms and 96 significant transcripts at FDR < 0.05 with different imputed expression in DLPFC between ADHD and ASD cases (Fig. 1 and Supplementary Data 4). The HIST1H2BD-201 isoform located in the chromosome 6 (HIST1) GWAS locus showed the strongest association (P = 2.08 × 10−9) with higher expression in ADHD compared to ASD cases (Supplementary Fig. 3b). The other genes/isoforms showed orders of magnitude less significant association, appointing HIST1H2BD-201 as the top-ranking causal candidate in the locus. The remaining 10 Bonferroni significant genes/isoforms were located in the chromosome 8 GWAS locus or in two loci on chromosome 3 (Supplementary Fig. 3c,d, respectively), where all except the gene encoding the TRAF interacting protein (TRAIP) were also genome-wide significant in gene-based analysis using MAGMA (Supplementary Fig. 4b and Supplementary Data 5).
Genetic correlations with other traits.
To examine the polygenic architecture of the identified shared and differentiating genetic risk for the disorders, we investigated the genetic correlations with 258 traits from a manually curated list of previously published GWAS and 597 traits from the UK Biobank, making use of LD Hub29 and LDSC30. Among the 258 previously reported GWAS, 30 (combined GWAS) and 32 (ADHDvsASD) traits showed significant correlations after Bonferroni correction for multiple testing (Supplementary Data 6 and Supplementary Fig. 7). The strongest correlations for the liability differentiating ADHDvsASD GWAS were observed for cognitive traits such as years of schooling (rG = −0.669, Pcorr = 3.68 × 10−85) and childhood IQ (rG = −0.609, Pcorr = 2.78 × 10−10), while the strongest correlations for the combined GWAS were with traits such as depressive symptoms (rG = 0.506, Pcorr = 2.08 × 10−19) and the PGC cross-disorder GWAS (rG = 0.433, Pcorr = 5.30 × 10−25).
Tissue and cell-type enrichment analyses.
We next tested whether genetic associations of shared and differentiating liabilities were enriched with respect to the transcriptomic profiles of human tissues. We found significant enrichment for the shared liability in several brain tissues, most significantly for the basal ganglia (Supplementary Fig. 8). Cell-type enrichment analyses revealed experiment-wide significant association (across all data sets tested) of the red nucleus (Supplementary Fig. 9c). Associations that were significant within one of the three tested data sets individually, but not overall, were observed for several cell types, including, e.g., dopaminergic and GABAergic neurons. For the disorder-differentiating analysis (ADHDvsASD), we observed no significant association with tissues or specific cell-types after correction for multiple testing (Supplementary Figs. 9 and 10). We also intersected our genetic associations with a recent multi-omics single-cell epigenetic catalog of the human brain31. Here both the combined and differentiating GWAS results showed significant enrichment for several neuronal cell populations (Supplementary Fig. 11 and Supplementary Data 8), including excitatory and inhibitory neurons. Interestingly, the only difference in terms of significant associations between the combined and differentiating GWAS was seen for oligodendrocytes (which were not significant in the combined GWAS but were significant in the ADHDvsASD GWAS). While aberrant myelination by oligodendrocytes resulting in disruption of white matter development has previously been reported in both ASD and ADHD32,33, the degree of severity of this alteration might be a distinct pathophysiological factor34.
Polygenic characterization of case subgroups.
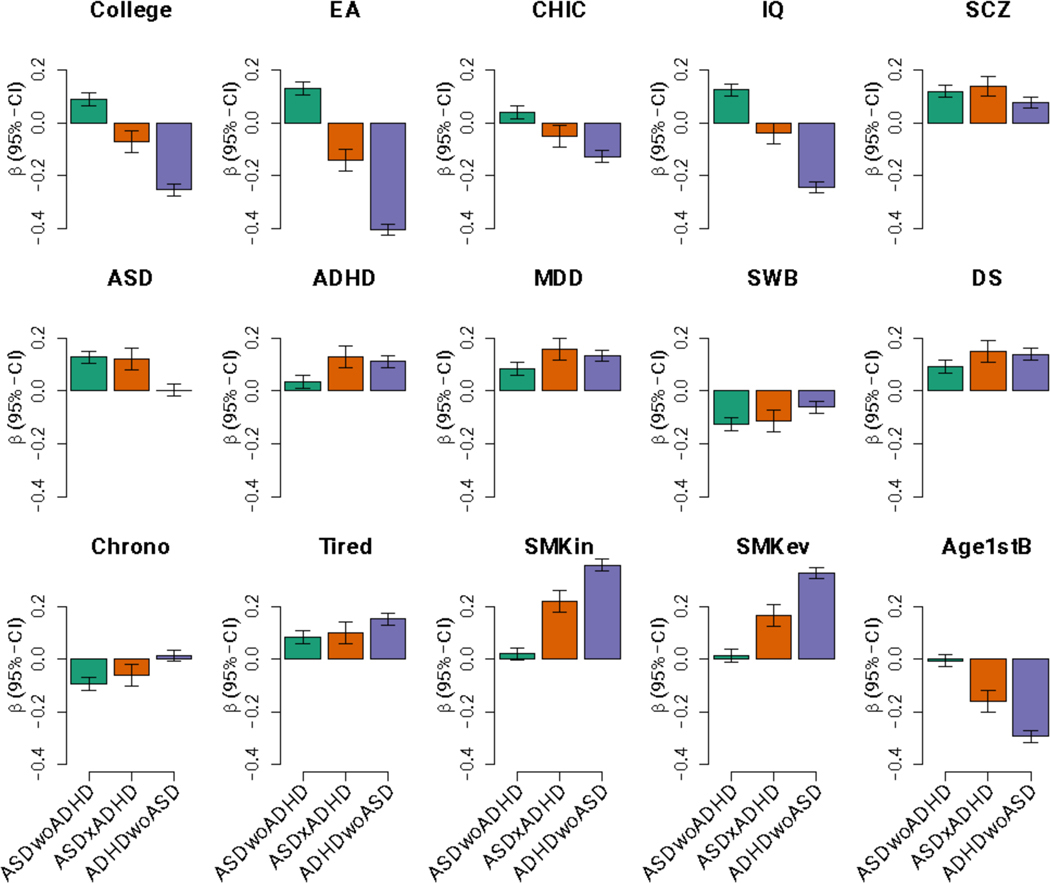
We used two complementary polygenic risk score (PRS) approaches to investigate differences in polygenic load for ADHD, ASD and related phenotypes in the iPSYCH data across the three phenotypic subgroups: ASD-only, ADHD-only and comorbid cases. The multivariate PRS framework showed, as expected, a significant association of the ASD-only subgroup with PRS for ASD (P = 6.89 × 10−26) and the ADHD-only subgroup with PRS for ADHD (P = 3.29 × 10−23; Fig. 2). Both scores were trained with PGC-only GWAS results5,35. Strikingly, the ASD-PRS load on comorbid ASD+ADHD cases was similar to that on ASD-only cases (P = 0.77), and likewise the ADHD-PRS load on the comorbid subgroup was similar to that on ADHD-only cases (P = 0.44; Fig. 2), demonstrating that the comorbid cases carry a load of both ADHD and ASD polygenic scores that are similar to the load carried by the single-disorder cases of their respective disorder PRS. In other words, comorbid cases are double-burdened with both ASD and ADHD PRS. In contrast, the ASD-PRS load on ADHD-only cases was not different from controls (P = 0.79) and the ADHD-PRS was only slightly increased in ASD-only cases compared to controls (P = 3.26 × 10−3; Fig. 2).
Figure 2 |. Comparison of PRS profiles across ADHD/ASD subtypes for 15 traits/phenotypes that have shown significant genetic correlation with ADHD and ASD in the past.
The bars display regression coefficients from a multivariate multivariable regression of the 15 normalized polygenic scores on ASD-ADHD comorbidity classes (n = 23,583) and controls as reference (n = 22,122, not shown). Green represents ASD-only cases (n = 9,315), orange depicts comorbid samples (n = 2,304), and purple for ADHD-only cases (n = 11,964). Error bars are 95% confidence intervals centered on the point estimate. ADHD, attention-deficit/hyperactivity disorder (PMID 20732625); ASD, autism spectrum disorder (PMID 30804558 without the iPSYCH sample); MDD, major depressive disorder (PMID 29700475 without DK or 23andMe); SWB, subjective well-being (PMID 27089181); DS, depressive symptoms (PMID 27089181); College, college completion (PMID 27046643); Edu, educational attainment (PMID 30038396); CHIC, childhood IQ (PMID 23358156); IQ, IQ (PMID 29942086); SCZ, schizophrenia (PGC3 without DK); Chrono, chronotype (PMID 30696823); Tired, self-reported tiredness (PMID 28194004); SMKos, smoking initiation (PMID 30643251); SMKev, ever smoker (PMID 30643258); Age1stB, age of first birth (PMID 20418890).
Results from our leave-one-out framework analysis (including only the iPSYCH data in the training GWAS) showed similar results (Table 3). We note that, in this analysis, the ASD-PRS load on ADHD-only cases as well as the ADHD-PRS load on ASD-only cases were increased compared to controls. Furthermore, secondary analysis in the leave-one-out framework suggested that ADHD cases with (n = 625) and without mild intellectual disability (ID) (n = 11,339) did not differ in terms of PRS for either ADHD or ASD. On the other hand, ASD cases with ID (n = 634) had lower PRSASD (OR = 0.89 (0.81–0.97), P = 0.0072) compared to those without mild ID (n = 8,681) but did not differ in terms of PRSADHD (Table 3).
Table 3 |.
Results of ADHD and ASD polygenic risk score analyses in the iPSYCH cohort using a leave-one-out analysis framework
| PRSADHD | PRSASD | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (coded as 1) | Comparison (coded as 0) | OR | LCI | UCI | P | OR | LCI | UCI | P |
|
|
|
|
|||||||
| ADHD-only | Controls | 1.45 | 1.41 | 1.48 | 1.3 • 10−207 | 1.08 | 1.06 | 1.11 | 7.5 • 10−12 |
| ASD-only | Controls | 1.10 | 1.07 | 1.13 | 3.1 • 10−13 | 1.21 | 1.18 | 1.24 | 1.2 • 10−48 |
| Comorbid | Controls | 1.32 | 1.25 | 1.39 | 2.8 • 10−25 | 1.22 | 1.16 | 1.29 | 3.5 • 10−14 |
| Comorbid | ADHD-only | 0.92 | 0.88 | 0.97 | 0.0015 | 1.13 | 1.08 | 1.19 | 4.7 • 10−7 |
| Comorbid | ASD-only | 1.22 | 1.16 | 1.28 | 6.4 • 10−16 | 1.01 | 0.96 | 1.06 | 0.68 |
| ASD-only | ADHD-only | 0.76 | 0.74 | 0.78 | 4.5 • 10−79 | 1.12 | 1.09 | 1.15 | 1.2 • 10−15 |
|
| |||||||||
| ADHD+ID | ADHD-no-ID | 0.97 | 0.88 | 1.06 | 0.46 | 0.94 | 0.86 | 1.03 | 0.19 |
| ASD+ID | ASD-no-ID | 1.03 | 0.93 | 1.12 | 0.58 | 0.89 | 0.81 | 0.97 | 0.0072 |
Results for per wave polygenic risk score analyses. PRSADHD, analyses using a PRS trained on an ADHD phenotype; PRSASD, analyses using a PRS trained on an ASD phenotype. Cases, group coded as 1 (cases) for the purpose of the analyses; Comparison, other group coded as 0 for the purpose of the analyses; OR, odds ratio; LCI, lower boundary for 95% confidence interval; UCI, upper boundary for 95% confidence interval; P, P-value (two-sided from regression model). Groups are as follows: ADHD-only, cases with ADHD diagnosis and without comorbid ASD diagnosis; ASD-only, cases with ASD diagnosis and without comorbid ADHD diagnosis; Comorbid, cases with comorbid ADHD and ASD diagnoses; Controls, individuals without ADHD and ASD diagnoses. P-values shown are without correction for multiple testing. Experiment-wide significant at 0.0042 (Bonferroni corrected for 2 × 6 tests). Additional secondary analyses also compare groups of individuals with ADHD or ASD with co-occurring mild intellectual disability (ADHD+ID and ASD+ID) to those without (ADHD-no-ID and ASD-no-ID).
To further dissect the genetic architecture across the ASD and ADHD subgroups, we examined the relative burden of PRS for phenotypes and traits that have shown significant genetic correlation with ADHD and ASD5,6,36. While PRS for schizophrenia and depression (and genetically related phenotypes) did not show substantially different loads across the subgroups, other traits showed compelling differences (Fig. 2). For instance, years of education, IQ, age at first birth, tiredness, and smoking showed differences between ADHD-only and ASD-only cases, with the comorbid cases at an intermediate level. An item-level analysis of neuroticism also revealed specific patterns of associations across the subgroups (Supplementary Fig. 12). On average, ADHD-only cases showed much stronger association than ASD-only cases with items belonging to the depressed affect cluster (e.g., the MOOD item) compared to the worry cluster. For comorbid cases, a distinct pattern was observed with PRS loads either ranking between the ADHD- and ASD-only cases (e.g., for the MOOD item) or even exceeding the two single-disorder groups (e.g., for the GUILT item).
Summarizing, we observe a genetic architecture of comorbid cases that presents itself in clear distinction from the ADHD and ASD single-disorder cases. Showing burden of both ASD and ADHD genetic risk, the comorbid cases also carry polygenic load profiles across other phenotypes that distinguishes them from their single-disorder cases, typically by carrying an intermediate load level but in some cases a load similar to just one of the single-disorder groups.
Genetic correlation and heritability across case subgroups.
We recently reported an LDSC genetic correlation of 0.36 between ASD and ADHD using the largest GWAS meta-analyses of the two disorders, including multiple cohorts and comorbid cases5. Here we investigated the correlations across diagnostic subgroups of the disorders in the iPSYCH sample using GCTA37. For ASD and ADHD overall, we found rG = 0.497 (s.e. = 0.054, P = 7.8 × 10−19). Excluding the comorbid cases reduced the correlation to rG = 0.397 (s.e. = 0.056, P = 6.3 × 10−12). After excluding cases with ID, the correlations between ASD and ADHD were even stronger: rG = 0.523 (s.e. = 0.054, P = 6.5 × 10−21) and rG = 0.425 (s.e. = 0.056, P = 1.7 × 10−13) with and without comorbid cases, respectively (Supplementary Data 9 and Supplementary Fig. 13).
Correlations between ADHD and ICD-10 diagnostic subcategories of childhood autism (F84.0), atypical autism (F84.1), Asperger’s syndrome (F84.5), and other/unspecified pervasive developmental disorders (other PDDs, F84.8–9) were similar to those for the ASD group overall, albeit with generally higher estimates for the groups with other PDDs and Asperger’s syndrome (Supplementary Data 9 and Supplementary Fig. 14).
Genetic liability in comorbid cases.
Guided by our results from the previously described analyses, we also performed a GWAS of the comorbid cases. Despite the small sample size (2,304 cases), we identified a genome-wide significant locus on chromosome 6 (rs1321614, P = 3.54 × 10−9, OR = 0.8190, MAF = 0.47 for the T allele). The lead SNP showed no association in the overall combined (ADHD+ASD) GWAS (P = 0.0261), the differentiating GWAS (P = 0.2883) or in GWASs of the ADHD-only and ASD-only cases (P = 0.7721 and P = 0.0086, respectively). The liability scale SNP-heritability for the GWAS using GCTA was 0.0557 (s.e. = 0.0088). Please see Supplementary Note for more information.
Discussion
This study dissects the genetic architecture for shared and differentiating genetic underpinnings of ADHD and ASD as well as across case subgroups. At the single variant level, we identified novel shared loci for the two disorders and five genome-wide significant loci differentiating the disorders, four of which are novel. Integration with DLPFC transcriptomic data identified and prioritized several possibly causal genes (see Supplementary Note). At the polygenic level, we revealed compelling differences across comorbid and single-disorder case groups.
The identified shared loci are generally highly pleiotropic and have previously been identified in GWAS of related disorders or cross-disorder studies including ADHD and/or ASD. However, considering only the eight major psychiatric disorders included in the most recent PGC cross-disorder study8, three of the loci (rs4916723, rs2391769, and rs227293) appear to be shared only between ADHD and ASD (Table 1 and Supplementary Data 2). For the other SNPs, only one (rs325506) shows support for involvement in more than one additional disorder. This is consistent with evidence from structural equation modeling of eight major psychiatric disorders, showing that ASD and ADHD cluster together in a group of early-onset neurodevelopmental disorders along with Tourette syndrome8.
In the ADHDvsASD GWAS, we identified five genome-wide significant loci, all showing opposite allelic directions in the separate GWAS of the two disorders, providing specific genetic clues to understanding the biology that drives the pathophysiology towards developing one or the other disorder. While one of the identified loci (rs3791033) supported the single ADHD-ASD differentiating locus reported previously24 (using CC-GWAS analysis on available summary statistics), the four novel loci all showed supportive (but not statistically significant) results in the CC-GWAS study, except the histone 1 locus at the MHC region, which was not included in the CC-GWAS (Supplementary Data 2). The yield of more significant loci in our study compared to the CC-GWAS could (in addition to methodological differences) be because we were able to remove comorbid ADHD+ASD cases, which were included in the GWAS results used in the CC-GWAS study, resulting in relatively stronger analytical power in our study.
The top-ranking differentiating TWAS gene/isoform was HIST1H2BD-201, which was two orders of magnitude more significant than the second-ranking (CAMKV-210) and the only Bonferroni significant transcript in the identified HIST1 GWAS locus. Deleterious de novo mutations in several histone modifying or interacting genes38–40 as well as in core histone genes39,41 have been associated with autism and developmental delay with autistic features. The haploinsufficiency resulting from these de novo mutations is consistent with our TWAS result showing reduced expression of HIST1H2BD-201 in ASD (relative to ADHD). Intriguingly, the ASD risk allele of the lead SNP in the locus is also associated with both increased educational performance25 and increased volume of the left globus pallidus27, while the opposite is the case for the ADHD risk allele. As part of the basal ganglia, globus pallidus is involved in several functions relating to phenotypic domains affected in ASD and/or ADHD such as cognition, social interactions, speech, repetitive behaviors and tics42. Taken together, our results suggest that the identified ADHD-ASD differentiating locus on chromosome 6 has downstream effects involving differential expression of the histone isoform HIST1H2BD-201 and volumetric changes of the left globus pallidus, which may contribute—as one weak-acting factor among many—to driving the pathophysiology towards either ASD or ADHD and impacting key phenotypic domains such as educational performance, social interaction, and motor impairments.
Previous studies found ASD and ADHD to display opposite genetic correlations with cognitive traits like educational attainment when assessing common variants genome-wide5,6,43. Corroborating these reports, we found that the ADHDvsASD GWAS showed the strongest correlations for cognitive traits (Supplementary Data 6 and Supplementary Fig. 7). Moreover, two of the identified differentiating loci (on chromosome 1 and 6) have lead SNPs that are genome-wide significant in educational attainment and show opposite allelic effects with increasing and decreasing educational performance for the ASD and ADHD risk alleles, respectively.
We note that the chromosome 1 locus (at position 44 Mb) was identified, counterintuitively, in both the shared and differentiating GWAS albeit with different lead SNPs (Tables 1 and 2). The locus covers a gene-rich 250-kb region of generally strong linkage disequilibrium (LD) but it also harbors variants with limited LD to the main haploblock (Supplementary Figs. 1a and 5d). The two lead SNPs are located 62 kb apart and show low pairwise LD (r2 = 0.1687; Table 2), indicating that the two SNPs are largely independent markers for association. This LD difference is also reflected in the different lists of other traits with previously reported associations for the lead SNPs or their LD proxies (Tables 1 and 2). Furthermore, this locus was the only locus showing significant heterogeneity across cohorts in the recent ADHD GWAS6, where the 23andMe sample provided no support for the otherwise consistently supported locus and, also in contrast to the other cohorts, exhibited limited genetic correlation with educational attainment.
Our analyses revealed enrichment of brain-expressed genes for the combined GWAS, implicating particularly the basal ganglia and cerebellum. Both structures have been found altered in both ASD42,44 and ADHD45–47, with evidence for reductions in basal ganglia volume the most robustly observed finding in the neuroimaging literature for both ASD and ADHD. The cell-type enrichment result implicating the red nucleus in midbrain is also consistent with our knowledge of phenotypic sharing between ASD and ADHD, as it relates to skilled movements and motor control in the limbs and jaw: both motor coordination and speech problems are frequent in both ASD and ADHD48,49. The red nucleus is strongly connected with many brain structures involved in ASD and ADHD, including the basal ganglia and the cerebellum50.
Dissecting the polygenic architecture using PRS approaches, we observed remarkable differences across comorbid and single-disorder (ADHD-only and ASD-only) case groups. The comorbid cases carry a double burden of ASD- and ADHD-PRS, whereas the single-disorder cases were largely just (single-) burdened for the respective disorder. Thus, cases diagnosed with both disorders have on average a similar level of genetic liability to each disorder as the single-disorder cases, providing strong biological support for the change in diagnostic guidelines from DSM-IV to DSM-5 allowing for diagnoses of both disorders in the same person. This is further highlighted by the identification of a genome-wide significant locus for comorbid cases (chromosome 6). It also supports pharmacological treatment of comorbid ADHD in individuals with ASD. In a recent meta-analysis, 25–32% of individuals with ASD also fulfill criteria for ADHD13, yet only 15–16% are treated with ADHD medications51,52, despite strong evidence of beneficial effects on the core symptoms of ADHD and potentially also reduced risk of injuries53, depression54, suicidal behavior55 and improved academic performance56. Moreover, it indicates that pharmacological treatment of symptoms like hyperactivity, inattention, impulsivity, aggression and tics in cases diagnosed with either ADHD or ASD may be guided by the individual symptomatology regardless of the given diagnosis.
We recently reported a significant genetic correlation of rG = 0.36 between ASD and ADHD, using LDSC and results from GWASs that included multiple cohorts and comorbid cases5. This was a considerable increase from the previous estimate of rG = 0.08 (s.e. = 0.10, P = 0.40), which was based on much smaller GWAS sample sizes without information on comorbid diagnoses57. Here we analyzed exclusively the iPSYCH cohort, which is relatively homogeneous and has information on all diagnoses given to each individual. We found a higher correlation (rG = 0.497), which remained substantial when excluding the comorbid cases (rG = 0.397), demonstrating that the genetic overlap between the disorders is not driven by comorbid cases alone. While we cannot exclude that under-diagnosis of comorbidity might exist, leading to an upwards bias of the correlation estimate between the single-disorder cases, our result is corroborated by data from Swedish twin studies that supports the distinction of ASD and ADHD, but also suggests considerable co-occurrence of symptoms of both disorders in individuals only fulfilling diagnostic criteria for one of the two disorders58,59.
In addition, the correlations increased when excluding cases with ID, indicating that cases with ID are more genetically heterogeneous in common variant risk between the two disorders than cases without ID. A recent exome sequencing study of ASD and ADHD (also in the iPSYCH cohort) showed that the disorders have substantial overlap in rare variant risk and that cases with ID carry a higher load of (ultra)rare damaging risk variants compared to cases without ID10. Consistent with this, our PRS analyses found lower ASD-PRS in the group of ASD cases with comorbid mild ID (IQ = 50–70) compared to those without mild ID. Taken together, these observations are consistent with the notion that the genetics differentiating the two disorders may be driven primarily by common variants (because the rare variant risk load is similar for the two disorders in the data available so far) and more extensively for cases with ID than without ID (because the common variant genetic correlation is lower for cases with ID). However, larger sample sizes for both GWAS and sequencing studies are needed to clarify this.
In conclusion, we have disentangled the shared and differentiating genetic liability underlying ASD and ADHD, identifying novel shared as well as disorder-specific risk variants informing on the pathophysiology. In addition, we have revealed specific patterns of polygenic architecture that are characteristic for comorbid cases compared to single-disorder cases. The results advance understanding of the complex etiologic basis and relationship between ASD and ADHD towards the long-term goals of better diagnosis and treatment of these disorders.
Methods
Ethics and overview.
The study was approved by the Regional Scientific Ethics Committee in Denmark and the Danish Data Protection Agency. We report results from different analyses all carried out in large-scale samples from the Psychiatric Genomics Consortium (PGC) and the Lundbeck initiative of integrative psychiatric research (iPSYCH). We used samples included in the most recent GWAS of ASD5 and ADHD6. For this manuscript, we will refer to individuals in the study cohort (most importantly in iPSYCH) that at the time of inclusion only had one of the two diagnoses registered (i.e., ADHD or ASD) as ADHD-only and ASD-only cases, respectively. We refer to individuals that during their lifetime and up to the time of inclusion had both an ADHD and ASD diagnosis registered as comorbid cases. Furthermore, we refer to these three groups of cases (i.e., ADHD-only, ASD-only, and comorbid) as ASD and ADHD subgroups.
Sample description and additional quality control.
Details about study specific case and control selection criteria and how individuals were drawn from the overall iPSYCH case-cohort sample60 can be found in the respective publications5,6. Here we focus on differences in selection criteria in the iPSYCH cohort and additional quality control (QC) procedures.
The majority of inclusion and exclusion criteria for the original studies were also used in this study. The only difference compared to the original studies was an additional exclusion criterion that removed individuals with a moderate to severe mental retardation (ICD10: F71-F79) from both the case and control cohorts. While this criterion was also used in the original ADHD GWAS6, it was not used in the original ASD GWAS5. The rationale for this decision lies in the interpretability of our results, where we treated ADHD and ASD consistently. We address the potential impact of this decision through different analyses (see Table 3, Supplementary Fig. 14b, and Supplementary Data 9).
Wave-wise pre-imputation QC and imputation of the iPSYCH case-cohort sample were taken from the original ADHD and ASD GWAS, respectively. Details about the respective steps and filters can be found elsewhere5,6. Since our analyses used a combined study cohort with samples from both the original ADHD and ASD GWAS, we performed some additional QC on the combined sample. Additional QC steps included the removal of related individuals across the original ADHD and ASD GWAS and a new principal component analysis (PCA) on the combined sample after exclusion of these related individuals. Following the same procedures as in the original studies, pairs of subjects were identified with pi-hat> 0.2 (using PLINK’s61 identity-by-state analysis) and one subject of each pair was excluded at random (with a preference for keeping cases). PCA was carried out using smartPCA in the EIGENSOFT software package62,63 using the Ricopili pipeline64. The original PGC datasets for ADHD and ASD did not include overlapping individuals and therefore the original datasets and summary statistics were used. The final combined dataset across all samples comprised 34,462 cases (i.e., individuals with an ADHD and/or ASD diagnosis) and 41,201 controls. We only included samples of European ancestry from the original ADHD and ASD GWAS. Among the cases in the iPSYCH cohort, 11,964 had an ADHD-only, 9,315 had an ASD-only, and 2,304 individuals had a comorbid diagnosis, respectively. Thus, the proportion of ADHD among ASD cases in the iPSYCH cohort was 19.8%, and the proportion of ASD among ADHD cases was 16.1%.
Genome-wide association analyses.
Like the original GWAS in ADHD and ASD, all processing and analyses for the individual GWAS and meta-analyses (see below) used the Ricopili pipeline64. More details on individual modules and steps can be found elsewhere5,6,64. We ran two main GWASs for our analyses. The first aimed to identify shared genetic risk for ADHD and ASD (combined GWAS) and the second aimed to identify differentiating genetic risk with an opposite direction of effects for ADHD and ASD (ADHD vs. ASD GWAS). All analyses of the iPSYCH sample and meta-analyses with the PGC samples were conducted at the secured national GenomeDK high-performance computing cluster in Denmark.
Combined GWAS.
We first ran an analysis in the combined dataset, i.e., on all 34,462 cases and 41,201 controls. The GWAS was conducted in each cohort (i.e. in the wave-wise iPSYCH samples and the individual PGC cohorts) using logistic regression with the imputed additive genotype dosages. The first 5 principal components (PCs) were included as covariates to correct for population stratification (Supplementary Note), and variants with imputation INFO score < 0.8 or minor allele frequency (MAF) < 0.01 were excluded. The resulting summary statistic files were then meta-analyzed using an inverse-variance weighted fixed effects model65. Post-processing of the summary statistics files through the Ricopili pipeline64 created Manhattan plots, individual regional associations plots, and forest plots. For a QQ-plot of the analysis, see Supplementary Fig. 14a.
ADHD vs. ASD GWAS.
To identify unique genetic risk loci or loci with opposite direction of effects for ADHD and ASD, we ran a case-only analysis for the ADHD-only (coded as 1, n = 11,964) against ASD-only cases (coded as 2, n = 9,315) in the iPSYCH cohort. This approach is in line with our recent study that compared the genetic risk to develop bipolar disorder and schizophrenia66. We excluded the comorbid cases from this GWAS, and the GWAS was conducted wave-wise using logistic regression with the imputed additive genotype dosages. The first 5 PCs were included as covariates to correct for population stratification, and variants with imputation INFO score < 0.8 or MAF < 0.01 were excluded. The resulting summary statistic files were then meta-analyzed using an inverse-variance weighted fixed effects model65 and visualization of results was achieved through the Ricopili pipeline64 (see above). For a QQ-plot of the analysis, see Supplementary Fig. 14b.
Identification of previously reported associations for top findings.
Different resources were used to identify previously reported associations of our top findings with other phenotypes and traits within and outside of psychiatry. We assessed associations reported in the open GWAS project database (https://gwas.mrcieu.ac.uk/about/, accessed 14 October 2020; see Supplementary Data 1 and 7 for results) and used the GWAS ATLAS website67 to visualize PheWAS analyses (see Supplementary Figs. 2 and 6). We also used results from the GWAS Catalog68 (see Table 2). Finally, we also compared our results with previous cross-disorder studies in the field. This included the recent analyses of the cross-disorder group in the PGC8, a study that used a new approach to study case-case associations in psychiatric disorders24, and a study that used conditional analyses to highlight associations that might be specific for individual psychiatric disorders69. Results are available in the Supplementary Note and Supplementary Data 2.
Transcriptomic imputation model construction and transcriptome-wide association study (TWAS).
Transcriptomic imputation models were constructed as previously described205 for dorso-lateral prefrontal cortex (DLPFC) transcript levels70. The genetic dataset of the PsychENCODE cohort was uniformly processed for QC steps before genotype imputation. We restricted our analysis to samples of European ancestry as previously described20. Genotypes were imputed using the University of Michigan server71 with the Haplotype Reference Consortium (HRC) reference panel72. Gene expression information (both at the level of gene and transcript) was derived from RNA-seq counts, which are adjusted for known and hidden confounds, followed by quantile normalization70. For the construction of the transcriptomic imputation models, we used EpiXcan20, an elastic net based method, which weighs SNPs based on available epigenetic annotation information73. EpiXcan was recently shown to increase power to identify genes under a causality model when compared to TWAS approaches that do not integrate epigenetic information74. We used this model (924 samples from DLPFC) due to power considerations20; in comparison, brain gene expression imputation models based on GTEx V875 are trained in 205 or fewer samples. Using only samples from DLPFC, we acknowledge that ADHD and ASD are both also associated with other brain regions and would like to highlight this as a potential limitation of our study. We performed the transcript-trait association analysis for the traits in this study as previously described20. Briefly, we applied the S-PrediXcan method20 to integrate the GWAS summary statistics and the transcriptomic imputation models constructed above to obtain association results at both the level of genes and transcripts.
Cell-type enrichment analysis.
A major portion of cell type specific enrichment is attributed to distal regulatory elements, as local regulatory events remain highly consistent across various tissues and cell types76. Therefore, we examined overlap of common genetic variants of investigated traits (see Supplementary Fig. 14 and Supplementary Data 8) and open chromatin from scATAC-seq study (single-cell assay for transposase accessible chromatin)31 using the LD-score partitioned heritability approach77. All regions of open chromatin were extended by 500 bp in either direction. The broad MHC-region (hg19 chr6:25–35Mb) was excluded due to its extensive and complex LD structure, but otherwise default parameters were used for the algorithm.
Additional functional characterization and annotation of main findings.
We used different approaches combining in-house scripts and data with those available via the FUMA v1.3.6a23 website (http://fuma.ctglab.nl) for downstream functional characterization and annotation of our findings. For FUMA, we uploaded our summary statistics from the individual analyses. We also used FUMA to perform tissue expression analyses on data available through their website. Finally, we used FUMA to perform cell-type specificity analyses78 based on our summary statistics. For all above-mentioned analyses, default settings were applied. More detailed information about the individual third-party datasets (available through FUMA) included in the analyses as well as individual aspects of the FUMA analyses can be found in the Supplementary Note. Supplementary Data 10 contains results from standard FUMA-based analyses, such as eQTL and chromatin interaction mapping.
Gene-based analysis.
We also used FUMA v1.3.6a23 to perform gene-based analysis. Genome-wide significance was assessed through Bonferroni correction for the number of genes tested. More detailed information about the individual third-party datasets (available through FUMA) included in the analyses as well as individual aspects of the gene-based analyses can be found in the Supplementary Note.
Our results in context of other findings.
Since the publication of the original ADHD and ASD results, a few studies have investigated the shared and unique risk architecture of these disorders. We compared our results with the findings of the Cross Disorder Working Group of the PGC8 and a recent analysis based on structural equation modelling of 11 major psychiatric disorders79. We also compared our results with recent analyses that aimed at identifying disorder-specific SNPs for psychiatric disorders24,69.
Polygenic risk score (PRS) analyses.
To examine potential polygenic heterogeneity across ADHD and ASD subtypes, we investigated how PRS trained on different phenotypes were distributed across ADHD-only, ASD-only and comorbid subgroups in the iPSYCH data through two complementary analysis frameworks: multivariate PRS and leave-one-out PRS. These two approaches have different strengths and limitations, allowing for robust interrogation of differences in ADHD and ASD subgroups in terms of polygenic burden for ADHD and ASD, as well as genetically related phenotypes.
Multivariate PRS analyses.
To examine the relative burden of PRS for phenotypes and traits that have shown significant genetic correlation with ADHD and ASD in the past5,6,36 across ADHD and ASD subgroups in the iPSYCH data, we ran a multivariate regression of the scores on these subgroups, adjusting for PCs and batch (for details, see Grove et al.5). In brief, this is a regression of multiple standardized PRSs variables and can superficially be viewed as running a linear regression for each score on the ADHD and ASD subgroups simultaneously. The regression coefficients can be interpreted as the mean value of the PRS relative to the value in controls. The framework allows us to compare the average PRS across subgroups for scores from several phenotypes while accounting for the inherent correlation between scores and adjusting for necessary covariates. This enables testing a whole array of hypotheses comparing both between subgroups and between PRSs. We can compare groups that are too small for GWAS and gauge genetic correlation with groups that are too small for LDSC, as is the case with the comorbid ASD-ADHD group. Polygenic scores were generated by clumping and thresholding employing standard Ricopili settings as explained5 and using summary statistics from the GWASs5,35,80–89.
Leave-one-out PRS analyses.
As a complementary approach, a leave-one-wave-out approach within the iPSYCH data was used to maximize power and maintain independent target and discovery samples for PRS analyses. Meta-analyses were run in METAL (using inverse-variance weighted fixed effects models with the STDERR scheme), including the per-wave GWAS summary results from all but one wave of data, for each combination of waves. Separate meta-analyses were run for GWAS of ADHD-only (excluding comorbid ASD or severe ID, defined as IQ ≤ 50) cases vs. controls and ASD-only (excluding comorbid ADHD or severe ID) cases vs. controls, using independent (split) controls. For each set of discovery results, LD-clumping was run in PLINK v.1.990 (with the parameters --clump-kb 500 --clump-r2 0.3) to obtain a relatively independent set of SNPs, while retaining the most significant SNP in each LD block. The SNP selection P-value threshold used was P < 0.5. Asymmetric/ambiguous SNPs (AT, TA, CG, GC), indels, multi-allelic and duplicate position SNPs were excluded. SNPs with MAF < 0.01, INFO < 0.8 or present in less than half of the sample were filtered out. PRS for ADHD and ASD were calculated by scoring the number of effect alleles weighted by the log(odds ratio [OR]) across the set of independent clumped, meta-analyzed SNPs in PLINK. PRS were derived in best guess imputed data after filtering out SNPs with MAF < 0.05 and INFO < 0.8. The PRS were standardized using z-score transformations; ORs can be interpreted as the increase in risk of the outcome, per standard deviation in PRS. Logistic regression analyses including 5 PCs were run to test for association of PRS with each of the outcomes within each wave, as follows: (a) ADHD-only cases vs. controls; (b) ASD-only cases vs. controls; (c) comorbid cases vs. controls; (d) ADHD-only cases vs. ASD-only cases; (e) ADHD-only cases vs. comorbid cases, and; (f) ASD-only cases vs. comorbid cases. Cases were coded as 1 and controls as 0, except that comorbid cases were coded as 1 in case-case comparisons and in analysis (d), the ASD-only cases were coded as 1. Overall meta-analyses of these per-wave analyses were performed in R using the ‘metafor’ package. As secondary tests, we stratified the ADHD-only and ASD-only cases by presence of mild ID (defined as IQ between 50–70). We also examined differences across several ASD hierarchical subtypes (childhood autism, atypical autism, Asperger’s, and pervasive developmental disorders mixed; see Grove et al.5 and Supplementary Data 9). Several sensitivity tests were also run (including sex as a covariate, excluding cases and controls with mild ID).
Genetic correlations (LD Hub).
The genetic correlations of our different datasets with other phenotypes were evaluated using LD Score regression (LDSC)30 and the LD Hub29 website (http://ldsc.broadinstitute.org/ldhub/). In brief, we re-reran analyses of the original GWAS of ADHD and ASD5,6 in the European-only datasets since new phenotypes have been added to LD Hub after publication of the original analyses. We also uploaded summary statistics for the two analyses described above, i.e., the combined GWAS and the ADHD vs. ASD GWAS, to assess correlation with the identified shared and differentiating genetic liability, respectively. We used all available phenotypes in LD Hub29 but performed analyses for the UKBB traits (n = 597) and the remaining individual phenotypes (n = 257) separately. For ADHD6 and ASD5, the most recent summary statistics replaced corresponding summary statistics in LD Hub as these had not been included at the date of analysis. The same was true for the summary statistics of major depressive disorder85 and bipolar disorder91. Levels of experiment-wide significance (Bonferroni correction for number of tests applied) were also established separately within the two groups, i.e., in the UKBB traits (P < 8.38 × 10−5) and the remaining individual phenotypes (P < 0.00019), respectively.
GCTA-GREML analyses across subgroups.
The additive variance explained by our GWAS dataset (SNP-based heritability; SNP-h2) was estimated in the iPSYCH sample using the GREML approach of GCTA37 for ADHD versus ASD and for ADHD versus each of the ASD sub-phenotypes (see below). The genetic relationship matrix (GRM) between all pairwise combinations of individuals was estimated using all case-control samples. The strict best-guess-genotypes (i.e., SNPs with INFO > 0.8, missing rate < 0.01 and MAF > 0.05, INDELs removed) were used for GRM estimation. GCTA-GREML accounts for linkage disequilibrium (LD)92, and the GRM estimation was performed on a non-LD-pruned dataset. Estimation of the phenotypic variance explained by the SNPs was performed for each of the sub-phenotypes listed in Supplementary Data 9, with PCs 1–20 included as continuous covariates and wave (1–23) as categorical dummy variables. ADHD prevalence of 0.05 and ASD prevalence of 0.01 was assumed to estimate the variance explained on the liability scale. Prevalence was estimated for hierarchical ASD phenotypes based on the estimate for the overall ASD phenotype and the proportion of each hierarchical phenotype over all ASD cases observed in our sample. Genetic covariance between pairs of traits (Supplementary Data 9) was estimated using the bivariate approach implemented in GCTA, by randomly splitting controls into two groups, one for each trait, in proportions corresponding to the proportion of the cases for each of the two traits in the total sample. PCs 1–20 and dummy variables for wave 1–23 were included as covariates in the bivariate analyses. Two-tailed P-values were obtained for rG point estimates based on the standard error estimated by GCTA using the approach by Altman and Bland93.
GCTA-GREML analyses were conducted for ADHD versus ASD main diagnosis (Supplementary Fig. 5a), by (1) excluding individuals with both phenotypes (comorbid) and (2) by randomly splitting comorbid cases into either ADHD or ASD. GCTA analyses were, in addition, conducted for ADHD versus four ASD sub-phenotypes, by (1) excluding individuals with both phenotypes (comorbid) and (2) by randomly splitting comorbid cases into either the ADHD or ASD sub-phenotype. These analyses were conducted both including and excluding individuals with intellectual disability. See Supplementary Data 9 and Supplementary Fig. 5 for an overview of comparisons.
Supplementary Material
Acknowledgements
The iPSYCH team was supported by grants from the Lundbeck Foundation (R102-A9118, R155-2014-1724, and R248-2017-2003), the EU H2020 Program (Grant No. 667302, “CoCA”), NIMH (1U01MH109514-01 to A.D.B.) and the Universities and University Hospitals of Aarhus and Copenhagen. The Danish National Biobank resource was supported by the Novo Nordisk Foundation. High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to A.D.B.). G.V. was funded by NIMH grant K08MH122911 and the 2020 NARSAD Young Investigator Grant #29350 from the Brain & Behavior Research Foundation. P.R. was funded by NIMH grants R01MH125246, U01MH116442, R01MH109677. Support for this research was further received by B.C. from the Spanish ‘Ministerio de Ciencia, Innovación y Universidades’ (RTI2018-100968-B-100), ‘Ministerio de Economía y Competitividad’, ‘AGAUR/Generalitat de Catalunya’ (2017-SGR-738), the European Union H2020 Program [H2020/2014-2020] under grant agreements n°667302 (CoCA), 643051 (MiND) and 728018 (Eat2beNICE), and the ECNP network “ADHD across the lifespan”. S.V.F. is supported by the European Union’s Horizon 2020 research and innovation programme under grant agreements No 667302 and 965381; NIMH grants U01MH109536-01, U01AR076092-01A1, R0MH116037 and 5R01AG06495502; Oregon Health and Science University, Otsuka Pharmaceuticals and Supernus Pharmaceutical Company. We deeply thank all participants in the cohorts included in this analysis.
B.F. has received educational speaking fees from Medice. In the past year, S.V.F. received income, potential income, travel expenses continuing education support and/or research support from Takeda, OnDosis, Tris, Otsuka, Arbor, Ironshore, Rhodes, Akili Interactive Labs, Sunovion, Supernus and Genomind. With his institution, S.V.F. has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. S.V.F. also receives royalties from books published by Guilford Press (Straight Talk about Your Child’s Mental Health), Oxford University Press (Schizophrenia: The Facts), and Elsevier (ADHD: Non-Pharmacologic Interventions). S.V.F. is Program Director of www.adhdinadults.com. B.M.N. is a member of the scientific advisory board at Deep Genomics and Neumora (formerly Neumora) and consultant for Camp4 Therapeutics, Takeda Pharmaceutical, and Biogen. M.J.D. is a founder of Maze Therapeutics and on the Scientific Advisory Board of RBNC Therapeutics.
Footnotes
Code availability
Please refer to individual sections of the methods above for published code (e.g., for EpiXcan or Ricopili, amongst other). Because the in-house scripts used for data processing and analysis of the iPSYCH data on the GenomeDK HPC infrastructure is highly dependent on that context, they can only be obtained from the authors upon request. This way we can ensure the proper context is explained in dialogue with the interested parties.
Competing interests
The other authors declare no competing interests.
Data availability
Summary statistics from this publication are available at http://ipsych.au.dk/downloads/. Summary statistics for the original ADHD and ASD GWAS analyses are also available at this site. For access to genotypes from the PGC samples and the iPSYCH sample, researchers should contact the lead PIs Elise Robinson / Anders Børglum (https://pgc.unc.edu/for-researchers/working-groups/autism-working-group/) and Anders Børglum for PGC-ASD and iPSYCH-ASD, respectively, and Benjamin Neale / Barbara Franke (https://pgc.unc.edu/for-researchers/working-groups/adhd-working-group/) and Anders Børglum for PGC-ADHD and iPSYCH-ADHD, respectively. Data used for brain transcriptome model generation are available from PsychENCODE (overview of available data sets at http://resource.psychencode.org/); genotypes are controlled data and access instructions are provided at https://www.synapse.org/#!Synapse:syn4921369/wiki/477467. Note that some datasets have been indirectly accessed at the respective analytical websites (e.g., GSE76381 through the FUMA website). Please refer to these websites (e.g., for FUMA https://fuma.ctglab.nl/links and https://fuma.ctglab.nl/tutorial#datasets) for availability of datasets used in the respective follow-up analyses / lookups (e.g., GSE76381).
References
- 1.Dalsgaard S. et al. Incidence rates and cumulative incidences of the full spectrum of diagnosed mental disorders in childhood and adolescence. JAMA Psychiatry 77, 155–164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faraone SV & Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry 24, 562–575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pettersson E. et al. Genetic influences on eight psychiatric disorders based on family data of 4 408 646 full and half-siblings, and genetic data of 333 748 cases and controls. Psychol Med 49, 1166–1173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandin S. et al. The heritability of autism spectrum disorder. JAMA 318, 1182–1184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grove J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demontis D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51, 63–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matoba N. et al. Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl Psychiatry 10, 265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482 e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin J. et al. Biological overlap of attention-deficit/hyperactivity disorder and autism spectrum disorder: evidence from copy number variants. J Am Acad Child Adolesc Psychiatry 53, 761–770 e26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satterstrom FK et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat Neurosci 22, 1961–1965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rommelse NN, Geurts HM, Franke B, Buitelaar JK & Hartman CA A review on cognitive and brain endophenotypes that may be common in autism spectrum disorder and attention-deficit/hyperactivity disorder and facilitate the search for pleiotropic genes. Neurosci Biobehav Rev 35, 1363–1396 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Zablotsky B, Bramlett MD & Blumberg SJ The co-occurrence of autism spectrum disorder in children with ADHD. J Atten Disord 24, 94–103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai MC et al. Prevalence of co-occurring mental health diagnoses in the autism population: a systematic review and meta-analysis. Lancet Psychiatry 6, 819–829 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Ottosen C. et al. Sex differences in comorbidity patterns of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 58, 412–422 e3 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Ghirardi L. et al. The familial co-aggregation of ASD and ADHD: a register-based cohort study. Mol Psychiatry 23, 257–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z. et al. Investigating shared genetic basis across Tourette syndrome and comorbid neurodevelopmental disorders along the impulsivity-compulsivity spectrum. Biol Psychiatry 90, 317–327 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabourdy F. et al. A MANBA mutation resulting in residual beta-mannosidase activity associated with severe leukoencephalopathy: a possible pseudodeficiency variant. BMC Med Genet 10, 84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W. et al. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat Commun 10, 3834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Leeuw CA, Mooij JM, Heskes T. & Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe K, Taskesen E, van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peyrot WJ & Price AL Identifying loci with different allele frequencies among cases of eight psychiatric disorders using CC-GWAS. Nat Genet 53, 445–454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JJ et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marzluff WF, Gongidi P, Woods KR, Jin J. & Maltais LJ The human and mouse replication-dependent histone genes. Genomics 80, 487–498 (2002). [PubMed] [Google Scholar]
- 27.Zhao B. et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet 51, 1637–1644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baselmans BML et al. Multivariate genome-wide analyses of the well-being spectrum. Nat Genet 51, 445–451 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Zheng J. et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan B. et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corces MR et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat Genet 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graciarena M, Seiffe A, Nait-Oumesmar B. & Depino AM Hypomyelination and oligodendroglial alterations in a mouse model of autism spectrum disorder. Front Cell Neurosci 12, 517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu ZM et al. White matter microstructural alterations in children with ADHD: categorical and dimensional perspectives. Neuropsychopharmacology 42, 572–580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki Y. et al. Association of white matter structure with autism spectrum disorder and attention-deficit/hyperactivity disorder. JAMA Psychiatry 74, 1120–1128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neale BM et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 49, 884–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagel M, Watanabe K, Stringer S, Posthuma D. & van der Sluis S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat Commun 9, 905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satterstrom FK et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584 e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duffney LJ et al. Epigenetics and autism spectrum disorder: A report of an autism case with mutation in H1 linker histone HIST1H1E and literature review. Am J Med Genet B Neuropsychiatr Genet 177, 426–433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Rubeis S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryant L. et al. Histone H3.3 beyond cancer: germline mutations in Histone 3 Family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients. Sci Adv 6, eabc9207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian K. et al. Basal ganglia and autism - a translational perspective. Autism Res 10, 1751–1775 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Clarke TK et al. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol Psychiatry 21, 419–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traut N. et al. Cerebellar volume in autism: literature meta-analysis and analysis of the Autism Brain Imaging Data Exchange Cohort. Biol Psychiatry 83, 579–588 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Hoogman M. et al. Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry 4, 310–319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw P. et al. A multicohort, longitudinal study of cerebellar development in attention deficit hyperactivity disorder. J Child Psychol Psychiatry 59, 1114–1123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfers T. et al. Individual differences v. the average patient: mapping the heterogeneity in ADHD using normative models. Psychol Med 50, 314–323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fliers E. et al. Motor coordination problems in children and adolescents with ADHD rated by parents and teachers: effects of age and gender. J Neural Transm (Vienna) 115, 211–220 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Franke B. et al. Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. Eur Neuropsychopharmacol 28, 1059–1088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basile GA et al. Red nucleus structure and function: from anatomy to clinical neurosciences. Brain Struct Funct 226, 69–91 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalsgaard S, Nielsen HS & Simonsen M. Five-fold increase in national prevalence rates of attention-deficit/hyperactivity disorder medications for children and adolescents with autism spectrum disorder, attention-deficit/hyperactivity disorder, and other psychiatric disorders: a Danish register-based study. J Child Adolesc Psychopharmacol 23, 432–439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg RE et al. Psychotropic medication use among children with autism spectrum disorders enrolled in a national registry, 2007–2008. J Autism Dev Disord 40, 342–351 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Dalsgaard S, Leckman JF, Mortensen PB, Nielsen HS & Simonsen M. Effect of drugs on the risk of injuries in children with attention deficit hyperactivity disorder: a prospective cohort study. Lancet Psychiatry 2, 702–709 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Chang Z, D’Onofrio BM, Quinn PD, Lichtenstein P. & Larsson H. Medication for attention-deficit/hyperactivity disorder and risk for depression: a nationwide longitudinal cohort study. Biol Psychiatry 80, 916–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang Z. et al. Medication for attention-deficit/hyperactivity disorder and risk for suicide attempts. Biol Psychiatry 88, 452–458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keilow M, Holm A. & Fallesen P. Medical treatment of attention deficit/hyperactivity disorder (ADHD) and children’s academic performance. PLoS One 13, e0207905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brainstorm Consortium et al. Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polderman TJ, Hoekstra RA, Posthuma D. & Larsson H. The co-occurrence of autistic and ADHD dimensions in adults: an etiological study in 17,770 twins. Transl Psychiatry 4, e435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronald A, Larsson H, Anckarsater H. & Lichtenstein P. Symptoms of autism and ADHD: a Swedish twin study examining their overlap. J Abnorm Psychol 123, 440–451 (2014). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 60.Pedersen CB et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry 23, 6–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patterson N, Price AL & Reich D. Population structure and eigenanalysis. PLoS Genet 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Lam M. et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics 36, 930–933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willer CJ, Li Y. & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715 e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watanabe K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet 51, 1339–1348 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Buniello A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Byrne EM et al. Conditional GWAS analysis to identify disorder-specific SNPs for psychiatric disorders. Mol Psychiatry 26, 2070–2081 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gandal MJ et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Das S. et al. Next-generation genotype imputation service and methods. Nat Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarthy S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao C. et al. Power analysis of transcriptome-wide association study: Implications for practical protocol choice. PLoS Genet 17, e1009405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Consortium GTEx. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X. et al. Functional architectures of local and distal regulation of gene expression in multiple human tissues. Am J Hum Genet 100, 605–616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe K, Umicevic Mirkov M, de Leeuw CA, van den Heuvel MP & Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat Commun 10, 3222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grotzinger AD et al. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis. Nat Genet 54, 548–559 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davies G. et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Mol Psychiatry 21, 758–767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okbay A. et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benyamin B. et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry 19, 253–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sniekers S. et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet 49, 1107–1112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wray NR et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okbay A. et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 48, 624–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones SE et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet 12, e1006125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deary V. et al. Genetic contributions to self-reported tiredness. Mol Psychiatry 23, 609–620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42, 441–447 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stahl EA et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang J, Lee SH, Wray NR, Goddard ME & Visscher PM GCTA-GREML accounts for linkage disequilibrium when estimating genetic variance from genome-wide SNPs. Proc Natl Acad Sci U S A 113, E4579–E4580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Altman DG & Bland JM How to obtain the confidence interval from a P value. BMJ 343, d2090 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics from this publication are available at http://ipsych.au.dk/downloads/. Summary statistics for the original ADHD and ASD GWAS analyses are also available at this site. For access to genotypes from the PGC samples and the iPSYCH sample, researchers should contact the lead PIs Elise Robinson / Anders Børglum (https://pgc.unc.edu/for-researchers/working-groups/autism-working-group/) and Anders Børglum for PGC-ASD and iPSYCH-ASD, respectively, and Benjamin Neale / Barbara Franke (https://pgc.unc.edu/for-researchers/working-groups/adhd-working-group/) and Anders Børglum for PGC-ADHD and iPSYCH-ADHD, respectively. Data used for brain transcriptome model generation are available from PsychENCODE (overview of available data sets at http://resource.psychencode.org/); genotypes are controlled data and access instructions are provided at https://www.synapse.org/#!Synapse:syn4921369/wiki/477467. Note that some datasets have been indirectly accessed at the respective analytical websites (e.g., GSE76381 through the FUMA website). Please refer to these websites (e.g., for FUMA https://fuma.ctglab.nl/links and https://fuma.ctglab.nl/tutorial#datasets) for availability of datasets used in the respective follow-up analyses / lookups (e.g., GSE76381).