Abstract
Surgical resection represents the standard of care for people with newly diagnosed diffuse gliomas, and the neuropathological and molecular profile of the resected tissue guides clinical management and forms the basis for research. The Response Assessment in Neuro-Oncology (RANO) consortium is an international, multidisciplinary effort that aims to standardise research practice in neuro-oncology. These recommendations represent a multidisciplinary consensus from the four RANO groups: RANO resect, RANO recurrent glioblastoma, RANO radiotherapy, and RANO/PET for a standardised workflow to achieve a representative tumour evaluation in a disease characterised by intratumoural heterogeneity, including recommendations on which tumour regions should be surgically sampled, how to define those regions on the basis of preoperative imaging, and the optimal sample volume. Practical recommendations for tissue sampling are given for people with low-grade and high-grade gliomas, as well as for people with newly diagnosed and recurrent disease. Sampling of liquid biopsies is also addressed. A standardised workflow for subsequent handling of the resected tissue is proposed to avoid information loss due to decreasing tissue quality or insufficient clinical information. The recommendations offer a framework for prospective biobanking studies.
Introduction
In patients with newly diagnosed diffuse glioma, maximal safe resection of the contrast-enhancing tumour on MRI represents the standard of care.1,2 Additional resection of non-contrast-enhancing tumour beyond the contrast-enhancing margins is increasingly being considered as a surgical goal if safely feasible.3,4 During the course of the disease, surgery is also often performed due to tumour progression or recurrence.5,6 Following resection, neuropathological evaluation of the tumour tissue allows accurate diagnosis to guide medical therapies.7 The resected tissue forms the basis for research, allowing insights into pathogenetic mechanisms; identification of biomarkers, including potential therapeutic targets; or pharmacodynamics and pharmacokinetics.8 However, there is no consensus about how the tumour tissue should be sampled during a resection in a standardised way to achieve representative insights into the tumour architecture. This concept of deliberate tissue sampling is particularly relevant given the considerable spatial intratumoural heterogeneity that has been shown at genetic, epigenetic, transcriptional, and cellular levels.9 Moreover, guidelines on sample handling, including labelling and storage, are yet to be defined, thus hampering comparative analyses among individual studies and different tumour tissue collection protocols.
Here, a multiprofessional group of experts from the fields of neurosurgery, neuropathology, medical neurooncology, radiation oncology, and neuroimaging (including representatives of the four Response Assessment in Neuro-Oncology [RANO] working groups: RANO resect, RANO recurrent glioblastoma, RANO radiotherapy, and RANO/PET) assembled to elaborate on the tissue volume and imaging-based tumour-specific regions from where the tumour should be sampled, the definition of how regions for sampling the non-contrast-enhancing presumed tumour on MRI should be identified, the consensus for blood and cerebrospinal fluid (CSF) sampling, the handling and storage recommendations to enable both accurate neuropathological diagnosis and in-depth research analyses, and the preferred sample labelling to avoid miscommunication across clinical specialities and research facilities and maximise the quality of clinical or technical information collected at the time of surgery.
The resulting suggestions for tissue sampling and handling during and after glioma resection aim to offer a framework to standardise the workflow and improve the quality of neuropathological diagnosis and prospective biobanking studies for basic and clinical research purposes.
Methods
Search strategy and selection criteria
An expert core group of representatives from the four RANO groups was formed (ie, the authors). We searched PubMed to identify current literature recommendations on tissue sampling, labelling, and storage using various combinations of the following search terms: “glioma”, “glioblastoma”, “tissue”, “MRI”, “PET”, “contrast enhancing”, “non-contrast-enhancing”, “heterogeneity”, “DNA”, “neuronavigation”, “spatial”, “therapy”, “resection”, “chemotherapy”, “radiotherapy”, “selection pressure”, “volume”, “diagnosis”, “neuropathology”, “frozen”, “storage”, “fixatives”, “formalin”, and “FFPE”. Google Scholar, the authors’ own files, and the references from relevant articles were also searched; only studies published in English-language journals between Jan 1, 2003, and July 1, 2023, were considered. The first and senior authors (PK and J-CT) screened papers for relevance, removed duplicates, and circulated information extracted from a preliminary reference list within the expert core group. The final reference list was then selected by all authors of the core group on the basis of originality and relevance to the topics covered in this paper. On the basis of the selected literature, a qualitative synthesis of the literature and recommendations on tissue sampling, labelling, and handling during glioma resection were formulated by the core group. Consensus between the authors was achieved through repeated circulation of the manuscript drafts.
Modified eDelphi survey: refinement of recommendations
For refinement of the recommendations, a panel of leading experts from the fields of surgical and medical neuro-oncology, neuropathology, radiation oncology, and neuroimaging were selected based on clinico-scientific expertise and previous academic contributions to the community. 50 experts were asked to participate, of whom 37 agreed to contribute as an Expert Rater Panel.
We used a two-stage eDelphi survey method, in which written questionnaires in English were provided to the panellists. In the first round of the eDelphi survey, each panellist received an instruction letter and a personalised file to score the importance of the individual recommendations. A 9-point Likert scale (ie, 1–3 points indicating limited importance, 4–6 points indicating importance but not critical importance, and 7–9 points indicating critical importance) was used for scoring, and an opportunity to explain the reasoning for the score was provided. Panellists were allowed to not rate individual recommendations if deemed outside their specific area of expertise. Outcomes of the first round of the eDelphi survey were reviewed by the expert core group. Each recommendation that was scored less than critical (ie, ≤6 points) by at least 20% of the panellists was revised by the expert core group on the basis of the provided feedback from the Expert Rater Panel. Only panellists who voted in the first round were allowed to proceed to the second round of the eDelphi survey. In the second round, the results for all recommendations were circulated and the revised recommendations (with highlighted changes) were open for re-scoring using the 9-point Likert scale. Beyond this final round, each recommendation that received 7 points or higher by 70% or more of the panellists was accepted as consensus, and each recommendation that received less than 7 points by more than 30% of the panellists was excluded. 36 panellists completed both stages of the eDelphi process (one panellist completed only the first round). Agreement for each recommendation is indicated as the number of experts who scored the individual recommendation as critical (ie, ≥7 points) out of the number of experts who responded to that recommendation.
Results
On the basis of the suggestions from the expert core group and the refinement from the eDelphi survey, recommendations for intraoperative tissue sampling and sample processing were formulated (panels 1, 2).
Panel 1: Consensus recommendations for intraoperative tissue sampling during diffuse glioma resection.
Preoperative imaging requirements for intraoperative neuronavigation
Recommendation 1: Intraoperative neuronavigation based on preoperatively obtained MRI should be used for anatomical orientation and distinguishing the various glioma imaging characteristics during resection.*
Recommendation 2: Minimum preoperative MRI datasets include T2-weighted sequences and contrast-enhanced T1-weighted sequences.*
Recommendation 3: MRI for neuronavigation should be obtained as close to resection as reasonably possible. As a benchmark, imaging should be obtained not more than 7 days before the resection. New clinical symptoms necessitate more proximate imaging.*
Recommendation 4: If available, data from physiological and metabolic MRI (eg, diffusion-weighted MRI, perfusion MRI, and magnetic resonance spectroscopy) and molecular imaging techniques (eg, amino acid PET) should be used to visualise metabolic hotspots and non-contrast-enhancing tumour. Visualisation of metabolic hotspots and non-contrast-enhancing tumour is especially important in the post-treatment recurrence setting when pseudoprogression is considered.
Recommendation 5: Whenever information from amino acid PET, physiological or metabolic MRI, or intraoperative tumour fluorescence is available, it should be noted for each individual sample taken. In case of intended removal, portions with or without the respective imaging signal should be labelled accordingly.*
Localisation of intraoperative sampling with respect to tumour appearance on MRI
Recommendation 6: Written patient (or caregiver) consent and institutional review board approval need to be obtained for storing tissue for research purposes beyond the clinical routine diagnostics.*
Recommendation 7: Although associated with uncertainty given the scarcity of prospective data, T2-hyperintense and FLAIR-hyperintense areas with less pronounced tumour cellularity (ie, oedema) might be indicated by the following MRI features: (1) strongly hyperintense on T2-weighted and FLAIR sequences (isointense or hyperintense compared with CSF on T2-weighted imaging; hyperintense compared with physiological white matter) and (2) respecting the integrity of the grey-white matter boundary without affecting the anatomical architecture of the cortex. More advanced MRI (including diffusion-weighted imaging) or PET imaging might provide additional insights.*
Recommendation 8: Although associated with uncertainty given the scarcity of prospective data, T2-hyperintense and FLAIR-hyperintense areas with high tumour cellularity (ie, non-contrast-enhancing tumour) might be indicated by the following MRI features: (1) mildly hyperintense on T2-weighted and FLAIR sequences (hypointense or isointense compared with CSF on T2-weighted imaging; hyperintense to physiological white matter) and (2) disrupting the anatomical architecture (ie, grey–white matter boundary) of the infiltrated tissue. More advanced MRI (including diffusion-weighted imaging) or PET imaging might provide additional insights.*
Recommendation 9: To identify regions of high tumour cellularity within T2-hyperintense and FLAIR-hyperintense abnormalities without contrast enhancement, closer proximity to contrast-enhancing tumour on MRI might increase the probability that that tissue contains a higher fraction of tumour cells. More advanced MRI or PET imaging might provide additional insights by showing metabolic or physiological changes associated with vital tumour.*
Recommendation 10: Separate samples should be taken from the tumour core (even when necrotic), the contrast-enhancing tumour portion, the surrounding non-contrast-enhancing presumed tumour portion, and the immediately adjacent macroscopically unaffected brain parenchyma, when clinically indicated and feasible.*
Recommendation 11: The precise location and presence or absence of contrast enhancement of the samples collected should be electronically documented with the intraoperative neuronavigation system or similarly adequate techniques (eg, stereotactic biopsy).*
Recommendation 12: Samples should be obtained early during resection to minimise anatomical distortions (ie, brain shift) on neuronavigation.*
Localisation of intraoperative sampling with respect to spatial tumour heterogeneity
Recommendation 13: When performing a large anatomical resection, such as an en bloc resection, specific documentation of sample orientation and margins should be performed to correlate samples with their respective locations on MRI. In cases where spatial mapping was not achieved, it should be explicitly disclosed in the information accompanying the samples.
Recommendation 14: In case of an intralesional piecemeal resection, at least two samples per respective MRI abnormality (ie, necrotic tumour core, contrast-enhancing tumour, presumed non-contrast-enhancing tumour, and macroscopically unaffected brain parenchyma if safe) should be obtained during debulking and documented with screenshots on neuronavigation. To account for spatial intratumoural heterogeneity, these two samples should be derived from regions as distant from each other as possible, if the surgical workflow safely allows.*
Sampling in the context of recurrent disease
Recommendation 15: Pretreatment information on systemic chemotherapy or localised treatment approaches (eg, radiotherapy or surgery) should be noted for the tumour area from which samples are being taken. Attention should be paid to whether the recurrent lesion is within the radiation field (ie, in-field vs out-of-field recurrence) or whether this particular area had previously undergone resection (ie, local vs distant recurrence).*
Recommendation 16: In the recurrent setting, samples should particularly be taken from actively growing tumour, as suspected from imaging, to enhance probability of sampling viable tumour.*
Recommendation 17: To ensure that stored research samples contain as much viable recurrent tumour as possible, close cooperation with the reporting neuropathologist (or pathologist) should be ensured. If biopsy alone is pursued for diagnostic purposes, intraoperative smear or frozen sections might be used to enhance probability of acquiring tissue with a high diagnostic yield.
Minimal volumes of tumour samples for further analyses
Recommendation 18: To accurately depict spatial intratumoural heterogeneity, separate samples might be defined as individual samples for research purposes if a minimum distance of 1 cm separates them.*
Recommendation 19: All resected, potentially viable tissue should be sampled for clinical or research purposes, and no tumour tissue should be discarded if local storage capacities allow for storage.*
Blood sampling as a source of germline DNA
Recommendation 20: The concurrent sampling of a peripheral blood sample as a source of matched, autologous constitutional DNA should be considered to distinguish whether DNA alterations in tumour tissue samples are of germline or somatic origin.
Recommendation 21: Patient consent must be obtained before large-scale sequencing of constitutional DNA due to the possible detection of germline alterations in genes predisposing for hereditary cancer syndromes (or even non-cancer diseases). Additionally, national requirements regarding genetic counselling have to be followed.
Blood and CSF sampling for scientific purposes, biomarker discovery, and immune phenotyping
Recommendation 22: If local storage capacities and institutional review board regulations permit, surgical interventions at initial glioma diagnosis and for presumed progression could allow for collection of peripheral blood specimens (ie, with EDTA [edetic acid] tubes, citrate tubes, and tubes without anticoagulants) for biobanking. This process could facilitate future translation of liquid biopsy approaches in neuro-oncology.
Recommendation 23: CSF can be considered for storage for future research purposes whenever it is sampled for clinical diagnostics or is surgically available, without additional risk, as part of routine procedures. These procedures might include lumbar punctures, but also clinically necessary steps of surgeries (eg, opening of basal cisterns or ventricles allowing drainage of CSF that is not contaminated by blood or brain tissue). A minimum target volume of 3 mL should be aimed for (but smaller volumes should not be discarded).
CSF=cerebrospinal fluid. FLAIR=T2-weighted fluid-attenuated inversion recovery. *Key recommendation.
Panel 2: Consensus recommendations for tissue processing after intraoperative sampling.
Sample processing in the operating room
Recommendation 24: Tissue samples should be processed in a timely manner and be cooled (not frozen) until final processing and fixation or deep freezing.*
Recommendation 25: Time of specimen acquisition and time of further processing should be recorded separately.
Final sample fixation for long-term storage
Recommendation 26: Samples should be fixed in buffered 4% formalin and embedded in paraffin for routine diagnostics.*
Recommendation 27: For biobanking purposes, samples should also be shock frozen in liquid nitrogen and stored as deep-frozen specimens in addition to formalin-fixed paraffin-embedded samples; and additional live tissue might be used to establish tumour-derived cell lines or organoids.*
Recommendation 28: Any excess tissue remaining after selection of samples to be fixed in formalin and embedded in paraffin for diagnostic purpose should not be discarded. Excess tissue should be either stored as deep-frozen samples or (only for short term) as wet tissue in formalin so that it can be used when needed at a later timepoint (eg, for diagnostic purposes when the original formalin-fixed paraffin-embedded samples are not sufficient to establish a definite diagnosis).*
Recommendation 29: Deep-frozen tissue specimens preserved in addition to formalin-fixed and paraffin-embedded tissue samples should not be exhausted for research purposes before the final diagnosis has been established.*
Recommendation 30: After establishment of diagnosis and storage of adequate tissue to allow future testing for clinical purposes (eg, to enter into a clinical trial), excess deep-frozen tissue that is not a part of the hospital archives might be indefinitely released for biobanking purposes. This process requires the patient’s consent and must be in accordance with local guidelines.
Labelling of the biobanking samples
Recommendation 31: Samples should be labelled with two unique identifiers and the date of surgery to allow back-tracking of clinical information, including re-identification of the patient’s identity, if national data protection laws allow. Legibility should be ensured by labelling in a manner that will endure storage conditions.*
Recommendation 32: Information on the exact sample position on MRI and other intraoperative information (eg, metabolic signal or intraoperative fluorescence) should be matched to a patient’s specific sample using an individual reference number.
*Key recommendation.
Intraoperative tissue sampling during diffuse glioma resection
Preoperative imaging requirements for intraoperative neuronavigation
A reliable visual distinction between the contrast-enhancing and non-contrast-enhancing tumour portions during surgery is morphologically not possible, but modern tools enable accurate visualisation of the surgeon’s intraoperative position.10 As such, intraoperative neuronavigation based on preoperatively obtained MRI (with a high resolution, such as 1×1×1 mm3) should be used when available to aid in distinguishing contrast-enhancing and non-contrast-enhancing tumour portions (recommendation 1: 33 [94%] of 35 experts agreed). On MRI, diffuse gliomas present as mostly hyperintense lesions on non-contrast-enhanced T2-weighted and T2-weighted fluid-attenuated inversion recovery (FLAIR) sequences, with variable signal on contrast-enhanced T1-weighted sequences (figure 1). As such, minimum preoperative MRI datasets include T2-weighted sequences and contrast-enhanced T1-weighted sequences (recommendation 2: 34 [92%] of 37 experts agreed).11 Among others, precontrast T1-weighted sequences and FLAIR sequences offer additional help to assess the tumour morphology. We recommend to use the standardised Brain Tumor Imaging Protocol, particularly in the setting of clinical trials.11
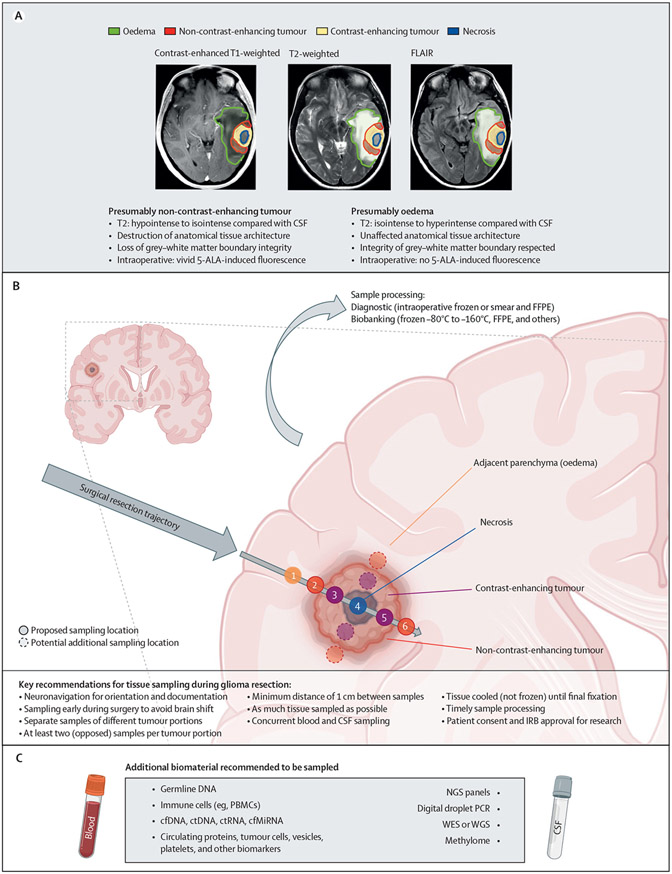
Figure 1: Proposed sampling recommendations.
(A) Tumour portions of a tempero-parietal glioblastoma as delineated on MRI. (B) Proposed sampling localisations respecting the surgical trajectory. Figure panel created with BioRender.com. (C) Additional biomaterials and potential applications. Figure panel created with BioRender.com. 5-ALA=5-aminolevulinic acid. cfDNA=cell-free DNA. cfMiRNA=cell-free microRNA. CSF=cerebrospinal fluid. ctDNA=circulating tumour DNA. ctRNA=circulating tumour RNA. FFPE=formalin-fixed paraffin-embedded. FLAIR=T2-weighted fluid-attenuated inversion recovery. IRB=institutional review board. NGS=next-generation sequencing. PBMCs=peripheral blood monocytes. WES=whole-exome sequencing. WGS=whole-genome sequencing.
Diffuse IDH-mutant gliomas of CNS WHO grade 2 often show little or no contrast-enhancement.2,4 Diffuse IDH-mutant gliomas of CNS WHO grade 3 and 4 might show at least focal contrast-enhancement.2 Most IDH-wildtype glioblastomas show vivid contrast-enhancement of the tumour core (often surrounding a central necrotic area without contrast-enhancement) accompanied by a non-contrast-enhancing rim.3,12 Such non-contrast-enhancing tissue abnormalities in the periphery of diffuse gliomas represent a combination of tumour cell invasion with an intact blood–brain barrier and oedema; however, brain infiltration by glioma cells extends beyond the tumour borders that are visible on MRI into brain parenchyma that appears morphologically unaffected on MRI.13
Contrast-enhancing gliomas (eg, IDH-wildtype glioblastomas of grade 4) can show changes in tumour size, configuration, and contrast-enhancing uptake within a few weeks. On the basis of pretherapeutic MRIs from 106 patients with glioblastoma, Stensjøen and colleagues estimated a growth rate of 1·4% per day and a radiographic volume-doubling time of 49·6 days.14 An even shorter doubling time of 21·1 days was reported by Ellingson and colleagues.15 To avoid discrepancies between intraoperative findings and preoperative imaging, MRI for neuronavigation should be obtained as close to the resection date as possible. As such, MRI should be obtained not more than 7 days preceding surgical tumour resection, according to expert consensus (recommendation 3: 22 [100%] of 22 experts agreed). Although we acknowledge the typically slower growth dynamics of non-contrast-enhancing tumours (eg, IDH-mutant gliomas of grade 2–3), imaging studies for such tumours should also be performed no more than 7 days preceding surgery for neuronavigation and because IDH-wildtype glioblastoma without contrast-enhancement cannot be excluded on the basis of imaging alone. Notably, the exactness of this 7-day time window is arbitrary and should serve only as a benchmark recommendation. If new neurological symptoms arise or major medication changes affecting tumour morphology are made, then new imaging should be obtained.
Beyond conventional MRI, physiological and metabolic MRI techniques (eg, diffusion-weighted imaging, perfusion MRI, and magnetic resonance spectroscopy) and advanced molecular imaging techniques involving radiolabelled amino acids (eg, [18F]fluoroethyl-L-tyrosine-PET, [11C]methionine-PET, or [18F]fluorodopa-PET) allow insights into tumour cellularity, perfusion, and metabolism of diffuse gliomas.16,17 Metabolic abnormalities, hyperperfusion, hypercellularity, and high uptake of amino acid tracer in diffuse gliomas often extend beyond areas of contrast enhancement on T1-weighted imaging.18-20 Furthermore, these imaging techniques depict hotspots in non-contrast-enhancing tumour portions of diffuse gliomas, which are presumably related to aggressive tumour growth.21-24 The visualisation of tumour hotspots is particularly relevant in the post-treatment setting to distinguish between tumour recurrence and treatment effects. Thus, these areas should be specifically targeted by integrating biological imaging (if available) into neuronavigation, allowing for intraoperative spatial co-registration with anatomical images (recommendation 4: 30 [88%] of 34 experts agreed). Additionally, tumour fluorescent-labelling techniques using 5-aminolevulinic acid (5-ALA) or fluorescein were established to improve the intraoperative delineation of the most proliferative areas in gliomas of CNS WHO grades 3 and 4.25,26 5-ALA is the most commonly used intraoperative fluorescent dye in glioma surgery. Vivid violet-red signal typically indicates the contrast-enhancing tumour bulk, pink signal extends into the non-contrast-enhancing tumour parts, and no fluorescence is shown for brain tissue with no or only minimal infiltration or for necrosis.25,27,28 5-ALA might be equally as helpful in supporting maximal safe resection of glioblastoma as intraoperative MRI.29 Accordingly, 5-ALA is recommended for intraoperatively distinguishing different tumour portions to be sampled in contrast-enhancing gliomas (particularly in post-treatment disease or situations when neuronavigation is less reliable than usual due to brain shift). Notably, accumulation of fluorescent dyes within samples can change their ex-vivo optical properties, which might affect intravital microscopy, immunofluorescence analysis, or flow cytometry studies.30 If known, the appearance of the individual tissue samples in these visualisation methods should be noted when stored for research purposes (recommendation 5: 30 [88%] of 34 experts agreed).
Localisation of intraoperative sampling with respect to tumour appearance on MRI
Patient (or caregiver) consent and institutional review board approval per local regulations need to be obtained in written form for storing tissue for research purposes beyond the clinical routine diagnostics (recommendation 6: 35 [95%] of 37 experts agreed). In emergency situations (eg, emergency craniotomies in cases of brain herniation due to tumour-related mass effects), tissue might be stored for routine diagnostics only and released for research at a later timepoint when approval is obtained.
The necrotic tumour core, contrast-enhancing tumour, non-contrast-enhancing presumed tumour, and macroscopically unaffected (but microscopically infiltrated) adjacent brain parenchyma might each possess distinct cellular, genetic, and epigenetic properties.9,31-33 Careful review of different MRI sequences might sometimes show characteristics that are suggestive of non-contrast-enhancing tumour with high tumour cell density (figure 1A).12 Although always associated with some degree of uncertainty, areas of less pronounced cellularity (ie, oedema) often have a strongly hyperintense signal on T2-weighted and FLAIR sequences (isointense or hyperintense compared with CSF on T2-weighted sequences; hyperintense compared with physiological white matter) and generally respect the grey–white matter boundary without affecting the cortex (recommendation 7: 25 [76%] of 33 experts agreed).3,34 By contrast, non-contrast-enhancing diffuse glioma parts with high cellularity might appear less hyperintense on T2-weighted and FLAIR sequences (hypointense or isointense compared with CSF on T2-weighted sequences; hyperintense compared with physiological white matter) than oedema and might destroy the anatomical structure of the infiltrated tissue, causing focal mass effect (recommendation 8: 26 [87%] of 30 experts agreed).3,34 Also, non-contrast-enhancing tumour is often located in closer proximity to contrast-enhancing tumour than are areas of oedema with only low tumour cell density (recommendation 9: 24 [71%] of 34 experts agreed).35,36 These MRI-based criteria will need to be correlated with neuropathological findings in prospective studies, and might eventually undergo refinement. In the current setting, these recommendations will help to establish a uniform nomenclature for sampling but might eventually also help to identify non-contrast-enhancing regions to be targeted by resection or radiotherapy. These imaging-based criteria need to be used cautiously in post-treatment situations, because therapy-induced changes might mimic tumour progression on conventional MRI.
We advocate that tissue should be sampled from each individual tumour portion that is visible on MRI to avoid undersampling: the tumour core, including necrosis with peri-necrotic tissue (if present), the contrast-enhancing tumour portion (if present), the surrounding non-contrast-enhancing tumour portion, and, when feasible (diagnostically or based on surgical trajectory), the immediately adjacent non-functioning and macroscopically unaffected brain parenchyma (recommendation 10: 30 [81%] of 37 experts agreed; figure 1B). Distant non-affected brain tissue might also be acquired along the surgical trajectory to the tumour, when feasible. Importantly, sampling refers only to areas that are clinically indicated to be resected for either diagnostic purposes or for cytoreduction.
The respective sampling position should be electronically documented with the intraoperative neuronavigation system so that these specimens are permanently linked to the image localisation from where they are taken (recommendation 11: 32 [91%] of 35 experts agreed). Minimal cauterisation should be used to preserve sample viability. During resection of hemispheric tumours, anatomical distortions (ie, brain shift) violating the rigid body assumption of neuronavigation might occur, which translate into differences between the reported virtual location and the actual location. Samples should therefore be obtained early during resection (recommendation 12: 28 [88%] of 32 experts agreed). In cases of uncertainty, navigated three-dimensional ultrasound or acquisition of intraoperative MRI can serve for orientation, adjustment of intraoperative neuronavigation, and definition of sampling localisation during later stages of resection.37,38
Localisation of intraoperative sampling with respect to spatial tumour heterogeneity
Diffuse gliomas of CNS WHO grade 4 in particular, but also diffuse gliomas of CNS WHO grades 2 or 3, exhibit considerable spatial intratumoural heterogeneity. In this context, tumour tissue sampled from spatially distinct areas (not necessarily of different appearance on preoperative diagnostic imaging) might have distinct molecular profiles indicating subclonal diversification in addition to joint clonal alterations linked to tumour initiation.9 Therefore, multiple samples would be required to craft an approximation of the true spatial cellular, molecular, and genetic landscape encountered within the tumour tissue.33 Although we do not advocate for a specific surgical technique, we acknowledge that there might be two general methods for glioma resection. The en bloc resection of gliomas by circumferential preparation of perilesional and lesional tissue preserves the entire tumour architecture,39 but such an approach requires specific documentation of sample orientation and margins to identify the exact MRI localisation of single tissue fragments from the sample later (recommendation 13: 30 [86%] of 35 experts agreed). Intralesional piecemeal resection might be the only feasible approach in about 60% of people with newly diagnosed glioblastoma undergoing resection due to crucial neurovascular or cerebral structures surrounding the tumour.39 In this setting, at least two samples per respective MRI abnormality (ie, necrotic tumour core; contrast-enhancing tumour; non-contrast-enhancing presumed tumour; and macroscopically unaffected, but presumably histologically infiltrated, adjacent brain parenchyma) should be taken from opposing regions as distant from each other as possible to assess spatial tumour heterogeneity (recommendation 14: 27 [84%] of 32 experts agreed; figure 1B). If geographical sample mapping cannot be achieved in some people with diffuse intracranial glioma, it should be explicitly disclosed in the information accompanying the samples.
Sampling in the context of recurrent disease
Previous therapies (eg, chemotherapy, radiotherapy, and surgery) might induce selective pressures on glioma cells and infiltration of non-tumorous immune cells, which shape the longitudinal subclonal and clonal tumour architecture.35,40-43 As such, clonal evolution of tumour cells during the natural history of the disease and as a result of therapeutic interventions might translate into relevant divergence in expression of potentially targetable alterations between paired untreated and treated recurrent tumour samples.44,45 Therefore, information regarding previous treatments should be collected or at least made retrospectively available (recommendation 15: 32 [86%] of 37 experts agreed).
Previous therapies might not only alter the mutational profile and (sub-)clonal composition of the tumour cells but also lead to marked reactive changes, such as reactive gliosis, inflammation, scar formation, and radionecrosis. Thus, particular care should be taken to select samples from active tumour tissue and not just areas with therapy-induced reactive changes (recommendation 16: 35 [95%] of 37 experts agreed). Specific imaging features (sometimes referred to as soap-bubble-like or Swiss-cheese-like appearance) and spatiotemporal patterns have been described for treatment-induced changes,46 and might allow distinction from true tumour recurrence on conventional MRI. In cases of remaining uncertainty, other imaging methods might be warranted, including advanced MRI sequences or amino acid PET. To ensure that stored research samples contain as much viable recurrent tumour as possible, fluorescent dyes should be used intraoperatively and the neurosurgeon should cooperate closely with the reporting neuropathologist (or pathologist). If biopsy alone is pursued for diagnostic purposes, intraoperative smear or frozen sections might be prepared to enhance the probability of acquiring tissue with a high diagnostic yield (recommendation 17: 24 [80%] of 30 experts agreed).
Minimal volumes of tumour samples for further analyses
With a minimum distance of 1 cm between each individual sample, distinct cellular and genetic phenotypes are encountered.32 Accordingly, a distance of 1 cm between each sample should be respected to capture spatial intratumoural heterogeneity (recommendation 18: [76%] of 33 experts agreed). Even very small viable tissue samples of 1 mm3 have proven sufficient for a reliable neuropathological diagnosis and advanced genetic and epigenetic analyses by experienced neuropathologists (or pathologists).47 However, larger total volumes of sampled glioma tissue were shown to correspond to increased histological accuracy, because the risk of sampling bias is ameliorated.48 Gutt-Will and colleagues49 reported on 111 IDH-wildtype gliomas, in which glioblastoma-like histopathological features were found in all surgical specimens larger than 10 cm3, but the rate of such histopathological findings decreased with smaller sample volumes. Although this study was conducted before the implementation of molecular biomarkers such as the presence or absence of IDH mutations, which can be reliably diagnosed in small tissue volumes, as an integral part of the final diagnosis according to The 2021 WHO Classification of Tumors of the Central Nervous System,7 these results emphasise that larger tissue volumes might be more suitable to allow for a representative diagnosis and reduce the risk of sampling bias. As such, all resected, potentially viable tissue should be stored, if local storage capacities allow (recommendation 19: 33 [94%] of 35 experts agreed), including the collected tissue fragments if ultrasound aspiration is used during tumour resection.50
Blood sampling as a source of germline DNA
Large-scale sequencing analysis of the sampled tissue provides an overview of the genetic tumour profile. The clinical benefit over targeted sequencing methods is yet to be shown for adults with diffuse gliomas, but a thorough identification of targetable genetic alterations might be possible (besides the inherent interest from a research perspective).51,52 Somatic versus germline origin can be identified with certainty only when the results obtained for tumour DNA are compared with autologous non-tumour (ie, constitutional) DNA samples.52 Furthermore, constitutional reference DNA might also facilitate diagnostic detection of somatic copy number lesions.7,53 Blood samples provide a simple source for extraction of matched constitutional DNA from peripheral leukocytes. Thus, a peripheral blood sample (eg, 5 mL blood in a tube containing EDTA [edetic acid]) should be obtained and stored whenever feasible in case of later need for diagnostic or research purposes (recommendation 20: 31 [91%] of 34 experts agreed).54 Notably, patient (or caregiver) consent must be obtained before large-scale sequencing of constitutional DNA due to the possible detection of germline alterations in genes predisposing for hereditary cancer (or even non-cancer diseases) per institutional and national guidelines (recommendation 21: 35 [97%] of 36 experts agreed).55
Blood and CSF sampling for scientific purposes, biomarker discovery, and immune phenotyping
Peripheral blood and CSF might potentially serve as valuable sources for biomarker discovery towards a liquid biopsy approach for tumour characterisation and monitoring treatment response in neuro-oncology.56,57 From CSF and blood samples, novel technologies can analyse cell-free nucleic acids, including circulating cell-free tumour DNA, circulating cell-free tumour RNA and microRNA, circulating proteins, extracellular vesicles, circulating tumour cells, tumour-educated platelets, and soluble and cellular immune biomarkers (figure 1C).58
If local storage capacities are available and institutional review board regulations permit collection of peripheral blood, then surgical interventions at initial glioma diagnosis and for presumed progression could allow for collection of peripheral blood specimens for biobanking (recommendation 22: 29 [91%] of 32 experts agreed). EDTA blood tubes, and citrate blood tubes to some degree, serve for direct whole blood analysis, enabling the assessment of all cellular fractions in the peripheral blood, subsequent isolation, and biobanking of peripheral blood mononuclear cells (excluding platelets and granulocytes) for immune phenotyping or separation of cell-free plasma for liquid biomarkers. Blood should ideally be processed within 3 h after collection for research purposes but can be stored in oscillating trays for up to 24 h at various temperatures depending on the intended use and the types of blood tubes being used. For collection of cell-free DNA from plasma, special blood collection tubes are commercially available that contain preservatives to stabilise nucleated blood cells and prevent release of nuclear DNA, and thereby allow for isolation of high-quality cell-free DNA.
CSF can be obtained during tumour resection (eg, when opening basal cisterns or ventricles), via placement of ventricular catheters, or during routine procedures (eg, lumbar puncture). CSF should be collected for research purposes only in the setting of clinically indicated procedures (recommendation 23: 28 [85%] of 33 experts agreed). The required volume for standard diagnosis (ie, 5 mL at least for cytological assessment of leptomeningeal seeding) should be secured before the collection of CSF for research purposes. CSF is usually collected in dedicated sterile CSF collection tubes and should ideally be processed within 30–60 min for research purposes. Collection of 3 mL of CSF should be aimed for, because advanced molecular tests, including next-generation sequencing panels, droplet digital PCR, and whole-exome sequencing or whole-genome sequencing, can be analysed at this volume (recommendation 23: 28 [85%] of 33 experts agreed).57,59 A separation of the cell pellet and supernatant by centrifugation is recommended.57,59 CSF samples can be deep-frozen (ie, stored at −80°C or lower) in dedicated CSF collection tubes for later use.
We recommend adhering to quality measurements proposed by the International Liquid Biopsy Standardisation Alliance.60 The types and number of tubes should be regularly assessed and updated depending on the exact research plans.
Tissue processing after intraoperative sampling
Sample processing in the operating room
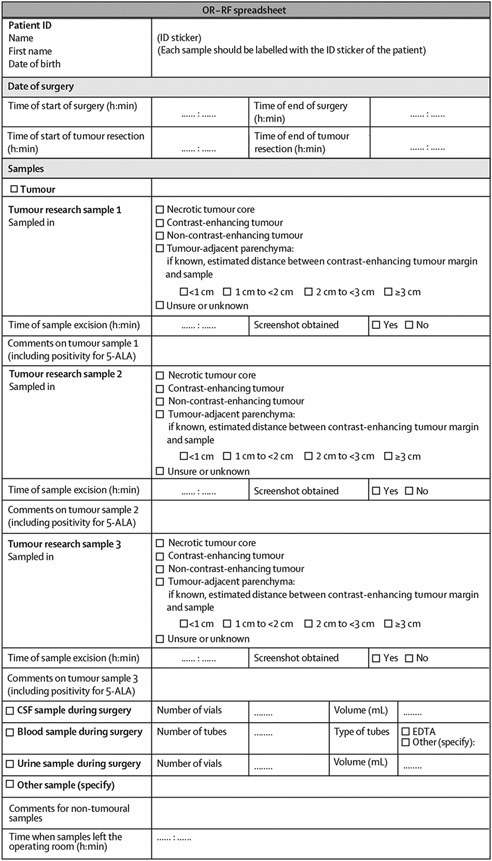
Following resection, glioma tissue should be immediately placed in a sterile vessel and cooled (not frozen) on wet ice to minimise ischaemic ex-vivo changes in tumour cell properties (ie, gene expression patterns) until final fixation or deep freezing (eg, until a designated technician arrives and cryotubes are labelled; recommendation 24: 28 [90%] of 31 experts agreed).61 Metabolite and protein detection by mass spectrometry provides insights into cancer metabolism and cell signalling, and freezing delay can severely affect collected data in terms of quality.62 Time of specimen acquisition and time of further processing, particularly deep freezing, should therefore be recorded separately (recommendation 25: 30 [91%] of 33 experts agreed), because the interval between resection and final fixation or deep freezing should be kept to a minimum, where fixation or deep freezing would ideally be done immediately within a 30-min interval following sampling. A standardised spreadsheet could be used to transfer the information that accompanies the samples (figure 2).
Figure 2: Template for a standardised spreadsheet for transfer of data accompanying surgical samples between the operating room and the research facility.
5-ALA=5-aminolevulinic acid. CSF=cerebrospinal fluid. EDTA=EDTA (edetic acid). ID=identifier. OR–RF=operating room and research facility communication.
Ideally, a neuropathologist (or a pathologist) should be involved in the macroscopic evaluation of the entire resected tissue and the decision as to which parts should be taken for routine diagnostics versus deep freezing or other research purposes, such as establishment of primary tumour cell cultures. If involvement of the pathologist or neuropathologist is not feasible, they should be informed that further tissue has been deep frozen for biobanking or specific research projects.
Shock freezing of tissue samples should be done in liquid nitrogen. For this process, tissue samples should be placed in appropriately labelled cryotubes, which are transferred directly into liquid nitrogen. For intraoperative frozen sections, native tissue samples are mounted on appropriate carrier plates using a specimen matrix that is suitable for cryosectioning. Then, the samples are typically shock frozen in isopentane precooled in liquid nitrogen to −160°C, which allows for a better preservation of histology than does direct placement in liquid nitrogen. In case a diagnosis cannot be made from the formalin-fixed paraffin-embedded samples (eg, due to bad tissue quality or insufficient viable tumour) or the samples are not representative of the neuroimaging abnormality, such deep-frozen specimens should still be available for diagnostic purposes, albeit yielding suboptimal morphology.63,64 Intraoperative frozen sections not only allow for an initial diagnostic impression but also assure that sufficient tumour tissue has been sampled for diagnosis. Sampling sufficient and viable tumour tissue is particularly important when freshly resected tumour specimens are subjected to procedures that destroy the original tumour histology (eg, establishment of primary cultures or immediate native tissue homogenisation). Tissue processing might be further tailored to the specific research project pursued.65
Final sample fixation for long-term storage
Tissue is routinely fixed in buffered 4% formalin and embedded in paraffin for histology; immuno-histochemistry; and DNA (or RNA) isolation for genetic and epigenetic studies according The 2021 WHO Classification of Tumors of the Central Nervous System,7 including DNA methylation profiling and next-generation sequencing approaches (recommendation 26: 27 [93%] of 29 experts agreed).66,67 If sufficient tissue is available, parts of the resected tumour tissue should also be put into appropriate cryovessels and shock frozen in liquid nitrogen as previously mentioned, ideally within 30 min after resection to ensure high quality of RNA, proteins, and metabolites for further dedicated molecular analyses.67 Deep-frozen samples should be stored at −80°C or even lower temperatures (eg, in a −140°C freezer or in liquid nitrogen) to ensure optimal long-term preservation of sensitive analytes, such as RNA, post-translational protein modifications, and metabolites, thereby limiting potential bias due to variations in preservation times across different samples. Variations in freezing temperature and speed as well as temperature for thawing of samples should be avoided. Shipment of deep-frozen tissue samples should be done on dry ice without interruption of the cold chain. High-molecular weight DNA that is extracted immediately after tissue resection from either native or shock-frozen samples can also serve for rapid DNA methylation and copy number profiling; for example, by use of long-read nanopore-based sequencing.68,69
Biobanking should include both frozen and formalin-fixed paraffin-embedded tissue (recommendation 27: 28 [93%] of 30 experts agreed), as formalin-fixed paraffin-embedded tissue quality is more suitable for histological or immunohistochemical analyses and less prone to physical damage than frozen tissue. Designated live (ie, unfixed and unprocessed) tissue might also be needed and could be transferred into culture media to establish patient-derived cell lines or organoids.70 Rapid histological assessment of frozen tissue sections is recommended to ensure that diagnostic tissue has been sampled before the tissue is dissected and transferred into cell cultures. Other chemical fixatives are also available for specific purposes, such as buffered glutaraldehyde for electron microscopy and alcohols for nucleic acid analyses.64 For other specific research purposes, dedicated equipment, special media or fixatives, and distinct workflows might be used (eg, for dissociation of tissue samples into single cell suspensions as required for flow cytometry, cloning, and single cell omics). Moreover, techniques for isolation of viable glioma cells, including ex-vivo electrophysiological analyses and single cell sequencing, and techniques for the generation of viable glioma models, such as organotypic slice cultures, have been reported.65,71,72
Any excess tissue should be kept and not discarded, so that it is available for additional diagnostic testing at a later timepoint. This might include situations where the original sample is of poor quality and not sufficient to establish a definitive diagnosis, when a patient wants to enter a clinical trial where dedicated tissue analysis is required, or to assess tumour properties along the disease trajectory (recommendation 28: 28 [90%] of 31 experts agreed). Such excess tissue should be stored either as deep-frozen samples or as formalin-fixed paraffin-embedded tissue. Deep-frozen storage has the advantage of retaining the natural protein structure but has a negative effect on the tissue morphology.73 Short-term storage (eg, up to a few weeks) of excess tissue in formalin is also possible and allows for later paraffin-embedding of additional tissue if required (eg, when a diagnosis cannot be made from the original formalin-fixed paraffin-embedded samples). Long-term storage in formalin as wet material, however, is not recommended due to tissue shrinkage, poor preservation of morphology, protein crosslinking with reduction or loss of immunostaining, and marked degradation of nucleic acids. As previously stated, the neuropathologist (or pathologist) should have access to samples in the biobanking collection, and deep-frozen tissue specimens preserved in addition to formalin-fixed and paraffin-embedded tissue samples should not be exhausted until a final diagnosis is established, if not otherwise specified in research projects (eg, methylation profiling as part of a research protocol, which then also allows diagnosis; recommendation 29: 32 [97%] of 33 experts agreed). In this setting, formalin-fixed paraffin-embedded samples should be used first before frozen tissue is retrieved, because frozen tissue ensures optimal long-term preservation of sensitive analytes. After establishment of the diagnosis and storage of adequate tissue to allow future testing for clinical purposes (eg, to match into a clinical trial), excess deep-frozen tissue that is not a part of the hospital archives might be indefinitely released for biobanking purposes. This process requires the patient’s consent and must be in accordance with local guidelines (recommendation 30: 33 [94%] of 35 experts agreed).
Labelling of the biobanking samples
Careful and informative labelling of deposited specimens is crucial not only to ensure the correct clinical context but also to retain the patient’s right to retract their specimens. As stated by the US National Cancer Institute Office of Biorepository and Biospecimens best practice guidelines, each biospecimen must have an “identifier or combination of identifiers that are firmly affixed to the container […] and able to endure storage conditions”.74 Many institutions have adopted a policy in which they require the use of two unique institutional identifiers together with the date of surgery to allow back-tracking to patient information.64,75 This practice should be encouraged because it helps to narrow the possibilities in case of mislabelling (recommendation 31: 33 [94%] of 35 experts agreed). Importantly, national and local data protection laws need to be respected as they might forbid using re-identifiable labelling and call for pseudonymisation via software-generated labels specifically for the purpose of biobanking. A label printer or barcoding system, rather than handwriting, should be used to preserve legibility and reduce human errors. The label and the two reference numbers on the label should be resistant to −80°C (or even lower temperatures if needed in selected cases).
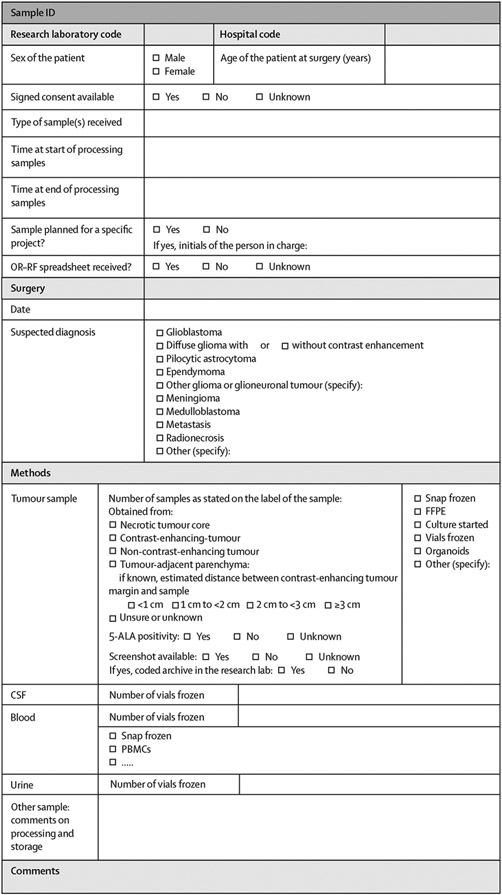
In addition to this labelling technique, which matches a specific sample to an individual patient and their respective operation, information on the exact tumour portion from which the tissue was sampled (eg, contrast-enhancing tumour),63 the sample number (eg, sample one of two from contrast-enhancing tumour samples), data for PET signal or intraoperative fluorescence, and clinical data should be stored in the biobank files (figure 3). The reference numbers could also be stored within the neuronavigation system, where the location of the sample is recorded, and on the technician’s datasheet, which should be filled out during the sampling and on which information on PET signal or intraoperative fluorescence can be deposited (recommendation 32: 33 [92%] of 36 experts agreed).
Figure 3: Template for a standardised spreadsheet for de-identified data storage in the tissue biobank.
5-ALA=5-aminolevulinic acid. CSF=cerebrospinal fluid. FFPE=formalin-fixed paraffin-embedded. ID=identifier. OR-RF=operating room and research facility communication. PBMCs=peripheral blood monocytes.
Conclusions
The 2021 WHO Classification of Tumors of the Central Nervous System established the assessment of molecular markers as a prerequisite for an accurate neuropathological diagnosis across the different entities of diffuse gliomas.7 As such, diagnosis and management need to follow an in-depth analysis of tumour tissue. Any scientific progress crucially relies on the tumour tissue. The RANO recommendations provide guidance for the ideal surgical workflow and biospecimen processing and labelling to achieve a genuine representation of the tumour architecture and standardisation among various tissue collections.
Supplementary Material
Footnotes
Declaration of interests
MS declares consultancy for Bracco and honoraria from GE Healthcare and AuntMinnie. ELR declares honoraria from AbbVie, Adastra, Daiichi Sankyo, LEO Pharma, Seagen, and Tocagen. BME declares advisory board participation and consulting for Medicenna, MedQIA, Servier Pharmaceuticals, Siemens, Janssen Pharmaceuticals, Imaging Endpoints, Kazia, Chimerix, Sumitomo Dainippon Pharma Oncology, ImmunoGenesis, Ellipses Pharma, Monteris, Neosoma, Alpheus Medical, Sagimet Biosciences, Sapience Therapeutics, and the Global Coalition for Adaptive Research. MMK declares consultancy for Blue Earth Diagnostics and research grants from EpicentRx and Blue Earth Diagnostics. NLA declares research grants from Novocure and honoraria from Novartis and Telix. MPM declares consulting for Karyopharm Therapeutics, Mevion Medical Systems, ZappRx, Sapience Therapeutics, and Xoft; being part of the board of directors for Oncoceutics; ownership of WARF patent 14/934,27, Topical Vasoconstrictor Preparations and Methods for Protecting Cells During Cancer Chemotherapy and Radiotherapy; and stock at Chimerix. MvdB declares consultancy for Celgene, BMS, Agios, Boehringer, AbbVie, Bayer, Carthera, Nerviano, and Genenta. MW declares research grants from Quercis and Versameb; honoraria from Novocure and Bayer; and consulting or advisory roles for CureVac, Medac, Novartis, Orbus Therapeutics, Philogen, and Bayer. MAV declares indirect equity and patent royalty interests from Infuseon Therapeutics; honoraria from Chimerix and Midatech; and research grants from DeNovo Pharma, Oncosynergy, Infuseon, and Chimerix. J-CT declares research grants from Novocure and Munich Surgical Imaging; participating on an advisory board for AAA Novartis; and royalties from Springer Publisher. PK, GR, NG, JTH, OS, PNH, MM, LvB, RYH, SMC, and MSB declare no competing interests.
Contributor Information
Philipp Karschnia, Department of Neurosurgery, Ludwig-Maximilians-University of Munich, Munich, Germany; German Cancer Consortium, Partner Site Munich, Munich, Germany.
Marion Smits, Department of Neuroradiology and Nuclear Medicine, Erasmus MC Cancer Institute, Rotterdam, Netherlands.
Guido Reifenberger, Institute of Neuropathology, Heinrich Heine University Medical Faculty and University Hospital Düsseldorf, Düsseldorf, Germany.
Emilie Le Rhun, Department of Neurosurgery, University Hospital of Zurich and University of Zurich, Zurich, Switzerland; Department of Neurology University Hospital of Zurich and University of Zurich, Zurich, Switzerland.
Benjamin M Ellingson, UCLA Brain Tumor Imaging Laboratory, Department of Radiological Sciences, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Norbert Galldiks, Department of Neurology, Faculty of Medicine, University of Cologne and University Hospital Cologne, Cologne, Germany; Research Center Juelich, Institute of Neuroscience and Medicine, Juelich, Germany.
Michelle M Kim, Department of Radiation Oncology, University of Michigan Hospital, Ann Arbor, MI, USA.
Jason T Huse, Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Oliver Schnell, Department of Neurosurgery, University of Freiburg, Freiburg, Germany.
Patrick N Harter, German Cancer Consortium, Partner Site Munich, Munich, Germany; Center for Neuropathology and Prion Research, Faculty of Medicine, Ludwig-Maximilians-University of Munich, Munich, Germany.
Malte Mohme, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Louisa von Baumgarten, Department of Neurosurgery, Ludwig-Maximilians-University of Munich, Munich, Germany.
Nathalie L Albert, Department of Nuclear Medicine, Ludwig-Maximilians-University of Munich, Munich, Germany.
Raymond Y Huang, Division of Neuroradiology, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA.
Minesh P Mehta, Department of Radiation Oncology, Miami Cancer Institute, Baptist Health South Florida, Miami, FL, USA.
Martin van den Bent, Department of Neurology, Erasmus MC Cancer Institute, Rotterdam, Netherlands.
Michael Weller, Department of Neurology, University Hospital of Zurich and University of Zurich, Zurich, Switzerland.
Michael A Vogelbaum, Department of NeuroOncology, Moffitt Cancer Center, Tampa, FL, USA.
Susan M Chang, Department of Neurosurgery and Division of NeuroOncology, University of California, San Francisco, CA, USA.
Mitchel S Berger, Department of Neurosurgery and Division of NeuroOncology, University of California, San Francisco, CA, USA.
Joerg-Christian Tonn, Department of Neurosurgery, Ludwig-Maximilians-University of Munich, Munich, Germany; German Cancer Consortium, Partner Site Munich, Munich, Germany.
References
- 1.Weller M, van den Bent M, Preusser M, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 2021; 18: 170–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JJ, Gonzalez Castro LN, McBrayer S, et al. Isocitrate dehydrogenase (IDH) mutant gliomas: a Society for Neuro-Oncology (SNO) consensus review on diagnosis, management, and future directions. Neuro Oncol 2023; 25: 4–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karschnia P, Young JS, Dono A, et al. Prognostic validation of a new classification system for extent of resection in glioblastoma: a report of the RANO resect group. Neuro Oncol 2022; 24 (suppl 7): vii255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hervey-Jumper SL, Zhang Y, Phillips JJ, et al. Interactive effects of molecular, therapeutic, and patient factors on outcome of diffuse low-grade glioma. J Clin Oncol 2023; 41: 2029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karschnia P, Dono A, Young JS, et al. Prognostic evaluation of re-resection for recurrent glioblastoma using the novel RANO classification for extent of resection: a report of the RANO resect group. Neuro Oncol 2023; 25: 1672–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ringel F, Pape H, Sabel M, et al. Clinical benefit from resection of recurrent glioblastomas: results of a multicenter study including 503 patients with recurrent glioblastomas undergoing surgical resection. Neuro Oncol 2016; 18: 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 2021; 23: 1231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Groot J, Penas-Prado M, Alfaro-Munoz K, et al. Window-of-opportunity clinical trial of pembrolizumab in patients with recurrent glioblastoma reveals predominance of immune-suppressive macrophages. Neuro Oncol 2020; 22: 539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klughammer J, Kiesel B, Roetzer T, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med 2018; 24: 1611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mert A, Buehler K, Sutherland GR, et al. Brain tumor surgery with 3-dimensional surface navigation. Neurosurgery 2012; 71 (2 suppl operative): ons286–94. [DOI] [PubMed] [Google Scholar]
- 11.Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized brain tumor imaging protocol in clinical trials. Neuro Oncol 2015; 17: 1188–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molinaro AM, Hervey-Jumper S, Morshed RA, et al. Association of maximal extent of resection of contrast-enhanced and non-contrast-enhanced tumor with survival within molecular subgroups of patients with newly diagnosed glioblastoma. JAMA Oncol 2020; 6: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkataramani V, Yang Y, Schubert MC, et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell 2022; 185: 2899–917.e31. [DOI] [PubMed] [Google Scholar]
- 14.Stensjøen AL, Solheim O, Kvistad KA, Håberg AK, Salvesen Ø, Berntsen EM. Growth dynamics of untreated glioblastomas in vivo. Neuro Oncol 2015; 17: 1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellingson BM, Nguyen HN, Lai A, et al. Contrast-enhancing tumor growth dynamics of preoperative, treatment-naive human glioblastoma. Cancer 2016; 122: 1718–27. [DOI] [PubMed] [Google Scholar]
- 16.Law I, Albert NL, Arbizu J, et al. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [18F]FDG: version 1.0. Eur J Nucl Med Mol Imaging 2019; 46: 540–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galldiks N, Langen KJ, Albert NL, et al. Investigational PET tracers in neuro-oncology—what’s on the horizon? A report of the PET/RANO group. Neuro Oncol 2022; 24: 1815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann P, Stavrinou P, Lipke K, et al. FET PET reveals considerable spatial differences in tumour burden compared to conventional MRI in newly diagnosed glioblastoma. Eur J Nucl Med Mol Imaging 2019; 46: 591–602. [DOI] [PubMed] [Google Scholar]
- 19.Wahl DR, Kim MM, Aryal MP, et al. Combining perfusion and high B-value diffusion MRI to inform prognosis and predict failure patterns in glioblastoma. Int J Radiat Oncol Biol Phys 2018; 102: 757–64. [DOI] [PubMed] [Google Scholar]
- 20.Kim MM, Sun Y, Aryal MP, et al. A phase 2 study of dose-intensified chemoradiation using biologically based target volume definition in patients with newly diagnosed glioblastoma. Int J Radiat Oncol Biol Phys 2021; 110: 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunz M, Albert NL, Unterrainer M, et al. Dynamic 18F-FET PET is a powerful imaging biomarker in gadolinium-negative gliomas. Neuro Oncol 2019; 21: 274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roodakker KR, Alhuseinalkhudhur A, Al-Jaff M, et al. Region-by-region analysis of PET, MRI, and histology in en bloc-resected oligodendrogliomas reveals intra-tumoral heterogeneity. Eur J Nucl Med Mol Imaging 2019; 46: 569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordova JS, Shu HK, Liang Z, et al. Whole-brain spectroscopic MRI biomarkers identify infiltrating margins in glioblastoma patients. Neuro Oncol 2016; 18: 1180–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avalos LN, Luks TL, Gleason T, et al. Longitudinal MR spectroscopy to detect progression in patients with lower-grade glioma in the surveillance phase. Neurooncol Adv 2022; 4: vdac175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stummer W, Pichlmeier U, Meinel T, Wiestler OD, Zanella F, Reulen HJ. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol 2006; 7: 392–401. [DOI] [PubMed] [Google Scholar]
- 26.Acerbi F, Broggi M, Schebesch KM, et al. Fluorescein-guided surgery for resection of high-grade gliomas: a multicentric prospective phase II study (FLUOGLIO). Clin Cancer Res 2018; 24: 52–61. [DOI] [PubMed] [Google Scholar]
- 27.Schucht P, Knittel S, Slotboom J, et al. 5-ALA complete resections go beyond MR contrast enhancement: shift corrected volumetric analysis of the extent of resection in surgery for glioblastoma. Acta Neurochir (Wien) 2014; 156: 305–12. [DOI] [PubMed] [Google Scholar]
- 28.Coburger J, Engelke J, Scheuerle A, et al. Tumor detection with 5-aminolevulinic acid fluorescence and Gd-DTPA-enhanced intraoperative MRI at the border of contrast-enhancing lesions: a prospective study based on histopathological assessment. Neurosurg Focus 2014; 36: E3. [DOI] [PubMed] [Google Scholar]
- 29.Roder C, Stummer W, Coburger J, et al. Intraoperative MRI-guided resection is not superior to 5-aminolevulinic acid guidance in newly diagnosed glioblastoma: a prospective controlled multicenter clinical trial. J Clin Oncol 2023; published online June 19. 10.1200/JCO.22.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R, Teich W, Frenzel F, et al. Optical characterization of sodium fluorescein in vitro and ex vivo. Front Oncol 2021; 11: 654300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravi VM, Neidert N, Will P, et al. T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat Commun 2022; 13: 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA 2013; 110: 4009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravi VM, Will P, Kueckelhaus J, et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 2022; 40: 639–55. [DOI] [PubMed] [Google Scholar]
- 34.Ellingson BM, Lai A, Nguyen HN, Nghiemphu PL, Pope WB, Cloughesy TF. Quantification of nonenhancing tumor burden in gliomas using effective T2 maps derived from dual-echo turbo spin-echo MRI. Clin Cancer Res 2015; 21: 4373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S, Hartzendorf S, Vogt P, et al. Progression patterns in non-contrast-enhancing gliomas support brain tumor responsiveness to surgical lesions. Pathol Oncol Res 2022; 28: 1610268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suchorska B, Jansen NL, Linn J, et al. Biological tumor volume in 18FET-PET before radiochemotherapy correlates with survival in GBM. Neurology 2015; 84: 710–19. [DOI] [PubMed] [Google Scholar]
- 37.Senft C, Bink A, Franz K, Vatter H, Gasser T, Seifert V. Intraoperative MRI guidance and extent of resection in glioma surgery: a randomised, controlled trial. Lancet Oncol 2011; 12: 997–1003. [DOI] [PubMed] [Google Scholar]
- 38.Prada F, Bene MD, Fornaro R, et al. Identification of residual tumor with intraoperative contrast-enhanced ultrasound during glioblastoma resection. Neurosurg Focus 2016; 40: e7. [DOI] [PubMed] [Google Scholar]
- 39.Al-Holou WN, Hodges TR, Everson RG, et al. Perilesional resection of glioblastoma is independently associated with improved outcomes. Neurosurgery 2020; 86: 112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barthel FP, Johnson KC, Varn FS, et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature 2019; 576: 112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoogstrate Y, Draaisma K, Ghisai SA, et al. Transcriptome analysis reveals tumor microenvironment changes in glioblastoma. Cancer Cell 2023; 41: 678–92.e7 [DOI] [PubMed] [Google Scholar]
- 42.Kocakavuk E, Anderson KJ, Varn FS, et al. Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer. Nat Genet 2021; 53: 1088–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knudsen AM, Halle B, Cédile O, et al. Surgical resection of glioblastomas induces pleiotrophin-mediated self-renewal of glioblastoma stem cells in recurrent tumors. Neuro Oncol 2022; 24: 1074–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schäfer N, Gielen GH, Rauschenbach L, et al. Longitudinal heterogeneity in glioblastoma: moving targets in recurrent versus primary tumors. J Transl Med 2019; 17: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karschnia P, Teske N, Thon N, et al. Chimeric antigen receptor T cells for glioblastoma: current concepts, challenges, and future perspectives. Neurology 2021; 97: 218–30. [DOI] [PubMed] [Google Scholar]
- 46.Winter SF, Loebel F, Loeffler J, et al. Treatment-induced brain tissue necrosis: a clinical challenge in neuro-oncology. Neuro Oncol 2019; 21: 1118–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katzendobler S, Do A, Weller J, et al. Diagnostic yield and complication rate of stereotactic biopsies in precision medicine of gliomas. Front Neurol 2022; 13: 822362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim BYS, Jiang W, Beiko J, et al. Diagnostic discrepancies in malignant astrocytoma due to limited small pathological tumor sample can be overcome by IDH1 testing. J Neurooncol 2014; 118: 405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutt-Will M, Murek M, Schwarz C, et al. Frequent diagnostic undergrading in isocitrate dehydrogenase wild-type gliomas due to small pathological tissue samples. Neurosurgery 2019; 85: 689–94. [DOI] [PubMed] [Google Scholar]
- 50.Kirby AJ, Lavrador JP, Bodi I, et al. Ex vivo ultrasonic samples of human brain tumors in the molecular era. Neurooncol Adv 2020; 2: vdaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blobner J, Dengler L, Blobner S, et al. Significance of molecular diagnostics for therapeutic decision-making in recurrent glioma. Neurooncol Adv 2023; 5: vdad060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capper D, Reifenberger G, French PJ, et al. EANO guideline on rational molecular testing of gliomas, glioneuronal, and neuronal tumors in adults for targeted therapy selection. Neuro Oncol 2023; 25: 813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woehrer A, Hainfellner JA. Molecular diagnostics: techniques and recommendations for 1p/19q assessment. CNS Oncol 2015; 4: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng S, Alfaro-Munoz K, Wei W, et al. Prospective clinical sequencing of adult glioma. Mol Cancer Ther 2019; 18: 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bunnik EM, Dondorp WJ, Bredenoord AL, de Wert G, Cornel MC. Mainstreaming informed consent for genomic sequencing: a call for action. Eur J Cancer 2021; 148: 405–10. [DOI] [PubMed] [Google Scholar]
- 56.Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat Rev Clin Oncol 2019; 16: 409–24. [DOI] [PubMed] [Google Scholar]
- 57.Soffietti R, Bettegowda C, Mellinghoff IK, et al. Liquid biopsy in gliomas: a RANO review and proposals for clinical applications. Neuro Oncol 2022; 24: 855–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov 2021; 11: 858–73. [DOI] [PubMed] [Google Scholar]
- 59.Friedman JS, Hertz CAJ, Karajannis MA, Miller AM. Tapping into the genome: the role of CSF ctDNA liquid biopsy in glioma. Neurooncol Adv 2022; 4 (suppl 2): ii33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Connors D, Allen J, Alvarez JD, et al. International liquid biopsy standardization alliance white paper. Crit Rev Oncol Hematol 2020; 156: 103112. [DOI] [PubMed] [Google Scholar]
- 61.Wan T, Zhu W, Zhao Y, et al. Astrocytic phagocytosis contributes to demyelination after focal cortical ischemia in mice. Nat Commun 2022; 13: 1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mock A, Rapp C, Warta R, et al. Impact of post-surgical freezing delay on brain tumor metabolomics. Metabolomics 2019; 15: 78. [DOI] [PubMed] [Google Scholar]
- 63.Darrigues E, Elberson BW, De Loose A, et al. Brain tumor biobank development for precision medicine: role of the neurosurgeon. Front Oncol 2021; 11: 662260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hojat A, Wei B, Olson MG, Mao Q, Yong WH. Procurement and storage of surgical biospecimens. Methods Mol Biol 2019; 1897: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Straehle J, Ravi VM, Heiland DH, et al. Technical report: surgical preparation of human brain tissue for clinical and basic research. Acta Neurochir (Wien) 2023; 165: 1461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018; 555: 469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sahm F, Brandner S, Bertero L, et al. Molecular diagnostic tools for the World Health Organization (WHO) 2021 classification of gliomas, glioneuronal and neuronal tumors; an EANO guideline. Neuro Oncol 2023; published online June 2. 10.1093/neuonc/noad100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Euskirchen P, Bielle F, Labreche K, et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol 2017; 134: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Djirackor L, Halldorsson S, Niehusmann P, et al. Intraoperative DNA methylation classification of brain tumors impacts neurosurgical strategy. Neurooncol Adv 2021; 3: vdab149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacob F, Salinas RD, Zhang DY, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 2020; 180: 188–204.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, Straehle J, Joseph K, et al. Isolation and profiling of viable tumor cells from human ex vivo glioblastoma cultures through single-cell transcriptomics. STAR Protoc 2023; 4: 102383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Das A, Gunasekaran A, Stephens HR, et al. Establishing a standardized method for the effective intraoperative collection and biological preservation of brain tumor tissue samples using a novel tissue preservation system: a pilot study. World Neurosurg 2022; 161: e61–74. [DOI] [PubMed] [Google Scholar]
- 73.Esteve-Codina A, Arpi O, Martinez-García M, et al. A comparison of RNA-Seq results from paired formalin-fixed paraffin-embedded and fresh-frozen glioblastoma tissue samples. PLoS One 2017; 12: e0170632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biorepositories and Biospecimen Research Branch of the National Cancer Institute, National Institutes of Health. NCI best practices for biospecimen resources. March 21, 2016. https://biospecimens.cancer.gov/bestpractices/2016-NCIBestPractices.pdf (accessed Aug 15, 2023). [Google Scholar]
- 75.Kay AB, Estrada DK, Mareninov S, et al. Considerations for uniform and accurate biospecimen labelling in a biorepository and research environment. J Clin Pathol 2011; 64: 634–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.