Abstract
Snakebite envenoming (SBE) is a global public health concern, primarily due to the lack of effective antivenom for treating snakebites inflicted by medically significant venomous snakes prevalent across various geographic locations. The rising demand for safe, cost-effective, and potent snakebite treatments highlights the urgent need to develop alternative therapeutics targeting relevant toxins. This development could provide promising discoveries to create novel recombinant solutions, leveraging human monoclonal antibodies, synthetic peptides and nanobodies. Such technologies as recombinant DNA, peptide and epitope mapping phage display etc) have the potential to exceed the traditional use of equine polyclonal antibodies, which have long been used in antivenom production. Recombinant antivenom can be engineered to target certain toxins that play a critical role in snakebite pathology. This approach has the potential to produce antivenom with improved efficacy and safety profiles. However, there are limitations and challenges associated with these emerging technologies. Therefore, identifying the limitations is critical for overcoming the associated challenges and optimizing the development of recombinant antivenoms. This review is aimed at presenting a thorough overview of diverse technologies used in the development of recombinant antivenom, emphasizing their limitations and offering insights into prospects for advancing recombinant antivenoms.
Keywords: Snake venom, Antivenom, Antibodies, Nanobodies, Recombinant antivenom, Traditional antivenom
Graphical abstract
1. Introduction
Snakes, belonging to the monophyletic group Squamata: Serpentes, comprise approximately 3600 extant species and are found across all continents except Antarctica [1,2]. In an evolutionary context, these reptiles are distinguished by their notable absence of limbs, elongated body structure, and strict carnivorous dietary habits [3]. Venomous snakes have specialized venom glands that produce a combination of toxins, which they utilize for defensive or survival functions. Using their fangs, these toxins are delivered by the snakes into other animals to incapacitate and aid in the digestion process [4,5]. Snake venom toxins can be classified into 63 distinct families, and among these, four families are identified as significant in clinical contexts. They are 3-finger toxins (3FTxs), phospholipases A2 (PLA2), snake venom metalloproteinases (SVMPs), and snake venom serine proteinases (SVSPs). Other protein families include cysteine-rich secretory proteins (CRISPs), l-amino acid oxidases (LAAOs), and C-type lectin-like proteins (CTLPs) [6].
Snakes release venom into humans as a means of self-defence, and the quantity of venom they inject into their prey or victims frequently surpasses the lethal dose by more than 100 times, as noted by Casewell et al. [5]. This has a detrimental impact on human victims. The diversity in venom composition at both inter- and intraspecific levels leads to a wide range of snakebite envenoming pathologies, posing a significant challenge in developing snakebite treatments that can effectively counteract various venom pathologies. The pathological effects of snakebites encompass a range of issues, such as nerve and muscle dysfunction (neurotoxicity), bleeding and clotting problems (hemotoxicity), as well as localized swelling, formation of blisters, and tissue death (cytotoxicity) in the area where the snakebite occurred [7]. According to the report by Gutierrez et al. [8], the most severe instances of snakebite envenomation typically involve snake species from the Elapidae family (like cobras, kraits, mambas, Australasian species, and sea snakes) and the Viperidae family (including rattlesnakes, lance-headed pit vipers, and true vipers).
Globally, there are around 5.4 million snakebite incidents each year, resulting in an estimated 1.8 to 2.7 million cases of envenomation. Furthermore, there are an estimated 81,410 to 137,880 snakebite-related deaths annually, with roughly three times as many cases of amputations and other enduring disabilities stemming from these incidents [9]. The majority of snakebite victims are impoverished agricultural labourers residing in rural areas [10]. In June 2017, snake envenoming was officially categorized as one of the neglected tropical diseases by the World Health Organization [11]. This constitutes a significant public health crisis worldwide due to the lack of effective antivenom for treating snakebites inflicted by medically significant venomous snakes prevalent across various regions. The most severe snakebite incidents are concentrated in Africa, Asia, and Latin America. In Asia, up to 2 million individuals experience snakebite envenomation annually, while in Africa, there are an estimated 435,000 to 580,000 snake bites requiring treatment each year [9]. The escalating rates of snakebite-related mortality and amputations in low- and middle-income countries can be attributed to the inadequate availability of antivenoms, substandard healthcare services, variations in antivenom efficacy, and limited ability to mitigate local damage caused by venom variation intra- and interspecifically. Additionally, adverse reactions to antivenom further complicate this issue [[12], [13], [14], [15], [16]].
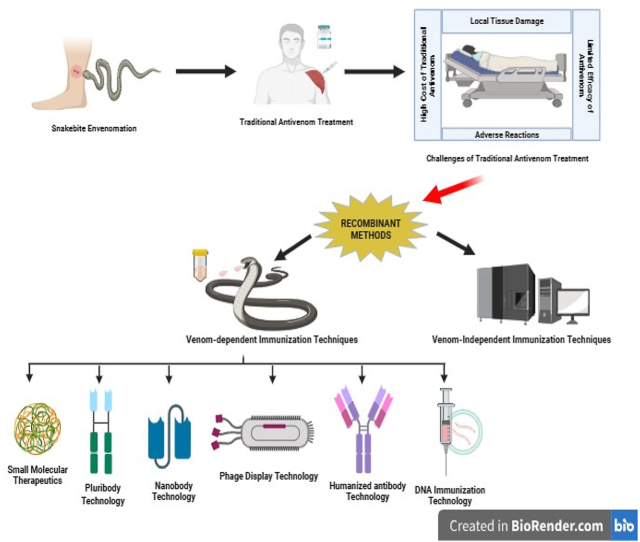
Currently, despite significant progress in technology and science, immunotherapy remains the primary treatment for snakebite envenoming. While existing antivenoms are playing a crucial role in saving lives, their limitations have spurred researchers to seek alternative neutralizing agents that can enhance or complement traditional antivenom treatment [4]. The development of next-generation antivenoms is actively underway, employing various approaches including phage display technology, humanized antibody technology, DNA immunization methods, small molecular therapeutics, and single-domain antibodies (nanobodies). Furthermore, the production of next-generation antivenom might not be dependent on venoms themselves, instead, it might depend on the understanding of venom composition and toxicity for specific snake species. This knowledge will be essential in formulating and dosing antivenom, ensuring more effective and targeted treatment [17].
This review is aimed at presenting a thorough overview of diverse recombinant technologies used in the development of antivenom, emphasizing their limitations and offering insights into prospects for advancing recombinant antivenom.
2. Traditional antivenom
One of the earliest experiments conducted using serum therapy was Sewall's experiments, which demonstrated that immunized pigeons could withstand up to six lethal doses of rattlesnake venom mixed with glycerine [18], making it the first evidence of serum therapy in snakebite management. Phisalix and Bertrand in 1894 established the antitoxic activity of animal blood immunized with European viper venom. Calmette, a disciple of Louis Pasteur, was responsible for developing antivenoms, with McFarland, Tidwell, and Venzor following his instructions, marking the beginning of antivenom development.
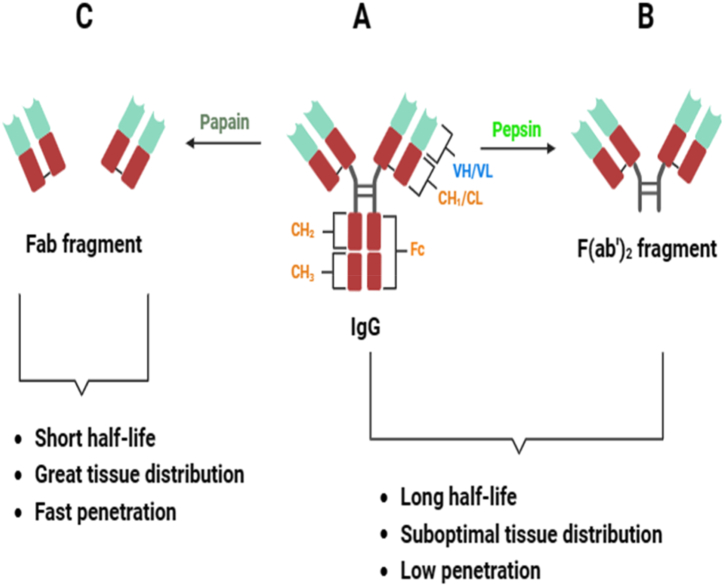
Traditional polyvalent antivenoms refer to a concentrated portion or fragment of immunoglobulins sourced from the plasma of animals hyperimmunized against specific snake venoms [4]. Traditional antivenom being the only available therapy for snake envenomation is currently produced in three formats by enzymatic digestion (papain and pepsin) as shown in Fig. 1 (A - C): Immunoglobulin Gs (150 kDa) as well as their derivatives like F (ab')2 fragments (100 kDa) and Fab fragments (50 kDa) [[19], [20], [21]].
Fig. 1.
Antibody formats. (A) Complete IgG (immunoglobulin G) made up of VH and VL (the heavy and light variable domains of antibodies respectively) as well as the CL (constant domain of light chains) and CHn (constant domain of heavy chains) (B) Pepsin digestion cleaves IgG at a distinct site within the hinge region, resulting in a bivalent fragment that retains both binding sites, known as F (ab')2. (C) Papain digestion cleaves disulfide bonds located near the hinge region, producing a crystallizable fragment composed of the constant segments of two H chains (Fc) and two antigen-binding fragments (Fab). The schematic diagram was created with biorender.com.
2.1. Limitations of traditional antivenoms
2.1.1. Local tissue damage
Immunoglobulin G and its fragments can effectively mitigate systemic damage by snakebites, but face limitations in treating localized tissue damage. These limitations arise from the pharmacodynamic properties of large molecules, preventing efficient antivenom penetration into affected local tissues. Consequently, this restriction can lead to enduring motor impairment [8,22,23]. The administration of F (ab')2 and Fab fragments addresses this issue by reducing limb disability and increasing bioavailability due to their smaller size, facilitating enhanced tissue penetration [24,25]. However, these fragments exhibit shorter serum half-lives and are rapidly cleared from the systemic circulation within an hour of administration.
2.1.2. Adverse reactions
Current venom immunization protocols lack a strategic approach to guide antibody specificities towards the most pathogenic venom proteins and also overlook the varying immunogenicity of distinct venom components. Consequently, conventional antivenoms contain varying antibodies, including those against weak antigens or non-toxic elements, reducing the potency of toxin-specific antibodies. Furthermore, these antivenoms often display inadequate antibody levels against highly pathogenic venom toxins [26]. The consequence of this untargeted production approach necessitates the administration of large volumes (20–200 mL) of antivenom for a curative impact, significantly increasing the likelihood of patients experiencing antivenom-induced adverse reactions [4,5].
These adverse reactions can be categorized into early and late adverse reactions. The early adverse reactions, such as anaphylactic and pyrogenic reactions, manifest within 24 h of antivenom administration and may include symptoms such as urticaria, nausea, colic, vomiting, generalized rash, chills, and, less commonly, hypotension, bronchospasm and angioneurotic oedema. Conversely, late reactions, identified as serum sickness, involve the humoral immune system response against the antibodies in the antivenom. This manifests as itching, fever, rashes, joint pain, proteinuria, and glomerulonephritis, typically occurring between 5 and 24 days after treatment [16,27]. In severe cases, antivenoms can trigger anaphylactic shock, which can potentially lead to death.
Reported incidences of adverse reactions following diverse antivenom treatments exhibit significant variability. Acute reactions were observed in 23–56 % of patients, with some cases showing high rates of serum sickness (up to 75 %). Conversely, certain studies reported a relatively low rate of approximately 10 %, for these complications [[28], [29], [30], [31], [32], [33]]. This inconsistency primarily arises from the fact that older immunoglobulin-based antivenoms had numerous side effects. In contrast, newer, refined equine antivenoms containing F (ab')2 or Fab fragments result in fewer adverse reactions due to their reduced immunogenicity and improved safety profiles [34].
Adulterated antivenoms present additional challenges, ranging from minimal efficacy to serious safety risks [35,36]. The effectiveness and safety of antivenoms are influenced by diverse factors, including the physicochemical properties of formulations, immunoglobulin fragments and antibody purity, presence of protein aggregates, endotoxin levels, preservative content, batch consistency, and the ability to neutralize key toxins in targeted venoms. Ensuring the efficacy and safety of antivenoms is crucial for manufacturers [37]. In response to these challenges, supplementary agents are administered with antivenom to mitigate adverse reactions. One example is hydrocortisone, although it has shown limited efficacy in reducing adverse reactions or delaying their onset in patients [38]. Despite these challenges, traditional antivenom remains the established and primary treatment for snakebites.
2.1.3. High cost of traditional antivenom
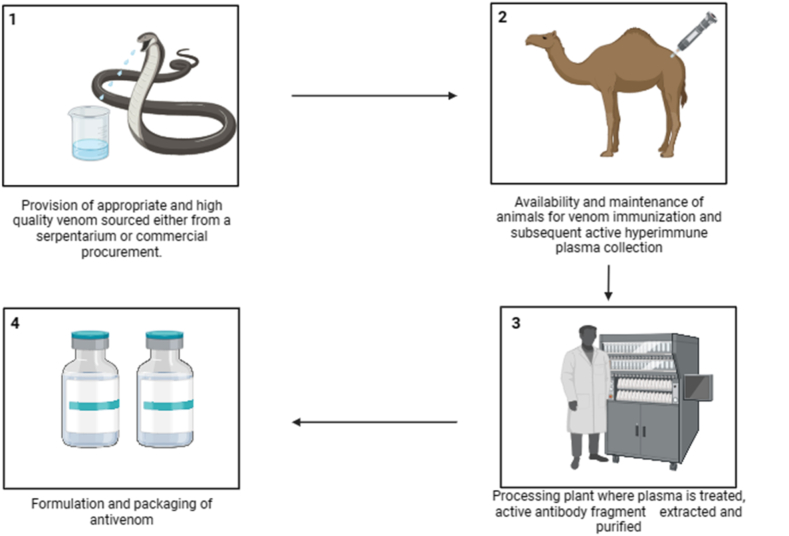
The socioeconomic impact of snakebite envenoming has been extensively documented but largely neglected by the global health community for many years. Every year, millions of individuals in low and middle-income countries experience the threat of death, disability, and societal disadvantage due to snakebite envenoming (SBE) with lack of access to suitable treatment. Health-economic considerations deeply influence all facets of this overlooked problem [39,40]. Currently, the global landscape comprises 46 antivenom producers [41], contributing to the low investment in the industry leading to the high cost of antivenom. The existing methods for antivenom production entail four key components, and each of these components contributes to the overall manufacturing cost of antivenom (Fig. 2).
Fig. 2.
Traditional antivenom production steps.
2.1.4. Limited efficacy of antivenom
The efficacy of antivenom is significantly influenced by the source of snake venoms used in horse immunization for antivenom production. This results in diverse venom composition among immunized species and those causing snakebite incidents across various geographic locations. Moreover, intraspecific variations stemming from geographic and ontogenetic differences also play a role in determining efficacy [37]. Additionally, different toxin categories exhibit varying levels of antigenicity, with low-molecular-weight toxins being less immunogenic than their high-molecular-weight counterparts. This discrepancy can lead to suboptimal antibody responses in the production animal, thereby limiting the antivenom's efficacy against toxic yet non-immunogenic venom components [17]. Furthermore, captivity significantly influences snake venom composition, resulting in substantial distinctions between wild and captive snakes. Venom production is influenced by factors such as size, sex, and age, with larger snakes and adult females, like Bothrops leucurus, displaying increased venom production compared to smaller snakes and adult males. Additionally, older animals may not be suitable for antivenom production due to their decreased ability to mount effective immune responses [42,43].
2.2. Alternative approach to using immunoglobulin G antibody
Although alternative and supplementary treatments for SBE are under development, commercializing them will require a significant amount of time. Hence, improving the current antivenom therapy is crucial for an immediate reduction in the global burden of SBE. Utilizing production hosts like chickens and ostriches for IgY-based antivenom production presents a non-invasive and cost-effective method [44]. IgY antivenoms offer advantages over traditional equine IgGs, including cost-effectiveness, as approximately 63 chickens can produce antibodies equivalent to a horse annually. Moreover, IgY antibodies do not activate the complement system, potentially allowing for larger doses to be administered to patients. However, comprehensive pre-clinical studies are necessary to validate the therapeutic efficacy of chicken IgY antivenoms [43,45].
3. Recombinant antivenom technology
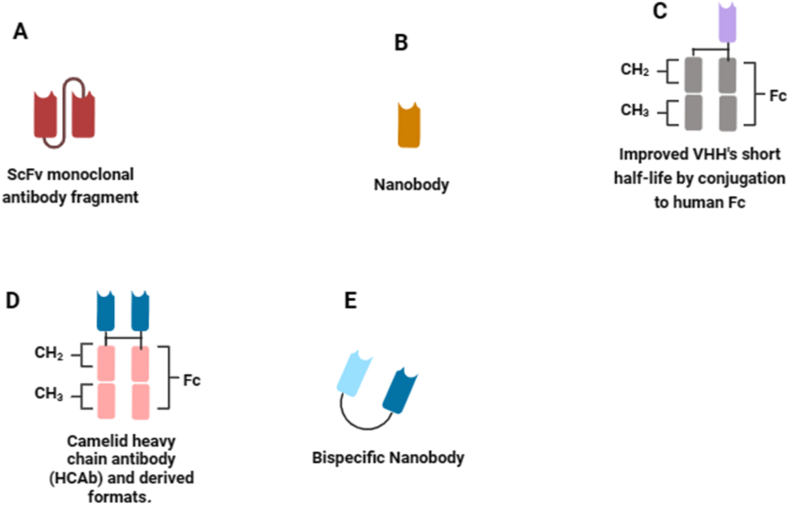
The evolutionary path of snake antivenom development, requires the integration of diverse knowledge levels into a unified framework. The integration of omics technologies and related disciplines has made significant qualitative and quantitative advancements, accelerating the journey towards recombinant antivenom technology [46]. However, given the advancements in modern biotechnological methods, the most critical toxins in the venom have been identified and characterized. Effective neutralization of such toxins, holds the potential to counter the effects of in vivo envenomation [47]. When selecting a specific snake venom for antitoxin discovery, the subsequent step involves identifying the medically most significant venom toxins for neutralization. In traditional antivenom production, this concern is not present as entire venoms are utilized for immunizing animals, allowing antibodies to be generated against all venom components in a polyclonal manner. Fig. 3 (A - E) provides an overview of the recombinant antibody fragments used in next generation antivenomunder development.
Fig. 3.
Immunoglobulin formats used in modern technologies. (A) scFv monoclonal antibody fragment is a fusion protein formed by combining the variable regions of heavy and light chains of immunoglobulins using a short linker peptide. (B) Nanobody is an antigen-binding VHH fragments originating from the heavy chain of IgG antibodies. (C) Nanobody fused to the Fc region of human immunoglobulin. (D) Heavy chain antibodies (HCAbs) recognize target antigens using a single chain containing a variable element referred to as camelid single domain antibody (VHH). (E) Bispecific nanobodies consist of two nanobodies designed to bind to distinct antigens simultaneously. VH and VL denote the heavy and light variable domains of antibodies. Other abbreviations include: CL (constant domain of light chains), CHn (constant domain of heavy chains), Fc (crystallizable fragment of antibodies), scFv (single-chain antibody fragment), VHH (heavy chain variable domain antibodies), HCAb (Camelid heavy chain antibody derived formats) and IgG (immunoglobulin G). The schematic diagram was created with biorender.com.
The adoption of recombinant technology offers significant advantages over conventional antivenom therapy, demonstrating enhanced efficacy and reduced adverse reactions in preclinical models [48]. Additionally, recombinant technology provides cost benefits, with studies indicating a reduction in production costs [40,49,50]. In comparison, animal-derived antivenoms typically range from USD 13 to $1120 per treatment, while recombinantly produced monovalent and polyvalent antivenoms can be manufactured at approximately USD 20 to $225 and USD 48 to $1354 per treatment, respectively [51]. Research suggests that producing antivenoms on a large scale using a combination of recombinant antibodies, which would require an estimated quantity of 500–2000 kg of therapeutically active antibodies, could be achieved at a cost of approximately US$55 to $65 per gram. This approach could effectively treat the approximately 1 million individuals bitten by snakes annually in sub-Saharan Africa [52,53]. However, before introducing recombinant antivenoms into clinical practice, it's essential to establish specific guidelines and classify them appropriately as either blood products or biotherapeutics [54].
3.1. Antivenom development using venom-independent immunization techniques
Eliminating the need for venom in antivenom production can significantly reduce the laborious and potentially risky tasks related to handling venomous animals. Nonetheless, venom remains essential for ensuring the quality and efficacy of antivenom through quality control and research validation [55]. One innovative approach involves a gene-based method that has been developed to create toxin-specific antivenom. This approach utilizes gene sequence information to differentiate venom toxins from non-toxins and employs bioinformatics tools to identify conserved regions (epitopes) among related toxins (derived from the same gene family). These epitopes are linked to form an ‘epitope-string,’ which, upon immunization, triggers the production of numerous toxin-specific antibodies capable of neutralizing venom-induced harm [5,56,57]. Another method involves using in-silico models to simulate systemic snakebite envenomation and treatment. By analyzing molecular size and key pharmacokinetic parameters, the model enables comparative simulations of antivenoms with different molecular formats. A case study using this model for simulated treatment of N. sumatrana envenomation highlights significant dose reductions that could be achieved through the adoption of recombinant antivenoms [20].
In proteomics, three primary strategies exist for identifying proteins in a sample: bottom-up, middle-down, and top-down [58]. The commonly used bottom-up approach involves enzymatic digestion of the sample into peptides, followed by mass spectrometry sequencing for protein identification. On the other hand, top-down proteomics is an emerging approach that directly analyzes intact proteins fragmented and sequenced using mass spectrometry. When top-down is not feasible, the middle-down approach is a compromise, involving enzymatic digestion into longer peptides for identification [59]. Regarding snake genomics, only 21 complete snake genomes are sequenced currently. Advances in sequencing technologies are anticipated to address challenges like venom diversification and repeat sequences during assembly. Third-generation sequencing technologies show promise in mitigating these challenges and enhancing accuracy. Sequencing more venomous snake genomes will deepen our insights into adaptation and venom evolution [3].
3.2. Alternative approaches utilizing venom dependency for immunization
In the field of antivenom development, various strategies have been employed to enhance efficacy and mitigate challenges. In the early 1980s, New and collaborators pioneered the use of liposomes made of sphingomyelin-cholesterol to encapsulate venom from Echis carinatus snakes, demonstrating its potential to elicit robust immune responses in mice, rabbits, and sheep [60]. Within venom-dependent approaches, innovative methods involving improved delivery systems, adjuvants, and detoxification techniques have been introduced to augment immune responses and enhance antibody neutralization of toxins. For instance, in Brazil, producing coral antivenom involves combining venoms from Micrurus frontalis and Micrurus corallinus, which can be challenging to obtain in adequate quantities. To address this limitation, researchers have explored alternative strategies, including the use of synthetic and recombinant antigens [61]. Additionally, nanoparticle adjuvant IMS3012 has emerged as a promising alternative to emulsion adjuvants, showing superior immunopotentiation capacity and safety in donor animals, particularly during the primary phase of antivenom immunization [62].
Innovative molecular engineering has led to the creation of a novel recombinant fusion protein, proficiently expressed in E. coli. This fusion protein combines crotamine and sphingomyelinase D and represents a promising immunogen to boost the reactivity of Mexican pit viper antivenoms against crotamine. Although initial tests were conducted in mice, further experiments employing ex vivo or in vitro models are essential for a comprehensive understanding of protamine neutralization. These findings underscore the potential of protamine-enriched venoms and the recombinant fusion protein as valuable immunogens to enhance neutralization potency against protamine, thus offering a pathway to improved Mexican antivenoms [63].
3.2.1. Pluribody technology
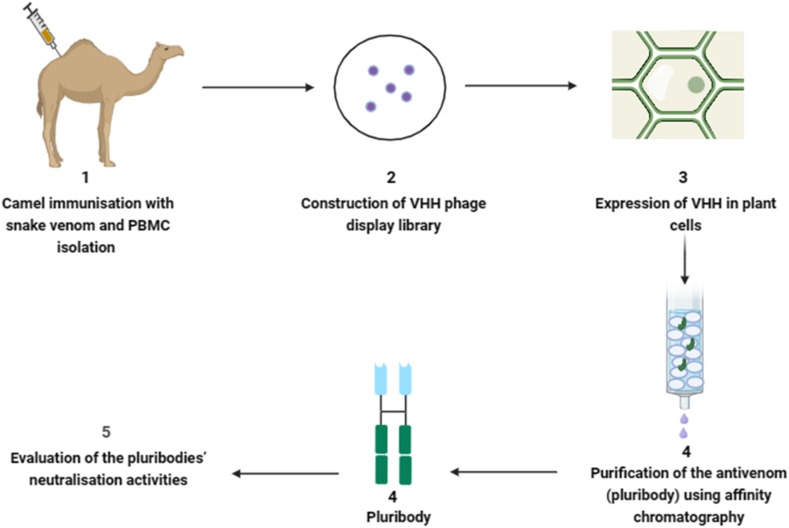
Pluribody technology is an innovative approach that utilizes genetically engineered plants to produce a diverse array of antibodies, referred to as pluribodies, that mimic those found in hyperimmunized camels. This technology leverages the natural defence mechanism of virus superinfection exclusion, resulting in diverse antibody-producing mosaics being produced consistently in plant leaves. The recombinant antivenom created through this process, using genetic data from immune cells in camels accurately mimics the toxin-binding properties of the immune response (Fig. 4). These properties resemble the venom-binding profile observed in equine antivenom [64]. Plant-based production systems offer distinct advantages, including ease of scalability, cost-efficiency, and an improved safety profile [43].
Fig. 4.
Major steps involved in pluribody technology. The numbers represent ordered steps involved. PBMC denote peripheral blood mononuclear cell and VHH is heavy chain variable domain antibodies. Schematic diagram was created with biorender.com.
Through a synthetic biological approach, pluribodies, the first plant-made recombinant polyclonal antibodies, were successfully created. Camels were immunized with venom fractions from Bothrops asper, resulting in the development of an immune VHH library. The pluribody-based formulation, derived from camels, exhibited the ability to neutralize toxin activities and provided effective protection to mice against lethal doses of venom [64]. Additionally, the study advocates for the utilization of plant cells to produce scFvBaP1, emphasizing its potential as a biotechnological alternative to horse immunization protocols for human therapy against snakebites [4,65].
Pluribodies show a promising potential in vitro model but exhibit limitations in effectively neutralizing small toxins or complex venoms. Parreno et al. [64] suggested leveraging plant biofactories for the production of recombinant polyclonal antibodies. However, utilizing plants as expression systems for antibodies poses challenges such as restricted protein yields, non-human glycosylation patterns, and the necessity of extracting and purifying antibodies from the plant matrix, characterized by abundant cellulose, lignin, and other polymeric macromolecules. Addressing these challenges is essential to enhance and optimize the viability of plant-based antibody production [66].
3.2.2. Phage display technology
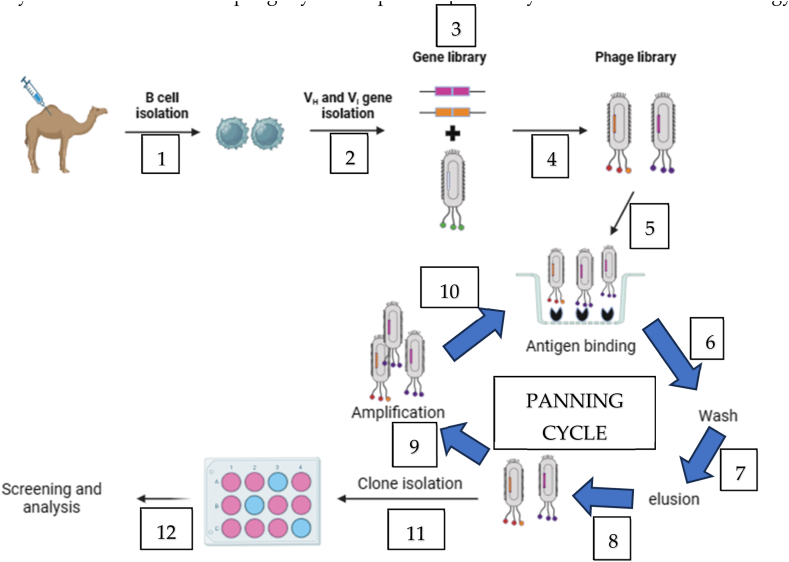
The phage display technology provides a promising approach in antivenom research by avoiding the need for extensive immunization and screening procedures. It effectively addresses challenges associated with high toxicity and low immunogenicity of the target protein. This technology capitalizes on the connection between the genetic blueprint (genotype) of antibodies and their observable characteristics (phenotype). This connection is established by integrating antibody genes, specifically single-chain variable fragments (scFvs) or single-domain antibody fragments (VHHs or Nanobodies) [15,67], into the DNA of bacteriophages. These modified bacteriophages display the antibody peptide sequence on their outer coat, facilitating the study and analysis of antibodies within a phage context [35]. The selection process in phage display involves constructing a library of proteins or peptides linked to the coat proteins of phage virions. During the in vitro selection, a target is affixed to a surface, and phages with strong affinity bind to it. These selected phages are then extracted, amplified, and subjected to further rounds of binding selection [68]. The resulting pool of phages is isolated, and characterized, and the proteins or peptides are then expressed for functional screening (Fig. 5). In this technology, deselection steps can be introduced to improve specificity by removing undesired binders from the pool. Various bacteriophage systems, including T4, lambda, and filamentous M13 bacteriophages, can be employed for phage display [69]. Each of these systems has its own set of advantages and limitations. In recent years, the M13 phage has been extensively utilized and is the sole phage system explored specifically within the field of toxicology [70].
Fig. 5.
Steps involved in phage display technology. VH and VL are variable heavy chain and variable light chain respectively. The schematic diagram was developed with biorender.com.
The biotinylation method proves effective for utilizing snake toxins as antigens in phage display selection experiments. It highlights the importance of achieving an optimal biotin-to-toxin ratio for successful outcomes in these experiments. Biotinylated antigens can be effectively presented by immobilizing them onto streptavidin using a linker, enabling precise control of concentration and preservation of their native conformation. However, this biotinylation process may introduce an additional step and potentially interfere with optimal selection sites. Utilizing biotinylation and streptavidin capture for the immobilization of phospholipase A2 may offer improved selection conditions for the discovery of toxin-binding single-chain variable fragments (scFvs) via phage display [68,71]. The hydrophobic nature of aliphatic linkers can cause biotinylated toxins to precipitate. To address this concern, the use of hydrophilic linkers like the polyethene glycol linker with four repeat units (PEG4-linker), particularly for smaller snake toxins, can help mitigate this issue. Furthermore, the length of the linker can influence antigen presentation and potentially affect phage display selection experiments. Biotinylation and streptavidin capture could potentially lead to toxin crowding, making parts or the entire toxin antigen inaccessible for effective binding interactions [68].
The outcomes of a phage display experiment are influenced by various factors, such as the incorporation of deselection steps to eliminate undesired antibody fragments, the affinity of the displayed antibody fragments, the extent of antibody display, antigen immobilization and presentation, and clonal variation. Clonal variation can impact processes related to antibody fragments like translation, folding, transport, and fusion stability. This can potentially cause amplification biases favouring phage virions that display unwanted antibody fragments [68,72]. Truncated phage virions, which are less metabolically straining on host cells [73], rapidly multiply, generating a larger quantity of non-antigen binding virions, which dominate the phage virion pool. In phage display technology, a challenge arises during amplification where antibody fragments possessing toxicity to bacteria may be eliminated due to adverse selection pressure [74,75]. This growth bias can be undesired and disrupt the successful isolation of high-affinity toxin binders in phage display experiments. Additionally, amber codons present in single-chain variable fragment (scFv) during phage display selection experiments may be misinterpreted as glutamate in certain organisms. Tomlinson libraries deliberately introduce amber codons following the myc-tag, enabling complete biosynthesis in TG1 cells while halting protein synthesis in non-suppressor strains. This deliberate design facilitates the straightforward expression of soluble scFvs following phage infection, streamlining the experimental process [68]. Amber codons have been identified as triggers for premature protein synthesis, as demonstrated in studies by Wu et al. [76] and Roncolato et al. [77]. Several strategies can be employed to mitigate this issue, including sequencing phagemid DNA, incorporating a codon that encodes glutamate, or synthesizing the complete scFv gene construct [78]. However, these approaches can be laborious and time-intensive. Another study by Laustsen [35] emphasizes the detrimental impact of amplification bias on phage display selection, favouring high-affinity binder phages over those with lower affinity or poorly assembled binders. It emphasizes the significance of maintaining optimal phage particle display levels during panning for efficient outcomes. Furthermore, the study reveals how different immobilization techniques can impact antigen presentation, potentially leading to the creation of suboptimal particles that exhibit increased binding capacity. Despite these limitations, integrating toxicogenomics and phage display in toxin analysis offers potential for advancing the development of synthetic or recombinant antivenom.
3.2.3. Humanized antibody technology
Humanized antibody technology is a potential strategy for improving the treatment of envenomation. Traditional methods relying on animal antisera have faced challenges due to impurities and the non-human origin of antibodies, leading to immunogenic reactions in patients [79]. The growing interest in utilizing human monoclonal antibodies (mAbs) to address animal envenomings stems from their flexibility, suitability for the human immune system, proven effectiveness across diverse applications, and the cost-efficiency associated with culturing mammalian cells [80]. Utilizing monoclonal antibody technology has shown promise in enhancing production efficiency and cost-effectiveness, paving the way for “next-generation antivenom”. This approach involves tailored mixtures of human monoclonal antibodies designed to target essential toxins in snake venoms [47,67,81]. The development of humanized or fully human monoclonal antibodies targeting venom toxins represents a significant advancement, enabling faster diffusion, improved tissue biodistribution, and less immunogenicity compared to traditional polyclonal preparations [67].
The process of humanizing antibodies, although aiming for therapeutic safety, does not assure complete safety due to potential CD4+ helper T cell epitopes within Complementarity Determining Regions (CDR) domains. Therefore, modifications to the CDRs of antibodies can be undertaken to decrease immunogenicity while preserving affinity and specificity for the antigen, as suggested by Harding et al. [82]. Antibody phage display technology can also be employed for the in vitro discovery of human antibodies [83], replicating the human immune system within a controlled laboratory environment using bacteriophages to display antibodies [25]. A phage display-based pipeline has been established to identify and enhance high-affinity, broadly-neutralizing human monoclonal antibodies. These antibodies effectively reduce lethality caused by elapid snake venom and significantly improve survival in mice exposed to specific venom types [84,85]. Furthermore, research by Ledsgaard et al. [86] and Ledsgaard et al. [54] has demonstrated the potential for complete neutralization of entire snake venom using a single recombinant monoclonal antibody, suggesting that a solitary monoclonal antibody holds promise in preventing lethality in venoms containing dominant toxin groups.
The potential of larger biomolecular therapies, including monoclonal human single chain variable fragments (scFvs) and complete monoclonal human IgGs show promise for inhibiting snake venom components in the future [15,[87], [88], [89]]. However, their size can impede tissue penetration, affecting efficacy against viper bites. Single-domain antibodies (sdAb) derived from heavy chain variable domains are being investigated for potential future antivenoms [90]. Another significant drawback of utilizing monoclonal antibodies lies in the necessity to address the diverse and varied toxic components present in most venom types. However, utilizing proteomics to identify venom toxin epitopes enables the selection of the most medically relevant toxins. This approach can lead to the development of more precisely targeted and safer products, reducing hypersensitivity reactions and potentially resulting in antivenoms that are more potent and effective [40].
3.2.4. DNA immunization approach
The DNA immunization approach involves innovative strategies for antivenom development. This method includes administering short or full-length toxins into animals based on amino acid sequences or introducing a plasmid encoding the relevant gene into animal cells using a Gene Gun. These approaches induce a toxin-specific immune response and subsequent antibody production by B lymphocytes [55]. Experimental exploration of immunization using cDNA encoding these pertinent toxins has shown promising results [55,91].
DNA synthesis presents a cost-effective option compared to peptides and recombinant proteins for antivenom development. By employing a computational pipeline, significant epitopes for antivenom production can be predicted using toxin sequences from medically relevant snakes. Through bioinformatics analysis involving sequence conservation and protein toxicity scoring, the most important conserved epitopes can be identified and incorporated into a DNA construct for immunization. Leveraging on in silico tools offers the advantage of analyzing sequence similarities between different species [92]. However, further bioinformatics ‘fine tuning’ may be necessary to eliminate sequences resembling human proteins in order to prevent cross-recognition [93]. Additionally, a broadly neutralizing antibody pool with cross-reactive properties can be generated using interspecies conserved toxins [43]. This method is particularly advantageous for venoms reliant on a limited number of toxins, such as the South American rattlesnake Crotalus durissus. Key toxins like neuro- and myotoxic PLA2, crotoxin, along with a thrombin-like serine proteinase, are essential for its venom toxicity. Immunization with these specific toxins is likely to yield an antivenom capable of effectively neutralizing the most significant toxic effects of the entire venom. However, antivenom production requires a substantial amount of venom for immunization and subsequent quality control processes. While obtaining venom from abundant snake species presents no challenges, collecting and maintaining venom sources for certain species is difficult due to national conservation policies or significant declines in their natural populations [94,95].
DNA immunization designed for human use encounters challenges in achieving high IgG titers, despite success in pre-clinical models. To address this shortcoming, studies have explored priming with DNA immunization followed by boosting with recombinant proteins, aiming to reduce the necessary protein antigen [96,97]. Another potential strategy involves combining DNA immunization with adjuvants, offering prospects for antivenom production. Future advancements may incorporate peptides or antibodies within liposomes to achieve targeted payload delivery, specifically targeting professional antigen-presenting cells or T helper cells using DNA sequences for immunization. The efficacy of this approach could further be enhanced by incorporating successful methodologies from gene therapy and DNA vaccination research [55].
3.2.5. Small molecular therapeutics (SMTs)
Small Molecular Therapeutics (SMTs) are designed to target specific enzymatic toxins, which make them potentially unsuitable as standalone therapeutic solutions. However, their potential lies in being adjunctive treatments, reinforcing agents for hybrid antivenoms, or aiding in initial medical care within healthcare [25]. Combinations of these inhibitors have the potential to effectively neutralize medically significant snake venoms across various species [[98], [99], [100]]. Synthetic compounds have been shown to inhibit snake venom toxins, particularly targeting snake venom metalloproteinases (SVMPs) and phospholipase A2 (PLA2). Inhibitors like chelating agents and hydroxamate peptidomimetics effectively hinder venom activity. For example, inhibitors like Varespladib and its derivatives have shown efficacy in inhibiting PLA2 activity in venom [101,102].
Snake venoms, particularly from the Viperidae family, contain enzymatic components with specific active sites relying on a minimal set of amino acids. Batimastat and Marimastat, categorized as small molecule inhibitors, appeared promising in mitigating the effects of venom from the snake species Echis ocellatus in mice [98]. These inhibitors have demonstrated efficacy in inhibiting various venom activities, especially hemorrhagic actions. The speed of administration influenced the degree of hemorrhage inhibition, with faster administration yielding better results, and Batimastat showed better effectiveness in inhibiting hemorrhagic activity compared to Marimastat [90,103,104].
Metal chelators like EDTA, categorized as small molecule enzymatic inhibitors, have also demonstrated the ability to mitigate toxic effects induced by specific snake venoms [105,106]. These inhibitors target enzymatically active toxins by mimicking natural substrates or scavenging vital co-factors essential for the toxin's function as reported by Laustsen [80]. However, it is important to note that there are currently no commercially available small molecule inhibitors designed specifically for treating snakebite envenoming [107].
3.2.6. Single-domain antibodies (nanobodies) technology
Single-domain antibodies (nanobodies) present a viable option for future antivenoms due to their small size, specificity, stability, and high epitope affinity. These nanobodies produced through microbial expression systems, exhibit strong and specific binding to target molecules, demonstrating high affinity, robustness, rapid distribution, and low immunogenicity in humans [79]. In comparison to intact monoclonal antibodies, single-chain variable fragments (scFvs) exhibit favourable characteristics, including a low molecular weight, which reduces their immunogenicity and enhances economic viability. However, scFvs face challenges such as lower avidity and issues related to stability, solubility, and affinity, necessitating ongoing research to optimize their production processes [67,108]. Conversely, VHHs, representing the smallest functional antibody fragments, resembling F (ab’) antibody domains, offer improved tissue penetration due to their small size, high solubility, tissue permeability, stability, interactions with inaccessible sites, and low immunogenicity. These properties make them promising for various biomedical applications such as research, diagnostics, and therapy [67]. Additionally, nanobodies have demonstrated therapeutic potentials against Bothrops atrox snake venom-induced envenoming, effectively neutralizing local tissue hemorrhage and myonecrosis, although their efficacy in preventing venom lethality requires further investigation [109].
Nanobodies, characterized by their low molecular weight, are rapidly eliminated primarily through renal clearance, with over 50 % elimination within 1 h [110]. However, their clinical effectiveness is impeded by challenges such as low stability, solubility, and affinity [111]. To address these issues, leveraging multivalent binding proteins as a technology platform has been proposed to enhance nanobodies' potency. This involves assembling multiple binding domains within a single molecule, augmenting both molecular size and functional affinity. This approach shows promise in improving the efficacy and pharmacokinetic properties of nanobodies, enabling effective targeting of complex toxin mixtures like snake venoms [112]. Despite this promise, these alternatives have not yet advanced to clinical application. Future research is anticipated to focus on nanobodies and human antibody formats, showcasing their potential to advance snakebite treatments [66,113].
4. Comparing the strengths and weaknesses of traditional and recombinant antivenom
Traditional antivenom, a long-standing century-old approach, and recombinant antivenom, a more recent development, each possess their own set of advantages and drawbacks, as presented in Table 1.
Table 1.
The strengths and weaknesses of traditional and recombinant antivenom.
| Aspect | Traditional Antivenom | Recombinant Antivenom | Reference(s) |
|---|---|---|---|
| Venom Diversity | May effectively neutralize a wide range of snake species and venom types due to the use of whole venoms for immunization. | May struggle to completely neutralize all snake species or venom types due to the specific targeting of venom components or toxins. | [17,27,42,114] |
| Regulatory Approval | Typically has a more straightforward and well-established regulatory approval process. | May face a more intricate and time-consuming regulatory approval process, potentially causing delays in availability. | [40,53,54] |
| Stability and Storage | Generally, has a longer shelf life, more stable storage conditions, and consistent dosing. | Could require specific stability and storage conditions, potentially having a shorter shelf life, dose variations, and potential renal elimination. | [77] |
| Adverse Reactions | Can elicit adverse reactions, including anaphylaxis, requiring medical supervision and monitoring. | Carries the potential for adverse reactions, necessitating thorough medical supervision and monitoring. | [16,27,81,115] |
| Clinical Efficacy and Safety | Well-established clinical efficacy and safety through extensive use and trials. | Clinical efficacy and safety may be less comprehensively established, with uncertainty about long-term effectiveness and safety. | [48,116,117] |
| Cost | Typically ranges from USD 13 to $1120 per treatment. | Potentially more cost-effective, with estimates of approximately USD 20 to $225 for monovalent and USD 48 to $1354 for polyvalent recombinant antivenoms per treatment. | [51] |
| Scalability | May be limited by the availability of venom sources and animal immunization. | Offers potential for large-scale production, providing cost-effective solutions for regions with high snakebite incidents. | [52,53] |
| Production Advancements | Rely on traditional methods and do not integrate modern biotechnological advances. | Benefit from modern biotechnological advancements and targeted toxin identification, potentially enhancing efficacy and reducing adverse reactions. | [35,46,47] |
5. Future perspectives
In the dynamic realm of snakebite research, scientists are exploring the development of recombinant antivenoms. This cutting-edge approach leverages the power of sequencing and "-omics” technologies to create adaptable therapeutic solutions capable of effectively countering a wide range of venom types. Although the proteomic profiles of over 200 snake species are now accessible, researchers are still diligently conducting functional studies to pinpoint the specific toxins responsible for snake envenomation [35,79].
Another promising avenue in this quest involves the use of short peptides with modified structural properties as lead molecules. This approach offers a significant advantage over traditional phage antibodies [118,119]. However, due to the cost implications of synthesizing these lead peptides, researchers have to focus on optimizing production processes to achieve larger yields [5,27,120].
The field of toxin neutralization for antivenom sera is witnessing the emergence of aptamers as a promising substitute for traditional antibodies. These aptamers show great potential for enhanced production, storage, and safety, despite facing inherent challenges [121]. To maximize the neutralization capacity of antivenoms, it is crucial that they possess molecular weights comparable to the target toxins. This characteristic ensures efficient bioavailability, even in deep tissues, thus, facilitating effective neutralization [67].
Moreover, in the battle against snakebite envenomation, regional collaboration, increased investment, and financial support are imperative. This collaborative approach would ensure that all snakebite victims receive suitable and timely antivenom treatments, ultimately saving lives [122]. The assessment of in vivo neutralization of snake venoms involves a protocol in which toxins and antibodies are pre-incubated in rodents before injection. While this method enhances reproducibility and comparability between antivenoms, it does not precisely simulate real-life envenomation and treatment situations. Therefore, evaluating the neutralizing capacity in separate experiments is more pertinent for a comprehensive assessment [81].
Looking ahead to the future of antivenom manufacturing, researchers are exploring the use of transgenic animals with a human antibody repertoire. This innovative approach holds the promise of enhancing safety and enabling early administration to victims, potentially improving treatment and recovery outcomes by reducing the risk of adverse reactions [55,123].
6. Conclusions
The journey to revolutionize snakebite care with novel antivenoms is marked by both groundbreaking breakthroughs and formidable barriers. The fusion of cutting-edge technologies has enabled the development of recombinant antivenoms, offering adaptable therapeutic remedies effective against diverse venom types. However, the complex task of identifying specific toxins in snake venoms remains a challenge. The use of short peptides with modified structural properties shows promise but is accompanied by cost considerations, which researchers are actively addressing. Aptamers are emerging as potential substitutes for traditional antibodies, offering advantages in production, storage, and safety. To optimize neutralization capacity, antivenoms must possess molecular weights similar to target toxins, ensuring efficient bioavailability, even in deep tissues. Enhanced regional collaboration and increased support are vital to ensure timely antivenom treatment for snakebite victims. While in vivo neutralization protocols enhance comparability, their limitations necessitate separate experiments for a comprehensive assessment. Looking to the future, the use of transgenic animals with human antibody repertoires holds promise for safer and more efficient antivenom manufacturing. Despite the barriers, the pursuit of novel antivenoms continues to advance snakebite care, offering hope for improved treatment and outcomes for snakebite victims worldwide.
Funding
This research received no external funding.
Data availability statement
Data is available upon request from the corresponding author.
CRediT authorship contribution statement
Samuel Odo Uko: Writing – original draft, Methodology, Investigation, Data curation, Conceptualization. Ibrahim Malami: Writing – review & editing. Kasimu Ghandi Ibrahim: Writing – review & editing. Nafiu Lawal: Writing – review & editing. Muhammad Bashir Bello: Writing – review & editing. Murtala Bello Abubakar: Writing – review & editing. Mustapha Umar Imam: Writing – review & editing, Supervision, Resources, Project administration, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The Authors acknowledge the staff and students of CAMRET for their contribution towards the idea that served as the foundation for this review.
References
- 1.Burbrink F., Pyron R. The taming of the skew: estimating proper confidence intervals for divergence dates. Syst. Biol. 2008;57:317–328. doi: 10.1080/10635150802040605. [DOI] [PubMed] [Google Scholar]
- 2.Wallach V., Williams K., Boundy J. first ed. CRC Press; Boca Raton: 2014. Snakes of the World: A Catalogue of Living and Extinct Species. [Google Scholar]
- 3.Rao W.Q., Kalogeropoulos K., Allentoft M.E., Gopalakrishnan S., Zhao W.N., Workman C.T., et al. The rise of genomics in snake venom research: recent advances and future perspectives. GigaScience. 2022:11. doi: 10.1093/gigascience/giac024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adriao A.A.X., Dos Santos A.O., de Lima E., Maciel J.B., Paz W.H.P., da Silva F.M.A., et al. Plant-derived toxin inhibitors as potential Candidates to complement antivenom treatment in snakebite envenomations. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.842576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casewell N.R., Wuster W., Vonk F.J., Harrison R.A., Fry B.G. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Tasoulis T., Isbister G. A review and database of snake venom proteomes. Toxins. 2017;9:290. doi: 10.3390/toxins9090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017;3 doi: 10.1038/nrdp.2017.79. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez J.M., Theakston R.D., Warrell D.A. Confronting the neglected problem of snake bite envenoming: the need for a global partnership. PLoS Med. 2006;3:e150. doi: 10.1371/journal.pmed.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO Snakebite envenoming. 2023. https://www.who.int/news-room/fact-sheets/detail/snakebite-envenoming>
- 10.Ooms G.I., van Oirschot J., Waldmann B., Okemo D., Mantel-Teeuwisse A.K., van den Ham H.A., et al. The burden of snakebite in rural communities in Kenya: a Household survey. Am. J. Trop. Med. Hyg. 2021;105:828–836. doi: 10.4269/ajtmh.21-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Who . 2017. Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins; pp. 197–388. [Google Scholar]
- 12.Armstrong L.J., Cynthia S., George M., Zachariah A. Comparing community and hospital data of snakebite in North Bihar: a community incidence survey and a parallel hospital-based clinical study. Trop. Doct. 2019;49:285–292. doi: 10.1177/0049475519865036. [DOI] [PubMed] [Google Scholar]
- 13.Calvete J.J. Next-generation snake venomics: protein-locus resolution through venom proteome decomplexation. Expert Rev. Proteomics. 2014;11:315–329. doi: 10.1586/14789450.2014.900447. [DOI] [PubMed] [Google Scholar]
- 14.Calvete J.J., Sanz L., Pla D., Lomonte B., Gutierrez J.M. Omics meets biology: application to the design and preclinical assessment of antivenoms. Toxins. 2014;6:3388–3405. doi: 10.3390/toxins6123388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva M.C.S., Pereira S.S., Gouveia M.P., Luiz M.B., Sousa R.M.O., Kayano A.M., et al. Anti-Metalloprotease P-I single-domain antibodies: tools for next-generation snakebite antivenoms. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/2748962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sriapha C., Rittilert P., Vasaruchapong T., Srisuma S., Wananukul W., Trakulsrichai S. Early adverse reactions to snake antivenom: Poison center data analysis. Toxins. 2022;14 doi: 10.3390/toxins14100694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casewell N.R., Jackson T.N.W., Laustsen A.H., Sunagar K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020;41:570–581. doi: 10.1016/j.tips.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sewall H. Experiments on the preventive Inoculation of rattlesnake venom. J. Physiol. 1887;8:203–210. doi: 10.1113/jphysiol.1887.sp000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keating G.M. Crotalidae polyvalent immune Fab: in patients with North American crotaline envenomation. BioDrugs. 2011;25:69–76. doi: 10.2165/11207250-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Morris N.M., Blee J.A., Hauert S. Developing a computational pharmacokinetic model of systemic snakebite envenomation and antivenom treatment. Toxicon. 2022;215:77–90. doi: 10.1016/j.toxicon.2022.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Zurbano B.N., Tavarone E., Viacava B.G., Dokmetjian J.C., Cascone O., Fingermann M. Critical aspects on traditional antivenom production processes and their optimization by factorial analysis. Biologicals. 2020;68:65–73. doi: 10.1016/j.biologicals.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes C.M., Teixeira CdFP., Leite A., Gutiérrez J.M., Rocha F.A.C. The snake venom metalloproteinase BaP1 induces joint hypernociception through TNF-α and PGE2-dependent mechanisms. Br. J. Pharmacol. 2007;151:1254–1261. doi: 10.1038/sj.bjp.0707351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimenes S., Sachett J., Colombini M., Sousa L., Ibiapina H., Costa A.G., et al. Observation of Bothrops atrox snake envenoming blister formation from five patients: pathophysiological insights. Toxins. 2021;12 doi: 10.3390/toxins13110800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerardo C.J., Quackenbush E., Lewis B., Rose S.R., Greene S., Toschlog E.A., et al. The efficacy of crotalidae polyvalent immune Fab (ovine) antivenom versus placebo plus optional rescue therapy on recovery from copperhead snake envenomation: a randomized, double-blind, placebo-controlled, clinical trial. Ann. Emerg. Med. 2017;70:233–244 e3. doi: 10.1016/j.annemergmed.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 25.Kini R.M., Sidhu S.S., Laustsen A.H. Biosynthetic oligoclonal antivenom (BOA) for snakebite and next-generation treatments for snakebite victims. Toxins. 2018;10 doi: 10.3390/toxins10120534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alomran N., Blundell P., Alsolaiss J., Crittenden E., Ainsworth S., Dawson C.A., et al. Exploring the utility of recombinant snake venom serine protease toxins as immunogens for generating experimental snakebite antivenoms. Toxins. 2022;14 doi: 10.3390/toxins14070443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alangode A., Rajan K., Nair B.G. Snake antivenom: challenges and alternate approaches. Biochem. Pharmacol. 2020;181 doi: 10.1016/j.bcp.2020.114135. [DOI] [PubMed] [Google Scholar]
- 28.Açikalin A., Gökel Y., Kuvandik G., Duru M., Köseoglu Z., Satar S. The efficacy of low-dose antivenom therapy on morbidity and mortality in snakebite cases. Am. J. Emerg. Med. 2008;26:402–407. doi: 10.1016/j.ajem.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Chippaux J., Lang J., Eddine S., Fagot P., Rage V., Peyrieux J., et al. Clinical safety of a polyvalent F(ab')2 equine antivenom in 223 African snake envenomations: a field trial in Cameroon. Trans. R. Soc. Trop. Med. Hyg. 1998;92:657–662. doi: 10.1016/s0035-9203(98)90802-1. [DOI] [PubMed] [Google Scholar]
- 30.Dart R., McNally J. Efficacy, safety, and use of snake antivenoms in the United States. Ann. Emerg. Med. 2001;37:181–188. doi: 10.1067/mem.2001.113372. [DOI] [PubMed] [Google Scholar]
- 31.Leon G., Segura A., Herrera M., Otero R., Franca F.O., Barbaro K.C., et al. Human heterophilic antibodies against equine immunoglobulins: assessment of their role in the early adverse reactions to antivenom administration. Trans. R. Soc. Trop. Med. Hyg. 2008;102:1115–1119. doi: 10.1016/j.trstmh.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 32.Stone S.F., Isbister G.K., Shahmy S., Mohamed F., Abeysinghe C., Karunathilake H., et al. Immune response to snake envenoming and treatment with antivenom; complement activation, cytokine production and mast cell degranulation. PLoS Neglected Trop. Dis. 2013;7:e2326. doi: 10.1371/journal.pntd.0002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tariang D., Philip P., Alexander G., Macaden S., Jeyaseelan L., Peter J., et al. Randomized controlled trial on the effective dose of anti-snake venom in cases of snake bite with systematic envenomation. JAPI. 1999;47:369–371. [PubMed] [Google Scholar]
- 34.Ha T.H., Hojer J., Trinh X.K., Nguyen T.D. A controlled clinical trial of a novel antivenom in patients envenomed by Bungarus multicinctus. J. Med. Toxicol. 2010;6:393–397. doi: 10.1007/s13181-010-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laustsen A. University of Copenhagen; 2016. Recombinant Antivenoms. [Google Scholar]
- 36.Sofyantoro F., Yudha D.S., Lischer K., Nuringtyas T.R., Putri W.A., Kusuma W.A., et al. Bibliometric analysis of literature in snake venom-related research worldwide (1933-2022) Animals (Basel) 2022:12. doi: 10.3390/ani12162058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patra A., Herrera M., Gutierrez J.M., Mukherjee A.K. The application of laboratory-based analytical tools and techniques for the quality assessment and improvement of commercial antivenoms used in the treatment of snakebite envenomation. Drug Test. Anal. 2021;13:1471–1489. doi: 10.1002/dta.3108. [DOI] [PubMed] [Google Scholar]
- 38.Kularatne S.A., Weerakoon K., Silva A., Maduwage K., Walathara C., Rathnayake I., et al. Efficacy of intravenous hydrocortisone administered 2-4 h prior to antivenom as prophylaxis against adverse drug reactions to snake antivenom in Sri Lanka: an open labelled randomized controlled trial. Toxicon. 2016;120:159–165. doi: 10.1016/j.toxicon.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Francis M.F., Vianney S.J., Heitz-Tokpa K., Kreppel K. Risks of snakebite and challenges to seeking and providing treatment for agro-pastoral communities in Tanzania. PLoS One. 2023;18 doi: 10.1371/journal.pone.0280836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Habib A.G., Brown N.I. The snakebite problem and antivenom crisis from a health-economic perspective. Toxicon. 2018;150:115–123. doi: 10.1016/j.toxicon.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Habib A.G., Musa B.M., Iliyasu G., Hamza M., Kuznik A., Chippaux J.P. Challenges and prospects of snake antivenom supply in sub-Saharan Africa. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braga R.M., Oitaven L.P.C., Rocha MMTd, Vieira S.E.M., Rúbio D.T., Sant’anna S.S., et al. Influence of size, sex and age on venom yield of Bothrops leucurus (Serpentes, Viperidae) under captivity conditions. Basic Appl. Herpetol. 2022;37:31–45. [Google Scholar]
- 43.Rathore A.S., Kumar R., Tiwari O.S. Recent advancements in snake antivenom production. Int. J. Biol. Macromol. 2023;240 doi: 10.1016/j.ijbiomac.2023.124478. [DOI] [PubMed] [Google Scholar]
- 44.Choraria A., Somasundaram R., Gautam M., Paray B.A., Al-sadoon M.K., Michael A. Experimental antivenoms from chickens and rabbits and their comparison with commercially available equine antivenom against the venoms of Daboia russelii and Echis carinatus snakes. Toxin Rev. 2020;1–12 [Google Scholar]
- 45.Bello C., Torrico F., enez J.C.J., Cepeda M.V., opez M.A.L., Rodríguez-Acosta A. A new approach of immunotherapy against crotalus snakes envenoming: ostrich (Struthio camelus) egg yolk antibodies (IgY-technology) Invest. Clin. 2022;63:57–69. [Google Scholar]
- 46.Calvete J.J. Venomics: integrative venom proteomics and beyond. Biochem. J. 2017;474:611–634. doi: 10.1042/BCJ20160577. [DOI] [PubMed] [Google Scholar]
- 47.Dias da Silva W., De Andrade S.A., Megale A.A.A., De Souza D.A., Sant'Anna O.A., Magnoli F.C., et al. Antibodies as snakebite antivenoms: past and future. Toxins. 2022;14 doi: 10.3390/toxins14090606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos-Filho N.A., Sousa T.S., Boldrini-Franca J., Santos-Silva L.K., Menaldo D.L., Henrique-Silva F., et al. rBaltMIP, a recombinant alpha-type myotoxin inhibitor from Bothrops alternatus (Rhinocerophis alternatus) snake, as a potential candidate to complement the antivenom therapy. Toxicon. 2016;124:53–62. doi: 10.1016/j.toxicon.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 49.Laustsen A.H., Johansen K.H., Engmark M., Andersen M.R. Recombinant snakebite antivenoms: a cost-competitive solution to a neglected tropical disease? PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu B.S., Jiang B.R., Hu K.C., Liu C.H., Hsieh W.C., Lin M.H., et al. Development of a broad-spectrum antiserum against cobra venoms using recombinant three-finger toxins. Toxins. 2021;13 doi: 10.3390/toxins13080556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins T.P., Laustsen A.H. Cost of manufacturing for recombinant snakebite antivenoms. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laustsen A.H. Snakebites: costing recombinant antivenoms. Nature. 2016;538:41. doi: 10.1038/538041e. [DOI] [PubMed] [Google Scholar]
- 53.Hammerschmidt N., Tscheliessnig A., Sommer R., Helk B., Jungbauer A. Economics of recombinant antibody production processes at various scales: industry-standard compared to continuous precipitation. Biotechnol. J. 2014;9:766–775. doi: 10.1002/biot.201300480. [DOI] [PubMed] [Google Scholar]
- 54.Ledsgaard L., Laustsen A.H., Pus U., Wade J., Villar P., Boddum K., et al. In vitro discovery of a human monoclonal antibody that neutralizes lethality of cobra snake venom. mAbs. 2022;14 doi: 10.1080/19420862.2022.2085536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bermudez-Mendez E., Fuglsang-Madsen A., Fons S., Lomonte B., Gutierrez J.M., Laustsen A.H. Innovative immunization strategies for antivenom development. Toxins. 2018;10 doi: 10.3390/toxins10110452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrison R.A. Development of venom toxin-specific antibodies by DNA immunisation: rationale and strategies to improve therapy of viper envenoming. Vaccine. 2004;22:1648–1655. doi: 10.1016/j.vaccine.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 57.Harrison RAea. Antibody from mice immunized with DNA encoding the carboxyl-disintegrin and cysteine-rich domain (JD9) of the haemorrhagic metalloprotease, Jararhagin, inhibits the main lethal component of viper venom. Clin. Exp. Immunol. 2000;121:358–363. doi: 10.1046/j.1365-2249.2000.01287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han X., Aslanian A., Yates rJ. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richard H.V., Carolina A.N., Jonas P., Ana GdCN-F. 2016. Snake Venom Proteopeptidomics: what Lies behind the Curtain; pp. 333–365. [Google Scholar]
- 60.New R.R.C., Theakston R.D.G., Zumbuehl O., Iddon D., Friend J. Liposomal immunisation against snake venoms. Toxicon. 1985;23:215–219. doi: 10.1016/0041-0101(85)90144-8. [DOI] [PubMed] [Google Scholar]
- 61.de Castro K.L.P., Lopes-de-Souza L., de Oliveira D., Machado-de-Avila R.A., Paiva A.L.B., de Freitas C.F., et al. A combined strategy to improve the development of a coral antivenom against Micrurus spp. Front. Immunol. 2019;10:2422. doi: 10.3389/fimmu.2019.02422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waghmare A., Deopurkar R.L., Salvi N., Khadilkar M., Kalolikar M., Gade S.K. Comparison of Montanide adjuvants, IMS 3012 (Nanoparticle), ISA 206 and ISA 35 (Emulsion based) along with incomplete Freund's adjuvant for hyperimmunization of equines used for production of polyvalent snake antivenom. Vaccine. 2009;27:1067–1072. doi: 10.1016/j.vaccine.2008.11.103. [DOI] [PubMed] [Google Scholar]
- 63.Ponce-Lopez R., Neri-Castro E., Olvera-Rodriguez F., Sanchez E.E., Alagon A., Olvera-Rodriguez A. Neutralization of crotamine by polyclonal antibodies generated against two whole rattlesnake venoms and a novel recombinant fusion protein. Toxicon. 2021;197:70–78. doi: 10.1016/j.toxicon.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parreno J.J.M., Huet E., Fernandez-Del-Carmen A., Segura A., Venturi M., Gandia A., et al. A synthetic biology approach for consistent production of plant-made recombinant polyclonal antibodies against snake venom toxins. Plant Biotechnol. J. 2018;16:727–736. doi: 10.1111/pbi.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gomes M., Alvarez M.A., Quellis L.R., Becher M.L., Castro J.M.A., Gameiro J., et al. Expression of an scFv antibody fragment in Nicotiana benthamiana and in vitro assessment of its neutralizing potential against the snake venom metalloproteinase BaP1 from Bothrops asper. Toxicon. 2019;160:38–46. doi: 10.1016/j.toxicon.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 66.Knudsen C., Laustsen A.H. Recent advances in next generation snakebite antivenoms. Trav. Med. Infect. Dis. 2018;3 doi: 10.3390/tropicalmed3020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fernandes C.F.C., Pereira S.S., Luiz M.B., Silva N., Silva M.C.S., Marinho A.C.M., et al. Engineering of single-domain antibodies for next-generation snakebite antivenoms. Int. J. Biol. Macromol. 2021;185:240–250. doi: 10.1016/j.ijbiomac.2021.06.043. [DOI] [PubMed] [Google Scholar]
- 68.Laustsen A.H., Lauridsen L.P., Lomonte B., Andersen M.R., Lohse B. Pitfalls to avoid when using phage display for snake toxins. Toxicon. 2017;126:79–89. doi: 10.1016/j.toxicon.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 69.Bazan J., Całkosi ński I., Gamian A. Phage display—a powerful technique for immunotherapy. Hum. Vaccines Immunother. 2012;8:1817–1828. doi: 10.4161/hv.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ledsgaard L., Kilstrup M., Karatt-Vellatt A., McCafferty J., Laustsen A.H. Basics of antibody phage display technology. Toxins. 2018;10 doi: 10.3390/toxins10060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richard G., Meyers A.J., McLean M.D., Arbabi-Ghahroudi M., MacKenzie R., Hall J.C. vol. 8. PLoS ONE; 2013. (Neutralization of A-Cobratoxin with High-Affinity Llama Single-Domain Antibodies (VHHs) and a VHH-Fc Antibody). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCafferty J. Academic Press Inc.; San Diego, CA: 1996. Phage Display: Factors Affecting Panning Efficiency. Phage Display of Peptides and Proteins: A Laboratory Manual; pp. 261–276. [Google Scholar]
- 73.Bruin Rd, Spelt K., Mol J., Koes R., Quattrocchio F. Selection of high-affinity phage antibodies from phage display libraries. Nat. Biotechnol. 1999;17:397–399. doi: 10.1038/7959. [DOI] [PubMed] [Google Scholar]
- 74.Umlauf B.J., McGuire M.J., Brown K.C. Introduction of plasmid encoding for rare tRNAs reduces amplification bias in phage display biopanning. Biotechniques. 2015;58:81–84. doi: 10.2144/000114256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schofield D.J., Pope A.R., Clementel V., Buckell J., Chapple S.D., Clarke K.F., et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu S., Ke A., Doudna J.A. A fast and efficient procedure to produce scFvs specific for large macromolecular complexes. J. Immunol. Methods. 2007;318:95–101. doi: 10.1016/j.jim.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roncolato E.C., Campos L.B., Pessenda G., Costa e Silva L., Furtado G.P., Barbosa J.E. Phage display as a novel promising antivenom therapy: a review. Toxicon. 2015;93:79–84. doi: 10.1016/j.toxicon.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Barderas R., Shochat S., Martínez-Torrecuadrada J., Altschuh D., Meloen R., Ignacio Casal J. A fast mutagenesis procedure to recover soluble and functional scFvs containing amber stop codons from synthetic and semisynthetic antibody libraries. J. Immunol. Methods. 2006;312:182–189. doi: 10.1016/j.jim.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 79.Alirahimi E., Kazemi-Lomedasht F., Shahbazzadeh D., Habibi-Anbouhi M., Hosseininejad Chafi M., Sotoudeh N., et al. Nanobodies as novel therapeutic agents in envenomation. Biochim. Biophys. Acta Gen. Subj. 2018;1862:2955–2965. doi: 10.1016/j.bbagen.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 80.Laustsen A.H. Guiding recombinant antivenom development by omics technologies. N Biotechnol. 2018;45:19–27. doi: 10.1016/j.nbt.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 81.Laustsen A.H., Maria Gutierrez J., Knudsen C., Johansen K.H., Bermudez-Mendez E., Cerni F.A., et al. Pros and cons of different therapeutic antibody formats for recombinant antivenom development. Toxicon. 2018;146:151–175. doi: 10.1016/j.toxicon.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 82.Harding F.A., Stickler M.M., Razo J., DuBridge R.B. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. mAbs. 2010;2:256–265. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCafferty J., Griffiths A.D., Winter G., Chiswell D.J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 84.Ledsgaard L., Wade J., Jenkins T.P., Boddum K., Oganesyan I., Harrison J.A., et al. Discovery and optimization of a broadly-neutralizing human monoclonal antibody against long-chain alpha-neurotoxins from snakes. Nat. Commun. 2023;14:682. doi: 10.1038/s41467-023-36393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ledsgaard L., Wade J., Boddum K., Oganesyan I., Harrison J., Jenkins T.P., et al. 2022. Discovery of a Broadly-Neutralizing Human Antibody that Can Rescue. [Google Scholar]
- 86.Ledsgaard L., Laustsen A.H., Pus U., Wade J., Villar P., Boddum K., et al. 2021. In Vitro Discovery and Optimization of a Human Monoclonal Antibody that Neutralizes Neurotoxicity and Lethality of Cobra Snake Venom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Silva L.C., Pucca M.B., Pessenda G., Campos L.B., Martinez E.Z., Cerni F.A., et al. Discovery of human scFvs that cross-neutralize the toxic effects of B. jararacussu and C. d. terrificus venoms. Acta Trop. 2018;177:66–73. doi: 10.1016/j.actatropica.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 88.Laustsen A.H., Karatt-Vellatt A., Masters E.W., Arias A.S., Pus U., Knudsen C., et al. In vivo neutralization of dendrotoxin-mediated neurotoxicity of black mamba venom by oligoclonal human IgG antibodies. Nat. Commun. 2018;9:3928. doi: 10.1038/s41467-018-06086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nazari A., Samianifard M., Rabie H., Mirakabadi A.Z. Recombinant antibodies against Iranian cobra venom as a new emerging therapy by phage display technology. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020;26 doi: 10.1590/1678-9199-JVATITD-2019-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Williams H.F., Layfield H.J., Vallance T., Patel K., Bicknell A.B., Trim S.A., et al. The urgent need to develop novel strategies for the diagnosis and treatment of snakebites. Toxins. 2019;11 doi: 10.3390/toxins11060363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Azofeifa-Cordero G., Arce-Estrada V., Flores-Díaz M., Alape-Girón A. Immunization with cDNA of a novel P-III type metalloproteinase from the rattlesnake Crotalus durissus durissus elicits antibodies which neutralize 69% of the hemorrhage induced by the whole venom. Toxicon. 2008;52:e8. doi: 10.1016/j.toxicon.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 92.Ros-Lucas A., Bigey P., Chippaux J., Gascón J., Alonso-Padilla J. Computer-aided analysis of west sub-Saharan Africa snakes venom towards the design of epitopebased poly-specific antivenoms. Toxins. 2022;14:418. doi: 10.3390/toxins14060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Melo PDVd, Lima SdA., Araújo P., Santos R.M., Gonzalez E., Belo A.A., et al. Immunoprotection against lethal effects of Crotalus durissus snake venom elicited by synthetic epitopes trapped in liposomes. Int. J. Biol. Macromol. 2020;161:299–307. doi: 10.1016/j.ijbiomac.2020.05.171. [DOI] [PubMed] [Google Scholar]
- 94.Gutierrez J.M., Leon G., Burnouf T. Antivenoms for the treatment of snakebite envenomings: the road ahead. Biologicals. 2011;39:129–142. doi: 10.1016/j.biologicals.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 95.Cardona-Ruda A., Rey-Súarez P., Núñez V. Anti-neurotoxins from Micrurus mipartitus in the development of coral Snake antivenoms. Toxins. 2022;14:265. doi: 10.3390/toxins14040265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ramos H.R., Junqueira-de-Azevedo I.D.L.M., Novo J.B., Castro K., Duarte C.G., Machado-de-Ávila R.A., et al. A heterologous multiepitope DNA prime/recombinant protein boost immunisation strategy for the development of an antiserum against Micrurus corallinus (coral snake) venom. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu S., Wang S., Lu S. DNA immunization as a technology platform for monoclonal antibody induction. Emerg. Microb. Infect. 2016;5:e33. doi: 10.1038/emi.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Albulescu L.O., Xie C., Ainsworth S., Alsolaiss J., Crittenden E., Dawson C.A., et al. A therapeutic combination of two small molecule toxin inhibitors provides broad preclinical efficacy against viper snakebite. Nat. Commun. 2020;11:6094. doi: 10.1038/s41467-020-19981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Albulescu L.-O., Melissa S.H., Stuart A., Jaffer A., Edouard C., Juan J.C., et al. Preclinical validation of a repurposed metal chelator as an early-intervention therapeutic for hemotoxic snakebite. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aay8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bulfone T.C., Samuel S.P., Bickler P.E., Lewin M.R. Developing small molecule therapeutics for the initial and adjunctive treatment of snakebite. J. Trop. Med. 2018;2018 doi: 10.1155/2018/4320175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gutierrez J.M., Albulescu L.O., Clare R.H., Casewell N.R., Abd El-Aziz T.M., Escalante T., et al. The search for natural and synthetic inhibitors that would complement antivenoms as therapeutics for snakebite envenoming. Toxins. 2021;13 doi: 10.3390/toxins13070451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Flipo M., Charton J., Hocine A., Dassonneville S., Deprez B., Deprez-Poulain R. Hydroxamates: relationships between structure and plasma stability. J. Med. Chem. 2009;52:6790–6802. doi: 10.1021/jm900648x. [DOI] [PubMed] [Google Scholar]
- 103.Lewin M.R., Samuel S., Merkel J., Bickler P. Varespladib (LY315920) appears to be a potent, broad-spectrum, inhibitor of snake venom phospholipase A2 and a possible pre-referral treatment for envenomation. Toxins. 2016;8:248. doi: 10.3390/toxins8090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arias A.S., Rucavado A., Gutiérrez J.M. Peptidomimetic hydroxamate metalloproteinase inhibitors abrogate local and systemic toxicity induced by Echis ocellatus (saw-scaled) snake venom. Toxicon. 2017;132:40–49. doi: 10.1016/j.toxicon.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 105.Ainsworth S., Slagboom J., Alomran N., Pla D., Alhamdi Y., King S.I., et al. The paraspecific neutralisation of snake venom induced coagulopathy by antivenoms. Commun. Biol. 2018;1:34. doi: 10.1038/s42003-018-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Escalante T., Ovadia M., Cury Y., Franceschi A., León G., Chaves F., et al. Inhibition of local hemorrhage and dermonecrosis induced by Bothrops asper snake venom: effectiveness of early in situ administration of the peptidomimetic metalloproteinase inhibitor batimastat and the chelating agent CaNa2EDTA. Am. J. Trop. Med. Hyg. 2017;63:313–319. [PubMed] [Google Scholar]
- 107.Knudsen C., Ledsgaard L., Dehli R.I., Ahmadi S., Sorensen C.V., Laustsen A.H. Engineering and design considerations for next-generation snakebite antivenoms. Toxicon. 2019;167:67–75. doi: 10.1016/j.toxicon.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 108.Castro J.M., Oliveira T.S., Silveira C.R., Caporrino M.C., Rodriguez D., Moura-da-Silva A.M., et al. A neutralizing recombinant single chain antibody, scFv, against BaP1, A P-I hemorrhagic metalloproteinase from Bothrops asper snake venom. Toxicon. 2014;87:81–91. doi: 10.1016/j.toxicon.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 109.Bailon C.H., Yaniro C.V.O., Caceres R.O.A., Colque A.E.G., Leiva D.W.J., Padilla R.C., et al. Development of nanobodies against hemorrhagic and myotoxic components of Bothrops atrox snake venom. Front. Immunol. 2020;11:655. doi: 10.3389/fimmu.2020.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pavlinkova G., Beresford G.W., Booth B.J., Batra S.K., Colcher D. Pharmacokinetics and biodistribution of engineered single-chain antibody constructs of MAb CC49 in colon carcinoma xenografts. J. Nucl. Med. 1999;40:1536–1546. [PubMed] [Google Scholar]
- 111.Wang R., Xiang S., Feng Y., Srinivas S., Zhang Y., Lin M., et al. Engineering production of functional scFv antibody in E. Coli by co-expressing the molecule chaperone Skp. Front. Cell. Infect. Microbiol. 2013;3:72. doi: 10.3389/fcimb.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wade J., Rimbault C., Ali H., Ledsgaard L., Rivera-de-Torre E., Abou Hachem M., et al. Generation of multivalent nanobody-based proteins with improved neutralization of long alpha-neurotoxins from elapid snakes. Bioconjugate Chem. 2022;33:1494–1504. doi: 10.1021/acs.bioconjchem.2c00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pucca M.B., Cerni F.A., Janke R., Bermudez-Mendez E., Ledsgaard L., Barbosa J.E., et al. History of envenoming therapy and current perspectives. Front. Immunol. 2019;10:1598. doi: 10.3389/fimmu.2019.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Antibody Society . 2022. Snakebite Antivenoms: Global Challenges and Progress toward Recombinant Antibody Therapeutics.https://www.antibodysociety.org/snakebite-antivenoms-global-challenges-and-progress-toward-recombinant-antibody-therapeutics/ [Google Scholar]
- 115.Hansel T.T., Kropshofer H., Singer T., Mitchell J.A., George A.J.T. The safety and side effects of monoclonal antibodies. Nat. Rev. Drug Discov. 2010;9:325–338. doi: 10.1038/nrd3003. [DOI] [PubMed] [Google Scholar]
- 116.Levi M., Levy J.H., Andersen H.F., Truloff D. Safety of recombinant activated factor VII in randomized clinical trials. N. Engl. J. Med. 2010;363:1791–1800. doi: 10.1056/NEJMoa1006221. [DOI] [PubMed] [Google Scholar]
- 117.Williams D.J., Habib A.G., Warrell D.A. Clinical studies of the effectiveness and safety of antivenoms. Toxicon. 2018;150:1–10. doi: 10.1016/j.toxicon.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 118.Almagro J.C., Pedraza-Escalona M., Arrieta H.I., Pérez-Tapia S.M. vol. 8. Antibodies; Basel, Switzerland: 2019. (Phage Display Libraries for Antibody Therapeutic Discovery and Development). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gomez-Betancur I., Gogineni V., Salazar-Ospina A., Leon F. vol. 24. Molecules; Basel, Switzerland: 2019. (Perspective on the Therapeutics of Anti-snake Venom). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Laustsen A.H., Dorrestijn N. Integrating engineering, manufacturing, and regulatory considerations in the development of novel antivenoms. Toxins. 2018;10 doi: 10.3390/toxins10080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ascoet S., De Waard M. Diagnostic and therapeutic value of aptamers in envenomation cases. Int. J. Mol. Sci. 2020:21. doi: 10.3390/ijms21103565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patikorn C., Ismail A.K., Zainal Abidin S.A., Othman I., Chaiyakunapruk N., Taychakhoonavudh S. Potential economic and clinical implications of improving access to snake antivenom in five ASEAN countries: a cost-effectiveness analysis. PLoS Neglected Trop. Dis. 2022;16 doi: 10.1371/journal.pntd.0010915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anderson V.E., Gerardo C.J., Rapp-Olsson M., Bush S.P., Mullins M.E., Greene S., et al. Early administration of Fab antivenom resulted in faster limb recovery in copperhead snake envenomation patients. Clin. Toxicol. 2019;57:25–30. doi: 10.1080/15563650.2018.1491982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is available upon request from the corresponding author.