Abstract
Common fragile sites (CFSs) are regions prone to chromosomal rearrangements, thereby contributing to tumorigenesis. Under replication stress (RS), CFSs often harbor under-replicated DNA regions at the onset of mitosis, triggering homology-directed repair known as mitotic DNA synthesis (MiDAS) to complete DNA replication. In this study, we identified an important role of DNA mismatch repair protein MutSβ (MSH2/MSH3) in facilitating MiDAS and maintaining CFS stability. Specifically, we demonstrated that MutSβ is required for the increased mitotic recombination induced by RS or FANCM loss at CFS-derived AT-rich and structure-prone sequences (CFS-ATs). We also found that MSH3 exhibits synthetic lethality with FANCM. Mechanistically, MutSβ is required for homologous recombination (HR) especially when DNA double-strand break (DSB) ends contain secondary structures. We also showed that upon RS, MutSβ is recruited to Flex1, a specific CFS-AT, in a PCNA-dependent but MUS81-independent manner. Furthermore, MutSβ interacts with RAD52 and promotes RAD52 recruitment to Flex1 following MUS81-dependent fork cleavage. RAD52, in turn, recruits XPF/ERCC1 to remove DNA secondary structures at DSB ends, enabling HR/break-induced replication (BIR) at CFS-ATs. We propose that the specific requirement of MutSβ in processing DNA secondary structures at CFS-ATs underlies its crucial role in promoting MiDAS and maintaining CFS integrity.
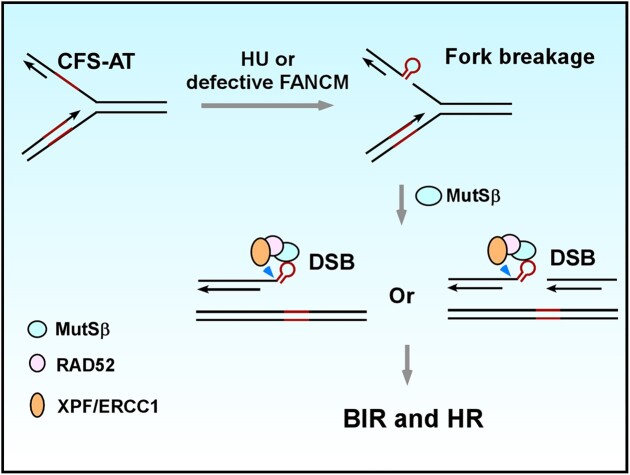
Graphical Abstract
Graphical Abstract.
Introduction
Common fragile sites (CFSs) are specific genomic loci that exhibit gaps or breaks on metaphase chromosomes when replication is partially impaired (1). These sites are known hot spots for chromosomal rearrangements such as deletions and translocations, particularly in cancer (2–4). CFSs exhibit a high susceptibility to DNA replication stress (RS) (5), which is often induced by the aberrant expression of oncogenes (6). As a result, CFS instability is believed to be a significant contributor to tumorigenesis (7–9). CFSs are ‘difficult-to-replicate’, which can be attributed to several intrinsic features. For instance, CFSs often contain clusters of AT-rich sequences that tend to form stable secondary structures (10–12). Additionally, they frequently harbor large genes that can lead to R-loop formation and collisions between replication and transcription (13,14). Furthermore, CFSs are typically late-replicating and have a shortage of replication origins (15).
Upon encountering replication disturbances, CFSs frequently fail to complete DNA replication and proceed into M-phase with under-replicated DNA (16–18). At the onset of mitosis, the structure-specific endonuclease (SSE) MUS81/EME1 cleaves under-replicated DNA regions, followed by induction of mitotic DNA synthesis (MiDAS) in the prophase of mitosis to complete DNA replication at CFSs (17,19,20). MiDAS exhibits a conservative DNA replication pattern and relies on the involvement of POLD3 and PIF1 (20–22), sharing characteristics with break-induced replication (BIR), a subtype of homologous recombination (HR) (23–25). Notably, MiDAS is independent of BRCA1 and RAD51 but requires RAD52 (21). However, the precise mechanism underlying MiDAS is still not fully understood.
Disturbed replication at CFSs leads to the formation of DNA secondary structures at AT-rich sequences on replication forks, causing replication stalling and double-strand break (DSB) formation, and these DSBs contribute to the fragility of CFSs (10,11,15,26,27). FANCM is important for dismantling these DNA secondary structures, thereby preventing DSB formation at CFS-ATs (11). HR, in the forms of both short tract gene conversion [STGC (28), often refer to general HR] and BIR, is employed to repair DSBs at CFS-ATs (22). HR is initiated by end resection, followed by invasion of the 3′ single-strand DNA (ssDNA) of one DSB end into its homologous template (29,30). When the second end homology is in close proximity, short DNA synthesis and strand displacement are followed by the annealing of the newly synthesized DNA strand to the second end homology, completing STGC [or general HR (28,29)]. However, if DSBs are single-ended or the second end is far away, BIR is activated, and BIR DNA synthesis requires POLD3 and PIF1, which are dispensable for STGC (25). In our previous study, we showed that after end resection at DSB ends, CFS-ATs form DNA secondary structures on the 3′ ssDNA overhangs (12), which require XPF/ERCC1 for removal before new DNA synthesis can initiate (31). Additionally, we demonstrated that RAD52 is specifically necessary for repairing DSBs with DNA secondary structures at the ends but is dispensable for HR at ‘clean ends’ (11,32).
DNA mismatch repair (MMR) plays a vital role in correcting DNA mismatches arising during DNA replication (33,34). The MutSα (MSH2/MSH6) and MutSβ (MSH2/MSH3) heterodimeric complexes, which are highly conserved, are involved in identifying mismatched nucleotides with distinct substrate specificities. MutSα binds to single-nucleotide mismatches and 1–2 nucleotide insertions, whereas MutSβ recognizes small DNA loops (35–39). Mismatch recognition triggers ATP-dependent activation of the MutLα endonuclease, generating a nick near the mismatched DNA to initiate MMR (38–40). Unlike MutSα, MutSβ can bind to various branched DNA structures (40). Previous studies have shown that MutSβ binds to hairpin structures formed in trinucleotide repeats (TNRs) and promotes pathogenic TNR expansion (41–44). MutSβ also interacts with hairpin loops in RPA-ssDNA and facilitates ATR activation (45). MMR proteins have also been implicated in HR. In yeast, Msh2 and Msh3 are important for gene conversion when DSB ends contain nonhomologous tails and are also required for single-strand DNA annealing (SSA) (46,47). Yeast MutSβ stimulates endonuclease activity of MutLα to cleave recombination intermediates in meiosis (48). In mammalian cells, MutSβ has also been shown to be involved in checkpoint activation and HR (45,49–51).
In this study, we demonstrated that MutSβ exhibits a synthetical lethal interaction with FANCM, an ATP-dependent DNA-remodeling translocase (52) that is required for preventing DSB formation at CFS-ATs (11). We showed that MutSβ plays a critical role in both HR and BIR of DSBs that contain structure-forming CFS-ATs (such as Flex1, Figure 7B). The MutSβ function in BIR is important for MiDAS at CFSs and the maintenance of CFS integrity. Mechanistically, MutSβ binds to DNA secondary structures formed at CFS-ATs and interacts with RAD52, a process that is enhanced by RS. RAD52 in turn recruits XPF/ERCC1 to CFS-ATs, enabling removal of DNA secondary structures at DSB ends and facilitating BIR at CFSs. These findings provide valuable insights into the intricate molecular interactions and repair mechanisms that underlie the maintenance of CFS integrity and highlight the role of MutSβ in promoting MiDAS to preserve genomic stability at CFSs.
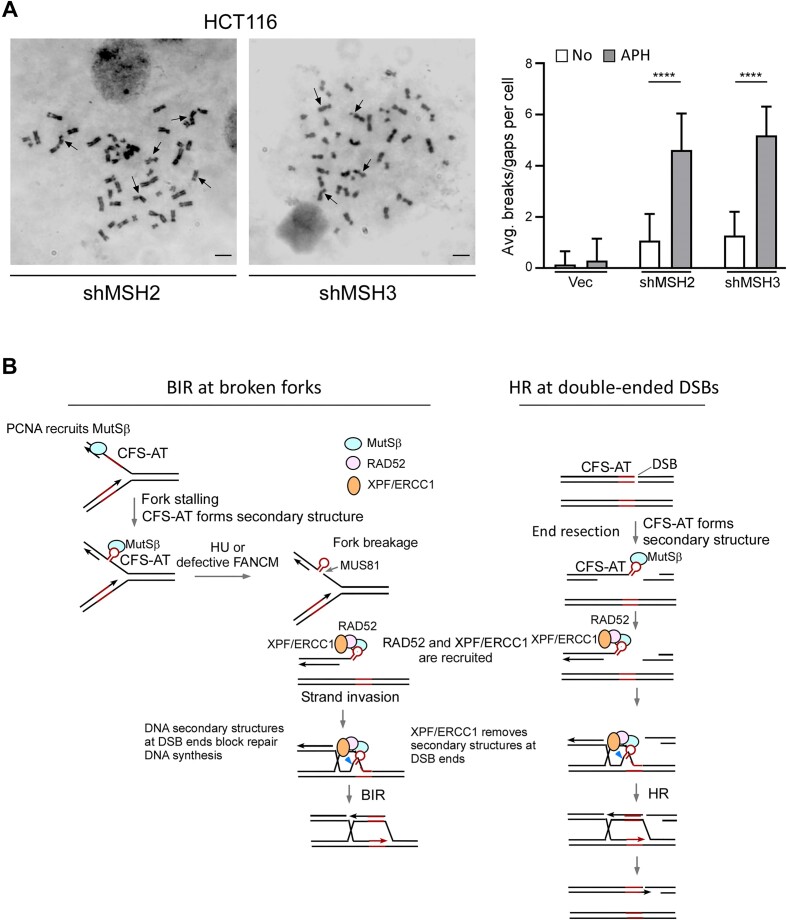
Figure 7.
MutSβ prevents CFS instability. (A) Metaphase spreads of HCT116 cells expressing shRNAs targeting MSH2 or MSH3, as well as the shRNA vector, were conducted before and after APH treatment (0.4 μM, 24 h). Representative images of metaphase spread of HCT116 cells expressing MSH2 and MSH3 shRNAs are shown and breaks and gaps on metaphase chromosomes are indicated by arrows (left). ∼100 metaphase spreads were analyzed in each sample. The overall chromosome gaps and breaks per cell were quantified (right). Scale bars, 6μm. (B) A working model to depict the involvement of MutSβ in facilitating BIR at stalled forks upon RS (left), and in promoting HR at double-ended DSBs (right) when DSB ends contain secondary structures such as CFS-ATs. See details in the main context.
Materials and methods
Cell culture and generation of MSH3-KO cell line by CRISPR
Human U2OS, HCT116, and 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum at 37°C and 5% CO2. The FANCM-knockout HCT116 cells used in this study have been previously described (53).
MSH3-knockout (KO) in EGFP-HR-Flex reporter (U2OS) cells was generated using CRISPR–Cas9 technology. A gRNA sequence (TGAACAAACAGTCTGTGAGT) targeting the fifth exon of human MSH3 was sub-cloned into PX459 (Addgene#62988) for making MSH3-KO. Transfected cells with this plasmid were subjected to puromycin selection for 48 hours, followed by isolation of single clones. The MSH3-KO clones were confirmed by Western blot analysis.
Plasmid construction and shRNAs
To construct expression vectors, MSH2, MSH3, MSH6, and XPF complementary DNAs (cDNAs) were sub-cloned into the pCDNA3.0-SFB vector containing the SFB (S protein-2xFlag-Streptavidin-binding peptide) tag (54). The MSH3 N-terminal deletion mutant (MSH3-ND: Δ1–200aa, defective in PCNA binding) was generated by PCR amplification with its nuclear localization signal added back to the construct (55). RAD52 cDNA was subcloned in the pCDNA3.0-HA vector for co-immunoprecipitation (co-IP) assay and in the NBLV0051-EGFP (Novo Bio) vector for monitoring repair foci.
Short hairpin RNAs (shRNAs) for FANCM and RAD52 were previously described (11). Other shRNA target sequences were obtained from Sigma-Aldrich and are listed below:
MSH2 shRNA #1: TCTGCAGATGTGCTTAG
MSH2 shRNA #2: CCAGTAATGGAATGAAGGTAA
MSH3 shRNA #1: GCCATTTAGATCACAACTTTA
MSH3 shRNA #2: GCACAGAAGGAATAAGGTCAT
MSH6 shRNA #1: GCCAGAAGAATACGAGTTGAA
MSH6 shRNA #2: TTCTGACAAAGGTGGTAAATT
MUS81 shRNA: CACGCGCTTCGTATTTCAGAA
XPF shRNA: GUAGGAUACUUGUGGUUGA
PCNA shRNA: GATGCTGTTGTAATTTCCTGT
Immunoblotting and RT-qPCR
Western blot analysis was performed using a standard protocol described previously (12). Briefly, cell lysates were boiled in 6X SDS loading buffer and separated using SDS-PAGE. Antibodies against FANCM were kindly provided by Dr. Weidong Wang (53). Commercially available antibodies used in this study included: MSH2 (HUABIO, EM1801, 1:2000), MSH3 (BD Biosciences, 611390, 1:2000), MSH6 (HUABIO, ET1604, 1:2000), β-Actin (Abmart, P30002, 1:5000), γH2AX (Abclonal, AP0099, 1:2000), H2AX (Cell Signaling #2595), MUS81 (Abclonal, A6818, 1:2000), XPF (Abmart, PA1397, 1:5000), PCNA (Millipore, 3428716, 1:2000), RAD52 (Abclonal, A5186, 1:2000), HA (Abmart, M20003, 1:5000) and Flag (Sigma, F1804, 1:2000).
Total RNA was extracted from cell lines using the RNeasyMini Kit (Qiagen). cDNA was synthesized through reverse transcription using the iScript cDNA synthesis kit (Bio-Rad). The iTaq Universal SYBR Green Supermix (Bio-Rad) was used for qPCR on C1000 Thermal Cycler (Bio-Rad). The primer sequences are listed below:
FANCM-F: 5′GCTTATTGTTCCGCTTGGTG
FANCM-R: 5′TCAAAGAACGAGCAAATGATTCC
MSH2-F: 5′ AGGCATCCAAGGAGAATGATTG
MSH2-R: 5′GGAATCCACATACCCAACTCCAA
MSH3-F: 5′CTACCAGCTATCTTCTGTGCATC
MSH3-R: 5′ CCTCTGTTTGCTCGGACAAG
MSH6-F: 5′GCAATGCAACGTGCAGATGAA
MSH6-R: 5′ACTTCGCCTAGATCCTTGTGT
Cell cycle analysis
For cell cycle analysis, the lentivirus-infected cell cells were collected by centrifugation at 4°C, and washed with cold PBS. Cells were resuspended in an appropriate amount of cold PBS, followed with addition of three volumes of cold anhydrous ethanol to achieve a final concentration of 75%. Cells were gently inverted, mixed well and incubated at 4°C for at least 18 hours. Subsequently, cells after fixing were centrifuged at 4°C and the supernatant was removed. After several rounds of PBS wash to remove ethanol, cells were treated with propidium iodide (PI, 50 μg/ml), along with RNaseA (50 μg/ml) at 37°C for 30 min. Cell cycle profiles were analyzed by FACS analysis (Attune Flow Cytometer, Thermo Fisher) and the proportion of cells in each phase was determined by FlowJo_v10.8.1.
Co-Immunoprecipitation (Co-IP)
293T cells were co-transfected with pCDNA3.0-SFB-MSH3 or SFB-MSH6 and pCDNA3.0-HA-RAD52 by linear polyethylenimine (PEI; Polysciences, 24314-2), and 8 hours later, medium was changed followed by additional 48-hour culturing. Co-IP was performed with or without HU treatment (2 mM HU, 2 h).
Whole cell lysis and co-IP were performed as previously described (56). Briefly, cells were lysed in NETN buffer [150 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl (pH 8.0), 0.5% NP-40] supplemented with protease inhibitors. IP was performed using anti-streptavidin magnetic beads (Abmart, A10005), followed by five washes with NETN buffer.
Chromatin immunoprecipitation (ChIP)
Streptavidin-ChIP was performed as previously described (57,58). Following the indicated treatment, the medium was removed, and the cells were washed with PBS. A cross-linking solution (1% formaldehyde in PBS) was added to the cells for 10 minutes at room temperature, then glycine was added to a final concentration of 125 mM to terminate cross-linking. After three PBS washes, the cells were resuspended in ultrasonic buffer [1% SDS, 50 mM Tris–HCl (pH 8.0), 10 mM EDTA] supplemented with protease inhibitors. Chromatin was fragmented by ultrasound until the DNA fragments were between 0.2 and 1.0 kb in length. The sonicated samples were diluted 10 times with IP buffer [0.01% SDS, 1.1% Trition X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl (pH 8.0), 167 mM NaCl] supplemented with protease inhibitors. From the diluted sample, 10% was retained as a total sample, while the remaining 90% was pre-cleared with Protein A Sepharose beads (GE HealthCare). The IP was performed using H2AX-S139p (Cell Signaling #2577) or anti-streptavidin magnetic beads. The IP samples were washed successively with 1 ml each of LS buffer (0.1% SDS, 20 mM Tris–HCl (pH 8.0), 1% Triton X-100, 2 mM EDTA, 150 mM NaCl), HS buffer (0.1% SDS, 20 mM Tris–HCl (pH 8.0), 1% Triton X-100, 2 mM EDTA, 500 mM NaCl), LiCl buffer (0.25 M LiCl, 10 mM Tris–HCl (pH 8.0), 1% NP-40, 1% deoxycholate, 1 mM EDTA), and TE buffer, all supplemented with protease inhibitors. The DNA-protein complexes were eluted with EL buffer (1% SDS, 0.1 M NaHCO3), and cross-linking was reversed by adding NaCl to a final concentration of 200 mM, followed by overnight incubation at 65°C. Proteinase K (10 mM EDTA, 20 mM Tris–HCl (pH 6.8)) digestion was performed for 2 h at 42°C, then the DNA was purified using the TIANquick Mini Purification Kit (TIANGEN) according to the manufacturer's instructions. For ChIP analysis, the recovered DNA was analyzed by qPCR and ΔΔCt was calculated using the GAPDH locus as an internal control. The primer sequences used have been previously published (11) and are listed below:
Flex-F: 5′CTCCAATTCGCCCTATAGTGAGTCGTATTA
Flex-R: 5′TTACTTGTACAGCTCGTCCATGC
FRA3B-F: 5′TTAGCCTACTTCAGGGTTTCT
FRA3B-R: 5′TGGAGAGGTTACTACTGGCA
GAPDH-F: 5′CCCTCTGGTGGTGGCCCCTT
GAPDH-R: 5′GGCGCCCAGACACCCAATCC
Cell proliferation and cell viability assay
Cell proliferation was determined by growth curves, as previously described (31). Briefly, HCT116 and its derived cell lines were seeded at a density of 1 × 105 cells in 6 cm cell culture dishes. Cell proliferation was assessed by counting trypsinized cells using a hemocytometer every 24 h. The cell number was normalized to day 1.
For the cell viability assay, cells were seeded in 96-well plates at a density of 2000 cells per well and treated with different concentrations of HU and aphidicolin (APH; Sigma, A4487) for 72 h. MTS reagent (Promega) was mixed with the medium in equal volumes and incubated at 37°C for at least 2 h. Cell viability was determined by measuring the emission at 490 nm using a Spectramax M5 reader (Molecular Devices).
Mitotic recombination assay and I-SceI-induced HR assay
For the HU-induced HR assay, reporter cells underwent initial double thymidine block (2 mM, two cycles of 16-hour in drug with a 12-hour interval of drug-free medium in between) to enrich the population in S-phase, followed by treatment with 2 mM HU for 24 h. For the I-SceI-induced HR assay, reporter cell lines were infected with lentiviruses encoding HA-I-SceI. Four days after HU or I-SceI expression, EGFP-positive events were quantified by FACS analysis using a BD Accuri C6 flow cytometer and accompanying data analysis software (CFlow, Becton-Dickinson).
Analysis of HR used in the HR-Flex/D-Flex reporter and HR-Luc/D-Luc reporter was performed as previously described (11,31). Briefly, genomic DNA was extracted four days after I-SceI viral infection, followed by PCR amplification. The PCR products were digested with or without BamHI and EcoRI and resolved by 1.5% agarose gel electrophoresis. The percentage of restriction enzyme digestible PCR products relative to the total DNA provided an estimation of HR efficiency, determined by quantifying the DNA bands using ImageJ.
Metaphase chromosome analysis
Metaphase chromosome analysis was performed according to standard protocols (12,59). HCT116 cells were treated with APH (0.4 μM) for 24 h, then nocodazole was added to a final concentration of 330 nM. The cells were then incubated for an additional 4 h to synchronize the population to the metaphase stage. Subsequently, the cells were collected and resuspended in a hypotonic solution (75 mM KCl) for 30 minutes at 37°C, then fixed in fixative solution (3:1 ratio of methanol and acetic acid) at room temperature for 30 min, with multiple changes of fixative solution. Fixed cells were dropped onto pre-cooled slides from a height and aged overnight at room temperature. Metaphase chromosome staining was performed using Giemsa. Breaks and gaps were detected and counted on Giemsa-stained metaphases.
Mitotic DNA synthesis (MiDAS)
MiDAS was performed as previously described (60,61). U2OS cells were treated with APH (0.4 μM) for 8 hours, then RO-3306 was added to a final concentration of 7 μM. The cells were then cultured for an additional 8 h to synchronize the population to the late G2 phase. The medium was removed, and the cells were washed three times with cold PBS, followed by releasing into fresh DMEM medium at 37°C and within 5 min, replacing with the medium containing 20 μM EdU and 0.1 μg/ml Colcemid for incubation at 37°C for 60 min.
Cells were shaken-off, then washed with cold PBS on ice. The cells were resuspended in 75 mM KCl for 20 min at 37°C. Swollen mitotic cells were collected and fixed in a fixative solution (3:1 ratio of methanol and acetic acid) at room temperature for at least 30 minutes, then dropped onto pre-cooled slides from a height. Fixed cells were aged overnight at room temperature.
Slides were blocked with blocking buffer (3% BSA in PBS) for 30 minutes then permeabilized with 0.5% Triton X-100 for 20 min. After the permeabilization buffer was removed, the cells were washed with blocking buffer three times. EdU incorporation was detected using the Click-IT EdU Alexa Fluor 488 Imaging Kit (Invitrogen). The Click-iT reaction was terminated with blocking buffer, and chromosome staining was performed with DAPI (0.25 μg/ml) for 3 mi. Images were analyzed using an Olympus IX81 FL microscope.
Immunofluorescence (IF) and in situ proximity ligation assay (PLA) on metaphase chromosomes
Cells were synchronized to mitosis following the MiDAS protocol, with the exception of being released into the medium containing only 0.1 μg/ml Colcemid for incubation at 37°C for 60 min. Mitotic cells were fixed and dropped onto slides, after which the slides were subjected to a 30-min blocking step (using 3% BSA in PBS) and subsequent permeabilization with 0.5% Triton X-100 for 20 min. This was followed by an overnight incubation at 4°C with the relevant primary antibodies. For IF, as previously described (62), following washing three times with blocking buffer, cells were incubated with appropriate secondary antibodies for 1 h at 37°C in the dark. PLA was performed using Duolink PLA technology (Sigma-Aldrich) according to the manufacturer's instructions. Anti-mouse PLUS and anti-rabbit MINUS PLA probes were coupled to the primary antibodies. After washing in buffer-A (0.01M Tris, 0.15M NaCl and 0.05% Tween-20), PLA probes were ligated for 45 min at 37°C then washed. Coverslips were washed in buffer-B (0.2M Tris and 0.1M NaCl) following amplification.
Finally, chromosome staining was performed with DAPI (0.25 μg/ml) for 3 min. Images were analyzed using an Olympus IX81 FL microscope.
Statistical analysis
Statistical analysis was performed using GraphPad Prism9 and Microsoft Excel. For sample sizes ≤30, significant differences were determined using an unpaired Student's t-test. For sample sizes greater than 30, a Mann–Whitney U test was used. The following notation was used to denote statistical significance: n.s. (not significant) (P> 0.05); *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
Results
MSH2 and MSH3 are synthetic lethal with FANCM
DNA secondary structures often arise upon RS (63–65). FANCM plays an important role in resolving DNA secondary structures and prevents RS-induced DSB formation at CFS-ATs (11,22). Interestingly, we found that depletion of MSH2 or MSH3 by shRNAs drastically induces cell death in FANCM-knockout (KO) but not in wildtype (WT) HCT116 cells (Figure 1A and Supplementary Figure S1A). Depleting MSH6 in FANCM-KO cells, however, does not result in significant cell death of FANCM-KO cells (Supplementary Figure S1B). These data suggest that MSH2/MSH3 (MutSβ) but not MSH2/MSH6 (MutSα) is synthetic lethal with FANCM.
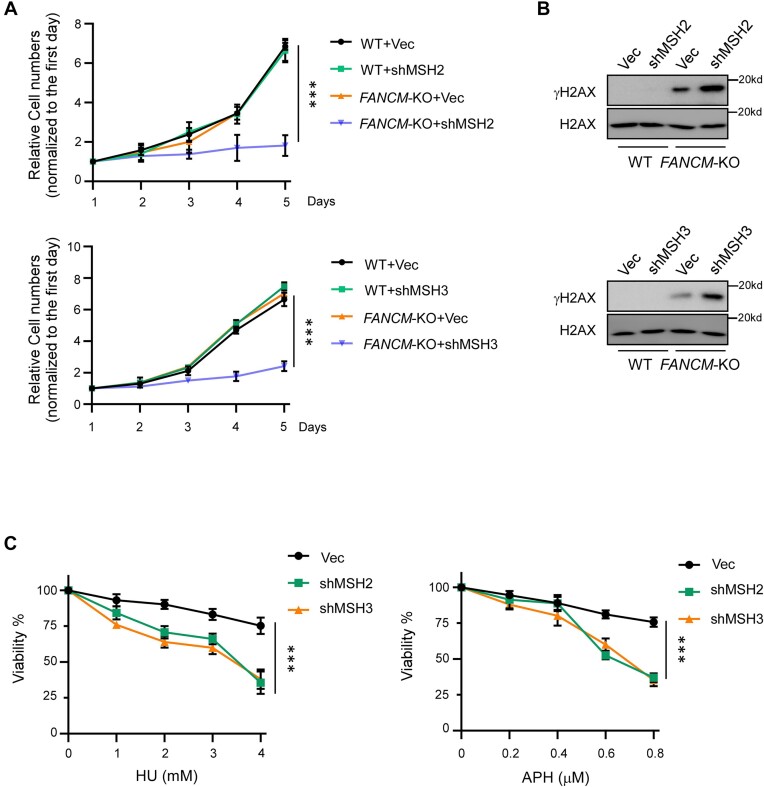
Figure 1.
MSH2 and MSH3 are synthetically lethal with FANCM. (A) The growth curves of HCT116 WT and FANCM-KO cells were plotted after infection with lentiviruses expressing shRNAs targeting MSH2 (top) or MSH3 (bottom), along with a vector control (Vec). (B) γ-H2AX Western blot analysis was conducted on HCT116 WT and FANCM-KO cells, which were infected with lentiviruses encoding shRNAs targeting MSH2 (top) or MSH3 (bottom), along with a vector control (Vec), after 4 days of infection. H2AX serves as the loading controls. (C) Cell viability was determined in HCT116 cells expressing shRNAs for MSH2, MSH3 or a vector (Vec) after treatment with the indicated concentrations of HU (left) and APH (right) for 48 hours.
When FANCM is deficient, DSB formation is increased at CFS-ATs (11). One possible mechanism could involve MutSβ’s role in repairing DSBs that accumulate in FANCM-deficient cells, thus contributing to the prevention of cell death. Indeed, we observed a significant increase of γH2AX signals in Western blot analysis when MSH2 or MSH3 is depleted in FANCM-KO cells, in comparison to MSH2 or MSH3 depletion in WT cells (Figure 1B), suggesting that MutSβ is actively involved in preventing and/or repairing DSBs accumulated due to FANCM deficiency. In addition, depletion of MSH2 or MSH3 by shRNAs leads to sensitivity of HCT116 cells to hydroxyurea (HU) and aphidicolin (APH) (Figure 1C and Supplementary Figure S1C). This suggests that MutSβ is important for cells to cope with RS, likely due to its role in minimizing DSB accumulation at structure prone DNA sequences on stalled replication forks.
MutSβ is required for mitotic recombination at Flex1 induced by FANCM deficiency and replication stress
We showed that the CFS-AT sequence, Flex1 derived from FRA16D induces spontaneous mitotic recombination (HR), and this effect is further elevated in FANCM-deficient cells or after HU treatment (11,12). As assayed by our EGFP-HR-Flex reporter (11,12) (Figure 2A), we found that FANCM depletion-induced mitotic recombination is suppressed by further depleting MSH3 (Figure 2B and Supplementary Figure S2A). In addition, HU-induced mitotic recombination at Flex1 is markedly reduced in MSH3-KO U2OS cells compared to WT cells (Figure 2C and Supplementary Figure S2B). Depletion of MSH2 or MSH3 but not MSH6 by shRNAs also inhibits HU-induced mitotic recombination at Flex1, without affecting cell cycle profiles (Figure 2D and Supplementary Figure S2C, S2D). ChIP analysis revealed that γH2AX is increased at Flex1 after APH treatment or after FANCM depletion, which is further elevated in MSH3-KO cells compared to WT cells (Figure 2E and Supplementary Figure S2E). We also performed ChIP at endogenous FRA3B locus in HCT116 cells and showed that γH2AX is accumulated at the vicinity of AT-rich sequences at FRA3B after APH treatment (Supplementary Figure S2F). These data suggest that MutSβ is required for promoting mitotic recombination to repair DSBs at Flex1 and other CFS-AT sequences when FANCM is deficient or upon RS, thereby preventing DSB accumulation at Flex1.
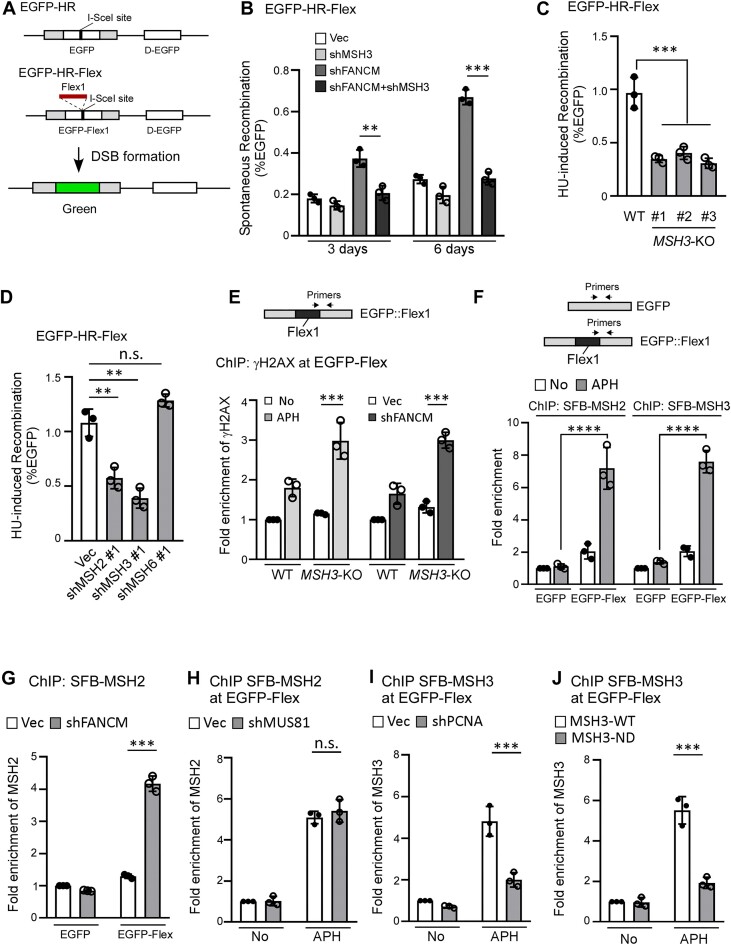
Figure 2.
MutSβ is required for promoting mitotic recombination at Flex1. (A) Schematic drawing of the EGFP-HR and EGFP-HR-Flex reporters described previously (11,89), and the HR-mediated repair product. D-EGFP: donor EGFP. (B) Mitotic recombination frequency was determined in U2OS (EGFP-HR-Flex) cells depleted for MSH3 by shRNAs with the vector (Vec) as a control, followed by depleting FANCM with shRNAs or expressing shRNA vector (Vec). FACS analysis was performed 3 and 6 days after MSH3 shRNA lentivirus infection. (C) U2OS (EGFP-HR-Flex) WT or MSH3-KO cells were synchronized to S-phase by double thymidine block, and subsequently were treated with HU (2 mM, 24 h). Mitotic recombination (HR) was assessed by FACS analysis 4 days after HU treatment. (D) U2OS (EGFP-HR-Flex) cells expressing shRNA#1 targeting MSH2, MSH3, MSH6, or a control vector (Vec) were synchronized to S-phase using double thymidine block, and subsequently treated with HU (2 mM, 24 h). HU-induced recombination was assessed by FACS analysis 4 days after HU treatment. (E) Anti-γH2AX ChIP analysis was performed in U2OS (EGFP-HR-Flex) WT and MSH3-KO cells at Flex1, using indicated primer sets before and after APH treatment (0.8 μM, 16 h, left) or after expressing FANCM shRNA and vector (Vec, right). The ChIP value in No treatment (left) and in Vec control (right) samples were set as 1 for normalization. (F) Anti-streptavidin ChIP analysis at Flex1 was performed in U2OS (EGFP-HR) and U2OS (EGFP-HR-Flex) cells expressing SFB-MSH2 (left) and SFB-MSH3 (right) before and after APH (0.8 μM, 16 h) treatment. Streptavidin beads recognize the SBP tag in SFB [S protein-2xFlag-Streptavidin-binding peptide (SBP)] (54). The ChIP value at EGFP in the EGFP-HR reporter with No treatment was set as 1 for normalization. (G) Anti-streptavidin ChIP analysis at Flex1 was performed in U2OS (EGFP-HR) and U2OS (EGFP-HR-Flex) cells expressing SFB-MSH2 after depleting FANCM with shRNAs or expressing shRNA vector (Vec). The ChIP value at EGFP in the EGFP-HR reporter with Vec control was set as 1 for normalization. (H and I). Anti-streptavidin ChIP analysis at Flex1 was performed in U2OS (EGFP-HR-Flex) cells expressing SFB-MSH2 with MUS81 silenced by shRNAs (H) or expressing SFB-MSH3 with PCNA silenced by shRNAs (I) using shRNA vector (Vec) as a control with or without APH treatment (0.8 μM, 16 h). The ChIP value in the sample of No treatment with Vec control was set as 1 for normalization. (J) Anti-streptavidin ChIP analysis at Flex1 was performed in U2OS (EGFP-HR-Flex) cells expressing SFB-MSH3-WT or SFB-MSH3-ND (Δ1–200aa, impaired in PCNA binding) before and after APH treatment (0.8 μM, 16 h). The ChIP value in the sample of No treatment with SBP-MSH3-WT expression was set as 1 for normalization.
We performed ChIP analysis of MSH2 and MSH3 at Flex1 in the EGFP-HR-Flex reporter using the EGFP-HR reporter, which does not contain Flex1, as a control. We found that the recruitment of MSH2 and MSH3 to Flex1 is significantly increased after APH treatment (Figure 2F). Recruitment of MSH2 to Flex1 is also increased after FANCM depletion (Figure 2G). We also showed that MSH3 is enriched at FRA3B close to AT-rich sequences in HCT116 cells after APH treatment (Supplementary Figure S2G). To test whether MutSβ is recruited before or after DSB formation at Flex1, we depleted MUS81, which cleaves stalled replication forks upon RS (66). MSH2 recruitment to Flex1 after APH is not altered when MUS81 is depleted, suggesting that MutSβ binds to Flex1 probably prior to fork breakage (Figure 2H and Supplementary Figure S2H). However, we found that APH-induced MSH3 recruitment is dependent on PCNA (Figure 2I and Supplementary Figure S2I). It has been shown that MSH3 directly binds to PCNA through a highly conserved PCNA-binding motif at the N-terminus of MSH3 and deleting the N-terminal 200 amino acids abolished the MSH3 and PCNA interaction (55). We found that the MSH3 N-terminus deletion mutant [MSH3-ND: Δ1–200 aa with insertion of its nuclear localization signal (NLS)] is strongly defective in recruitment to Flex1 after APH (Figure 2J and Supplementary Figure S2J). We propose that MutSβ is loaded by PCNA onto forks, enabling it to bind to DNA secondary structures (such as Flex1 and other CFS-ATs) which are formed on stalled replication forks when ssDNA is accumulated upon RS prior to DSB formation (Figure 7B, left).
MutSβ interacts with RAD52 and recruits RAD52 to Flex1 in a manner dependent on MUS81
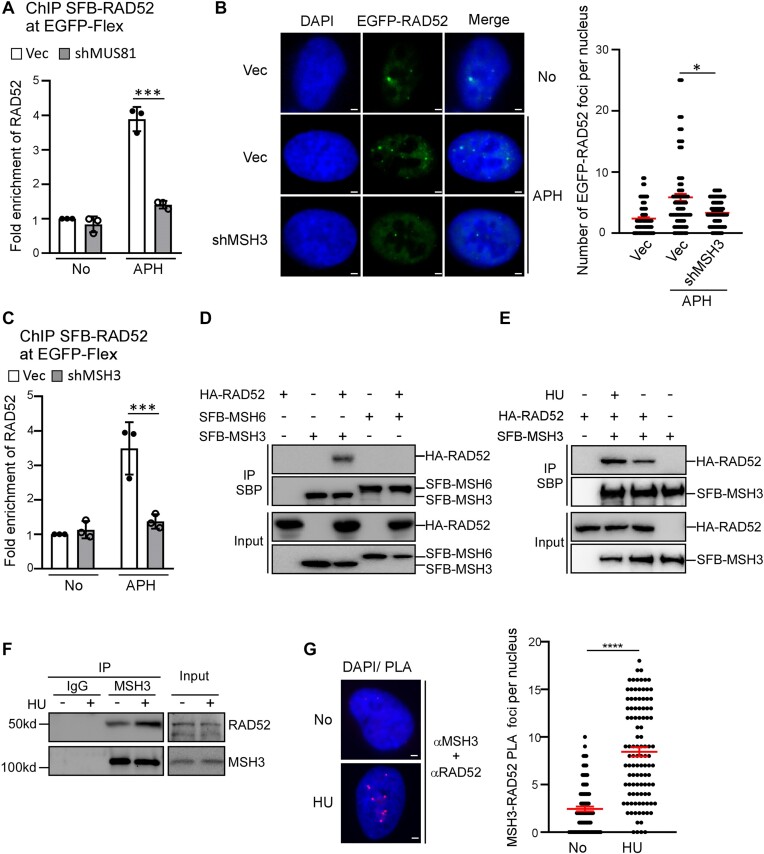
We showed previously that elevated mitotic recombination at Flex1 in FANCM deficient cells is dependent on RAD52 (11). We performed ChIP analysis and further showed that RAD52 is recruited to Flex1 after APH treatment in a manner dependent on MUS81 (Figure 3A and Supplementary Figure S3A), suggesting that RAD52 recruitment to Flex1 is likely after DSB formation. Since MutSβ recruitment to Flex1 is independent of MUS81 and probably prior to DSB formation, we asked whether MutSβ is required for RAD52 binding to DNA damage sites upon RS. We showed that APH-induced RAD52 foci are significantly reduced when MSH3 is depleted by shRNA (Figure 3B and Supplementary Figure S3B). ChIP analysis also revealed that APH-induced RAD52 binding to Flex1 is significantly reduced when MSH3 is deficient (Figure 3C and Supplementary Figure S3C). In addition, we found that RAD52 interacts with MSH3 but not MSH6 as revealed by co-immunoprecipitation (co-IP) of tagged RAD52 and MSH3 or MSH6 (Figure 3D) and the interaction of RAD52 and MSH3 is further enhanced after HU treatment (Figure 3E). Using co-IP and PLA, we verified that endogenous RAD52 and MSH3 interact with each other, and their interactions are enhanced following HU treatment (Figure 3F and 3G). These results suggest that RAD52 is recruited to DSBs at Flex1 or other structure-prone DNA sequences in a manner dependent on MutSβ upon RS, and RAD52 recruitment to damage sites is possibly enhanced by its physical interaction with MutSβ.
Figure 3.
MutSβ recruits RAD52 to Flex1 after DSB formation. (A) Anti-streptavidin ChIP analysis at Flex1 was performed in U2OS (EGFP-HR-Flex) cells expressing SFB-RAD52 with MUS81 silenced by shRNA or expressing shRNA vector (Vec) with or without APH treatment (0.8 μM, 16 h). The ChIP value in the sample of No treatment with Vec control was set as 1 for normalization. (B) EGFP-RAD52 foci were monitored in U2OS cells expressing MSH3 shRNAs or shRNA vector (Vec), and representative EGFP-RAD52 foci are displayed along with DAPI staining before and after APH treatment (0.4 μM, 16 h, left). The quantification of EGFP-RAD52 foci per nucleus is presented (right). ∼100 nuclei were analyzed in each sample. Scale bars, 2 μm. (C) Anti-streptavidin ChIP analysis at Flex1 was performed in U2OS (EGFP-HR-Flex) cells expressing SFB-RAD52 with MSH3 silenced by shRNA using the shRNA vector (Vec) as a control. The ChIP value in the sample of No treatment with Vec control was set as 1 for normalization. (D) HA-RAD52 and SFB-MSH3 or SFB-MSH6 were expressed alone or together in 293T cells and co-IP was performed using anti-streptavidin magnetic beads, recognizing the SBP tag in SFB [S protein-2xFlag-Streptavidin-binding peptide (SBP)] (54). Anti-HA (for HA-RAD52) and anti-Flag (for SFB-MSH3 and SFB-MSH6) Western blot analysis was performed. (E). HA-RAD52 and SFB-MSH3 were expressed alone or together in 293T cells and co-IP using anti-streptavidin magnetic beads, recognizing the SBP (SBP: streptavidin-binding peptide) tag in SFB-MSH3, was performed in the presence or absence of HU (2 mM, 2 h). Anti-HA (for HA-RAD52) and anti-Flag (for SFB-MSH3) western blot analysis was performed. (F) Endogenous interaction of MSH3 and RAD52 was performed by IP of MSH3 in U2OS cells with or without HU treatment (2 mM, 2 h), followed by Western blot analysis of RAD52 and MSH3. (G) PLA analysis of RAD52 with MSH3 was performed in U2OS cells, treated with or without HU (2 mM, 24 h). A representative image of RAD52 and MSH3 PLA is shown (left) and the PLA foci of RAD52 with MSH3 were quantified (right). ∼100 nuclei were analyzed in each sample. Scale bars, 2 μm.
MutSβ is important for removing Flex1 at DSB ends to complete HR
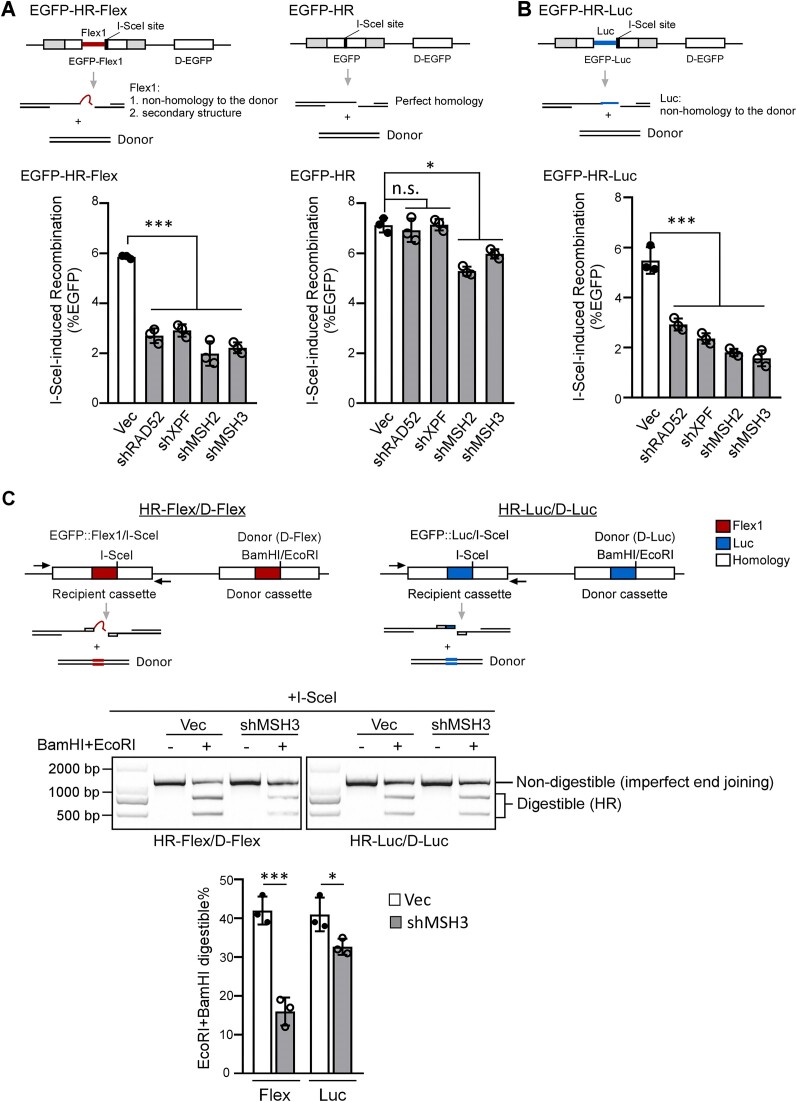
Since DSBs are accumulated at Flex1 when MSH2 or MSH3 is depleted in FANCM deficient cells (Figure 1B), we asked whether MutSβ plays a role in repairing DSBs at Flex1. We used I-SceI to generate DSBs at the 3′ end of Flex1 (0.3 kb) in the EGFP-HR-Flex reporter (Figure 4A, top left). Depletion of MSH2 or MSH3 strongly inhibits HR after I-SceI cleavage of the EGFP-HR-Flex reporter (Figure 4A, left and Supplementary Figure S4A). However, HR reduction, assayed by the general EGFP-HR reporter, is much weaker in MSH2- or MSH3-depleted cells although it is still statistically significant compared to the vector expressing cells (Figure 4A, right and Supplementary Figure S4B).
Figure 4.
MutSβ is important for removing Flex1 at DSB ends to promote HR. (A and B). HR frequency was determined by FACS analysis in U2OS (EGFP-HR-Flex) (A, left), U2OS (EGFP-HR) (A, right) and U2OS (EGFP-HR-Luc) (B) cells, expressing shRNAs for RAD52, XPF, MSH2 and MSH3 or a control vector (Vec), 4 days after I-SceI lentiviral infection. (C) HR reporters HR-Flex/D-Flex and HR-Luc/D-Luc were used as described previously (11). Schematic drawing of HR reporters HR-Flex/D-Flex and HR-Luc/D-Luc is shown on top. U2OS (HR-Flex/D-Flex) or U2OS (HR-Luc/D-Luc) cells expressing MSH3 shRNAs or a control vector were infected with I-SceI lentiviruses. 4 days later, genomic DNA was extracted and digested with I-SceI, followed by PCR using indicated primers. The PCR products with or without BamHI and EcoRI digestion were resolved on an agarose gel (middle), and the percentage of BamHI- and EcoRI-digestible PCR products relative to the total DNA was calculated (bottom).
Different from the EGFP-HR reporter, the EGFP-HR-Flex reporter contains an inserted 0.3 kb Flex1, which does not have homology to the donor and would become a nonhomologous tail at the left side of the DSB after I-SceI cleavage. In addition, Flex1 would form DNA secondary structures in the 3′ ssDNA overhangs after end resection (Figure 4A, top). After strand invasion, DNA secondary structures, like nonhomologous tails, could block DSB ends from initiating new DNA synthesis for HR (Figure 7B). In yeast, it has been shown that Msh2/Msh3 is critical for removing 3′ nonhomologous tails (3′ flaps) to support SSA and HR (47,67,68). We thus examined the role of MutSβ in HR using the EGFP-HR-Luc reporter which contains a 0.3 kb luciferase (Luc) fragment at the same position as Flex1 but does not form secondary structures after end resection (Figure 4B, top). Inhibition of MSH2 or MSH3 also strongly suppresses HR in the EGFP-HR-Luc reporter (Figure 4B and Supplementary Figure S4C), suggesting that MutSβ is also required for HR when a DSB end contains a nonhomologous tail in mammalian cells.
Given that MutSβ is required for HR when Luc, acting as a nonhomologous tail, and Flex1, a nonhomologous tail with secondary structure, are present at DSB ends, further investigation is needed to determine whether HR would still be dependent on MutSβ when DSB ends contain perfect homology to the donor but are prone to forming DNA secondary structures. We used the previously established DSB repair reporters (HR-Flex/D-Flex and HR-Luc/D-Luc, Figure 4C, top), of which Flex1 and Luc are inserted to both the donor template and the recipient cassette (containing the I-SceI cleavage site) (11). In this design, the DSB ends in both reporters possess perfect homology to the donors. However, after end resection, Flex1 in HR-Flex/D-Flex would form a secondary structure, whereas Luc in HR-Luc/D-Luc would not (Figure 4C, top). Because of the insertion of Flex1 and Luc in the donor templates, HR would not produce green cells, and we used PCR analysis to score the repair events. Four days after I-SceI lentiviral infection, genomic DNA was extracted and digested with I-SceI to remove the parental EGFP recipient cassettes (including DNA uncut by I-SceI or perfect end joining products after I-SceI cut), followed by PCR with primers specific for the EGFP recipient cassette (Figure 4C, top). BamHI and EcoRI sites were introduced to the donor templates, corresponding to the I-SceI position in the recipient cassettes. If HR is used, BamHI and EcoRI sites would be copied from the donor cassettes to the recipient cassettes in the HR repair products, while end joining products would not contain BamH1 and EcoRI sites in the recipient cassettes. Thus, the ratio of BamHI and EcoRI digestible and non-digestible PCR products reflects the ratio of the repair products by HR or by imperfect end joining (Figure 4C, middle). We observed that after MSH3 depletion, the percentage of using HR in U2OS (HR-Flex/D-Flex) cells is substantially reduced, but the reduction in U2OS (HR-Luc/D-Luc) cells is minor (Figure 4C, middle and bottom, and Supplementary Figure S4D). These data suggest that even with perfect homology of DSB ends to the donor templates, MutSβ is still required for HR when DNA secondary structures are formed at DSB ends.
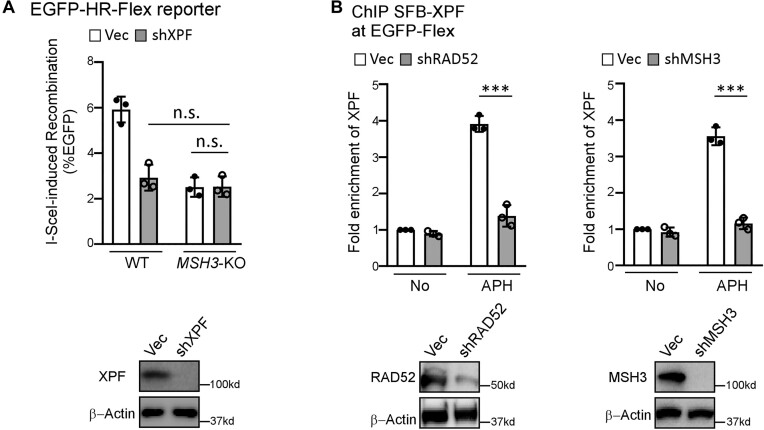
MutSβ and RAD52 are required for XPF recruitment to Flex1
As we described previously (31), XPF/ERCC1 and RAD52 are required for HR when DSB ends contain nonhomologous tails and/or DNA secondary structures such as Luc and Flex1 (EGFP-HR-Luc and EGFP-HR-Flex reporters, Figure 4A, left and 4B), but not at the clean DSB ends (EGFP-HR, Figure 4A, right), suggesting that XPF/ERCC1 and RAD52 are important for removing nonhomologous tails and DNA secondary structures at DSB ends to facilitate HR. Interestingly, depleting XPF by shRNA in MSH3-KO cells does not further decrease HU-induced recombination (Figure 5A), suggesting that XPF and MSH3 are in the same pathway to promote HR at Flex1 upon RS. By ChIP analysis, we showed that the recruitment of XPF to Flex1 after APH is dependent on RAD52 and MSH3 (Figure 5B). Given that RAD52 interacts with XPF (69), we propose that the role of MutSβ in recruiting RAD52, which in turn recruits XPF/ERCC1 to DSBs at Flex1, is important for DNA secondary structure removal at DSB ends to facilitate HR.
Figure 5.
MutSβ and RAD52 are required for XPF recruitment to Flex1. (A) HR frequency was determined by FACS analysis in WT or MSH3-KO U2OS (EGFP-HR-Flex) cells expressing XPF shRNAs or a control vector (Vec) 4 days after I-SceI lentiviral infection (top). XPF depletion was shown by Western blot analysis using β-Actin as a loading control (bottom). (B) Anti-streptavidin ChIP analysis at Flex1 was performed in U2OS (EGFP-HR-Flex) cells expressing SFB-XPF with RAD52 (left) or MSH3 (right) silenced by shRNAs using the shRNA vector as a control (Vec) before and after APH treatment (0.8 μM, 16 h). The ChIP value in the sample of No treatment with Vec control was set as 1 for normalization. Depletion of RAD52 or MSH3 was shown by Western blot analysis using β-Actin as a loading control (bottom).
MutSβ is important for MiDAS and CFS stability
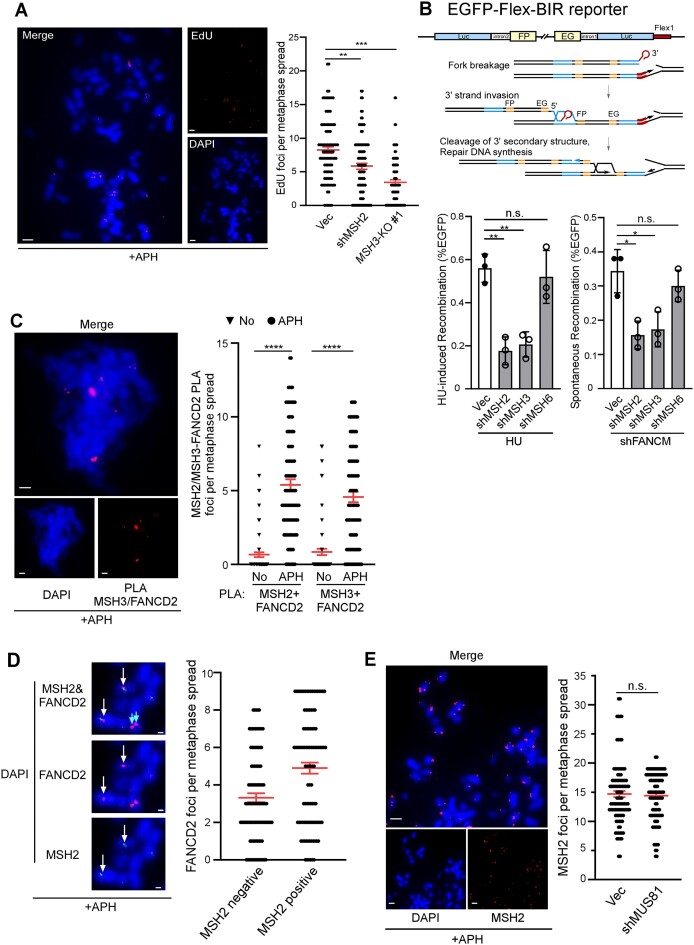
CFSs frequently harbor DNA sequences that are susceptible to forming DNA secondary structures (15,70–74), with Flex1 being one of these sequences derived from the CFS FRA16D (10). We hypothesized that MutSβ would play an important role in safeguarding CFS integrity due to its critical function in processing DNA secondary structures at DSB ends to promote homology-directed repair. When replication is partially disturbed, DNA replication often cannot be completed at CFSs, triggering the activation of MiDAS to ensure replication completion during mitosis (20). Interestingly, we observed that MiDAS is much reduced in MSH3-KO cells and in MSH2-depleted cells (Figure 6A and Supplementary Figure S5A). Since MiDAS exhibits BIR features (20), we used our previously established BIR reporter cell line U2OS (EGFP-Flex-BIR) containing Flex1 (22) (Figure 6B, top) to determine the BIR efficiency after depleting MSH2 and MSH3. Indeed, HU- and FANCM deficiency-induced BIR at Flex1 is dependent on MSH2 and MSH3, but not MSH6 (Figure 6B, bottom and Supplementary Figure S5B).
Figure 6.
MutSβ is important for MiDAS. (A) MiDAS analysis was performed in WT U2OS cells expressing shRNA for MSH2 or the shRNA vector (Vec) and in MSH3-KO U2OS cells. Cells were treated with APH (0.4 μM, 8 h), followed by addition of RO-3306 (7 μM, 8 h), and then released into fresh medium containing EdU (20 μM) and Colcemid (0.1 μg/ml) as described in Materials and Methods. A representative image of EdU incorporation on metaphase chromosomes is shown (left) and EdU foci formed on each metaphase spread were quantified (right). ∼100 metaphase spreads were analyzed in each sample. Scale bars, 5 μm. (B) Schematic drawing of the EGFP-Flex-BIR reporter (22) is shown on top. U2OS (EGFP-Flex-BIR) cells expressing shRNAs targeting MSH2, MSH3 or MSH6 with the vector (Vec) as a control were synchronized to S-phase using double thymidine block followed by HU treatment (2 mM, 24 h, bottom left) or infected with lentiviruses encoding shRNAs for FANCM (bottom right). BIR frequency was assessed by FACS 6 days after. (C) PLA analysis of MSH2 or MSH3 with FANCD2 was performed in U2OS cells, treated with or without APH (0.4 μM, 8 h), followed by addition of RO-3306 (7 μM, 8 h), and then released into fresh medium containing Colcemid (0.1 μg/ml) as described in Materials and Methods. A representative image of MSH3 and FANCD2 PLA on metaphase chromosomes is shown (left) and the PLA foci of MSH2 or MSH3 with FANCD2 on metaphase spread were quantified (right). ∼100 metaphase spreads were analyzed in each sample. Scale bars, 5 μm. (D) U2OS cells were treated with APH (0.4 μM, 8 h), followed by addition of RO-3306 (7 μM, 8 h), and subsequently released into fresh medium containing Colcemid (0.1 μg/ml), and immunostaining of MSH2 and FANCD2 was performed. Representative immunofluorescence images of MSH2 foci (green) that co-localize with FANCD2 foci (red) indicated by white arrows or not (indicated with blue arrows) on metaphase chromosomes are shown on the left, and the quantification is present on the right. ∼100 metaphase spreads were analyzed in each sample. Scale bars, 2 μm. (E) U2OS cells expressing MUS81 shRNAs or vector (Vec) were treated with APH (0.4 μM, 8 h), followed by addition of RO-3306 (7 μM, 8 h), and released into fresh medium containing Colcemid (0.1 μg/ml), and immunostaining of MSH2 was performed. Representative immunofluorescence images of MSH2 foci on metaphase chromosomes are shown on the left, and the quantification is present on the right. ∼100 metaphase spreads were analyzed in each sample. Scale bars, 5 μm.
In response to RS, FANCD2 forms foci that persist into mitosis and serves as a surrogate marker for the location of CFSs (20,75). Using PLA, we found that MSH3 and FANCD2 are localized in proximity after APH treatment (Figure 6C). We also showed that MSH2 form foci on metaphase chromosomes after APH treatment, and majority of FANCD2 foci are overlapping with MSH2 foci (Figure 6D). Collectively, these data suggest that MutSβ is recruited to CFSs upon RS. Furthermore, similar to FANCD2 foci (76), MSH2 foci formed during mitosis are independent of MUS81 (Figure 6E and Supplementary Figure S5C), indicating that either MutSβ recruitment to CFSs is independent of MUS81 or occurs before MUS81-mediated cleavage of under-replicated DNA regions.
To examine whether MutSβ is important for preventing CFS instability, we treated HCT116 cells with low dose of APH (0.4 μM), the condition that causes CFS expression (5) and examined gaps and breaks on metaphase chromosomes. We observed a significant increase of chromosomal gaps and breaks when MSH2 and MSH3 are depleted (Figure 7A and Supplementary Figure S5D), supporting the notion that MutSβ is important for maintaining CFS stability.
Discussion
A growing body of evidence indicates that besides its role in MMR, MutSβ is actively engaged in the cellular response to DSBs. It has been shown that following laser microirradiation in human cells, MSH2, MSH3, MSH6 and MLH1 are rapidly recruited to DSB sites (77). MSH2 and MSH3 are also important for checkpoint activation in response to ionizing radiation and contribute to cellular resistance against agents that induce DSBs (45,78–80). Moreover, chromatid breaks are significantly increased in MSH2 and MSH3 null mouse embryotic fibroblasts (81). Our findings provide compelling evidence that MutSβ plays a distinctive role in processing DNA secondary structures at DSB ends to facilitate HR and BIR. This unique function is crucial for safeguarding structure-prone DNA sequences within the genome, which holds particular significance for maintaining the stability of CFSs.
Our study showed that MutSβ is enriched at the CFS-AT sequence Flex1 after RS such as APH treatment or when FANCM is deficient. We propose that MutSβ binds to DNA secondary structures formed in the ssDNA regions at stalled replication forks (Figure 7B, left) and also at 3′ ssDNA overhangs at double ended DSB ends following end resection (Figure 7B, right). Flex1, along with other CFS-AT sequences, is predicted to contain multiple stem loops (72), and previous study has shown that MutSβ directly interacts with stem loops formed in TNRs and in RPA-bound ssDNA regions (41,42,45). Additionally, MSH2 recruitment to laser-generated DSBs is dependent on CtIP and MRE11 (45), suggesting that end resection is required for MSH2 to bind to double ended DSB ends. Thus, DNA loops formed on ssDNA likely represent a common structural feature to which MutSβ binds. We have further demonstrated that APH-induced MSH2 binding to Flex1 occurs independently of MUS81 (Figure 2H), implying that MutSβ’s interaction with Flex1 at stalled forks does not require the presence of MUS81 and probably takes place before fork breakage and DSB formation. Moreover, our study indicated that APH-induced MutSβ binding to Flex1 relies on the presence of PCNA and the association of MSH3 with PCNA (Figure 2I and J). One possibility is that MutSβ species, loaded onto forks by PCNA during normal replication, are recruited to stalled forks to bind to DNA secondary structures formed in the ssDNA regions accumulated upon RS.
It has been suggested that MutSβ plays a role in HR in mammalian cells (45,49–51). In our study, we observed that when DSB ends are homologous to the donor, deficiency in MSH2 or MSH3 results in a relatively minor yet significant defect in HR. However, when DSB ends contain secondary structures or nonhomologous tails, HR is strongly impaired in MSH2- or MSH3-deficient cells (Figure 4A and B). This is reminiscent of the observations in yeast that MutSβ, together with Rad1/Rad10 (yeast homolog of XPF/ERCC1), is required for removing nonhomologous tails at DSBs during gene conversion, but is dispensable for gene conversion when DSB ends are homologous to the donor (46,47,82–85). MutSβ is also required for SSA in yeast when the annealing homology is relatively short (47). In mammalian cells, the role of MutSβ in HR appears to be multifaceted. One study showed that MutSβ aids in EXO1 recruitment, thereby promoting end resection and facilitating HR (51). However, another study indicated that end resection remains unaffected when MSH2 or MSH3 is depleted after CPT treatment (45). Moreover, MutSβ binds to Holliday junctions (HJs), stimulating the activity of SLX4-binding SSEs (SLX1, MUS81-EME1 and XPF-ERCC1) to resolve recombination intermediates, and deficiency in MutSβ results in a reduction of sister chromatid exchange and an increase in ultrafine bridges in mitosis (50). Despite the various roles of MutSβ in relation to HR, based on our study, we propose that MutSβ is most crucially required when DSB ends contain DNA secondary structures or nonhomologous tails that block new DNA synthesis for HR/BIR.
In yeast, removing nonhomologous tails at DSBs for HR requires Rad1/Rad10 (84,85). We showed that this mechanism is conserved in mammalian cells, where XPF/ERCC1 is essential for HR when DSB ends contain either nonhomologous tails or DNA secondary structures (31). Additionally, RAD52 is specifically required for HR when DSB ends are obstructed by nonhomologous tails or DNA secondary structures (11). We also showed that RAD52 recruits XPF/ERCC1 to DSB ends containing G-quadruplexes, possibly through a direct interaction (32). In this study, we presented evidence that the recruitment of XPF to DSBs at Flex1 (AT-rich and secondary structure prone) is dependent on both RAD52 and MutSβ (Figure 5B). Notably, we showed that RAD52 interacts with MSH3 and this interaction is enhanced after HU treatment (Figure 3D and E). One plausible scenario is that MutSβ directly binds to DNA secondary structures in Flex1 after RS, and upon DSB formation at Flex1, RAD52 is recruited to the proximity of Flex1 by binding to RPA-ssDNA. The local accumulation of MutSβ and RAD52 would strengthen their interactions, consequently retaining more RAD52 at Flex1. Alternatively, RS stimulates the interaction between MutSβ and RAD52, although its exact mechanism remains unknown. This increased association could potentially lead to the recruitment of more RAD52 to Flex1, given that MutSβ binds to Flex1. As a result of the enrichment of RAD52 at Flex1, XPF/ERCC1 is recruited through the interaction with RAD52, acting to cleave DNA secondary structures at Flex1 to promote homology-directed repair. Besides the MutSβ/RAD52 axis, there is also a possibility that SLX4 offers an alternative route for recruiting XPF/ERCC1 to DNA secondary structures recognized by MutSβ, as SLX4 has been found to form a complex with both MutSβ and XPF/ERCC1 (50).
Collectively, based on this study, we propose a working model illustrating the role of MutSβ in fork repair under RS (Figure 7B, left). At stalled replication forks, ssDNA accumulates, leading to formation of DNA secondary structures, such as DNA stem loops at Flex1 and other CFS-ATs. MutSβ is likely loaded onto the replication forks through interacting with PCNA during replication, and as DNA secondary structures (such as CFS-ATs) are formed subsequently at stalled forks due to RS, MutSβ is recruited to bind these structures. MUS81 is likely responsible for cleaving stalled forks, and following that, RAD52 is recruited to Flex1, facilitated at least in part through its interaction with MutSβ. Subsequently, RAD52 recruits XPF/ERCC1, which removes DNA secondary structures at DSB ends, allowing new DNA synthesis that is required for BIR. This coordinated recruitment of MutSβ, RAD52 and XPF/ERCC1 at Flex1 and also other CFS-ATs highlights their crucial roles in addressing challenging DNA secondary structures under RS conditions and underscores their significance in maintaining CFS stability. Additionally, at double-ended DSBs, DNA secondary structures (such as CFS-ATs) could form in the ssDNA regions after end resection and similar mechanisms involving MutSβ, RAD52 and XPF/ERCC1 to remove DNA secondary structures at DSB ends are used to promote HR (Figure 7B, right).
FANCM plays a crucial role in safeguarding Flex1, and its deficiency results in the accumulation of DSBs at Flex1 (11). RAD52, XPF and PIF1, which are essential for HR or BIR to repair DSBs at Flex1, exhibit a synthetic lethal interaction with FANCM (11,22). MSH2 and MSH3 are also synthetic lethal with FANCM, consistent with our findings that MutSβ functions together with RAD52 and XPF to process DNA secondary structures at DSB ends to promote HR/BIR-mediated repair at Flex1.
CFSs are intrinsically difficult to replicate, and when faced with RS, they frequently fail to complete replication before entering mitosis, resulting in the formation of breaks/gaps on metaphase chromosomes, which is visualized as fragility at these loci, commonly referred to as ‘CFS expression’ (5). Upon entry into mitosis, the MUS81/EME1 complex, in association with SLX4 and XPF/ERCC1, cleaves DNA regions at under-replicated CFS loci to activate MiDAS to complete DNA replication (20). The dependence of MiDAS on POLD3 and PIF1 indicates that BIR mechanism is used for MiDAS to complete DNA synthesis at CFSs (20). In this study, we demonstrated that MSH2 and MSH3 are in proximity to FANCD2 in mitotic cells and MSH2 foci colocalize with FANCD2 foci on metaphase chromosomes, suggesting that MutSβ is localized to CFSs as FANCD2 is commonly used as a surrogate marker for CFSs (20,75). Structure-prone AT-rich sequences are commonly present at CFSs, and these sequences could form stable secondary structures to hinder fork progression (10,15,72) and become targets for endonuclease-mediated cleavage to initiate MiDAS. We propose that upon MUS81 cleavage CFSs, DNA secondary structures formed in the ssDNA regions need be excised to enable new DNA synthesis for MiDAS, similar to the mechanism we described for promoting BIR at Flex1/CFS-ATs in our reporter upon RS (Figure 7B, left). In support of this idea, we showed that MSH3 is required for efficient MiDAS, and MSH3 deficiency leads to an increased number of gaps and breaks on metaphase chromosomes when replication is disturbed.
MMR proteins have been implicated in CFS protection. Notably, the tumor suppressor genes FHIT and WWOX, located within CFS FRA3B and FRA16D, respectively, are frequently found to be mutated in colon cancer cases with MMR deficiency (86). Moreover, MMR-deficient cells exhibit increased sensitivity to chromosomal damage induced by APH especially at CFSs (87). In our study, we provide direct evidence that MutSβ is involved in maintaining CFS integrity, and propose that its role in directing the removal of DNA secondary structures at DSBs to promote BIR/MiDAS underlies its function in CFS protection. Importantly, CFS instability is further exacerbated upon oncogene expression due to oncogene-induced RS (88). Therefore, revealing the role of MutSβ in MiDAS and its connection to CFS stability could provide valuable insights into the processes involved in maintaining genomic integrity and preventing oncogene-driven genomic instability.
Supplementary Material
Acknowledgements
We thank Dr Weidong Wang for providing anti-FANCM antibody and Dr Catherine Freudenreich (Tufts University) for sharing the Flex1-containing plasmids.
Contributor Information
Youhang Li, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China; Department of Molecular Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
Yunkun Zhang, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Sameer Bikram Shah, Department of Molecular Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
Chia-Yu Chang, Department of Molecular Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
Hailong Wang, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Xiaohua Wu, Department of Molecular Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health (NIH) [CA187052, GM141868, CA244912 to X.W.]; National Natural Science Foundation of China [31971221 to H.W.]. Funding for open access charge: National Natural Science Foundation of China [31971221].
Conflict of interest statement. None declared.
References
- 1. Glover T.W. Common fragile sites. Cancer Lett. 2006; 232:4–12. [DOI] [PubMed] [Google Scholar]
- 2. Arlt M.F., Durkin S.G., Ragland R.L., Glover T.W.. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst.). 2006; 5:1126–1135. [DOI] [PubMed] [Google Scholar]
- 3. Burrow A.A., Williams L.E., Pierce L.C., Wang Y.H.. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009; 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilson T.E., Arlt M.F., Park S.H., Rajendran S., Paulsen M., Ljungman M., Glover T.W.. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015; 25:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glover T.W., Berger C., Coyle J., Echo B.. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984; 67:136–142. [DOI] [PubMed] [Google Scholar]
- 6. Macheret M., Halazonetis T.D.. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015; 10:425–448. [DOI] [PubMed] [Google Scholar]
- 7. Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A. Jr, Kastrinakis N.G., Levy Bet al.. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005; 434:907–913. [DOI] [PubMed] [Google Scholar]
- 8. Tsantoulis P.K., Kotsinas A., Sfikakis P.P., Evangelou K., Sideridou M., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G.et al.. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008; 27:3256–3264. [DOI] [PubMed] [Google Scholar]
- 9. Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C.et al.. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005; 434:864–870. [DOI] [PubMed] [Google Scholar]
- 10. Zhang H., Freudenreich C.H.. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell. 2007; 27:367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang H., Li S., Oaks J., Ren J., Li L., Wu X.. The concerted roles of FANCM and Rad52 in the protection of common fragile sites. Nat. Commun. 2018; 9:2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H., Li Y., Truong L.N., Shi L.Z., Hwang P.Y., He J., Do J., Cho M.J., Li H., Negrete A.et al.. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 2014; 54:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Helmrich A., Ballarino M., Tora L.. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011; 44:966–977. [DOI] [PubMed] [Google Scholar]
- 14. Le Tallec B., Millot G.A., Blin M.E., Brison O., Dutrillaux B., Debatisse M. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 2013; 4:420–428. [DOI] [PubMed] [Google Scholar]
- 15. Ozeri-Galai E., Lebofsky R., Rahat A., Bester A.C., Bensimon A., Kerem B.. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol. Cell. 2011; 43:122–131. [DOI] [PubMed] [Google Scholar]
- 16. Letessier A., Millot G.A., Koundrioukoff S., Lachages A.M., Vogt N., Hansen R.S., Malfoy B., Brison O., Debatisse M.. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011; 470:120–123. [DOI] [PubMed] [Google Scholar]
- 17. Naim V., Wilhelm T., Debatisse M., Rosselli F.. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 2013; 15:1008–1015. [DOI] [PubMed] [Google Scholar]
- 18. Le Beau M.M., Rassool F.V., Neilly M.E., Espinosa R. 3rd, Glover T.W., Smith D.I., McKeithan T.W.. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 1998; 7:755–761. [DOI] [PubMed] [Google Scholar]
- 19. Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D.. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009; 11:753–760. [DOI] [PubMed] [Google Scholar]
- 20. Minocherhomji S., Ying S., Bjerregaard V.A., Bursomanno S., Aleliunaite A., Wu W., Mankouri H.W., Shen H., Liu Y., Hickson I.D.. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015; 528:286–290. [DOI] [PubMed] [Google Scholar]
- 21. Bhowmick R., Minocherhomji S., Hickson I.D.. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol. Cell. 2016; 64:1117–1126. [DOI] [PubMed] [Google Scholar]
- 22. Li S., Wang H., Jehi S., Li J., Liu S., Wang Z., Truong L., Chiba T., Wang Z., Wu X.. PIF1 helicase promotes break-induced replication in mammalian cells. EMBO J. 2021; 40:e104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu X., Malkova A.. Break-induced replication mechanisms in yeast and mammals. Curr. Opin. Genet. Dev. 2021; 71:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu L., Malkova A.. Break-induced replication: unraveling each step. Trends Genet. 2022; 38:752–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anand R.P., Lovett S.T., Haber J.E.. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 2013; 5:a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang H., Li S., Zhang H., Wang Y., Hao S., Wu X.. BLM prevents instability of structure-forming DNA sequences at common fragile sites. PLoS Genet. 2018; 14:e1007816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Irony-Tur Sinai M., Salamon A., Stanleigh N., Goldberg T., Weiss A., Wang Y.H., Kerem B. AT-dinucleotide rich sequences drive fragile site formation. Nucleic Acids Res. 2019; 47:9685–9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson R.D., Jasin M.. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000; 19:3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wright W.D., Shah S.S., Heyer W.D.. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018; 293:10524–10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cejka P., Symington L.S.. DNA end resection: mechanism and Control. Annu. Rev. Genet. 2021; 55:285–307. [DOI] [PubMed] [Google Scholar]
- 31. Li S., Lu H., Wang Z., Hu Q., Wang H., Xiang R., Chiba T., Wu X.. ERCC1/XPF is important for repair of DNA double-strand breaks containing secondary structures. iScience. 2019; 16:63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S., Wang Z., Shah S.B., Chang C.Y., Ai M., Nguyen T., Xiang R., Wu X.. DNA repair protein RAD52 is required for protecting G-quadruplexes in mammalian cells. J. Biol. Chem. 2023; 299:102770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006; 7:335–346. [DOI] [PubMed] [Google Scholar]
- 34. Kunkel T.A., Erie D.A.. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015; 49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Genschel J., Littman S.J., Drummond J.T., Modrich P.. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 1998; 273:19895–19901. [DOI] [PubMed] [Google Scholar]
- 36. Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol. Cell. Biol. 1996; 16:5604–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drummond J.T., Li G.M., Longley M.J., Modrich P.. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995; 268:1909–1912. [DOI] [PubMed] [Google Scholar]
- 38. Habraken Y., Sung P., Prakash L., Prakash S.. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr. Biol. 1996; 6:1185–1187. [DOI] [PubMed] [Google Scholar]
- 39. Palombo F., Iaccarino I., Nakajima E., Ikejima M., Shimada T., Jiricny J.. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 1996; 6:1181–1184. [DOI] [PubMed] [Google Scholar]
- 40. Surtees J.A., Alani E.. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J. Mol. Biol. 2006; 360:523–536. [DOI] [PubMed] [Google Scholar]
- 41. Owen B.A., Yang Z., Lai M., Gajec M., Badger J.D. 2nd, Hayes J.J., Edelmann W., Kucherlapati R., Wilson T.M., McMurray C.T.. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005; 12:663–670. [DOI] [PubMed] [Google Scholar]
- 42. Tian L., Hou C., Tian K., Holcomb N.C., Gu L., Li G.M.. Mismatch recognition protein MutSbeta does not hijack (CAG)n hairpin repair in vitro. J. Biol. Chem. 2009; 284:20452–20456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tome S., Holt I., Edelmann W., Morris G.E., Munnich A., Pearson C.E., Gourdon G.. MSH2 ATPase domain mutation affects CTG*CAG repeat instability in transgenic mice. PLoS Genet. 2009; 5:e1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keogh N., Chan K.Y., Li G.M., Lahue R.S.. MutSbeta abundance and Msh3 ATP hydrolysis activity are important drivers of CTG*CAG repeat expansions. Nucleic Acids Res. 2017; 45:10068–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burdova K., Mihaljevic B., Sturzenegger A., Chappidi N., Janscak P.. The mismatch-binding factor MutSbeta can mediate ATR activation in response to DNA double-strand breaks. Mol. Cell. 2015; 59:603–614. [DOI] [PubMed] [Google Scholar]
- 46. Colaiacovo M.P., Paques F., Haber J.E.. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics. 1999; 151:1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sugawara N., Paques F., Colaiacovo M., Haber J.E.. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rogacheva M.V., Manhart C.M., Chen C., Guarne A., Surtees J., Alani E.. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J. Biol. Chem. 2014; 289:5664–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bennardo N., Gunn A., Cheng A., Hasty P., Stark J.M.. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009; 5:e1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Young S.J., Sebald M., Shah Punatar R., Larin M., Masino L., Rodrigo-Brenni M.C., Liang C.C., West S.C.. MutSbeta Stimulates Holliday Junction Resolution by the SMX Complex. Cell Rep. 2020; 33:108289. [DOI] [PubMed] [Google Scholar]
- 51. Oh J.M., Kang Y., Park J., Sung Y., Kim D., Seo Y., Lee E.A., Ra J.S., Amarsanaa E., Park Y.U.et al.. MSH2-MSH3 promotes DNA end resection during homologous recombination and blocks polymerase theta-mediated end-joining through interaction with SMARCAD1 and EXO1. Nucleic Acids Res. 2023; 51:5584–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xue X., Sung P., Zhao X.. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 2015; 29:1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Y., Leung J.W., Jiang Y., Lowery M.G., Do H., Vasquez K.M., Chen J., Wang W., Li L.. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol. Cell. 2013; 49:997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J.. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007; 131:901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kleczkowska H.E., Marra G., Lettieri T., Jiricny J.. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001; 15:724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W.et al.. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013; 9:e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J.. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:7480–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kolodziej K.E., Pourfarzad F., de Boer E., Krpic S., Grosveld F., Strouboulis J.. Optimal use of tandem biotin and V5 tags in ChIP assays. BMC Mol. Biol. 2009; 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Casper A.M., Nghiem P., Arlt M.F., Glover T.W.. ATR regulates fragile site stability. Cell. 2002; 111:779–789. [DOI] [PubMed] [Google Scholar]
- 60. Garribba L., Wu W., Ozer O., Bhowmick R., Hickson I.D., Liu Y.. Inducing and detecting mitotic DNA synthesis at difficult-to-replicate loci. Meth. Enzymol. 2018; 601:45–58. [DOI] [PubMed] [Google Scholar]
- 61. Li S., Wang H., Jehi S., Li J., Liu S., Wang Z., Truong L., Chiba T., Wang Z., Wu X.. PIF1 helicase promotes break-induced replication in mammalian cells. EMBO J. 2021; 40:e104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hao S., Wang Y., Zhao Y., Gao W., Cui W., Li Y., Cui J., Liu Y., Lin L., Xu X.et al.. Dynamic switching of crotonylation to ubiquitination of H2A at lysine 119 attenuates transcription-replication conflicts caused by replication stress. Nucleic Acids Res. 2022; 50:9873–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li S., Wu X.. Common fragile sites: protection and repair. Cell Biosci. 2020; 10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Irony-Tur Sinai M., Kerem B. DNA replication stress drives fragile site instability. Mutat. Res. 2018; 808:56–61. [DOI] [PubMed] [Google Scholar]
- 66. Osman F., Whitby M.C.. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst.). 2007; 6:1004–1017. [DOI] [PubMed] [Google Scholar]
- 67. Goldfarb T., Alani E.. Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair and nonhomologous tail removal. Genetics. 2005; 169:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Paques F., Haber J.E.. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997; 17:6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Motycka T.A., Bessho T., Post S.M., Sung P., Tomkinson A.E.. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J. Biol. Chem. 2004; 279:13634–13639. [DOI] [PubMed] [Google Scholar]
- 70. Lukusa T., Fryns J.P.. Human chromosome fragility. Biochim. Biophys. Acta. 2008; 1779:3–16. [DOI] [PubMed] [Google Scholar]
- 71. Limongi M.Z., Pelliccia F., Rocchi A.. Characterization of the human common fragile site FRA2G. Genomics. 2003; 81:93–97. [DOI] [PubMed] [Google Scholar]
- 72. Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B.. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 2003; 23:7143–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ried K., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D.et al.. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000; 9:1651–1663. [DOI] [PubMed] [Google Scholar]
- 74. Arlt M.F., Miller D.E., Beer D.G., Glover T.W.. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer. 2002; 33:82–92. [DOI] [PubMed] [Google Scholar]
- 75. Ozer O., Bhowmick R., Liu Y., Hickson I.D.. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget. 2018; 9:15836–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Naim V., Wilhelm T., Debatisse M., Rosselli F.. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 2013; 15:1008–1015. [DOI] [PubMed] [Google Scholar]
- 77. Hong Z., Jiang J., Hashiguchi K., Hoshi M., Lan L., Yasui A.. Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J. Cell Sci. 2008; 121:3146–3154. [DOI] [PubMed] [Google Scholar]
- 78. Franchitto A., Pichierri P., Piergentili R., Crescenzi M., Bignami M., Palitti F.. The mammalian mismatch repair protein MSH2 is required for correct MRE11 and RAD51 relocalization and for efficient cell cycle arrest induced by ionizing radiation in G2 phase. Oncogene. 2003; 22:2110–2120. [DOI] [PubMed] [Google Scholar]
- 79. Pichierri P., Franchitto A., Piergentili R., Colussi C., Palitti F.. Hypersensitivity to camptothecin in MSH2 deficient cells is correlated with a role for MSH2 protein in recombinational repair. Carcinogenesis. 2001; 22:1781–1787. [DOI] [PubMed] [Google Scholar]
- 80. Park J.M., Huang S., Tougeron D., Sinicrope F.A.. MSH3 mismatch repair protein regulates sensitivity to cytotoxic drugs and a histone deacetylase inhibitor in human colon carcinoma cells. PLoS One. 2013; 8:e65369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van Oers J.M., Edwards Y., Chahwan R., Zhang W., Smith C., Pechuan X., Schaetzlein S., Jin B., Wang Y., Bergman A.et al.. The MutSbeta complex is a modulator of p53-driven tumorigenesis through its functions in both DNA double-strand break repair and mismatch repair. Oncogene. 2014; 33:3939–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Evans E., Sugawara N., Haber J.E., Alani E.. The Saccharomyces cerevisiae Msh2 mismatch repair protein localizes to recombination intermediates in vivo. Mol. Cell. 2000; 5:789–799. [DOI] [PubMed] [Google Scholar]
- 83. Bertrand P., Tishkoff D.X., Filosi N., Dasgupta R., Kolodner R.D.. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fishman-Lobell J., Haber J.E.. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992; 258:480–484. [DOI] [PubMed] [Google Scholar]
- 85. Ivanov E.L., Haber J.E.. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995; 15:2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jahid S., Sun J., Gelincik O., Blecua P., Edelmann W., Kucherlapati R., Zhou K., Jasin M., Gumus Z.H., Lipkin S.M.. Inhibition of colorectal cancer genomic copy number alterations and chromosomal fragile site tumor suppressor FHIT and WWOX deletions by DNA mismatch repair. Oncotarget. 2017; 8:71574–71586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vernole P., Muzi A., Volpi A., Terrinoni A., Dorio A.S., Tentori L., Shah G.M., Graziani G.. Common fragile sites in colon cancer cell lines: role of mismatch repair, RAD51 and poly(ADP-ribose) polymerase-1. Mutat. Res. 2011; 712:40–48. [DOI] [PubMed] [Google Scholar]
- 88. Miron K., Golan-Lev T., Dvir R., Ben-David E., Kerem B.. Oncogenes create a unique landscape of fragile sites. Nat. Commun. 2015; 6:7094. [DOI] [PubMed] [Google Scholar]
- 89. Wang H., Shao Z., Shi L.Z., Hwang P.Y., Truong L.N., Berns M.W., Chen D.J., Wu X.. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 2012; 287:21471–21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.